Abstract
Despite recent progress, the origin of the eukaryotic cell remains enigmatic. It is now known that the last eukaryotic common ancestor was complex and that endosymbiosis played a crucial role in eukaryogenesis at least via the acquisition of the alphaproteobacterial ancestor of mitochondria. However, the nature of the mitochondrial host is controversial, although the recent discovery of an archaeal lineage phylogenetically close to eukaryotes reinforces models proposing archaea-derived hosts. We argue that, in addition to improved phylogenomic analyses with more comprehensive taxon sampling to pinpoint the closest prokaryotic relatives of eukaryotes, determining plausible mechanisms and selective forces at the origin of key eukaryotic features, such as the nucleus or the bacterial-like eukaryotic membrane system, is essential to constrain existing models.
Keywords: eukaryogenesis, archaea, symbiosis, origin of nucleus, metabolism, syntrophy, membrane
A long-lasting query
The origin of the eukaryotic cell was a major evolutionary event that led to a wide diversification of lineages displaying very different morphologies, several of which independently evolved towards multicellularity [1]. Compared to the average prokaryotic cell, the average early eukaryotic cell represented a considerable increase in structural complexity, typified by the presence of an endomembrane system delimiting a hallmark eukaryotic feature, the nucleus, and membrane-bound organelles, notably mitochondria. In the past decades progress in cell and molecular biology, microbial diversity studies and, most of all, comparative genomics and molecular phylogeny, have helped to better constrain the nature of that transition. Several facts related to the early evolution of eukaryotes have been unambiguously established including, among others, a mixed archaeal-bacterial legacy in eukaryotic genomes (see Glossary; Box 1). It is now clear that the last eukaryotic common ancestor (LECA) was a rather complex organism already possessing major idiosyncratic features associated with extant eukaryotes (Box 2). It is also recognized that endosymbiosis did play a crucial role in eukaryogenesis and that the evolution of the alphaproteobacterial endosymbiont at the origin of mitochondria contributed the basics of energy metabolism [2,3] and largely shaped the eukaryotic genome [4], leading to innovations (Box 3). While more detailed knowledge about the last common eukaryotic ancestor and the alphaproteobacterial ancestor of mitochondria is still needed, the most fundamental open query relates to the nature of the host that acquired the mitochondrial ancestor and the eukaryogenic process itself. This is the point upon which most models for the origin of eukaryotes have traditionally diverged [5-8]. The recent discovery of an archaeal lineage, the Lokiarchaeota, sharing more, and seemingly more closely related, genes with eukaryotes [9] represents a significant advance towards the understanding of eukaryotic origins. From the phylogenomics perspective, this observation supports an archaeon as (or an archaeal contribution to) the host of mitochondria [10]. However, from the mechanistic perspective, this solution renovates fundamental open questions that relate to the specific evolutionary process and the underlying selective drivers at the origin of key eukaryotic features. In the following, we very briefly recapitulate the general types of current eukaryogenic models, exposing key mechanistic questions that need to be answered in order to constrain and/or test those hypotheses.
Box 1. Established facts about eukaryotic origins.
Two major cell structural types, prokaryotic and eukaryotic, exist. The eukaryotic cell is, on average, structurally more complex, possessing an endomembrane system with Golgi apparatus, lysosomes or peroxisomes, and endoplasmic reticulum (continuous with the nuclear membrane). Transcription and translation are thought to be generally coupled in prokaryotes (as mRNA is being synthesized, ribosomes start protein synthesis [61]); however, whereas eukaryotic transcription occurs in the nucleus, translation takes place in the cytoplasm.
There are three classically recognized phylogenetic domains of life: Archaea and Bacteria (both prokaryotic), and Eucarya [62]. Established in the 1970s with the first universal molecular phylogenetic analyses [63], this tripartite division has been validated over the years with thousands of genes and genomes from cultured and environmental lineages [64]. Although the three domains share basic biochemistry, genetic code and some universally conserved molecular complexes (ribosome, membrane ATPase) [65], each has distinctive characteristics: a different DNA replication machinery for bacteria [66], unique ether-linked isoprenoid membrane phospholipids in archaea [57], and various complex cellular components and processes in eukaryotes.
In addition to universal and domain-specific traits, eukaryotes specifically share exclusive characteristics with either bacteria or archaea, suggesting some kind of chimeric heritage. Eukaryotic machineries involved in informational processes (replication, transcription, translation) are more similar to, or share homologs only with, archaea [51,67,68]. Genes involved in energy and carbon metabolism, and membrane phospholipids are bacterial-like [57].
Historically, prokaryotes predate eukaryotes, as supported by two independent sources of evidence: the fossil record and the universal occurrence of mitochondria (or derivatives) in extant eukaryotes. The oldest unambiguous eukaryotic microfossils date back to ~2 Ga [69], compatible with molecular-dating inferences for LECA [70,71]. By contrast, geochemical isotope records support the likely occurrence of bacterial and/or archaeal metabolisms (methanogenesis, sulfate reduction, nitrogen fixation) much earlier (3.2-3.4 Ga) [72-74], and the 2.4-Ga atmospheric oxygen hike, clearly attests to the prior evolution of cyanobacterial oxygenic photosynthesis [75]. Consequently, microbial communities at 3.5 Ga [76] were exclusively prokaryotic; eukaryotes appeared >1 Ga later. Likewise, compelling evidence shows that parasitic and anaerobic protists lacking typical mitochondria lost them secondarily: they possess genes of mitochondrial origin in nuclear genomes and mitochondria-related organelles [77]. Therefore, LECA already possessed mitochondria. Because mitochondria evolved from alphaproteobacteria, itself a derived bacterial lineage, bacteria had diversified well before divergence of current eukaryotic lineages.
Finally, symbiosis did play a crucial role in the evolution of the eukaryotic cell, at least via the mitochondrial acquisition from an alphaproteobacterial endosymbiont [78].
Box2. The nature of the last common eukaryotic ancestor (LECA).
Ultrastructural and phylogenetic studies have provided overwhelming support to the idea that all eukaryotes are monophyletic: namely, that they derive from a singleLECA. Ultrastructural and genomic characters widespread in contemporary eukaryotic lineages are likely to have been inherited vertically and can be used as source of information to infer ancestral characteristics. Using this comparative approach, many studies have contributed elements to reconstruct a detailed portrait of LECA [52,79]. It possessed all the paradigmatic eukaryotic features, including the nucleus (with nuclear lamina and nuclear pores), a complex endomembrane system, and a sophisticated tubulin-actin-based cytoskeleton. In relation with the endomembrane system, LECA possessed developed endocytic and exocytic pathways and concomitant vesicle trafficking networks (including Golgi apparatus, lysosomes and autophagosomes) which also involved the cytoskeleton. The cytoskeleton was also essential for phagocytosis, and the presence of this mechanism indicates that LECA was most likely heterotrophic and fed on organic matter, perhaps as predator of other cells. Its metabolism was most likely aerobic because it possessed oxygen-respiring mitochondria. The cytoskeleton had also a key role in mitosis, cell cytokinesis, and cell motility (probably by several flagella). Meiosis was likely present, opening the possibility for some form of sexual reproduction. The genome contained introns, making necessary the existence of a splicing system, likely integrated is a sophisticated gene regulation machinery that also included the activity of small non-coding RNAs and RNA interference. This list is not exhaustive as many other processes, such as ubiquitination and proteasome-mediated degradation, were also present. Coding for all those characters, some of them based on the participation of hundreds of different proteins, requires a very large number of genes. Conservative estimates suggest that the eukaryotic ancestor had a genome with at least 4,000-5,000 genes [52,79]. This implies that LECA was complex, fully comparable to many modern eukaryotes, and that the toolkit for eukaryotic cell components was established very early. The subsequent evolution of eukaryotic lineages involved mostly a fine-tuning of those components rather than major evolutionary innovations (two major exceptions are the endosymbiotic acquisition of photosynthesis and the multiple origins of multicellularity, both at the origin of massive eukaryotic evolutionary radiations).
Box3. Symbiosis in evolution: the case of mitochondria.
The idea that certain membrane-bound organelles derive from endosymbiotic bacteria dates back to the early twentieth century, when Konstantin Mereschkowsky proposed such an evolutionary origin for chloroplasts (but also the nucleus) [21]. Several decades later, Lynn Margulis (Sagan) revived the idea that chloroplasts evolved from endosymbiotic cyanobacteria and further proposed that mitochondria derived from purple bacteria [80]. Highly controversial at the time, final proof came from the first molecular phylogenies of organelle genes that placed chloroplasts within cyanobacteria and mitochondria within alphaproteobacteria [81]. Since then, phylogenetic analysis not only of mitochondrial genes from organellar genomes but also of those that were transferred to nuclear genomes corroborated those affiliations showing, in addition the respective monophyly (hence, the unique origin) of extant mitochondria and chloroplasts [4,78].
Mitochondrial evolution illustrates particularly well the magnitude of the impact that symbiosis can have in evolution. From a metabolic point of view, mitochondria conferred the energy-efficient ability of aerobic respiration to early eukaryotes, allowing the colonization of new ecological niches and opening up the potential to evolve much more complex, including multicellular, structures [2,3]. Thus, even if many mitochondria display (facultative) anaerobic respiration and if many eukaryotes (including some animals) thrive in strictly anoxic conditions, the contribution of oxygen respiration to the evolution and diversification of eukaryotes is beyond any doubt. From the genomic perspective, mitochondrial evolution exemplifies a general trend that is observed in obligatory symbioses. These become definitely stabilized when one partner (usually the endosymbiont) transfers essential genes to the other partner while losing them from its genome. A dependency relation is then established and endosymbiotic gene transfer can further proceed, leading to a progressive genome reduction and, eventually, extinction[82,83]. Genome reduction is manifest in mitochondria. Alphaproteobacterial genomes range from ~1.3 Mbp (parasitic Rickettsia and free-living Pelagibacter spp.) to >9 Mbp, but the largest mitochondrial genomes have only ~100 genes (some excavate protists) and the smallest, animal and apicomplexan mitochondrial genomes, have only 13 and 3 protein-coding genes, respectively. Genome extinction was achieved in many hydrogenosomes and other mitochondria-related organelles (MROs) found in some parasitic and/or anaerobic protists [4,77,82]. Some genes transferred to the nucleus encode proteins that have retained their original mitochondrial function and need to be targeted back to the organelle via signal peptides. However, many of the original mitochondrial genes were lost, their functions being redundant or no longer necessary; others that were transferred to the nucleus either replaced host genes or, accelerating their evolutionary rate, led to brand-new functions.
Mechanistic types of eukaryogenesis models
Many different hypotheses for the origin of eukaryotes have been proposed (for some reviews, see [5-8]). Here, we briefly highlight mechanisms proposed by the two main general types of eukaryogenesis models. Many models exclusively focus on the phylogenetic ancestry of eukaryotes, looking for their closest prokaryotic relatives, or on particular mechanistic details, but only a few models provide combined phylogenetic and mechanistic frameworks.
Autogenous models
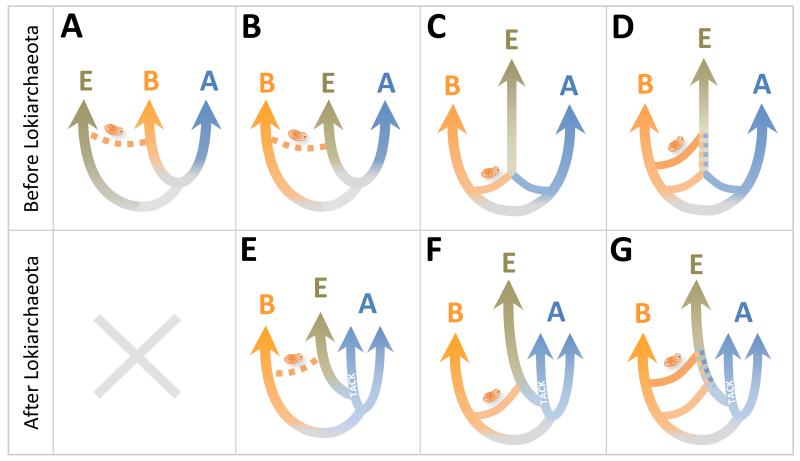
For autogenous (or endogenous) hypotheses, eukaryotes descend from a proto-eukaryotic lineage in which most typical eukaryotic properties had already evolved prior to the acquisition of mitochondria, notably a complex endomembrane system, including the nucleus, and a developed cytoskeleton. Those two components were essential for the evolution of phagocytosis, a critical property that subsequently allowed the engulfment of the mitochondrial ancestor. Although for some authors that proto-eukaryotic lineage was ancestral and retained primitive characters [11,12] (Figure 1.A), for the majority, it was sister to the archaea (Figure 1.B). This would explain the archaeal-like features of ‘informational’ processes in eukaryotes [5,13]; the bacterial-like ‘operational’ genes would derive from mitochondria. This was, until recently, the most popular model. A radical variant is the Neomura hypothesis, proposing that the protoeukaryotic lineage sister to archaea formed a clade with Gram positive bacteria (Actinobacteria and Firmicutes) nested within the remaining, Gram negative bacteria [14]. The emergence of a proto-eukaryotic lineage from within bacteria and, more specifically, Planctomycetes, has also been proposed based on the presence of endomembranes in members of this lineage [15]. However, this hypothesis fails to explain the archaeal-like features found in eukaryotes. In addition, many different bacteria have endomembranes, generally to compartmentalize metabolic regions in the cytoplasm [16]. A very well-known case is that of cyanobacterial thylakoids. Indeed, prior to the general acceptance of the endosymbiotic theory for chloroplasts and mitochondria, cyanobacteria were evoked as eukaryotic ancestors precisely because of their ability to generate endomembranes [17]. As for Planctomycetes[18], these are cases of analogy showing that producing endomembranes is feasible and common among prokaryotes.
Figure 1.
Schematic summary of general types of hypotheses on the phylogenetic origin of eukaryotes in the tree of life before and after the discovery of Lokiarchaeota and related deep-branching archaea. A, archaea; B, bacteria; E, eukaryotes. The schematic organelle drawing represents the mitochondrial acquisition.
Symbiogenetic models
A large category of hypotheses of increasing popularity contends that eukaryotes derive directly from symbiosis between archaea and bacteria, refusing the existence of a third, independent proto-eukaryotic lineage for which direct evidence is missing. Thus, there would be only two primary phylogenetic domains (bacteria and archaea), eukaryotes being mergers (a secondary domain). Originally judged mechanistically unlikely based on the lack of phagocytosis in prokaryotes (hence, the impossibility to acquire endosymbionts), the discovery of endosymbionts within bacteria [19], together with the discovery of membrane remodeling in bacteria and archaea [16,20], made this endosymbiotic process plausible. Margulis recovered the idea of an endosymbiotic origin of organelles from Mereschkowski’s pioneer proposal [21] and further hypothesized that eukaryotic flagella derive from symbiotic spirochetes (serial endosymbiotic theory) [22]. The rest of symbiogenetic models can be generally classed in two categories. One states that the endosymbiosis of the alphaproteobacterial ancestor of mitochondria within one archaeon triggered eukaryogenesis (Figure 1.C). Searcy suggested a metabolic symbiosis mediated by sulfur transfer in a wall-less Thermoplasma-like (belonging to the Euryarchaeota, one of the two major archaeal branches) archaeon [23]. The hydrogen hypothesis is the most elaborate of these models; here, the symbiosis was driven by interspecies hydrogen transfer to a methanogenic archaeon (Euryarchaeota) [24]. Later, different archaeal hosts for the mitochondrial ancestor have been proposed based on gene content and molecular phylogenetic analyses of subsequently discovered archaea, including members of the recently recognized TACK superphylum (Thaumarchaeota, Aigarchaeota, Crenarchaeota, Korarchaeota) [25,26] or deep-branching archaea [27]. The second group of symbiogenetic models posits the endosymbiotic origin of the nucleus, which would derive from the endosymbiosis of one archaeon within a bacterium. The acquisition of mitochondria would constitute a second, independent symbiotic event (Figure 1.D). Some models posit that the nucleus derived from an endosymbiotic eocyte archaeon within a Gram negative bacterium [28] or from one archaeon within a gammaproteobacterium [29] based on gene content and phylogenetic analyses, but fail to provide any clear selective advantage for the nuclear endosymbiosis. The syntrophy hypothesis proposes a highly detailed mechanism suggesting that eukaryotes evolved from a two-step symbiosis. A methanogen would have first established a hydrogen-mediated metabolic symbiosis with a fermentative ancestral myxobacterium (Deltaproteobacteria); mitochondria would derive from metabolically versatile, facultative aerobic alphaproteobacteria [30,31].
Lokiarchaeota come into play
Until recently, attempts to test the different hypotheses and pinpoint the nature of the mitochondrial host based on phylogenomic analysis with increasingly improved methods and taxonomic sampling led to contradictory and/or inconclusive results [32,33]. More recent analyses raised serious suspicions that eukaryotes might branch within one of the two major archaeal branches [34] and be a secondary domain[35] resulting from the merging of archaeal and bacterial lineages. At the same time, many environmental lineages in the two major archaeal clades, the Euryarchaeota and the TACK superphylum or Proteoarchaeota [36], were known by their 16S rRNA sequences, but their genomes remained largely inaccessible until the advent of metagenomics and single cell genomics. Recently, genomic information assembled from seemingly eukaryote-free deep-sea sediment metagenomes for one environmental lineage branching deeply within the TACK superphylum, the Lokiarchaeota, reveals a somewhat larger repertoire of genes in common with eukaryotes than the rest of known archaea, in particular genes involved in membrane remodeling and the cytoskeleton [9]. Eukaryotes branch as sister of this archaeal group in phylogenomic analyses, suggesting the eukaryogenic involvement of an ancestral archaeon belonging to this lineage or of a yet-to-discover archaeal lineage with an even richer eukaryotic-like gene content.
Relatively little is still known from this environmental lineage. Previous microbial ecology and geochemical analyses show that members of this clade (also known as Deep Sea Archaeal Group –DSAG– or marine benthic group B) are diverse and occur in various anoxic settings, often dominating in marine deep-subsurface sediments [37-39]. They appear to be very small (<1 μm diameter) as shown by fluorescence in-situ hybridization and their presence in sediment cores correlates with that of typical ether-linked isoprenoid lipids, strongly suggesting typical archaeal membranes [37,38]. Stable isotope analyses suggest that they are involved in the anaerobic degradation of sedimentary organic matter, a process in which intricate syntrophic interactions involving fermentation of complex organics and subsequent reactions leading to CH4 or CO2, depending on the syntrophic partners. It has been suggested that these archaea might participate in dissimilatory CH4 oxidation [37] or iron or manganese reduction[38].
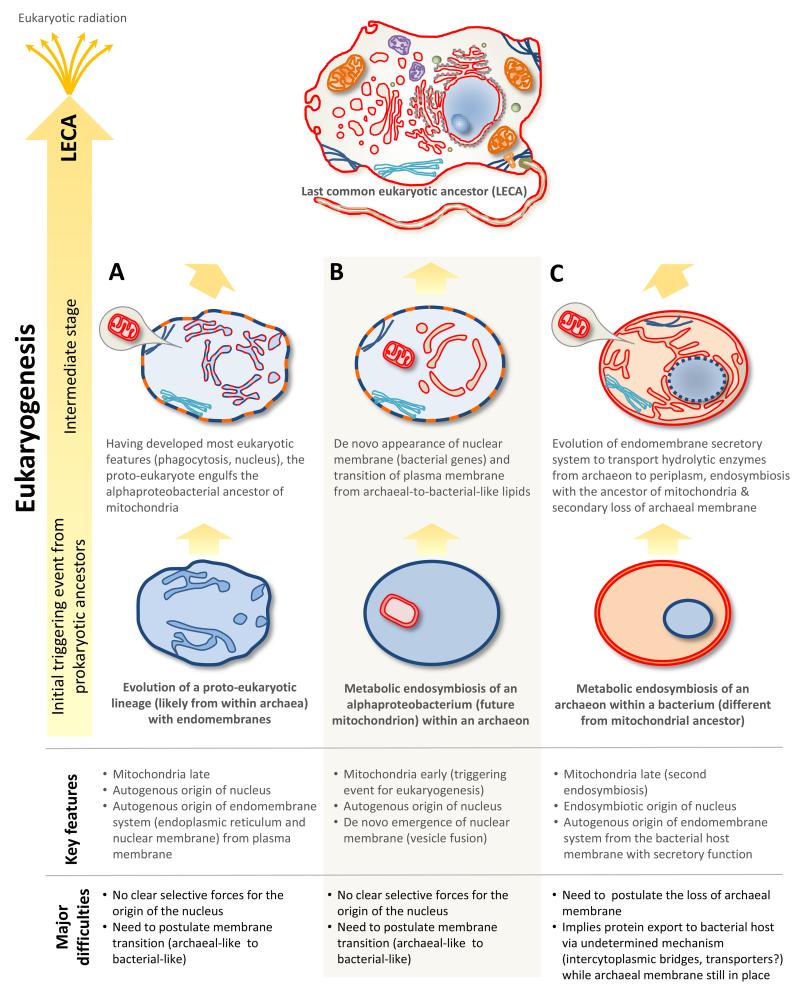
While the discovery of Lokiarchaeota rules out the existence of a third proto-eukaryotic lineage independent of bacteria and archaea (Figure 1.A-B), further supporting the existence of only two primary domains [10], it is still compatible with three different models for the origin of eukaryotes (Figure 1.E-G) that imply three distinct mechanisms (Figure 2). One corresponds to the classical autogenous model invoking a proto-eukaryotic lineage transposed to one specific subgroup of the archaea, and is the view that seems favored by several authors [9,10]. Here, an extensive endomembrane system and cytoskeleton would have evolved in an archaeon, leading to the emergence of a proto-eukaryotic lineage of archaeal origin (Figure 2.A). The two other models would imply endosymbiosis of the mitochondrial ancestor within a member of this archaeal lineage (Figure 2.B) or endosymbiosis of the archaeon within a bacterium before mitochondrial symbiosis took place (Figure 2.C). Therefore, if the discovery of Lokiarchaeota can help in refining the phylogenetic ancestry of eukaryotes, it also renovates old open questions.
Figure 2.
Three hypothetical mechanisms to explain the evolution of the eukaryotic cell. Focus is made on key eukaryotic cellular structures (nucleus, mitochondria, membrane system). (A) Evolution of a proto-eukaryotic lineage before the mitochondrial acquisition event; it is compatible with the classical model proposing a third primary proto-eukaryotic lineage (Figure 1.A, B) and with models proposing that the eukaryotic host derived from within archaea (Figure 1.E). (B) Endosymbiosis of mitochondrial ancestor within one archaeon as the triggering event for eukaryogenesis (Figure 1.C, F). (C) Endosymbiotic origin of nucleus and secondary symbiosis with ancestor of mitochondria (Figure 1.D, G).
Open questions on the origin of eukaryotes
Details about the more specific nature of the symbiotic partners that originated eukaryotes remain to be specified. Which are the closest living archaeal relatives of eukaryotes? Many deep-branching archaeal lineages other than the Lokiarchaeota exist that might contain more eukaryotic-like genes [9]. What are these archaea like? Do they have eukaryotic-like features (actin cytoskeleton, vesicular-trafficking and membrane-remodeling capabilities, endocytosis and/or phagocytosis) as their genome content would predict or are those genes involved in other functions? Having tiny cells and thriving in energy-challenging environments seems at odds with high cellular complexity, but only studies on Lokiarchaeota ultrastructure and biology will provide an answer.
There are also open questions about the nature of the alphaproteobacterial ancestor of mitochondria. The parasitic Rickettsiales or the free-living Pelagibacter/SAR11 group have been proposed as closest relatives to mitochondria, but recent phylogenomic analyses with improved sequence evolution models suggest that those results were affected by long-branch attraction artifacts and compositional (high A+T content) biases, leaving the question unresolved [40].
Some models propose the involvement of another bacterial symbiont different from the ancestor of mitochondria as host of the archaeon, which would become the future nucleus [28,29], sometimes of very specific nature (e.g., myxobacteria) [30,31]. Was it the case and, if so, what was that bacterium like? Phylogenomic analyses generally show a variable contribution of different bacteria (other than alphaproteobacteria, for which phylogenetic signal is strong) to eukaryotes, which is interpreted as ‘noise’ derived from bacteria-to-eukaryote horizontal gene transfer (HGT) and as lack of evidence for any particular bacterial lineage [33]. Although this might well be the case, there are alternative explanations. First, many of the non-alphaproteobacterial genes in eukaryotes might have been acquired via the archaeal host, given that mesophilic archaeal lineages seem to have massively imported genes from bacteria [41], including the Lokiarchaeota [9], or via the mitochondrial ancestor, as has been proposed for myxobacterial genes present in eukaryotes [42]. Second, if such a bacterial host ever existed, it would be extremely difficult to unveil its phylogenetic signal because i) it would imply an older event than the mitochondrial symbiosis, such that the phylogenetic signal would have eroded, and ii) such bacterial host underwent a very important functional shift, in contrast to mitochondria (which essentially conserve the energy metabolism of their free-living relatives) and the archaeal component (which kept the ‘informational’ functions).
However, as phylogenomic information improves, the more open and deeper questions do no longer relate to the phylogenetic origin of symbiotic partners but to the mechanisms and selective forces underlying the eukaryogenic process. We highlight four key questions below; answering them will be crucial to constrain existing models.
What kind of metabolic symbiosis between eukaryogenic partners?
Regardless the type of eukaryogenic model considered (Figure 2), metabolic interspecies interactions must have been fundamental for the evolution of the eukaryotic cell. These are widespread in nature, and particularly extended in anoxic environments, where specialized microorganisms are most often the source and/or sink of electron donors and acceptors [43,44]. Because mitochondria are still responsible for the essential of eukaryotic cell energetics, some type of syntrophic relationship must have been established between the ancestor of mitochondria and its host. Traditional views postulate a strict aerobic ancestor of mitochondria [45], while most symbiogenetic models postulate a facultative aerobe, suggesting that eukaryotes first evolved in anoxic or microaerophilic environments [24,30,31]. The latter view would be consistent with the low-oxygen conditions prevailing not only in Proterozoic (2.5-0.54 Ga) oceans [46] but also in contemporary oceanic (and freshwater) sediments, where Lokiarchaeaota and related archaeal lineages thrive [9]. Indeed, many mitochondria across the eukaryotic tree (e.g. animals, fungi, euglenids, ciliates, algae, diatoms, foraminifera) produce ATP using electron acceptors different from oxygen (e.g., nitrate, nitrite, or fumarate) [2]. This might suggest an anaerobic and metabolically versatile ancestor for mitochondria. However, whereas the phylogeny of some mitochondrial proteins involved in anaerobic metabolism suggests common ancestry, that of others suggests HGT and secondary adaptation to anaerobiosis [47]. Having access to mitochondrial genes of a broader sampling of anaerobic protists would allow testing whether anaerobic respiration was ancestral and/or gained independently several times.
Thus, the original metabolic interaction between the mitochondrial ancestor and its host remains undetermined. Lokiarchaeota might participate in syntrophic interactions involving fermentation [9,37]. If the mitochondrial host was an archaeon or an archaea-derived proto-eukaryote (Figure 2.A-B), it might have produced hydrogen and/or intermediate fermentation products. Was the mitochondrial ancestor an original sink for those metabolites? If the archaeon was an endosymbiont (Figure 2.C), the bacterial host might have been that metabolic sink. At any rate, interspecies hydrogen-transfer might have been originally involved in the original eukaryogenic symbiosis. Hydrogen-mediated syntrophy is widespread in anoxic settings; the best known examples involving fermentative or sulfate-reducing deltaproteobacteria with, respectively, methanogenic or methanotrophic archaea [43,44]. The hydrogen and the syntrophy hypotheses converged in proposing such a metabolic interaction [48], where the archaeon was methanogenic (using H2 to reduce CO2 to CH4) and the bacterium involved in the primary symbiosis (the mitochondrial ancestor in the hydrogen hypothesis, the deltaproteobacterial host in the syntrophy hypothesis) was fermentative. Therefore, the archaeal partners were hydrogen sinks, not sources. While the possibility of a methanogenic ancestor for eukaryotes cannot be completely ruled out (although Lokiarchaeota seem to lack methanogenesis, other deep-branching archaea might have possessed this pathway, especially if the last common archaeal ancestor was a methanogen, as some phylogenomic analyses seem to suggest [49]), this now seems unlikely. However, the mechanistic essence of the two symbiogenetic models (Figure 2.B-C) remains valid under a reversed metabolic interaction, where the archaeon is the hydrogen producer and the bacterium, the hydrogen sink. In the case of the syntrophy hypothesis, the bacterial host might have been a sulfate-reducer (the original hypothesis indeed proposed a facultative sulfate reducer ancestral to myxobacteria [30,48]).
Mitochondria: early or late?
Before the discovery of the Lokiarchaeota, symbiogenetic models proposing a direct endosymbiosis of the mitochondrial ancestor within one archaeon had gained popularity [24,26,50] (Figure 2.B). They imply a triggering effect of eukaryogenesis for the mitochondrial endosymbiosis. However, the fact that the newly discovered archaea possess several homologs to membrane modeling and cytoskeleton-related eukaryotic proteins has opened the possibility for a proto-eukaryotic lineage endowed with phagocytosis to evolve from within archaea, which seems the currently favored hypothesis [9,10]. In this case, mitochondria would incorporate later (Figure 2.A). The difference between the two situations is far from irrelevant because the driving forces underlying eukaryogenesis and the predictions of the two models are very different. In the former case, the mitochondrial symbiosis would be the cause of eukaryogenesis; in the latter, the consequence. In the former case, amitochondriate eukaryotes would have never existed; in the latter, the possibility that archaea-derived amitochondriate proto-eukaryotes exist is real. In models invoking a bacterial host for the archaeon (Figure 2.C), the mitochondrial symbiosis is a second, independent event; mitochondria would therefore come later but, as in the case of autogenous models, the time between the start of eukaryogenesis and the mitochondrial stabilization is difficult to determine. The strong alphaproteobacterial signal in eukaryotic genomes [33,51] would rather suggest a relatively late mitochondrial incorporation event.
Which selective forces for the evolution of the nucleus?
The origin of the nucleus remains mysterious. While it was present in LECA and evolved as part of the endomembrane system involving many protein components of archaeal and bacterial ancestry [52], most models do not provide any (or any convincing) selective force for the evolution of this defining character. Two driving forces have been evoked by autogenous models. Cavalier-Smith proposed that the nucleus evolved to prevent DNA damage caused by cytoskeletal pulling [53], an idea adopted by others [26]. However, eukaryotic chromosomes are able to overcome mechanical stresses during mitosis, when the nuclear envelope disintegrates in many protists, even in species where chromosomes are permanently uncondensed. In addition, eukaryotes (as do prokaryotes) have efficient DNA repair systems to cope with single and double-strand breaks occurring during the mechanically-challenging DNA-dependent processes (replication, transcription, recombination), and many eukaryotes have genomes with several dozens of chromosomes, which reduces the individual size of DNA molecules, diminishing breakage probability. Jékely proposed that the nucleus appeared to safeguard ribosome biogenesis, preventing the formation of chimeric ribosomes during mitochondrial endosymbiosis [54]. However, the formation of chimeric ribosomes could have been more simply achieved by retaining the ribosomal protein genes in the mitochondrial genome, as is the case with the ribosomal RNA and other protein genes for which the cytosolic synthesis and transport back to the mitochondrion poses a problem.
In the framework of the hydrogen hypothesis, the nucleus was proposed to appear de novo through the synthesis of bacterial-like lipids that would form vesicles in the archaeal cytoplasm (Figure 2.B). The nuclear compartment would have appeared to decouple transcription and translation, thus preventing the synthesis of aberrant proteins as introns appeared [55]. However, this explanation is at odds with the fact that a single intron in an essential gene is deleterious if transcription and translation are not already uncoupled and a splicing system is not in place. Thus, the decoupling of transcription and translation by the nuclear membrane must precede intron invasion. Such an idea was put forward in the framework of the syntrophy hypothesis, which proposes the endosymbiotic origin of the nucleus [31] (Figure 2.C). Endosymbiotic models for the origin of the nucleus easily account for the presence of a different compartment [21,28,31], but the difficulty here lies in how to explain what drives the endosymbiosis and the origin of the endomembrane system. In the syntrophy hypothesis, the endoplasmic reticulum and the nuclear membrane evolve with a primary secretory role from the internalization of the inner plasma membrane of the host bacterium, and in a way that is analogous to that proposed by autogenous models (and that would be most similar to the membrane internalization seen in other Gram negative bacteria, with the external membrane retaining its cell-boundary role). This endomembrane system would secure the distribution of hydrolytic enzymes synthesized by the archaeon towards the periplasm which, progressively, would be internalized in form of hydrolysis-related vesicles (lysosomes, peroxisomes). The formation of a proto-nuclear membrane to which ribosomes attached to deliver their products would have allowed intron expansion. Once ribosomes migrated out towards the cytoplasm (interestingly, ribosomal subunits are still assembled in the nucleus), with attachment to the endoplasmic reticulum, the proto-nuclear membrane was exapted to prevent the synthesis of aberrant proteins [31]. Though these explanations seem plausible, this hypothesis is not without problems. First, endosymbiotic origins for the nucleus are often disregarded as less parsimonious than models involving only two symbionts, despite the fact that dual and multiple symbioses are frequent in natural environments [56]. Second, the transition from archaeal endosymbiont to nucleus involves the loss of the archaeal membrane and related mechanistic shifts that are difficult to explain.
How did the bacterial-like eukaryotic membranes evolve?
Archaeal membranes are radically different from bacterial and eukaryotic membranes, having phospholipids of different composition and stereochemistry [57]. If currently favored models based on an archaea-derived host are correct, they entail a transition from archaeal to bacterial membranes (Figure 2.A-B). However, this poses two challenges. First, the plasma membrane is not only a lipid bilayer but a dynamic cell-environment interface, so that the driving force leading to such transition from an already optimized system is obscure. Second, it has been proposed that a change of membrane composition would be feasible because mixed liposomes of archaeal and bacterial lipids are stable [8]. However, while this argument might apply to primitive, not yet optimized membranes, such as those of the last common ancestor [57], it is more unrealistic for complex membranes harboring many transmembrane proteins, which are specifically adapted to their lipid environment [58]. A change in membrane phospholipids must therefore be accompanied by a pervasive change in membrane proteins. Is that major shift possible? While there is no direct evidence for an archaea-to-bacteria membrane transition, that possibility cannot be completely ruled out. Indeed, recent studies show that mesophilic archaeal lineages for which genomic data are available have undergone extensive and convergent HGT from bacteria. Several of those convergently acquired genes relate to fatty acid biosynthesis and membrane-associated electron transfer carriers and transporters [41]. However, even if important lineages of mesophilic archaea (e.g. haloarchaea, marine Thaumarchaeota and Euryarchaeota, mesophilic methanogens) have imported bacterial fatty acid genes, their fate in the cell is uncertain since their membrane lipids remain typically archaeal (e.g. tetraether lipids) and based on isoprenoid moieties and glycerol-1 (instead of 3)-phosphate [59,60]. Therefore, in the absence of observable transitional membrane stages, the archaea-to-bacteria membrane shift remains the Achilles’ heel for these models. Models proposing an archaeal endosymbiont (Figure 2C) within a bacterial host offer, by contrast, a straightforward explanation for the bacterial-like nature of eukaryotic membranes.
Conclusions
The long-inscrutable quest for the origin of the eukaryotic cell has become increasingly tractable with recent progress in genome sequencing, phylogenomic analyses and the exploration of microbial diversity in natural ecosystems. The discovery of new lineages of uncultured archaea in anoxic sediments that appear more related to eukaryotes than other archaea constrains existing models for the origin of eukaryotes. However, if the prokaryotic ancestry of eukaryotes begins to be refined, fundamental questions on the eukaryogenic process remain fully open. Among them are the number and type of metabolic symbioses at the origin of the eukaryotic cell, whether mitochondria evolved early or late and, most critically, how and why the nucleus evolved, and how the eukaryotic membrane lipids came to be (see Outstanding Questions). The devil is in the detail; thus the elaboration of detailed, plausible and, if possible, testable mechanistic models accounting for all eukaryotic features is necessary to constrain existing models and unravel the transition to eukaryotic complexity.
Outstanding questions.
What was the last common ancestor of eukaryotes like? What was its level of complexity?
Which is the closest alphaproteobacterial relative to mitochondria? What were the metabolic properties of the mitochondrial ancestor? Was it a facultative anaerobe?
What was the phylogenetic affiliation of the mitochondrial host? Was it a bona fide archaeon? Was it a member of a proto-eukaryotic lineage derived from within archaea? Was it a bacterium-archaeon consortium?
What was the phenotypic nature of the mitochondrial host? Were major eukaryotic features (endomembrane system, phagocytosis, complex cytoskeleton) present in that ancestor or were those features developed during symbiogenesis?
What type of metabolic symbiosis or symbioses led to eukaryogenesis?
When did the mitochondrial symbiosis establish? Was it an early event triggering eukaryogenesis? Was it a later event and mitochondria established in a proto-eukaryotic lineage containing all major eukaryotic features except mitochondria?
What were the selective forces promoting the origin of the eukaryotic nucleus? Was the origin of the nucleus autogenous? Was it endosymbiotic?
How did the bacterial-like membrane of the eukaryotic cell evolve if the mitochondrial host was an archaeon?
Trends.
Eukaryotes arose from the endosymbiosis of an alphaproteobacterium in an unknown host. Eukaryogenic models diverge in the hypothetical host proposed
Recent phylogenomic analyses and the discovery of archaea with seemingly more eukaryotic-like genes suggest that the mitochondrial host was an archaeon or had a vital archaeal contribution, excluding a third hypothetical proto-eukaryotic lineage different from archaea
As the phylogenetic origin of eukaryotes gets clearer, mechanistic questions remain open: the type of metabolic symbioses involved, the timing of mitochondrial acquisition and, most importantly, the origin of the eukaryotic nucleus and bacterial-like membranes
Plausible driving forces and processes for the evolution of the eukaryotic nucleus are missing
If the mitochondrial host was an archaeon, a difficult-to-explain archaeal-to-bacterial membrane transition is required
Acknowledgements
We apologize for not being able to cite the work of many authors due to space limitations. We acknowledge funding from the European Research Council under the European Union’s Seventh Framework Program ERC Grant Agreement 322669 ‘ProtistWorld’.
Glossary
- Alphaproteobacteria
highly diversified and metabolically versatile class of bacteria within the phylum Proteobacteria from which the ancestor of mitochondria evolved.
- Archaea
one of the three classically recognized domains of life and one of the two primary phylogenetic domains. Archaea exhibit prokaryotic cell structure. They are traditionally divided in two main branches, the Euryarchaeota and the TACK (Thaumarchaeota, Aigarchaeota, Crenarchaeota, Korarchaeota) superphylum or Proteoarchaeota.
- Autogenous models
hypotheses postulating that the endomembrane system and the nucleus result from the invagination of the plasma membrane in a proto-eukaryotic or a prokaryotic ancestor. Historically, before the general acceptance of the endosymbiotic origin of mitochondria and chloroplasts, the term also referred to the internal development of these organelles from endogenous endomembranes.
- Deltaproteobacteria
proteobacterial class encompassing predominantly anaerobic, sulfate-reducing bacteria, fermentative syntrophic bacteria (e.g. Syntrophomonas) and myxobacteria.
- Endosymbiosis
symbiotic relationship in which one partner (endosymbiont) is within the other (host).
- Endosymbiotic gene transfer
transfer of genes from the genome of the endosymbiont to that of the host; in some cases it can lead to complete genome extinction.
- Eocyte
term historically applied to the archaeal clade that was more similar to eukaryotes based on ribosomal proteins (originally, the Crenarchaeota); by extension, some authors apply it today to the TACK superphylum.
- Eukaryogenesis
process that led to the evolution of the eukaryotic cell from prokaryotic ancestors.
- Hydrogenosome
hydrogen-producing organelle that evolved from mitochondria.
- Mitosome
a type of MRO, genome-lacking simple mitochondrial remnant where Fe-S clusters are assembled.
- MRO
mitochondria-related organelle; includes hydrogenosomes, mitosomes and, in general, organelles derived from mitochondria.
- Mutualism
symbiosis beneficial for the interacting partners.
- Myxobacteria
also known as ‘slime’ or ‘social’ bacteria, group of Deltaproteobacteria characterized by complex genomes, gliding motility, coordinated multicellularity and cell differentiation.
- Neomura
hypothetic clade composed by Archaea + Eucarya that would have evolved from single-membrane-bound, Gram-positive bacteria.
- Planctomycetes
bacterial phylum containing some members with more-or-less developed endomembrane systems that sometimes resemble nuclear-like membranes (e.g., Gemmata obscuriglobus).
- Phylogenomics
molecular phylogenetic analyses of genome-derived information.
- Serial endosymbiotic theory
evolutionary eukaryogenic scenario proposed by Lynn Margulis stating that mitochondria and chloroplasts derived from endosymbiotic bacteria subsequent to merging symbiosis between a wall-less archaeon and motility-providing spirochetes.
- Symbiogenesis
evolutionary process mediated by symbiosis at the origin of the eukaryotic cell.
- Symbiosis
intimate and durable inter-species interaction; generally applied to parasitic, mutualistic and commensal interactions, but sometimes applied more restrictively to mutualism.
- Syntrophy
symbiosis mediated by metabolic interactions.
References
- 1.Adl SM, et al. The revised classification of eukaryotes. J Eukaryot Microbiol. 2012;59:429–493. doi: 10.1111/j.1550-7408.2012.00644.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mentel M, Martin W. Energy metabolism among eukaryotic anaerobes in light of Proterozoic ocean chemistry. Philos Trans R Soc Lond B Biol Sci. 2008;363:2717–2729. doi: 10.1098/rstb.2008.0031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lane N, Martin W. The energetics of genome complexity. Nature. 2010;467:929–934. doi: 10.1038/nature09486. [DOI] [PubMed] [Google Scholar]
- 4.Timmis JN, et al. Endosymbiotic gene transfer: organelle genomes forge eukaryotic chromosomes. Nat Rev Genet. 2004;5:123–135. doi: 10.1038/nrg1271. [DOI] [PubMed] [Google Scholar]
- 5.Embley TM, Martin W. Eukaryotic evolution, changes and challenges. Nature. 2006;440:623–630. doi: 10.1038/nature04546. [DOI] [PubMed] [Google Scholar]
- 6.Poole AM, Penny D. Evaluating hypotheses for the origin of eukaryotes. Bioessays. 2007;29:74–84. doi: 10.1002/bies.20516. [DOI] [PubMed] [Google Scholar]
- 7.Keeling PJ. The impact of history on our perception of evolutionary events: endosymbiosis and the origin of eukaryotic complexity. Cold Spring Harb Perspect Biol. 2014;6:a016196. doi: 10.1101/cshperspect.a016196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McInerney JO, et al. The hybrid nature of the Eukaryota and a consilient view of life on Earth. Nat Rev Microbiol. 2014;12:449–455. doi: 10.1038/nrmicro3271. [DOI] [PubMed] [Google Scholar]
- 9.Spang A, et al. Complex archaea that bridge the gap between prokaryotes and eukaryotes. Nature. 2015 doi: 10.1038/nature14447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Embley TM, Williams TA. Evolution: Steps on the road to eukaryotes. Nature. 2015 doi: 10.1038/nature14522. [DOI] [PubMed] [Google Scholar]
- 11.Sogin ML. Early evolution and the origin of eukaryotes. Curr. Opin. Genet. Dev. 1991;1:457–463. doi: 10.1016/s0959-437x(05)80192-3. [DOI] [PubMed] [Google Scholar]
- 12.Poole A, et al. Early evolution: prokaryotes, the new kids on the block. Bioessays. 1999;21:880–889. doi: 10.1002/(SICI)1521-1878(199910)21:10<880::AID-BIES11>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 13.Doolittle WF, Brown JR. Tempo, mode, the progenote, and the universal root. Proc. Natl. Acad. Sci. USA. 1994;91:6721–6728. doi: 10.1073/pnas.91.15.6721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cavalier-Smith T. The neomuran revolution and phagotrophic origin of eukaryotes and cilia in the light of intracellular coevolution and a revised tree of life. Cold Spring Harb Perspect Biol. 2014;6:a016006. doi: 10.1101/cshperspect.a016006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fuerst JA, Sagulenko E. Keys to eukaryality: planctomycetes and ancestral evolution of cellular complexity. Front Microbiol. 2012;3:167. doi: 10.3389/fmicb.2012.00167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Diekmann Y, Pereira-Leal JB. Evolution of intracellular compartmentalization. Biochem J. 2013;449:319–331. doi: 10.1042/BJ20120957. [DOI] [PubMed] [Google Scholar]
- 17.Cavalier-Smith T. The origin of nuclei and of eukaryotic cells. Nature. 1975;256:463–468. doi: 10.1038/256463a0. [DOI] [PubMed] [Google Scholar]
- 18.McInerney JO, et al. Planctomycetes and eukaryotes: a case of analogy not homology. Bioessays. 2011;33:810–817. doi: 10.1002/bies.201100045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.von Dohlen CD, et al. Mealybug beta-proteobacterial endosymbionts contain gamma-proteobacterial symbionts. Nature. 2001;412:433–436. doi: 10.1038/35086563. [DOI] [PubMed] [Google Scholar]
- 20.Godde JS. Breaking through a phylogenetic impasse: a pair of associated archaea might have played host in the endosymbiotic origin of eukaryotes. Cell Biosci. 2012;2:29. doi: 10.1186/2045-3701-2-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mereschkowsky C. Über natur und usprung der chromatophoren im pflanzenreiche. Biologisches Centralblatt. 1905;25:593–604. [Google Scholar]
- 22.Margulis L. Archaeal-eubacterial mergers in the origin of Eukarya: phylogenetic classification of life. Proc Natl Acad Sci U S A. 1996;93:1071–1076. doi: 10.1073/pnas.93.3.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Searcy DG. Origins of mitochondria and chloroplasts from sulfur-based symbioses. In: Hartman H, Matsuno K, editors. The origin and evolution of the cell. World Scientific; 1992. pp. 47–78. [Google Scholar]
- 24.Martin W, Muller M. The hydrogen hypothesis for the first eukaryote. Nature. 1998;392:37–41. doi: 10.1038/32096. [DOI] [PubMed] [Google Scholar]
- 25.Guy L, Ettema TJ. The archaeal ‘TACK’ superphylum and the origin of eukaryotes. Trends Microbiol. 2011;19:580–587. doi: 10.1016/j.tim.2011.09.002. [DOI] [PubMed] [Google Scholar]
- 26.Martijn J, Ettema TJ. From archaeon to eukaryote: the evolutionary dark ages of the eukaryotic cell. Biochem Soc Trans. 2013;41:451–457. doi: 10.1042/BST20120292. [DOI] [PubMed] [Google Scholar]
- 27.Makarova KS, et al. Evolution of diverse cell division and vesicle formation systems in Archaea. Nat Rev Microbiol. 2010;8:731–741. doi: 10.1038/nrmicro2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gupta RS, Golding GB. The origin of the eukaryotic cell. Trends Biochem Sci. 1996;21:166–171. [PubMed] [Google Scholar]
- 29.Horiike T, et al. Origin of eukaryotic cell nuclei by symbiosis of Archaea in Bacteria is revealed by homology-hit analysis. Nat Cell Biol. 2001;3:210–214. doi: 10.1038/35055129. [DOI] [PubMed] [Google Scholar]
- 30.Moreira D, López-García P. Symbiosis between methanogenic archaea and delta-Proteobacteria as the origin of eukaryotes: The syntrophic hypothesis. J. Mol. Evol. 1998;47:517–530. doi: 10.1007/pl00006408. [DOI] [PubMed] [Google Scholar]
- 31.López-García P, Moreira D. Selective forces for the origin of the eukaryotic nucleus. Bioessays. 2006;28:525–533. doi: 10.1002/bies.20413. [DOI] [PubMed] [Google Scholar]
- 32.Gribaldo S, et al. The origin of eukaryotes and their relationship with the Archaea: are we at a phylogenomic impasse? Nat Rev Microbiol. 2010;8:743–752. doi: 10.1038/nrmicro2426. [DOI] [PubMed] [Google Scholar]
- 33.Rochette NC, et al. Phylogenomic test of the hypotheses for the evolutionary origin of eukaryotes. Mol Biol Evol. 2014;31:832–845. doi: 10.1093/molbev/mst272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Williams TA, Embley TM. Archaeal “dark matter” and the origin of eukaryotes. Genome Biol Evol. 2014;6:474–481. doi: 10.1093/gbe/evu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Williams TA, et al. An archaeal origin of eukaryotes supports only two primary domains of life. Nature. 2013;504:231–236. doi: 10.1038/nature12779. [DOI] [PubMed] [Google Scholar]
- 36.Petitjean C, et al. Rooting the domain archaea by phylogenomic analysis supports the foundation of the new kingdom proteoarchaeota. Genome Biol Evol. 2014;7:191–204. doi: 10.1093/gbe/evu274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Biddle JF, et al. Heterotrophic Archaea dominate sedimentary subsurface ecosystems off Peru. Proc Natl Acad Sci U S A. 2006;103:3846–3451. doi: 10.1073/pnas.0600035103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jorgensen SL, et al. Correlating microbial community profiles with geochemical data in highly stratified sediments from the Arctic Mid-Ocean Ridge. Proc Natl Acad Sci U S A. 2012;109:E2846–2855. doi: 10.1073/pnas.1207574109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jorgensen SL, et al. Quantitative and phylogenetic study of the Deep Sea Archaeal Group in sediments of the Arctic mid-ocean spreading ridge. Front Microbiol. 2013;4:299. doi: 10.3389/fmicb.2013.00299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rodriguez-Ezpeleta N, Embley TM. The SAR11 group of Alpha-Proteobacteria is not related to the origin of mitochondria. PLoS ONE. 2012;7:e30520. doi: 10.1371/journal.pone.0030520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.López-García P, et al. Bacterial gene import and mesophilic adaptation in archaea. Nat Rev Microbiol. 2015;13:447–456. doi: 10.1038/nrmicro3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schluter A, et al. Phylogenomic evidence for a myxococcal contribution to the mitochondrial fatty acid beta-oxidation. PLoS ONE. 2011;6:e21989. doi: 10.1371/journal.pone.0021989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Orphan VJ. Methods for unveiling cryptic microbial partnerships in nature. Curr Opin Microbiol. 2009;12:231–237. doi: 10.1016/j.mib.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 44.Kouzuma A, et al. Microbial interspecies interactions: recent findings in syntrophic consortia. Front Microbiol. 2015;6:477. doi: 10.3389/fmicb.2015.00477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kurland CG, Andersson SG. Origin and evolution of the mitochondrial proteome. Microbiol Mol Biol Rev. 2000;64:786–820. doi: 10.1128/mmbr.64.4.786-820.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scott C, et al. Tracing the stepwise oxygenation of the Proterozoic ocean. Nature. 2008;452:456–459. doi: 10.1038/nature06811. [DOI] [PubMed] [Google Scholar]
- 47.Hug LA, et al. Phylogenetic distributions and histories of proteins involved in anaerobic pyruvate metabolism in eukaryotes. Mol Biol Evol. 2010;27:311–324. doi: 10.1093/molbev/msp237. [DOI] [PubMed] [Google Scholar]
- 48.López-García P, Moreira D. Metabolic symbiosis at the origin of eukaryotes. Trends Biochem Sci. 1999;24:88–93. doi: 10.1016/s0968-0004(98)01342-5. [DOI] [PubMed] [Google Scholar]
- 49.Raymann K, et al. The two-domain tree of life is linked to a new root for the Archaea. Proc Natl Acad Sci U S A. 2015;112:6670–6675. doi: 10.1073/pnas.1420858112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yutin N, et al. The origins of phagocytosis and eukaryogenesis. Biol Direct. 2009;4:9. doi: 10.1186/1745-6150-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thiergart T, et al. An evolutionary network of genes present in the eukaryote common ancestor polls genomes on eukaryotic and mitochondrial origin. Genome Biol Evol. 2012;4:466–485. doi: 10.1093/gbe/evs018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Koumandou VL, et al. Molecular paleontology and complexity in the last eukaryotic common ancestor. Crit Rev Biochem Mol Biol. 2013;48:373–396. doi: 10.3109/10409238.2013.821444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cavalier-Smith T. Origin of the cell nucleus, mitosis and sex: roles of intracellular coevolution. Biol Direct. 2010;5:7. doi: 10.1186/1745-6150-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jekely G. Origin of eukaryotic endomembranes: a critical evaluation of different model scenarios. Adv Exp Med Biol. 2007;607:38–51. doi: 10.1007/978-0-387-74021-8_3. [DOI] [PubMed] [Google Scholar]
- 55.Martin W, Koonin EV. Introns and the origin of nucleus-cytosol compartmentalization. Nature. 2006;440:41–45. doi: 10.1038/nature04531. [DOI] [PubMed] [Google Scholar]
- 56.Dubilier N, et al. Endosymbiotic sulphate-reducing and sulphide-oxidizing bacteria in an oligochaete worm. Nature. 2001;411:298–302. doi: 10.1038/35077067. [DOI] [PubMed] [Google Scholar]
- 57.Lombard J, et al. The early evolution of lipid membranes and the three domains of life. Nat Rev Microbiol. 2012;10:507–515. doi: 10.1038/nrmicro2815. [DOI] [PubMed] [Google Scholar]
- 58.Pogozheva ID, et al. Structural adaptations of proteins to different biological membranes. Biochim Biophys Acta. 2013;1828:2592–2608. doi: 10.1016/j.bbamem.2013.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Koga Y, Morii H. Biosynthesis of ether-type polar lipids in archaea and evolutionary considerations. Microbiol Mol Biol Rev. 2007;71:97–120. doi: 10.1128/MMBR.00033-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lincoln SA, et al. Planktonic Euryarchaeota are a significant source of archaeal tetraether lipids in the ocean. Proc Natl Acad Sci U S A. 2014;111:9858–9863. doi: 10.1073/pnas.1409439111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Martin W, Koonin EV. A positive definition of prokaryotes. Nature. 2006;442:868. doi: 10.1038/442868c. [DOI] [PubMed] [Google Scholar]
- 62.Woese CR, et al. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc. Natl. Acad. Sci. USA. 1990;87:4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Woese CR, Fox GE. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc. Natl. Acad. Sci. USA. 1977;74:5088–5090. doi: 10.1073/pnas.74.11.5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.López-García P, Moreira D. Tracking microbial biodiversity through molecular and genomic ecology. Res Microbiol. 2008;159:67–73. doi: 10.1016/j.resmic.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 65.Mirkin BG, et al. Algorithms for computing parsimonious evolutionary scenarios for genome evolution, the last universal common ancestor and dominance of horizontal gene transfer in the evolution of prokaryotes. BMC Evol Biol. 2003;3:2. doi: 10.1186/1471-2148-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Leipe DD, et al. Did DNA replication evolve twice independently? Nucleic Acids Res. 1999;27:3389–3401. doi: 10.1093/nar/27.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rivera MC, et al. Genomic evidence for two functionally distinct gene classes. Proc. Natl. Acad. Sci. USA. 1998;95:6239–6244. doi: 10.1073/pnas.95.11.6239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ribeiro S, Golding GB. The mosaic nature of the eukaryotic nucleus. Mol Biol Evol. 1998;15:779–788. doi: 10.1093/oxfordjournals.molbev.a025983. [DOI] [PubMed] [Google Scholar]
- 69.Knoll AH. Paleobiological perspectives on early eukaryotic evolution. Cold Spring Harbor Perspectives in Biology. 2014:6. doi: 10.1101/cshperspect.a016121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Parfrey LW, et al. Estimating the timing of early eukaryotic diversification with multigene molecular clocks. Proc Natl Acad Sci U S A. 2011;108:13624–13629. doi: 10.1073/pnas.1110633108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Eme L, et al. On the age of eukaryotes: evaluating evidence from fossils and molecular clocks. Cold Spring Harb Perspect Biol. 2014;6:a016139. doi: 10.1101/cshperspect.a016139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ueno Y, et al. Evidence from fluid inclusions for microbial methanogenesis in the early Archaean era. Nature. 2006;440:516–519. doi: 10.1038/nature04584. [DOI] [PubMed] [Google Scholar]
- 73.Shen Y, et al. Isotopic evidence for microbial sulphate reduction in the early Archaean era. Nature. 2001;410:77–81. doi: 10.1038/35065071. [DOI] [PubMed] [Google Scholar]
- 74.Stueken EE, et al. Isotopic evidence for biological nitrogen fixation by molybdenum-nitrogenase from 3.2 Gyr. Nature. 2015;520:666–669. doi: 10.1038/nature14180. [DOI] [PubMed] [Google Scholar]
- 75.Farquhar J, et al. Geological constraints on the origin of oxygenic photosynthesis. Photosynth Res. 2011;107:11–36. doi: 10.1007/s11120-010-9594-0. [DOI] [PubMed] [Google Scholar]
- 76.Allwood AC, et al. Stromatolite reef from the Early Archaean era of Australia. Nature. 2006;441:714–718. doi: 10.1038/nature04764. [DOI] [PubMed] [Google Scholar]
- 77.Hjort K, et al. Diversity and reductive evolution of mitochondria among microbial eukaryotes. Philos Trans R Soc Lond B Biol Sci. 2010;365:713–727. doi: 10.1098/rstb.2009.0224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gray MW. Mitochondrial evolution. Cold Spring Harb Perspect Biol. 2012;4:a011403. doi: 10.1101/cshperspect.a011403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Koonin EV. Preview. The incredible expanding ancestor of eukaryotes. Cell. 2010;140:606–608. doi: 10.1016/j.cell.2010.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sagan L. On the origin of mitosing cells. J Theor Biol. 1967;14:255–274. doi: 10.1016/0022-5193(67)90079-3. [DOI] [PubMed] [Google Scholar]
- 81.Schwartz RM, Dayhoff MO. Origins of prokaryotes, eukaryotes, mitochondria, and chloroplasts. Science. 1978;199:395–403. doi: 10.1126/science.202030. [DOI] [PubMed] [Google Scholar]
- 82.Dyall SD, et al. Ancient invasions: from endosymbionts to organelles. Science. 2004;304:253–257. doi: 10.1126/science.1094884. [DOI] [PubMed] [Google Scholar]
- 83.McCutcheon JP, Moran NA. Extreme genome reduction in symbiotic bacteria. Nat Rev Microbiol. 2012;10:13–26. doi: 10.1038/nrmicro2670. [DOI] [PubMed] [Google Scholar]