Abstract
Background
Early aerial senescence in switchgrass (Panicum virgatum) can significantly limit biomass yields. WRKY transcription factors that can regulate senescence could be used to reprogram senescence and enhance biomass yields.
Methods
All potential WRKY genes present in the version 1.0 of the switchgrass genome were identified and curated using manual and bioinformatic methods. Expression profiles of WRKY genes in switchgrass flag leaf RNA-Seq datasets were analyzed using clustering and network analyses tools to identify both WRKY and WRKY-associated gene co-expression networks during leaf development and senescence onset.
Results
We identified 240 switchgrass WRKY genes including members of the RW5 and RW6 families of resistance proteins. Weighted gene co-expression network analysis of the flag leaf transcriptomes across development readily separated clusters of co-expressed genes into thirteen modules. A visualization highlighted separation of modules associated with the early and senescence-onset phases of flag leaf growth. The senescence-associated module contained 3000 genes including 23 WRKYs. Putative promoter regions of senescence-associated WRKY genes contained several cis-element-like sequences suggestive of responsiveness to both senescence and stress signaling pathways. A phylogenetic comparison of senescence-associated WRKY genes from switchgrass flag leaf with senescence-associated WRKY genes from other plants revealed notable hotspots in Group I, IIb, and IIe of the phylogenetic tree.
Conclusions
We have identified and named 240 WRKY genes in the switchgrass genome. Twenty three of these genes show elevated mRNA levels during the onset of flag leaf senescence. Eleven of the WRKY genes were found in hotspots of related senescence-associated genes from multiple species and thus represent promising targets for future switchgrass genetic improvement. Overall, individual WRKY gene expression profiles could be readily linked to developmental stages of flag leaves.
Electronic supplementary material
The online version of this article (doi:10.1186/s12864-015-2057-4) contains supplementary material, which is available to authorized users.
Background
Switchgrass (Panicum virgatum) is a temperate, warm-season perennial that is being developed as a cellulosic biofuel crop [1, 2]. Tetraploid switchgrass populations and cultivars have higher yields as compared to octaploid populations [3]. Thus, most current breeding efforts are focused on improving biomass yields and quality in tetraploid lines [4, 5]. Tetraploid populations can occur as upland and lowland ecotypes, with the lowland plants significantly out-yielding the upland lines across several locations [5]. However, the latitudinal adaptation of these different ecotypes presents challenges, since most of southerly-adapted, high-yielding, lowland germplasm suffers from significant winter-kill at more northern sites of the USA [6]. Some crosses of upland x lowland plants show heterosis for yields [7], and this approach appears to hold promise in the continuing development of switchgrass as a biomass crop [8]. Nevertheless, extending the period of carbon assimilation by delaying aerial senescence could be a means to significantly improve yields, as long as other plant attributes, such as dormancy onset and nutrient remobilization are not impaired [6].
Senescence is a genetically programmed trait that can potentially be reprogrammed by several molecular breeding strategies such as marker-assisted selection. To develop switchgrass cultivars with delayed senescence, it is critical to determine the key molecular events that occur during senescence to identify the regulators that trigger this process. Senescence is the final stage of plant development and is tightly controlled to increase the fitness of the whole plant [9]. Transcriptome analysis of Arabidopsis thaliana (A. thaliana; At) leaf senescence suggests that several families of transcription factors play major roles in the cellular reprogramming associated with senescence. The major transcription factor families associated with A. thaliana leaf senescence are NACs, WRKYs, C2H2 zinc finger proteins, AP2/ERFs, MYBs, homeobox proteins, bZIPs, bHLHs, and C3H zinc finger proteins. WRKY TFs were the second largest TF family to be induced during senescence in this study [10].
WRKY transcription factors are key regulators of many plant processes, including responses to biotic and abiotic stresses, wounding, senescence, seed dormancy, and seed germination [11]. They are components of intracellualar signaling webs, for example many are phosphorylated by MAP kinase cascades [12]. The defining feature of WRKY transcription factors is their DNA binding domain referred to as the WRKY domain, which is named after the almost invariant WRKY amino acid sequence within the N-terminal region [13]. The WRKY domain is about 60 residues in length and also possesses a Cx4–5Cx22–23HxH or Cx7Cx23HxC zinc-finger structure at the C-terminus [11]. Structural determination of the WRKY domain bound to its W box cis-acting element revealed that part of a four-stranded β-sheet enters the major groove of DNA almost perpendicular to the DNA helical axis in a β-wedge. Amino acids in the conserved WRKYGQK signature motif contact the W box DNA bases [14].
Functional genomic studies of individual WRKY transcription factors have provided clear evidence that specific WRKY proteins are regulators of senescence, although some of these transcription factors play multiple roles in planta [15–17]. The first evidence supporting a role of WRKY transcription factors in the senescence process came from studies of A.thaliana AtWRKY6 [18, 19]. One target gene for AtWRKY6 is FLG22-induced receptor-like kinase 1 (FRK1 previously called SIRK) whose expression is strongly induced during leaf senescence. Senescing leaves of wrky6 knockout mutants showed a drastic reduction in FRK1 transcript levels and green leaves of WRKY6 overexpression lines showed clearly elevated FRK1 transcript levels. In A.thaliana, AtWRKY54 and AtWRKY70 appear to have cooperative and partly redundant functions in senescence, as revealed by single and double mutant studies [20]. AtWRKY54 and AtWRKY70 are both negative regulators of senescence and interact independently with AtWRKY30 which is expressed during developmental leaf senescence [20]. Another member of the WRKY family in A.thaliana, AtWRKY53, acts as a convergence node between senescence and pathogen responses [21]. The AtWRKY53-interacting protein UPL5 is a HECT domain E3 ubiquitin ligase that regulates leaf senescence in A.thaliana through degradation of AtWRKY53, demonstrating that targeted breakdown of AtWRKY53 is a feature of senescence in A.thaliana. Recently it has been shown that AtWRKY18 represses AtWRKY53 activity and acts as a positive regulator of senescence [22]. AtWRKY22 has also been implicated in regulating dark-induced leaf senescence and appears to share cross-regulation with AtWRKY6, AtWRKY53, and AtWRKY70 [23]. Other WRKY transcription factors that have been implicated in regulating senescence include rice OsWRKY42 [24], OsWRKY80 [25], and OsWRKY23 [26]. Taken together, studies of both specific WRKY transcription factors and the WRKY family as a whole demonstrate that WRKY proteins play important roles in regulating the process of senescence.
In this study, the members of the WRKY gene family that are present in Version 1.1 of the genome sequence of switchgrass (www.phytozome.org) were identified. The names and genomic locations were enumerated for 191 full-length WRKY genes, together with 49 partial WRKY genes where complete sequence of the gene was lacking. Some of these 49 genes are likely pseudogenes due to the presence of nonsense mutations within the ORF and missing portions of the WRKY domain. Switchgrass also contains a Group RW5 R protein-WRKY gene, consisting of a domain structure of B3-LRR-NB ARC-LRR-WRKY. The presence of a B3 DNA-binding domain together with a WRKY domain and domains from intracellular resistance proteins suggest that this R protein has at least two different DNA-binding domains with different cis-acting element specificities that could be responsive to both biotic and abiotic stresses. We also show that the switchgrass genome contains a second R protein-WRKY gene. The PviWRKY174 protein is a Group RW6 protein orthologous to sorghum SbRWRKY2 and SbRWRKY3. Existing RNA-Seq datasets [27] from flag leaves obtained from field grown switchgrass plants at distinct stages of development were used to understand the relationships between WRKY gene expression and leaf developmental state. Using a range of bioinformatic analyses, distinct modules of co-expressed genes were found to be associated with specific flag leaf developmental stages. A co-expressed cluster of 3000 genes containing 23 WRKYs were specifically associated with the onset of senescence.
Methods
Sequence data sets
The sequences of the complete WRKY gene family from switchgrass were taken from v1.1 genome sequence at Phytozome (http://www.phytozome.org/) [28]. These sequence data were produced by the US Department of Energy Joint Genome Institute. Senescence associated WRKY genes from other species were obtained from the Leaf Senescence Database (http://www.eplantsenescence.org/) by performing a text search with the term “WRKY”.
Identification and manual curation of the switchgrass WRKY transcription factor family
To identify the WRKY family in switchgrass, a modification of the TOBFAC pipeline was used [29]. Tblastn searches were performed against the JGI release v1.1 of the switchgrass genome sequence using a representative WRKY domain from each of the flowering plant subfamilies of WRKY transcription factors (I, IIa, IIb, IIc, IId, IIe, and III) [29]. The e-value was set to 10 to ensure that all potential WRKY domain-encoding sequences, however diverse or fragmentary, were discovered. All hits were pooled into a single data set and duplicate sequences were then removed. For each positive genomic sequence, about 20 kb of genomic sequence around the WRKY domain-encoding region (if available) was used in the gene prediction program FGENESH (http://www.softberry.com/) with the monocot plant setting and the resultant amino acid prediction compared to the gene model (if present). Positive genomic sequences were also analyzed by Hidden Markov Model analyses using the protein sequence vs profile-HMM 624 database tool at Janelia.org (http://hmmer.janelia.org). For this analysis the default settings of the program were used to search the Pfam, Gene3D, and Superfamily databases. The R protein-WRKY genes were further investigated by blastp, PSI-BLAST and tblastn searches at NCBI (http://www.ncbi.nlm.nih.gov/) [30, 31].
Phylogenetic analyses
The amino acid sequences of the WRKY domains or the complete amino acid sequences of the R protein-WRKYs [32] were used for phylogenetic analyses. Alignments were constructed using MUSCLE [33] with the following parameters; Gap Penalties: Gap open −2.9, Gap Extended 0, Hydrophobicity multiplier 1.2 Memory/Iterations: Max Memory in MB 4095, Max Iterations 8; Clustering Method Iteration 1, 2 (UPGMB), Clustering Method (Other Iterations (UPGMB), Min. Diag. Length (Lambda) 24. For the Neighbor Joining tree [34], the percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) were determined [35]. The evolutionary distances were computed using the Poisson correction method [36] and are in the units of the number of amino acid substitutions per site. All ambiguous positions were removed for each sequence pair. Evolutionary analyses were conducted in MEGA6 [37]. All positions containing alignment gaps and missing data were eliminated in pairwise sequence.
Switchgrass Flag Leaf RNA-Seq Data
Previously published [27] RNA-Seq data from switchgrass was used to analyze WRKY expression (SRA Accession SRX481052). Briefly, flag leaves from field grown cv Summer plants were collected in 2012 at five time points: heading (July 3), anthesis (July 27), seed set (August 16), mature seed (August 31), and senescence onset (September 19). At each time point, three pools of 10 flag leaves each were collected from randomly selected plants. RNA was isolated from all samples and 100 bp single-end sequencing was performed using an Illumina HiSeq2000 instrument with five samples per lane, yielding an average of 45 million reads per sample.
Mapping and differential gene expression analysis
HiSeq2000 100-bp reads were mapped to version 1.1 of the switchgrass genome (www.phytozome.org). Tophat2 (version 2.0.11) [38] was used with default parameters for mapping and reads with multiple alignments were discarded prior to counting gene expression, whereby only uniquely mapped reads were used for all subsequent analyses. Expression values were calculated using the featureCounts function in the Subread (version 1.4.4) analysis program [39], along with the version 1.1 gene annotation file which was modified to include WRKY genes identified as already described. Differentially expressed genes (FDR < 0.05) across the time series were identified using the likelihood ratio test in DESeq2 (version 1.6.3) [40, 41] in R [42].
NMDS and Hierarchical Clustering Analysis
Raw read counts were normalized and subjected to the variance stabilization transformation from DESeq2 to enable comparisons of expression levels between WRKY genes and to correct for differences in gene lengths. Standardized counts of WRKY genes were analyzed via NMDS using the ‘metaMDS’ function from the vegan package [43] in R (version 3.1.1 for Linux) to determine if WRKY expression profiles changed over the course of development time in coordinated manner and to ascertain whether flag leaves collected at the same developmental time points displayed similar WRKY expression patterns.
To identify clusters of WRKY genes that were activated during each flag leaf developmental stage, variance stabilized counts of differentially expressed WRKY genes that were obtained above were also subjected to agglomerative hierarchical analysis as follows: Variance-stabilized read counts obtained in the previous section were log transformed and Z-scores were computed. A compositional Euclidean dissimilarity matrix was computed and clustering analysis was performed using Ward’s method on z-scores derived from the average of the replicates collected at each time point in JMP® Version 9.0 (SAS Institute Inc, Cary, NC, 1989–2007).
Co-expression Network Analysis
Weighted Gene Co-expression Network Analysis (WGCNA, version 1.43), an R package, was used to identify groups of genes having similar expression patterns across the flag leaf time series [44, 45]. Differentially expressed genes with a log2 fold change of 1.5 or greater were used for network analysis (19,049 total genes). A soft threshold (β) value of 12 was used in the transformation of the adjacency matrix in order to meet the scale-free topology criteria. Co-expression modules were created with the blockwiseModules function using the following parameters: TOMType=”unsigned”, maxBlockSize = 20000, mergeCutHeight = 0.4, minModuleSize = 15. The expression pattern of the resulting modules is represented by the module eigengene (ME), which corresponds to the first principal component of a given module.
Module Visualization
Cytoscape (version 3.2.0) [46] was used to visualize co-expression networks identified using WGCNA. The topological overlap measure (TOM) calculated by WGCNA was used as a measure of co-expression for pairs of genes. Prior to visualization, the overall network size was reduced in two ways. First, all gene pairs were filtered by requiring one of the two genes to be a transcription factor. Putative switchgrass transcription factors were identified by PFAM [47] annotations and following the family assignment rules detailed in the Plant Transcription Factor Database v3 [48]. A total of 901 putative transcription factors (including WRKYs) were identified in the flag leaf time series differentially expressed gene list. Second, the top 2.5 % of the TOM values for each gene pair were retained. The final network contained 13,405 nodes (genes) connected by 428,378 edges (TOM values). The network was drawn in Cytoscape using the AllegroLayout plugin with an edge-weighted Allegro Fruchterman-Reingold algorithm.
Promoter Analysis of Genes of Interest
To identify potential cis-regulatory elements in WRKY genes induced during senescence, the promoter regions of the senescence-related WRKY genes, defined as 1000 bp upstream of the start codon, were scanned for putative regulatory motifs using Place Web Signal Scan (https://sogo.dna.affrc.go.jp/cgi-bin/sogo.cgi?lang=en&pj=640&action=page&page=newplace) [49, 50]. While promoter regions could not be identified for several of the WRKY genes, which were directly adjacent to scaffold boundaries or were located on unplaced contigs, we were able to extract full 1000 bp promoter regions for 17 of the 23 WRKY genes that were activated during flag leaf senescence.
Availability of supporting data
The data used in this manuscript are available as part of the short-reads archive depository within the NCBI at http://www.ncbi.nlm.nih.gov/sra/SRX481052/
Results
The WRKY transcription factor family in switchgrass
To identify all members of the WRKY gene family in the switchgrass (v1.1) genome sequence, the same modified version of the TOBFAC pipeline used previously to identify WRKY genes in Brachypodium distachyon [29] was employed. Briefly, tblastn searches [30] were performed against the Panicum virgatum v1.1 genome sequence using a representative WRKY domain from each of the subfamilies of WRKY transcription factors found in flowering plants (I, IIa, IIb, IIc, IId, IIe, and III). These multiple searches employed a cut off e-value of 10 in order to identify all possible WRKY domain encoding sequences even if these sequences were incomplete. All of the positive sequences were combined into a single dataset and redundant sequences were removed. Finally, every sequence was manually curated to ensure that each sequence contained at least part of a WRKY domain and could therefore be regarded as a switchgrass WRKY gene. This pipeline enabled us to produce a data set of switchgrass WRKY genes that is more complete than the predicted gene models in the v1.1 genome sequence. For each positive genomic sequence, about 20 kb of genomic sequence around the WRKY domain-encoding region (if available) was used in the gene prediction program FGENESH with the monocot plant setting and the resultant amino acid prediction was compared to the gene model (if present in phytozome). Positive genomic sequences were also analyzed by Hidden Markov Model analyses using the protein sequence vs profile-HMM 624 database tool at Janelia.org (http://hmmer.janelia.org). Using this pipeline we identified 191 full length WRKY genes and named them PviWRKY1-PviWRKY191 (Additional file 1: Table S1). We gave the genes PviWRKY names to avoid confusion with Phaseolus vulgaris, the common bean, whose genome sequence is publically available. In addition, we found 49 WRKY domain-containing sequences that did not encode a full length gene. Inspection of these incomplete sequences revealed that many were located on short contigs and were therefore lacking the complete genomic sequences. Several others were present on chromosomal sequences but coding regions were interrupted by regions of “Ns” and therefore have missing portions of the gene. This situation is not surprising as version 1.1 of the switchgrass genome contains 636.1 Mb of sequence localized to chromosomes with an additional 593.5 Mb which is not localized. The Panicum virgatum Genome Sequencing Project expects that there will be significant future movement of genes as they integrate direct sequence from the clone based genome improvement project (http://phytozome.jgi.doe.gov) into future assemblies. The total number of WRKY sequences identified in the P.virgatum genome assembly was 240. Of the 191 complete WRKY genes, nine were completely lacking a gene model in phytozome and three had gene models that incorrectly predicted the WRKY domain. The forty nine incomplete WRKY genes were named PartialWRKY1-PartialWRKY49 and will be added to the list of complete genes or pseudogenes when additional sequence data become available. Of these 49 sequences, 27 had a partial gene model, suggesting that they could be functional. RNA-Seq data collected from switchgrass flag leaves detected transcripts from 37 partial WRKYs and 169 full-length WRKYs indicating a final number of at least 206 expressed WRKY genes in switchgrass leaves (Additional file 1: Table S1).
Resistance protein-WRKY genes in the switchgrass genome
One key feature of the WRKY gene family in many, but not all, species of flowering plants is the existence of chimeric proteins comprising domains typical for both resistance (R) proteins and WRKY transcription factors. An atlas of R protein-WRKY genes has been assembled and the proteins classified into eight groups [32]. These groups of R protein-WRKYs have been named RW1-8 and new groups can be expected to be discovered as more plant genome sequences become available. The switchgrass WRKY transcription factor PviWRKY178 encodes a Group RW5 protein of the domain structure B3-LRR-NB ARC-LRR-WRKY (Fig. 1) that we have also previously called PvRWRKY1 [32]. This gene was expressed in flag leaves (see below). This gene is a distinct type of R protein-WRKY because it contains two different types of DNA-binding domain (a WRKY domain and a B3 domain). The B3 domain has previously been identified in three major classes of transcription factors, ABI3/VP1-like factors, the RAV-like family, and auxin response factors (ARFs) [51] but Group RW5 proteins are the first reported proteins that combine B3 domains and WRKY domains. The B3 domain proteins play roles in the responses to abscisic acid and auxin [52, 53] but the role of PviWRKY178/PvRWRKY1 is currently unknown.
Fig. 1.
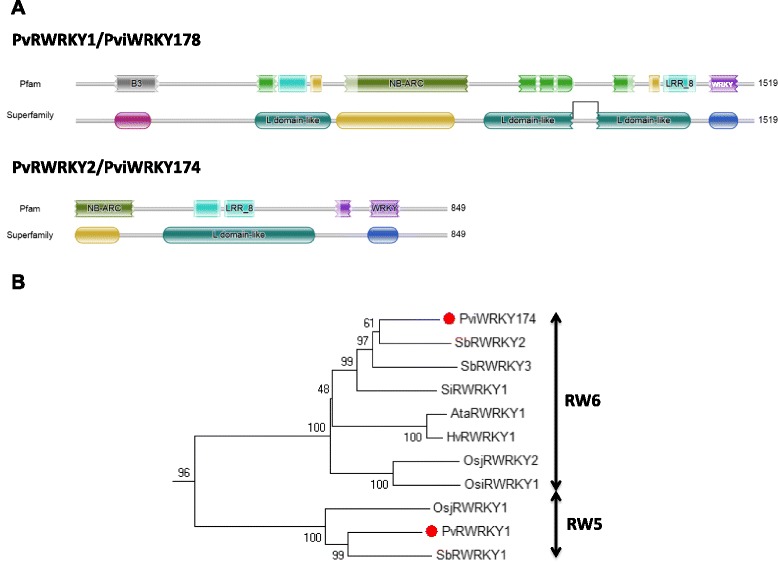
Resistance protein – WRKY (RW) genes in switchgrass. a Domain structures of PvRWRKY1/PviWRKY178 and PvRWRKY2/PviWRKY174. Hidden Markov Model analyses were performed with the complete amino acid sequences using the protein sequence vs profile-HMM database tool at Janelia.org (http://hmmer.janelia.org) and searching the Pfam and Superfamily databases. b Neighbor Joining phylogenetic tree derived from a MUSCLE alignment of full length R protein-WRKYs from Groups RW5 and RW6. Numbers indicate bootstrap values from 1000 replicates. Red dots denote switchgrass proteins
Detailed searches also revealed that PviWRKY174 also encodes an R protein-WRKY and that it had previously been overlooked [32] as it is located on a short contig of only 6.13 kb (Additional file 1: Table S1). Nevertheless, HMMER analysis suggests that the complete coding sequence may be present and phylogenetic analyses demonstrate that the PviWRKY174 protein is a Group RW6 protein similar to proteins encoded by sorghum SbRWRKY2 and SbRWRKY3 (Fig. 1). Therefore the switchgrass genome contains two expressed RW genes, one belonging to Group RW5 and one to group RW6. Both sorghum (Sorghum bicolor) and rice (Oryza sativa) also contain RW genes from these two groups and this may be a feature of some, but not all monocot genomes. For example, the Brachypodium distachyon genome appears to lack RW genes entirely.
This comprehensive analysis of the WRKY family of genes and proteins in switchgrass permitted a deeper enquiry of existing transcriptomic datasets to quantitate specific WRKY gene expression during flag leaf development in field grown switchgrass plants. Our intent was to (a) describe the overall patterns of WRKY gene expression at specific stages of leaf and plant development; and (b) to analyze gene networks associated with leaf senescence with the goal of discovering WRKYs that could activate flag leaf senescence.
The dataset used in the current study was obtained from flag leaves [27] collected from plants at heading (7/3/12); anthesis (7/27/12); early seed set (8/16/12); seeds at hard seed set (8/31/12); and at physiological maturity when flag leaf senescence was visually obvious (9/19/12). Global aspects of these data sets have been described in Palmer et al. (2014). A total of 110 WRKYs were differentially expressed across all harvest dates.
Non-metric multidimensional scaling (NMDS) analysis of WRKY gene expression
To determine whether the WRKY expression profiles were correlated with flag leaf development, NMDS ordination based on a Bray-Curtis dissimilarity matrix of the standardized expression levels of the differentially expressed (DE) WRKY genes in switchgrass flag leaves was performed. This analysis (Fig. 2) demonstrated that WRKY gene expression levels in flag leaves collected at each developmental stage (harvest date) were distinct and that individual replicates within each time point were quite similar to each other, indicating that the expression levels of WRKY genes were highly consistent in flag leaves at the same stages of development. Furthermore, the DE-WRKY expression profiles of young expanding flag leaves (red circles, Fig. 2) were separated from the DE-WRKY expression profiles of flag leaves collected at other harvest times. DE-WRKY expression patterns were also more similar in flag leaves as they transitioned to source leaves (blue circles, Fig. 2) compared to other harvest dates when flag leaves were expected to be fully functional in terms of carbon assimilation and transport (green and black circles, Fig. 2). The first and last harvest dates were most differentiated by the NMDS axis 1, when flag leaves were elongating or had started to senesce respectively. These results indicated that the WRKY expression profile changed in a coordinated manner over the course of flag leaf development and WRKYs contributing to senescence could be identified.
Fig. 2.
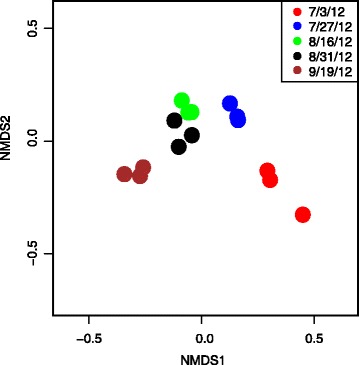
Non-metric multidimensional scaling (NMDS) analysis of WRKY expression patterns in switchgrass flag leaves collected across five different developmental states. The three biological replicates at each harvest date are shown in the same color
Hierarchical Clustering Analysis
Because the NMDS analysis indicated that activation of specific WRKY genes may stimulate senescence in flag leaves, a hierarchical clustering to identify groups of senescence-related WRKY genes was conducted (Fig. 3). Two distinct clusters of WRKY genes whose expression levels were elevated during senescence were observed (Additional file 1: Table S1). The first cluster (red) included 16 genes whose expression levels were elevated on the 9/19/12 collection date compared to other time points, indicating that their induction is likely linked to senescence in flag leaves. The second cluster (blue) included 10 genes (Additional file 2: Table S2) whose expression levels were highest on 8/16/12 and 9/19/12 time points. A third cluster (orange) included 8 WKRY genes whose expression levels were elevated at both the 7/13/12 and 9/19/12 time points, although their expression levels were significantly increased on 9/19/12 relative to 7/13/12, potentially linking them to leaf senescence. A fourth cluster (cyan) included 8 WRKY genes with variable patterns of expression levels. Of significance are three WRKY genes whose expression levels were elevated on 8/31/12 and 9/19/12 (Fig. 3). Notably, a large cluster containing 39 highly expressed WRKY genes was also observed during early flag leaf development and may play roles in regulating developmental processes involved in leaf expansion and functionality (Additional file 2: Table S2).
Fig. 3.

Hierarchical clustering analysis of 110 differentially expressed WRKY genes. The Z-score heatmap was derived from the average of the replicates collected at each time point (displayed in Fig. 2). Yellow is low expression; Magenta is high expression
WRKY genes within co-expression modules
Unsigned weighted gene co-expression network analysis (WGCNA) was utilized to place genes that were identified as differentially expressed over the course of flag leaf development (FDR < 0.05, log2 fold change > 1.5) into thirteen co-expression modules. Gene membership in the modules ranged from 5682 genes in module 1 to 21 genes in module 13. The expression profile of each module is summarized by a module eigengene, which is analogous to the first principal component of the module expression data. Comparison of the module eigengenes (Fig. 4, Additional file 3: Figure S1, Additional file 4: Figure S2) revealed four related module sets that displayed similar expression profiles across flag leaf development: (1) modules 1, 8, and 13; (2) modules 4, 5, 6, and 12; (3) modules 2, 10, and 11; (4) modules 3, 7, and 9. Key expression characteristics within each module set included high expression at (a) anthesis for set 1, (b) during seed development (seed set and mature seed) for set 2, (c) at heading for set 3, and (d) at the onset of senescence for set 4.
Fig. 4.
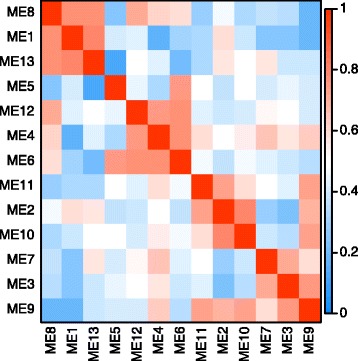
Module eigengene adjacency heatmap. Module-eigengenes (ME) in this analysis are defined as the first principal component of a coexpression module matrix. The heatmap shows the relatedness of the 13 co-expression modules (ME1-ME13) identified by WGCNA with red being highly related and blue being not related
A closer inspection of WRKY genes within the context of co-expression modules indicated that 79 out of 85 WRKYs included in WGCNA were assigned to five co-expression modules (Fig. 5). Module 2, whose expression peaked at heading (7/3/12) when flag leaves were still expanding [27], contained 39 WRKYs. Module 1 was the largest co-expression module and contained 5682 genes whose expression peaked at 7/27/12 coincident with anthesis and yet only contained six WRKYs. Nine WRKYs were found in module 5, whose expression levels peaked at seed set (8/16/12). This was a transition stage for flag leaves and plants as documented by the down regulation of genes linked to chlorophyll biosynthesis along with the appearance of a new sink tissue (seeds) respectively [27].
Fig. 5.
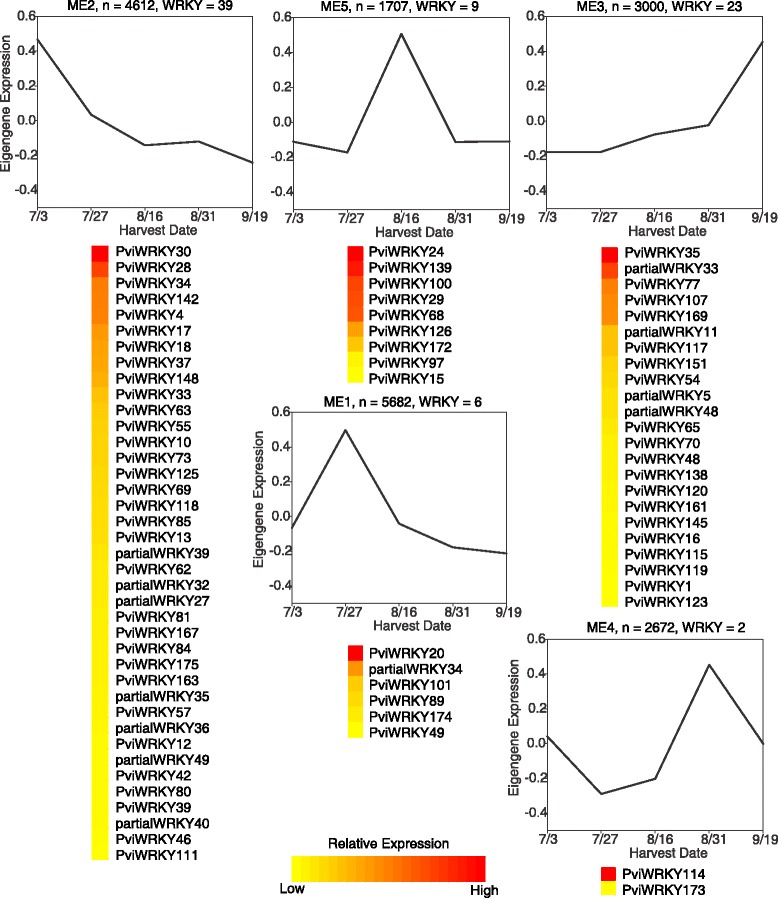
Select WRKY containing co-expression modules. Module eigengene expression profiles across the time series are shown, along with relative expression levels of individual WRKY genes within each select co-expression module. Red indicates high relative expression and yellow indicates low relative expression
Two WRKYs were present in module 4, both with maximum expression at or around the time seeds attained physiological maturity [27]. Genes assigned to module 3, including 23 WRKYs (Fig. 5), had highest expression coinciding with senescence onset (9/19/12). These 23 WRKYs may be associated with senescence related processes.
Senescence associated WRKY genes from switchgrass
Figure 6a shows a combined phylogentic tree of the WRKY gene families from A.thaliana and switchgrass. As an example, the senescence-associated co-expression module 3 switchgrass genes are indicated in red. The WRKY genes from module 3 had no representatives from Groups IIa and only a single gene from Group IId and IIe. To verify if these patterns of WRKY associations were common to other plants as well, a number of senescence associated WRKY genes from rice, banana (Musa acuminata), and Medicago truncatula taken from the Leaf Senescence Database 2.0 (http://www.eplantsenescence.org/) were used for further analysis (Fig. 6b). Notably there are clusters of closely-related senescence associated WRKY genes in Group I, IIb, and IIc. Evidence supporting the possible involvement of these WRKY genes in regulating senescence comes from various data including expression analyses, mutants, and overexpression/knock down lines in several systems [11]. It is also possible that other WRKYs (for example module 5) are associated with senescence.
Fig. 6.
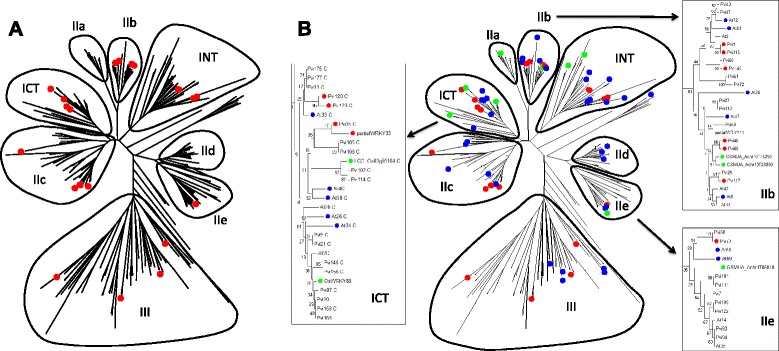
Senescence associated WRKY genes from switchgrass and other plants. a A combined Neighbor-Joining phylogenetic tree of all switchgrass and A.thaliana WRKY domain containing genes. The senescence-associated co-expression module 3 switchgrass genes are indicated in red. b A combined phylogenetic tree of all switchgrass and Arbabidopsis WRKY domains and several other senescence-inducible genes from other plants. The senescence-associated module 3 switchgrass genes are indicated in red, senescence-associated WRKY genes from A. thaliana (blue), switchgrass (red) and other plants (green) are shown with inset close ups of clusters of senescence associated WRKY genes. The other plants include rice, banana, and Medicago truncatula. Higher plants groups of WRKY genes (I-III) are shown. The evolutionary history was inferred using the Neighbor-Joining method. The optimal tree with the sum of branch length = 29.93 is shown. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree
It is clear from Fig. 6b that there are local hot spots in the phylogeny where closely-related WRKY genes from multiple species are associated with senescence. This observation is particularly apparent for WRKYs belonging to Group I, IIb, and IIe. One hot spot in Group IIb contains three switchgrass WRKY genes (PviWRKY48, PviWRKY65, and PviWRKY117) together with two banana genes and two A. thaliana WRKY genes including the well-characterized regulator of senescence AtWRKY6 [18, 19]. This suggests that AtWRKY6-like genes may regulate aspects of the senescence process across flowering plants, particularly as Fig. 6b shows representatives from both monocots and dicots. Group IIe is completely devoid of senescence-associated genes except for four very similar genes (AtWRKY65, AtWRKY69, PviWRKY77, and the banana gene Achr1T05010). This suggests that hot spots of senescence-associated genes are localized in the phylogenetic tree. However, it also suggests that possible function cannot be predicted from phylogenetic position alone as there are many instances such as PviWRKY77 and PviWRKY58 where apparent paralogs do not share the same association with senescence (Fig. 6b).
Taken together, the expression and phylogenetic studies have identified 23 switchgrass WRKY genes that show significantly enhanced mRNA levels during senescence of flag leaves under field conditions. Of these 23 genes, several are similar to senescence associated WRKY genes from other species and may represent conserved nodes in senescence signaling. Therefore, these genes represent potential targets for increasing biomass yields in switchgrass (and other flowering plants) by delaying senescence.
Network visualizations across five different developmental states
To better understand and identify co-expression clusters of genes regulated by the WRKYs, transcriptional networks were visualized. A transcription factor centered selection of 13,405 genes from the flag leaf expression data set [27] was used for these network analyses. These networks are shown in Fig. 7. The overall network patterns indicate that dynamic restructuring of the flag leaf transcriptomes was associated with key developmental events occurring both in the flag leaves as well as in the plant.
Fig. 7.

Edge-weighted network visualizations. a Each node represents an individual gene 13,405) and edges (428,378) connecting nodes are weighted by the topological overlap measure (TOM) as calculated by WGCNA. Nodes are colored based on the time point where each gene showed peak expression. Red = 7/3/12, Orange = 7/27/12, Yellow = 8/16/12, Green = 8/31/21, and Magenta = 9/19/12. b Only genes identified as transcript factors are shown. c Only WRKY transcription factors are shown. d Only WRKY genes and genes directly connected to WRKYs by an edge are shown. These are the genes with the highest degree of co-expression with the differentially expressed WRKYs
At the first harvest date, flag leaves were still expanding and had not yet transitioned into source leaves [27]. Genes with high expression during this time point (red; Fig. 7a) are linked to a cluster of genes associated with anthesis through a smaller network of genes (orange). At anthesis flag leaves had transitioned into source leaves [27], and the cluster of genes observed at this leaf developmental stage (orange; Fig. 7a) shares more connections to all the other network clusters (Fig. 7a). However, genes overexpressed in flag leaves at around the time of seed set (yellow; 8/16/12) were well separated from the central network hub, possibly because it was in response to an abiotic or biotic stress experienced by these plant/flag leaves around the collection date. Flag leaf gene networks when seeds were at the hard-seed stage (green; 8/31/12) were more closely aligned to the central network observed at anthesis (Fig. 7a) suggesting that transcriptional networks had possibly recalibrated after a stress-event observed the earlier stage. Genes with peak expression during senescence onset (magenta; 9/19/12) were distantly associated with the anthesis hub, although there were a few connections to the networks originating at around the time of seed set.
The 901 transcription factors forming the backbone of this entire network are highlighted (Fig. 7b) along with just WRKY transcription factors (Fig. 7c). The association patterns for the WRKYs are consistent with the involvement of specific WRKYs with specific stages of switchgrass flag leaf development. Interestingly, two WRKYs (yellow circles; Fig. 7c), namely PviWRKY29 and PviWRKY97 were more closely aligned to the WRKYs upregulated at the time of senescence onset (magenta circles; Fig. 7c). Genes directly connected to WRKYs by high topological overlap measure (TOM) value are depicted in Fig. 7d. Many of these genes could serve as direct targets for each respective WRKY.
To further investigate the relationships between WRKY expression patterns and cellular processes associated with flag leaf development, specific gene sets arising from the network analysis were performed to (a) evaluate the types and numbers of genes associated with senescence-related WRKYs and (b) searched for cis-acting elements in the available promoter sequences to identify putative W boxes that could serve as direct targets for WRKY transcription factors.
As described in Fig. 7c, although PviWRKY29 and PviWRKY97 were part of the genes overexpressed at the time of seed set, they were more closely aligned to the gene networks occurring at the time of senescence. PviWRKY29 is found in the Group IIb senescence-associated hot spot and is a potential paralog of PviWRKY117. PviWRKY29 and PviWRKY117 are also the two apparent switchgrass orthologs of the well-known senescence regulator AtWRKY6 (Fig. 6b) [18, 19]. PviWRKY97 is a Group I protein similar to the senescence-related WRKY protein encoded by the OsWRKY80 gene (Fig. 6b).
There were 33 genes found with direct connections to PviWRKY29 and PviWRKY97. Many of these genes encoded proteins destined to the cell-wall regions or were related to phosphate metabolism. Others appeared to be functioning in regulation/signal transduction. Of these genes directly linked to PviWRKY29 and PviWRKY97, 23 had sufficient 5' upstream (putative promoter) sequence available. Nineteen of these 23 genes (Table 1) had one or more putative W boxes in these upstream sequences, suggesting they could be directly regulated by PviWRKY29 and/or PviWRKY97. The genes where promoter regions are not currently available may also contain one or more W boxes, and the final number of genes that may be regulated by WRKYs could be higher.
Table 1.
Potential W box containing genes in module 5 that were directly connected to PviWRKY29 and PviWRKY97 through co-expression analysis. Switchgrass gene ID, number of putative W boxes, the highest blast match to A.thaliana and functional description are given
| Gene ID | No. W Box | Best match | Function description |
|---|---|---|---|
| Pavir.J02056 | 1 | AT2G26190 | calmodulin-binding family protein |
| Pavir.Da01490 | 2 | AT5G47750 | D6 protein kinase like 2 |
| Pavir.J40688 | 1 | AT4G39150 | DNAJ heat shock N-terminal domain protein |
| Pavir.J24695 | 1 | AT1G78060 | Glycosyl hydrolase family protein |
| Pavir.Bb00101 | 2 | AT4G05190 | kinesin 5 |
| Pavir.J13678 | 3 | AT3G09220 | laccase 7 |
| Pavir.Cb01890 | 3 | AT5G49760 | Leucine-rich repeat protein kinase family |
| Pavir.Fa01734 | 3 | AT1G56130 | Leucine-rich repeat transmembrane kinase |
| Pavir.Fb01478 | 3 | AT1G78130 | Major facilitator superfamily protein |
| Pavir.J34655 | 1 | AT2G37770 | NAD(P)-linked oxidoreductase superfamily |
| Pavir.J38308 | 1 | AT2G37770 | NAD(P)-linked oxidoreductase superfamily |
| Pavir.Eb02929 | 1 | AT5G11670 | NADP-malic enzyme 2 |
| Pavir.Ib03917 | 4 | AT5G08030 | PLC-like phosphodiesterases superfamily |
| Pavir.Ab01298 | 3 | AT3G02040 | senescence-related gene 3 |
| Pavir.Da00281 | 3 | AT2G20850 | STRUBBELIG-receptor family 1 |
| Pavir.Ba02108 | 2 | AT2G01770 | vacuolar iron transporter 1 |
| Pavir.Cb01458 | 1 | N/A | N/A |
| Pavir.J21234 | 1 | N/A | N/A |
| Pavir.J09086 | 2 | N/A | N/A |
In-Silico Promoter Analysis of Senescence-Related WRKY Genes
To determine potential environmental triggers that stimulate the expression of module 3 WRKY genes, the putative promoter region 1000 bp upstream of the start codon was scanned for cis-regulatory elements. All 17 senescence related WRKY genes with a putative promoter region from module 3, had a putative W box (TTGACY), suggesting auto and cross-regulation of WRKY genes. Additionally, putative ABA-responsive elements were present in multiple copies in the promoters of the majority of these module 3 WRKY genes, supporting their roles in activating cascades of genes involved in senescence. Common ABA-responsive motifs detected in the promoter regions of these WRKY genes included ACGTG, MACGYGB, ACGT, CCACGTGG, and ACGTSSSC. These sequences, if functional, would represent potential binding sites for several classes of transcription factors including bZIP and bHLHs. In addition to their proposed roles in triggering senescence, these WRKY genes also likely have roles in responding to abiotic stressors that serve as common triggers of senescence because motifs involved in drought responsiveness (DRE-1 and DRE-2), cold responsiveness (LTRE domains), phosphate starvation (P Starv), and sulfur responsiveness (Sulfur RE) were frequently detected in the promoter regions.
Further, the promoters of these WRKY genes contained putative regulatory elements for pathogen responsiveness (GT1GMSCAM4 and GCC box). These WRKY genes could represent a convergence point between the pathogen response and senescence pathways in switchgrass. WRKY transcription factors are known to play roles in both biotic and abiotic stress responses [11, 54, 55] and therefore the presence of putative W boxes in several WRKY gene promoters (Fig. 8) suggests that many of the senescence-associated WRKY genes represent common nodes between senescence and stress signaling pathways.
Fig. 8.

Promoter features of module 3 WRKYs. Seventeen of the 23 WRKYs in Module 3 had putative promoter regions which were further analyzed for the presence of additional motifs. Fourteen motifs (see methods for more details) were detected and the number of occurrences of each is shown along with the relative expression level of each WRKY (magenta being high, yellow being low). ABRE = Abscisic acid response element, Low Temp = Low temperature responsiveness, P Starve = Phosphate starvation response element, DRE-1 = Dehydration response element, Amm RE = Ammonium response element, DRE-2 = Dehydration responsive element, Salt, Path RE = Salt and Pathogen responsiveness element, Path RE = Pathogen responsiveness element, AntiOx RE = Antioxidant response element, Ethylene RE = Ethylene response element, HD BS = Homeodomain transcription factor binding site, Myb BS = Myb transcription factor binding site, MYC BS = MYC binding site, Sulfur RE = Sulfur responsive element
Our results present some evidence to support these common nodes between senescence and stress signaling pathways. Figure 7c shows that several WRKY genes from 8/16/12 (yellow) are more closely aligned to the gene networks occurring at the time of senescence (magenta). The yellow genes in Fig. 7 had been subjected to stress and this suggests that the yellow genes that map more closely with the senescence-associated genes are responsive to both stress and senescence. This includes PviWRKY29 an apparent switchgrass ortholog of the well-characterized senescence regulator AtWRKY6. AtWRKY6 is up-regulated by both pathogen attack and senescence and the results with PviWRKY29 are therefore what would have been predicted for AtWRKY6.
Discussion
The WRKY gene family in switchgrass
WRKY transcription factors were identified in the whole-genome sequence of the tetraploid switchgrass clone AP13 (Version 1.1). Alamo, the cultivar from which AP13 originated, is extensively distributed throughout switchgrass breeding programs and is a heterozygous tetraploid with two sub-genomes designated A and B [56]. An identified total of 240 WRKY genes in tetraploid switchgrass is consistent with the total number of genes in rice, the most extensively characterized monocot species. Diploid rice contains approximately 125 WRKY genes [57] which is about half the number identified in switchgrass. Examination of the switchgrass WRKY phylogenetic tree depicted in Fig. 7 shows clearly that the majority of the genes are present in pairs that presumably represent A and B genes. Over the entire phylogenetic tree, about two thirds of senescence-associated switchgrass WRKY genes are present as A/B pairs, suggesting extensive conservation of function between similar genes in the A and B sub-genomes. Many of these pairs of genes also show similar senescence-associated expression characteristics (for example PviWRKY1/PviWRKY115, PviWRKY120/PviWRKY123, PviWRKY35/PartialWRKY33 and PviWRKY48/PviWRKY65), although there are exceptions to this observation (PviWRKY58/PviWRKY77, PviWRKY50/PviWRKY145, and PviWRKY29/PviWRKY117).
Switchgrass contains both a Group RW6 resistance protein-WRKY (PviWRKY174) and a Group RW5 protein (PviWRKY178) [32]. R protein-WRKY genes appear to have evolved recently in flowering plants and each class appears to be restricted to specific flowering plant lineages [32], which suggests that RW5 and RW6 genes may have been present in the last common ancestor of switchgrass, sorghum and rice. Brachypodium distachyon appears to completely lack RW genes [29].
Co-expression modules during leaf senescence
The role of WRKYs in a number of different plant developmental processes, especially in defense and senescence, is well established [11]. Senescence is a complex process and is influenced by a number of events both internal and external to the leaf [58]. Not unsurprisingly, many transcription factors, including WRKYs and NACs, can impact these processes. Often there is redundancy in the molecular events controlled by groups of transcription factors, suggesting that there is a dynamic balance in the interactions between these transcriptional regulators with each other and with the entire transcriptional machinery. More often than not, the specific functions of all the individual WRKYs sharing similar expression profiles are not known. However, high throughput expression analyses can provide some measure of understanding of these interactions.
The transcriptional datasets used in the current study were collected from switchgrass flag leaves at five different plant developmental stages from field grown plants [27]. Using NMDS, it was first established that WRKY gene expression within this dataset was consistent between replicates, indicating that flag leaves collected at each harvest dates were similar physiologically. In addition, NMDS analysis also established that the WRKYs were well differentiated at each harvest date, indicating that specific WRKYs were associated with cellular metabolism in flag leaves at different developmental stages. These differences were confirmed by the hierarchical clustering of the DE-WRKYs, which showed that specific WRKYs were up/down regulated in a manner that followed previously described changes in leaf physiology [27].
Through analysis of co-expression modules, it became possible to link changes in flag leaf transcription to specific physiological stages of flag leaf development. Of significance was the discovery of modules that were associated with leaf expansion (module 2), mature leaves (modules 1 and 5), pre-senescence (module 4) and senescence onset (module 3). Most WRKY genes were associated with early and late leaf development stages, consistent with their deduced roles in other species [11, 29, 59].
Module 4 which contained genes that had peak expression levels just before the onset of senescence only contained two WRKYs (PviWRKY173 and PviWRKY114). Although PviWRKY173 was not highly expressed at this time point, PviWRKY114 was. A closer inspection of PviWRKY114 (Pavir.J00850) indicated that it has an unusual structure with an N-terminal DUF domain protein (domain of unknown function: DUF3598), and a C-terminal WRKY domain. This large fusion gene appears to be present in a few other plant genomes (for example, Setaria italica and rice). However, in many instances the two domains are located on independent genes. RNA-Seq mapping data indicated that reads were confined only to the N-terminal DUF3598 domain of Pavir.J00850 (data not shown), suggesting the WRKY domain is not expressed. These findings suggest that WRKY transcription factors may not be regulators of genes assigned to module 4.
Senescence associated WRKY genes from switchgrass
A number of previous studies have shown the importance of WRKY transcription factors in regulating senescence [19, 60]. A recent study in cotton identified 3624 senescence-associated genes that showed differential expression during the process of senescence [61]. Of these genes, 519 encode transcription factors and the WRKY family had the most members associated with senescence (54) followed by bHLH (44), and C3H (42). Many members of the WRKY gene family were up-regulated early during the onset of senescence [61]. Unfortunately, only the raw Illumina reads from Lin et al. [61] are currently available and therefore it is not possible to include these data in our analysis. Nevertheless, it is clear that WRKY transcription factors are regulators of senescence [19, 60]. The 23 WRKY genes in module 3 are likely to include major regulators of senescence in switchgrass and represent excellent candidate genes for increasing switchgrass biomass by delaying senescence in the field.
PviWRKY117 which was strongly associated with module 3 is an apparent switchgrass ortholog of AtWRKY6 (Fig. 6b). AtWRKY6 has been shown to positively regulate both senescence- and pathogen defense-associated genes [18]. One target gene of AtWRKY6 is a receptor-like protein kinase (FRK1) whose expression is strongly induced during leaf senescence and is activated by AtWRKY6 binding to W boxes in the FRK1 promoter. PviWRKY117 had increased mRNA levels of over 13-fold during flag leaf development. The peak of its induction occurred at the last harvest date (9/19/12) when flag leaves were beginning to senesce. This expression profile is consistent with a role as a possible master regulator of senescence in switchgrass.
Inspection of the PviWRKY117 demonstrated that its promoter contained known transcription factor binding sites, including a cluster of putative W boxes (underlined), a G box, and an S box (TTGACCCCATTGACC, CACGTGG, and AGCCACC). These are potential binding sites for WRKY, ERF, bZIP, and bHLH transcription factors. The presence of these sequences in the promoter of PviWRKY117 suggests possible auto-regulation and previously AtWRKY6 has been shown to suppress its own transcription [18].
Network analyses provide robust evidence for the role of WRKYs during leaf development
Inferring functional relationships through co-expression and network analyses has already been a useful tool for the analysis of WRKY transcription factors [15–17]. Our network analyses provided a remarkable visual representation of the dynamic changes in flag leaf transcriptomes over time. Both the complexity of the connections as well as the apparent distinct molecular signatures at each major point in leaf development could be distinguished. Similar scenarios have been described for other monocot species [62–67].
Transcriptomic networks at early and late stages were especially well resolved and added to the overall interpretation of the changes in WRKY gene expression discussed above. As anticipated each developmental stage (harvest date) was linked to a number of different transcription factors, while WRKY-associated networks were particularly abundant during leaf expansion and the onset of senescence. Additionally, these analyses implied that specific WRKY transcription factors were strongly associated with specific flag leaf growth stages, and WRKY-controlled networks especially at an early and late stage of flag leaf development were generally independent. However, WRKYs present within module 5 associated with the time of seed set, presented an interesting profile possibly linking different cellular processes to the first molecular signatures for the onset of leaf senescence.
This specific module 5 subnetwork associated with the senescence cluster (see Fig. 7) contained only two WRKYs, namely PviWRKY29 and PviWRKY97, which had direct connections to at least 19 genes that fell into two major categories: cell wall/defense and phosphate responsiveness. The role of WRKYs in plant defense is well established [11]. It is possible that the defense genes such as LRRs, a laccase, and a putative wall-bound xylosidase upregulated at this harvest date (8/16/12), (Pavir.J24695), were activated in response to an undetermined stress. Two other switchgrass genes, classified as NADP-linked oxidoreductase superfamily proteins, were found in this cluster. Related proteins have been indicated to have a direct role in the detoxification of stress-related accumulation of reactive carbonyls [68].
Recently, A. thaliana AtWRKY45 [69] was shown to directly influence plant P levels through control of a phosphate transporter. A related WRKY, A.thaliana AtWRKY42 exerted a greater influence on plant P status [70], and was suggested to impact plant P homeostasis. PviWRKY29 is most similar to A.thaliana AtWRKY6 and AtWRKY42, which both appear to impact plant P nutrition [70, 71], and AtWRKY6 plays a role in leaf senescence [18], suggesting a potential link between early sensing of P status to the onset of senescence in switchgrass flag leaves. Pavir.Ab01298 (Table 1) is the switchgrass ortholog of the A.thaliana senescence associated gene 3 (SAG 3), and encodes a phosphodiesterase. A similar gene plays a key role in maintaining plastid/cellular P homeostasis, especially under P starvation in A.thaliana [72].
Several other SAGs, including NADP-Malic enzyme 2, also associated with module 5 [27] providing evidence for a link between PviWRKY29 and PviWRKY97 and initiation of the senescence process. This expression network also contained a switchgrass ortholog of a vacuolar iron transporter gene. In rice, knockdowns of two leaf iron transporters led to increased iron translocation to the seeds [73]. However, switchgrass contains two sink tissues towards the end of a growing season, seeds and rhizomes [6]. Generally seeds are physiologically mature prior to completion of flag leaf senescence and rhizome dormancy [6, 27]. These observations raise the possibility that nutrient remobilization is staggered in switchgrass to meet the sink demands of different tissues.
Transcriptomic evidence for a causal relationship between WRKY expression and the onset of leaf senescence was seen at the last harvest date of flag-leaf sampling. Transcripts for 23 WRKYs were associated with the senescence-associated module 3, and the putative promoter regions for many of these WRKYs were populated with cis-elements known to confer response to both biotic and abiotic stresses. Several module 3 WRKYs were enriched for ABREs, DREs, low temperature, MYB, MYC, and sulfur-responsive elements, suggesting that module 3 WRKY gene expression was reflective of the internal and external environment during senescence onset. Further, these WRKYs were part of a network consisting of 3000 genes.
Among the SAGs within these networks were transcription factors, genes coding for enzymes in the chlorophyll degradation pathway, and several nutrient transporters with known roles during leaf senescence see Table 2) [27, 74–77]. Among these module 3 genes was a NAC transcription factor encoded by Pavir.J16651. This specific NAC (called PvNAC1) was demonstrated to impact leaf senescence in switchgrass [78], and is most similar to AtNAC29, which has been implicated in leaf senescence in A. thaliana [79].
Table 2.
Potential Module 3 SAGs. SAGs were identified as described previously [27]. Other descriptions are as given for Table 1
| Gene ID | Nearest At | At Description | Phase 4 RPKM |
|---|---|---|---|
| Pavir.Ba03899 | AT1G26870 | NAC domain containing protein 9 | 45 |
| Pavir.Fb00689 | AT2G33480 | NAC domain containing protein 41 | 659 |
| Pavir.Hb00869 | AT3G04070 | NAC domain containing protein 47 | 106 |
| Pavir.J16651 | AT1G69490 | NAC domain containing protein 29 | 103 |
| Pavir.Ca02775 | AT3G12977 | NAC domain containing protein | 1270 |
| Pavir.Ba01244 | AT4G22920 | non-yellowing 1 | 1280 |
| Pavir.Gb00362 | AT5G13800 | pheophytinase | 542 |
| Pavir.Hb02058 | AT5G13800 | pheophytinase | 387 |
| Pavir.J04787 | AT3G02040 | senescence-related gene 3 | 367 |
| Pavir.J37002 | AT5G45890 | senescence-associated gene 12 | 692 |
| Pavir.Bb01489 | AT5G45890 | senescence-associated gene 12 | 1300 |
| Pavir.Ab02441 | AT5G24380 | YELLOW STRIPE like 2 | 116 |
| Pavir.Gb01191 | AT5G53550 | YELLOW STRIPE like 3 | 159 |
| Pavir.Cb00745 | AT2G03530 | ureide permease 2 | 6913 |
| Pavir.Ea02698 | AT5G11670 | NADP-malic enzyme 2 | 565 |
| Pavir.J06980 | AT3G45140 | lipoxygenase 2 | 36 |
| Pavir.J32181 | AT2G42490 | Copper amine oxidase family protein | 54 |
| Pavir.J16835 | AT4G35090 | catalase 2 | 163 |
| Pavir.Fb00414 | AT5G60360 | aleurain-like protease | 71 |
| Pavir.Ia01427 | AT4G36220 | ferulic acid 5-hydroxylase 1 | 95 |
In addition to WRKY genes, Fischer-Kilbienski et al. have reported a protein containing the DUF548 domain (AtS40-3; AT4G18980) was targeted to the nucleus and regulated senescence via an AtWRKY53-dependent or independent route [80]. DUF548 is now recognized as a “senescence regulator" domain. The present version of the switchgrass genome contains 29 genes coding for proteins with the senescence regulator domain. Transcripts for 19 of these 29 genes were found in the flag leaf dataset, and only four of these genes were present in module 3 (Additional file 5: Figure S3). All four genes appeared to belong to group 1 proteins [80] (data not shown) indicating their importance to switchgrass flag leaf senescence. AtWRKY53 is a group III protein and of the large number of similar switchgrass genes, only PviWRKY119 appears to be associated with senescence (Additional file 2: Table S2). However, it is possible that WRKYs could be part of the regulatory cascade influenced by the four switchgrass proteins containing the senescence regulator domain (Additional file 5: Figure S3).
Overall our analyses have classified all of the available full-length and partial switchgrass WRKY genes into specific protein clades and have placed their expression profiles within a framework of flag leaf development. More notably, it was possible to identify expression networks and expression modules that serve to integrate WRKY gene expression with specific genes. Many of these genes have known functions during leaf senescence in other plants. These findings provide a good platform for future analysis of specific genes and their ability to serve as markers for the continued improvement of switchgrass as a biofuel crop.
Availability of supporting data
The data used in this manuscript are available as part of the short-reads archive depository within the NCBI at http://www.ncbi.nlm.nih.gov/sra/SRX481052/.
Acknowledgements
This work was supported in part by the Office of Science (BER), U. S. Department of Energy Grant Number DE-AI02-09ER64829, USDA-NIFA Grant Number 2011-67009-30096, and by the USDA-ARS CRIS projects 3042-21000-030-00D and 3042-21220-032-00D. The U.S. Department of Agriculture, Agricultural Research Service, is an equal opportunity/affirmative action employer and all agency services are available without discrimination. Mention of commercial products and organizations in this manuscript is solely to provide specific information. It does not constitute endorsement by USDA-ARS over other products and organizations not mentioned. The authors would like to thank Mani Kant Choudhary, Marissa Miller, Naveen Kumar, Malini Rao, Deena Rushton, Ying-Sheng Huang and Nikhil Kesarla in the Rushton lab. This project was supported in part by National Research Initiative grants 2008-35100-04519 and 2008-35100-05969 from the USDA National Institute of Food and Agriculture. This research was supported by funding from the North Central Regional Sun Grant Center at South Dakota State University through a grant provided by the US Department of Agriculture under award number 2010-38502-21861.
Additional files
The WRKY gene family in switchgrass. (XLSX 25 kb)
WRKY genes in the pro-senescence and early season clusters. (XLSX 9 kb)
Module Eigengene Dendrogram of the 13 modules (ME1-ME13) identified by WGCNA. (PDF 21 kb)
Expression profile of Module Eigengenes. (PDF 31 kb)
Expression heatmap of switchgrass genes coding for proteins containing the “senescence- regulator” domain. (PDF 27 kb)
Footnotes
Charles I. Rinerson, Erin D. Scully and Nathan A. Palmer contributed equally to this work.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
GS, JR, NP, ES, TDR, CIR, RCR, PT, QJS, and PJR conceived the experiments. CIR, PJR, ES, NP, and TDR analyzed data. GS, ES, NP, SS, JR, and PJR wrote the paper. All authors read and approved the final manuscript.
Contributor Information
Charles I. Rinerson, Email: crinerson@gmail.com
Erin D. Scully, Email: Erin.Scully@ars.usda.gov
Nathan A. Palmer, Email: nathan.palmer@ars.usda.gov
Teresa Donze-Reiner, Email: TDonze-Reiner@wcupa.edu.
Roel C. Rabara, Email: roel.rabara@tamu.edu
Prateek Tripathi, Email: tprateek@dornsife.usc.edu.
Qingxi J Shen, Email: jeffery.shen@unlv.edu.
Scott E. Sattler, Email: Scott.Sattler@ars.usda.gov
Jai S. Rohila, Email: jai.rohila@sdstate.edu
Gautam Sarath, Email: Gautam.Sarath@ars.usda.gov.
Paul J. Rushton, Email: prushton@xxiicentury.com
References
- 1.Sarath G, Mitchell RB, Sattler SE, Funnell D, Pedersen JF, Graybosch RA, Vogel KP. Opportunities and roadblocks in utilizing forages and small grains for liquid fuels. J Ind Microbiol Biotechnol. 2008;35(5):343–54. doi: 10.1007/s10295-007-0296-3. [DOI] [PubMed] [Google Scholar]
- 2.Mitchell R, B, Vogel, K.P, Schmer M.R. Switchgrass (Panicum virgatum) for biofuel production. . In: Sustainable Ag Energy Community of Practice, eXtension. (http://extensionorg/pages/switchgrass-for-Biofuel-Production). 2012.
- 3.Vogel KP, Sarath G, Saathoff AJ, Mitchell RB. Switchgrass. Rsc Energy Environ S. 2011;3:341–80. [Google Scholar]
- 4.Casler MD. Switchgrass breeding, genetics and genomics. In: Monti A, editor. Switchgrass, Green Energy and Technology. London, U.K: Springer; 2012. pp. 29–53. [Google Scholar]
- 5.Bartley L, Wu Y, Saathoff A, Sarath G. Switchgrass Genetics and Breeding Challenges. Ames, IA: John Wiley and Sons; 2013. pp. 7–31. [Google Scholar]
- 6.Sarath G, Baird LM, Mitchell RB. Senescence, dormancy and tillering in perennial C4 grasses. Plant Sci. 2014;217–218:140–51. doi: 10.1016/j.plantsci.2013.12.012. [DOI] [PubMed] [Google Scholar]
- 7.Martinez-Reyna JM, Vogel KP. Heterosis in switchgrass: Spaced plants. Crop Science. 2008;48(4):1312–20. doi: 10.2135/cropsci2007.12.0695. [DOI] [Google Scholar]
- 8.Vogel K, Mitchell R, Sarath G, Casler MD. Registration of 'Liberty' switchgrass. J Plant Regist. 2014;8:242–7. doi: 10.3198/jpr2013.12.0076crc. [DOI] [Google Scholar]
- 9.Lin JF, Wu SH. Molecular events in senescing Arabidopsis leaves. Plant J. 2004;39(4):612–28. doi: 10.1111/j.1365-313X.2004.02160.x. [DOI] [PubMed] [Google Scholar]
- 10.Guo Y, Cai Z, Gan S. Transcriptome of Arabidopsis leaf senescence. Plant Cell and Environment. 2004;27(5):521–49. doi: 10.1111/j.1365-3040.2003.01158.x. [DOI] [Google Scholar]
- 11.Rushton PJ, Somssich IE, Ringler P, Shen QJ. WRKY transcription factors. Trends Plant Sci. 2010;15(5):247–58. doi: 10.1016/j.tplants.2010.02.006. [DOI] [PubMed] [Google Scholar]
- 12.Popescu SC, Popescu GV, Bachan S, Zhang Z, Gerstein M, Snyder M, Dinesh-Kumar SP. MAPK target networks in Arabidopsis thaliana revealed using functional protein microarrays. Genes Dev. 2009;23(1):80–92. doi: 10.1101/gad.1740009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rushton PJ, Torres JT, Parniske M, Wernert P, Hahlbrock K, Somssich IE. Interaction of elicitor-induced DNA-binding proteins with elicitor response elements in the promoters of parsley PR1 genes. Embo Journal. 1996;15(20):5690–700. [PMC free article] [PubMed] [Google Scholar]
- 14.Yamasaki K, Kigawa T, Watanabe S, Inoue M, Yamasaki T, Seki M, Shinozaki K, Yokoyama S. Structural Basis for Sequence-specific DNA Recognition by an Arabidopsis WRKY Transcription Factor. J. Biol Chem. 2012;287(10):7683–91. doi: 10.1074/jbc.M111.279844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berri S, Abbruscato P, Faivre-Rampant O, Brasileiro AC, Fumasoni I, Satoh K, Kikuchi S, Mizzi L, Morandini P, Pe ME, et al. Characterization of WRKY co-regulatory networks in rice and Arabidopsis. Bmc Plant Biol. 2009;9:120. doi: 10.1186/1471-2229-9-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Choura M, Rebaï A, Masmoudi K. Unraveling the WRKY transcription factors network in Arabidopsis Thaliana by integrative approach. Network Biology.2015; 5(2).
- 17.Wanke D, Hahn A, Kilian J, Harter K, Berendzen KW. Regulatory Networks: Inferring Functional Relationships through Co-Expression. Qtm Prob Wht Noi. 2010;26:405–24. [Google Scholar]
- 18.Robatzek S, Somssich IE. Targets of AtWRKY6 regulation during plant senescence and pathogen defense. Genes Dev. 2002;16(9):1139–49. doi: 10.1101/gad.222702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robatzek S, Somssich IE. A new member of the Arabidopsis WRKY transcription factor family, AtWRKY6, is associated with both senescence- and defence-related processes. Plant J. 2001;28(2):123–33. doi: 10.1046/j.1365-313X.2001.01131.x. [DOI] [PubMed] [Google Scholar]
- 20.Besseau S, Li J, Palva ET. WRKY54 and WRKY70 co-operate as negative regulators of leaf senescence in Arabidopsis thaliana. J Exp Bot. 2012;63(7):2667–79. doi: 10.1093/jxb/err450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miao Y, Zentgraf U. A HECT E3 ubiquitin ligase negatively regulates Arabidopsis leaf senescence through degradation of the transcription factor WRKY53. Plant J. 2010;63(2):179–88. doi: 10.1111/j.1365-313X.2010.04233.x. [DOI] [PubMed] [Google Scholar]
- 22.Potschin M, Schlienger S, Bieker S, Zentgraf U. Senescence Networking: WRKY18 is an Upstream Regulator, a Downstream Target Gene, and a Protein Interaction Partner of WRKY53. J. Plant Growth Regul. 2014;33(1):106–18. doi: 10.1007/s00344-013-9380-2. [DOI] [Google Scholar]
- 23.Zhou X, Jiang YJ, Yu DQ. WRKY22 transcription factor mediates dark-induced leaf senescence in Arabidopsis. Mol. Cells. 2011;31(4):303–13. doi: 10.1007/s10059-011-0047-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Han M, Kim CY, Lee J, Lee SK, Jeon JS. OsWRKY42 Represses OsMT1d and Induces Reactive Oxygen Species and Leaf Senescence in Rice. Mol Cells. 2014;37(7):532–9. doi: 10.14348/molcells.2014.0128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ricachenevsky FK, Sperotto RA, Menguer PK, Fett JP. Identification of Fe-excess-induced genes in rice shoots reveals a WRKY transcription factor responsive to Fe, drought and senescence. Mol Biol Rep. 2010;37(8):3735–45. doi: 10.1007/s11033-010-0027-0. [DOI] [PubMed] [Google Scholar]
- 26.Jing SJ, Zhou X, Song Y, Yu DQ. Heterologous expression of OsWRKY23 gene enhances pathogen defense and dark-induced leaf senescence in Arabidopsis. Plant Growth Regul. 2009;58(2):181–90. doi: 10.1007/s10725-009-9366-z. [DOI] [Google Scholar]
- 27.Palmer NA, Donze-Reiner T, Horvath D, Heng-Moss T, Waters B, Tobias C, Sarath G. Switchgrass (Panicum virgatum L) flag leaf transcriptomes reveal molecular signatures of leaf development, senescence, and mineral dynamics. Funct Integr Genomics. 2014;15(1):1–16. doi: 10.1007/s10142-014-0393-0. [DOI] [PubMed] [Google Scholar]
- 28.Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, et al. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 2012;40(Database issue):D1178–86. doi: 10.1093/nar/gkr944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tripathi P, Rabara RC, Langum TJ, Boken AK, Rushton DL, Boomsma DD, Rinerson CI, Rabara J, Reese RN, Chen X, et al. The WRKY transcription factor family in Brachypodium distachyon. Bmc Genomics. 2012;13:270. doi: 10.1186/1471-2164-13-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 31.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rinerson CI, Rabara JC, Tripathi P, Shen QJ, Rushton PJ. The evolution of WRKY transcription factors. Bmc Plant Biol. 2015;15:66. doi: 10.1186/s12870-015-0456-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–7. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4(4):406–25. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 35.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;783–791. [DOI] [PubMed]
- 36.Zuckerkandl E, Pauling L. Molecules as documents of evolutionary history. J. Theor. Biol. 1965;8(2):357–66. doi: 10.1016/0022-5193(65)90083-4. [DOI] [PubMed] [Google Scholar]
- 37.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. J. Therm. Biol. 2013;30(12):2725–9. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14(4):R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liao Y, Smyth GK, Shi W. FeatureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30(7):923–30. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 40.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11(10). [DOI] [PMC free article] [PubMed]
- 41.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Team RC.R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2012. Open access available at: http://cranr-projectorg2011.
- 43.Oksanen J, Kindt R, Legendre P, O'Hara B, Stevens M, Oksanen M, et al. The vegan package. Community ecology package. 2007.
- 44.Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat. Appl. Genet. Mol. Biol. 2005;4:Article17. doi: 10.2202/1544-6115.1128. [DOI] [PubMed] [Google Scholar]
- 45.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saito R, Smoot ME, Ono K, Ruscheinski J, Wang PL, Lotia S, Pico AR, Bader GD, Ideker T. A travel guide to Cytoscape plugins. Nat methods. 2012;9(11):1069–76. doi: 10.1038/nmeth.2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, et al. Pfam: the protein families database. Nucleic Acids Res. 2014;42(Database issue):D222–30. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jin J, Zhang H, Kong L, Gao G, Luo J. PlantTFDB 3.0: a portal for the functional and evolutionary study of plant transcription factors. Nucleic Acids Res. 2014;42(Database issue):D1182–7. doi: 10.1093/nar/gkt1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Higo K, Ugawa Y, Iwamoto M, Korenaga T. Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 1999;27(1):297–300. doi: 10.1093/nar/27.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Prestridge DS. Signal Scan - a Computer-Program That Scans DNA-Sequences for Eukaryotic Transcriptional Elements. Comput Appl Biosci. 1991;7(2):203–6. doi: 10.1093/bioinformatics/7.2.203. [DOI] [PubMed] [Google Scholar]
- 51.Riechmann JL, Heard J, Martin G, Reuber L, Jiang C, Keddie J, Adam L, Pineda O, Ratcliffe OJ, Samaha RR, et al. Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science. 2000;290(5499):2105–10. doi: 10.1126/science.290.5499.2105. [DOI] [PubMed] [Google Scholar]
- 52.Mccarty DR, Carson CB, Stinard PS, Robertson DS. Molecular Analysis of Viviparous-1 - an Abscisic Acid-Insensitive Mutant of Maize. Plant Cell. 1989;1(5):523–32. doi: 10.1105/tpc.1.5.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ulmasov T, Hagen G, Guilfoyle TJ. ARF1, a transcription factor that binds to auxin response elements. Science. 1997;276(5320):1865–8. doi: 10.1126/science.276.5320.1865. [DOI] [PubMed] [Google Scholar]
- 54.Eulgem T, Rushton PJ, Robatzek S, Somssich IE. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000;5(5):199–206. doi: 10.1016/S1360-1385(00)01600-9. [DOI] [PubMed] [Google Scholar]
- 55.Rushton DL, Tripathi P, Rabara RC, Lin J, Ringler P, Boken AK, Langum TJ, Smidt L, Boomsma DD, Emme NJ, et al. WRKY transcription factors: key components in abscisic acid signalling. Plant Biotechnol J. 2012;10(1):2–11. doi: 10.1111/j.1467-7652.2011.00634.x. [DOI] [PubMed] [Google Scholar]
- 56.Casler MD, Tobias CM, Kaeppler SM, Buell CR, Wang ZY, Cao PJ, Schmutz J, Ronald P. The Switchgrass Genome: Tools and Strategies. Plant Genome. 2011;4(3):273–82. doi: 10.3835/plantgenome2011.10.0026. [DOI] [Google Scholar]
- 57.Shen QXJ, Yu DQ, Jeon JS, Piffanelli P, Abbruscato P, Guo ZJ, et al. Nomenclature report on rice WRKY's - Conflict regarding gene names and its solution. Rice. 2012; 5. [DOI] [PMC free article] [PubMed]
- 58.Thomas H. Senescence, ageing and death of the whole plant. New Phytol. 2013;197(3):696–711. doi: 10.1111/nph.12047. [DOI] [PubMed] [Google Scholar]
- 59.Li Z, Peng J, Wen X, Guo H. Gene Network Analysis and Functional Studies of Senescence-associated Genes Reveal Novel Regulators of Arabidopsis Leaf Senescence. J Integr Plant Biol. 2012. [DOI] [PubMed]
- 60.Miao Y, Laun T, Zimmermann P, Zentgraf U. Targets of the WRKY53 transcription factor and its role during leaf senescence in Arabidopsis. Plant Mol Biol. 2004;55(6):853–67. doi: 10.1007/s11103-005-2142-1. [DOI] [PubMed] [Google Scholar]
- 61.Lin M, Pang CY, Fan SL, Song MZ, Wei HL, Yu SX. Global analysis of the Gossypium hirsutum L. Transcriptome during leaf senescence by RNA-Seq. Bmc Plant Biol. 2015; 15. [DOI] [PMC free article] [PubMed]
- 62.Tausta SL, Li P, Si Y, Gandotra N, Liu P, Sun Q, Brutnell TP, Nelson T. Developmental dynamics of Kranz cell transcriptional specificity in maize leaf reveals early onset of C4-related processes. J Exp Bot. 2014;65(13):3543–55. doi: 10.1093/jxb/eru152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shakoor N, Nair R, Crasta O, Morris G, Feltus A, Kresovich S. A Sorghum bicolor expression atlas reveals dynamic genotype-specific expression profiles for vegetative tissues of grain, sweet and bioenergy sorghums. Bmc Plant Biol. 2014;14:35. doi: 10.1186/1471-2229-14-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang P, Kelly S, Fouracre JP, Langdale JA. Genome-wide transcript analysis of early maize leaf development reveals gene cohorts associated with the differentiation of C-4 Kranz anatomy. Plant J. 2013;75(4):656–70. doi: 10.1111/tpj.12229. [DOI] [PubMed] [Google Scholar]
- 65.Liu WY, Chang YM, Chen SCC, Lu CH, Wu YH, Lu MYJ, Chen DR, Shih ACC, Sheue CR, Huang HC, et al. Anatomical and transcriptional dynamics of maize embryonic leaves during seed germination. P Natl Acad Sci USA. 2013;110(10):3979–84. doi: 10.1073/pnas.1301009110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gregersen PL, Holm PB, Krupinska K. Leaf senescence and nutrient remobilisation in barley and wheat. Plant Biol (Stuttg) 2008;10(Suppl 1):37–49. doi: 10.1111/j.1438-8677.2008.00114.x. [DOI] [PubMed] [Google Scholar]
- 67.Liu L, Zhou Y, Zhou G, Ye R, Zhao L, Li X, Lin Y. Identification of early senescence-associated genes in rice flag leaves. Plant Mol Biol. 2008;67(1–2):37–55. doi: 10.1007/s11103-008-9300-1. [DOI] [PubMed] [Google Scholar]
- 68.Yamauchi Y, Hasegawa A, Taninaka A, Mizutani M, Sugimoto Y. NADPH-dependent reductases involved in the detoxification of reactive carbonyls in plants. J Biol Chem. 2011;286(9):6999–7009. doi: 10.1074/jbc.M110.202226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang H, Xu Q, Kong YH, Chen Y, Duan JY, Wu WH, Chen YF. Arabidopsis WRKY45 transcription factor activates PHOSPHATE TRANSPORTER1;1 expression in response to phosphate starvation. Plant Physiol. 2014;164(4):2020–9. doi: 10.1104/pp.113.235077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Su T, Xu Q, Zhang FC, Chen Y, Li LQ, Wu WH, et al. WRKY42 Modulates Phosphate Homeostasis through Regulating Phosphate Translocation and Acquisition in Arabidopsis. Plant Physiol. 2015. [DOI] [PMC free article] [PubMed]
- 71.Chen YF, Li LQ, Xu Q, Kong YH, Wang H, Wu WH. The WRKY6 transcription factor modulates PHOSPHATE1 expression in response to low Pi stress in Arabidopsis. Plant Cell. 2009;21(11):3554–66. doi: 10.1105/tpc.108.064980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cheng Y, Zhou W, El Sheery NI, Peters C, Li M, Wang X, Huang J. Characterization of the Arabidopsis glycerophosphodiester phosphodiesterase (GDPD) family reveals a role of the plastid-localized AtGDPD1 in maintaining cellular phosphate homeostasis under phosphate starvation. Plant J. 2011;66(5):781–95. doi: 10.1111/j.1365-313X.2011.04538.x. [DOI] [PubMed] [Google Scholar]
- 73.Zhang Y, Xu YH, Yi HY, Gong JM. Vacuolar membrane transporters OsVIT1 and OsVIT2 modulate iron translocation between flag leaves and seeds in rice. Plant J. 2012;72(3):400–10. doi: 10.1111/j.1365-313X.2012.05088.x. [DOI] [PubMed] [Google Scholar]
- 74.Hortensteiner S, Krautler B. Chlorophyll breakdown in higher plants. Biochim Biophys Acta. 2011;1807(8):977–88. doi: 10.1016/j.bbabio.2010.12.007. [DOI] [PubMed] [Google Scholar]
- 75.Hortensteiner S. Stay-green regulates chlorophyll and chlorophyll-binding protein degradation during senescence. Trends Plant Sci. 2009;14(3):155–62. doi: 10.1016/j.tplants.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 76.Waters BM, Uauy C, Dubcovsky J, Grusak MA. Wheat (Triticum aestivum) NAM proteins regulate the translocation of iron, zinc, and nitrogen compounds from vegetative tissues to grain. J Exp Bot. 2009;60(15):4263–74. doi: 10.1093/jxb/erp257. [DOI] [PubMed] [Google Scholar]
- 77.Gregersen PL, Culetic A, Boschian L, Krupinska K. Plant senescence and crop productivity. Plant Mol Biol. 2013;82(6):603–22. doi: 10.1007/s11103-013-0013-8. [DOI] [PubMed] [Google Scholar]
- 78.Yang J, Worley E, Torres-Jerez I, Miller R, Wang M, Fu C, Wang ZY, Tang Y, Udvardi M. PvNAC1 and PvNAC2 Are Associated with Leaf Senescence and Nitrogen Use Efficiency in Switchgrass. Bioenergy Res. 2014;8:868–80. doi: 10.1007/s12155-014-9566-x. [DOI] [Google Scholar]
- 79.Guo Y, Gan S. AtNAP, a NAC family transcription factor, has an important role in leaf senescence. Plant J. 2006;46(4):601–12. doi: 10.1111/j.1365-313X.2006.02723.x. [DOI] [PubMed] [Google Scholar]
- 80.Fischer-Kilbienski I, Miao Y, Roitsch T, Zschiesche W, Humbeck K, Krupinska K. Nuclear targeted AtS40 modulates senescence associated gene expression in Arabidopsis thaliana during natural development and in darkness. Plant Mol Biol. 2010;73(4–5):379–90. doi: 10.1007/s11103-010-9618-3. [DOI] [PubMed] [Google Scholar]


