Abstract
Matrix metalloproteinase-13 (MMP-13) plays a critical role in parathyroid hormone (PTH)-induced bone resorption. PTH acts via protein kinase A (PKA) to phosphorylate and stimulate the transactivation of Runx2 for MMP-13 promoter activation. We show here that PTH stimulated Runx2 phosphorylation in rat osteoblastic cells. Runx2 was phosphorylated on serine 28 and threonine 340 after 8-bromo cyclic adenosine mono phosphate (8-Br-cAMP) treatment. We further demonstrate that in the presence of 8-Br-cAMP, the wild-type Runx2 construct stimulated MMP-13 promoter activity, while the Runx2 construct having mutations at three phosphorylation sites (S28, S347 and T340) was unable to stimulate MMP-13 promoter activity. Thus, we have identified the Runx2 phosphorylation sites necessary for PKA stimulated MMP-13 promoter activation and this event may be critical for bone remodeling.
Keywords: Runx2, Matrix metalloproteinase-13, Collagenase-3, Cyclic adenosine mono phosphate, Parathyroid hormone
1. Introduction
Parathyroid hormone (PTH) is an essential regulator of calcium homeostasis. The hormone has multiple actions, including indirect activation of the osteoclast through osteoblastic production of RANKL [1] resulting in increased bone resorption, as well as many direct changes in the functions of the osteoblast. Besides the stimulation of the production of RANKL, bone resorbing hormones such as PTH increase the production of proteases such as matrix metalloproteinase-13 (MMP-13; collagenase-3) which appear to be required for bone and cartilage matrix degradation and the later action of osteoclasts [2–4]. MMP-13 appears to play a critical role in PTH-induced bone resorption and calcemic responses and endochondral bone formation [5,6].
In the rat osteoblastic cell line, UMR 106-01, PTH induces MMP-13 gene transcription through a protein kinase A (PKA)-dependent pathway requiring de novo protein synthesis [7,8]. Investigation of the regulatory region of the MMP-13 gene identified the PTH-response elements as being the runt domain (RD at −132/−126) and the activator protein-1 (AP-1 at −48/−42) binding sites in the MMP-13 promoter. We have demonstrated a PTH-dependent cooperative interaction between the sites and the proteins (Runx2/Fos/Jun) binding to them [9]. However, there was no significant change in the abundance of Runx2 protein binding to the RD site and there was also no change in the levels of Runx2 mRNA or protein in UMR cells after PTH treatment [8]. Using a Gal4 fusion construct of the activation domain 3 (AD3) of Runx2, and in vitro reactions with PKA, we have shown that PTH acts via PKA to phosphorylate and stimulate the transactivation of Runx2 through a PKA consensus site in AD3 [8].
Runx2 has been found to be an essential transcription factor for inducing osteoblast differentiation [9–13]. Runx2 regulates expression of several genes including alkaline phosphatase, types I and II collagen,RANKL,TGF-β type I receptor,C/EBPdelta,nuclear matrix associated proteins, and TWIST, a basic helix–loop–helix transcription factor [14]. Runx2 seems to be involved in the balance between bone formation and bone resorption and participates in several bone and bone related diseases [14–18]. Runx2 activity can be modulated in several ways, including direct regulation of its gene expression, post-translational modifications, and protein–protein interactions. In this study we investigated how Runx2 is modified in response to PTH treatment, and how this modification is biologically significant for MMP-13 promoter activation. We identified Runx2 phosphorylation sites in vivo and their requirement for PKA stimulation of MMP-13 promoter activity in osteoblastic and non-osteoblastic cells.
2. Materials and methods
2.1. Reagents and cells
Tissue culture media and reagents were obtained from Invitrogen. The Runx2, c-myc tag and phospho tyrosine antibodies were purchased from Santa Cruz Biotechnology, CA. The phospho threonine antibody was obtained from Calbiochem, CA. Other chemicals were obtained from Sigma, St. Louis. UMR 106-01 cells were maintained in culture in Eagle’s minimal essential medium (EMEM) supplemented with 10% fetal bovine serum (FBS) at 37 °C in a humidified atmosphere of 5% CO2 and 95% air. COS-7 and C3H10T1/2 cells were maintained in DMEM medium plus 10% FBS.
2.2.Immunoprecipitation
Cells were lysed in immunoprecipitation buffer containing protease inhibitors and phenylmethylsulfonyl fluoride (PMSF). Protein A/G beads were added to the extracts and allowed to pre-absorb for 1 h, after which primary antibody and protein A/G bead mixture were added. Incubation was continued at 4 °C with agitation for 2 h or overnight. After washing, the beads were suspended in 2x SDS sample buffer, boiled for 5 min and centrifuged. The proteins in the resulting supernatant were separated by SDS–PAGE and subjected to Western blotting (8, 9).
2.3.Western blot analysis
The proteins were transferred electrophoretically to polyvinylidene difluoride membrane (Bio-Rad). After blocking in Tris-buffered saline containing 5% (w/v) non-fat dry milk, the membrane was exposed to primary antibody overnight at 4 °C. The membrane was washed and exposed to horseradish peroxidase-conjugated secondary antibody. The immunoreactive signals were visualized using an enhanced chemiluminescence detection kit (Amersham Biosciences).
2.4. Metabolic labeling of Runx2 protein
COS-7 cells were transiently transfected with pCMV–c-myc– Runx2 construct [15] for 24 h. The cells were then incubated with 100 μCi of [32P]phosphate (Phosphorus-32; Amersham BioSciences) in phosphate free DMEM medium containing 0.1% FBS for 2 h, followed by control or 8-Br-cAMP treatment. Whole cell lysates were prepared and immunoprecipitated with antibody to the c-myc tag. The immunoprecipitated proteins were fractionated by 12% SDS–PAGE, followed by autoradiography.
2.5. Phosphoamino acid analysis
The radioactive band (Runx2) as visualized by autoradiography was cut out and incubated for 45 min at 95 °C in the presence of 6 N HCl. The resulting samples were dried and redissolved in a solution containing unlabeled phosphorylated serine, threonine, and tyrosine standards (Sigma Company, St. Louis). Separation of the amino acids by thin layer chromatography (TLC) electrophoresis and staining of the amino acid standards with ninhydrin were done as described previously [19]. Visualization of Runx2-derived 32P-phosphorylated amino acid(s) was accomplished by autoradiography.
2.6. Identification of phosphorylation sites by mass spectrometry
COS-7 cells were transiently transfected with pCMV–c-myc– Runx2 construct [15] for 24 h. The cells were then treated with control or 8-Br-cAMP containing media. Whole cell lysates were prepared and immunoprecipitated with antibody to the c-myc tag. The immunoprecipitated proteins were fractionated by 12% SDS–PAGE, followed by silver staining (Amersham Biosciences). The gel bands corresponding to the Runx2 proteins based on their sizes were excised and washed with 30% ACN in 50 mM ammonium bicarbonate prior to 7 μg DTT reduction and 35 μg iodoacetamide alkylation. Trypsin (0.2 μg) was used for digestion at 37 °C overnight. The resulting peptides were extracted with 30 μl of 1% trifluoracetic acid (TFA) followed by C18 Ziptip (Millpore Corporation, Billerica, MA) desalting according to manufacturer’s protocol. The peptides were dried in a Speedvac, and re-suspended in 10 μl of solvent A (2% acetonitrile (ACN), 0.1% formic acid (FA)) for LC– MS/MS analysis. In brief, the peptides were first separated by Reversed Phase Liquid Chromatography (RPLC, capillary PepMap100 column (75 μm × 150 mm, 3 μm, 100 Å, C18), Dionex, Sunnyvale, CA, USA) in a 60 min linear gradient from 10% solvent A to 40% solvent B (95% ACN, 0.1% FA). The RPLC eluent was directly introduced into a nano-ESI source on an API-US QTOF tandem MS system (Waters Corporation, Milford, MA). The ESI capillary voltage was set at 3000 V. The MS spectra (m/z 400–1900) were acquired in the positive ion mode. Argon was used as the collision gas and the collision energy was set within a range between 17 and 55 V, depending on the charge states and the m/z values of the ions analyzed. MS/MS spectra were acquired in data-dependent mode, in which the top five most abundant precursors with two to five charges from each MS survey scan were selected for fragmentation. Protein identification was performed by searching the MS/MS spectra against mammalian NCBI database using a local MASCOT search engine (V. 1.9). Oxidized methionine, carbamidomethyl labeled-cysteine and serine/threonine phosphorylation were set as variable modifications as the search parameters. The MS/MS spectra of phosphopeptides were manually confirmed.
2.7. Site-directed mutagenesis and transient transfections
The mutations in the Runx2 constructs were carried using the Quick Change site-directed mutagenesis kit (Stratagene, La Jolla, CA). The serine and threonine amino acids were converted to alanine. The constructs were verified by sequencing at the UMDNJRWJMS-core DNA automatic sequencing facility. The plasmid DNAs were transiently transfected into cells using GeneJammer (Stratagene). Briefly, cells were plated at 2–4 × 105/well in sixwell plates in 10% FBS-containing medium. The following day, the cells were transfected with 1 μg DNA and 5 μl GeneJammer per well in 1 ml of serum-free medium. After 3 h, 1 ml of 10% FBS-containing medium were added. After 24 h, the cells were treated with either control or 8-Br-cAMP (10−3M)-containing media for 24 h. CAT activity was measured by reacting 50 5 μl of cell lysate in duplicate in a 100 μl reaction volume consisting of final concentrations of 250 μM n-butyryl-coenzyme A and 23 mM [14C]-chloramphenicol (0.125 μCi/assay). The values were normalized to protein as determined by the Bradford dye binding (BioRad, Hercules, CA) method. A standard curve using purified CAT was performed every experiment to determine the linear range of the enzyme assay. The Renilla luciferase construct was co-transfected to normalize the transfection efficiency. The Renilla luciferase assay was carried out using the Renilla luciferase assay kit from Promega [8,9].
2.8. Real-time reverse transcriptase PCR
Total RNA was isolated by a kit from Qiagen (Valencia, CA) and subjected to real-time RT-PCR (15). Reverse transcriptase reaction was carried out using TaqMan Reverse Transcription reagents (Roche, Indianapolis, IN). PCR reactions were performed according to the realtime thermocycler machine manufacturer’s instructions (DNA Engine Opticon, MJ Research, MA, USA), which allow realtime quantitative detection of the PCR product by measuring the increase in SYBR green fluorescence caused by binding of SYBR green to double-stranded DNA. The SYBR green kit for PCR reactions was purchased from Perkin Elmer Applied Biosystems (Wellesley, MA). Primers used in this study were designed using the PrimerExpress software (Perkin–Elmer Applied Biosystems). For PCR amplification, the following sets of mouse specific MMP-13 primers were used:
Sense 5́ GCC CTG ATG TTT CCC ATC TA 3́
Antisense 5́ TTT TGG GAT GCT TAG GGT TG 3́
3. Results
3.1. PTH stimulates Runx2 phosphorylation in vitro
We have previously demonstrated that PTH stimulates the transactivation of AD3 in Runx2 through a consensus PKA site [8]. We also showed that the purified PKA catalytic subunit could phosphorylate this portion of Runx2 in vitro but not if the PKA site at amino acid 347 in AD3 was mutated. In this paper, to identify PKA-dependent transactivation of Runx2 for MMP-13 promoter activation, we first examined PTH stimulation of Runx2 phosphorylation in vivo. Whole cell lysates were prepared from control or PTH-treated UMR 106-01 cells and subjected to immunoprecipitation with either IgG or Runx2 antibody, followed by Western blot analysis using antibodies for phosphorylated serine, threonine, and tyrosine. As shown in Fig. 1A, PTH stimulated Runx2 phosphorylation within 5 min in UMR 106-01 cells and this occurred mostly on serine residues with a small amount at threonine and tyrosine residues. PTH stimulation of Runx2 phosphorylation at serine, threonine, and tyrosine residues was seen even out to 30 min after PTH treatment (Fig. 1B).
Fig. 1.
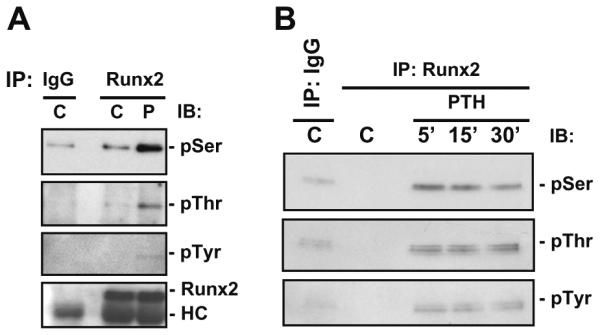
PTH stimulation of Runx2 phosphorylation in vivo. (A) UMR 106-01 cells were treated with control or PTH (10−8M)-containing media for 5 min. (B) UMR 106-01 cells were treated with control or PTH (10−8M)-containing media for 5, 15 and 30 min. Whole cell lysates were prepared and immunoprecipitated with either IgG or Runx2 antibody and immunoblotted with antibodies as indicated followed by detection by ECL kit (C, control; P, PTH; IP, immunoprecipitation; IB, immunoblot; HC, heavy chain; p, phosphorylated).
3.2. Identification of Runx2 phosphorylation in vivo
Since osteoblastic cells contain several Runx isoforms (8) and overexpression of a Runx2 construct may downregulate its own promoter activity (15), in this study we utilized an expression plasmid of c-myc-tagged Runx2 (type II) to specifically identify type II Runx2 phosphorylation, and transfected this into COS-7 cells since these cells do not have endogenous Runx2. The cells were then metabolically labeled with radioactive free orthophosphate, followed by 8-Br-cAMP treatment and immunoprecipitation with c-myc antibody (Fig. 2A). The effect of PTH is mimicked by using 8-Br-cAMP, a cell permeable form of cAMP which will activate protein kinase A. The immunoprecipitated Runx2 protein was subjected to acid hydrolysis and TLC analysis as described in Section2. The results indicate that there was Runx2 phosphorylation on serines in both control and 8-Br-cAMP treated lysates and there appeared to be more serine phosphorylation after 5 min of 8-Br-cAMP treatment (Fig. 2B). This is consistent with our previous result (Fig. 1) that Runx2 was more greatly phosphorylated on serine residues than threonine and tyrosine residues in response to PTH treatment.
Fig. 2.
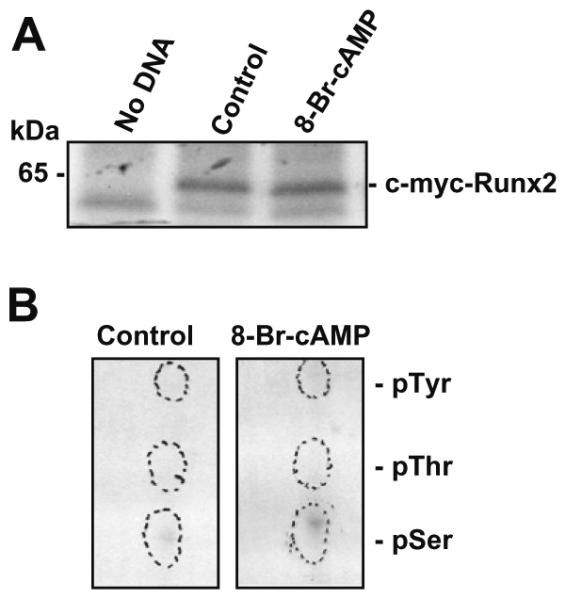
(A) Metabolic labeling of Runx2 protein. The c-myc-tagged Runx2 construct (pCMV–c-myc–Runx2) was transiently transfected into COS-7 cells. The cells were then metabolically labeled with free orthophosphate (100 μCi) for 2 h, followed by 8-Br-cAMP (10−3M) treatment for 5 min. Whole cell lysates were prepared and immunoprecipitated with antibody to the c-myc tag, fractionated by 12% SDS–PAGE and autoradiographed. (B) Phosphoamino acid analysis of Runx2 protein. The labeled Runx2 protein from the gel was excised and digested with 6 N HCl, followedby TLC analysis. The cold phosphorylated serine, threonine and tyrosine amino acids were included as standards. The cold amino acids were visualized with Ninhydrin spray and UV light. The labeled phosphorylated amino acids were visualized by autoradiography.
3.3. Identification of Runx2 phosphorylation sites in vivo
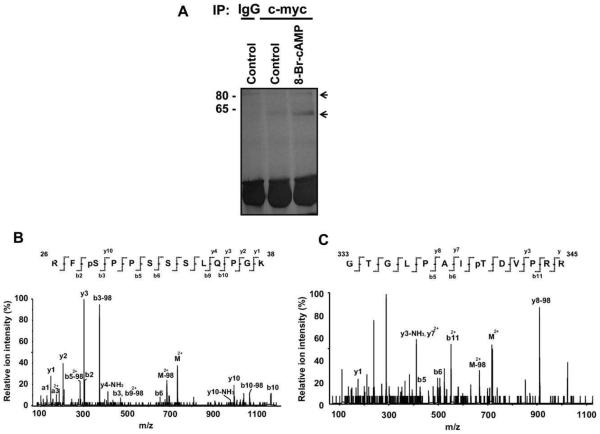
To precisely identify Runx2 phosphorylation sites in vivo, COS-7 cells were transiently transfected with c-myc-tagged Runx2 and treated with control or 8-Br-cAMP-containing media. Whole cell lysates were immunoprecipitated with either IgG or c-myc tag antibody. This was followed by SDS–PAGE fractionation and silver staining. The two bands indicated by arrows (Fig. 3A) were excised and subjected to matrix-assisted laser desorption/ionization (MAL-DI) MS analysis. The results identi?ed the bottom band as Runx2 and that it was phosphorylated on serines and threonines and identified that these were serine 28 (Fig. 3B) and threonine 340 after 8-Br-cAMP treatment (Fig. 3C).
Fig. 3.
MALDI/MS analysis of Runx2 phosphorylation sites in vivo. (A) The c-myc-tagged Runx2 construct (pCMV–c-myc–Runx2) was transiently transfected into COS-7 cells. The cells were then treated with control or 8-Br-cAMP (10−3M)-containing media for 5 min. Whole cell lysates were prepared, immunoprecipitated with antibody to the c-myc tag, fractionated by 12% SDS–PAGE and silver stained. The two protein bands corresponding to 65 kDa and 80 kDa were excised from the gel and subjected to MALDI/MS analysis. (B) MS/MS spectrum of a doubly-charged ion corresponds to RFSPPSSSLQPGK plus one phosphorylation site (m/z 734.42). The observed y- and b-ion series confirmed the peptide sequence and localized the phosphorylation site at S28. (C) MS/MS spectrum of a doubly-charged ion corresponds to GTGLPAITDVPRR plus one phosphorylation site (m/z 716.87). The observed y- and b-ion series tentatively confirmed the peptide sequence and localized the phosphorylation site at T340.
3.4. Functional characterization of Runx2 phosphorylation sites for MMP-13 promoter activation
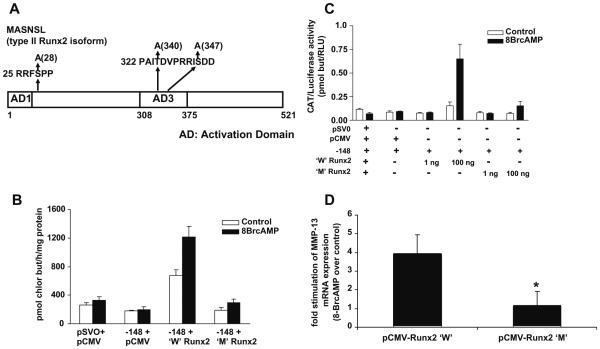
In order to assess the biological significance of the Runx2 phosphorylation sites for MMP-13 promoter activation, the amino acids were mutated in single or double sites by site-directed mutagenesis. The wild-type Runx2 and the Runx2 mutated at these phosphorylation sites were transiently transfected along with the rat MMP-13 promoter construct into either COS-7 cells or C3H10T1/2 cells since the latter are of mesenchymal lineage. The cells were then treated with control or 8-Br-cAMP-containing media and CAT activity was measured. Both the wild-type Runx2 and Runx2 constructs having mutations of either single or double phosphorylation sites still stimulated MMP-13 promoter activity after 8-Br-cAMP treatment (data not shown). We previously demonstrated the requirement of serine amino acid at 347 in the PKA site of AD3 of the type II Runx2 for MMP-13 promoter activation (8). Hence, we mutated this site and used the triple mutant for transfection experiments. Fig. 4A indicates the changes of serine (28, 347) and threonine (340) amino acids in the wild-type Runx2 construct into alanine amino acids in the mutant-type Runx2. The wild-type Runx2 and the Runx2 mutated at these phosphorylation sites were transiently transfected along with the rat MMP-13 promoter construct into either COS-7 cells or C3H10T1/2 cells. The Runx2 construct having mutations of all three phosphorylation sites was unable to stimulate MMP-13 promoter activity in either of the cell lines (Fig. 4B and C). To determine the importance of Runx2 phosphorylation sites for endogenous MMP-13 gene expression, the wild-type and the mutated Runx2 constructs were transiently transfected into C3H10T1/2 cells, followed by 8-BrcAMP treatment. Real-time RT-PCR analysis shows that the wild-type Runx2 construct stimulated MMP-13 mRNA expression in the presence of 8-BrcAMP; while the mutated Runx2 construct was unable to increase MMP-13 mRNA expression in the presence of 8-BrcAMP (Fig. 4D), suggesting the importance of Runx2 phosphorylation sites for endogenous MMP-13 gene activation in response to 8-BrcAMP treatment.
Fig. 4.
8-Br-cAMP stimulation of MMP-13 promoter activation. (A) Schematic diagram of 8-Br-cAMP stimulation of phosphorylated residues in Runx2 (type II). The serine (S) residues at 28 and 347, and threonine (T) residue at 340 in the Runx2 construct were mutated to alanine residues. (B) The wild-type Runx2 or the Runx2 construct with the three phosphorylation sites mutated were transiently transfected into COS-7 cells along with the rat MMP-13 promoter for 24 h. (C) The wild-type Runx2 or the Runx2 construct with the three phosphorylation sites mutated, at two different concentrations (1 ng and 100 ng), were transiently transfected into C3H10T1/2 cells along with the rat MMP-13 promoter for 24 h. The cells were then treated with control or 8-Br-cAMP (10−3M)-containing media for 24 h. Whole cell lysates were prepared and assayed for CAT activity (pmoles of chloramphenicol butyrylated per hour per mg protein). Data represent mean ± S.E. of three replicate plates. (D) The wild-type Runx2 or the Runx2 construct with the three phosphorylation sites mutated were transiently transfected into C3H10T1/2 cells for 24 h. The cells were then treated with control or 8-Br-cAMP (10−3M)-containing media for 4 h. Total RNA was isolated and real-time RT-PCR was carried out using the mouse MMP-13 and GAPDH primers. The MMP-13 mRNA expression was normalized with GAPDH mRNA expression, and the fold stimulation was calculated over control. The experiment was carried out in triplicate. The asterisks represent P < 0.05 compared with the wild-type Runx2 (W, wild-type; M, mutant).
To determine whether the phosphorylation site mutations affect expression of Runx2 protein, the wild-type and triple-mutant c-myc-tagged Runx2 constructs were transiently transfected into C3H10T1/2 cells. Whole cell lysates were prepared and subjected to Western blot analysis using the c-myc and b-actin antibodies. Expression of c-myc-tagged wild-type Runx2 and c-myc-tagged mutant-type Runx2 proteins was seen in C3H10T1/2 cells in response to transfection with the c-myc-tagged wild-type Runx2 construct and the c-myc-tagged mutant Runx2 construct, respectively (Fig. 5). There was no endogenous expression of c-myc-tagged Runx2 proteins in either C3H10T1/2 cells or UMR 106-01 cells (Fig. 5).
Fig. 5.
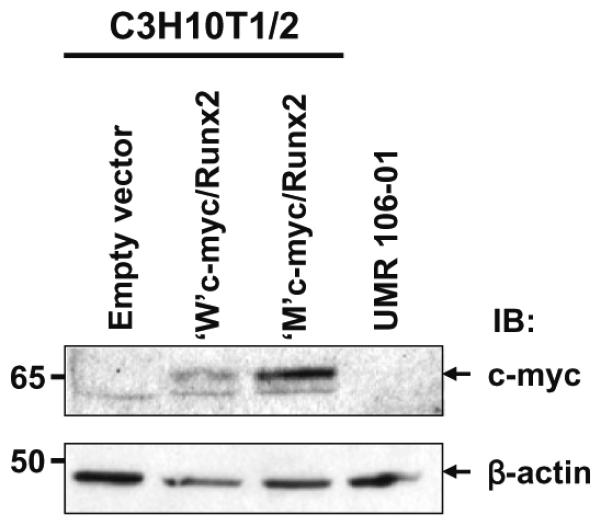
The wild-type and the mutant Runx2 protein expression. C3H10T1/2 cells were transiently transfected with either the c-myc-tagged wild-type Runx2 construct or the c-myc-tagged mutant Runx2 construct for 24 h. As negative controls, cells (C3H10T1/2 and UMR 106-01) were transiently transfected with empty vector. Whole cell lysates were prepared and Western blot analysis was carried out using the antibodies as indicated. β-Actin has been used as loading control. IB, immunoblot.
4. Discussion
Runx proteins are subject to post-translational modifications that affect their activity, in particular phosphorylation [20–22]. In this study we report that PTH stimulated Runx2 phosphorylation in rat osteoblastic cells and this occurred mostly on serine residues with a small amount at threonine and tyrosine residues (Fig. 1). The phosphorylation that occurs at different amino acids within the Runx2 protein may control or regulate its transactivation differently. Even though Runx2 protein level has been reported to be decreased by prolonged cAMP treatment via ubiquitination and the proteasomal pathway [23], we found that the total amount of Runx2 protein after PTH treatment in the time period we used was apparently unchanged (Fig. 1) [8,9].
In the TLC analysis (Fig. 2B) we could not detect phosphorylated threonine residues but the MALDI/MS study revealed phosphorylation of Runx2 at threonine residues (Fig. 3C) in addition to serine residues. This could be due to either less sensitivity of the TLC technique due to less incorporation of label or lower loading of the acid hydrolyzed samples. Even though PTH stimulated Runx2 phosphorylation at serine, threonine, and tyrosine residues (Fig. 1), 8-Br-cAMP treatment of COS-7 cells identified Runx2 phosphorylation only at serine (28) and threonine (340) residues by MALDI/ MS analysis (Fig. 3B and C), suggesting that there might be activation of other intermediate signaling pathway(s) by PTH. We were not able to detect serine phosphorylation at 347 by MALDI/MS analysis and that could be due to either hindrance effect of two amino acids closely located with each other (340, 347) or rapid dephosphorylation at this amino acid, or technical problems, perhaps related to dephosphorylation. The phosphorylated threonine (340) and serine (347) residues in the Runx2 protein are located in the AD3 region and this region has been previously shown to transactivate Runx2 through its PKA site [8]. Consistent with these data, Krishnan et al. showed PKA-dependent increases in endogenous Runx2 activity mediated PTH action [23]. It should be noted that the labeling and immunoprecipitations for MALDI/MS had to be done in COS-7 cells because of the poor transfection efficiency in UMR 106-01 cells and the need to use tagged Runx2 to immunoprecipitate sufficient pure protein for MALDI/MS.
During osteoblastic differentiation of human bone marrow stromal cells, there is increased Runx2 activity which could be due to, at least in part, phosphorylation of key residues in the Runx2 protein [24]. Runx2 has been shown to be a substrate for the MAPK pathway and this pathway can be stimulated by a variety of signals including those initiated by extracellular matrix, osteogenic growth factors like BMPs and fibroblast growth factor-2, NELL1 and mechanical loading [25–27]. In these studies, an increase in Runx2 phosphorylation is coincident with enhanced Runx2 transcriptional activity. Runx2 activity has been reported to be negatively regulated by phosphorylation of two serine residues [28]. Dexamethasone was shown to decrease Runx2 phosphorylation levels on a serine residue, thereby inducing osteogenesis [29]. Recently it has been reported that ERK-MAPK activation stimulates osteoblast differentiation and skeletal development through a pathway involving Runx2 [30]. It appears that the putative signaling cascades or effector molecules could regulate Runx2 activity in opposite ways by phosphorylating it at different amino acids at different positions. There is a confiict of results indicating transcriptional activation or repression of Runx2 by its phosphorylation status [24–30], and our studies provide evidence that Runx2 phosphorylation at serine 28, 347 and threonine 340 residues after 8-Br-cAMP treatment may be correlated with its transcriptional activation of the MMP-13 gene (Fig. 4B–D). This transcriptional activation could result from recruitment or sequestration of factors and cofactors by phosphorylated Runx2 on the MMP-13 promoter. Thus, we have identified for the first time the Runx2 phosphorylation sites necessary for PKA stimulated MMP-13 promoter activation and this event may be critical for bone remodeling.
Acknowledgements
We thank Dr. Gerard Karsenty for providing the rat Runx2 (type II) construct. This work was supported by a research grant (DK 47420) from the National Institutes of Health (to N.C.P.).
Abbreviations
- PTH
parathyroid hormone
- PKA
protein kinase A
- 8-Br-cAMP
8-bromo cyclic adenosine mono phosphate
- MMP-13
matrix metalloproteinase-13
- MALDI
matrix-assisted laser desorption/ionization
- TLC
thin layer chromatography
- CAT
chloramphenicol acetyl transferase
- GAPDH
glyceraldehyde 3-phosphate dehydrogenase
References
- [1].Yasuda H, Shima N, Nakagawa N, Yamaguchi K, Kinosaki M, Mochizuki S, Tomoyasu A, Yano K, Goto M, Murakami A, Tsuda E, Morinaga T, Higashio K, Udagawa N, Takahashi N, Suda T. Osteoclast differentiation factor is a ligand for osteoprotegerin/osteoclastogenesis-inhibitory factor and is identical to TRANCE/RANKL. Proc. Natl. Acad. Sci. USA. 1998;95:3597–3602. doi: 10.1073/pnas.95.7.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Partridge NC, Jeffrey JJ, Ehlich LS, Teitelbaum SL, Fliszar C, Welgus HG, Kahn AJ. Hormonal regulation of the production of collagenase and a collagenase inhibitor activity by rat osteogenic sarcoma cells. Endocrinology. 1987;120:1956–1962. doi: 10.1210/endo-120-5-1956. [DOI] [PubMed] [Google Scholar]
- [3].Witty JP, Foster SA, Stricklin GP, Matrisian LM, Stern PH. Parathyroid hormone-induced resorption in fetal rat limb bones is associated with production of the metalloproteinases collagenase and gelatinase B. J. Bone Miner. Res. 1996;11:72–78. doi: 10.1002/jbmr.5650110111. [DOI] [PubMed] [Google Scholar]
- [4].Delaisse JM, Eeckhout Y, Vaes G. Bone-resorbing agents affect the production and distribution of procollagenase as well as the activity of collagenase in bone tissue. Endocrinology. 1988;123:264–276. doi: 10.1210/endo-123-1-264. [DOI] [PubMed] [Google Scholar]
- [5].Zhao W, Byrne MH, Boyce BF, Krane SM. Bone resorption induced by parathyroid hormone is strikingly diminished in collagenase-resistant mutant mice. J. Clin. Invest. 1999;103:517–524. doi: 10.1172/JCI5481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Inada M, Wang Y, Byrne MH, Rahman MU, Miyaura C, Lopez-Otin C, Krane SM. Critical roles for collagenase-3 (Mmp13) in development of growth plate cartilage and in endochondral ossification. Proc. Natl. Acad. Sci. USA. 2004;101:17192–17197. doi: 10.1073/pnas.0407788101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Scott DK, Brakenhoff KD, Clohisy JC, Quinn CO, Partridge NC. Parathyroid hormone induces transcription of collagenase in rat osteoblastic cells by a mechanism using cyclic adenosine 3′,5′-monophosphate and requiring protein synthesis. Mol. Endocrinol. 1992;6:2153–2159. doi: 10.1210/mend.6.12.1337147. [DOI] [PubMed] [Google Scholar]
- [8].Selvamurugan N, Pulumati MR, Tyson DR, Partridge NC. Parathyroid hormone regulation of the rat collagenase-3 promoter by protein kinase A-dependent transactivation of core binding factor alpha1. J. Biol. Chem. 2000;275:5037–5042. doi: 10.1074/jbc.275.7.5037. [DOI] [PubMed] [Google Scholar]
- [9].Selvamurugan N, Chou WY, Pearman AT, Pulumati MR, Partridge NC. Parathyroid hormone regulates the rat collagenase-3 promoter in osteoblastic cells through the cooperative interaction of the activator protein-1 site and the runt domain binding sequence. J. Biol. Chem. 1998;273:10647–10657. doi: 10.1074/jbc.273.17.10647. [DOI] [PubMed] [Google Scholar]
- [10].Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, Shimizu Y, Bronson RT, Gao YH, Inada M, Sato M, Okamoto R, Kitamura Y, Yoshiki S, Kishimoto T. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997;89:755–764. doi: 10.1016/s0092-8674(00)80258-5. [DOI] [PubMed] [Google Scholar]
- [11].Otto F, Thornell AP, Crompton T, Denzel A, Gilmour KC, Rosewell IR, Stamp GW, Beddington RS, Mundlos S, Olsen BR, Selby PB, Owen MJ. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell. 1997;89:765–771. doi: 10.1016/s0092-8674(00)80259-7. [DOI] [PubMed] [Google Scholar]
- [12].Ducy P, Zhang R, Geoffroy V, Ridall AL, Karsenty G. Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell. 1997;89:747–754. doi: 10.1016/s0092-8674(00)80257-3. [DOI] [PubMed] [Google Scholar]
- [13].Banerjee C, McCabe LR, Choi JY, Hiebert SW, Stein JL, Stein GS, Lian JB. Runt homology domain proteins in osteoblast differentiation: AML3/CBFA1 is a major component of a bone-specific complex. J. Cell Biochem. 1997;66:1–8. doi: 10.1002/(sici)1097-4644(19970701)66:1<1::aid-jcb1>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- [14].Lian JB, Javed A, Zaidi SK, Lengner C, Montecino M, van Wijnen AJ, Stein JL, Stein GS. Regulatory controls for osteoblast growth and differentiation: role of Runx/Cbfa/AML factors. Crit. Rev. Eukar. Gene Exp. 2004;14:1–41. [PubMed] [Google Scholar]
- [15].Selvamurugan N, Jefcoat SC, Kwok S, Kowalewski R, Tamasi JA, Partridge NC. Overexpression of Runx2 directed by the matrix metalloproteinase-13 promoter containing the AP-1 and Runx/RD/Cbfa sites alters bone remodeling in vivo. J. Cell Biochem. 2006;99:545–557. doi: 10.1002/jcb.20878. [DOI] [PubMed] [Google Scholar]
- [16].Coffman JA. Runx transcription factors and the developmental balance between cell proliferation and differentiation. Cell Biol. Int. 2003;27:315–324. doi: 10.1016/s1065-6995(03)00018-0. [DOI] [PubMed] [Google Scholar]
- [17].Westendorf JJ, Hiebert SW. Mammalian runt-domain proteins and their roles in hematopoiesis, osteogenesis, and leukemia. J. Cell Biochem. 1999;(Suppl.):51–58. doi: 10.1002/(sici)1097-4644(1999)75:32+<51::aid-jcb7>3.3.co;2-j. [DOI] [PubMed] [Google Scholar]
- [18].Javed A, Barnes GL, Pratap J, Antkowiak T, Gerstenfeld LC, van Wijnen AJ, Stein JL, Lian JB, Stein GS. Impaired intranuclear trafficking of Runx2 (AML3/CBFA1) transcription factors in breast cancer cells inhibits osteolysis in vivo. Proc. Natl. Acad. Sci. USA. 2005;102:1454–1459. doi: 10.1073/pnas.0409121102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Namchuk M, Lindsay L, Turck CW, Kanaani J, Baekkeskov S. Phosphorylation of serine residues 3, 6, 10, and 13 distinguishes membrane anchored from soluble glutamic acid decarboxylase 65 and is restricted to glutamic acid decarboxylase 65alpha. J. Biol. Chem. 1997;272:1548–1557. doi: 10.1074/jbc.272.3.1548. [DOI] [PubMed] [Google Scholar]
- [20].Tanaka T, Kurokawa M, Ueki K, Tanaka K, Imai Y, Mitani K, Okazaki K, Sagata N, Yazaki Y, Shibata Y, Kadowaki T, Hirai H. The extracellular signal-regulated kinase pathway phosphorylates AML1, an acute myeloid leukemia gene product, and potentially regulates its transactivation ability. Mol. Cell Biol. 1996;16:3967–3979. doi: 10.1128/mcb.16.7.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Tintut Y, Parhami F, Le V, Karsenty G, Demer LL. Inhibition of osteoblast-specific transcription factor Cbfa1 by the cAMP pathway in osteoblastic cells. Ubiquitin/proteasome-dependent regulation. J. Biol. Chem. 1999;274:28875–28879. doi: 10.1074/jbc.274.41.28875. [DOI] [PubMed] [Google Scholar]
- [22].Huang G, Shigesada K, Ito K, Wee HJ, Yokomizo T, Ito Y. Dimerization with PEBP2beta protects RUNX1/AML1 from ubiquitin–proteasome-mediated degradation. EMBO J. 2001;20:723–733. doi: 10.1093/emboj/20.4.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Krishnan V, Moore TL, Ma YL, Helvering LM, Frolik CA, Valasek KM, Ducy P, Geiser AG. Parathyroid hormone bone anabolic action requires Cbfa1/Runx2-dependent signaling. Mol. Endocrinol. 2003;17:423–435. doi: 10.1210/me.2002-0225. [DOI] [PubMed] [Google Scholar]
- [24].Shui C, Spelsberg TC, Riggs BL, Khosla S. Changes in Runx2/Cbfa1 expression and activity during osteoblastic differentiation of human bone marrow stromal cells. J. Bone Miner. Res. 2003;18:213–221. doi: 10.1359/jbmr.2003.18.2.213. [DOI] [PubMed] [Google Scholar]
- [25].Selvamurugan N, Kwok S, Alliston T, Reiss M, Partridge NC. Transforming growth factor-beta 1 regulation of collagenase-3 expression in osteoblastic cells by cross-talk between the Smad and MAPK signaling pathways and their components, Smad2 and Runx2. J. Biol. Chem. 2004;279:19327–19334. doi: 10.1074/jbc.M314048200. [DOI] [PubMed] [Google Scholar]
- [26].Bokui N, Otani T, Igarashi K, Kaku J, Oda M, Nagaoka T, Seno M, Tatematsu K, Okajima T, Matsuzaki T, Ting K, Tanizawa K, Kuroda S. Involvement of MAPK signaling molecules and Runx2 in the NELL1-induced osteoblastic differentiation. FEBS Lett. 2008;582:365–371. doi: 10.1016/j.febslet.2007.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Franceschi RT, Xiao G, Jiang D, Gopalakrishnan R, Yang S, Reith E. Multiple signaling pathways converge on the Cbfa1/Runx2 transcription factor to regulate osteoblast differentiation. Conn. Tissue Res. 2003;44:109–116. [PMC free article] [PubMed] [Google Scholar]
- [28].Wee HJ, Huang G, Shigesada K, Ito Y. Serine phosphorylation of RUNX2 with novel potential functions as negative regulatory mechanisms. EMBO Rep. 2002;3:967–974. doi: 10.1093/embo-reports/kvf193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Phillips JE, Gersbach CA, Wojtowicz AM, Garcia AJ. Glucocorticoid-induced osteogenesis is negatively regulated by Runx2/Cbfa1 serine phosphorylation. J. Cell. Sci. 2006;119:581–591. doi: 10.1242/jcs.02758. [DOI] [PubMed] [Google Scholar]
- [30].Ge C, Xiao G, Jiang D, Franceschi RT. Critical role of the extracellular signal-regulated kinase-MAPK pathway in osteoblast differentiation and skeletal development. J. Cell. Biol. 2007;176:709–718. doi: 10.1083/jcb.200610046. [DOI] [PMC free article] [PubMed] [Google Scholar]