Abstract
Enrichment analysis has been widely applied in the genome-wide association studies (GWAS), where gene sets corresponding to biological pathways are examined for significant associations with a phenotype to help increase statistical power and improve biological interpretation. In this work, we expand the scope of enrichment analysis into brain imaging genetics, an emerging field that studies how genetic variation influences brain structure and function measured by neuroimaging quantitative traits (QT). Given the high dimensionality of both imaging and genetic data, we propose to study Imaging Genetic Enrichment Analysis (IGEA), a new enrichment analysis paradigm that jointly considers meaningful gene sets (GS) and brain circuits (BC) and examines whether any given GS-BC pair is enriched in a list of gene-QT findings. Using gene expression data from Allen Human Brain Atlas and imaging genetics data from Alzheimer’s Disease Neuroimaging Initiative as test beds, we present an IGEA framework and conduct a proof-of-concept study. This empirical study identifies 12 significant high level two dimensional imaging genetics modules. Many of these modules are relevant to a variety of neurobiological pathways or neurodegenerative diseases, showing the promise of the proposal framework for providing insight into the mechanism of complex diseases.
Keywords: Imaging genetics, enrichment analysis, genome wide association study, quantitative trait
1 Introduction
Brain imaging genetics is an emerging field that studies how genetic variation influences brain structure and function. Genome-wide association studies (GWAS) have been performed to identify genetic markers such as single nucleotide polymorphisms (SNPs) that are associated with brain imaging quantitative traits (QTs) [15, 16]. Using biological pathways and networks as prior knowledge, enrichment analysis has also been performed to discover pathways or network modules enriched by GWAS findings to enhance statistical power and help biological interpretation [5]. For example, numerous studies on complex diseases have demonstrated that genes functioning in the same pathway can influence imaging QTs collectively even when constituent SNPs do not show significant association individually [13]. Enrichment analysis can also help identify relevant pathways and improve mechanistic understanding of underlying neurobiology [6,10,11,14].
In the genetic domain, enrichment analysis has been widely studied in gene expression data analysis and has recently been modified to analyze GWAS data. GWAS-based enrichment analysis first maps SNP-level scores to gene-level scores, and then test whether a pre-defined gene set S (e.g., a pathway) is enriched in a set of significant genes L (e.g., GWAS findings). Two strategies are often used to compute enrichment significance: threshold-based [3, 4, 8, 19] and rank-based [17]. Threshold-based approaches aim to solve an independence test problem (e.g., chi-square test, hypergeometric test, or binomial z-test) by treating genes as significant if their scores exceed a threshold. Rank-based methods take into account the score of each gene to determine if the members of S are randomly distributed throughout L.
In brain imaging genetics, the above enrichment analysis methods are applicable only to genetic findings associated with each single imaging QT. Our ultimate goal is to discover high level associations between meaningful gene sets (GS) and brain circuits (BC), which typically include multiple genes and multiple QTs. To achieve this goal, we propose to study Imaging Genetic Enrichment Analysis (IGEA), a new enrichment analysis paradigm that jointly considers sets of interest (i.e., GS and BC) in both genetic and imaging domains and examines whether any given GS-BC pair is enriched in a list of gene-QT findings.
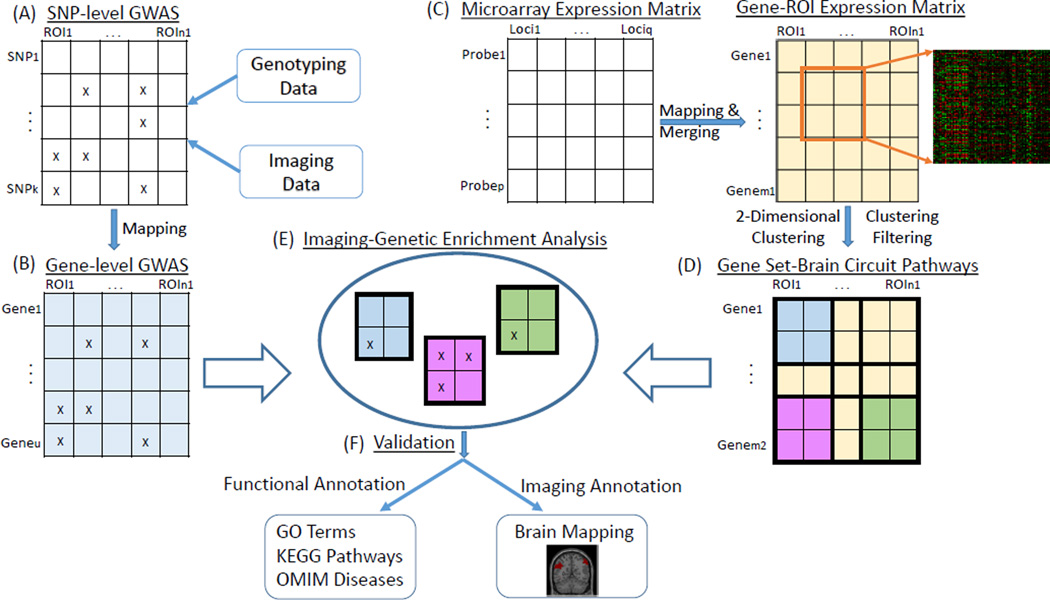
Using whole brain whole genome gene expression data from Allen Human Brain Atlas (AHBA) and imaging genetics data from Alzheimer’s Disease Neuroimaging Initiative (ADNI) as test beds, we present a novel IGEA framework and conduct a proof-of-concept study to explore high level imaging genetic associations based on brain-wide genome-wide association study (BWGWAS) results. For consistency purpose, in this paper, we use GS to indicate a set of genes and BC to indicate a set of regions of interest (ROIs) in the brain. The proposed framework consists of the following steps (see also Figure 1): (1) use AHBA to identify meaningful GS-BC modules, (2) conduct BWGWAS on ADNI amyloid imaging genetics data to identify SNP-QT and gene-QT associations, (3) perform IGEA to identify GS-BC modules significantly enriched by gene-QT associations using threshold-based strategy, and (4) visualize and interpret the identified GS-BC modules.
Fig. 1.
Overview of the proposed Imaging Genetic Enrichment Analysis (IGEA) framework. (A) Perform SNP-level GWAS of brain wide imaging measures. (B) Map SNP-level GWAS findings to gene-level. (C) Construct gene-ROI expression matrix from AHBA data. (D) Construct GS-BC modules by performing 2D hierarchical clustering, and filter out non-significant 2D clusters. (E) Perform IGEA by mapping gene-level GWAS findings to identified GS-BC modules. (F) For each enriched GS-BC module, examine the GS using GO terms, KEGG pathways, and OMIM disease databases, and map the BC to the brain.
2 Methods and Materials
2.1 Brain Wide Genome Wide Association Study (BWGWAS)
The imaging and genotyping data used for BWGWAS were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu). One goal of ADNI has been to test whether serial magnetic resonance imaging (MRI), positron emission tomography (PET), other biological markers, and clinical and neuropsychological assessment can be combined to measure the progression of mild cognitive impairment (MCI) and early AD. For up-to-date information, see www.adni-info.org. Preprocessed [18F]Florbetapir PET scans (i.e., amyloid imaging data) were downloaded from adni.loni.usc.edu, then aligned to each participant’s same visit scan and normalized to the Montreal Neurological Institute (MNI) space as 2 × 2 × 2 mm voxels. ROI level amyloid measurements were further extracted based on the MarsBaR AAL atlas. Genotype data of both ADNI-1 and ADNI-GO/2 phases were also downloaded, and then quality controlled, imputed and combined as decribed in [9]. A total of 980 non-Hispanic Caucasian participants with both complete amyloid measurements and genome-wide data were studied. Associations between 105 (out of a total 116) baseline amyloid measures and 5,574,300 SNPs were examined by performing SNP-based GWAS using PLINK [12] with sex, age and education as covariates. To facilitate the subsequent enrichment analysis, a gene-level p-value was determined as the smallest p-value of all SNPs located in ±50K bp of the gene.
2.2 Constructing GS-BC Modules using AHBA
There are many types of prior knowledge that can be used to define meaningful GS and BC entities. In the genomic domain, the prior knowledge could be based on Gene Ontology or functional annotation databases; in the imaging domain, the prior knowledge could be neuroanatomic ontology or brain databases. In this work, to demonstrate the proposed IGEA framework, we use gene expression data from the Allen Human Brain Atlas (AHBA, Allen Institute for Brain Science, Seattle, WA; available from http://www.brain-map.org/) to extract GS and BC modules such that genes within a GS share similar expression profiles and so do ROIs within a BC. We hypothesize that, given these similar co-expression patterns across genes and ROIs, each GS-BC pair forms an interesting high level imaging genetic entity that may be related to certain biological function and can serve as a valuable candidate for two-dimensional IGEA.
The AHBA includes genome-wide microarray-based expression covering the entire brain through systematic sampling of regional tissue. Expression profiles for eight health human brains have been released, including two full brains and six right hemispheres. One goal of AHBA is to combine genomics with the neuroanatomy to better understand the connections between genes and brain functioning. As an early report indicated that individuals share as much as 95% gene expression profile [21], in this study, we only included one full brain (H0351.2001) to construct GS-BC modules. First all the brain samples (~ 900) were mapped to MarsBaR AAL atlas, which included 116 brain ROIs. Due to many-to-one mapping from brain samples to AAL ROIs, there are > 1 samples for each ROI. Following [20], samples located in the same ROI were merged using the mean statistics. Probes were then merged to genes using the same strategy. Finally the preprocessed gene-ROI profiles were normalized for each ROI. As a result, the expression matrix contained 16,097 genes over 105 ROIs.
We performed a 2D cluster analysis on the gene-ROI expression matrix to identify interesting GS-BC modules. First, we calculated the distance matrices for both genes and ROIs, respectively. In other words, we computed the dissimilarity between each pair of genes, and the dissimilarity between each pair of ROIs, using Equation (1).
| (1) |
Two dendrograms were constructed by applying hierarchical clustering to two distance matrices separately, using the UPGMA (Unweighted Pair Group Method with Arithmetic Mean) algorithm. As most enrichment analyses placed constraints on genetic pathways of sizes from 10 to 400 [13], we cut the dendrogram at half of its height to build genetic clusters (i.e., GSs) whose sizes are mostly within the above range. For the imaging domain, we also employed the same parameter to construct ROI clusters (i.e., BCs).
We tested the statistical significance of each GS-BC pair based on a null hypothesis that the expression level of a gene is independent from the expression level of other genes across relevant brain ROIs in the same GS-BC module, assuming that the average Pearson’s correlation coefficients (PCCs) of gene expression levels for genes in the GS-BC module are higher than the ones from random GS-BC modules. Thus, for each GS-BC module, we constructed another GS-BC module with the same number of randomly selected genes and ROIs, and calculated its average PCC (avgPCC), This procedure was repeated N = 1000 times and the empirical p-value of original GS-BC module was calculated using the following equation, where I is an indicator function [7].
| (2) |
2.3 Imaging Genetic Enrichment Analysis (IGEA)
Pathway enrichment analysis has been extensively employed to genomic domain to analyze the genetic findings associated with a specific imaging QT. In this study, our goal is to identify high level associations between gene sets and brain circuits, which typically include multiple genes and multiple QTs.
In this study, we propose the threshold-based IGEA by extending the existing threshold-based enrichment analysis. SNP level findings have been mapped to gene level findings in Section 2.1. The GWAS findings are a list L of N = NG × NB gene-QT associations, where we have a set Gd of NG = |Gd| genes and a set Bd of NB = |Bd| QTs in our analysis. From Section 2.2, significant GS-BC modules, where relevant genes share similar expression profiles across relevant ROIs, have been constructed. Given an interesting GS-BC module with gene set Gk and QT set Bk, IGEA aims to determine whether the target GS-BC module T = {(g, b)|g ∈ Gd ∩ Gk, b ∈ Bd ∩ Bk} is enriched in L.
Now we describe our threshold-based IGEA method. We have N gene-QT pairs from GWAS. Out of these, n = |A| pairs (the set A) are significant ones with GWAS p-value passed a certain threshold. We also have m = |P| (the set P) gene-QT pairs from a given GS-BC module, and k significant pairs are from P. Using Fisher’s exact test for independence, the enrichment p-value for the given GS-BC module is calculated as:
| (3) |
2.4 Evaluation of the Identified GS-BC Modules
For evaluation purpose, we tested the statistical significance of the IGEA results. We hypothesize that the gene-QT associations from BWGWAS of the original data should be overrepresented in certain GS-BC modules, and the BWGWAS results on permuted data should not be enriched in a similar number of GS-BC modules. We performed IGEA analyses on 50 permuted BWGWAS data sets, and estimated the distribution of the number of significant GS-BC modules. The distribution appeared to be normal. Using this normal distribution, we estimated an empirical p-value for the number of significant GS-BC modules discovered from the original data.
To determine the functional relevance of the enriched GS-BC modules, we also tested whether genes from each module are overrepresented for specific neurobiological functions, signaling pathways or complex neurodegenerative diseases. We performed pathway enrichment tests using gene ontology (GO) biological process terms, KEGG pathways and OMIM (Online Mendelian Inheritance in Man) database.
3 Results and Discussions
3.1 Significant GS-BC Modules from AHBA
By performing hierarchical clustering on both genetic and imaging domains, 275 out of 357 genetic clusters (only those with size ranging from 10 to 400) and 8 imaging clusters (with size ranging from 4 to 23, no clusters are excluded) were identified. 2200 GS-BC modules were generated by combining each pair of genetic and ROI clusters. After performing 1000 permutation tests, 610 modules were kept with a p-value ≤ 0.05. We did not use extremely stringent statistical thresholds for the selection, to avoid the exclusion of potentially interesting candidates. For the BWGWAS results, we obtained 21, 028 × 105 = 2, 207, 940 gene-QT associations after mapping SNP-based p-values to genes. Out of these, 1679 gene-QT associations passed the BWGWAS p-value of 1.0e-5.
All 610 constructed GS-BC modules were tested for whether they could be enriched by BWGWAS results using IGEA, and 12 of them turned out to be significant after Bonferroni correction (see Table 1). We also tested the significance of the number of identified GS-BC modules. Compared to permuted results, the analysis on the original data yielded a significantly larger number of enriched GS-BC modules with empirical p-value = 2.6e-2, indicating that imaging genetic associations existed in these enriched GS-BC modules.
Table 1.
Twelve significantly enriched GS-BC modules from IGEA. See also Table 2 and Figure 2 for details about relevant GSs and BCs respectively.
| Module ID | BC ID | # of ROIs | GS ID | # of genes | Corrected P-value |
|---|---|---|---|---|---|
| 01 | BC02 | 13 | GS01 | 59 | 1.83E-12 |
| 02 | BC01 | 14 | GS01 | 59 | 8.46E-07 |
| 03 | BC05 | 12 | GS02 | 18 | 1.58E-02 |
| 04 | BC02 | 13 | GS03 | 107 | 1.08E-04 |
| 05 | BC06 | 11 | GS03 | 107 | 9.17E-09 |
| 06 | BC07 | 10 | GS03 | 107 | 6.99E-07 |
| 07 | BC03 | 18 | GS03 | 107 | 2.96E-09 |
| 08 | BC04 | 23 | GS03 | 107 | 7.42E-06 |
| 09 | BC06 | 11 | GS04 | 50 | 4.51E-02 |
| 10 | BC07 | 10 | GS04 | 50 | 2.88E-02 |
| 11 | BC03 | 18 | GS04 | 50 | 4.56E-03 |
| 12 | BC02 | 13 | GS05 | 52 | 1.25E-07 |
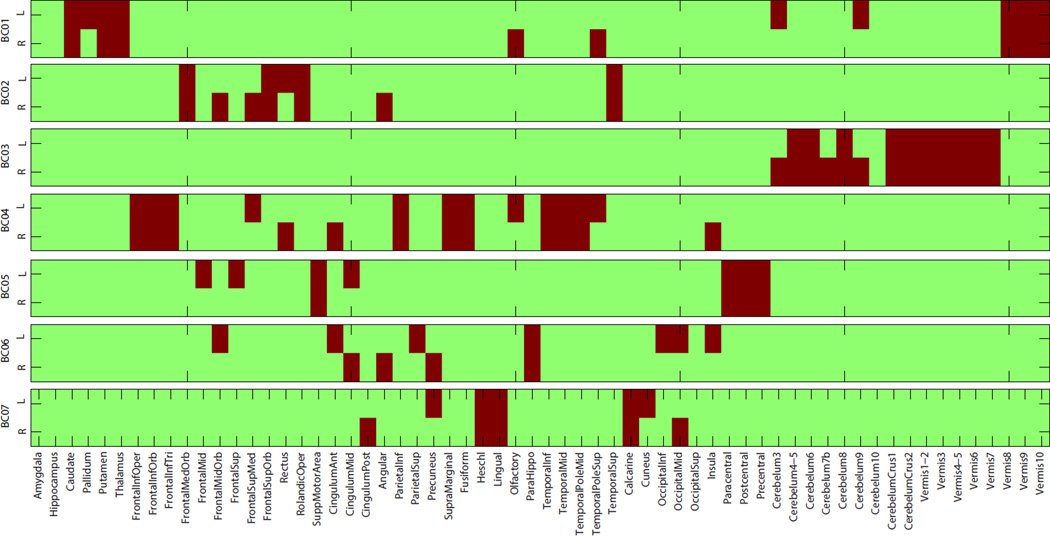
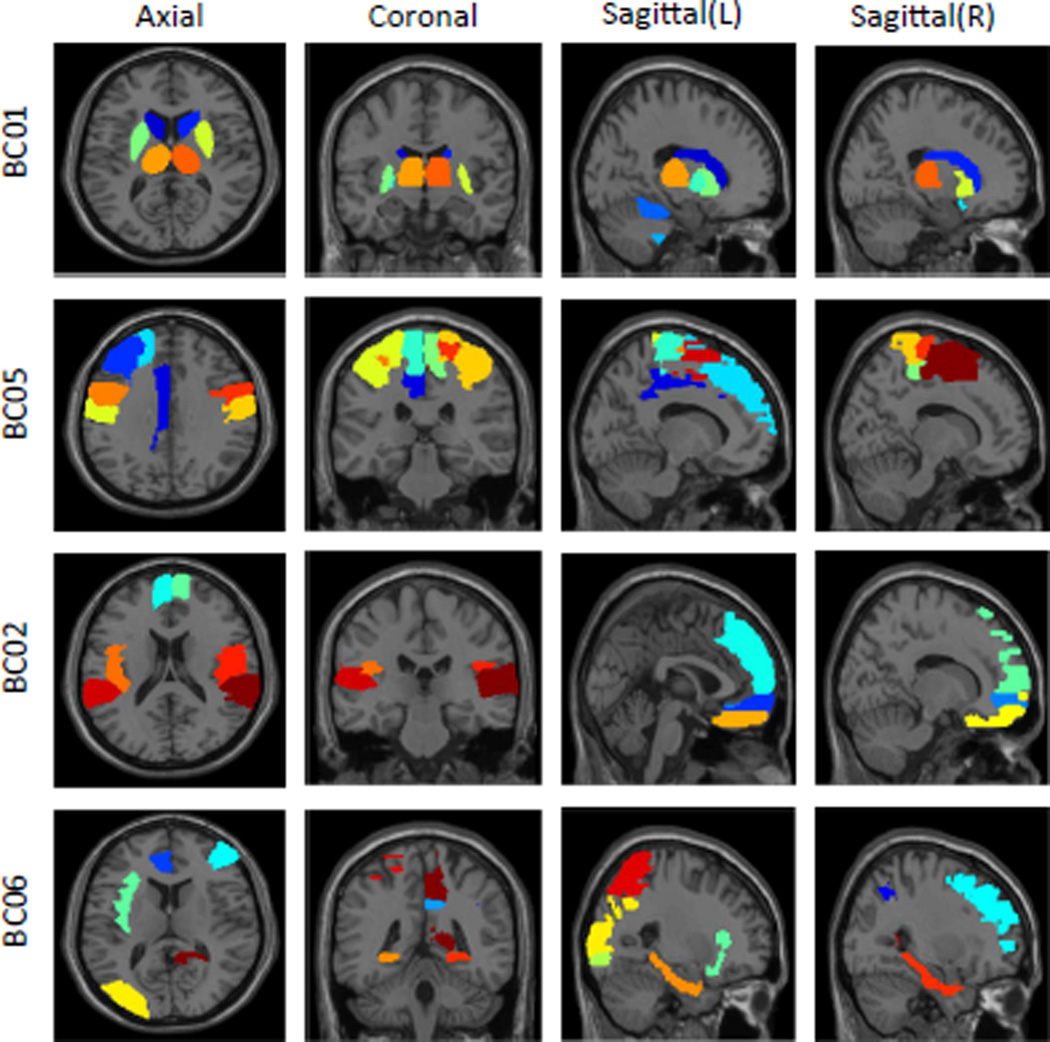
Across all 12 identified modules, there are 5 and 7 unique GS and BC entities respectively. Table 2 lists the 5 unique GSs with gene symbols. Figure 2 shows the 7 unique BCs with corresponding ROI names, and Figure 3 maps four of those onto the brain. For example, BC01 involves structures responsible for motivated behaviors (e.g., caudate, pallidum, putamen) and sensory information processing (e.g., thalamus). BC02 involves various frontal regions responsible for executive functions. BC06 includes structures that are major spots for amyloid accumulation in AD (e.g., cingulum, precuneus). Details of all 12 modules are listed in Table 1. We can find that some modules share common gene sets with different brain circuits, and some share the same brain circuits with different gene sets. This illustrates the complex associations among multiple genes and multiple brain ROIs.
Table 2.
Five unique gene sets (GSs) identified from IGEA.
| GS ID | Gene symbols |
|---|---|
| GS01 | AASDHPPT, APOC1, APOC4, ARHGAP1, ARMC2, BCR, C16orf74, CHST11, COL25A1, DCLK3, DNAH6, DNAI1, DOCK4, DRD1, ELMOD1, ERLIN1, EXD2, EYA1, FAM107A, FAM118B, FZD2, GNAL, GPR6, GPR88, GSTM3, HTR4, HYDIN, IL17D, ITPK1, KLF5, MFN2, MIPEP, MLLT3, MMD2, MTHFD1, MYB, NCKAP1, NSUN3, PALM, PDE1B, PDYN, PFDN6, PHF21B, PPP1R1A, RAP1GAP, RGS14, RGS20, RNF44, SLC17A8, SLC2A4, STYXL1, TRIM69, UBQLN4, UBR7, VAX1, WNT8A, ZC3HAV1L, ZMAT2, ZNF883 |
| GS02 | ACSS1, CPLX1, CSMD2, DDX4, ITGB3, LRIG1, METTL7A, NDRG2, NPAS4, PDIA6, PLA2G5, POTED, PPARGC1A, PSD2, PXDNL, TFCP2L1, USH2A, VEGFA |
| GS03 | ADPRHL2, ADRM1, AKR1A1, ANAPC2, AP4M1, AP5Z1, APBB1, APBB3, ARFGAP3, ARMC6, ASL, ATP5B, AUP1, AURKAIP1, B4GALT3, C17orf59, CAPN1, CCS, CCT3, CDIPT, COG4, CPNE1, CSNK2B, CSTF1, DAPK3, DDX21, DECR2, DEPDC5, DHPS, DHX16, DHX38, DNAJC30, EIF2B4, EIF2B5, ELOF1, ERAL1, FAM50B, FAM96B, FLII, GALK1, GGNBP2, GPAA1, GPATCH3, GPI, GPR137, HEXIM2, HN1, HOOK2, HPS6, IFFO1, KAT5, KHK, KLHL22, LDLR, LRSAM1, LZTR1, MAF1, MAGEF1, MFSD10, MMS19, MRPL38, MRPL54, NARFL, NCAPH2, NCLN, NISCH, NRBP1, NTHL1, PHB2, PI4KB, PIH1D1, POLD2, POLG, PPOX, PRDX2, PRMT1, PRPF31, PTBP2, PTOV1, RABGGTA, RALY, RPS19BP1, RRN3, SH2B1, SLC25A42, SLC41A3, SMPD1, SNRPA, SSNA1, STK19, STUB1, SULT1A1, TCEA2, TCEB3, TMED3, TMEM106C, TMEM161A, TOMM40, TRMU, TRPC4AP, TTC27, TUSC1, TXN2, TXNL4B, UBR4, YTHDF2, ZFAND2B |
| GS04 | AIPL1, AP1M2, ARRDC5, CD1C, CST1, DEFB113, DEFB126, EPGN, FBP2, FGF19, FNDC7, FRG2, GPRC5D, IL22, INMT, KCNK18, KIF18A, KLRG2, KRT79, MBD3L2, MMP7, MS4A1, MS4A3, MSMB, NEIL3, OR13C3, OR1M1, OR4F15, OR51I2, OR5AN1, OR5AR1, OR5M1, OR7G3, PDZK1IP1, PRAMEF8, PTCHD3, RLN1, SIRPD, TBX20, TEDDM1, TGM3, TIAF1, TIMD4, TM4SF19, TM4SF20, TMC1, TPD52L3, WFDC13, XDH, ZFP42 |
| GS05 | ALDH9A1, ANKFN1, APOE, ATP6V0A4, BIN1, C11orf65, C15orf52, C1orf64, CD81, CDH1, CNN3, ECSCR, EDNRB, ENG, FAM84B, GGT5, GIMAP5, GPR137B, GREM1, GSTM2, GTF2F2, HRASLS2, ID1, LMO2, MAPKAPK3, MARCH10, PARP4, PAWR, PGR, PHF10, PLSCR4, PMAIP1, POLI, PRDX1, RAB13, RGS22, SDS, SLC2A1, SLC40A1, SMAD9, STX18, SULT1C4, SVOPL, TIE1, TM4SF18, TMEM204, TRIP6, TST, WASF2, WFDC3, WRB, WWC2 |
Fig. 2.
Seven unique brain circuits (BCs) identified from IGEA. ROIs belonging to each BC are colored in red.
Fig. 3.
Brain maps of four brain circuits (BCs) identified from IGEA.
3.2 Pathway Analysis of Identified GS-BC Modules
To explore and analyze functional relevance of our identified GS-BC modules, we performed pathway analysis from three aspects including biological processes, functional pathways and diseases using Gene Ontology, KEGG pathways and OMIM diseases databases, respectively.
Most identified GSs have a significant functional enrichment, and several can be related to the neurodegenerative disease and its development. For instance, calcium signaling pathway (from Module #01 and #02) playing key role in short- and long-term synaptic plasticity, has shown abnormality in many neurodegenerative disorders including AD, Parkinson’s disease, amyotrophic lateral sclerosis, Huntington’s disease, spinocerebellar ataxias and so on [1]. There are also several enriched pathways related to oxidative stress, which is a critical factor for a range of neurodegenerative disorders. For example, DNA polymerase (from Module #04-08) deficiency can lead to neurodegeneration and exacerbates AD phenotypes by reducing repair of oxidative DNA damage [18]; glycolysis and gluconeogenesis (from Module #04-08) are associated with hypoxia, ischemia, and AD [2]; others like adherens junction (from Module #12) and focal adhesion (from Module #03) have also been shown disorder-related by indirectly affecting oxidative stress. For the enriched disease results, we also find some neurodegeneration-related (like anomalies from Module #01 and #02, neuropathy from module #04-08), while a large part of them are cancer-related (like prostate cancer from Module #09, #10 and #11). A large body of studies have focused on investigating the relationship between cancer and neurodegeneration, with abnormal cell growth and cell loss in common. For the GO Biological Process enrichment, various Biological Process terms are enriched and can be grouped to 5 categories including cellular process, cell cycle, metabolic process, neurological system process and response to stimulus. Most of these terms have direct or indirect relationships with neurodegenerative diseases or phenotypes.
4 Conclusions
We have presented a two dimensional imaging genetic enrichment analysis (IGEA) framework to explore the high level imaging genetic associations by integrating whole brain genomic, transcriptomic and neuroanatomic data. Traditional pathway enrichment analysis focused on investigating genetic findings of a single phenotype one at a time, and relationships among imaging QTs could be ignored. Such approach could be inadequate to provide insights into the mechanisms of complex diseases that involve multiple genes and multiple QTs. In this paper, we have proposed a novel enrichment analysis paradigm IGEA to detect high level associations between gene sets and brain circuits. By jointly considering the complex relationships between interlinked genetic markers and correlated brain imaging phenotypes, IGEA provides additional power for extracting biological insights on neurogenomic associations at a systems biology level.
Acknowledgments
This work was supported by NIH R01 LM011360, U01 AG024904 (details available at http://adni.loni.usc.edu), RC2 AG036535, R01 AG19771, P30 AG10133, and NSF IIS-1117335 at IU, and by NIH R01 LM011360, R01 LM009012, and R01 LM010098 at UPenn.
References
- 1.Bezprozvanny I. Calcium signaling and neurodegenerative diseases. Trends Mol Med. 2009;15(3):89–100. doi: 10.1016/j.molmed.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Butterfield D, Lange M. Multifunctional roles of enolase in Alzheimer’s disease brain: beyond altered glucose metabolism. J Neurochem. 2009;111(4):915–933. doi: 10.1111/j.1471-4159.2009.06397.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Draghici S, Khatri P, et al. Global functional profiling of gene expression. Genomics. 2003;81(2):98–104. doi: 10.1016/s0888-7543(02)00021-6. [DOI] [PubMed] [Google Scholar]
- 4.Draghici S, Khatri P, et al. Onto-tools, the toolkit of the modern biologist: Onto-express, onto-compare, onto-design and onto-translate. Nucleic Acids Res. 2003;31(13):3775–3781. doi: 10.1093/nar/gkg624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hirschhorn JN. Genomewide association studies–illuminating biologic pathways. N Engl J Med. 2009;360(17):1699–1701. doi: 10.1056/NEJMp0808934. [DOI] [PubMed] [Google Scholar]
- 6.Hong MG, Alexeyenko A, et al. Genome-wide pathway analysis implicates intracellular transmembrane protein transport in Alzheimer disease. J Hum Genet. 2010;55(10):707–709. doi: 10.1038/jhg.2010.92. [DOI] [PubMed] [Google Scholar]
- 7.Jin D, Lee H. A computational approach to identifying gene-microRNA modules in cancer. PLoS Comput Biol. 2015;11(1):e1004042. doi: 10.1371/journal.pcbi.1004042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Khatri P, Draghici S. Ontological analysis of gene expression data: current tools, limitations, and open problems. Bioinformatics. 2005;21(18):3587–3595. doi: 10.1093/bioinformatics/bti565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim S, Swaminathan S, et al. Influence of genetic variation on plasma protein levels in older adults using a multi-analyte panel. Plos One. 2013;8(7) doi: 10.1371/journal.pone.0070269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lambert JC, Grenier-Boley B, et al. Implication of the immune system in Alzheimer’s disease: evidence from genome-wide pathway analysis. J Alzheimers Dis. 2010;20(4):1107–1118. doi: 10.3233/JAD-2010-100018. [DOI] [PubMed] [Google Scholar]
- 11.O’Dushlaine C, Kenny E, et al. Molecular pathways involved in neuronal cell adhesion and membrane scaffolding contribute to schizophrenia and bipolar disorder susceptibility. Mol Psychiatry. 2011;16(3):286–292. doi: 10.1038/mp.2010.7. [DOI] [PubMed] [Google Scholar]
- 12.Purcell S, Neale B, et al. Plink: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ramanan V, Shen L, et al. Pathway analysis of genomic data: concepts, methods, and prospects for future development. Trends Genet. 2012;28(7):323–332. doi: 10.1016/j.tig.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ramanan VK, Kim S, et al. Genome-wide pathway analysis of memory impairment in the Alzheimer’s Disease Neuroimaging Initiative (ADNI) cohort implicates gene candidates, canonical pathways, and networks. Brain Imaging Behav. 2012;6(4):634–648. doi: 10.1007/s11682-012-9196-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saykin AJ, Shen L, et al. Alzheimer’s Disease Neuroimaging Initiative biomarkers as quantitative phenotypes: Genetics core aims, progress, and plans. Alzheimers Dement. 2010;6(3):265–273. doi: 10.1016/j.jalz.2010.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shen L, Thompson PM, et al. Genetic analysis of quantitative phenotypes in AD and MCI: imaging, cognition and biomarkers. Brain Imaging Behav. 2014;8(2):183–207. doi: 10.1007/s11682-013-9262-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Subramanian A, Tamayo P, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sykora P, Misiak M, et al. Dna polymerase beta deficiency leads to neurodegeneration and exacerbates Alzheimer disease phenotypes. Nucleic Acids Res. 2015;43(2):943–959. doi: 10.1093/nar/gku1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ulitsky I, Maron-Katz A, et al. Expander: from expression microarrays to networks and functions. Nature Protocols. 2010;5(2):303–322. doi: 10.1038/nprot.2009.230. [DOI] [PubMed] [Google Scholar]
- 20.Yan J, Du L, et al. Transcriptome-guided amyloid imaging genetic analysis via a novel structured sparse learning algorithm. Bioinformatics. 2014;30(17):i564–i571. doi: 10.1093/bioinformatics/btu465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zeng H, Shen EH, et al. Large-scale cellular-resolution gene profiling in human neocortex reveals species-specific molecular signatures. Cell. 2012;149(2):483–496. doi: 10.1016/j.cell.2012.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]