Abstract
A model is proposed for the evaluation of dispersive forces in a continuum solvent representation for use in large-scale computer simulations. It captures the short and long-range effects of water-exclusion in conditions of partial and anisotropic hydration. The model introduces three parameters, one of which represents the degree of hydration (water occupancy) at any point in the system, which depends on the solute conformation, and two that represent the strength of water-water and water-solute dispersive interactions. It is optimized for proteins, using hydration data of a sub-optimally hydrated binding site and results from dynamics simulations in explicit water. The model is applied to a series of aliphatic-alcohol/protein complexes and a set of binary and ternary complexes of various sizes. Implications for weak and ultra-weak protein-protein association and for simulation in crowded media are discussed.
I. Introduction
When a solute approaches another solute in an aqueous solution the water displaced is reorganized structurally and dynamically. The overall effect of water removal and reorganization can be divided into electrostatic and non-electrostatic forces. A continuum solvent representation for use in large-scale (i.e., long-time, big-size) simulations requires both effects to be properly incorporated into the force field, and to be computationally efficient as well. To this end, effective potentials have been developed that represent the solvent effects implicitly,1–5 but these have focused mainly on the treatment of electrostatics. Non-electrostatic effects include solvent-induced forces (SIF) and dispersion forces (DF), both essential components of the hydration process. SIF are short-ranged, non-pairwise forces that originate in the rearrangements of the solute-water and water-water hydrogen-bond network in the solute hydration shells,6–9 and thus modulate molecular interactions at close proximity. In purely nonpolar solutes, SIF are hydrophobic forces, for which several continuum theories have been proposed.10–13 For polar/charged solutes the treatment of SIF is more complicated, especially if the surfaces are topographically irregular, as most proteins are.14,15 SIF make important contributions to the intermolecular potentials of mean force,16 and partially determine the height of the desolvation barriers and the strength of hydrogen bonds (HB) between hydrated groups.5 A model has been proposed to account for HB modulation by SIF in protein simulations,5,17 and an algorithm developed to incorporate the non-pairwise nature of SIF in Langevin dynamics.18
The focus here is on the implicit treatment of DF. These forces are weak but pervasive,19 and their importance in implicit solvation has long been recognized.20–23 The fundamental role of DF in protein hydration energies,21,24 protein-ligand interactions,21,25–28 thermal stability of proteins,29,30 and preferential hydration/binding underlying protein denaturation by cosolutes31 have been probed experimentally and computationally. Increasing awareness of the importance of water-mediated DF has led to a number of extensions of implicit solvent models (ISMs) to incorporated DF in protein simulations.32,33
Because DF are attractive at all distances, failure to account for solvent DF may lead to overly compact structures of peptides and proteins, over-stabilization of non-native conformers, unphysical orientations and reduced fluctuations of side chains at protein surfaces, and stronger protein/ligand binding. In the latter case the problem is typically circumvented by neglecting the direct protein-protein dispersive energy altogether. This ad hoc solution is based on the notion that proteins and the water they displace upon association make equal contributions to the dispersive energy, and thus cancel out. This assumption is not always justified because the density of interfacial water varies substantially throughout the protein surface, and range from mild over-hydration,34 to partial dewetting,35 to significant dehydration in crevices and narrow pockets.26 Indeed, it has been shown that dispersive interactions are the main contributions to the binding energy in sub-optimally hydrated binding sites as a result of uncompensated dispersive attraction upon binding.26,27 In addition, DF between large solutes have a long-range, shape-dependent component36 (e.g., ~1/L for spheres, and ~1/L2 for planes, where L is the separation between the surfaces), which may result in an effective attraction or repulsion between the solutes in an aqueous solution,28,37 depending on their densities and materials, as determined by the relative values of the Hamaker constants.38 The strength of protein-water dispersion interactions has been estimated by computer simulations,21 and it was shown that a significant contribution is made by atoms in the interior of the protein (for ubiquitin, a rather small protein, the protein-water van der Waals energy was estimated at ~45 kcal/mol21). Other simulation studies have shown similar results.24 Although water and proteins are arguably similar organic substances, small differences in dispersive energies could rapidly add up as the system size increases, and could conceivably make a measurable contribution in multimeric complexes and assemblies. Such effective attraction may even exist between proteins and small molecules, as shown in μs-length simulations of urea binding to hen lysozyme.31 The implications for the behavior of crowded, anisotropic media, such as those in the interior of a living cell could be far reaching, possibly affecting subcellular organization and molecular translocation.
The model introduced here corrects the aforementioned limitations of current ISMs, and is designed for large-scale simulations. Parameterization is based on results from dynamic simulations of barnase/barstar and cytochrome C peroxidase/cytochrome C complexes in explicit water, and on calorimetric data of a sub-optimally hydrated protein binding site. Results are presented for a set of binary and ternary complexes of different sizes, and for a series of aliphatic alcohol/MUP-I complexes.
II. Model
The simplest treatment of electrostatic and non-electrostatic forces in a continuum solvent representation is through a local, position-dependent permittivity ε and density ρ.18,39 The dependence of these functions at the solute/liquid interface is complicated, but suitable approximations based on classical theories of dielectrics40 and liquid structure41 can provide practical expressions for both.3,18,39 In this case each element of liquid exerts a force on the solute, and the total force is obtained by integration over the space occupied by the liquid.18 Improvements can be introduced empirically by incorporating non-local and other non-linear effects in ε and ρ.39,42,43 Most ISMs used in macromolecular simulations assume ρ = 0, so the solute-solute DF tend to be stronger, especially for atoms close to the interface, where uncompensated forces are more apparent. The model proposed here adopts an intermediate approach, in which ρ ≠ 0 only at critical points, thus correcting for solvent DF at a fraction of computational cost.
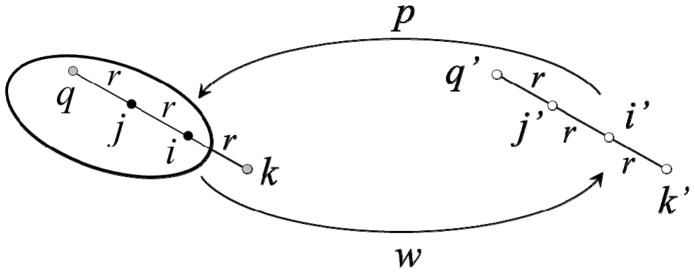
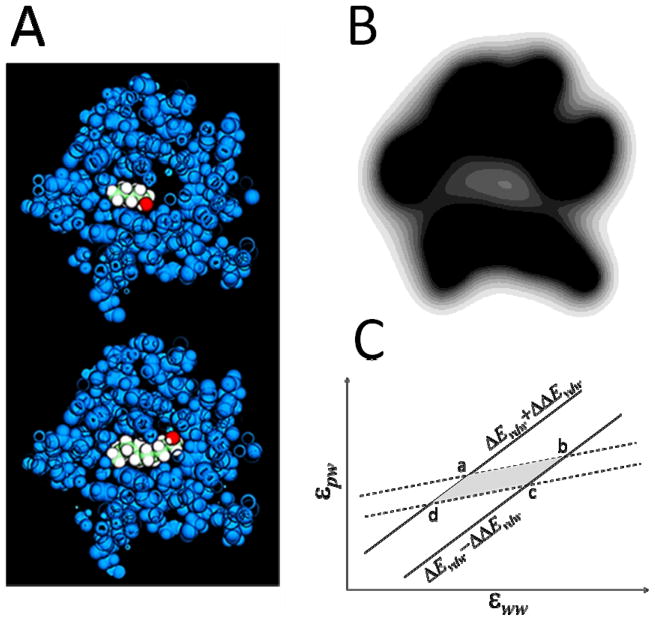
A solute atom i in a liquid is subject to an average dispersion force f = |f|r/r towards atoms within a volume dv at a position r (relative to i); an element dv at −r exerts an average force f′ = −|f′|r/r in the opposite direction. If both elements of volume are in average occupied by the same kind of substance, both forces cancel out. An imbalance of forces can result from different substances occupying r and −r or by different water densities in either position. In the absence of explicit water a major imbalance of forces will systematically occur throughout the system, which must be corrected. In addition, when a solute is assembled in solution the (dispersion) work necessary to move an atom i from bulk water to its final position in the solute is partially compensated by the work of removing water from the site in the opposite direction. These two independent processes are summarized in the cycle of fig. 1. A solute element p is moved from i′ in bulk water to i in the solute, whereas a water element w is moved from i to i′. The position j is occupied by another solute element, and site k is occupied by water with an occupancy sk dependent on the solute conformation (s → 1 in bulk water, and s → 0 within the solute); positions j′ and k′ are both occupied by bulk water. If the dispersive energy between two elements of volume separated a distance r is Vd = −2εσ6/r6 (with ε > 0) the energy change upon transfer, , is given by
Figure 1.
Formation of a solute in water. A solute element p is transferred from bulk water (site i′) to its final position (i) in the growing solute, while water w is moved from i to i′. Site j at a distance r from i is occupied by solute; sites k and k′ contain water with occupancies sk ≤ 1 and sk′ = 1; likewise for q and q′; si and sj are assumed to be unity.
| (1) |
which is probably the simplest way to incorporate the dependence of the solvent dispersion energy on the solute conformation. All the σ’s are here set equal to σij, so water-exclusion effects are incorporated only through ε’s. By setting εik = εiw sk, εwj = εwj si, and εwk = εww sk si, and because εwj′ = εwk′ = εww and εi′j′ = εi′k′ = εiw, the term in parenthesis in eq 1 can be cast into a single effective dispersive factor , where
| (2) |
is the solvent-corrected strength of the direct (εij) dispersive interaction between the two solute elements (si was set to unity, i.e., the site is assumed to be occupied by water prior to transferring the atom). Similar calculation for j in the presence of i yields (cf. fig. 1)
| (2′) |
where sq is the water occupancy at site q. To preserve the symmetry of the potential the effective strength of the pair (ε̃ij) is obtained as the arithmetic mean of and , i.e.,
| (3) |
where the solvent-corrected interaction is determined by ε̃′ij = [ε′ij + ε′ij]/2. Because ε̃ij is symmetrical upon permutation of indexes i and j, the total van der Waals energy of a solute composed of N atoms is
| (4) |
where is the direct solute-solute van der Waals energy and Vdisp is the correction to the dispersive energy due to the solvent. The dependence on the solute conformation, represented by r ≡ {r1, r2, …, rN}, where ri is the position of atom i, is made explicit in the last term.
An obvious choice for s(r) in eqs 2 and 2′ is a function proportional to the local water density ρ(r), for which any of the common approximations in liquid theory, including a simple barometric law probably suffices. For computational efficiency the approach used here is based on a contact model similar to that used in the development of the SCP implicit model of electrostatics,5,44 namely,
| (5) |
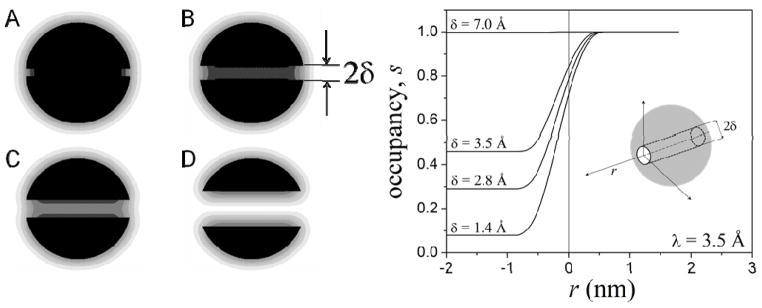
where rkl ≡|rl + rj − 2ri| is the distance between an atom l of the solute and the off-solute site rk = 2ri − rj determined by the (i, j) pair; a similar equation holds for sq, with rql = |rl + ri − 2rj|. Thus, s → 1 for a point far from the solute (bulk water), and s0 is chosen so that s → 0 for a point in the interior of the solute. For proteins, s0 can be estimated analytically5 as s0 ≈ 3(ζ + 2ζ2 + 2ζ3)exp(−1/ζ), where ζ = λ/pa and pa = (3Vp/4πNp)1/3 ~1.4 Å is the average radius of a protein atom, and Vp ~183 Å3 and NP ~15 are the average volume and average number of atoms per residue, respectively, estimated from the PDB. The behavior of eq 5 is illustrated in fig. 2 for a sphere with a radius of 2 nm containing a cylindrical cavity of radius δ along its major axis. The sphere is composed of atoms distributed uniformly in a cubic lattice, with a number density NP/residue. To illustrate the effects of cavity size on water occupancy, δ is varied from 1.4 Å (radius of a water molecule) to 7 Å (effective diameter of an amino acid). Figure 2 shows a grey-scale representation of s(r) on a plane containing the cavity axis (left panel), along with the values of water occupancy along the axis (right panel) for λ = 3.5 Å. For a narrow cavity only its openings are partially accessible to water, while the interior remains fully dehydrated (s ~ 0). As the cavity size increases the level of hydration increases from sub-optimal (0 < s < 1) to optimal (s ~1). In the current implementation the level of hydration is represented by the single parameter λ, which must be optimized using experimental or simulation data (cf. Section III). The extent of hydration inside the cavity depends not only on the thermodynamic conditions, the size and the topology of the pocket, but also on the chemical properties of the lining atoms. Improvement of the parameterization (not pursued here) would then require at least two λ’s, one for non-polar groups and one for polar/charged groups.
Figure 2.
Left panel: gray-scale representation of water occupancy s (eq 5) in a sphere containing a cylindrical cavity of radius δ along one of its major axes (black: s = 0; white: s = 1). The system mimics a typical globular protein with a diameter of ~4 nm, containing a binding site with varying degrees of water accessibility. The sphere is formed by identical atoms distributed in a cubic lattice with a constant of 1.4 Å; λ in eq 5 is 3.5 Å. (A) δ = 1.4 Å (radius of a water molecule); only the openings of the cavity are accessible to water; (B) δ = 2.8 Å; hydration at the center of the cavity remains significantly reduced; (C) δ = 3.5 Å (effective radius of an amino acid in a protein); hydration reaches half the value of bulk water; (D) δ = 7 Å; near optimal hydration. Right panel: water occupancy as a function of the distance r along the cavity axis for different cavity radii δ; r = 0 is at the entrance of the cavity (inset).
Introducing the expression of sk and sq into eq 3 yields ε̃′ij = aij + biΣexp(−rkl/λ) + bjΣexp(−rql/λ) where aij = εww − εiw − εwj; bi = (εww − εiw)/2s0; and bj = (εww − εwj)/2s0. For proteins, an additional simplification is introduced in which a single dispersion strength is assigned to all the atoms (represented by a generic index p), i.e., εiw = εpw. Given the known sensitivity of simulation results to Lennard-Jones parameters, as evidenced by the continuous effort in force field parameterizations based on chemical atom types,45 this simplification may require revision. In addition, water occupancy at a given off-solute site is largely determined by the solute atoms close to the site, so the summation in eq 5 can be restricted to a subset Nk of atoms l within a distance dc from k; likewise for a subset Nq around q. Finally, the parameter λ is made independent of k, so the final expression for ε̃′ij is
| (6) |
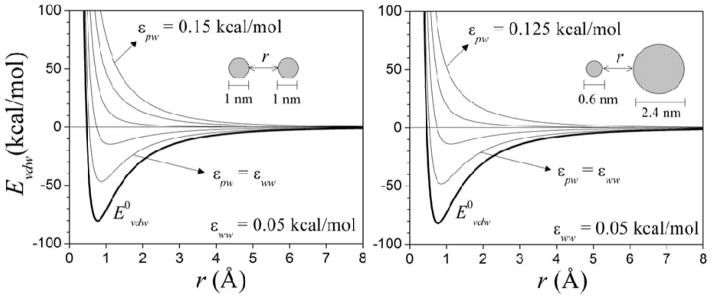
This expression contains three parameters to be determined, either a, b, and λ; or εww, εpw, and λ. In the latter approach, which is followed here, εww and εpw have a physical meaning. In particular, these are positive quantities, and their values are expected to be close to those of LJ parameters in the protein force field. Their relative values determine whether the effective interactions between hydrated solutes are attractive or repulsive, as illustrated in fig. 3. All the potentials show a long-range decay.
Figure 3.
van der Waals interaction energy Evdw (eq 4) between two spheres in water, as a function of the distance r between their surfaces, and for different values εww and εpw (eq 6). The spheres are composed of identical atoms distributed uniformly on a cubic lattice with a constant of 1.4 Å. Lennard-Jones parameters are εij = 0.15 kcal/mol and σij = 2.5 Å (LB mixing rules used). The energies at equilibrium are similar (within ~2 kcal/mol) in both systems for the same values of εww and εpw. The direct potentials (E0vdw; for εww = εpw = 0) corresponding to the uncorrected interactions are also shown.
The van der Waals force Fvdw,i on an atom i is calculated as Fvdw,i = −∇iEvdw, where the gradient is taken with respect to ri while keeping all the off-solute positions rk fixed in the calculation of the partial derivatives. From eq 4 , where the first term is the direct van der Waals force ( ) and the second term is the correction of the total dispersion force on i due to the solvent, given by
| (7) |
where r̂ij = (rj − ri)/|rj − ri| is the unit vector in the direction determined by the pair (i, j), and r̂ik = (rk − ri)/|rk − ri| is the unit vector determined by i and the off-solute site k. The first term in eq 7 has the usual pairwise form of a dispersion force determined by ε̃′ij, and originates in the presence of solvent at sites k and q. The quantity η in the second sum of eq 7 is given by
| (8) |
and originates in the changes of solvent occupancy at site k due to changes in the position of atom i; θ is the Heaviside step function, and the index k is determined by the corresponding (m, n) atom pairs, i.e., rk = 2rm − rn, and rik = rk − ri, so σk ≡ σnm and rk ≡ rnm. In practice, the cutoff distance dc can be made rather small, so most of the N(N−1) terms in eq 7 vanish.
III. Results
Isothermal titration calorimetry (ITC) of a series of aliphatic alcohols bound to the major urinary protein (MUP-I) showed that the binding enthalpy is determined mainly by ligand-protein dispersive interactions.26,27 This is an example of suboptimal hydration, hence an ideal system for optimization of λ in eq 5. The crystal structures of the complexes (cf. caption fig. 7) show a narrow binding pocket located deep into MUP-I (fig. 4A) and lined mostly with nonpolar residues. These structures are used here to calculate the average water occupancy within the binding pocket and the binding enthalpies of the complexes. For computational efficiency the summation in eq 5 is restricted to Cα atoms only, so s0 is determined by pa = 3.5 Å. This simplification was used previously to treat long-range water-exclusion effects on electrostatics.5 The protein is immersed into a cubic lattice with a cell length of 1.4 Å, and s calculated with eq 5 at each point of the lattice. The density of water within the binding pocket was estimated26 at ~0.21 g/cm3. An average occupancy <s> ~0.2 is well reproduced by eq 5 with λ = 2 Å, although s is heterogeneous throughout the pocket (fig. 4B). In general, increasing λ lowers s. This value of λ was estimated with no cutoffs in eq 5, but <s> remains unchanged with dc as small as ~12 Å.
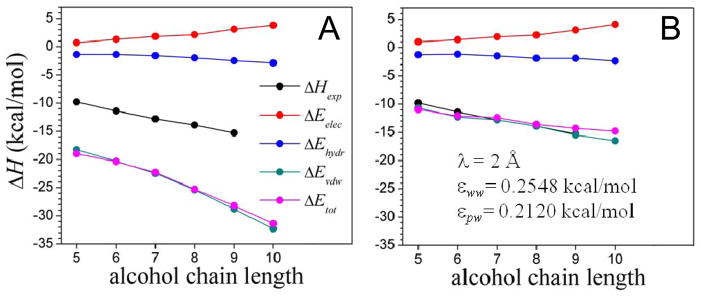
Figure 7.
Energy contributions to the binding energy of the MUP-I protein to a series of aliphatic alcohols as a function of the chain length, calculated with the implicit solvent model (Eelec given by eq 10; Evdw by eq 4, and Ehydr is the cavity-formation energy). (A) Uncorrected dispersion energy ( in eq 4); and (B) solvent-dispersion corrected energy ( ). The structures of the complexes are taken from the PDB; accession codes 1znd, 1zne, 1zng, 1znh, 1znk, 1znl, for pentan-, hexan-, heptan-, octan-, nonan-, and decan-1-ol, respectively. The experimental binding enthalpy26 (ΔHexp) is also shown (ITC data for decan-1-ol not available). Binding is dominated by dispersive forces, as determined by the extent of sub-optimal hydration of the binding pocket.
Figure 4.
(A) Cross section of the pentan-1-ol/MUP-I (upper) and decan-1-ol/MUP-I (lower) complexes, showing the buried cavity and the alcohol molecules filling the space (PDB 1znd used). (B) Gray-scale representation of water occupancy s on a (arbitrary) plane across the sub-optimally hydrated binding cavity. To be compared with fig. 2. Water occupancy was calculated with λ = 2 Å in eq 5, which yields <s> ~0.2 inside the binding pocket, based on reported results.27 (C) Iso-energy lines of the van der Waals dissociation energies of barnase/barstar (dashed lines) and cytochrome C peroxidase/cytochrome C (solid) complexes in the εww−εpw plane, determined by the statistical errors (~2 kcal/mol) estimated from MD simulation in explicit water. The overlapping section is defined by the following (εww, εpw) pairs: a = (0.2252, 0.2501); b = (0.4313, 0.4074); c = (0.2824, 0.2898); d = (0.0763, 0.1325); all in kcal/mol.
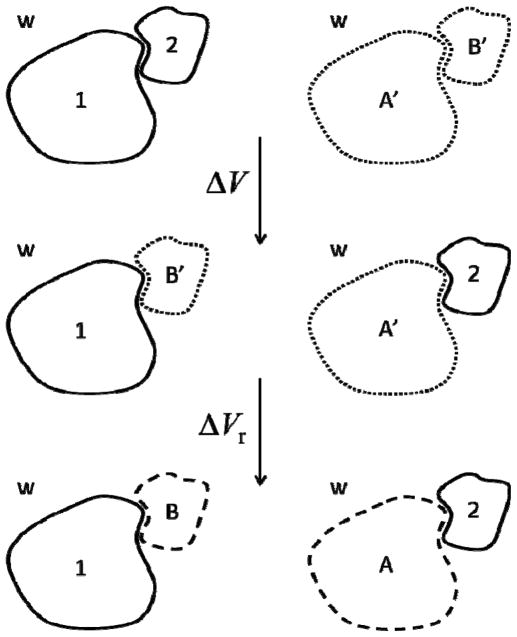
The optimized λ is used next to obtain εww and εpw in eq 6. Figure 5 depicts a generalization of the cycle of fig. 1, in which two proteins (1 and 2) are dissociated from their bound state. The van der Waals contribution to the total dissociation energy is ΔEvdw = ΔV + ΔVr, where ΔV is the energy required to separate the proteins without affecting the behavior of water (an ideal process), and ΔVr is the reorganization energy of water in contact with the protein. A′ and B′ are regions of bulk water with the same shapes and volumes as proteins 1 and 2, whereas A and B are the same regions of (non-bulk) water in contact with the proteins. After evaluating ΔV and ΔVr the energy change can be approximated as
Figure 5.
Thermodynamic cycle used to estimate the total van der Waals contribution to protein-protein binding in water used in the molecular dynamics simulations. A′ and B′ are regions of bulk water with the same sizes and shapes of proteins 1 and 2, respectively; whereas A and B are the same regions of water in contact with the proteins 2 and 1 upon structural and dynamic reorganization (see text).
| (9) |
where EIJ is the van der Waals interaction energy between subsystems I and J, and the terms with I = J are self-energies; all the terms corresponding to interactions of the proteins with bulk water (E1w and E2w) cancel out, and the differences (EAw − EA′w) and (EBw − EB′w) are small compared to the other terms in eq 9, thus neglected. A system in which the protein interfaces are well hydrated prior to binding appears to be the barnase/barstar complex, for which the binding enthalpy at 25 °C and pH 7 has been measured46 at ~19.3 kcal/mol. Each term of eq 9 is here calculated for this complex using molecular dynamic simulations in explicit water, so ΔEvdw and the individual contributions can be estimated directly. The all-atom representation of the CHARMM protein force field45 (version c35b5) with default protonation states (pH 7) was used, resulting in an electrically neutral system. The simulations were carried out in the canonical ensemble at 25 °C and 1 atm, using periodic boundary conditions and the CHARMM-optimized TIP3P water model. A cubic simulation cell with a side length of ~9.33 nm was used, large enough to accommodate both bulk and non-bulk (interfacial) water around the complex and the unbound proteins. Long-range electrostatics was treated with PME summations,47 with specifications reported previously.16 Bond lengths and angles were constrained with SHAKE, and a 2-fs time step was used (leapfrog integrator). Pressure was maintained with the Langevin piston method,48 with a mass of 400 amu and a collision frequency of 20 ps−1; temperature was fixed with the Hoover thermostat,49 using a mass of 103 kcal mol−1 ps2. NPT-equilibration of pure water yielded an average density of ~0.993 g/cm3.
The conformations of the proteins were taken from the crystal structure of the complex (PDB id 1brs). To calculate E12 (1 = barnase; 2 = barstar) the complex was initially centered in the simulation cell and water molecules overlapping protein atoms were removed. To calculate E1B, E2A, EAA, and EBB the unbound proteins were positioned within the simulation cell in the same positions they occupied in the simulation of the complex. Throughout the simulations all the Cα atoms were constrained at fixed positions, and the system equilibrated for 5 ns. To calculate EA′A′, EB′B′ and EA′B′ only the dynamics of water was simulated. The regions of non-bulk (A, B) and bulk (A′, B′) water were defined by the molecular surfaces of the corresponding proteins, using the Lee-Richards algorithm (probe radius set to zero). The error in the estimate of ΔEvdw is chosen to be ΔΔEvdw ~2 kcal/mol, which is the expected accuracy of the force field. Therefore, each term in eq 9 was evaluated with a statistical error ΔE = σ/n1/2 ~0.25 kcal/mol, where σ is the standard deviation of the corresponding term EIJ obtained from the fluctuating energy EIJ(t), and n is the number of uncorrelated steps given by n = τ/τc, where τ is the total simulation time and τc the correlation time estimated from the autocorrelation function C(t) ~ ∫EIJ(t′)EIJ(t+t′)dt′. To decrease the errors to the required values each term in eq 9 required a different simulation length τ, ranging from 10 to 20 ns. For the calculation of the energies from the trajectories PME was abandoned, and all the non-bonded terms were calculated directly without truncation. All the energies reported below are in kcal/mol. The changes in water self-energies upon reorganization were practically identical despite the different amounts of water in each region: EAA − EA′A′ = 12.5 ± 0.8 and EBB − EB′B′ = 12.7 ± 0.8. Both energies are also positive, implying unfavorable rehydration. The similarity of self-energies appears to be related to the number of water molecules “in contact” with the protein surfaces, which are the ones affected the most by the reorganization at the interfaces. Thus, region A has a number of water molecules in contact with protein 2, which is similar to the number of water molecules in B in contact with 1. However, this similarity is not expected to be a general feature, and larger differences would likely be observed in the presence of more complex surfaces. There is also a slight increase in the average number of water molecules within each region: nA = 668 ± 10 and nA′ = 663 ± 10; and nB = 544 ± 9 and nB′ = 542 ± 9, where Δn are now standard deviations. From a thermodynamic standpoint, these changes are related to the differences in chemical potential (μ) induced by the surfaces. Thus, μ of water in A′ cannot be equal to μw of the bulk water, but μ in A becomes equal to μw once all the structural rearrangements (including water absorption) set in; similar for B and B′. The changes in self-energies within each region can be partially explained by an increase in water packing induced by the protein surfaces. The interaction terms are: E2A = −38.5 ± 0.1, E1B = −41.2 ± 0.1, EA′B′ = 7.3 ± 0.1, and E12 = −62.9 ± 0.1. The resulting van der Waals contribution is thus ΔEvdw = 1.1 ± 2 kcal/mol, a small attraction but within the statistical error of the calculation. Therefore, in this case where water reorganization at the interface is modest and the proteins are relatively small, ΔEvdw ~ 0 is justified.
The parameterization requires a second protein complex. To partially incorporate the effect of protein size, the complex is chosen on the basis of the following criteria: One of the proteins must be similar in size to barnase (or barstar) and the second protein must be larger than barstar; in addition, the proteins must be globular and relatively rigid, with a topographically simple protein/protein interface to also expect near-optimal pre-binding hydration. Cytochrome C peroxidase contains the same number of residues than barnase; and its ligand, cytochrome C, is ~3.5 times larger than barstar. The binding enthalpy of this complex (PDB id: 2pcc) has been measured at ~2.6 kcal/mol. Dynamic simulations were performed using the same protocol described above. The van der Waals contribution to dissociation was estimated at ΔEvdw = 0.5 ± 2 kcal/mol, also weakly attractive but still within the statistical error.
The dispersive contributions to the dissociation energies are now calculated using eqs 4 and 6 with the optimized λ. Robust methods exist to evaluate the relative binding affinities of small ligands for a rigid binding site,50 including free energy perturbation and thermodynamic integration. Accurate estimates of absolute free energies are more challenging, even for small system, and no reliable methods are yet available for proteins, although progress is being made.51 The method employed here is simple, yet robust enough for the purpose of parameterization. The conformations of the complexes are first relaxed using canonical MC simulations of the proteins in the bound state at 25 °C. The average energy is calculated52 as <Eb> = Z−1Σi Ei exp(−Ei/kT) ≈ Σi Ei/n, where Ei and n in the last sum are the energy of the accepted conformation i and the number of accepted conformations, respectively. The energy is given by Ei = Eelec,i + Evdw,i + Ehydr,i, each term corresponding, respectively, to electrostatic, van der Waals, and hydrophobic contributions. Electrostatic interactions are calculated with the SCP model.5,53 In this model the electrostatic energy Eelec of a solute composed of N atoms is given by,
| (10) |
where r denotes the conformation of the system, and q are the atomic charges. In the partition of eq 10 the first sum is the interaction energy term, and the second sum is the self-energy term. The dependence of Eelec on the conformation is through the screening functions D and the effective radii R, as both of these quantities account for solvent-exclusion effects at different length scales.5 The effects of SIF on HB energies are introduced5 through a suitable modification of R, whereas the hydrophobic energy is given by a cavity term Ehydr = aγ, where γ is the solute solvent-accessible surface-area, and a is a parameter obtained from hydration energies of small alkanes.53 The functions D, R, and γ are all estimated with a contact model similar to eq 5.
Optimization of the model requires adjusting εww and εpw in such a way that the association energy ΔEd = <Eb> − E∞ equals the experimental binding enthalpy at same temperature. This procedure would require a fully-converged MC simulation for each parameter set over the course of the optimization. To simplify the computational demand only one structure in the bound state is used, which is chosen from a MC simulation carried out with the non-corrected model ( in eq 4). A member k of the canonical ensemble is selected such that Ek is closest to the average <Eb> of the ensemble. The representative structure k is subjected to a short minimization to relax it with the dispersion-corrected force field. For each values of εww and εpw the dissociation energy is calculated simply as ΔEd = Ek − E∞, a major simplification but good enough for the optimization. Then, for each (εww, εpw) pair, the van deer Waals dissociation energy ΔEvdw = Evdw,k − Evdw,∞ is compared to ΔEvdw obtained from the explicit-water simulations. Calculations are also carried out with the all-atom CHARMM force field,45 using standard protonation states, and no truncations of the non-bonded interactions.
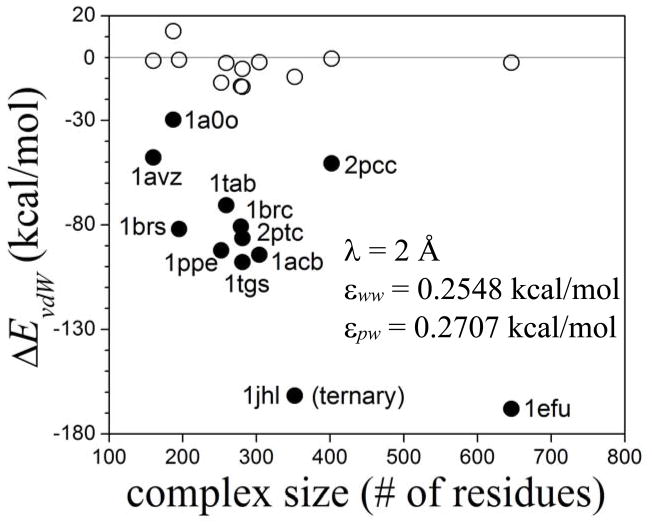
Figure 4C shows the region in the εww−εpw plane within which ΔEvdw (in kcal/mol) equals the dissociation energies. For each complex the iso-energy contour plots are straight lines, and lines corresponding to different complexes intersect, forming an extended rhomboid-shaped region. Any combination of εww and εpw within this region can be chosen as optimized parameters; in particular the values at the center, namely, εww ~0.2548 kcal/mol and εpw ~0.2707 kcal/mol. These values make physical sense as they are within the order of magnitude of the LJ parameters of the CHARMM force field. These values are used to estimate the van der Waals contributions to the binding energies of a set of binary and ternary protein complexes of varying sizes. The procedure is the same as described above for 1brs and 2pcc; for the ternary complex the dissociated state corresponds to the three components separated from one another. Results are shown in fig. 6. The uncorrected energies (solid circles) are large in all cases, but correction by water dispersion destabilize the complexes significantly (open circles). Yet, in all but one of the complexes the corrected van der Waals contributions are attractive. The degree of stabilization does not correlate with complex size (at least within this range of sizes considered here) but appears to be related more closely to the structural details of the protein/protein interfaces. For the binary complexes the contribution is negative (ΔEvdW < − 2 kcal/mol) for complexes in which a specificity-conferring residue (Arg or Lys) in one protein is buried in the other protein (1tgs, 2pts, 1brc, 1ppe). The interfaces of the other complexes are less tight, including 1a0o, which shows a rather low degree of complementarity. This reinforces the notion that dispersion in the presence of topographically complex interfaces characterized by crevices and narrow pockets could make important contributions to binding. This is illustrated by the aliphatic-alcohols/MUP-I protein complexes. Binding energies are calculated with the ISM at T = 27 °C, using the same protocol used for the protein complexes. Figure 7A shows the components of the binding energy without corrections for water dispersion effects. The hydrophobic attraction increases with chain length, as expected, but it is compensated by an unfavorable electrostatic energy. This leaves only the direct van der Waals interaction ( in eq 4) as the main contribution to the binding energy. This energy, however, is almost twice as large as the experimental enthalpy27 ΔHexp, a consequence of uncompensated dispersive interaction. The partial weakening of the binding energy is due to the solvent dispersion correction (Vdisp in eq 4), as shown in fig. 7B. To reproduce the experimental enthalpy within ~2 kcal/mol accuracy the parameter εpw for the alcohol atoms has to be adjusted to ~0.2120 kcal/mol. Using the same value of εpw obtained for proteins, the van der Waals contribution is underestimated by ~6 kcal/mol, which demonstrates the sensitivity of the results to the LJ parameters; nonetheless, the linear behavior with the chain length is still reproduced. As mentioned, a single εpw is unlikely to be optimal for all chemical species, and different values are needed for amino acids, nucleic acids, alcohols, fatty acids, and other basic units. Ultimately, an atom-type-based parameterization would probably be required, in line with the parameterization of the LJ potential in the force field. Another reason to recalibrate εwp for alcohols is that the Cα-based calculation of s can no longer be used; in this case the sum in eq 5 included all the C atoms of the aliphatic chain, which underestimates the values of s if the same λ is used.
Figure 6.
Water-dispersion effects of the binding energies of a number of binary and ternary protein complexes of varying sizes (identified by PDB id) as estimated with the ISM. Van der Waals energy of binding (ΔEvdw); solid symbols: uncorrected dispersive interactions ( in eq 4); open: corrected dispersion ( ). Barnase/barstar (1brs) and cytochrome C peroxidase/cytochrome C (2pcc) used in the optimization are also shown. The proteins include enzymes bound to substrates, inhibitors, or activators; and an antigen-antibody complex.
IV. Discussion and Conclusions
Dispersion forces have been shown to play a role in protein-ligand interactions,25–28 the conformational stability of proteins,29,30 and the preferential hydration/binding mechanism underlying denaturation by cosolutes.31 The importance of incorporating DF in an implicit solvent model has been established.20,22,23 Progress has been made in recent years, and models have been proposed that incorporate solvent DF empirically.32,33 The treatment of long-range effects and the dependence on the solute conformation are important physical features that still need careful consideration.
In this paper a model was proposed which incorporates solvent-corrected DF through effective interactions between the solute atoms. The model captures the short- and long-range effects of water-exclusion in conditions of partial and anisotropic hydration. It was developed in the context of a classical (molecular mechanics) force field, where electrostatics and van der Waals interactions are treated independently36 (although both are electrostatic in origin). The solvent correction is introduced through the LJ coefficients ε (eq 6). The extent of water exclusion is represented in ε through a conformation-dependent water-occupancy function s(r) (eq 5), using a contact model similar to that used earlier in the treatment of water-exclusion in electrostatics.5,44 In the current implementation ε are made independent of temperature; such dependence can eventually be incorporated through s. The model is simple yet general, as its formulation does not depend on the nature of the solute. In combination with the implicit electrostatic model developed previously5,44,53 (which includes effects of SIF) it forms a complete ISM, as it captures the main physical effects of partial, anisotropic hydration common in real biological systems. Improvements are probably needed both in the treatment of s and in the parameterization. The current implementation uses only three parameters, one of which (λ) represents the degree of hydration and two others that represent the strength of water-water (εww) and solute-water (εpw) dispersive interactions. These parameters were optimized for proteins, using hydration data of a sub-optimally hydrated binding site (MUP-I), and results from explicit-water MD simulations of two medium-sized complexes (barnase/barstar and cytochrome C peroxidase/cytochrome C).
The model was applied to a series of aliphatic-alcohol/MUP-I complexes for which ITC data suggest dispersive interactions to be the major contributions to the binding enthalpy. The model reproduces the linear increase of affinity with chain length. It was found, however, that (favorable) hydrophobic and (unfavorable) electrostatic energies are not negligible, but compensate each other for all the chain lengths, thus leaving dispersion as the main contribution. The model was also used to estimate the binding energies of several protein complexes of various sizes, demonstrating that a significant reduction of the dispersive attraction is achieved with the current implementation. Need for improvements can only be determined by MD simulations in explicit water, and accomplished through atom-type-based parameterization of εpw.
To account for long-range interactions no cutoffs were used to evaluate either eq 4 or eq 5, so the computational cost of the current implementation makes the model more suitable for peptides and small proteins. However, several strategies can be used to speed up computation of the long-range contributions, in line with the treatment of long-range electrostatics in the SCP model.5 These include suitable truncations of eqs 4 and 5; the use of Hamaker-like potentials between distant residues; infrequent updates of eq 3 during Langevin dynamics; and neglect of the 3-body terms in eq. 7. Depending on how these approximations are implemented the CPU time can be improved substantially, to a maximum of about ~2–4 times the computational cost required in the gas phase; these approximations, however, require careful re-parameterization, and will be the topic of a future study.
In general, aqueous interfaces display non-bulk behavior that can extend up to a few nanometers into the bulk, depending on the surface size, topography, and charge distribution.5 In particular, the density of interfacial water can vary from mild over-hydration,34 to partial dewetting,35 to significant dehydration in crevices and narrow pockets.26 The imbalance of DF introduced by density variations at the interface could affect the association/dissociation mechanism. Furthermore, dispersive interactions have a long-range component that may lead to effective attraction/repulsion between proteins in large complexes and assemblies. Small differences in the strength of protein-protein and protein-water DF could make significant contributions in weak and ultra-weak protein association.52,54 The implications in crowded, anisotropic environments, such as those in the interior of a living cell, can be far reaching. Evidence suggests that a large proportion of biologically active molecules in the cell do not diffuse freely in the bulk medium despite their high solubility, but are transiently bound to one another, to membranes, or to the cytoskeleton.55 In addition to electrostatics, long-range uncompensated DF may play a role in such non-specific interactions, possibly affecting subcellular organization and molecular translocation.
Acknowledgments
This study utilized the high-performance computer capabilities of the Biowulf PC/Linux cluster at the NIH, and was supported by the NIH Intramural Research Program through CIT.
References
- 1.Gilson MK, Davis ME, Luty BA, McCammon JA. Computation of Electrostatic Forces on Solvated Molecules Using the Poisson-Boltzmann Equation. J Phys Chem. 1993;97:3591. doi: 10.1063/1.1924448. [DOI] [PubMed] [Google Scholar]
- 2.Cramer CJ, Truhlar DG. Implicit Solvation Models: Equilibria, Structure, Spectra, and Dynamics. Chem Revs. 1999;1999:2161. doi: 10.1021/cr960149m. [DOI] [PubMed] [Google Scholar]
- 3.Tomasi J, Mennucci B, Cammi R. Quantum mechanical continuum solvation models. Chem Rev. 2005;105:2999. doi: 10.1021/cr9904009. [DOI] [PubMed] [Google Scholar]
- 4.Feig M, Brooks CL. Recent Advances in the Development and Application of Implicit Solvent models in Biomolecule Simulations. Curr Op Struc Biol. 2004;14:217. doi: 10.1016/j.sbi.2004.03.009. [DOI] [PubMed] [Google Scholar]
- 5.Hassan SA, Steinbach PJ. Water-Exclusion and Liquid-Structure Forces in Implicit Solvation. J Phys Chem B. 2011;115:14668. doi: 10.1021/jp208184e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ben-Naim A. Solvent-Induced Forces in Protein Folding. J Phys Chem. 1990;94:6893. [Google Scholar]
- 7.Bruge F, Fornilli SL, Malenkov GG, Palma-Vittorelli MB, Palma MU. Solvent-Induced Forces on a Molecular Scale: Non-Additivity, Modulation and Causal Relation to Hydration. Chem Phys Lett. 1996;254:283. [Google Scholar]
- 8.Durell SR, Brooks BR, Ben-Naim A. Solvent-Induced Forces Between Two Hydrophilic Groups. J Phys Chem. 1994;98:2198. [Google Scholar]
- 9.Hassan SA. Amino Acid Side Chain Interactions in the Presence of Salts. J Phys Chem B. 2005;109:21989. doi: 10.1021/jp054042r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lum K, Chandler D, Weeks JD. Hydrophobicity at Small and Large Length Scales. J Phys Chem B. 1999;103:4570. [Google Scholar]
- 11.Chandler D. Interfaces and the Driving Force of Hydrophobic Assembly. Nature. 2005;437:640. doi: 10.1038/nature04162. [DOI] [PubMed] [Google Scholar]
- 12.Pratt LR. Molecular theory of Hydrophobic Effects: She is too mean to have her name repeated. Annu Rev Phys Chem. 2002;53:409. doi: 10.1146/annurev.physchem.53.090401.093500. [DOI] [PubMed] [Google Scholar]
- 13.Hummer G, Garde S, Garcia AE, Pratt EA. New Perspectives on Hydrophobic Effects. Chem Phys. 2000;258:349. [Google Scholar]
- 14.Tan ML, Cendagorta JR, Ichiye T. Effects of Microcomplexity on Hydrophobic hydration in Amphiphiles. J Amer Chem Soc. 2013;135:4918. doi: 10.1021/ja312504q. [DOI] [PubMed] [Google Scholar]
- 15.Giovambattista N, Lopez CF, Rossky PJ, Debenedetti PG. Hydrophobicity of protein surfaces: Separating geometry from chemistry. Proc Nat Acad Sci (USA) 2008;105:2274. doi: 10.1073/pnas.0708088105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hassan SA. Intermolecular Potentials of Mean Force of Amino Acid Side Chain Interactions in Aqueous Medium. J Phys Chem B. 2004;108:19501. [Google Scholar]
- 17.Hassan SA, Guarnieri F, Mehler EL. Characterization of Hydrogen Bonding in a Continuum Solvent Model. J Phys Chem B. 2000;104:6490. [Google Scholar]
- 18.Hassan SA. Liquid-structure Forces and Electrostatic Modulation of Biomolecular Interactions in Solution. J Phys Chem B. 2007;111:227. doi: 10.1021/jp0647479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parsegian VA. Van der Waals Forces: A Handbook for Biologists, Chemists, Engineers, and Physicists. Cambridge University Press; New York: 2005. [Google Scholar]
- 20.Floris F, Tomasi J. Evaluation of the Dispersion Contribution to the Solvation Energy: a Simple Computational Model in the Continuum Approximation. J Comput Chem. 1989;10:616. [Google Scholar]
- 21.Pitera JW, van Gunsteren WF. The importance of solute-solvent van der Waals Interactions with interior atoms of biopolymers. J Amer Chem Soc. 2001;123:3163. doi: 10.1021/ja0057474. [DOI] [PubMed] [Google Scholar]
- 22.Zacharias M. Continuum Solvent Modeling of Nonpolar Solvation: Improvement by separating Surface Area dependent Cavity and Dispersion Contributions. J Phys Chem A. 2003;107:3000. [Google Scholar]
- 23.Wagoner JA, Baker NA. Assessing Implicit Models for Nonpolar Mean Solvation Forces: the Importance of Dispersion and Volume Terms. Proc Nat Acad Sci (USA) 2006;103:8331. doi: 10.1073/pnas.0600118103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Levy RM, Zhang LY, Gallicchio E, Felts AK. On the nonpolar hydration free energy of proteins: surface area and continuum solvent models for the solute-solvent interaction energy. J Amer Chem Soc. 2003;125:9523. doi: 10.1021/ja029833a. [DOI] [PubMed] [Google Scholar]
- 25.Chapman KT, Still WC. A Remarkable Effect of Solvent Size on the Stability of a Molecular Complex. J Amer Chem Soc. 1989;111:3075. [Google Scholar]
- 26.Barrat E, Bingham RJ, Warner DJ, Laughton CA, Phillips SEV, Homans SW. Van der Waals interactions Dominate Ligand-Protein Association in a Protein Binding Site Occluded from Solvent Water. J Amer Chem Soc. 2005;127:11827. doi: 10.1021/ja0527525. [DOI] [PubMed] [Google Scholar]
- 27.Malham R, Johnstone S, Bingham RJ, Barratt E, Phillips SEV, Laughton CA, Homans SW. Strong Solute-Solute Dispersive Interactions in a Protein-Ligand Complex. J Amer Chem Soc. 2005;127:17061. doi: 10.1021/ja055454g. [DOI] [PubMed] [Google Scholar]
- 28.Janshoff A, Neitzert M, Oberdorfer Y, Fuchs H. Force Spectroscopy of Molecular Systems. Single Molecule Spectroscopy of Polymers and Biomolecules. Angew Chem Int Ed. 2000;39:3212. doi: 10.1002/1521-3773(20000915)39:18<3212::aid-anie3212>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 29.Vondrasek J, Kubaz T, Jenney FE, Adams MWW, Kozisek M, Cerny J, Sklenar V, Hobza P. Dispersion Interactions Govern the Strong Thermal Stability of a Protein. Chem Eur J. 2007;13:9022. doi: 10.1002/chem.200700428. [DOI] [PubMed] [Google Scholar]
- 30.Berka K, Hobza P, Vondrasek J. Analysis of Energy Stabilization inside the Hydrophobic Core of Rubredoxin. Chem Phys Chem. 2009;10:543. doi: 10.1002/cphc.200800401. [DOI] [PubMed] [Google Scholar]
- 31.Hua L, Zhou R, Thirumalai D, Berne BJ. Urea Denaturation by Stronger Dispersion Interactions with Proteins than Water Implies a 2-Stage Unfolding. Proc Nat Acad Sci (USA) 2008;105:16928. doi: 10.1073/pnas.0808427105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gallicchio E, Levy RM. AGBNP: an analytic implicit solvent model suitable for molecular dynamics simulations and high-resolution modeling. J Comp Chem. 2004;25:479. doi: 10.1002/jcc.10400. [DOI] [PubMed] [Google Scholar]
- 33.Dzubiella J, Swanson JMJ, McCammon JA. Coupling hydrophobicity, dispersion, and electrostatics in continuum solvent models. Phys Rev Lett. 2006;96:087802. doi: 10.1103/PhysRevLett.96.087802. [DOI] [PubMed] [Google Scholar]
- 34.Merzel F, Smith JC. Is the First Hydration Shell of Lysozyme of Higher Density than Bulk Water? Proc Nat Acad Sci (USA) 2002;99:5378. doi: 10.1073/pnas.082335099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jensen TR, Ostergaard M, Reitzel N, Balashev K, Peters GH, Kjaer K, Bjornholm T. Water in Contact with Extended Hydrophobic Surfaces: Direct Evidence of Weak Dewetting. Phys Rev Lett. 2003;90:086101. doi: 10.1103/PhysRevLett.90.086101. [DOI] [PubMed] [Google Scholar]
- 36.Israelachvili JN. Intermolecular and Surfaces Forces. 3. Academic Press; San Diego, CA: 2011. [Google Scholar]
- 37.Hamaker HC. The London-van der Waals attraction between spherical particles. Physica. 1937;10:1058. [Google Scholar]
- 38.Hunter CA. Quantifying Intermolecular Interactions: Guidelines for the Molecular Recognition Toolbox. Angew Chem Int Ed. 2004;43:5310. doi: 10.1002/anie.200301739. [DOI] [PubMed] [Google Scholar]
- 39.Hassan SA. Self-consistent treatment of the local dielectric permittivity and electrostatic potential in solution for polarizable macromolecular force fields. J Chem Phys. 2012;137:074102. doi: 10.1063/1.4742910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bottcher CJF. Theory of Electric Polarisation: Dielectrics in Static Fields. 2. Vol. 1 Elsevier; 1973. [Google Scholar]
- 41.Hansen J-P, McDonald IR. Theory of Simple Liquids. 2. Elsevier; London: 1986. [Google Scholar]
- 42.Liszi J, Meszaros L, Ruff I. The field dependence of the Kirkwood factor and the nonlinear dielectric behavior of some liquids. J Chem phys. 1981;74:6896. [Google Scholar]
- 43.Rubinstein A, Sherman S. Influence of the Solvent Structure on the Electrostatic Interactions in Proteins. Biophys J. 2004;87:1544. doi: 10.1529/biophysj.103.038620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hassan SA, Mehler EL, Zhang D, Weinstein H. Molecular Dynamics Simulations of Peptides and Proteins with a Continuum Electrostatic Model Based on Screened Coulomb Potentials. Proteins. 2003;51:109. doi: 10.1002/prot.10330. [DOI] [PubMed] [Google Scholar]
- 45.Brooks BR, Brooks CL, Mackerell AD, Nilsson L, Petrella RJ, Roux B, Won Y, Archontis G, Bartels C, Boresch S, Caflisch A, Caves L, Cui Q, Dinner AR, Feig M, Fischer S, Gao J, Hodoscek M, Im W, Kuczera K, Lazaridis T, Ma J, Ovchinnikov V, Paci E, Pastor RW, Post CB, Pu JZ, Schaefer M, Tidor B, Venable RM, Woodcock HL, Wu X, Yang W, York DM, Karplus M. CHARMM: The Biomolecular Simulation Program. J Comp Chem. 2009;30:1545. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Frisch C, Schreiber G, Johnson CM, Fersht AR. Thermodynamics of the interaction of Barnase and Barstar: changes in free energy versus changes in enthalpy on mutation. J Mol Biol. 1997;267:696. doi: 10.1006/jmbi.1997.0892. [DOI] [PubMed] [Google Scholar]
- 47.Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG. A Smooth Particle Mesh Ewald Method. J Chem Phys. 1995;103 [Google Scholar]
- 48.Feller SE, Zhang Y, Pastor RW, Brooks BR. Constant Pressure Molecular Dynamics Simulations: the Langevin Piston Method. J Chem Phys. 1995;103:4613. [Google Scholar]
- 49.Hoover WG. Canonical Dynamics: Equilibrium Phase-Space Distributions. Phys Rev A. 1985;31:1695. doi: 10.1103/physreva.31.1695. [DOI] [PubMed] [Google Scholar]
- 50.Gilson MK, Given JA, Bush BL, McCammon JA. The Statistical-Thermodynamic Basis for Computation of Binding Affinities: A Critical Review. Biophys J. 1997;72:1. doi: 10.1016/S0006-3495(97)78756-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gilson MK, Zhou HX. Calculation of protein-ligand binding affinities. Annu Rev Biophys Biomol Struct. 2007;36:21. doi: 10.1146/annurev.biophys.36.040306.132550. [DOI] [PubMed] [Google Scholar]
- 52.Cardone A, Pant H, Hassan SA. Specific and non-specific protein association in solution: computation of solvent effects and prediction of first-encounter modes for efficient configurational bias Monte Carlo simulations. J Phys Chem B. 2013;117:12360. doi: 10.1021/jp4050594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hassan SA, Guarnieri F, Mehler EL. A General Treatment of Solvent Effects Based on Screened Coulomb Potentials. J Phys Chem B. 2000;104:6478. [Google Scholar]
- 54.Clore GM, Tang C, Iwahara J. Elucidating transient macromolecular interactions using paramagnetic relaxation enhancement. Curr Op Struc Biol. 2007;17:603. doi: 10.1016/j.sbi.2007.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Luby-Phelps K. Cytoarchitecture and Physical Properties of Cytoplasm: Volume, Viscosity, Diffusion, Intracellular Surface Area. Int Rev Cytol. 2000;192:189. doi: 10.1016/s0074-7696(08)60527-6. [DOI] [PubMed] [Google Scholar]