A redesigned production process enables expanded molecular variation and altered bioactivity for the antibiotic erythromycin.
Keywords: erythromycin, antibiotic, E. coli, Polyketide, heterologous biosynthesis, analogue
Abstract
Type I modular polyketide synthases are responsible for potent therapeutic compounds that include avermectin (antihelinthic), rapamycin (immunosuppressant), pikromycin (antibiotic), and erythromycin (antibiotic). However, compound access and biosynthetic manipulation are often complicated by properties of native production organisms, prompting an approach (termed heterologous biosynthesis) illustrated in this study through the reconstitution of the erythromycin pathway through Escherichia coli. Using this heterologous system, 16 tailoring pathways were introduced, systematically producing eight chiral pairs of deoxysugar substrates. Successful analog formation for each new pathway emphasizes the remarkable flexibility of downstream enzymes to accommodate molecular variation. Furthermore, analogs resulting from three of the pathways demonstrated bioactivity against an erythromycin-resistant Bacillus subtilis strain. The approach and results support a platform for continued molecular diversification of the tailoring components of this and other complex natural product pathways in a manner that mirrors the modular nature of the upstream megasynthases responsible for aglycone polyketide formation.
INTRODUCTION
Natural products are well recognized for their medicinal impact. Environmentally derived compounds have altered worldwide historical events and revolutionized modern medicine through the introduction of numerous clinical agents (1–5). Balancing the beneficial applications of final natural product therapies is the challenge and complexity associated with individual compound formation.
At one level, harnessing the potential of natural products is made challenging by the native environmental cellular sources responsible for biosynthesis. Plants, filamentous fungi and bacteria, and other microbial sources responsible for the breadth of natural products found in the environment are often unculturable within a laboratory setting and are always less tractable relative to model bacterial hosts (6). This situation introduced an approach termed heterologous biosynthesis, in which the native genetic pathway for a desired natural product is transferred to a surrogate host, which provides advantages in the directed production of the encoded product (7, 8). Although many model systems have been used in this capacity, few offer the innate growth kinetics and genetic tractability of Escherichia coli, making this host a prime candidate for heterologous biosynthetic efforts.
However, the lack of native natural product biosynthetic pathways within E. coli and the complexity associated with the enzymatic machinery needed for compound formation pose substantial hurdles to accomplishing heterologous biosynthesis with this host system. Without innate natural product formation capabilities, metabolic routes must be introduced or engineered within E. coli in support of biosynthesis. Furthermore, the foreign biosynthetic pathways pose significant challenges in the form of numerous and large genes that must be coordinately expressed and successfully translated to active protein products. These issues are more pronounced when attempting to generate compounds derived from type I modular polyketide synthase systems (9, 10). As an illustrative example highlighted in this study, the macrolactone core of the antibiotic erythromycin is the result of three large modular polyketide synthase enzymes (each ≥330 kD) that produce a cyclized product termed 6-deoxyerythronolide B (6dEB) (11, 12). A combination of deoxysugar biosynthetic, hydroxylase, methyltransferase, and resistance enzymes (17 in total) enable conversion of 6dEB to the final erythromycin compound (fig. S1) (13, 14). A previous work by our group accomplished the heterologous biosynthesis of the final erythromycin A compound by systematically confirming individual and coordinated pathway expression and activity through a metabolically engineered strain of E. coli termed BAP1 (15, 16).
This synthesis opens up new opportunities for compound production and pathway engineering. Type I modular polyketide synthases, in particular, suggest a myriad of diversification opportunities through a combination of altered biosynthetic design and the recombinant tools afforded by a heterologous host (17). However, regardless of the chemical variability imparted to the aglycone polyketide product when using this approach, biological activity will be significantly reduced or altogether eliminated without accompanying compound tailoring, highlighted by the biosynthesis and attachment of two deoxysugars (l-mycarose and d-desosamine) in the case of erythromycin (18–21). A key question is whether these downstream tailoring pathways offer the same potential for compound diversification as their upstream polyketide synthase counterparts. The information accumulating for natural product deoxysugar pathways, coupled with the advantages afforded by the E. coli heterologous host, allows for a systematic assessment of tailoring diversification. Specifically, this study features the construction and introduction of glycosylation patterns to vary the native mycarose component of erythromycin (Fig. 1). Key aspects of the work include unique deoxysugar pathways built from putative individual enzymatic steps and notable flexibility of the tailoring pathway enzymes to generate new compounds. Success emphasizes the engineering advantages of the E. coli host system and the plasticity of downstream tailoring reactions to accommodate molecular variation. The results also underscore a counterbalance to the previous efforts to engineer compound variation through manipulation of the modular nature of the upstream polyketide synthase associated with erythromycin and similarly derived natural products. Our results suggest a similar modularity associated with the downstream tailoring reactions that offer as much or more opportunity for compound variation and altered bioactivity given the necessity of these steps in endowing therapeutic properties.
Fig. 1. Modular engineering of tailoring pathways for glycosylation diversification.
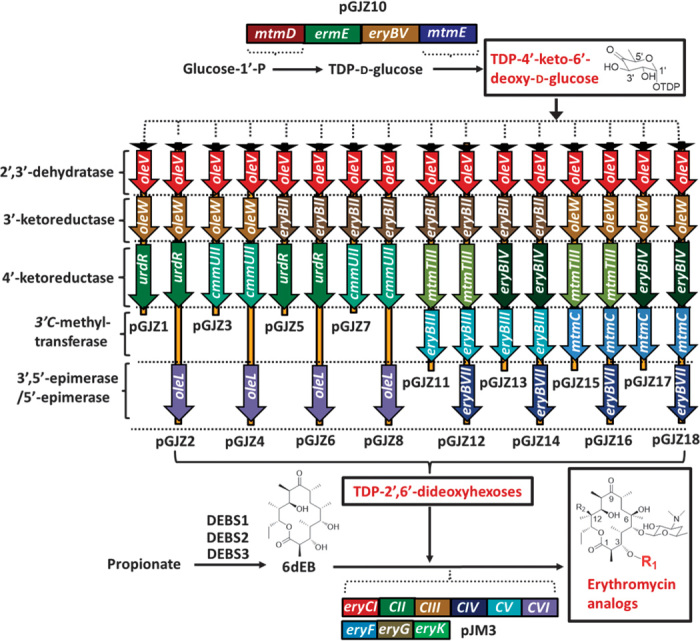
Indicated are the pathways, enzymes, intermediates, and final products of the heterologous system used to generated erythromycin analogs. Black triangles represent the T7 promoter, and homologous genes are grouped by dashed lines. R1, diversified glycosylation as a result of the introduced pathways to generate TDP-2′,6′-dideoxyhexoses; R2, H or OH as a result of EryK activity. DEBS, deoxyerythronolide B synthase.
RESULTS
Metabolic engineering for improved deoxysugar formation
E. coli has the native metabolic ability to convert d-glucose-1′-phosphate to TDP (thymidine 5′-diphosphate)–4′-keto-deoxy-d-glucose, which provides a starting intermediate for the dideoxysugar pathways to be described below, through the activity of glucose-1′-phosphate:TTP (thymidine 5′-triphosphate) thymidylyl transferase (RmlA) and TDP-d-glucose 4′,6′-dehydratase (RmlB). However, although this native capability can support erythromycin formation when using E. coli (16), it is likely that the process could be improved through engineering steps that accompany the remaining erythromycin heterologous pathway. Namely, expression was increased for these two steps via the introduction of analogous pathway genes, mtmD and mtmE, native to Streptomyces argillaceus (Fig. 1) (22).
The impact on product formation was then first tested for heterologous erythromycin with a threefold titer improvement observed (Fig. 2A). The approach was next applied to one of the analog pathways (from pGJZ1), and a similar threefold improvement in product titer was achieved (Fig. 2B). As a result of the improvements observed for both the original erythromycin compound and an analog, the use of MtmD and MtmE was included in all subsequent attempts to generate analog compounds.
Fig. 2. Improved erythromycin production by engineering early deoxysugar pathway steps.
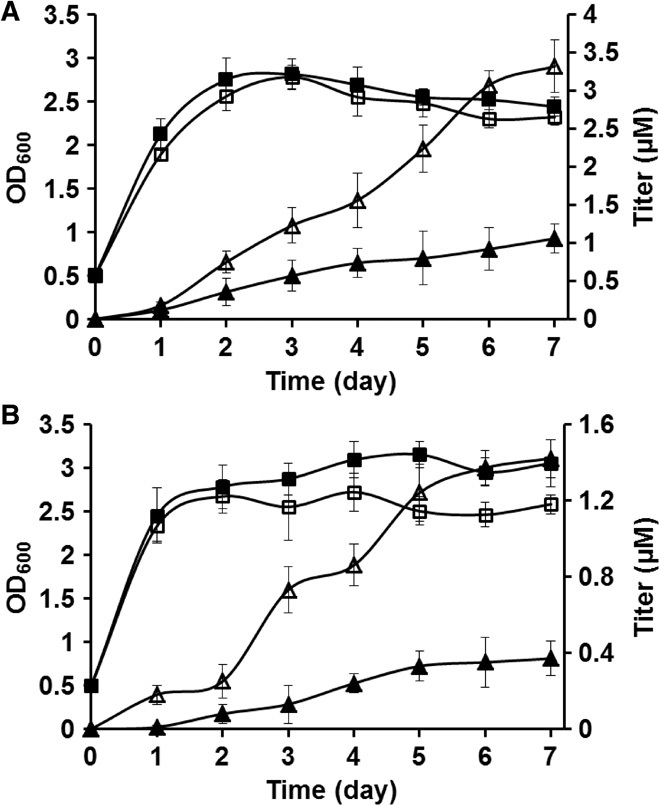
(A and B) Growth and production comparisons with and without MtmD and MtmE for erythromycin (A) and d-olivosyl-erythromycin (B). Squares represent OD600 values; triangles represent titer values; closed markers are without MtmD/MtmE; and open markers are with MtmD/MtmE.
Pathway construction for systematic mycarose replacement
The deoxysugar tailoring pathways introduced for analog formation were constructed as described in Materials and Methods (figs. S2 and S3). Then, the pathways were divided into two generations. In the first generation, enzymes from previously studied natural product systems (described further below) were designed for eight pathways (pGJZ1-8; Fig. 1), with one pair of 3′-ketoreductases (OleW and EryBII) controlling the stereochemistry at C3′, one pair of 4′-ketoreductases (UrdR and CmmUII) controlling the stereochemistry at C4′, and one 3′,5′-epimerase (OleL) responsible for configuration conversion at the C5′ position, thus leading to four chiral partner deoxysugars (fig. S4). However, only one of these deoxysugars had been previously tested in the context of attachment to 6dEB (23), and the symmetry established between chiral deoxysugar pairs enabled a systematic assessment of pathway flexibility and final compound structural diversity. Hence, the set provided a basis to test the capabilities of both the deoxysugar enzymes within hypothetical tailoring pathways and the EryBV glycosyltransferase to accept the eight resulting deoxysugars.
The approach was extended through the introduction of an extra methyltransferase (EryBIII or MtmC; pGJZ11-18; Fig. 1), resulting in a second generation of eight deoxysugar pathways and four additional chiral partner substrates (fig. S5). Five of these constructed routes, in particular, had no precedent in available pathway architecture. Instead, they were built from hypothetical activity of individual pathway enzymes. In addition, seven of the resulting eight deoxysugars (excluding the native l-mycarose) had not been tested for EryBV-based transfer to 6dEB. In summary, the pathway design used in this study for deoxysugar-based analog production is both comprehensive relative to previous studies (23–36) and unique due to the use of a type I modular polyketide system established in E. coli.
Once isolated, the individual genes used to build the deoxysugar pathways described above were first tested for expression by SDS–polyacrylamide gel electrophoresis (SDS-PAGE) analysis (fig. S6). Soluble protein formation was observed for each expressed gene. In each case, expression derived from genes cloned within the pET28a vector, which includes an N-terminal 6× His tag and has been previously demonstrated to aid erythromycin pathway gene expression during heterologous reconstitution through E. coli (16). Upon confirming expression, operons were constructed from the successfully expressed individual gene cassettes.
Expanded erythromycin analog formation
Production of new analogs was tested in a two-stage process in which 6dEB was first generated and isolated for addition to a second culture of E. coli containing combinations of operons required for 6dEB conversion. Successful analog formation was observed in each of the 16 deoxysugar pathways engineered in this study. Representative examples are presented in Fig. 3, and a comprehensive list of produced analogs is provided in table S2. Product titers varied between introduced pathways, and, like the native erythromycin pathway, derivatives associated with analogs were observed on the basis of the tailoring activities of EryK and EryG (fig. S1). Although analogs resulting from some pathways (derived from pGJZ6, pGJZ7, and pGJZ12) showed reduced final titers, seven (from pGJZ3, pGJZ4, pGJZ13, pGJZ14, pGJZ15, pGJZ17, and pGJZ18) demonstrated levels that approached or surpassed the original erythromycin heterologous levels (table S2).
Fig. 3. Production analysis of erythromycin analogs.
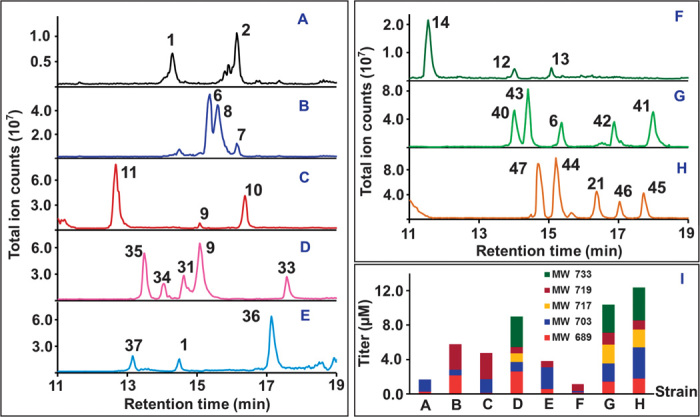
(A to I) LC-MS profiles from representative heterologous systems harboring pGJZ1 (A), pGJZ3 (B), pGJZ4 (C), pGJZ14 (D), pGJZ15 (E), pGJZ5 (F), pGJZ17 (G), and pGJZ18 (H) and corresponding compound distributions (I) as a function of EryG and/or EryK activity.
Finally, new analogs were tested for preliminary antibiotic activity (fig. S14). Three different antibiotic-resistant strains of Bacillus subtilis were used to assess activity, with most of the analogs tested matching the effectiveness of the original compound against erythromycin-sensitive B. subtilis strains (Table 1; analogs produced at reduced titers were excluded from this analysis). In addition, three of the analog pathways [two from this study and one previously described by our group (23)] produced compounds demonstrating activity against an erythromycin-resistant strain of B. subtilis, providing preliminary demonstration of the therapeutic potential of this diversification strategy.
Table 1. MIC values (μg/ml) for erythromycin analog pathways tested against antibiotic-resistant (+) B. subtilis strains.
| B. subtilis strain | pGJZ1 | pGJZ2 | pGJZ3 | pGJZ4 | pGJZ5 | pGJZ8 | Erythromycin A |
| Erythromycin+ | 0.20 | — | 0.20 | 0.20 | — | — | — |
| Chloramphenicol+ | 0.15 | 0.10 | 0.10 | 0.15 | 0.10 | 0.40 | 0.16 |
| Streptomycin+ | 0.15 | 0.30 | 0.10 | 0.10 | 0.10 | 0.40 | 0.24 |
| B. subtilis strain | pGJZ11 | pGJZ13 | pGJZ14 | pGJZ15 | pGJZ16 | pGJZ17 | pGJZ18 |
| Erythromycin+ | — | — | — | — | — | — | — |
| Chloramphenicol+ | 0.10 | 0.20 | 0.15 | 0.10 | 0.10 | 0.20 | 0.20 |
| Streptomycin+ | 0.20 | 0.20 | 0.20 | 0.20 | 0.10 | 0.20 | 0.40 |
DISCUSSION
The successful production of erythromycin analogs for each newly introduced deoxysugar pathway and comparable titers to the original erythromycin compound in numerous cases demonstrate the remarkable flexibility of the various deoxysugar enzymes to function broadly across engineered pathways. In addition, insight was gained into the distribution of derivatives associated with analog compounds as a by-product of the activities of EryK (C12 hydroxylase) and EryG (C3′-O-methyltransferase). As is the case for the original erythromycin compound, the activity of these enzymes will dictate the distribution of four distinct A, B, C, and D derivatives, and the established reaction order is EryK [acting on erythromycin D; molecular weight (MW) 703] followed by EryG (acting on erythromycin C; MW 719) (37, 38) (fig. S1). However, production analysis indicated that the action of EryK and EryG is variable depending on the analog substrate encountered, allowing for the biosynthesis of 42 new structures (table S1 and figs. S7 to 12).
Compounds without a C3′ methyl group resulting from the first-generation analog pathways (fig. S7) prompt the activity of EryG, which appears to then enable EryK activity (in all cases but pathway 1) (fig. S13). Specifically, substrates with a C3 methoxy group tailored by EryG (compounds 4, 7, 10, 13, 16, 19, and 22; MW 703; fig. S8) can be further processed by EryK to generate products with MW 719 (compounds 5, 8, 11, 14, 17, 20, and 23; fig. S11). To further support this result, we removed the EryG step from our heterologous system (by replacing pJM3 with pGJZ9) (fig. S13B). Without EryG, the heterologous system failed to generate an MW 719 product but instead accumulated the nonmethoxylated starting compound, indicating a preferential activity of EryG relative to EryK toward first-generation analogs. The observed reaction order contradicts the previously established sequence of EryK and then EryG, but also emphasizes an additional level of plasticity by downstream enzymes in providing fully tailored final products.
In the case of the second generation of analogs (which all contain a C3′ methyl group), EryK activity was observed in every case but the pathway associated with pGJZ12 (which was difficult to interpret due to low production levels) (table S1). However, in four pathways (from pGJZ11, pGJZ13, pGJZ15, and pGJZ16), EryG activity was not observed. Results from the second-generation pathways reestablished a reaction order of EryK and EryG and, when combined with data from the first-generation analogs, support the selective activity of EryK on MW 703 compounds (fig. S13A). More direct biochemical and structural studies need to be dedicated to better understand the reasons for variable EryK/G activity, but the approach presented here, in lieu of sufficient quantities of the substrates that would be needed for more in-depth characterization, provides a complementary route to traditional enzymatic assessment.
The analog production results also help to culminate research to identify and characterize both the deoxysugar pathways associated with complex polyketide products and their mechanisms of attachment. The efforts in identifying the olivose, oliose, digitoxose, and boivinose pathways associated with the oleandomycin, mithramycin, chromomycin, and urdamycin A polyketide products guided the design of the first-generation analog pathways (34, 39–44). Likewise, assessment of the original l-mycarose pathway prompted the inclusion of an alternative methylation step and the resulting second-generation pathways. Similar tools and engineering approaches associated with E. coli are expected to expand on the diversification capabilities presented here, for example, by separately engineering alterations of the desosamine addition to 6dEB, combinatorially varying both deoxysugars associated with erythromycin, or combining tailoring pathway engineering with precursor-directed biosynthesis (45–47). Furthermore, additional metabolic engineering and synthetic biology tools will be available to enable and improve the outcomes from the diversification studies, just as the inclusion of mtmD and mtmE was used to assist the objectives of this study.
The structural modifications resulting from the three analog pathways (resulting from pGJZ1, pGJZ3, and pGJZ4) responsible for rescued antibiotic activity include both macrolactone and mycarose alterations. Although minor, such changes likely altered ribosomal binding affinity enough to restore bioactivity though direct structural assessment is needed to confirm the results observed here. Similarly minor structural variations have also been observed to enable semisynthetic erythromycin analogs to effectively combat resistance mechanisms (48). It is also interesting that antibiotic activity was restored to analogs modified at the mycarosyl unit as opposed to the desosamine moiety, which has direct interaction with the common erythromycin resistance mechanism (methylation of A2058 in the 23S ribosomal RNA subunit) (49–51). This suggests that even more pronounced resistance recovery could be accomplished by similarly engineering the desosamine portion of erythromycin. Also of note is the discrepancy between general analog production levels and restored antibiotic potency. The second-generation analogs include a C3′ methylation step similar to the native l-mycarose pathway, and this similarity in structure may have allowed for stronger production levels (Fig. 3 and table S1). However, antibiotic effectiveness against an erythromycin-resistant B. subtilis was achieved with first-generation analogs lacking this C3′ methyl functionality. The more pronounced deviation from the native l-mycarose may have caused increased pathway incorporation challenges during intermediate and final product formation for the first-generation analogs, thus resulting in reduced product titers relative to second-generation compounds. However, this same structural variation may have prompted the bioactivity observed within the first analog set.
The approach also presents another option to the renewed interest in natural product discovery (52). As opposed to isolating a completely new compound, a suitably complex natural product biosynthetic pathway lends itself to directed randomization strategies such as the one presented in this work. Theoretically, the combination of modularity presented in the upstream polyketide synthase and similarly proposed modularity in the downstream reactions, as supported by this work, allows for an extensive means of broadening the molecular space associated with an initial structure to the point where new chemical diversity begins to allow for new bioactivity. Such potential relies on precise, efficient, and rapid engineering of the target biosynthetic system, which is a key advantage afforded by heterologous reconstitution through a host such as E. coli.
MATERIALS AND METHODS
Experimental design
Deoxysugar pathways were designed, engineered, and introduced to E. coli to systematically diversify the erythromycin antibiotic. The process was facilitated by a metabolically engineered strain of E. coli capable of supporting complex natural product biosynthesis. In addition to new analog formation, insight was gained into the flexibility of tailoring reactions required for final compound assembly and activity.
Materials
Polymerase chain reaction (PCR) primers were purchased from Eurofins Genomics and are listed in table S2. The primers were used to amplify genes provided by J. Rohr (University of Kentucky), C. Méndez and J. Salas (University of Oviedo), K. Yang (Chinese Academy of Sciences), and B.-G. Kim (Seoul National University). Selection antibiotics, culture medium components, isopropyl β-d-1-thiogalactopyranoside (IPTG), ethyl acetate, and buffer components were purchased from Fisher Chemical, whereas erythromycin and roxithromycin were obtained from Sigma-Aldrich. Restriction endonucleases, T4 DNA ligase, and Phusion High-Fidelity PCR Master Mix were purchased from New England Biolabs. The GroEL/ES chaperonin plasmid pGro7 was purchased from Takara.
Strains and plasmids
Strain BAP1 was used in the biosynthesis of 6dEB and has been engineered to (i) enable polyketide posttranslational modification through the activity of an Sfp 4′-phosphopantetheinyl transferase and (ii) convert exogenously fed propionate to propionyl–coenzyme A (CoA) through the activity of a propionyl-CoA synthetase. Plasmids pBP130 and pBP144 were added to this strain to introduce the propionyl-CoA carboxylase and deoxyerythronolide B synthase enzymes needed to generate (2S)-methylmalonyl-CoA and produce 6dEB, respectively (fig. S1).
The pGJZ vectors were designed in an operon fashion with one T7 promoter and one T7 terminator and a ribosomal binding site preceding each gene. Operons were derived from PCR products first individually inserted into pET28a. Individual gene expression was confirmed by SDS-PAGE (fig. S6) before operon construction began. Operons were built according to the general design presented in fig. S2. Each gene contained a 5′ restriction site and a series of 3′ restriction sites (introduced during PCR amplification) that enabled successive gene introduction based on the compatible cohesiveness of Xba I (X) and Spe I (S) in a procedure similar to that described earlier (16). In this case, however, additional restriction sites (Avr II, Pst I, Bst BI, Mun I, Sna BII, and Hind III) were included to allow interchange of genes as needed in future cloning steps. Plasmids pJM2 [l-mycarose biosynthesis and attachment; erythromycin resistance (ermE)] and pJM3 (d-desosamine biosynthesis and attachment; eryF, eryG, and eryK) have been described previously (53) and enable erythromycin A production (fig. S1). Plasmid pGJZ9 is constructed similar to pJM3 but without eryG. Final plasmid maps are provided in fig. S3. Deoxysugar pathways associated with constructed plasmids are presented in figs. S4 and S5.
Once completed, plasmid combinations (as outlined in Fig. 1) were introduced to strain TB3 [a derivative of BAP1 (54)] via sequential electroporation. For example, TB3 would contain pGJZ10 (ampicillin-resistant), pJM3 (or pGJZ9, as further described below; streptomycin-resistant), and pGJZ1 (or another pathway operon; kanamycin-resistant). pGro7 (chloramphenicol-resistant) would also be introduced to these strains to provide chaperonin support. Once selected on antibiotic-containing LB agar, glycerol stocks were prepared for subsequent analog production studies.
Analog production cultures
Strain BAP1/pBP130/pBP144 was first used to generate 6dEB. A starter culture incubated at 37°C with shaking was initiated from a glycerol stock of the strain in 3 ml of LB medium [tryptone (10 g/liter), yeast extract (5 g/liter), and NaCl (10 g/liter)] containing selection antibiotics. The starter culture was used to inoculate (1% v/v) a 50-ml LB seed culture, which was incubated at 37 °C with shaking in a 125-ml flask until reaching an OD600 value of 0.6 to 0.8. At which point, the entire culture was added to a 3-liter bioreactor vessel containing 2 liters of production medium [tryptone (15 g/liter), glycerol (25 g/liter), yeast extract (5 g/liter), NaCl (10 g/liter), and 100 mM Hepes; adjusted to pH 7.6 using NaOH]. The bioreactor (Applikon 1030) was then operated at an agitation rate of 500 rpm, a filtered air flow rate of 3 liter/min, and pH 7.6. The system was maintained at 37 °C until 0.6 to 0.8 OD600 and then shifted to 22 °C and induced with 0.1 mM IPTG. After continued operation for 5 days, the bioreactor contents were extracted once with 2 liters of ethyl acetate. The extract was concentrated under vacuum, and the 6dEB content was quantified by liquid chromatography–mass spectrometry (LC-MS) for subsequent addition to analog conversion strains.
Analog-producing strains (using BAP1 derivative TB3) contained combinations of pGJZ10, pJM3 (or pGJZ9), pGro7, and the pathway-specific pGJZ plasmids outlined in Fig. 1. For comparison to the original erythromycin pathway, plasmids pJM2 and pJM3, which contain the native eryB and eryC operons to generate l-mycarose and d-desosamine, respectively, were used (53) (fig. S1). Glycerol stocks of these strains were used to initiate 3 ml of overnight LB starter cultures, incubated as described above, which were then inoculated (1% v/v) into 30-ml production medium cultures, with both cultures including selection antibiotics as needed. Production cultures were first incubated with shaking at 37 °C until 0.6 to 0.8 OD600 and then shifted to 22 °C for IPTG induction (0.1 mM). After 3 hours, the cultures were supplemented with 30 μM 6dEB, continued for 7 days, and extracted three times with 30 ml of ethyl acetate each time before concentration under vacuum. OD measurements were made at 600 nm and used to monitor cell growth over time (Fig. 2). Similarly, product quantification was carried out using LC-MS.
Production analysis and assessment
LC-MS analysis was performed using an API 3000 Triple Quad LC-MS with a Turbo Ion Spray source (PE Sciex) coupled with a Shimadzu Prominence LC system. All MS analyses were conducted in positive ion mode, and chromatography was performed on a Waters XTerra C18 column (5 μm, 2.1 × 250 mm). After an injection of 3 μl of crude extract, a linear gradient of 30% buffer A (95% water/5% acetonitrile/0.1% formic acid) to 100% buffer B (5% water/95% acetonitrile/0.1% formic acid) was used at a flow rate of 0.2 ml/min. In the absence of authentic standards, analog concentrations were estimated using erythromycin A as an external standard and roxithromycin as an internal standard. Known amounts of erythromycin A were added to completed TB3 cultures (without plasmids) grown under the same conditions described for analog production but using only 2-ml cultures. After ethyl acetate extraction and air drying, the samples were subjected to MS analysis to prepare a calibration curve. Both calibration and analog extracts were dissolved in 100 μl of methanol containing roxithromycin (0.1 mg/liter). The ratio of the erythromycin A and roxithromycin standard peak areas was correlated with erythromycin A concentrations to quantify experimental analog production titers with a suitable calibration curve made before every experimental analysis. Results are presented in table S1, and compound assignments per MW are summarized in figs. S7 to 12. HR-MS (high-resolution mass spectrometry) analysis was completed on an Agilent 6538 HRESI QTOF (quadrupole time-of-flight) MS for analogs resulting from pathways encoded in pGJZ1, pGJZ3, and pGJZ4. MS/MS (tandem mass spectrometry) and HR-MS data are presented in a dedicated section at the end of the Supplementary Materials (beneath fig. S15). Analog production levels were compared to erythromycin A produced through the use of pJM2 and pJM3 (table S1). In addition, the presence or absence of EryG was tested through the inclusion of pJM3 or pGJZ9, respectively (table S1).
A filter disc zone of inhibition assay was first performed to qualitatively test antibiotic activity. In this assay, filter paper discs were placed onto the surface of a freshly prepared LB agar plate seeded with 2.5% (v/v) of an overnight B. subtilis culture and subsequently loaded with 4 μl of analog extracts. Analogs produced at trace levels (from pGJZ6, pGJZ7, and pGJZ12; table S1) were not tested in either this or the subsequent minimum inhibitory concentration (MIC) assay. B. subtilis strains resistant to chloramphenicol, streptomycin, and erythromycin were included in the antibiotic assessment studies (with both the chloramphenicol- and erythromycin-resistant strains also resistant to streptomycin; strains provided by L. Sonenshein (Tufts University)]. After overnight incubation at 37°C, the inhibition zones of the erythromycin analogs were compared with discs containing control extracts (prepared exactly the same as for the erythromycin analogs), control antibiotics (1 μl added per disk from stock concentrations of 500 μg/ml for chloramphenicol and 100 μg/ml for streptomycin and erythromycin A), 6dEB (1 μl from a stock concentration of 100 μg/ml), and methanol.
A liquid-phase MIC assay was completed for the erythromycin analogs in a 96-well plate format. The protocol used was identical to that recommended by the National Committee for Clinical Laboratory Standards. Resulting OD600 values of each well were recorded after 12 hours, and results were compared to erythromycin A standard.
Statistical analysis
Error bars represent SD values generated from three independent experiments.
Supplementary Material
Acknowledgments
The authors recognize material contributions from J. Rohr (University of Kentucky; mtmTIII and mtmC); C. Méndez and J. Salas (University of Oviedo) and K. Yang (Chinese Academy of Sciences) (mtmD, mtmE, cmmUII, and urdR); B.-G. Kim (Seoul National University; Streptomyces fradiae genomic DNA used to isolate oleV and oleW); and L. Sonenshein (Tufts University; B. subtilis strains). Author contributions: G.Z. designed the study and cowrote the draft with B.A.P. Y.L. and L.F. assisted with culturing and antibiotic assessment studies, respectively. Competing interests: The authors declare that they have no competing interests.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/1/4/e1500077/DC1
Fig. S1. Biosynthetic pathway for erythromycin A emphasizing deoxysugar biosynthesis and attachment and the enzymatic steps catalyzed by EryK and EryG.
Fig. S2. General genetic design used in constructing plasmids and operons.
Fig. S3. Maps of the pGJZ plasmids used to generate erythromycin analogs.
Fig. S4. Deoxysugar pathways associated with first-generation pGJZ plasmids.
Fig. S5. Deoxysugar pathways associated with second-generation pGJZ plasmids.
Fig. S6. SDS-PAGE confirmation of gene expression.
Fig. S7. Compounds associated with MW 689.
Fig. S8. Compounds associated with MW 703.
Fig. S9. Compounds associated with MW 705.
Fig. S10. Compounds associated with MW 717.
Fig. S11. Compounds associated with MW 719.
Fig. S12. Compounds associated with MW 733.
Fig. S13. MW correlation with EryK activity.
Fig. S14. (A and B) Filter disk antibiotic activity assessment of first-generation (A) and second-generation (B) deoxysugar pathway analogs against three B. subtilis strains resistant (+) to chloramphenicol, streptomycin, and erythromycin.
Fig. S15. Common fragmentation patterns in the MS/MS spectra of erythromycin analogs.
Table S1. Production analysis of erythromycin analogs from first (pGJZ1 to pGJZ8) and second (pGJZ11 to pGJZ18) generation deoxysugar pathways.
Table S2. PCR oligonucleotide primers.
REFERENCES AND NOTES
- 1.Cragg G. M., Newman D. J., Natural products: A continuing source of novel drug leads. Biochim. Biophys. Acta 1830, 3670–3695 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Newman D. J., Cragg G. M., Natural products as sources of new drugs over the 30 years from 1981 to 2010. J. Nat. Prod. 75, 311–335 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Demain A. L., From natural products discovery to commercialization: A success story. J. Ind. Microbiol. Biotechnol. 33, 486–495 (2006). [DOI] [PubMed] [Google Scholar]
- 4.Demain A. L., Sanchez S., Microbial drug discovery: 80 years of progress. J. Antibiot. 62, 5–16 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kardos N., Demain A. L., Penicillin: The medicine with the greatest impact on therapeutic outcomes. Appl. Microbiol. Biotechnol. 92, 677–687 (2011). [DOI] [PubMed] [Google Scholar]
- 6.Davies J., Ryan K. S., Introducing the parvome: Bioactive compounds in the microbial world. ACS Chem. Biol. 7, 252–259 (2012). [DOI] [PubMed] [Google Scholar]
- 7.Zhang H., Boghigian B. A., Armando J., Pfeifer B. A., Methods and options for the heterologous production of complex natural products. Nat. Prod. Rep. 28, 125–151 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ongley S. E., Bian X., Neilan B. A., Müller R., Recent advances in the heterologous expression of microbial natural product biosynthetic pathways. Nat. Prod. Rep. 30, 1121–1138 (2013). [DOI] [PubMed] [Google Scholar]
- 9.Weissman K. J., Leadlay P. F., Combinatorial biosynthesis of reduced polyketides. Nat. Rev. Microbiol. 3, 925–936 (2005). [DOI] [PubMed] [Google Scholar]
- 10.Khosla C., Harbury P. B., Modular enzymes. Nature 409, 247–252 (2001). [DOI] [PubMed] [Google Scholar]
- 11.Donadio S., Staver M. J., McAlpine J. B., Swanson S. J., Katz L., Modular organization of genes required for complex polyketide biosynthesis. Science 252, 675–679 (1991). [DOI] [PubMed] [Google Scholar]
- 12.Cortes J., Haydock S. F., Roberts G. A., Bevitt D. J., Leadlay P. F., An unusually large multifunctional polypeptide in the erythromycin-producing polyketide synthase of Saccharopolyspora erythraea. Nature 348, 176–178 (1990). [DOI] [PubMed] [Google Scholar]
- 13.Summers R. G., Donadio S., Staver M. J., Wendt-Pienkowski E., Hutchinson C. R., Katz L., Sequencing and mutagenesis of genes from the erythromycin biosynthetic gene cluster of Saccharopolyspora erythraea that are involved in l-mycarose and d-desosamine production. Microbiology 143, 3251–3262 (1997). [DOI] [PubMed] [Google Scholar]
- 14.Salah-Bey K., Doumith M., Michel J. M., Haydock S., Cortés J., Leadlay P. F., Raynal M. C., Targeted gene inactivation for the elucidation of deoxysugar biosynthesis in the erythromycin producer Saccharopolyspora erythraea. Mol. Gen. Genet. 257, 542–553 (1998). [DOI] [PubMed] [Google Scholar]
- 15.Pfeifer B. A., Admiraal S. J., Gramajo H., Cane D. E., Khosla C., Biosynthesis of complex polyketides in a metabolically engineered strain of E. coli. Science 291, 1790–1792 (2001). [DOI] [PubMed] [Google Scholar]
- 16.Zhang H., Wang Y., Wu J., Skalina K., Pfeifer B. A., Complete biosynthesis of erythromycin A and designed analogs using E. coli as a heterologous host. Chem. Biol. 17, 1232–1240 (2010). [DOI] [PubMed] [Google Scholar]
- 17.Khosla C., Harnessing the biosynthetic potential of modular polyketide synthases. Chem. Rev. 97, 2577–2590 (1997). [DOI] [PubMed] [Google Scholar]
- 18.Weymouth-Wilson A. C., The role of carbohydrates in biologically active natural products. Nat. Prod. Rep. 14, 99–110 (1997). [DOI] [PubMed] [Google Scholar]
- 19.Thorson J. S., Hosted T. J. Jr, Jiang J., Biggins J. B., Ahlert J., Natures carbohydrate chemists the enzymatic glycosylation of bioactive bacterial metabolites. Curr. Org. Chem. 5, 139–167 (2001). [Google Scholar]
- 20.Rix U., Fischer C., Remsing L. L., Rohr J., Modification of post-PKS tailoring steps through combinatorial biosynthesis. Nat. Prod. Rep. 19, 542–580 (2002). [DOI] [PubMed] [Google Scholar]
- 21.Olano C., Mendez C., Salas J. A., Post-PKS tailoring steps in natural product-producing actinomycetes from the perspective of combinatorial biosynthesis. Nat. Prod. Rep. 27, 571–616 (2010). [DOI] [PubMed] [Google Scholar]
- 22.Lombó F., Siems K., Braña A. F., Méndez C., Bindseil K., Salas J. A., Cloning and insertional inactivation of Streptomyces argillaceus genes involved in the earliest steps of biosynthesis of the sugar moieties of the antitumor polyketide mithramycin. J. Bacteriol. 179, 3354–3357 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang M., Zhang H., Park S. H., Li Y., Pfeifer B. A., Deoxysugar pathway interchange for erythromycin analogues heterologously produced through Escherichia coli. Metab. Eng. 20, 92–100 (2013). [DOI] [PubMed] [Google Scholar]
- 24.Rodríguez L., Aguirrezabalaga I., Allende N., Braña A. F., Méndez C., Salas J. A., Engineering deoxysugar biosynthetic pathways from antibiotic-producing microorganisms. A tool to produce novel glycosylated bioactive compounds. Chem. Biol. 9, 721–729 (2002). [DOI] [PubMed] [Google Scholar]
- 25.Pérez M., Baig I., Braña A. F., Salas J. A., Rohr J., Méndez C., Generation of new derivatives of the antitumor antibiotic mithramycin by altering the glycosylation pattern through combinatorial biosynthesis. Chembiochem 9, 2295–2304 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shepherd M. D., Liu T., Méndez C., Salas J. A., Rohr J., Engineered biosynthesis of gilvocarcin analogues with altered deoxyhexopyranose moieties. Appl. Environ. Microbiol. 77, 435–441 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lombó F., Gibson M., Greenwell L., Braña A. F., Rohr J., Salas J. A., Méndez C., Engineering biosynthetic pathways for deoxysugars: Branched-chain sugar pathways and derivatives from the antitumor tetracenomycin. Chem. Biol. 11, 1709–1718 (2004). [DOI] [PubMed] [Google Scholar]
- 28.Han A. R., Park J. W., Lee M. K., Ban Y. H., Yoo Y. J., Kim E. J., Kim E., Kim B. G., Sohng J. K., Yoon Y. J., Development of a Streptomyces venezuelae-based combinatorial biosynthetic system for the production of glycosylated derivatives of doxorubicin and its biosynthetic intermediates. Appl. Environ. Microbiol. 77, 4912–4923 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Han A. R., Park S. R., Park J. W., Lee E. Y., Kim D. M., Kim B. G., Yoon Y. J., Biosynthesis of glycosylated derivatives of tylosin in Streptomyces venezuelae. J. Microbiol. Biotechnol. 21, 613–616 (2011). [PubMed] [Google Scholar]
- 30.Han A. R., Shinde P. B., Park J. W., Cho J., Lee S. R., Ban Y. H., Yoo Y. J., Kim E. J., Kim E., Park S. R., Kim B. G., Lee D. G., Yoon Y. J., Engineered biosynthesis of glycosylated derivatives of narbomycin and evaluation of their antibacterial activities. Appl. Microbiol. Biotechnol. 93, 1147–1156 (2012). [DOI] [PubMed] [Google Scholar]
- 31.Hong J. S., Park S. H., Choi C. Y., Sohng J. K., Yoon Y. J., New olivosyl derivatives of methymycin/pikromycin from an engineered strain of Streptomyces venezuelae. FEMS Microbiol. Lett. 238, 391–399 (2004). [DOI] [PubMed] [Google Scholar]
- 32.Blanco G., Patallo E. P., Braña A. F., Trefzer A., Bechthold A., Rohr J., Méndez C., Salas J. A., Identification of a sugar flexible glycosyltransferase from Streptomyces olivaceus, the producer of the antitumor polyketide elloramycin. Chem. Biol. 8, 253–263 (2001). [DOI] [PubMed] [Google Scholar]
- 33.Schell U., Haydock S. F., Kaja A. L., Carletti I., Lill R. E., Read E., Sheehan L. S., Low L., Fernandez M. J., Grolle F., McArthur H. A., Sheridan R. M., Leadlay P. F., Wilkinson B., Gaisser S., Engineered biosynthesis of hybrid macrolide polyketides containing d-angolosamine and d-mycaminose moieties. Org. Biomol. Chem. 6, 3315–3327 (2008). [DOI] [PubMed] [Google Scholar]
- 34.Pérez M., Lombó F., Baig I., Braña A. F., Rohr J., Salas J. A., Méndez C., Combinatorial biosynthesis of antitumor deoxysugar pathways in Streptomyces griseus: Reconstitution of “unnatural natural gene clusters” for the biosynthesis of four 2,6-d-dideoxyhexoses. Appl. Environ. Microbiol. 72, 6644–6652 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Melançon C. E. III, Liu H. W., Engineered biosynthesis of macrolide derivatives bearing the non-natural deoxysugars 4-epi-d-mycaminose and 3-N-monomethylamino-3-deoxy-d-fucose. J. Am. Chem. Soc. 129, 4896–4897 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Melançon C. E. III, Yu W. L., Liu H. W., TDP-mycaminose biosynthetic pathway revised and conversion of desosamine pathway to mycaminose pathway with one gene. J. Am. Chem. Soc. 127, 12240–12241 (2005). [DOI] [PubMed] [Google Scholar]
- 37.Lambalot R. H., Cane D. E., Aparicio J. J., Katz L., Overproduction and characterization of the erythromycin C-12 hydroxylase, EryK. Biochemistry 34, 1858–1866 (1995). [DOI] [PubMed] [Google Scholar]
- 38.Paulus T. J., Tuan J. S., Luebke V. E., Maine G. T., DeWitt J. P., Katz L., Mutation and cloning of eryG, the structural gene for erythromycin O-methyltransferase from Saccharopolyspora erythraea, and expression of eryG in Escherichia coli. J. Bacteriol. 172, 2541–2546 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aguirrezabalaga I., Olano C., Allende N., Rodriguez L., Braña A. F., Méndez C., Salas J. A., Identification and expression of genes involved in biosynthesis of l-oleandrose and its intermediate l-olivose in the oleandomycin producer Streptomyces antibioticus. Antimicrob. Agents Chemother. 44, 1266–1275 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.González A., Remsing L. L., Lombó F., Fernández M. J., Prado L., Braña A. F., Künzel E., Rohr J., Méndez C., Salas J. A., The mtmVUC genes of the mithramycin gene cluster in Streptomyces argillaceus are involved in the biosynthesis of the sugar moieties. Mol. Gen. Genet. 264, 827–835 (2001). [DOI] [PubMed] [Google Scholar]
- 41.Wang G., Kharel M. K., Pahari P., Rohr J., Investigating mithramycin deoxysugar biosynthesis: Enzymatic total synthesis of TDP-d-olivose. Chembiochem 12, 2568–2571 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang G., Pahari P., Kharel M. K., Chen J., Zhu H., Van Lanen S. G., Rohr J., Cooperation of two bifunctional enzymes in the biosynthesis and attachment of deoxysugars of the antitumor antibiotic mithramycin. Angew. Chem. Int. Ed. Engl. 51, 10638–10642 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hoffmeister D., Ichinose K., Domann S., Faust B., Trefzer A., Dräger G., Kirschning A., Fischer C., Künzel E., Bearden D., Rohr J., Bechthold A., The NDP-sugar co-substrate concentration and the enzyme expression level influence the substrate specificity of glycosyltransferases: Cloning and characterization of deoxysugar biosynthetic genes of the urdamycin biosynthetic gene cluster. Chem. Biol. 7, 821–831 (2000). [DOI] [PubMed] [Google Scholar]
- 44.Menéndez N., Nur-e-Alam M., Braña A. F., Rohr J., Salas J. A., Méndez C., Biosynthesis of the antitumor chromomycin A3 in Streptomyces griseus: Analysis of the gene cluster and rational design of novel chromomycin analogs. Chem. Biol. 11, 21–32 (2004). [DOI] [PubMed] [Google Scholar]
- 45.Harvey C. J., Puglisi J. D., Pande V. S., Cane D. E., Khosla C., Precursor directed biosynthesis of an orthogonally functional erythromycin analogue: Selectivity in the ribosome macrolide binding pocket. J. Am. Chem. Soc. 134, 12259–12265 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee H. Y., Harvey C. J., Cane D. E., Khosla C., Improved precursor-directed biosynthesis in E. coli via directed evolution. J. Antibiot. 64, 59–64 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee H. Y., Khosla C., Bioassay-guided evolution of glycosylated macrolide antibiotics in Escherichia coli. PLOS Biol. 5, e45 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tu D., Blaha G., Moore P. B., Steitz T. A., Structures of MLSBK antibiotics bound to mutated large ribosomal subunits provide a structural explanation for resistance. Cell 121, 257–270 (2005). [DOI] [PubMed] [Google Scholar]
- 49.Katz L., Brown D., Boris K., Tuan J., Expression of the macrolide-lincosamide-streptogramin-B-resistance methylase gene, ermE, from Streptomyces erythraeus in Escherichia coli results in N6-monomethylation and N6,N6-dimethylation of ribosomal RNA. Gene 55, 319–325 (1987). [DOI] [PubMed] [Google Scholar]
- 50.Schlünzen F., Harms J. M., Franceschi F., Hansen H. A., Bartels H., Zarivach R., Yonath A., Structural basis for the antibiotic activity of ketolides and azalides. Structure 11, 329–338 (2003). [DOI] [PubMed] [Google Scholar]
- 51.Schlünzen F., Zarivach R., Harms J., Bashan A., Tocilj A., Albrecht R., Yonath A., Franceschi F., Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria. Nature 413, 814–821 (2001). [DOI] [PubMed] [Google Scholar]
- 52.Grushkin D., Natural products emergent. Nat. Med. 19, 390–392 (2013). [DOI] [PubMed] [Google Scholar]
- 53.Jiang M., Fang L., Pfeifer B. A., Improved heterologous erythromycin a production through expression plasmid re-design. Biotechnol. Prog. 29, 862–869 (2013). [DOI] [PubMed] [Google Scholar]
- 54.Zhang H., Boghigian B. A., Pfeifer B. A., Investigating the role of native propionyl-CoA and methylmalonyl-CoA metabolism on heterologous polyketide production in Escherichia coli. Biotechnol. Bioeng. 105, 567–573 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/1/4/e1500077/DC1
Fig. S1. Biosynthetic pathway for erythromycin A emphasizing deoxysugar biosynthesis and attachment and the enzymatic steps catalyzed by EryK and EryG.
Fig. S2. General genetic design used in constructing plasmids and operons.
Fig. S3. Maps of the pGJZ plasmids used to generate erythromycin analogs.
Fig. S4. Deoxysugar pathways associated with first-generation pGJZ plasmids.
Fig. S5. Deoxysugar pathways associated with second-generation pGJZ plasmids.
Fig. S6. SDS-PAGE confirmation of gene expression.
Fig. S7. Compounds associated with MW 689.
Fig. S8. Compounds associated with MW 703.
Fig. S9. Compounds associated with MW 705.
Fig. S10. Compounds associated with MW 717.
Fig. S11. Compounds associated with MW 719.
Fig. S12. Compounds associated with MW 733.
Fig. S13. MW correlation with EryK activity.
Fig. S14. (A and B) Filter disk antibiotic activity assessment of first-generation (A) and second-generation (B) deoxysugar pathway analogs against three B. subtilis strains resistant (+) to chloramphenicol, streptomycin, and erythromycin.
Fig. S15. Common fragmentation patterns in the MS/MS spectra of erythromycin analogs.
Table S1. Production analysis of erythromycin analogs from first (pGJZ1 to pGJZ8) and second (pGJZ11 to pGJZ18) generation deoxysugar pathways.
Table S2. PCR oligonucleotide primers.


