Abstract
Leptospirosis, caused by pathogenic Leptospira, is a worldwide zoonotic infection. The genus Leptospira includes at least 21 species clustered into three groups—pathogens, non-pathogens, and intermediates—based on 16S rRNA phylogeny. Research on Leptospira is difficult due to slow growth and poor transformability of the pathogens. Recent identification of extrachromosomal elements besides the two chromosomes in L. interrogans has provided new insight into genome complexity of the genus Leptospira. The large size, low copy number, and high similarity of the sequence of these extrachromosomal elements with the chromosomes present challenges in isolating and detecting them without careful genome assembly. In this study, two extrachromosomal elements were identified in L. borgpetersenii serovar Ballum strain 56604 through whole genome assembly combined with S1 nuclease digestion following pulsed-field gel electrophoresis (S1-PFGE) analysis. Further, extrachromosomal elements in additional 15 Chinese epidemic strains of Leptospira, comprising L. borgpetersenii, L. weilii, and L. interrogans, were successfully separated and identified, independent of genome sequence data. Southern blot hybridization with extrachromosomal element-specific probes, designated as lcp1, lcp2 and lcp3-rep, further confirmed their occurrences as extrachromosomal elements. In total, 24 plasmids were detected in 13 out of 15 tested strains, among which 11 can hybridize with the lcp1-rep probe and 11 with the lcp2-rep probe, whereas two can hybridize with the lcp3-rep probe. None of them are likely to be species-specific. Blastp search of the lcp1, lcp2, and lcp3-rep genes with a nonredundant protein database of Leptospira species genomes showed that their homologous sequences are widely distributed among clades of pathogens but not non-pathogens or intermediates. These results suggest that the plasmids are widely distributed in Leptospira species, and further elucidation of their biological significance might contribute to our understanding of biology and infectivity of pathogenic spirochetes.
Author Summary
Leptospirosis, caused by a diversity of pathogenic species within the genus Leptospira, is a worldwide public health problem affecting both developed and developing countries. In 2003, the whole genome sequencing of pathogenic Leptospira interrogans strain 56601 opened the genomic research of this specific pathogen and accelerated genomic sequencing speed afterwards. The availability of whole genome sequences of the pathogenic species L. interrogans and L. borgpetersenii, saprophyte L. biflexa and the intermediate L. licerasiae, which represented three phylogenetic groups of Leptospira spp., has undoubtedly facilitated the understanding of the genetic complexity of this organism. Genome drafts of more than 300 strains have been released recently. The analysis of these sequences will provide invaluable information for understanding the evolution and adaption to various environment of the Leptospiraceae. Extrachromosomal replicons are important for communication of genetic information between bacteria. Despite first leptospiral genome was sequenced in 2003, these small auto-replicons, however, were not identified in the whole genome sequence of pathogenic L. interrogans until 2014. How to effectively identify the presence of small auto-replicons in Leptospira cell is a technical challenge which hinders researchers in fully understanding the complexity and diversity of Leptospira genomes. In this study, we report identification of small auto-replicons in 15 Leptospira strains endemic in China by S1-PFGE analysis, independent of genome sequence assembly. Further analysis suggested that these plasmids are widely presented in Leptospira species. The study, combined with the recently available genomic sequencing data, will help us to elucidate the diversity of Leptospira genomes and a deeper insight into evolutionary perspective of this unique clade of organism.
Introduction
Leptospires are thin, spiral, highly motile bacteria that belong to the order Spirochaetales, an early branch of eubacteria. The genus Leptospira includes at least 21 species based on 16S rRNA phylogeny, further distinguished into three clades: pathogens, non-pathogens and intermediates[1]. Pathogenic Leptospira are comprised of at least 14 species, which share a common branch in evolution, genetically distinct from non-pathogens. Leptospires are also serologically classified into serovars, including more than two hundred that are pathogenic in human and animals[1].
Pathogenic Leptospira is known to cause the widespread water-related zoonosis, called leptospirosis. Hosts usually become infected through direct contact with soil or water contaminated by the urine of infected animals [2]. Infection produces a wide spectrum of clinical manifestations, ranging in severity from a mild influenza-like disease to an acute, potentially lethal infection. Pathogenic Leptospira species, such as L. interrogans, L. borgpetersenii, L. kirschneri, L. noguchii, and L.weillii [3–5], are the causative pathogens of leptospirosis, among which L. interrogans and L. borgpetersenii are most prevalent globally. Although infection with L. interrogans and L. borgpetersenii cause similar clinical symptoms, the transmission modes are different: for example, L. interrogans is commonly waterborne, whereas L. borgpetersenii is transmitted via direct host-to-host contact [6]. Currently, the completed genome sequences include five pathogenic strains of L. interrogans, two pathogenic strains of L. borgpetersennii, and two strains of saprophytic L. biflexa [6–12]. Comparison of the L. interrogans and L. borgpetersenii genome sequences revealed genome reduction in the L. borgpetersenii genome that is supposedly IS-mediated [6].
Plasmids are regarded as one of the most effective vehicles for bacterial communication of genetic information [13], promoting the rapid evolution and adaptation abilities of bacteria [14]. Because of the diverse genetic information they carry, plasmids often play specific biological roles in the host bacterium. Also, they can potentially be engineered as efficient genetic tools for microbial genetic manipulation and analysis through the introduction, modification, or removal of target genes [15]. In fact, many plasmids were found in spirochaetes, with a majority identified in the genus Borrelia [13]. In addition to its linear chromosome, Borrelia contains multiple circular and linear plasmids within a single cell [16]. As for the genus Leptospira, plasmid P74 and bacteriophage LE1 were first reported within the saprophytic L. biflexa serovar Patoc strain Patoc I genome, and then a LE1-like prophage was found in the intermediate species L. licerasiae [9,17]. It was long believed that pathogenic Leptospira species contain only two chromosomes [5]. Recently, a 54-kb genomic island, LaiGI, was confirmed as an extrachromosomal replicon stable within the first sequenced pathogenic strain of L. interrogans strain Lai [18,19]. Two plasmids, pGui1 and pGui2, in L. interrogans pathogenic strain Gui44 and three plasmids, lcp1, lcp2, and lcp3, in L. interrogans pathogenic strain 56609 were reported in succession, which significantly contribute to revealing the diversity of the pathogenic Leptospira genome[10,12]. Based on the plasmids identified, L. interrogans–Escherichia coli shuttle vectors with the predicted replication rep gene or rep combined with parAB loci from the three plasmids of L. interrogans pathogenic strain 56609 were reported to be successfully transformed into both saprophytic and pathogenic Leptospira species, which is considered as a new milestone in research efforts involving pathogenic Leptospira [12,16].
Although the plasmids have originally been identified in L. interrogans, the plasmid sequence in other infectious strains, including L. borgpetersenii is still unknown. In this study, two extrachromosomal circular elements of L. borgpetersenii serovar Ballum strain 56604 were detected and estimated by S1 nuclease digestion following pulsed-field gel electrophoresis (S1-PFGE), which allowed detection of low-copy replicons [20]. These two plasmids were further characterized in details through whole genome sequencing. We subsequently used S1-PFGE to identify the plasmids in the remaining 14 reference strains of Leptospira in China belonging to species L. borgpetersenii, L. interrogans, and L. weilii. These efforts will contribute to a better understanding the genetic complexity of this bacterium, delivering the most effective genomic information, and accelerating the process of complete genomic sequencing of the genus Leptospira.
Materials and Methods
Bacterial strains and culture conditions
Leptospira borgpetersenii serogroup Ballum serovar Ballum strain 56604 was obtained from the Institute for Infectious Disease Control and Prevention (CDC), Beijing, China. The remaining 14 domestic reference strains of Leptospira were also obtained from the Chinese CDC and used in this study: Leptospira interrogans serogroup Icterohaemorrhagiae serovar Lai strain 56601, serogroup Canicola serovar Canicola strain 56603, serogroup Pyrogenes serovar Pyrogenes strain 56605, serogroup Autumnalis serovar Autumnalis strain 56606a and 56606v, serogroup Australis serovar Australis strain 56607, serogroup Pomona serovar Pomona strain 56608, serogroup Grippotyphosa serovar Linhai strain 56609, serogroup Hebdomadis serovar Hebdomadis strain 56610, serogroup Bataviae serovar Paidjan strain 56612, serogroup Sejroe serovar Wolffi strain 56635; Leptospira borgpetersenii serogroup Javanica serovar Javanica strain 56602, serogroup Tarassovi serovar Tarassovi strain 56613, serogroup Mini serovar Mini strain 56655; Leptospira weilii serogroup Manhao serovar Qingshui strain 56615. Of note, 56606a is avirulent strain derived from strain 56606v and has lost its virulence after long time in vitro passages in our laboratory. The strains were grown in liquid Ellinghausen–McCullough–Johnson–Harris (EMJH) medium under aerobic conditions at 28°C to mid-log phase and then collected at an optical density of 1.3–2.0 at 600 nm.
Next-generation DNA sequencing and assembly
The genome of L. borgpetersenii serovar Ballum strain 56604 was sequenced using a 454 GS20 system (454 Life Sciences). Six micrograms of 56604 genomic DNA was prepared to create sequencing libraries according to the manufacturer’s protocol (454 Life Sciences). A total of 421,952 reads (average read length 282bp) were generated and 421,410 reads of high quality (99.8%) were selected for genome assembly, providing 29.5 fold coverage. 179 contigs (144 contigs >500bp) were yielded and the N50 size of the contigs was 28,039bp. In the finishing process, the reference genome sequences of L. borgpetersenii serovar Hardjo strain L550 and JB197 were used to determine the suppositional contig order of 56604. In the following physical gap closing, PCR was performed thousands of times based on the following conditions: 5 min at 95°C, followed by 35 cycles of 30 sec at 95°C, 30 sec at 58°C, and 30 sec at 72°C. After that, sequence assembly was accomplished using Phred, Phrap, and Consed programs[21–23].
Genome annotation and sequence similarity analysis
The sequence alignments were performed using BLAST (http://blast.st-va.ncbi.nlm.nih.gov/Blast.cgi). The open reading frames (ORFs) were predicted and manually checked by the combined use of GLIMMER, GeneMark, and Z-curve programs[24,25]. Clusters of orthologous groups (COG) functional annotation for each gene was performed through RPS-BLAST in the NCBI Conserved Domain Database (CDD), and conserved domains were analyzed to further verify and supplement the annotation by searching the Pfam database[26,27]. Transfer RNA genes were identified with tRNAscan-SE (http://selab.janelia.org/tRNAscan-SE/). Insertion sequence (IS) elements were determined using the IS-finder online tool (https://www-is.biotoul.fr/). Orthologous proteins were identified by performing BLAST searches against the NCBI non-redundant protein database of L. borgpetersenii serovar Hardjo strain L550 and strain JB197 genomes and L. borgpetersenii serogroup Ballum serovar Ballum strain 56604 and subsequently checked manually. Whole-genome sequence comparison was performed at the nucleotide level using the program BLASTn (with a cutoff E value of 1e-10) and visualized with EasyFig[28].
Preparation of agarose gel plugs
Bacteria were grown in EMJH medium under aerobic conditions at 28°C to mid-log phase. Each culture was suspended in 100 μl cell suspension buffer (50mM Tris-HCl pH7.2, 25M NaCl, 50mM EDTA) to a turbidity of 1.3–2.0 OD600, as specified in S1 Table. An equal volume of plasmid-harboring bacteria was embedded in agarose plugs (1%) (SeaKemGold Agarose gels), immediately dispensed into wells of plug molds, then incubated at 55°C with proteinase K solution (2% (w/v) Na-deoxycholate, 10% (w/v) Na-lauroyl sarcosine, 0.5M EDTA pH 8.0, 0.5 M NaCl, 0.5% proteinase K) overnight. The plugs were washed twice in tubes shaking in a 54°C water bath for 15 min each time with both pre-warmed double distilled water and TE buffer to inactivate the proteinase K, then used immediately or stored in TE buffer at 4°C.
S1 nuclease and restriction enzyme digestion of DNA in plugs and pulsed-field gel electrophoresis (PFGE)
S1 nuclease, a specific enzyme that can convert supercoiled plasmids into full-length linear molecules, was used to digest the 2-mm gel plugs. For different Leptospira species, the digestion reaction was similar in time but different in the enzyme added (S1 nuclease, Thermo Scientific; NotI and PstI, New England Biolabs), as shown in S1 Table. The standard strain Salmonella enterica serotype Braenderup (H9812), kindly provided by the Department of Clinical Microbiology, Ruijin Hospital (Shanghai, China), was digested with XbaI and used as a molecular weight marker. Gel plugs were subjected to PFGE immediately after completion of the digestion reaction using a contour-clamped homogeneous electric field machine (CHEF-DR III; Bio-Rad). Electrophoresis with linear ramp time from 5 to 65 s at a gradient of 6 V/cm and an included angle of 120° was performed for 20 h to separate the DNA fragments, and gels were cooled continuously at 14°C during the running process. Gels were then stained in 1μg/mL ethidium bromide for 40 min and visualized in a gel image acquisition and analysis system.
Preparation of probe and southern hybridization
The plasmid DNA was transferred and cross-linked to positively charged nylon membranes (Roche Diagnostics) and hybridized against digoxigenin-labeled probes generated through polymerase chain reaction amplification (PCR DIG Probe Synthesis Kit, Roche) according to the manufacturer’s instructions with some modifications. Briefly, the membrane was washed in 2X SSC twice for 5 min each time at 25°C, then washed twice in 0.1X SSC for 15 min at 68°C. After blocking in 1% blocking reagent for 30 min, the membrane with DIG-labeled probe was detected with anti-Digoxigenin-AP, Fab fragments (Roche), and CDP-Star (Roche). All primers used are listed in S2 Table.
Nucleotide sequence accession number
The complete genomic sequences of L. borgpetersenii strain 56604 have been deposited in GenBank under the following accession numbers CP012029, CP012030, CP012031 and CP012032.
Results
General features of L. borgpetersenii strain 56604
The genome of strain L. borgpetersenii strain 56604 consists of two circular chromosomes and two circular extrachromosomal replicons. Chromosome CI and CII are 3,550,837 bp and 361,762 bp, respectively, with GC content of 40.2%. Table 1 summarizes the genome features of strains 56604, L550 and JB197. The genome of strain 56604 is smaller than L. interrogans, similar to that of L. borgpetersenii strains L550 and JB197[6].
Table 1. General features of the L. borgpetersenii serovar Ballum and serovar Hardjo genome.
| Genome Features | Ballum 56604 | Hardjo-bovis L550 | Hardjo-bovis JB197 | |||||
|---|---|---|---|---|---|---|---|---|
| CI | CII | lbp1 | lbp2 | CI | CII | CI | CII | |
| Genome Size(bp) | 3.55Mb | 362Kb | 65Kb | 59Kb | 3.61Mb | 317Kb | 3.58Mb | 300Kb |
| G+C content (%) | 40.2 | 40.2 | 41 | 39.7 | 40.23 | 40.16 | 40.23 | 40.43 |
| Protein coding (%) | 75.94 | 75.95 | 75.81 | 75.5 | 74 | 73.9 | 73.8 | 74.8 |
| Total CDSs | 2,360 | 258 | 39 | 35 | 2,607 | 235 | 2,540 | 230 |
| CDSs with assigned function | 1,828 | 196 | 26 | 24 | 1,647 | 134 | 1,594 | 131 |
| CDSs without assigned function | 532 | 62 | 13 | 11 | 960 | 101 | 946 | 99 |
| Average CDS length(bp) | 1,142 | 1,065 | 1,272 | 1,285 | 1,026 | 998 | 1,039 | 975 |
| Total IS element | 45 | 8 | 1 | 0 | 114 | 7 | 111 | 5 |
| IS1533 | 26 | 5 | 0 | 0 | 72 | 5 | 81 | 3 |
| Transfer RNA | 37 | 0 | 1 | 0 | 37 | 0 | 35 | 0 |
| 23S Ribosomal RNA | 2 | 0 | 0 | 0 | 2 | 0 | 2 | 0 |
| 16S Ribosomal RNA | 2 | 0 | 0 | 0 | 2 | 0 | 2 | 0 |
| 5S Ribosomal RNA | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 0 |
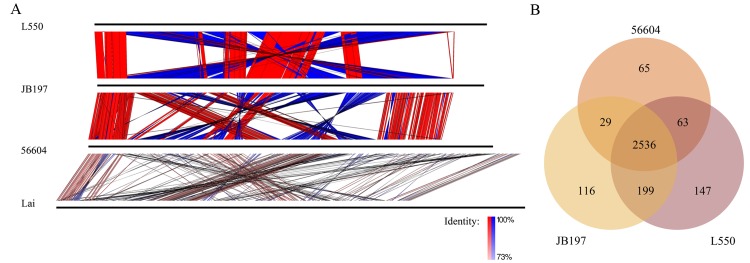
The genome sequence of strain 56604, L550 and JB197 shared extensive collinear disrupted by few rearrangements, but not L. interrogans strain Lai (Fig 1A).The numbers of conserved genes amongst the three strains are 2,536 as shown in Fig 1B. Two additional circular extrachromosomal replicons, designated lbp1 and lbp2, which are 65,435 bp with GC content of 41% and 59,545 bp with GC content 39.7%, respectively (detailed in Table 1 and S1 Fig) were present in strain 56604. Several insertion sequence (IS) elements have been described in pathogenic Leptospira, including IS1500, IS1501, IS1533, and ISLin1[5,29]. We identified a total of approximately 54 ISs scattered in chromosomes of strain 56604, including 31 copies of IS1533, 15 copies of ISLin1, 4 copies of IS1502, 2 copies of IS1500, 1 copy of IS1501, and 1 copy of ISLin2(S3 Table). The number of IS copies is apparently less than those in L. borgpetersenii strain L550 and JB197 [6,30,31].
Fig 1. Comparison of replicons and the shared and unique orthologs of L. borgpetersenii.
(A) The replicons of L. borgpetersenii serovar Hardjo strains L550 and JB197, serovar Ballum strains 56604 and L. interrogans serovars Lai were aligned by BLASTn (with a cutoff E value of 1e-10) and visualized with EasyFig[28]. Colored lines drawn between two adjacent linearized chromosomes (horizontal black lines) showed the location of homologous regions and vertical blocks between sequences indicated regions of shared similarity in the same (red) or opposite (blue) direction. (B) Venn diagram showing the distribution of shared and unique proteins by L. borgpetersenii species (BLASTp with a cutoff E value of 1e-5, identity of ≥50% and coverage of ≥50%).
Two extrachromosomal replicons, designated lbp1 and lbp2, share similar replication sequence with lcp1 and lcp2
Two circular extrachromosomal replicons, designated lbp1 and lbp2, were identified in strain 56604 through whole genome sequencing. According to sequence-reads coverage of each assembly level, the two plasmids were estimated to share equal copy numbers with the chromosomes. The GC content is higher than that of plasmids lcp1, lcp2, and lcp3 (35%, 35%, 39%) in L. interrogans strain 56609 as well as pGui1 (34.63%) and pGui2 (33.33%) in L. interrogans strain Gui44 (Table 2)[10,12]. Sequence comparisons showed low similarity amongst them (less than 20% coverage). BLASTp analysis showed that plasmid lbp1 has 1, 3, 0, 10 and 1 orthologs in plasmid lcp1, lcp2, lcp3, pGui1and pGui2 respectively; that plasmid lbp2 has 1, 1, 0, 1 and 3 orthologs in plasmid lcp1, lcp2, lcp3, pGui1 and pGui2 respectively.
Table 2. General features of the plasmids in sequenced pathogenic Leptospira genome.
| Features | L. borgpetersenii serovar Ballum strain 56604 | L. interrogans serovar Linhai strain 56609 | L. interrogans serovar Canicola strain Gui44 | ||||
|---|---|---|---|---|---|---|---|
| lbp1 | lbp2 | lcp1 | lcp2 | lcp3 | pGui1 | pGui2 | |
| Size (bp) | 65,435 | 59,545 | 67,282 | 56,757 | 54,986 | 74,981 | 66,851 |
| G+C content (%) | 41 | 39.7 | 35.91 | 34.67 | 39.43 | 34.63 | 33.33 |
| Protein coding (%) | 75.81 | 75.5 | 75.4 | 68.8 | 83.9 | 78.32 | 72.5 |
| Protein-coding genes | 39 | 35 | 71 | 56 | 77 | 62 | 63 |
| Hypothetical genes | 13 | 11 | 31 | 30 | 30 | 35 | 52 |
| Gene density (bp per gene) | 1,272 | 1,285 | 714 | 698 | 599 | 947 | 769 |
| Transposases | 1 | 1 | 6 | 7 | 0 | 9 | 4 |
| t RNA | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| r RNA | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
99% identity with 100% coverage with lcp2-rep. parAB genes were also found to be located immediately upstream of rep genes in lbp1 and lbp2 respectively. The parAB genes are less conserved and they showed low similarities between each other.
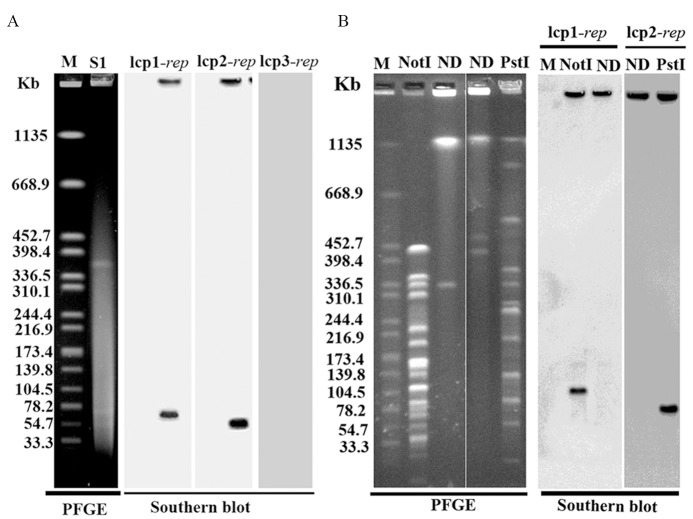
S1-PFGE separation followed by Southern blotting with lcp1-rep, lcp2-rep, and lcp3-rep specific probes confirmed that lbp1 could hybridize with lcp1-rep sequence and lbp2 with lcp-2-rep sequence (Fig 2). Genomic DNA digested with selected restriction enzyme followed by in situ PFGE-based Southern blot analysis also confirmed that lbp1 and lbp2 were not integrated into chromosomes. lbp1, cutting with a single restriction enzyme NotI in lbp1was confirmed by Southern blotting using lcp1-rep. lbp2, cutting with a single restriction enzyme PstI in lbp2 was confirmed by Southern blotting using lcp2-rep. The obtained DNA bands were same as above (Fig 2). These data showed that lbp1 contained same rep sequence with lcp1 and lbp2 contained same rep sequence with lcp2, both of which are extrachromosomal. Further bioinformatics analysis showed that the rep of lbp1 shared 87% identity with 100% coverage with lcp1-rep, and the rep of lbp2 shared
Fig 2. Detection of lbp1 and lbp2 plasmids by PFGE.
(A) S1-PFGE-based Southern blot analysis. (B) Single restriction enzyme digestion of the plasmids followed by Southern blot analysis. M represents the standard strain Salmonella enterica serotype Braenderup (H9812) digested with XbaI electrophoresed under pulsed-field conditions as a molecular weight marker. ND, undigested L. borgpetersenii serovar Ballum strain 56604; L. borgpetersenii serovar Ballum strain 56604 were digested by S1 nuclease, NotI and PstI. For Southern blot analysis, the genomic DNA of L. borgpetersenii serovar Ballum strain 56604 was blotted to a nylon membrane and hybridized by lcp1-rep and lcp2-rep. Probes were generated by PCR of T-vector plasmid DNA containing rep genes with primer pairs of lcp1-rep-probe FR, lcp2-rep-probe-FR and lcp3-rep-probe-FR, respectively (S2 Table).
S1-PFGE detection of plasmids within the 15 Chinese epidemic Leptospira strains
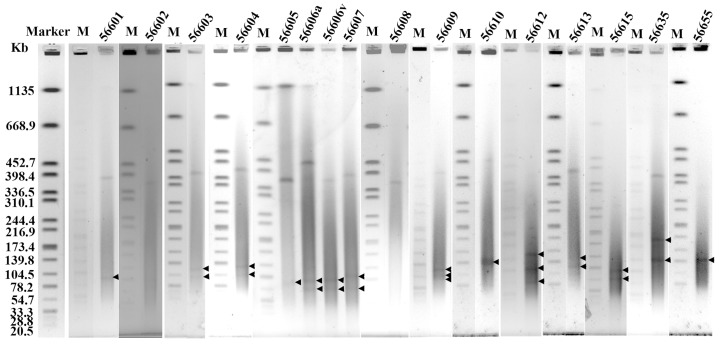
The large size and low copy of plasmids within Leptospira cells presents a challenge for their isolation[12]. Moreover, the high proportion of homologous sequence also makes it difficult to differentiate them from the chromosome and affects the assembly process, even during whole genome sequencing[12]. S1 nuclease treatment can convert the supercoiled plasmids into full-length linear molecules. When the bacteria harboring plasmid embedded in agarose are digested with S1 nuclease followed by pulsed-field gel electrophoresis (PFGE), plasmids can be detected and their sizes can be estimated with appropriate linear DNA markers [20]. In this study, 15 Chinese epidemic Leptospira strains were subjected to S1-PFGE, a distinct approach to detect the presence and the size of plasmids within Leptospira cells. According to the results of S1-PFGE, plasmids can be directly detected in 13 out of 15 tested Leptospira strains, with the exception of L. borgpetersenii strain 56602 and L. interrogans strain 56608 (Fig 3 and Table 3). The sizes ranged from 50 kb to 150 kb (Fig 3), and one Leptospira cell can contain up to three plasmids. It showed that the plasmids have been detected in all tested species, including L. borgpetersenii, L. weilii, and L. interrogans.
Fig 3. Plasmids detection in 15 Chinese epidemic Leptospira strains by S1-PFGE separation.
Leptospira strains were embedded in agarose, lysed and digested with S1 nuclease and electrophoresed under pulsed-field conditions. Marker represents the pattern of the standard strain Salmonella enterica serotype Braenderup (H9812) digested with XbaI electrophoresed under pulsed-field conditions as a molecular weight marker. M represents the marker electrophoresed with each sample. 56601–56655 represent 15 Chinese epidemic Leptospira strains as detailed in Table 3.
Table 3. Homology sequence of lcp1-rep, lcp2-rep and lcp3-rep detected in 15 Chinese epidemic Leptospira strains.
| Strain | Species | Serovar | lcp1-rep | lcp2-rep | lcp3-rep |
|---|---|---|---|---|---|
| 56601 | L.interrogans | Lai | 0 | 1 | 0 |
| 56602 | L.borgpetersenii | Javanica | 0 | 0 | 0 |
| 56603 | L.interrogans | Canicola | 1 | 1 | 0 |
| 56604 | L.borgpetersenii | Ballum | 1 | 1 | 0 |
| 56605 | L.interrogans | Pyrogenes | 1 | 0 | 0 |
| 56606a | L.interrogans | Autumnalis | 1 | 1 | 0 |
| 56606v | L.interrogans | Autumnalis | 1 | 1 | 0 |
| 56607 | L.interrogans | Australis | 1 | 0 | 1 |
| 56608 | L.interrogans | Pomona | 0 | 0 | 0 |
| 56609 | L.interrogans | Linhai | 1 | 1 | 1 |
| 56610 | L.interrogans | Hebdomadis | 0 | 1 | 0 |
| 56612 | L.interrogans | Paidjan | 2 | 1 | 0 |
| 56613 | L.borgpetersenii | Tarassovi | 1 | 1 | 0 |
| 56615 | L.interrogans | Qingshui | 1 | 1 | 0 |
| 56635 | L.borgpetersenii | Wolffi | 1 | 1 | 0 |
| 56655 | L.weilii | Mini | 0 | 1 | 0 |
Number represents the band detected by Southern blot hybridized by specific rep sequence probes following S1-PFGE separation.
S1-PFGE-based Southern blot analysis of rep gene homology among Leptospira with lcp1, lcp2, and lcp3-rep specific probes
In our previous study, the whole genome of L. interrogans serovar Linhai strain 56609 was sequenced, and three extrachromosomal replicons, designated lcp1, lcp2, and lcp3, were in the cell[12]. The homology of rep genes in the plasmids detected by S1-PFGE was tested by Southern blot hybridization using lcp1, lcp2, and lcp3-rep specific probes. As shown in Fig 4 and Table 3, each of the plasmids detected by S1-PFGE in 13 Chinese epidemic strains can hybridize with one of three known rep specific probes. Eleven plasmids distributed in 10 strains can hybridize with lcp1-specific probes (Fig 4A). Of note, two plasmids in strain 56612 can hybridize with lcp1-specific probes. Eleven plasmids distributed in 11 strains can hybridize with lcp2-specific probes (Fig 4B). Two plasmids distributed in two strains can hybridize with lcp3-specific probes (Fig 4C). Plasmid partitioning system encoded two partitioning proteins ParAB protein and a replication protein rep protein. Plasmids with the same replication control are “incompatible”, whereas the plasmids with different replication control are “compatible”[32]. rep has been frequently used to classify plasmids[33]. It seems that the plasmids in Chinese epidemic strains can be divided into three types: lcp1, lcp2, and lcp3.
Fig 4. Identification of homologous sequence of lcp1-rep, lcp2-rep and lcp3-rep in 15 Chinese epidemic Leptospira strains.
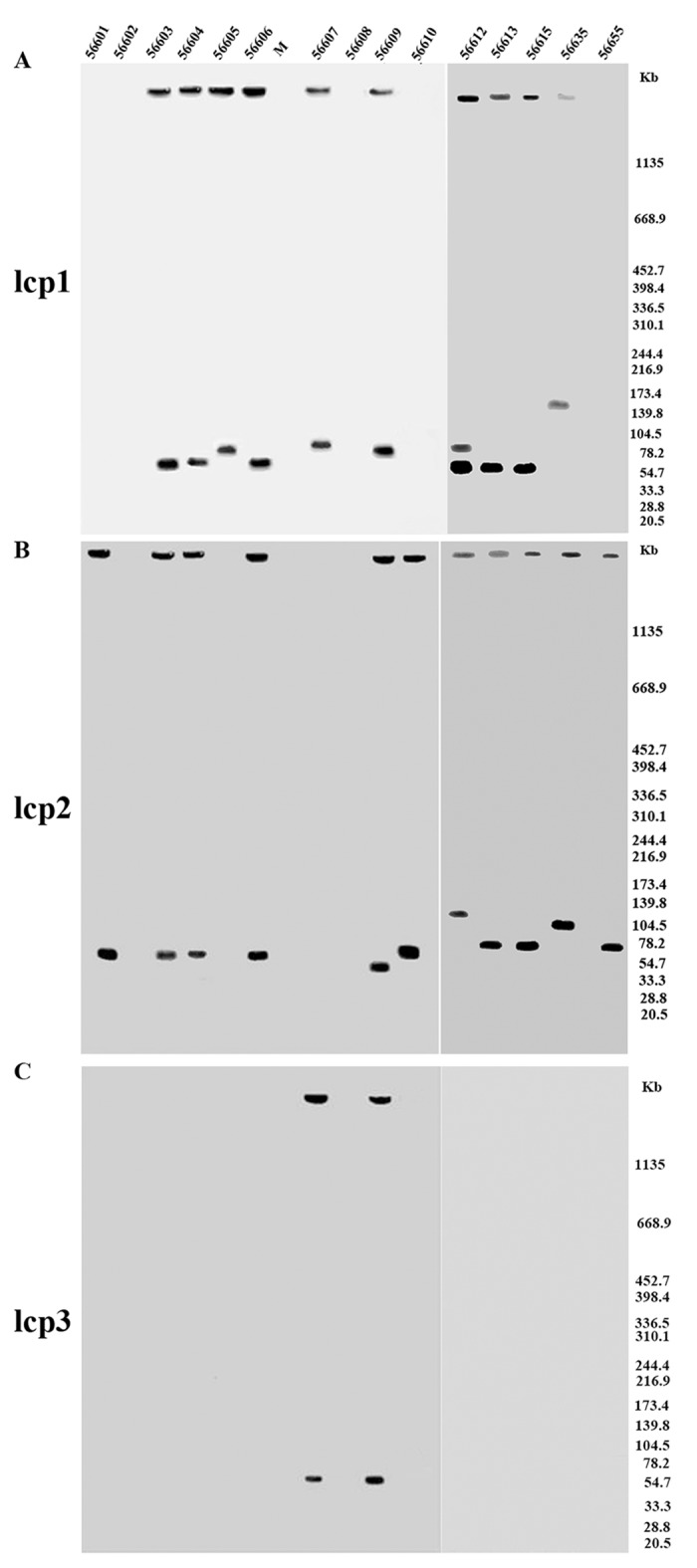
Leptospira strains were embedded in agarose, lysed and digested with S1 nuclease and electrophoresed under pulsed-field conditions, blotted to a nylon membrane and then hybridized with the probes of lcp1-rep(A), lcp2-rep(B) and lcp3-rep(C). Probes were generated by PCR of T-vector plasmid DNA containing rep genes with primer pairs of lcp1-rep-probe FR, lcp2-rep-probe-FR and lcp3-rep-probe-FR, respectively (S2 Table). Markers are shown on the right. 56601–56655 represent 15 Leptospira Chinese epidemic strains as detailed in Table 3.
When compared with the previous results to test for presence of the rep gene in the same 15 Chinese epidemic strains, more plasmids were detected in this study[12]. The only difference is that in the previous study, the embedded bacterial cells were digested with the selected restriction enzyme, whereas in this study, the cells were digested with S1 nuclease. In the previous study, some plasmids without cleavage sites for the selected restriction enzyme may have gone undetected because of being supercoiled. S1-PFGE is a general method independent of knowing restriction enzyme cleavage sites and thus can detect all the plasmids in a cell[20].
lcp1, lcp2, and lcp3-rep homologous sequence widely distributed in pathogens (Group I) of genus Leptospira
The genus Leptospira is comprised of 21 species clustered into following groups, pathogen (Group I), intermediate pathogen (Group II), and non-pathogen based on 16S rRNA phylogeny[1]. The infectious group includes at least 14 species, nine species of pathogens, and five species of intermediate pathogens. The genomes of 319 Leptospira strains belonging to 20 species have been sequenced and deposited in the GenBank database. Blastp search of the lcp1, lcp2, and lcp3-rep genes against the sequenced Leptospira genomes showed that 6 species (299 strains sequenced) including 77 strains harbored homologous sequence with lcp1-rep, 6 species (299 strains sequenced) including 80 strains harbored homologous sequence with lcp2-rep, and 7 species (301 strains sequenced) including 71 strains harbored homologous sequence with lcp3-rep (Table 4). All of the 7 species belonged to pathogens (Group I). Of the 319 sequenced strains, 304 strains belonged to pathogens (Group I); only 8 strains belonged to intermediate pathogens, and 7 strains belonged to the non-pathogens group. Obviously, more genome data of intermediate pathogens and non-pathogens are needed to understand the relationship between extrachromosomal replicons and their host range.
Table 4. The distribution of lcp1-rep, lcp2-rep and lcp3-rep in sequenced Leptospira strains.
| Group | Species | Sequenced strain | lcp1-rep strain | lcp2-rep strain | lcp3-rep strain |
|---|---|---|---|---|---|
| pathogens | L. alexanderi | 1 | - | - | - |
| L. weilii | 9 | 6 | 7 | 7 | |
| L. borgpetersenii | 22 | 10 | 10 | 10 | |
| L. santarosai | 27 | 7 | 1 | 7 | |
| L. meyeri | 2 | - | - | - | |
| L. alstonii | 2 | - | - | 1 | |
| L. interrogans | 206 | 39 | 51 | 34 | |
| L. kirschneri | 26 | 9 | 9 | 9 | |
| L. noguchii | 9 | 6 | 2 | 3 | |
| Intermediate group | L. licerasiae | 4 | - | - | - |
| L. wolffii | 1 | - | - | - | |
| L. fainei | 1 | - | - | - | |
| L. inadai | 1 | - | - | - | |
| L. broomii | 1 | - | - | - | |
| non-pathogens | L. idonii | - | - | - | |
| L. vanthielii | 1 | - | - | - | |
| L. biflexa | 2 | - | - | - | |
| L. wolbachii | 1 | - | - | - | |
| L. terpstrae | 1 | - | - | - | |
| L. yanagawae | 1 | - | - | - | |
| L. kmetyi | 1 | - | - | - |
“-” represents zero.
Discussion
Heterogeneity within the genus Leptospira is well confirmed based on recent studies adopting DNA-DNA hybridization, comparative genomics, and whole genome sequencing efforts [2,5,12]. There are currently 21 species within the genus Leptospira, 14 of which are supposedly pathogenic. However, based on prevalence and pathogenicity, L. interrogans and L. borgpetersenii are the two largest species, containing about half of the known 230 pathogenic serovars, although the latter strain is less well characterized [6]. L. borgpetersenii genomes are 700 kb smaller than that of L. interrogans [6,30] and its genome reduction may be IS-mediated, because there are more copy numbers of IS elements in sequenced L. borgpetersenii serovar Hardjo compared with in L. interrogans serovars Lai and Copenhageni [6]. IS sequence, as a mobile genetic element, varies between serovars. Several IS elements, including IS1500, IS1501, IS1502, IS1533, and ISLin1, were identified in Leptospira [2,5,6,29,30]. Although the genome size of L. borgpetersenii serovar Ballum strain 56604 showed a similar trend of genome reduction as L. borgpetersenii serovar Hardjo, the type and copy number of IS elements in serovar Ballum were apparently less than those in serovar Hardjo, with the exception of two single copy extrachromosomal elements.
Plasmids are important genetic vehicles playing an important role in the processes of genomic organization, adaptation, evolution, and virulence for bacterial pathogens [13]. Also, plasmids are very important genetic tools to manipulate and analyze microorganisms through introduction, modification, or removal of target genes [13]. However, in some cases due to their occurrence in low copy numbers and containment of repeat sequences (homologous to chromosomes), the method to detect plasmids in Leptospira is primarily dependent on whole genome sequencing and assembly[10,12]. Even using whole genome sequencing, plasmid could be miss annotated and assembled into chromosomes [19]. Based on the plasmids identified, shuttle vectors constructed with the rep genes can be successfully transformed into some pathogenic Leptospira[12,16]. Although genetic incompatibility is supposed to hinder successful transformation, the precise mechanism is still unknown[12]. Identification of more replicons in different strains will help to understand the mechanism and facilitate the manipulation of genetic tools. In this study, S1-PFGE was applied as an effective method to detect the plasmids within Leptospira cells, which yielded accurate bands and sizes of plasmids in all tested Leptospira strains, although despite our best efforts bands were smeared to some degree. We speculate that despite S1-PFGE method is sensitive, the low copy number of plasmids in Leptospira probably render them more difficult for detection.
We tested the plasmid distribution among 15 Chinese epidemic Leptospira strains, including L. borgpetersenii, L. weilii, and L. interrogans using S1-PFGE, which enable us to detect potentially all plasmids in sequenced strains 56601, 56603(Gui44), and 56609. The band numbers (corresponding to one, two, and three bands, respectively) and size values were consistent with previously identified plasmids. Furthermore, Southern blotting using known lcp1, lcp2, and lcp3 specific probes further confirmed them as extrachromosomal elements. Interesting, it seems that plasmids detected in Chinese epidemic strains can be divided into three types: lcp1, lcp2, and lcp3. They can exist together or separately within Leptospira cells. Moreover, same type plasmids (lcp1 in strain 56612) can also coexist in one cell.
Further analysis of the plasmid-encoded genes did not reveal any unique gene cluster but confirmed the distribution and diversity of plasmids in the genus Leptospira. Blastp search of lcp1, lcp2, and lcp3-rep homologous sequence with 319 sequenced Leptospira genomes further showed they are widely distributed in pathogens (Group I) of Leptospira. Based on the information provided in this study, we speculate that additional assessment of plasmid contents in additional globally prevalent Leptospira strains, should shed new light on significance of these intriguing extrachromosomal DNA elements in leptospiral biology and evolution.
Supporting Information
Circles are numbered from outer to inner and are designated as follows. (A-B) Large chromosome (CI) and small chromosome (CII), (C-D) Plasmid lbp1 and lbp2. Circles 1 and 2 denote forward and reverse strand genes (colors represent functional categories according to COGs). Circles 3, tRNA genes and rRNA genes. The two inner circles for the chromosomes display GC content and GC skew calculated using a 1,000 bp (CI) / 600 bp (CII) window sliding 500 bp (CI) /300 bp (CII) at a time. The two inner circles for the plasmids display GC content and GC skew calculated using a 1,000 bp window sliding 900 bp at a time.
(TIF)
(DOCX)
(DOCX)
(DOCX)
Acknowledgments
We would like to thank Faith Kung and Weinan Zhu for their assistance with the study.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by grants: the National Natural Science Foundation of China (grant numbers 81261160321, 81171587, 81271793, 81201334, 81471908) to XG, KD and PH; http://www.nsfc.gov.cn/; Research Fund for the Doctoral Program of Higher Education of China (grant number 201200731100719) to XG; http://www.cutech.edu.cn/; the Natural Science Foundation of Shanghai (grant number 13ZR1423000) to PH; http://www.stcsm.gov.cn/. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Levett PN (2015) Systematics of leptospiraceae. Curr Top Microbiol Immunol 387: 11–20. 10.1007/978-3-662-45059-8_2 [DOI] [PubMed] [Google Scholar]
- 2. Lehmann JS, Matthias MA, Vinetz JM, Fouts DE (2014) Leptospiral pathogenomics. Pathogens 3: 280–308. 10.3390/pathogens3020280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Vijayachari P, Sugunan AP, Shriram AN (2008) Leptospirosis: an emerging global public health problem. J Biosci 33: 557–569. [DOI] [PubMed] [Google Scholar]
- 4. Bharti AR, Nally JE, Ricaldi JN, Matthias MA, Diaz MM, et al. (2003) Leptospirosis: a zoonotic disease of global importance. The Lancet Infectious Diseases 3: 757–771. [DOI] [PubMed] [Google Scholar]
- 5. Picardeau M (2015) Genomics, proteomics, and genetics of leptospira . Curr Top Microbiol Immunol 387: 43–63. 10.1007/978-3-662-45059-8_4 [DOI] [PubMed] [Google Scholar]
- 6. Bulach DM, Zuerner RL, Wilson P, Seemann T, McGrath A, et al. (2006) Genome reduction in Leptospira borgpetersenii reflects limited transmission potential. Proc Natl Acad Sci U S A 103: 14560–14565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ren SX, Fu G, Jiang XG, Zeng R, Miao YG, et al. (2003) Unique physiological and pathogenic features of Leptospira interrogans revealed by whole-genome sequencing. Nature 422: 888–893. [DOI] [PubMed] [Google Scholar]
- 8. Nascimento AL, Ko AI, Martins EA, Monteiro-Vitorello CB, Ho PL, et al. (2004) Comparative genomics of two Leptospira interrogans serovars reveals novel insights into physiology and pathogenesis. J Bacteriol 186: 2164–2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Picardeau M, Bulach DM, Bouchier C, Zuerner RL, Zidane N, et al. (2008) Genome sequence of the saprophyte Leptospira biflexa provides insights into the evolution of Leptospira and the pathogenesis of leptospirosis. PLoS One 3: e1607 10.1371/journal.pone.0001607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhu WN, Huang LL, Zeng LB, Zhuang XR, Chen CY, et al. (2014) Isolation and characterization of two novel plasmids from pathogenic Leptospira interrogans serogroup Canicola Serovar Canicola strain Gui44. PLoS Negl Trop Dis 8: e3103 10.1371/journal.pntd.0003103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jahn K, Sudek H, Stoye J (2012) Multiple genome comparison based on overlap regions of pairwise local alignments. BMC Bioinformatics 13 Suppl 19: S7 10.1186/1471-2105-13-S19-S7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhu W, Wang J, Zhu Y, Tang B, Zhang Y, et al. (2015) Identification of three extra-chromosomal replicons in Leptospira pathogenic strain and development of new shuttle vectors. BMC Genomics 16: 90 10.1186/s12864-015-1321-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shintani M, Sanchez ZK, Kimbara K (2015) Genomics of microbial plasmids: classification and identification based on replication and transfer systems and host taxonomy. Front Microbiol 6: 242 10.3389/fmicb.2015.00242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Aminov RI (2011) Horizontal gene exchange in environmental microbiota. Front Microbiol 2: 158 10.3389/fmicb.2011.00158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Raz Y, Tannenbaum ED (2014) Repression/depression of conjugative plasmids and their influence on the mutation-selection balance in static environments. PLoS One 9: e96839 10.1371/journal.pone.0096839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pappas CJ, Benaroudj N, Picardeau M (2015) A replicative plasmid vector allows efficient complementation of pathogenic Leptospira strains. Appl Environ Microbiol 81: 3176–3181. 10.1128/AEM.00173-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ricaldi JN, Fouts DE, Selengut JD, Harkins DM, Patra KP, et al. (2012) Whole genome analysis of Leptospira licerasiae provides insight into leptospiral evolution and pathogenicity. PLoS Negl Trop Dis 6: e1853 10.1371/journal.pntd.0001853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bourhy P, Salaun L, Lajus A, Medigue C, Boursaux-Eude C, et al. (2007) A genomic island of the pathogen Leptospira interrogans serovar Lai can excise from its chromosome. Infect Immun 75: 677–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Huang L, Zhu W, He P, Zhang Y, Zhuang X, et al. (2014) Re-characterization of an extrachromosomal circular plasmid in the pathogenic Leptospira interrogans serovar Lai strain 56601. Acta Biochim Biophys Sin (Shanghai) 46: 605–611. [DOI] [PubMed] [Google Scholar]
- 20. Barton BM, Harding GP, Zuccarelli AJ (1995) A general method for detecting and sizing large plasmids. Anal Biochem 226: 235–240. [DOI] [PubMed] [Google Scholar]
- 21. Ewing B, Green P (1998) Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res 8: 186–194. [PubMed] [Google Scholar]
- 22. Ewing B, Hillier L, Wendl MC, Green P (1998) Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res 8: 175–185. [DOI] [PubMed] [Google Scholar]
- 23. Gordon D, Abajian C, Green P (1998) Consed: a graphical tool for sequence finishing. Genome Res 8: 195–202. [DOI] [PubMed] [Google Scholar]
- 24. Delcher AL, Phillippy A, Carlton J, Salzberg SL (2002) Fast algorithms for large-scale genome alignment and comparison. Nucleic Acids Res 30: 2478–2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhang CT, Wang J (2000) Recognition of protein coding genes in the yeast genome at better than 95% accuracy based on the Z curve. Nucleic Acids Res 28: 2804–2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Marchler-Bauer A, Anderson JB, Cherukuri PF, DeWeese-Scott C, Geer LY, et al. (2005) CDD: a Conserved Domain Database for protein classification. Nucleic Acids Res 33: D192–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, et al. (2014) Pfam: the protein families database. Nucleic Acids Res 42: D222–230. 10.1093/nar/gkt1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sullivan MJ, Petty NK, Beatson SA (2011) Easyfig: a genome comparison visualizer. Bioinformatics 27: 1009–1010. 10.1093/bioinformatics/btr039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zuerner RL, Huang WM (2002) Analysis of a Leptospira interrogans locus containing DNA replication genes and a new IS, IS1502. FEMS Microbiol Lett 215: 175–182. [DOI] [PubMed] [Google Scholar]
- 30. Zhong Y, Chang X, Cao XJ, Zhang Y, Zheng H, et al. (2011) Comparative proteogenomic analysis of the Leptospira interrogans virulence-attenuated strain IPAV against the pathogenic strain 56601. Cell Res 21: 1210–1229. 10.1038/cr.2011.46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nascimento AL, Verjovski-Almeida S, Van Sluys MA, Monteiro-Vitorello CB, Camargo LE, et al. (2004) Genome features of Leptospira interrogans serovar Copenhageni. Braz J Med Biol Res 37: 459–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Couturier M, Bex F, Bergquist PL, Maas WK (1988) Identification and classification of bacterial plasmids. Microbiol Rev 52: 375–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, et al. (2005) Identification of plasmids by PCR-based replicon typing. J Microbiol Methods 63: 219–228. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Circles are numbered from outer to inner and are designated as follows. (A-B) Large chromosome (CI) and small chromosome (CII), (C-D) Plasmid lbp1 and lbp2. Circles 1 and 2 denote forward and reverse strand genes (colors represent functional categories according to COGs). Circles 3, tRNA genes and rRNA genes. The two inner circles for the chromosomes display GC content and GC skew calculated using a 1,000 bp (CI) / 600 bp (CII) window sliding 500 bp (CI) /300 bp (CII) at a time. The two inner circles for the plasmids display GC content and GC skew calculated using a 1,000 bp window sliding 900 bp at a time.
(TIF)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.