The voltage-gated proton channel, HV1, is notoriously unique among ion channels (1), and plays key roles in the health and disease of diverse tissues and species (2). Li et al. (3) combine biochemical, computational, and electron paramagnetic resonance (EPR) spectroscopic approaches to shed light on structural aspects of the human proton channel, hHV1. Their results advance the field in several key areas, culminating in a bold new model for gating.
The voltage-sensing domain (VSD) is the part of voltage-gated ion channels that senses the electrical potential across the cell membrane where the channel resides. Most such channels open upon membrane depolarization, which is accomplished mainly by cationic amino acids located in the fourth transmembrane helical segment (S4) moving outward when the inside of the cell is made more positive. This movement is transduced from the VSD (S1–S4) to the pore region (S5–S6), opening a conduction pathway. In addition to biologically important voltage-gated K+, Na+, Ca2+, and H+ channels, other classes of membrane proteins with VSDs exist that are not channels at all. One example is a voltage-sensing phosphatase (VSP), an enzyme whose activity is regulated by membrane potential. In a landmark study in 2014 (4), the Li–Perozo group reported crystal structures for CiVSP in both “down” and “up” conformations, the first VSD-containing molecule to have structures determined in both states. Voltage-gated ion channels are closed at negative voltages, and open upon depolarization as the S4 helix moves “up” through the membrane electrical field. Because the VSP is not a channel, “closed” and “open” become “down” and “up.” The gene for hHV1 was identified only in 2006 (5). To the astonishment of everyone, the gene product bore a striking resemblance to the VSD of other voltage-gated ion channels, so much so that the simultaneously identified mouse (mHV1) and Ciona intestinalis (CiHV1) gene products were dubbed VSOP or “voltage sensor-only protein” (6). HV1 has only four membrane-spanning helices (S1–S4); these sense voltage but also contain the proton conduction pathway (7).
Crystal structures are great, up to a point. They provide tremendously detailed information about molecules, but they have limitations that are sometimes overlooked. They tell us about structure, but only the structure of whatever exists in the crystal. Proteins have many conformations, and one must determine or guess which one was captured during crystallization, and hope it is a native conformation and not a broken one. Forming crystals of membrane proteins is challenging, and often the protein is modified to facilitate crystallization. Ligands or chaperone-like proteins are included, parts of the molecule are truncated, chimerae are produced: whatever it takes to get a good crystal. HV1 has not been successfully crystallized in its entirety. First came crystal structures of the C terminus alone that lacked the entire transmembrane region (8, 9). Then in 2014, the first exciting glimpse of HV1 appeared. Well, not quite HV1, but a chimera of the mouse proton channel, mHV1, with the C terminus replaced by a leucine zipper transcriptional activator GCN4 from Saccharomyces cerevisiae, and with the cytoplasmic ends of S2–S3 replaced by the corresponding section of CiVSP. Nevertheless, this three-species chimera functions as a proton channel and thus retains essential features. A protein in a crystal senses no membrane potential, and is assumed to be in a state occupied at 0 mV. This means HV1 is closed, although with no pH gradient, the channel begins to open within 20 mV (10). HV1 exhibits complex gating kinetics (10–12), suggesting it has multiple closed states. Because 0 mV is close to the “threshold” voltage where channels first open, the crystal may have captured a shallow closed state.
EPR provides useful information not available from other approaches, but has its own limitations. Significantly, the protein could be studied in situ in its native environment, a lipid bilayer, in contrast to a crystal in which interactions with the membrane are lost. The greatest limitation may be the necessity to introduce a bulky spin label whose presence inside the protein may perturb the native structure at least locally. Li et al. (3) replaced the lone native cysteine (Cys) of hHV1 and then introduced Cys at each of 149 locations encompassing the entire VSD. A spin label was attached to each construct and the molecules reconstituted into liposomes. Both the full-length 273-amino acid protein and a VSD-only construct were purified and shown to mediate proton conduction in liposomes; VSD-only constructs were used in all EPR measurements.
Three parameters obtained from EPR are mobility, O2 accessibility (which indicates proximity to lipid), and NiEDDA (Ni2+ ethylenediaminediacetic acid) accessibility, which reports aqueous exposure. Both mobility and O2 accessibility are larger for hHV1 than for other VSD-containing molecules, revealing a dynamic molecule, deficient in tertiary contacts, that historically has resisted all attempts to obtain a crystal structure. NiEDDA accessibility defines the boundaries of the transmembrane regions (S1–S4), and reveals that the channel pore has a short isthmus between two aqueous vestibules, reminiscent of the VSDs of K+ and Na+ channels, which also focus the electric field over a short distance (13–15). This “hydrophobic gasket” forms a dielectric barrier to water and ion permeation and separates “in” from “out.” The hydrophobic region (yellow in Fig. 1) encompasses the outer two arginines of the S4 helix (R1 and R2), but the third (R3) is accessible internally. Homology models of hHV1 (16) and CiHV1 (17) indicate a ∼10 Å long hydrophobic region that in hHV1 includes the selectivity filter Asp112 (18) and Phe150 that is conserved universally in VSD-containing molecules (19).
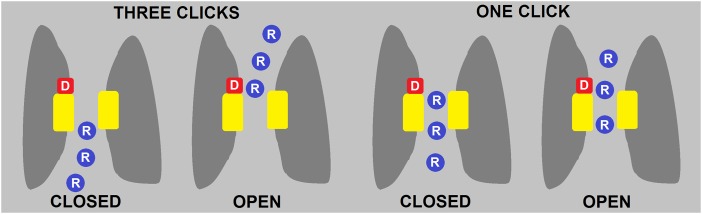
Fig. 1.
The “three-click” model (Left) is a logical extrapolation to HV1 of S4 movement during K+ or Na+ channel opening. The three Arg (R) in the S4 helix all move outward past the yellow hydrophobic gasket, from intracellular to extracellular aqueous vestibules. In the new “one-click” model (Right), Li et al. (3) propose that S4 moves only one turn of the helix upwards.
Mammalian HV1 are dimers, although each protomer has its own conduction pathway and can function independently (20, 21). EPR measurements at each of 149 positions reveal the distance between each pair in the dimer, unambiguously defining the dimer interface and resolving internecine disputes on this point (22, 23). The EPR data resoundingly establish the dimer interface to be at the top (extracellular end) of S1 and the lower part of S4. Dimerization was thought to result mainly from extensive coiled-coil interactions between the C termini (20–22). Surprisingly, Li et al. (3) found that their VSD-only construct of hHV1, which lacks the C terminus, spontaneously associated as a dimer with Kd 3 μM.
Previous homology models of HV1 used VSDs from crystal structures of other channels as templates. Li et al. (3) generated models based on structures of KVAP, KV1.2, NaVAb, or CiVSP, and tuned these using molecular dynamics. EPR solvent accessibility data from hHV1 favored the CiVSP-based model. This is reasonable, because phylogenetically, HV1 are related more closely to VSP than to other channels (24). Although the closed hHV1 model agrees well with the structure of the closed mHV1 chimera, parts of S2 and S3 from the CiVSP-based model must be shifted up or down to match the crystal structure. A consequence of this mismatch is seen in Fig. 2. The residues in hHV1 corresponding to those forming the hydrophobic gasket in CiVSP (4), V109, F150, and V178 (yellow in Fig. 2), align horizontally in CiVSP and in our homology model of hHV1 (16). However, in the mHV1 structure, V174 in the S3 helix is too low. Perhaps CiVSP is not an ideal template, but it seems more than coincidental that parts of both S2 and S3 were replaced in the chimera. Shifting S3 of mHV1 up by one “click” to match CiVSP would align the gasket nicely.
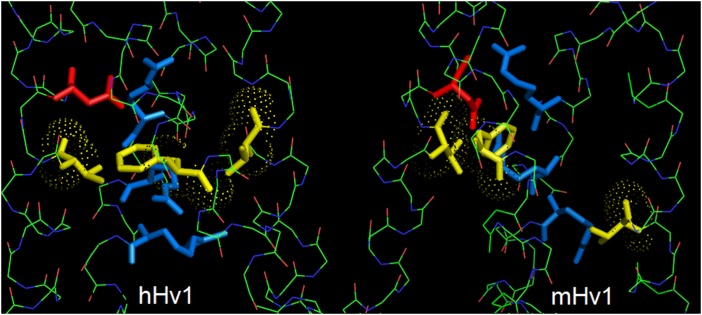
Fig. 2.
The hydrophobic gasket (three yellow amino acids) is aligned in the new hHV1 model based on the CiVSP crystal structure (Left), but not in the chimeric mHV1 crystal structure (Right). In each closed channel viewed from the side, the three S4 Arg are blue and the Asp in S1 that produces proton selectivity is red. The apparent misalignment in mHV1 could reflect a disturbance due to the spliced-in S2–S3 segment from CiVSP, which ends two positions below V174, the anomalous residue in mHV1. Alternatively, a different amino acid might complete the gasket in mHV1.
The most far-reaching conclusion of Li et al. (3) is that S4 moves much less during gating than most envisioned. Seduced by the surprising resemblance of HV1 to other VSDs, everyone initially assumed that its S4 would move just like it does in other channels: basic amino acids spaced every three positions along S4 move past the hydrophobic gasket. In other channels, three to four charges move past the gasket during opening, so it was expected that all three Arg in S4 of HV1 would move from the intracellular to the extracellular side (Fig. 1). However, replacing the innermost Arg with histidine (R211H) resulted in inhibition by internal Zn2+ even when the channel was open, arguing that S4 movement was far more restricted in hHV1 than in other VSDs (16). Given overall agreement of the EPR data with the mHV1 structure, the fact that CiVSP is the only VSD with structures of both down and up conformations, and the phylogenetic proximity of CiVSP and hHV1 (24), Li et al. (3) propose a gating model for hHV1 (Fig. 1) based on their “one-click” model for CiVSP (4). The S4 segment moves up just one turn of the helix, in contrast to the three or four clicks of other VSDs. If this model survives the test of time, it will add one more distinctive feature to an already unique channel.
Acknowledgments
This work was supported by National Science Foundation Award MCB-0943362 and National Institutes of Health Grant R01-GM087507.
Footnotes
The author declares no conflict of interest.
See companion article on page E5926.
References
- 1.DeCoursey TE. The voltage-gated proton channel: A riddle, wrapped in a mystery, inside an enigma. Biochemistry. 2015;54(21):3250–3268. doi: 10.1021/acs.biochem.5b00353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.DeCoursey TE. Voltage-gated proton channels: Molecular biology, physiology, and pathophysiology of the HV family. Physiol Rev. 2013;93(2):599–652. doi: 10.1152/physrev.00011.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li Q, et al. Resting state of the human proton channel dimer in a lipid bilayer. Proc Natl Acad Sci USA. 2015;112:E5926–E5935. doi: 10.1073/pnas.1515043112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li Q, et al. Structural mechanism of voltage-dependent gating in an isolated voltage-sensing domain. Nat Struct Mol Biol. 2014;21(3):244–252. doi: 10.1038/nsmb.2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ramsey IS, Moran MM, Chong JA, Clapham DE. A voltage-gated proton-selective channel lacking the pore domain. Nature. 2006;440(7088):1213–1216. doi: 10.1038/nature04700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sasaki M, Takagi M, Okamura Y. A voltage sensor-domain protein is a voltage-gated proton channel. Science. 2006;312(5773):589–592. doi: 10.1126/science.1122352. [DOI] [PubMed] [Google Scholar]
- 7.Lee SY, Letts JA, MacKinnon R. Functional reconstitution of purified human Hv1 H+ channels. J Mol Biol. 2009;387(5):1055–1060. doi: 10.1016/j.jmb.2009.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li SJ, et al. The role and structure of the carboxyl-terminal domain of the human voltage-gated proton channel Hv1. J Biol Chem. 2010;285(16):12047–12054. doi: 10.1074/jbc.M109.040360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fujiwara Y, Takeshita K, Nakagawa A, Okamura Y. Structural characteristics of the redox-sensing coiled coil in the voltage-gated H+ channel. J Biol Chem. 2013;288(25):17968–17975. doi: 10.1074/jbc.M113.459024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cherny VV, Markin VS, DeCoursey TE. The voltage-activated hydrogen ion conductance in rat alveolar epithelial cells is determined by the pH gradient. J Gen Physiol. 1995;105(6):861–896. doi: 10.1085/jgp.105.6.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeCoursey TE, Cherny VV. Voltage-activated hydrogen ion currents. J Membr Biol. 1994;141(3):203–223. doi: 10.1007/BF00235130. [DOI] [PubMed] [Google Scholar]
- 12.Villalba-Galea CA. Hv1 proton channel opening is preceded by a voltage-independent transition. Biophys J. 2014;107(7):1564–1572. doi: 10.1016/j.bpj.2014.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang N, George AL, Jr, Horn R. Molecular basis of charge movement in voltage-gated sodium channels. Neuron. 1996;16(1):113–122. doi: 10.1016/s0896-6273(00)80028-8. [DOI] [PubMed] [Google Scholar]
- 14.Starace DM, Bezanilla F. Histidine scanning mutagenesis of basic residues of the S4 segment of the Shaker K+ channel. J Gen Physiol. 2001;117(5):469–490. doi: 10.1085/jgp.117.5.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Starace DM, Bezanilla F. A proton pore in a potassium channel voltage sensor reveals a focused electric field. Nature. 2004;427(6974):548–553. doi: 10.1038/nature02270. [DOI] [PubMed] [Google Scholar]
- 16.Kulleperuma K, et al. Construction and validation of a homology model of the human voltage-gated proton channel hHV1. J Gen Physiol. 2013;141(4):445–465. doi: 10.1085/jgp.201210856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chamberlin A, Qiu F, Wang Y, Noskov SY, Larsson HP. Mapping the gating and permeation pathways in the voltage-gated proton channel Hv1. J Mol Biol. 2015;427(1):131–145. doi: 10.1016/j.jmb.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Musset B, et al. Aspartate 112 is the selectivity filter of the human voltage-gated proton channel. Nature. 2011;480(7376):273–277. doi: 10.1038/nature10557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tao X, Lee A, Limapichat W, Dougherty DA, MacKinnon R. A gating charge transfer center in voltage sensors. Science. 2010;328(5974):67–73. doi: 10.1126/science.1185954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koch HP, et al. Multimeric nature of voltage-gated proton channels. Proc Natl Acad Sci USA. 2008;105(26):9111–9116. doi: 10.1073/pnas.0801553105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tombola F, Ulbrich MH, Isacoff EY. The voltage-gated proton channel Hv1 has two pores, each controlled by one voltage sensor. Neuron. 2008;58(4):546–556. doi: 10.1016/j.neuron.2008.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee SY, Letts JA, Mackinnon R. Dimeric subunit stoichiometry of the human voltage-dependent proton channel Hv1. Proc Natl Acad Sci USA. 2008;105(22):7692–7695. doi: 10.1073/pnas.0803277105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Musset B, et al. Zinc inhibition of monomeric and dimeric proton channels suggests cooperative gating. J Physiol. 2010;588(Pt 9):1435–1449. doi: 10.1113/jphysiol.2010.188318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith SME, et al. Voltage-gated proton channel in a dinoflagellate. Proc Natl Acad Sci USA. 2011;108(44):18162–18167. doi: 10.1073/pnas.1115405108. [DOI] [PMC free article] [PubMed] [Google Scholar]