Abstract
The advent of sensitive and robust quantitative proteomics techniques has been emerging as a vital tool for deciphering complex biological puzzles that would have been challenging to conventional molecular biology methods. The method here describes the use of two isotope labeling techniques – isobaric tags for relative and absolute quantification (iTRAQ) and redox isotope-coded affinity tags (ICAT), to elucidate the cardiovascular redox-proteome changes and thioredoxin 1 (Trx1)-regulated protein network in cardiac-specific Trx1 transgenic mouse models. The strategy involves the use of an amine-labeling iTRAQ technique, gauging the global proteome changes in Trx1 transgenic mice at the protein level, while ICAT, labeling redox-sensitive cysteines, reveals the redox-status of cysteine residues. Collectively, these two quantitative proteomics techniques not only can quantify global changes of the cardiovascular proteome, but also pinpoint specific redox sensitive cysteine sites that are subjected to Trx1-catalzyed reduction.
Keywords: Quantitative proteomics, liquid chromatography, tandem mass spectrometry, iTRAQ, Redox-ICAT, hypertrophy, thioredoxin 1
1. Introduction
Chemical labeling of peptides/proteins with isotope-coded reagents (1), rendering peptide/proteins with mass differences that are readily discernible in mass spectrometers, enable the comparative proteome quantitation from multiple biological samples. One advantage of the chemical labeling technique is its versatile applicability to all sources of proteins (cells, tissues, serum, bone, hair etc.). Unlike stable isotope labeling with amino acid in cell culture (SILAC) technique (2), which incorporates stable isotopic amino acids during cell culture, but is limited to proteins that can be retrieved from cultured cells that undergo rapid protein turnover. Here we will introduce the application of two distinct chemical labeling approaches - iTRAQ (1, 3) and ICAT (4, 5), to quantify the proteome changes in left ventricular tissues of wild type and a cardiac-specific Trx1 transgenic mouse (6, 7).
iTRAQ reagents label the primary amines on the N-terminus and lysine residues of peptides and can accommodate the quantification of up to 8 different samples simultaneously (8-plex iTRAQ (8)). The isobaric nature of iTRAQ reagents does not add to the complexity of chromatography and the mass spectrum (MS) and only releases signature fragments (m/z 114 to 117 for 4-plex and 113 to 121 for 8-plex) of individual tags upon collision-induced dissociation, that can be observed in tandem mass (MS/MS) spectra for peptide identification and quantification (Fig. 1A). On the other hand, ICAT reagents, available in light and heavy versions, label free thiol groups of cysteine residues. ICAT reagents incorporate a biotin tag to enable selective avidin-based enrichment of ICAT-labeled peptides from non-cysteine containing peptides, therefore reducing sample complexity. The light and heavy ICAT labeled peptides appear as doublets in MS spectra, within which the peak intensity/integrated chromatographic peak area of the doublet are used for peptide quantitation. Peptide sequence is obtained from the MS/MS spectra of either the light or heavy ICAT-labeled peptide (Fig. 1B). Many derivatives of the ICAT technique were created to gauge the redox status of cysteines in peptides by introducing different reduction agents and workflows (5, 9, 10).
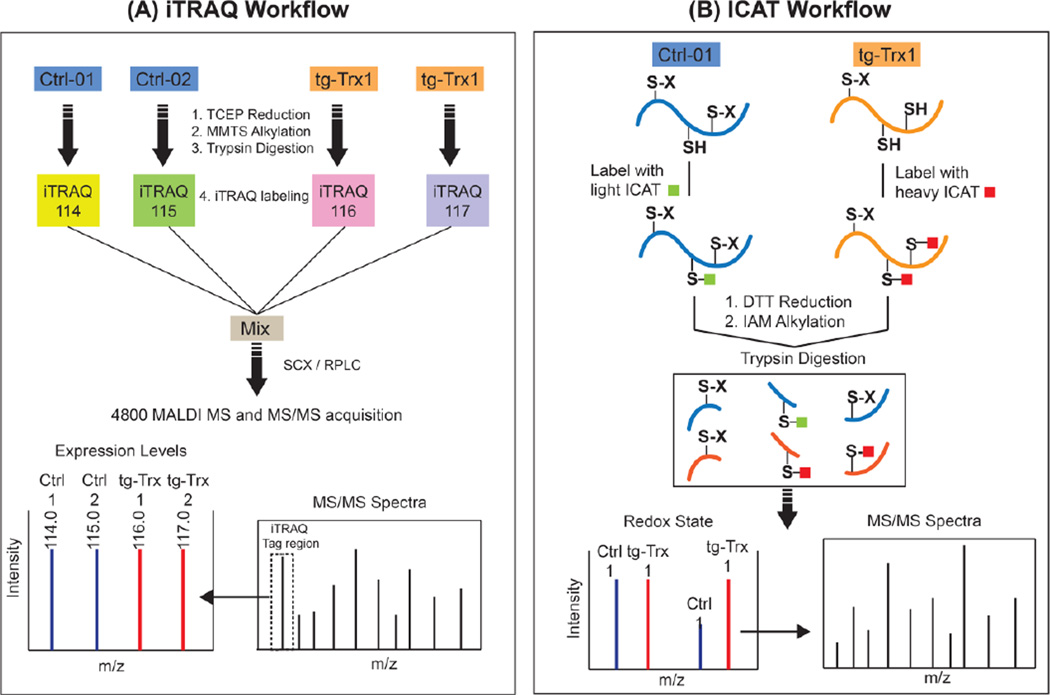
Figure 1. Typical workflow for ICAT and iTRAQ quantitation.
A) In the iTRAQ workflow, protein samples are first subjected to TCEP reduction, MMTS alkylation and trypsin digestion. The resulting tryptic peptides are then labeled by designated iTRAQ reagents separately. After quenching the reaction, labeled peptides are mixed and separated by multidimensional chromatography. Finally, MS data is acquired on a 4800 MALDI TOF/TOF in a data dependent acquisition mode. B) In the ICAT workflow, protein thiols are first labeled by either the light ICAT (control) or heavy ICAT (Trx1-overexpressing tissue) reagents. Protein disulfide bonds are then reduced by DTT and alkylated with IAM, sequentially. The labeled proteins are mixed, digested with trypsin and separated sequentially using SCX, avidin affinity and RPLC separations. ICAT-labeled peptides are identified and quantified by a 4800 MALDI-TOF/TOF mass spectrometer. Peptides containing Trx1-reduced cysteines had an ICAT H/L ratio larger than one, and can be quantified by the precursor peak intensity and identified by the MS/MS spectrum (Modified from Molecular & Cellular Proteomics, 2009 (8), 1674–1687 with permission).
A general shotgun proteomics approach commonly deals with a massive number of tryptic peptides (20,000 to 100,000) in a single liquid chromatography coupled with a tandem mass spectrometrer (LC/MS/MS) experiment (11). To maximize proteome coverage and discovery of low abundant proteins, multiple chromatographic separations are routinely applied in conjunction with these chemical labeling techniques for peptide fractionation and enrichment. Some of the most popular multidimensional chromatographic methods include multidimensional protein identification technology (MUDPIT) (12), OFFGEL (13, 14), strong cation exchange coupled with reversed phase liquid chromatography (SCX-RPLC) (3, 15, 16) and SCX-affinity chromatography-RPLC (5, 7). Here we will describe the application of the latter two techniques for the preparation of iTRAQ and ICAT-labeled peptides for LC/MS/MS identification and quantification of peptides and their reduction by Trx1.
Many lines of evidence (17, 18) have established Trx1, an 11 kDa antioxidant protein, as a negative regulator of oxidative stress-induced hypertrophy. Here we demonstrate a detailed proteomics method involving the use of two complementary stable isotope labeling proteomics techniques to identify the cardiac Trx1 targeted protein network in a Trx1 transgenic mouse model. By use of this protocol, we were able to identify 78 putative Trx1 reductive sites in 55 proteins (7), including many metabolic enzymes within the protein networks regulating the tricarboxylic acid (TCA) cycle and oxidative phosphorylation pathways that have been shown previously to be regulated by Trx1 (17). Some novel target protein networks, including the creatine-phosphocreatine shuttle, the mitochondrial permeability transition pore complex, and the cardiac contractile apparatus were observed for the first time. By using the two comparative proteomics methods including iTRAQ and redox ICAT, we were able to find that Trx1 plays not only a conventional role as an antioxidant, but also a role in remodeling the cardiovascular system to regulate cardiac energy dynamics and muscle contraction.
2. Materials
2.1. Tissue Homogenization and Protein Extraction
The left ventricular heart tissues from control and Trx1-overexpressed mice (three in each group) are diced into ~ 2 × 2 mm cubes and rinsed thoroughly by ice cold PBS (3×) to remove blood content.
ICAT Lysis Buffer: 6 M urea, 2% CHAPS, 1% Triton X-100, and 30 mM Tris-HCl at pH 7.5 and 0.1% (v/v) of protease inhibitor cocktail (Sigma, cat no. P8340, St Louis, MO, USA). (See Note 1)
Omni Tissue Homogenizer: (Omni International Inc., Marietta, GA, USA).
BCA Protein Assay Kit: (Pierce, cat #. 23225, Rockford, IL. USA).
Spectra MAX 190 microplate spectrophotometer (Molecular Devices, Sunnyvale, CA, USA).
iTRAQ Lysis Buffer: 150 mM TEAB, 1.0 % Igepal CA630 (NP-40), 1.0 % Triton X-100, 0.1% v/v protease inhibitor cocktail.
2.2 ICAT Labeling
Cleavable ICAT® Reagent - 10 Assay kit (Sciex Cat# 4339036, Forster City, CA. USA)
Accessory for ICAT: Cartridge holder (4326688), Needleport adaptor (4326689), Outlet connector (4326690), Avidin affinity cartridges (4326694), Cation exchange cartridges (4326695)
Cysteine Reducing Reagent: 50 mM Dithiothreitol (DTT, BioRad Cat #161-0611, Hercules, CA. USA). Cysteine
Alkylation Reagent: 50 mM Iodoacetamide (IAM, BioRad Cat # 163-2109, Hercules, CA. USA).
Eppendorf Vacufuge concentrator 5301 (Eppendorf North America, Inc. Westbury, NY, USA).
2.3 iTRAQ Labeling
Reducing Reagent: 50 mM Tris-(2-carboxyethyl) phosphine (TCEP)
Cysteine Blocking Reagent: 200 mM methyl methanethiosulfonate (MMTS)
HPLC grade ethanol
HPLC grade water
Trypsin (20 µg/vial, Promega, cat no. V5111, Madison, WI, USA)
iTRAQ™ reagents: 114, 115, 116, 117, (Applied Biosystems Inc., ABI, Forster City, CA. USA)
Eppendorf Vacufuge concentrator 5301 (Eppendorf North America, Inc. Westbury, NY, USA)
2.4. Liquid Chromatography Systems
2.4.1. Strong Cation Exchange liquid chromatography (SCX-LC)
Mobile Phase A: 10 mM KH2PO4 and 20% acetonitrile (ACN), pH 3.0.
Mobile Phase B: 600 mM KCl, 10 mM KH2PO4 and 20% ACN, pH 3.0.
BioCAD Sprint™ perfusion chromatography system (PerSeptive BioSystems).
Column: Polysulfoethyl-A column (4.6 × 200 mm, 5 µm, 300 Å, Poly LC Inc., Columbia, MD. USA).
2.4.2. Peptide Desalting
PepClean C18 spin columns (Pierce, cat #. 89870, Rockford, IL, USA).
Loading Solution: 5% ACN containing 0.5% trifluoroacetic acid (TFA, Pierce, cat # 28904, Rockford, IL, USA).
Activation Solution: 50% ACN containing 0.5% TFA.
Elution Solvent: 70% ACN.
Eppendorf Vacufuge concentrator 5301 (Eppendorf North America, Inc. Westbury, NY, USA).
2.4.3. Reversed-phase Liquid Chromatography (RPLC)
Mobile Phase A: 5% ACN containing 0.1% TFA.
Mobile Phase B: 95% ACN containing 0.1% TFA.
LC-Packings Ultimate Chromatography System equipped with a Probot MALDI spotting device (Dionex, Sunnyvale, CA, USA).
C18 PepMap trapping column (0.3 × 5 mm, 5 µm, 100 Å, Dionex, P/N 160454).
C18 PepMap capillary column (0.1 × 150 mm, 3 µm, 100 Å, Dionex, P/N 160321).
Matrix Assisted Laser Desorption Ionization (MALDI) Matrix Solution: 7 mg/ml α-cyano-4-hydroxycinnamic acid (Sigma, cat #. 476870, St Louis, MO, USA) in 60% ACN, 5 mM ammonium monobasic phosphate and internal peptide calibrants (50 fmol/ml each of (Glu1)-fibrinopeptide B (GFP, m/z 1570.677, Sigma, cat #. F3261) and adrenocorticotropic hormone 18–39 (ACTH 18–39, m/z 2465.199, Sigma, cat #. A8346)).
2.5. Mass Spectrometry
4800 Proteomics Analyzer (ABI).
MALDI plates (ABI).
Mass Standards Kit containing 6 peptides mixture (ABI, cat# 4333604).
2.6. Data Analysis Software
4000 Series Explorer (ABI).
GPS Data Explorer v3.5 (ABI).
Mascot Search Engine v1.9 (Matrix Science Ltd. London, UK).
3. Methods
3.1. Protein Extraction
Mouse left ventricular tissues (~100 mg) are diced into 2 × 2 mm cubes and wash thoroughly by ice cold PBS (repeat twice) to remove blood content in a 2 ml Eppendorf tube.
Spin down heart tissues and remove supernatant.
Add 500 µl of either ICAT or iTRAQ lysis buffer to each sample tube. (See Note 2)
Perform heart tissue homogenization on an Omni Tissue Homogenizer at 4°C. Six strike cycles (15 seconds each) were carried out with 2 minutes cooling interval to avoid overheating. (See Note 3)
Remove tissue debris in the homogenates by centrifugation for 30 min at 14,000 × g at 4 °C in a bench-top centrifuge. Transfer supernatants into a fresh 1.5 ml Eppendorf tube and keep it on ice.
Measure protein concentrations for all six samples using the BCA protein assay with bovine serum albumin (BSA) diluted in the Lysis Buffer as standards. Protein yield will be in the range of 4 – 10 mg/ml depending on the lysis buffer of choice.
Adjust protein concentration of each sample to the same level with either ICAT or iTRAQ lysis buffer.
3. 2. ICAT Labeling
Pipette 120 µg protein from each sample into separate tubes (See Note 4).
Precipitate protein in cold acetone (5:1 ratio at −20° C) overnight (See Note 5).
Pellet protein content by high speed centrifugation for 15 min at 14,000 × g at 4 °C.
Remove supernatant and wash the pellets three time with cold acetone (−20° C).
Solubilize protein with 80 µl of ICAT labeling buffer: 6 M urea, 2% CHAPS, 0.01% SDS, and 30 mM Tris-HCl at pH 8.3. (See Note 6)
Bring the ICAT reagent tubes to room temperature and briefly spin down the powder to the bottom of the tubes.
Add 20 µl of ACN to each ICAT tube and vortex the solution.
Spin down the solution to the bottom of the tubes and transfer the entire content to designated sample tubes for protein labeling.
Incubate the mixture for 2 h at 37 °C.
Briefly centrifuge to bring all solution to the bottom of each tube.
Quench excess ICAT reagents by adding 10 mM DTT (final concentration) and incubate for 15 min. (See Note 7)
Alkylate newly generated sulfyhydryls by 15 mM IAM (final concentration) and incubate for 15 min at room temperature in dark. (See Note 8)
Mix light and heavy ICAT labeled sample pairs.
Dilute the sample volume at least 6 times with 20 mM ammonium bicarbonate buffer. (See Note 9)
Add trypsin solution to a final 1:50 ratio (enzyme: protein) and digest overnight at 37 °C. (See Note 8)
Dry the digested peptide samples in a speedvac.
3. 3. SCX-LC
The combined peptide mixture was separated by SCX-LC on a polysulfoethyl-A column to remove excess ICAT reagents and unwanted detergent (SDS and CHAPS), prior to fractionation of the peptides.
Reconstitute the ICAT labeled peptides by adding ~ 500 µl of SCX mobile phase A. Adjust pH to 2.5–3.0 with phosphoric acid if necessary.
Centrifuge the sample at 20,000 × g for 10 min to remove any particulates.
Equilibrate the SCX column with Mobile Phase A, and then inject the ICAT labeled peptides onto the SCX column through a 500 µl sample loading loop.
The gradient profile of SCX consisted of 10 min of 100% mobile phase A followed by 30 min of 0–25% mobile phase B and 20 min of 25–100% B at 1 ml/min. Collect peptide fractions at 2 min/fraction after the elution of neutral and anionic interference.
Dry all the SCX fractions in a speedvac for subsequent desalting steps.
3.4. Peptide Desalting Using C18 Spin Columns
Reconstitute each dried SCX fraction in 150 µl of the Loading Solution (See Note 10).
Add 200 µl of the Activation Solution into a C18 spin column and centrifuge at 1,500 × g for 1 min. Repeat this step once.
Equilibrate the spin column with 200 µl of the Loading Solution, centrifuge the column at 1,500 × g for 1 min. Repeat this step twice.
For each SCX fraction, load 150 µl of the peptides in the Loading Solution onto the spin column and centrifuge at 1,000 × g for 1 min and collect the flow through. Reload the flow through materials onto the spin column.
Wash the bound peptides with 200 µl of the Loading Solution and centrifuge at 1,500 × g for 1 min to remove salts. Repeat this step twice.
Elute the peptides using 100 µl of the Elution Solution by centrifugation at 1,500 × g for 1 min and repeat twice. Collect all three eluants into the same eppendorf tube.
Dry the peptides solution in a speedvac.
3.5. Enrichment of ICAT labeled peptides by Avidin Affinity Chromatography
Reconstitute each dried peptide fraction in 500 µl of the Affinity-load buffer, vortex to mix the solution. Confirm the pH of solution is ~7.0. (See Note 11)
Briefly centrifuge to bring all solution to the bottom of the tubes.
Assemble avidin cartridge system.
Load 2 ml Affinity-Elution Buffer to the cartridge and discard the eluate.
Load 2 ml Affinity-Load Buffer to the cartridge and discard the eluate.
Slowly load (drop by drop) the peptide samples in 500 µl of the Affinity-Load Buffer and collect the flow through. (See Note 12)
Re-load the flow on the avidin cartridge and collect the flow through.
Wash the avidin cartridge with 1 ml of Wash1, divert the eluate to waste.
Wash the avidin cartridge with 1 ml of Wash2, divert the eluate to waste.
Wash the avidin cartridge with 1 ml of Milli-Q water, divert the eluate to waste.
Load 800 µl of Affinity-Elution Buffer into syringe and inject slowly to the cartridge (~1 drop/5 s), discard the first 50 µl of eluate. Collect the remaining 750 µl of eluate into a glass vial.
Repeat steps 1–11 for remaining peptide fractions.
3.6. TFA Cleavage of Biotin Moiety from ICAT Peptides
Speedvac the affinity eluates to complete dryness. (See Note 13)
Prepare cleavage mixture of 95 µl of cleavage reagent A with 5 µl of cleavage reagent B and mix them with dry peptide samples.
Vortex the reaction mixture and incubate at 37 °C for 2 h.
Centrifuge and dry the reaction mixtures.
3.7. RPLC and Spotting MALDI Plate
Reconstitute cleaved ICAT peptide samples in 20 µl of RPLC Mobile Phase A (MPA), vortex vigorously, then centrifuge at 10,000 × g for 5 min.
Equilibrate the RPLC column with 5% RPLC Mobile Phase B for at least 15 min for stable column pressure.
Each fraction (6.4 µl) will be loaded onto a C18 trapping column using a Microliter Pickup method at a flow rate of 20 µl/min. Online desalting step was carried out by a 5-min MPA wash.
- Peptides bound to the trapping column are subsequently resolved on a C18 capillary PepMap column with the following gradient profiles at a flow rate of 400 nl/min:
Time (min) Solvent A Solvent B 0 95 5 2 95 5 75 70 30 90 10 90 100 10 90 105 95 5 115 95 5 Mix the RPLC eluants in line with MALDI matrix in a 1:2 ratio through a 30 nl mixing tee, and deposited onto a MALDI plate using the Probot, at 12 sec per spot.
Repeat the RPLC steps for each SCX fraction.
3.8. Mass Spectrometry
Mix 50 fmol of 6-peptide calibrants at a ratio of 1:1 with MALDI matrix solution. Deposit the freshly prepared calibrant solution on the calibration spots on the MALDI plate.
Create a new spot set, load and align the ICAT sample plates. Load the sample plates into the plate loader of the 4800 Proteomics Analyzer.
Acquire and update the MS calibration file using the six peptides mixture in the Mass Standard Kit. The MS/MS calibration file needs be updated using GFP MS/MS ions (m/z 1570.677).
Acquire MS spectra for each spot in the positive ion mode with a laser intensity of 3,200 and mass range of 850 – 3,500 and sum of the first 1,500 laser shots. In the MS processing method, use internal calibration standards [GFP (m/z 1570.677) and ACTH 18–39 (m/z 2465.199)] to achieve a mass accuracy better than 50 ppm.
After MS analysis, identify and extract ICAT ion pairs (Δ m/z 9.03 ± 0.03 Da) by GPS Explorer software (v3.5 ABI). Compute the relative ICAT ratios with the integrated chromatographic areas of each ICAT ion pair. Submit ICAT pair with over 20% change ratio and Signal/Noise ratio (S/N) > 50 to MS/MS acquisition. (Note 14)
In the MS/MS acquisition method, spectra are accumulation of 2,000 laser shots at a laser intensity of 3,200 using 2-keV collision energy and 5 × 10−7 torr collision gas pressure. Generate peak lists with 4000 Series Explorer with the following settings: set S/N threshold to 10, local noise window width at 250 m/z, and minimum peak width bin size was 2.9; set resolution at 22,000 at m/z 2,400 for MS and 8,000 at m/z 2,000 for MS/MS. Smooth MS/MS spectra with the Savitsky-Golay algorithm (FWHM = 9, polynomial order = 4).
3.9. Database Search
Perform peptide identification on a MASCOT search engine (v1.9) integrated in the GPS Explorer software with the following search parameters: one missed tryptic cleavage, 50 ppm for MS mass error tolerance and 0.3 Da for MS/MS mass error tolerance, variable modifications included ICAT L/H modifications, carbamidomethylation of cysteines, and methionine oxidation.
Unique peptides with confidence interval (C. I.) values above 95% from the MS/MS search are considered significant. (Note 15)
3.10. Trypsin Digestion and iTRAQ labeling
Pipette 100 µg of protein from the four samples into four separate tubes (see Note 16). To each sample, add 2 µl of reducing reagent and vortex. Bring down the contents with a brief centrifugation. Incubate the sample tubes at 60 °C for 1 h. Spin briefly to settle the liquid to the bottom of each tube.
To each sample, carefully add 1 µl of the cysteine blocking reagent. Mix by vortexing and centrifuge briefly to collect the solutions at the bottom of the tube. Incubate at room temperature for 10 min.
Reconstitute two vials of trypsin (20 µg/vial) with 25 µl each of HPLC grade water. Vortex briefly.
To each sample tube, add 10 µl of the trypsin solution, vortex and centrifuge briefly to collect the solution at the bottom of the tube. Incubate at 37 °C for 12 to 16 h. Spin briefly to bring the sample solution to the bottom of the tubes (see Note 17).
Bring the iTRAQ reagents to room temperature. Add 70 µl of ethanol into each reagent vial, cap the vial and vortex vigorously, then centrifuge briefly to settle the iTRAQ reagents to the bottoms of the vials (see Note 18).
Transfer the entire content of one iTRAQ reagent vial into each of the four sample tubes, and vortex to mix thoroughly. Spin briefly to collect the liquid at the bottom of the tubes. Peptides derived from the two control samples are labeled with iTRAQ Reagents 114 and 115 whereas peptides obtained from the two Trx1 over-expressed samples are labeled with iTRAQ Reagents 116 and 117. Incubate the reaction vials at room temperature for 1 h.
Carefully combine the entire contents of all four iTRAQ labeled samples into one tube, mix thoroughly by vortexing, then centrifuge briefly.
3.11. 2D-LC separation and MS analysis
The combined peptide mixture will be first separated by SCX-LC to remove excess iTRAQ reagents. In order to remove both TEAB and the organic solvent from the sample, dry the combined sample completely in a vacuum concentrator (see Note 19). Follow the same steps in 3.3. to fractionate the peptides.
- Follow the same steps in 3.4. to desalt the peptides. Dry the resulting peptides by Speedvac and reconstitut the peptides with 10 μl Solvent A. Use Nano-RPLC for peptide separations following the same steps in 3.7., except using the following gradient:
Time (min) Solvent A Solvent B 0 95 5 4 92 8 34 82 18 57 62 38 64 5 95 69 5 95 70 95 5 85 95 5 Mix the RPLC eluants in line with MALDI matrix and deposited onto a MALDI plate. Follow steps 1 to 3 in 3.8. to caliberate the 4800 MALDI TOF/TOF analyzer.
Create an acquisition, a processing and a job-wide interpretation method for both MS and MS/MS analyses. Use the MS acquisition method in a positive MS reflector with a mass range of 850–3,000 (in Da) and a focus mass of 1950 Da. Set the laser intensity to 3,000 and the detector voltage multiplier at 0.90. Averaged MS spectrum over 1,000 laser shots. In the processing method, GluFib (m/z 1570.677) and ACTH 18–39 (m/z 2465.199) masses are used as the internal calibrants.
For the interpretation method, the precursor selection is based on a minimum S/N filter of 50, precursor mass tolerance of 200 ppm and from weakest to strongest peaks as an MS/MS acquisition order.
Use a 2 KV positive MS/MS method. Set the laser intensity to 4,000 and detector voltage multiplier at 0.90. Specify the metastable suppression as “on” and the precursor mass window at relative 400 resolution (FWHM). Each MS/MS spectrum is accumulated over 4,000 laser shots. In the MS/MS processing method, each spectrum is smoothed using the Savitsky-Golay algorithm with points across the peak set at 3 and polynomial order set at 4. Set the medium CID gas recharge pressure to medium with a threshold of 5.0 × 107 torr.
3. 12. Bioinformatics Analysis
Peptide identification is performed by searching the MS/MS spectra against Swissprot mouse database (see Note 20), using a local MASCOT search engine (v. 1.9) on a GPS (v. 3.5, ABI) server. The following search parameters are used: trypsin with one missed cleavage, mass tolerance of 50 ppm for the precursor ions, and 0.3 Da for the MS/MS fragment. iTRAQ labeled N-terminal and lysine, and cysteine methanethiolation were selected as fixed modifications, while methionine oxidation and iTRAQ-labeled tyrosine were considered as variable modifications. Only peptides identified with confidence interval (C. I.) values greater than 95% should be used for protein identification and quantitation.
Extract the iTRAQ reporter ions cluster areas using GPS explorer. Only ion counts greater than 5,000 are used for quantification analysis. The individual reporter ion peak areas for each iTRAQ channel are normalized by the population median.
For each peptide, the ratio of normalized reporter ion peak areas at 115, 116 and 117 are divided by the normalized reporter ion peak areas at 114. Such ratios are then transformed into log2 values. In cases of multiple MS/MS spectra matched to the same peptide sequence, the peptide ratio is calculated and weighted based on the relative proportion of each spectrum.
- The mean of all peptide ratios from the same protein are calculated. The relative protein expression between Tg-Trx and Control samples are computed based on the following equation.
Pi: the pooled protein log2 ratio of the ith protein. (i = 1, 2, 3, … N, where N is the total number of identified proteins). The p-values in Student’s t-tests are calculated by comparing each protein log2 ratio in the control group (P114 and P115) to those in the Tg-Trx group (P116 and P117) using Microsoft Excel. Anti-log2 of Pi values are calculated to produce the exact protein fold change values.
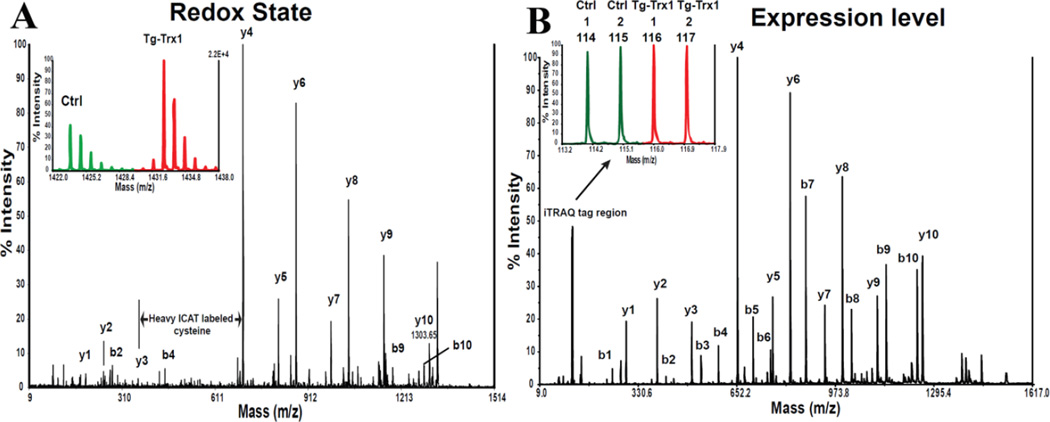
Figure 2. Peptide identification and quantitation by ICAT and iTRAQ.
Panel A) Sequence of cysteine containing peptide is identified by matching MS2 fragment ion mass to the predicted theoretical fragments. The addition of 227 Da (236 Da for the heavy ICAT tag) to a cysteine residue is a hallmark of an ICAT labeled peptide (mass difference between y3 and y4 ions). Quantitation of light and heavy labeled peptides (redox states of cysteines in this case) are computed from the precursor intensities acquired from the MS1 scan (left insert) or from the integrated extracted ion chromatogram. B) iTRAQ quantitation is carried out on the reporter region (mass range 114 –117 for 4-plex iTRAQ) in the MS2 spectra (see insert of panel B). The individual peak intensity of each mass reporter ion reflects the corresponding peptide/protein level in each sample. The peptide sequence is identified in a similar manner as ICAT technology (Modified from Molecular & Cellular Proteomics, 2009 (8), 1674–1687 with permission).
Acknowledgements
The authors are grateful for the funding support from NIH/NINDS grant NS046593 to H.L for the support of a NINDS NeuroProteomics Core Facility at UMDNJ-New Jersey Medical School.
Footnotes
It is recommended to use freshly prepared lysis buffer. Selection of proper detergents is discretional upon protein of interest (e.g. Membrane proteins)
To preserve the native redox states of protein cysteines, it is highly recommended to minimize sample exposure to air, keep samples on ice during sample preparation if compatible and purge with high-purity nitrogen for extended incubation (e. g. trypsin digestion step).
Depending on the detergents of choice, excessive bubbles may be formed in the lysis process. A quick spin-down of sample tubes at 4°C will help to remove bubbles.
Estimate free protein thiol content in the samples. For example, for 100 µg protein with an average mass of 50 kDa and 6 cysteines per protein, the total cysteine content can be estimated as 100×10−6g/50,000g/mol*6=12 nmol. Each ICAT tube contains 175 nmol of labeling reagent to maintain excessive reagent/free cysteines ratio >10 times for complete labeling.
Whole cell lysates contain many small molecules that could have adverse effects on protein ICAT labeling. For example, glutathione and other cysteine-containing antioxidants are observed at high levels (mM) and will consume ICAT reagents at much faster reaction rates than protein thiols. Alternative precipitation methods (such as TCA precipitation and Methanol/Chloroform precipitation) and buffer exchange methods (membrane ultrafiltration) can be implemented to remove interfering molecules.
To avoid protein loss and sample variation, complete solubilization of proteins is a key step. Mild agitation with eppendorf pipet tips and sonication in a water bath will facilitate protein solubilization. Increasing SDS concentration to 0.05% can also enhance the degree of protein solubilization.
In addition to scavenging excess ICAT reagents; DTT also reduces disulfide bonds and other reversible cysteine modifications. This is important in the forward redox ICAT labeling scheme, since the reduction of disulfide bonds (omit before ICAT labeling) will facilitate tryptic digestions and improve protein and peptide identifications.
Aliquot 1 µl of solution from each reaction mixture (Step 13), and 1 µl of solution before and after tryptic digestion for as a quality control step. Load the samples in separate lanes of 1D-SDS PAGE. After electrophoresis and protein staining; evaluate the initial sample loading and digestion efficiency. In the second QC test, mix 1 µl each of tryptic heavy and light ICAT labeled peptides, desalt with ZipTip and spot the sample at 1:1 ratio with MALDI matrix solution on a MALDI plate. Acquire MS spectrum of the peptides, evaluate the abundance and relative ratio of ICAT pairs with 9 Da (or multiplier) mass differences.
Reducing urea concentration to <1M is important for effective trypsin digestion. It is also important to adjust the pH of the solution to the range of 8.0 – 8.5 for optimum trypsin activity.
Combining adjacent fractions with less peptide abundance according to the SCX chromatogram, greatly reduces the sample processing time without much loss of total protein identification and quantification. Late eluted fractions might have more salts and require additional loading buffer for full dissolution.
The loading capacity of the avidin cartridge is ~ 10 µg of peptide. A new avidin cartridge can be used up to 50 times with proper usage and storage.
Using a syringe pump (e.g. Standard Infusion Only Pump 11 Elite Syringe Pumps, Harvard Apparatus, Holliston, MA. USA) for solvent delivery yields more consistent and robust results.
Residual water content will have an adverse effect on the cleavage reaction. It is critical to dry the peptide samples completely before TFA cleavage. Perform the reaction in a hood with proper ventilation. Since TFA is reactive and corrosive to a broad range of materials, it is suggested to use glass vials and glass syringe with metal plunger to transfer and hold the reaction mixtures.
Peptides containing multiple cysteines can result in multiple ICAT labeling, thus the mass difference for these peptides may be observed in multipliers of (Δ m/z 9.03 ± 0.03 Da). This can be addressed by setting allowance of multiple ICAT labeling in the GPS Explorer software.
One-hit-wonder is one of the caveats in using ICAT labeled peptides for protein identification and quantifications, given the low observation frequency of cysteines (< 3.3%) in vertebrate proteomes. It is not uncommon to find only one ICAT labeled peptide for a given protein. Careful inspection of MS and MS/MS spectra for positive identification and removal of potential interference is important to reduce false positive results. One confirmative hallmark for ICAT labeled peptide is the signature mass differences of 339.1 for heavy ICAT labeled, and 330.1 for light ICAT labeled, between yn and yn2212;1 ions, where n indicates the location of ICAT labeled cysteine (Figure 2).
Based on iTRAQ instructions from ABI, each protein sample should be between 5 to 100 µg for each iTRAQ labeling reaction. To ensure maximum labeling efficiency, sample volumes should be less than 50 µl each. If the sample volume is larger than 50 µl, a speedvac can be used to reduce the sample volume before iTRAQ labeling.
It is important to check the protein digestion efficiency before iTRAQ labeling. Take 1 µl of each digested sample and desalt it using a C18 ZipTip (Millipore, Billerica, MA). Mix the eluted peptides with the MALDI matrix solution in a 1:1 ratio and spot them onto a MALDI plate. Acquire MS spectra to check if the peptide ion signals are comparable.
To maximize labeling efficiency, the concentration of organic reagents (ethanol and iTRAQ reagents) in iTRAQ labeling reactions should be larger than 60% (v/v).
To remove all of the TEAB, reconstitute the combined iTRAQ labeled samples in 100 µl of HPLC grade water and dry the sample in a vacuum concentrator. Repeat this step twice to ensure all the TEAB is evaporated.
It is important to use the latest version of the protein database to ensure comprehensive peptide identification. Swissprot, IPI, NCBI protein database or EST (6 frame translation into protein sequences) can be used, with an increasing number of entries and database size. Generally speaking, using bigger databases will likely increase one’s chance to match a spectrum to a peptide sequence. However, it will also increase the odds for random matching. We chose the Swissprot database for our study, because of its high protein sequence accuracy and low redundancy.
References
- 1.Ross PL, Huang YN, Marchese JN, Williamson B, Parker K, Hattan S, Khainovski N, Pillai S, Dey S, Daniels S, Purkayastha S, Juhasz P, Martin S, Bartlet-Jones M, He F, Jacobson A, Pappin DJ. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol Cell Proteomics. 2004;3:1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- 2.Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 3.Liu T, Donahue KC, Hu J, Kurnellas MP, Grant JE, Li H, Elkabes S. Identification of Differentially Expressed Proteins in Experimental Autoimmune Encephalomyelitis (EAE) by Proteomic Analysis of the Spinal Cord. Journal of Proteome Research. 2007;6:2565–2575. doi: 10.1021/pr070012k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gygi SP, Rist B, Gerber SA, Turecek F, Gelb MH, Aebersold R. Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat Biotechnol. 1999;17:994–999. doi: 10.1038/13690. [DOI] [PubMed] [Google Scholar]
- 5.Fu C, Hu J, Liu T, Ago T, Sadoshima J, Li H. Quantitative analysis of redox-sensitive proteome with DIGE and ICAT. J Proteome Res. 2008;7:3789–3802. doi: 10.1021/pr800233r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yamamoto M, Yang G, Hong C, Liu J, Holle E, Yu X, Wagner T, Vatner SF, Sadoshima J. Inhibition of endogenous thioredoxin in the heart increases oxidative stress and cardiac hypertrophy. J Clin Invest. 2003;112:1395–1406. doi: 10.1172/JCI17700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fu C, Wu C, Liu T, Ago T, Zhai P, Sadoshima J, Li H. Elucidation of thioredoxin target protein networks in mouse. Mol Cell Proteomics. 2009;8:1674–1687. doi: 10.1074/mcp.M800580-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choe L, D'Ascenzo M, Relkin NR, Pappin D, Ross P, Williamson B, Guertin S, Pribil P, Lee KH. 8-Plex quantitation of changes in cerebrospinal fluid protein expression in subjects undergoing intravenous immunoglobulin treatment for Alzheimer's disease. Proteomics. 2007;7:3651–3660. doi: 10.1002/pmic.200700316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sethuraman M, McComb ME, Huang H, Huang S, Heibeck T, Costello CE, Cohen RA. Isotope-coded affinity tag (ICAT) approach to redox proteomics: identification and quantitation of oxidant-sensitive cysteine thiols in complex protein mixtures. J Proteome Res. 2004;3:1228–1233. doi: 10.1021/pr049887e. [DOI] [PubMed] [Google Scholar]
- 10.Leichert LI, Gehrke F, Gudiseva HV, Blackwell T, Ilbert M, Walker AK, Strahler JR, Andrews PC, Jakob U. Quantifying changes in the thiol redox proteome upon oxidative stress in vivo. Proceedings of the National Academy of Sciences. 2008;105:8197–8202. doi: 10.1073/pnas.0707723105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Michalski A, Cox J, Mann M. More than 100,000 Detectable Peptide Species Elute in Single Shotgun Proteomics Runs but the Majority is Inaccessible to Data-Dependent LC−MS/MS. Journal of Proteome Research. 2011;10:1785–1793. doi: 10.1021/pr101060v. [DOI] [PubMed] [Google Scholar]
- 12.Washburn MP, Wolters D, Yates JR., 3rd Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 13.Chenau J, Michelland S, Sidibe J, Seve M. Peptides OFFGEL electrophoresis: a suitable pre-analytical step for complex eukaryotic samples fractionation compatible with quantitative iTRAQ labeling. Proteome Science. 2008;6:9. doi: 10.1186/1477-5956-6-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Obad S, dos Santos CO, Petri A, Heidenblad M, Broom O, Ruse C, Fu C, Lindow M, Stenvang J, Straarup EM, Hansen HF, Koch T, Pappin D, Hannon GJ, Kauppinen S. Silencing of microRNA families by seed-targeting tiny LNAs. Nat Genet. 2011;43:371–378. doi: 10.1038/ng.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ralhan R, DeSouza LV, Matta A, Chandra Tripathi S, Ghanny S, Datta Gupta S, Bahadur S, Siu KWM. Discovery and Verification of Head-and-neck Cancer Biomarkers by Differential Protein Expression Analysis Using iTRAQ Labeling, Multidimensional Liquid Chromatography, and Tandem Mass Spectrometry. Molecular & Cellular Proteomics. 2008;7:1162–1173. doi: 10.1074/mcp.M700500-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keshamouni VG, Michailidis G, Grasso CS, Anthwal S, Strahler JR, Walker A, Arenberg DA, Reddy RC, Akulapalli S, Thannickal VJ, Standiford TJ, Andrews PC, Omenn GS. Differential Protein Expression Profiling by iTRAQ−2DLC−MS/MS of Lung Cancer Cells Undergoing Epithelial-Mesenchymal Transition Reveals a Migratory/Invasive Phenotype. Journal of Proteome Research. 2006;5:1143–1154. doi: 10.1021/pr050455t. [DOI] [PubMed] [Google Scholar]
- 17.Ago T, Yeh I, Yamamoto M, Schinke-Braun M, Brown JA, Tian B, Sadoshima J. Thioredoxin1 upregulates mitochondrial proteins related to oxidative phosphorylation and TCA cycle in the heart. Antioxid Redox Signal. 2006;8:1635–1650. doi: 10.1089/ars.2006.8.1635. [DOI] [PubMed] [Google Scholar]
- 18.Yang Y, Ago T, Zhai P, Abdellatif M, Sadoshima J. Thioredoxin 1 negatively regulates angiotensin II-induced cardiac hypertrophy through upregulation of miR-98/let-7. Circ Res. 2011;108:305–313. doi: 10.1161/CIRCRESAHA.110.228437. [DOI] [PMC free article] [PubMed] [Google Scholar]