Abstract
Background
The human heart consists of several cell types with distinct lineage origins. Interactions between these cardiac progenitors are very important for heart formation. The muscle segment homeobox gene family plays a key role in the cell morphogenesis and growth, controlled cellular proliferation, differentiation, and apoptosis, but the relationships between the genetic abnormalities and CHD phenotypes still remain largely unknown. The aim of this work was to evaluate variations in MSX1 and MSX2 for their possible associations with CHD.
Methods
We sequenced the MSX1 and MSX2 genes for 300 Chinese Han CHD patients and 400 normal controls and identified the variations. The statistical analyses were conducted using Chi-Square Tests as implemented in SPSS (version 19.0). The Hardy-Weinberg equilibrium test of the population was carried out using the online software OEGE.
Results
Six variations rs4647952, rs2048152, rs4242182, rs61739543, rs111542301 and rs3087539 were identified in the MSX2 gene, but the genetic heterozygosity of those SNPs was very low. In contrast, the genetic heterozygosity of two variations rs3821949 near the 5’UTR and rs12532 within 3’UTR of the MSX1 gene was considerably high. Statistical analyses showed that rs3821949 and rs12532 were associated with the risk of CHD (specifically VSD).
Conclusions
The SNPs rs3821949 and rs12532 in the MSX1 gene were associated with CHD in Chinese Han populations.
Introduction
Congenital heart diseases (CHD) are a group of complex congenital anatomic malformations worldwide with high morbidity and mortality. The incidence of the illness is about 7.5% in newborns [1] and 1% of the patients required clinical intervention [2]. There are numerous types of this disease, including ventriculap septal defect, pulmonary stenosis, tetralogy of Fallot, patent ductus arteriosus, mitral valve insufficiency, etc. [3], which are often complicated with arrhythmias and heart failure [4]. So far, many gene mutations and chromosomal variants have been identified in familiar and sporadic CHD cases [5–7]. However, the relationships between those genetic abnormalities and CHD phenotypes still remain largely unknown.
The mammalian heart is a complex and also one of the first formed organs during embryogenesis [3], and the formation is strictly regulated and controlled by gene regulatory networks, consisting of signaling pathways, transcription factors, epigenetic factors and miRNAs [8, 9]. Among the regulatory networks, the Nodal/TGF-βsignaling pathway has a key role in early stages of human embryonic stem (HES) cell differentiation, directing the cells to develop into different embryonic lineages. Any malfunctions in the pathway may lead to errors in the transformation of the embryonic lineages [10–12]. For example, defects in the transforming growth factors LEFTY in the Nodal/TGF-β signaling pathway may affect the signaling of NODAL and TGF-β[3, 13]. In a previous study, we found that single nucleotide polymorphisms (SNPs) of the Lefty genes are associated with the risk of CHD [3]. The Nodal gene in the Nodal/TGF-β signaling pathway can initiate a series of signal transduction events in the later stages of embryonic development [13, 14]. However, no variations in the Nodal gene so far have been associated with the risk of CHD [3]. Therefore, it is not clear whether it is the Nodal/TGF-β signaling pathway or only Lefty that is associated with the risk of CHD. Additionally, SMAD3, an intracellular regulating factor in the Nodal/TGF-β signaling pathway to modulate the transcription of many genes [15, 16], together with LEFTY plays central roles in the signaling pathway [15, 17]. Our previous work has demonstrated that the variant rs2289263 before 5’UTR of the SMAD3 gene is associated with increased risk of VSD in the Chinese Han population [18]. Additionally, as the process of HES cell differentiation during embryonic development is very important for the heart development, it may also be involved in the pathogenesis of CHDs.
During embryonic development, HES cells differentiate to various cell types of ectoderm, endoderm and mesoderm, and the cardiomyocytes are generated and differentiated in the mesoderm [19]. As the heart consists of several cell types with distinct lineage origins [20], such as myocardium cells, cardiac neural crest (NC) cells, aorticopulmonary septum cells and membranous ventricular septum cells, etc. [21], interactions between these cardiac progenitors are very important for the cardiac development, and any mistakes may result in congenital heart malformations [1]. The muscle segment homeobox gene family is an important transcriptional regulator during embryonic development and has an important role in cell morphogenesis and growth [22]. Muscle segment homeobox 1 (MSX1) and Muscle segment homeobox 2 (MSX2) are members of the muscle segment homeobox gene family that encode transcription factors, playing important roles in the organogenesis and tissue–tissue interactions during vertebrate embryonic development [23], and mutations in MSX1 or MSX2 have been associated with impaired development of cranial neural crest-derived structures, oral clefts, and nonsyndromic oligodontia [23–27]. Studies with animal models also identified MSX1 and MSX2 double mutants in a broad range of heart malformations, such as tetralogy of Fallot and persistent truncus arteriosus [20, 23].
In this study, we analyzed the transcribed regions and splicing sites of the MSX1 and MSX2 genes and compared the sequences between 300 Chinese Han CHD patients and 400 controls to validate the possible associations of MSX1 and MSX2 with CHDs,. We found that variations rs3821949 near the 5’UTR and rs12532 within the 3’UTR of the MSX1 gene were closely associated with the risk of CHD (specifically, VSD).
Results
Patients
Clinical diagnosis of all recruited members was confirmed at the Fourth or the Second Affiliated Hospitals of Harbin Medical University. The CHD patients had no history or manifestations of any other systemic abnormalities. We also established that their mothers did not take medicines or attract infections during gestation, because these factors had been found to be associated with heart malformation in pregnancy [28, 29].
A total of 300 CHD patients (male 136, female 164, the min and max age were 0.2 and 61.0 respectively, and the average age was 15.47 years) and 400 unrelated controls (male 173, female 227, the min and max age were 0.3 and 60.0 respectively, and the average age was 13.68 years) were recruited for this study, and there was no statistical differences of the gender composition and age between the two groups (Table 1). The 300 CHD patients contained 128 with ventricular septal defects (VSD), 107 with atrial septal defects (ASD), 44 with patent ductus arteriosus (PDA), 6 with tetralogy of Fallot, 4 with pulmonary stenosis, and 11 with other types of congenital heart defects.
Table 1. Clinical characteristics and analysis of the study population.
| Parameter | CHD | Control | F | t | P | 95%CI-Up | 95%CI-Low |
|---|---|---|---|---|---|---|---|
| Sample (n) | 300 | 400 | - | - | - | - | - |
| Male/Female (n) | 136/164 | 173/227 | - | - | 0.583 | - | - |
| Age (years) | 15.47±17.47 | 13.68±10.22 | 147.419 | -1.579 | 0.115 | -4.00676 | 0.43695 |
Data are shown as mean±SD; between the two groups, there were no statistical differences of the age and gender composition.
MSX1 and MSX2 gene analysis
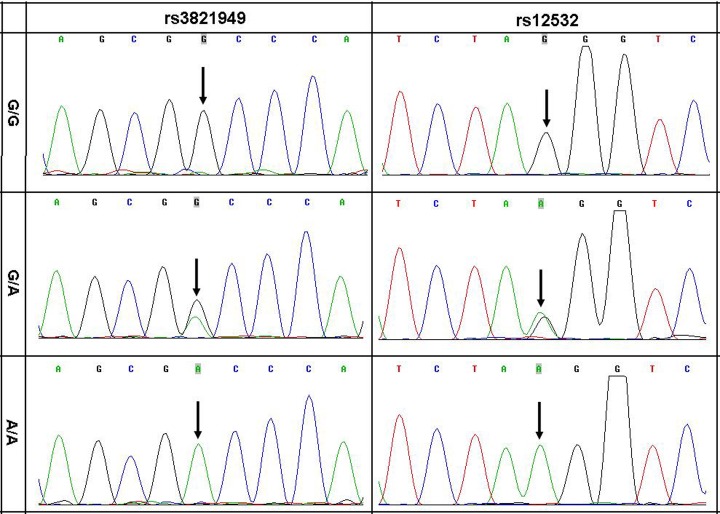
We sequenced the MSX1 and MSX2 genes to test the hypothesis that germline common genetic variants in MSX1 or MSX2 may confer susceptibility to CHD. Upon analyzing the transcribed regions and splicing sites of MSX1 and MSX2, we identified rs4242182, rs61739543 and rs111542301 within the translated region, and rs2048152 within an intron, and rs4647952 and rs3087539 within 5’UTR and 3’UTR respectively of the MSX2 gene, but the genetic heterozygosity of all these SNPs loci was low. On the other hand, we identified rs3821949 near 5’UTR and rs12532 within 3’UTR of the MSX1 gene, and the genetic heterozygosity of the two SNPs was considerably high (Fig 1).
Fig 1. Three genotypes of DNA sequence chromatograms of rs3821949 and rs12532.
SNP rs3821949 and rs12532 genotyping statistical analysis
To test possible associations between MSX1 and CHD, we conducted SNP analyses and found that both rs3821949 and rs12532 were associated with the risk of CHD (specifically VSD) in Chinese Han population (Tables 2 and 3).The Hardy-Weinberg equilibrium test for the CHD and controls were conducted and it was in line with the equilibrium.
Table 2. The genotype and allele frequency of SNP rs3821949 and rs12532 in 300 Chinese Han CHD patients and 400 non-CHD controls.
| SNP | Group | Genotype frequency (%) | Allele frequency (%) | ||||
|---|---|---|---|---|---|---|---|
| rs3821949 | Genotype | G/G | G/A | A/A | G | A | |
| CHD | 300 | 189(63.0) | 74(24.7) | 37(12.3) | 452(75.3) | 148(24.7) | |
| VSD | 128 | 81(63.3) | 31(24.2) | 16(12.5) | 193(75.4) | 63(24.6) | |
| ASD | 107 | 63(58.9) | 29(27.1) | 15(14.0) | 155(72.4) | 59(27.6) | |
| Controls | 400 | 292(73.0) | 72(18.0) | 36(9.0) | 656(82.0) | 144(18.0) | |
| rs12532 | Genotype | G/G | G/A | A/A | G | A | |
| CHD | 300 | 121(40.3) | 139(46.3) | 40(13.3) | 381(63.5) | 219(36.5) | |
| VSD | 128 | 55(43.0) | 62(48.4) | 11(8.6) | 172(67.2) | 84(32.8) | |
| ASD | 107 | 31(29.0) | 64(59.8) | 12(11.2) | 126(58.9) | 88(41.1) | |
| Controls | 400 | 126(31.5) | 219(54.8) | 55(13.8) | 471(58.9) | 329(41.1) | |
Table 3. SNP rs3821949 and rs12532 within MSX1 gene associated with the risk of congenital heart diseases in Chinese populations.
| Title | Pearson Chi-square | Pearson’s R | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Genotyped SNP | Disease Type | Statistical Types | Value | Min count a | df | Asymp. Sig. (2-sided) | Value | Asymp. Std. error b | Approx. T c | Approx. Sig |
| rs3821949 | CHD-Control | Genotype | 7.974 | 31.29 | 2 | 0.019 | -0.098 | 0.038 | -2.606 | 0.009 d |
| Allele | 9.231 | 125.14 | 1 | 0.002 | -0.081 | 0.027 | -3.046 | 0.002 d | ||
| VSD-Control | Genotype | 4.425 | 12.61 | 2 | 0.109 | -0.086 | 0.045 | -1.976 | 0.049 d | |
| Allele | 5.376 | 50.18 | 1 | 0.020 | -0.071 | 0.032 | -2.322 | 0.020 d | ||
| ASD-Control | Genotype | 8.029 | 10.76 | 2 | 0.018 | -0.118 | 0.047 | -2.661 | 0.008 d | |
| Allele | 9.657 | 42.84 | 1 | 0.002 | -0.098 | 0.034 | -3.119 | 0.002 d | ||
| rs12532 | CHD-Control | Genotype | 6.187 | 40.71 | 2 | 0.045 | 0.069 | 0.038 | 1.825 | 0.068 d |
| Allele | 3.079 | 234.86 | 1 | 0.079 | 0.047 | 0.027 | 1.755 | 0.079 d | ||
| VSD-Control | Genotype | 6.509 | 16.00 | 2 | 0.039 | 0.110 | 0.042 | 2.536 | 0.012 d | |
| Allele | 5.627 | 100.12 | 1 | 0.018 | 0.073 | 0.030 | 2.376 | 0.018 d | ||
| ASD-Control | Genotype | 0.972 | 14.14 | 2 | 0.615 | 0.000 | 0.043 | 0.001 | 0.999 d | |
| Allele | 0.000 | 88.01 | 1 | 0.999 | 0.000 | 0.031 | 0.001 | 0.999 d | ||
a: The minimum expected count
b: Not assuming the null hypothesis
c: Using the asymptotic standard error assuming the null hypothesis
d: Based on normal approximation
Discussion
In this study, we analyzed the transcribed regions and splicing sites of the MSX1 and MSX2 genes in a large cohort of CHD patients and controls, and found that the variations rs3821949 and rs12532 in the MSX1 gene were associated with the risk of CHD in the Chinese Han population, demonstrating the involvement of the MSX1 gene in the CHD etiology.
The mammalian heart is a complex organ, starting to form in the mesoderm 18 or 19 days after fertilization, and many genes with strict temporal, spatial, and sequential expression are involved in the formation [3]. The genes MSX1 and MSX2 encode the transcription factors that play key roles in the survival and differentiation of secondary heart field precursors [20, 30]. MSX1 and MSX2 double mutants in mice show malposed, elongated or spiral rotated heart outflow tract [20, 31, 32], and MSX1 and MSX2 null mutants in embryos can increase apoptosis in the secondary heart field, hence leading to many heart malformations [33]. Mutations in MSX1 and MSX2 may perturb the differentiation of heart field precursors and myocardial cells in the heart outflow tract [34–36], while mice with mutation in only MSX1 or MSX2 did not show obvious cardiac defects [24, 25], suggesting that there may be some complementary roles of the MSX1 and MSX2 transcription factors in heart development. However, in this study, the genetic heterozygosity of the SNPs located within the MSX2 gene was very low in the Chinese Han population.
LEFTY and SMAD3 play important roles in the Nodal/TGF-Lefty signaling pathway [3, 13, 15–18] and we have previously found that SNPs in Lefty and SMAD3 genes are associated with the risk of CHD [3, 18]. Normal functions of the Nodal/TGF-β signaling pathway are very important for the early stages of HES cell differentiation into different embryonic lineages [10, 12]. During embryonic development the HES cells differentiate to various cell types including cardiomyocytes in the mesoderm [19]. The heart consists of several cell types with distinct lineage origins [20, 21], and interactions between these cells are very important for the cardiac development [1]. The muscle segment homeobox gene family controls cell morphogenesis, growth, proliferation, differentiation, and apoptosis during embryonic development [22]. So the differentiation of HES cells and cardiac progenitor cell interactions during embryonic development are important for the heart development and any defects or mistakes may cause CHDs.
MSX1 seems to be an active gene in the HES cells as implicated by its up-regulated expression when the HES cells are co-cultured with PA6 cells [37], and, conversely, the expression levels of MSX1 become lower when the HES cells are treated with dopamine [38]. In a genome-wide methylation-gene expression study between the epigenetic modifications of retinoic acid treated and undifferentiated HES cells, the author uncovered 166 differentially methylated CpG sites and 2,013 differentially expressed genes, in which 19 genes including MSX1 are highly correlated with each other [39].
The 3’UTR and 5’UTR sequences play important roles in regulating the expression of genes [40] [41]. Of great significance, variations within 3’UTR may be associated with human tumorigenesis and survival of the patient [42, 43]. The 5′UTR region of gene is a structural complex, which can bind with some miRNAs and may be involved in gene expression, protein translation and disease pathogenesis [44, 45]. In a recently study, we demonstrated that the variant rs2289263 before the 5’UTR of SMAD3 gene is associated with increased risk of VSD in the Chinese Han population[18]. Probably, the SNPs in 3’UTR or 5′UTR may affect the binding of the untranslated regions with regulatory factors such as miRNAs and eventually hamper the gene expression and function. Findings in this study with rs3821949 and rs12532 of the MSX1 gene have updated our understanding on 5’UTR and r 3’UTR and may lead to new insights into the pathogenesis of CHDs.
Materials and Methods
The study population
From the Fourth and the Second Affiliated Hospitals of Harbin Medical University, Harbin, China, we collected specimens of 300 CHD patients and 400 normal controls for this study (Table 1); specimens of 90 of the 300 CHD patients were overlapping with those used for another study [3, 18]. All the CHD patients and normal controls were given comprehensive physical examination, electrocardiogram and ultrasonic echocardiogram examinations. None of the patients showed any other cardiac or systematic abnormalities, and the normal controls did not show any defects in the heart or other body parts. For this work, we obtained a written informed consent from each participant or their parents on behalf of minors, and the Ethics Committee of the Harbin Medical University approved this work, consistent with the 1975 Declaration of Helsinki.
DNA analysis
Using standard protocols, we extracted the genomic DNA from the peripheral blood leukocytes of the participants. The human MSX1 and MSX2 genes each consist of two exons located on 6p16.2 and 5q35.2, respectively. To determine the SNP genotypes in the genes, we amplified the four exons and splicing sites of the genes using polymerase chain reaction (PCR) method, and sequenced the products using standard protocols [46]. After that, the genotypes of the SNP were determined using PCR and gene sequencing methods [3].
Rs3821949 and rs12532 SNP genotyping analysis and Statistical methods
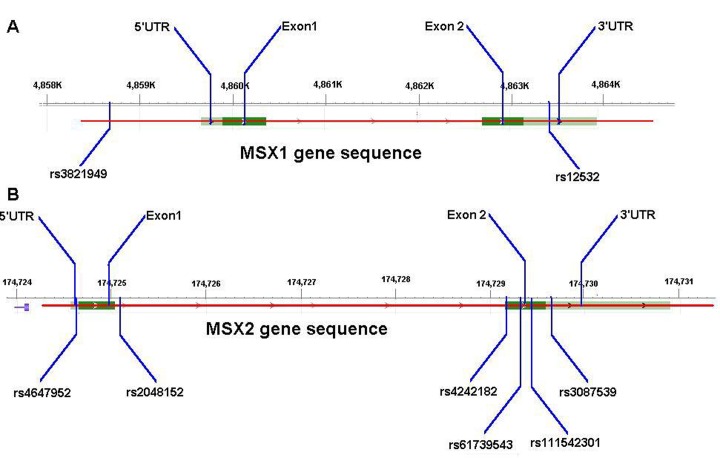
We determined genotypes of the rs3821949, rs12532 and rs4647952, rs2048152, rs4242182, rs61739543, rs111542301, rs3087539 in the MSX1 and MSX2 genes (Fig 2), and all the measurements were conducted by two independent researchers (Table 4). And then overall CHD meta-analysis was conducted according to the types of CHD and sample sizes.
Fig 2. Schematic diagrams of SNP in the genes.
A: locations of rs3821949 and rs12532 in the MSX1 gene; B: locations of rs4647952, rs2048152, rs4242182, rs61739543, rs111542301 and rs3087539 in the MSX2 gene.
Table 4. PCR primers used for SNP statistical analysis.
| Gene | SNPs | Directions | primers | Size | Tm (°C) |
|---|---|---|---|---|---|
| MSX1 | Rs3821949 | Forward | CTCCCCTGACCCCAACTC | 388bp | 52.8 |
| Reverse | CCACCCTCGCTCTGAACT | ||||
| Rs12532 | Forward | CTCCGAAGTCTGATCCCT | 215 bp | 51.0 | |
| Reverse | CTTTTCTTGCCTGGTGT | ||||
| MSX2 | Rs4647952 | Forward | TTCGGGAAGAGCCAATCA | 376 bp | 59.4 |
| Reverse | TTCTTGTCGGACATGAGCG | ||||
| Rs2048152 | Forward | CAGAGGGTGTCTTGGATGCG | 249 bp | 59.3 | |
| Reverse | AGATGGAGCGGCGTGGAT | ||||
| Rs4242182 | Forward | AGAGGGGAGGCCCGAAAG | 203 bp | 53.9 | |
| Reverse | GCGAGGAGCTGGGATGTG | ||||
| Rs61739543 | Forward | ACGCCCTTTACCACATCC | 542 bp | 55.3 | |
| Reverse | CCTCCGCCTACAGAACAA | ||||
| Rs111542301 | Forward | ACCACATCCCAGCTCCTC | 346 bp | 56.8 | |
| Reverse | GGTACATGCCATATCCCACT | ||||
| Rs3087539 | Forward | GCATGTACCACCTGTCCT | 268 bp | 52.5 | |
| Reverse | CCATCAGAGCCAATCTTT |
The continuous variable (measurement data, such as age) statistical analyses were conducted using independent-samples T test and the discrete variable (enumeration data, such as gender composition and genotype frequency) statistical analyses were conducted using Chi-Square Tests to calculate odds ratios and P value as implemented in SPSS (version 19.0). P values less than 0.05 were considered statistically significant. The Hardy-Weinberg equilibrium test of the CHD and control population was conducted with the online software OEGE.
Acknowledgments
The authors thank the patients and family members for their cooperation and participation in this study; the physicians for the specimens collection and clinical examinations.
Patient consent: Obtained.
Ethics approval: Ethics Committee of Harbin Medical University.
Data Availability
All relevant data are within the paper.
Funding Statement
This work was supported by a grant from Heilongjiang Innovation Research Foundation for Graduate Studies (YJSCX2014-10HYD), and grants of National Natural Science Foundation of China (NSFC81271786, 81110378, 30970119, 81030029) to SLL. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Hoffman JI, Kaplan S. The incidence of congenital heart disease. J Am Coll Cardiol. 2002;39(12):1890–900. Epub 2002/06/27. doi: S0735109702018867 [pii]. . [DOI] [PubMed] [Google Scholar]
- 2. Hoffman JI, Kaplan S, Liberthson RR. Prevalence of congenital heart disease. Am Heart J. 2004;147(3):425–39. Epub 2004/03/05. 10.1016/j.ahj.2003.05.003 S0002870303007294 [pii]. . [DOI] [PubMed] [Google Scholar]
- 3. Deng X, Zhou J, Li FF, Yan P, Zhao EY, Hao L, et al. Characterization of Nodal/TGF-Lefty Signaling Pathway Gene Variants for Possible Roles in Congenital Heart Diseases. PLoS One. 2014;9(8):e104535 Epub 2014/08/12. 10.1371/journal.pone.0104535 PONE-D-14-16633 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. van der Bom T, Zomer AC, Zwinderman AH, Meijboom FJ, Bouma BJ, Mulder BJ. The changing epidemiology of congenital heart disease. Nat Rev Cardiol. 2011;8(1):50–60. Epub 2010/11/04. 10.1038/nrcardio.2010.166 nrcardio.2010.166 [pii]. . [DOI] [PubMed] [Google Scholar]
- 5. Bruneau BG. The developmental genetics of congenital heart disease. Nature. 2008;451(7181):943–8. Epub 2008/02/22. 10.1038/nature06801 nature06801 [pii]. . [DOI] [PubMed] [Google Scholar]
- 6. Richards AA, Garg V. Genetics of congenital heart disease. Curr Cardiol Rev. 2010;6(2):91–7. Epub 2011/05/03. 10.2174/157340310791162703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pierpont ME, Basson CT, Benson DW Jr., Gelb BD, Giglia TM, Goldmuntz E, et al. Genetic basis for congenital heart defects: current knowledge: a scientific statement from the American Heart Association Congenital Cardiac Defects Committee, Council on Cardiovascular Disease in the Young: endorsed by the American Academy of Pediatrics. Circulation. 2007;115(23):3015–38. Epub 2007/05/24. doi: CIRCULATIONAHA.106.183056 [pii] 10.1161/CIRCULATIONAHA.106.183056 . [DOI] [PubMed] [Google Scholar]
- 8. Buckingham M, Meilhac S, Zaffran S. Building the mammalian heart from two sources of myocardial cells. Nat Rev Genet. 2005;6(11):826–35. Epub 2005/11/24. doi: nrg1710 [pii] 10.1038/nrg1710 . [DOI] [PubMed] [Google Scholar]
- 9. van Weerd JH, Koshiba-Takeuchi K, Kwon C, Takeuchi JK. Epigenetic factors and cardiac development. Cardiovasc Res. 2011;91(2):203–11. Epub 2011/05/25. 10.1093/cvr/cvr138 cvr138 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Postovit LM, Margaryan NV, Seftor EA, Kirschmann DA, Lipavsky A, Wheaton WW, et al. Human embryonic stem cell microenvironment suppresses the tumorigenic phenotype of aggressive cancer cells. Proc Natl Acad Sci U S A. 2008;105(11):4329–34. Epub 2008/03/13. 10.1073/pnas.0800467105 0800467105 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Costa FF, Seftor EA, Bischof JM, Kirschmann DA, Strizzi L, Arndt K, et al. Epigenetically reprogramming metastatic tumor cells with an embryonic microenvironment. Epigenomics. 2009;1(2):387–98. Epub 2010/05/25. 10.2217/epi.09.25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Malchenko S, Galat V, Seftor EA, Vanin EF, Costa FF, Seftor RE, et al. Cancer hallmarks in induced pluripotent cells: new insights. J Cell Physiol. 2010;225(2):390–3. Epub 2010/06/23. 10.1002/jcp.22280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tabibzadeh S, Hemmati-Brivanlou A. Lefty at the crossroads of "stemness" and differentiative events. Stem Cells. 2006;24(9):1998–2006. Epub 2006/05/27. doi: 2006–0075 [pii] 10.1634/stemcells.2006-0075 . [DOI] [PubMed] [Google Scholar]
- 14. Dvash T, Sharon N, Yanuka O, Benvenisty N. Molecular analysis of LEFTY-expressing cells in early human embryoid bodies. Stem Cells. 2007;25(2):465–72. Epub 2006/10/14. doi: 2006–0179 [pii] 10.1634/stemcells.2006-0179 . [DOI] [PubMed] [Google Scholar]
- 15. van de Laar IM, Oldenburg RA, Pals G, Roos-Hesselink JW, de Graaf BM, Verhagen JM, et al. Mutations in SMAD3 cause a syndromic form of aortic aneurysms and dissections with early-onset osteoarthritis. Nat Genet. 2011;43(2):121–6. Epub 2011/01/11. 10.1038/ng.744 ng.744 [pii]. . [DOI] [PubMed] [Google Scholar]
- 16. Liying J, Yuchun T, Youcheng W, Yingchen W, Chunyu J, Yanling Y, et al. A SMAD3 gene polymorphism is related with osteoarthritis in a Northeast Chinese population. Rheumatol Int. 2013;33(7):1763–8. Epub 2013/01/08. 10.1007/s00296-012-2593-z . [DOI] [PubMed] [Google Scholar]
- 17. van der Linde D, van de Laar IM, Bertoli-Avella AM, Oldenburg RA, Bekkers JA, Mattace-Raso FU, et al. Aggressive cardiovascular phenotype of aneurysms-osteoarthritis syndrome caused by pathogenic SMAD3 variants. J Am Coll Cardiol. 2012;60(5):397–403. Epub 2012/05/29. 10.1016/j.jacc.2011.12.052 S0735-1097(12)01238-7 [pii]. . [DOI] [PubMed] [Google Scholar]
- 18. Li FF, Zhou J, Zhao DD, Yan P, Li X, Han Y, et al. Characterization of SMAD3 Gene Variants for Possible Roles in Ventricular Septal Defects and Other Congenital Heart Diseases. PLoS One. 2015;10(6):e0131542 Epub 2015/06/26. 10.1371/journal.pone.0131542 PONE-D-14-42761 [pii]. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Schuldiner M, Benvenisty N. Factors controlling human embryonic stem cell differentiation. Methods Enzymol. 2003;365:446–61. Epub 2003/12/31. . [DOI] [PubMed] [Google Scholar]
- 20. Chen YH, Ishii M, Sun J, Sucov HM, Maxson RE Jr. Msx1 and Msx2 regulate survival of secondary heart field precursors and post-migratory proliferation of cardiac neural crest in the outflow tract. Dev Biol. 2007;308(2):421–37. Epub 2007/07/03. doi: S0012-1606(07)01107-4 [pii] 10.1016/j.ydbio.2007.05.037 . [DOI] [PubMed] [Google Scholar]
- 21. Cheng G, Litchenberg WH, Cole GJ, Mikawa T, Thompson RP, Gourdie RG. Development of the cardiac conduction system involves recruitment within a multipotent cardiomyogenic lineage. Development. 1999;126(22):5041–9. Epub 1999/10/26. . [DOI] [PubMed] [Google Scholar]
- 22. Han J, Ishii M, Bringas P Jr., Maas RL, Maxson RE Jr., Chai Y. Concerted action of Msx1 and Msx2 in regulating cranial neural crest cell differentiation during frontal bone development. Mech Dev. 2007;124(9–10):729–45. Epub 2007/08/19. doi: S0925-4773(07)00106-2 [pii] 10.1016/j.mod.2007.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ishii M, Han J, Yen HY, Sucov HM, Chai Y, Maxson RE Jr. Combined deficiencies of Msx1 and Msx2 cause impaired patterning and survival of the cranial neural crest. Development. 2005;132(22):4937–50. Epub 2005/10/14. doi: dev.02072 [pii] 10.1242/dev.02072 . [DOI] [PubMed] [Google Scholar]
- 24. Satokata I, Maas R. Msx1 deficient mice exhibit cleft palate and abnormalities of craniofacial and tooth development. Nat Genet. 1994;6(4):348–56. Epub 1994/04/01. 10.1038/ng0494-348 . [DOI] [PubMed] [Google Scholar]
- 25. Satokata I, Ma L, Ohshima H, Bei M, Woo I, Nishizawa K, et al. Msx2 deficiency in mice causes pleiotropic defects in bone growth and ectodermal organ formation. Nat Genet. 2000;24(4):391–5. Epub 2000/03/31. 10.1038/74231 . [DOI] [PubMed] [Google Scholar]
- 26. Souza LT, Kowalski TW, Collares MV, Felix TM. MSX1 gene and nonsyndromic oral clefts in a Southern Brazilian population. Braz J Med Biol Res. 2013;46(7):555–8. Epub 2013/08/02. 10.1590/1414-431X20133054 S0100-879X2013005033054 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kimura M, Machida J, Yamaguchi S, Shibata A, Tatematsu T, Miyachi H, et al. Novel nonsense mutation in MSX1 in familial nonsyndromic oligodontia: subcellular localization and role of homeodomain/MH4. Eur J Oral Sci. 2014;122(1):15–20. Epub 2013/12/18. 10.1111/eos.12105 . [DOI] [PubMed] [Google Scholar]
- 28. van Driel LM, Smedts HP, Helbing WA, Isaacs A, Lindemans J, Uitterlinden AG, et al. Eight-fold increased risk for congenital heart defects in children carrying the nicotinamide N-methyltransferase polymorphism and exposed to medicines and low nicotinamide. Eur Heart J. 2008;29(11):1424–31. Epub 2008/04/29. 10.1093/eurheartj/ehn170 ehn170 [pii]. . [DOI] [PubMed] [Google Scholar]
- 29. Kebed KY, Bishu K, Al Adham RI, Baddour LM, Connolly HM, Sohail MR, et al. Pregnancy and Postpartum Infective Endocarditis: A Systematic Review. Mayo Clin Proc. 2014;89(8):1143–52. Epub 2014/07/06. doi: S0025-6196(14)00388-7 [pii] 10.1016/j.mayocp.2014.04.024 . [DOI] [PubMed] [Google Scholar]
- 30. Chan-Thomas PS, Thompson RP, Robert B, Yacoub MH, Barton PJ. Expression of homeobox genes Msx-1 (Hox-7) and Msx-2 (Hox-8) during cardiac development in the chick. Dev Dyn. 1993;197(3):203–16. Epub 1993/07/01. 10.1002/aja.1001970305 . [DOI] [PubMed] [Google Scholar]
- 31. Barbosky L, Lawrence DK, Karunamuni G, Wikenheiser JC, Doughman YQ, Visconti RP, et al. Apoptosis in the developing mouse heart. Dev Dyn. 2006;235(9):2592–602. Epub 2006/08/02. 10.1002/dvdy.20885 . [DOI] [PubMed] [Google Scholar]
- 32. Yelbuz TM, Waldo KL, Kumiski DH, Stadt HA, Wolfe RR, Leatherbury L, et al. Shortened outflow tract leads to altered cardiac looping after neural crest ablation. Circulation. 2002;106(4):504–10. Epub 2002/07/24. . [DOI] [PubMed] [Google Scholar]
- 33. Ward C, Stadt H, Hutson M, Kirby ML. Ablation of the secondary heart field leads to tetralogy of Fallot and pulmonary atresia. Dev Biol. 2005;284(1):72–83. Epub 2005/06/14. doi: S0012-1606(05)00274-5 [pii] 10.1016/j.ydbio.2005.05.003 . [DOI] [PubMed] [Google Scholar]
- 34. Lo CW, Waldo KL, Kirby ML. Gap junction communication and the modulation of cardiac neural crest cells. Trends Cardiovasc Med. 1999;9(3–4):63–9. Epub 1999/12/01. doi: S1050-1738(99)00015-8 [pii]. . [DOI] [PubMed] [Google Scholar]
- 35. Vorotnikova E, McIntosh D, Dewilde A, Zhang J, Reing JE, Zhang L, et al. Extracellular matrix-derived products modulate endothelial and progenitor cell migration and proliferation in vitro and stimulate regenerative healing in vivo. Matrix Biol. 2010;29(8):690–700. Epub 2010/08/28. 10.1016/j.matbio.2010.08.007 S0945-053X(10)00132-0 [pii]. . [DOI] [PubMed] [Google Scholar]
- 36. Miller KA, Davidson S, Liaros A, Barrow J, Lear M, Heine D, et al. Prediction and characterisation of a highly conserved, remote and cAMP responsive enhancer that regulates Msx1 gene expression in cardiac neural crest and outflow tract. Dev Biol. 2008;317(2):686–94. Epub 2008/03/25. 10.1016/j.ydbio.2008.02.016 S0012-1606(08)00135-8 [pii]. . [DOI] [PubMed] [Google Scholar]
- 37. Chan AA, Hertsenberg AJ, Funderburgh ML, Mann MM, Du Y, Davoli KA, et al. Differentiation of human embryonic stem cells into cells with corneal keratocyte phenotype. PLoS One. 2013;8(2):e56831 Epub 2013/02/26. 10.1371/journal.pone.0056831 PONE-D-12-16042 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Belinsky GS, Sirois CL, Rich MT, Short SM, Moore AR, Gilbert SE, et al. Dopamine receptors in human embryonic stem cell neurodifferentiation. Stem Cells Dev. 2013;22(10):1522–40. Epub 2013/01/05. 10.1089/scd.2012.0150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cheong HS, Lee HC, Park BL, Kim H, Jang MJ, Han YM, et al. Epigenetic modification of retinoic acid-treated human embryonic stem cells. BMB Rep. 2010;43(12):830–5. Epub 2010/12/30. 10.5483/BMBRep.2010.43.12.830 . [DOI] [PubMed] [Google Scholar]
- 40. Lou F, Ma HN, Xu L, Chen M, Zhu YB. Two polymorphisms of CD44 3'UTR weaken the binding of miRNAs and associate with naso-pharyngeal carcinoma in a Chinese population. Eur Rev Med Pharmacol Sci. 2014;18(17):2444–52. Epub 2014/10/01. doi: 7755 [pii]. . [PubMed] [Google Scholar]
- 41. Fujimuro T, Matsui T, Nitanda Y, Matta T, Sakumura Y, Saito M, et al. Hes7 3'UTR is required for somite segmentation function. Sci Rep. 2014;4:6462 Epub 2014/09/25. 10.1038/srep06462 srep06462 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chen S, He Y, Ding J, Jiang Y, Jia S, Xia W, et al. An insertion/deletion polymorphism in the 3' untranslated region of beta-transducin repeat-containing protein (betaTrCP) is associated with susceptibility for hepatocellular carcinoma in Chinese. Biochem Biophys Res Commun. 2010;391(1):552–6. Epub 2009/11/26. 10.1016/j.bbrc.2009.11.096 S0006-291X(09)02273-6 [pii]. . [DOI] [PubMed] [Google Scholar]
- 43. Christensen BC, Moyer BJ, Avissar M, Ouellet LG, Plaza SL, McClean MD, et al. A let-7 microRNA-binding site polymorphism in the KRAS 3' UTR is associated with reduced survival in oral cancers. Carcinogenesis. 2009;30(6):1003–7. Epub 2009/04/22. 10.1093/carcin/bgp099 bgp099 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tosetto E, Casarin A, Salviati L, Familiari A, Lieske JC, Anglani F. Complexity of the 5'UTR region of the CLCN5 gene: eleven 5'UTR ends are differentially expressed in the human kidney. BMC Med Genomics. 2014;7:41 Epub 2014/07/09. 10.1186/1755-8794-7-41 1755-8794-7-41 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Orom UA, Nielsen FC, Lund AH. MicroRNA-10a binds the 5'UTR of ribosomal protein mRNAs and enhances their translation. Mol Cell. 2008;30(4):460–71. Epub 2008/05/24. 10.1016/j.molcel.2008.05.001 S1097-2765(08)00328-6 [pii]. . [DOI] [PubMed] [Google Scholar]
- 46. Tan ZX, Li FF, Qu YY, Liu J, Liu GR, Zhou J, et al. Identification of a known mutation in Notch 3 in familiar CADASIL in China. PLoS One. 2012;7(5):e36590 Epub 2012/05/25. 10.1371/journal.pone.0036590 PONE-D-11-24676 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.