Abstract
Gap junctions play important roles in auditory function and skin biology; mutations in the Cx26 (connexin26) gene are the predominant cause of inherited non-syndromic deafness and cause disfiguring skin disorders. Mass spectrometry (MS) was used to identify PTMs (post-translational modifications) of Cx26 and to determine whether they occur at sites of disease-causing mutations. Cx26 was isolated from transfected HeLa cells by sequential immunoaffinity and metal chelate chromatography using a tandem C-terminal haemagglutinin epitope and a (His-Asn)6 sequence. In-gel and in-solution enzymatic digestions were carried out in parallel with trypsin, chymotrypsin and endoproteinase GluC. Peptides were fractionated using a reversed-phase matrix by stepwise elution with increasing concentrations of organic solvent. To improve detection of low-abundance peptides and to maximize sequence coverage, MALDI–TOF-MS (matrix-assisted laser desorption/ionization–time-of-flight mass spectrometry; MS) and MALDI–TOF/TOF-MS/MS (matrix-assisted laser desorption/ionization-time-of-flight/time-of-flight tandem mass spectrometry; MS/MS) spectra were acquired from each elution step using an Applied Biosystems 4800 tandem mass spectrometer. Acquisition, processing and interpretation parameters were optimized to improve ionization and fragmentation of hydrophobic peptides. MS and MS/MS coverage of Cx26 was significantly above that reported for other membrane proteins: 71.3 %by MS, with 29.9 %by MS/MS. MS coverage was 92.6 % if peptides resulting from in-source collisions and/or partial enzymatic cleavages were considered. A variety of putative PTMs of Cx26 were identified, including acetylation, hydroxylation, γ-carboxyglutamation, methylation and phosphorylation, some of which are at sites of deafness-causing mutations. Knowledge of the PTMs of Cx26 will be instrumental in understanding how alterations in the cellular mechanisms of Cx26 channel biogenesis and function lead to losses in auditory function and disfiguring skin disorders.
Keywords: connexin, deafness, mass spectrometry, mutation, post-translational modification, skin disease
INTRODUCTION
At least four connexin isoforms are expressed in the cochlea. Prominent among those, Cx26 (connexin26) is found in the supporting cells of the organ of Corti, the basal cell region of the stria vascularis and type I fibrocytes of the spiral ligament, which are important components for auditory function [1]. Mutations in the Cx26 gene (GJB2) are the predominant cause of inherited non-syndromic sensorineural deafness in humans [2,3]. Knockout mouse studies have provided insights into the role of Cx26 in auditory function and have demonstrated that Cx26 is essential for cochlear function and survival of the sensory epithelium of the inner ear [4,5]. Although Cx26 mutations that cause complete loss of functional junctional channels lead to non-syndromic deafness, which is principally recessive, mutations that cause altered channel function can lead to dominant deafness syndromes that include severe and disfiguring skin pathologies [6]. These mutations are found in all topological domains of Cx26; connexin proteins have four transmembrane domains (M1–M4), cytoplasmic N-terminal (NT) and C-terminal (CT) domains, one cytoplasmic loop (CL) between M2 and M3, and two extracellular loops (E1, between M1 and M2, and E2, between M3 and M4).
Cx26deaf (mutations of Cx26 that cause non-syndromic or syndromic human deafness) disrupt the intercellular molecular/ionic signalling pathway by affecting connexin channel function, assembly and/or trafficking [7]. In other proteins, these processes can be modulated or controlled by PTMs (post-translational modifications) [8]. With the exception of phosphorylation, predominantly studied in Cx43 (connexin43), modulation of connexin channel properties and cellular dynamics by PTMs have not been studied. Furthermore, the potential role of disrupted PTMs in Cx26deaf pathologies is unexplored. Knowledge of Cx26 PTMs will be instrumental in guiding experiments to understand how cellular mechanisms of connexin channel biogenesis and function can become altered, and how Cx26deaf mutation can lead to losses in auditory function and to skin pathology.
Unfortunately, analysis of PTMs of membrane proteins by mass spectrometry (MS) remains challenging. Sequence coverage of membrane proteins is typically lower than that of soluble proteins, largely because of proteolytic resistance, and poor ionization of hydrophobic peptides makes detection of low-abundance peptides in complex spectra and PTM identification by tandem MS (MS/MS) difficult [9,10].
In the present study, significant methodological improvements were made to existing methods for MALDI–TOF-MS (matrix-assisted laser desorption/ionization–time-of-flight mass spectrometry) and were used to identify Cx26 PTMs. Cx26 was isolated by sequential immunoaffinity and metal chelate chromatography using a pair of tandem C-terminal tags. In-gel and in-solution digestions were carried out in parallel with three enzymes, with peptides recovered with and fractionated from a reversed-phase matrix to improve the recovery of low-abundance peptides in complex digestion mixtures, and thereby maximize sequence coverage. MS acquisition, processing and interpretation parameters were also optimized to improve ionization and MS/MS fragmentation of hydrophobic connexin peptides.
The results reveal a variety of PTMs of Cx26, some of which are at Cx26deaf sites. The findings are interpreted in the context of what is known about the effects of mutations at those sites on connexin channel biogenesis and function.
EXPERIMENTAL
Materials
Components of the Tet-On connexin expression system were from BD Biosciences. DMEM (Dulbecco’s modified Eagle’s medium), G418 sulfate, hygromycin and doxycycline were from Life Technologies. Agarose-conjugated anti-HA (haemagglutinin) clone HA-7 mouse IgG was from Sigma, as were other reagents unless stated. Talon superflow metal (cobalt) affinity resin was from Clontech. The detergent OG (n-octyl-β-D-glucopyranoside; 99.5 %purity) was from Glycon Biochemicals, and DoDM (n-dodecyl-β-D-maltoside; ≥95 % purity) was from EMD Biosciences. Water was LC (liquid chromatography)-MS grade or quartz-distilled reverse-osmosis-purified.
Expression and purification of Cx26 channels
Bidirectional tetracycline-responsive expression vectors (Clontech) were used to express Cx26 channels in HeLa cells, which have virtually no endogenous connexin expression [11]. The rat Cx26 coding sequence was subcloned in-frame with a sequence coding for a 3.3 kDa CT domain ‘tag’, consisting of a thrombin cleavage site followed by an HA epitope and six His-Asn repeats. Tet-On cells were maintained in 200 μg/ml hygromycin and 100 μg/ml G418. Cells were induced for Cx26 expression when approx. 35 % confluent with 1 μg/ml doxycycline for 48 h [12].
Tagged homomeric Cx26 hemichannels were purified from confluent Tet-On HeLa cells (8000–10000 cm2), solubilized in 50 mM NaH2PO4, 50 mM NaCl, 5 mM EDTA, 5 mM EGTA, 1 %(w/v) DoDM, 1 mM 2-mercaptoethanol, 0.5 mg/ml azolectin and 0.5 mM iPr2P-F (di-isopropyl fluorophosphate; Calbiochem), pH 7.4, with added protease and phosphatase inhibitors (cocktails I and II; Sigma), for 2 h at 4 °C with rocking. The supernatant (100 000 gav for 30 min at 4 °C) was incubated with 0.25 ml of agarose-immobilized anti-HA mouse IgG overnight at 4 °C with shaking. The antibody matrix was collected (700 gav for 1 min at 4 °C), and washed in a fritted column with 10 ml of 0.1 M PBS, 1 M NaCl and 0.5 % DoDM, pH 7.4, followed by 15 ml of the same buffer containing 138 mM NaCl. Bound Cx26 was eluted with 50 mM sodium acetate, 0.5 M NaCl, 10 mM KCl and 0.2 % DoDM, pH 4.0, and 0.6 ml fractions were collected into 0.05 ml of 1 M NaHCO3, 10 mM KCl and 0.2 % DoDM, pH 9.0. The final pH was approx. 7.4. Solubilization of gap junctions with the glucoside detergents DoDM and OG yields hemichannels [13].
Elution fractions containing Cx26 (as determined by Western blot analysis with a monoclonal anti-HA antibody) were combined and diluted 5-fold with 0.1 M PBS, pH 7.4, containing 0.2 % DoDM and EDTA-free protease and phosphatase inhibitors (Roche). Cx26 was repurified by immobilized metal affinity chromatography using 0.5 ml of Talon superflow cobalt affinity resin. After incubation for 2 h at 4 °C, the resin was washed in a fritted column with 5 ml of 0.1 M PBS, pH 7.4, 10 mM imidazole and 0.2 % DoDM, then Cx26 was eluted by 0.1 M PBS, pH 7.4, 0.1 M imidazole and 0.2 % DoDM. After 0.1 % TFA (trifluoroacetic acid) was added to the eluate, purified Cx26 was concentrated by centrifugation (3800 gav for 20 min at 25 °C) using an AmiconUltra4 30 kDa cut-off filter (Millipore).
SDS/PAGE, Western blot analysis and gold staining
Cx26 was resolved by SDS/PAGE on 12.5 % (w/v) acrylamide Tris/HCl gels (Bio-Rad Laboratories) under denaturing and reducing conditions. For Western blot analysis, proteins were transferred on to PVDF membranes and stained with specific antibodies to Cx26 (Zymed) or the HA epitope of the CT domain tag. Reactive species were detected with the relevant phosphatase-conjugated secondary antibodies using NBT/BCIP (Nitro Blue Tetrazolium/5-bromo-4-chloroindol-3-yl phosphate; Pierce Endogen). For gold staining, nitrocellulose membranes were used, blocked (0.1 M PBS, pH 7.4, and 0.3 % Tween 20; 30 min for 37 °C), washed and stained with colloidal gold total protein stain (Bio-Rad Laboratories). The blots were washed with quartz-distilled reverse-osmosis-purified water and air-dried.
In-gel digestion
Cx26, resolved by SDS/PAGE, was visualized with E-Zinc stain (Pierce Endogen). The Cx26 band was excised with a sterile scalpel blade and cut into 1 mm3 pieces, washed with 50 % (v/v) acetonitrile, destained according to the manufacturer’s instructions and dehydrated (10 min) in 100 % acetonitrile. Gel pieces were rehydrated (10 min) in 0.1 M NH4HCO3, dehydrated (10 min) in 100 % acetonitrile, and reduced with 0.1 M TCEP [tris-(2-carboxyethyl)phosphine] in 0.1 M NH4HCO3 (30 min at 37 °C). Following ‘washing’ by dehydration–rehydration–dehydration as above, alkylation was performed with 50 mM iodoacetamide in 0.1 M NH4HCO3 [1 h at room temperature (25 °C) in the dark]. Gel pieces were then ‘washed’ as described previously.
Sequencing-grade Tryp (trypsin; 1 μg/μl in 50 mM acetic acid; Stratagene), modified to resist autodigestion, and Chymo (chymotrypsin; 1 μg/μl in 50 mM acetic acid; Roche) were diluted 1/20 with 50 mM NH4HCO3. Gel pieces were rehydrated in a minimal volume of 50 mM NH4HCO3 and digested overnight at 37 °C with 50–100 ng of Tryp or Chymo. Sequencing-grade GluC (endoproteinase GluC; 1 μg/μl in water; Roche) was diluted 1/20 with 50 mM NH4HCO3. Gel pieces were rehydrated in a minimal volume of 50 mM NH4HCO3 and digested overnight at 25°C with 50–100 ng of GluC.
After overnight digestion, Cx26 peptides from each digestion were extracted with 10–20 μl suspensions of 2.5 % (v/v) Poros-20R2 reversed-phase beads (Applied Biosystems) in 5 % (v/v) acetic acid, 0.2 % TFA and 0.1 % OG by vortex-mixing (1000 rev./min at 4 °C) overnight.
Poros-20R2 beads were back-loaded on to micro-C18 or micro-SCX (strong cation exchange) ziptips (Millipore) and were washed according to the manufacturer’s instructions. Cx26 peptides were recovered by step gradient elution: for C18 ziptips, elution conditions were 5, 15, 25, 35, 60 and 80 % (v/v) acetonitrile, followed by 50 % (v/v) methanol/20 % (v/v) acetonitrile (all in 0.1 %TFA); for SCX ziptips, elution conditions were 1, 5 and 10 %(v/v) NH4OH in 30 % (v/v) methanol [13].
Solution volumes were reduced by brief drying under vacuum before each elution fraction was mixed with matrix/standard solution {7 mg/ml α-cyanohydroxycinnamic acid in 50 % (v/v) methanol, 20 % (v/v) acetonitrile and 0.1 % TFA containing 50 fmol/μl each of proteomics grade Glu-fibrinogen peptide [m/z 1570.677 a.m.u. (atomic mass units)] and ACTH18–39 (m/z 2465 a.m.u.) (both from AnaSpec)}. Aliquots (0.5–1.0 μl) were spotted on to a stainless steel MALDI plate and were analysed with a 4800 proteomics analyser tandem mass spectrometer (Applied Biosystems).
In-solution digestion
Purified concentrated Cx26 was reduced with 0.1 M TCEP (for 30 min at 37 °C in the dark), and then immediately alkylated with 50 mM iodoacetamide (1 h at room temperature in the dark), both in 0.1 M NH4HCO3. Cx26 samples (1 vol.) were precipitated by phase separation using 2 vols of ice-cold methanol, and 1 vol. of ice-cold chloroform, with vortex-mixing after each addition. For phase separation, 1.5 vols of ice-cold water were added, the samples were vortex-mixed and centrifuged (10 000 gav for 5 min at 4 °C). The upper phase was discarded and 1.5 vols of cold methanol was added to the lower phase and protein/Cx26 at the interface. The samples were vortex-mixed and centrifuged (14 000 gav for 5 min at 4 °C) to pellet protein/Cx26. Excess solution was removed and the pellet was dried briefly under a vacuum.
Cx26 was resuspended with 10 μl of 50 mM NH4HCO3, 5 % (v/v) acetonitrile and 2 M urea and incubated for 30 min at 37 °C. After dilution to approx. 0.5 M urea with 50 mM NH4HCO3 and 5 % (v/v) acetonitrile, Cx26 was digested overnight with 50–100 ng of Tryp, Chymo (both at 37 °C) or GluC (at 25 °C). Digestions were stopped with an equal volume of 10 % (v/v) methanol, 60 %(v/v) acetonitrile, 1 %(v/v) TFA and 0.1 %OG, and then dried under vacuum. Cx26 peptides were resuspended in 0.2 %TFA buffer compatible for desalting by micro-C18 or micro-SCX ziptips. Alternatively, peptides from each digestion were recovered with 2.5 % (v/v) Poros-20R2 reversed-phase beads, as described above.
MS
Spectra were acquired using an ABI4800 mass spectrometer (Applied Biosystems) in a positive-ion mode with internal mass calibration using the Glu-fibrinogen peptide, ACTH18–39 and Tryp autodigestion products (m/z 842.502 and 2211.096 a.m.u.) if present. MS spectra (m/z 800–3600) were processed by standard methods, including noise removal (2.0 S.D.), baseline correction and de-isotoping. Acquisition, processing and interpretation parameters were optimized further ‘on-the-fly’ to improve ionization and fragmentation of hydrophobic and low-abundance Cx26 peptides. The eight most and least intense ions per MALDI spot, with signal/noise ratios >25, were selected for subsequent MS/MS analysis in both 1 and 2 keV modes. MS/MS spectra were acquired using heavy CID (collision-induced dissociation) and higher gas pressure (1×10−6 compared with 1×10−7 Pa), laser intensity (for MS, 3200–3500; and for MS/MS, 3875 or higher), and 3125–5000 subspectra were collected to increase the possibility of hydrophobic peptide fragmentation and improve peptide signal intensity. Each spectrum was averaged with 3000 laser shots and smoothed with the Savitsky–Golay algorithm [FWHM (full width at half-maximum) = 9, polynomial order = 4].
GPS Explorer v3.5 (Applied Biosystems) was used to process MS and MS/MS spectra to submit the peak lists to the MASCOT v1.9 search engine (http://www.matrixscience.com; Matrix Science) for peptide identification against the MSDB v20060831, SwissProt v57.7 and NCBI (National Center for Biotechnology Information) v20090912 databases. Typically, the total ion score CI (confidence interval) was greater than 95 %. The peptide sequences were additionally matched against a connexin-only database. For MS analyses, the precursor mass error tolerance was set to 20 p.p.m., whereas the MS/MS mass error tolerance was set to 0.3 Da, three missed cleavages were allowed, and carbamidomethyl [CAM-Cys (carboxyami-domethylcysteine)] and oxidation [Oxi-Met (methionine oxidation)] were set as variable modifications.
Other artifactual modifications potentially introduced during sample preparation, such as methylated glutamate, acetylated aspartate or C-terminus and formylated N-terminus, as well as contamination from human keratin, were excluded in peak lists used for database searches (FindPept; http://expasy.org/tools/findpept.html). In separate database searches, to identify non-tryptic and semi-tryptic cleavages in Tryp digestions, the enzyme parameter was set as ‘none’ or ‘semi-Tryp’.
Peak lists were also matched against the FindMod (http://au.expasy.org/tools/findmod/) and/or ProFound (http://prowl.rockefeller.edu) databases as ‘unknowns’ (parameters: rodentia 20–40 kDa, pI 0–14, mass tolerance 50 p.p.m.), and subsequently against the expected fragment masses of digested Cx26 to determine m/z peaks corresponding to modified Cx26 peptides. The Delta mass tables listing PTMs were also consulted (http://prowl.rockefeller.edu/aainfo/deltamassv2.html).
Parameters for individual Cx26 PTM analyses were set up separately in different searches and, in each case, the PTMs were set up as variable modifications. Suspected PTMs of Cx26 were rejected if the modified peptides were not found unmodified in the same digestion, there was lack of recurrence in different digestions, or when the error was greater than 20 p.p.m. Putative Cx26 PTMs in each spectrum were matched further by percentage CI or confirmed manually by peak assignment using Data Explorer v4.6 (Applied Biosystems).
RESULTS
Identification of peptides from Cx26 by MALDI–TOF/TOF-MS/MS
Enzymatic digestion and MS analysis of soluble proteins generally yields 40–60 % sequence coverage [9,10]. Sequence coverage of membrane proteins is typically lower due to the hydrophobicity of transmembrane domains, which are resistant to proteolysis, and because the resulting hydrophobic peptides are difficult to extract from in-gel digestions, do not ionize well or are difficult to solubilize in buffers compatible with MS [9]. To address these problems, several preparative and analytic methods were applied to optimize Cx26 sequence coverage.
To maximize sequence coverage, in-gel and in-solution enzymatic digestions of Cx26 were carried out in parallel with Tryp, GluC and Chymo. After stepwise elution of peptides from the reversed-phase bead matrix, both MS and MS/MS spectra were acquired for peptides in each elution fraction. The matched m/z lists of Cx26 enzyme digestions are shown in Table 1. MS sequence coverage was 62.6 % for Tryp (Table 1A), 7.9 % for GluC (Table 1B) and 29.9 % for Chymo (Table 1C). Total MS sequence coverage was 71.3 %, with 29.9 %of the Cx26 sequence confirmed by MS/MS sequencing (Figure 1), values significantly higher than typically observed in membrane protein analysis [10].
Table 1. Sequence coverage of Cx26.
Sequence coverage of Cx26 purified from HeLa cells by Tryp digestion was 62.6% (A), by GluC digestion was 7.9% (B) and by Chymo digestion was 29.9% (C). Overall MS sequence coverage was 71.3%, with 29.9% sequence confirmed by MS/MS (✓). The cytoplasmic, membrane and extracellular domain boundaries in all of the Tables follow those described by Maeda et al. [15]. The modified residue is in bold, and the specific modification and amino acid position from the N-terminus of the peptide are given. CAM-Cys is the result of alkylation with 2-iodoacetamide.
| (A)
| ||||||
|---|---|---|---|---|---|---|
| Confirmed by MS/MS | Calculated m/z | Observed m/z | Δ(p.p.m.) | Missed cuts | Peptide, observed modification | Domain |
| ✓ | 1618.8257 | 1618.8197 | − 4 | 0 | M1DWGTLQSILGGVNK15 | NT |
| ✓ | 1634.8207 | 1634.8186 | − 1 | 0 | M1DWGTLQSILGGVNK15, Oxi-Met at position 1 | NT |
| 2353.0222 | 2353.0203 | − 1 | 0 | E42VWGDEQADFVCNTLQPGCK61, 2× CAM-Cys at positions 12 and 19 | E1 | |
| ✓ | 1820.8536 | 1820.8643 | 6 | 0 | N62VCYDHYFPISHIR75, CAM-Cys at position 3 | E1/M2 |
| 2596.4507 | 2596.4451 | − 2 | 0 | L76WALQLIMVSTPALLVAMHVAYR98 | M2 | |
| ✓ | 1691.8856 | 1691.8792 | − 3 | 2 | G109EIKNEFKDIEEIK122 | CL |
| 2049.0862 | 2049.0811 | − 2 | 3 | G109EIKNEFKDIEEIKTQK125 | CL | |
| ✓ | 1264.6425 | 1264.6384 | − 3 | 1 | N113EFKDIEEIK122 | CL |
| ✓ | 1621.8431 | 1621.8359 | − 4 | 2 | N113EFKDIEEIKTQK125 | CL |
| 2863.3333 | 2863.2498 | − 29 | 0 | V144IFEAVFMYVFYIMYNGFFMQR165, 3× Oxi-Met at positions 8,14 and 20 | M3/E2 | |
| 2452.0842 | 2452.0789 | − 2 | 0 | C169NAWPCPNTVDCFISRPTEK188, 3× CAM-Cys at position 1, 6 and 12 | E2 | |
| ✓ | 1051.6735 | 1051.7043 | 29 | 1 | S222KRPVLVPR230 | CT/Tag |
| ✓ | 836.5464 | 836.5432 | − 4 | 0 | R224PVLVPR230 | CT/Tag |
| ✓ | 2866.2214 | 2866.2271 | 2 | 0 | G231SYPYDVDPYALHNHNHNHNHNHN254 | Tag |
| (B)
| ||||||
|---|---|---|---|---|---|---|
| Confirmed by MS/MS | Calculated m/z | Observed m/z | Δ (p.p.m.) | Missed cuts | Peptide, modification | Domain |
| 1167.6666 | 1167.6754 | 8 | 0 | K102KRKFMKGE110, Oxi-Met at position 6 | CL | |
| 1635.9363 | 1635.9386 | 1 | 1 | K102KRKFMKGEIKNE114 | CL | |
| 1651.9312 | 1651.925 | − 4 | 1 | K102KRKFMKGEIKNE114, Oxi-Met at position 6 | CL | |
| 1114.6942 | 1114.6963 | 2 | 0 | I121KTQKVRIE129 | CL | |
| (C)
| ||||||
|---|---|---|---|---|---|---|
| Confirmed by MS/MS | Calculated m/z | Observed m/z | Δ (p.p.m.) | Missed cuts | Peptide, modification | Domain |
| 2877.4637 | 2877.4177 | − 16 | 3 | R32IMILVVAAKEVWGDEQADFVCNTL56, CAM-Cys at position 22 | M1/E1 | |
| 1233.5714 | 1233.5955 | 20 | 1 | C53NTLQPGCKNV63, 1× CAM-Cys at position 1 or 8 | E1 | |
| 2447.1746 | 2447.1919 | 7 | 2 | Q57PGCKNVCYDHYFPISHIRL76, 1× CAM-Cys at position 4 or 8 | E1/M2 | |
| 1859.0544 | 1859.0139 | − 22 | 0 | R98RHEKKRKFMKGEI111, Oxi-Met at position 10 | CL | |
| 2543.1451 | 2543.1438 | − 1 | 3 | F162MQRLVKCNAWPCPNTVDCF181, 3× CAM-Cys at positions 8, 13 and 19 | E2 | |
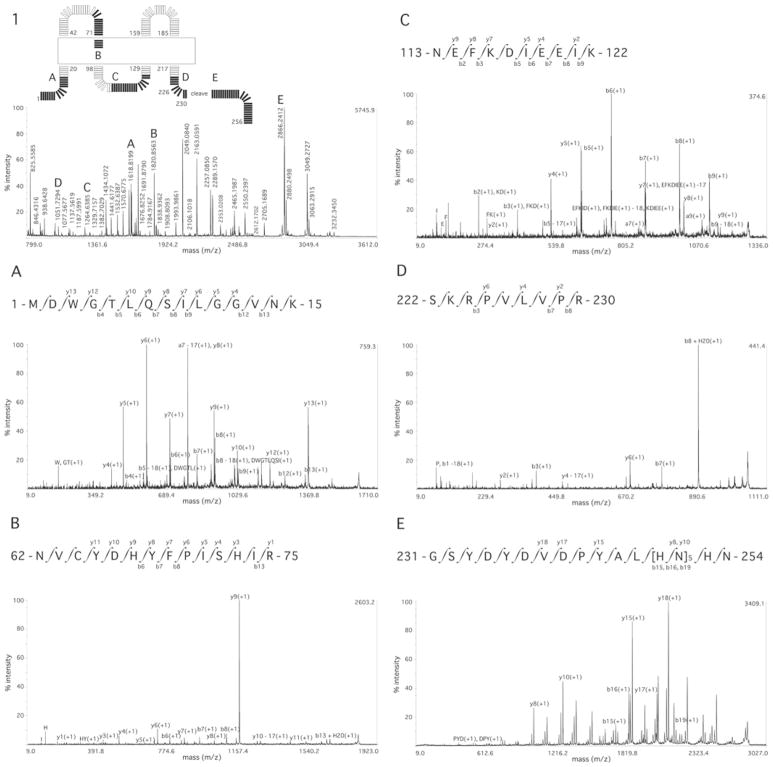
Figure 1. MS and MS/MS sequence coverage of Cx26.
(1) MS spectrum of Cx26 Tryp digestion, with letters A–E indicating the peptides for which MS/MS spectra are subsequently shown. Inset, the exact location of these peptides in a schematic diagram of Cx26 showing the cytoplasmic, membrane (boxed) and extracellular domain boundaries as described by Maeda et al. [15]. (A) NT domain; (B) E1; (C) CL; (D) CT domain; (E) CT domain purification tag. Labelling of b and y ions in MS/MS spectra was according to Biemann et al. [14].
Figure 1 (panel 1) shows the MS spectra of one elution fraction from a Tryp digestion (showing approx. 32 %sequence coverage; peaks with m/z of approx. 1691, 2049 and 2353 a.m.u. are also from Cx26), and selected MS/MS spectra of peptides from each of the Cx26 domains: the NT domain (Figure 1A), E1 (Figure 1B), CL (Figure 1C), CT domain (Figure 1D) and the CT domain purification tag (Figure 1E).
Sequences not covered by MS analyses were only from the transmembrane domains: a region of M1 proximal to the NT domain, a region of M3 proximal to CL and almost all of M4.
Non-tryptic and semi-tryptic cleavages of peptides
To increase the possibility of hydrophobic peptide fragmentation and improve peptide ion signal intensities, MS and MS/MS spectra were acquired, as needed, using CID, higher gas pressure (1×10−6 Pa) and laser intensity (>3875). Such changes can lead to better secondary fragmentation of peptides by in-source collisions. Additionally, in tryptic digestions some cleavages may not occur specifically at arginine or lysine residues due to residual chymotryptic activity, result from pseudotrypsin activity (Tryp autodigestion), partial enzymatic cleavage (i.e. one end of the peptide arises from non-specific enzymatic cleavage) or can occur by degradation during protein purification itself. Thus a number of other peptides were observed (Table 2), including those ‘missing’ from M1, M3 and M4, which were not predicted by an in silico digestion of Cx26. These were very low abundance in the spectra and not amenable to analysis that could reveal PTMs. However, if included, MS sequence coverage would rise to 92.6 % of Cx26.
Table 2. Peptides from non-tryptic and semi-tryptic cleavages of Cx26.
Format designating modification and abbreviations are as in the legend to Table 1.
| Calculated m/z | Observed m/z | Δ(p.p.m.) | Peptide, modification | Domain |
|---|---|---|---|---|
| 1054.6118 | 1054.6056 | − 6 | V27LFIFRIM34, Oxi-Met at position 8 | M1 |
| 1136.5470 | 1136.5457 | − 1 | N113EFKDIEEI121 | CL |
| 1168.5747 | 1168.5813 | 6 | A148VFMYVFYI156, Oxi-Met at position 4 | M3 |
| 1433.6300 | 1433.6211 | − 6 | L56QPGCKNVCYDH67, 1× CAM-Cys at position 5 or 9 | E1 |
| 1440.6543 | 1440.6287 | − 18 | P234YDVDPYALHNH245 | Tag |
| 1452.7151 | 1452.7059 | − 6 | T177VDCFISRPTEK188, CAM-Cys at position 4 | E2 |
| 1490.7308 | 1490.7432 | 8 | M1DWGTLQSILGGVN14 | NT |
| 1552.6882 | 1552.6636 | 16 | D50FVCNTLQPGCKN62, 2× CAM-Cys at positions 4 and 11 | E1 |
| 1638.8639 | 1638.8557 | − 5 | K125VRIEGSLWWTYT137 | CL/M3 |
| 1650.8019 | 1650.8164 | 9 | N206ITELCYLFIRYC218 | M4/CT |
| 1656.8137 | 1656.8007 | − 8 | G160FFMQRLVKCNAW172, CAM-Cys at position 10 | E2 |
| 1666.808 | 1666.8086 | 0 | L210CYLFIRYCSGKS222, 2× CAM-Cys at positions 2 and 9 | M4/CT |
| 1676.8036 | 1676.8248 | 12 | Y158NGFFMQRLVKCN170, CAM-Cys at position 12 | E2 |
| 1697.8760 | 1697.8760 | 0 | I140FFRVIFEAVFMY152, Oxi-Met at position 12 | M3 |
| 1784.9080 | 1784.9152 | 4 | S139IFFRVIFEAVFMY152, Oxi-Me at position 13 | M3 |
| 1993.9921 | 1993.9855 | − 3 | F141FRVIFEAVFMYVFY− 155, Oxi-Met at position 11 | M3 |
| 2111.9094 | 2111.9385 | 14 | F51VCNTLQPGCKNVCYDH− 67, 3× CAM-Cys at positions 3,10 and 14 | E1 |
| 2185.0139 | 2185.0464 | 15 | F161FMQRLVKCNAWPCPNT177, 2× CAM-Cys at positions 9 and 14, and Oxi-Met at position 3 | E2 |
| 2364.0178 | 2364.03 | 5 | G231SYPYDVDPYALHNHNHNHN250 | Tag |
| 2367.0745 | 2367.064 | − 4 | K41EVWGDEQADFVCNTLQPGCK61 | E1 |
| 2435.0916 | 2435.0681 | − 10 | M157YNGFFMQRLVKCNAWPCP175, 2× CAM-Cys at positions 13 and 18, and 1× Oxi-Met at position 1 or 7 | E2 |
| 2447.1746 | 2447.1985 | 10 | L56QPGCKNVCYDHYFPISHIR75, 1× CAM-Cys at position 5 or 9 | E1/M2 |
| 2447.1746 | 2447.1985 | 10 | Q57PGCKNVCYDHYFPISHIRL76, 1× CAM-Cys at position 4 or 8 | E1/M2 |
| 2451.1228 | 2451.1394 | 7 | I156MYNGFFMQRLVKCNAWPC174, 2× CAM-Cys at positions 14 and 19, and 1× Oxi-Met at position 2 or 8 | E2 |
| 2501.2202 | 2501.2551 | 14 | P173CPNTVDCFISRPTEKTVFTVF194 | E2/M4 |
| 2510.1626 | 2510.1379 | − 10 | K168CNAWPCPNTVDCFISRPTEKT189 | E2 |
| 2550.1938 | 2550.2397 | 18 | L166VKCNAWPCPNTVDCFISRPTE187, 1× CAM-Cys at position 4, 9 or 15 | E2 |
| 2566.2866 | 2566.292 | 2 | C202ILLNITELCYLFIRYCSGKS222, 2× CAM-Cys at two of positions 1, 10 or 17 | M4/CT |
| 2566.3018 | 2566.292 | − 4 | I128EGSLWWTYTTSIFFRVIFEA148 | M3 |
| 2615.1196 | 2615.1445 | 10 | G231SYPYDVDPYALHNHNHNHNHN252 | Tag |
| 2705.1582 | 2705.1624 | 2 | A49DFVCNTLQPGCKNVCYDHYFP70, 3× CAM-Cys at positions 5, 12 and 16 | E1 |
| 2894.2703 | 2894.2478 | − 8 | M157YNGFFMQRLVKCNAWPCPNTVDC180, 1× CAM-Cys at position 13, 18 or 24 | E2 |
| 3264.5427 | 3264.5261 | − 5 | I145FEAVFMYVFYIMYNGFFMQRLVKC169, CAM-Cys at position 25, and 3× Oxi-Met at positions 7, 13 and 19 | M3/E2 |
| 3227.4392 | 3227.468 | 9 | Y155IMYNGFFMQRLVKCNAWPCPNTVDC180, 2× CAM-Cys at two of positions 15, 20 or 26 | E2 |
PTMs of Cx26
The PTMs of Cx26 identified by MS and by MS/MS analysis are listed in Table 3. All MS/MS sequence assignments that passed acceptance criteria after PTM database searches were manually verified, as most algorithms used for automated peptide identification are potentially less reliable in identifying spectra when the precursor ion is in the + 1 charge state characteristic of MALDI. The peaks in each MS/MS spectrum were assigned as b and y ions [14], resulting, respectively, from cleavage of amide bonds from the N-terminal or C-terminal of the peptide. Where identification was by MS only, suspected PTMs were rejected if the modified peptides were not found unmodified in the same digestion, there was a lack of recurrence of observation in different digestions or the error was >20 p.p.m. (see the Experimental section). Several putative PTMs of Cx26 were identified, including acetylation, hydroxylation, γ-carboxyglutamation, methylation and phosphorylation.
Table 3. PTMs of Cx26.
Suspected PTMs were rejected if (i) the modified peptides were not found unmodified in the same digestion, (ii) there was insufficient recurrence of observation in different digestions, (iii) the error was >20 p.p.m., or (iv) the amino acids involved were cysteine residues that were CAM-Cys-modified. Formats designating modifications are as in the legend to Table 1. Acetyl-Lys, acetylation of lysine; Acetyl-Met, acetylation of methionine; GGLU-Glu, glutamate γ-carboxylation; Hydroxyl-Asn, hydroxylation of asparagine; Methyl-Lys, methylation of lysine; Methyl-Argo, methylation of arginine; Phospho-Tyr, tyrosine phosphorylation; Phospho-Thr, threonine phosphorylation. Information is also shown schematically in Figure 4 (and its legend).
| Enzyme | Confirmed by MS/MS | Calculated m/z | Observed m/z | Δ (p.p.m.) | Missed cuts | Peptide, modification | Domain |
|---|---|---|---|---|---|---|---|
| Tryp | 1650.8156 | 1650.8195 | 2 | 0 | M1DWGTLQSILGGVNK15, Oxi-Met at position 1, and Hydroxyl-Asn at position 14 | NT | |
| Tryp | ✓ | 1660.8363 | 1660.8247 | − 7 | 0 | M1DWGTLQSILGGVNK15, Acetyl-Met at position 1 | NT |
| Tryp | 1676.8312 | 1676.8179 | − 8 | 0 | M1DWGTLQSILGGVNK15, Oxi-Met at position 1, and Acetyl-Lys at position 15 | NT | |
| Tryp | 2367.0381 | 2367.064 | 11 | 0 | E42VWGDEQADFVCNTLQPGCK61, 2× CAM-Cys at positions 12 and 19, and Methyl-Lys at position 20 | E1 | |
| Tryp | 2383.9805 | 2383.9521 | − 12 | 0 | E42VWGDEQADFVCNTLQPGCK61, 2× CAM-Cys at positions 12 and 19, and 2× GGLU-Glu at positions 1 and 6 | E1 | |
| Tryp | 1777.8479 | 1777.8655 | 10 | 0 | N62VCYDHYFPISHIR75, Methyl-Arg at position 14 | E1/M2 | |
| Tryp | ✓ | 1834.8694 | 1834.8735 | 2 | 0 | N62VCYDHYFPISHIR75, CAM-Cys at position 3 and Methyl-Arg at position 14 | E1/M2 |
| Chymo | 1602.8976 | 1602.8877 | − 6 | 1 | A96YRRHEKKRKF106, 2× Acetyl-Lys at two of positions 7, 8 or 10 | M2/CL | |
| GluC | 2252.1743 | 2252.1506 | − 11 | 2 | K102KRKFMKGEIKNEFKD− 117, Oxi-Met at position 6, and 5× Acetyl-Lys at five of positions 1, 2, 4, 7, 11 or 15 | CL | |
| Chymo | 909.4676 | 909.4496 | − 20 | 0 | K112NEFKDI118, Hydroxy-Asn at position 2 | CL | |
| Chymo | 937.4625 | 937.4501 | − 13 | 1 | K112NEFKDI118, GGLU-Glu at position 3 | CL | |
| GluC | 1565.8298 | 1565.8145 | − 10 | 2 | I118EEIKTQKVRIE129, Phospho-Thr at position 6 | CL | |
| Tryp | 2694.2837 | 2694.2505 | − 12 | 2 | L166VKCNAWPCPNTVDCFISRPTEK188, 1× CAM-Cys at position 4, 9 or 15, and 1× Hydroxy-Asn at position 5 or 11 | E2 | |
| Tryp | 3032.2307 | 3032.2522 | 7 | 1 | L166VKCNAWPCPNTVDCFISRPTEK188, 3× CAM-Cys at positions 4, 9 and 15, and 3× Phospho-Thr at positions 12, 18 and 21 | E2 | |
| Tryp | 843.4028 | 843.3899 | − 15 | 1 | Y217CSGKSK223, CAM-Cys at position 2, and 1× Methyl-Lys at position 5 or 7 | CT | |
| Tryp | ✓ | 2880.2375 | 2880.2437 | 2 | 0 | G231SYPYDVDPYALHNHNHNHNHNHN254, 1× Methyl-Asn at position 22 | Tag |
| Tryp | 2946.1877 | 2946.1924 | 2 | 0 | G231SYPYDVDPYALHNHNHNHNHNHN254, 1× Phospho-Tyr at position 3, 5 or 10 | Tag |
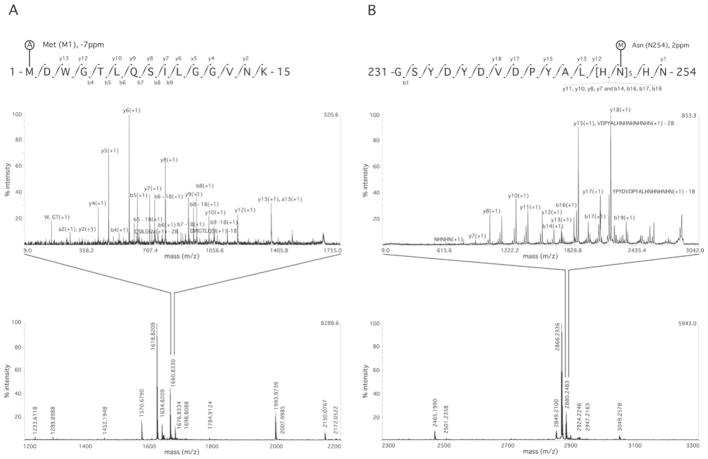
Figure 2 shows two MS/MS spectra identifying PTMs. These are the acetylation of the N-terminal methionine (Met1) (Figure 2A) and methylation of asparagine at position 254 (Asp254) (Figure 2B). These PTMs correspond to mass additions of 42.01 and 14.01 a.m.u. respectively, to the amino acids.
Figure 2. MS and MS/MS spectra showing PTMs of Cx26.
All modified peptides were also found in their unmodified state in the same digestions. (A) MS spectrum of the Cx26 Tryp digestion (lower panel) and MS/MS spectrum of the NT domain showing N-terminal acetylation (A) of methionine (position Met1). (B) MS spectrum of Cx26 Tryp digestion (lower panel) and MS/MS spectrum of the CT domain purification tag showing N-terminal methylation (M) of asparagine (position Asp254). See Table 1 for full details of the sequence coverage, and Table 3 for other PTMs. Labelling of b and y ions in MS/MS spectra was according to Biemann et al. [14].
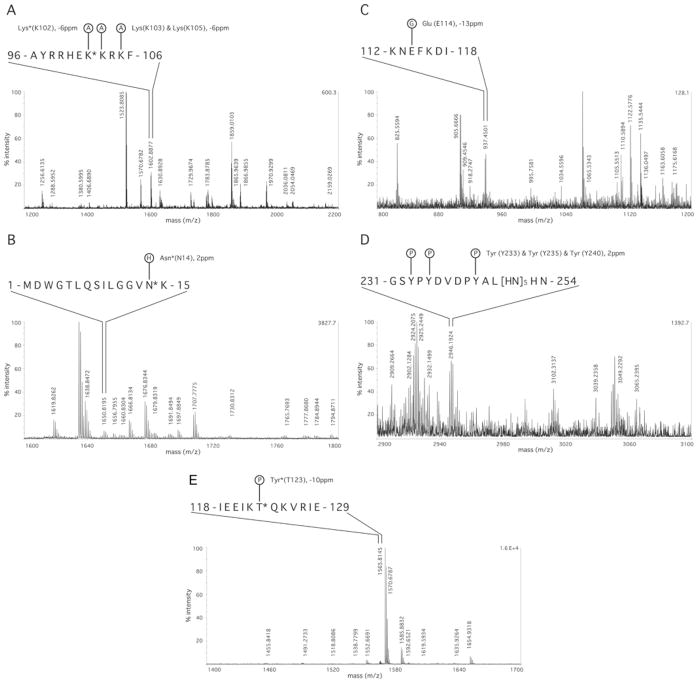
Many more PTMs of Cx26 were indicated by MS spectra only. Their acceptance was subject to the stringent criteria listed above. These PTMs include multiple acetylations in the NT and CL domains, hydroxylation in the NT, CL and E2 domains, γ-carboxylation of glutamate in the E1 and CL domains, methylation in the E1, M2 and CT domains, and phosphorylation in the CL and E2 domains, as well as PTMs (phosphorylation and methylation) in the CT domain purification tag. Representative MS spectra for these PTMs are shown in Figure 3.
Figure 3. MS spectra suggesting PTMs of Cx26 for which high-quality MS/MS spectra were not obtained.
Representative MS spectra for the indicated modifications. See the Experimental section and the legend to Table 3 for acceptance criteria. (A) Acetylation (A) in the CL proximal to the second transmembrane domain. (B) hydroxylation (H) in the NT domain proximal to the first transmembrane domain. (C) glutamate γ-carboxylation (G) in the central part of the CL. (D) phosphorylation (P) of the CT domain purification tag. (E) phosphorylation (P) in the central part of the CL. Amino acids marked with asterisks (*) are sites of Cx26deaf mutation. See Table 1 for details of sequence coverage and Table 3 for other PTMs.
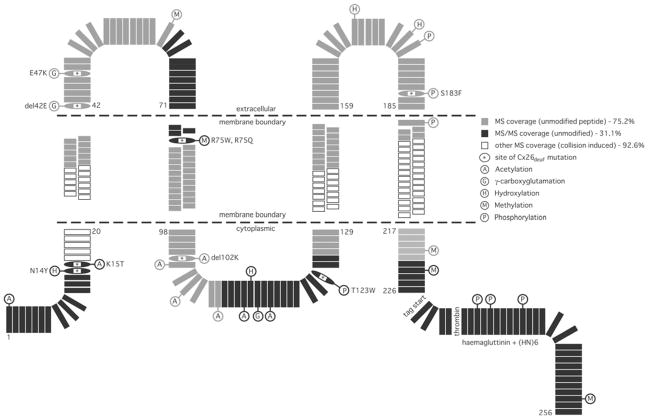
All of the PTMs identified by MS or MS/MS are presented schematically in Figure 4. To determine whether the PTMs identified were at sites of Cx26deaf, databases for mutations (http://davinci.crg.es/deafness and www.ncbi.nlm.nih.gov/omim) were consulted. A significant fraction of the PTMs identified by MS and MS/MS are at Cx26deaf sites (Table 4). These were hydroxylation at Asn14, γ-carboxylation at Glu42 and Glu47, methylation at Arg75, acetylation at Lys15 and Lys102, and phosphorylation at Thr123 and Ser183.
Figure 4. Schematic representation of the identified PTMs of Cx26.
Each amino acid of Cx26, and of the tandem CT domain epitopes used for protein purification, is shown individually. Filled black rectangles indicate regions for which MS/MS confirmation was achieved, with sequences for which only MS data were available is shown by filled grey rectangles. If rectangles are not filled (i.e. white), then no sequence information/peptides were obtained by MALDI–TOF (see Tables 1 and 2 for peptide lists and the text for discussion of their absence). The PTMs (see Table 3 and 4, and their legends) are identified using the following abbreviations: acetylation (A), hydroxylation (H), γ-carboxyglutamation (G), methylation (M) and phosphorylation (P). Sites of Cx26deaf disease-causing mutation, where PTMs were identified, are designated by single-letter amino acid code, are oval-shaped and marked inset by an asterisk (*). The positions of the cytoplasmic, membrane (stippled lines) and extracellular domain boundaries are as described by Maeda et al. [15].
Table 4. PTMs at sites of disease-causing mutations.
This Table summarizes information regarding the disease-causing mutations of Cx26 that are at sites of PTMs identified in the present study. Abbreviations: n, non-syndromic; s, syndromic; r, recessive; d, dominant. Information is also shown schematically (see Figure 4 and its legend).
| Mutation (type) | Domain | Mechanism (if known) | Observed PTM |
|---|---|---|---|
| Asn14Tyr (s, d*) and Asn14Lys (s, d†) | NT | Asn14Tyr: Keratitis-Ichthyosis-Deafness syndrome*, forms functional junctional channels, reduced junctional conductance, change to local structural flexibility of the NT domain [30]; Asn14Lys: Clouston syndrome†, caused open hemichannels and loss of voltage-dependence [51,52]. | Hydroxylation |
| Lys15Thr (n, r) | NT | No information on functional effect [53]. | Acetylation |
| del-Glu42 (s, d) | E1 | Forms junctional plaques but not functional junctional channels, behaves as dominant-negative with Cx26, Cx30 and Cx43 [35,36]. | γ-Carboxyglutamation |
| Glu47Lys (n, r) | E1 | Normal trafficking, but does not form functional hemichannels or junctional channels [37]. | γ-Carboxyglutamation |
| Arg75Gln (n, d) and Arg75Trp (s, d‡) | E1 | Forms junctional plaques in some reports, not in others, but no functional junctional channels, forms hemichannels with altered voltage-dependence and reduced permeability, behaves as dominant-negative with Cx26, Cx30 and Cx43 [42,54,55]. (Arg75Trp for Palmoplantar keratoderma† [55]). | Methylation |
| del-Lys102 (n, r) | CL | No information on functional effects§ [56]. | Acetylation |
| Thr123Asn (n, d§) | CL | No information on functional effects [57,58]. | Phosphorylation |
| Ser183Phe (s, d‡) | E2 | Behaves as dominant-negative with Cx26, impaired formation of junctional channels with those forming being Lucifer yellow permeable [48]. | Phosphorylation |
DISCUSSION
PTMs of proteins are diverse and play important modulatory roles in trafficking, biogenesis and function [8]. In the present study, MALDI–TOF/TOF-MS was employed to identify PTMs of Cx26 expressed in HeLa cells. Protocols for sample preparation for MS and MS/MS analysis were optimized to improve sequence coverage, ionization and fragmentation of hydrophobic regions of the protein, since they present difficulties for MS analysis. These refinements included the use of highly pure connexin, parallel in-gel and in-solution digestion by three proteases with distinct cleavage specificities, peptide fractionation with a reversed-phase matrix, and adjustment of the MS/MS and collision parameters to increase fragmentation. This resulted in 71.3 % confirmed sequence coverage by MS and 29.9 % confirmed by MS/MS sequencing. Low-abundance peptides, not amenable to MS/MS analysis and arising from other sources, such as in-source collisions, partial enzymatic cleavage, pseudotrypsin or chymotryptic activity in tryptic digestions, were observed. If these are included, MS sequence coverage was 92.6 %. The results indicate a variety of PTMs of Cx26 distributed over the protein sequence, several of which are at sites that when mutated in Cx26 (and other connexins) give rise to human pathologies.
Analysis of Cx26 PTMs using MALDI-MS/MS
According to the recent crystal structure of Cx26 [15], approx. 47 % of the sequence is in transmembrane regions. This high proportion of hydrophobic sequence presents difficulties for digestion by proteases and subsequent peptide analysis by MS, as indicated by the absence in Table 1 of peptides from short regions of M1 and M3, and the entire M4 domain. The two transmembrane helices assigned in the crystal as forming the outer wall of the pore (i.e. regions exposed to lipid for almost all of their lengths that are within the membrane) are part of M3 and principally M4, whereas M1, the principle pore-lining helix, is atypically hydrophobic [using ProtParam (http://au.expasy.org/tools/protparam.html), the average hydropathy of M1 is 1.85 compared with 1.20, 1.05 and 1.31 for M2, M3 and M4 respectively]; M1 does contain amino acids involved in hydrophobic intra-protomer interactions.
MALDI–TOF-MS typically has a detection limit of low femtomole to high attomole, but for membrane protein and peptides with hydrophobic character, the detection limit is much higher and signal intensity (peptide ionization) is usually much lower than for water-soluble peptides. This may be a consequence of membrane proteins having a greater tendency than soluble proteins to remain folded in the gas phase; additionally, MALDI–TOF/TOF-MS/MS sequencing using CID depends on protonation of amide groups, and folded hydrophobic regions are poorly ionized. With our optimized MALDI parameters, the detection sensitivity was improved but was still insufficient to detect low-abundance PTMs, resulting in incomplete coverage by MS/MS sequencing.
In theory, ESI-LC-QTOF-MS [electrospray-coupled LC with QTOF (quadrupole TOF)-MS] would provide improved and/or complementary ionization for MS/MS sequencing of peptides with hydrophobic character, since peptides can carry multiple charges, rather than single charges (here, + 1) as in MALDI–TOF-MS. Furthermore, this technology might allow more sensitive detection of, in particular, sites of phosphorylation of Cx26. The dominant neutral loss of phosphoric acid from phosphorylated peptides when using MALDI and a TOF analyser in reflector mode (‘post-source decay’) typically results in insufficient fragmentation of such phosphopeptides during MALDI–TOF/TOF-MS/MS. However, even by ESI-LC-QTOF-MS there may still be some element of uncertainty for assignment, especially where there is more than one potential location in the peptide for PTM. In this case, confirmation requires MS3 of product–phosphoric acid ions using an ‘ion trap’, which is not possible with the MALDI–TOF/TOF-MS employed in the present study. However, ESI-LC-QTOF-MS suffers from other technical challenges, notably being less tolerant than MALDI–TOF-MS to non-peptide contaminants such as salt, and especially lipid or detergent that may be likely to remain bound post-digestion to hydrophobic peptides.
One previous investigation of connexin PTMs by proteomic methods (Cx44 and Cx49) utilized HPLC for desalting, isolation and concentration of peptides, a custom-built MALDI-QTOF system, custom peptide identification software, two proteases and in-solution digestion, but achieved substantially less MS and MS/MS sequence coverage than in the present study, and allowed a greater margin of error for assigning PTMs [16]. Aside from N-terminal acetylation, several sites of phosphorylation were found in the CL and long CT domains of these connexins. The sites were identified, as outlined above, by neutral loss of phosphoric acid from MS/MS fragment ions. Peptides were not recovered from the transmembrane and extracellular domains, the proximal portions of the CT domain, or from substantial portions of the NT domain (Cx44) or the CL domain (Cx49).
The greater sequence coverage in Cx26 MS and MS/MS sequencing in the present study can be attributed to the various technical improvements in MALDI–TOF-MS and MALDI–TOF/TOF-MS/MS spectra acquisition described above and to the purity of the starting material (in the study of Cx44 and Cx49, only 10–18 out of 600–900 MS peaks were from the connexins). The purification of Cx26 employed in the present study used tandem CT domain tags and yielded a protein of high purity. This contributes to greater sensitivity in peptide and PTM detection, since non-connexin/contaminating peptides absorb ‘energy’ and suppress ionization of peptides of interest. Previous work using less optimized purifications and MS running conditions identified only a small number of PTMs of Cx26, which were also found in the present study (hydroxylation at Asn14 and γ-carboxylation at Glu114 [13]).
PTMs of Cx26
Several different PTMs of Cx26 were identified, including acetylation, hydroxylation, γ-carboxyglutamation, methylation and phosphorylation. The PTMs confirmed by MALDI–TOF/ TOF-MS/MS sequencing can be viewed with high confidence. The others were identified by mass consistency and accuracy of expected peptide fragments. The limitations of using single m/z peaks to identify PTMs are acknowledged; however, strict criteria were applied to minimize the chance of erroneous PTM identification (see the Experimental section). In particular, assignment of the PTMs was made using criteria that each was supported by the observation of re-occurrence of the modified peptide in different digestions along with identification of the unmodified peptide in the same digestion. The PTMs identified from MS by these criteria are highly likely, but not certain, to be correct. For simplicity, below, PTMs may be referred to as ‘identified’, although this is not, in the strictest sense, true for those resulting from MS analysis alone.
Acetylation of Cx26
Acetylation was identified at the N-terminal methionine (Met1, by MS/MS), elsewhere in the NT domain and at several lysine residues in the CT domain (Lys15, two among Lys102, Lys103 and Lys105, and all of Lys108, Lys112 and Lys116; by MS). N-terminal acetylation is a common irreversible cotranslational modification. Proteins with the Met1-Asp2 sequence, such as Cx26, undergo an obligatory N-terminal acetylation. This can affect a variety of factors, most prominently protein stability due to the blocked N-terminus, but also protein function and subcellular targeting [17].
Acetylation of the N-terminus eliminates its positive charge. In connexins, the NT domain folds into the aqueous pore to act as a voltage sensor and gate [15,18–20]. Thus charge elimination at the N-terminus may have a profound impact on channel gating. It was reported that the N-terminal methionine residue in Cx44 and Cx49 is endogenously removed and the newly exposed N-terminal glycine residue becomes acetylated [16], so N-terminal acetylation may be a widespread connexin PTM.
Unlike N-terminal acetylation, acetylation of internal lysine residues is post-translational and reversible. An internal lysine acetylation in the NT domain (Lys15) was identified, as were several other lysine residue acetylations in a short segment of the CL domain proximal to M2. Internal lysine residue acetylation can regulate diverse functions, including protein–protein interaction and protein stability [21]. Lysine residue acetylation can also affect other PTMs, e.g. lysine residues are targets for ubiquitination and acetylation can block this modification [22].
According to the Cx26 crystal structure, Lys15 is at the outer edge of the cytoplasmic entrance/vestibule of the channel pore, so charge removal by acetylation could affect charge selectivity to permeants. Perhaps more important, Lys15 is near the ‘hinge’ region at which the NT domain turns into the pore to form the voltage sensor/gate [15,18], so a structural or charge modification at this site could affect voltage gating as well.
Lys15Thr is a Cx26deaf mutation. The acetylation and this mutation both remove the positive charge. In Cx32, this position is an arginine residue, and the Arg15Gln mutation, which also eliminates positive charge, causes a peripheral neuropathy. Interestingly, Arg15Gln channels are functional, but have altered voltage sensitivity [23,24], consistent with the structural inferences. There is no functional information about Cx26deaf Lys15Thr channel function, but, extrapolating from Cx32 Arg15Gln, one could propose that acetylation/charge removal at this site in Cx26 has a similar effect on channel voltage sensitivity, which results in non-syndromic deafness.
Several internal lysine residues are potentially acetylated in the Cx26 CL near M2. The connexin CL is involved in gating by pH and related intramolecular interactions [25,26], so dynamic modulation of the charge of this domain by multiple acetylations could modulate these processes, as occurs in other proteins [22].
In light of this, it is notable that deletion of Lys102 is a Cx26deaf mutation. Acetylation or del-Lys102 eliminates a negative charge at this position. There is no functional information about Cx26deaf del-Lys102; however, in the CL of Cx32, Glu102Gly eliminates the positive charge, as would acetylation, alters pH sensitivity and causes a mild neuropathy [27]. Thus it appears that a lack of charge at this position has a pathological effect on pH gating for both connexins.
Hydroxylation of Cx26
Hydroxylation was identified at Asn14 in the NT domain, at Asn113 in the CL, and at either Asn170 or Asn176 in E2. Protein hydroxylation is an irreversible modification that occurs at aromatic residues in a wide variety of proteins, but is less common at asparagine residues. Hydroxylated asparagine is found in several proteins, such as hypoxia-inducible transcription factors, epidermal growth factor and vitamin K clotting factors, where they are associated with high-affinity Ca2+-binding sites [28]. Cx26 does not contain the putative consensus sequence for hydroxylation by 2-oxoglutarate-dependent dioxygenases, but it presumes a characteristic β-fold [29].
There are two Cx26deaf mutations at Asn14: Asn14Tyr and Asn14Lys. Asn14 is at the ‘hinge’ region that is, as noted above, where the NT domain folds into the pore [15,18], so its modification would be expected to affect (voltage) gating. Indeed, the Asn14Tyr mutation alters local peptide flexibility [30] and leads to reduced junctional conductance, consistent with altered gating, whereas the introduction of positive charge by Asn14Lys mutation generates open hemichannels, as well as functional gap junction channels, but they lack voltage-dependence. Asn14Tyr does not introduce a charge, so could be considered the mutation that better approximates the lack of hydroxylation.
Asn176 is one of the residues in the Cx26 crystal structure suggested to be directly involved in the docking interaction between apposed hemichannels [15]. The Asn175Asp mutation in Cx32 (corresponding to Asn176 in Cx26) causes a peripheral neuropathy, but the effect on channel function has not been characterized [31].
Glutamate γ-carboxylation of Cx26
Glutamate γ-carboxylation was identified at Glu42 and Glu47 in E1, and at Glu114 in the CL. This irreversible modification generates high-affinity Ca2+-binding sites; addition of the negatively charged carboxy group forms a Ca2+-chelation site with the existing glutamate side-chain carboxy group [8,32]. In this context, the position of these modifications in E1 is intriguing as, according to the crystal structure of Cx26 [15], they are in a 310 helix involved in Ca2+-sensitive ‘loop gating’ of connexin hemichannels, i.e. the gate that keeps unapposed hemichannels closed [33,34].
Intriguingly, both Glu42 and Glu47 are sites of Cx26deaf mutations. Deletion of Glu42 or Glu47Lys allows formation of junctional channels, but they are non-functional [35–37], perhaps indicating a loop gate malfunction, e.g. remaining closed in spite of hemichannel docking to form junctional channels. Other studies have identified Glu42 as a major determinant of the charge selectivity of the channel [38]. The Cx26deaf Glu47Lys mutation eliminates the possibility of γ-carboxyglutamation, but imposes the same charge that γ-carboxyglutamation would, suggesting that it is the PTM itself and not the charge alteration itself that is the key factor for normal channel function.
Glutamate γ-carboxylation was identified in the CL, which contains a high density of glutamate resides (four between residues 114 and 120), although only one was detected so modified in the present study. As γ-carboxylated glutamate residues can mediate strong Ca2+-dependent interactions with membranes [32], these could mediate Cx26 sensitivity to cytosolic Ca2+ (proposed previously for Cx26 [13]). The Glu114Gly mutation apparently has no dramatic effect on dye permeability (pH sensitivity untested) [39]. Introduction of an increased negative charge into the CL may also modulate the various gating processes and related intramolecular interactions involving it, as mentioned above.
Methylation of Cx26
Methylation was identified at Lys61 in E1, at Arg75 at the E1/M2 border (by MS/MS) and at either Lys221 or Lys223 in the CT domain. Methylation at arginine and lysine residues is relatively widespread, irreversible and implicated in a wide spectrum of cellular processes, including protein stability, activity and protein–protein interactions [40]. In most cases, its primary effect follows from increased side-chain bulk and hydrophobicity.
The most intriguing methylation identified is at Arg75, a site of two Cx26deaf mutations that inhibit junctional coupling in Cx26 as well as Cx32 [41,42]. Arg75Trp, unlike Arg75Gln, forms somewhat functional hemichannels [43]. These mutations both eliminate the positive charge, as well as the possibility of methylation, so the consequences of methylation itself cannot be inferred. However, it should be noted that Arg75 is at the E1/M2 border (i.e. the aqueous–lipid interface) and so its increased side-chain bulk and/or hydrophobicity after methylation may influence the positioning of M2 and E1 relative to the membrane.
In Cx32, the site that corresponds to Cx26 Lys221 is Arg220, which conserves the charge and the potential for methylation. The Arg220Gly mutation in Cx32 forms junctional plaques and causes neuropathy, but the function of the junctional channels has not been assessed [41].
Phosphorylation of Cx26
Phosphorylation was identified at Thr123 in the CL, at Thr177 in E2, at Ser183 and Thr186 near the E2/M4 border, and at one of Tyr233, Tyr235 or Tyr240 in the epitope tag. To date, Cx26 has not been definitively reported to be a phosphoprotein. Our present findings indicate potential phosphorylation in the CL, a domain in which at least two other connexins (Cx36 and Cx56) are phosphorylated [44,45]. In contrast with CT domain phosphorylation, the effects of CL phosphorylation are underexplored. Thr123 is a possible Cx26deaf mutation, but there is no information on the functional effects of mutation at this site.
Several phosphorylation sites are indicated in E2. Such modifications could occur by intracellular kinase activity, but could also be introduced by ecto-kinases found on many cells, including HeLa cells, which modulate other membrane proteins [46, 47]. According to the Cx26 crystal structure [15], one of the suspected sites in E2 (Thr177) forms a docking interaction with the apposed hemichannel.
Interestingly, the Cx26deaf Ser183Phe mutation significantly reduces the number of junctional channels formed [48], but those formed retain normal dye permeability. It is therefore unknown if the primary defect is in trafficking, junctional formation or greater internalization of undocked hemichannels. The corresponding Ser182Thr mutation in Cx32 that causes neuropathy also results in non-functional junctional channels [24]. Given the biological reversibility of phosphorylation, this might imply a role for it in regulating hemichannel docking at this site in both connexins.
Phosphorylation (and methylation; by MS/MS) was also indicated in the HA epitope, which may be relevant to evaluation of the function or cell biology of connexins carrying this tag.
Cx26 PTMs and deafness mutations
Mutations in the Cx26 gene (GJB2) account for over 50 % of autosomal recessive non-syndromic congenital deafness, the most common form of genetic deafness, and 10–30 % of sporadic cases. More than 50 mutations in the coding region of GJB2 have been reported to be associated with autosomal-recessive (DFNB1) and autosomal-dominant (DFNA3) forms of hearing impairment [2,3].
The mechanism(s) by which deafness is caused by mutations that delete or alter the function of Cx26 is unknown. It has been proposed that disruption of intercellular K+ recycling in the cochlea is involved [5], leading to reversed operation of a glutamate transporter and accumulation of extracellular glutamate, which may inhibit synthesis of glutathione, causing death from oxidative stress [4].
Previous evidence has suggested that disruption of intercellular molecular signalling, rather than K+ recycling, is the important factor. Some Cx26deaf mutations produce channels with unaltered K+ permeability, but with disrupted molecular permeability [49,50]. In addition, when Cx26 is genetically deleted, the remaining Cx30 junctional channels, found in the same locations, are highly permeable to K+ [4]. Thus changes in the character, degree or regulation of connexin channel molecular permeability, as opposed to ionic conductance, appear to be sufficient to cause deafness and/or skin pathology.
The location of PTMs identified in the present study at which there are known deafness-causing mutations are indicated in Figure 4. Information about the functional effects of mutation at these sites is summarized in Table 4. Of the eight Cx26deaf sites at locations of PTMs, five are dominant mutations and four have syndromic phenotypes.
At this point it is difficult to make mechanistic links between the presence of specific PTMs normally at these sites and disruptions of normal Cx26 function when these sites have Cx26deaf mutations. Each Cx26deaf mutation alters the sequence, which can have its own effects, in addition to eliminating the (proper) PTM. It is unknown whether the detected PTMs are introduced as part of normal channel maturation, degradation or modulatory processes. However, if a particular PTM is part of a degradatory process and does not occur due to mutation at the site, one would expect the result to be increased channel function rather than decreased or compromised function. Such an effect is not observed, so one can presume that the observed irreversible PTMs at Cx26deaf sites are to promote proper channel maturation and function, rather than proper degradation. Of the observed PTMs, only phosphorylation and acetylation are reversible, so one cannot infer a priori whether their presence or absence favours channel function.
It is notable that most of the identified PTMs, and all of those at Cx26deaf locations (with the possible exception of Ser183, noted above), are in regions known or strongly suspected from biophysical work and/or the recent Cx26 crystal structure to be crucial for proper channel function: Asn14 and Lys15 are involved in voltage gating, Glu42 and Glu47 are involved in loop gating and Arg75 interacts directly with residues in M1, the pore-lining helix. Although there is little functional information about specific PTM sites in the CL loop (Lys102, Asn113 and Glu114), it is recognized that the CL is a primary site of connexin channel functional modulation. One would anticipate that in most cases lack of proper PTMs would substantially affect trafficking, but the available evidence does not indicate this; many other Cx26deaf mutations do affect trafficking or channel assembly, but these were not identified in the present study as sites of PTM. Of the five PTM sites that are also sites of Cx26deaf mutation for which there are relevant data, all form junctional channels and, of those, three form functional junctional channels. Thus a case can be made that disease-causing disruption of PTMs are more likely to affect mature channel function than biogenesis.
Several of the disease mutations that do form functional channels are at sites at which PTMs have been identified. These are particularly interesting targets for future investigation.
Conclusions
The overall high sequence coverage of Cx26 achieved using MALDI–TOF and MS/MS made it possible to identify several new sites for PTM of Cx26. Many of the sites of PTM are at sites of Cx26 disease-causing mutation. Mutation at these sites could preclude these PTMs from playing important roles in connexin channel biogenesis and/or function. Such PTMs are promising targets to study the regulatory functions of connexin channel gating and permeation properties.
Acknowledgments
We thank Jade Liu (UMDNJ) for technical support.
FUNDING
This work was supported by the National Institutes of Health [grant numbers NS046593 (NeuroProteomics Core Grant) (to H. L.), and DC7470, NS56509 and GM36044 (to A. L. H.). The funding agency had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Abbreviations used
- a.m.u
atomic mass units
- CAM-Cys
carboxyamidomethylcysteine
- Chymo
chymotrypsin
- CI
confidence interval
- CID
collision-induced dissociation
- CL
cytoplasmic loop
- CT
C-terminal domain
- Cx26
connexin26
- Cx26deaf
mutations of Cx26 that cause non-syndromic or syndromic human deafness
- DoDM
n-dodecyl-β-D-maltoside
- E1 (etc.)
extracellular loop 1 domain (etc.)
- ESI-LC-QTOF
electrospray ionization liquid chromatography coupled with quadrupole time-of-flight
- GluC
endoproteinase GluC
- HA
haemagglutinin
- LC
liquid chromatography
- M1 (etc.)
transmembrane 1 domain (etc.)
- MALDI
matrix-assisted laser desorption/ionization
- MS
mass spectrometry
- MS/MS
tandem MS
- NT
N-terminal domain
- OG
n-octyl-β-D-glucopyranoside
- Oxi-Met
methionine oxidation
- PTM
post-translational modification
- QTOF
quadrupole time-of-flight
- SCX
strong cation exchange
- TCEP
tris-(2-carboxyethyl)phosphine
- TFA
trifluoroacetic acid
- TOF
time-of-flight
- Tryp
trypsin
Footnotes
AUTHOR CONTRIBUTION
Darren Locke conceived and designed the experiments; Shengjie Bian and Darren Locke performed the experiments; Shengjie Bian, Andrew Harris and Darren Locke analysed the data; Hong Li contributed reagents/materials/analysis tools; Andrew Harris and Darren Locke wrote the paper.
References
- 1.Forge A, Becker D, Casalotti S, Edwards J, Marziano N, Nevill G. Gap junctions in the inner ear: comparison of distribution patterns in different vertebrates and assessement of connexin composition in mammals. J Comp Neurol. 2003;467:207–231. doi: 10.1002/cne.10916. [DOI] [PubMed] [Google Scholar]
- 2.Kelsell DP, Dunlop J, Stevens HP, Lench NJ, Liang JN, Parry G, Mueller RF, Leigh IM. Connexin 26 mutations in hereditary non-syndromic sensorineural deafness. Nature. 1997;387:80–83. doi: 10.1038/387080a0. [DOI] [PubMed] [Google Scholar]
- 3.Petersen MB. Non-syndromic autosomal-dominant deafness. Clin Genet. 2002;62:1–13. doi: 10.1034/j.1399-0004.2002.620101.x. [DOI] [PubMed] [Google Scholar]
- 4.Cohen-Salmon M, Ott T, Michel V, Hardelin JP, Perfettini I, Eybalin M, Wu T, Marcus DC, Wangemann P, Willecke K, Petit C. Targeted ablation of connexin26 in the inner ear epithelial gap junction network causes hearing impairment and cell death. Curr Biol. 2002;12:1106–1111. doi: 10.1016/s0960-9822(02)00904-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kudo T, Kure S, Ikeda K, Xia AP, Katori Y, Suzuki M, Kojima K, Ichinohe A, Suzuki Y, Aoki Y, et al. Transgenic expression of a dominant-negative connexin26 causes degeneration of the organ of Corti and non-syndromic deafness. Hum Mol Genet. 2003;12:995–1004. doi: 10.1093/hmg/ddg116. [DOI] [PubMed] [Google Scholar]
- 6.Gerido DA, DeRosa AM, Richard G, White TW. Aberrant hemichannel properties of Cx26 mutations causing skin disease and deafness. Am J Physiol Cell Physiol. 2007;293:C337–C345. doi: 10.1152/ajpcell.00626.2006. [DOI] [PubMed] [Google Scholar]
- 7.Martinez AD, Acuña R, Figueroa V, Maripillan J, Nicholson B. Gap-junction channels dysfunction in deafness and hearing loss. Antioxid Redox Signaling. 2009;11:309–322. doi: 10.1089/ars.2008.2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walsh CT, Garneau-Tsodikova S, Gatto GJ. Protein posttranslational modifications: the chemistry of proteome diversifications. Angew Chem. 2005;44:7342–7372. doi: 10.1002/anie.200501023. [DOI] [PubMed] [Google Scholar]
- 9.Wu CC, MacCoss MJ, Howell KE, Yates JR. A method for the comprehensive proteomic analysis of membrane proteins. Nat Biotechnol. 2003;21:532–538. doi: 10.1038/nbt819. [DOI] [PubMed] [Google Scholar]
- 10.Wu CC, Yates JR. The application of mass spectrometry to membrane proteomics. Nat Biotechnol. 2003;21:262–267. doi: 10.1038/nbt0303-262. [DOI] [PubMed] [Google Scholar]
- 11.Mesnil M, Krutovskikh V, Piccoli C, Elfgang C, Traub O, Willecke K, Yamasaki H. Negative growth control of HeLa cells by connexin genes: connexin species specificity. Cancer Res. 1995;55:629–639. [PubMed] [Google Scholar]
- 12.Koreen IV, Elsayed WA, Liu YJ, Harris AL. Tetracycline-regulated expression enables purification and functional analysis of recombinant connexin channels from mammalian cells. Biochem J. 2004;383:111–119. doi: 10.1042/BJ20040806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Locke D, Koreen IV, Harris AL. Isoelectric points and post-translational modifications of connexin26 and connexin32. FASEB J. 2006;20:1221–1223. doi: 10.1096/fj.05-5309fje. [DOI] [PubMed] [Google Scholar]
- 14.Biemann K. Contributions of mass spectrometry to peptide and protein structure. Biomed Environ Mass Spectrom. 1988;16:99–111. doi: 10.1002/bms.1200160119. [DOI] [PubMed] [Google Scholar]
- 15.Maeda S, Nakagawa S, Suga M, Yamashita E, Oshima A, Fujiyoshi Y, Tsukihara T. Structure of the connexin 26 gap junction channel at 3.5Å resolution. Nature. 2009;458:597–602. doi: 10.1038/nature07869. [DOI] [PubMed] [Google Scholar]
- 16.Shearer D, Ens W, Standing K, Valdimarsson G. Posttranslational modifications in lens fiber connexins identified by off-line-HPLC MALDI-quadrupole time-of-flight mass spectrometry. Invest Ophthalmol Visual Sci. 2008;49:1553–1562. doi: 10.1167/iovs.07-1193. [DOI] [PubMed] [Google Scholar]
- 17.Meinnel T, Peynot P, Giglione C. Processed N-termini of mature proteins in higher eukaryotes and their major contribution to dynamic proteomics. Biochimie. 2005;87:701–712. doi: 10.1016/j.biochi.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 18.Purnick PE, Benjamin DC, Verselis VK, Bargiello TA, Dowd TL. Structure of the amino terminus of a gap junction protein. Arch Biochem Biophys. 2000;381:181–190. doi: 10.1006/abbi.2000.1989. [DOI] [PubMed] [Google Scholar]
- 19.Oh S, Rivkin S, Tang Q, Verselis VK, Bargiello TA. Determinants of gating polarity of a connexin 32 hemichannel. Biophys J. 2004;87:912–928. doi: 10.1529/biophysj.103.038448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oshima A, Tani K, Hiroaki Y, Fujiyoshi Y, Sosinsky GE. Three-dimensional structure of a human connexin26 gap junction channel reveals a plug in the vestibule. Proc Natl Acad Sci USA. 2007;104:10034–10039. doi: 10.1073/pnas.0703704104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kouzarides T. Acetylation: a regulatory modification to rival phosphorylation? EMBO J. 2000;19:1176–1179. doi: 10.1093/emboj/19.6.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang XJ, Seto E. Lysine acetylation: codified crosstalk with other posttranslational modifications. Mol Cell. 2008;31:449–461. doi: 10.1016/j.molcel.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abrams CK, Freidin MM, Verselis VK, Bennett MV, Bargiello TA. Functional alterations in gap junction channels formed by mutant forms of connexin 32: evidence for loss of function as a pathogenic mechanism in the X-linked form of Charcot-Marie-Tooth disease. Brain Res. 2001;900:9–25. doi: 10.1016/s0006-8993(00)03327-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang HL, Chang WT, Yeh TH, Wu T, Chen MS, Wu CY. Functional analysis of connexin-32 mutants associated with X-linked dominant Charcot-Marie-Tooth disease. Neurobiol Dis. 2004;15:361–370. doi: 10.1016/j.nbd.2003.11.005. [DOI] [PubMed] [Google Scholar]
- 25.Manthey D, Banach K, Desplantez T, Lee CG, Kozak CA, Traub O, Weingart R, Willecke K. Intracellular domains of mouse connexin26 and -30 affect diffusional and electrical properties of gap junction channels. J Membr Biol. 2001;181:137–148. doi: 10.1007/s00232-001-0017-1. [DOI] [PubMed] [Google Scholar]
- 26.Shibayama J, Lewandowski R, Kieken F, Coombs W, Shah S, Sorgen PL, Taffet SM, Delmar M. Identification of a novel peptide that interferes with the chemical regulation of connexin43. Circ Res. 2006;98:1365–1372. doi: 10.1161/01.RES.0000225911.24228.9c. [DOI] [PubMed] [Google Scholar]
- 27.Abrams CK, Freidin M, Bukauskas F, Dobrenis K, Bargiello TA, Verselis VK, Bennett MV, Chen L, Sahenk Z. Pathogenesis of X-linked Charcot-Marie-Tooth disease: differential effects of two mutations in connexin 32. J Neurosci. 2003;23:10548–10558. doi: 10.1523/JNEUROSCI.23-33-10548.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Semenza GL. Hypoxia-inducible factor 1 (HIF-1) pathway. Sci Stke. 2007:cm8. doi: 10.1126/stke.4072007cm8. [DOI] [PubMed] [Google Scholar]
- 29.Coleman ML, McDonough MA, Hewitson KS, Coles C, Mecinovic J, Edelmann M, Cook KM, Cockman ME, Lancaster DE, Kessler BM, et al. Asparaginyl hydroxylation of the Notch ankyrin repeat domain by factor inhibiting hypoxia-inducible factor. J Biol Chem. 2007;282:24027–24038. doi: 10.1074/jbc.M704102200. [DOI] [PubMed] [Google Scholar]
- 30.Arita K, Akiyama M, Aizawa T, Umetsu Y, Segawa I, Goto M, Sawamura D, Demura M, Kawano K, Shimizu H. A novel N14Y mutation in Connexin26 in keratitis-ichthyosis-deafness syndrome: analyses of altered gap junctional communication and molecular structure of N terminus of mutated connexin26. Am J Pathol. 2006;169:416–423. doi: 10.2353/ajpath.2006.051242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Silander K, Meretoja P, Juvonen V, Ignatius J, Pihko H, Saarinen A, Wallden T, Herrgård E, Aula P, Savontaus ML. Spectrum of mutations in Finnish patients with Charcot-Marie-Tooth disease and related neuropathies. Hum Mutat. 1998;12:59–68. doi: 10.1002/(SICI)1098-1004(1998)12:1<59::AID-HUMU9>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 32.Nelsestuen GL, Shah AM, Harvey SB. Vitamin K-dependent proteins. Vitam Horm. 2000;58:355–389. doi: 10.1016/s0083-6729(00)58031-5. [DOI] [PubMed] [Google Scholar]
- 33.Verselis VK, Trelles MP, Rubinos C, Bargiello TA, Srinivas M. Loop gating of connexin hemichannels involves movement of pore-lining residues in the first extracellular loop domain. J Biol Chem. 2009;284:4484–4493. doi: 10.1074/jbc.M807430200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tang Q, Dowd TL, Verselis VK, Bargiello TA. Conformational changes in a pore-forming region underlie voltage-dependent ‘loop gating’ of an unapposed connexin hemichannel. J Gen Physiol. 2009;133:555–570. doi: 10.1085/jgp.200910207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thomas T, Aasen T, Hodgins M, Laird DW. Transport and function of Cx26 mutants involved in skin and deafness disorders. Cell Commun Adhes. 2003;10:353–358. doi: 10.1080/cac.10.4-6.353.358. [DOI] [PubMed] [Google Scholar]
- 36.Rouan F. Trans-dominant inhibition of connexin-43 by mutant connexin-26: implications for dominant connexin disorders affecting epidermal differentiation. J Cell Sci. 2001;114:2105–2113. doi: 10.1242/jcs.114.11.2105. [DOI] [PubMed] [Google Scholar]
- 37.Stong BC, Chang Q, Ahmad S, Lin X. A novel mechanism for connexin 26 mutation linked deafness: cell death caused by leaky gap junction hemichannels. Laryngoscope. 2006;116:2205–2210. doi: 10.1097/01.mlg.0000241944.77192.d2. [DOI] [PubMed] [Google Scholar]
- 38.Oh S, Rubin JB, Bennett MV, Verselis VK, Bargiello TA. Molecular determinants of electrical rectification of single channel conductance in gap junctions formed by connexins 26 and 32. J Gen Physiol. 1999;114:339–364. doi: 10.1085/jgp.114.3.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Choung YH, Moon SK, Park HJ. Functional study of GJB2 in hereditary hearing loss. Laryngoscope. 2002;112:1667–1671. doi: 10.1097/00005537-200209000-00026. [DOI] [PubMed] [Google Scholar]
- 40.Lee YH, Stallcup MR. Protein arginine methylation of nonhistone proteins in transcriptional regulation. Mol Endocrinol. 2009;23:425–433. doi: 10.1210/me.2008-0380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yum SW, Kleopa KA, Shumas S, Scherer SS. Diverse trafficking abnormalities of connexin32 mutants causing CMTX. Neurobiol Dis. 2002;11:43–52. doi: 10.1006/nbdi.2002.0545. [DOI] [PubMed] [Google Scholar]
- 42.Deng Y, Chen Y, Reuss L, Altenberg GA. Mutations of connexin 26 at position 75 and dominant deafness: essential role of arginine for the generation of functional gap-junctional channels. Hear Res. 2006;220:87–94. doi: 10.1016/j.heares.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 43.Chen Y, Deng Y, Bao X, Reuss L, Altenberg GA. Mechanism of the defect in gap-junctional communication by expression of a connexin 26 mutant associated with dominant deafness. FASEB J. 2005;19:1516–1518. doi: 10.1096/fj.04-3491fje. [DOI] [PubMed] [Google Scholar]
- 44.Berthoud VM, Beyer EC, Kurata WE, Lau AF, Lampe PD. The gap-junction protein connexin 56 is phosphorylated in the intracellular loop and the carboxy-terminal region. Eur J Biochem. 1997;244:89–97. doi: 10.1111/j.1432-1033.1997.00089.x. [DOI] [PubMed] [Google Scholar]
- 45.Urschel S, Höher T, Schubert T, Alev C, Söhl G, Wörsdörfer P, Asahara T, Dermietzel R, Weiler R, Willecke K. Protein kinase A-mediated phosphorylation of connexin36 in mouse retina results in decreased gap junctional communication between AII amacrine cells. J Biol Chem. 2006;281:33163–33171. doi: 10.1074/jbc.M606396200. [DOI] [PubMed] [Google Scholar]
- 46.Redegeld FA, Caldwell CC, Sitkovsky MV. Ecto-protein kinases: ecto-domain phosphorylation as a novel target for pharmacological manipulation? Trends Pharm Sci. 1999;20:453–459. doi: 10.1016/s0165-6147(99)01399-1. [DOI] [PubMed] [Google Scholar]
- 47.Wirkner K, Stanchev D, Köles L, Klebingat M, Dihazi H, Flehmig G, Vial C, Evans RJ, Fürst S, Mager PP, et al. Regulation of human recombinant P2 × 3 receptors by ecto-protein kinase C. J Neurosci. 2005;25:7734–7742. doi: 10.1523/JNEUROSCI.2028-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.de Zwart-Storm EA, van Geel M, van Neer PA, Steijlen PM, Martin PE, van Steensel MA. A novel missense mutation in the second extracellular domain of GJB2, p. Ser183Phe, causes a syndrome of focal palmoplantar keratoderma with deafness. Am J Pathol. 2008;173:1113–1119. doi: 10.2353/ajpath.2008.080049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Beltramello M, Piazza V, Bukauskas FF, Pozzan T, Mammano F. Impaired permeability to Ins(1,4,5)P3 in a mutant connexin underlies recessive hereditary deafness. Nat Cell Biol. 2005;7:63–69. doi: 10.1038/ncb1205. [DOI] [PubMed] [Google Scholar]
- 50.Zhang Y, Tang W, Ahmad S, Sipp JA, Chen P, Lin X. Gap junction-mediated intercellular biochemical coupling in cochlear supporting cells is required for normal cochlear functions. Proc Natl Acad Sci USA. 2005;102:15201–15206. doi: 10.1073/pnas.0501859102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van Steensel MA, Steijlen PM, Bladergroen RS, Hoefsloot EH, van Ravenswaaij-Arts CM, van Geel M. A phenotype resembling the Clouston syndrome with deafness is associated with a novel missense GJB2 mutation. J Invest Dermatol. 2004;123:291–293. doi: 10.1111/j.0022-202X.2004.23204.x. [DOI] [PubMed] [Google Scholar]
- 52.Lee JR, Derosa AM, White TW. Connexin mutations causing skin disease and deafness increase hemichannel activity and cell death when expressed in Xenopus oocytes. J Invest Dermatol. 2009;129:870–878. doi: 10.1038/jid.2008.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lim LH, Bradshaw JK, Guo Y, Pilipenko V, Madden C, Ingala D, Keddache M, Choo DI, Wenstrup R, Greinwald JH. Genotypic and phenotypic correlations of DFNB1-related hearing impairment in the Midwestern United States. Arch Otolaryngol Head Neck Surg. 2003;129:836–840. doi: 10.1001/archotol.129.8.836. [DOI] [PubMed] [Google Scholar]
- 54.Marziano NK, Casalotti SO, Portelli AE, Becker DL, Forge A. Mutations in the gene for connexin 26 (GJB2) that cause hearing loss have a dominant negative effect on connexin 30. Hum Mol Genet. 2003;12:805–812. doi: 10.1093/hmg/ddg076. [DOI] [PubMed] [Google Scholar]
- 55.Richard G, White TW, Smith LE, Bailey RA, Compton JG, Paul DL, Bale SJ. Functional defects of Cx26 resulting from a heterozygous missense mutation in a family with dominant deaf-mutism and palmoplantar keratoderma. Hum Genet. 1998;103:393–399. doi: 10.1007/s004390050839. [DOI] [PubMed] [Google Scholar]
- 56.Mueller RF, Nehammer A, Middleton A, Houseman M, Taylor GR, Bitner-Glindzciz M, Van Camp G, Parker M, Young ID, Davis A, et al. Congenital non-syndromal sensorineural hearing impairment due to connexin 26 gene mutations: molecular and audiological findings. Int J Ped Otorhinolaryngol. 1999;50:3–13. doi: 10.1016/s0165-5876(99)00242-6. [DOI] [PubMed] [Google Scholar]
- 57.Park HJ, Hahn SH, Chun YM, Park K, Kim HN. Connexin26 mutations associated with nonsyndromic hearing loss. Laryngoscope. 2000;110:1535–1538. doi: 10.1097/00005537-200009000-00023. [DOI] [PubMed] [Google Scholar]
- 58.Snoeckx RL, Djelantik B, Van Laer L, Van de Heyning P, Van Camp G. GJB2 (connexin 26) mutations are not a major cause of hearing loss in the Indonesian population. Am J Med Genet A. 2005;135:126–129. doi: 10.1002/ajmg.a.30726. [DOI] [PubMed] [Google Scholar]