Abstract
The NLR family apoptosis inhibitory proteins (NAIPs) bind conserved bacterial ligands, such as the bacterial rod protein PrgJ, and recruit NLR family CARD-containing protein 4 (NLRC4) as the inflammasome adapter to activate innate immunity. We found that the PrgJ-NAIP2-NLRC4 inflammasome is assembled into multisubunit disk-like structures through a unidirectional adenosine triphosphatase polymerization, primed with a single PrgJ-activated NAIP2 per disk. Cryo–electron microscopy (cryo-EM) reconstruction at subnanometer resolution revealed a ~90° hinge rotation accompanying NLRC4 activation. Unlike in the related heptameric Apaf-1 apoptosome, in which each subunit needs to be conformationally activated by its ligand before assembly, a single PrgJ-activated NAIP2 initiates NLRC4 polymerization in a domino-like reaction to promote the disk assembly. These insights reveal the mechanism of signal amplification in NAIP-NLRC4 inflammasomes.
The nucleotide-binding domain (NBD) and leucine-rich repeat (LRR)–containing protein (NLR) family participates in the formation of inflammasomes that activate caspase-1 for cell death induction and cy-tokine maturation. NLR family apoptosis inhibitory proteins (NAIPs) are so far the only NLR family members with specifically defined ligands (1–4). NAIP2 detects the inner rod protein of the bacterial type III secretion system, including Salmonella typhimurium PrgJ, whereas NAIP5 and NAIP6 detect bacterial flagellin such as Salmonella typhimurium FliC (2, 4, 5). NLR family caspase recruitment domain (CARD)– containing protein 4 (NLRC4) was initially found to participate in caspase-1 activation and inter-leukin (IL)–1β secretion in response to cytoplasmic flagellin (6) and was only recently shown to be the common adapter for NAIPs (2, 4). The NAIP-NLRC4 inflammasomes perform effector functions against intracellular bacteria (7, 8), play protective roles in mouse models of colitisassociated colorectal cancer (9, 10), and serve as a potential strategy in tumor immunotherapy (11). Mutations in NLRC4 also induce auto-inflammatory diseases in humans (10, 12–14).
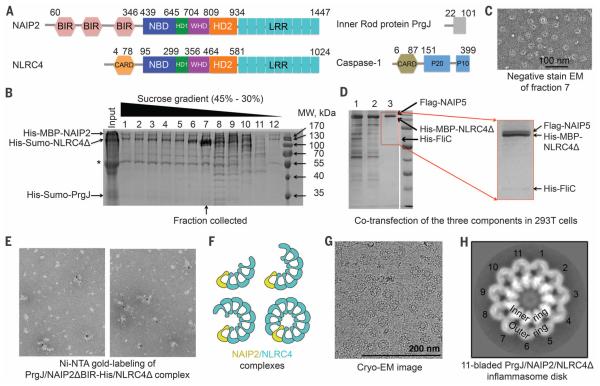
We assembled the FliC-activated NAIP5-NLRC4 complex and the PrgJ-activated NAIP2-NLRC4 complex with the use of CARD-deleted NLRC4 (NLRC4Δ) to avoid potential CARD-mediated aggregation (Fig. 1A). Either full-length NAIP2 or N-terminal baculovirus inhibitor of apoptosis protein repeat (BIR) domain–deleted NAIP2 (NAIP2ΔBIR) was used. During purification, sucrose gradient fractions showed variable molar ratios between NAIP2 and NLRC4Δ (Fig. 1B). The fraction with the highest amount of NLRC4Δ relative to NAIP2, by approximately one order of magnitude, contained mostly single, complete disk-like particles (Fig. 1C and fig. S1A). Similarly, overstoichiometry of NLRC4Δ to NAIP5 was observed in the reconstituted FliC-NAIP5-NLRC4Δ complex (Fig. 1D), also with mostly complete disks under electron microscopy (fig. S1B). In contrast, our earlier PrgJ-NAIP2-NLRC4Δ preparations exhibited much lower NLRC4Δ/NAIP2 molar ratios (fig. S1C), with mostly incomplete disks (fig. S1D). Labeling of NAIP2 with 5-nm Ni-NTA gold particles in the PrgJ-NAIP2ΔBIR–His-NLRC4Δ inflammasome revealed singularly labeled complexes (Fig. 1E). These data, together with the previous report that NAIP5 did not oligomerize in the presence of flagellin (15), demonstrated that a single NAIP exists in each complex, whether in a full disk or a partially assembled disk (Fig. 1F).
Fig. 1. Preparation and characterization of NAIP-NLRC4Δ complexes.
(A) Domain organizations of Salmonella typhimurium PrgJ, mouse NAIP2, mouse NLRC4, and mouse caspase-1. Domain size is drawn approximately to scale; residue numbers are labeled. (B) SDS–polyacrylamide gel electrophoresis (PAGE) of different fractions of the sucrose gradient ultracentrifugation during the purification of the PrgJ-NAIP2-NLRC4Δ complex. Locations of the three component proteins are labeled. The asterisk indicates a contaminating band. (C)A representative negative-stain EM image from fraction 7 in (B). (D) SDS-PAGE of amylose resin elution (lane 1), anti-Flag flow-through (lane 2), and anti-Flag elution (lane 3) fractions during the purification of the coexpressed His-FliC– Flag-NAIP5–His-MBP-NLRC4Δ complex. An enlarged image of lane 3 is shown. (E) Ni-NTA gold labeling (5 nm) of purified His-Sumo-PrgJ–NAIP2ΔBIR-His– His-Sumo-NLRC4Δ complex upon removal of the His-Sumo tag. (F) Schematic diagram of partial and complete inflammasome particles that contain variable ratios between NAIP2 (yellow) and NLRC4 (cyan). (G) Representative cryo-EM micrograph of PrgJ-NAIP2-NLRC4Δ particles. (H) An averaged 2D class of the 11-bladed PrgJ-NAIP2-NLRC4Δ inflammasome complex. The dimensions of the image are 43.5 nm × 43.5 nm.
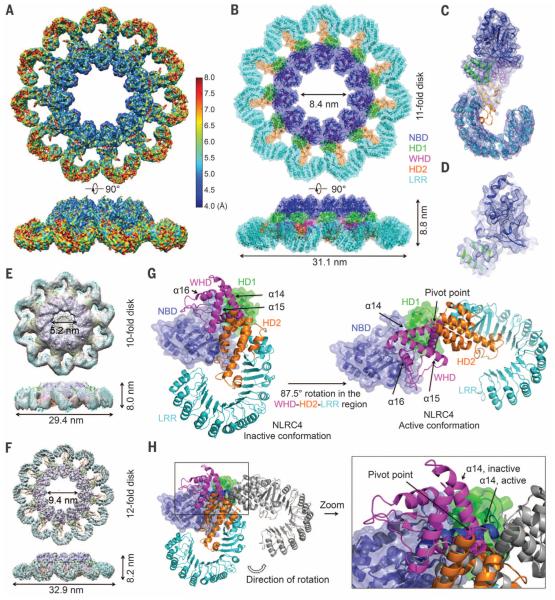
We collected cryo–electron microscopy (cryo-EM) data on the PrgJ-NAIP2-NLRC4Δ complex (Fig. 1G and figs. S2 and S3A). Reference-free two-dimensional (2D) classification revealed mostly 11-bladed, but also 12- and 10-bladed inflammasome complexes (fig. S3B), implying conformational flexibility. From the top or bottom view, an inflammasome disk comprises an inner ring and an outer ring (Fig. 1H); 3D classification yielded models with apparent C10, C11, and C12 symmetries (fig. S2). The individual blades did not show observable differences to indicate the single PrgJ-NAIP2 complex in each disk, probably due to similar domain organizations of NAIP2 and NLRC4 (Fig. 1A). Upon imposing the apparent symmetry, the C11, C12, and C10 reconstructions were refined to resolutions of 4.7 Å, 7.5 Å, and 12.5 Å, respectively (Fig. 2, A to F, and fig. S3, C to G). Local resolution estimation of the C11 reconstruction suggests that the inner ring possesses resolutions of 4.0 to 6.0 Å (Fig. 2A and fig. S4), with secondary structural features consistent with a resolution of at least 6.0 Å (Fig. 2, A to D, and fig. S5). Using the crystal structure of NLRC4Δ in the inactive conformation (PDB ID 4KXF) (16), we built and refined an atomic model of the active NLRC4Δ (table S1). All structures have a domed center and a prominent inner hole (Fig. 2, B, E, and F). The inner ring of the disk contains the NBD, helical domain 1 (HD1), and the winged helix domain (WHD); the outer ring comprises helical domain 2 (HD2) and the LRR domain (Fig. 1A and Fig. 2B).
Fig. 2. Cryo-EM structure determination and conformational activation of NLRC4.
(A) Cryo-EM map of the C11 PrgJ-NAIP2-NLRC4Δ complex colored with local resolution calculated by ResMap using two separately refined half maps. (B) Superimposed ribbon diagram and transparent surface of the C11 NLRC4Δ structure. (C) Cryo-EM density superimposed with one NLRC4Δ subunit. (D) A close-up view of the structure of the NBD of NLRC4Δ superimposed with the cryo-EM density. (E and F) Cryo-EM maps and fitted NLRC4Δ models for the C10 reconstruction at 12.5 Å resolution (E) and the C12 reconstruction at 7.5 Å resolution (F). (G) The WHD-HD2-LRR domain of NLRC4 swings 87.5° to transit from the inactive conformation (left, PDB ID 4KXF) to the active conformation (right). NBD and HD1 are shown in superimposed ribbon diagram and transparent surface, and the WHD-HD2-LRR module is shown in ribbon diagram. (H) Superimposed inactive (colored) and active (gray, except for a14 helix, which is in dark blue) conformations of NLRC4Δ. The α14 helices in the two conformations are labeled to show the relative rotations and the rotational pivot point.
We focus our further discussions on the 11-bladed structure with the highest resolution. The conformation of active NLRC4Δ in the 11-bladed inflammasome exhibits differences from that of inactive NLRC4Δ (16). When the NBD-HD1 regions of NLRC4 in the two states are aligned, the WHD-HD2-LRR module needs to rotate 87.5° along an axis at the junction between HD1 and WHD to turn from the inactive state to the active state (Fig. 2, G and H). The pivot point of the long-range hinge motion is where the inactive and active conformations of the a14 helix of WHD intercept (Fig. 2H). The rotation rearranges the intramolecular interactions between WHD and the NBD-HD1 module. Several missense mutations in human NLRC4 that are associated with auto-inflammatory conditions (12–14)—Thr337 → Ser, Val341 → Ala, and His443 → Pro—localize to this highly dynamic region (fig. S6A).
We do not know whether NLRC4 conformational transition is accompanied by exchange of adenosine diphosphate (ADP) in the inactive state to adenosine triphosphate (ATP) in the active state, as observed for apoptosome assembly by the related adenosine triphosphatase (ATPase) Apaf-1 (17, 18). Lack of nucleotide density in the cryo-EM map, local conformational changes, a modified Walker B motif, and absence of a conserved Arg in the sensor I motif (19) may all suggest alternative mechanisms (fig. S6, B and C). Consistently, the NLRC4 Walker A motif mutant Lys175 → Arg induced cell killing almost as effectively as did the wild type when coexpressed with NAIP5, flagellin, and caspase-1 (2).
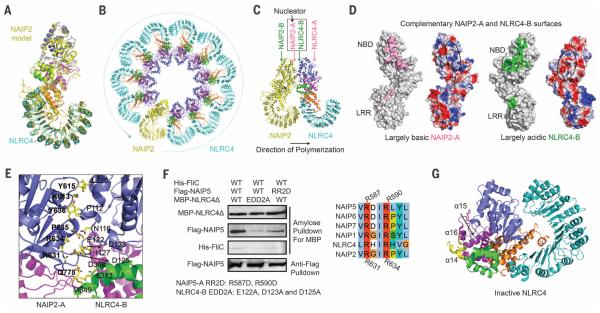
To facilitate analysis on NAIP2-NLRC4 interactions, we generated a homology model of NAIP2ΔBIR based on the NLRC4Δ structure (20) (Fig. 3A). By replacing one of the NLRC4 molecules in the initially fitted C11 structure, we generated a NAIP2-NLRC4 inflammasome model with one NAIP2 and 10 NLRC4 molecules, in which a single PrgJ-activated NAIP2 initiates NLRC4 activation and polymerization in a domino-like reaction (Fig. 3B). This mechanism differs from other oligomeric ATPases such as apoptosome proteins Apaf-1, CED-4, and DARK, which need to be activated before assembly, either conformationally (by its ligand cytochrome c) or by removal of the inhibitory protein CED-9 (21–23).
Fig. 3. Conformational activation of NLRC4 by activated NAIP2.
(A) Superposition of the NLRC4Δ structure and the NAIP2ΔBIR homology model in the active conformations. (B) A ribbon diagram of the 11-bladed NAIP2-NLRC4 inflammasome disk, with the single NAIP2 molecule in yellow and NLRC4 molecules in cyan. (C) Locations of A and B surfaces, in particular NAIP2-A and NLRC4-B, in the PrgJ-NAIP2-NLRC4Δ inflammasome. (D) Mapped interactions at the NAIP2-A surface (pink) and the NLRC4-B surface (green) and their surface electrostatic potentials. Dotted ovals show the approximate locations of the interface on the electrostatic surfaces. (E) Detailed interactions between the NAIP2-A surface and the NLRC4-B surface. Those on the A surface are labeled in boldface. (F) Mutations at the NAIP5-A and NLRC4-B surfaces impaired complex formation. The mutated residues in NAIP5 are completely conserved in NAIP2 and NLRC4. The three proteins were coexpressed in 293T cells; the MBP tag was used to pull down the complex, and the component proteins were detected using Western blots. (G) Ribbon diagram of NLRC4Δ in inactive conformation. The region of WHD at the tip of the α14, α15, and α16 helices, in slight clash with an interacting NAIP2, is shown in yellow. Amino acid abbreviations: D, Asp; E, Glu; H, His; I, Ile; K, Lys; L, Leu; M, Met; N, Asn; P, Pro; Q, Gln; R, Arg; S, Ser; V, Val; Y, Tyr.
The NAIP2-NLRC4 interactions are extensive, with a total surface area of ~1000 Å2 per subunit per interaction, and contain a mixture of hydrophobic, hydrophilic, and charged interactions. For brevity, we named one surface as A and the opposing surface as B (Fig. 3C), each comprising a large patch near the NBD and a small patch at the LRR (Fig. 3D). The LRR does appear to play a minor role in strengthening the oligomerization interactions because an LRR-deleted NLRC4 showed attenuation, not abrogation, of killing and oligomerization in comparison with ligand-activated wild-type NLRC4 (2). Calculation of surface electrostatics shows that the NAIP2-A surface, formed by NBD and WHD, is largely basic, whereas the NLRC4-B surface, formed by NBD, HD1, and WHD, is largely acidic; this would suggest that the NAIP2-A surface interacts with the NLRC4-B surface to initiate the directional formation of an inflammasome disk (Fig. 3D). Consistently, the NAIP2-B surface is largely incompatible with either NLRC4-A (fig. S7, A and B) or NAIP2-A (fig. S7, C and D). We thus named NAIP2-A the nucleating surface.
Interaction analysis revealed that residues Lys613, Tyr615, Arg631, Arg634, Pro635, Tyr636, and Gln778 of NAIP2-A and residues Pro112, Asn116, Glu122, Asp123, Asp125, Ile127, Leu220, Leu313, Met349, and Asp368 of NLRC4-B bury a large surface area at the interface (Fig. 3E). To test this interface, we used the His-FliC–Flag-NAIP5–His-MBP-NLRC4Δ inflammasome system because of the availability of differentially tagged constructs for coexpression in 293T cells and because of the sequence similarity between NAIP2 and NAIP5 (Fig. 3F). For NAIP5-A, we mutated Arg587 and Arg590, which correspond to Arg631 and Arg634 of NAIP2-A, to Asp (mutation RR2D). For NLRC4-B, we mutated the three acidic residues, Glu122, Asp123, and Asp125 to Ala (mutation EDD2A). Whereas the wild-type constructs showed robust copurification of NAIP5 and FliC by NLRC4, mutations on either NAIP5-A or NLRC4-B reduced the amount of copurified NAIP5 and FliC (Fig. 3F).
For NAIP2 to initiate NLRC4 polymerization, we hypothesized that it must have weak affinity with the inactive NLRC4. Mapping NLRC4-A and NLRC4-B residues onto the inactive NLRC4 conformation showed that NLRC4-B, but not NLRC4-A, is already largely formed, with only a small part of the WHD in clash with an interacting NAIP2-A (Fig. 3G and fig. S7E). The bound NAIP2-A at this site would push the WHD of NLRC4 exactly at a14, the hinge for conformational changes to occur (Fig. 2, G and H). We propose that the active NAIP2-A surface makes an initial encounter with the NLRC4-B surface in the inactive conformation to initiate the activating conformational change (fig. S7F).
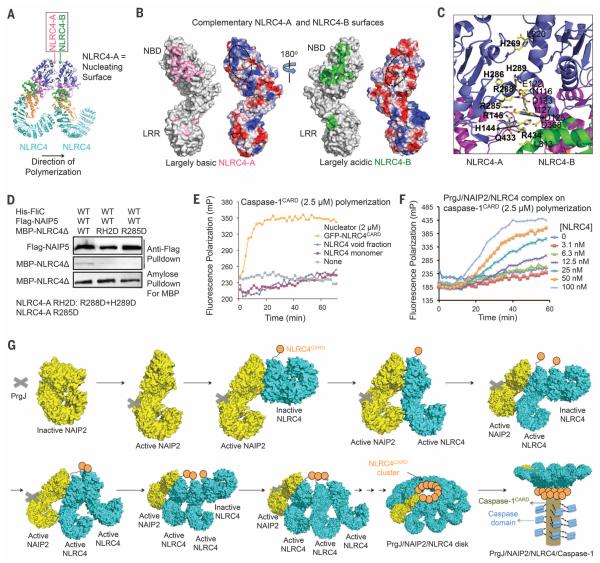
Calculation of surface electrostatics revealed charge complementarity between the mostly basic NLRC4-A surface and the opposing, largely acidic NLRC4-B surface (Fig. 4, A and B); this finding supports unidirectional polymerization nucleated by NLRC4-A. Structural analysis identified interfacial residues including His144, Arg145, His269, Arg285, His286, Arg288, His289, Gln433, and Arg434 of NLRC4-A, and Asn116, Glu122, Asp123, Asp125, Ile127, Leu220, Leu313, and Asp368 of the opposing NLRC4-B (Fig. 4C). We mutated NLRC4-A residues Arg288 and His289 to Asp (mutation RH2D) and Arg285 to Asp (mutation R285D) and tested their interactions using the same 293T cell coexpression system of His-FliC, Flag-NAIP5, and His-MBP-NLRC4Δ. Both RH2D and R285D mutations reduced the amount of NLRC4Δ in the inflammasome complex (Fig. 4D). Therefore, like a ligand-activated NAIP2, a newly activated NLRC4 triggers activation of another NLRC4 molecule by inducing conformational changes (fig. S7F). The NBD-NBD interactions between adjacent NLRC4 subunits differ from those in the heptameric apoptosome, with distinct angular relationships that may have explained the existence of more subunits in each NLRC4 disk (21, 22) (fig. S8, A to C).
Fig. 4. NLRC4 polymerization and caspase-1 activation.
(A) Locations of NLRC4-A and NLRC4-B surfaces. (B) Mapped interactions at NLRC4-A (pink) and NLRC4-B (green) surfaces and the surface electrostatic potentials. (C) Detailed interactions between two neighboring NLRC4 molecules. Those on the A surface are labeled in boldface. (D) Mutations at the NLRC4-A surface impaired NLRC4 recruitment. Three proteins were coexpressed in 293T cells. The Flag tag was used to pull down the complex; component proteins were detected using Western blots. (E) NLRC4CARD, instead of NLRC4FL, nucleates filament formation of labeled caspase-1CARD, as shown by increase in fluorescence polarization. (F) The PrgJ-NAIP2-NLRC4FL inflammasome nucleates filament formation of labeled caspase-1CARD at substoichiometric ratios, as shown by increase in fluorescence polarization. (G) Schematic diagram for mechanism of PrgJ-NAIP2–nucleated polymerization of NLRC4, followed by caspase-1 dimerization and activation.
ASC, like NLRC4, is an inflammasome adapter protein. We showed previously that ASC-dependent inflammasomes activate caspase-1 by ASCCARD-mediated caspase-1CARD polymerization (24). To examine whether the PrgJ-NAIP2-NLRC4 inflammasome may directly activate caspase-1 through the CARD in NLRC4, we adopted the same fluorescence polarization assay of caspase-1CARD (24). We reconstituted the full-length PrgJ-NAIP2-NLRC4 inflammasome, which showed stacked disk-like structures similar to the previously reconstituted FliC-NAIP5-NLRC4 complex (15) (fig. S8D), and expressed NLRC4CARD fused to green fluorescent protein (GFP-NLRC4CARD), which was filamentous under EM (fig. S8E). GFP-NLRC4CARD augmented the rate of caspase-1CARD polymerization, whereas full-length NLRC4 from either the monomeric or the void fraction had little effect (Fig. 4E). The PrgJ-NAIP2-NLRC4 inflammasome robustly promoted filament formation of caspase-1CARD even at a 1:100 substoichiometric ratio (Fig. 4F).
Given that NLRC4CARD alone is filamentous, we expect that the ~10 molecules of NLRC4CARD in each NAIP2-NLRC4 inflammasome form a short filament at the center of the disk. The observed stacking in full-length NAIP-NLRC4 inflammasomes (15) is likely due to interactions between unengaged NLRC4 CARDs. In the presence of caspase-1, the inflammasome may change into single disks, just like the transition of the Drosophila apoptosome from double rings to single rings in the presence of the caspase (21). Curiously, the central hole of the inflammasome has a diameter just a bit smaller than that of CARD filaments at ~9 nm, which may provide a perfectly sized “basin” to cradle the protruded CARD filament. These studies demonstrate that ASC-independent NAIP-NLRC4 inflammasomes make use of a similar mechanism for caspase-1 activation, as shown for ASC-dependent inflammasomes (24).
Our studies suggest that activation of NAIP-NLRC4 inflammasomes may proceed through the following steps (Fig. 4G), a conclusion also reached independently by the accompanying study (25): (i) Because of the domain similarity of NAIPs to NLRC4, we propose that the NAIP resting state is similar to the NLRC4 inactive conformation. After a cell is infected and bacterial products appear in the cytosol, a NAIP recognizes its specific bacterial ligand, likely through a surface on the HD1, WHD, and HD2 region (26). The specific ligand drives the NAIP into the open, activated conformation. (ii) The ligand-bound NAIP uses its nucleating surface to interact with the adapter NLRC4 that is yet to be activated. The interaction forces the WHD and its linked C-terminal region to change into the activated conformation, overcoming NLRC4 auto-inhibition. The activated NLRC4 uses its newly exposed nucleating surface to repeat recruitment and activation of additional NLRC4 molecules, until a complete disk is formed or until the NLRC4 concentration falls below the dissociation constant of the interaction. (iii) NLRC4 clustering induces oligomerization of the CARD of NLRC4, enabling the recruitment of caspase-1 through CARD-CARD interactions and triggering caspase-1 dimerization, autoproteolysis, and activation. The activation mechanism ensures signal amplification from the receptor to the adapter, and then to the effector.
As the most abundant energy source in living organisms, ATP is used widely in enzymes to mediate force generation, conformation change, oligomerization, and transport. The ATPase-mediated nucleated polymerization through a domino-like chain reaction identified here adds an important, elegant mechanism to this universal and already complex enzyme family. Nucleated polymerization in NAIP-NLRC4 inflammasomes also presents yet another mode of higher-order oligomerization, which may play a role in facilitating proximityinduced enzyme activation, threshold response, and prion-like propagation in immune signaling (27–30).
Supplementary Material
ACKNOWLEDGMENTS
We thank G. Bozkurt for technical assistance; H. Ploegh for providing engineered, Ca2+-independent sortase and the peptide-fluorophore conjugate Gly-Gly-Gly-TAMRA (GGG-TAMRA); and M. Ericsson for use of the Harvard Medical School EM facility. The data presented in this manuscript are tabulated in the main paper and in the supplementary materials. Cryo-EM maps and atomic coordinates have been deposited in the Electron Microscopy Data Bank (EMDB IDs EMD-6458, EMD-6459, and EMD-6460 for C11, C12, and C10 reconstructions, respectively) and Protein Data Bank (PDB ID 3JBL for the C11 reconstruction). Supported by NIH K99 grant AI108793 (Q.Y.), a Cancer Research Institute postdoctoral fellowship (L.D.), an Intel academic grant, research funds at Peking University, and NIH grant 1DP1HD087988. The cryo-EM facility was funded through the NIH grant AI100645 Center for HIV/AIDS Vaccine Immunology and Immunogen Design (CHAVI-ID). The experiments were performed in part at the Center for Nanoscale Systems at Harvard University, a member of the National Nanotechnology Infrastructure Network (NNIN), which is supported by NSF award ECS-0335765.
Footnotes
SUPPLEMENTARY MATERIALS
www.sciencemag.org/content/350/6259/404/suppl/DC1
Materials and Methods
Figs. S1 to S8
Table S1
References (31–48)
REFERENCES
- 1.Growney JD, Dietrich WF. Genome Res. 2000;10:1158–1171. doi: 10.1101/gr.10.8.1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kofoed EM, Vance RE. Nature. 2011;477:592–595. doi: 10.1038/nature10394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Diez E, et al. Nat. Genet. 2003;33:55–60. doi: 10.1038/ng1065. [DOI] [PubMed] [Google Scholar]
- 4.Zhao Y, et al. Nature. 2011;477:596–600. doi: 10.1038/nature10510. [DOI] [PubMed] [Google Scholar]
- 5.Yang J, Zhao Y, Shi J, Shao F. Proc. Natl. Acad. Sci. U.S.A. 2013;110:14408–14413. doi: 10.1073/pnas.1306376110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miao EA, et al. Nat. Immunol. 2006;7:569–575. doi: 10.1038/ni1344. [DOI] [PubMed] [Google Scholar]
- 7.Miao EA, et al. Nat. Immunol. 2010;11:1136–1142. doi: 10.1038/ni.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Franchi L, et al. Nat. Immunol. 2012;13:449–456. doi: 10.1038/ni.2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu B, et al. Proc. Natl. Acad. Sci. U.S.A. 2010;107:21635–21640. doi: 10.1073/pnas.1016814108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Janowski AM, Kolb R, Zhang W, Sutterwala FS. Front. Immunol. 2013;4:370. doi: 10.3389/fimmu.2013.00370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garaude J, Kent A, van Rooijen N, Blander JM. Sci. Transl. Med. 2012;4:120ra16. doi: 10.1126/scitranslmed.3002868. [DOI] [PubMed] [Google Scholar]
- 12.Canna SW, et al. Nat. Genet. 2014;46:1140–1146. doi: 10.1038/ng.3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kitamura A, Sasaki Y, Abe T, Kano H, Yasutomo K. J. Exp. Med. 2014;211:2385–2396. doi: 10.1084/jem.20141091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Romberg N, et al. Nat. Genet. 2014;46:1135–1139. doi: 10.1038/ng.3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Halff EF, et al. J. Biol. Chem. 2012;287:38460–38472. doi: 10.1074/jbc.M112.393512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu Z, et al. Science. 2013;341:172–175. doi: 10.1126/science.1236381. [DOI] [PubMed] [Google Scholar]
- 17.Reubold TF, Wohlgemuth S, Eschenburg S. J. Biol. Chem. 2009;284:32717–32724. doi: 10.1074/jbc.M109.014027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bao Q, Lu W, Rabinowitz JD, Shi Y. Mol. Cell. 2007;25:181–192. doi: 10.1016/j.molcel.2006.12.013. [DOI] [PubMed] [Google Scholar]
- 19.Proell M, Riedl SJ, Fritz JH, Rojas AM, Schwarzenbacher R. PLOS ONE. 2008;3:e2119. doi: 10.1371/journal.pone.0002119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martí-Renom MA, et al. Annu. Rev. Biophys. Biomol. Struct. 2000;29:291–325. doi: 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- 21.Yuan S, Akey CW. Structure. 2013;21:501–515. doi: 10.1016/j.str.2013.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yuan S, et al. Structure. 2011;19:128–140. doi: 10.1016/j.str.2010.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qi S, et al. Cell. 2010;141:446–457. doi: 10.1016/j.cell.2010.03.017. [DOI] [PubMed] [Google Scholar]
- 24.Lu A, et al. Cell. 2014;156:1193–1206. doi: 10.1016/j.cell.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu Z, et al. Science. 2015;350:399–404. doi: 10.1126/science.aac5489. [DOI] [PubMed] [Google Scholar]
- 26.Tenthorey JL, Kofoed EM, Daugherty MD, Malik HS, Vance RE. Mol. Cell. 2014;54:17–29. doi: 10.1016/j.molcel.2014.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu H. Cell. 2013;153:287–292. doi: 10.1016/j.cell.2013.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Franklin BS, et al. Nat. Immunol. 2014;15:727–737. doi: 10.1038/ni.2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cai X, et al. Cell. 2014;156:1207–1222. doi: 10.1016/j.cell.2014.01.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baroja-Mazo A, et al. Nat. Immunol. 2014;15:738–748. doi: 10.1038/ni.2919. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.