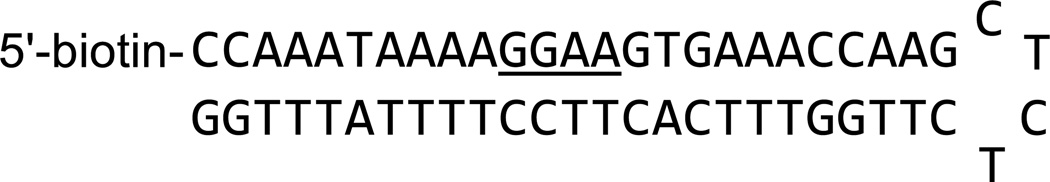Abstract
Biosensor-surface plasmon resonance (SPR) technology has emerged as a powerful label-free approach for the study of nucleic acid interactions in real time. The method provides simultaneous equilibrium and kinetic characterization for biomolecular interactions with minimal materials and without an external probe. A detailed and practical guide for protein-DNA interaction analyses using biosensor-SPR methods is presented. Details of the SPR technology and basic fundamentals are described with recommendations on the preparation of the SPR instrument, sensor chips and samples, as well as extensive information on experimental design, quantitative and qualitative data analyses and presentation. A specific example of the interaction of a transcription factor with DNA is shown with results evaluated by both kinetic and steady-state SPR methods.
Keywords: Biosensor, surface plasmon resonance, protein-nucleic acid interaction, kinetics, steady state analysis, mass transfer, transcription factor
1. Introduction
1.1 Surface Plasmon Resonance
During the past twenty years, commercial biosensors using surface plasmon resonance (SPR) detection have been introduced to the scientific community and have emerged as a major and powerful approach for characterizing biomolecular interactions with high quality kinetic and thermodynamic information (1–4). From the initial development on protein-protein interactions, the applications of SPR have been significantly extended to diverse biomolecule complexes, including protein-nucleic acid, protein-small molecules, and nucleic acid-small molecules (5–8). In the SPR method one component of an interaction is immobilized on a sensor chip to create the biosensor interaction surface. The other component(s) of the interaction is then injected over the sensor surface in a solution at the desired ionic strength and pH. Upon complex formation on the sensor surface between these species, the refractive index changes are converted into SPR responses. The real time responses allow the extent and rates of complex formation to be quantitatively determined. The unique SPR detection mode has numerous advantages over conventional interaction analyses, such as optical methods for systems involving strong interactions and/or low fluorescence and absorbance. In addition, SPR generally requires only picomole to nanomole quantities of material which is minimal compared to other techniques generally used for evaluation of biomolecular interactions.
Despite the widespread and long-standing use of SPR in characterizing protein-ligand and protein-protein interactions, evaluation of protein-DNA complexes with such instruments has been less extensive. Protein-DNA interactions present specific challenges for SPR characterization due to their generally high binding affinity and strong electrostatic nature. These features give rise to several practical complications: i) mass transfer limits on kinetics, where the rates of transfer of components from the injected solution to the immobilized component is slower than the association reaction, ii) very slow dissociation rates with potential rebinding during the dissociation phase, and iii) limited time for the association reaction due to volume limitations in the injection syringe. These limitations have restricted use of these instruments with protein-DNA complex. Several approaches have been devised to mitigate these challenges, including the use of high flow rates, low immobilization densities, and the addition of DNA in the flow solution (9–12).
To illustrate the utility of SPR in protein-DNA interactions, and methods to optimize experimental conditions for overcoming the limitations for some challenging systems, the interaction between the transcription factor PU.1 (Spi-1) with a specific DNA sequence will be used as an example in this protocol. PU.1 is a member of the ETS-family of proteins which comprise an evolutionarily conserved family of transcription factors (13). ETS-family proteins regulate the expression of a functionally diverse array of genes throughout the Metazoan kingdom (14–16). Aberrent expression of ETS-regulated genes are frequently implicated in human and veterinary cancers (17–20). All ETS proteins share a structurally conserved DNA binding domain (known as the ETS domain) that recognizes DNA sequences containing a central 5’-GGAA/T-3’ consensus (21). Structurally, ETS/DNA complexes are universally characterized by the insertion of an essential recognition helix in the major groove at the core consensus, while the flanking bases are recognized via backbone contacts (22). Since ETS factors share only limited interchangeability in vivo (23–25), the mechanism by which they achieve biological specificity continues to be an active area of inquiry. Factor-specific interactions with their target DNA represent major specificity determinants, as evidenced by the strong correspondence of sequence preference of ETS domains in vitro to genomic occupancy of native ETS proteins in vivo (21, 26). In the case of PU.1, recent thermodynamic and kinetic studies of DNA sequence recognition have shed light on the mechanism of DNA recognition (27–29), and established a molecular paradigm against which other ETS proteins may be quantitatively compared. Sequence-specific PU.1 ETS-DNA interactions, thus, represent an excellent model system for evaluating protein-DNA complex formation by SPR in terms of both equilibrium and kinetic signatures. The techniques described for ETS/DNA interactions should be highly transferrable to the investigation of other sequence-specific protein/DNA and peptide/DNA interactions.
1.2 Basic principles for biosensor-SPR method
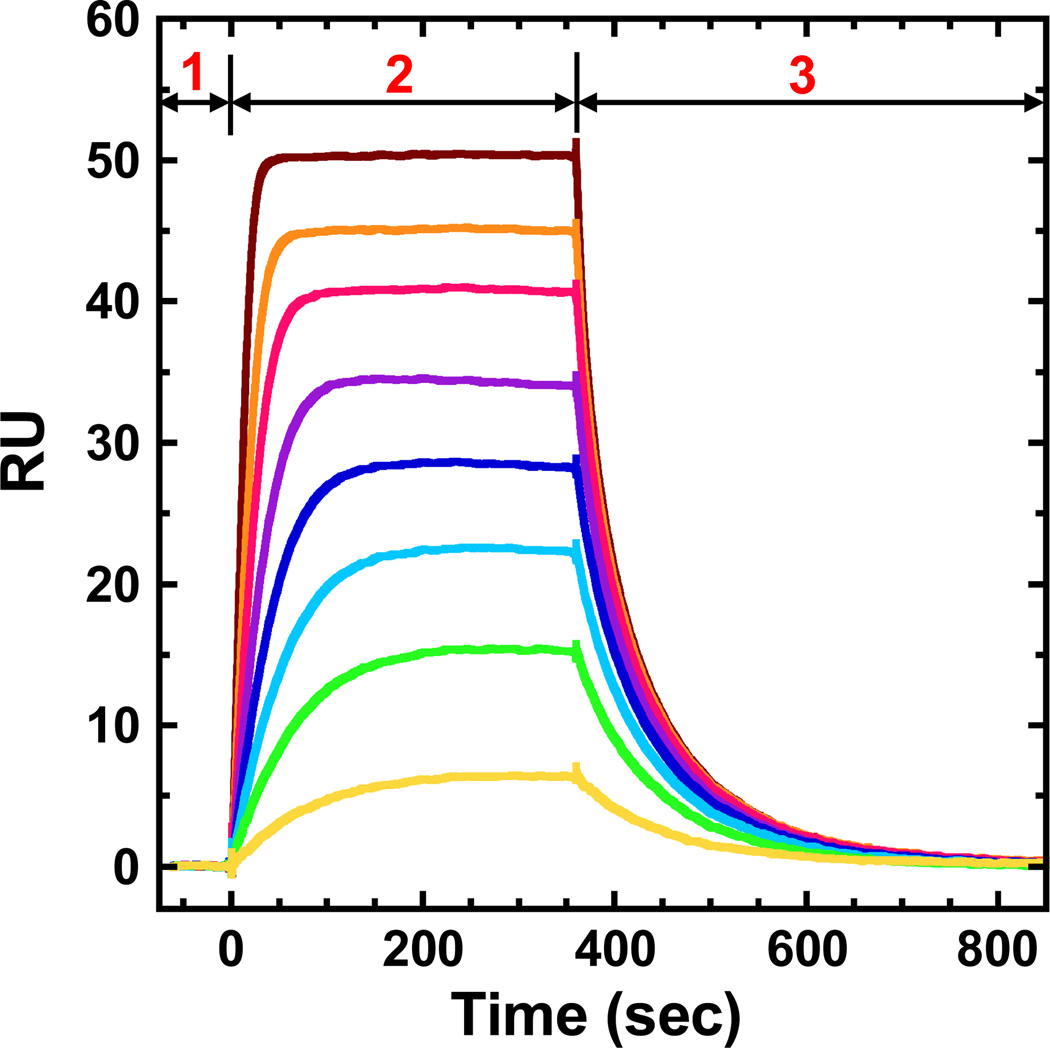
The results of a biosensor-SPR experiment are typically presented as a series of sensorgrams, which shows the SPR response units (RU) as a function of time (Figure 1). With a DNA sequence immobilized on the chip surface, first, initial buffer flow gives a reference baseline. Second, a protein solution is injected and as the solution flows over the surface, protein binding to DNA is monitored by SPR changes. With sufficient time a steady-state plateau is established where association and dissociation of protein are occurring at an equal rate. Third, buffer flow (without protein) is restarted and the dissociation of the complex can be monitored as a function of time (Figure 1).
Figure 1.
Representative SPR sensorgrams with three major stages: (1) initial buffer flow for a stable reference baseline; (2) ligand association phase with sample injections over the sensor chip surface; (3) ligand dissociation phase with buffer flow over the surface.
For a protein (P) binding to a DNA sequence and forming a single complex (C), the interaction is given by:
| (1) |
and the equilibrium binding affinity for this interaction is:
| (2) |
where [P] is the concentration of the protein at the sensor surface, [DNA] is the concentration of the immobilized DNA which is not bound to protein, the free DNA concentration, and [C] is the concentration of the complex; KA is the equilibrium binding constant, ka is the association rate constant and kd is the dissociation rate constant.
For association:
| (3) |
and for dissociation:
| (4) |
Both the association and dissociation phases of the sensorgram can be simultaneously fit to a desired binding model with several sensorgrams at different protein concentrations using a global fitting routine (30, 31). Global fitting allows the most robust determination of the kinetic constants and the calculation of equilibrium constant, KA, from the ratio of kinetic constants (Equation 2). Steady-state binding results from SPR experiments, where the SPR response reaches a plateau region, can be fitted with the following model:
| (5) |
(limit of r as Cfree → +∞, r → = n)
where n is the stoichiometry and is one for the single-site model, r represents the moles of bound protein per mole of DNA and Cfree is the free protein concentration in equilibrium with the complex. When Cfree is very high, r equals n. RUobs is the observed (experimental) response in the plateau region and RUmax is the predicted maximum response for a monomer protein binding to a DNA site (8). RUmax can be calculated and determined experimentally at the RU for saturation of the DNA binding sites, or used in Equation 5 as a fitting parameter such that KA, r and RUmax are determined by fitting RUobs versus Cfree. Dividing the observed steady-state response RUobs by RUmax at saturation yields the binding stoichiometry.
If the complex dissociates slowly, the surface can be regenerated before the complete dissociation occurs with a solution that causes rapid dissociation of the complex without irreversible damage to the immobilized DNA (31, 32). For example, a solution at low or high pH (pH ≤ 2.5 or pH ≥10) can unfold DNA and cause the complex to completely dissociate. Additional injections of buffer at pH near 7 allow the immobilized DNA to refold for additional steps in the binding experiment. After the dissociation and regeneration phase, a stable baseline is re-established. Then another sample containing a different concentration of protein can be injected to generate another sensorgram. With a series of sensorgrams generated with a broad range of concentrations, both the kinetics and equilibrium constant can be determined as discussed above.
1.3 Critical factors for protein-DNA interaction evaluation by biosensor-SPR methods
1.3.1 Concentration range and binding affinity KD
For the determination of an equilibrium constant by any method, the selected set of experimental concentrations must provide both free and bound concentrations of reactants. In the biosensor-SPR method with DNA bound to the surface, the protein concentrations should go from below to above the KD to determine the KD so that a range of bound states of DNA is obtained. The initial concentrations have little bound protein but as the concentration of protein injected is increased, the fraction of DNA saturation approaches one. In this way the most accurate equilibrium constant can be obtained. The sensorgrams will go from a very low RU and will approach a saturation value at high protein concentration, relatively to KD. If too low a set of protein concentrations is used, all sensorgrams will be low and it will be hard to determine the RUmax. If too high a set of concentrations is used, all sensorgrams will be near the saturation limit and global fitting will not be useful. Some experimentation is needed in any new protein-DNA binding system to get an approximate KD so that an appropriate set of protein concentration can be prepared.
1.3.2 Mass transport and rebinding in dissociation
For interactions on a sensor surface, the reaction component in sample solution is injected over the flow cell surface and must be transported from the bulk solution to the surface, which is called mass transport. In the reaction of interest, a protein, such as PU.1, is transported to the immobilized DNA for complex formation. This is a diffusion-controlled process, and the transport rate can directly influence the binding kinetics if it is slower than the binding reaction. A key requirement in accurate determination of kinetic constants by the SPR method is that the concentration of free protein in the matrix quickly equilibrates with the flow solution. If the association reaction is much faster than mass transport, the observed binding will be limited by the mass transport processes. Conversely, if transport is fast and association is slow, the observed binding will represent the true interaction kinetics (33). Therefore, the mass transport rate is a critical factor that must be considered in biosensor experimental design and in evaluating kinetic constants from biosensor-SPR methods.
A similar factor that affects kinetics is the rebinding induced slow dissociation. After the protein-DNA complex formation reaches the injection time limit, the running buffer flows over the sensor surface to dissociate the bound protein from the immobilized DNA. For proteins with very high binding affinities, they can frequently rebind DNA after they dissociate from DNA but before leaving the matrix. When rebinding occurs the rate of protein transport away from the surface is slow compared to the real dissociation rate at the surface, which results in a very slow apparent dissociation. He et al. presented a theoretical and experimental approach to deal with the mass transfer effect on strong binding of proteins to DNA (9). They used the lac repressor-operator interaction as a test system and compared their results with the previous SPR data that did not agree with filter binding results (10, 11). They clearly showed that, with the target DNA immobilized and lac repressor protein in the injection solution, strong mass transport effects and rebinding in dissociation were observed at low flow rates and high density of DNA immobilization. As the flow rate increased, the observed association rate constants were significantly increased in agreement with elimination of mass transport effects. In the dissociation phase of the sensorgram, the rebinding of the dissociated protein markedly restricted the determination of the true dissociate rate. The authors designed a clever method to deal with the rebinding that includes excess lac DNA in the flowing buffer for dissociation. By this method, the dissociated lac repressor protein bound to the DNA sequence in the flowing buffer, instead of the immobilized DNA on the surface, and consequently the rebinding was relieved and the real dissociation rate was determined (9).
Overall, for kinetic measurements, it is generally recommended to use low surface densities of the immobilized DNA and high protein flow rate (≥ 50 µL/min) to minimize the limitations on binding rates by mass transport processes. In addition, the dissociation phase can be set up for several hours or even longer with Biacore SPR which allows at least 50% of bound protein to dissociate and a reliable kinetic fit can be performed, even with very slow dissociation.
1.3.3 Limited time for the association due to volume limitations in the injection syringe
The volume of the injection syringe of a biosensor SPR system is limited, and thus the sample injection and association time is consequently limited. For example, the Biacore T200 instrument has an injection volume of 350 µL (syringe size), which obviously means that the association time can only be set to 7 min maximum with a flow rate of 50 µL/min, or 3.5 min with a flow rate of 100 µL/min. These limited association times are not sufficient for some interactions to reach the binding equilibrium necessary for steady state analyses. Myszka et al. showed that this problem can be resolved by immobilizing the target DNA sequence and placing the protein at different concentrations in the running buffer (9). Then, protein samples are able to be injected for hours and the kinetics can be determined with limited effects from mass transfer and rebinding. This method also allows the steady-state plateau to be reached and the KA to be determined directly (Equation 5) without any possible mass transfer issues.
In our biosensor-SPR evaluation of the interaction of PU.1 ETS domain with DNA, both mass transfer and rebinding are carefully evaluated and minimized in the experimental protocol which is described in detail below. The incorporation of optimally designed flow cells in current instrumentation and optimized experimental protocols and sensor chips have qualified biosensor-SPR as an excellent method for quantitative analysis of protein-DNA interactions, especially for strong binding systems. Here, we show that careful use of ionic conditions allows useful data collection over a broad range of conditions without limiting mass transfer or rebinding problems.
2 Materials
2.1 Instrument Cleaning
These materials are for the Biacore T200 research instrument but similar materials are required for other instruments.
Maintenance chip with a glass flow cell surface for cleaning without damage to experimental sensor chips.
0.5% (w/v) sodium dodecyl sulphate (SDS, Biacore Desorb solution 1).
50 mM glycine pH 9.5 (Biacore Desorb solution 2). (see Note 1)
1% (v/v) acetic acid solution
0.2 M sodium bicarbonate solution.
6 M guanidine HCl solution.
10 mM HCl solution. (see Note 2)
2.2 Sensor Chip Preparation Solutions for DNA Immobilization
A CM4 or CM5 sensor chip that has been at room temperature for at least 30 min prior to use (all sensor chips are available from GE Healthcare Inc.). (see Note 3)
NHS: 100 mM N-hydroxysuccinimide freshly prepared in water.
EDC: 400 mM 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride freshly prepared in water.
Immobilization buffer: 10 mM sodium acetate buffer pH 4.5.
200 µg/ml streptavidin solution in immobilization buffer.
1 M ethanolamine hydrochloride in water pH 8.5.
Running buffer (HBS-EP): 10 mM HEPES pH7.4, 150 mM NaCl, 3 mM EDTA, 0.05% (v/v) surfactant P20.
2.3 DNA Immobilization Solutions for a SA chip
A streptavidin–coated sensor chip (SA chip or prepared as outlined above) that has been at room temperature for at least 30 min.
Running buffer (HBS–EP): 10 mM HEPES pH7.4, 150 mM NaCl, 3 mM EDTA, 0.05% (v/v) surfactant P20. (as in Section 2.2).
Activation buffer: 1 M NaCl in 50 mM NaOH.
Biotin-labeled nucleic acid solution (20 nM of 5’-biotinylated single strand or hairpin DNA dissolved in HBS-EP buffer.
2.4 Flow Solutions: General Buffers
HBS-EP (see Note 4): 10 mM HEPES pH 7.4, 150 mM NaCl, 3 mM EDTA, 0.05% (v/v) surfactant P20.
10 mM Tris pH 7.4, 100 mM NaCl, 1 mM EDTA, 0.05% (v/v) surfactant P20.
25 mM Na2HPO4 pH 7.4, 400 mM NaCl, 1 mM EDTA, 0.05% (v/v) surfactant P20.
2.5 Regeneration Solution
Generally used regeneration solutions are listed in Table 2. In general, milder conditions are initially used, and more harsh conditions are applied as needed. Some other regeneration solutions for special samples are available from the Biacore website. In our studies, 1 M NaCl solution is typically used as a gentle but efficient regeneration solution to remove protein from the DNA immobilized sensor chip surface. (see Note 5)
Table 2.
Regeneration solutions.
| Interaction Strength |
Acidic | Basic | Hydrophobic | Ionic |
|---|---|---|---|---|
| Weak | pH > 2.5 | pH < 9 | pH < 9 | |
| 10 mM Glycine/HCl | 10 mM HEPES/NaOH | 50% ethylene glycol | 1 M NaCl | |
| HCl | ||||
| formic acid | ||||
| Intermediate | pH 2–2.5 | pH 9–10 | pH 9–10 | |
| 10 mM Glycine/HCl | 10 mM Glycine/NaOH | 50% ethylene glycol | 2 M MgCl2 | |
| formic acid | ||||
| HCl | NaOH | |||
| H3PO4 | ||||
| Strong | pH < 2 | pH > 10 | pH > 10 | |
| 10 mM Glycine/HCl | NaOH | 25–50% ethylene glycol | 4 M MgCl2 | |
| HCl | ||||
| formic acid | 6 M guanidine-chloride | |||
| H3PO4 |
3 Methods
3.1 Sensor Chip Preparation for DNA Immobilization
Dock the CM4 or CM5 chip, then Prime with running buffer (HBS-EP). Start a sensorgram in all flow cells with a flow rate of 5 µL/ min until baseline is stable (drifting < 1 response unit (RU) / min). “Dock” and “Prime” are Biacore control software commands that instruct the instrument to carry out specific operations. The commands and corresponding functions are listed in Table 1.
Mix 100 µL of NHS and 100 µL of EDC into one vial.
Inject the mixture of NHS/EDC for 10 min (50 µL) to activate the carboxymethyl surface to reactive esters.
Use Manual Inject to inject streptavidin solution over all flow cells with a flow rate of 5 µL/min for 20 min (100 µL). Track the RU immobilized, which is available in real time readout, and stop the injection after the desired level is reached (typically 2500 ~ 3000 RU for CM5 chip and 1000 ~ 1500 RU for CM4 chip).
Inject ethanolamine hydrochloride for 10 min (50 µL) to deactivate any remaining reactive esters.
Prime several times to ensure surface stability.
Then the sensor chip is ready for DNA immobilization as described under Section 3.2. (see Note 6)
Table 1.
Biacore instrument commands*.
| Biacore Control Software commands |
Function |
|---|---|
| Desorb | Removes adsorbed materials from the flow system |
| Sanitize | Removes disinfects from the flow system |
| Prime | Strongly flushes the flow system with running buffer |
| Dock | Docks the sensor chip into the instrument |
| Undock | Undocks the sensor chip from the instrument |
These commands are for Biacore instruments, but other commands with the same functions might be used with other instruments.
3.2 DNA Immobilization on a SA chip
Dock a streptavidin–coated chip (SA chip) and start a sensorgram with a 25 µL/min flow rate.
Inject activation buffer for 1 min (25 µL) five to seven times to remove any unbound streptavidin from the sensor chip.
Prime several times to ensure surface stability.
Allow buffer to flow at least 5 min before immobilizing the nucleic acids.
Start a new sensorgram with a flow rate of 1 µL/min by choosing only one flow cell under “flow path” (e.g., flow cell 2 (FC2)) on which to immobilize the nucleic acid. Note not to immobilize nucleic acid on the flow cell chosen as the control flow cell. Generally, flow cell 1 (FC1) is used as a control and is left blank for subtraction, and different nucleic acids are immobilized on the remaining three flow cells (FC2-4).
Wait for the baseline to stabilize which usually takes a few minutes. Use Manual Inject, load the injection loop with ~100 µL of a 20 nM nucleic acid solution and inject over the current flow cell.
- The amount of DNA to immobilize on the sensor chip depends on the relative molecular weights of the target DNA and protein and on the sensitivity of the biosensor system. Since the SPR response is directly proportional to the mass concentration of material at the surface, the theoretical protein binding capacity for a 1:1 interaction of a given surface is related to the amount of DNA immobilized (12):
For example, the molecular weight of DNA in Figure 2 is 16962 Daltons and the molecular weight of the PU.1 ETS domain is 12000 Daltons, immobilizing 100 RU of DNA will give a theoretical PU.1 binding capacity of 70 RU assuming that the PU.1 is 100% bound in a 1:1 complex. (see Note 7)(6) Track the RU immobilized and stop the injection after the desired level is reached (typically ~ 300 RU for hairpin nucleic acid in ~20–30 bases in length, and ~100 RU for hairpin nucleic acid containing ~ 50–60 bases for kinetic experiments to minimize the mass transport effects).
At the end of the injection and after the baseline has stabilized, use the instrument’s crosshair to determine the RUs of nucleic acid immobilized. The amount of nucleic acid immobilized is required to determine the theoretical moles of protein binding sites for the current flow cell.
Repeat steps 5 to 9 for other flow cells (e.g., FC3 and FC4).
Stop the sensorgram and change the immobilization buffer for the experimental buffer and Prime the system four times.
Before the first experiment, leave the sensorgram running overnight at 10 µL/min in order to stabilize the baseline of the newly prepared sensor chip.
Figure 2.
5’-biotin-labeled target hairpin DNA sequence for PU.1.
3.3 Sample preparation
The sample solution must be prepared in the same buffer used to establish the baseline (the running buffer). If the protein requires the presence of a reducing agent, such as DTT (dithiothreitol) or TCEP (Tris(2-carboxyethyl)phosphine), to prevent oxidation of free cysteines, or nonspecific DNA (NS DNA) to reduce the nonspecific interaction with immobilized DNA, the same amount of agent or NS DNA must be added to the running buffer to minimize the refractive index difference.
The sample concentration depends on the magnitude of the binding constant (KA). With a single binding site, for example, concentrations at least 10 times above and below 1/ KA should be used (i.e., a 100 fold difference between the lowest and highest concentrations). A larger concentration range above and below 1/ KA will yield a more complete binding curve. For binding constants of 107–109 M−1, as observed with many nucleic acid/protein complexes, protein concentrations from 0.01 nM to 10 µM in the flow solution allow accurate determination of binding constants (see Section 1.3.1). Injecting samples from low to high concentration is useful for eliminating artifacts in the data from adsorption or carry over (see Note 8).
Possible problems at high sample concentrations: poor sensorgrams and nonspecific binding may be obtained. For proteins that self-associate, it is important to maintain concentrations well below levels at which oligomerization occurs. For example, the PU.1 ETS domain is known to dimerize at above 10 µM and above 1 µM in the DNA bound-state (29). Therefore, the sample concentrations of PU.1 in this protocol are maintained well below this level (< 0.4 µM) to ensure that the protein presents as a monomer in both free and bound states.
3.4 Regeneration
Regeneration is the process of removing bound analyte from the sensor chip surface after analysis of a sample, in preparation for the next analysis cycle.
Regeneration conditions should remove the bound analyte completely from the surface without destroying the immobilized reagent. Generally used regeneration solutions are listed in Table 2. In general, milder conditions are initially used, and more harsh conditions are applied as needed. Some other regeneration solutions for special samples are available from the Biacore website. In our studies, 1 M NaCl solution is typically used as a gentle but efficient regeneration solution to remove protein from the DNA immobilized sensor chip surface. (see Note 5)
Injections of 30 ~ 60 sec of regeneration solution are usually sufficient. Longer exposure to regeneration conditions involves greater risks of lose of binding activity on the surface, and often does not improve regeneration.
After injection of regeneration solution, three 1-min injections of running buffer are recommended to reduce the remaining regeneration solution.
At the end of each cycle, 5 min running with buffer flowing is also set to ensure that the chip surface is re–equilibrated for binding (i.e., the dextran matrix is re-equilibrated with running buffer) and the baseline has stabilized before the next sample injection.
3.5 Data Collection and Processing
The Biacore control software allows users to write a method or to use a method wizard to set up experiments. Several key factors, such as flow rate, flow path, association and dissociation time, injection order, surface regeneration and post-regeneration re-equilibration, must be considered in setting up experiments. An example of the method used to collect PU.1 binding data on DNA surface is shown below.
A Biacore T200 instrument (GE Healthcare Inc.) is used as an example in this protocol. This is one of the most sensitive biosensor-SPR instruments but other instruments are excellent and the application will determine what sensitivity is needed. Binding small molecules to a protein or DNA requires high sensitivity while the interaction of two proteins, a protein with DNA or other macromolecule complexes, requires less sensitivity.
Streptavidin is immobilized on a CM4 sensor chip as described in Section 3.1, and then three biotin-labeled hairpin DNAs are immobilized in different flow cells as described in Section 3.2. The biotin-labeled target DNA sequence discussed in this example is shown in Figure 2. This DNA is based on the λB motif of the lg2-4 enhancer (34), 5’-AAAGGAAGTG-3’, a native high-affinity cognate site for PU.1 (35, 36).
1 M NaCl is used as regeneration solution.
The ETS domain of murine PU.1 (residues 167–262 from the murine sequence) is overexpressed in Escherichia coli and purified as previously described (28).
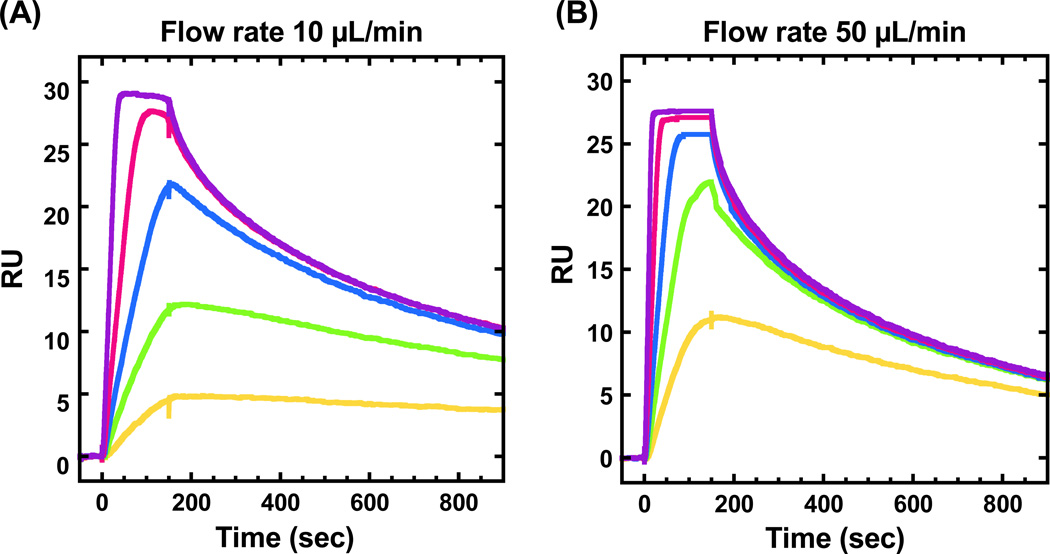
Na2HPO4 buffer (25 mM Na2HPO4, 1 mM EDTA and 0.05% (v/v) surfactant P20, pH 7.4) with different concentrations of NaCl have been used as experimental and running buffer in search of the optimum ionic conditions. An example of mass transport effects on PU.1-DNA interaction with 300 mM NaCl is shown in Figure 3. It is clear to see that the shapes of sensorgram and apparent kinetics are strongly dependent on flow rates (see Section 1.3.2). Sensorgrams with the same samples at other flow rates, such as 75 and 100 µL/min, have also been run with 300 mM and 400 mM NaCl and they are very alike (data not shown) which suggests that by increasing the flow rate the mass transport effects could be minimized and finally removed. Therefore, a high flow rate (100 µL/min) is employed in this protocol and Na2HPO4 buffer containing 400 mM NaCl, with and without 300 µM/base pair of salmon sperm DNA as non-specific DNA, is used as the experimental buffer. (see Note 9)
A series of concentrations (concentration range is from 1 nM to 400 nM) of PU.1 is prepared with the experimental buffer to cover the concentration range around the KD at any salt concentration. (see Section 1.3.1)
The flow rate is set to 100 µL/min to minimize mass transport effects.
Several buffer samples are injected at the start of each experiment as a baseline stabilization step. At the beginning of each sample injection cycle, experimental buffer flows over the sensor chip surface for 5 minutes to give a very stable baseline that is essential for accurate binding analysis.
Inject 250 µL of each concentration of PU.1 solution and set 600 seconds as the dissociation time (see Note 10). Protein samples are injected from low to high concentration to eliminate artifacts in the data from adsorption carry-over on the instrument flow system.
Inject 1 min regeneration solution (1 M NaCl) in the end of the dissociation phase, followed by three 1-min experimental buffer flow to provide a stable baseline for the next sample cycle.
After the data are collected, open the experimental sensorgrams in the Biacore evaluation software for data processing (see Note 11). Zero the baselines on the response (y-) and the time (x-) axes by choosing a small region of a few seconds for averaging prior to sample injection on both the sample and control flow cells.
Subtract the response of the reference flow cell (FC1) from the reaction flow cell (i.e. FC2-1, FC3-1, and FC4-1). This can remove the effects from any bulk shift contribution on the changes of RUs.
Subtract a buffer injection, or an average of several buffer injections from a series of ligand injections at different concentrations on the same reaction flow cell. The reference correction and the buffer correction are known as double subtraction and can eliminate specific baseline irregularities (8, 37). At this stage, the data are ready for analysis as discussed below.
Figure 3.
Sensorgrams of PU.1 binding with target DNA sequence with 300 mM NaCl at flow rate of (A) 10 µL/min and (B) 50 µL/min. The PU.1 concentrations from bottom to top are 1, 2, 3, 5 and 10 nM in both plots.
3.6 Data Analysis
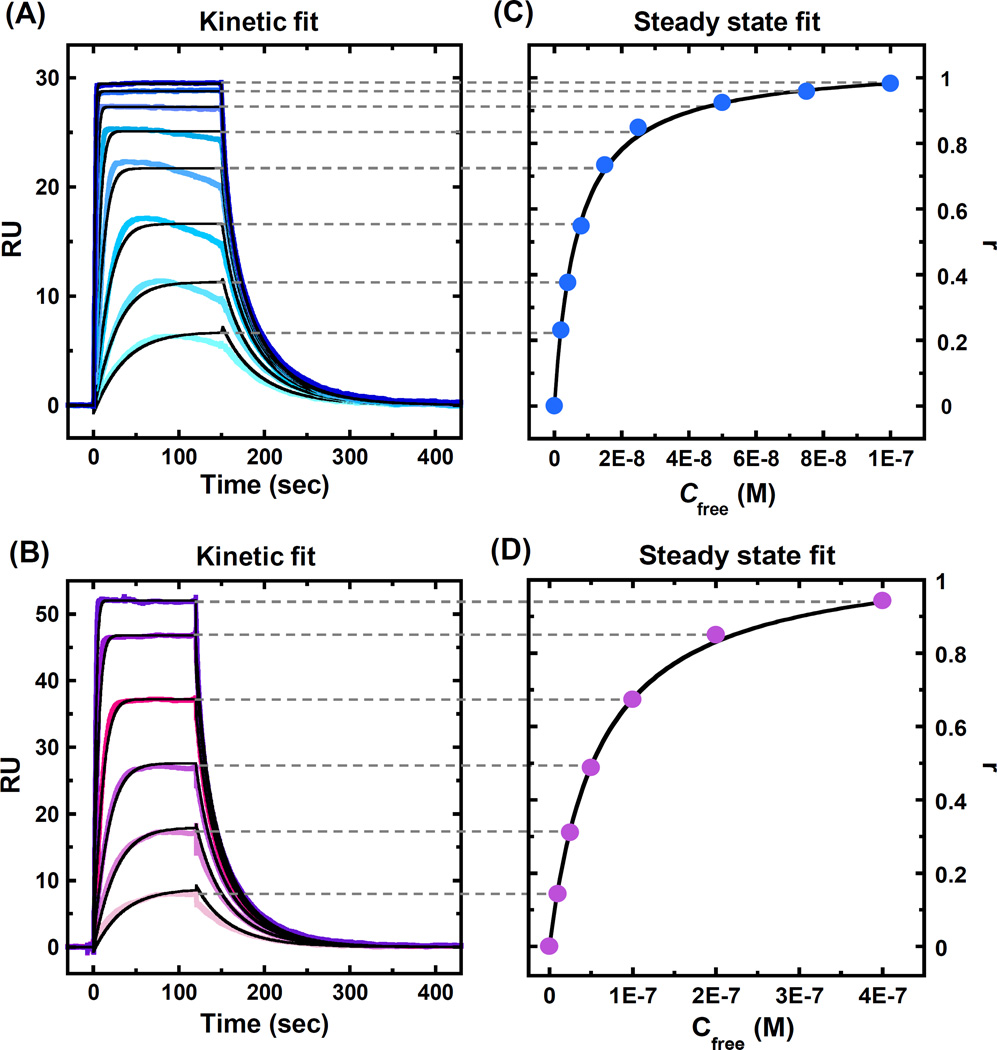
After the sensorgrams are processed as described above, kinetic and/or steady state analysis is performed. Both kinetic and steady state fitting can be done in Biacore evaluation software or with the Scrubber-2 package written by Myszka and collaborators (http://www.biologic.com.au). As can be seen in Figures 4A–B, PU.1 binding reaches a steady state plateau in the injection period so that both kinetic and steady state analysis can be used. In this case the binding rate is not limited by mass transfer and the association and dissociation rate constants can be determined. The average of the data over a selected time period in the steady state region of each sensorgram can be obtained, converted to r (r = RUobs/RUmax, Equation 5) and plotted as a function of protein concentration in the flow solution, as shown in Figures 4C–D. (see Note 12)
Equilibrium constants can be obtained by fitting the sensorgrams to the equivalent site model in Equation 5.
The RU on the surface is directly indicating the amount of PU.1 bound. Based on the RU at saturation we can determine that PU.1 forms a 1:1 complex with target DNA as expected.
A global kinetic fit in a 1:1 model with mass transport is applied for the sensorgrams in Figures 4A–B to determine the binding kinetics and affinity, and the results are listed in Table 3. With 400 mM NaCl, PU.1-DNA interaction has a very fast association [ka= (3.1 ± 0.1) × 107 M−1 s−1] and an overall binding affinity of 7.0 nM. In the presence of non-specific DNA, the dissociation of PU.1 is barely affected, while the association rate is decreased around 10-fold resulting in a 10-time weaker binding affinity (KD= 73 ± 1 nM) compared to PU.1 binding without non-specific DNA. The steady state plateau is obtained for every injection in Figures 4A–B, which allows the steady state fits to be performed (Figures 4C–D). The binding affinity values are in excellent agreement to the results determined by kinetic rate constants (KD= kd / ka, Table 3). This suggests that the mass transfer effect is not significant and does not dominate the kinetics evaluation in these experiments. (see Note 13 and Note 14)
Comments on more complex binding models can be input through Biacore or other evaluation software.
Figure 4.
Sensorgrams (color) and global kinetic fitting (black overlays) in a 1:1 binding model for PU.1 binding to the target DNA sequence with (A) 400 mM NaCl and (B) 400 mM NaCl in the presence of 300 µM salmon sperm DNA as non-specific DNA. The PU.1 concentrations from bottom to top are (A) 2, 4, 8, 15, 25, 50, 75 and 100 nM, and (B) 10, 25, 50, 100, 200 and 400 nM. (C) and (D) Steady state fitting of the sensorgrams in Figures 4A and 4B, respectively. RU values from the steady state region were converted to r (r = RUobs/RUmax) and are plotted as a function of unbound protein concentration with equilibrium in the complex.
Table 3.
Binding affinities and kinetics for PU.1 with and without non-specific (NS) DNA at 400 mM NaCl*.
|
Ka (× 106 M−1 S−1) |
kd (S−1) |
KD (nM) | ||
|---|---|---|---|---|
| Kinetic fit | Steady-state fit | |||
| No NS DNA | 31 ± 1 | 0.22 ± 0.01 | 7.0 ± 0.2 | 6.8 ± 0.3 |
| 300 µM NS DNA | 2.4± 0.1 | 0.18± 0.01 | 73± 1 | 63 ± 2 |
Errors listed in this table are standard errors for the fit of 1:1 binding model.
Acknowledgments
We gratefully thank the NIH (AI064200) for the support for biosensor-SPR studies on DNA complexes, and the Georgia Research Alliance for funding of the Biacore instruments. We thank Carol Wilson for manuscript proofreading.
Footnotes
Maintenance chips are available from GE Healthcare Inc. “Desorb” is a Biacore software command that instructs the instrument to remove adsorbed proteins from the flow system. A detailed list of commands and operations are shown in Table 1. Make sure that the analysis and sample compartment temperatures are not below 20 °C, since SDS in Desorb solution 1 may precipitate at low temperature. For biological samples such as protein, the Sanitize method should also be used after Desorb to insure that there is no protein left for microorganisms to grow in the liquid injection and flow system.
After running the regular Desorb, if the baseline is still not stable with ±1.0 RU/min, an additional super clean method may be used. First, run Desorb using 1% (v/v) acetic acid in place of desorb solutions 1 and 2 with deionized water at 50 °C as running buffer, followed by one Prime to wash out the residual acetic acid. Then, run Desorb using 0.2 M sodium bicarbonate followed by one Prime to wash out the sodium bicarbonate residuals. Last, run Desorb using 6 M guanidine HCl in place of SDS (solution 1) and 10 mM HCl for glycine (solution 2). Prime the instrument a few times to thoroughly clean all residuals and flow buffer to stabilize the instrument.
The choice of sensor chip depends on the nature and demands on the application. For general purposes, a Biacore CM5 sensor chip, which carries a matrix of carboxymethylated (CM) dextran covalently attached to the gold surface, can be used. It has a high surface capacity for immobilizing a wide range of ligands from protein to nucleic acids and carbohydrates. For protein-DNA interaction investigation, the Biacore CM4 sensor chip is another good choice because it is similar to sensor chip CM5 but has a lower degree of carboxymethylation (~ 30% of that of CM5 chip) and charge that helps to reduce non-specific binding of the highly positively charged molecules, such as protein, to the surface. Sensor chip SA has a surface carrying a dextran matrix to which streptavidin has been covalently attached. Streptavidin has a very high binding affinity with biotin (KD ≈ 10−15 M), so that the surface provides a high capture of biotinylated ligands. The SA chip is particularly suited to work with nucleic acids since the 5’ or 3’ terminal biotinylation of nucleic acid is a well-established procedure.
The selection of experimental buffer depends on the nature of the target protein and DNA sequence. Salt concentration can be adjusted based on the experimental requirement. With the increasing of salt concentration, the binding affinity for protein-DNA interaction typically decreases for positively charged protein.
Regeneration conditions must be harsh enough to break the complex and remove the bound reagent but mild enough to keep the DNA strand intact. It is highly recommended to start with the mildest conditions and short surface contact times since regeneration solutions can cause an undesired effect on DNA or immobilized matrix.
The same procedure can be applied to immobilize other protein or DNA that is labeled with a terminal amino group. Other molecules with a free amino group can also be captured by this method.
The Biacore T200 is good for experiments with less than 10 RU, while the Biacore 3000 and X100 have around 1/10th sensitivity of Biacore T200. Another way to calculate the amount of DNA immobilized is by using a standard ligand with known binding stoichiometry and binding affinity such as DNA minor groove binder netropsin to titrate the amount of DNA on the surface (31). This is especially useful as sensor chips are used numerous times and begin to lose immobilized DNA.
To estimate an unknown KA, it is necessary to conduct a preliminary experiment with several samples in concentrations spreading over a broad range. Then a more focused set of concentrations covering the binding range is run to determine the KA accurately.
As mentioned above (Note 4), the salt concentration in buffer can be adjusted for experimental requirements. Here, the strong binding of PU.1 with its native high-affinity DNA needs at least 400 mM NaCl to be evaluated by SPR without mass transport effects. By conducting the experiments at different salt concentrations, a linear log KD vs. log [Na+] plot is obtained that can be extrapolated to less salt concentrations (27).
A sufficient association phase with a plateau region is needed for steady state analysis. For the most accurate fitting of the dissociation phase it is highly recommended to allow sufficient time for the protein to dissociate at least 50% from the complex.
Other software programs such as Scrubber 2 and CLAMP are available for processing Biacore data. The results can also be exported and presented in graphing software such as KaleidaGraph. For the Biacore T200 user, data processing can be automatically performed with the Biacore T200 evaluation software, which is sufficient for most routine analyses and much more convenient for new users.
In some instances at low concentrations where the response does not reach the steady state, the equilibrium responses can be obtained from kinetic fits of the sensorgrams using the known RUmax from the higher concentration sensorgrams. This extrapolation method works well with sensorgrams where the observed response is at least 50% of the equilibrium RU.
The different RUmax values at PU.1 binding saturation in Figures 4A–B are due to the different amounts of immobilized DNA left on the experimental sensor chips. As mentioned in Note 7, through appropriate calibration of the amount of immobilized DNA on the flow cell, binding kinetics and affinity analysis can be well performed.
Reference
- 1.Homola J. Surface plasmon resonance sensors for detection of chemical and biological species. Chem Rev. 2008;108:462–493. doi: 10.1021/cr068107d. [DOI] [PubMed] [Google Scholar]
- 2.Rich RL, Myszka DG. Advances in surface plasmon resonance biosensor analysis. Curr Opin Biotechnol. 2000;11:54–61. doi: 10.1016/s0958-1669(99)00054-3. [DOI] [PubMed] [Google Scholar]
- 3.Wilson WD. Analyzing biomolecular interactions. Science. 2002;295:2103–2105. doi: 10.1126/science.295.5562.2103. [DOI] [PubMed] [Google Scholar]
- 4.Piliarik M, Vaisocherova H, Homola J. Surface Plasmon Resonance Biosensing. In: Rasooly A, Herold KE, editors. Methods in Molecular Biology. Vol. 503. New York City: Humana Press; 2009. pp. 65–88. [DOI] [PubMed] [Google Scholar]
- 5.Davis TM, Wilson WD. Surface plasmon resonance biosensor analysis of RNA-small molecule interactions. Methods Enzymol. 2001;340:22–51. doi: 10.1016/s0076-6879(01)40416-2. [DOI] [PubMed] [Google Scholar]
- 6.Papalia GA, Giannetti AM, Arora N, Myszka DG. Thermodynamic characterization of pyrazole and azaindole derivatives binding to p38 mitogen-activated protein kinase using Biacore T100 technology and van't Hoff analysis. Anal Biochem. 2008;383:255–264. doi: 10.1016/j.ab.2008.08.010. [DOI] [PubMed] [Google Scholar]
- 7.Liu Y, Wilson WD. Quantitative analysis of small molecule-nucleic acid interactions with a biosensor surface and surface plasmon resonance detection. In: Fox KR, editor. Methods in Molecular Biology. Vol. 613. New York City: Humana Press; 2010. pp. 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nanjunda R, Munde M, Liu Y, Wilson WD. Real-time monitoring of nucleic acid interactions with biosensor-surface plasmon resonance. In: Wanunu M, Tor Y, editors. Methods for studying nucleic acid/drug interactions. Boca Raton: CRC Press; 2011. pp. 91–122. [Google Scholar]
- 9.He X, Coombs D, Myszka DG, Goldstein B. A theoretical and experimental study of competition between solution and surface receptors for ligand in a Biacore flow cell. Bull Math Biol. 2006;68:1125–1150. doi: 10.1007/s11538-006-9093-9. [DOI] [PubMed] [Google Scholar]
- 10.Bondeson K, Frostellkarlsson A, Fagerstam L, Magnusson G. Lactose repressor-operator DNA interactions: kinetic analysis by a surface plasmon resonance biosensor. Anal Biochem. 1993;214:245–251. doi: 10.1006/abio.1993.1484. [DOI] [PubMed] [Google Scholar]
- 11.Goeddel DV, Yansura DG, Caruthers MH. Binding of synthetic lactose operator DNAs to lactose represessors. Proc Natl Acad Sci USA. 1977;74:3292–3296. doi: 10.1073/pnas.74.8.3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davis TM, Wilson WD. Determination of the refractive index increments of small molecules for correction of surface plasmon resonance data. Anal Biochem. 2000;284:348–353. doi: 10.1006/abio.2000.4726. [DOI] [PubMed] [Google Scholar]
- 13.Degnan BM, Degnan SM, Naganuma T, Morse DE. The ETS multigene family is conserved throughout the metazoa. Nucleic Acids Res. 1993;21:3479–3484. doi: 10.1093/nar/21.15.3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oikawa T, Yamada T. Molecular biology of the Ets family of transcription factors. Gene. 2003;303:11–34. doi: 10.1016/s0378-1119(02)01156-3. [DOI] [PubMed] [Google Scholar]
- 15.Sharrocks AD. The ETS-domain transcription factor family. Nat Rev Mol Cell Bio. 2001;2:827–837. doi: 10.1038/35099076. [DOI] [PubMed] [Google Scholar]
- 16.Sementchenko VI, Watson DK. Ets target genes: past, present and future. Oncogene. 2000;19:6533–6548. doi: 10.1038/sj.onc.1204034. [DOI] [PubMed] [Google Scholar]
- 17.Hsu T, Trojanowska M, Watson DK. Ets proteins in biological control and cancer. J Cell Biochem. 2004;91:896–903. doi: 10.1002/jcb.20012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gilliland DG. The diverse role of the ETS family of transcription factors in cancer. Clin Cancer Res. 2001;7:451–453. [PubMed] [Google Scholar]
- 19.Oikawa T. ETS transcription factors: possible targets for cancer therapy. Cancer Sci. 2004;95:626–633. doi: 10.1111/j.1349-7006.2004.tb03320.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Galang CK, Muller WJ, Foos G, Oshima RG, Hauser CA. Changes in the expression of many Ets family transcription factors and of potential target genes in normal mammary tissue and tumors. J Biol Chem. 2004;279:11281–11292. doi: 10.1074/jbc.M311887200. [DOI] [PubMed] [Google Scholar]
- 21.Wei G-H, Badis G, Berger MF, Kivioja T, Palin K, Enge M, et al. Genome-wide analysis of ETS-family DNA-binding in vitro and in vivo. EMBO J. 2010;29:2147–2160. doi: 10.1038/emboj.2010.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hollenhorst PC, McIntosh LP, Graves BJ. Genomic and biochemical insights into the specificity of ETS transcription factors. In: Kornberg RD, Raetz CRH, Rothman JE, Thorner JW, editors. Annu Rev Biochem. Vol. 80. 2011. pp. 437–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.DeKoter RP, Singh H. Regulation of B lymphocyte and macrophage development by graded expression of PU.1. Science. 2000;288:1439–1441. doi: 10.1126/science.288.5470.1439. [DOI] [PubMed] [Google Scholar]
- 24.Ross IL, Yue X, Ostrowski MC, Hume DA. Interaction between PU.1 and another Ets family transcription factor promotes macrophage-specific basal transcription initiation. J Biol Chem. 1998;273:6662–6669. doi: 10.1074/jbc.273.12.6662. [DOI] [PubMed] [Google Scholar]
- 25.Kopp JL, Wilder PJ, Desler M, Kim JH, Hou J, Nowling T, et al. Unique and selective effects of five Ets family members, Elf3, Ets1, Ets2, PEA3, and PU.1, on the promoter of the type II transforming growth factor-beta receptor gene. J Biol Chem. 2004;279:19407–19420. doi: 10.1074/jbc.M314115200. [DOI] [PubMed] [Google Scholar]
- 26.Pham TH, Minderjahn J, Schmidl C, Hoffmeister H, Schmidhofer S, Chen W, et al. Mechanisms of in vivo binding site selection of the hematopoietic master transcription factor PU.1. Nucleic Acids Res. 2013;41:6391–6402. doi: 10.1093/nar/gkt355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Munde M, Poon GM, Wilson WD. Probing the electrostatics and pharmacological modulation of sequence-specific binding by the DNA-binding domain of the ETS family transcription factor PU.1: a binding affinity and kinetics investigation. J Mol Biol. 2013;425:1655–1669. doi: 10.1016/j.jmb.2013.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poon GM. Sequence discrimination by DNA-binding domain of ETS family transcription factor PU.1 is linked to specific hydration of protein-DNA interface. J Biol Chem. 2012;287:18297–18307. doi: 10.1074/jbc.M112.342345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poon GM. DNA binding regulates the self-association of the ETS domain of PU.1 in a sequence-dependent manner. Biochemistry. 2012;51:4096–4107. doi: 10.1021/bi300331v. [DOI] [PubMed] [Google Scholar]
- 30.Myszka DG. Kinetic, equilibrium, and thermodynamic analysis of macromolecular interactions with BIACORE. Methods Enzymol. 2000;323:325–340. doi: 10.1016/s0076-6879(00)23372-7. [DOI] [PubMed] [Google Scholar]
- 31.Nguyen B, Tanious FA, Wilson WD. Biosensor-surface plasmon resonance: quantitative analysis of small molecule-nucleic acid interactions. Methods. 2007;42:150–161. doi: 10.1016/j.ymeth.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 32.Tanious FA, Nguyen B, Wilson WD. Biosensor-surface plasmon resonance methods for quantitative analysis of biomolecular interactions. In: Correia JJ, Detrich HW, editors. Methods Cell Biol. Vol. 84. 2008. pp. 53–77. [DOI] [PubMed] [Google Scholar]
- 33.Karlsson R. Affinity analysis of non-steady-state data obtained under mass transport limited conditions using BIAcore technology. J Mol Recognit. 1999;12:285–292. doi: 10.1002/(SICI)1099-1352(199909/10)12:5<285::AID-JMR469>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 34.Eisenbeis CF, Singh H, Storb U. PU-1 is a component of a multiprotein complex which binds an essential site in the murine immunoglobulin lambda-2-4 enhancer. Mol Cell Biol. 1993;13:6452–6461. doi: 10.1128/mcb.13.10.6452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Poon GM, Gross P, Macgregor RB. The sequence-specific association of the ETS domain of murine PU.1 with DNA exhibits unusual energetics. Biochemistry. 2002;41:2361–2371. doi: 10.1021/bi015572q. [DOI] [PubMed] [Google Scholar]
- 36.Poon GM, Macgregor RB. Base coupling in sequence-specific site recognition by the ETS domain of murine PU.1. J Mol Biol. 2003;328:805–819. doi: 10.1016/s0022-2836(03)00362-0. [DOI] [PubMed] [Google Scholar]
- 37.Myszyka DG. Improving biosensor analysis. J Mol Recognit. 1999;12:279–284. doi: 10.1002/(SICI)1099-1352(199909/10)12:5<279::AID-JMR473>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]