Summary
Apicomplexa possess a complex pellicle that is composed of a plasma membrane and a closely apposed inner membrane complex (IMC) that serves as a support for the actin-myosin motor required for motility and host cell invasion. The IMC consists of longitudinal plates of flattened vesicles, fused together and lined on the cytoplasmic side by a subpellicular network of intermediate filament-like proteins. The spatial organization of the IMC has been well described by electron microscopy, but its composition and molecular organization is largely unknown. Here, we identify a novel protein of the IMC cytoskeletal network in Toxoplasma gondii, called TgSIP, and conserved among apicomplexan parasites. To finely pinpoint the localization of TgSIP, we used structured illumination super resolution microscopy and revealed that it likely decorates the transverse sutures of the plates and the basal end of the IMC. This suggests that TgSIP might contribute to the organization or physical connection among the different components of the IMC. We generated a T. gondii SIP deletion mutant and showed that parasites lacking TgSIP are significantly shorter than wild-type parasites and show defects in gliding motility, invasion and reduced infectivity in mice.
Keywords: Apicomplexa, Toxoplasma gondii, Plasmodium, inner membrane complex, motility
Introduction
Dinoflagellates, Ciliates and Apicomplexa, jointly form the superphylum Alveolata. This very large group of unicellular eukaryotic organisms (protists) includes parasites of tremendous clinical importance such as the causative agents of malaria, cryptosporidiosis and toxoplasmosis. Alveolates are highly diverse and gather organisms with very different environmental niches, lifestyles and morphologies. The unifying ultrastructural feature of the disparate alveolates is the presence of flattened vesicles underlying the plasma membrane. Consequently, the pellicle surrounding the cytoplasm of alveolates is a triple bilayer. This highly-specialized structure is known as alveoli in ciliates, amphiesmal vesicles in dinoflagellates, or inner membrane complex (IMC) in apicomplexan parasites (Reger, 1967). The alveoli seem to have evolved independently towards specific functions in each taxon. In ciliates they are thought to serve as calcium store, in dinoflagellates they can contain cellulose plates and thus form a remarkable coat of armor (Ladenburger et al., 2009). In Apicomplexa they anchor the actin-myosin motor complex, a key component of the molecular machinery required for the motility and invasion processes of these parasitic organisms (for a review see (Morrissette et al., 2013)). The IMC also provides structural stability to the parasite and has an important role during the development of daughter cells during parasite division (for reviews see (Francia et al., 2014, Morrissette et al., 2013)).
In Apicomplexa, this endomembranous system is also associated with a network of 8–10 nm intermediate filament-like filaments (also called subpellicular network) (Mann et al., 2001). Beneath the IMC vesicles and the cytoskeleton network lie 22 microtubules that emanate from the apical polar ring and extend two-thirds of the length of the parasite (Nicholset al., 1987). The cortical cytoskeleton of Apicomplexa is therefore composed of two main components: the microtubules and the inner membrane complex (IMC), which itself is formed by the membrane cisternae lined by the subpellicular network (Mann et al., 2001). The IMC plates together with the subpellicular network will be hereafter referred to as IMC.
The IMC is organized according to a characteristic pattern conserved in most apicomplexan zoites. In Toxoplasma tachyzoites, it is composed of a single cone-shaped plate of approximately 1 μm at the apex of the parasite (called the apical cap), followed by 3 or 4 rows of six rectangular plates. The plates are contiguous longitudinally and horizontally, leading to longitudinal and transversal junction lines also called sutures. The IMC covers almost the entire length of the tachyzoite leaving openings at the apex above the conoid and at the base of the cell, where the plates narrow in a helical fashion (Porchet et al., 1977). In certain stages of Plasmodium spp., the pattern may be simplified to a single spherical plate (in merozoites (Hanssen et al., 2013)) or a single longitudinal plate (in sporozoites (Dubremetz et al., 1979)). In Plasmodium falciparum gametocytes, the IMC is organized in rectangular plates running around the girth of the banana shaped gametocyte (Hanssen et al., 2012, Konoet al., 2012, Meszoely et al., 1987).
Freeze-fracture studies have shown that the cytoplasmic faces of the IMC plates are covered with longitudinal lines of intramembranous particles (IMPs) (Porchet et al., 1977), organized into a Triton X-100 resistant 32 nm lattice (Morrissette et al., 1997). Regularly spaced double lines of IMPs with a periodicity reminiscent of the underlying 22 subpellicular microtubules have been suggested to contribute to the stability of the pellicle potentially by providing a link to the subpellicular microtubules (Morrissette et al., 1997). However, the IMP lines extend across the entire length of the parasite, while the microtubules run only two-thirds of the parasite. To date, the molecular composition and the functional significance of these IMPs are unknown. However, a novel family of six-pass transmembrane proteins called GAPMs, are likely IMP candidates and are proposed to play a role in tethering the gliding motor to the cytoskeleton network. Indeed, these proteins localize to the IMC and were shown to co-purify with cytoskeleton proteins and the acto-myosin motor (Bullen et al., 2009).
Over the past decade, an increasing number of proteins associated with the IMC have been identified (for reviews see (Anderson-White et al., 2012, Harding et al., 2014). Most of them are conserved across the Apicomplexa. One group contains detergent soluble proteins that are mainly associated with the membrane of the IMC plates through coordinated myristoylation and palmitoylation (Beck et al., 2010, Frenal et al., 2010, de Miguel et al., 2008, De Napoli et al., 2013, Fung et al., 2012, Gaskins et al., 2004). They include a family of proteins termed IMC sub-compartment proteins (ISPs) (Beck et al., 2010, Poulin et al., 2013). Some are distributed over the entire length of the IMC, while others are restricted to sub-compartments, such as the apical cap to which ISP1 is localized exclusively at and the middle region where ISP2 is found. Proteins of the gliding machinery GAP45, 40, 50, 70, are also associated with the IMC plates and are also detergent soluble (Frenal et al., 2010, Gaskins et al., 2004). In contrast, proteins associated with the subpellicular network of the IMC are highly resistant to deoxycholate (DOC) detergent extraction (Mann et al., 2001, Gilket al., 2006, Gubbels et al., 2004, Hu et al., 2006). These include the articulin-like proteins, also named IMC proteins in Toxoplasma, and conserved in most Apicomplexa (Mann et al., 2001, Anderson-White et al., 2011, Khater et al., 2004, Tremp et al., 2008, Tremp et al., 2011). These proteins have structural similarity to mammalian intermediate filament proteins. A recent study characterized 14 articulin proteins in Toxoplasma (Anderson-White et al., 2011) and revealed that, as for the ISP proteins, the articulins have different subcellular localizations. Some proteins localize to the cortical cytoskeleton and others such as IMC5, 8, 9, and 13 are exclusively associated with the basal cytoskeleton or basal complex. Subtle differences in the localization of IMC proteins within the basal complex have been recently described by conventional immune-microscopy (Anderson-White et al., 2011). Two layers of IMC proteins were found: i) a widest structure containing IMC9, and 13, as well as the membrane occupation and recognition nexus protein 1 (MORN1), which is recruited early to the edge of the growing IMC, where it is thought to be important for the interaction between IMC and microtubules, and to play a role in IMC biogenesis; and ii) a more distal structure composed of IMC5 and 8, partially co-localizing with centrin 2 (Hu et al., 2006). Whether these two layers correspond to the two sub-structures identified by electron microscopy at the basal complex (Anderson-White et al., 2011) remains to be demonstrated.
Apicomplexa undergo various complex division processes, unified by the presence of a growth phase during which the parasite replicates its genome by successive rounds of DNA replication and mitosis, and a budding phase (for a review see (Francia et al., 2014)). The later corresponds to the assembly of daughter cell structures. In Toxoplasma, the internal budding is synchronous with nuclear division and is called endodyogeny, by contrast to most Apicomplexa, such as Plasmodium in which the budding of daughter cells occurs after several rounds of nuclear divisions (schizogony). In both cases, the budding phase is a synchronous step and therefore tightly regulated in time and space. During this step, the parasite orchestrates the sequential expression and assembly of IMC proteins, microtubules, as well as the packaging of specific organelles dedicated to the invasion process, called micronemes and rhoptries. The rhoptries and IMC associated proteins are expressed and assembled within a narrow window of time (Behnke et al., 2010), while microneme proteins are synthesized and packaged later. In the final stages of endodyogeny the mother's cytoskeleton disassembles and the mother's plasma membrane is transferred to the daughters. The budding step is therefore a highly complex process in which the parasite needs to control the correct scaffolding of the components of the cortical structure. This includes the differential targeting of proteins to sub-compartments within the IMC, but also the correct assembly of the different IMC plates and their association with the gliding machinery, the subpellicular network and the microtubules.
Here, we present the characterization of a new protein associated with the cytoskeleton of the IMC with a localization reminiscent of the junctions between IMC plates. This protein, called TgSIP (Toxoplasma stripes IMC protein), is not essential for parasite survival but appears crucial for maintaining the shape of the parasite and for optimal motility, host cell invasion and virulence.
Results
Identification of SIP, a conserved protein in Apicomplexa parasites, present at the edges of the IMC plates
While searching for new proteins that shared the expression profile of rhoptry proteins and were conserved among apicomplexan parasites (Behnke et al., 2010), we selected a predicted open reading frame, TGME49_267500, which encodes a 230 amino acid protein. Genome mining and multiple sequence alignment with TGME49_267500 revealed orthologs in all Apicomplexa (Supplementary Fig. 1). The gene is present as a single copy in Coccidia and Cryptosporidium parasites; while two paralogs are found in Babesia, Theileria and Plasmodium species. TGME49_267500 also possesses two orthologs in Perkinus marinus, a dinoflagellate closely related to Apicomplexa. The highest degree of conservation lies in the central part of the protein. TGME49_267500 lacks putative transmembrane domains or other domains and is predicted to be a 25.7 kDa protein with a pI of 6.5.
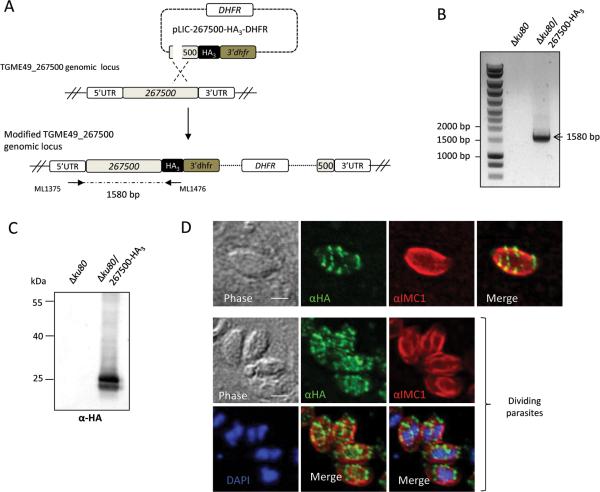
To explore the localization of TGME49_267500, a sequence coding for a triple HA tag was inserted in the open reading frame by single homologous recombination at the endogenous TGME49_267500 locus in the RH-ku80ko strain (ΔKu80) (Huynh et al., 2009) (Fig. 1A). Correct integration of the targeting plasmid was verified by PCR (Fig. 1B) and the expression of the C-terminal epitope-tagged protein 267500-HA3 was assessed by Western blot (Fig. 1C). A specific doublet migrating at the expected size was detected in the tagged-strain with anti-HA antibodies. In immunofluorescence assays (IFA), epitope-tagged protein 267500-HA3 displayed a peculiar banding pattern not previously observed (Fig. 1D). Transversal discontinuous lines are visible along the longitudinal axis of the tachyzoite. The banding pattern is also visible in the daughter cells during internal budding, as shown by co-staining with IMC1, a protein of the articulin family associated with the subpellicular network of the IMC (Fig. 1D). IMC1 labels both the subpellicular network of the mother and of the daughter cells, and is thought to provide stability to the structure (Mann et al., 2001). We thereafter named this protein SIP (for Stripes IMC Protein).
Figure 1. Expression and localization of TgSIP (TGME49_267500) in Toxoplasma tachyzoites.
A. Schematic representation of the generation a transgenic parasite expressing a triple HA-tag at the C -terminal end of TGME49_267500. The 3’ end of the TGME49_267500 gene was cloned in frame with the HA3 tag in pLIC-HA3-DHFR vector, giving pLIC-267500-HA3-DHFR. Single homologous recombination at the endogenous locus allowed the endogenous tagging of TGME49_267500. The modified TGME49_267500 genomic locus is depicted.
B. Vector integration at the 3’ end of TGME49_267500 (knock-in Δku80 strain) was confirmed by the PCR amplification of a 1580 bp fragment, as indicated by arrows. The corresponding non transfected line was used as control (no PCR amplification).
C. Immunoblot carried out on Δku80 or Δku80 expressing TGME49_267500-HA3 parasite lysates probed with anti-HA antibodies showed that TGME49_267500-HA3 is detected at the expected size (25 kDa).
D. IFA performed on intracellular transgenic parasites using anti-HA antibodies and anti-IMC1 to detect the IMC of the mother and of daughter cells. The nuclei were stained with Hoechst. TGME49_267500 is present on transverse lines, producing a clownfish pattern. Note that the banding pattern is also detected in daughter cells during endodyogeny (bottom panel). Scale bars represent 2 μm.
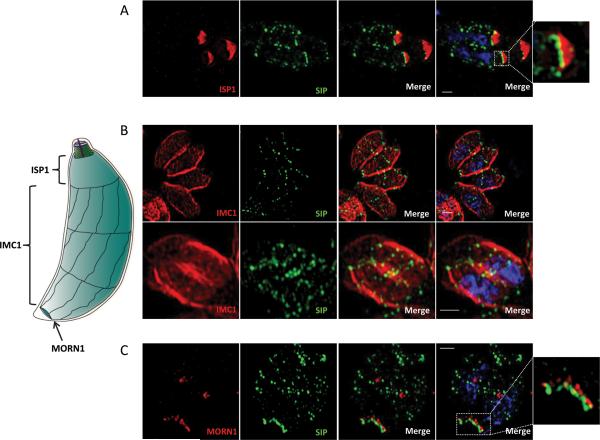
To gain more insight into the localization of TgSIP, we next used structured illumination super resolution microscopy (SIM-SR). We performed SIM-SR on HA-tagged parasites labelled with different markers. First we used ISP1, which localizes to the apical cap of the parasite and can be observed early upon the initiation of the daughter IMC (Beck et al., 2010). Our analysis revealed the juxtaposition of TgSIP at the base of the IMC cap, in both the mother and the daughter cells (Fig. 2A). (Hu et al., 2006). We therefore performed co-localization of TgSIP with TgCen2 (Supplementary Fig. 2). A sequence coding for YFP was inserted in the open reading frame by single homologous recombination at the endogenous cen2 locus in parasites that expressed HA-tagged TgSIP. As shown previously, TgCen2 fluorescence is visible at the pre-conoidal ring, the centriole, at the basal pole and as distinct dots positioned at the lower border of the apical cap. Remarkably, while TgSIP co-localized with the annuli of centrin 2, TgSIP was consistently found just apical to TgCen2 at the posterior end of the parasite.
Figure 2. Refining TgSIP localization using super-resolution microscopy.
On the left, schematic representation of the IMC structure of the Toxoplasma tachyzoites. The conical apical cap and three bands of longitudinal plates are drawn. The ISP1 marker associates exclusively with the apical cap. The IMC1 protein delineates the entire length of longitudinal plates, and MORN1 the basal complex. MORN1 labels the ring shaped structure at the basal end of the IMC.
A. Structured illumination Super Resolution (SIM-SR) microscopy of parasites stained using anti-HA (TgSIP) and anti-ISP1 antibodies. Maximum intensity projection shows that in daughter cells and mother cells, TgSIP localized at the posterior edge of the ISP1 label. The squared framed apical structure is an enlargement of the right side of the image. The nuclei were stained with DAPI (blue). Scale bars represent 1 μm.
B. Structured illumination Super Resolution (SIM-SR) analysis of parasites in S phase (upper panel) or undergoing division (bottom panel) using anti-HA and anti-IMC1 antibodies. Upper panel: single plane of a Z-stack acquisition. Lower panel: maximal projection. The nuclei were stained with DAPI (blue). TgSIP showed a striped organization delineating the apical and posterior edges of the peripheral IMC1staining together with 2 interspersed discrete rows of puncta crossing the width of the parasite. Scale bars represent 1 μm.
C. Co-localization with MORN1 defines a very distal structure decorated by TgSIP. Scale bars represent 1 μm. The nuclei were stained with DAPI (blue). The squared frame distal structure is enlarged on the right of the figure.
Dual labelling with MORN1, a protein that delineates the basal complex (Gubbels et al., 2006, Hu et al., 2006), showed that TgSIP was also present at the basal edge of the IMC, where it localized just beneath MORN1 (Fig. 2C). The lines visible by confocal microscopy (Fig. 1D) appeared as rows of punctate spots regularly interspaced across the width of the parasite when observed using super-resolution (Fig. 2B and C). The organization and localization of these rows is reminiscent of the junctions between the plates of the IMC. To define the subcellular localization of TgSIP during cell division, we also performed SIM-SR microscopy on dividing parasites labeled with anti-IMC1 (Mann et al., 2001). Super-resolution confirmed that TgSIP is associated with transversal lines in the daughter cells (Fig. 2B). Maximal projections (lower panel) showed that the lines encircled the parasite, which suggests that TgSIP is associated with the periphery of the parasite.
Finally, in order to pinpoint the localization of TgSIP, we endogenously tagged ISP3 with YFP in HA-tagged TgSIP parasites. ISP3 is present from the border of the apical cap (no overlap with ISP1) to the posterior end of the IMC. Co-localizations of TgSIP with ISP3 (Supplementary Fig. 3) revealed that the apical ring of TgSIP always coincided with the apical edge of the ISP3 signal, supporting the notion that SIP is located at the junction between the apical cap and the first row of IMC plates. As expected, we also found TgSIP associated with the posterior edge of the ISP3 signal.
Altogether, our observations suggest that TgSIP is located at the seams that connect IMC plates perpendicular to the axis of the parasite. This label is already found during daughter cell formation and is subsequently maintained in mature parasites.
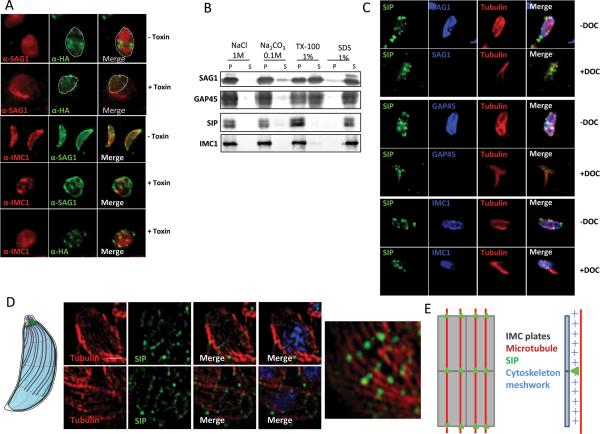
TgSIP is associated with the cytoskeleton network
The presence of TgSIP in internal daughter cells suggested that it may associate with the IMC rather than with the plasma membrane, as the later only encapsulates the daughter cells late in the division process when the two daughter cells emerge from the mother. This prompted us to analyze in more detail which part of the pellicle TgSIP is associated with. The distance between the plasma membrane and the IMC is very small (approximately 15 nm), which precludes discriminating between them by light microscopy. However, treatment with alpha-toxin induces the separation of the plasma membrane from the IMC, enabling differential localization of plasma membrane versus IMC associated proteins (Gaskins et al., 2004). Alpha-toxin binds to GPI-anchored proteins and forms pores that induce swelling of the plasma membrane, while the IMC remains unchanged. After treatment with the alpha-toxin aerolysin, swelling of the plasma membrane was visible with anti-SAG1 staining, while TgSIP remained associated with IMC1 (Fig. 3A), TgSIP is thus not associated with the plasma membrane. Next we performed detergent extractions of proteins from extracellular parasites to discriminate whether TgSIP is associated with the soluble part of the IMC, the insoluble part of the IMC, or with the microtubular cytoskeleton. Parasites expressing HA-tagged TgSIP were re-suspended in buffers varying in their detergent and salt composition, followed by centrifugation and western-blot using anti-HA antibodies to detect the presence of TgSIP in the soluble (supernatant, S) or insoluble (pellet, P) fraction (Fig. 3B). TgSIP remained in the insoluble pellet after treatment with 1% Triton X-100, 0.1 M sodium carbonate, or 1 M NaCl while it was fully soluble in 1% SDS, an extraction profile shared by IMC1 and characteristic of the cytoskeleton proteins that underlie the IMC plates (Fig. 3B). In contrast proteins anchored to the plasma membrane (SAG1) and to the IMC through lipid modification (GAP45) are solubilized in the presence of Triton X-100. To confirm the association of TgSIP with the cytoskeleton, we then performed IFA on extracellular parasites treated with 1% DOC. In this condition, the plasma membrane and the IMC membranes are removed by extraction; in contrast, the IMC network and the underlying microtubules remain intact. DOC-extracted parasites expressing HA-tagged TgSIP were triple-labeled with anti-HA and anti-tubulin antibodies, and either with antibodies directed against SAG1 (plasma membrane), GAP45 (IMC membrane) or IMC1 (subpellicular cytoskeleton) (Fig. 3C). In extracted parasites, TgSIP was detected in the absence of SAG1 and GAP45 labeling. This result showed that TgSIP was associated with the peripheral cytoskeleton. However we noted that the characteristic banding pattern was not as well defined when compared to untreated parasites.
Figure 3. TgSIP is associated with the detergent resistant skeleton of the parasite pellicle.
A. Incubation of TgSIP-HA3 parasites with aerolysin alpha-toxin revealed that TgSIP is associated with the IMC. In untreated cells, the transversal lines of TgSIP occupied the width of the parasite labeled with the plasma membrane marker SAG1. Following alpha-toxin treatment, the dramatic swelling of the plasma membrane separates it from the underlying IMC. In this condition, TgSIP followed the IMC1 staining and became distant from the plasma membrane, demonstrating the association of TgSIP with the IMC.
B. Detergent extraction profile of TgSIP after solubilisation of HA-tagged TgSIP parasites with 1% Triton X-100, 1% SDS, 1M NaCl or 0.1M Na2CO3. Extracts were subjected to SDS-PAGE, blotted and probed with antibodies as indicated. As expected, the GPI-anchored plasma membrane SAG1 and the acylated-GAP45 protein were solubilized in 1% Triton X-100. In contrast, the detergent resistant IMC protein meshwork containing IMC1 remained in the pellet upon Triton-X-100 solubilization. As IMC1, TgSIP was only solubilized in 1% SDS, demonstrating that it is probably embedded in the meshwork of the IMC.
C. Localization by IFA of TgSIP after DOC-extraction. As expected, in DOC-extracted parasites, SAG1 and GAP45 were not detected by IFA, while the tubulin and IMC1 proteins remained in the cytoskeleton ghost. IFA with anti-HA revealed the presence of dots of SIPHA3 associated with the cytoskeleton ghost, indicating that TgSIP associates with the IMC network.
D. Structured illumination Super Resolution (SIM-SR) microscopy of parasites using anti-HA and anti-tubulin antibodies. Scale bars represent 1 μm. Upper panel: single plane of a Z-stack acquisition. Lower panel and right panel: maximum intensity projection. The nuclei were stained with DAPI (blue). TgSIP showed a periodic row organization matching in most cases the periodicity of one of the microtubules. On the left, schematic representation of the IMC structure and of the subpellicular microtubules of Toxoplasma tachyzoites.
E. Schematic representation of TgSIP localization on IMC sutures. The plates are represented by rectangles and the subpellicular microtubules are in red; TgSIP is indicated by green dots located on the transverse sutures at the intersections with microtubules; lateral view (right) shows that TgSIP is located in the subpellicular network.
Our analysis of TgSIP's localization by super-resolution microscopy (Fig. 2) revealed a punctate signal with a distinct periodicity along the transversal lines spanning the parasite's length. To test if the periodicity of the dots followed the path of sub-pellicular microtubules, we performed a dual staining of TgSIP with anti-tubulin. As shown in figure 3D, some of the labeling indeed looked like dots of TgSIP on the microtubules, while some other looked more like bars linking microtubules. This suggested an association of TgSIP with the microtubules, and/or with the double lines of IMPs within the cytoplasmic face of the IMC that overlay the path of the microtubules (Morrissette et al., 1997) (Fig. 3E).
TgSIP is required to maintain the shape of the parasite
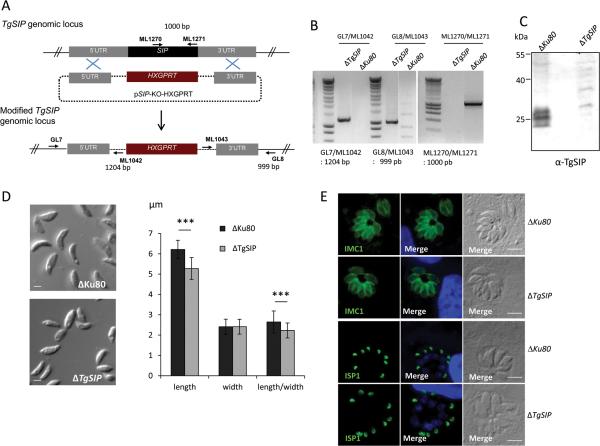
To investigate the function of TgSIP, we deleted the TgSIP gene by double homologous recombination in ΔKu80 tachyzoites as described in Figure 4A. Stable clones that had successfully integrated the targeting vector were obtained after mycophenolic acid-xanthine selection and clones were isolated by limiting dilution. Genomic PCR analysis confirmed the replacement of the open reading frame of TgSIP with the HXGPRT resistance cassette in a clone named hereafter ΔTgSIP (Fig. 4B). The loss of TgSIP protein was then confirmed by Western blot (Fig. 4C) using a TgSIP specific antiserum (Supplementary Fig. 5). The anti-TgSIP antibodies did not react by IFA, precluding further characterization of the loss of TgSIP by IFA (data not shown).
Figure 4. Ablation of the TgSIP gene and phenotypic characterization of ΔTgSIP parasites reveal an abnormal parasite shape.
A. Schematic drawing of the replacement strategy used for the TgSIP knock-out. The pSIPKO_HXGPRT plasmid was inserted at the TgSIP endogenous locus by double homologous recombination at the 5’ and 3’ sides of TgSIP, removing the TgSIP gene. Primers used to verify the correct vector integration are indicated as arrows.
B. PCR verification of the correct integration of pSIP-KO_HXGPRT at the 5’ (primers GL7 and ML1042) and 3’ (primers ML1043 and GL8) loci. Primers ML1270 and ML1271 were used to verify the absence of the TgSIP gene in the parasite genome.
C. Immunoblot of lysates from control (ΔKu80) and ΔTgSIP parasites using specific anti-TgSIP antibodies demonstrating that TgSIP is missing from null (KO) lines.
D. DIC images illustrating the difference of morphology of ΔTgSIP compared to control ΔKu80. Measurement of the length and the width, showed that the ΔTgSIP parasites were statistically shorter than ΔKu80 parasites, while the width was unchanged. Student's t test ***p < 0.001; mean ± s.e.m (n=50). Scale bars represent 2 μm.
E. IFA analysis of ΔTgSIP and ΔKu80 parasites using anti-IMC1 and anti-ISP1 antibodies. No apparent changes in the distribution of IMC1 and ISP1 between the control and the mutant were observed. Scale bars represent 5 μm.
The phenotypic consequences of TgSIP disruption were first evaluated by light microscopy using extracellular parasites. The ΔTgSIP parasites had a stumpy appearance and were about 16% shorter than wild-type parasites (Fig. 4D) indicating significant morphological changes. In contrast parasite width remained unchanged. We did not observe any disturbance in the localization of the proteins ISP1 and IMC1 (Fig. 4E).
ΔTgSIP parasites show defects in motility and invasion
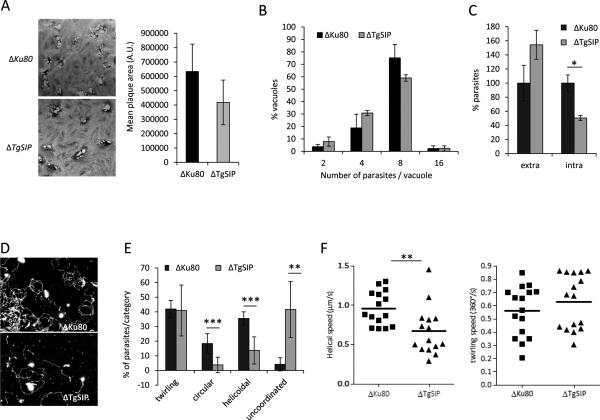
To investigate the phenotypic consequences of the loss of TgSIP on the lytic cycle of the parasite, we performed plaque assays. Fresh monolayers of HFF cells were infected with wild-type or ΔTgSIP parasites for 7 days and the size of the plaques were measured. As shown in Figure 5A, the ΔTgSIP formed plaques similar in size to ΔKu80 parasites, indicating that the mutant is not severely affected in the lytic cycle. This assay reflects the global ability of the parasite to perform invasion, several rounds of replication and egress, but does not allow the detection of subtle defects in each of these successive steps. Yet, we quantified individually each step of the lytic cycle. Further analysis established that removal of TgSIP did not change the replication of the parasite (Fig. 5B). To examine the ability of the ΔTgSIPto invade host cells, we used an assay to differentially detect between intracellular from extracellular parasites, as described previously (Huynh et al., 2006). The ΔTgSIP parasites exhibited a reproducible decrease of around 50% in invasion (Fig. 5C). A defect in invasion might be the consequence of a defect of the parasite to glide, a step that involves the secretion of microneme proteins (MICs) and their relocalization to the surface of the parasite coupled to their association with an actin-myosin motor complex anchored to the IMC plates (Opitz et al., 2002). To monitor if parasites depleted in TgSIP are affected in gliding motility, trails deposited by moving parasites on poly-lysine coated cover slips were visualized by IFA (Hakansson et al., 1999). Gliding was induced by treating the parasites with the calcium ionophore A23187. The density and shape of the trails deposited by ΔTgSIP were different from those deposited by the control strain (Fig. 5D), pointing to an overall defect in gliding motility in ΔTgSIP parasites. To gain further insight into this defect, we used time-lapse video microscopy to examine gliding of extracellular parasites. A significant decrease (P<0.001) in the number of gliding parasites was observed. 80.4% of wild-type parasites showed gliding during a fixed time-frame of 60 seconds, while only 40.9% of ΔTgSIP showed gliding during the same period (Supplementary movies 1 and 2). On a two-dimensional (2D) surface, wild-type parasites exhibit three types of motility (Hakansson et al., 1999): 1) twirling, where the parasite is attached to the substrate by its posterior end and rotates clockwise; 2) circular gliding, where the parasite lies on its side and moves in counterclockwise circles; and finally 3) a helical gliding, where the parasite glides in a clockwise arc, a motion similar to horizontal twirling, resulting in forward movement that follows the path of a corkscrew. ΔTgSIP parasites exhibited apparent normal twirling motion, with a similar speed (Fig. 5E, F). For both strains, the average time required to complete a 360° rotation was measured to be around 2.0 s. However, ΔTgSIP parasites were defective in productive forward circular and helical motions (Fig. 5E). Indeed, most of the observed ΔTgSIP parasites were unable to accomplish a complete circular movement and their average net helical motility speed was significantly decreased (Fig. 5F). Most ΔTgSIP initiated circular and helical movements but their movements stopped or could not be clearly classified, leading to no net translocation (we scored such patterns as uncoordinated movement in figure 5E) (Supplementary movies 3, 4 and 5). In conclusion, removal of TgSIP significantly affected the behavior and speed of motility.
Figure 5. Loss of TgSIP affects parasite motility and host cell invasion.
A. Plaque formation of indicated parasites was analyzed after 7 days. Plaques represent areas where host cells have been lysed and are visible as oval white spots on the background HFF staining. Quantification of the size of the lysis plaques is shown in the graph. The size difference between control (ΔKu80) and ΔTgSIP is not statistically significant.
B. Intracellular growth of Δku80 and ΔTgSIP parasites assessed by counting the number of parasites per vacuole. Only vacuoles containing at least two parasites were counted. Values represent means ± SEM, n = 3, from a representative experiment out of 3 independent assays. No significant difference was found.
C. Host cell invasion assay. Control and ΔTgSIP parasites were allowed to invade for 5 min before fixation and differential staining of attached extracellular and intracellular parasites. Results were normalized to ΔKu80. Values represent means ± SEM, n = 3, from a representative experiment out of 3 independent assays. Student's t test *p < 0.05.
D. Trail deposition staining after gliding of control (ΔKu80) and ΔTgSIP parasites. Parasites were allowed to glide on polyL-lysine coated coverslips for 15 min after induction with 1 μM of calcium ionophore A23187, and trails were stained with anti-SAG1 antibody. Scale bars in red represent 2 μm.
E. Quantification of parasite motility by time-lapse video microscopy. The types of motility were categorized in four groups, including the three well-described circular, helicoidal and twirling movements. A fourth group of unconventional movements highly represented in ΔTgSIP was added (see Materials and Methods). Data are plotted as a percentage of the total movement observed. Values represent mean ± s.e.m, n = 7, from 3 independent assays. Student's t test **p < 0.01, ***p < 0.001.
F. Effect of TgSIP suppression on the speed of gliding. Scatter plots depicting the speeds of helical (left) and twirling (right) gliding motility in ΔKu80 and ΔTgSIP parasites. The gliding speeds (μm/s for helical and 360°/s for twirling) were calculated from following the movements of at least 15 parasites for each strain. Bars represent median values. Student's t test **p < 0.01.
Disruption of the TgSIP gene impairs parasite virulence in mice
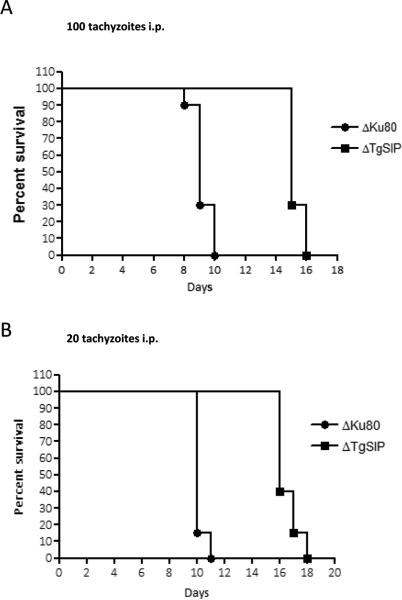
We tested the virulence of ΔTgSIP parasites in a mouse model. The control ΔKu80 parasite and its derivative ΔTgSIP originate from the RH strain, a type 1 strain, which kills mice 8-10 days after i.p. injection of a single tachyzoite. We infected mice with 20 or 100 tachyzoites and monitored them daily. The mice infected with 20 or 100 tachyzoites of the control ΔKu80 strain died within 10-11 days and 8-10 days, respectively (Fig. 6). In contrast, mice infected with ΔTgSIP parasites survived longer, between 15-19 and 14-16 days for infection with 20 and 100 tachyzoites, respectively. This delay in killing was consistently observed in two independent experiments demonstrating that the lack of TgSIP impacted the virulence in mice.
Figure 6. Deletion of TgSIP gene attenuates parasite virulence in a mouse model.
20 tachyzoites (A) or 100 tachyzoites (B) of the indicated strains were injected i.p. into Swiss female mice (n=20) and mouse survival was monitored daily. The ΔTgSIP strain is statistically less virulent than wild-type (p-value <0.001, by Logrank test).
Discussion
We have identified a new protein in T. gondii, TgSIP that is associated with the cytoskeletal network that lies beneath the IMC plates. This is the first protein known to show a particular pattern suggesting that it might correspond to the transversal sutures joining the edges of the IMC plates. SIP is conserved in all Apicomplexa and present in another parasite of the Alveolate superphylum, Perkinsus marinus. Functional analysis of TgSIP in Toxoplasma tachyzoites showed that its removal altered the shape of the parasites, with a significant decrease of their length while the width remained unchanged, suggesting a change in the tensile strength of the IMC. Altogether, these modifications affected the motility of the parasite, its ability to invade cells and its infectivity in mice. During revision of this manuscript an independent characterization of TgSIP has been published (Tilley et al., 2014). In this publication the protein has been named TgCBAP (Conserved Basal Apical Peripheral protein). These data on protein localization and on the modification of parasite shape in absence of the protein corroborate ours. All infectious stages of Apicomplexa share the same basic pellicle, including a plasma membrane that completely encloses the parasite, and the IMC, a patchwork of flattened vesicles underlying the whole plasma membrane except at both ends. The organization of the plates of the IMC and of their sub-structures was initially revealed by freeze fracture electron microscopy. In T. gondii tachyzoites, the IMC is made of an apical trunk-conical plate (apical cap) followed by 3 or 4 parallel rows of 6 rectangular plates narrowing posteriorly in a helical fashion at the basal ring. The IMC is continuous, the plates being joined by a characteristic patchwork of transverse and longitudinal sutures. The recent descriptions of proteins specifically associated with IMC subdomains (Anderson-White et al., 2011, Beck et al., Gilk et al., 2006, Lorestani et al., 2012) have shed light on the complex molecular organization of the IMC and on the existence of specific mechanisms of assembly and targeting during IMC biogenesis. Until now it is poorly understood, how the IMC plates are joined together. A proteinaceous material in P. falciparum gametocytes has been identified by electron microscopy (Kaidoh et al., 1993) and cryo-electron tomography (Dearnley et al., 2012) running perpendicular to the microtubule network and corresponding probably to the transverse sutures encircling the gametocytes (Meszoely et al., 1987). It was recently suggested that these proteinaceous sutures might provide a means of stabilizing the IMC and the sub-pellicular microtubule network (Dearnley et al., 2012). The localization of TgSIP might be consistent with a role for this protein in building and maintaining IMC sutures. Indeed, TgSIP localizes to four transversal bands that match the number and likely positions of the IMC plate sutures in T. gondii. For the first suture we have robust molecular markers. First, using the apical cap protein ISP1 (Beck et al., 2010) TgSIP precisely delineates the basal boundary of the ISP1 compartment (Fig. 2). Second, this boundary is also delineated by the annuli of TgCen2 (Beck et al., 2010, Hu et al., 2006) and accordingly TgSIP co-localizes with this ring of TgCen2 annuli (Supplementary Fig. 2). Finally, ISP3 that labels a compartment that begins at the base of the ISP1 apical cap, also starts at the apical ring of TgSIP (Supplementary Fig. 3).
The IMC1 and ISP3 staining at the posterior end terminates with a line of TgSIP staining, showing that TgSIP localizes at the very basal end of the IMC. Recent studies identified three substructures in the basal complex (Anderson-White et al., 2011). The widest and most apical structure contains MORN1, IMC9, IMC13, and IMC15. It is followed by a structure comprising IMC5 and IMC8, which is then followed at the very basal end by the smallest structure containing TgCen2. The TgSIP label appears more distal than MORN1 but less than TgCen2, a situation that might correspond to the structure decorated by IMC5 and IMC8. Definitive validation of this structure will require additional studies.
Interestingly, SIM-SR microscopy resolved the TgSIP bands into rows of individual dots that appear to follow the general patterning of subpellicular microtubules. Whether this is due to direct interaction with microtubules or reflects the patterning of the intramembranous particles (IMPs) that follows the microtubule path is not fully distinguished by our experiments. However, we note that TgSIP staining is also found in the basal portion of the parasite IMC that is typically devoid of microtubules. We therefore deem it is most likely that TgSIP localizes to the intersection of transverse sutures with the double lines of IMPs found within the IMC plates because these align with the microtubules, but extend across the entire length of the parasite (Morrissette et al., 1997). TgSIP does not contain predicted transmembrane domains and is resistant to detergent extraction, suggesting that it is associated to the equally detergent resistant subpellicular network underlining the IMC plates. Whether TgSIP directly acts as a physical link between the subpellicular network and the transverse sutures of the IMC plates is unknown at this point. In addition, how TgSIP associates with the subpellicular network remains unsolved. TgSIP has neither a transmembrane domain nor a microtubule binding domain, nonetheless it is found in the insoluble fraction (this study and in (Tilley et al., 2014)) and in a conoid-enriched cytoskeletal fraction (Hu et al., 2006). It is likely that the association of TgSIP with the subpellicular network requires cytoskeleton partners that remain to identify. Further investigations are now required to determine the precise localization of TgSIP with respect to the subpellicular network and the molecular mechanisms of this link.
ΔTgSIP parasites are shorter than normal. We do not know if ΔTgSIP tachyzoites display an abnormal number of rows of longitudinal plates. We could only verify that the apical cap is present in this mutant, well delineated by the ISP1 specific antibody. The hypothesis of a defect in the number of plates or its patchwork organization could only be tested by freeze fracture. The phenotypic characterization of ΔTgSIP also revealed defects in motility, invasion and virulence, the two later being likely the consequence of abnormal motility patterns. We showed in this study that TgSIP is not essential for tachyzoite motility in vitro, but that its disruption does adversely affect the ability of the parasites to perform circular and helical gliding. As the curved trajectories of both circular and helical gliding are likely dependent of the crescent shape of the parasite (Hakansson et al., 1999), the gliding phenotype of ΔTgSIP could be a result of the reduced cell length and of the loss of it typical crescent morphology. Interestingly there is another mutant in T. gondii, ΔTgPhiL1, that causes a similar morphological change (Barkhuff et al., 2011) resulting in changes in the precise spatial control of gliding (Leung et al., 2014). While the loss of TgPhiL1 function was only observable using 3D-matrigel-based gliding assays (Leung et al., 2014), the loss of TgSIP function was readily detected using classical 2D assays. Thus the reduction of motility of ΔTgSIP parasites in 2D motility assay is therefore unlikely to be solely attributable to a modification of its cell shape. One of the important roles of the IMC is to act as anchor for the actin-myosin motor (Opitz et al., 2002). Parasite gliding results from an anterior-posterior translocation of surface exposed MICs interacting with substrates while being coupled to the submembranous actin-myosin motor which is rigidly anchored to the IMC plates. MyoA allows the movement of short actin filament from the anterior to the posterior of the tachyzoite in association with the cytoplasmic tail of surface exposed MICs. Based on this model, it is tempting to speculate that if the absence of TgSIP disturbs the IMC organization at some points, it would induce the arrest of the forward translocation of MICs and would impact gliding.
The IMC is composed of a mosaic of proteins of varying evolutionary origin; some are ancient with apparent homologs throughout the eukaryota whereas others are restricted to the Apicomplexa (Kono et al., 2012). Most IMC proteins display a great diversity in their spatiotemporal distribution even within a single family. A Plasmodium IMC-membrane protein (MAL13P1.228) that highlights the transversal structures in gametocytes has been recently identified (Kono et al., 2012). In contrast to most of the IMC proteins, MAL13P1.228 is restricted to Plasmodium sp., indicating a unique function within the Plasmodium genus. In contrast to MAL13P1.228, SIP is conserved in all Apicomplexa. Interestingly, SIP appears not to be transcribed in the P. falciparum blood stages (www.plasmoDB.org), where the merozoite IMC consists of a single sub-spherical plate (Hanssen et al., 2013). In contrast, the protein is expressed in gametocytes (www.plasmoDB.org), which share a patchwork of IMC plates and subpellicular microtubules with T. gondii (Kono et al., 2012, Meszoely et al., 1987). SIP is also expressed in ookinetes (www.plasmoDB.org), which again show a more complex IMC organization similar to T. gondii tachyzoites (Raibaud et al., 2001). Lastly, the alveolate Perkinsus spp. also encodes a SIP homolog, however the structure of its IMC, and whether it is organized into plates, are not known. It is interesting to note that other Alveolates, such as the ciliates, do not possess a SIP homolog. In this case, however, the organization of the alveolar network is very distinct from the one occurring in Apicomplexa. The exclusive expression of SIP in stages harboring organized rows of IMC plates and a link between SIP and the structure and function of IMC remain to be validated.
Experimental procedures
Ethics statement
This study was conducted according to the European Union guidelines for the handling of laboratory animals and the immunization protocol for antibody production in rabbits was conducted at the CRBM animal house (Montpellier) and approved by the Committee on the Ethics of Animal Experiments (Languedoc-Roussillon, Montpellier) (Permit Number: D34-172-4, delivered on 20/09/2009).
All mice protocols were approved by the Institutional Animal Care and Utilization Committee (IACUC) of the American University of Beirut (IACUC Permit Number #14-3-278). All animals were housed in approved pathogen-free housing. Eye pricks were done following deep anesthesia with isoflurane.
Host cells and parasite cultures
Tachyzoites from RH-ΔKu80 strain (deleted for the ku80 gene (Huynh et al., 2009)) and referred to hereafter as ΔKu80 were used throughout in this study. Parasites were grown in human foreskin fibroblast (HFFs) in Dulbecco's Modified Eagle's Medium (DMEM; GIBCO, Invitrogen) supplemented with 5% of fetal calf serum (FCS), 1% penicillin-streptomycin and 1% glutamine.
Generation of transgenic parasites
For transfection, 20.106 parasites were re-suspended in 800μL of cytomix (10mM KPO4, 120mM KCl, 0.15mM CaCl2, 5mM MgCl2, 25mM Hepes, 2mM EDTA) complemented with 3mM ATP and 3mM of reduced glutathione and electroporated with 30-50μg DNA as described previously (Kim et al., 1993). Selections of the transgenic parasites were performed with mycophenolic acid (20μg/mL) and xanthine (50μg/mL) for HXGPRT selection or pyrimethamine (1μM) for DHFR-TS selection and chloramphenicol (20μM) for chloramphenicol acetyltransferase selection. The isolation of clonal transgenic populations was performed using limiting dilution in 96-well plates.
Antibodies
The antibodies used and their dilution for western blot (WB) and IFA were as follows:
-mouse monoclonal antibodies (MAb) T4 1E5 anti-SAG1, 1:2000 (IFA), (1/2000) (WB) (Couvreur et al., 1988)
-rabbit anti-ROP1, 1:500 (WB), 1:3000 (IFA) (Dubremetz&Puijalon, unpublished)
-rat MAb anti-HA 3F10 (Roche, clone 3F10), 1:1000 (WB) 1:1000 (IFA)
-rabbit anti-SAG1 1:2000 (IFA) (Harning et al., 1996)
-rabbit anti-GAP45, 1:1000 (WB), 1:1000 (IFA) (Mann et al., 2001)
-rabbit anti-IMC1, 1:1000 (WB), 1:1000 (IFA) (Mann et al., 2001)
-mouse mAb 7E8 anti-ISP1, 1:4000 (IFA) (Beck et al., 2010)
-mouse mAb 12G10 anti-alpha tubulin, 1:1000 (IFA) (a gift of Jacek Gaertig, University of Georgia, USA)
-rabbit anti-MORN1, 1:250 (IFA) (Gubbels et al., 2006)
For immunofluorescence, the secondary antibodies used were AlexaFluor 488 (Sigma), AlexaFluor 594 (Sigma), and AlexaFluor 546 (Invitrogen), at dilution recommended by the manufacturer. For immunoblot, the secondary rat, mouse or rabbit antibodies used were coupled to alkaline phosphatase (Promega).
Molecular cloning
All the primers are listed below.
To verify the predicted cDNA of TGME49_267500, total RNA was extracted from ΔKu80 tachyzoites using the NucleoSpin RNAII extraction kit (MACHEREY-NAGEL GmbH & Co.). Following RT-PCR using an oligo-dT and the Superscript first strand synthesis kit (Invitrogen), the specific full-length cDNA of TGME49_267500 gene was amplified using the PrimeSTAR HS DNA polymerase (Takara Bio Inc.) and primers ML1527 and ML1468. The obtained fragment was cloned in the pCR-Blunt II-TOPO vector (Invitrogen) and sequenced. The complete cDNA sequence matched the one predicted by ToxoDB (www.toxodb.org).
Plasmid pET24b-SIP-His6 was designed to produce a recombinant TgSIP protein. The cDNA sequence encoding the full-length TgSIP gene was PCR amplified using the PrimeSTAR HS DNA polymerase (Takara Bio Inc.) and primers ML1527 and ML1468. After sequencing, the full-length cDNA was cloned into the NdeI and XhoI restriction sites of pET24b vector (Merck Biosciences) allowing expression of C-terminal His-tagged recombinant protein.
Plasmid pLIC-267500-HA3 was designed to add a sequence coding for a HA3 at the endogenous locus of TGME49_267500 ORF. A 1000bp genomic fragment corresponding to the open reading frame upstream the stop codon of TGME49_267500 was PCR amplified using the primers ML1270/ML1271 and cloned in pLIC-DHFR-HA3 as described previously (Huynh et al., 2009). Plasmid pLIC-267500-HA3 was sequenced and linearized with BstBI prior to transfection to allow single homologous recombination in the ΔKu80 strain. After transfection, parasites resistant to pyrimethamine were isolated by limiting dilution cloning and correct integration of pLIC-267500-HA3 was verified by PCR using the primers ML1476 and ML1375.
Plasmids pLIC-CEN2-YFP-CAT and pLIC-ISP3-YFP-CAT were designed to add a YFP sequence at the endogenous locus of TgCen2 (TGME49_250340) and TgISP3 (TGME49_316540) respectively. A 1500bp and 1771 bp genomic fragment corresponding to TgCen2 and Tgisp3 respectively were PCR amplified using the primers ML1914/ML1915 and ML1912/ML1913 respectively and cloned in pLIC-CAT-YFP as described previously (Huynhet al., 2009). Plasmids pLIC-CEN2-YFP-CAT and pLIC-ISP3-YFP-CAT were sequenced and linearized with EcoRV and NruI respectively prior to transfection to allow single homologous recombination in HA-tagged TgSIP strain. After transfection and selection of the parasites with chloramphenicol, correct integration of the plasmids pLIC-CEN2-YFP-CAT and pLICISP3-YFP-CAT were verified by PCR using the primers ML1952/ML1950 and ML1951/ML1950 respectively.
pSIP-KO-HXGPRT was designed to generate the knockout construct 5’TgSIPHXGPRT-3’TgSIP. 1000pb and 850pb of the 5’-UTR and 3’-UTR respectively of TgSIP gene were amplified from genomic DNA using the primers GL3-GL4 and GL5-GL6 respectively. The amplified fragments were sub-cloned in pCR-Blunt-II-TOPO vector (Invitrogen) and sequenced. Both fragments were then cloned XhoI/HindIII for the 5’UTR fragment and BamHI/XbaI for the 3’UTR fragment, in the 2854.HX plasmid vector. After linearization by XhoI/XbaI and transfection in ΔKu80 strain, parasites resistant to mycophenolic acid in presence of xanthine were isolated by limiting dilution cloning and were subsequently screened by PCR for the 5’ and 3’ integration using the couples of primers GL7/ML1042 and GL8/ML1043 respectively, and for the loss of TgSIP gene using primers ML1270/ML1271.
Production of a recombinant SIP protein and of a specific serum
Escherichia coli BL21 competent cells were transformed with pET24b-SIP-His6 plasmid. Expression of TgSIP-His6 recombinant protein was induced by adding 0.5 mM IPTG at 37°C for 3 hours. Cells from 1L culture were harvested at 4000g for 20 min at 4°C and the pellet was re-suspended in 100 mL of buffer A (Hepes 100mM pH 7.5, NaCl 200mM, 2-mercaptoethanol 2mM, TX-100 0.01%, lysozyme 100μg/mL and a complete protease inhibitor cocktail (Roche)). After sonication and centrifugation of the lysate at 17000 rpm during 30 min at 4°C, TgSIP-His6 recombinant protein was abundantly recovered in the insoluble fraction. TgSIP-His6 protein was electroeluted after SDS-PAGE under denaturing conditions. Recombinant protein SIP-His6 (100μg) was used to raise polyclonal antibodies in rabbit as described previously (Besteiro et al., 2009).
Detergent extraction
For detergent extraction experiments, 100.106 freshly egressed parasites from ΔKu80 and TgSIP-HA3 strains were harvested at 3,000 rpm, 10 min at room temperature. The pellets were re-suspended in 1mL of Tris-HCl 50mM, pH7.5 with a complete protease inhibitor cocktail (Roche) and sonicated twice 15 sec at 30% amplitude. The intact parasites were removed by centrifugation at 500 rpm for 10 min. The supernatants containing lysed parasites were separated in 5 samples of 200μL and centrifuged at 100,000g for 30 min. Then, the pellets were re-suspended in Tris-HCl 50mM pH7.5 containing either 1% Triton X-100, 1% SDS, 1M NaCl or 0.1M Na2CO3). After 1H at 4°C on a rotative wheel, the samples were centrifuged at 100,000g during 30 min. After TCA protein precipitation of the soluble fraction, equivalent amounts of supernatant (soluble) and pellet (insoluble) fractions were separated on a 12% gel, transferred to nitrocellulose and blotted using anti-IMC1, anti-SAG1, anti-HA, and anti-GAP45 antibodies.
Aeromonas hydrophila aerolysin experiments
Separation of the parasite plasma membrane and the IMC was achieved using aerolysin treatment as previously described (Gaskins et al., 2004). Prior to use, purified recombinant aerolysin was activated for 20 min at 37°C with trypsin (1mg/ml) in HBS (140mM NaCl, 2.7mM KCl, 20mM Hepes, pH7. Freshly egressed parasites from the control (ΔKu80) or the ΔTgSIP strains were treated or not in presence of activated toxin diluted at 100 ng/mL in phosphate buffered saline (PBS) during 3 hours at 37°C. After treatment, parasites were settled onto cover slips coated with Poly-L-Lysine and fixed with 4% PFA-PBS for 10 min at room temperature. The parasites were permeabilized with 0.1% Triton X-100 in PBS and IFAs were performed using rabbit anti-IMC1, mouse mAb T4 1E5 anti-SAG1, and rat monoclonal anti-HA as described below.
IFA and super-resolution
For IFAs, host cells (HFF) were plated onto coverslips and infected with parasites. Coverslips were fixed with 4% formaldehyde in PBS and permeabilized with 0.2% Triton X-100 in PBS/3% BSA. For IFAs on extracellular parasites treated with 1% DOC for 25min or with aerolysin (see below), the parasites were settled onto coverslips coated with Poly-L-Lysine and fixed with 4% PFA-PBS for 10 min at room temperature. The parasites were then permeabilized with 0.1% Triton X-100 in PBS. Coverslips were blocked in PBS 10% FCS (3% bovine serum albumin (BSA) for super-resolution) and proceeded further for IFA as previously described (El Hajj et al., 2008). Samples were observed with a Zeiss Axioimager epifluorescence microscope equipped with an apotome and a Zeiss Axiocam MRmCCD camera driven by the Axiovision software (Zeiss), at the Montpellier RIO imaging facility. Super-Resolution images were acquired using the Zeiss ELYRA S1 (SR-SIM) microscope. Images were collected and processed using Zeiss Zen software.
Immunoblots
Freshly released tachyzoites were harvested, washed in PBS and resuspended directly in SDS sample buffer. Polyacrylamide gels were run under non-reducing conditions. Separated proteins were transferred onto nitrocellulose membranes using a semi-dry electroblotter and probed with primary antibodies diluted in 5% non-fat milk powder in TNT buffer (50 mM Tris pH 8.0; 150 mM NaCl; 0.05% Tween20). After several washes, the nitrocellulose membrane was incubated for 1h with secondary antibodies coupled to alkaline phosphatase. Bound secondary conjugated antibodies were visualized using an alkaline phosphatase kit according to manufacturer's instructions (Promega).
Morphology analysis
Freshly egressed parasites from control (ΔKu80) and ΔTgSIP parasites were resuspended in HBSS and allowed to settle on coverslips previously coated with poly-L-lysine. After fixation with 4% PAF in HBSS containing 10mM HEPES pH.7.2, the parasites were visualized by differential interference contrast (DIC) microscopy and images were collected. The length and the width of 50 parasites were determined for each strain using Zeiss Zen software.
Invasion assays
Two-color invasion assays were performed as previously (Cerede et al., 2005). Briefly, to synchronize invasion, 5.106 freshly egressed tachyzoites were first sedimented on ice for 20 min on HFF monolayer grown on coverslips in 24-well plates. Invasion was allowed during 5 min at 37°C and stopped by fixation in 4% PAF in Hank's Balanced Salt Solution (HBSS). A first immuno-detection with the mouse mAb T4 1E5 anti-SAG1 in 2% FCS/HBSS was performed to detect extracellular parasites. After permeabilization with 0.01% saponin for 15min, a second IFA was performed using rabbit anti-ROP1 to label the parasitophorous vacuole of intracellular parasites. Extracellular and intracellular parasites were counted from 20 fields per coverslip from three coverslips.
Plaque assays
HFF monolayer grown in 24-well plates were infected with parasites from the control (ΔKu80) or the ΔTgSIP strains. After 7 days, HFF monolayers were fixed with 4% PFA-PBS and stained with Giemsa. Images were obtained with an Olympus MVX10 macro zoom microscope equipped with an Olympus XC50 camera. Plaque area measurements were performed with CellA software (Olympus).
Intracellular growth assay
HFF monolayers grown on coverslips in a 24-well plate were infected with 2.105 freshly egressed parasites of the ΔKu80 or ΔTgSIP strains. After 2 hours of incubation à 37°C, the remaining extracellular parasites were removed by 5 washings in HBSS. Then, the infected cells were incubated for 22 hours at 37°C and fixed with 4% PFA-PBS. Intracellular parasites were stained by IFA using mouse mAb T4 1E5 anti-SAG1, and the number of parasites per vacuole was counted.
Gliding assay
5.106 freshly egressed parasites of the control (ΔKu80) or the ΔTgSIP strains were harvested and resuspended in 500μL of Ringer's medium (155 mM NaCl, 3 mM KCl, 2 mM CaCl2, 1 mM MgCl2, 3 mM NaH2PO4, 10 mM HEPES, 10 mM glucose) containing 1μM of the calcium ionophore A23187 (Calbiochem, EMD Chemicals, USA). Immediately, 100μL of the parasite suspension was placed on poly-L-lysine coated slides and incubated at 37°C in a wet environment. After 15 minutes, unattached parasites were removed by washing with PBS and IFA were performed on parasites fixed with 4% PFA, using mouse mAb T4 1E5 anti-SAG1 to visualize the trails of SAG1formed by gliding.
Video microscopy and analysis of gliding motility
Time-lapse video microscopy was conducted using a Leica DM-IRB phase-contrast microscope. The immersion objective and condenser are temperature-controlled to maintain the sample at 37°C on an area of 1 cm2 (TC344B, Warner Instrument Corporation). Pictures were collected under low-light illumination, using a 100X APO oil immersion objective and a Miro LC310 (Vision Research) camera. Freshly egressed parasites were harvested, resuspended in Ringer's medium containing 1μM of the calcium ionophore A23187 (Calbiochem, EMD Chemicals, USA) and immediately added to glass-bottom culture dishes previously coated with BSA at 500μg/ml. Once parasites were settled, recording was started. Time-lapse movies were recorded with an exposure time of 6ms per frame and at 24 frames per second for a total of 5 minutes. The video was then processed using ImageJ.
The motility of the parasites and the type of motility (helical, twirling and circular) defined previously (Hakansson et al., 1999) were scored by the observers. Non motile parasites corresponded to parasites that remained attached to the substrat without any change in their position over the time period analyzed. Incomplete circular movement, very short helical movement without net movement, side-to-side rocking behavior, and under-clocked twirling movement were classified as uncoordinated movements. Data presented the values of at least 200 parasites for each strain from seven independent movies, taken from three independent experiments. An average of 25 parasites was counted per video. The speed of motility was calculated by manual tracking by recording the position of each parasite every 0.5sec along 50sec. Data presented the values of at least 15 parasites for each type of movement from at least five independent movies.
In vivo experiments
12-16 week-old Swiss mice (Charles River, France) were infected by i.p. injection of 20 or 100 tachyzoites freshly harvested from cell culture. Invasiveness of the parasites was evaluated by simultaneous plaque assay of a similar dose of parasites on HFFs. Mouse survival was monitored daily until their death, endpoint of all experiments. The immune response of surviving animals was tested following eye pricks performed on day 9 post-infection. Sera were tested by Western blotting against tachyzoite lysates. Data were represented as Kaplan and Meier plots using GraphPad.
Statistics
All results are presented as mean values ±SEM. Two-tailed Student's t tests were used using GraphPad to determine the statistical significance of differences observed between indicated groups for parametric comparisons. For in vivo experiments, the level of significance was determined with the Logrank test using GraphPad.
Oligonucleotides used in this study
ML1527 5’-TACCATATGTCTGACAGAACAGGAACAGAC-3’
ML1468 5’-CCGCTCGAGCTGGTAGACGGGACTGTAAAG-3’
ML1270 5’- TACTTCCAATCCAATTTAATGCCTGGCTGCGAGCAGG-3’
ML1271 5’- TCCTCCACTTCCAATTTTAGCCTGGTAGACGGGACTGTAAAG-3’
ML 1375 5’-CTGGGAAAGGCAATTGTCGG-3’
ML 1476 5’-CAGCGTAGTCCGGGACGTCGTAC-3’
GL3 5’-TATACTCGAGGCCGCGAAAGTCACGCACAC-3’
GL4 5’-AAGCTTACGTCAACTAGAAGACTGACGCG-3’
GL5 5’-TATAGGATCCAGCTCCAAGAGCCGTTGGGTTT-3’
GL6 5’- TATATCTAGAAAACGTACTTTTCAGGCTTCGGACTC-3’
GL7 5’-AGGCAAGCAGGCTTTCCCTGTATA-3’
GL8 5’-CGGTCGTGTGCAGAAGTGAACT-3’
ML1042 5’-GCCGACAGGACGCTACTGG-3’
ML1043 5’-CAGTGACACCGCGGTGGAGG-3’
ML1912 5’- TACTTCCAATCCAATTTAATGCACTGCAGTCGCGTTAACTTCT-3’
ML1913 5’- TCCTCCACTTCCAATTTTAGCGTTTCTGTCGTTGCTTGCTG-3’
ML1914 5’- TACTTCCAATCCAATTTAATGCAAGCTGGTGAGAAAGTGTTTG-3’
ML1915 5’- TCCTCCACTTCCAATTTTAGCCGGGAAAGTCTTCTTGGTC -3’
ML 1950 5’- GGTGGTGCAGATGAACTTCAG-3’
ML1951 5’- TGCTAGACGTATCCAGATGAGTG-3’
ML1952 5’- ATGCAGCGAGGAGCACTG -3’
Supplementary Material
Acknowledgements
We are grateful to Michael White and Olivier Lucas (University of Florida, US) for the bioinformatic screen based on Toxoplasma mRNA abundance over the cell cycle and for providing a list of proteins that have a cell cycle profile that matched with the periodic expression pattern of rhoptry proteins. We thank Dominique Soldati-Favre for providing the purified Aeromonas hydrophyla aerolysin and for plasmid 2854.HX, Peter Bradley for the anti-ISP1 antibodies, Con Beckers for the anti-IMC1 and anti-GAP45 antibodies, Jacek Gaertig for mouse mAb 12G10 anti-alpha tubulin, and Vern Carruthers for sharing the RH-ku80ko strain and pLIC-DHFR-3HA plasmid. We thank Manouk Abkarian for time-lapse video assistance. We are grateful to Marina Lavigne and Eliane Rubio for her technical assistance in production of recombinant protein. We are also grateful to Montpellier RIO imaging facility at the University of Montpellier 2 and the Biomedical Microscopy Core at the University of Georgia. This research was supported by the Laboratoire d'Excellence (LabEx) (ParaFrap ANR-11-LABX-0024) and by the Fondation pour la Recherche Médicale (Equipe FRM DEQ20130326508) to M.L., by the Cèdre France-Lebanon program (Project N21774) to H. E. H and M.L. and by grants from the National Institutes of Health to B.S. who is a Georgia Research Alliance distinguished investigator.
Abbreviations
- IMC
inner membrane complex
- ISP1
IMC sub-compartment protein 1
- MORN1
Membrane Occupation and Recognition Nexus Protein1
- IMPs
Intra-Membranous Particles
References
- Anderson-White B, Beck JR, Chen CT, Meissner M, Bradley PJ, Gubbels MJ. Cytoskeleton assembly in Toxoplasma gondii cell division. Int Rev Cell Mol Biol. 2012;298:1–31. doi: 10.1016/B978-0-12-394309-5.00001-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson-White BR, Ivey FD, Cheng K, Szatanek T, Lorestani A, Beckers CJ, et al. A family of intermediate filament-like proteins is sequentially assembled into the cytoskeleton of Toxoplasma gondii. Cell Microbiol. 2011;13:18–31. doi: 10.1111/j.1462-5822.2010.01514.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkhuff WD, Gilk SD, Whitmarsh R, Tilley LD, Hunter C, Ward GE. Targeted disruption of TgPhIL1 in Toxoplasma gondii results in altered parasite morphology and fitness. PLoS One. 2011;6:e23977. doi: 10.1371/journal.pone.0023977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck JR, Rodriguez-Fernandez IA, Cruz de Leon J, Huynh MH, Carruthers VB, Morrissette NS, Bradley PJ. A novel family of Toxoplasma IMC proteins displays a hierarchical organization and functions in coordinating parasite division. PLoS Pathog. 6 doi: 10.1371/journal.ppat.1001094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck JR, Rodriguez-Fernandez IA, de Leon JC, Huynh MH, Carruthers VB, Morrissette NS, Bradley PJ. A novel family of Toxoplasma IMC proteins displays a hierarchical organization and functions in coordinating parasite division. PLoS Pathog. 2010;6:e1001094. doi: 10.1371/journal.ppat.1001094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behnke MS, Wootton JC, Lehmann MM, Radke JB, Lucas O, Nawas J, et al. Coordinated progression through two subtranscriptomes underlies the tachyzoite cycle of Toxoplasma gondii. PLoS One. 2010;5:e12354. doi: 10.1371/journal.pone.0012354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besteiro S, Michelin A, Poncet J, Dubremetz JF, Lebrun M. Export of a Toxoplasma gondii rhoptry neck protein complex at the host cell membrane to form the moving junction during invasion. PLoS Pathog. 2009;5:e1000309. doi: 10.1371/journal.ppat.1000309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullen HE, Tonkin CJ, O'Donnell RA, Tham WH, Papenfuss AT, Gould S, et al. A novel family of Apicomplexan glideosome-associated proteins with an inner membrane-anchoring role. J Biol Chem. 2009;284:25353–25363. doi: 10.1074/jbc.M109.036772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerede O, Dubremetz JF, Soete M, Deslee D, Vial H, Bout D, Lebrun M. Synergistic role of micronemal proteins in Toxoplasma gondii virulence. J. Exp. Med. 2005;201:453–463. doi: 10.1084/jem.20041672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couvreur G, Sadak A, Fortier B, Dubremetz JF. Surface antigens of Toxoplasma gondii. Parasitology. 1988;97(Pt 1):1–10. doi: 10.1017/s0031182000066695. [DOI] [PubMed] [Google Scholar]
- de Miguel N, Lebrun M, Heaslip A, Hu K, Beckers CJ, Matrajt M, et al. Toxoplasma gondii Hsp20 is a stripe-arranged chaperone-like protein associated with the outer leaflet of the inner membrane complex. Biol Cell. 2008;100:479–489. doi: 10.1042/BC20080004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Napoli MG, de Miguel N, Lebrun M, Moreno SN, Angel SO, Corvi MM. N-terminal palmitoylation is required for Toxoplasma gondii HSP20 inner membrane complex localization. Biochim Biophys Acta. 2013;1833:1329–1337. doi: 10.1016/j.bbamcr.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dearnley MK, Yeoman JA, Hanssen E, Kenny S, Turnbull L, Whitchurch CB, et al. Origin, composition, organization and function of the inner membrane complex of Plasmodium falciparum gametocytes. J Cell Sci. 2012;125:2053–2063. doi: 10.1242/jcs.099002. [DOI] [PubMed] [Google Scholar]
- Dubremetz JF, Prensier G, Torpier G, Maurois P, Sinden RE. Structure de la pellicule du sporozoite de Plasmodium yoelii: étude en cryofracture. C R Acad Sci D. 1979;228:623–626. [Google Scholar]
- El Hajj H, Papoin J, Cerede O, Garcia-Reguet N, Soete M, Dubremetz JF, Lebrun M. Molecular signals in the trafficking of Toxoplasma gondii protein MIC3 to the micronemes. Eukaryot Cell. 2008;7:1019–1028. doi: 10.1128/EC.00413-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francia ME, Striepen B. Cell division in apicomplexan parasites. Nat Rev Microbiol. 2014;12:125–136. doi: 10.1038/nrmicro3184. [DOI] [PubMed] [Google Scholar]
- Frenal K, Polonais V, Marq JB, Stratmann R, Limenitakis J, Soldati-Favre D. Functional dissection of the apicomplexan glideosome molecular architecture. Cell Host Microbe. 2010;8:343–357. doi: 10.1016/j.chom.2010.09.002. [DOI] [PubMed] [Google Scholar]
- Fung C, Beck JR, Robertson SD, Gubbels MJ, Bradley PJ. Toxoplasma ISP4 is a central IMC sub-compartment protein whose localization depends on palmitoylation but not myristoylation. Mol Biochem Parasitol. 2012;184:99–108. doi: 10.1016/j.molbiopara.2012.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaskins E, Gilk S, DeVore N, Mann T, Ward G, Beckers C. Identification of the membrane receptor of a class XIV myosin in Toxoplasma gondii. J Cell Biol. 2004;165:383–393. doi: 10.1083/jcb.200311137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilk SD, Raviv Y, Hu K, Murray JM, Beckers CJ, Ward GE. Identification of PhIL1, a novel cytoskeletal protein of the Toxoplasma gondii pellicle, through photosensitized labeling with 5-[125I]iodonaphthalene-1-azide. Eukaryot Cell. 2006;5:1622–1634. doi: 10.1128/EC.00114-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubbels MJ, Vaishnava S, Boot N, Dubremetz JF, Striepen B. A MORN-repeat protein is a dynamic component of the Toxoplasma gondii cell division apparatus. J Cell Sci. 2006;119:2236–2245. doi: 10.1242/jcs.02949. [DOI] [PubMed] [Google Scholar]
- Gubbels MJ, Wieffer M, Striepen B. Fluorescent protein tagging in Toxoplasma gondii: identification of a novel inner membrane complex component conserved among Apicomplexa. Mol Biochem Parasitol. 2004;137:99–110. doi: 10.1016/j.molbiopara.2004.05.007. [DOI] [PubMed] [Google Scholar]
- Hakansson S, Morisaki H, Heuser J, Sibley LD. Time-lapse video microscopy of gliding motility in Toxoplasma gondii reveals a novel, biphasic mechanism of cell locomotion. Mol Biol Cell. 1999;10:3539–3547. doi: 10.1091/mbc.10.11.3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanssen E, Dekiwadia C, Riglar DT, Rug M, Lemgruber L, Cowman AF, et al. Electron tomography of Plasmodium falciparum merozoites reveals core cellular events that underpin erythrocyte invasion. Cell Microbiol. 2013;15:1457–1472. doi: 10.1111/cmi.12132. [DOI] [PubMed] [Google Scholar]
- Hanssen E, Knoechel C, Dearnley M, Dixon MW, Le Gros M, Larabell C, Tilley L. Soft X-ray microscopy analysis of cell volume and hemoglobin content in erythrocytes infected with asexual and sexual stages of Plasmodium falciparum. Journal of structural biology. 2012;177:224–232. doi: 10.1016/j.jsb.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding CR, Meissner M. The inner membrane complex through development of Toxoplasma gondii and Plasmodium. Cell Microbiol. 2014 doi: 10.1111/cmi.12285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harning D, Spenter J, Metsis A, Vuust J, Petersen E. Recombinant Toxoplasma gondii surface antigen 1 (P30) expressed in Escherichia coli is recognized by human Toxoplasma-specific immunoglobulin M (IgM) and IgG antibodies. Clin Diagn Lab Immunol. 1996;3:355–357. doi: 10.1128/cdli.3.3.355-357.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu K, Johnson J, Florens L, Fraunholz M, Suravajjala S, DiLullo C, et al. Cytoskeletal components of an invasion machine--the apical complex of Toxoplasma gondii. PLoS Pathog. 2006;2:e13. doi: 10.1371/journal.ppat.0020013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh MH, Carruthers VB. Toxoplasma MIC2 is a major determinant of invasion and virulence. PLoS Pathog. 2006;2:e84. doi: 10.1371/journal.ppat.0020084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh MH, Carruthers VB. Tagging of endogenous genes in a Toxoplasma gondii strain lacking Ku80. Eukaryot Cell. 2009;8:530–539. doi: 10.1128/EC.00358-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaidoh T, Nath J, Okoye V, Aikawa M. Novel structure in the pellicular complex of Plasmodium falciparum gametocytes. J Eukaryot Microbiol. 1993;40:269–271. doi: 10.1111/j.1550-7408.1993.tb04916.x. [DOI] [PubMed] [Google Scholar]
- Khater EI, Sinden RE, Dessens JT. A malaria membrane skeletal protein is essential for normal morphogenesis, motility, and infectivity of sporozoites. J Cell Biol. 2004;167:425–432. doi: 10.1083/jcb.200406068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Soldati D, Boothroyd JC. Gene replacement in Toxoplasma gondii with chloramphenicol acetyltransferase as selectable marker. Science. 1993;262:911–914. doi: 10.1126/science.8235614. [DOI] [PubMed] [Google Scholar]
- Kono M, Herrmann S, Loughran NB, Cabrera A, Engelberg K, Lehmann C, et al. Evolution and architecture of the inner membrane complex in asexual and sexual stages of the malaria parasite. Mol Biol Evol. 2012;29:2113–2132. doi: 10.1093/molbev/mss081. [DOI] [PubMed] [Google Scholar]
- Ladenburger EM, Sehring IM, Korn I, Plattner H. Novel types of Ca2+ release channels participate in the secretory cycle of Paramecium cells. Mol Cell Biol. 2009;29:3605–3622. doi: 10.1128/MCB.01592-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung JM, Rould MA, Konradt C, Hunter CA, Ward GE. Disruption of TgPHIL1 alters specific parameters of Toxoplasma gondii motility measured in a quantitative, three-dimensional live motility assay. PLoS One. 2014;9:e85763. doi: 10.1371/journal.pone.0085763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorestani A, Ivey FD, Thirugnanam S, Busby MA, Marth GT, Cheeseman IM, Gubbels MJ. Targeted proteomic dissection of Toxoplasma cytoskeleton sub-compartments using MORN1. Cytoskeleton (Hoboken) 2012;69:1069–1085. doi: 10.1002/cm.21077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann T, Beckers C. Characterization of the subpellicular network, a filamentous membrane skeletal component in the parasite Toxoplasma gondii. Mol Biochem Parasitol. 2001;115:257–268. doi: 10.1016/s0166-6851(01)00289-4. [DOI] [PubMed] [Google Scholar]
- Meszoely CA, Erbe EF, Steere RL, Trosper J, Beaudoin RL. Plasmodium falciparum: freeze-fracture of the gametocyte pellicular complex. Exp Parasitol. 1987;64:300–309. doi: 10.1016/0014-4894(87)90040-3. [DOI] [PubMed] [Google Scholar]
- Morrissette N, Gubbels MJ. Weiss LM, Kim K, editors. The Toxoplasma Cytoskeleton: Structures, Proteins and Processes. Toxoplasma gondii: The Model Apicomplexan-Perspectives and Methods. 2013:455–503. [Google Scholar]
- Morrissette NS, Murray JM, Roos DS. Subpellicular microtubules associate with an intramembranous particle lattice in the protozoan parasite Toxoplasma gondii. J Cell Sci. 1997;110(Pt 1):35–42. doi: 10.1242/jcs.110.1.35. [DOI] [PubMed] [Google Scholar]
- Nichols BA, Chiappino ML. Cytoskeleton of Toxoplasma gondii. The Journal of protozoology. 1987;34:217–226. doi: 10.1111/j.1550-7408.1987.tb03162.x. [DOI] [PubMed] [Google Scholar]
- Opitz C, Soldati D. ‘The glideosome’: a dynamic complex powering gliding motion and host cell invasion by Toxoplasma gondii. Mol. Microbiol. 2002;45:597–604. doi: 10.1046/j.1365-2958.2002.03056.x. [DOI] [PubMed] [Google Scholar]
- Porchet E, Torpier G. [Freeze fracture study of Toxoplasma and Sarcocystis infective stages (author's transl)]. Z Parasitenkd. 1977;54:101–124. doi: 10.1007/BF00380795. [DOI] [PubMed] [Google Scholar]
- Poulin B, Patzewitz EM, Brady D, Silvie O, Wright MH, Ferguson DJ, et al. Unique apicomplexan IMC sub-compartment proteins are early markers for apical polarity in the malaria parasite. Biology open. 2013;2:1160–1170. doi: 10.1242/bio.20136163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raibaud A, Lupetti P, Paul RE, Mercati D, Brey PT, Sinden RE, et al. Cryofracture electron microscopy of the ookinete pellicle of Plasmodium gallinaceum reveals the existence of novel pores in the alveolar membranes. Journal of structural biology. 2001;135:47–57. doi: 10.1006/jsbi.2001.4396. [DOI] [PubMed] [Google Scholar]
- Reger JF. The fine structure of the gregarine Pyxinoides balani parasitic in the barnacle Balanus tintinnabulum. The Journal of protozoology. 1967;14:488–497. doi: 10.1111/j.1550-7408.1967.tb02034.x. [DOI] [PubMed] [Google Scholar]
- Tilley LD, Krishnamurthy S, Westwood NJ, Ward GE. Identification of TgCBAP, a novel cytoskeletal protein that localizes to three distinct subcompartments of the Toxoplasma gondii pellicle. PLoS One. 2014;9:e98492. doi: 10.1371/journal.pone.0098492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremp AZ, Dessens JT. Malaria IMC1 membrane skeleton proteins operate autonomously and participate in motility independently of cell shape. J Biol Chem. 2011;286:5383–5391. doi: 10.1074/jbc.M110.187195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremp AZ, Khater EI, Dessens JT. IMC1b is a putative membrane skeleton protein involved in cell shape, mechanical strength, motility, and infectivity of malaria ookinetes. J Biol Chem. 2008;283:27604–27611. doi: 10.1074/jbc.M801302200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.