Radical revision of tiger taxonomy for a pragmatic and scientifically sound approach to tiger conservation management.
Keywords: Felidae, Management Units, One Plan Approach, Subspecies, Taxonomy
Abstract
Although significantly more money is spent on the conservation of tigers than on any other threatened species, today only 3200 to 3600 tigers roam the forests of Asia, occupying only 7% of their historical range. Despite the global significance of and interest in tiger conservation, global approaches to plan tiger recovery are partly impeded by the lack of a consensus on the number of tiger subspecies or management units, because a comprehensive analysis of tiger variation is lacking. We analyzed variation among all nine putative tiger subspecies, using extensive data sets of several traits [morphological (craniodental and pelage), ecological, molecular]. Our analyses revealed little variation and large overlaps in each trait among putative subspecies, and molecular data showed extremely low diversity because of a severe Late Pleistocene population decline. Our results support recognition of only two subspecies: the Sunda tiger, Panthera tigris sondaica, and the continental tiger, Panthera tigris tigris, which consists of two (northern and southern) management units. Conservation management programs, such as captive breeding, reintroduction initiatives, or trans-boundary projects, rely on a durable, consistent characterization of subspecies as taxonomic units, defined by robust multiple lines of scientific evidence rather than single traits or ad hoc descriptions of one or few specimens. Our multiple-trait data set supports a fundamental rethinking of the conventional tiger taxonomy paradigm, which will have profound implications for the management of in situ and ex situ tiger populations and boost conservation efforts by facilitating a pragmatic approach to tiger conservation management worldwide.
INTRODUCTION
Fewer than 4000 tigers inhabit the forests of Asia—a historically low number (1). These tigers occupy only 7% of their estimated former distribution range (2), and 70% of them inhabit 42 source sites, which occupy only 0.5% of their historical range (3). Almost US$50 million is spent annually by range countries, non-governmental organizations, and private donors on the conservation of wild tigers (3). This amount has massively increased since the launch of the World Bank’s Global Tiger Initiative (GTI) in 2008. No other threatened species commands such resources and attention from the international community.
A precondition for a successful tiger recovery and global tiger management (including global captive breeding programs and conservation management of free-ranging meta-populations) is a consensus on the number of tiger conservation units [that is, subspecies, evolutionarily significant units (ESUs), ecotypes, or management units (MUs)] because active interventions, such as translocations or releases of captive-bred tigers, will presumably become more important in the future for reversing the decline of wild tigers.
Despite repeated previous efforts to unravel tiger diversity and define conservation units in terms of subspecies (4, 5), results were often contradictory and no consensus has been reached so far. Up to nine subspecies of tigers are currently recognized (6) (Fig. 1A), three of which are already extinct and a fourth probably survives only in captivity (7) (Table 1). All but one subspecies were first described on the basis of morphological characters measured in only one or a few specimens (4) (Table 1). Only the Malayan tiger was identified on the basis of molecular markers, and it has not been diagnosed in terms of morphological traits (8). Although a type specimen was designated, the formal scientific description, according to the International Code of Zoological Nomenclature, is still lacking, rendering this taxon a nomen nudum (9). Even so, the proposed subspecies name “Panthera tigris jacksoni” is widely used, for example, by The IUCN/SSC Red List of Threatened Species (6).
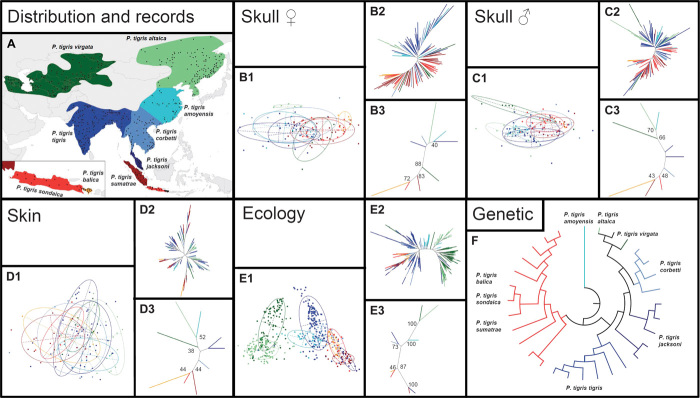
Fig. 1. Variation among and phenotypic space across all nine putative subspecies of tigers.
(A) Former distribution of the nine putative subspecies and occurrence records used for the ecological analysis. (B1 to E3) Multivariate analyses of skull traits [(B1 to B3) females; (C1 to C3) males], pelage (D1 to D3) and ecological preferences (E1 to E3). (B1 to E1) The 1.5 inertia ellipse for all nine putative subspecies displayed on the plane defined by the first two principal components; (B2 to E2) unrooted binary neighbor-joining trees based on the matrices of Euclidean distance between individuals in the multivariate space defined by all principal components; (B3 to E3) same as in (B2 to E2) but with distances being measured between the centroids of each putative subspecies with bootstrap values of groupings. (F) Radial tree of the maximum likelihood analysis of 3968 bp of mtDNA (see Fig. 2).
Table 1. Current tiger P. tigris subspecies classification based on the IUCN/SSC Red List of Threatened Species.
(6). Subspecies are chronologically ordered by the year of their description. † indicates subspecies recognized as extinct under the current IUCN/SSC Red List of Threatened Species (6).
| Subspecies | Common English name | Abbreviation | No. of specimens in type description | Authority |
| P. t. tigris | Bengal tiger | TIG | 0 | (L., 1758) |
| P. t. virgata† | Caspian tiger | VIR | 1 skin | (Illiger, 1815) |
| P. t. altaica | Amur tiger | ALT | 1 skin | (Temminck, 1844) |
| P. t. sondaica† | Javan tiger | SON | 1 skin | (Temminck, 1844) |
| P. t. amoyensis | South Chinese tiger | AMO | 5 skulls | (Hilzheimer, 1905) |
| P. t. balica† | Balinese tiger | BAL | 1 skin; 1 skull | (Schwarz, 1912) |
| P. t. sumatrae | Sumatran tiger | SUM | 1 skull; 1 live animal | Pocock, 1929 |
| P. t. corbetti | Indochinese tiger | COR | 13 skins; 19 skulls | Mazák, 1968 |
| P. t. jacksoni | Malayan tiger | JAC | 22 genetic (mtDNA and microsatellite) | Luo et al., 2004 |
Whereas some molecular analyses supported the distinction of multiple subspecies (5, 8, 10), others did not (11, 12) and observed that differences may have been overemphasized simply owing to fragmentary sampling along a more or less complex cline of variation (4, 13). This was recently illustrated for Bengal tigers, where seemingly clear molecular differences among current Bengal tiger populations (14) vanished after museum specimens from extinct Bengal tiger populations were included in analyses (15). There are minimal molecular differences between the extinct Caspian tiger and the extant Amur tiger, suggesting that they belong to the same subspecies (16). Similarly, a recent molecular study showed that the two other extinct subspecies, the Javan and the Balinese tigers, have a close genetic affinity to Sumatran tigers (17), whereas morphological studies had suggested that these two are distinct from all other putative tiger subspecies (18–20). In summary, previous studies often used small sample sizes and used either only morphological characters or only molecular markers to characterize subspecies. The two approaches have only occasionally been combined in a superficial way (4, 5, 8, 18, 19). Consequently, the validity of some or all subspecies remains a matter of debate.
The contradictory views on tiger taxonomy highlight a more general issue about the taxonomic implications of newly collected phylogenetic data and their consequences for conservation management priorities. In recent years, there has been a considerable “taxonomic inflation” among birds and mammals (21, 22) because the liberal application of the phylogenetic species concept (PSC) was combined with new molecular methods and statistical tools to facilitate the distinction of even closely related populations with high statistical support. In tigers, this led to the wide acceptance of several subspecies and even attempts to raise some of these to species level (18). Therefore, the tiger is an instructive example to illustrate how the tension between an increase in phylogenetic information and the resolution of taxonomic disputes may affect and impede conservation management plans and priorities.
Here, we present the most comprehensive analysis to date of molecular, morphological (craniodental and pelage data), and ecological (climate, habitat, and prey data) characteristics of all nine putative tiger subspecies. We show that with a comprehensive multi-trait data set, only two subspecies (one consisting of two management units) receive robust support. We conclude with a discussion of appropriate conservation strategies, and propose a scientifically sound and more pragmatic approach to reverse the tiger’s population decline.
RESULTS AND DISCUSSION
Evolutionary history of tigers
Whereas saving the tiger as a species is the main and unanimous goal, a key aim of current conservation efforts is the protection of tiger diversity (2) to maintain the species’ evolutionary potential. For a better understanding of the tiger’s evolutionary history and its current intraspecific diversity, we conducted a molecular analysis of all nine putative tiger subspecies. For this, we completed a previously used data set of 4 kb of mitochondrial DNA (mtDNA) (8, 16) with additional sequences generated in this study from the extinct Javan and Balinese tigers. We focused on this data set because nuclear autosomal, X-linked, and Y-chromosome markers showed no variation within tigers (10) and even major histocompatibility complex and autosomal microsatellite variability were low (8). Consistent with nuclear data, mitochondrial data indicated that modern tigers contain substantially lower genetic diversity than other pantherine (table S16) or Southeast Asian cats (10). The demographic reconstruction revealed a population decline around 80 thousand years ago (kya), followed by a Late Pleistocene expansion (Fig. 2C). This suggests that modern tigers succumbed to rapid environmental changes after the Toba super-volcanic eruption (~73.5 kya) in northern Sumatra (23). The impact of this eruption has been linked to population bottlenecks in other mammals such as humans (24), orangutans (25), and clouded leopards (26). Thus, tigers may have only survived in a single refugium outside the region of ash-cloud fallouts. The ancestral position of haplotype AMO1 (Fig. 2B) suggests the presence of such a refugium in or around southern China (8, 16, 27).
Fig. 2. Phylogenetic analyses of all nine putative subspecies using 3968 bp of mtDNA.
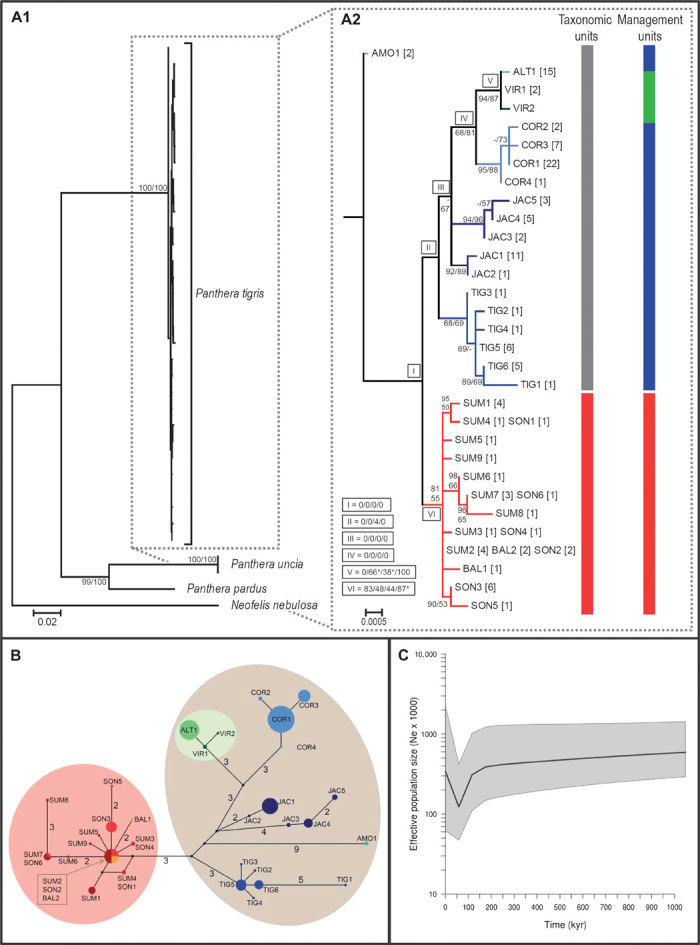
(A1 and A2) Maximum likelihood tree of intraspecific variation among all putative tiger subspecies in relation to three pantherine cat species [snow leopard (Panthera uncia), leopard (Panthera pardus), and clouded leopard (Neofelis nebulosa)]. Values above or below branches show maximum likelihood and Bayesian inference bootstrap supports. (A1) Maximum likelihood tree including three pantherine cat species as outgroups. (A2) Enlargement of the maximum likelihood tree part showing the tigers. Roman numerals indicate bootstrap supports of nodes for skull {females} / skull {males} / skin / ecological preferences. Abbreviations for putative subspecies are given in Table 1. * indicates that one additional putative subspecies clusters with this group. (B) Haplotype network. The size of the circles is proportional to haplotype frequency. Connecting lines between haplotypes represent one mutational step unless indicated otherwise by numbers. (C) Bayesian demographic skyline reconstruction of tigers for the last 1 million years.
Although hints of such a bottleneck had already been reported (8, 17), two reasons prevented a broader discussion of the Late Pleistocene population decline: (i) the widely accepted view that the tiger’s high morphological variation across its wide geographical distribution was an ancient adaptation to local habitats, and (ii) the presence of tiger fossils dating back ca. 2.55 million years (28) from throughout the current tiger’s range from northern China to Java, Sri Lanka, and Japan. It was thus assumed that the tiger’s geographical distribution had essentially remained unchanged for more than 2 million years (4).
However, neither argument contradicts a Late Pleistocene bottleneck. (i) After the bottleneck, the tiger’s population expansion was accompanied by a large range expansion, which probably led to rapid local adaptations, allowing for survival and reproduction in different habitats as varied as cold subarctic taiga and tropical rainforests. (ii) If the wide distribution of tiger fossils were to indicate continuous multiregional evolution throughout the entire Pleistocene (29), we would expect molecular data to show substantial genotypic differentiation over time (30). Therefore, the low molecular as well as morphological (see below) variation observed strongly favors the scenario that ancestral Pleistocene tigers became extinct in large parts of their former distributional range and were replaced by modern tigers recolonizing large parts of Asia as recently as the Late Pleistocene.
Multi-trait tiger taxonomy
Our combined approach of molecular, morphological (skull and pelage), and ecological data revealed that different traits supported different intraspecific groupings (Fig. 1). Although some single measures distinguished one or a few of the nine putative subspecies by the 75% rule (31) (fig. S4), the combination of traits rejected their validity. The multi-trait approach only supported the distinction of two subspecies (Fig. 3, A1 to A5), continental tigers and Sunda tigers. These were consistently supported by bootstrap analyses, could be strictly differentiated according to the 75% rule (31), and were demonstrably significantly different (Fig. 4 and tables S18 to S21).
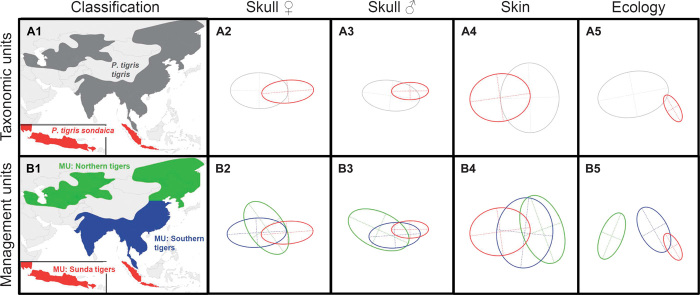
Fig. 3. Tiger variation for the two taxonomic units (subspecies) and three management units recognized by the current study.
(A1 and B1) Former distribution of the (A1) taxonomic units and (B1) management units. (A2 to B5) The 1.5 inertia ellipses of the multivariate analyses displayed on the plane defined by the first two principal components for (A2 to A5) two taxonomic units and (B2 to B5) management units for the different tiger traits.
Fig. 4. Pairwise comparison of the two tiger subspecies P. tigris tigris and P. tigris sondaica recognized by the current study.
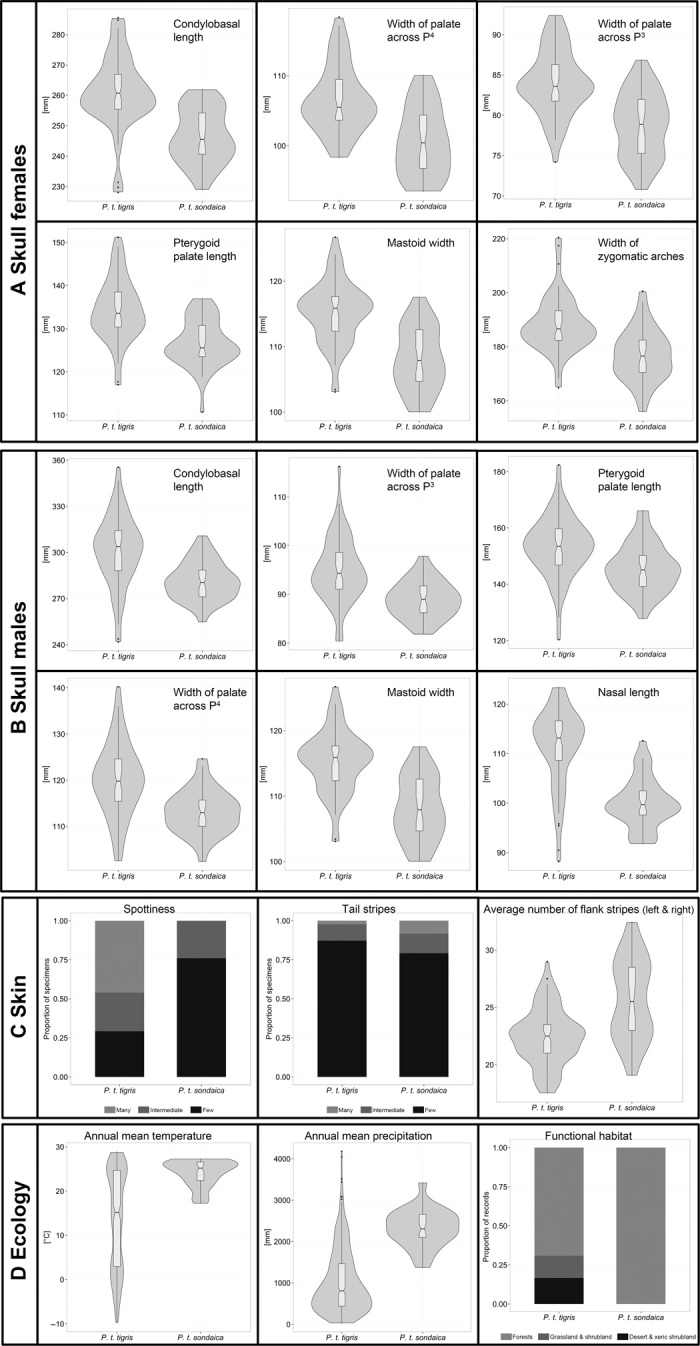
(A and B) For skulls [(A) females; (B) males], the six variables that explained most of the variation in tigers in the multivariate analyses are plotted as violin plots. (C and D) For pelage (C) and ecological preferences (D), the three variables that explained most of the variation in tigers are shown. Ordinal and categorical data are shown as bar plots.
Craniodental morphology
Analysis of 201 tiger skulls (102 males and 99 females) for 41 characters distinguished Sunda tigers from those of continental tigers with a high bootstrap support (BS for females, 83%; BS for males, 48%; see Materials and Methods and table S11 for details on the interpretation of the BS values; Fig. 1, B3 and C3), and the majority of the 41 characters was significantly different between Sunda and continental tigers for both males and females (table S18).
Pelage morphology
Bootstrap analyses of pelage markings of 114 tiger skins (males and females considered jointly) supported the differentiation (BS = 44%; Fig. 1, D3) of Sunda and continental tigers, which differed significantly in their number of flank stripes and spots (table S19).
Ecology
We used two different approaches to summarize the ecological preferences of each subspecies—ecological niche similarity analysis, on the basis of an ecological envelope modeling framework (fig. S2 and table S12), and factor analysis for mixed data (FAMD; Fig. 1, E1). The results of both analyses were highly consistent with each other. Ecologically, Sunda tigers were significantly distinct from continental tigers (BS = 87%; Fig. 1, E3, and tables S20 and S21), except that Malayan tigers grouped ecologically with Sunda tigers (Fig. 1, E1 and E3). The latter was expected because of the highly similar climatic and environmental conditions in peninsular Malaysia and Sumatra.
Molecular population structure
Sunda tigers formed a supported group [BS = 81% maximum likelihood (ML), 55% Bayesian inference (BI); Fig. 2, A2, red], with several diagnostic sites separating Sunda tigers from continental tigers (Fig. 2B and Table 2). Despite the young evolutionary age of modern tiger populations—most variation found in today’s wild tigers probably evolved within the last ~80,000 years (Fig. 2C)—molecular data revealed two groups (continental tigers and Sunda tigers) starting to diverge around (continental) or soon after (Sunda) the population decline (table S17).
Table 2. Diagnosis of continental tigers [P. tigris tigris (L., 1758)] and Sunda tigers [P. tigris sondaica (Temminck, 1844)].
| P. tigris tigris | P. tigris sondaica | |
| Principal synonyms: virgata (Illiger, 1815); altaica (Temminck, 1844); amoyensis (Hilzheimer, 1905); corbetti Mazák, 1968; jacksoni Luo et al., 2004 |
Principal synonyms: balica (Schwarz, 1912); sumatrae Pocock, 1929 |
|
| Skulls (Fig. 4, A and B) | ||
| ~75% of males distinguishable from ~75% of sondaica males |
Condylobasal length ≥288 mm; interorbital width ≥71 mm; nasal length ≥105 mm; ventral posterior length ≥81 mm; width of zygomatic arches ≥218 mm; mastoid width ≥126 mm; canine height ≥56 mm |
Condylobasal length <288 mm; interorbital width <70.5 mm; nasal length <105 mm; ventral posterior length <81 mm; width of zygomatic arches <218 mm; mastoid width <126 mm; canine height <56 mm |
| ~75% of females distinguishable from ~75% of sondaica |
Condylobasal length ≥255 mm; interorbital width ≥60 mm; nasal length ≥91.5 mm; ventral posterior length ≥72.5 mm; width of zygomatic arches ≥182 mm; mastoid width ≥112.5 mm; canine height ≥47 mm |
Condylobasal length <255 mm; interorbital width <60 mm; nasal length <91.5 mm; ventral posterior length <72.5 mm; width of zygomatic arches <182 mm; mastoid width <112.5 mm; canine height <47 mm |
| Pelage (Fig. 4C) | ||
| Ground color | Variable but averages lighter than sondaica | Darker than tigris |
| Mean number of flank stripes (distinguishes 75% of tigris from 75% of sondaica) |
<23 | >24 |
| Stripes | Thicker and less dense, especially on hindquarters | Thin and particularly dense on hindquarters |
| Pale ventral coloration | Extends further up flanks (one-third to halfway), tending to be white in color |
Extends minimally up flanks, tending to be yellow in color |
| Brow patches | Whiter and more extensive than in sondaica | Smaller than in tigris and tending to be yellow cranially |
| Ecological preferences (Fig. 4D) | ||
| Climate (~75% of tigris occurrence records are distinguishable from sondaica) |
More seasonal; cooler and drier climates than sondaica, but occurrence records from parts of its range (for example, Peninsular Malaysia) are indistinguishable from sondaica |
Less seasonal; warmer and wetter climates than tigris |
| Molecular markers (mtDNA) | ||
| SNPs that distinguish all tigris from sondaica [see also Luo et al. (8)] |
Position of the cat mtDNA genome (71) 5608 ND2: C in tigris 15743 Cytb: A in tigris |
Position of the cat mtDNA genome (71) 5608 ND2: T in sondaica 15743 Cytb: G in sondaica |
| SNPs that distinguish most tigris from sondaica |
Position of the cat mtDNA genome (71) 4442 ND1: T in most tigris 5155 ND2: T in most tigris 13029 ND5: G in most tigris 14118 ND5: C in most tigris |
Position of the cat mtDNA genome (71) 4442 ND1: C in all sondaica 5155 ND2: C in all sondaica 13029 ND5: A in most sondaica 14118 ND5: T in all sondaica |
Our analysis of this multiple-trait data set supports recognition of two distinct evolutionary groups of tigers and thus two subspecies: continental tigers and Sunda tigers (Table 2).
Continental tigers
This group includes six currently recognized subspecies: P. tigris tigris (Bengal tiger, TIG), P. tigris virgata† (Caspian tiger, VIR), P. tigris altaica (Amur tiger, ALT), P. tigris amoyensis (South Chinese tiger, AMO), P. tigris corbetti (Indochinese tiger, COR), and P. tigris jacksoni (Malayan tiger, JAC).
Craniodental morphology. Analysis of tiger skulls revealed a high degree of overlap in phenotypic space between putative continental subspecies for both males and females (Fig. 1, B1 and C1), and almost no separation or distinction between continental subspecies (Fig. 1, B2 and C2). Only the skulls of male tigers from Northern and East Asia (ALT, VIR, and AMO) were grouped together and distinct from those of other continental tigers (BS = 66%; Fig. 1, C3). The high degree of overlap between putative continental subspecies in our results is largely in agreement with those of other studies (18, 19).
Pelage morphology. Analysis of tiger pelages showed a large overlap between all putative continental subspecies (Fig. 1, D1), with no apparent significant differentiation (Fig. 1, D2). Only Northern tigers (ALT and VIR) were grouped distinctly from other continental tigers, whose pelage characteristics were largely overlapping. Our results are in agreement with previous observations of a North-South cline in several pelage characters (4).
Ecology. Owing to the wide geographical distribution of tigers across several distinct climatic regions, the disjunct Northern tigers (ALT and VIR) were clearly separated from all other tigers (Fig. 1, E3; BS = 100%). Compared with other traits, the overlap between Caspian and Amur tigers was lower, reflecting the natural distribution gap in large parts of Mongolia. Southern continental tigers, except Malayan tigers, clustered together. The high similarity in ecological preferences of Malayan and Sumatran tigers (Fig. 1, E3; BS = 100%) contrasts with other Malayan tiger traits, which showed a high degree of similarity with those of continental tigers. However, this result emphasizes the high degree of ecological adaptability of tigers that originate from two different phylogenetic clades.
Molecular population structure. On the basis of mtDNA, the populations of mainland tigers (Fig. 2, A2, gray) were structured, with Amur and Caspian tigers grouping together (Fig. 2, A2, green; BS = 94% ML, 87% BI), and Indochinese (BS = 95% ML, 88% BI) and Bengal (BS = 68% ML, 69% BI) tigers forming distinct groups. However, these are not all reciprocally monophyletic. Malayan tigers, although genetically separated from other continental tigers, were polyphyletic, forming two well-supported subclades (Fig. 2, A2; JAC clade 1, BS = 94% ML, 96% BI; JAC clade 2, BS = 92% ML, 89% BI). Few, if any, mitochondrial single-nucleotide polymorphisms (SNPs) separated continental tiger subspecies (Table 2), and no nuclear SNPs were found in Y- and X-linked genes (10). Currently, sampling is still incomplete across the tiger’s wide geographical distribution and relies on the inclusion of captive specimens. Therefore, it remains uncertain whether the observed mtDNA-based phylogenetic patterns within continental tigers have resulted from genuine evolutionary differences between isolated groups or just from incomplete sampling along a cline (4).
A robust intraspecific taxonomy should rely on well-established, multiple, independent characters (32, 33), especially if there are uncertainties in the available molecular data. None of the six putative continental subspecies could be consistently distinguished by morphological or ecological characters, because these overlapped greatly among the putative subspecies Fig. 1, B1 to E1 and B2 to E2. In contrast to tigers, similar data for the Sunda clouded leopard, Neofelis diardi, revealed significant differences between populations in Sumatra and Borneo, contributing to their recognition as two subspecies N. diardi diardi and N. diardi borneensis (26). Therefore, our data reject the taxonomic division of continental tigers into six subspecies. Instead, they should be merged into a single subspecies, which shows only minor local ecological adaptation and differentiation by distance.
Sunda tigers
This group includes three currently recognized subspecies: P. tigris sondaica† (Javan tigers, SON), P. tigris balica† (Bali tigers, BAL), and P. tigris sumatrae (Sumatran tigers, SUM).
Craniodental morphology, pelage morphology, ecology, and molecular population structure. The skulls of male and female Sunda tigers (SON, BAL, and SUM; Fig. 1, B1 and B2, C1 and C2), as well as their pelage markings, overlapped in their traits (Fig. 1, D1 and D2). Although Javan and Balinese tigers were more ecologically distinct from Sumatran tigers than in the other traits, their ecological niches still substantially overlapped (Fig. 1, E1 and E2). Molecular analysis showed that Javan and Balinese tigers cannot be distinguished from each other or from Sumatran tigers. Although a recent study found a single diagnostic SNP (a position not used in our study) between Javan/Balinese and Sumatran tigers (17), all three putative subspecies shared haplotypes in our analysis (Fig. 2B). This indicates recent gene flow between the tiger populations of these islands, which was facilitated by land bridges around the time of the Last Glacial Maximum, when lower sea levels exposed the continental shelf until as recently as 10 kya (34). Our data demonstrate that all Sunda tigers constitute a single subspecies.
Conservation management units (ecotypes) of modern tigers
In addition to the taxonomy proposed here, which supports the recognition of only two subspecies, our data also support the recognition of two management units (ecotypes) within continental tigers: northern tigers (ecotype 1: ALT and VIR) and southern tigers (ecotype 2: AMO, COR, TIG, and JAC). These management units are justified because northern continental tigers show molecular, skull (only males), pelage, and ecological characters distinct from those of southern continental tigers (Fig. 3, B2 to B5). However, the recognition of a separate northern tiger subspecies is inappropriate, because (i) within the phylogenetic tree, northern tigers are a subclade within the continental group, and thus, a southern continental tiger subspecies would be paraphyletic, and (ii) the absence of tigers in northern China was most probably caused by ancient-to-modern human overexploitation and overemphasizes the distinctiveness of the northern end of what was probably a continuous cline. In other words, this sampling bias, exacerbated by a recent population bottleneck, has led to the inappropriate recognition of a northern tiger subspecies, which in isolation shows significant local adaptations to a temperate climate and habitats with their particular arrays of prey species. Owing to these adaptations and the current large distribution gap between northern and southern continental tigers—which exceeds 1000 km and is greater than the maximum dispersal distance of tigers (13)—the treatment of northern and southern continental tigers as separate management units is justified.
Management implications
With only ca. 1000 breeding females in the wild (35), the traits of all tiger individuals will probably be of importance for long-term conservation because the remaining variation will be the key to maintaining the highest possible adaptability to changing environments. Therefore, to achieve the goal of doubling the number of free-ranging tigers by 2022 (36), all remaining wild tiger populations are essential, and all tiger range countries share equal responsibility for restoring the geographical spread and ecological diversity of tigers. Basing current conservation strategies on numerous subspecies, for which there is little or no scientific support, may actually hinder the tiger’s survival by preventing large-scale cooperative conservation management programs, such as conservation captive breeding, reintroduction initiatives, or trans-boundary projects, which will be restricted to the currently recognized putative subspecies. “Subspecific hybrid” tigers or animals of supposedly different subspecies, even if bred in reputable conservation breeding programs and not “tiger farms,” are therefore excluded from many conservation campaigns. This greatly reduces the number of available animals, unnecessarily restricting gene pools and thus impeding the success of conservation measures.
Recent coalescent simulations highlighted that, in the absence of gene flow between currently recognized subspecies, today’s levels of genetic diversity in tigers cannot be maintained (37). Our analyses of and recommendations on the taxonomic status of all currently recognized subspecies now provide a solid foundation for a more pragmatic approach to tiger conservation worldwide. It is worth noting that reducing the number of recognized tiger subspecies to two and the number of management units to three will not hinder cooperative conservation efforts, nor will they be impeded by people and governments of range countries labeling their “own” tiger populations with vernacular names such as “Indian,” “Chinese,” or “Malayan” tigers as an expression of pride in their local population, because this will be of paramount importance for local or regional conservation success. If local conservation activities for wild tiger populations are well planned, and law and policy enforcement is effective, tiger populations can rapidly recover (38). In contrast to overarching, coordinated international initiatives, the success of local conservation activities is largely independent of the tiger’s evolutionary history, its subspecies status, or management unit assignment.
With the population of wild tigers dwindling to a worryingly low number, the success of future conservation management is likely to depend more on intensive modes of conservation intervention to boost the diversity of genetically depleted populations or reestablish populations after local extinctions. Such active interventions may include translocations or the release of captive-bred individuals by internationally recognized conservation breeding programs, which may also arise from assisted reproduction techniques such as artificial insemination or (in future) embryo transfer. Therefore, the successful conservation management of the tiger requires an integrated conservation management approach, which simultaneously considers and coordinates in situ and ex situ conservation management activities. This is embodied in the IUCN Captive Breeding Specialist Group’s One Plan Approach to species conservation planning, which aims to bridge the gap between captive and wild populations for more integrated species conservation (39).
Our results demonstrate that only Sumatran tigers should be used for interventions in Sumatra, Java, and Bali—even if there is currently a lack of sufficient tiger habitats that are devoid of tiger populations on these islands. For northeastern Asia and the Caspian region, our data support previous results that suggested the use of Amur tigers for reintroduction programs to their former Caspian range and elsewhere (40). For conservation management interventions in southern continental Asia, we recommend giving highest priority to the transfer of either wild tigers or captive-bred tigers with known origin from reputable conservation breeding institutions to repopulate areas with currently no tigers. These tigers should originate from less than 1000 km away, which is consistent with the maximum distance recorded for tiger dispersal and hence possible gene flow (13). Only if such individuals are not available should tigers from other parts of the southern continental Asian range be used.
Our results mostly support current conservation management of captive tigers. A previous study (41) genotyped a significant proportion of captive tigers. Many were of unknown origin and others were classified as hybrids of former putative subspecies, with the result that “hybrid” individuals were often removed from their respective breeding programs. This highlights the importance of a scientifically sound and more pragmatic approach to the management of captive tigers. Zoos have spent significant amounts of money to have their tigers genotyped, and currently focus their breeding efforts on “pure-bred” tigers as defined by the conventional definition of nine subspecies. Our results demonstrate that the current breeding programs accredited by the World Association of Zoos and Aquariums (WAZA) could be extended by inclusion of many of these supposed hybrids because southern continental tigers do not need to be managed according to their hitherto conventional subspecific classification. Such a conservation management approach would enhance the persistence of alleles that may only survive in supposedly “subspecific hybrid” tigers, which substantially outnumber the so-called pure captive tigers (41).
Our recommendations assist in the maintenance of genetic diversity and are consistent with recent findings, according to which every tiger, both wild and captive, is important as a potential reservoir of genetic diversity for the species (37). The separate breeding of Indochinese, South Chinese, and Malayan tigers is likely to be unsustainable because only few founder individuals were available—for instance, only six individuals for South Chinese tigers (42). In contrast, both the northern continental tiger management units and the Sunda tiger subspecies have been traditionally maintained in separate breeding programs. Their numbers of founders were much higher; several hundred individuals are currently part of international breeding programs. For Sumatran tigers, the international zoo community led by WAZA has even successfully established a Global Species Management Plan. The separate conservation breeding and management of these units is supported by our results and should be continued.
CONCLUSION
The previous traditional tiger intraspecific taxonomy was often based on small sample sizes and arbitrary morphological characters and lacked a comprehensive approach so that subspecies names were little more than labels for local populations. By integrating morphological, molecular, and ecological data with biogeography and geological events, we obtained robust support for two subspecies that have differentiated in isolation, with one of them consisting of two conservation management units. Our rationalization provides a solid foundation to boost both in situ and ex situ conservation management of the appropriate evolutionary, taxonomic, and management units at a global level. This will assist an integrated conservation management approach to reverse the tiger’s decline and thus to achieve the key challenge—saving the species, Panthera tigris, as a whole.
MATERIALS AND METHODS
Experimental design
Craniodental and mandibular morphology
Craniodental and mandibular diversity within tigers were assessed using a sample of 201 tiger adult skulls [102 males (table S1) and 99 females (table S2)]. For each specimen, 41 craniomandibular and dental measurements were analyzed (table S3).
Pelage morphology
Altogether, we analyzed photographs of 114 tigers, which were either photographs of museum skin specimens or photographs taken in the wild (for example, photographs taken in camera-traps) or in zoos (known-origin animals only) so that the origin was known (table S4). For each photograph, we scored six pelage characters (tables S5 and S6). The number of stripes on flanks was the mean count of the number of stripes on the right and left flanks. The other five characters were categorized between 1 (none or few) and 3 (many) because some specimens or photographs were not always complete and thus only comparable on a semiquantitative scale.
Ecological preferences
Occurrence records
In total, we collated 482 occurrence records of tigers (table S7). Most tiger records were extracted from Mazák (43) and Kitchener and Dugmore (13), which are based on historical records. We allocated each of these historical records a latitude/longitude value with an uncertainty of about 50 km because of the low resolution of the used maps (13, 43). These historical records are valuable because they predate the current distribution, which we judge to be biased as a consequence of tiger persecution and habitat loss. In addition to the historical records, we selected random points from the Tiger Conservation Landscapes (3) to include locations of the modern distribution not covered by historical sampling. For the putative Javan and Bali subspecies, we included localities recorded by Yamaguchi et al. (20) and used localities from where museum specimens were originally collected, which we had used for the skull, pelage, and molecular analyses. Because of the low number of records for these two putative subspecies, we selected additionally three (Java) and nine (Bali) random points from these islands to reach a minimum number of 15 occurrence records per putative subspecies (table S7).
Digital environmental information
We used a set of 19 global climate data maps with a spatial resolution of about 1 km2 (30 arc sec). They comprise temperature and precipitation parameters (listed in table S10) calculated from long-term time series of monthly values (44) (www.worldclim.org/bioclim). We also used available information about the world’s terrestrial ecoregions (45) (http://worldwildlife.org/publications/terrestrial-ecoregions-of-the-world). We reclassified the 16 ecoregions within the tiger’s range into four functional habitat classes (table S8). Because we expected that the availability of prey species would also influence the occurrence of tigers, we reviewed the existing literature to obtain information about tiger prey species. We only included those genera as potential tiger prey species, for which we could find at least two independent references, and we grouped prey genera into functional prey groups according to their ecological similarity (table S9). For all prey species, we used the former distributional ranges provided by the IUCN/SSC Red List of Threatened Species (www.iucnredlist.org/). The distributional ranges of all potential prey species within one prey group were merged to obtain the presence/absence layer for each prey group.
All maps were clipped and resampled to the same extent and cell size. All spatial analyses used ArcInfo 9.3 and ArcGIS v. 9.3.1 (ArcView, Spatial Analyst, 3D Analyst, Geostatistical Analyst) by ESRI, and R 3.0.1 (46).
Genetic samples
We obtained epithelial tissue (from skulls or skins) or maxillo-turbinal bone samples from P. tigris sondaica (n = 22) and P. tigris balica (n = 3) voucher specimens from natural history museums. Ten Javan archival DNA samples did not provide amplicons for all of the fragments (see below) and thus were excluded from further analyses (table S13). DNA extraction of all samples was performed following an established protocol (26) in a separate clean pre-PCR (polymerase chain reaction) laboratory solely used for the extraction of DNA of museum samples. PCR amplification was performed with eight known sets of mitochondrial primers (16) and four new sets of mitochondrial primers (table S14), which covered regions shown to be variable and informative for the distinction of Sumatran tigers (8). In total, we sequenced 1772 base pairs (bp) of the 4078 bp previously analyzed (8). Our sequenced regions were selected to cover all variable positions within Sumatran and between Sumatran and continental tigers. All PCR reactions and sequencing analyses were done following the established protocol (26). Sequences were assembled, aligned, and edited using the software Clustal X2 (47) and analyzed together with published sequences (8, 16) and GenBank accession no. JF357969 (11) (SUM9).
Data analyses
Multivariate analysis of skulls, skins, and ecological data
For each trait, the phenotypic space was summarized by performing appropriate ordination methods (Figs. 1 and 3 and fig. S1). Such a multivariate approach ensured that the variation within each trait was presented in an identical manner across traits, efficiently summarized the total phenotypic space in a limited number of dimensions or components, and therefore also permitted a useful geometrical representation of the total phenotypic space. We preferred an ordination approach because this does not require a prior assignment of each individual to a specific subspecies and allows the number of true groups/subspecies to emerge from the analysis rather than being set before analysis, which other multivariate approaches, such as discriminant analysis, would require.
Specifically, for skull data, we performed a scaled principal components analysis (PCA), that is, a PCA on data that were transformed into z-scores (mean, 0; SD, 1) to avoid an overemphasis of the larger measurements. PCA is a well-established technique for efficiently summarizing phenotypic space in skull measurements for taxonomic purposes (48). We performed separate PCAs for males and females owing to sexual dimorphism in tigers. PCAs were computed in R using the function “dudi.pca” from the package ade4 1.5-2 (49). For skin data, one variable was quantitative and others were ordinal with three levels. We performed a scaled PCA on these data after coding the ordinal variables by their corresponding numerical values (1–3) because our ordinal records reflect continuous variation. Although PCA assumes a multivariate normal distribution of variables, PCA has been shown to be very robust when performed on standardized semiquantitative variables (50). For ecological data, we used FAMD (51), a method designed to combine both categorical and continuous variables within the same analysis by combining a scaled PCA on quantitative variables and a multiple correspondence analysis (MCA), a nonparametric ordination technique, on qualitative variables. The FAMD was computed in R using the function “FAMD” from the package FactoMineR 1.25 (52). Figure 1 (B1 and E1) shows the first two principal components and the 1.5 inertia ellipses for all traits; the third and fourth components are shown in fig. S3. Tables S3, S6, and S10 provide the relative contribution of each trait to the first four principal components. We illustrate the phenotypic space first with respect to the nine putative subspecies (Fig. 1 and fig. S3) and then with respect to the two subspecies and three management units recognized by the results of our study (Fig. 3 and fig. S3).
We then used the results of our multivariate analyses to generate unrooted binary trees to visualize how individuals and the nine putative subspecies were related to each other as a function of each set of traits. Trees were constructed by applying a neighbor-joining algorithm onto Euclidean distance matrices derived from coordinates of individuals on all components in the phenotypic space. Trees were computed and displayed using the package ape 3.0-9 in R (53). To assess the bootstrap support of nodes of interest in trees (putative subspecies groupings), we built 1000 bootstrap samples, reran the multivariate analysis, and built a new neighbor-joining tree for each of these samples. Then, bootstrap values resembled directly the frequency of trees that presented the node under consideration. We generated each bootstrap sample by drawing at random a number of individuals (with replacement) within each group equal to the sample size of the corresponding group. This bootstrap analysis was performed independently for each of the four subspecies trees initially obtained, that is, two for skulls (males and females), one for pelage, and one for ecological traits. We then assessed whether the nodes of the mtDNA tree (Fig. 2) were supported by the other three character sets of skull, pelage, and ecological characters. To compare the strength of obtained bootstrap supports to reference values in the absence of any population structure, we permuted subspecies in the Euclidean distance matrices for each of 1000 bootstrap runs and calculated the mean, median, and 95% confidence intervals (CIs) of nodes pooling two, three, or four putative subspecies in the neighbor-joining trees. This simulation study showed that all bootstrap values higher than 40% can be regarded as high in our analysis because such a support was much higher than the upper 95% CI (table S11). All computations for multivariate analyses were performed using R 3.0.1 (46).
Ecological niche modeling
The tiger occurrence records, the environmental data, and the data on the distribution of tiger prey were used to model the ecological niches for each of the nine putative subspecies and for all subspecies combined, using a maximum entropy algorithm implemented in MaxEnt v.3.3.3a (54) with the default settings (replicated run type, “cross-validate”; regularization multiplier, 1; maximum number of background points, 10,000). We ran 10 replicates and used the mean relative probability of occurrence for further analyses.
First, we used all occurrence records to predict the historical (pre-Anthropocene) distribution of tigers (fig. S1). We used a presence/absence map based on the 10-percentile training presence logistic threshold (10P threshold) of this model, together with published maps of the historical tiger distribution (13, 43, 55), to create a predictive presence/absence historical distributional range map of tigers (fig. S1). This map was used, together with information from Kitchener and Dugmore (13) and Luo et al. (8), to delimit the ranges of the nine putative subspecies (Fig. 1A).
Subsequently, we performed MaxEnt models for all nine putative subspecies (fig. S2, B to I), followed by a pairwise comparison of all the resulting putative subspecies models with ENMTools (56). Here, we used the niche overlap tool, which assesses the similarity between predictions of habitat suitability. For each possible pair of putative subspecies, the respective MaxEnt habitat suitability maps were restricted to the combined ranges of the two subspecies, and the niche overlap was calculated by two measures, I and D, as implemented in ENMTools (table S12).
Subspecies diagnosis
To visualize the subspecies diagnosis, we plotted the main traits (those that contributed most in the multivariate analysis) for the two tiger subspecies as violin plots, that is, a combination of a box plot and a kernel density plot. Nonoverlapping quartile boxes (in fig. S3) can be regarded as reliably differentiating more than 75% of individuals between the subspecies. Violin plots were plotted in R using the package ggplot2 (57). Furthermore, we performed a pairwise comparison between the two tiger subspecies, which we finally recognized using Mann-Whitney U tests for all continuous and ordinal variables (skull, skin, and climate). Categorical count variables (functional prey group presence and functional habitats) were compared using Fisher’s exact test.
Molecular analysis
All analyses described below were performed on two data sets, 1772 and 3968 bp in length [primer sequences were trimmed off the 4078 bp of Luo et al. (8)], from 121 specimens. Because both data sets gave similar results (table S15), we only presented the results of the ~4-kb fragment to be compared with previous studies (8, 16, 17). All sequences represented conspecifics of P. tigris, except for sequences from two Panthera uncia [accession nos. NC010638 and EF551004 (58)], one Panthera pardus [accession no. EF551002 (58)], and one N. nebulosa [accession no. DQ257669 (59)], which were included as outgroups in phylogenetic and Bayesian skyline analyses. The known divergence times from these outgroups to P. tigris were used for molecular clock calibration [6.37 ± 0.3185 Ma for the most recent common ancestor (MRCA) of Neofelis and Panthera (60), 3.94 ± 0.197 Ma for the MRCA of the different Panthera species (61), and 3.19 ± 0.1595 Ma for the MRCA of P. uncia and P. tigris (61)]. Summary statistics were computed using DnaSP 5.10.01 (62), grouping all P. tigris sequences in a single unit, or splitting sequences into two units according to their geographical distribution on the continent and in the Sunda region (table S15). For phylogenetic analyses and Bayesian skylines, we haplotyped each data set using FaBox v.1.41 (63), resulting in 35 haplotypes. Furthermore, we used jModelTest v.0.1.1 and the Bayesian Information Criterion (64) to select HKY+Γ8 as the best model of molecular evolution. We then performed maximum likelihood and Bayesian phylogenetic inferences using PhyML v.3.0 (65) and MrBayes v.3.2.2 (66) (Fig. 2, A1 and A2). In maximum likelihood analyses, node support was estimated on the basis of 100 bootstrap pseudo-replicates, whereas in Bayesian inference, node support was based on the posterior distribution of 75,000 trees (100,000,000 generations; sampling frequency, 1/1000; burn in, 25,000) sampled from two parallel runs with four chains each. Finally, we used BEAST v.1.7.0 (67) to estimate divergence times at three subclades of the tree (which were forced to monophyly in agreement with maximum likelihood and Bayesian phylogenetic analyses): (i) the MRCA of all P. tigris sequences; (ii) the MRCA of all P. tigris, disregarding sequence AMO1, which branched outside of all other P. tigris sequences; and (iii) the MRCA of all P. tigris sequences from the Sunda region. This was performed under the HKY+Γ8 and the Bayesian Skyline models, assuming either a strict constant molecular clock or a relaxed molecular clock (with an uncorrelated lognormal distribution). A total of 2.5 × 108 generations was performed in each analysis, sampling 1 generation for every 1000, and the best-fit model was selected by means of Bayes factor (BF) calculations as provided by Tracer v.1.4 (68) after convergence and using 25% of sampled generations as burn in. For both sequence data sets, the relaxed clock model received significant support (log10 BF = 0.985 and 4.103 for the 1772- and 3968-bp sequence data sets, respectively). Therefore, we based divergence times (table S17) and demographic skyline reconstructions (Fig. 2C) on the posterior distributions from this model. We assumed an average biological generation time of 5 years for tigers (69). To display all possible maximum parsimonious relationships among haplotypes, a haplotype median-joining network (Fig. 2B) was constructed using Network v.4.6.0.0 (70).
Supplementary Material
Acknowledgments
We thank D. Mörike, F. Mayer, G. Lenglet, N. Mascarenhas, T. M. Kaiser, and C. Stefen for supplying samples of Javan and Bali specimens on which the molecular work is based. We also thank R. Redel for his assistance with the laboratory analyses, and D. Hills, P. Jenkins, L. Tomsett, and R. Portela-Miguez (Mammal Section, Natural History Museum, London), C. Smeenk (Naturalis, Leiden), and F. Mayer (Museum für Naturkunde, Berlin) for access to tiger specimens in their care. Funding: This work was funded by the German Research Foundation (DFG grant Fi-698/5-1) and the Federal Ministry of Education and Research (BMBF FKZ: 01LN1301A). Author contributions: A.W., J.F., and A.C.K. conceived the idea and wrote the manuscript; A.C., P.C., J.N., A.K.S., L.O., N.B., H.H., and S.K.-S. edited the manuscript; A.W., L.O., N.B., and J.F. performed the molecular analysis; P.C. collected the skull data; A.C.K. collected the pelage data; A.C. analyzed the skull, pelage, and ecological data; A.W., J.N., A.K.S., and S.K.-S. performed the ecological modeling and the GIS analyses. Competing interests: The authors declare that they have no competing interests. Data and materials availability: GenBank accession no. KR349728-KR349907.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/1/5/e1400175/DC1
Fig. S1. Summary of the work flow to predict the historical distribution of tigers.
Fig. S2. Predicted historical distribution of tigers P. tigris (top left) and MaxEnt analysis for the nine putative subspecies.
Fig. S3. Phenotypic space across all nine putative subspecies of tigers.
Fig. S4. Comparison of the nine putative tiger subspecies.
Table S1. Sources and details of male specimens included in the craniodental analysis.
Table S2. Sources and details of female specimens included in the craniodental analysis.
Table S3. The 41 craniodental characters and their contribution to the first four principal components.
Table S4. Sources and details of the specimens or photographs included in the pelage analysis.
Table S5. Means and SDs for the six pelage characters across the nine putative subspecies.
Table S6. The six pelage characters and their contribution to the first four principal components.
Table S7. Number of occurrence records including the references for each of the nine putative subspecies used for the PCA and for the species distribution modeling (see Materials and Methods for details).
Table S8. Definitions of the four functional habitats and the eco-regions assigned to them.
Table S9. List of the tiger prey species.
Table S10. The 29 ecological variables and their contribution to the first four principal components.
Table S11. Simulation of bootstrap values of nodes pooling two, three, or four putative subspecies in the neighbor-joining tree.
Table S12. Niche overlap between putative subspecies as computed by ENMTools.
Table S13. Sources and details of specimens included in the molecular analysis in addition to the sequences from Luo et al. (8), Driscoll et al. (16), and GenBank accession no. JF357969 (11) (SUM9).
Table S14. Details of mitochondrial primer sets used in addition to the eight primer sets described by Driscoll et al. (16).
Table S15. Population summary statistics calculated for the short and long mtDNA data sets.
Table S16. Nucleotide diversity of pantherine cats.
Table S17. Divergence dates in years before present derived from the short and long mtDNA data sets.
Table S18. Pairwise comparison between the two tiger subspecies P. tigris tigris and P. tigris sondaica for the craniodental characteristics using the Mann-Whitney U test (♂: ntigris = 64, nsondaica = 38; ♀: ntigris = 55, nsondaica = 44).
Table S19. Pairwise comparison between the two tiger subspecies P. tigris tigris and P. tigris sondaica for the pelage characteristics using the Mann-Whitney U test (ntigris = 89, nsondaica = 25).
Table S20. Pairwise comparison between the two tiger subspecies P. tigris tigris and P. tigris sondaica for the continuous ecological variables (climate) using the Mann-Whitney U test (ntigris = 428, nsondaica = 50).
Table S21. Pairwise comparison between the two tiger subspecies P. tigris tigris and P. tigris sondaica for the categorical ecological variables (functional prey groups and functional habitat) using Fisher’s exact test (ntigris = 428, nsondaica = 50).
REFERENCES AND NOTES
- 1.Seidensticker J., Saving wild tigers: A case study in biodiversity loss and challenges to be met for recovery beyond 2010. Integr. Zool. 5, 285–299 (2010). [DOI] [PubMed] [Google Scholar]
- 2.E. W. Sanderson, J. Forrest, C. Loucks, J. Ginsberg, E. Dinerstein, J. Seidensticker, P. Leimgruber, M. Songer, A. Heydlauff, T. O’Brien, G. Bryja, S. Klenzendorf, E. Wikramanayake, in Tigers of the World, R. Tilson, P. J. Nyhus, Eds. (Academic Press, London, ed. 2, 2010), pp. 144–161.
- 3.Walston J., Robinson J. G., Bennett E. L., Breitenmoser U., da Fonseca G. A., Goodrich J., Gumal M., Hunter L., Johnson A., Karanth K. U., Leader-Williams N., Mackinnon K., Miquelle D., Pattanavibool A., Poole C., Rabinowitz A., Smith J. L. D., Stokes E. J., Stuart S. N., Vongkhamheng C., Wibisono H., Bringing the tiger back from the brink-the six percent solution. PLOS Biol. 8, e1000485 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.A. C. Kitchener, N. Yamaguchi, in Tigers of the World, R. Tilson, P. J. Nyhus, Eds. (Academic Press, London, ed. 2, 2010), pp. 54–84.
- 5.S.-J. Luo, W. E. Johnson, J. L. D. Smith, S. J. O’Brien, in Tigers of the World, R. Tilson, P. J. Nyhus, Eds. (Academic Press, London, ed. 2, 2010), pp. 36–51.
- 6.R. S. Chundawat, B. Habib, U. Karanth, K. Kawanishi, J. Ahmad Khan, T. Lynam, D. Miquelle, P. J. Nyhus, S. Sunarto, R. Tilson, S. Wang, Panthera tigris IUCN 2013. IUCN Red List Threat. Species (2013); www.iucnredlist.org/details/15955/0.
- 7.J. Seidensticker, B. Gratwicke, M. Shrestha, in Tigers of the World, R. Tilson, P. J. Nyhus, Eds. (Academic Press, London, ed. 2, 2010), pp. 295–299.
- 8.Luo S. J., Kim J. H., Johnson W. E., van der Walt J., Martenson J., Yuhki N., Miquelle D. G., Uphyrkina O., Goodrich J. M., Quigley H., Tilson R., Brady G., Martelli P., Subramaniam V., McDougal C., Hean S., Huang S. Q., Pan W., Karanth U. K., Sunquist M., Smith J. L. D., O’Brien S. J., Phylogeography and genetic ancestry of tigers (Panthera tigris). PLOS Biol. 2, e442 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.International Commission on Zoological Nomenclature (ICZN), International Code for Zoological Nomenclature (The International Trust for Zoological Nomenclature, London, ed. 4, 1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luo S.-J., Zhang Y., Johnson W. E., Miao L., Martelli P., Antunes A., Smith J. L. D., O’Brien S. J., Sympatric Asian felid phylogeography reveals a major Indochinese–Sundaic divergence. Mol. Ecol. 23, 2072–2092 (2014). [DOI] [PubMed] [Google Scholar]
- 11.Kitpipit T., Tobe S. S., Kitchener A. C., Gill P., Linacre A., The development and validation of a single SNaPshot multiplex for tiger species and subspecies identification—Implications for forensic purposes. Forensic Sci. Int. Genet. 6, 250–257 (2012). [DOI] [PubMed] [Google Scholar]
- 12.J. Wentzel, J. C. Stephens, W. E. Johnson, M. Menotti-Raymond, J. Pecon-Slattery, N. Yuhki, M. Carrington, H. B. Quigley, D. G. Miquelle, R. Tilson, J. Manansang, G. Brady, L. Zhi, U. Karanth, S. J. O’Brien, in Riding the Tiger: Tiger Conservation in Human Dominated Landscapes, J. Seidensticker, S. Christie, P. Jackson, Eds. (Cambridge Univ. Press, Cambridge, UK, 1999), pp. 20–49.
- 13.Kitchener A. C., Dugmore A. J., Biogeographical change in the tiger, Panthera tigris. Anim. Conserv. 3, 113–124 (2000). [Google Scholar]
- 14.Sharma S., Stuckas H., Bhaskar R., Rajput S., Khan I., Goyal S. P., Tiedemann R., mtDNA indicates profound population structure in Indian tiger (Panthera tigris tigris). Conserv. Genet. 10, 909–914 (2009). [Google Scholar]
- 15.Mondol S., Bruford M. W., Ramakrishnan U., Demographic loss, genetic structure and the conservation implications for Indian tigers. Proc. Biol. Sci. 280, 20130496 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Driscoll C. A., Yamaguchi N., Kahila Bar-Gal G., Roca A. L., Luo S.-J., Macdonald D., O’Brien S. J., Mitochondrial phylogeography illuminates the origin of the extinct Caspian tiger and its relationship to the Amur tiger. PLOS One 4, e4125 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xue H.-R., Yamaguchi N., Driscoll C. A., Han Y., Bar-Gal G. K., Zhuang Y., Mazak J. H., Macdonald D. W., O’Brien S. J., Luo S.-J., Genetic ancestry of the extinct Javan and Bali tigers. J. Hered. 106, 247–257 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mazak J., Groves C., A taxonomic revision of the tigers (Panthera tigris) of Southeast Asia. Mamm. Biol. 71, 268–287 (2006). [Google Scholar]
- 19.Mazak J. H., Craniometric variation in the tiger (Panthera tigris): Implications for patterns of diversity, taxonomy and conservation. Mamm. Biol. 75, 45–68 (2010). [Google Scholar]
- 20.Yamaguchi N., Driscoll C. A., Werdelin L., Abramov A. V., Csorba G., Cuisin J., Fernholm B., Hiermeier M., Hills D., Hunter L., Itakura H., Johansson U. S., Kascheev V., Krohmann K., Martin T., Nowak-Kemp M., Pavlinov I. Y., Renoud F., Tomsett L., Van Der Mije S., Zholnerovskaya E., Groves C., Kitchener A. C., Nijman V., Macdonald D. W., Locating specimens of extinct tiger (Panthera tigris) subspecies: Javan tiger (P. T. sondaica), Balinese tiger (P. T. balica), and Caspian tiger (P. T. virgata), including previously unpublished specimens. Mammal Study 38, 187–198 (2013). [Google Scholar]
- 21.Isaac N. J. B., Mallet J., Mace G. M., Taxonomic inflation: Its influence on macroecology and conservation. Trends Ecol. Evol. 19, 464–469 (2004). [DOI] [PubMed] [Google Scholar]
- 22.Zachos F. E., Apollonio M., Bärmann E. V., Festa-Bianchet M., Göhlich U., Habel J. C., Haring E., Kruckenhauser L., Lovari S., McDevitt A. D., Pertoldi C., Rössner G. E., Sánchez-Villagra M. R., Scandura M., Suchentrunk F., Species inflation and taxonomic artefacts—A critical comment on recent trends in mammalian classification. Mamm. Biol. 78, 1–6 (2013). [Google Scholar]
- 23.Rampino M. R., Self S., Volcanic winter and accelerated glaciation following the Toba super-eruption. Nature 359, 50–52 (1992). [Google Scholar]
- 24.Ambrose S. H., Late Pleistocene human population bottlenecks, volcanic winter, and differentiation of modern humans. J. Hum. Evol. 34, 623–651 (1998). [DOI] [PubMed] [Google Scholar]
- 25.Nater A., Nietlisbach P., Arora N., van Schaik C. P., van Noordwijk M. A., Willems E. P., Singleton I., Wich S. A., Goossens B., Warren K. S., Verschoor E. J., Perwitasari-Farajallah D., Pamungkas J., Krützen M., Sex-biased dispersal and volcanic activities shaped phylogeographic patterns of extant orangutans (genus: Pongo). Mol. Biol. Evol. 28, 2275–2288 (2011). [DOI] [PubMed] [Google Scholar]
- 26.Wilting A., Christiansen P., Kitchener A. C., Kemp Y. J. M., Ambu L., Fickel J., Geographical variation in and evolutionary history of the Sunda clouded leopard (Neofelis diardi) (Mammalia: Carnivora: Felidae) with the description of a new subspecies from Borneo. Mol. Phylogenet. Evol. 58, 317–328 (2011). [DOI] [PubMed] [Google Scholar]
- 27.S. J. Herrington, in Tigers of the World, R. L. Tilson, U. S. Seal, Eds. (Noyes Publishing, Park Ridge, NJ, 1987), pp. 51–61.
- 28.Mazák J. H., Christiansen P., Kitchener A. C., Oldest known pantherine skull and evolution of the tiger. PLOS One 6, e25483 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hooijer D., Pleistocene remains of Panthera tigris (Linnaeus) subspecies from Wanhsien, Szechuan, China, compared with fossil and recent tigers from other localities. Am. Mus. Novit. 1346, 1–17 (1947). [Google Scholar]
- 30.Turelli M., Barton N. H., Coyne J. A., Theory and speciation discuss recent examples of progress in each of these areas. Trends Ecol. Evol. 16, 330–343 (2001). [DOI] [PubMed] [Google Scholar]
- 31.E. Mayr, P. D. Ashlock, Principles of Systematic Zoology (McGraw-Hill, New York, ed. 2, 1991). [Google Scholar]
- 32.Wilson E. O., Brown W. L. J., The subspecies concept and its taxonomic application. Syst. Zool. 2, 97–111 (1953). [Google Scholar]
- 33.Haig S. M., Beever E. A., Chambers S. M., Draheim H. M., Dugger B. D., Dunham S., Elliott-Smith E., Fontaine J. B., Kesler D. C., Knaus B. J., Lopes I. F., Loschl P., Mullins T. D., Sheffield L. M., Taxonomic considerations in listing subspecies under the U.S. Endangered Species Act. Conserv. Biol. 20, 1584–1594 (2006). [DOI] [PubMed] [Google Scholar]
- 34.Voris H., Maps of Pleistocene sea levels in Southeast Asia: Shorelines, river systems and time durations. J. Biogeogr. 27, 1153–1167 (2000). [Google Scholar]
- 35.U. Karanth, J. M. Goodrich, S. Vaidyanathan, V. G. Reddy, Landscape Scale, Ecology-Based Management of Wild Tiger Populations (Global Tiger Initiative, World Bank, and Wildlife Conservation Society, Washington, DC, 2010). [Google Scholar]
- 36.Wikramanayake E., Dinerstein E., Seidensticker J., Lumpkin S., Pandav B., Shrestha M., Mishra H., Ballou J., Johnsingh A. J. T., Chestin I., Sunarto S., Thinley P., Thapa K., Jiang G., Elagupillay S., Kafley H., Pradhan N. M. B., Jigme K., Teak S., Cutter P., Aziz M. A., Than U., A landscape-based conservation strategy to double the wild tiger population. Conserv. Lett. 4, 219–227 (2011). [Google Scholar]
- 37.Bay R. A., Ramakrishnan U., Hadly E. A., A call for tiger management using “reserves” of genetic diversity. J. Hered. 105, 295–302 (2013). [DOI] [PubMed] [Google Scholar]
- 38.Rabinowitz A., Hunter L., Smith J., Tiger conservation: Trust tradition. Science 333, 1824 (2011). [DOI] [PubMed] [Google Scholar]
- 39.Byers O., Lees C., Wilcken J., Schwitzer C., The One Plan Approach: The philosophy and implementation of CBSG’s approach to integrated species conservation planning. WAZA Mag. 14, 2–5 (2013). [Google Scholar]
- 40.Driscoll C. A., Chestin I., Jungius H., Pereladova O., Darman Y., Dinerstein E., A postulate for tiger recovery: The case of the Caspian Tiger. J. Threat. Taxa 4, 2637–2643 (2012). [Google Scholar]
- 41.Luo S.-J., Johnson W. E., Martenson J., Antunes A., Martelli P., Uphyrkina O., Traylor-Holzer K., Smith J. L. D., O’Brien S. J., Subspecies genetic assignments of worldwide captive tigers increase conservation value of captive populations. Curr. Biol. 18, 592–596 (2008). [DOI] [PubMed] [Google Scholar]
- 42.K. Traylor-Holzer, X. Zhong, Y. Yuzhong, in Tigers of The World, R. Tilson, P. J. Nyhus, Eds. (Academic Press, London, ed. 2, 2010), pp. 457–461.
- 43.V. Mazák, Der Tiger (A. Ziemsen Verlag, Wittenberg Lutherstadt, Germany, 1965). [Google Scholar]
- 44.Hijmans R. J., Cameron S. E., Parra J. L., Jones P. G., Jarvis A., Very high resolution interpolated climate surfaces for global land areas. Int. J. Climatol. 25, 1965–1978 (2005). [Google Scholar]
- 45.Olson D. M., Dinerstein E., Wikramanayake E. D., Burgess N. D., Powell G. V. N., Underwood E. C., D’amico J. A., Itoua I., Strand H. E., Morrison J. C., Loucks C. J., Allnutt T. F., Ricketts T. H., Kura Y., Lamoreux J. F., Wettengel W. W., Hedao P., Kassem K. R., Terrestrial ecoregions of the world: A new map of life on earth. Bioscience 51, 933–938 (2001). [Google Scholar]
- 46.R Core Development Team, R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, Vienna, Austria, 2013). [Google Scholar]
- 47.Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., Higgins D. G., Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 (2007). [DOI] [PubMed] [Google Scholar]
- 48.P. H. A. Sneath, R. R. Sokal, Numerical Taxonomy: The Principles and Practice of Numerical Classification (Freeman, San Francisco, CA, 1973). [Google Scholar]
- 49.Dray S., Dufour A. B., The ade4 package: Implementing the duality diagram for ecologists. J. Stat. Softw. 22, 1–20 (2007). [Google Scholar]
- 50.D. Borcard, F. Gillet, P. Legendre, Numerical Ecology with R (Springer, New York, 2011). [Google Scholar]
- 51.Pages J., Analyse factorielle de donnees mixtes. Rev. Stat. Appl. LII, 93–111 (2004). [Google Scholar]
- 52.Lê S., Josse J., Husson F., FactoMineR: An R package for multivariate analysis. J. Stat. Softw. 25, 1–18 (2008). [Google Scholar]
- 53.Paradis E., Claude J., Strimmer K., APE: Analyses of phylogenetics and evolution in R language. Bioinformatics 20, 289–290 (2004). [DOI] [PubMed] [Google Scholar]
- 54.Philips S. J., Anderson R. P., Schapire R. E., Maximum entropy modeling of species geographic distributions. Ecol. Model. 190, 231–259 (2006). [Google Scholar]
- 55.Panthera, Tiger range map (2009).
- 56.Warren D. L., Glor R. E., Turelli M., ENMTools: A toolbox for comparative studies of environmental niche models. Ecography 33, 607–611 (2010). [Google Scholar]
- 57.H. Wickham, ggplot2: Elegant Graphics for Data Analysis (Springer, New York, 2009). [Google Scholar]
- 58.Wei L., Wu X., Zhu L., Jiang Z., Mitogenomic analysis of the genus Panthera. Sci. China Life Sci. 54, 917–930 (2011). [DOI] [PubMed] [Google Scholar]
- 59.Wu X., Zheng T., Jiang Z., Wei L., The mitochondrial genome structure of the clouded leopard (Neofelis nebulosa). Genome 50, 252–257 (2007). [DOI] [PubMed] [Google Scholar]
- 60.Johnson W. E., Eizirik E., Pecon-Slattery J., Murphy W. J., Antunes A., Teeling E., O’Brien S. J., The late Miocene radiation of modern Felidae: A genetic assessment. Science 311, 73–77 (2006). [DOI] [PubMed] [Google Scholar]
- 61.Davis B. W., Li G., Murphy W. J., Supermatrix and species tree methods resolve phylogenetic relationships within the big cats, Panthera (Carnivora: Felidae). Mol. Phylogenet. Evol. 56, 64–76 (2010). [DOI] [PubMed] [Google Scholar]
- 62.Librado P., Rozas J., DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452 (2009). [DOI] [PubMed] [Google Scholar]
- 63.Villesen P., FaBox: An online toolbox for fasta sequences. Mol. Ecol. Notes 7, 965–968 (2007). [Google Scholar]
- 64.Posada D., jModelTest: Phylogenetic model averaging. Mol. Biol. Evol. 25, 1253–1256 (2008). [DOI] [PubMed] [Google Scholar]
- 65.Guindon S., Dufayard J.-F., Lefort V., Anisimova M., Hordijk W., Gascuel O., New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321 (2010). [DOI] [PubMed] [Google Scholar]
- 66.Ronquist F., Teslenko M., van der Mark P., Ayres D. L., Darling A., Höhna S., Larget B., Liu L., Suchard M. A., Huelsenbeck J. P., MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Drummond A. J., Suchard M. A., Xie D., Rambaut A., Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 29, 1969–1973 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.A. Rambaut, A. J. Drummond, Tracer v1.4 (2007).
- 69.Smith J. L. D., McDougal C. W., The contribution of variance in lifetime reproduction to effective population size in tigers. Conserv. Biol. 5, 484–490 (1991). [Google Scholar]
- 70.Bandelt H., Forster P., Röhl A., Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 16, 37–48 (1999). [DOI] [PubMed] [Google Scholar]
- 71.Lopez J. V, Cevario S., O’Brien S. J., Complete nucleotide sequences of the domestic cat (Felis catus) mitochondrial genome and a transposed mtDNA tandem repeat (Numt) in the nuclear genome. Genomics 33, 229–246 (1996). [DOI] [PubMed] [Google Scholar]
- 72.V. G. Heptner, A. A. Sludskii, in Mammals of the Soviet Union, R. S. Hoffmann, Ed. (Vysshaya Shkola Publisher, Moscow, 1992), vol. 2, pp. 95–183.
- 73.Mazak V., Groves C. P., van Bree J. H., On a skin and skull of the Bali tiger, and a list of preserved specimens of Panthera tigris balica (Schwarz, 1912). Z. Saeugetierkunde 43, 108–113 (1978). [Google Scholar]
- 74.M. E. Sunquist, F. C. Sunquist, in Handbook of the Mammals of the World, D. E. Wilson, R. A. Mittermeier, Eds. (Lynx Edicions, Barcelona, 2009), pp. 54–168.
- 75.J. Seidensticker, C. W. McDougal, in Symposium of the Zoological Society of London, N. Dunstone, M. L. Gorman, Eds. (The Zoological Society of London, Oxford, UK, 1993), vol. 65, pp. 105–125.
- 76.Barber-Meyer S. M., Jnawali S. R., Karki J. B., Khanal P., Lohani S., Long B., MacKenzie D. I., Pandav B., Pradhan N. M. B., Shrestha R., Subedi N., Thapa G., Thapa K., Wikramanayake E., Influence of prey depletion and human disturbance on tiger occupancy in Nepal. J. Zool. 289, 10–18 (2013). [Google Scholar]
- 77.Johnson A., Vongkhamheng C., Hedemark M., Saithongdam T., Effects of human–carnivore conflict on tiger (Panthera tigris) and prey populations in Lao PDR. Anim. Conserv. 9, 421–430 (2006). [Google Scholar]
- 78.Andheria A. P., Karanth K. U., Kumar N. S., Diet and prey profiles of three sympatric large carnivores in Bandipur Tiger Reserve, India. J. Zool. 273, 169–175 (2007). [Google Scholar]
- 79.C. M. Francis, A Guide to the Mammals of Southeast Asia (Princeton Univ. Press, Oxford, UK, 2008), 392 pp. [Google Scholar]
- 80.O’Brien T. G., Kinnaird M. F., Wibisono H. T., Crouching tigers, hidden prey: Sumatran tiger and prey populations in a tropical forest landscape. Anim. Conserv. 6, 131–139 (2003). [Google Scholar]
- 81.Tilson R., Defu H., Muntifering J., Nyhus P. J., Dramatic decline of wild South China tigers Panthera tigris amoyensis: Field survey of priority tiger reserves. Oryx 38, 40–47 (2004). [Google Scholar]
- 82.Ramakrishnan U., Coss R. G., Pelkey N. W., Tiger decline caused by the reduction of large ungulate prey: Evidence from a study of leopard diets in southern India. Biol. Conserv. 89, 113–120 (1999). [Google Scholar]
- 83.Reddy H. S., Srinivasulu C., Rao K. T., Prey selection by the Indian tiger (Panthera tigris tigris) in Nagarjunasagar Srisailam tiger reserve, India. Mamm. Biol. 69, 384–391 (2004). [Google Scholar]
- 84.Biswas S., Sankar K., Prey abundance and food habit of tigers (Panthera tigris tigris) in Pench National Park, Madhya Pradesh, India. J. Zool. 256, 411–420 (2002). [Google Scholar]
- 85.Eizirik E., Kim J.-H., Menotti-Raymond M., Crawshaw P. G., O’Brien S. J., Johnson W. E., Phylogeography, population history and conservation genetics of jaguars (Panthera onca, Mammalia, Felidae). Mol. Ecol. 10, 65–79 (2001). [DOI] [PubMed] [Google Scholar]
- 86.Uphyrkina O., Johnson W. E., Quigley H. B., Miquelle D. G., Marker L., Bush M. E., O’Brien S. J., Phylogenetics, genome diversity and origin of modern leopard, Panthera pardus. Mol. Ecol. 10, 2617–2633 (2001). [DOI] [PubMed] [Google Scholar]
- 87.Antunes A., Troyer J. L., Roelke M. E., Pecon-Slattery J., Packer C., Winterbach C., Winterbach H., Hemson G., Frank L., Stander P., Siefert L., Driciru M., Funston P. J., Alexander K. A., Prager K. C., Mills G., Wildt D., Bush M., O’Brien S. J., Johnson W. E., The evolutionary dynamics of the lion Panthera leo revealed by host and viral population genomics. PLOS Genet. 4, e1000251 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/1/5/e1400175/DC1
Fig. S1. Summary of the work flow to predict the historical distribution of tigers.
Fig. S2. Predicted historical distribution of tigers P. tigris (top left) and MaxEnt analysis for the nine putative subspecies.
Fig. S3. Phenotypic space across all nine putative subspecies of tigers.
Fig. S4. Comparison of the nine putative tiger subspecies.
Table S1. Sources and details of male specimens included in the craniodental analysis.
Table S2. Sources and details of female specimens included in the craniodental analysis.
Table S3. The 41 craniodental characters and their contribution to the first four principal components.
Table S4. Sources and details of the specimens or photographs included in the pelage analysis.
Table S5. Means and SDs for the six pelage characters across the nine putative subspecies.
Table S6. The six pelage characters and their contribution to the first four principal components.
Table S7. Number of occurrence records including the references for each of the nine putative subspecies used for the PCA and for the species distribution modeling (see Materials and Methods for details).
Table S8. Definitions of the four functional habitats and the eco-regions assigned to them.
Table S9. List of the tiger prey species.
Table S10. The 29 ecological variables and their contribution to the first four principal components.
Table S11. Simulation of bootstrap values of nodes pooling two, three, or four putative subspecies in the neighbor-joining tree.
Table S12. Niche overlap between putative subspecies as computed by ENMTools.
Table S13. Sources and details of specimens included in the molecular analysis in addition to the sequences from Luo et al. (8), Driscoll et al. (16), and GenBank accession no. JF357969 (11) (SUM9).
Table S14. Details of mitochondrial primer sets used in addition to the eight primer sets described by Driscoll et al. (16).
Table S15. Population summary statistics calculated for the short and long mtDNA data sets.
Table S16. Nucleotide diversity of pantherine cats.
Table S17. Divergence dates in years before present derived from the short and long mtDNA data sets.
Table S18. Pairwise comparison between the two tiger subspecies P. tigris tigris and P. tigris sondaica for the craniodental characteristics using the Mann-Whitney U test (♂: ntigris = 64, nsondaica = 38; ♀: ntigris = 55, nsondaica = 44).
Table S19. Pairwise comparison between the two tiger subspecies P. tigris tigris and P. tigris sondaica for the pelage characteristics using the Mann-Whitney U test (ntigris = 89, nsondaica = 25).
Table S20. Pairwise comparison between the two tiger subspecies P. tigris tigris and P. tigris sondaica for the continuous ecological variables (climate) using the Mann-Whitney U test (ntigris = 428, nsondaica = 50).
Table S21. Pairwise comparison between the two tiger subspecies P. tigris tigris and P. tigris sondaica for the categorical ecological variables (functional prey groups and functional habitat) using Fisher’s exact test (ntigris = 428, nsondaica = 50).