Abstract
Escherichia coli amine oxidase (ECAO), encoded by the tynA gene, catalyzes the oxidative deamination of aromatic amines into aldehydes through a well-established mechanism, but its exact biological role is unknown. We investigated the role of ECAO by screening environmental and human isolates for tynA and characterizing a tynA-deletion strain using microarray analysis and biochemical studies. The presence of tynA did not correlate with pathogenicity. In tynA+ Escherichia coli strains, ECAO enabled bacterial growth in phenylethylamine, and the resultant H2O2 was released into the growth medium. Some aminoglycoside antibiotics inhibited the enzymatic activity of ECAO, which could affect the growth of tynA+ bacteria. Our results suggest that tynA is a reserve gene used under stringent environmental conditions in which ECAO may, due to its production of H2O2, provide a growth advantage over other bacteria that are unable to manage high levels of this oxidant. In addition, ECAO, which resembles the human homolog hAOC3, is able to process an unknown substrate on human leukocytes.
Introduction
Primary amine oxidases (EC 1.4.3.21, PrAOs, also known as copper-containing amine oxidases, CAOs) are expressed in mammals, plants, and bacteria. These enzymes catalyze the oxidation of primary amines into aldehyde through the reaction R-CH2-NH2 + O2 + H2O → R-CHO + H2O2 + NH3. The preferred amine substrate depends on the specific amine oxidase, and the aldehyde product depends on the substrate [1]. The by-products of the reaction, hydrogen peroxide and ammonia, are invariably released.
In Escherichia coli (E. coli), a copper amine oxidase (ECAO) is encoded by the tynA gene (Entrez Gene ID: 945939, previously known as maoA, [2]). ECAO is a soluble, periplasmic, homodimeric protein with one copper ion and post-translationally derived topaquinones at both of its active sites [3]. Aromatic amines are the preferred substrates of ECAO, as compared to aliphatic amines and polyamines; therefore, it is also called 2-phenylethylamine oxidase or tyramine oxidase (http://www.expasy.org, [3]). ECAO was originally identified in E. coli K-12 [4], and its expression is detected only when the cells are growing in a medium supplemented with tyramine or phenylethylamine (PEA); it is not expressed under anaerobic conditions [5]. The expression of ECAO is regulated by FeaR, an AraC family regulator that also activates a phenylacetaldehyde dehydrogenase–encoding gene, feaB [5]. Because a transposon inserted into the tynA gene causes a constitutive SOS response [6], which protects against severe DNA damage caused by ultraviolet light or DNA-damaging chemicals [7], it has been suggested that tynA could be involved in catabolizing toxic compounds [8].
The crystal structure of ECAO has been solved [9]. It is a prototypic homodimer that has a catalytic section consisting of the D2–D4 domains and an additional D1 domain that is absent from other solved copper amine oxidase (CAO) structures. Although ECAO (as well as PrAOs from Arthrobacter globiformis and Hansenula polymorpha) has been used as a model enzyme to elucidate the enzymatic mechanism of amine oxidases [10–17], its known biological functions have so far been limited to enabling the bacteria to grow on different carbon or nitrogen sources [5,9,18]. In mammals, the corresponding primary amine oxidase, hAOC3, is involved in processes such as leukocyte trafficking and glucose metabolism [19].
In this study, we determined how widely this amine oxidase is found within the bacterial kingdom and in what circumstances it is expressed. We assessed ECAO’s biological role by performing microarray analysis on a deletion strain of E. coli K-12, ΔtynA. We found that ECAO can utilize a human leukocyte cell surface molecule(s) as a substrate and is inhibited by a hAOC3 inhibitor, which suggests that it shares similarities to its human counterpart. These results broaden our knowledge about this enzyme in bacteria and provide us with new insights into its role in E. coli.
Materials and Methods
Reagents
All the reagents were from Sigma-Aldrich unless otherwise stated.
Bacterial strains and DNA
The E. coli strains used were DH5α [20], SY327 λpir [21], S17-1 λpir [22], and wild-type (wt) E. coli strains isolated from different environmental and human sources. Extended-spectrum beta-lactamase (ESBL) strains and wastewater samples were obtained from the National Institute for Health and Welfare, Turku, Finland. Bacterial pathogens were obtained from the Department of Microbiology, University of Turku, Finland. The DNAs of Finnish enterohemorrhagic E. coli (EHEC) strains were from the National Institute for Health and Welfare, Helsinki, Finland. The strains of different E. coli pathotypes were from the Institute for Molecular Infection Biology, University of Würzburg, Germany. E. coli strains isolated from well waters were obtained from the Finnish Water Protection Association, South-Western Finland, Turku, Finland. Strains from natural gut microbiota were collected from the fecal samples of healthy volunteers, and were only used for the development of the cultivation method. Stool samples analyzed for tynA and ECAO activity were from routine clinical microbiology laboratories. The vector pUC19 was from Fermentas (Waltham, MA, USA). E. coli (pKK233-3-tynA) [11] and pRV1 [23] have been characterized before.
The presence of semicarbazide-sensitive amine oxidases in various bacteria
The presence of PrAO enzymes in bacteria was studied using two strategies: bioinformatics and functional assays. In the bioinformatics approach, we searched for bacteria with PrAO enzymes by querying published bacterial sequences (NCBI The Entrez Protein database) for established PrAO PROSITE motifs, namely a topaquinone signature (PS01164, LVVrwisTvgNYDY) and a copper-binding site signature (PS01165, TtGttHVaraEEwP). In the functional assay, we cultured 85 different E. coli strains in minimal medium with a specified amine as the only carbon source and tested the bacterial lysate for the presence of tynA by polymerase chain reaction (PCR, see below).
Culture conditions
Bacteria were cultured in lysogeny broth (LB, [24]) or on Luria agar plates (LA) supplemented with the appropriate antibiotics (ampicillin, 25 μg/mL; chloramphenicol, 20 μg/mL; kanamycin, 100 μg/mL in the plates and 20 μg/mL in liquid media). Selected strains were screened for the expression of tynA by culturing the bacteria in an M9 minimal medium [25] supplemented with 2 g/L lactose and 5 mM PEA (M9-lactose-PEA medium). To induce ECAO activity, an overnight culture of wt E. coli in M9-lactose medium was diluted 1:20 in M9-lactose-PEA with 5 μM CuSO4. The bacteria were grown aerobically at 30°C with 250 rpm shaking. The induction of ECAO was assayed by measuring its activity (see below). To overexpress ECAO, we cultured E. coli (pKK233-3-tynA) in LB supplemented with 5 μM CuSO4 at 37°C and 250 rpm shaking to early log phase and induced the expression of ECAO by adding isopropyl β-1-D-thiogalactopyranoside (IPTG) to a final concentration of 2 mM.
PCR screening for tynA
For the tynA screening, we took E. coli samples from fresh LA plates, suspended the bacteria in water, and incubated the samples for 5 min at 95°C. A total of 1 μL of bacterial lysate was used for PCR. We used forward primer EAO1 (5`-gacggggcatctttaccac-3`) and reverse primer EAO2 (5`-cacgccgctgaccgtagg-3`) to detect a 500–base pair (bp) fragment of the tynA gene. LACYfor (5`-ggtacttcaaacatatgcagcg-3`) and LACYrev (5`-ggctacatgacatcaaccatatc-3`), which amplify a 900-bp fragment of the E. coli lacY gene, were used as positive controls. Primers were from Operon (Cologne, Germany), and the Dynazyme polymerase was from Thermo Scientific (Espoo, Finland). The reaction products were subjected to agarose gel electrophoresis (Qbiogene QA-agarose, Qbiogene, Montreal, Canada), stained with ethidium bromide (Bio-Rad, Hercules, CA, USA), and visualized under ultraviolet light.
Sequencing tynA
The tynA genes from three tynA+ strains that grew in the M9-lactose-PEA medium and five tynA+ strains that did not grow (indicating the enzyme was inactive) were cloned into the pCDNA3.1 plasmid and sequenced (Turku Centre for Biotechnology, University of Turku and Åbo Akademi University, Finland, primer sequences are available on request).
tynA in the E. coli genome
The genomes of tynA+ and tynA- strains were compared using the Artemis Comparison Tool (ACT) online (http://www.webact.org/, [26]).
Construction of a tynA knockout strain (ΔtynA)
The tynA gene and 600 bp of flanking sequences on either side were amplified by PCR, using E. coli K-12 as a template, with the forward primer 5`- GTCAGAGAATTCCACATATGGATAAGATTGCTG -3`(EcoRI site in bold) and the reverse primer 5`- TCTGACGGATCCGACACGCTGGTTAGTGG -3`(BamHI site in bold). The primers were designed according to the published sequence of the tynA locus in the E. coli genome [27]. The 3740-bp PCR product was cloned into the EcoRI/BamHI site of pUC19. A tynA fragment of 2250 bp was removed from the plasmid by inverse PCR (forward primer 5`-CTCAAGATGCATTCAGGTTGTTTTACGGGCAGAATAC-3`and reverse primer 5`-GTCAGAATGCATGACGAAACGCCAACGCTAG-3`, NsiI sites in bold). The tynA gene was replaced with a 1200-bp kanamycin resistance cassette (kmGB), which was a PstI cleavage product from the pUC-4K plasmid (GE Healthcare, Little Chalfont, UK). The tynA ΔkmGB was excised from pUC19 using PvuII, and the blunt ends were ligated into the EcoRV sites of the pRV1 suicide vector. The restriction enzymes EcoRI, BamHI, and NsiI were from NEB (New England Biolabs, Beverly, Massachusetts, USA), and EcoRV and PvuII were from Promega (Madison, Wisconsin, USA). Digestion products were purified from an agarose gel using the Qiagen Gel Extract Kit (Hilden, Germany), and the T4-ligation kit (NEB) was used for ligations.
The pRV1 vector containing the tynA knockout cassette (pRV1-TK) was electroporated first into SY327 λpir and then into S17-1 λpir. The plasmid was conjugated from S17-1 λpir into wt ESBL E. coli 201. The transconjugant bacteria, now having the suicide vector integrated into their genomes by homologous recombination, were subjected to a cycloserine enrichment to select for clones resistant to kanamycin and ampicillin, and sensitive to chloramphenicol. In these clones, a second homologous recombination event had eliminated the suicide vector and the wild-type allele. The constructed mutants were verified by PCR using the primers 200bpFS (5`-GCAACGGCGAATAGTTGC-3`) and 200bpFSrev (5`-CGGTGTGGGTAAACAGCC-3`), and by Southern blotting.
Southern blotting
Genomic DNA from wt and ΔtynA E. coli strains was isolated using the NucleoSpin Tissue Kit (Macherey-Nagel, Dueren, Germany). DNA was digested with HindIII (NEB), and Southern blotting was performed as described previously [28]. In the wt, the 3400-bp fragment encompassing tynA and 600-bp of flanking sequences was labeled using [α-32P]dCTP (50 μCi, 3000 Ci/mmol), according to the Prime-a-Gene Labeling System (Promega, Madison, WI, USA). The probe was purified using a ProbeQuant G-50 Micro Column, and the hybridization was performed in ULTRAhyb hybridization buffer (Ambion, Foster City, CA USA) at 42°C for 14–16 h. Probe detection was performed using X-ray film. The exposure time was 3 h.
Preparation of cell lysates
Bacterial cultures were centrifuged at 3200 × g at 4°C for 15 min, and cell pellets were lysed in phosphate-buffered saline (PBS) containing 1 mM phenylmethylsulfonyl fluoride (PMSF), 1% aprotinin, and 0.1% nonyl phenoxypolyethoxylethanol (NP-40). The cells were lysed by three freeze-thaw cycles using liquid nitrogen and a 37°C water bath. The crude lysates were further treated with short ultrasound pulses (2 × 15 s).
Determination of protein concentration
The concentration of protein within the lysates was determined using the Bio-Rad DC Protein Assay, with bovine serum albumin as the standard. The concentration of a purified protein was determined by a Bradford assay (Bio-Rad).
Amine oxidase activity assay
To determine the amine oxidase activity in the lysates, we utilized the Amplex Red assay (Molecular Probes Europe BV), using 10 μg of bacterial lysate per well. PEA (0.5 mM) was used as a substrate. To specifically determine semicarbazide-sensitive amine oxidase activity, we used 1 mM semicarbazide (SC) in the control wells. ECAO activity was measured as the difference in fluorescence between SC-inhibited and uninhibited samples. To test two new substrates, isoamylamine and propylamine, we used the same assay and identical amounts of substrate. The activity assays were performed at 30°C.
To test if ECAO has a substrate on human granulocytes, we used purified recombinant protein ECAO, whose production and purification has been described [9]. To collect human neutrophils, we extracted granulocytes from the fresh peripheral blood of healthy volunteers using Ficoll (GE Healthcare). We collected buffy coats according to standard procedure, lysed red blood cells with Pharm Lyse (BD Biosciences, Franklin Lakes, NJ, USA), washed with PBS, counted the cells, and used the cells immediately. The assay was conducted in 25 mM HEPES and 150 mM NaCl (pH 7.4), and we used 30 nM ECAO and 200,000 viable granulocytes per well. PEA was used as a positive control for the substrate, and the specific activity was determined using parallel wells with SC, as described above. The assay was conducted at 37°C. Three independent assays (different blood donors) were performed with duplicates.
ECAO inhibitor tests
To determine the effect of different inhibitors on ECAO activity, we used purified ECAO protein and a spectroscopic amine oxidase activity assay, which is not as sensitive to additives as the Amplex Red assay [29]. In short, 21 ng of ECAO protein was incubated with differing amounts of inhibitors in Krebs-Ringer phosphate glucose buffer for 20 min at 30°C. After incubation, we added substrate (0.5 mM PEA) and a mixture of 1 mM vanillic acid, 0.5 mM 4-aminopyridine, and 16 U of horseradish peroxidase, and measured the A492nm immediately and hourly thereafter.
To assess the effect of inhibitors on bacterial growth, discs with different concentrations of SC were placed on LA plates freshly plated with either tynA+ E. coli (three different strains) or tynA- E. coli (five strains). The inhibition zones were measured the next day. For the growth inhibition assay in a liquid medium, the same strains were cultured in tryptic soy broth (TSB) medium for 14–16 h. Then, 103 cells were inoculated into TSB medium with different concentrations (0–100 mg/mL) of SC and cultured further at 37°C with shaking at 100 rpm until the following day. Then, 10 μL of bacterial mixture was plated on lactose medium plates, and the plates were incubated at 37°C for 14–16 h before counting the colonies.
The inhibition of growth by a selective hydrazine-based human amine oxidase inhibitor, BTT-2052 [30], and SC on tynA+ and tynA- strains was tested similarly in M9-lactose-PEA medium using inhibitor concentrations of 0–0.5 mM.
RNA extraction
To extract RNA from the bacteria cultured in M9-lactose-PEA medium, we added two volumes of RNA Protect Bacteria (Qiagen) immediately to samples taken from the E. coli cultures at 0 h, 1 h, and 4 h. Total RNA was isolated using the RNeasy Mini-Kit (Qiagen), and DNA was removed using the RNase-Free DNase Set (Qiagen). The concentration of the extracted RNA was determined by a NanoDrop 2000 (Thermo Scientific), and the quality of RNA was quantified by Bio-Rad’s Experion electrophoresis station.
cDNA preparation and array hybridization
Microarray studies were performed at the Finnish DNA Microarray Centre at Turku Centre for Biotechnology, Turku, Finland. Standard microarray methods were used for cDNA synthesis, fragmentation, and end-terminus biotin labeling, and were based on Affymetrix protocols. Equal amounts of total RNA from seven different samples were pooled to yield the required 10 μg of starting material, and 2.4 μg of fragmented and labeled cDNA was used in the preparation of the hybridization cocktail. The cocktail was prepared according to the Midi format. Each sample was hybridized to the GeneChip E. coli 2.0 Array. A GeneChip Fluidics Station 450 was used to wash and stain the arrays, and a GeneChip Scanner 3000 with Autoloader was used to scan the arrays. Affymetrix GeneChip Command Console (AGCC) software version 1.0 was used to control the Fluidic Station and Scanner. CEL files were automatically generated by AGCC. After the hybridization, quality control was done with the Affymetrix Expression Console.
Data analysis
All the microarray data analysis was done using the R language and environment for statistical computing and Bioconductor [31,32]. The array data was normalized with the gcRMA method [33]. The resulting data was filtered using a threshold of >2.5 and <-2.5 for the fold-change of up- and down-regulated genes, respectively. The microarray data discussed here has been deposited in NCBI's Gene Expression Omnibus [34] and are accessible through GEO Series accession number GSE65385 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE65385).
Production of hydrogen peroxide in the medium by ECAO
Wt ECAO was induced in M9-lactose-PEA medium cultures as described above. The ΔtynA strain was used as a negative control, and the cultures were grown for 6 h, as described. The cells were separated by centrifugation (1.5 min at 15300 × g), and the supernatant was assayed for activity by measuring the amount of hydrogen peroxide in the medium using the Amplex Red assay and by measuring the fluorescence using a fluorometer (TECAN Ultra, Tecan, Zürich, Switzerland).
Ethics
For the cultivation of tynA+ strains, as well as for measuring ECAO activity, we used collections of bacterial specimens that are investigated in routine clinical microbiology laboratories in Finland. In 2006, when the experiments were done, no ethical approval for the usage of these bacterial samples was needed, as all of them were from anonymous donors. For the development of a method to cultivate specimens from stool under specific conditions, we collected a few stool samples from the authors, who gave their oral informed consent for the usage of their samples for this purpose. One of the authors (T.K.) collected, stored, and cultivated the samples anonymously. We did not collect any identifying information from the donors, and none of the experiments or results were ever connected to their identities.
Statistical analyses
The data are shown as the mean ± the standard error of the mean (SEM). Statistical tests, unless otherwise mentioned, were conducted using the Student’s t-test, and a value of p < 0.05 was considered statistically significant.
Results
Amine oxidases in various bacteria
To analyze which bacteria could harbor a homolog of PrAO, we conducted a database search using two PROSITE motifs that defined the active sites and the copper-binding sites of amine oxidases. The database search revealed that the amine oxidase gene exists in many environmental bacterial species (isolated from soil, sea water, pesticide- and metal-contaminated soil), as well as in opportunistic pathogens, but it exists in only a few clinically important species (S1 Table).
ECAO in E. coli
The existence of tynA in different E. coli strains was assayed first by screening 38 Finnish wastewater and well water samples for the presence of tynA using PCR (Table 1, S1 Fig). More than half of the well water samples (57%) and about one third of the wastewater samples (35%) were tynA+. The subsequent screening of different human isolates and E. coli pathotypes indicated that this gene could be frequently detected in diarrhea-causing pathotypes, such as non-O157:H7 EHECs (82% tynA+), enterotoxigenic E. coli (ETEC; 69%), and enteroinvasive E. coli (EIEC; 67%), as well as in fecal isolates (38%, Table 2). Of note, tynA was less frequently detected in enteropathogenic E. coli (EPEC; 17%). Among extra-intestinal pathogenic isolates, tynA was most frequently detected in sepsis isolates (27%). Out of the few tested pus (7), wound (3), and blood (8) samples, 71%, 67%, and 50% were tynA+, respectively. The tynA screening was negative in newborn meningitis (11) and in O157:H7 EHEC (28) isolates.
Table 1. Frequency of E. coli in environmental isolates. a .
| Waste water | Well water | |
|---|---|---|
| Total | 20 | 18 |
| tynA+ | 7 | 12 |
| % | 35 | 57 |
atested by PCR.
Table 2. Frequency of E. coli in human isolates. a .
| Wound | Pus | Blood | Sepsis | Feces | EIEC | EPEC | UPEC | ETEC | NBM | EHEC O157:H7 | EHEC Non-O157:H7 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total | 3 | 7 | 8 | 15 | 99 | 3 | 6 | 45 | 13 | 11 | 28 | 49 |
| tynA+ | 2 | 5 | 4 | 4 | 38 | 2 | 1 | 3 | 9 | 0 | 0 | 40 |
| % | 67 | 71 | 50 | 27 | 38 | 67 | 17 | 7 | 69 | 0 | 0 | 82 |
atested by PCR.
Genome comparisons
To gain further insight into the evolution of tynA in E. coli, we compared the genomes of a tynA-positive (K-12) and a tynA-negative (UPEC CFT073) strain with the ACT [26], which revealed that a 33-kb area flanking the tynA gene is missing from CFT073 (S2 Fig). When another tynA-negative strain, IHE3034, was included, we found the rearrangement of 930 kb of flanking sequence by an inversion.
Natural tynA mutations
When screening for culture conditions that induced tynA expression, we found that ECAO activity was induced only when tynA+ strains were grown with PEA as the carbon source (data not shown). However, when we tested the amine oxidase activity of several strains that were tynA+, according to our PCR screening, some did not grow in M9-lactose-PEA medium. Only a subset (22 out of 39 tynA+ strains) both grew and had amine oxidase activity, as determined in our standard Amplex Red assay (S2 Table). The sequencing of tynA genes from seven strains, both inactive and active, demonstrated that four strains, including DH5α, had mutations in tynA when compared to the tynA of wt E. coli K-12, but none of the mutations were in the conserved residues essential for amine oxidase activity (S3 Table). Therefore, we propose that the presence of tynA does not necessarily reflect the existence of the functional enzyme.
ECAO expression and activity depends on the presence of both lactose and PEA in the medium
To study the biological role of tynA in E. coli K-12, we constructed a tynA-deletion strain (ΔtynA) through allelic exchange. The successful allelic exchange was verified by PCR, and the deletion of tynA was confirmed by Southern blot (S3 Fig).
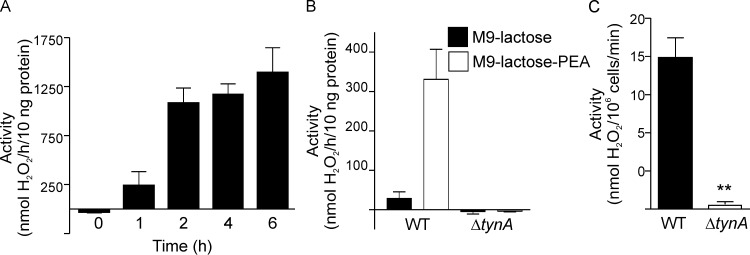
We assayed the expression and activity of the tynA gene product, ECAO, on PEA in different media. When culturing wt bacteria in M9-lactose-PEA medium, we detected activity on PEA (5 mM) after 1 h, and the activity reached a maximum at 2 h after induction (Fig 1A). No significant activity was detected in M9-PEA medium supplemented with another sugar, such as glucose, arabinose, rhamnose, sucrose, or maltose, as a carbon source (data not shown). To analyze the relationship between ECAO expression and the presence of both lactose and PEA in the medium, we tested the amine oxidase activity of wt and ΔtynA strains in minimal medium M9 supplemented with lactose, with or without 5 mM PEA. We detected a clear dependence of activity on the presence of lactose and PEA for wt, but the ΔtynA strain had no activity in either (Fig 1B), which has been shown before with a different medium [18]. To measure the amount of H2O2 released into the medium by ECAO, we measured the H2O2 concentrations of stationary phase cultures of wt and ΔtynA strains. Again, the ΔtynA strain failed to produce any H2O2 in the medium, whereas the wt strain had concentrations of up to 1 μM H2O2 (Fig 1C), similar to earlier findings [18]. To test the endurance of ECAO expression, a total of nine different strains (and our ΔtynA strain) were cultured in similar conditions for 7 d, during which the cells continued to grow without entering stationary phase. ECAO activity remained at a similar level, although it differed among the strains. The ΔtynA strain was constantly inactive (data not shown).
Fig 1. The time and medium dependence of ECAO expression and activity.
(A) The kinetics of ECAO induction in E. coli K-12 (wt) cultured in M9-lactose medium supplemented with 5 mM PEA (n = 4). The 0 h time point corresponds to the time when the cells were diluted into PEA-containing culture medium. (B) The activity of ECAO is induced in E. coli K-12 (wt), but not in our ΔtynA strain, by the presence of both lactose and PEA 6 h after diluting the bacterial cultures in M9-lactose-PEA media (n = 3). (C) The H2O2 production of wt K-12 E. coli and ΔtynA bacteria in stationary phase in M9-lactose-PEA medium (** p < 0.01). The means ± SEMs are shown.
Inhibition of ECAO with semicarbazide
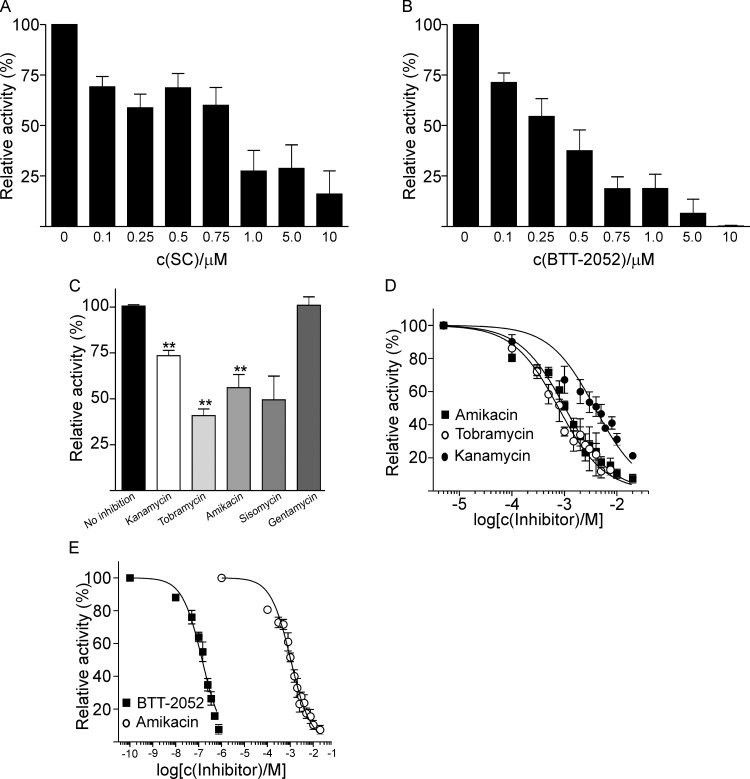
ECAO preferentially processes aromatic amine substrates such as PEA, tyramine, and tryptamine, whereas the human homolog of ECAO, hAOC3, is most active towards benzylamine (BA) and methylamine [3,35]. In our experiments, we confirmed the preferential activity of ECAO on PEA, tyramine, and tryptamine over BA (S4 Fig). To our knowledge, the effects of different amine oxidase inhibitors on ECAO have not been tested; therefore, we compared the effects of a general PrAO inhibitor, SC, and a selective hAOC3 inhibitor, BTT-2052, on ECAO activity on PEA (Fig 2). Among seven different inhibitor concentrations (0.1–10 μM), we observed a half maximal inhibitory concentration (IC50) of 0.7 μM for SC and 0.24 μM for BTT-2052.
Fig 2. Different ECAO inhibitors.
(A) The relative inhibition of ECAO by semicarbazide (SC) and (B) the hAOC3 inhibitor BTT-2052. (C) The effect of different aminoglycosides (1 mM) on the activity of ECAO (** p < 0.01). (D) The inhibition curves of different aminoglycosides on ECAO activity. (E) The pairwise comparison of inhibition curves between BTT-2052 and amikacin.
We tested the effect of the SC and BTT-2052 inhibitors on the growth of the wt and ΔtynA strains on plates and in liquid media. When we used SC concentrations lower than 5–10 mM, we detected no significant effect on growth in liquid rich media (S5 Fig). As expected, SC had no effect on the growth of the ΔtynA strain (S5 Fig). On LA plates, SC had no effect on bacterial growth under the concentration of 100 mg/mL, and even at 100 mg/mL, the effect was minimal (data not shown). BTT-2052 had no effect on the growth of either wt or ΔtynA strains on plates at the investigated concentrations (0.1–0.5 mM, data not shown).
Certain aminoglycosides are able to inhibit ECAO activity
Aminoglycosides, which consist of branched primary amine groups attached to a ring structure (tetrahydropyran or cyclohexane), are antibiotics against Gram-negative bacteria and are reported to inhibit bovine amine oxidase [36]. We tested the effect of aminoglycosides on ECAO activity; kanamycin, tobramycin, amikacin, and sisomycin (p = 0.06) decreased activity by 23–61% (Fig 2C). Gentamycin had no effect. To quantify the inhibition kinetics of amikacin, tobramycin, and kanamycin, we measured the activity of ECAO on PEA in different aminoglycoside concentrations (10 μM–10 mM). The half maximal inhibitory concentrations (IC50) were 1 mM for amikacin, 0.8 mM for tobramycin, and 3.8 mM for kanamycin (Fig 2D). The inhibition kinetics of amikacin and BTT-2052 were compared across nine different inhibitor concentrations (Fig 2E). The shape of the curves is similar, but the IC50 values differ by an order of magnitude.
Knocking out tynA interferes with several pathways in E. coli
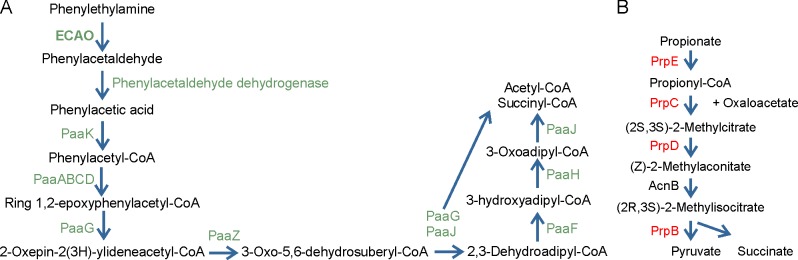
To assay the biological role of tynA in E. coli, we performed microarray analysis using RNA from wt and ΔtynA cell lysates. After 4 h of growth in M9-lactose-PEA medium, the most differentially expressed gene between ΔtynA and wt was tynA, as expected (214-fold down-regulated, Table 3). In addition, the enzymes involved in the aerobic catabolic pathway of phenylacetate (paaABCDEFGHIJKXY and maoC [paaZ]) and phenylacetaldehyde dehydrogenase (feaB) were down-regulated (FC = -108 –-2.9, Fig 3A). Furthermore, we noticed a decrease in the expression of oxyS, grxA, and ahpCF, which are regulated by the oxidative stress regulator OxyR [37]; OxyS is a regulatory sRNA, GrxA is a glutaredoxin, and AphCF is an alkyl hydroperoxide reductase [38–40]. Other genes, which were expressed with less than a -2.5-fold difference, were hypothetical proteins c0693 and c4090, and S-adenosyl-L-methionine-dependent methyltransferase (smtA, FC -2.8, -2.8, and -2.9, respectively).
Table 3. Differentially down-regulated (FC < -2.50) genes in ΔtynA. a .
| Gene symbol | Gene | FC |
|---|---|---|
| tynA | Tyramine oxidase, copper-requiring | -213.77 |
| pad | Predicted multicomponent oxygenase/reductase subunit for phenylacetic acid degradation | -108.30 |
| paaC | Predicted multicomponent oxygenase/reductase subunit for phenylacetic acid degradation | -86.95 |
| maoC | Fused aldehyde dehydrogenase/enoyl-CoA hydratase | -83.89 |
| paaA | Predicted multicomponent oxygenase/reductase subunit for phenylacetic acid degradation | -76.72 |
| paaB | Predicted multicomponent oxygenase/reductase subunit for phenylacetic acid degradation | -67.16 |
| paaG | Acyl-CoA hydratase | -55.67 |
| paaE | Predicted multicomponent oxygenase/reductase subunit for phenylacetic acid degradation | -53.42 |
| paaF | Enoyl-CoA hydratase-isomerase | -49.38 |
| feaB | Phenylacetaldehyde dehydrogenase | -39.84 |
| paaI | Predicted thioesterase | -24.85 |
| paaJ | Predicted beta-ketoadipyl CoA thiolase | -22.80 |
| paaK | Phenylacetyl-CoA ligase | -13.82 |
| paaH | 3-Hydroxybutyryl-CoA dehydrogenase | -13.09 |
| oxyS | OxyS RNA | -10.16 |
| grxA | Glutaredoxin 1 | -6.30 |
| paaX | DNA-binding transcriptional repressor of phenylacetic acid degradation, aryl-CoA responsive | -4.81 |
| smtA | Putative metallothionein | -2.88 |
| pay | Predicted hexapeptide repeat acetyltransferase | -2.87 |
| ahpF | Alkyl hydroperoxide reductase subunit F | -2.85 |
| c4090 | Hypothetical protein | -2.79 |
| ahpC | Alkyl hydroperoxide reductase subunit C | -2.76 |
| c0693 | Hypothetical protein | -2.75 |
aComparisons to wt bacteria were made at the 4 hr time point
Fig 3. Pathways altered by tynA knockout.
(A) The phenylethylamine metabolic pathway is down-regulated (green) in the ΔtynA strain when compared to the wt strain in M9-lactose-PEA medium. (B) The oxidation of propionate to pyruvate is up-regulated (red) in the ΔtynA strain. Pathways are depicted according to Brock et al. and Zeng et al. [5,41].
The most up-regulated genes in ΔtynA were those encoding the enzymes involved in oxidizing propionate to pyruvate: prpBCDE with FC 9.38 to 3.39 (Table 4, Fig 3B). Notably, there were fewer genes up-regulated than down-regulated, and the magnitude of up-regulation was lower (the highest fold-change was 9.38, whereas the lowest was -213.77). Two other pairs of up-regulated genes were those of the nitrogen regulatory protein C (NtrC)–activated polar amino acid transporter family yhdXZ [42], with FC = 6.09 and 3.44, respectively, and the respiratory nitrate reductase genes narUW [43], with FC = 2.97 and 3.14 respectively. See Table 4 for the other up-regulated genes.
Table 4. Differentially up-regulated (FC > 2.50) genes in ΔtynA. a .
| Gene symbol | Gene name | FC |
|---|---|---|
| prpE | Putative propionyl-CoA synthetase | 9.38 |
| prpD | 2-Methylcitrate dehydratase | 8.50 |
| prpC | Methylcitrate synthase | 7.99 |
| yhdX | Putative transport system permease protein | 6.09 |
| gabT | 4-Aminobutyrate aminotransferase | 3.53 |
| yhdZ | Hypothetical amino-acid ABC transporter ATP-binding protein YhdZ | 3.44 |
| prpB | 2-Methylisocitrate lyase | 3.39 |
| narW | Cryptic nitrate reductase 2 delta | 3.14 |
| narU | Nitrite extrusion protein 2 | 2.97 |
| aldB | Aldehyde dehydrogenase B (lactaldehyde dehydrogenase) | 2.83 |
| pqiA | Paraquat-inducible protein A | 2.71 |
| hemN | Coproporphyrinogen-III oxidase | 2.60 |
| ygaM | Hypothetical protein | 2.53 |
| glgS | Glycogen synthesis protein GlgS | 2.53 |
| ldhA | D-Lactate dehydrogenase | 2.50 |
aComparisons to wt E. coli were done at the 4 h time point.
ECAO activity on granulocytes
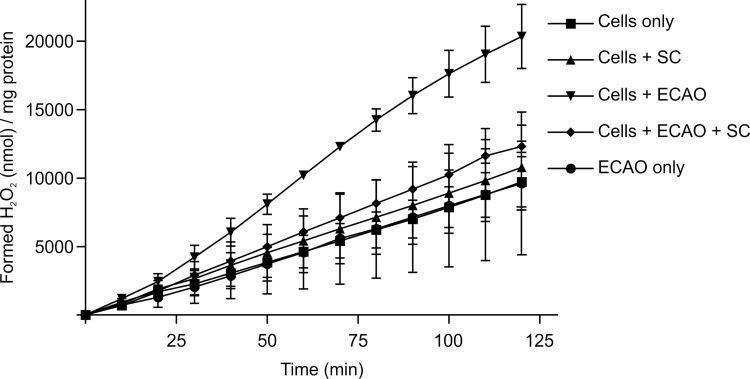
To determine whether bacterial ECAO could function like its human counterpart, hAOC3, by acting on human leukocytes, we tested the activity of purified, recombinant ECAO protein [9] with fresh human granulocytes as the sole source of substrate. We detected significantly more H2O2 production when granulocytes were added to ECAO than in the condition without cells, without ECAO, or with SC as an inhibitor (Fig 4). Clorgylin was added to prevent monoamine oxidase activity.
Fig 4. ECAO is able to use human granulocytes as a substrate.
We detected the production of H2O2 by ECAO when granulocytes were the only source of substrate (Cells + ECAO), and the activity was inhibited by semicarbazide (Cells + ECAO + SC). The activity is significantly higher than without ECAO (Cells only, p = 0.0003; cells with SC, p = 0.0004; without cells, p = 0.017, and with added SC inhibitor [Cells+ECAO+SC], p = 0.004). The mean of three independent experiments is shown ± SEM. Statistical significance was calculated with paired t-test.
Discussion
We found that the gene encoding the primary amine oxidase, tynA, is present in various bacteria. We screened several environmental and human isolates for both tynA presence and activity, and demonstrated that some strains possess the gene, but do not have its activity. We replicated the previous results that ECAO enables bacteria to grow on a rare nitrogen or carbon source, such as PEA, and that the activity of the tynA gene product in E. coli periplasm drives the production and release of hydrogen peroxide into the environment [18]. As new findings, we demonstrated that ECAO is inhibited by certain aminoglycosides and report that ECAO can utilize human leukocytes as substrates. In addition, ECAO was inhibited by a human amine oxidase inhibitor suggesting that the bacterial protein mimics the enzymatic activity of the human homolog.
tynA is a reserve gene for harsh culture conditions
Our database search for the existence of tynA revealed that opportunistic pathogens, human and plant pathogens, antibiotic-producing bacteria, and bacteria resistant to antibiotics harbor an amine oxidase homolog of ECAO (S1 Table). The environmental bacteria included isolates from harsh conditions, such as the soil, pesticide- or metal-contaminated soil or seawater, suggesting that tynA serves as a reservoir gene for circumstances when nutrients are not abundantly available. This was supported by our detecting more tynA-positive E. coli strains in well water than in wastewater (Table 1). On the other hand, the release of hydrogen peroxide into the immediate environment may be bactericidal to those bacteria that do not express peroxidases, such as catalase in E. coli K-12 [44,45]. As has been shown previously [18], we also measured hydrogen peroxide at a concentration of 1 μM in the medium of tynA+ bacterial cultures in stationary phase, and this was tynA dependent (Fig 1). ECAO is a periplasmic enzyme and the hydrogen peroxide formed by its enzymatic reaction is secreted into the environment, because the flux of the formed hydrogen peroxide is larger through the outer membrane than through the inner membrane [18]. Even small amounts of hydrogen peroxide may give a growth advantage to ECAO-producing bacteria in their immediate surroundings.
O157:H7 E. coli are tynA-negative
We observed the absence of tynA in serotype O157:H7 EHEC (Table 2). Some O157:H7 bacteria lose O157 antigenicity or survive poorly in water cultures [46,47], suggesting that growth conditions favoring the presence of tynA could be specifically useless or harmful for O157:H7 pathogenicity. In addition, PEA inhibits cell division and reduces O157:H7 E. coli counts in beef [48], which further supports our finding.
ECAO processes aromatic amines and is affected by a human PrAO inhibitor and aminoglycosides
As a semicarbazide-sensitive amine oxidase, ECAO is able to process PEA, tyramine, and tryptamine, and its activity is inhibited by SC, hydroxylamine, phenelzine, iproniazid, and tranycypromine [3]. We found that the hAOC3 inhibitor BTT-2052 also inhibited ECAO with an efficient inhibitor concentration lower than that of SC (0.24 μM vs. 0.7 μM, respectively, Fig 2A and 2B). This is within the same range of IC50 values for recombinant hAOC3, mouse and rat AOC3, and purified porcine AOC1 (0.05–0.18 μM, [30]). In addition, the IC50 of BTT-2052 for ECAO was lower than that for mouse PrAO, hAOC2, and for recombinant human MAO-A [30]. We also demonstrated that the aminoglycosides tobramycin, amikacin, and kanamycin inhibited ECAO activity (IC50 of 0.8–3.8 mM, Fig 2C–2E), which may have some clinical relevance. Although observed IC50 concentrations are one order of magnitude higher than the maximum concentration recommended for these antibiotics in human serum (2–72 μM), concentrations 100 times higher than in serum can be locally achieved, as when antibiotics accumulate in the kidney during their excretion [49].
ECAO is a metabolic enzyme
To analyze the role of ECAO in E. coli K-12, we performed a microarray analysis on the wt and ΔtynA strain. Most of the down-regulated genes in ΔtynA were either part of the paaABCDEFGHIJKXYZ operon, which produces enzymes to convert phenylacetic acid to acetyl-CoA and succinyl-CoA, or were related to it (tynA and feaB) and were active in utilizing PEA as a carbon source (Fig 3A, [50,51]). Another down-regulated group of genes were those under the control of OxyR (oxyS, aphCF, and grxA), which is present in its inactive, reduced form in vivo [52]. OxyR is activated by hydrogen peroxide, and once oxidized, drives the transcription of oxyS, ahpCF, and grx, among other genes [52,53]. Furthermore, ΔoxyR cells are unable to grow on PEA, but grow on phenylacetic acid [18]. Because we observed no down-regulation of the other members of the regulon [54], the down-regulation of oxyS, aphCF, and grxA most likely results from the activation of OxyR by the hydrogen peroxide produced by ECAO. In addition, there was no significant difference in the levels of the cyclic adenosine monophosphate (cAMP) or the cAMP regulatory protein (CRP) genes (cyaA, FC 1.21 and crp FC 1.11, respectively), despite paaABCDEFGHIJK and paaZ being regulated by CRP [55–57]. This is supported by the recent results that paaXY, which were both down-regulated in our work, form an auto-regulated operon, and PaaY regulates the transcription of other paa genes [58]. The roles of the rest of the down-regulated genes in ΔtynA remain to be elucidated.
In the ΔtynA strain, the prpBCDE operon was up-regulated. Its proteins are involved in degrading propionate to pyruvate in the 2-methylcitrate pathway ([41], Fig 3B). The prpBCDE operon is activated by carbon catalytic repression, which ensures the utilization of the most energy-rich sugar (glucose) in the growth environment via regulation by cAMP, CRP, and σ54 ([55,59–61]). We believe that when tynA+ E. coli strains are grown in PEA-lactose minimal medium, this operon is down-regulated, enabling the utilization of PEA. When PEA cannot be utilized by the ΔtynA strain, the repression of the operon is not active, and the genes are up-regulated compared to the wt strain. Previously, it was shown that prpBCDE is down-regulated by 2 g/L lactose in rich culture medium [62], and we found this is also true for minimal medium. The fourth enzyme involved in the 2-methylcitrate pathway, acnB, was only slightly up-regulated (FC 1.6), but it is expressed in all growth conditions, independent of the energy source [41]. Two other genes related to the propionate pathway were also up-regulated in ΔtynA cells: ldhA, whose gene product, lactate dehydrogenase, converts pyruvate to lactate [63] and aldB, which encodes for an enzyme that converts propanal to propionate [64].
Nitrogen starvation can induce tynA expression [5]. Therefore, the up-regulation of genes in ΔtynA could result from the absence of ammonia produced by ECAO in wt cells. Of note, narWU and yhdXZ genes were up-regulated in the ΔtynA strain. Both yhdX and yhdZ are activated by nitrogen limitation [42,65]. In addition, narU, which is involved in nitrate uptake [66], and narW, which is needed in the formation of active respiratory nitrate reductase complex NRZ from the narZYWV operon [43], were up-regulated. The NarY subunit of the complex contains an iron-sulfur cluster, and the formation of such clusters are catalyzed, in anaerobic conditions, by an oxygen-independent coproporphyrinogen-III oxidase, a gene product of hemN [67], which was also up-regulated in the ΔtynA strain. Moreover, prpBCDE, yhdXZ, and hemN are all under the control of the RNA polymerase subunit σ54, which is important in nitrogen assimilation [61].
Finally, glgS was also up-regulated. It encodes for a glycogen synthesis molecule that is induced during stationary phase and may be a significant repressor of bacterial adhesion and biofilm formation [68]. Its up-regulation in ΔtynA cells could simply be a consequence of more stringent culture conditions. In conclusion, the microarray analysis of up-regulated genes in the ΔtynA strain identified new genes involved in PEA-dependent nitrogen utilization and confirm the metabolic pathways affected by the lack tynA.
It has been known for a long time that PrAOs are involved in amine metabolism. It is an advantage for bacteria to be able to grow on different nutrients. For example, Klebsiella oxytoca, E. coli, and Arthrobacteria use oxidases to grow on, preferably, aromatic amines [3,69,70]. Arthobacter globiformis has two different oxidases, enabling it to grow on both PEA and histamine [71,72], and Arthrobacter P1 is able to grow on methylamine [73]. This capability is not, however, restricted to bacteria. Yeasts have been proposed to use these enzymes in their metabolism [74,75]. Eukaryotic PrAOs, in general, have markedly different substrate specificity than prokaryotes, as they process BA and methylamine over other substances like tyramine or PEA [76–78], but their biological functions differ from those of prokaryotes [77,79].
ECAO can use human granulocytes as a substrate
The human counterpart of ECAO, hAOC3, is involved in the adhesion and transmigration of leukocytes from blood vessels to the sites of inflammation [80]. hAOC3 utilizes lymphocyte surface molecules as a substrate, and Siglec-9 and -10 (Sialic acid–binding immunoglobulin-like lectins) are its leukocyte counter-receptors [81–83]. The activity of hAOC3 is important for its biological role in leukocyte trafficking [84]. Because ECAO is inhibited by a hAOC3 inhibitor and has a similar molecular structure, we tested ECAO’s activity on human leukocytes. Indeed, purified recombinant ECAO was able to process human granulocytes, as determined by the production of H2O2 (Fig 4), even though we and others have demonstrated different substrate specificities for these two proteins [3,76]. Although it is possible that purified PrAOs from other bacteria, plants, and yeast can also process molecules on human granulocytes, we demonstrated that E. coli ECAO behaves in a similar manner as hAOC3 under these experimental conditions, yet the in vivo relevance remains to be seen.
Supporting Information
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Acknowledgments
We thank our collaborators for the bacterial strains used in ECAO testing: Director Jari Jalava, PhD, from the Unit of Bacterial Infections, National Institute for Health and Welfare, Turku, Finland, Erkki Eerola, PhD, from the Department of Medical Microbiology and Immunology, University of Turku, Turku, Finland, as well as Sanna Nurmela and Katja Eriksson from Water and Environment Research of Southwest Finland, Turku, Finland. We are grateful for Maritta Pohjansalo and Sari Mäki for excellent technical work, Anne Sovikoski-Georgieva for indispensable secretarial help, as well as MSc. Ruth Fair-Mäkelä for reviewing the language of the manuscript.
Data Availability
All Micro array data files are available from the NCBI's Gene Expression Omnibus database (accession number GSE65385).
Funding Statement
This work was supported by the Academy of Finland (www.aka.fi) #132701 (to HE), by the Academy of Finland (www.aka.fi) #141136 (to MS and SJ), by the Turku Graduate School of Biomedical Sciences (www.tubs.utu.fi, to TH), by the University of Turku foundation (https://www.utu.fi/fi/yksikot/yliopistosaatio/en/Sivut/home.aspx, to HE), and by the Maud Kuistila foundation (http://www.maudkuistilanmuistosaatio.fi/index.php?lang=en, to HE). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Klinman JP, Mu D (1994) Quinoenzymes in biology. Annu Rev Biochem 63: 299–344. 10.1146/annurev.bi.63.070194.001503 [DOI] [PubMed] [Google Scholar]
- 2. Azakami H, Yamashita M, Roh J, Hyeob H, Suzuki H, Kumagai H, et al. (1994) Nucleotide sequence of the gene for monamine oxidase (maoA) from Escherichia coli. J Ferment Bioeng 77: 315–319. [Google Scholar]
- 3. Roh JH, Suzuki H, Azakami H, Yamashita M, Murooka Y, Kumagai H (1994) Purification, characterization, and crystallization of monoamine oxidase from Escherichia coli K-12. Biosci Biotechnol Biochem 58: 1652–1656. [DOI] [PubMed] [Google Scholar]
- 4. Murooka Y, Higashiura T, Harada T (1978) Genetic mapping of tyramine oxidase and arylsulfatase genes and their regulation in intergeneric hybrids of enteric bacteria. J Bacteriol 136: 714–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zeng J, Spiro S (2013) Finely tuned regulation of the aromatic amine degradation pathway in Escherichia coli. J Bacteriol 195: 5141–5150. 10.1128/JB.00837-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. O'Reilly EK, Kreuzer KN (2004) Isolation of SOS constitutive mutants of Escherichia coli. J Bacteriol 186: 7149–7160. 10.1128/JB.186.21.7149-7160.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Janion C (2008) Inducible SOS response system of DNA repair and mutagenesis in Escherichia coli. Int J Biol Sci 4: 338–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rankin LD, Bodenmiller DM, Partridge JD, Nishino SF, Spain JC, Spiro S (2008) Escherichia coli NsrR regulates a pathway for the oxidation of 3-nitrotyramine to 4-hydroxy-3-nitrophenylacetate. J Bacteriol 190: 6170–6177. 10.1128/JB.00508-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Parsons MR, Convery MA, Wilmot CM, Yadav KD, Blakeley V, Corner AS, et al. (1995) Crystal structure of a quinoenzyme: copper amine oxidase of Escherichia coli at 2 A resolution. Structure 3: 1171–1184. [DOI] [PubMed] [Google Scholar]
- 10. Wilmot CM, Murray JM, Alton G, Parsons MR, Convery MA, Blakeley V, et al. (1997) Catalytic mechanism of the quinoenzyme amine oxidase from Escherichia coli: exploring the reductive half-reaction. Biochemistry 36: 1608–1620. 10.1021/bi962205j [DOI] [PubMed] [Google Scholar]
- 11. Murray JM, Saysell CG, Wilmot CM, Tambyrajah WS, Jaeger J, Knowles PF, et al. (1999) The active site base controls cofactor reactivity in Escherichia coli amine oxidase: x-ray crystallographic studies with mutational variants. Biochemistry 38: 8217–8227. 10.1021/bi9900469 [DOI] [PubMed] [Google Scholar]
- 12. Wilmot CM, Hajdu J, McPherson MJ, Knowles PF, Phillips SE (1999) Visualization of dioxygen bound to copper during enzyme catalysis. Science 286: 1724–1728. [DOI] [PubMed] [Google Scholar]
- 13. Mure M, Kurtis CR, Brown DE, Rogers MS, Tambyrajah WS, Saysell C, et al. (2005) Active site rearrangement of the 2-hydrazinopyridine adduct in Escherichia coli amine oxidase to an azo copper(II) chelate form: a key role for tyrosine 369 in controlling the mobility of the TPQ-2HP adduct. Biochemistry 44: 1583–1594. 10.1021/bi0479860 [DOI] [PubMed] [Google Scholar]
- 14. Mure M, Brown DE, Saysell C, Rogers MS, Wilmot CM, Kurtis CR, et al. (2005) Role of the interactions between the active site base and the substrate Schiff base in amine oxidase catalysis. Evidence from structural and spectroscopic studies of the 2-hydrazinopyridine adduct of Escherichia coli amine oxidase. Biochemistry 44: 1568–1582. 10.1021/bi047988k [DOI] [PubMed] [Google Scholar]
- 15. Saysell CG, Tambyrajah WS, Murray JM, Wilmot CM, Phillips SE, McPherson MJ, et al. (2002) Probing the catalytic mechanism of Escherichia coli amine oxidase using mutational variants and a reversible inhibitor as a substrate analogue. Biochem J 365: 809–816. 10.1042/BJ20011435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Murray JM, Kurtis CR, Tambyrajah W, Saysell CG, Wilmot CM, Parsons MR, et al. (2001) Conserved tyrosine-369 in the active site of Escherichia coli copper amine oxidase is not essential. Biochemistry 40: 12808–12818. [DOI] [PubMed] [Google Scholar]
- 17. Kurtis CR, Knowles PF, Parsons MR, Gaule TG, Phillips SE, McPherson MJ (2011) Tyrosine 381 in E. coli copper amine oxidase influences substrate specificity. J Neural Transm 118: 1043–1053. 10.1007/s00702-011-0620-y [DOI] [PubMed] [Google Scholar]
- 18. Ravindra Kumar S, Imlay JA (2013) How Escherichia coli tolerates profuse hydrogen peroxide formation by a catabolic pathway. J Bacteriol 195: 4569–4579. 10.1128/JB.00737-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Salmi M, Jalkanen S (2012) Ectoenzymes controlling leukocyte traffic. Eur J Immunol 42: 284–292. 10.1002/eji.201142223 [DOI] [PubMed] [Google Scholar]
- 20. Hanahan D (1985) Techniques for transformation of E. coli In: Glover D, editor. DNA cloning: A practical approach: IRL Press, Washington DC: pp. 109–135. [Google Scholar]
- 21. Miller VL, Falkow S (1988) Evidence for two genetic loci in Yersinia enterocolitica that can promote invasion of epithelial cells. Infect Immun 56: 1242–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. de Lorenzo V, Timmis KN (1994) Analysis and construction of stable phenotypes in gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods Enzymol 235: 386–405. [DOI] [PubMed] [Google Scholar]
- 23. Skurnik M, Venho R, Toivanen P, al-Hendy A (1995) A novel locus of Yersinia enterocolitica serotype O:3 involved in lipopolysaccharide outer core biosynthesis. Mol Microbiol 17: 575–594. [DOI] [PubMed] [Google Scholar]
- 24. Bertani G (2004) Lysogeny at mid-twentieth century: P1, P2, and other experimental systems. J Bacteriol 186: 595–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Maniatis T, Fritch EF, Sambrook J (1982) Molecular cloning: a laboratory manual: Cold Spring Harbor, NY: Cold Spring Harbor Laboratory. [Google Scholar]
- 26. Carver TJ, Rutherford KM, Berriman M, Rajandream MA, Barrell BG, Parkhill J (2005) ACT: the Artemis Comparison Tool. Bioinformatics (Oxford, England) 21: 3422–3423. 10.1093/bioinformatics/bti553 [DOI] [PubMed] [Google Scholar]
- 27. Blattner FR, Plunkett G 3rd, Bloch CA, Perna NT, Burland V, Riley M, et al. (1997) The complete genome sequence of Escherichia coli K-12. Science 277: 1453–1462. [DOI] [PubMed] [Google Scholar]
- 28. Sambrook J, Fritsch E, Maniatis T (1989) Molecular cloning: a laboratory manual: Cold Spring Harbor, NY: Cold Spring Harbor Laboratory. [Google Scholar]
- 29. Holt A, Sharman DF, Baker GB, Palcic MM (1997) A continuous spectrophotometric assay for monoamine oxidase and related enzymes in tissue homogenates. Anal Biochem 244: 384–392. 10.1006/abio.1996.9911 [DOI] [PubMed] [Google Scholar]
- 30. Marttila-Ichihara F, Smith DJ, Stolen C, Yegutkin GG, Elima K, Mercier N, et al. (2006) Vascular amine oxidases are needed for leukocyte extravasation into inflamed joints in vivo. Arthritis Rheum 54: 2852–2862. 10.1002/art.22061 [DOI] [PubMed] [Google Scholar]
- 31. Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, et al. (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5: R80 10.1186/gb-2004-5-10-r80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. R Development Core Team R (2006) R: A language and environment for statistical computing: R Foundation for Statistical Computing; Vienna, Austria: ISBN 3-900051-07-0, URL http://www.R-project.org. [Google Scholar]
- 33.Hovatta I, Kimppa K, Lehmussola A, Pasanen T, Saarela J, Saarikko I, et al. (2006) DNA Microarray Data Analysis; Tuimala J, Laine MM, editors.
- 34. Edgar R, Domrachev M, Lash AE (2002) Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30: 207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Smith D, Fulop F, Pihlavisto M, Lazar L, Alaranta S, Vainio P (2003) Carbocyclic hydrazino inhibitors of copper-containing amine oxidases. Int Pat Appl; Publ. No.: WO 03/006003 A1; Chem Abstr. 138, 57890. [Google Scholar]
- 36. Olivieri A, Tipton KF, O'Sullivan J (2012) Characterization of the in vitro binding and inhibition kinetics of primary amine oxidase/vascular adhesion protein-1 by glucosamine. Biochim Biophys Acta 1820: 482–487. 10.1016/j.bbagen.2011.12.009 [DOI] [PubMed] [Google Scholar]
- 37. Chiang SM, Schellhorn HE (2012) Regulators of oxidative stress response genes in Escherichia coli and their functional conservation in bacteria. Arch Biochem Biophys 525: 161–169. 10.1016/j.abb.2012.02.007 [DOI] [PubMed] [Google Scholar]
- 38. Kehres DG, Janakiraman A, Slauch JM, Maguire ME (2002) Regulation of Salmonella enterica serovar Typhimurium mntH transcription by H(2)O(2), Fe(2+), and Mn(2+). J Bacteriol 184: 3151–3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tao K (1997) oxyR-dependent induction of Escherichia coli grx gene expression by peroxide stress. J Bacteriol 179: 5967–5970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Morgan RW, Christman MF, Jacobson FS, Storz G, Ames BN (1986) Hydrogen peroxide-inducible proteins in Salmonella typhimurium overlap with heat shock and other stress proteins. Proc Natl Acad Sci U S A 83: 8059–8063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Brock M, Maerker C, Schutz A, Volker U, Buckel W (2002) Oxidation of propionate to pyruvate in Escherichia coli. Involvement of methylcitrate dehydratase and aconitase. Eur J Biochem 269: 6184–6194. [DOI] [PubMed] [Google Scholar]
- 42. Zimmer DP, Soupene E, Lee HL, Wendisch VF, Khodursky AB, Peter BJ, et al. (2000) Nitrogen regulatory protein C-controlled genes of Escherichia coli: scavenging as a defense against nitrogen limitation. Proc Natl Acad Sci U S A 97: 14674–14679. 10.1073/pnas.97.26.14674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Blasco F, Pommier J, Augier V, Chippaux M, Giordano G (1992) Involvement of the narJ or narW gene product in the formation of active nitrate reductase in Escherichia coli. Mol Microbiol 6: 221–230. [DOI] [PubMed] [Google Scholar]
- 44. Selva L, Viana D, Regev-Yochay G, Trzcinski K, Corpa JM, Lasa I, et al. (2009) Killing niche competitors by remote-control bacteriophage induction. Proc Natl Acad Sci U S A 106: 1234–1238. 10.1073/pnas.0809600106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Jansen WT, Bolm M, Balling R, Chhatwal GS, Schnabel R (2002) Hydrogen peroxide-mediated killing of Caenorhabditis elegans by Streptococcus pyogenes. Infect Immun 70: 5202–5207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hara-Kudo Y, Miyahara M, Kumagai S (2000) Loss of O157 O antigenicity of verotoxin-producing Escherichia coli O157:H7 surviving under starvation conditions. Appl Environ Microbiol 66: 5540–5543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wang G, Doyle MP (1998) Survival of enterohemorrhagic Escherichia coli O157:H7 in water. J Food Prot 61: 662–667. [DOI] [PubMed] [Google Scholar]
- 48. Lynnes T, Horne SM, Pruss BM (2014) ss-Phenylethylamine as a novel nutrient treatment to reduce bacterial contamination due to Escherichia coli O157:H7 on beef meat. Meat Sci 96: 165–171. 10.1016/j.meatsci.2013.06.030 [DOI] [PubMed] [Google Scholar]
- 49. Edwards CQ, Smith CR, Baughman KL, Rogers JF, Lietman PS (1976) Concentrations of gentamicin and amikacin in human kidneys. Antimicrob Agents Chemother 9: 925–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Teufel R, Mascaraque V, Ismail W, Voss M, Perera J, Eisenreich W, et al. (2010) Bacterial phenylalanine and phenylacetate catabolic pathway revealed. Proc Natl Acad Sci U S A 107: 14390–14395. 10.1073/pnas.1005399107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Song F, Zhuang Z, Finci L, Dunaway-Mariano D, Kniewel R, Buglino JA, et al. (2006) Structure, function, and mechanism of the phenylacetate pathway hot dog-fold thioesterase PaaI. J Biol Chem 281: 11028–11038. 10.1074/jbc.M513896200 [DOI] [PubMed] [Google Scholar]
- 52. Pomposiello PJ, Demple B (2001) Redox-operated genetic switches: the SoxR and OxyR transcription factors. Trends Biotechnol 19: 109–114. [DOI] [PubMed] [Google Scholar]
- 53. Imlay JA (2013) The molecular mechanisms and physiological consequences of oxidative stress: lessons from a model bacterium. Nat Rev Microbiol 11: 443–454. 10.1038/nrmicro3032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Baez A, Shiloach J (2013) Escherichia coli avoids high dissolved oxygen stress by activation of SoxRS and manganese-superoxide dismutase. Microb Cell Fact 12: 23 10.1186/1475-2859-12-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Shimada T, Fujita N, Yamamoto K, Ishihama A (2011) Novel roles of cAMP receptor protein (CRP) in regulation of transport and metabolism of carbon sources. PLoS One 6: e20081 10.1371/journal.pone.0020081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ferrandez A, Garcia JL, Diaz E (2000) Transcriptional regulation of the divergent paa catabolic operons for phenylacetic acid degradation in Escherichia coli. J Biol Chem 275: 12214–12222. [DOI] [PubMed] [Google Scholar]
- 57. Fic E, Bonarek P, Gorecki A, Kedracka-Krok S, Mikolajczak J, Polit A, et al. (2009) cAMP receptor protein from escherichia coli as a model of signal transduction in proteins—a review. J Mol Microbiol Biotechnol 17: 1–11. 10.1159/000178014 [DOI] [PubMed] [Google Scholar]
- 58. Fernandez C, Diaz E, Garcia JL (2014) Insights on the regulation of the phenylacetate degradation pathway from Escherichia coli. Environ Microbiol Rep 6: 239–250. 10.1111/1758-2229.12117 [DOI] [PubMed] [Google Scholar]
- 59. Gorke B, Stulke J (2008) Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nat Rev Microbiol 6: 613–624. 10.1038/nrmicro1932 [DOI] [PubMed] [Google Scholar]
- 60. Lee SK, Newman JD, Keasling JD (2005) Catabolite repression of the propionate catabolic genes in Escherichia coli and Salmonella enterica: evidence for involvement of the cyclic AMP receptor protein. J Bacteriol 187: 2793–2800. 10.1128/JB.187.8.2793-2800.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Reitzer L, Schneider BL (2001) Metabolic context and possible physiological themes of sigma(54)-dependent genes in Escherichia coli. Microbiol Mol Biol Rev 65: 422–444, table of contents. 10.1128/MMBR.65.3.422-444.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Park JM, Vinuselvi P, Lee SK (2012) The mechanism of sugar-mediated catabolite repression of the propionate catabolic genes in Escherichia coli. Gene 504: 116–121. 10.1016/j.gene.2012.04.074 [DOI] [PubMed] [Google Scholar]
- 63. Bunch PK, Mat-Jan F, Lee N, Clark DP (1997) The ldhA gene encoding the fermentative lactate dehydrogenase of Escherichia coli. Microbiology 143 (Pt 1): 187–195. [DOI] [PubMed] [Google Scholar]
- 64. Ho KK, Weiner H (2005) Isolation and characterization of an aldehyde dehydrogenase encoded by the aldB gene of Escherichia coli. J Bacteriol 187: 1067–1073. 10.1128/JB.187.3.1067-1073.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Jiang H, Shang L, Yoon SH, Lee SY, Yu Z (2006) Optimal production of poly-gamma-glutamic acid by metabolically engineered Escherichia coli. Biotechnol Lett 28: 1241–1246. 10.1007/s10529-006-9080-0 [DOI] [PubMed] [Google Scholar]
- 66. Jia W, Tovell N, Clegg S, Trimmer M, Cole J (2009) A single channel for nitrate uptake, nitrite export and nitrite uptake by Escherichia coli NarU and a role for NirC in nitrite export and uptake. Biochem J 417: 297–304. 10.1042/BJ20080746 [DOI] [PubMed] [Google Scholar]
- 67. Layer G, Kervio E, Morlock G, Heinz DW, Jahn D, Retey J, et al. (2005) Structural and functional comparison of HemN to other radical SAM enzymes. Biol Chem 386: 971–980. 10.1515/BC.2005.113 [DOI] [PubMed] [Google Scholar]
- 68. Rahimpour M, Montero M, Almagro G, Viale AM, Sevilla A, Canovas M, et al. (2013) GlgS, described previously as a glycogen synthesis control protein, negatively regulates motility and biofilm formation in Escherichia coli. Biochem J 452: 559–573. 10.1042/BJ20130154 [DOI] [PubMed] [Google Scholar]
- 69. Hacisalihoglu A, Jongejan JA, Duine JA (1997) Distribution of amine oxidases and amine dehydrogenases in bacteria grown on primary amines and characterization of the amine oxidase from Klebsiella oxytoca. Microbiology 143 (Pt 2): 505–512. 10.1099/00221287-143-2-505 [DOI] [PubMed] [Google Scholar]
- 70. Sugino H, Sasaki M, Azakami H, Yamashita M, Murooka Y (1992) A monoamine-regulated Klebsiella aerogenes operon containing the monoamine oxidase structural gene (maoA) and the maoC gene. J Bacteriol 174: 2485–2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Shimizu E, Ichise H, Yorifuji T (1990) Phenylethylamine oxidase, a novel enzyme of Arthrobacter globiformis may be a quinoprotein. Agric Biol Chem 54: 851–853. [Google Scholar]
- 72. Choi YH, Matsuzaki R, Fukui T, Shimizu E, Yorifuji T, Sato H, et al. (1995) Copper/topa quinone-containing histamine oxidase from Arthrobacter globiformis. Molecular cloning and sequencing, overproduction of precursor enzyme, and generation of topa quinone cofactor. J Biol Chem 270: 4712–4720. [DOI] [PubMed] [Google Scholar]
- 73. Zhang X, Fuller JH, McIntire WS (1993) Cloning, sequencing, expression, and regulation of the structural gene for the copper/topa quinone-containing methylamine oxidase from Arthrobacter strain P1, a gram-positive facultative methylotroph. J Bacteriol 175: 5617–5627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Cai D, Klinman JP (1994) Evidence of a self-catalytic mechanism of 2,4,5-trihydroxyphenylalanine quinone biogenesis in yeast copper amine oxidase. J Biol Chem 269: 32039–32042. [PubMed] [Google Scholar]
- 75. Peter C, Laliberte J, Beaudoin J, Labbe S (2008) Copper distributed by Atx1 is available to copper amine oxidase 1 in Schizosaccharomyces pombe. Eukaryot Cell 7: 1781–1794. 10.1128/EC.00230-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Lyles GA (1995) Substrate-specificity of mammalian tissue-bound semicarbazide-sensitive amine oxidase. Prog Brain Res 106: 293–303. [DOI] [PubMed] [Google Scholar]
- 77. Stolen CM, Marttila-Ichihara F, Koskinen K, Yegutkin GG, Turja R, Bono P, et al. (2005) Absence of the endothelial oxidase AOC3 leads to abnormal leukocyte traffic in vivo. Immunity 22: 105–115. 10.1016/j.immuni.2004.12.006 [DOI] [PubMed] [Google Scholar]
- 78. Mu D, Medzihradszky KF, Adams GW, Mayer P, Hines WM, Burlingame AL, et al. (1994) Primary structures for a mammalian cellular and serum copper amine oxidase. J Biol Chem 269: 9926–9932. [PubMed] [Google Scholar]
- 79. Salmi M, Jalkanen S (2014) Ectoenzymes in leukocyte migration and their therapeutic potential. Semin Immunopathol 36: 163–176. 10.1007/s00281-014-0417-9 [DOI] [PubMed] [Google Scholar]
- 80. Salmi M, Jalkanen S (2005) Cell-surface enzymes in control of leukocyte trafficking. Nat Rev Immunol 5: 760–771. [DOI] [PubMed] [Google Scholar]
- 81. Salmi M, Yegutkin GG, Lehvonen R, Koskinen K, Salminen T, Jalkanen S (2001) A cell surface amine oxidase directly controls lymphocyte migration. Immunity 14: 265–276. [DOI] [PubMed] [Google Scholar]
- 82. Kivi E, Elima K, Aalto K, Nymalm Y, Auvinen K, Koivunen E, et al. (2009) Human Siglec-10 can bind to vascular adhesion protein-1 and serves as its substrate. Blood 114: 5385–5392. 10.1182/blood-2009-04-219253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Aalto K, Autio A, Kiss EA, Elima K, Nymalm Y, Veres TZ, et al. (2011) Siglec-9 is a novel leukocyte ligand for vascular adhesion protein-1 and can be used in PET imaging of inflammation and cancer. Blood 118: 3725–3733. 10.1182/blood-2010-09-311076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Koskinen K, Vainio PJ, Smith DJ, Pihlavisto M, Yla-Herttuala S, Jalkanen S, et al. (2004) Granulocyte transmigration through the endothelium is regulated by the oxidase activity of vascular adhesion protein-1 (VAP-1). Blood 103: 3388–3395. 10.1182/blood-2003-09-3275 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
All Micro array data files are available from the NCBI's Gene Expression Omnibus database (accession number GSE65385).