SUMMARY
The reprogramming factors that induce pluripotency have been identified primarily from embryonic stem cell (ESC)-enriched, pluripotency-associated factors. Here we report that during mouse somatic cell reprogramming, pluripotency can be induced with lineage specifiers that are pluripotency rivals to suppress ESC identity, most of which are not enriched in ESCs. We found that OCT4 and SOX2, the core regulators of pluripotency, can be replaced by lineage specifiers that are involved in mesendodermal (ME) specification and in ectodermal (ECT) specification, respectively. OCT4 and its substitutes attenuated the elevated expression of a group of ECT genes whereas SOX2 and its substitutes curtailed a group of ME genes during reprogramming. Surprisingly, the two counteracting lineage specifiers can synergistically induce pluripotency in the absence of both OCT4 and SOX2. Our study suggests a “seesaw model,” in which a balance that is established using pluripotency factors and/or counteracting lineage specifiers can facilitate reprogramming.
INTRODUCTION
Understanding the establishment of cellular identity in programming and reprogramming is a major goal of modern biology. For years, pluripotency-associated factors and their rivals, lineage specifiers, have generally been considered to determine the identities of pluripotent and differentiated cells, respectively (Jaenisch and Young, 2008; Young, 2011). Accordingly, the reprogramming factors that induce pluripotency have been identified primarily from embryonic stem cell (ESC)-enriched factors, pluripotency-associated factors, or maternal factors, such as Oct4, Sox2, Klf4, c-Myc, Nanog, PRDM14, Sall4, Esrrb, Utf1, Tet2, and Glis1 (Aoi et al., 2008; Buganim et al., 2012; Chia et al., 2010; Doege et al., 2012; Maekawa et al., 2011; Maherali et al., 2008; Takahashi and Yamanaka, 2006; Yu et al., 2007). Likewise, the direct reprogramming of differentiated cells into other differentiated cell types has been successfully demonstrated by several lineage specifiers, such as Gata4 and Hnf4a (Vierbuchen and Wernig, 2011). Thus, the perspective that the direct conversion of cell state A to cell state B should be realized by a set of master regulatory factors of cell type B has been a prevailing strategy (Graf and Enver, 2009; Jopling et al., 2011); however, whether this is the only strategy for cell fate conversion is unclear.
Recent data indicate that the most critical reprogramming factor, Oct4, which inhibits the expression of differentiation-related genes in ESCs (Kim et al., 2008; Pardo et al., 2010), is sufficient to direct the reprogramming of somatic cells into induced pluripotent stem cells (iPSCs) (Li et al., 2010b; Zhu et al., 2010). Therefore, it is of great significance to find substitutes for Oct4 to elucidate its physiological role and gain a better understanding of the reprogramming mechanisms, which remain largely unknown. The ESC-enriched factor Nr5a2 has been identified as an Oct4 substitute (Heng et al., 2010). However, the physiological role of Oct4 remains unclear because Nr5a2 directly regulates Oct4 and binds to the upstream promoter region of Oct4 (Gu et al., 2005; Guo and Smith, 2010). Therefore, extensively screening for novel Oct4 substitutes among factors including, but not limited to, ESC-related factors may cast light on the molecular mechanisms that underlie reprogramming and pluripotency, thereby facilitating the development of safer and more efficient reprogramming strategies.
Here, we identified eight lineage specifiers as OCT4 substitutes, including GATA3, GATA6, and SOX7, which are involved in mesendodermal (ME) lineage specification. OCT4 and its substitutes attenuated the elevated expression of a group of ectodermal (ECT) genes, such as the ECT lineage specifier Dlx3, that were collectively triggered by SOX2, KLF4, and c-MYC (SKM). Knockdown of Dlx3 enhanced reprogramming in the absence of OCT4, rather than in the absence of SOX2. In addition, SOX2 can be replaced by lineage specifiers involved in ECT lineage specification, such as GMNN. Surprisingly, the two counteracting lineage specifiers can synergistically induce pluripotency in the absence of both OCT4 and SOX2. We suggest a “seesaw model,” in which the balance that is established by pluripotency factors and/or lineage specifiers facilitates the induction of pluripotency in somatic cells. This model could shed light on fundamental questions regarding the establishment of cellular identity during programming and reprogramming.
RESULTS
Induction of pluripotency in mouse somatic cells with GATA3
In our study, we screened an initial plasmid library of 10,080 human genes for their ability to replace OCT4 when introduced together with virally expressed SKM to direct the reprogramming of mouse embryonic fibroblasts (MEFs) containing a green fluorescent protein (GFP) reporter driven by an Oct4 promoter and enhancer. Reprogramming efficiency was evaluated by determining the number of Oct4-driven GFP-positive colonies. We found that GATA3 had the most significant effect in the primary hits (Figure 1A and Table S1). Interestingly, GATA3 is not enriched in ESCs and is an important regulator of development and differentiation (Figure S1D and Table S4) (Ting et al., 1996).
Figure 1. GATA3 Can Substitute for OCT4 to Induce Pluripotency in Mouse Somatic Cells.
(A) Schematic representation of the gene screening process. For the screen, a plasmid library of 10,080 human genes was analyzed. An OCT4 plasmid and empty vectors (EV) were used as positive and negative controls in the first and second column, respectively.
(B) Reprogramming assay that determines the ability of the lineage specifier GATA3 to enhance reprogramming in the absence of OCT4, SOX2, KLF4, or c-MYC. The Oct4-GFP-positive colonies were counted at 9 days post-infection (dpi). SKM, OKM, OSM, OSK, and SK plus EV were used as negative controls. OSKM was used as a positive control. Error bars indicate the SD (n = 3).
(C) The kinetics of reprogramming using different combinations of genes. For the 50,000 MEFs seeded per well, the GFP fluorescence was monitored every 24 hours. The orange bars indicate that no Oct4-GFP-positive cells were observed, whereas the green bars indicate the emergence of Oct4-GFP-positive cells. Two representative independent experimental sets are shown.
(D) The Oct4-GFP-positive colonies generated by dox-inducible GATA3 and constantly expressed SKM were counted at 10 dpi. Dox was added to the culture medium for various periods of time to induce the expression of GATA3. SKM plus EV was used as a negative control. Error bars indicate the SD (n = 3).
(E) The generation of iPS colonies with G3SKM from Oct4-GFP MEFs. Phase and GFP images of a primary iPS colony, GFP images of passaged iPS colonies, and the expression of NANOG in iPS colonies are depicted from left to right. Scale bars represent 100 m.
(F) An adult chimeric mouse (left) generated from G3SKM-induced iPSCs (#1). Phase (middle) and GFP (right) images of the male gonads dissected from an E13.5 G3SKM-iPS-#1 chimeric embryo.
(G) Germline transmission mice (agouti) from #1 are depicted.
(H) The flow cytometric analysis of GFP in G3SKM-secondary MEFs without (left panel) and with (middle panel) dox. RT-qPCR analysis of the relative expression of endogenous Oct4 in G3SKM-secondary MEFs in the process of dox induction. Representative results from three independent experiments are shown. Error bars indicate the SD (n = 3).
See also Figures S1, S2 and Tables S1, S2.
We further validated the ability of GATA3 to replace OCT4 during the reprogramming of MEFs, mouse adult dermal fibroblasts (MDFs), mouse gastric epithelial cells (GECs), and mouse keratinocytes using viral vectors (Figures 1B, S1A, and S1B). The expression of exogenous genes was verified (Figure S1E). We found that GATA3 achieved a reprogramming efficiency that was comparable to, or even higher than, that of OCT4. We subsequently evaluated the ability of GATA3 to enhance reprogramming in the absence of SOX2, KLF4, or c-MYC. We observed that GATA3 was also able to enhance reprogramming in the absence of c-MYC, but it was unable to substitute for SOX2 or KLF4 (Figure 1B). Next, we monitored the kinetics of GATA3- and OCT4-induced reprogramming. We found that Oct4-GFP-positive cells emerged at 6–7 days post-infection (dpi) during both GATA3- and OCT4-induced reprogramming (Figure 1C). We found that GATA3 may mainly function at 4–7 dpi (Figure 1D), which corresponds to the period during which the pluripotency circuitry is reconstructed (Polo et al., 2012).
iPSCs generated with GATA3 are pluripotent
The iPSCs generated using GATA3, SOX2, KLF4, and c-MYC (G3SKM) had morphology similar to mouse ESCs (Figures 1E and S2A). The G3SKM-induced iPSCs were stable during long-term passaging and stained positive for alkaline phosphatase (AP), SSEA-1, UTF1, and NANOG (Figures 1E and S2B). The methylation levels of the Nanog and Oct4 promoters were similar to the methylation levels in mouse ESCs (Figure S2C). Genomic integrations of the viruses into the genomic DNA were confirmed in iPSCs, teratomas, and tissues from chimeric mice, and showed no OCT4 transgene integration (Figure S2D). The expression of endogenous pluripotency-associated genes was activated, and the expression of exogenous GATA3, SOX2, KLF4, and c-MYC was silenced in these cells (Figure S2E), which indicates that they were fully reprogrammed. G3SKM-induced iPSCs produced germline-competent chimeras (Figures 1F and 1G), and these iPSCs were further validated by the characterization of teratoma formation, gene expression profiles, and other assays (Figure S2F and S2G and Tables S2).
GATA3 has little effect on the events noted in previous studies
To identify the potential mechanisms by which GATA3 could replace OCT4, we examined several pathways that have been reported to facilitate or inhibit reprogramming.
To test whether GATA3 could activate endogenous Oct4 to a high level shortly after induction so that it was the activated endogenous Oct4 plus SKM that induced pluripotency, we monitored the endogenous Oct4 expression levels using doxycycline (dox)-inducible G3SKM-secondary MEFs (Wernig et al., 2008), 20% to 40% of which could be reprogrammed. We did not detect significant Oct4 activation until Oct4-GFP-positive cells had begun to emerge (Figure 1H). There was also no significant activation of the pluripotency-associated factor Nanog until Oct4-GFP-positive cells began to emerge (Figure S1C).
We found that GATA3 had little effect on mesenchymal-epithelial transition (MET) (Figure 2A) (Li et al., 2010a; Samavarchi-Tehrani et al., 2010). In addition, p53 and p21 expression was not affected by GATA3 (Figure 2A) (Zhao et al., 2008). Moreover, we did not observe any significant changes in cell proliferation (Figure 2B), which suggests that GATA3 does not contribute to reprogramming by accelerating cell proliferation. These results suggest that GATA3 functions in a manner different from the mechanisms noted in previous studies to promote reprogramming.
Figure 2. OCT4 and GATA3 Can Inhibit a Group of ECT Genes that are Elevated by SKM during Reprogramming.
(A) RT-qPCR analysis of the expression of endogenous Cdh1, Cdh2, Snai1, p53, and p21 relative to their expression in MEFs. The samples were tested at 4 dpi. Error bars indicate the SD (n = 3).
(B) Cell proliferation curves. SKM was used as a control, and 50,000 MEFs were seeded per well. Three wells were harvested every 24 hours to count the number of cells. The cell populations were analyzed using a paired two-tailed Student's t-test. No significant difference was observed between G3SKM- and SKM-infected MEFs (P = 0.7803).
(C) A Venn diagram illustrating the overlap between the expression changes identified in cells induced with G3SKM and OSKM. The blue circle represents the DE lists (expression change > two fold, P < 0.05, FDR < 0.01) in G3SKM-induced cells compared with SKM-induced cells. The purple circle represents the DE lists of OSKM-induced cells compared with SKM-induced cells.
(D) Gene Ontology (GO) analysis of the overlapping genes in (C). The GO analysis was based on the DE list. The P values represent the Bonferroni-corrected EASE score.
(E) GO analysis of genes regulated by SKM, G3SKM, and OSKM at 7 dpi. The GO analysis was based on the DE list. The P values represent the Bonferroni-corrected EASE score.
(F) Heatmaps depicting the relative fold change of gene expression at 7 dpi based on the ECT-related genes by RNA-seq. Red and green indicate increased and decreased expression, respectively.
OCT4 and GATA3 inhibit the ECT-related genes that are elevated by SKM
To further determine the roles of OCT4 and GATA3 in inducing pluripotency, we performed RNA-seq assays at 7 dpi based on our kinetics test (Figures 1C and 1D). The shared targets of OCT4 and GATA3 were involved in several important processes, such as cell adhesion and the regulation of transcription, based on Gene Ontology (GO) function enrichment analysis (Figures 2C and 2D). Interestingly, we observed that compared with MEFs, the up-regulated genes that were most significantly enriched were related to ectoderm and epidermis development following the introduction of SKM. When OCT4 and GATA3 were introduced along with SKM, the most significantly enriched down-regulated genes were also related to ectoderm and epidermis development (Figures 2E and 2F). These results suggest that SKM may have the potential to direct cells toward an ECT state and that the inhibition of ECT-related genes by OCT4 and GATA3 may facilitate the induction of pluripotency.
Screening of other lineage specifiers as substitutes for OCT4
The current model proposes that ESCs are maintained by a shield of pluripotency factors that collaboratively prohibit differentiation into any lineage to preserve an undifferentiated state (Boyer et al., 2005; Silva et al., 2009). A more recent and provocative perspective is that pluripotency factors function as classical lineage-specifying factors to direct the differentiation of ESCs into a specific lineage while inhibiting their commitment to mutually exclusive lineages (Loh and Lim, 2011). In ESCs, Oct4 promotes ME and primitive endoderm differentiation and suppresses ECT differentiation (Niwa et al., 2000; Thomson et al., 2011; Wang et al., 2012). Interestingly, the overexpression of Gata3 in ESCs directs primitive endodermal differentiation, and, similar to Oct4, Gata3 also functions in some ME lineages (Nishiyama et al., 2009). The similarity of OCT4 and GATA3 in lineage specification suggests that other lineage specifiers may also replace OCT4. To test this possibility, we screened several additional lineage specifiers using viral overexpression (Table S3). The results indicated that the lineage specifiers GATA6, SOX7, PAX1, GATA4, CEBPa, HNF4a, and GRB2 were able to substitute for OCT4 to induce pluripotency (Figures 3A, 3B, and S1E). These lineage specifiers have been shown to function mainly during multiple stages of ME differentiation and in early embryonic patterning (Table S4). Most of these substitutes for OCT4 are not enriched in ESCs (Figure S1D). However, the lineage specifiers that are primarily involved in ectodermal lineage specification (e.g., SOX1 and ASCL1) were unable to replace OCT4 (Table S3).
Figure 3. Identification of Other Substitutes for OCT4 that also Inhibit ECT-related Genes Elevated by SKM during Reprogramming.
(A) Identification of additional lineage specifiers that are related to the ME lineage that can replace OCT4 during reprogramming. The Oct4-GFP-positive colonies were counted at 9 dpi. SKM plus EV was used as a control. Error bars indicate the SD (n = 3).
(B) GFP images of iPS colonies generated with SKM+GATA6 (G6SKM), SKM+GATA3 (G3SKM), SKM+SOX7 (S7SKM), SKM+PAX1 (P1SKM), SKM+GATA4 (G4SKM), SKM+CEBPa (CaSKM), SKM+HNF4a (H4SKM), and SKM+GRB2 (GrSKM). Scale bars represent 100 m.
(C) Heatmaps depicting the relative fold change of gene expression at 7 dpi based on the ECT-related genes by microarray. Red and green indicate increased and decreased expression, respectively.
(D) RT-qPCR analysis of the expression of endogenous Dlx3 (upper panel) and Lhx5 (lower panel) relative to the expression in SKM. The samples were tested at 7 dpi after removal of Oct4-GFP-positive cells. Significance was assessed compared with the controls using a one-tailed Student’s t-test. ***, P < 0.001. Error bars indicate the SD (n = 3).
(E) RT-qPCR analysis of the expression of endogenous Dlx3 (upper panel) and Lhx5 (lower panel) relative to the expression in SKM from a single colony; three colonies from each condition were tested. Significance was assessed compared with the controls using a one-tailed Student's t-test. * P < 0.05. Error bars indicate the SD (n = 3).
See also Figures S3, S4 and Tables S2, S3, S4.
Other OCT4 substitutes also inhibit ECT-related genes elevated by SKM
Because both OCT4 and GATA3 attenuated the elevated expression of the group of ECT genes that was collectively triggered by SKM, we asked whether the other substitutes for OCT4 could also attenuate their expression. We performed microarray assays to analyze the roles of other OCT4 substitutes. In addition, we analyzed another lineage specifier, MIXL1, as a negative control, which could not replace OCT4, despite its involvement in ME lineage specification. Consistent with our expectations, the other OCT4 substitutes, but not MIXL1, attenuated the up-regulation of ECT genes by SKM (Figure 3C). GO analysis also showed that the most significantly enriched down-regulated genes after the introduction of OCT4 substitutes other than MIXL1 were associated with ectoderm and epidermis development and other terms related to ECT specification (Figure S3).
Next, to validate the tendency apparent from the RNA-seq and microarray analyses (Figures 2F and 3C), we performed RT-qPCR assays. We observed that typical ECT marker genes, such as Dlx3 and Lhx5, were up-regulated by SKM and significantly down-regulated after the introduction of OCT4 and its substitutes (Figures 3D). To further validate these findings, we performed single-colony assays. We acquired AP-positive, Oct4-GFP-negative colonies in different conditions, including controlled SKM conditions. Then, we mechanically split each colony into two sub-colonies. We analyzed one sub-colony for gene expression while tracing the other half sub-colony in culture to monitor whether it became Oct4-GFP-positive and dox-independent (Figure S4). In this case, we could analyze those reprogramming-competent colonies that were headed toward pluripotency. AP-positive colonies grown in SKM conditions failed to become Oct4-GFP-positive, suggesting that they are reprogramming refractory. We found a concomitant down-regulation of Dlx3 and Lhx5 after the introduction of OCT4 and its substitutes (Figure 3E). This finding is consistent with the results from mixed cell populations, supporting the lineage specification tendency observed in the bulk population. These results suggest that OCT4 and its substitutes can repress the up-regulation of ECT-related genes that are triggered by SKM.
Suppression of Dlx3, a master ECT-related gene, facilitates reprogramming in the presence of SKM
To test whether the inhibition of ECT-related genes by OCT4 and its substitutes correlated with the promotion of reprogramming, we further evaluated whether the knockdown of the master ECT lineage-related genes that are up-regulated by SKM could also promote reprogramming in the absence of OCT4. We found that the knockdown of the key ECT marker Dlx3, which subsequently resulted in the down-regulation of several other ECT genes, promoted reprogramming in the absence of OCT4, rather than in the absence of SOX2 (Figures 4 and Figure S5), functionally supporting the data observed by gene profiling (Figures 2E, 2F, and 3C-3E). Furthermore, iPSCs generated with Dlx3 shRNAs were proven to be pluripotent by stringent characterization assays (Figures 4E, 4F, and Table S2). We found no significant binding of Oct4 and Gata3 to the Dlx3 promoter region in published ChIP data from “hmChIP,” which suggests that Oct4 and Gata3 may indirectly regulate Dlx3 (Figure S5D). Together, our data indicate that OCT4 and its substitutes attenuate the up-regulation of ECT-related genes that is triggered by the SKM combination; this inhibition correlates with facilitating the induction of pluripotency.
Figure 4. Inhibition of the Master ECT Gene Dlx3 Facilitates Reprogramming in the Presence of SKM.
(A) shRNAs targeting Dlx3 were introduced into MEFs with SKM or OKM. Scrambled shRNA (control RNAi) was used as a control. The Oct4-GFP-positive colonies were counted at 9 dpi. Error bars indicate the SD (n = 3).
(B) RT-qPCR analysis of the expression of endogenous Dlx3 relative to the expression in control. SKM plus scramble shRNA (control RNAi) was used as a control. The samples were tested at 7 dpi. Error bars indicate the SD (n = 3).
(C) Western blot analysis of DLX3 in control and knockdown cells in the presence of a SKM virus cocktail. GAPDH was used as a loading control.
(D) Phase and GFP images of iPS colonies generated with Dlx3 RNAi 1 and Dlx3 RNAi 2 in the presence of SKM. Scale bars represent 100 µm.
(E) Adult chimeric mice generated from Dlx3 RNAi 2-induced iPSCs (#1).
(F) A germline transmission mouse (agouti) from #1 is depicted. See also Figure S5.
SOX2 and its substitutes attenuate the expression of ME-related genes elevated by OKM
Similar to Oct4, Sox2 also regulates lineage specification in ESCs. Sox2 inhibits mesendodermal differentiation and promotes neural ectodermal differentiation (Thomson et al., 2011; Wang et al., 2012). Intuitively, we asked whether the lineage specifiers involved in ECT lineage specification could substitute for SOX2 during reprogramming. Indeed, the previously identified SOX2 substitutes, Sox1, Sox3, and RCOR2, are regulators of ECT development or are particularly expressed in neural tissues (Nakagawa et al., 2007; Tontsch et al., 2001; Yang et al., 2011; Zeng et al., 2010). In addition, we demonstrated that GMNN, which is involved in ECT lineage specification (Seo, 2005), could substitute for SOX2 to promote reprogramming (Figures 5A, 5B, S1E, and Table S4).
Figure 5. SOX2 and Its Substitutes Attenuate the Expression of ME Genes that are Elevated by OKM during Reprogramming.
(A) Identification of lineage specifiers related to the ECT lineage that can facilitate reprogramming in the presence of OKM. The Oct4-GFP-positive colonies were counted at 9 dpi. OKM plus EV was used as a control. Error bars indicate the SD (n = 3).
(B) GFP images of iPS colonies generated with OKM+SOX1 (OS1KM), OKM+SOX3 (OS3KM), and OKM+GMNN (OGmKM). Scale bars represent 100 m.
(C) Heatmaps depicting the relative fold change of gene expression at 7 dpi based on the ME-related genes by microarray. Red and green indicate increased and decreased expression, respectively.
(D) RT-qPCR analysis of the expression of endogenous T (left panel) and Eomes (right panel) relative to the expression in OKM. Significance was assessed compared with the controls using a one-tailed Student's t-test. *** P < 0.001. The samples were tested at 7 dpi. Error bars indicate the SD (n = 3).
(E) The expression of endogenous T (upper panel) and Eomes (lower panel) relative to the expression in OKM from a single colony; three colonies from each condition were tested. Significance was assessed compared with the controls using a one-tailed Student's t-test. * P < 0.05, *** P < 0.001. Error bars indicate the SD (n = 3).
To assess whether SOX2 and its substitutes could attenuate the up-regulation of the ME-related genes that are induced by OCT4, KLF4, and c-MYC (OKM), we performed microarray and RT-qPCR analyses. The expression of a group of classical ME markers was elevated after the induction of OKM; these elevated expression levels were attenuated by SOX2 and its substitutes (Figures 5C and 5D). Furthermore, we performed similar single-colony assays as previously mentioned to test this tendency in reprogramming-competent cells. We observed that SOX2 and its substitutes inhibited the expression of T and Eomes in these colonies (Figure 5E). These data indicate that SOX2 and its substitutes attenuate the up-regulation of ME-related genes that is induced by OKM during the reprogramming process.
A “seesaw model” suggests that the balancing of counteracting forces facilitates the induction of pluripotency
A two-node model of cell fate determination has been studied in various instances of pluripotent stem or progenitor cells that undergo a binary cell fate decision (e.g., GATA1 and PU.1, RUNX2 and PPARg) (Huang, 2009). This circuit has already hinted at the concept of “balanced pluripotent state” (Figure S6A and S6B). To better understand the induction of pluripotency by lineage specifiers, we propose a “seesaw model” (Figures 6A and 7) based on previous studies (Hanna et al., 2009; Loh and Lim, 2011; Niakan et al., 2010; Niwa et al., 2000; Polo et al., 2012; Rosa and Brivanlou, 2010; Teo et al., 2011; Thomson et al., 2011; Wang et al., 2012) and our current data (Figures 2–5 and S3–S5). This model consists of two coupled modules: the pluripotency module and the differentiation module. The pluripotency module is represented by the mutual activation of Oct4 and Sox2, while the differentiation module is modeled by mutually inhibiting the ME and ECT genes (Figure 6A).
Figure 6. Balanced Equilibrium Facilitates the Induction of Pluripotency.
(A) A model for the coupled pluripotency module (self-activation of Oct4 and Sox2) and for the differentiation module (mutual antagonism between the ME and ECT lineages).
(B) The cell-state landscape obtained from the density of the trajectories. The blue color indicates deep “attractors” near which the cell states tend to stay. The x-axis represents the difference between the two groups of lineage inducers, and the y-axis indicates the "pluripotency" of the cell.
(C and D) The phase space without (box i)/with KM (box ii) under no input, respectively (C). Different reprogramming strategies with KM and the trajectory starting from the ME state are shown (D). The blue circuit indicates the ME state where the trajectories start. Yellow dots indicate stable cell states. The changing directions of cell states are marked by arrows. The trajectories that end up in the ME state, the ECT state, and the pluripotent state are colored by blue, green, and red, respectively. Cyan indicates the region where the reprogramming is possible. Dark lines are the nullclines of the differentiation module when the pluripotency-module is OFF. When the stable crossing point falls into the cyan reprogramming region, pluripotency can be restored.
(E) The generation of mouse iPSCs in the absence of both OCT4 and SOX2. KM plus EV was used as a control. The Oct4-GFP-positive colonies were counted at 9 dpi. Error bars indicate the SD (n = 3).
(F) GFP images of iPS colonies generated with KM+GATA3+SOX1 (G3S1KM), KM+GATA3+SOX3 (G3S3KM), KM+GATA6+SOX1 (G6S1KM), KM+GATA6+SOX3 (G6S3KM), KM+GATA6+ GMNN (G6GmKM), KM+PAX1+SOX1 (P1S1KM), KM+PAX1+SOX3 (P1S3KM), and OSKM. Scale bars represent 100 m.
See also Figures S6, S7 and Table S5, S6.
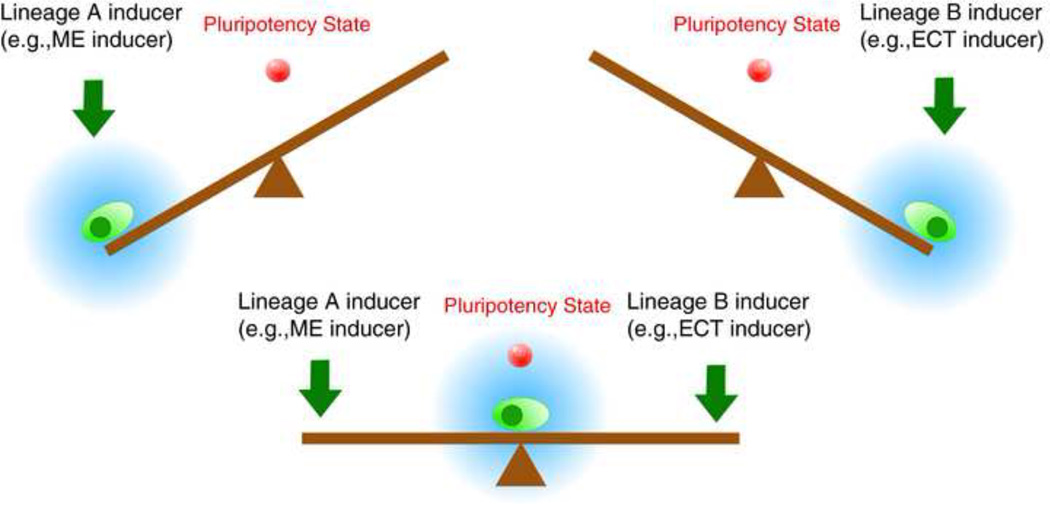
Figure 7. The “seesaw model”.
A diagram of the “seesaw model.” Blue clouds indicate the regions that the cell states are likely to sample with noise. The pluripotency state (red ball) is located near the balanced region. When the seesaw is balanced between the two differentiation potentials, the cell has a higher probability of entering the pluripotent state.
Simulations of the model produced three cell states: the pluripotency state, the ME state, and the ECT state (Figures 6B and S6C). In somatic states, the pluripotency state is unreachable (Figure 6C, box i). However, overexpression of KLF4 and c-MYC (KM) may act as a driving force that reduces reprogramming barriers (Polo et al., 2012), so the pluripotency state becomes a local attractor that is reachable from a somatic state if properly perturbed (Figure 6C, box ii). Note that the pluripotent state is most easily reached (high pluripotency potential) if the expression strengths of the ME and ECT genes are forced to be in the blue region, i.e., if they are balanced.
A key result of this model is that pluripotency and reprogramming can be achieved by a variety of perturbations to the system, as illustrated in Figure 6D. For example, the overexpression of SOX2 alone will switch on the ECT genes that inhibit the pluripotency module in turn, bestowing the ECT state onto the cell, regardless of the initial cell state (Figures 6D, box i; S6I; and S6N). The overexpression of OCT4 (or the ME gene GATA3) in combination with SOX2 counteracts the inhibition of the ECT genes to the pluripotency module, which permits the cell to move into the pluripotency attractor region (Figures 6D, boxes ii and iii; S6D; and S6E). More strategies for restoring pluripotency as suggested by the model are shown in Figures 6D, S6 and Table S5, S6.
Some of the results from our model are consistent with previous studies (Table S6). Deviation from the balanced equilibrium for pluripotency directs cells to flow into divergent differentiated states. For example, the enforced expression of OCT4 results in cellular transdifferentiation into mesodermal hematopoietic cells (Szabo et al., 2010). SOX2, SKM with neural lineage specifiers, and reduced Oct4 expression in the OSKM cocktail can result in neuroectodermal lineage transdifferentiation from fibroblasts (Han et al., 2012; Ring et al., 2012; Thier et al., 2012). Other strategies identified by our model call for further investigation to validate or falsify the model.
One of the striking predictions of the model is that when expressed together, an ME specifier and an ECT specifier can synergistically enhance reprogramming in the absence of both OCT4 and SOX2. The mutually antagonistic nature of the differentiation module makes the system behave like an easily tilted “seesaw;” when the system tends to fall into either the ME or ECT state, in turn suppressing the pluripotency module, the cell is left with no chance to enter the pluripotency state. However, when both ME and ECT specifiers are exogenously expressed at appropriate levels, their external expression may be unable to compensate for the decrease in their endogenous expression due to the mutual inhibition. The differentiation module may end up in the balanced region, wherein neither ME specifiers nor ECT specifiers are present at levels that are sufficiently high to suppress the pluripotency module. Then, a self-activating pluripotency module may switch on by itself, eliciting the serial activation of a pluripotency network to realize successful reprogramming (Figures 6D, box v; S6G; and S6R-6T).
To test the feasibility of this idea, we screened different combinations of lineage specifiers. We found that either (1) GATA3, GATA6, PAX1, and either SOX1 or SOX3 or (2) GATA6 plus GMNN, can promote successful reprogramming without the two most critical pluripotency factors, OCT4 and SOX2 (Figures 6E, 6F, and S1E). Furthermore, these iPSCs were proven to be pluripotent (Figure S7). These lineage specifiers are not enriched in ESCs (Figure S1D), which indicates that the induction and maintenance of pluripotency may be two different scenarios.
DISCUSSION
Our data present the first evidence that the lineage specifiers GATA3, GATA6, SOX7, PAX1, GATA4, CEBPa, HNF4a, GRB2, and GMNN, which are generally considered as pluripotency rivals, can unexpectedly facilitate reprogramming and replace reprogramming factors of a corresponding lineage-specifying potential. We suggest a “seesaw model,” in which lineage specifiers facilitate reprogramming when they are balanced with other mutually exclusive lineage specifiers. There are multiple ways to reach the pluripotency state, all involving balancing the forces that would otherwise drive the cell to a differentiated fate. Deviation from the balance directs cells to divergent differentiated states, reducing the possibility of restoring pluripotency. This hypothesis is complementary to previous studies demonstrating that lineage specifiers, such as Gata6, a reported repressor of Nanog, hinder the reprogramming process (Chazaud et al., 2006; Mikkelsen et al., 2008). We suggest that pluripotency factors may direct lineage specification and that lineage specifiers may in turn influence the induction of pluripotency; this model provides the conceptually new perspective that one can reach the pluripotency “destination” by getting on cars that do not normally run at the destination.
Our “seesaw model” is in agreement with the recent perspective that pluripotency factors act as classical lineage-specifying factors to direct the differentiation of ESCs into a specific lineage while cross-inhibiting commitment to mutually exclusive lineages (Loh and Lim, 2011). The collaborative result is the temporary inhibition of all lineages to preserve an undifferentiated state. The self-activating pluripotency module could be activated when all the lineage specifying forces are counteracted, i.e., at the dynamic balanced point of the “seesaw”, wherein no particular lineage specifying activity is dominant to inhibit the pluripotency module. Once the pluripotency module is activated, the ME and ECT lineage fates are blocked by SOX2 and OCT4, respectively, and the pluripotency state is maintained.
Most of these substitutes are usually depicted as typical ME markers (GATA6, GATA4, etc.) or ECT markers (SOX1, SOX3, etc.). To make our model succinct, these lineage specifiers are classified as either ME or ECT. However, this classification is not absolute, as indicated by our “seesaw model.” The ME and ECT lineages are the two major counteracting lineages, but in the reprogramming symphony, an enormously intricate self-conflicted coalition of different lineage specifiers that are not limited to the ME and ECT lineages orchestrates the concerted induction of pluripotency.
The “seesaw model” focuses on one major aspect in reprogramming: understanding the implications of the reprogramming landscape that is shaped by the interactions among the different cell states. Needless to say, reprogramming is not merely a “seesaw.” Several other important events, such as epigenetic changes, are also involved in this process. Furthermore, there are several important barriers to overcome for successful reprogramming. The various reprogramming factors execute their corresponding functions to facilitate the induction of pluripotency. Concomitantly, these factors may induce side effects, such as the stimulation of lineage specification. In addition to their reported functions in the activation of the pluripotency circuitry and in epigenetic and cell cycle regulation, KLF4 and c-MYC may also promote stimuli for particular lineages (Cao et al., 2012; Smith et al., 2010; Sridharan et al., 2009) to concert an equilibrium symphony. The function of these lineage specifiers may be involved in other processes and warrants further in-depth study.
We hope that our model can serve as a starting point to decipher the mysterious reprogramming code in a novel scenario. Our results suggest that a chemical screen for small molecules that can substitute for OCT4 and SOX2 in directing the corresponding lineage specification would be a novel and feasible strategy to generate iPSCs. Our findings also demonstrate the need for further investigation of the conventional perspectives regarding the functions of pluripotency factors and lineage specifiers and on the manner in which the pluripotency regulatory circuitry is established during reprogramming.
EXPERIMENTAL PROCEDURES
Screen
The 10,080 cDNAs that were screened for their ability to replace OCT4 were obtained from Origene Co., Ltd. Oct4-GFP MEFs were infected with SKM (SOX2, KLF4 and c-MYC) lentiviral supernatant (MOI: 4) supplemented with 10 ng/ l polybrene (Sigma) for 12 hours. Details are provided in the Extended Experimental Procedures.
Cell Culture
Primary MEFs, mouse GECs, keratinocytes and MDFs were derived from ICR×Oct4-GFP transgenic mice. Mouse GECs were isolated and cultured as previously described (Aoi et al., 2008). Details are provided in the Extended Experimental Procedures.
iPSCs Generation
The dox-inducible lentiviral system used has been previously described (Maherali et al., 2008). Further details are provided in the Extended Experimental Procedures.
Time Course of GATA3 Induction
A dox-inducible lentiviral system was used, so the expression of tet-GATA3, but not pll3.7- U6 GFP-SOX2, pll3.7- U6 GFP-KLF4, or pll3.7- U6 GFP-c-MYC (Zhao et al., 2008), could be induced by dox. Dox was added to the culture medium for different periods of time, and the number of Oct4-GFP-positive colonies was counted at 10 dpi.
Characterization of iPSCs
The teratoma formation and chimera experiments were performed as previously described (Li et al., 2010b). The primers used for RT-PCR, genomic PCR, and RT-qPCR are listed in Table S7. Bisulfite treatment of DNA was performed using the CpGenome™ Fast DNA Modification Kit (Millipore). The primers used for promoter fragment amplification have been previously described (Takahashi and Yamanaka, 2006). The details for Immunofluorescence and the detection of AP activity are provided in the Extended Experimental Procedures.
Single colony analysis
Five days after infection with different reprogramming cocktails, Alkaline Phosphatase Live Stain (Invitrogen) was used for the detection of AP activity. AP-positive colonies were then picked and each colony was divided into two parts. One part was kept culturing in induction medium for 2–4 days. The other part was frozen at −80°C. If Oct4-GFP turned on in further 2–4 days induction and became dox-independent in 4 days without dox, total RNA was isolated from the frozen part using the RNeasy micro kit (Qiagen) and then used for RT-qPCR analysis.
Knockdowns
Dlx3 knockdown was achieved using shRNA lentiviral vectors (Sigma) containing a puromycin resistance gene. Further details are provided in the Extended Experimental Procedures.
Western Blotting
Cells were washed with PBS, treated with 1x SDS sample buffer, and then boiled. Additional details are provided in the Extended Experimental Procedures.
DNA Microarray
Total mRNA from iPSCs, MEFs, MEFs at 7 dpi, and R1 were labeled with Cy5, hybridized to a mouse Oligo Microarray (Phalanx Mouse Whole Genome OneArray; Phalanx Biotech). The data were analyzed according to the manufacturer’s protocol.
RNA-seq
RNA sequencing libraries were constructed using the Illumina mRNA-seq Prep Kit. The fragmented and randomly primed 200-bp paired-end libraries were sequenced using Illumina HiSeq 2000. Details are included in the Extended Experimental Procedures.
Statistical Analysis
Cell proliferation curves were analyzed using a paired two-tailed Student's t-test. The unpaired one-tailed t-test was used for statistical analysis of other experiments. The standard deviation (SD) was used to quantify variability.
Model Construction
A coarse-grained model was built based on our results and the results of previous studies. The interactions involved in this model are illustrated in Figure 6A. Additional details are provided in the Extended Experimental Procedures.
Supplementary Material
HIGHLIGHTS.
OCT4 and SOX2 can be replaced by lineage specifiers during reprogramming
OCT4 substitutes and SOX2 substitutes inhibit some ECT and ME genes, respectively
A seesaw model suggests that balancing counteracting forces facilitates reprogramming
ACKNOWLEDGEMENTS
We thank Chengyan Wang and Xiaolei Yin for helpful discussions. We also thank Haisong Liu, Jiancheng Wang, Zhiyuan Hou, Ren Li, Fangfang Zhu, Dongyi Xu, and Xing Zhang for technical assistance. J.S., C.W., and Y.T.W. conducted most of the experiments in this study. Z.Y.L. performed the computer analysis and mathematical model analysis. S.D.S., W.H.Z., X.T., H.Y., LJ.S., X.H.Z., M.L.Q., W.F.Y., Y.S., and G.G. performed the experimental work. H.K.D., Y.Z., and C.T. conceived of and supervised the study. This work was supported by grants from the National Basic Research Program of China (973 program Grant Nos. 2012CB966401, 2009CB941101, 2010CB945204, and 2009CB918500), the Key New Drug Creation and Manufacturing Program (Grant Nos. 2011ZX09102-010-03), the National Natural Science Foundation of China (Grant Nos. 90919031 and 10721403), the 111 project, and NSF (CMMI-0941355).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBERS
Microarray and RNA-seq data are available in the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/) under the accession number GSE43995.
SUPPLEMENTAL INFORMATION
The supplemental information includes the Extended Experimental Procedures, seven figures, and seven tables.
REFERENCES
- Aoi T, Yae K, Nakagawa M, Ichisaka T, Okita K, Takahashi K, Chiba T, Yamanaka S. Generation of Pluripotent Stem Cells from Adult Mouse Liver and Stomach Cells. Science. 2008;321:699–702. doi: 10.1126/science.1154884. [DOI] [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, et al. Core Transcriptional Regulatory Circuitry in Human Embryonic Stem Cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buganim Y, Faddah Dina A, Cheng Albert W, Itskovich E, Markoulaki S, Ganz K, Klemm Sandy L, van Oudenaarden A, Jaenisch R. Single-Cell Expression Analyses during Cellular Reprogramming Reveal an Early Stochastic and a Late Hierarchic Phase. Cell. 2012;150:1209–1222. doi: 10.1016/j.cell.2012.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Q, Zhang X, Lu L, Yang L, Gao J, Gao Y, Ma H, Cao Y. Klf4 is required for germ-layer differentiation and body axis patterning during Xenopus embryogenesis. Development. 2012;139:3950–3961. doi: 10.1242/dev.082024. [DOI] [PubMed] [Google Scholar]
- Chazaud C, Yamanaka Y, Pawson T, Rossant J. Early Lineage Segregation between Epiblast and Primitive Endoderm in Mouse Blastocysts through the Grb2-MAPK Pathway. Developmental Cell. 2006;10:615–624. doi: 10.1016/j.devcel.2006.02.020. [DOI] [PubMed] [Google Scholar]
- Chia N-Y, Chan Y-S, Feng B, Lu X, Orlov YL, Moreau D, Kumar P, Yang L, Jiang J, Lau M-S, et al. A genome-wide RNAi screen reveals determinants of human embryonic stem cell identity. Nature. 2010;468:316–320. doi: 10.1038/nature09531. [DOI] [PubMed] [Google Scholar]
- Doege CA, Inoue K, Yamashita T, Rhee DB, Travis S, Fujita R, Guarnieri P, Bhagat G, Vanti WB, Shih A, et al. Early-stage epigenetic modification during somatic cell reprogramming by Parp1 and Tet2. Nature. 2012;488:652–655. doi: 10.1038/nature11333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf T, Enver T. Forcing cells to change lineages. Nature. 2009;462:587–594. doi: 10.1038/nature08533. [DOI] [PubMed] [Google Scholar]
- Gu P, Goodwin B, Chung ACK, Xu X, Wheeler DA, Price RR, Galardi C, Peng L, Latour AM, Koller BH, et al. Orphan Nuclear Receptor LRH-1 Is Required To Maintain Oct4 Expression at the Epiblast Stage of Embryonic Development. Molecular and Cellular Biology. 2005;25:3492–3505. doi: 10.1128/MCB.25.9.3492-3505.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo G, Smith A. A genome-wide screen in EpiSCs identifies Nr5a nuclear receptors as potent inducers of ground state pluripotency. Development. 2010;137:3185–3192. doi: 10.1242/dev.052753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Dong W, Tapia N, Hermann A, Hemmer K, Höing S, Araúzo-Bravo Marcos J, Zaehres H, Wu G, Frank S, Moritz S, et al. Direct Reprogramming of Fibroblasts into Neural Stem Cells by Defined Factors. Cell Stem Cell. 2012;10:465–472. doi: 10.1016/j.stem.2012.02.021. [DOI] [PubMed] [Google Scholar]
- Hanna J, Saha K, Pando B, van Zon J, Lengner CJ, Creyghton MP, van Oudenaarden A, Jaenisch R. Direct cell reprogramming is a stochastic process amenable to acceleration. Nature. 2009;462:595–601. doi: 10.1038/nature08592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heng J-CD, Feng B, Han J, Jiang J, Kraus P, Ng J-H, Orlov YL, Huss M, Yang L, Lufkin T, et al. The Nuclear Receptor Nr5a2 Can Replace Oct4 in the Reprogramming of Murine Somatic Cells to Pluripotent Cells. Cell Stem Cell. 2010;6:167–174. doi: 10.1016/j.stem.2009.12.009. [DOI] [PubMed] [Google Scholar]
- Huang S. Reprogramming cell fates: reconciling rarity with robustness. BioEssays. 2009;31:546–560. doi: 10.1002/bies.200800189. [DOI] [PubMed] [Google Scholar]
- Jaenisch R, Young R. Stem Cells, the Molecular Circuitry of Pluripotency and Nuclear Reprogramming. Cell. 2008;132:567–582. doi: 10.1016/j.cell.2008.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jopling C, Boue S, Belmonte JCI. Dedifferentiation, transdifferentiation and reprogramming: three routes to regeneration. Nature Reviews Molecular Cell Biology. 2011;12:79–89. doi: 10.1038/nrm3043. [DOI] [PubMed] [Google Scholar]
- Kim J, Chu J, Shen X, Wang J, Orkin SH. An Extended Transcriptional Network for Pluripotency of Embryonic Stem Cells. Cell. 2008;132:1049–1061. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Liang J, Ni S, Zhou T, Qing X, Li H, He W, Chen J, Li F, Zhuang Q, et al. A Mesenchymal-to-Epithelial Transition Initiates and Is Required for the Nuclear Reprogramming of Mouse Fibroblasts. Cell Stem Cell. 2010a;7:51–63. doi: 10.1016/j.stem.2010.04.014. [DOI] [PubMed] [Google Scholar]
- Li Y, Zhang Q, Yin X, Yang W, Du Y, Hou P, Ge J, Liu C, Zhang W, Zhang X, et al. Generation of iPSCs from mouse fibroblasts with a single gene, Oct4, and small molecules. Cell Research. 2010b;21:196–204. doi: 10.1038/cr.2010.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh Kyle M, Lim B. A Precarious Balance: Pluripotency Factors as Lineage Specifiers. Cell Stem Cell. 2011;8:363–369. doi: 10.1016/j.stem.2011.03.013. [DOI] [PubMed] [Google Scholar]
- Maekawa M, Yamaguchi K, Nakamura T, Shibukawa R, Kodanaka I, Ichisaka T, Kawamura Y, Mochizuki H, Goshima N, Yamanaka S. Direct reprogramming of somatic cells is promoted by maternal transcription factor Glis1. Nature. 2011;474:225–229. doi: 10.1038/nature10106. [DOI] [PubMed] [Google Scholar]
- Maherali N, Ahfeldt T, Rigamonti A, Utikal J, Cowan C, Hochedlinger K. A High-Efficiency System for the Generation and Study of Human Induced Pluripotent Stem Cells. Cell Stem Cell. 2008;3:340–345. doi: 10.1016/j.stem.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Hanna J, Zhang XL, Ku MC, Wernig M, Schorderet P, Bernstein BE, Jaenisch R, Lander ES, Meissner A. Dissecting direct reprogramming through integrative genomic analysis. 2008;454:49. doi: 10.1038/nature07056. 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]; Nature. 454:794–794. [Google Scholar]
- Nakagawa M, Koyanagi M, Tanabe K, Takahashi K, Ichisaka T, Aoi T, Okita K, Mochiduki Y, Takizawa N, Yamanaka S. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nature Biotechnology. 2007;26:101–106. doi: 10.1038/nbt1374. [DOI] [PubMed] [Google Scholar]
- Niakan KK, Ji H, Maehr R, Vokes SA, Rodolfa KT, Sherwood RI, Yamaki M, Dimos JT, Chen AE, Melton DA, et al. Sox17 promotes differentiation in mouse embryonic stem cells by directly regulating extraembryonic gene expression and indirectly antagonizing self-renewal. Genes & Development. 2010;24:312–326. doi: 10.1101/gad.1833510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama A, Xin L, Sharov AA, Thomas M, Mowrer G, Meyers E, Piao Y, Mehta S, Yee S, Nakatake Y, et al. Uncovering Early Response of Gene Regulatory Networks in ESCs by Systematic Induction of Transcription Factors. Cell Stem Cell. 2009;5:420–433. doi: 10.1016/j.stem.2009.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H, Miyazaki J, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nature Genetics. 2000;24:372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- Pardo M, Lang B, Yu L, Prosser H, Bradley A, Babu MM, Choudhary J. An Expanded Oct4 Interaction Network: Implications for Stem Cell Biology, Development, and Disease. Cell Stem Cell. 2010;6:382–395. doi: 10.1016/j.stem.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo Jose M, Anderssen E, Walsh Ryan M, Schwarz Benjamin A, Nefzger Christian M, Lim Sue M, Borkent M, Apostolou E, Alaei S, Cloutier J, et al. A Molecular Roadmap of Reprogramming Somatic Cells into iPS Cells. Cell. 2012;151:1617–1632. doi: 10.1016/j.cell.2012.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ring Karen L, Tong Leslie M, Balestra Maureen E, Javier R, Andrews-Zwilling Y, Li G, Walker D, Zhang William R, Kreitzer Anatol C, Huang Y. Direct Reprogramming of Mouse and Human Fibroblasts into Multipotent Neural Stem Cells with a Single Factor. Cell Stem Cell. 2012 doi: 10.1016/j.stem.2012.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa A, Brivanlou AH. A regulatory circuitry comprised of miR-302 and the transcription factors OCT4 and NR2F2 regulates human embryonic stem cell differentiation. The EMBO Journal. 2010;30:237–248. doi: 10.1038/emboj.2010.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samavarchi-Tehrani P, Golipour A, David L, Sung H-k, Beyer TA, Datti A, Woltjen K, Nagy A, Wrana JL. Functional Genomics Reveals a BMP-Driven Mesenchymal-to-Epithelial Transition in the Initiation of Somatic Cell Reprogramming. Cell Stem Cell. 2010;7:64–77. doi: 10.1016/j.stem.2010.04.015. [DOI] [PubMed] [Google Scholar]
- Seo S. Geminin regulates neuronal differentiation by antagonizing Brg1 activity. Genes & Development. 2005;19:1723–1734. doi: 10.1101/gad.1319105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva J, Nichols J, Theunissen TW, Guo G, van Oosten AL, Barrandon O, Wray J, Yamanaka S, Chambers I, Smith A. Nanog Is the Gateway to the Pluripotent Ground State. Cell. 2009;138:722–737. doi: 10.1016/j.cell.2009.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KN, Singh AM, Dalton S. Myc Represses Primitive Endoderm Differentiation in Pluripotent Stem Cells. Cell Stem Cell. 2010;7:343–354. doi: 10.1016/j.stem.2010.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridharan R, Tchieu J, Mason MJ, Yachechko R, Kuoy E, Horvath S, Zhou Q, Plath K. Role of the Murine Reprogramming Factors in the Induction of Pluripotency. Cell. 2009;136:364–377. doi: 10.1016/j.cell.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo E, Rampalli S, Risueño RM, Schnerch A, Mitchell R, Fiebig-Comyn A, Levadoux-Martin M, Bhatia M. Direct conversion of human fibroblasts to multilineage blood progenitors. Nature. 2010;468:521–526. doi: 10.1038/nature09591. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of Pluripotent Stem Cells from Mouse Embryonic and Adult Fibroblast Cultures by Defined Factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Teo AKK, Arnold SJ, Trotter MWB, Brown S, Ang LT, Chng Z, Robertson EJ, Dunn NR, Vallier L. Pluripotency factors regulate definitive endoderm specification through eomesodermin. Genes & Development. 2011;25:238–250. doi: 10.1101/gad.607311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thier M, Wörsdörfer P, Lakes Yenal B, Gorris R, Herms S, Opitz T, Seiferling D, Quandel T, Hoffmann P, Nöthen MM, et al. Direct Conversion of Fibroblasts into Stably Expandable Neural Stem Cells. Cell Stem Cell. 2012;10:473–479. doi: 10.1016/j.stem.2012.03.003. [DOI] [PubMed] [Google Scholar]
- Thomson M, Liu Siyuan J, Zou L-N, Smith Z, Meissner A, Ramanathan S. Pluripotency Factors in Embryonic Stem Cells Regulate Differentiation into Germ Layers. Cell. 2011;145:875–889. doi: 10.1016/j.cell.2011.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting CN, Olson MC, Barton KP, Leiden JM. Transcription factor GATA-3 is required for development of the T-cell lineage. Nature. 1996;384:474–478. doi: 10.1038/384474a0. [DOI] [PubMed] [Google Scholar]
- Tontsch S, Zach O, Bauer H-C. Identification and localization of M-CoREST (1A13), a mouse homologue of the human transcriptional co-repressor CoREST, in the developing mouse CNS. Mechanisms of Development. 2001;108:165–169. doi: 10.1016/s0925-4773(01)00477-4. [DOI] [PubMed] [Google Scholar]
- Vierbuchen T, Wernig M. Direct lineage conversions: unnatural but useful? Nature Biotechnology. 2011;29:892–907. doi: 10.1038/nbt.1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Oron E, Nelson B, Razis S, Ivanova N. Distinct Lineage Specification Roles for NANOG, OCT4, and SOX2 in Human Embryonic Stem Cells. Cell Stem Cell. 2012;10:440–454. doi: 10.1016/j.stem.2012.02.016. [DOI] [PubMed] [Google Scholar]
- Wernig M, Lengner CJ, Hanna J, Lodato MA, Steine E, Foreman R, Staerk J, Markoulaki S, Jaenisch R. A drug-inducible transgenic system for direct reprogramming of multiple somatic cell types. Nature Biotechnology. 2008;26:916–924. doi: 10.1038/nbt1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P, Wang Y, Chen J, Li H, Kang L, Zhang Y, Chen S, Zhu B, Gao S. RCOR2 Is a Subunit of the LSD1 Complex That Regulates ESC Property and Substitutes for SOX2 in Reprogramming Somatic Cells to Pluripotency. Stem Cells. 2011;29:791–801. doi: 10.1002/stem.634. [DOI] [PubMed] [Google Scholar]
- Young Richard A. Control of the Embryonic Stem Cell State. Cell. 2011;144:940–954. doi: 10.1016/j.cell.2011.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, et al. Induced Pluripotent Stem Cell Lines Derived from Human Somatic Cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- Zeng W, Kong Q, Li C, Mao B. Xenopus RCOR2 (REST corepressor 2) interacts with ZMYND8, which is involved in neural differentiation. Biochemical and Biophysical Research Communications. 2010;394:1024–1029. doi: 10.1016/j.bbrc.2010.03.115. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Yin X, Qin H, Zhu F, Liu H, Yang W, Zhang Q, Xiang C, Hou P, Song Z, et al. Two Supporting Factors Greatly Improve the Efficiency of Human iPSC Generation. Cell Stem Cell. 2008;3:475–479. doi: 10.1016/j.stem.2008.10.002. [DOI] [PubMed] [Google Scholar]
- Zhu S, Li W, Zhou H, Wei W, Ambasudhan R, Lin T, Kim J, Zhang K, Ding S. Reprogramming of Human Primary Somatic Cells by OCT4 and Chemical Compounds. Cell Stem Cell. 2010;7:651–655. doi: 10.1016/j.stem.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.