Abstract
Biocuration is a time-intensive process that involves extraction, transcription, and organization of biological or clinical data from disjointed data sets into a user-friendly database. Curated data is subsequently used primarily for text mining or informatics analysis (bioinformatics, neuroinformatics, health informatics, etc.) and secondarily as a researcher resource. Biocuration is traditionally considered a Ph.D. level task, but a massive shortage of curators to consolidate the ever-mounting biomedical “big data” opens the possibility of utilizing biocuration as a means to mine today’s data while teaching students skill sets they can utilize in any career. By developing a biocuration assembly line of simplified and compartmentalized tasks, we have enabled biocuration to be effectively performed by a hierarchy of undergraduate students. We summarize the necessary physical resources, process for establishing a data path, biocuration workflow, and undergraduate hierarchy of curation, technical, information technology (IT), quality control and managerial positions. We detail the undergraduate application and training processes and give detailed job descriptions for each position on the assembly line. We present case studies of neuropathology curation performed entirely by undergraduates, namely the construction of experimental databases of Amyotrophic Lateral Sclerosis (ALS) transgenic mouse models and clinical data from ALS patient records. Our results reveal undergraduate biocuration is scalable for a group of 8–50+ with relatively minimal required resources. Moreover, with average accuracy rates greater than 98.8%, undergraduate biocurators are equivalently accurate to their professional counterparts. Initial training to be completely proficient at the entry-level takes about five weeks with a minimal student time commitment of four hours/week.
Keywords: biocuration, text mining, database, biomedical informatics, bioinformatics, neuroinformatics, health informatics, data science, lab management, big data, undergraduate research
Defined by the International Society for Biocuration, biocuration involves the translation and integration of information relevant to biology or medicine into a database or resource that enables integration of the scientific literature as well as large data sets. The primary goal of biocuration is to accurately and comprehensively present integrated data as a user-friendly resource for working scientists and as a basis for computational analysis. There has been rapid expansion of published literature, experimental data, electronic clinical records, and continuous health monitoring data. The curation of biomedical data has become a necessity to explore new problem domains using informatics analysis, a field more recently referred to as “big data.” In fact, because of the magnitude and breadth of available data and the development of innovative informatics analysis techniques, biocuration is quickly becoming one of the most invaluable research aids.
Curation of any kind of biomedical data or literature is typically considered a Ph.D. level task due to the integrated level of knowledge required to classify complex information (Burge et al., 2012). While automated tools exist to expedite the process, biocuration currently remains a largely manual and time-consuming task. The level of education and the required manual labor and time are critical reasons why there is a massive shortage of biocurators (Burge et al., 2012). On the other hand, the number of undergraduates wishing to obtain research experience and crucial career-enhancing data analytical skills is nearly unlimited. To this end, we have developed a biocuration process that enables undergraduates to successfully curate biomedical data at accuracy and productivity rates that equal professional curators.
Our approach includes partitioning the complex biocuration process into digestible steps that collectively encompass a serial assembly line, which can be operated by a “small army” of undergraduate biocurators, from 8–50+ per semester. By dispersing the workload, each undergraduate curator works a reasonable but effective four hours/week, thereby preventing burnout. Another important part of undergraduate biocuration success is the creation of an undergraduate lab environment that closely resembles a business structure with a hierarchy of curator, technical, managerial, and administrative positions. Curation is the foundational layer of the hierarchy. However, the business structure creates additional opportunities for undergraduate skill set development for both academic research and traditional industry careers while simultaneously alleviating the managerial time commitment of the primary investigator (PI)/professor.
In this article, we outline the process utilized to initiate and maintain an undergraduate biocuration assembly line. This method is ideally suited to informatics/theoretical labs or primary investigators/programs who wish to establish undergraduate research positions suitable for students headed either into academic research or industry. Smaller-scale versions could also be envisioned in wet/experimental/clinical labs that have their own large data sources that they wish to curate. Compared to many research projects, biocuration requires very little start-up financial or physical resources while still offering amazing career-transforming opportunity for undergraduates. Moreover, the developed curated database products and enabled informatics analysis is invaluable to biomedical data science.
Because our lab’s primary area of expertise is computational analysis of neurophysiology and neuropathology, all of our biocuration has focused on neuropathology of disease (Amyotrophic Lateral Sclerosis, Alzheimer’s Disease, Spinal Cord Injury). However, this method is adaptable to any experimental (in vitro, in vivo, physiology or pathology) or clinical data set.
METHODS
Biocuration involves first establishing a data path as well as developing a human workflow. Below we summarize the required resources and the data path process and go into detail on how an assembly line of curators and hierarchical undergraduate research positions can be utilized as a “undergraduate curation corporation.”
Required resources
Other than a data source and eager undergraduate students, the physical resources needed include basic computers with standard office productivity software, database software, a database server, and a secure local area network from which the database server runs such that users can simultaneously enter data into a remote database. A quiet purpose-specific environment is preferable. Security of the database and the environment must be assessed depending on the type of data being curated (e.g., clinical data curation requires many more security protections compared to experimental data). For our ALS informatics project, curation was done on project-specific computers in a room with a closed door with multiple layers of environmental (controlled access) and computational security (multiple logins, firewalls, etc.). Resources required for automated and/or manual database back up must also be in place.
Establishing a data path
Establishing the data path includes instituting the process from accessing the data to database development to curating the data to the ability to analyze the data. Note that, while initially serial, a data path is an iterative loop. That is, there are always more refinements as the curation project progresses. Bicourators of different sub-fields each have their own personal preferences for establishing a data path. Here we summarize the activities for establishing a data path that we believe work best for undergraduates.
Data source. The first step to developing undergraduate biocuration is to establish one or more large data sources to begin curating. Data sources could be clinical medical records, unpublished experimental data, or published experimental data. When establishing a data source, simply keep in mind that the point of biocuration is to pool and organize data sets into a form that is both useable to other researchers and can be used to perform bioinformatics analysis. Also, determine that all protocols for permission and approval to data access (e.g., Internal Review Board for clinical data, etc.) are in place.
Data pool. The next step is to establish what parts of the data source will be curated. Being as inclusive as possible is usually the best option. More metrics allow more analytical options and more complex research questions to be pursued. Quantitative metrics are the most preferred, but parametric data is also valuable. Any data that has pervasive commonality (e.g., a survey that all patients have in common) or can be standardized or normalized (e.g., a biomedical experiment that includes a wild type or control) should definitely be included.
Alpha curation. Before the development of a database, a group of alpha testers employ individualized manual methods to collect entries from the data pool. Collecting data in a database program or even a simple spreadsheet is acceptable. Having multiple testers helps to include multiple points of view. These test curators, with the PI, establish the data fields and the initial anticipated workflow.
Establish database. Based on the alpha curation, a relational database is created that allows entry, review, and easy export of curated data for statistical analysis. We use the Filemaker (Filemaker, Inc.) database software, because, in our experience, it is more friendly and intuitive for novice database users, both in terms of layouts (graphical user interfaces) and in scripting language. However, Access (Microsoft, Inc.) or any other preferred database platform could be utilized.
Beta curation. Before the database is released for official curation by a large group, beta testers not involved in the construction of the database use the database to identify bugs, suggest edits in design, or suggest addition of automation/user tools to reduce error. The database is refined based on beta curator input.
Workflow. The order in which the data is curated and how it is formatted must be explicitly specified. The development of written protocols is key to insure data homogeneity and integrity.
Primary curation. Primary curation is the bulk of curation performed as part of the main project or database construction.
Data quality control. Establishing manual and automated data quality control procedures is critical for insuring data integrity. These procedures will vary slightly depending on the curated data type. More detail is given in the Quality Control section under Positions.
Classification. After a substantive amount of primary curation, classifications (such as functional ontologies) can be imposed to assist in searching, aggregating, and analyzing data. We recommend designing for both a universal ontology, to be used by any database user, and user-specified ontologies, to be used and customized by specific tech teams (see Positions) as part of their analysis.
Prioritization. For very large biocuration projects, developing a prioritization scheme can be helpful to insuring that the most important or time-sensitive data or data relevant to the pilot projects’ goals is curated first.
Meta-data analysis. Meta-data simply means “data on the data.” Meta-data of curated data in the database is used to determine sample sizes, which specify the feasibility of a pilot project’s goals.
Feasibility study. A feasibility study is the process of evaluating potential pilot project alternatives based on meta-data, scientific or clinical significance or user need.
Pilot project. A pilot project is a research project that utilizes the curated database for exploratory or hypothesis driven research or to develop a product. Depending on available sample sizes and timelines, it may or may not be a full-size study. Some pilot projects can develop into full-size studies once proof-of-concept has been obtained.
Applicants
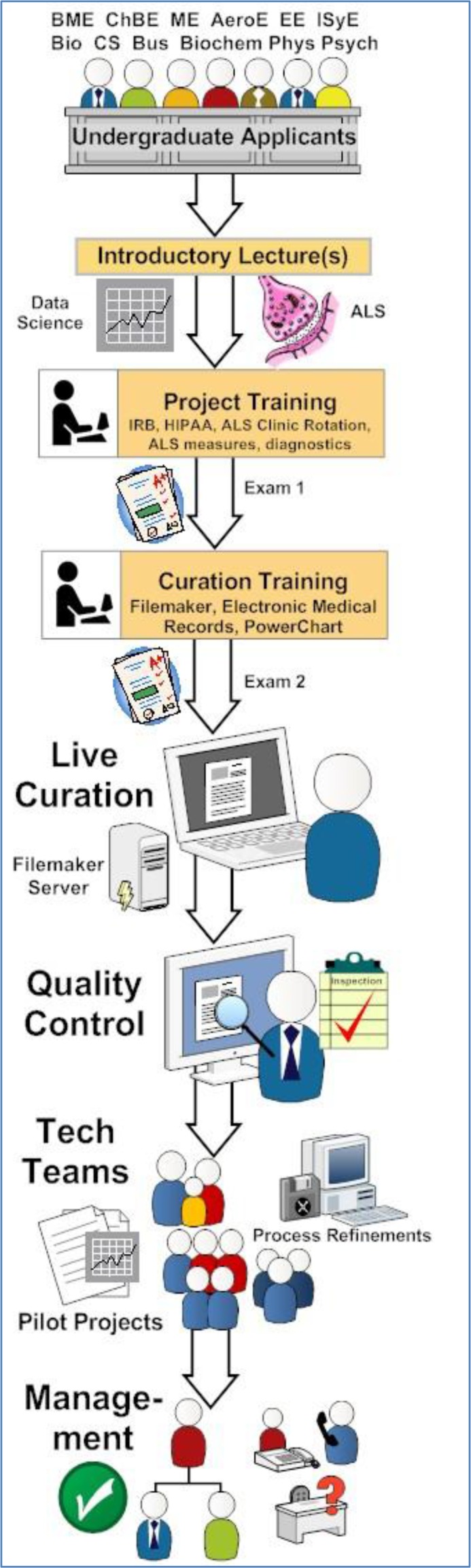
Applicants range from high school juniors through senior undergraduate students. As shown in Figure 1, our applicants come from a variety of majors, including all different types of engineering, biological science, chemisty, computer science, business/finance majors, and many others. Irrespective of major, most applicants seek us out due to their perceived interest in neuroscience or medicine. Because we are in the biomedical engineering (BME) department, the majority of applicants are BME majors. Many of the BME majors and especially the non-BME majors have a pre-health track, which means they have had requisite exposure to basic biology knowledge and quantitative skills necessary for curation. Applicants complete our lab’s official application and submit a one-page resume. The application is a combination of questions, including questions to assess personality, attention to detail, interests/extracurricular activities, classwork, special skill sets, future career plans, and perhaps most importantly, a paragraph written by the applicant stating why they desire a position. An in-person interview by the PI is utilized to assess intangible qualities.
Figure 1.
Application, training, and job positions. ALS clinical informatics project used as a detailed example.
Unlike many labs seeking undergraduates, we don’t simply target “the crème de la crème” with the highest GPA, etc. In fact, we have found that GPA is not even a good predictor of success in a biocuration and informatics environment. Desire, commitment, consistency, attention to detail, and fundamental biological and quantitative knowledge (e.g., how to read a graph or an experimental protocol, etc.) appear to be the essential basic applicant qualities at the entry-level. Every person starts at the same position so the students sort themselves out in the hierarchy.
Initial Training
As shown in Figure 1, there are three phases of initial training: introductory lectures, project training and curation training. Introductory lectures are classroom-style teaching sessions where associates are given a primer on biocuration, an overview of basic neuroscience, and fundamental scientific and/or clinical knowledge on the topic/condition/pathology being assessed. Interactive or hands-on project training teaches the applicants everything they specifically need to know about the project, especially the types of measures being curated and their definitions, and any ethical or research protocols for handling the data. For example, most of our clinical ALS informatics project associates had at least a one-day rotation at an ALS Clinic to help them better understand the information that was being curated in the chart. Interactive or hands-on curation training teaches the applicant everything they need to know in terms of data transcription—how to access the data, how to use the database software, how the database is organized, and ultimately how to find/search, view, edit and add new data
Exams testing concept recall immediately follow each training session. The curation training session also has a practical exam where students curate specified data into their own individual practice database for a period of one week. A score of 95% is required to pass the practical exam. Individualized tutoring and up to two retakes are allowed before dismissal. Upon passing the practical, students undergo a competency assessment, which is a four-week period of live curation in the actual project database. If the associate shows good promise in meeting the quality control productivity and accuracy standards and has consistently worked their specified schedule, they are finally offered a contracted position.
Positions
Curators: Curation is our foundational and entry task/position. Curators collect/transcribe data and enter it into the project-specific database. Whether an associate is a senior in high school or a senior in college, he/she will begin as a curator. The amount of expertise required for curation will vary by project. For very complex data paths and/or to lower the experience barrier for applicants, curation can be divided into different “levels” that form a serial assembly line. Curators are promoted to each subsequent level as they gain the necessary experience and training and prove their desire and skill. For example, our SOD1 G93A ALS transgenic mouse informatics projects has multiple levels of curators: 1) document capture—Level 1 associates access and save the data files (in this case, published articles) in an organized, appropriate format for subsequent transcription; 2) figure capture—associates transcribe figure captions by panel. 3) data series capture—associates transcribe noninterpretative experimental parameters names, experimental methods, mouse genotypes, etc.; 4) response value capture—associates transcribe quantitative data, which may or may not require subjective interpretation by reading data from a figure, etc.; 5) informatics capture—capturing additional project-specific data, applying organization or ontological schemes, or even doing meta-data analysis. In essence, these curators serve as consultants to the technical teams.
Despite detailed initial training, there are alwa ys questions that occasionally surface in regards to specific entries. We developed a “curator que” that uses a freely available online interactive classroom website (Piazza), which we have adapted for posting questions/answers, reminders, and tips. Quality control managers respond to posted questions within 24 hours.
Most curators work a reasonable but effective 4–6 hours/week. This amount of workload is perfect for associates who are assessing their interest. Moreover, in an assessment of curator workload, we have determined that 4–6 hours/week results in optimal productivity and accuracy. Curation is a very detail-oriented task, and this must be considered when determining the workload (hours worked and required productivity). In fact, we prefer that associates work in multiple two-hour intervals versus a continuous four or six hours. Online scheduling is utilized to manage computer access, which optimizes computer resource requirements and insures curators always have a computer when they come in to the lab to work their scheduled time periods. Our typical curator tenure is about three semesters, with a percentage of students being promoted to tech teams at the end of semester two. However, many students work as a curator much longer than the average simply because they enjoy the task or the time required fits their schedule while still satisfying their desire to participate in research.
Quality Control (QC): Just like any manufacturing process or company, the purpose of quality control is to ensure that the product (i.e., the database) and performed service (i.e., biocuration) adheres to a defined set of quality criteria. Our QC personnel quantify curator productivity and accuracy, and provide weekly feedback and required corrections. Quality control personnel are typically highly skilled former curators who wish to obtain real-world management experience. As such, many have aspirations to do industry research and development project management. Most QC personnel work eight hours/week, although the minimum is four hours/week.
A written quality control protocol specifies how automated script checks and visual inspection of entries should proceed for each data source and curation type. This insures consistency among QC personnel. The lead QC manager also monitors the efficacy and consistency of QC personnel via random visual inspection and automated statistical analysis. The training of the quality control team is similar to the biocuration training: it consists of a hands-on workshop, a concept exam testing QC protocol knowledge, and a competency exam assessing their performance of entries. The QC personnel to biocurator ratio ranges from 1:5 to 1:10, depending on the degree of complexity of the curation and the amount of available automation as part of the QC process.
The quality control process, itself, consists of the QC personnel using a combination of automation and visual inspection to check curated entry accuracy and curator productivity. For every single entry in the curated database, there is an associated scribe ID, creation time stamp, and modified time stamp so that we can track when the entry was made, modified, and who completed it. Each week, quality control personnel are randomly assigned a list of associates’ work to check. Automation works well for finding most missing entries or fields, typos/mis-spellings, assessing qualitative entries, and checking for formatting. Automation also works well for calculating productivity (entries completed over a specific time period). However, to determine if quantitative data is entered correctly or if all possible data has been gathered from the data source, QC personnel must visually inspect and compare the curator’s entry to the data source. The QC personnel complete a feedback form for each curator detailing their mistakes and positively commenting about good performances (e.g., accurately entering a very complex figure, etc.) as appropriate. The feedback cites the database field keys for each entry that requires modification. The curators have one week to fix identified mistakes before re-checking by QC personnel.
For each curation level, there is an associated requirement for productivity and accuracy. Prior to corrections, we set the required curator weekly accuracy standard to be 97.5%. This rate was based on accuracy standards referenced by other academic and industrial curation projects using Ph.D. biocurators (see Results). Nonetheless, our actual accuracy rate prior to correction is 98.8%. Productivity requirements vary by curation level and project. We utilize beta tests during data process development to determine productivity standards. The general rule of thumb is that the productivity standard is 2 standard deviations below the beta test average. This method ensures that the productivity standard provides ample motivation to be efficient but is also sufficiently low so as to not rush or penalize a curator for doing a thorough job or taking a little time to learn about data along the way.
A point-based system is used to track productivity and accuracy for both penalization and reward. Penalty points for not meeting quality control standards are used for performance grading and determining readiness of promotion (see Performance Review).
Technical Teams: About 60% of our curators go on to join a technical or “tech” team. Tech teams consist of 2–5 students who perform small pilot data science projects using curated data. Projects may be exploratory or hypothesis-driven. All teams start by assessing meta-data and creating any necessary classification or ontologies. Teams are typically paired with one or two curator consultants to expedite getting the data in the form the team needs for their specific project. In our lab, exploratory studies utilize complex systems-based techniques to identify relationships within the data and testable hypotheses. Traditional hypothesis-driven studies typically consist of large-scale meta-analyses or the use of standard statistics to answer a specific question. Tech teams, on average, require three semesters to complete a published article although we have had several extremely motivated teams submit an article or conference proceeding after two semesters. A few teams also develop data science tools to improve and automate our curation process.
Tech team project penultimate deliverables or outcomes can vary based on both the PI’s desires (ongoing projects and deadlines) and the team members’ desires (long-term career goals like authoring an article in preparation for graduate school, development of specific skills for industry, planned tenure in the lab, school schedule, etc.). Typically, we have had the best success if the penultimate deliverable was a useable product/tool by the lab or public, a published conference proceeding, or a peer-reviewed journal article. Such deliverables encourage and externally motivate not only the tech teams, but also the curators. Some continuity within teams (i.e., one or more team members seeing the tech team project to completion) is preferred but is not necessarily a requirement with proper documentation and, if possible, a hand-off or transition period that includes start-up training and/or an apprenticeship by at least one of the new team’s members.
The primary investigator provides one or more broad topics, questions, or product goals from which the team can pursue to choose. However, it is up to the tech team to shape the chosen path into successful project. A senior-level experienced undergraduate technical team manager handles day-to-day management of tech teams, which have pre-set weekly intermediate deliverables set by the PI. Initially, tech teams are given tasks that include project-specific curation combined with intense literature searches or the equivalent thereof to become proficient on the background of their project’s topic. Common technical skills (how to use reference software, export data, make a figure, scientific writing, etc.) are taught in sessions led by tech team managers. After ascertaining a knowledge background, the focus then shifts to activities beyond the primary curation data process (see Establishing the data process). The first semester concludes with a feasibility study assessing pilot project alternatives, one of which the tech team ultimately pursues in the next semester.
After semester one, a tech team is analogous to a graduate student fulfilling his/her research proposal. Tech team managers are still available to teach specific common statistical/analytical skills (meta-analysis, ANOVA, cox proportional hazards, Kaplan-Meir, etc.) and to oversee day-to-day productivity and intermediate deliverables. However, the PI/professor oversees the tech team project direction as the research becomes more project-specific, especially during the construction and formatting of the penultimate deliverable.
Information Technology (IT) Team. The IT team is really just another specialized form of a tech team whose focus is on enhancing IT aspects of biocuration rather than performing analytical informatics on a particular research question. Thus, some of our associates with high-end computer skills or IT-related career plans choose to join our IT team instead of a traditional research tech team. The IT team refines the database and develops automation to enhance curation workflow, quality control, and pilot project informatics analysis. They may also serve as consultants to tech teams by assisting in writing scripts or other programming tasks. Finally, the IT team is responsible for all IT maintenance, including weekly backups, login accounts, servers, hardware and software.
Management. As noted in the above sections, there are undergraduate managers for curation quality control, IT, and tech teams. Technical team managers, who are senior-level students, oversee the daily activities of the technical teams; lead skills training sessions (using reference software, making figures, scientific writing tips) and provide consulting for statistical and informatics analysis. QC managers direct the curator que (online system for asking curation questions), maintain our training and QC protocols, and oversee the curator and quality control personnel master schedules. Our IT managers coordinate and oversee all of the IT team’s activities.
Our managers are mostly advanced students that have been with us for the majority of their undergraduate tenure. Minimally, they must have completed all levels of curation. In fact, QC or IT managers are only required to have previously completed curation. However, undergraduate tech team managers must have completed curation and at least one and preferably two semesters as a tech team member. Managers may also be technicians or graduate students. The commonality among all of the managers is that they oversee the day-to-day activity of their core group and the submission of weekly and monthly productivity reports. They are also the primary point-of-contact to the PI/professor. Managers are mostly trained through an apprenticeship system, although our lab does have specific written protocols in place regarding the responsibilities of each managerial role.
Performance Review
As noted in the quality control section, a point-based infraction system is used to grade curator accuracy and productivity. A similar system is used to grade technical team and manager productivity via the use of weekly or biweekly intermediate deliverables. All students are given contracts at the beginning of each semester that outline the expectations for their position(s) and the equivalent points which translate to a specific performance grade. Students are given weekly feedback on their performance, so there are no surprises. Students must maintain a “B” to have their contract renewed the following semester. Consistently poor performance during a semester results in demotion to a lesser position (for example, a Level 2 curator may be demoted to Level 1). If productivity standards for the demoted position are not met, the student’s contract is ultimately rescinded. A total of three communicated warnings precede dismissal. Dismissal is done in an in-person meeting with the PI.
On a more positive note, Biocurator-of-the-Month awards, which include a small prize and a publicly displayed certificate on the lab door, are given to the most productive (e.g., entries/hour) biocurator(s) who also did not incur penalty points for excessive errors. Analogously, technical-associate-of-the-month awards recognize outstanding performance by tech team members or managers.
Compensation
During the initial training and competency assessment period (approximately six weeks), students are uncompensated. For their first three semesters with a position, most students take optional research course credit at the rate of 3–4 working hours per credit hour, depending on position type. For subsequent semesters, managers and exceptional technical team members may receive hourly pay if PI funds are available. Many of the exceptional student researchers apply for and receive independent research funding through the university’s undergraduate research program (Georgia Tech President’s Undergraduate Research Award) or other outside similar programs. In general, curators are not given authorship on articles, although in special cases, Level 5 curators who serve as consultants may be considered. Authorship is generally reserved for tech team members, and is discussed as part of the initial tech team establishment.
Primary Investigator Commitment
As one might expect, leading an “undergraduate biocuration corporation” can be a substantive time commitment to the primary investigator. Nonetheless, the commitment is manageable with PI scheduling forethought. The greatest amount of time is spent with the initial set-up (writing of the curation and management protocols, determining data sources and possible technical project topics, and interviewing/training the first batch of students). This phase temporarily requires a full-time commitment by the PI; for example, a summer semester might be an ideal time to start a program. After initial set-up, the steady-state operation of the overall program is most directly proportional to the number of advanced technical teams, which require the most input on behalf of the PI. As a point of reference, approximately two advanced technical teams, properly and personally mentored by the PI, take the equivalent PI time investment as a full-time graduate student. PI oversight of data source biocuration fluctuates as a function of project phase and especially the undergraduate quality control management experience. On average, eight fully trained and mentored curators require approximately the same PI time commitment as one full-time graduate student.
RESULTS AND DISCUSSION
We have developed our process using three different experimental disease models: spinal cord injury (Mitchell and Lee, 2008), ALS (e.g., Mitchell and Lee, 2012; Irvin et al., 2015; Kim et al., 2015; Mitchell et al., 2015; Coan and Mitchell, 2015; Pfohl et al., 2015), and Alzheimer’s disease (Foley et al., 2015). As part of our biocuration protocol and database development, we have utilized over 350 different undergraduate curators and 10 high school curators. One of the major pros of an undergraduate biocuration program is that it is scalable. We started with a team of three alpha curators. We went through four major scale-ups, about one per year. We currently maintain a total team of 50+ per semester, which includes about 30–40 primary curators and 15–20 tech team members and managers. Based on our experience, to run and easily maintain a true curation assembly line with both curators and full-time quality control, at least eight students are necessary. The addition of a couple of tech/IT teams and managers brings the minimum total for a “curation corporation” to be around 15 students. The maximum number of students is simply a function of student interest, physical and data resources, and of course PI/professor time.
Case Study: Clinical health informatics
By curating ALS Clinic medical records, we developed a novel clinical ALS database, which consists of over 300 different quantitative and qualitative measures, including pre-ALS health, ALS progression metrics, clinical treatments, diagnostic tests, and autopsy reports. The pilot project included 300 patients, and the completed database includes an astounding 1,587 patients, the largest and most comprehensive ALS data set available to date. Such databases make way for epidemiological studies of demographics, disease progression and treatment. For example, our resultant comprehensive assessment of antecedent disease, which found that ALS patients have substantially less other disease compared to matched non-ALS controls, has resulted in novel hypotheses regarding possible neuroprotective mechanisms (Mitchell et al., 2015). Other examples of published related tech team projects include the identification of novel autopsy pathological marker relationships (Coan and Mitchell, 2015). Currently, an additional five tech teams have ongoing projects utilizing the clinical ALS database.
Case Study: Experimental model informatics
Our largest database is our ALS transgenic mouse database, which curates quantitative data from 3,500+ articles and 35,000+ figure panels into ∼50,000 different metrics and treatments assessed over 160,000 time points. Since the inception of tech teams a couple of years ago, 12 ALS tech teams and two AD tech teams have published six peer-reviewed journal articles and eight conference proceedings to date. Numerous other articles are in review or in preparation. Examples of hypothesis-driven tech team projects include: meta-analysis examining the relationship between amyloid beta and mouse cognition (Foley et al., 2015) and sex-dependent progression patterns in SOD1 G93A mice (Pfohl et al., 2015). Examples of exploratory data science tech team projects include: informatics-based analysis of the SOD1 G93A field topics to develop a functional ontology (Kim et al., 2015) and assessing novel homeostatic instabilities in ALS metabolism (Irvin et al., 2015). Finally, the curated products (i.e., the databases), themselves, are an invaluable researcher resource. Our first release of the searchable SOD1 G93A ALS mouse figure database is available on our website: http://www.pathology-dynamics.org. Ongoing work continues.
Undergraduate curators are productive
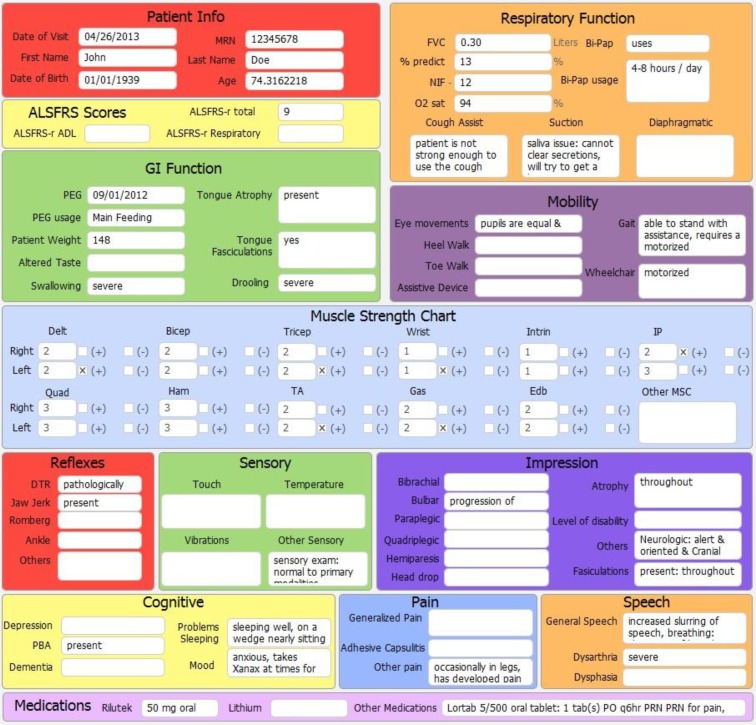
Eager undergraduates are very productive. Because curation consists of much more than copy and paste transcription, the required capacity and opportunity to learn about the topic being curated maintains interest. The quantified productivity of biocuration obviously depends on the complexity of the data being curated. As a reference, an undergraduate curator reading through paragraphs of unorganized dictated text from medical records of standard clinic visits can transcribe, on average, about 10 layouts per hour of about the size and complexity shown in Figure 2 (ALS Clinical Informatics layout).
Figure 2.
Example curation data entry layout from ALS clinical informatics project. This is one of the four data entry layouts used as part of our ALS clinical informatics project. The layout above shows some of the parametric and non-parametric data that is recorded during a standard ALS clinic patient visit. Additional separate layouts (not shown) exist for cognitive testing, patient history (medical and family history, onset symptom timeline, diagnostic and genetic testing), and autopsy and pathological reports. If no data was present in the medical record or survey for a particular field, the field is simply left blank. Note that patient name and MRN fields are only shown for reference as to how data is obtained from data source; curated data is ultimately de-identified to protect patient privacy.
For an experimental data capture project, Table 1 illustrates the curation rates for different levels of data capture from published primary experimental data articles (SOD1 G93A ALS mouse). The actual rate of productivity varies by the amount and type of data in each article. Generally, capturing all qualitative and quantitative data from an article takes less than two hours.
Table 1.
Biocuration capture type descriptions and entry times for published experimental model (e.g., SOD1 G93A ALS mouse).
| Capture Type | Average Time (per article) | Curation Description |
|---|---|---|
| Document | 2 to 3 min. | Download full-text PDF document from GT library or PubMed Central |
| Figure | 10 to 25 min. | Extract figure/table captions, panel description, experimental type |
| Data Series | 30 to 60 min. | Extract data series types: mouse strain & attributes, treatments, significance |
| Response Values | 60 to 120 min. | Extract experimental parameters & numeric data from quantifiable figures |
| Full Capture (total) | 100 to 200 min. | All of the above. |
Undergraduate curators are accurate
Our biocuration accuracy requirement is 97.5% and is based on published tolerance of error in similar projects by professional biocurators (Keseler et al., 2014; Wu et al., 2014a; Wu et al., 2014b). However, for our SOD1 G93A transgenic ALS mouse experimental database, which curates published in vivo and in vitro data, our actual accuracy (based on 4+ years of biocuration quality control calculations) is an astounding 98.8% with a per-semester standard deviation of 0.2%.
Of the average 1.2% of entries recognized by quality control as erroneous, only 10.5% are classified as “critical errors,” which meaningfully compromise the integrity of the data. Thus, our effective accuracy is 99.9%. Discounting the fact that the ALS clinical informatics database does not require reading quantitative values from graphs, the accuracy of biocuration in that database is very comparable.
Our average error rate of 1.2% is in line with other similar databases that employ professional curators. For example, the Candida Genome Database (CGD) and the EcoCyc Escherichia Coli Database employ manual biocurators that are Ph.D. biologists. The CGD has an overall error rate average of 1.82% (Keseler et al., 2014). The EcoCyc Escherichia coli database has an overall error rate average of 1.40% (Keseler et al., 2013). Thus, utilizing a curation assembly line, undergraduates are very capable of doing professional quality biocuration.
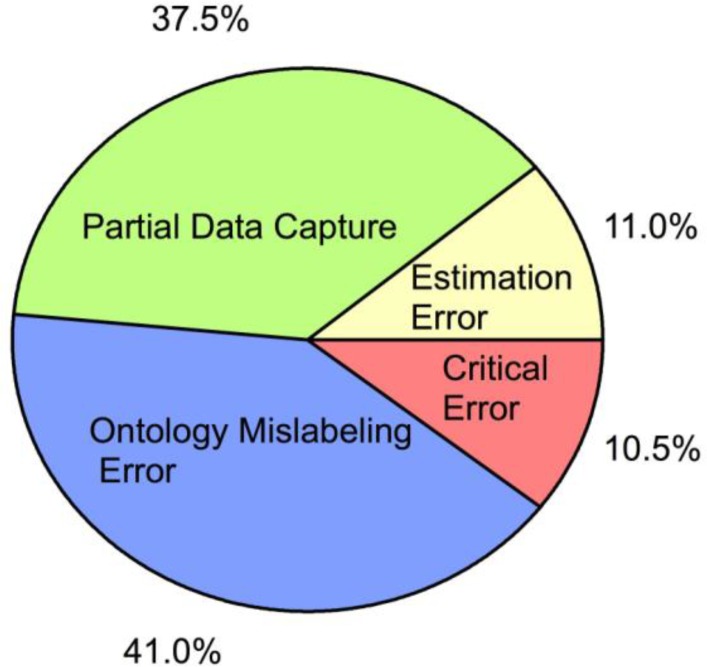
Figure 3 shows the breakdown of error types for the SOD1 G93A ALS mouse database for full data capture (aggregated curation for levels, 1–4). The largest percentage of errors is ontological labeling errors (the placement of tags to make finding data easier). Labeling of ontological terms requires the most knowledge of the SOD1 G93A ALS pathophysiology. Ontological labeling is also the only subjective or interpretative entry in the database. Thus, it is not surprising that ontological labeling has the highest error rate. Fortunately, ontological labeling errors do not in any way affect the integrity of the curated data, itself. Partial data capture errors are the second-most common error. Finally, about 11% of all errors are estimation errors, which are quantitative interpretative errors from reading a graph. Almost all critical errors, which affect data integrity, are the direct result of over- or under-estimation of quantitative values.
Figure 3.
Pie chart illustrating curator error types for Full Data Capture (Curation Levels 1–4, see curation section in Positions) for our SOD1 G93A transgenic mouse ALS experimental database as identified by quality control personnel. On average, trained curators commit errors on less than 1.2% of their entries with a standard deviation of ± 1%. The pie chart represents the breakdown of error type of this 1.2% of total errors. Partial Data Capture (green): curator fails to collect all data from a figure or table. For example, trendline data was recaptured only for G93A and not for G93A + treatment. Ontology mislabeling error (blue): curator assigns response value entry to wrong ontological classification. Estimation error (yellow): captured data point value (typically from a graph) is visually estimated as greater than ± 5% from the actual value. Note that value estimation off by more than 10% is defined as a critical error. Critical error (red): incorrectly entered data that compromises data integrity.
Automation and Future Directions
Recently, we have been utilizing automated ontological scripts to assign ontological tags established on the presence of certain key words in relevant fields. Based on one semester of data, this method appears to be substantively reducing this error type. To reduce partial data capture errors, we have also recently added automated field checks, which appear to be very effective in reminding curators to fill in required fields. We are also in the process of testing freeware to estimate values in-between tick marks on graphs embedded in pdf files. Finally, automation is also being employed to calculate productivity using computerized time stamps. Our IT team continues to pursue projects to enhance automation in the entire data path process, including curation, quality control, and analysis.
Biocuration enhances career opportunities
One of the major benefits of biocuration in comparison to more traditional undergraduate research opportunities is that it opens up research to a greater number of students with more varied skill and/or career desires. Curation teaches basic data organization and analytical skills necessary for any career. It also serves as an equalizer, giving all students uniform opportunity to climb the hierarchy. Technical teams favor students that intend to pursue graduate school, academia or research-focused jobs. Management and IT tend to favor students headed into industry or project management. Formal end-of-semester forms are used to track the students’ undergraduate research and post-graduate career plans throughout their undergraduate tenure in the lab. Additionally, informal exit surveys are utilized to track postgraduation positions and acceptance. Based on this data for about 350 students, we have determined that a tenure of three or more semesters was analogous to a 0.4-point GPA boost in the very competitive biomedical engineering industry, including student co-op/internships and postgraduation job offers. A management position is analogous to 0.6-point GPA boost for biomedical industry, and about one-third of our multi-semester managers were offered industry project management positions. To date, 80% of students who authored a research publication or proceeding and applied to graduate school or professional school have been admitted.
Conclusions
Undergraduate biocuration can be successfully utilized to develop large, powerful databases and analyze corresponding informatics data. Undergraduate biocurators using the assembly line curation method described have accuracy and productivity comparable to professional Ph.D. biocurators. Moreover, biocuration provides invaluable research experience to a broader population of students who may not otherwise obtain a research position or hands-on experience. Because of the breadth of positions involved in biocuration, it utilizes many different skill sets which are applicable to both research and industry jobs.
Footnotes
This work was supported by National Institute of Health grants (NS081426 and NS069616) to CSM.
REFERENCES
- Burge S, Attwood TK, Bateman A, Berardini TZ, Cherry M, O'donovan C, Xenarios L, Gaudet P. Database. Oxford: 2012. 2012. Biocurators and biocuration: surveying the 21st century challenges; p. bar059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coan G, Mitchell CS. An assessment of possible neuropathology and clinical relationships in 46 sporadic amyotrophic lateral sclerosis patient autopsies. Neurodegener Dis. 2015;15:301–312. doi: 10.1159/000433581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley AM, Ammar ZM, Lee RH, Mitchell CS. Systematic review of the relationship between amyloid-beta levels and measures of transgenic mouse cognitive deficit in Alzheimer’s disease. J Alzheimers Dis. 2015;44:787–795. doi: 10.3233/JAD-142208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irvin CW, Kim RB, Mitchell CS. Seeking homeostasis: temporal trends in respiration, oxidation, and calcium in the SOD1 G93A Amyotrohpic Lateral Sclerosis mouse. Front Cell Neurosci. 2015;9:248. doi: 10.3389/fncel.2015.00248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keseler IM, Mackie A, Peralta-Gil M, Santos-Zavaleta A, Gama-Castro S, Bonavides-Martinez C, Fulcher C, Huerta AM, Kothari A, Krummenacker M, Latendresse M, Muniz-Rascado L, Ong Q, Paley S, Schroder I, Shearer AG, Subhraveti P, Travers M, Weerasinghe D, Weiss V, Collado-Vides J, Gunsalus RP, Paulsen I, Karp PD. EcoCyc: fusing model organism databases with systems biology. Nucleic Acids Res. 2013;41:D605–612. doi: 10.1093/nar/gks1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keseler IM, Skrzypek M, Weerasinghe D, Chen AY, Fulcher C, Li GW, Lemmer KC, Mladinich KM, Chow ED, Sherlock G, Karp PD. Database. Oxford: 2014. 2014. Curation accuracy of model organism databases; p. bau058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim RB, Irvin CW, Tilva KR, Mitchell CS. State of the field: an informatics-based systematic review of the SOD1-G93A amyotrophic lateral sclerosis transgenic mouse model. Amyotroph Lateral Scler Frontotemporal Degener. 2015;2015:1–14. doi: 10.3109/21678421.2015.1047455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell CS, Hollinger SK, Goswami SD, Polak MA, Lee RH, Glass JD. Antecedent disease is less prevalent in amyotrophic lateral sclerosis. Neurodegener Dis. 2015;15:109–113. doi: 10.1159/000369812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell CS, Lee RH. Pathology dynamics predict spinal cord injury therapeutic success. J Neurotrauma. 2008;25:1483–1497. doi: 10.1089/neu.2008.0658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell CS, Lee RH. Dynamic meta-analysis as a therapeutic prediction tool for amyotrophic lateral sclerosis. In: Maurer M, editor. Amyotrophic lateral sclerosis. InTech; 2012. pp. 59–80. www.intechopen.com/books/amyotrophic-lateral-sclerosis/dynamic-metaanalysis-as-a-therapeutic-prediction-tool-foramyotrophic-lateral-sclerosis. [Google Scholar]
- Pfohl S, Halicek M, Mitchell CS. Characterization of genetic background and sex on disease progression in the SOD1 G93A transgenic Amyotrophic Lateral Sclerosis mouse model: a meta-analysis. J Neuromuscular Dis. 2015;2:137–150. doi: 10.3233/JND-140068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu HY, Chiang CW, Li L. Text mining for drug-drug interaction. Methods Mol Biol. 2014a;1159:47–75. doi: 10.1007/978-1-4939-0709-0_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu TJ, Shamsaddini A, Pan Y, Smith K, Crichton DJ, Simonyan V, Mazumder R. Database. Oxford: 2014b. 2014. A framework for organizing cancer-related variations from existing databases, publications and NGS data using a High-performance Integrated Virtual Environment (HIVE) p. bau022. [DOI] [PMC free article] [PubMed] [Google Scholar]