Abstract
Introduction:
Porphyromonas gingivalis is associated with periodontitis and exhibit a wide array of virulence factors, including fimbriae which is encoded by the FimA gene representing six known genotypes.
Objetive:
To identify FimA genotypes of P. gingivalis in subjects from Cali-Colombia, including the co-infection with Aggregatibacter actinomycetemcomitans, Treponema denticola, and Tannerella forsythia.
Methods:
Subgingival samples were collected from 151 people exhibiting diverse periodontal condition. The occurrence of P. gingivalis, FimA genotypes and other bacteria was determined by PCR.
Results:
P. gingivalis was positive in 85 patients. Genotype FimA II was more prevalent without reach significant differences among study groups (54.3%), FimA IV was also prevalent in gingivitis (13.0%). A high correlation (p= 0.000) was found among P. gingivalis, T. denticola, and T. forsythia co-infection. The FimA II genotype correlated with concomitant detection of T. denticola and T. forsythia.
Conclusions:
Porphyromonas gingivalis was high even in the healthy group at the study population. A trend toward a greater frequency of FimA II genotype in patients with moderate and severe periodontitis was determined. The FimA II genotype was also associated with increased pocket depth, greater loss of attachment level, and patients co-infected with T. denticola and T. forsythia.
Keywords: Porphyromonas gingivalis, bacterial fimbriae, polymerase chain reaction, periodontitis, gingivitis
Abstract
Introducción:
Porphyromonas gingivalis es una bacteria asociada con la periodontitis. Expresa una amplia gama de factores de virulencia, incluyendo las fimbrias, las cuales están codificadas por el gen FimA que representa seis genotipos conocidos.
Objetivo:
Identificar los genotipos de FimA de P. gingivalis en pacientes de Cali - Colombia, incluyendo la co -infección con Aggregatibacter actinomycetemcomitans, Treponema denticola y Tannerella forsythia .
Métodos:
Se obtuvieron muestras subgingivales de 151 individuos con diferentes diagnósticos periodontales. La ocurrencia de P. gingivalis, los genotipos de FimA y otras bacterias se determinó por PCR.
Resultados:
Porphyromonas gingivalis fue positiva en 85 pacientes. El genotipo FimA II fue más prevalente, pero no hubo diferencias significativas entre los grupos de estudio (54.3%) , FimA IV fue el más frecuente en la gingivitis (13.0%). Una alta correlación (p= 0.000 ) se encontró entre P. gingivalis , T. denticola y T. forsythia. El genotipo FimA II estuvo correlacionado con la detección de T. denticola y T. forsythia .
Conclusiones:
Porphyromonas gingivalis tuvo una alta frecuencia incluso en el grupo de individuos sanos. Se encontró una tendencia hacia una mayor frecuencia de FimA II en pacientes con periodontitis moderada y severa. El genotipo FimA II también se asoció con una mayor profundidad de la bolsa, una mayor pérdida de nivel de inserción, y con los pacientes en los que se detectó co - infección con T. denticola y T. forsythia.
Introduction
Periodontitis is an inflammatory disease that affects supporting tissue around teeth 1. One of the triggering factors of the disease is the persistence of the biofilm formed on the dental surfaces and the gingival margin 2.
In Colombia, according to the Third National Study on Oral Health in 1999, periodontal insertion loss is 50.2% and this generates increasing awareness to the health system because its high prevalence.
Porphyromonas gingivalis is associated to the onset and progression of chronic and aggressive periodontitis 3-5, frequently present in patients with periodontal disease and is detected even in healthy subjects 6. Porphyromona gingivalis carry multiple virulence factors 4,7-8 and fimbriae is considered a key factor 7,9 composed by subunits of fimbrillin 10, located on the cell surface, which gives the bacteria the capacity to adhere to the supporting periodontal tissue, the acquired pellicle and to other bacterial species during biofilm formation and consolidation. The gene encoding the fimbriae is denominated FimA and six genotypes are known (I, Ib, II, III, IV, V) based on their nucleotide sequences 10.
In periodontitis subjects FimA II and FimA IV genotypes have been the more frequently identified in contrast to healthy subjects in whom the FimA I genotype is the most prevalent 11-14.
This cross-sectional descriptive study seeks to determine the prevalence of FimA genotypes from P. gingivalis isolated from subgingival samples in health, gingivitis and chronic periodontitis subjects and determine its association to other periodontopathic microorganisms like Aggregatibacter actinomycetemcomitans, Treponema denticola, and Tannerella forsythia.
Materials and Methods
Subgingival samples were taken from 151 subjects, selected by convenience. Sixty seven men and 84 women with age range 22-84 yrs old, (43.4 ±10.6 yrs), coming from 3 clinics: The Dental School at Valle University, The Dental Clinics of Santiago de Cali University, and one private dental clinic in Cali-Colombia. Subjects had different periodontal diagnoses (25 healthy, 77 gingivitis and 49 chronic periodontitis). The patients authorized participation. The study was approved an IRB committee at the Faculty of Health -Valle University-, protocol No. 012-010. Selection was made of subjects who had more than 15 teeth, who had not taken antibiotics, corticoids, and/or AINES at least three months prior to be included in the study, without systemic diseases and who had not received periodontal treatment and/or professional dental cleaning at least six months prior to participating in the study.
Periodontal examination and sampling
A full-mouth periodontal probing chart was performed by two calibrated periodontist accordingly to the diagnostic criteria of the AAP 1999. The measurements of probing pocket depth and clinical attachment loss was recorded at six points around each tooth. The variables considered for clinical diagnosis were probing depth, clinical attachment loss, the rate of bleeding and the number of teeth. Healthy subjects were considered had not signs of inflammation, probing depth sites equal or less than 3 mm, no bleeding on probing without clinical attachment loss. Gingivitis patients presented clinical signs of gingival inflammation and edema, bleeding on probing without clinical attachment loss. Periodontitis patients presented swelling, bleeding on probing and probing depth of 4 mm in at least one tooth (localized) or more than 50% of the teeth (generalized). After removal of the supragingival plaque with sterile gauzes, two sterile paper tips were introduced in the deepest pockets (>4 mm) in patients with periodontitis and in the mesial vestibular surface of the first four molars in healthy subjects and patients with gingivitis, leaving them in the sulcus for one minute. Thereafter, the paper tips were deposited in Eppendorf tubes and cryopreserved at -20° C until processing 14,17
DNA extraction and PCR
Bacterial DNA extraction from the paper tips was performed through adsorption to silica particles, according to Boom's 1989 protocol, and the P. gingivalis genotyping procedure was carried out via the Polymerase Chain Reaction (PCR) technique, using published reports 16-18. First, DNA extraction was confirmed by using specific generic primers for 16S rRNA; then, the presence of DNA from P. gingivalis was confirmed with specific primers 16-18 and, thereafter PCR was performed with the specific primers for each FimA I, II, III, IV, V, and Ib genotype, as widely reported in literature 11,14-17.
DNA from P. gingivalis ATCC33227 (FimA I), W83 (FimA IV), ATCC33279 (FimA Ib) strains was used as positive controls 14. For positive controls of FimA II and FimA III genotypes, two clinical isolates were used numbered 486 and 723, which were typified and donated by the Microbiology laboratory at Bosque University 13.
The PCR tests to study the FimA genotype were conducted in a thermocycler (AXYGEN) with the following amplification program: initial denaturing at 95° C for 5 min, followed by 36 cycles at 94° C for 30 s, 60° C for 30 s, and 72° C for 30 s, and a final extension at 72° C for 7 min. Besides the FimA genotypes from P. gingivalis, the same samples were also used to identify the presence of other periodontopathic bacteria, like A. actinomycetemcomitans, T. denticola and T. forsythia through a single round PCR technique using specific primers for the 16S rRNA gene from each of the three microorganisms before mentioned. The PCR products were separated electrophoretically in agarose gels and the DNA bands were stained with SYBR safe and visualized via UV light in a trans-illuminator (Invitrogen). The results were documented through photographic registration. Identification of FimA genotypes was carried out according to the molecular size of the amplification bands obtained, compared to the respective positive control 14.
Standardization and analytic sensitivity for the PCR for P. gingivalis Fim A gene detection
To determine the limit of detection of the FimA gene from P. gingivalis by PCR technique,
a serial and tittered dilutions of known bacterial genotypes coming from clinical isolates of FimA II and FimA III, or control strains ATCC33279 (FimA Ib), W83 (FimA IV), and ATCC 33227 (FimA I) were done. Likewise, PCR sensitivity for FimA genotypes in clinical samples was determined by experimental inoculation of GCFs negative for P. gingivalis using diverse dilutions of prototype strains in spike samples.
PCR specificity for the P. gingivalis FimA gene
Primers and PCR specificity of FimA genotypes (I, II, II, IV, and Ib) was probed against other periodontopathic bacteria like A. actinomycetemcomitans (D11-s1), T. denticola (ATCC. 43056), and T. forsythia (ATCC 43037).
Statistical analyses
Pearson´s Chi squared statistical test was used to compare the frequencies of the FimA genotypes. To analyze the frequency of other periodontopathogens, Fisher's exact test was applied taking in account that sample size was below 20. Clinical attachment loss or CAL was analyzed by Student's T test and quantitative variables like probing pocket depth - PPD, and Bleeding on Probing - BOP, number of teeth, and age were analyzed by using Wilcoxon's test and Mann-Whitney test based on previous testing of Normal distribution.
Results
PCR sensitivity and specificity for the P. gingivalis FimA gene
The detection limit of the FimA gene was determined to detect up to 50 bacterial cells in the dilutions carried out from a pure culture from each P. gingivalis strain available. However, the detection limit increases up to 500 cells when the bacterial dilutions were inoculated in GCF possibly by the action of PCR inhibitors or DNAases. High specificity was found between the primers used and the representative strains of each of the FimA genotypes studied.
Frequency of P. gingivalis
A total of 85 subjects were positive for P. gingivalis (56.3%) being the bacteria more prevalent in women 57.6% of that in men 42.3%. Frequency of P. gingivalis by diagnoses was 52.0% for healthy subjects, 59.7% for gingivitis patients and 53.1% for chronic periodontitis (Table 1).
Table 1. Prevalence and Distribution of FimA genotypes of P. gingivalis according to periodontal diagnosis (AAP 1999).
| Genotypes | Periodontal Diagnosis | ||||||
|---|---|---|---|---|---|---|---|
| Healthy | Gingivitis | Periodontitis | p value* | ||||
| **n | (%) | n | (%) | n | (%) | >0.050 | |
| P. gingivalis | 13 | 52.0 | 46 | 59.7 | 26 | 53.1 | 0.681 |
| FimA I | 2 | 15.4 | 12 | 26.1 | 3 | 11.5 | 0.301 |
| FimA II | 6 | 46.2 | 25 | 54.3 | 15 | 57.7 | 0.792 |
| FimA III | 2 | 15.4 | 2 | 4.3 | 2 | 7.7 | 0.386 |
| FimA IV | 1 | 7.7 | 6 | 13.0 | 0 | 0.0 | 0.154 |
| FimA Ib | 3 | 12.0 | 15 | 19.5 | 2 | 4.1 | 0.057 |
| FimA V | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | |
| Not tipificable | 5 | 38.0 | 10 | 21.7 | 8 | 30.7 | 0.428 |
| fimA I, II, Ib | 1 | 7.7 | 8 | 17.4 | 0 | 0.0 | 0.066 |
| fimA III, Iv, Ib | 1 | 7.7 | 0 | 0.0 | 0 | 0.0 | 0.079 |
| fim A I, III, Ib | 1 | 7.7 | 0 | 0.0 | 0 | 0.0 | 0.079 |
| fimA II, IV | 0 | 0.0 | 2 | 4.3 | 0 | 0.0 | 0.377 |
| fimA I, II | 0 | 0.0 | 1 | 2.1 | 2 | 4.3 | 0.406 |
| fim A III, Ib | 0 | 0.0 | 0 | 0.0 | 1 | 2.1 | 0.350 |
| fim A III, IV | 0 | 0.0 | 1 | 2.1 | 0 | 0.0 | 0.616 |
| fim A I, Ib | 0 | 0.0 | 4 | 8.7 | 1 | 3.8 | 0.375 |
* Pearson´s Chi squared statistical test.
**The values of n not add up to total shown since the same subject can express various genotypes time.
Frequency and distribution of P. gingivalis FimA genotypes
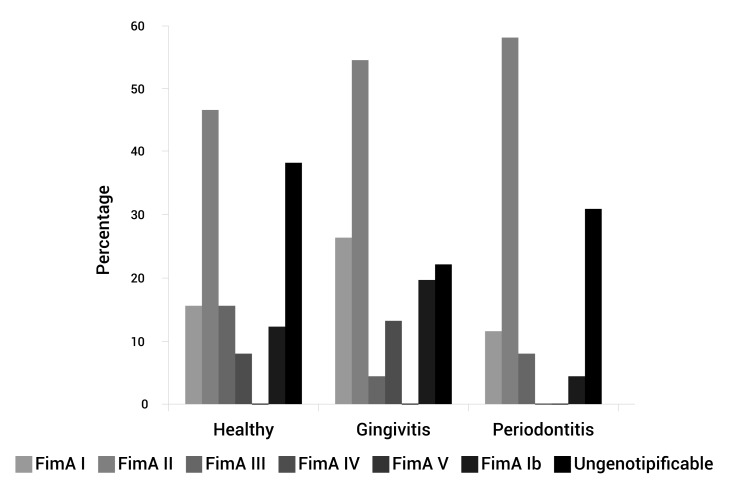
The most frequent genotype was FimA II (54.3%), followed by FimA Ib (23.5%), FimA I (20%), FimA IV (8.2%), and FimA III (7.1%). No positive samples were found for the FimA V genotype. To the 85 patients positive for P. gingivalis, 23 (27%) were negative for the six genotypes studied hence, assigned as the unknown genotype or non/tipificable (Table 1). In the three groups: healthy, gingivitis, and periodontitis patients, the percentages of fimA II genotypes were 46.2%, 54.3%, and 57.7%, respectively (Table 1, Fig. 1).
Figure 1. Genotypes FimA P. gingivalis frequency according periodontal diagnosis.
The FimA II genotype was more frequent as increasing the severity of periodontitis (AAP 1999), although no statistically significant difference was reached (p= 0.813). We also analyzed the prevalence and distribution of the different FimA genotypes according to the clinical variables analyzed and it was found that FimA II and FimA III genotypes were linked to a high PD per site. The FimA II was related to a higher CAL and tooth loss. FimA III genotype was related to higher BOP (Table 2).
Table 2. Relation between FimA genotypes and clinical variables.
| Genotypes | Clinical variables* | |||
|---|---|---|---|---|
| PD | CAL | BOP | NoTe | |
| FimA I | 4.3 | 4.2 | 55.8 | 25.1 |
| FimA II | 4.6 | 4.5 | 53.4 | 23.2 |
| FimA III | 4.6 | 4.0 | 63.5 | 24.0 |
| FimA IV | 4.4 | 4.3 | 61.8 | 23.4 |
| FimA Ib | 4.2 | 4.0 | 56.0 | 24.0 |
*All data are mean
PD: probing pocket depth site (mm)
CAL: clinical attachment loss site (mm)
BOP Bleeding on Probing (%)
NoTe: number of teeth
Eleven patients were positive for three FimA genotypes and 12 patients harbored two fimA genotypes, meaning that coinfection is a not rare event (Table 1).
Prevalence of other periodontopathogens
It was a high prevalence of T. forsythia (65.3%) in the periodontitis patients while its prevalence in healthy patients and gingivitis patients was 56.0% and 68.8% respectively. The prevalence of T. denticola in healthy, gingivitis and periodontitis patients was 40.0%, 58.4% and 40.8% respectively and for A. actinomycetencomitans was 8.0% in healthy patients, 20.8% in gingivitis and 14.2% in periodontitis (Table 3).
Table 3. Positive co-infection with other periodontopathogens and FimA genotypes.
| Species* | ||||
|---|---|---|---|---|
| A. actinomyc** | T. denticola | T. forsythia | ||
| Periodontal diagnosis | Healthy | 8.0 | 40.0 | 56.0 |
| Gingivitis | 20.8 | 58.4 | 68.8 | |
| Periodontitis | 14.2 | 40.8 | 65.3 | |
| P .gingivalis | 64.0 | 74.7 | 72.7 | |
| Genotypes FimA | ||||
| FimA I | 31.2 | 21.4 | 22.2 | |
| FimA II | 68.8 | 55.3 | 52.8 | |
| FimA III | 6.7 | 7.1 | 5.5 | |
| FimA IV | 6.7 | 8.9 | 6.94 | |
*Percentage
**A. actinomycetemcomitans
Co-infection of P. gingivalis and FimA genotypes with A. actinomycetemcomitans, T. denticola, and T. forsythia
A high correlation was observed between P. gingivalis, T. denticola, and T. forsythia, (p= 0.000). The co-infection of these microorganisms was observed in higher percentages for the FimA II genotype with prevalence in the samples positive for A. actinomycetemcomitans (68.7%), T. denticola (55.3%), and T. forsythia (52.8%), although this relationship was not statistically significant (Table 3).
Discussion
Frequency of P. gingivalis
The results of this study in the P. gingivalis frequency differ from those reported by Amano et al., 14,15, Guo et al., 19, Wu et al., 22, and, Zhao et al., 23, but are similar to those reported by Beikler 20 and Missailidis et al 16.
High P. gingivalis prevalence in healthy patients is common in the Colombian population, according to that reported by Botero et al., 24, Lafaurie et al., 25, and Mayorga et al., 26. This finding could be influenced by the ethnicity, low sanitation and poverty. Most of the patients in our study live on wages below $400- $500 US dollars per month. This hinders access to dental services, worsening periodontitis, causing delay or lack on dental treatment and favoring perhaps horizontal and vertical transmission of important periodontal pathogens.
Frequency and distribution of P. gingivalis FimA genotypes
In general, the most frequent genotype was FimA II, which was detected in 46 samples (54.1%).
Healthy patients revealed prevalence for FimA II followed by FimA I These results largely differ from studies published in literature, that reported lower frequencies of P. gingivalis in healthy patients 16,20,21, as well as low frequencies of the FimA II genotype and high frequencies of the FimA I, FimA III, and FimA V genotypes 14,16,22,23, considering these genotypes as low pathogens (Table 1, Fig. 1).
It is also worth considering the importance of conducting studies on genotype expression through, for example, Real Time-PCR and to determine the relationship between the phenotype and the genotype. Another important issue to bear in mind is that possibly the most virulent genotypes are present during initial stages of the periodontal disease, where the process of colonization and infection begins, as well as the stimulus of the immune and inflammatory response, which over time will generate damage and destruction of tissue.
The difference between the gingivitis and healthy groups may be associated to the bacterial load or the combination of virulence factor of diverse organisms. It might be either important to do a follow up of the populations to unveil if genotypes remain constant or varies over the time and after treatment. Further studies are required to prove these hypotheses.
Results in the gingivitis and periodontitis groups with respect to higher prevalence (FimA II genotype), specifically in generalized gingivitis moderate periodontitis, and severe periodontitis coincide with studies from Japan 15,16,18, China 19,23, Germany 20, Norway, the US, Canada, and The Netherlands 21, Brazil 11,16, Mexico 17, and, Colombia 13. The second most common genotype in these groups (FimA I), which in different studies has been associated to healthy individuals, revealed values of 26.1 and 11.5%, respectively, (Table 1) which is quite similar to that reported by Missailidis et al., 16 and Davila et al., 17, who reported this genotype as the second most prevalent in patients with gingivitis and periodontitis.
In our study there were no statistically significant differences in the distribution of genotypes for periodontal diagnosis. However a main reason could be the sample size. Moon et al., reported the finding that primers specific for fimA II could cross react with FimA Ib genotype leading to errors 27. The same research group reported the FimA II prevalence in Koreans reporting 44.5% in healthy subjects and 67.3% in periodontitis and concluding that results of previous studies should be reviewed as entailed to an overestimation of the relative risk for FimA II genotype. The authors suggest that healthy subjects carry genotype II have increased risk of developing periodontal disease 28. Interestingly, these results are very similar to our findings.
All the positive samples for P. gingivalis were negative for the FimA V genotype (Table 1). This result coincides with that reported by Missailidis et al., 16 and by Enersen et al., 29, but differs from the results from Japan 18, and from China 23 who reported prevalence of 16 to 29%; however, very low prevalence of this genotype has been reported (1.0-3.9%) in studies published in Germany 20 and Norway 21.
It is worth highlighting that in developing this study P. gingivalis FimA V control strain was not available and although PCR was performed using specific primers for this genotype, no sample turned out positive. The lack of the FimA V strain for the positive control is a limitation in our study.
Upon comparing the clinical variables, it was found that the FimA III genotype was most related to increasing PD, increasing CAL and bleeding on probing. Nonetheless, when comparing the relationship of the genotypes with the probing depth and insertion level loss at sampling sites (Table 2), it was found that the FimA II genotype was related to greater PD and greater CAL being similar to previous reports 11,14,29.
The presence of more than one genotype has been widely reported in literature 16,30. The current study presented 23 samples, mostly distributed in the gingivitis group with frequencies at 9.4% of the total for two genotypes and 9.4% for three genotypes (Table 1). This heterogeneous distribution of P. gingivalis genotypes on the population could be associated to ethnic differences.
Prevalence of periodontopathogens: A. actinomycetemcomitans, T. denticola, and T. forsythia
This study found high prevalence of the three microorganisms in the whole sample, being most prevalent in the gingivitis and periodontitis group as compared to healthy individuals, which agrees with other studies in Colombia done by Botero et al., 24 and Lafaurie et al 25.
Co-infection of P. gingivalis genotypes with T. denticola, T. forsythia, and A. actinomycetemcomitans
The current study found positive association among the three microorganisms from the red complex described Socransky et al., 31 and corroborated by others 23-25 (Table 3). The study also found a high percentage of positive samples for P. gingivalis and A. actinomycetemcomitans; these two microorganisms have been considered important etiological factors during the onset and progression of periodontal disease 32. Of the samples positive for P. gingivalis and the other three microorganisms studied, the majority were from the FimA II genotype, which could indicate a higher co-infection ratio of this genotype with T. denticola, T. forsythia, and A. actinomycetemcomitans. These results are similar to those obtained by Zhao et al., 23.
Conclusions
FimA II of P. gingivalis was detected in gingivitis, periodontitis and healthy subjects, and was also associated with periodontal disease severity. Porphyromonas gingivalis was associated with T. denticola and T. forsythia and FimA II genotype was frequent in patients that presented co-infection with these periodontal pathogens.
Acknowledgments:
This study was funded by the Research Award from the University of the Valle, 2012. Código 1695
References
- 1.Darveau R. Periodontitis: a polymicrobial disruption of host homeostasis. Nat Rev Microbiol. 2010;8:481–490. doi: 10.1038/nrmicro2337. [DOI] [PubMed] [Google Scholar]
- 2.Zijnge V, van Leeuwen MB, Degener JE, Abbas F, Thurnheer T, Gmür R, et al. Oral Biofilm Architecture on Natural Teeth. PLoS One. 2010;5(2):e9321. doi: 10.1371/journal.pone.0009321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dornelles F, Santos D, Van Dyke T, Calixto J, Batista E, Campos M. In vivo up-regulation of kinin B1 receptors after treatment with Porphyromonas gingivalis lipopolysaccharide in rat paw. J Pharmacol Exp Ther. 2009;33(3):756–763. doi: 10.1124/jpet.109.155762. [DOI] [PubMed] [Google Scholar]
- 4.Yilmaz O. The chronicles of Porphyromonas gingivalis: the microbium, the human oral epithelium and their interplay. Microbiology. 2008;154:2897–2903. doi: 10.1099/mic.0.2008/021220-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Holt SC, Kesavalu L, Walker S, Genco CA. Virulence factors of Porphyromonas gingivalis. Periodontol 2000. 1999;20:168–238. doi: 10.1111/j.1600-0757.1999.tb00162.x. [DOI] [PubMed] [Google Scholar]
- 6.Slots J, Ting M. Actinobacillus actinomycetemcomitans and Porphromonas gingivalis in human periodontal disease: occurrence and treatment. Periodontol 2000. 1999;20:82–121. doi: 10.1111/j.1600-0757.1999.tb00159.x. [DOI] [PubMed] [Google Scholar]
- 7.Brunner J, Scheres N, El Idrissi NB, Deng DM, Laine ML, van Winkelhoff AJ, et al. The capsule of Porphyromonas gingivalis reduces the immune response of human gingival fibroblasts. BMC Microbiol. 2010;10:5–5. doi: 10.1186/1471-2180-10-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stathopoulou PG, Galicia JC, Benakanakere MR, Garcia CA, Potempa J, Kinane DF. Porphyromonas gingivalis induce apoptosis in human gingival epithelial cells through a gingipain-dependent mechanism. BMC Microbiol. 2009;9:107–119. doi: 10.1186/1471-2180-9-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. M Wang, Liang S, Hosur KB, Domon H, Yoshimura F, Amano A, et al. Differential virulence and innate immune interactions of Type I and II fimbrial genotypes of Porphyromonas gingivalis. Oral Microbiol Immunol. 2009;24(6):478–484. doi: 10.1111/j.1399-302X.2009.00545.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Inaba H, Nakano K, Kato T, Nomura R, Kawai S, Kuboniwa M, et al. Heterogenic virulence and related factors among clinical isolated of Porphyromonas gingivalis with type II fimbriae. Oral Microbiol Immunol. 2008;23:29–35. doi: 10.1111/j.1399-302X.2007.00386.x. [DOI] [PubMed] [Google Scholar]
- 11.Teixeira SR, Mattarazo F, Feres M, Figueiredo LC, de Faveri M, Simionato MR, et al. Quantification of Porphyromonas gingivalis and FimA genotypes in smoker chronic periodontitis. J Clin Periodontol. 2009;36:482–487. doi: 10.1111/j.1600-051X.2009.01411.x. [DOI] [PubMed] [Google Scholar]
- 12.Nakano K, Inaba H, Nomura R, Nemoto H, Takeuchi H, Yoshioka H, et al. Distribution of Porphyromonas gingivalis FimA genotypes in cardiovascular specimens from Japanese patients. Oral Microbiol Immunol. 2008;23:170–172. doi: 10.1111/j.1399-302X.2007.00406.x. [DOI] [PubMed] [Google Scholar]
- 13.Pérez P, Lafaurie G, Gracieux P, Meuric V, Tamanai-Shacoori Z, Castellanos JE, et al. Distribution of Porphyromonas gingivalis FimA genotypes in isolates from subgingival plaque and blood sample during bacteremia. Biomédica. 2009;29:298–306. [PubMed] [Google Scholar]
- 14.Amano A, Nakagawa I, Kataoka K, Morisaki I, Amada S. Distribution of Porphyromonas gingivalis strains with FimA genotypes in periodontitis patients. J Clin Microbiol. 1999;37:1426–1430. doi: 10.1128/jcm.37.5.1426-1430.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Amano A, Kuboniwa I, Nakagawa, Akiyama S, Morisaki I, Hamada S. Prevalence of specific genotypes of Porphyromonas gingivalis FimA and periodontal health status. J Dent Res. 2000;79:1664–1668. doi: 10.1177/00220345000790090501. [DOI] [PubMed] [Google Scholar]
- 16.Missailidis CG, Umeda JE, Ota-Tsuzuki C, Anzai D, Mayer MP. Distribution of FimA genotypes of Porphyromonas gingivalis in subjects with various periodontal conditions. Oral Microbiol Immunol. 2004;19:224–229. doi: 10.1111/j.1399-302X.2004.00140.x. [DOI] [PubMed] [Google Scholar]
- 17.Davila-Pérez C, Amano A, Alpuche-Solis AG, Patiño-Marin N, Pontigo-Loyola AP, Hamada S, et al. Distribution of genotypes of Porphyromonas gingivalis in type 2 diabetic patients with periodontitis in Mexico. J Clin Periodontol. 2007;34:25–30. doi: 10.1111/j.1600-051X.2006.01011.x. [DOI] [PubMed] [Google Scholar]
- 18.Nakagawa I, Amano A, Kimura RK, Nakamura T, Kawabata S, Hamada S. Distribution and molecular characterization of Porphyromonas gingivalis carrying a new type of FimA gene. J Clin Microbiol. 2000;38:1909–1914. doi: 10.1128/jcm.38.5.1909-1914.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo YH, Wu YF, Liu TJ, Xiao XR, Zhou B, Zhou XP. The distribution of FimA genotype of Porphyromonas gingivalis in chronic periodontitis patients. Hua Xi Kou Qiang Yi Xue Za Zhi. 2005;33:99–102. [PubMed] [Google Scholar]
- 20.Beikler T, Peters U, Prajaneh S, Prior K, Ehmke B, Flemmig TF. Prevalence of Porphyromonas gingivalis FimA genotypes in caucasians. Eur J Oral Sci. 2003;111:390–394. doi: 10.1034/j.1600-0722.2003.00065.x. [DOI] [PubMed] [Google Scholar]
- 21.Van der Ploeg J, Giertsen E, Ludin B, Mörgeli C, Zinkernagel AS, Gmür R. Quantitative detection of Porphyromonas gingivalis FimA genotypes in dental plaque. FEMS Microbiol Lett. 2004;232:31–37. doi: 10.1016/S0378-1097(04)00064-3. [DOI] [PubMed] [Google Scholar]
- 22.Wu Y-F, Guo YH, Liu TJ, Xiao XR, Zhao L, Meng S, et al. Distribution of FimA genotype of Porphyromonas gingivalis in Chinese periodontitis patients and its relationship with chronic periodontitis. Sichuan Da Xue Xue Bao Yi Xue Ban. 2006;37:101–108. [PubMed] [Google Scholar]
- 23.Zhao L, Wu Y-F, Meng S, Yang H, OuYang YL, Zhou XD. Prevalence of FimA genotypes of Porphyromonas gingivalis and periodontal health status in chinese adults. J Periodontal Res. 2007;42:511–517. doi: 10.1111/j.1600-0765.2007.00975.x. [DOI] [PubMed] [Google Scholar]
- 24.Botero JE, Contreras A, Lafaurie G, Jaramillo A, Betancourt M, Arce RM. Occurrence of periodontopathic and superinfecting bacteria in chronic and aggressive periodontitis subjects in a Colombian population. J Periodontol. 2007;78(4):696–704. doi: 10.1902/jop.2007.060129. [DOI] [PubMed] [Google Scholar]
- 25.Lafaurie G, Contreras A, Barón A, Botero J, Mayorga-Fayad I, Jaramillo A, et al. Demographic, clinical, and microbial aspects of chronic and aggressive periodontitis in Colombia: a multicenter study. J Periodontol, 2007;78:629–639. doi: 10.1902/jop.2007.060187. [DOI] [PubMed] [Google Scholar]
- 26.Mayorga-Fayad I, Lafaurie GI, Contreras A, Castillo DM, Barón A, Aya Mdel R. Subgingival microbiota in chronic and aggressive periodontitis in Bogotá, Colombia: an epidemiological approach. Biomedica. 2007;27(1):21–33. [PubMed] [Google Scholar]
- 27.Moon JH, Shin SI, Chung JH, Lee SW, Amano A, Lee JY. Development and evaluation of new primers for PCR-based identification of type II fimA of Porphyromonas gingivalis. FEMS Immunol Med Microbiol. 2012;64:425–428. doi: 10.1111/j.1574-695X.2011.00889.x. [DOI] [PubMed] [Google Scholar]
- 28.Moon JH, Herr Y, Lee HW, Shin SI, Kim C, Amano A, et al. Genotype analysis of Porphyromonas gingivalis fimA in Korean adults using new primers. J Med Microbiol. 2013;62:1290–1294. doi: 10.1099/jmm.0.054247-0. [DOI] [PubMed] [Google Scholar]
- 29.Enersen M, Olsen I, Kvalheim O, Caugant DA. FimA genotypes and multilocus sequence types of Porphyromonas gingivalis from patients with periodontitis. J Clin Microbiol. 2008;46:31–42. doi: 10.1128/JCM.00986-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakagawa I, Amano A, Kuboniwa M, Nakamura T, Kawabata S, Hamada S. Functional differences among FimA variants of Porphyromonas gingivalis and their effects on adhesion to and Invasion of human epithelial cells. Infect Immun. 2002;70:277–285. doi: 10.1128/IAI.70.1.277-285.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Socransky SS, Hafajee AD. Dental biofilms: difficult therapeutic targets. Periodontol 2000. 2002;28:12–55. doi: 10.1034/j.1600-0757.2002.280102.x. [DOI] [PubMed] [Google Scholar]
- 32.Jaramillo A, Arce RM, Herrera D, Betancourth M, Botero JE, Contreras A. Clinical and microbiological characterization of periodontal abscesses. J Clin Periodontol. 2005;32(12):1213–1218. doi: 10.1111/j.1600-051X.2005.00839.x. [DOI] [PubMed] [Google Scholar]