The architecture of the pore membrane can control translocation of a growing polysaccharide across the membrane and alter the final polymer structure.
Keywords: mixed linkage glucan; (1-3,1-4) beta-glucan; CslF6; polysaccharide biosynthesis; membrane pore; protein stucture; cellulose synthase like
Abstract
The cereal cell wall polysaccharide (1-3,1-4)-β-glucan is a linear polymer of glucose containing both β1-3 and β1-4 bonds. The structure of (1-3,1-4)-β-glucan varies between different cereals and during plant growth and development, but little is known about how this is controlled. The cellulose synthase–like CslF6 protein is an integral membrane protein and a major component of the (1-3,1-4)-β-glucan synthase. I show that a single amino acid within the predicted transmembrane pore domain of CslF6 controls (1-3,1-4)-β-glucan structure. A new mechanism for the control of the polysaccharide structure is proposed where membrane pore architecture and the translocation of the growing polysaccharide across the membrane control how the acceptor glucan is coordinated at the active site and thus the proportion of β1-3 and β1-4 bonds within the polysaccharide.
INTRODUCTION
Many plant cell wall polysaccharides such as (1-4)-β-glucan (cellulose) and (1-3,1-4)-β-glucan (also known as mixed linkage glucan or simply β-glucan), mannans, and xyloglucans are synthesized by large integral membrane proteins with a cytoplasmic active site and a membrane pore through which the polysaccharides are transported to exit the cell (1). The biosynthetic genes belong to the cellulose synthase (CesA) (2) and cellulose synthase–like (Csl) gene families (3). The CslF6 protein is a major component of the (1-3,1-4)-β-glucan synthase of cereals because knockout mutants of this gene have essentially no (1-3,1-4)-β-glucan (4–6). The (1-3,1-4)-β-glucan is a major cell wall component of grasses and is important for human nutrition because of the cholesterol-lowering properties of soluble (1-3,1-4)-β-glucan found at high levels in oat and barley grains (7, 8). It is a linear polymer of glucose linked by about 75% β1-4 and 25% β1-3 bonds arranged in an irregular but nonrandom sequence such that no consecutive β1-3 bonds occur (Fig. 1A) (9). The structure of (1-3,1-4)-β-glucan can be interrogated by digestion with a bacterial enzyme lichenase (10), which only cleaves a β1-4 bond if it is preceded by a β1-3 bond, producing a series of oligosaccharides of degree of polymerization (DP) 3–9, with DP3 and DP4 making up about 90% of the polymer (Fig. 1, A and B) (9, 10). The irregular arrangement of the glycosidic linkages limits the ability of the polymer to form aggregates and increases solubility (11). The structure of (1-3,1-4)-β-glucan as measured by the DP3/DP4 ratio varies between cereal grains and during plant growth and development (12–15), affecting both the solubility and viscosity of the polymer in solution as well as the cholesterol-lowering properties (7, 8). Oat grain (1-3,1-4)-β-glucan has a low DP3/DP4 ratio around 2 or less and is very soluble, whereas wheat (1-3,1-4)-β-glucan has a higher ratio (>2.8) and is insoluble (15). Despite its simple structure, little is known about the biosynthesis of (1-3,1-4)-β-glucan other than the involvement of the cellulose synthase–like CslF and CslH protein families (16–18). Recently, crystal structures of a related protein, the bacterial cellulose synthase (BcsA and BcsB) complex, were determined, showing that cellulose is synthesized at a single intracellular active site and that transport of the polymer across the membrane occurs through a pore simultaneously with elongation of the glucan chain (19, 20). Additionally, it has been speculated that the transmembrane helices (TMHs) of the membrane pore of such polysaccharide synthases may affect enzyme processivity by holding the glucan receptor in place during catalysis (21). To define regions of the CslF6 protein that control the (1-3,1-4)-β-glucan structure, I created chimeric CslF6 genes from cereals that produce (1-3,1-4)-β-glucan with different structures and expressed these in a heterologous tobacco leaf system, which has no endogenous (1-3,1-4)-β-glucan background (4, 22).
Fig. 1. Characterization of (1-3,1-4)-β-glucan structure in Nicotiana benthamiana leaves.
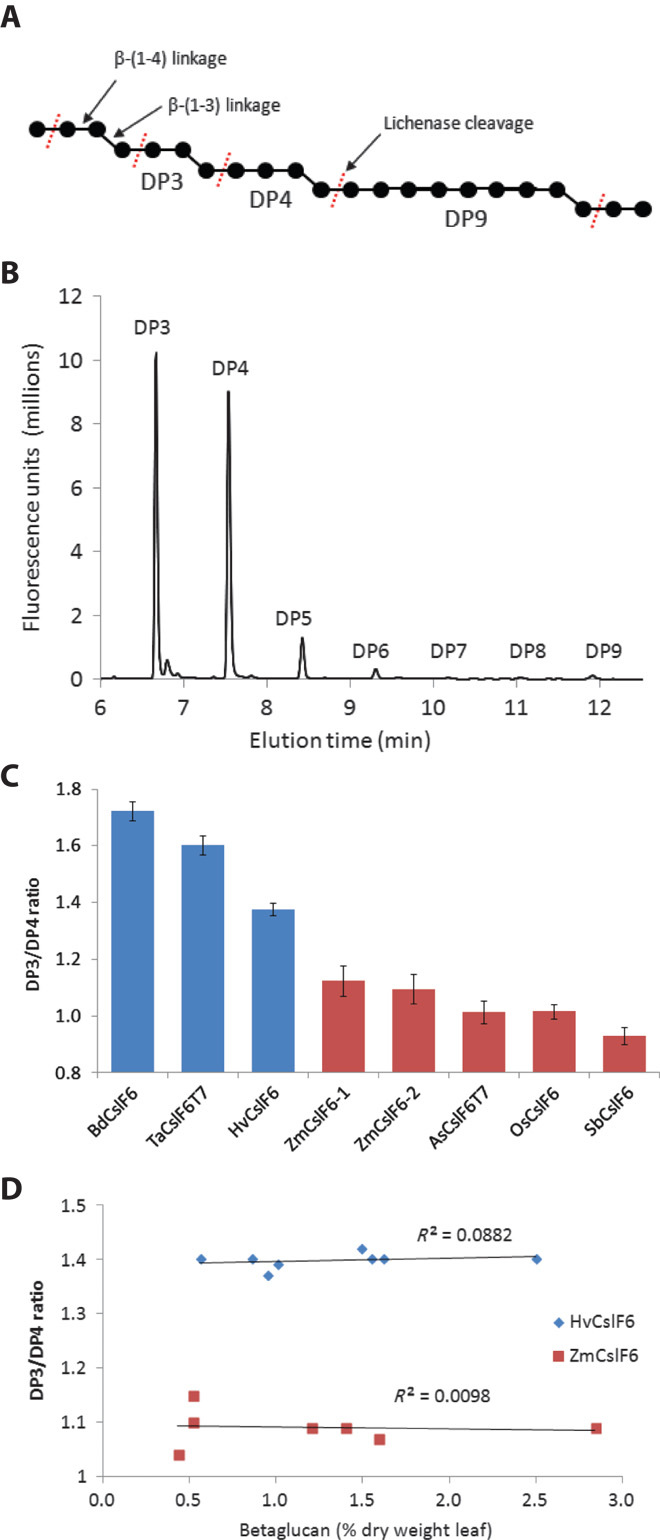
(A) Structure of (1-3,1-4)-β-glucan. Glucose molecules are represented by black dots with β1-4 linkages (straight lines) or β1-3 linkages (angled lines). The bacterial enzyme lichenase specifically cleaves (1-3,1-4)-β-glucan at a β1-4 linkage after a β1-3 linkage as indicated by dashed red lines, producing short oligosaccharides with a DP from DP3 to DP9. (B) Oligosaccharides released by lichenase digestion of betaglucan were fluorescently labeled with APTS and separated by capillary electrophoresis. (C) The source of the CslF6 gene affects the DP3/DP4 ratio of the (1-3,1-4)-β-glucan produced in N. benthamiana leaves. The CslF6 proteins can be classified into two groups: those that produce a (1-3,1-4)-β-glucan with a high (>1.3) DP3/DP4 ratio (Bd, Brachypodum distachyon; Ta, Triticum aestivum; Hv, Hordeum vulgare; shown in blue) or a low (<1.1) DP3/DP4 ratio (Zm, Zea mays; As, Avena sativum; Os, Oryza sativa; Sb, Sorghum bicolor; shown in red). T7 is an 11–amino acid epitope tag at the N terminus of some of the CslF6 proteins—it has no effect on the amount or structure of the (1-3,1-4)-β-glucan. Results are averages ± SD from replicate measurements of between 2 and 11 independent experiments with each gene. (D) The DP3/DP4 ratio of the (1-3,1-4)-β-glucan produced by the HvCslF6 or ZmCslF6 proteins is very stable over a wide range of (1-3,1-4)-β-glucan concentrations in independent experiments. There is no correlation between the DP3/DP4 ratio and the amount of (1-3,1-4)-β-glucan produced.
RESULTS
The structure of (1-3,1-4)-β-glucan is an intrinsic property of the CslF6 protein
The DP3/DP4 ratio of (1-3,1-4)-β-glucan in grain varies and is characteristic of the different cereal species (table S1). The surveyed cereal species may be divided into two groups. One group (Brachypodium, wheat, and barley) has grain (1-3,1-4)-β-glucan with a relatively high DP3/DP4 ratio, and the CslF6 genes from this group produce (1-3,1-4)-β-glucan with a relatively high DP3/DP4 ratio (>1.4) when transiently expressed in N. benthamiana leaf (Fig. 1C). Conversely, the group of species (maize, oat, rice, and sorghum) with low DP3/DP4 ratio (1-3,1-4)-β-glucan in their grain have CslF6 genes that produce (1-3,1-4)-β-glucan with a relatively low DP3/DP4 ratio (about 1.0) when expressed in N. benthamiana leaf (Fig. 1C). The DP3/DP4 ratios of the (1-3,1-4)-β-glucan produced from each CslF6 protein are very stable and did not vary over a wide range of (1-3,1-4)-β-glucan concentrations observed in a large number of independent experiments (Fig. 1D and table S2). Western blotting with a multiepitope antibody against the HvCslF6 protein showed a broad protein, perhaps doublet band, of around the expected size in membrane preparations from N. benthamiana leaf (Fig. 2A, lanes 3 to 5) and that this protein comigrated with the HvCslF6 protein from barley leaf tissue (Fig. 2A, lane 1). The cause of the apparent doublet has recently been explained as incomplete reduction of disulphide bonds during sample denaturation (23). A band of the expected size for the BdCslF6 protein can be seen in Fig. 2A (lane 5), but this band is very faint because the BdCslF6 protein does not react well against the HvCslF6 antibody. The expression level of each protein, however, can be compared using an antibody against the T7 epitope tag present on the N terminus of each protein. The Western blot in Fig. 2B shows that the BdCslF6 protein is actually expressed at higher levels than the TaCslF6 or HvCslF6 proteins but not as high as the HvCslF4 T7–tagged protein (which migrates faster because of its smaller size, 98 versus 106 kD). Specificity of the antibodies was demonstrated by a lack of reaction in control lanes (Fig. 2A, lane 2, containing an HvCslF4 T7–tagged protein, or Fig. 2B, lane 1, containing proteins extracted from a barley leaf).
Fig. 2. The CslF proteins are found in the membrane fraction.
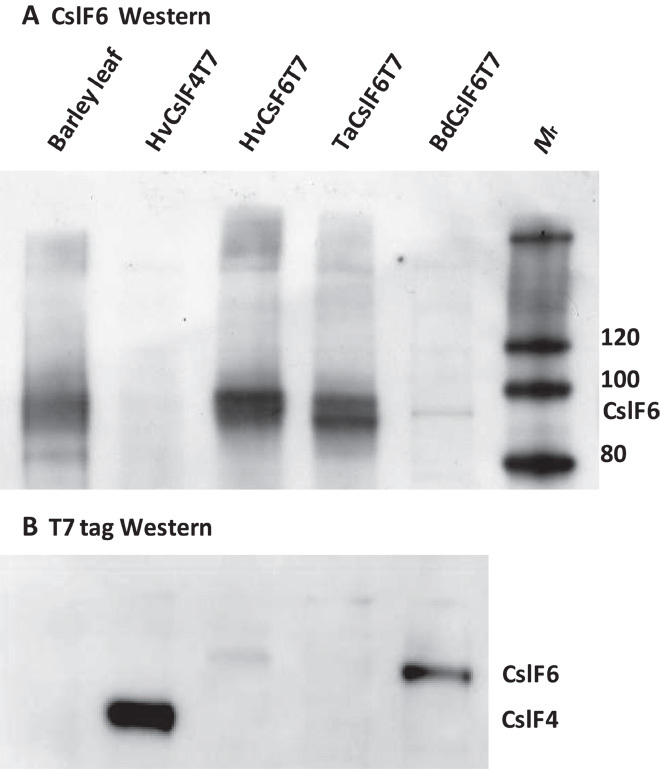
(A and B) Proteins from membrane preparations from barley leaf or N. benthamiana leaf expressing the indicated CslF protein probed with (A) multiepitope HvCslF6 antibody or (B) T7 tag antibody. Molecular mass markers (Mr) and the positions of CslF6 and CslF4 are shown at the side.
The predicted membrane pore region of CslF6 controls (1-3,1-4)-β-glucan structure
Because the DP3/DP4 ratio of (1-3,1-4)-β-glucan produced in N. benthamiana leaf appeared to be an intrinsic property of the expressed CslF6 protein, I hypothesized that it may be possible to determine the region(s) of the CslF6 protein that controls (1-3,1-4)-β-glucan structure by expressing chimeric proteins in this transient expression system. I selected barley (HvCslF6) and maize (ZmCslF6-2) genes as representative from the groups producing (1-3,1-4)-β-glucan with either a high or a low DP3/DP4 ratio, respectively, to make chimeric constructs based on conservation of restriction sites within the genes (fig. S1). Reciprocal chimeras were made using the Bgl II–Eco RI fragments, and other chimeras were made as restrictions digests allowed. Swapping the cytoplasmic active-site domain of ZmCslF6 protein with the HvCslF6 sequence (Sac I to Bgl II fragment) had no effect on (1-3,1-4)-β-glucan structure (Fig. 3). In contrast, the C-terminal one-third of the protein encompassing six predicted TMHs had the majority of control over the DP3/DP4 ratio because all proteins containing this region from barley produced (1-3,1-4)-β-glucan with a high ratio (>1.4), whereas proteins with this region from maize produced (1-3,1-4)-β-glucan with a low DP3/DP4 ratio (1.1 to 1.2) (Fig. 3). The importance of the C-terminal domain containing the six predicted TMHs in controlling the (1-3,1-4)-β-glucan structure was confirmed by swapping this region among several different CslF6 proteins. Chimeric proteins containing this region from BdCslF6 or HvCslF6 produced (1-3,1-4)-β-glucan with high ratios (>1.4), whereas chimeric proteins containing this region from AsCslF6 or ZmCslF6 produced (1-3,1-4)-β-glucan with a low DP3/DP4 ratio (1.11 to 1.18) (Fig. 4). All of the chimeric proteins produced large amounts of (1-3,1-4)-β-glucan, and there was no relationship between the structure of the (1-3,1-4)-β-glucan and the amount produced, as was the case for the native proteins (Fig. 4 and table S1).
Fig. 3. The effect of chimeric HvCslF6/ZmCslF6 genes on (1-3,1-4)-β-glucan structure.
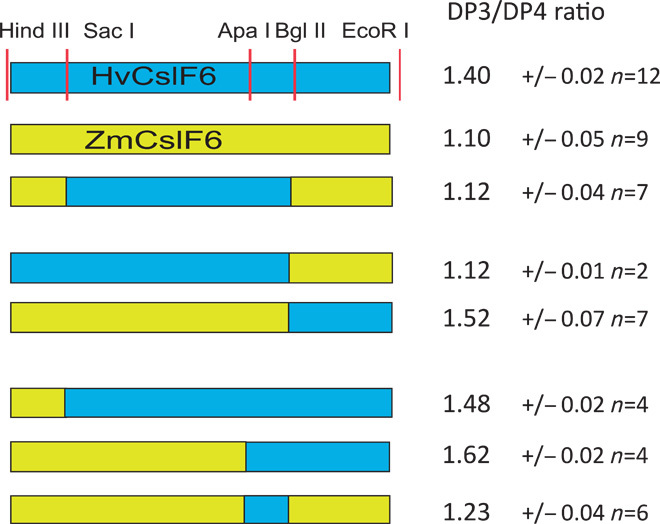
Restriction sites used in cloning are shown as vertical lines. The HvCslF6 gene is represented by blue rectangle, and the ZmCslF6-2 gene as a yellow rectangle. (Right) DP3/DP4 ratio of the lichenase digested (1-3,1-4)-β-glucan averaged from the indicated number of independent transformation experiments with SDs shown.
Fig. 4. The Bgl II–Eco RI fragment of CslF6 genes controls (1-3,1-4)-β-glucan structure.

Restriction sites used in cloning are shown as vertical lines. The blue or red rectangles represent the CslF6 genes that produce (1-3,1-4)-β-glucan with a high DP3/DP4 ratio, and the yellow or gray rectangles represent those that produce a (1-3,1-4)-β-glucan with a low DP3/DP4 ratio as indicated on the right. All chimeric constructs show a high (>1.4) or a low (<1.2) DP3/DP4 ratio depending on the source of the Bgl II–Eco RI fragment. SDs were less than 0.014 for all samples. The amount of (1-3,1-4)-β-glucan (BG) produced by each construct is shown in the last column as a percentage of dry weight of the freeze-dried leaf.
Further chimeric constructs were made within the C-terminal region containing the six predicted TMHs to narrow down the region controlling (1-3,1-4)-β-glucan structure. Using synthetic DNA blocks, an Xba I site was introduced into both the ZmCslF6 and HvCslF6 genes at a conserved Leu-Asp codon pair between predicted TMH6 and TMH7 without changing the amino acid sequence of the CslF6 proteins (Fig. 5A). Using this new Xba I site, reciprocal swaps of the two halves of this region were made, and expression of these genes in N. benthamiana leaf showed that the region encoding the predicted TMH3 to TMH6 controlled the (1-3,1-4)-β-glucan structure (Fig. 5B). Within this region, there are 13 amino acid differences between the maize and barley proteins (Fig. 5C and fig. S2). Further synthetic DNA Bgl II–Xba I fragments were synthesized, and expression of the chimeric proteins in N. benthamiana leaf demonstrated that it was the N-terminal half of this region with six amino acid differences that controlled the (1-3,1-4)-β-glucan structure (Fig. 5C). A further chimeric construct exchanging the Pst I–Eco RI fragment between the maize and barley proteins showed that the two amino acid differences N-terminal of the predicted TMH3 had no effect on the (1-3,1-4)-β-glucan structure, indicating that it was the four amino acid differences in predicted TMH4 that controlled the (1-3,1-4)-β-glucan structure (Fig. 5C).
Fig. 5. The predicted TMH4 of CslF6 controls (1-3,1-4)-β-glucan structure.
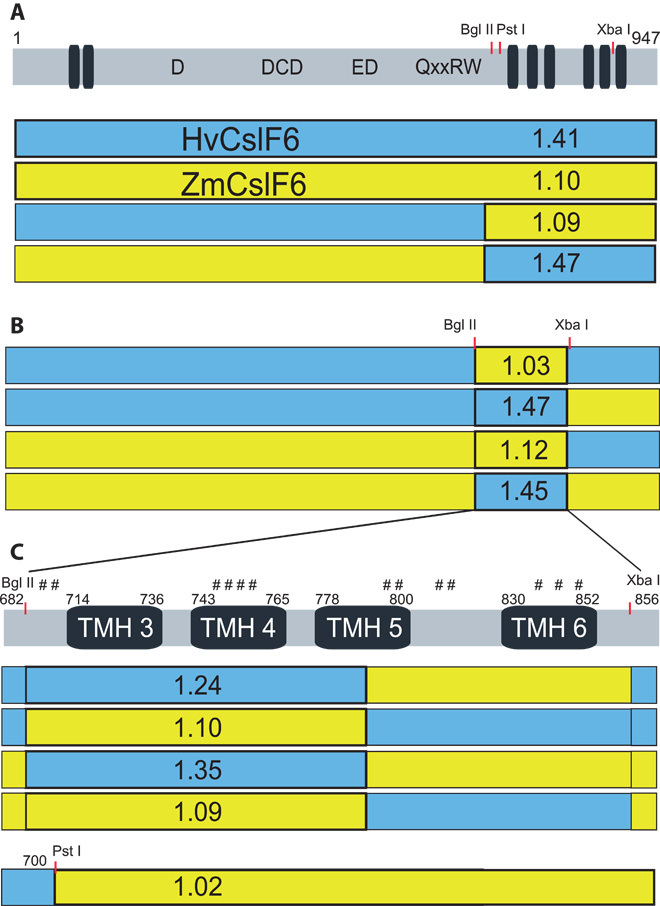
(A) Diagrammatic representation of the full-length CslF6 protein sequence (gray rectangle; 1 to 947 amino acids; numbers refer to the HvCslF6 protein), with predicted TMHs shown as thin dark barrels and the approximate positions of the conserved amino acids of the active site in single-letter code. Red lines indicate approximate positions of restriction sites used in construction of chimeric genes shown below as blue or yellow rectangles (representing barley HvCslF6 and maize ZmCslF6, respectively). The numbers within the rectangles are the DP3/DP4 ratios of the (1-3,1-4)-β-glucan produced by the corresponding chimeric proteins in N. benthamiana leaf, and the boxed area indicates the region controlling this ratio (that is, blue area has a high and yellow area has a low DP3/DP4 ratio). Genes were fused at the Bgl II site. (B) The region controlling (1-3,1-4)-β-glucan structure was further defined to be between the Bgl II and Xba I sites encompassing TMH3 to TMH6 of the CslF6 protein. (C) Expanded view of this region with the protein shown as a gray rectangle; numbers refer to the boundaries of the predicted TMHs of the HvCslF6 protein; and the positions of the Bgl II, Pst I, and Xba I sites are shown on top. The hash marks (#) represent the position of the 13 amino acid differences between the HvCslF6 and ZmCslF6 proteins in this region. By comparing the four Bgl II–Xba I chimeras and the single Pst I chimera, the region controlling the DP3/DP4 ratio is further limited to the four amino acid differences in TMH4 of the CslF6 protein.
A single amino acid change in predicted TMH4 controls the (1-3,1-4)-β-glucan structure
Each of the single amino acid differences in the HvCslF6 protein was then individually changed to the corresponding amino acid of the ZmCslF6 protein, and the effect on the (1-3,1-4)-β-glucan structure was assessed (Fig. 6). Two amino acid changes had no effect on the (1-3,1-4)-β-glucan structure, but when Ile757 of the HvCslF6 protein was changed to leucine (HvCslF6I757L), the structure of the (1-3,1-4)-β-glucan produced was indistinguishable from that produced from the ZmCslF6 protein, whereas that of HvCslF6S752G showed a DP3/DP4 ratio intermediate between the two species. All single amino acid variants were active and produced at least as much (1-3,1-4)-β-glucan as did the native CslF6 protein (Fig. 6). A reciprocal experiment was then performed where the equivalent amino acid of the ZmCslF6 protein was changed from leucine to isoleucine (ZmCslF6L757I), and this increased the DP3/DP4 ratio of the (1-3,1-4)-β-glucan produced significantly, such that it was similar to that produced from the native HvCslF6 protein (fig. S3). Similarly, the single amino acid change in the ZmCslF6 protein did not affect the amount of (1-3,1-4)-β-glucan produced (fig. S3).
Fig. 6. Single amino acid differences in the predicted TMH4 of CslF6 control (1-3,1-4)-β-glucan structure.
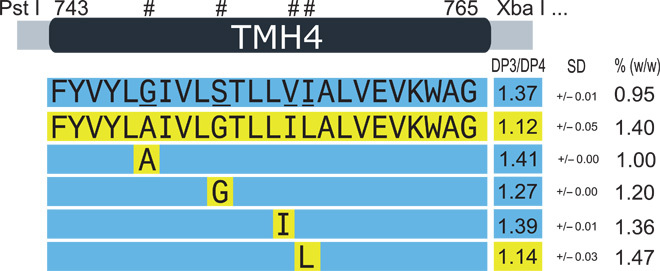
The amino acid sequence of the TMH4 region of HvCslF6 and ZmCslF6 proteins is shown. Each of the four different single amino acids (underlined) in the HvCslF6 protein was changed to the corresponding ZmCslF6 amino acid, and the effect on the DP3/DP4 ratio and the amount (% dry weight of leaf) of the (1-3,1-4)-β-glucan are shown at the right. Results are means ± SD from replicates of at least two transformation experiments.
Further evidence for the role of these two amino acids (Ile757 and Ser752) in controlling the (1-3,1-4)-β-glucan structure was obtained by making the same isoleucine for leucine change in the Brachypodium (BdCslF6) and oat (AsCslF6) proteins (see fig. S1 for an alignment of this region in all the CslF6 proteins), as well as combining the two single amino acid changes in the HvCslF6 protein. When expressed in tobacco leaf, the BdCslF6 protein produces a (1-3,1-4)-β-glucan with the highest DP3/DP4 ratio among all CslF6 proteins (1.72), whereas the BdCslF6I571L substitution reduces this ratio substantially down to 1.39 (Fig. 7). In contrast, the AsCslF6 protein produces a (1-3,1-4)-β-glucan with a low DP3/DP4 ratio of 1.01, and this ratio was further decreased to 0.89 when the modified AsCslF6I759L protein was expressed. In the double substitution protein HvCslF6S752G,I757L, a further decrease in the DP3/DP4 ratio from 1.14 to 1.05 was noted. The position of HvCslF6 Ile757 in relation to the bacterial and plant cellulose synthase and CslD proteins is shown in fig. S4 for comparison.
Fig. 7. The effect of amino acid changes in predicted TMH4 on (1-3,1-4)-β-glucan structure across species.
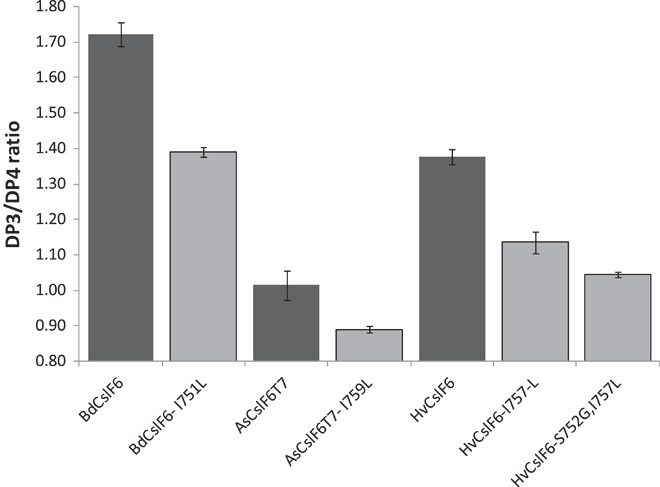
The DP3/DP4 ratio of (1-3,1-4)-β-glucan produced in N. benthamiana leaf from wild-type CslF6 proteins with an isoleucine in predicted TMH4 is shown as black bars. When the indicated isoleucine is changed to a leucine, the DP3/DP4 ratio of the (1-3,1-4)-β-glucan is decreased (light gray bars). A further decrease in the DP3/DP4 ratio is observed when a second amino acid substitution is made in the HvCslF6 TMH4 region (HvCslF6S752G,I757L). Shown are averages and SDs of replicates from at least two transformation experiments. The amount of (1-3,1-4)-β-glucan produced for the respective constructs was 1.8, 2.2, 0.4, 0.6, 1.8, 2.4, and 1.8% of the dry weight of the freeze-dried leaf tissue.
The isoleucine-to-leucine change alters the fine structure of the (1-3,1-4)-β-glucan
Calculations from the complete chain length distribution (DP3-DP9, table S2) show that the proportion of β1-4 bonds is increased in the (1-3,1-4)-β-glucan produced from those CslF6 proteins that have the isoleucine-to-leucine change in TMH4 (Fig. 8A, compare dark blue and red bars), approaching the proportion of β1-4 bonds in the (1-3,1-4)-β-glucan produced from those CslF6 proteins that have a native leucine in the equivalent position (Fig. 8A, light blue bars). An additional difference in the (1-3,1-4)-β-glucan structure produced from those CslF6 proteins with a native isoleucine is the higher level of short DP3 and DP4 oligosaccharides (>~92% of the total DP3-DP9; Fig. 8B, dark blue bars) compared to those CslF6 proteins with a leucine at the same position in TMH4 (Fig. 8B, red and light blue bars). These differences can largely be explained by the lower proportion of DP5 and DP6 in the (1-3,1-4)-β-glucan produced from those CslF6 proteins with an isoleucine in TMH4 (Fig. 8C).
Fig. 8. The isoleucine-to-leucine change alters the fine structure of (1-3,1-4)-β-glucan.
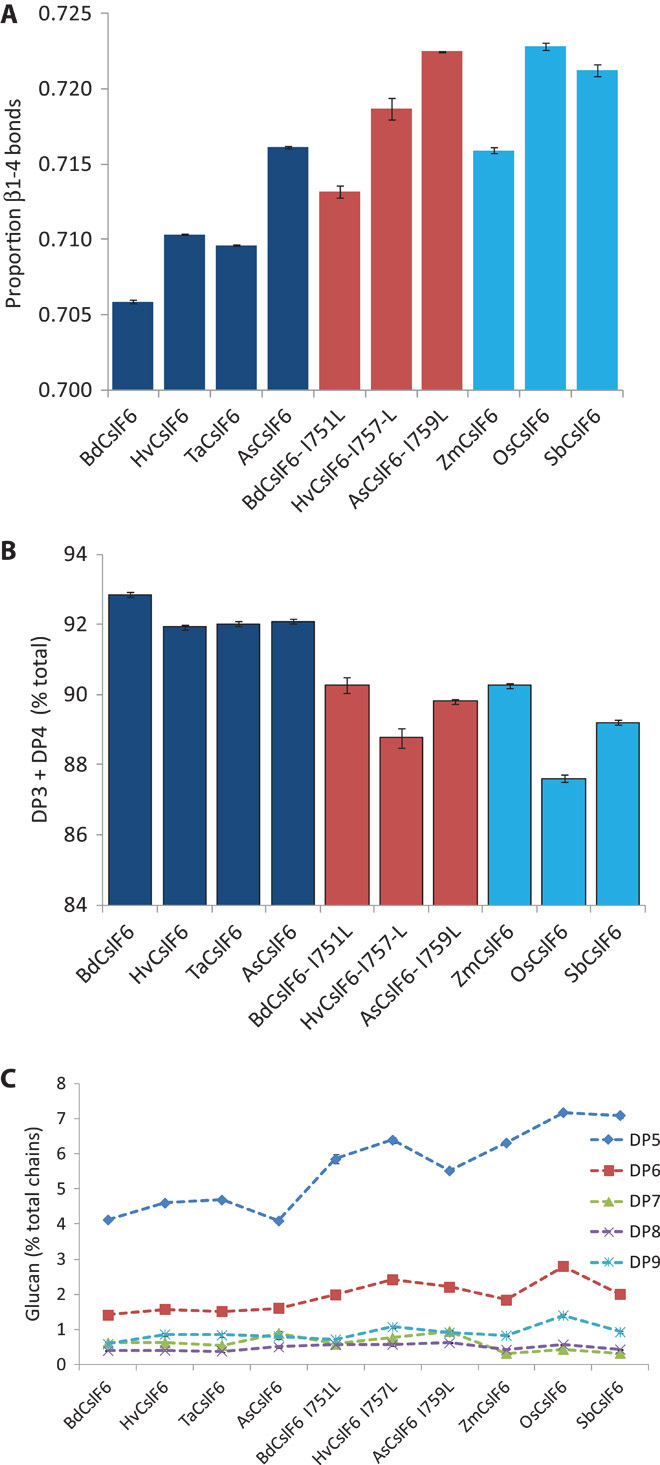
(A) Changing the native isoleucine in TMH4 of CslF6 protein (dark blue bars) to a leucine (red bars) increases the proportion of β1-4 bonds in (1-3,1-4)-β-glucan, similar to that produced from those native CslF6 proteins that have a leucine at the same position (light blue bars). (B) The increase in β1-4 bond frequency changes the (1-3,1-4)-β-glucan structure, decreasing the proportion of DP3 and DP4 oligosaccharides. (C) The increase in longer-chain oligosaccharides is largely explained by an increase in DP5 and DP6. Dashed lines are shown to indicate the trends in change of oligosaccharide profiles between the independent experimental measurements. Error bars from duplicate measurements are indicated.
DISCUSSION
The wide occurrence of (1-3,1-4)-β-glucan in vegetative tissues and grain of the Poaceae grasses suggests an important function for this linear glucan polymer perhaps related to the heterogeneous nature of the arrangement of the β1-3 and β1-4 bonds and the consequent effects on biophysical properties (11). Recent work has highlighted the importance of the CslF6 protein in (1-3,1-4)-β-glucan biosynthesis; however, how or if this protein contributes to the heterogeneity is not known. I have addressed this question by isolating the CslF6 gene from a wide range of grasses and by expressing them individually and as chimeras in a heterologous plant system that has no endogenous (1-3,1-4)-β-glucan background.
The CslF6 proteins fall into two groups: one group (Brachypodium, wheat, and barley) produces a (1-3,1-4)-β-glucan in N. benthamiana leaf with a high DP3/DP4 ratio, and the other (oat, maize, rice, and sorghum) produces a (1-3,1-4)-β-glucan with a low DP3/DP4 ratio, reflecting the structure of the (1-3,1-4)-β-glucan found in the respective grains. The structure of the (1-3,1-4)-β-glucan appears to be an intrinsic property of the CslF6 protein, whereas the amount of (1-3,1-4)-β-glucan produced varies between experiments wherein the DP3/DP4 ratio is remarkably controlled for each protein. All of the chimeric proteins are active and produce high levels of (1-3,1-4)-β-glucan, sometimes exceeding that of the native CslF6 proteins, suggesting that the CslF6 protein is a robust enzyme and can tolerate changes to the amino acid sequence while still maintaining enzyme activity. Swapping the cytoplasmic active site between the HvCslF6 and ZmCslF6 proteins showed that this region alone had little control over the (1-3,1-4)-β-glucan structure. A series of further chimeras narrowed down the region controlling the (1-3,1-4)-β-glucan structure to the predicted transmembrane pore region and specifically to one of the eight predicted TMHs—TMH4. Within the predicted TMH4, one of the four amino acid differences (Ile757 of HvCslF6) exerted the majority of the control because changing this to leucine (as found in the ZmCslF6 protein) produced a (1-3,1-4)-β-glucan with a DP3/DP4 ratio very similar to that produced by the ZmCslF6 protein. This same single amino acid change in the BdCslF6 and AsCslF6 proteins also reduced the DP3/DP4 ratio of the (1-3,1-4)-β-glucan significantly, and changing the leucine of the ZmCslF6 protein into isoleucine produced a reciprocal change in the (1-3,1-4)-β-glucan structure, indicating the general nature of this mechanism controlling the (1-3,1-4)-β-glucan structure.
Although Ile757 of HvCslF6 exerts the majority of control over the (1-3,1-4)-β-glucan structure, a comparison of the sequences from all of the CslF6 proteins (figs. S2 and S4) and the results from other chimeras suggests that other regions of the protein can also have some effects. For instance, the amino acid sequences of predicted TMH4 of BdCslF6 and two of the AsCslF6 proteins are identical (including the Ile, fig. S2), whereas the structure of the (1-3,1-4)-β-glucan produced by these proteins is very different. The AsCslF6 protein has many differences compared to the other CslF6 proteins, especially in the predicted TMH5 and TMH6 domains, and this may be the reason for the differences in the (1-3,1-4)-β-glucan structure; however, this needs to be experimentally determined. Also, some of the HvCslF6/ZmCslF6 chimeras produced (1-3,1-4)-β-glucans, with DP3/DP4 ratios significantly higher than that produced by the native HvCslF6 protein (Fig. 3), whereas swapping the C-terminal region (containing TMH3 to TMH8) of the AsCslF6 and ZmCslF6 proteins with that of BdCslF6 increased the DP3/DP4 ratio of the (1-3,1-4)-β-glucan, but it was not as high as that produced from the native BdCslF6 protein (Fig. 4). This suggests interactions between the predicted TMH domains of the transmembrane pore and other regions of the CslF6 protein, and to understand what these may be and how they might control (1-3,1-4)-β-glucan structure, a three-dimensional structure of the CslF6 protein or closely related protein is needed.
Although no crystal structure is available for a full-length plant cellulose synthase–related protein, the cytoplasmic domain of the cotton CesA and rice CesA8 proteins has been modeled (24, 25). These models include the interfacial domain 2 (IF2; containing the conserved QxxRW motif), which forms part of the base of the transmembrane pore. TMH3 and TMH4 immediately follow this domain (fig. S2), and these are equivalent to TMH5 and TMH6 of the BcsA crystal structure, which form the core of the membrane pore (fig. S4) (20). The newly synthesized cellulose chain crosses the membrane parallel to TMH5 and TMH6 of BcsA, making contact with the amino acid side chains of these helices (20). A recent x-ray structure of the BcsA-BcsB in complex with either the bacterial activator cyclic di guanosine monophosphate (c-di-GMP) or competitive inhibitor uridine diphosphate has identified sequence motifs such as the small loop and finger helix preceding IF2 and the gating loop between IF3 and TMH7, which are important for catalysis and membrane transport of cellulose (19). Critical amino acids within these motifs are essentially invariant among bacterial and eukaryotic cellulose synthase enzymes (20), suggesting that the overall topology/architecture in this region is the same. These amino acids are also conserved in the related CslD and CslF6 proteins (fig. S4); thus, it is likely that TMH3 and TMH4 of HvCslF6 also form part of the membrane pore and that some of the side chains may also interact with (1-3,1-4)-β-glucan and affect its translocation through the membrane pore.
The single amino acid difference (isoleucine or leucine) in TMH4 of CslF6 has a major and reproducible impact on the structure of (1-3,1-4)-β-glucan, demonstrating that small changes to the membrane pore architecture can strongly affect (1-3,1-4)-β-glucan biosynthesis. It is difficult to predict with certainty the corresponding position of Ile757 within the BcsA crystal structure because of the limited homology within the TMHs. However, when the alignment includes the CslD proteins, which are intermediate between CesA and CslF proteins, there is a strong indication that Ile757 of HvCslF6 is equivalent to Gln463 of BcsA toward the base of the membrane pore (fig. S4) (19, 20). In the c-di-GMP–bound BcsA structure (19), the adjacent region just inside the entrance to the transmembrane channel is involved with the coordination of the acceptor glucan chain along with the Trp383 of the QxxRW motif, so it is conceivable that Ile757 of HvCslF6 could affect movement of the growing β-glucan chain within the membrane channel, as well as glucan acceptor coordination and consequently the frequency of β1-3 or β1-4 bond formation during (1-3,1-4)-β-glucan biosynthesis. Although the single amino acid Ile/Leu757 position has the largest effect on the (1-3,1-4)-β-glucan structure, it is important to stress that it is probably the overall shape of the membrane pore that is important (that is, the membrane pore architecture) and that this will be affected by other regions of the protein that make up the membrane pore. Previously, it had been speculated that the TMHs of the membrane pore of a polysaccharide synthase may affect enzyme processivity by holding the glucan receptor in place during catalysis (21), but the results presented here are the first experimental evidence that this mechanism may also affect the type of bond incorporated at the active site.
Recently, a mutation in the predicted TMH4 region of an Arabidopsis thaliana CesA protein (CESA1A903V) was shown to affect the polymerization rate of the CesA protein and cellulose microfibril crystallinity (26), and this mutant is equivalent to Leu755 of HvCslF6 only two amino acids away from Ile757 (fig. S4). The predicted TMH4 region of the CesA protein is much more variable than the equivalent region of the CslF6 protein (12 of 21 variant amino acids in CesA versus 4 of 21 in CslF6), suggesting that this region in CslF6 is under stronger evolutionary constraint, perhaps not only by controlling the translocation of the growing polymer but also with the added function of controlling β1-3 or β1-4 bond formation at the active site.
At present, there are two models for (1-3,1-4)-β-glucan biosynthesis: one where the CslF6 protein can introduce both β1-3 and β1-4 bonds and the other (a two-stage model) where the CslF6 protein only makes short β1-4 glucans, and an additional protein at the plasma membrane links these short chains with a β1-3 bond to create the final (1-3,1-4)-β-glucan polymer at the extracellular surface. This latter model was proposed to explain the discrepancy that (1-3,1-4)-β-glucan could only be detected in the cell wall and not at the Golgi apparatus, the expected site of synthesis. Several lines of evidence now suggest that the control of (1-3,1-4)-β-glucan structure is an intrinsic property of the CslF6 protein—first, each CslF6 protein produces a (1-3,1-4)-β-glucan of defined structure, and second, these DP3/DP4 ratios are very stable across all experiments with a wide range of (1-3,1-4)-β-glucan levels. If (1-3,1-4)-β-glucan biosynthesis relies on a component supplied by the host plant (here N. benthamiana), which could vary in different physiological states or growing conditions, then it is difficult to see how this stability in the (1-3,1-4)-β-glucan structure would be maintained. A recent study also found that CslF6 expression in Pichia yeast produced (1-3,1-4)-β-glucan, suggesting that the CslF6 protein alone can make (1-3,1-4)-β-glucan (27). The large structural changes in (1-3,1-4)-β-glucan as a result of a single amino acid change in a transmembrane helix of the CslF6 protein are also highly suggestive that the CslF6 protein can make both β1-3 and β1-4 bonds. The two-stage model is unnecessarily complex in that a stop-start mechanism would be needed for making the individual short β1-4 linked oligosaccharides; there is no mechanism to explain what would control the proportion of the oligosaccharides [and hence the structure of the (1-3,1-4)-β-glucan], and it is difficult to envisage what the energy source would be for driving the short oligosaccharides through the membrane pore. The two-stage model has no direct experimental evidence, and a further difficulty, given the results presented here, is that there would be no physical connection between the acceptor glucan at the active site and the glucan interacting with the amino acid residues identified in predicted TMH4 as controlling the (1-3,1-4)-β-glucan structure. I conclude along with Kim et al. (27) that the CslF6 protein can make both β1-3 and β1-4 linkages, and further propose that it is the translocation of the (1-3,1-4)-β-glucan across the membrane and associated coordination of the acceptor glucan at the active site that controls the proportion of β1-3 and β1-4 bonds in (1-3,1-4)-β-glucan and that this is governed largely by the membrane pore architecture.
Because changes in only one or a few amino acids can profoundly affect the (1-3,1-4)-β-glucan structure, it may be possible, using new gene editing technologies, to alter the solubility of (1-3,1-4)-β-glucan and thus perhaps generate wheats with cholesterol-lowering properties or with altered cell wall digestibility for biofuel applications. Finally, the findings described here may also have implications for the large number of other CesA or Csl proteins and the polysaccharides they synthesize.
MATERIALS AND METHODS
Cloning of CslF6 genes
Full-length CslF6 complementary DNAs (cDNAs) were isolated from barley, wheat, Brachypodium, oat, rice, maize, and sorghum as follows. RNA was isolated from frozen tissue (about 100 mg of 1-week-old seedlings) using a NucleoSpin RNA Plant extraction kit as per the manufacturer’s instructions (Macherey-Nagel). Five micrograms of RNA (without deoxyribonuclease treatment) was reverse-transcribed for 1 hour at 50°C in a 20-μl reaction using 5 pmol of the RoRidT17 primer and SuperScript III as per the manufacturer’s instructions (Invitrogen). The reaction was diluted with tris-EDTA (pH 8.0) to 200 μl and heat-inactivated at 70°C for 15 min. Two microliters of this diluted seedling cDNA was amplified with gene-specific primers (table S3) using Phusion Hot Start polymerase [New England Biolabs (NEB)] in a 20-μl polymerase chain reaction (PCR) including 7 to 10% (w/v) dimethyl sulfoxide according to the manufacturer’s instructions with cycling conditions of 98°C for 30 s, followed by 36 cycles of 98°C for 7 s, 15 s at 62°C, and 72°C for 90 s with a final 5-min extension at 72°C. PCR products around 3 kb in size were separated on a 1.0% tris-borate-EDTA agarose gel, purified using an illustra kit (GE Healthcare), A-tailed with a nonproofreading Taq polymerase, and ligated into Xcm I–cut pCXSN vector as described using NEB blunt-T/A ligase (28). Inserts were screened for orientation and fully sequenced. The forward primers were either in the 5′ untranslated region or around the ATG start codon. Primer SJ116 anneals to barley, wheat, oat, and Brachypodium genes, and the equivalent primer SJ277 has an 11–amino acid T7 epitope tag before the initiating methionine. The reverse primers were generally gene-specific. Maize (Z. mays) has two CslF6 genes here designated as CslF6-1 and CslF6-2.
Chimeric gene constructs
The unique Hind III and Eco RI sites upstream of the CaMV 35 promoter and downstream of the NOS polyA in the pCXSN vector were used in conjunction with internal restriction sites to swap regions of the HvCslF6 and ZmCslF6-2 genes (fig. S1). The Bgl II site is conserved in all CslF6 genes. The Pst I site upstream of the CaMV 35 promoter was destroyed by cutting with Sbf I and repairing the ends with T4 DNA polymerase, leaving the Pst I site within the CslF6 gene as unique. An Xba I site was introduced into the CslF6 cDNA sequence (TCTCGA changed to TCTAGA) as a Bgl II–Afl II gBlock fragment (IDT Technologies) at the conserved Leu-Asp codon pair between TMH6 and TMH7 without changing the amino acid sequence. This enabled the creation of HvCslF6 and ZmCslF6 chimeras at the Xba I site. Further chimeras were created by using Bgl II–Xba I gBlocks fused within the conserved TMH5 domain. The two amino acid differences between the Bgl II and Pst I sites were shown not to affect the (1-3,1-4)-β-glucan structure by swapping the Pst I–Eco RI fragment of the two genes. Last, synthetic gBlock fragments with single amino acid changes in TMH4 were cloned as Pst I–Xba I fragments into the HvCslF6 cDNA. Similarly, Pst I and Xba I sites were introduced into the BdCslF6 gene as a Bgl II–Afl II gBlock fragment (IDT Technologies) without changing the amino acid sequence.
The C-terminal region of the CslF6 protein was swapped between all CslF6 genes using the Bgl II–Eco RI restriction sites. Subsequent amino acid changes in TMH4 of CslF6 were created using synthetic Pst I–Xba I gBlock fragments. All plant transformation vectors were transferred into Agrobacterium tumefaciens AGL1 by electroporation and selection on kanamycin (50 mg/liter) and rifampicin (25 mg/liter).
Transient expression in N. benthamiana leaves
Transient expression in N. benthamiana leaves was performed as described in (4, 22) using Agrobacterium cultures containing the CslF6 expression vector at an optical density (OD) of 0.4 along with the P19 viral suppressor at 0.2 OD. The top three leaves of 5- to 6-week-old plants were used for infiltration. There is a slight effect of leaf age on the amount and structure of (1-3,1-4)-β-glucan produced, so to minimize this effect, each Agrobacterium was infiltrated into a minimum of six half leaves, which were combined in further analysis. Four or 5 days later, leaves were harvested, the midrib was removed, and leaves were combined before freeze-drying them for 24 hours. Samples were then ball-milled to a fine powder. It was noted that the AsCslF6 gene always gave the lowest and the SbCslF6 gene the highest level of (1-3,1-4)-β-glucan compared to the other genes. The expression of the CslF6 genes was consistent between experiments and generally resulted in visible chlorosis of the leaf tissue, which was more severe in the younger leaves.
Membrane preparation
Fresh plant material (1.5 g) was ground in liquid nitrogen and resuspended in 6 ml of extraction buffer [25 mM tris-HCl (pH 7.5), 0.25 M sucrose, 2 mM EDTA, 20 mM KCl, 10 mM dithiothreitol (DTT), and 10% glycerol containing protease inhibitor mix (4 μl/ml) (Sigma) added just before use], filtered through Miracloth, and clarified by centrifugation for 10 min at 8000g at 4°C. Membranes were pelleted from the supernatant by centrifugation for 1 hour at 100,000g at 4°C, and the pellet was resuspended in 0.1 ml of extraction buffer.
Western blotting
Proteins from the membrane preparation were solubilized in sample buffer [0.125 M tris-HCl (pH 6.8), 15% glycerol, 3% SDS, 0.2 M DTT, 0.05% bromophenol blue] at 40°C for 1 hour, and about 20 μg of protein was run on a Novex (Life Technologies) 4 to 12% polyacrylamide gel, blotted onto nitrocellulose (Pall), and used in Western blots with either a T7 epitope tag antibody (Novagen, 1:10,000 dilution) or an HvCslF6 antibody (1:1000 dilution) and chemiluminescence detection (ECL Amersham). The rabbit polyclonal HvCslF6 antibody (Life Research) was raised against a multiepitope HvCslF6 antigen expressed in Escherichia coli and His tag–purified (GRVRSNEPVA SLDMDIVAMG QIGAVNDESW ESGAAVDDRP PRLAGLFAKT KYEKPGLEMT PKKTYGKSDA) corresponding to amino acids 9 to 18, 52 to 61, 62 to 71, 84 to 93, 526 to 535, 536 to 545, and 566 to 575 of the full-length HvCslF6 protein. Note that there is at least one amino acid mismatch in each of the homologous seven 10-mer epitopes in the BdCslF6 protein.
Quantification and analysis of (1-3,1-4)-β-glucan structure
The (1-3,1-4)-β-glucan content of the leaves was assayed as described (4). Briefly, a crude cell wall preparation was made from 20 mg of ground leaf material by heating for 30 min at 80°C in 1.8 ml of 80% ethanol in a 2-ml Eppendorf tube with mixing. The supernatant was removed after centrifugation at 10,000 rpm for 5 min, and the residue was reextracted in the same volume of 80% ethanol at 80°C for 10 min. After centrifugation, the pellet was washed at room temperature for 10 min in 50% ethanol with a final 5-min wash in 20 mM sodium phosphate buffer (pH 6.5). The pellet was resuspended in 0.5 ml of the same buffer, and the material was solubilized by heating at 90°C for 30 min with mixing. The sample was cooled to 50°C, and (1-3,1-4)-β-glucan was assayed with a Megazyme kit. Briefly, the sample was incubated for 2 hours with 20 μl (1 U) of lichenase (Megazyme) to digest the (1-3,1-4)-β-glucan into oligosaccharides of DP3-DP9 and centrifuged at 10,000 rpm for 5 min; triplicate 10-μl samples were removed for (1-3,1-4)-β-glucan assay by further digestion with β-glucosidase; and the glucose released was measured spectrophotometrically against glucose standards as described in the Megazyme kit protocol. To determine the structure of (1-3,1-4)-β-glucan, an additional 50 μl of sample of the lichenase digestion was dried in a SpeedVac and the oligosaccharides were fluorescently labeled by reductive amination with 8-amino-1,3,6-pyrenetrisulfonic acid (APTS) and then separated by fluorophore-assisted capillary electrophoresis (FACE) with laser-induced fluorescence detection as described (29). The advantage of this method is that each oligosaccharide has a single fluorophore attached, and the signal response from the detector will be independent of the oligosaccharide length, unlike in high-performance anion exchange chromatography methods with a pulsed amperometric detector where each oligosaccharide has a different response factor depending on the length. The area under each peak was calculated and expressed either as a DP3/DP4 ratio or as a percentage of the total DP3-DP9 area for each peak.
Supplementary Material
Acknowledgments
I would like to acknowledge the excellent technical assistance of J. Yang and R. Chapple for β-glucan analysis and O. Larroque for analysis of lichenase digests of β-glucan by FACE. Funding: This work was funded by the Commonwealth Industrial Research Organisation Agriculture Flagship. Competing interests: The work described here forms part of a patent application PCT/AU2014/050173. Data and materials availability: Sequences of full-length CslF6 cDNAs have been deposited in the National Center for Biotechnology Information under accession numbers KP260637 to KP260644.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/1/5/e1500069/DC1
Fig. S1. Plasmid map of the Agrobacterium transformation vector pSJ226 and pSJ195 used for transient expression studies in N. benthamiana.
Fig. S2. Amino acid sequence alignment of the C-terminal region of the CslF6 proteins.
Fig. S3. The Leu-Ile amino acid change in ZmCslF6 TMH4 increases DP3/DP4 ratio.
Fig. S4. Sequence alignment of the C-terminal region of Rhodobacter sphaeroides BcsA and Hordeum vulgare CslF6 with Oryza sativa CesA and CslD proteins.
Table S1. Comparison of (1-3,1-4)-β-glucan abundance and structure in N. benthamiana leaf and in cereal wholegrain.
Table S2. Abundance of DP3-DP9 from Fig. 8.
Table S3. Primers.
REFERENCES AND NOTES
- 1.Doblin M. S., Pettolino F., Bacic A., Plant cell walls: The skeleton of the plant world. Funct. Plant Biol. 37, 357–381 (2010). [Google Scholar]
- 2.Pear J. R., Kawagoe Y., Schreckengost W. E., Delmer D. P., Stalker D. M., Higher plants contain homologs of the bacterial celA genes encoding the catalytic subunit of cellulose synthase. Proc. Natl. Acad. Sci. U.S.A. 93, 12637–12642 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hazen S. P., Scott-Craig J. S., Walton J. D., Cellulose synthase-like genes of rice. Plant Physiol. 128, 336–340 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Taketa S., Yuo T., Tonooka T., Tsumuraya Y., Inagaki Y., Haruyama N., Larroque O., Jobling S. A., Functional characterization of barley betaglucanless mutants demonstrates a unique role for CslF6 in (1,3;1,4)-β-d-glucan biosynthesis. J. Exp. Bot. 63, 381–392 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tonooka T., Aoki E., Yoshioka T., Taketa S., A novel mutant gene for (1-3,1-4)-β-d-glucanless grain on barley (Hordeum vulgare L.) chromosome 7H. Breed. Sci. 59, 47–54 (2009). [Google Scholar]
- 6.Vega-Sánchez M. E., Verhertbruggen Y., Christensen U., Chen X., Sharma V., Varanasi P., Jobling S. A., Talbot M., White R. G., Joo M., Singh S., Auer M., Scheller H. V., Ronald P. C., Loss of cellulose synthase-like F6 function affects mixed-linkage glucan deposition, cell wall mechanical properties, and defense responses in vegetative tissues of rice. Plant Physiol. 159, 56–69 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collins H. M., Burton R. A., Topping D. L., Liao M. L., Bacic A., Fincher G. B., Variability in fine structures of noncellulosic cell wall polysaccharides from cereal grains: Potential importance in human health and nutrition. Cereal Chem. 87, 272–282 (2010). [Google Scholar]
- 8.Othman R. A., Moghadasian M. H., Jones P. J. H., Cholesterol-lowering effects of oat β-glucan. Nutr. Rev. 69, 299–309 (2011). [DOI] [PubMed] [Google Scholar]
- 9.Burton R. A., Fincher G. B., (1,3;1,4)-β-D-glucans in cell walls of the poaceae, lower plants, and fungi: A tale of two linkages. Mol. Plant 2, 873–882 (2009). [DOI] [PubMed] [Google Scholar]
- 10.Simmons T. J., Uhrin D., Gregson T., Murray L., Sadler I. H., Fry S. C., An unexpectedly lichenase-stable hexasaccharide from cereal, horsetail and lichen mixed-linkage β-glucans (MLGs): Implications for MLG subunit distribution. Phytochemistry 95, 322–332 (2013). [DOI] [PubMed] [Google Scholar]
- 11.Burton R. A., Gidley M. J., Fincher G. B., Heterogeneity in the chemistry, structure and function of plant cell walls. Nat. Chem. Biol. 6, 724–732 (2010). [DOI] [PubMed] [Google Scholar]
- 12.Gibeaut D. M., Pauly M., Bacic A., Fincher G. B., Changes in cell wall polysaccharides in developing barley (Hordeum vulgare) coleoptiles. Planta 221, 729–738 (2005). [DOI] [PubMed] [Google Scholar]
- 13.Wilson S. M., Burton R. A., Doblin M. S., Stone B. A., Newbigin E. J., Fincher G. B., Bacic A., Temporal and spatial appearance of wall polysaccharides during cellularization of barley (Hordeum vulgare) endosperm. Planta 224, 655–667 (2006). [DOI] [PubMed] [Google Scholar]
- 14.Izydorczyk M. S., Dexter J. E., Barley β-glucans and arabinoxylans: Molecular structure, physicochemical properties, and uses in food products—A review. Food Res. Int. 41, 850–868 (2008). [Google Scholar]
- 15.Burton R. A., Fincher G. B., Current challenges in cell wall biology in the cereals and grasses. Front. Plant Sci. 3, 130 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burton R. A., Jobling S. A., Harvey A. J., Shirley N. J., Mather D. E., Bacic A., Fincher G. B., The genetics and transcriptional profiles of the cellulose synthase-like HvCslF gene family in barley. Plant Physiol. 146, 1821–1833 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burton R. A., Wilson S. M., Hrmova M., Harvey A. J., Shirley N. J., Stone B. A., Newbigin E. J., Bacic A., Fincher G. B., Cellulose synthase-like CslF genes mediate the synthesis of cell wall (1,3;1,4)-β-d-glucans. Science 311, 1940–1942 (2006). [DOI] [PubMed] [Google Scholar]
- 18.Doblin M. S., Pettolino F. A., Wilson S. M., Campbell R., Burton R. A., Fincher G. B., Newbigin E., Bacic A., A barley cellulose synthase-like CSLH gene mediates (1,3;1,4)-β-d-glucan synthesis in transgenic Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 106, 5996–6001 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morgan J. L. W., McNamara J. T., Zimmer J., Mechanism of activation of bacterial cellulose synthase by cyclic di-GMP. Nat. Struct. Mol. Biol. 21, 489–496 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morgan J. L., Strumillo J., Zimmer J., Crystallographic snapshot of cellulose synthesis and membrane translocation. Nature 493, 181–186 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Davis J. K., Combining polysaccharide biosynthesis and transport in a single enzyme: Dual-function cell wall glycan synthases. Front. Plant Sci. 3, 138 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wood C. C., Petrie J. R., Shrestha P., Mansour M. P., Nichols P. D., Green A. G., Singh S. P., A leaf-based assay using interchangeable design principles to rapidly assemble multistep recombinant pathways. Plant Biotechnol. J. 7, 914–924 (2009). [DOI] [PubMed] [Google Scholar]
- 23.Wilson S. M., Ho Y. Y., Lampugnani E. R., Van de Meene A. M. L., Bain M. P., Bacic A., Doblin M. S., Determining the subcellular location of synthesis and assembly of the cell wall polysaccharide (1,3; 1,4)-β-d-glucan in grasses. Plant Cell 27, 754–771 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Olek A. T., Rayon C., Makowski L., Kim H. R., Ciesielski P., Badger J., Paul L. N., Ghosh S., Kihara D., Crowley M., Himmel M. E., Bolin J. T., Carpita N. C., The structure of the catalytic domain of a plant cellulose synthase and its assembly into dimers. Plant Cell 26, 2996–3009 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sethaphong L., Haigler C. H., Kubicki J. D., Zimmer J., Bonetta D., DeBolt S., Yingling Y. G., Tertiary model of a plant cellulose synthase. Proc. Natl. Acad. Sci. U.S.A. 110, 7512–7517 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harris D. M., Corbin K., Wang T., Gutierrez R., Bertolo A. L., Petti C., Smilgies D. M., Estevez J. M., Bonetta D., Urbanowicz B. R., Ehrhardt D. W., Somerville C. R., Rose J. K. C., Hong M., DeBolt S., Cellulose microfibril crystallinity is reduced by mutating C-terminal transmembrane region residues CESA1A903V and CESA3T942I of cellulose synthase. Proc. Natl. Acad. Sci. U.S.A. 109, 4098–4103 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim S.-J., Zemelis S., Keegstra K., Brandizzi F., The cytoplasmic localization of the catalytic site of CSLF6 supports a channeling model for the biosynthesis of mixed-linkage glucan. Plant J. 81, 537–547 (2015). [DOI] [PubMed] [Google Scholar]
- 28.Chen S. B., Songkumarn P., Liu J. L., Wang G. L., A versatile zero background T-vector system for gene cloning and functional genomics. Plant Physiol. 150, 1111–1121 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O’Shea M. G., Samuel M. S., Konik C. M., Morell M. K., Fluorophore-assisted carbohydrate electrophoresis (FACE) of oligosaccharides: Efficiency of labelling and high-resolution separation. Carbohydr. Res. 307, 1–12 (1998). [Google Scholar]
- 30.Hall T. A., BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98 (1999). [Google Scholar]
- 31.Thompson J. D., Higgins D. G., Gibson T. J., CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Slabaugh E., Davis J. K., Haigler C. H., Yingling Y. G., Zimmer J., Cellulose synthases: New insights from crystallography and modeling. Trends Plant Sci. 19, 99–106 (2014). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/1/5/e1500069/DC1
Fig. S1. Plasmid map of the Agrobacterium transformation vector pSJ226 and pSJ195 used for transient expression studies in N. benthamiana.
Fig. S2. Amino acid sequence alignment of the C-terminal region of the CslF6 proteins.
Fig. S3. The Leu-Ile amino acid change in ZmCslF6 TMH4 increases DP3/DP4 ratio.
Fig. S4. Sequence alignment of the C-terminal region of Rhodobacter sphaeroides BcsA and Hordeum vulgare CslF6 with Oryza sativa CesA and CslD proteins.
Table S1. Comparison of (1-3,1-4)-β-glucan abundance and structure in N. benthamiana leaf and in cereal wholegrain.
Table S2. Abundance of DP3-DP9 from Fig. 8.
Table S3. Primers.


