Abstract
FOXP3+ Tregs are central for the maintenance of self-tolerance and can be defective in autoimmunity. In multiple sclerosis and type-1 diabetes, dysfunctional self-tolerance is partially mediated by a population of IFNγ-secreting Tregs. It was previously reported that increased NaCl concentrations promote the induction of proinflammatory Th17 cells and that high-salt diets exacerbate experimental models of autoimmunity. Here, we have shown that increasing NaCl, either in vitro or in murine models via diet, markedly impairs Treg function. NaCl increased IFNγ secretion in Tregs, and reducing IFNγ — either by neutralization with anti-IFNγ antibodies or shRNA-mediated knockdown — restored suppressive activity in Tregs. The heightened IFNγ secretion and loss of Treg function were mediated by the serum/glucocorticoid-regulated kinase (SGK1). A high-salt diet also impaired human Treg function and was associated with the induction of IFNγ-secreting Tregs in a xenogeneic graft-versus-host disease model and in adoptive transfer models of experimental colitis. Our results demonstrate a putative role for an environmental factor that promotes autoimmunity by inducing proinflammatory responses in CD4 effector cells and Treg pathways.
Introduction
Tregs are central in mediating peripheral tolerance and are involved in the maintenance of tolerance to self-antigens (1, 2). Tregs employ Foxp3 as a key transcription factor and are characterized by a Treg-specific demethylated region (TSDR) (3). Tregs depend on IL-2 for survival and suppress ongoing immune responses through direct and indirect mechanisms involving secretion of the cytokines TGF-β and IL-10, in addition to cell-to-cell contact–dependent mechanisms (4, 5). Despite normal frequency of Tregs in patients with autoimmune diseases such as multiple sclerosis (MS), their suppressive function is substantially decreased in comparison to healthy individuals (6, 7). Similar to CD4 effector T cells, Tregs possess functional plasticity and are capable of secreting the proinflammatory cytokines IFNγ and IL-17, among others (1, 8–14). Secretion of IFNγ by Foxp3+ Tregs has been observed in patients with relapsing-remitting MS and type 1 diabetes (T1D) (13, 14). The mechanisms that trigger Treg plasticity and dysfunction in autoimmune diseases are not well understood.
Autoimmune diseases are mediated by genetic variants lowering immune cell activation thresholds, resulting in heightened inflammatory responses (15–17). Understanding the unfavorable interactions between genes and environment are critical in understanding the pathophysiology of autoimmune diseases (18, 19). Insufficient vitamin D intake, increased body mass index, and smoking are environmental risk factors for a number of autoimmune diseases. In this regard, diet is emerging as a potential environmental trigger (20–23). We have previously observed that higher physiologic salt concentrations, similar to concentrations that could be detected in the interstitium after sodium-rich diets in vivo, have direct effects on the induction of Th17 cells with initiation of a proinflammatory signature in vitro, suggesting a role for salt in adaptive immune function (24–27). Increases in dietary salt intake worsen animal models of MS (experimental autoimmune encephalomyelitis [EAE]), and is associated with increased in vivo induction of proinflammatory Th17 cells (24, 25). Of note, recent studies in patients with autoimmune diseases indicated that higher salt intake is associated with increased clinical and radiological disease activity in MS patients and with increased risk of developing rheumatoid arthritis (28, 29). Moreover, a recent longitudinal study under highly controlled conditions in humans indicated that changes in dietary salt levels could significantly impact cellularity of cells of the innate immune system and secreted levels of pro- and antiinflammatory cytokines such as IL-6, IL-23, and IL-10 in vivo (30). Besides raising interstitial sodium content, high-salt diets potentially exert direct effects on lymphocytes in gut-associated tissues and within the intestine (25). Finally, recent data have demonstrated that the gut microbiota may have a profound impact on Th17 cells and Tregs (31). As such, it is likely that high-salt diets also alter the gut microbiota, thereby having an indirect impact on CD4 T cells, further linking dietary salt intake to development of autoimmune disease (11).
Serum/glucocorticoid-regulated kinase 1 (SGK1) plays a key role in cellular stress response and downstream activation of potassium, sodium, and chloride channels (32, 33). While levels of SGK1 expression contribute to hypertension and renal disease, transcriptional network analysis identified Sgk1 as a key node for the induction of Th17 cells (25, 34). Moreover, recently SGK1 was shown to have an effect on other Th cellular subsets (35). SGK1-mediated induction of proinflammatory Th17 cells occurs in tandem with activation of p38/MAPK and NFAT5 pathways, leading to a FOXO1-dependent increase in expression of the IL-23 receptor and heightened signaling for the IL-17 inflammatory cascade (24, 25). Blocking the renin-angiotensin system also modulates immune responses and induction of EAE, associated with suppression of autoreactive Th17 cells and enhanced Treg responses (36, 37). Furthermore, excess NaCl affects the innate immune system via skin-residing macrophages, providing important implications for blood pressure regulation (26, 27), and it has been recently demonstrated that high-salt diets could boost the function of proinflammatory macrophages (38, 39).
Here, we report that physiologically elevated levels of sodium chloride block the suppressive capacity of human and murine Foxp3+ Tregs in vitro and in vivo. Under high-salt in vitro conditions, IFNγ blockade by either monoclonal antibodies or shRNA-mediated knockdown of IFNG restores Treg function. Additionally, shRNA-mediated knockdown or chemical inhibition of SGK1 in human Tregs inhibits IFNγ secretion while similarly restoring suppressive function. Thus, high-salt induces a proinflammatory Treg phenotype associated with an SGK1-dependent increase in IFNγ secretion. Finally, high-salt diets blocked Treg function in a murine model of colitis and in a humanized xenogeneic graft-versus-host disease model (x-GvHD model), and they were associated with increased Treg IFNγ secretion in vivo. These data provide evidence that, in addition to the increased induction of proinflammatory Th17 cells, excess dietary sodium intake can impact autoimmunity by inhibiting the function of Tregs. Thus, our data provide a mechanistic link between an environmental factor and immune dysregulation in autoimmune disease.
Results
High salt blocks suppressive function of Tregs in vitro.
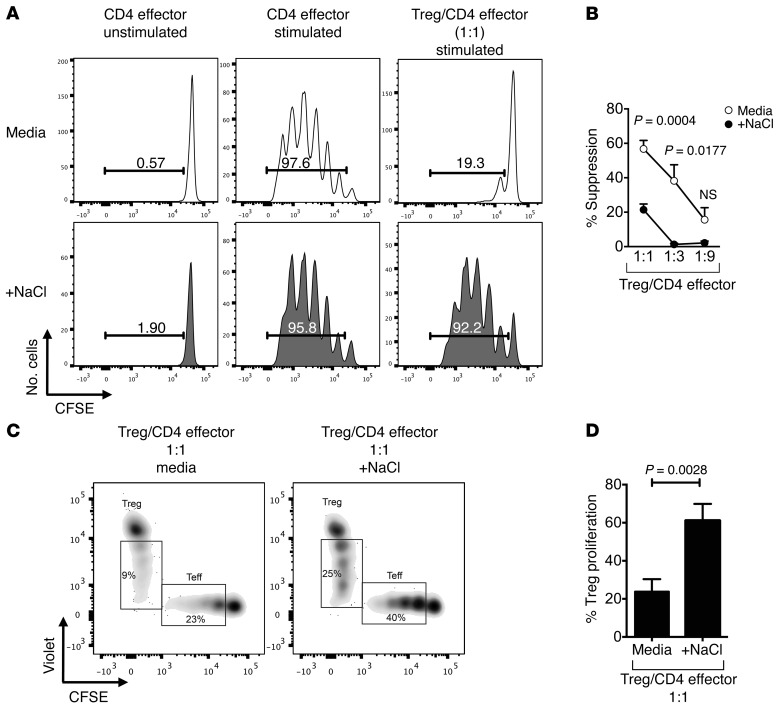
We used an in vitro coculture suppression assay system to study CD4+CD25hiCD127loFoxp3+ Treg functionality in the presence of high-salt conditions (13). While entry into cell cycle by CD4+CD25–CD45RA+CD45RO– naive T effector cells (CD4 effector cells) was inhibited by Tregs in standard media, the addition of physiologically relevant 40 mM NaCl (high salt) prevented efficient in vitro suppressor function of human Tregs, indicating that either the effector cells escaped suppression or that Tregs are acquiring a nonsuppressive phenotype (Figure 1, A and B, and refs. 24–27). In order to determine whether the loss of Treg function was related to cell death, we examined viability and Annexin V reactivity. There was no significant increase in the number of dead cells or evidence of increased apoptosis via Annexin V under high-salt conditions (Supplemental Figure 1, A and B; supplemental material available online with this article; doi:10.1172/JCI81151DS1). It was of interest to determine whether high-salt conditions induced increased Treg entry into cell cycle as measured by cell trace dye. Increases in both Treg and CD4 effector cell proliferation were observed in the high-salt condition (Figure 1, C and D). This was accompanied by stable Foxp3 expression (Supplemental Figure 2) in addition to an increase in CD25 expression on and IL-2 secretion by Tregs in the coculture system (Supplemental Figure 3, A and B). Using a transwell plate, diminished suppressor function in high-salt in vitro conditions was not altered by lack of cell contact (data not shown). Treg suppressor function was also dependent on NaCl concentration between 10–40 mM (Supplemental Figure 4). To explore if these findings were the result of hypertonic shifts in the culture medium, a suppression assay was performed in media enriched with 80 mM mannitol (Supplemental Figure 5, A and B). In line with our previous observations on Th17 cells, no change was observed between standard media and mannitol conditions, indicating that the effect was not simply dependent on hypertonicity (24, 25).
Figure 1. High-salt conditions block suppressive function in vitro.
(A) CD4+ naive effector T cells (CD4 effector) were labeled with CFSE, stimulated with αCD2/αCD3/αCD28-coated beads (at 2 beads/cell) and cultured alone or cocultured with CD4+CD25hiCD127lo Tregs at ratios as indicated. Cells were cultured either in media (white) or with an additional 40 mM NaCl (gray) added. CFSE dilution was measured by flow cytometry after 5 days. Histograms depict cellular proliferation and are gated on viable cells. (B) The line graph depicts a summary of experiments at Treg/CD4 effector cell ratios as indicated (n = 10). (C) Tregs were stained with Celltrace Violet and CD4 effector cells with CFSE to track proliferation within the coculture system either in normal media or in media enriched with 40 mM NaC. Cultures were stimulated with αCD2/αCD3/αCD28-coated beads (at 2 beads/cell). (D) Summary of findings (n = 5). Statistical analyses were performed using paired Student’s t test.
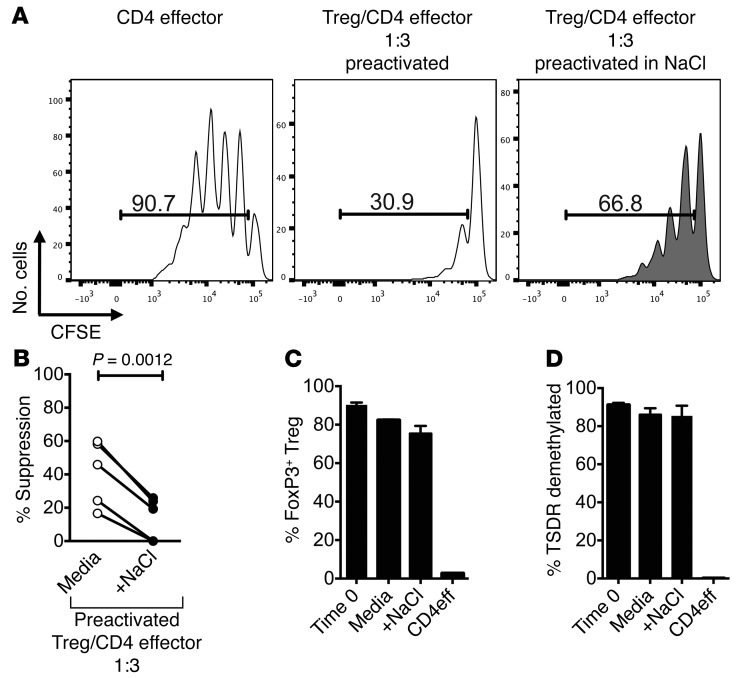
To confirm the direct influence of the high-salt condition on human Tregs and to exclude that the loss of suppression is perhaps mediated indirectly by heightened effector T cell responses, Tregs were activated either in standard media or in high-salt media for 72 hours, as previously reported (40). After the incubation period, Tregs were plated in a suppression assay with the standard NaCl concentration. As activated Tregs are more suppressive than ex vivo isolated Tregs, we used a lower Treg/CD4 effector cell ratio of 1:3 (40). Tregs preincubated in high-salt conditions similarly lost in vitro suppressor function, demonstrating that a direct effect of high-salt on Tregs is the main contributor to the observed effect (Figure 2, A and B). We next examined whether the high-salt conditions decreased either Foxp3 expression or changes in methylation status of the TSDR (3). There was no significant change in either Foxp3 expression or in the demethylation status of the TSDR region after 72 hours of activation, indicating that neither loss of FOXP3 expression nor loss of TSDR demethylation are accounting for the observed loss of suppression (Figure 2, C and D). There was also no observed change in cell viability (Supplemental Figure 6, A and B).
Figure 2. High-salt conditions directly influence Treg function.
(A) Tregs were activated with plate-bound αCD3 and soluble αCD28 at 1 μg/ml either in normal media (media, white) or media enriched with 40 mM NaCl (gray) for 72 hours with IL-2 (25 U/ml). Following incubation, Tregs were washed, counted, and plated in a suppression assay in normal media with freshly sorted allogeneic CD4 effector cells stained with CFSE. CFSE dilution was measured via flow cytometry after 5 days. (B) Line graph depicts a summary of experiments (n = 5). At onset of preactivation and following 72-hour preactivation, an aliquot of Tregs was analyzed. (C and D) We confirmed that Foxp3 expression (C) (n = 5) and percent demethylation of the TSDR (D) (n = 3) did not change with activation and that these are human Tregs. Statistical analyses were performed using paired Student’s t test.
High-salt promotes a Th1-type effector phenotype in Tregs.
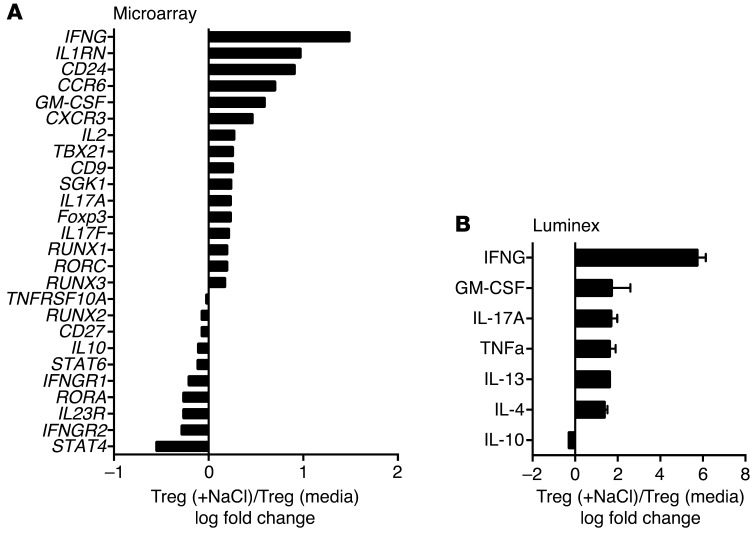
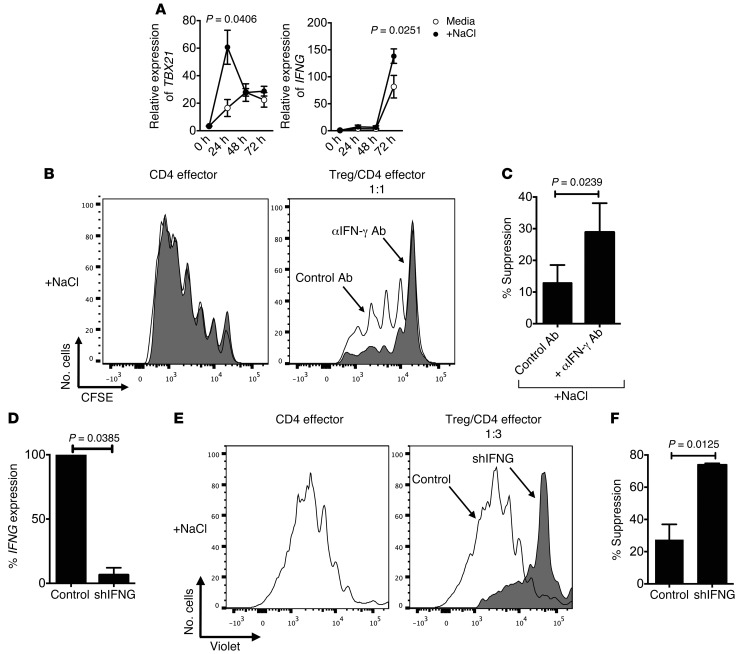
To examine the mechanism for the loss of Treg function and increased proliferation, we performed a microarray analysis of Tregs under standard and high-salt conditions. Purified Tregs were activated with anti-CD3/CD28 in the presence of IL-2 for 72 hours, and mRNA expression was determined using Affymetrix gene chip analysis (Figure 3A). The expression analysis indicated a shift toward a Th1-type proinflammatory Treg phenotype with a dominant increase in IFNG, TBX21, and CXCR3 expression. We also detected increases in Th17-related mRNA expression — including RORC, CCR6, IL17A, and IL17F — in line with what we previously observed in naive CD4 cells cultured under Th17 conditions in the presence of high salt (24, 25), although to much lower levels compared with IFNG. As SGK1 is a central node associated with induction of the Th17 CD4 program by high salt, it was of interest that this kinase was also induced in Tregs under similar conditions (Figure 3A).
Figure 3. High-salt conditions induce a Th1-type phenotype.
(A) Tregs were stimulated for 72 hours with plate-bound αCD3 and soluble αCD28 at 1 μg/ml with IL-2 (25 U/ml) either in the presence (+NaCl) or absence (media) of an additional 40 mM NaCl prior to being analyzed via Affymetrix array (U133 Plus 2.0) (n = 2). (B) Supernatants from activated Tregs either in normal media (media) or in media enriched with 40 mM NaCl were harvested and assayed via Luminex. Fold change was calculated based on +NaCl condition (n = 4).
To analyze protein expression of cytokines, we performed a Luminex assay on Treg culture supernatants induced under standard and high-salt conditions. Six-fold increases in secreted IFNγ were detected, thereby confirming the observed dominant changes of IFNG on mRNA levels (Figure 3B). Of note, in line with the expression data, the increase in IFNγ production was more pronounced than that of IL-17. Analysis of surface markers associated with suppression — such as CTLA4, GITR, PD-1, and LAP-TGFβ — did not reveal any significant high-salt–mediated changes in the Treg population (data not shown). Together, these data suggest that Tregs exposed to high-salt conditions are predominantly adopting a Th1-type effector phenotype, as evidenced by increased IFNγ, TBX21, and CXCR3 expression.
High-salt diet impairs the suppressive function of Tregs in vivo.
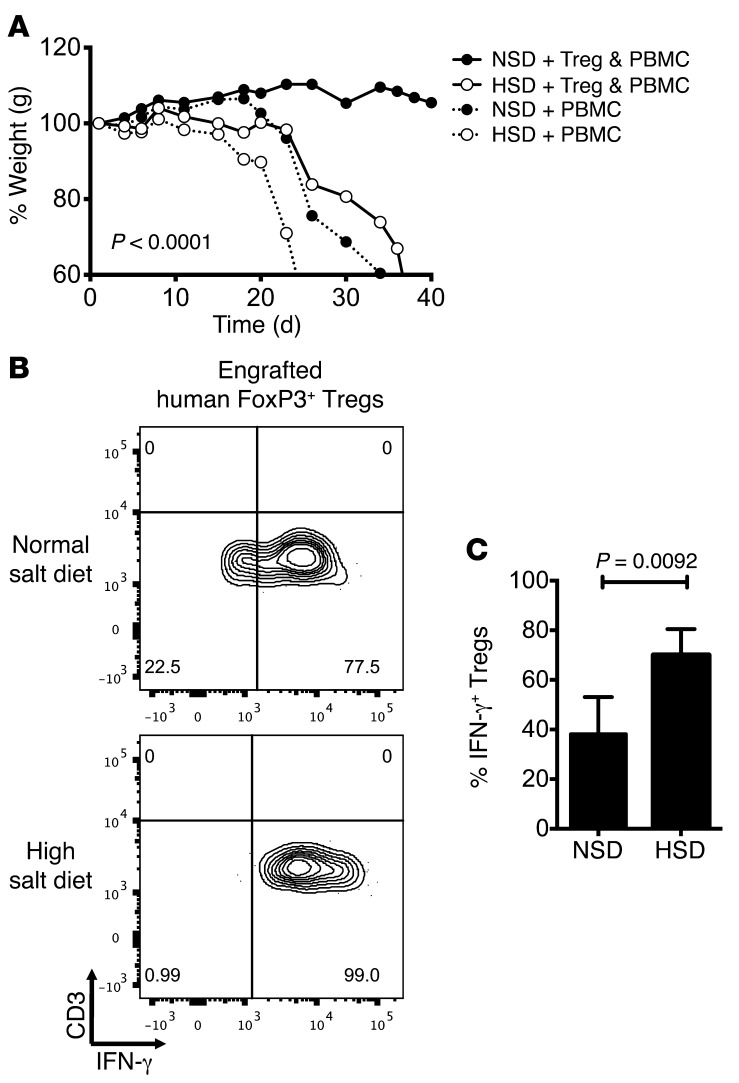
The significant in vitro effects of high-salt conditions on Treg function and IFNγ production prompted us to study the effects of increased dietary salt in vivo. We first used a model of acute x-GvHD to directly study human Treg functionality in an in vivo setting (41). Immune-deficient NOD-scid IL2Rgnull (NSG) mice were fed either a normal salt diet (NSD), containing 0.4% of NaCl or a high-salt diet, containing 8% NaCl and 1% of NaCl in the drinking water for 2 weeks prior to x-GvHD being induced through adoptive transfer of CD25-depleted human peripheral blood mononuclear cells (PBMCs). In this model, development of acute x-GvHD could be followed by weight loss and measurements of the general appearance in animals. Disease severity could be significantly delayed or prevented by the cotransfer of syngeneic human Tregs (41, 42). Animals on an NSD cotransferred with enriched Tregs and CD25-depleted PBMCs (Supplemental Figure 7A) had delayed onset of disease and were symptom free following a 40-day time course, whereas their high-salt diet counterparts developed significant disease (Figure 4A). Mice on a high-salt diet receiving only human PBMCs developed a more severe x-GvHD with earlier onset in comparison to those animals on normal diet (Figure 4A). ANOVA analysis between the NSD and high-salt diet groups receiving either Tregs and PBMCs, or PBMCs alone, revealed significance across subsets. The high-salt diet did not induce weight changes in animals that received no human cells compared with animals on an NSD (Supplemental Figure 7B). Analysis of total splenocytes revealed an increase in proinflammatory cytokines in the CD3+CD8– subset, and animals transferred only with CD25-depleted PBMC showed no significant induction of FOXP3-expressing cells (data not shown). Notably, engrafted human Tregs (CD3+CD8–Foxp3bright) isolated from spleens of animals receiving high-salt diets produced significantly higher levels of IFNγ in comparison to Tregs from NSD animals (Figure 4, B and C).
Figure 4. High-salt intake inhibits Treg function in vivo.
Six- to 10-week-old NSG mice received normal chow and tap water ad libitum (NSD) or sodium-rich chow containing 8% NaCl and tap water containing 1% NaCl ad libitum (high-salt diet, HSD). Animals were put on HSD 2 weeks prior to engraftment of either CD25– human PBMCs alone or Tregs and PBMCs. (A) Length of time to development of GvHD was observed in animals as proxy for Treg function and annotated by percent weight across donor-matched HSD and NSD animals (n = 8 per diet and condition). (B) At time of development of GvHD symptoms, NSD mice and HSD mice were sacrificed in diet-matched and donor-matched pairings. Splenocytes were isolated prior to being analyzed by FACS. Cells were stained for viability, CD3, CD8, Foxp3, and IFNγ. Tregs were gated as CD3+CD8–Foxp3bright. (C) Intracellular protein levels of IFNγ were calculated and depicted across donor-matched animals (n = 5 per diet and condition). Statistical analyses were performed using one-way ANOVA between respective dietary and adoptive transfer groups. A paired Student’s t test was used for FACS analyses.
In order to evaluate the function of murine Tregs under high-salt conditions, an in vitro suppression assay was used; GFP+ natural Tregs were isolated from Foxp3-GFP reporter mice and cultured with or without 40 mM NaCl in the presence of anti-CD3 and anti-CD28 for 3 days, and then cultured in suppression assays for 5 days. In line with the human data, murine Tregs activated in high-salt conditions exhibited lower suppressive capacity and were less efficient in inhibiting the proliferation of WT naive T effector cells (Supplemental Figure 8A). Given these findings, we placed Foxp3-GFP reporter mice on a high-salt diet for 3 weeks prior to harvesting Tregs from the lamina propria and mesenteric lymph nodes and analyzed those Tregs for gene expression. In both regions, we observed an increase in Sgk1 and Ifng, whereas Tbx21 expression was only significantly upregulated in Tregs isolated from the lamina propria in animals on a high-salt diet (Supplemental Figure 9). This data is in line with our previous findings on high-salt–inducing Th17 cells (25) and suggests that the primary site of action for inducing this phenotype in vivo is the small intestine. Since murine Tregs seemed to act similarly to human Tregs, this enabled us to directly test the impact of high-salt conditions on murine Tregs in an experimental animal model in vivo. The compromised suppressive function of murine Tregs exposed to high-salt was confirmed in a T cell cotransfer colitis model. Either natural (Foxp3+/GFP+) Tregs preactivated in high-salt conditions or preactivated under standard media conditions were transferred together with WT CD4+CD62L+CD44– naive T cells (as in Supplemental Figure 8A) at a ratio of 1:5 into Rag2–/– mice. Natural Tregs activated under standard conditions efficiently suppressed the development of colitis as measured by weight changes, while natural Tregs exposed to high-salt conditions showed reduced protection (Supplemental Figure 8B). Together, these data suggest that high-salt conditions decreased Treg suppressive function both in vitro and in vivo. Consistent with our in vitro data, the combined observations indicate that exposure to high-salt conditions causes functional loss in the Treg population and results in initiation of a Th1-type effector-like signature.
Neutralizing IFNγ recovers suppressive function under high-salt conditions.
Given our findings in vitro and in vivo, it was of interest to determine the mechanism by which the high-salt conditions led to increases in IFNγ with loss of suppressor function. To elucidate transcriptional changes across time, we performed a kinetic analysis and observed that most prominent increases in TBX21 at 24 hours precede a rise in IFNG expression at 72 hours (Figure 5A). It was demonstrated before that loss of in vitro function of Th1-type Tregs is mediated by IFNγ and could be restored by neutralizing IFNγ (13). In order to examine whether increased IFNγ levels similarly contribute to high-salt–mediated functional loss of suppression, we neutralized IFNγ within the coculture system. Notably, a human IFNγ neutralizing antibody added at the start of the coculture assay resulted in significant recovery of suppression under high-salt conditions (Figure 5, B and C). To confirm our findings, we added recombinant IFNγ to the start of the culture and observed that it resulted in a deficit in suppressive function similar to that observed secondary to high salt (data not shown). Given these observations, we utilized IFNG-specific shRNA (shIFNG) to directly target IFNG expression in Tregs. Lentiviral particles containing constructs of GFP-shRNA specific for IFNG were added to activated Tregs that were then sorted based on viability and GFP expression prior to being plated in a suppression assay with high-salt media. Transduction efficiency was controlled by GFP expression (data not shown), and IFNG knockdown was confirmed via quantitative PCR (qPCR) (Figure 5D). Silencing of IFNG in Tregs resulted in recovery of functional suppression in the setting of high-salt (Figure 5, E and F).
Figure 5. Neutralizing IFNγ restores Treg suppression in high-salt conditions.
(A) Tregs were stimulated for 24 hours, 48 hours, and 72 hours with plate-bound αCD3 and soluble αCD28 at 1 μg/ml respectively with IL-2 (25 U/ml) either in the presence (+NaCl) or absence (media) of an additional 40 mM NaCl prior to being analyzed via qPCR for changes in TBet (TBX21) and IFNG (n = 5). (B) CD4 effector cells were labeled with CFSE, stimulated with αCD2/αCD3/αCD28-coated beads, and cultured alone or cocultured with Tregs. A neutralizing monoclonal antibody for IFNγ was added to the start of the culture at onset at 10 μg/ml (αIFNγ Ab, gray). A normal goat IgG Control was added to the start of culture to account for any nonspecific changes. CFSE dilution was measured by flow cytometry after 5 days in both cell populations. (C) The bar graph depicts a summary of independent experiments (n = 5). Statistical analyses were performed using paired Student’s t test. (D–F) Tregs were stimulated overnight with plate-bound αCD3 (1 μg/ml) and soluble αCD28 (1 μg/ml). The following day, a GFP-tagged viral construct containing shRNA specific for IFNG (shIFNG, gray) and a nontarget viral (control, white) construct were added to the culture at MOI = 5 with IL-2 (25 U/ml). The culture was left for 5 days, and transduced live cells were sorted by GFP+ and propidium iodide. IFNG knockdown was confirmed via qPCR (D). Transduced GFP+ Tregs were cultured in a CFSE suppression assay at a Treg/CD4 effector cell ratio of 1:3. CFSE dilution was measured by flow cytometry after 5 days (E). The bar graphs depict a summary of experiments demonstrating recovery of suppression in shIFNG in relation to control Tregs (n = 4) (F).
High-salt–induced Treg dysfunction is dependent on SGK1.
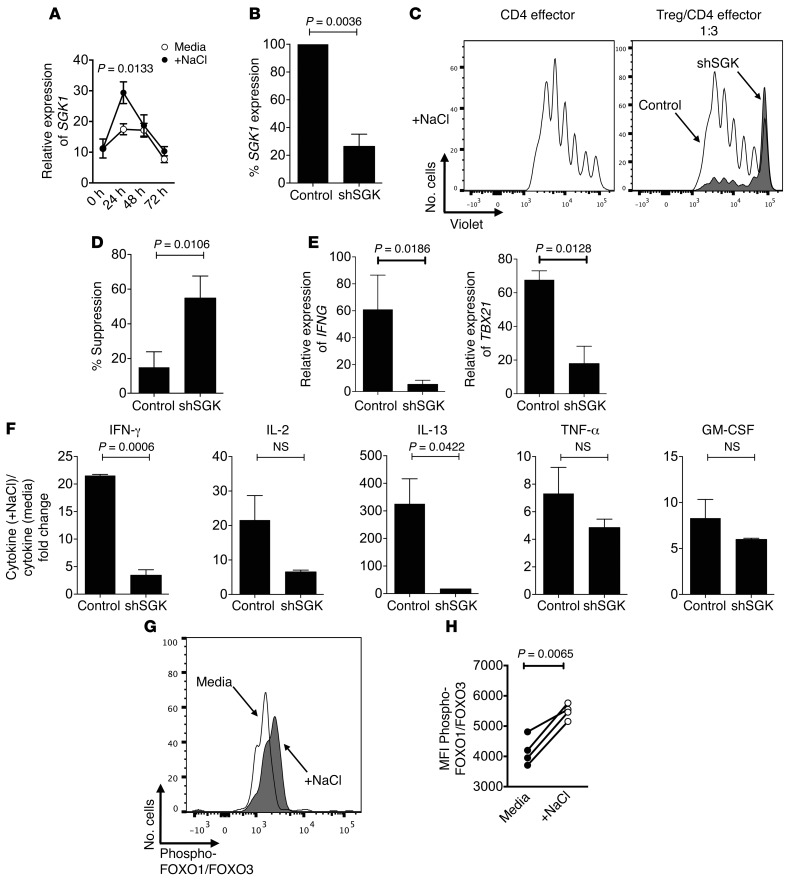
Because SGK1 is a central node in high-salt–mediated Th17 induction, it was of interest to study whether it is responsible for high-salt–mediated loss of suppression in Tregs (25, 33, 34). In line with published data, changes in SGK1 expression as an immediate early response gene followed a similar kinetic profile compared with TBX21 expression, and we observed a more prominent increase in SGK1 at 24 hours over a 72-hour kinetic profile (Figure 6A) (25). Pharmacologic blockade of SGK1 resulted in recovery of suppressive function in Tregs exposed to high-salt conditions (Supplemental Figure 10, A and B). Furthermore, we then utilized SGK1-specific shRNA to directly silence SGK1 expression in Tregs. Silencing of SGK1 recovered Treg suppression in the high-salt condition (Figure 6, B–D). Of note, shRNA-SGK1–transduced Tregs expressed less IFNG and TBX21 than those transduced with the control vector, suggesting that induction of a Th1–type program and suppressive activity under high-salt conditions depends on SGK1 (Figure 6E). Luminex analysis of supernatants of the coculture confirmed that the knockdown of SGK1 in Tregs efficiently restored suppression and blocked cytokine secretion (Figure 6F). Thus, SGK1 silencing both recovered Treg suppressive capacity in high-salt conditions and decreased IFNγ secretion.
Figure 6. High-salt–induced Treg dysfunction is dependent on SGK1.
(A) Tregs were stimulated for 24 hours, 48 hours, and 72 hours with plate-bound αCD3 and soluble αCD28 at 1 μg/ml with IL-2 (25 U/ml) either in the presence (+NaCl) or absence (media) of an additional 40 mM NaCl prior to being analyzed via qPCR for changes in SGK1 (n = 5). (B–F) Tregs were stimulated overnight with plate-bound αCD3 (1 μg/ml) and soluble αCD28 (1 μg/ml). The following day, a GFP-tagged viral construct containing shRNA specific for SGK1 (shSGK, gray) and a nontarget viral (control, white) construct were added to the culture at MOI = 5 with IL-2 (25 U/ml). The culture was left for 5 days, and transduced live cells were sorted by GFP+ and propidium iodide. (B) SGK1 knockdown was confirmed via qPCR. (C) Transduced GFP+ Tregs were cultured in a CFSE suppression assay at a Treg/CD4 effector cell ratio of 1:3. CFSE dilution was measured by flow cytometry after 5 days. (D) The bar graphs depict a summary of experiments demonstrating recovery of suppression in shSGK in relation to control Tregs (n = 3). (E) Transduced Tregs were stimulated for 4 hours with PMA and Ionomycin prior to analyzing changes in SGK1, IFNG, and TBX21 expression via qPCR (n = 3). (F) Luminex analysis of coculture supernatants. Bar graph depicts fold change in cytokine production (n = 4). (G) Tregs were activated with αCD2/αCD3/αCD28-coated beads (at 2 beads/cell) for 4 hours either in the presence (+NaCl) or absence (media) of an additional 40 mM NaCl prior to being harvested, fixed, and made permeable. Phosphorylation analysis was conducted by staining for phospho-FOXO1/FOXO3 and ascertaining changes via flow cytometry. (H) Graph depicts a summary of changes in phosphorylation (n = 4). Statistical analyses were performed using paired Student’s t test.
Downstream targets of SGK1 include FOXO1, which — in conjunction with FOXO3 — stabilizes the Foxp3 locus (25, 33, 43). Given this observation, we examined whether increases in phosphorylation of FOXO1/FOXO3 occurred following exposure to high-salt conditions in vitro. We observed an increase in phosphorylation of FOXO1/FOXO3 with high-salt conditions, implicating SGK1 engagement and suggesting that changes in Foxp3 stability may be an underlying mechanism for loss in Treg function (Figure 6, G and H). Overall, these findings identify parallel mechanisms between salt-mediated changes in Treg functional regulation and proinflammatory induction of CD4+ lymphocytes, though further experiments are necessary to directly implicate a role for FOXO1/FOXO3 phosphorylation in salt-mediated loss of Treg suppression.
Discussion
SGK1 is a key nodal point in a number of immunologically relevant systems pertaining to proinflammatory cascades and regulatory mechanisms. Here, we demonstrate that engagement of the SGK1 axis by elevated NaCl in CD4+CD25hiCD127loFoxp3+ Tregs results in functional loss of suppression. Exposing Tregs to standard media enriched with physiologically relevant 40 mM NaCl results in a dysregulated nonsuppressive phenotype. This phenotype is salt-specific and is accompanied by dominant initiation of a Th1-type signature both in vitro and in vivo, with increased mRNA expression of IFNG, TBX21, and CXCR3, in addition to marked increases in protein levels of IFNγ (44). Silencing of SGK1 in Tregs resulted in recovery of suppressive function in high-salt conditions, in addition to attenuation of the Th1-type phenotype. Moreover, blocking IFNγ either via neutralizing antibody within the coculture system or via shIFNG recovered suppressor function. High-salt diets in vivo similarly induce IFNγ-secreting Tregs with associated losses of function. These data link a newly discovered environmental risk factor with a central immunologic dysfunction associated with human autoimmune diseases.
We and others have shown that Tregs can exhibit plasticity and are able to adopt various functional signatures (8, 11, 13). That is, Tregs can acquire Th1-type, Th2-type, and Th17-type phenotypes marked by effector-like functions (11). The loss of in vitro Treg function is restored in IL-12–induced Th1-type Tregs by neutralizing IFNγ (13). Similarly, in high-salt conditions, downregulating IFNG expression also restores suppressive function to Tregs. While Tregs exposed to high-salt conditions have a slight increase in expression of IL17a, IL17f, RORC, and CCR6, neutralizing IL-17 in the coculture setting did not restore their suppressive function (data not shown). Moreover, silencing SGK1 did not change IL-17 production in the coculture, yet it resulted in decreases in IFNG expression, suggesting that the Th1-type phenotype is dominant in the high-salt Treg population. While there were only modest increases in IL-13 under high-salt conditions, it was of interest that SGK1 shRNA knockdown decreased IL-13 secretion, as well. However, this decrease in IL-13 does not appear to be related to loss of Treg function. Thus, these data suggest that the Th1-type signature is dominant in Tregs exposed to a high-salt condition.
Th1-type Tregs expressing high levels of TBX21, IFNγ, and the surface marker CXCR3 have been identified in numerous disease settings, including colitis, infection, and autoimmune disease (13, 45–47). Tregs isolated ex vivo from patients with MS and T1D secrete more IFNγ and possess a functionally impaired phenotype; they do not suppress as effectively as Tregs isolated from healthy individuals (13, 14). Neutralizing IFNγ restores suppressive capacity to Tregs isolated from MS patients, indicating a central role for IFNγ in mediating loss of Treg function. Both Tregs isolated from MS patients and Tregs exposed to high-salt conditions have loss of suppressive function with no significant changes in methylation of the FOXP3 locus, FOXP3 protein, or mRNA expression (13). Increases in Treg proliferation under high-salt conditions further suggest that Tregs can adopt specific phenotypes and effector-like functions. That is, high-salt conditions induced IFNγ+-secreting Tregs that more rapidly enter cell cycle, indicating a memory- and effector-like phenotype as evidenced by loss of anergy, increased growth, enhanced cytokine secretion, and decreased suppressive function (48).
Having observed that exposure to high salt in vitro induced IFNγ in Tregs, it was important to determine whether a high-salt diet in vivo similarly induced the same phenotype with functional loss of suppression, considering the potential influence of dietary salt in the gut and immune function. Thus, we examined both a x-GvHD model that is induced in immune-deficient mice through the transfer of human CD25– PBMCs and an adoptive transfer colitis model induced by the transfer of murine WT T cells in immune-deficient mice (41, 42, 49). Under normal dietary conditions, the addition of human CD25– PBMCs alone results in x-GvHD, and coadministration of autologous human Tregs (CD25+ PBMCs) markedly decreases disease onset (41, 50). However, in those animals exposed to a high-salt diet, the human Tregs were not able to halt onset of GvHD. Similar to what we observed under high-salt conditions in vitro, Foxp3bright Tregs in the humanized x-GvHD model exposed to a high-salt diet in vivo secreted higher amounts of IFNγ in comparison to those fed an NSD. In experimental colitis, Tregs exposed in vitro to high-salt conditions were unable to suppress development of transfer-colitis, compared with Tregs not exposed to high-salt conditions. Moreover, when Tregs are isolated ex vivo from the lamina propria of mice fed a high-salt diet for 3 weeks, they exhibit higher levels of Sgk1, Ifng, and Tbx21 than those animals on an NSD. Thus, we observed that a high-salt diet worsened the severity and onset of disease in the humanized x-GvHD model and in the murine adoptive transfer colitis models, with the site of action likely being the small intestine. These data provide evidence that our in vitro experiments model the in vivo administration of a high-salt diet.
We examined the mechanism by which high-salt conditions mediate loss of Treg function. SGK1 activation leads to downstream phosphorylation of FOXO1, which alongside FOXO3 are known placeholders of the Foxp3 locus (43). Thus, NaCl-induced activation and phosphorylation of FOXO1/FOXO3 by SGK1 could lead to their degradation and may affect Foxp3 stability (25). Overall, our data raise the question as to whether the IFNγ+ Treg signature observed in autoimmune disease may be mediated environmentally in a genetically susceptible host via exposure to high dietary salt conditions.
Large parts of the industrialized world currently consume a diet rich in processed foods that contains high amounts of salt, sugar, fat, and cholesterol (31, 51). This Western diet has widespread implications in renal, cardiovascular, and endocrine homeostasis. SGK1 has been established as a key response element in these systems. Changes in SGK1 function mediate adverse physiologic events through monitoring sodium transport, salt maintenance, and osmotic responses (52, 53). Although, SGK1 has not been directly studied in human autoimmune disease, investigations in the EAE model demonstrated that SGK1–/– mice have a diminished inflammatory response and attenuated induction of the disease (25). Our findings support that this may be due to heightened Treg suppressive function in the setting of decreased Th17 cell induction (24, 25).
Gastrointestinal changes in salt concentrations and sodium storage in muscle, skin, and organs without commensurate water retention may explain how increases in dietary salt are able to sustain long-term influences on the immune system, given effective clearance of excess salt through the renal system (54). Within the gastrointestinal tract, increased exposure to dietary salt causes an increased inflammatory milieu (25). Dietary changes may also influence both the function and composition of the gut microbiome, which in turn could impact both innate and adaptive immunity through inducing increases in inflammatory cells while causing loss of Treg function (55, 56). Moreover, it has been reported that caloric and dietary shifts manipulate the architecture and absorptive capacity of the gut epithelium, which may influence immune function via prolonged exposure to metabolites (57–59). Recent studies have also demonstrated that sodium retention can occur in skin, muscle, and organs without added water retention or gross changes (60). Buffering of sodium occurs via macrophages but also by CD4+ cells present in these microenvironments, further demonstrating the relationship between CD4 cells and salt balance (26, 61).
In summary, we identify a mechanism by which engagement of SGK1, secondary to increased sodium chloride exposure, results in a Th1-type effector signature in Foxp3+ Tregs. This phenotype is marked by loss of suppressor function, increased proliferation, and increased expression of IFNG, TBX21, and CXCR3. shRNA-mediated knockdown of IFNG and SGK1 recovers suppressor function. Utilizing a humanized mouse model of x-GvHD and a murine model of experimental colitis, we demonstrate that feeding a high-salt diet worsens disease marked by increases in IFNγ and nonfunctional Tregs. Our findings add further evidence that control of dietary salt intake may be efficacious in patients with autoimmune disease. In this regard, we have begun clinical trials in patients with MS to investigate whether inflammation can be attenuated by manipulating dietary salt intake. Finally, these findings mark SGK1 as a potential therapeutic target for autoimmune disease in human Tregs.
Methods
CD4+ T cell isolation.
Peripheral blood was obtained from healthy control volunteers. PBMCs were separated by Ficoll-Paque PLUS (GE Healthcare) gradient centrifugation. Untouched total CD4+ T cells were isolated from PBMCs by negative selection via the CD4+ T cell isolation kit (StemCell Technologies Inc.). CD25-enriched and -depleted CD4+ cells were isolated via CD25 microbeads II (Miltenyi Biotec). Tregs and naive effector T cells (CD4 effector cells) were then sorted by high-speed flow cytometry with a FACS Aria (BD Biosciences) to a purity >98% as verified by post-sort analysis using CD4 PerCP-Cy5.5 (clone RPA-T4), CD25 PE (clone M-A251), CD127 Alexa647 (clone HIL-7R-M21), CD45RA V500 (clone H100), and CD45RO APC-H7 (clone UCHL1) (BD Biosciences). Cells were isolated with the following parameters: Tregs, CD4+CD25hiCD127low; CD4 effector, CD4+CD25–CD45RA+CD45RO–.
Suppression assay.
Naive CD4+ T effectors (5 × 105) were stained with CellTrace CFSE (CD4 effector cells) at 2.5 μM and cultured with Tregs (5 × 105, titrated down as described) in 96-well round-bottom plates (Corning Inc.) in X-VIVO15 medium (BioWhittaker Inc.) with 5% human serum (Gemini Bio-Products). Where specified, Tregs were stained with CellTrace Violet (ThermoFisher Scientific) at 2.5 μM to detect proliferation in the coculture system. Cells were stimulated with Treg Inspector Beads (αCD2/αCD3/αCD28-coated beads at 2 beads/cell (Miltenyi Biotec) and left in culture for 5 days before being analyzed by FACS. Media was supplemented with an additional 40 mM NaCl or 80 mM Mannitol (both from Sigma-Aldrich) as indicated.
Blocking antibodies/pharmacologic inhibitors.
A neutralizing monoclonal antibody for IFNγ was added to the start of the culture at onset (AF-285-NA, R&D Systems) at 10 μg/ml (13). A normal goat IgG control was also added to the start of culture (AB-108-C, R&D Systems) to account for any nonspecific changes. Where indicated, a pharmacologic inhibitor for SGK1 (GSK650394, Tocris) was added to the start of the culture at 1 μM. A DMSO control was added to account for any nonspecific changes.
High-salt activation of Tregs.
Tregs (1 × 105/well) were stimulated in X-VIVO15 medium (BioWhittaker Inc.) with 5% human serum (Gemini Bio-Products) for 72 hours with plate-bound αCD3 (1 μg/ml), soluble αCD28 (1 μg/ml), and IL-2 (25 U/ml) in the presence or absence of an additional 40 mM NaCl. Following the 72-hour culture, Tregs were removed, washed, and plated in a suppression assay with allogeneic CD4 effector cells for 5 days to ascertain salt specific influences on the Treg population.
shRNA-mediated gene silencing.
Tregs and CD4 effector cells (1 × 105/well) were stimulated in X-VIVO15 medium (BioWhittaker Inc.) with 5% human serum (Gemini Bio-Products) overnight with plate-bound αCD3 (1 μg/ml) and soluble αCD28 (1 μg/ml). The following day, a GFP-tagged viral construct containing shRNA specific for IFNG (TRCN0000058499) or SGK1 (TRCN0000040175) and a control viral construct were added to the culture at multiplicity of infection equal to 5 with IL-2 (25 U/ml) (24). Media with IL-2 were replenished following 3 days, and was culture left for 5 days, at which point transduced live cells were sorted by GFP+ and prodidium iodide. Transduced cells were then either cultured in a CFSE suppression assay for 5 days with allogeneic CD4 effector cells or stimulated with phorbal myristate acetate (PMA, 50 ng/ml) and ionomycin (250 ng/ml; both from Sigma-Aldrich) for 4–5 hours prior to qPCR analysis.
Flow cytometry.
Cells were analyzed by flow cytometry if not specified elsewhere after a culture period of 5 days. Cells were first stained with the LIVE/DEAD cell kit (Invitrogen) to exclude dead cells. For surface staining, cells were stained with the respective antibodies for 20 minutes in PBS. For intracellular staining, cells were fixed and made permeable (Fix/Perm, eBioscience) according to the manufacturer’s instructions, and stained with Foxp3–Alexa Fluor 700, PE-Cyanine 7 (PCH101, eBioscience), or IFNγ–PE-Cyanine 7 (4S.B3, eBioscience) for intracellular Foxp3 expression and cytokine detection for 30–45 minutes. Data were acquired on a FACS Fortessa or LSRII (BD Biosciences) and analyzed with FlowJo software (TreeStar).
Phosphorylation analysis.
Tregs were stimulated with Treg Inspector Beads (αCD2/αCD3/αCD28-coated beads) at 2 beads/cell (Miltenyi Biotec) and left in culture for 4 hours prior to being harvested. Fixation (Fixation Buffer, BD Biosciences) and methanol permeabilization (Perm Buffer III, BD Biosciences) was performed following manufacturer’s instructions. A primary antibody for Phospho-FOXO1 (Thr24)/FOXO3a (Thr32) (9464, Cell Signaling Technology) and a secondary antibody for Fluorescein (FITC) anti-rabbit were used (Jackson ImmunoResearch Laboratories Inc.) to detect changes in phosphorylation of FOXO1/FOXO3 via flow cytometry. Data were acquired on a FACS Fortessa (BD Biosciences) and analyzed with FlowJo software.
qPCR.
Cells for RNA isolation were harvested following a 5-day culture if not specified elsewhere, and RNA was isolated using the Absolutely RNA 96 Microprep kit (Agilent Technologies) or RNeasy micro kit (QIAGEN) and converted to cDNA via reverse transcriptase by random hexamers and Multiscribe RT (TaqMan Gold RT–PCR kit, Applied Biosystems). All primers were purchased from Applied Biosystems (IFNG–Hs00982921_m1, SGK1–Hs00178612_m1, TBX21–Hs00203436_m1, FOXP3–Hs01085834_m1, B2M–4326319E, Gapdh–Mm03302249_g1, Sgk1–Mm00441387_g1, Tbx21–Mm00450960_m1, and Ifng–Mm01168134_m1). All reactions were performed on a StepOnePlus Real-Time PCR System (Applied Biosystems). The values are represented as the difference in CT values normalized to β2-microglobulin for each sample as per the following formula: relative RNA expression = (2–ΔCt) × 103.
Affymetrix microarray.
RNA was isolated from cells as indicated above. Samples were prepared for Affymetrix gene arrays (U133 Plus 2.0) as per manufacturer’s instructions and system. RNA samples were amplified using the NuGEN Ovation Pico WTA System prior to running gene array. These data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE63327 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE63327).
FOXP3 methylation analysis.
Genomic DNA was isolated from sorted Treg (CD4+CD25+CD127–) and CD4 effector cell populations, and DNA methylation analysis of the FOXP3 locus was handled by Epiontis.
Luminex.
Multiplex assays for cytokine secretion were performed on supernatants from stimulated Tregs, CD4 effector cells, and coculture CFSE assays using Milliplex Human Th17 Cytokine/Chemokine Magnetic Bead Panel (Millipore).
Mice; x-GvHD induction.
Six- to 10-week-old male NSG mice were purchased from The Jackson Laboratory or were bred and housed at the in-house animal care facility of the Yale University under standardized conditions. Mice received normal chow and tap water ad libitum (control group) or sodium-rich chow containing 8% NaCl (TestDiet) and tap water containing 1% NaCl ad libitum (high-salt group). Animals were put on high-salt diets 10–14 days before cell transfer. For induction of x-GvHD, each animal received i.v. either 2.5 × 107 CD25–-depleted PBMC alone or together with 0.5 × 107 CD25+-enriched Tregs as indicated. The weight and clinical symptoms of the mice were determined over the entire period of the experiment; clinical signs of the disease were scored according to weight, and general appearance (ruffled fur, hunched posture) and mobility (impaired movement) was monitored. Engraftment of human PBMCs was analyzed by FACS analysis of murine blood obtained by punctuation of the tail vein or by FACS analysis of spleen cells at different time points. All animal experimentation was performed in accordance to the US animal protection law and Yale University guidelines.
Mice; adoptive transfer colitis, suppression assay.
For suppression assay and sorting, methods were followed as outlined in (62). For T cell-induced colitis, CD4+CD62L+CD44– naive T cells (7 × 105) were injected i.p. with or without Foxp3+ Treg cells (Foxp3-GFP reporter mice) preactivated with 40 mM NaCl (1 × 105) into 6- to 10-week-old male Rag2–/– mice, and weight loss was monitored. Regarding assessment of intestinal inflammation, tissues were graded semiquantitatively from 0–5 in a blinded fashion. A score of 0 indicated no changes observed; 1 indicated minimal scattered mucosal inflammatory cell infiltrates, with or without minimal epithelial hyperplasia; 2 indicated mild scattered to diffuse inflammatory cell infiltrates, sometimes extending into the submucosa and associated with erosions, with minimal to mild epithelial hyperplasia and minimal to mild mucin depletion from goblet cells; 3 indicated mild to moderate inflammatory cell infiltrates that were sometimes transmural, often associated with ulceration, with moderate epithelial hyperplasia and mucin depletion; 4 indicated marked inflammatory cell infiltrates that were often transmural and associated with ulceration, with marked epithelial hyperplasia and mucin depletion; and 5 indicated marked transmural inflammation with severe ulceration and loss of intestinal glands.
Mice; ex vivo qPCR.
GFP+ Treg cells were isolated from the lamina propria and mesenteric lymph nodes of Foxp3-GFP reporter mice. Mice either received normal chow and tap water ad libitum (control group) or sodium-rich chow containing 8% NaCl (TestDiet) and tap water containing 1% NaCl ad libitum (high-salt group) for 3 weeks. Following this 3-week period, Tregs were isolated from the aforementioned tissues and analyzed via qPCR for gene expression.
Statistics.
Statistical analysis was run using Prism (Graph Pad Software). Unless otherwise indicated, standard paired 2-tailed t tests were used for analyses, with P values equal to or less than 0.05 being considered significant. For murine studies, one-way ANOVA analysis was performed. For all figures, error bars show mean ± SEM, unless indicated elsewhere.
Study approval.
All animal studies were performed in accordance with and approved by Yale University and Harvard University animal care protocol and Institutional Review Boards. All human subjects provided informed consent prior to inclusion in this project. All studies were conducted in compliance with Institutional Review Board (Yale University and Harvard University) protocols.
Supplementary Material
Acknowledgments
We thank L. Devine and C. Wang for their assistance with cell sorting and FACS. We thank M. Mitrovic and C. Cotsapas for their assistance with microarray samples. We thank A. Peters for assistance with performing nanostring. This research was supported by grants from the National Institute of Allergy and Infectious Disease (AI045757, AI046130, AI070352, and AI039671), the National Institute of Neurological Disorders and Stroke (NS067305 and F31NS086434), the National Institute of General Medical Sciences (GM093080), the National Multiple Sclerosis Society (CA1061-A-18), the Penates Foundation, and the Nancy Taylor Foundation for Chronic Disease.
Footnotes
Conflict of interest: David A. Hafler has been on Scientific Advisory Boards for the following companies over the past two years: Bristol-Myers Squibb, EMD Serono, Genzyme, Sanofi-Aventis, US Inc., MedImmune, Novartis Pharmaceuticals, Roche, and Teva Neuroscience.
Reference information:J Clin Invest. 2015;125(11):4212–4222. doi:10.1172/JCI81151.
Markus Kleinewietfeld’s present address is: Translational Immunology, Medical Faculty Carl Gustav Carus, TU Dresden, and DFG-Center for Regenerative Therapies Dresden (CRTD), Dresden, Germany.
See the related Commentary beginning on page 4002.
References
- 1.Kleinewietfeld M, Hafler DA. Regulatory T cells in autoimmune neuroinflammation. Immunol Rev. 2014;259(1):231–244. doi: 10.1111/imr.12169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sakaguchi S, et al. Foxp3+ CD25+ CD4+ natural regulatory T cells in dominant self-tolerance and autoimmune disease. Immunol Rev. 2006;212:8–27. doi: 10.1111/j.0105-2896.2006.00427.x. [DOI] [PubMed] [Google Scholar]
- 3.Baron U, et al. DNA demethylation in the human FOXP3 locus discriminates regulatory T cells from activated FOXP3(+) conventional T cells. Eur J Immunol. 2007;37(9):2378–2389. doi: 10.1002/eji.200737594. [DOI] [PubMed] [Google Scholar]
- 4.Ohkura N, Kitagawa Y, Sakaguchi S. Development and maintenance of regulatory T cells. Immunity. 2013;38(3):414–423. doi: 10.1016/j.immuni.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 5.Wing JB, Sakaguchi S. Multiple treg suppressive modules and their adaptability. Front Immunol. 2012;3: doi: 10.3389/fimmu.2012.00178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Viglietta V, Baecher-Allan C, Weiner HL, Hafler DA. Loss of functional suppression by CD4+CD25+ regulatory T cells in patients with multiple sclerosis. J Exp Med. 2004;199(7):971–979. doi: 10.1084/jem.20031579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kumar M, et al. CD4+CD25+FoxP3+ T lymphocytes fail to suppress myelin basic protein-induced proliferation in patients with multiple sclerosis. J Neuroimmunol. 2006;180(1–2):178–184. doi: 10.1016/j.jneuroim.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 8.Beriou G, et al. IL-17-producing human peripheral regulatory T cells retain suppressive function. Blood. 2009;113(18):4240–4249. doi: 10.1182/blood-2008-10-183251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Voo KS, et al. Identification of IL-17-producing FOXP3+ regulatory T cells in humans. Proc Natl Acad Sci U S A. 2009;106(12):4793–4798. doi: 10.1073/pnas.0900408106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koch MA, Tucker-Heard G, Perdue NR, Killebrew JR, Urdahl KB, Campbell DJ. The transcription factor T-bet controls regulatory T cell homeostasis and function during type 1 inflammation. Nat Immunol. 2009;10(6):595–602. doi: 10.1038/ni.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kleinewietfeld M, Hafler DA. The plasticity of human Treg and Th17 cells and its role in autoimmunity. Semin Immunol. 2013;25(4):305–312. doi: 10.1016/j.smim.2013.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sakaguchi S, Miyara M, Costantino CM, Hafler DA. FOXP3+ regulatory T cells in the human immune system. Nat Rev Immunol. 2010;10(7):490–500. doi: 10.1038/nri2785. [DOI] [PubMed] [Google Scholar]
- 13.Dominguez-Villar M, Baecher-Allan CM, Hafler DA. Identification of T helper type 1-like, Foxp3+ regulatory T cells in human autoimmune disease. Nat Med. 2011;17(6):673–675. doi: 10.1038/nm.2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McClymont SA, et al. Plasticity of human regulatory T cells in healthy subjects and patients with type 1 diabetes. J Immunol. 2011;186(7):3918–3926. doi: 10.4049/jimmunol.1003099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wellcome Trust Case Control Consortium. et al. Association scan of 14,500 nonsynonymous SNPs in four diseases identifies autoimmunity variants. Nat Genet. 2007;39(11):1329–1337. doi: 10.1038/ng.2007.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.International Multiple Sclerosis Genetics Consortium. The expanding genetic overlap between multiple sclerosis and type I diabetes. Genes Immun. 2009;10(1):11–14. doi: 10.1038/gene.2008.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cotsapas C, et al. Pervasive sharing of genetic effects in autoimmune disease. PLoS Genet. 2011;7(8): doi: 10.1371/journal.pgen.1002254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller FW, et al. Epidemiology of environmental exposures and human autoimmune diseases: findings from a National Institute of Environmental Health Sciences Expert Panel Workshop. J Autoimmun. 2012;39(4):259–271. doi: 10.1016/j.jaut.2012.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ascherio A, Munger KL. Environmental risk factors for multiple sclerosis. Part II: Noninfectious factors. Ann Neurol. 2007;61(6):504–513. doi: 10.1002/ana.21141. [DOI] [PubMed] [Google Scholar]
- 20.Munger KL, et al. Vitamin D intake and incidence of multiple sclerosis. Neurology. 2004;62(1):60–65. doi: 10.1212/01.WNL.0000101723.79681.38. [DOI] [PubMed] [Google Scholar]
- 21.Arnson Y, Shoenfeld Y, Amital H. Effects of tobacco smoke on immunity, inflammation and autoimmunity. J Autoimmun. 2010;34(3):J258–J265. doi: 10.1016/j.jaut.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 22.de Carvalho JF, Pereira RM, Shoenfeld Y. The mosaic of autoimmunity: the role of environmental factors. Front Biosci (Elite Ed) 2009;1:501–509. doi: 10.2741/e46. [DOI] [PubMed] [Google Scholar]
- 23.Croxford AL, Waisman A, Becher B. Does dietary salt induce autoimmunity? Cell Res. 2013;23(7):872–873. doi: 10.1038/cr.2013.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kleinewietfeld M, et al. Sodium chloride drives autoimmune disease by the induction of pathogenic TH17 cells. Nature. 2013;496(7446):518–522. doi: 10.1038/nature11868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu C, et al. Induction of pathogenic TH17 cells by inducible salt-sensing kinase SGK1. Nature. 2013;496(7446):513–517. doi: 10.1038/nature11984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Machnik A, et al. Macrophages regulate salt-dependent volume and blood pressure by a vascular endothelial growth factor-C-dependent buffering mechanism. Nat Med. 2009;15(5):545–552. doi: 10.1038/nm.1960. [DOI] [PubMed] [Google Scholar]
- 27.Wiig H, et al. Immune cells control skin lymphatic electrolyte homeostasis and blood pressure. J Clin Invest. 2013;123(7):2803–2815. doi: 10.1172/JCI60113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Farez MF, Fiol MP, Gaitan MI, Quintana FJ, Correale J. Sodium intake is associated with increased disease activity in multiple sclerosis. J Neurol Neurosurg Psychiatry. 2015;86(1):26–31. doi: 10.1136/jnnp-2014-307928. [DOI] [PubMed] [Google Scholar]
- 29.Sundstrom B, Johansson I, Rantapaa-Dahlqvist S. Interaction between dietary sodium and smoking increases the risk for rheumatoid arthritis: results from a nested case-control study. Rheumatology (Oxford) 2015;54(3):487–493. doi: 10.1093/rheumatology/keu330. [DOI] [PubMed] [Google Scholar]
- 30.Yi B, et al. Effects of dietary salt levels on monocytic cells and immune responses in healthy human subjects: a longitudinal study. Transl Res. 2015;166(1):103–110. doi: 10.1016/j.trsl.2014.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Manzel A, Muller DN, Hafler DA, Erdman SE, Linker RA, Kleinewietfeld M. Role of “Western diet” in inflammatory autoimmune diseases. Curr Allergy Asthma Rep. 2014;14(1): doi: 10.1007/s11882-013-0404-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lang F, Shumilina E. Regulation of ion channels by the serum- and glucocorticoid-inducible kinase SGK1. FASEB J. 2013;27(1):3–12. doi: 10.1096/fj.12-218230. [DOI] [PubMed] [Google Scholar]
- 33.Binger KJ, Linker RA, Muller DN, Kleinewietfeld M. Sodium chloride, SGK1, Th17 activation. Pfluger Arch. 2015;467(3):543–550. doi: 10.1007/s00424-014-1659-z. [DOI] [PubMed] [Google Scholar]
- 34.Yosef N, et al. Dynamic regulatory network controlling TH17 cell differentiation. Nature. 2013;496(7446):461–468. doi: 10.1038/nature11981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heikamp EB, et al. The AGC kinase SGK1 regulates TH1 and TH2 differentiation downstream of the mTORC2 complex. Nat Immunol. 2014;15(5):457–464. doi: 10.1038/ni.2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Platten M, et al. Blocking angiotensin-converting enzyme induces potent regulatory T cells and modulates TH1- and TH17-mediated autoimmunity. Proc Natl Acad Sci U S A. 2009;106(35):14948–14953. doi: 10.1073/pnas.0903958106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stegbauer J, et al. Role of the renin-angiotensin system in autoimmune inflammation of the central nervous system. Proc Natl Acad Sci U S A. 2009;106(35):14942–14947. doi: 10.1073/pnas.0903602106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ip WK, Medzhitov R. Macrophages monitor tissue osmolarity and induce inflammatory response through NLRP3 and NLRC4 inflammasome activation. Nat Commun. 2015;6: doi: 10.1038/ncomms7931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jantsch J, et al. Cutaneous Na+ storage strengthens the antimicrobial barrier function of the skin and boosts macrophage-driven host defense. Cell Metab. 2015;21(3):493–501. doi: 10.1016/j.cmet.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gautron AS, Dominguez-Villar M, de Marcken M, Hafler DA. Enhanced suppressor function of TIM-3 FoxP3 regulatory T cells. Eur J Immunol. 2014;44(9):2703–2711. doi: 10.1002/eji.201344392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mutis T, et al. Human regulatory T cells control xenogeneic graft-versus-host disease induced by autologous T cells in RAG2–/–γc–/– immunodeficient mice. Clin Cancer Res. 2006;12(18):5520–5525. doi: 10.1158/1078-0432.CCR-06-0035. [DOI] [PubMed] [Google Scholar]
- 42.Kleinewietfeld M, et al. CD49d provides access to “untouched” human Foxp3+ Treg free of contaminating effector cells. Blood. 2009;113(4):827–836. doi: 10.1182/blood-2008-04-150524. [DOI] [PubMed] [Google Scholar]
- 43.Ouyang W, Beckett O, Ma Q, Paik JH, DePinho RA, Li MO. Foxo proteins cooperatively control the differentiation of Foxp3+ regulatory T cells. Nat Immunol. 2010;11(7):618–627. doi: 10.1038/ni.1884. [DOI] [PubMed] [Google Scholar]
- 44.Antonelli A, Ferrari SM, Giuggioli D, Ferrannini E, Ferri C, Fallahi P. Chemokine (C-X-C motif) ligand (CXCL)10 in autoimmune diseases. Autoimmun Rev. 2014;13(3):272–280. doi: 10.1016/j.autrev.2013.10.010. [DOI] [PubMed] [Google Scholar]
- 45.Feng T, Cao AT, Weaver CT, Elson CO, Cong Y. Interleukin-12 converts Foxp3+ regulatory T cells to interferon-gamma-producing Foxp3+ T cells that inhibit colitis. Gastroenterology. 2011;140(7):2031–2043. doi: 10.1053/j.gastro.2011.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Oldenhove G, et al. Decrease of Foxp3+ Treg cell number and acquisition of effector cell phenotype during lethal infection. Immunity. 2009;31(5):772–786. doi: 10.1016/j.immuni.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Astier AL, Meiffren G, Freeman S, Hafler DA. Alterations in CD46-mediated Tr1 regulatory T cells in patients with multiple sclerosis. J Clin Invest. 2006;116(12):3252–3257. doi: 10.1172/JCI29251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang R, Green DR. Metabolic checkpoints in activated T cells. Nat Immunol. 2012;13(10):907–915. doi: 10.1038/ni.2386. [DOI] [PubMed] [Google Scholar]
- 49.Asseman C, Mauze S, Leach MW, Coffman RL, Powrie F. An essential role for interleukin 10 in the function of regulatory T cells that inhibit intestinal inflammation. J Exp Med. 1999;190(7):995–1004. doi: 10.1084/jem.190.7.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Issa F, Hester J, Goto R, Nadig SN, Goodacre TE, Wood K. Ex vivo-expanded human regulatory T cells prevent the rejection of skin allografts in a humanized mouse model. Transplantation. 2010;90(12):1321–1327. doi: 10.1097/TP.0b013e3181ff8772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McGuire S. Institute of Medicine. 2010. Strategies to Reduce Sodium Intake in the United States. Washington, DC: The National Academies Press. Adv Nutr. 2010;1(1):49–50. doi: 10.3945/an.110.1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McCormick JA, Bhalla V, Pao AC, Pearce D. SGK1: a rapid aldosterone-induced regulator of renal sodium reabsorption. Physiology (Bethesda) 2005;20:134–139. doi: 10.1152/physiol.00053.2004. [DOI] [PubMed] [Google Scholar]
- 53.Lang F, Artunc F, Vallon V. The physiological impact of the serum and glucocorticoid-inducible kinase SGK1. Curr Opin Nephrol Hypertens. 2009;18(5):439–448. doi: 10.1097/MNH.0b013e32832f125e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Titze J. Sodium balance is not just a renal affair. Curr Opin Nephrol Hypertens. 2014;23(2):101–105. doi: 10.1097/01.mnh.0000441151.55320.c3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vieira SM, Pagovich OE, Kriegel MA. Diet, microbiota and autoimmune diseases. Lupus. 2014;23(6):518–526. doi: 10.1177/0961203313501401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Honda K, Littman DR. The microbiome in infectious disease and inflammation. Annu Rev Immunol. 2012;30:759–795. doi: 10.1146/annurev-immunol-020711-074937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, Gordon JI. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med. 2009;1(6): doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Muegge BD, et al. Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science. 2011;332(6032):970–974. doi: 10.1126/science.1198719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yilmaz OH, et al. mTORC1 in the Paneth cell niche couples intestinal stem-cell function to calorie intake. Nature. 2012;486(7404):490–495. doi: 10.1038/nature11163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dahlmann A, et al. Magnetic resonance-determined sodium removal from tissue stores in hemodialysis patients. Kidney Int. 2015;87(2):434–441. doi: 10.1038/ki.2014.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Titze J, Muller DN, Luft FC. Taking another “look” at sodium. Can J Cardiol. 2014;30(5):473–475. doi: 10.1016/j.cjca.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 62.Joller N, et al. Treg cells expressing the coinhibitory molecule TIGIT selectively inhibit proinflammatory Th1 and Th17 cell responses. Immunity. 2014;40(4):569–581. doi: 10.1016/j.immuni.2014.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.