Significance
Studies examining paternal exposure to diverse environmental stimuli propose that epigenetic marks in germ cells, including small noncoding RNAs such as microRNA (miR), transmit experience-dependent information from parent to offspring. However, these nongenetic mechanisms of transgenerational inheritance are poorly understood, specifically how these germ-cell marks may act postfertilization to enact long-term changes in offspring behavior or physiology. In this study, through zygote microinjection of nine specific sperm miRs previously identified in our paternal stress mouse model, we demonstrate that sperm miRs function to reduce maternal mRNA stores in early zygotes, ultimately reprogramming gene expression in the offspring hypothalamus and recapitulating the offspring stress dysregulation phenotype.
Keywords: transgenerational, epigenetic, stress, microRNA, paternal
Abstract
Epigenetic signatures in germ cells, capable of both responding to the parental environment and shaping offspring neurodevelopment, are uniquely positioned to mediate transgenerational outcomes. However, molecular mechanisms by which these marks may communicate experience-dependent information across generations are currently unknown. In our model of chronic paternal stress, we previously identified nine microRNAs (miRs) that were increased in the sperm of stressed sires and associated with reduced hypothalamic–pituitary–adrenal (HPA) stress axis reactivity in offspring. In the current study, we rigorously examine the hypothesis that these sperm miRs function postfertilization to alter offspring stress responsivity and, using zygote microinjection of the nine specific miRs, demonstrated a remarkable recapitulation of the offspring stress dysregulation phenotype. Further, we associated long-term reprogramming of the hypothalamic transcriptome with HPA axis dysfunction, noting a marked decreased in the expression of extracellular matrix and collagen gene sets that may reflect an underlying change in blood–brain barrier permeability. We conclude by investigating the developmental impact of sperm miRs in early zygotes with single-cell amplification technology, identifying the targeted degradation of stored maternal mRNA transcripts including sirtuin 1 and ubiquitin protein ligase E3a, two genes with established function in chromatin remodeling, and this potent regulatory function of miRs postfertilization likely initiates a cascade of molecular events that eventually alters stress reactivity. Overall, these findings demonstrate a clear mechanistic role for sperm miRs in the transgenerational transmission of paternal lifetime experiences.
Evidence that offspring behavior and physiology can be shaped by parental life experiences has stimulated new consideration of the factors that underlie disease risk and resilience. Notably, perturbations such as parental stress, malnutrition, infection, or advanced age have been associated with an increased incidence of neurodevelopmental disease in offspring (1), and alterations in their hypothalamic–pituitary–adrenal (HPA) stress axis response may be central to increased disease predisposition (2, 3). Studies in diverse animal models have demonstrated similar outcomes, particularly that of offspring HPA axis dysregulation, following either maternal or paternal stress exposure (4–7); yet mechanisms by which parental lifetime stress experience modify offspring development and adult phenotypes remain unclear. In particular, transgenerational transmission via the maternal lineage likely relies on the complex maternal–fetal/neonatal interaction, whereas transmission through the paternal lineage suggests germ-line reprogramming (8).
Germ cell epigenetic marks, vulnerable to environmental stimuli and capable of directing profound developmental change, may mediate the effects of parental lifetime environmental exposures on offspring behavior and physiology (9, 10). Rodent models examining paternal transmission have identified epigenetic signatures in mature sperm as possible substrates of transgenerational programming, namely patterns of retained histone modifications, DNA methylation, and/or populations of small noncoding RNAs (11–21). RNA populations are of primary interest, as they may be altered through intercellular communication via epididymosomes even in transcriptionally inert mature sperm, where DNA condensation impedes other epigenetic change (22–24). Studies in which manipulation of total RNA content in postfertilization zygotes reproduced aspects of a paternally transmitted phenotype highlight the critical importance of RNA as a germ cell epigenetic mark and support the potential role of small RNAs, including microRNAs (miRs), in trait transmission (20, 25). However, neither the identity of specific sperm miRs responsive to environmental challenge, nor mechanisms by which they may function to impact offspring development, have been determined.
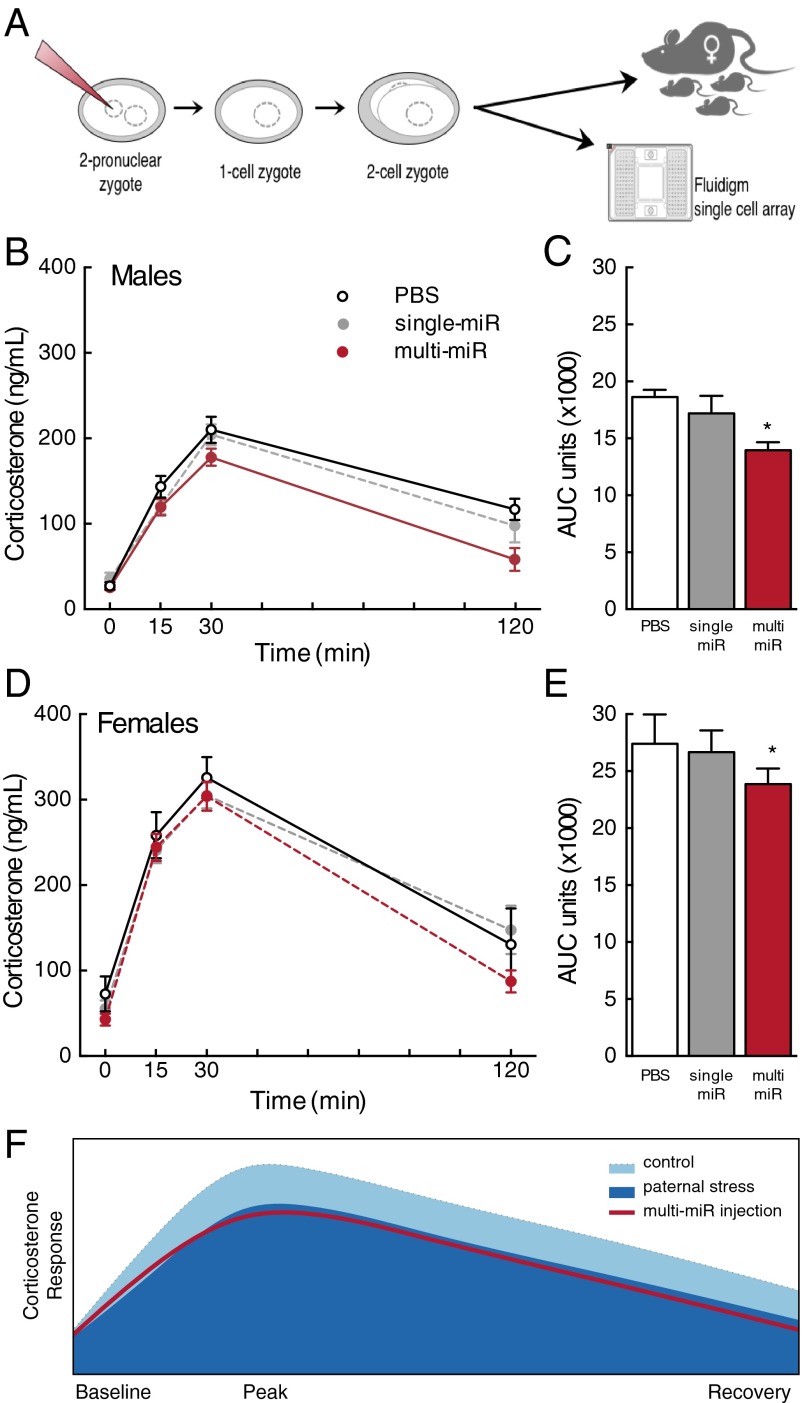
We previously established that the adult offspring of male mice exposed to chronic stress before breeding exhibit a significantly blunted HPA stress axis response and reprogramming of relevant gene sets within the paraventricular nucleus (PVN) of the hypothalamus. In this model, we identified nine specific miRs in the sires’ sperm as the potential germ cell mark sensitive to paternal stress experience (11). Here, to confirm sperm miR content as a mechanism of epigenetic transmission, we microinjected the nine miRs into single-cell zygotes (multi-miR injection). Zygotes were then implanted into surrogate females, reared normally, and examined for adult HPA stress axis sensitivity and long-term reprogramming of PVN gene expression as a recapitulation of the paternal stress phenotype (Fig. 1A). Further, to elucidate the currently unknown function of sperm miRs following fertilization, we evaluated the potential targeting of stored maternal RNA in early zygotes.
Fig. 1.
Zygote microinjection of nine sperm miRs recapitulated the HPA axis phenotype. (A) Schematic of experimental design in which single-cell mouse zygotes were injected with nine miRs (multi-miR; 1 ng/μL final concentration of miR-29c, miR-30a, miR-30c, miR-32, miR-193-5p, miR-204, miR-375, miR-532-3p, and miR-698) or with one of two control conditions: an injection control (PBS only) or a control for the total amount of injected miRs (single miR; 1 ng/μL final concentration miR-193-5p alone) before implantation into surrogate females or preparation for later analyses. (B and D) Corticosterone levels in male and female adult mice were significantly reduced in the multi-miR group relative to PBS controls in response to a 15-min restraint from t = 0–15 min. (C and E) Total AUC of corticosterone production over 120 min was significantly decreased in the multi-miR group compared with PBS controls, and expected sex differences were observed. Data are presented as mean ± SEM n = 6–10 mice per group. *P < 0.05 multi-miR vs. PBS. (F) Model demonstrating the similarity in the shape of the male corticosterone response curve in the multi-miR–injected group with the outcome previously reported in offspring following paternal stress (11).
Results
Recapitulation of Blunted HPA Axis Response.
To determine if the multi-miR injection elicited changes in HPA stress axis responsivity similar to that previously observed in our paternal stress model, we examined plasma corticosterone levels following a brief restraint stressor in adult male and female mice derived from injected zygotes. To ensure that observed outcomes resulted from the action of the nine miRs (multi-miR injection: miR-29c, miR-30a, miR-30c, miR-32, miR-193-5p, miR-204, miR-375, miR-532-3p, and miR-698) independent of experimental techniques and the potential effect of concentration, control conditions included both a PBS injection and a single-miR injection using miR-193-5p (randomly selected from the nine used in the composite) at the same total miR concentration. Corticosterone response was significantly reduced by miR injection [treatment (F2,43 = 3.32, P = 0.046)] and this effect did not vary across time (no interaction, Wilks’ lambda = 0.83, F6,82 = 1.31, P = 0.26), (Fig. 1 B and D). Expected changes in corticosterone levels across time were observed (F3,41 = 32.03, P < 0.0001), as well as expected basal sex differences in corticosterone (F1,43 = 63.82, P < 0.0001). The main effect of sex significantly interacted with time (F3,41 = 33.51, P < 0.0001), but not treatment condition (no interaction, F2,43 = 0.005, P = 0.99). There was no interaction of time, sex, and treatment (Wilks’ lambda = 0.90, F6,82 = 0.73, P = 0.62). Assessment of total corticosterone production in 120 min, measured as area under the curve (AUC), confirmed the significant and noninteracting effects of zygote injection and sex [treatment (F2,37 = 3.44, P = 0.043); sex (F1,37 = 45.02, P < 0.0001); and interaction (F2,37 = 0.33, P = 0.72)]. Total corticosterone was reduced in the multi-miR–injected group compared with PBS (P < 0.05) (Fig. 1 C and E). Animals were otherwise normal in weight and appearance (Table S1).
Programmatic Changes in PVN Transcriptome in Response to Zygote miR Injection.
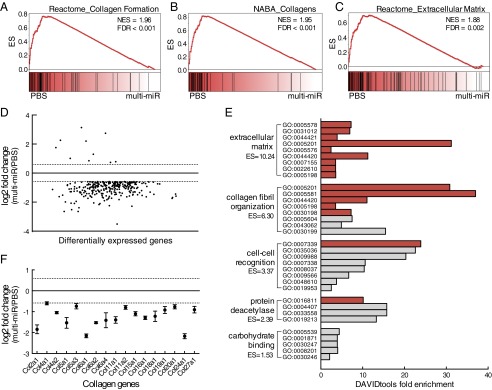
Gene expression patterns in the adult PVN were analyzed using RNA sequencing (RNA-seq) to investigate long-term reprogramming of the HPA axis. Gene set enrichment analysis (GSEA) of the PVN transcriptome in PBS vs. multi-miR groups revealed the robust enrichment of curated gene sets for collagen formation and extracellular matrix organization in the PBS group (26, 27), indicating that the expression of member genes was decreased in the multi-miR–injected group (Fig. 2 A–C). No gene sets were significantly increased in the multi-miR group (Table S2). Differential gene expression analysis using stringent threshold and significance criteria of P < 0.05, false discovery rate (FDR) < 0.05, and log2 fold change ≥ |0.585| identified 298 genes with altered expression in the PVN following correction for multiple comparisons (Table S3), and the majority of these genes (288 of 298) exhibited decreased expression (Fig. 2D). Gene ontology (GO) analysis of the 298 dysregulated PVN genes using DAVID tools functional annotation clustering similarly identified an enrichment of collagen and extracellular matrix-related terms (28, 29), driven in part by the decreased expression of 17 collagen genes (Fig. 2 E and F). Notably, expression of the nine injected miRs was unchanged in the adult PVN of the multi-miR group (Fig. S1).
Fig. 2.
Altered transcriptome profile in adult PVN from multi-miR–injected zygotes supports a lasting epigenetic modification. (A–C) GSEA of RNA-seq data identified three significantly enriched C2 gene sets with normalized enrichment score (NES) > 1.8 and FDR < 0.05, illustrating a decrease in collagen formation and extracellular matrix genes in the multi-miR–injected group. (D) Differential expression analysis identified 298 (10 up, 288 down) genes significantly changed by at least 1.5 fold (log2 fold change ≥ |0.585|, dotted lines). Genes are presented in ranked order of expression from low to high. P < 0.05, FDR < 0.05. (E) Functional annotation clustering of dysregulated genes in the multi-miR group according to GO annotation (DAVID tools, ES = 1.3 equivalent to α = 0.05, gray bar indicates P < 0.05, and FDR < 0.05 for unique GO terms). (F) Significantly decreased expression of 17 collagen genes found in top extracellular matrix-enriched cluster. P < 0.05, FDR < 0.05. Data are shown relative to PBS-injected zygotes and presented as mean ± SEM, n = 6 biological replicates per group. ES, cluster enrichment score.
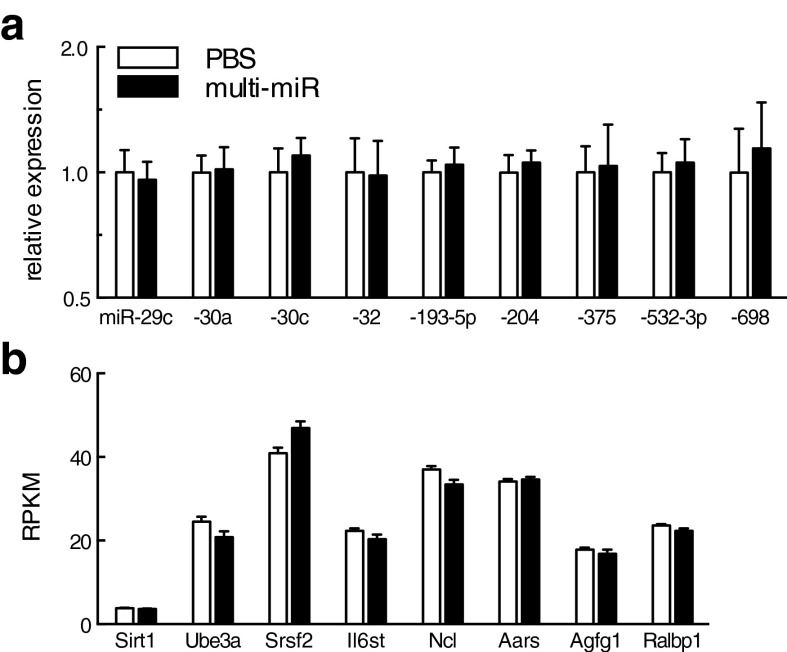
Fig. S1.
No change in PVN expression of the nine injected miRs or their mRNA targets. (A) qRT-PCR analysis of microRNA in the adult PVN in the multi-miR–injected group revealed no difference in the expression of miR-29c, miR-30a, miR-32, miR-193-5p, miR-204, miR-375, miR-532-3p, or miR-698. (B) PVN expression of the eight targeted maternal mRNA was similarly unchanged in the adult PVN. Expression in RPKM for Sirt1, Ube3a, Srsf2, IL6st, Ncl, Aara, Agfg1, and Ralbp1 was obtained by RNA-seq analysis. All data are presented as mean ± SEM, n = 6 mice per group. RPKM, reads per kilobase of transcript per million.
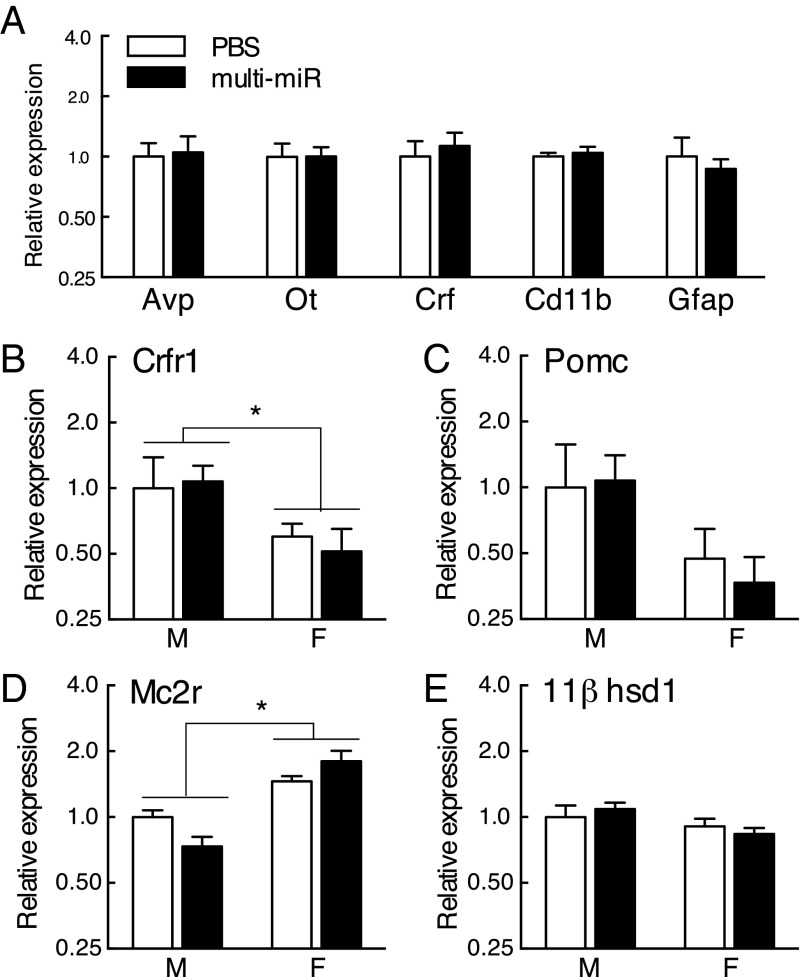
Further, to ensure zygote miR injection did not impact overall development of the HPA axis, gene expression in the PVN, pituitary gland, and adrenal gland was examined. Markers for primary and dominant PVN cell populations were unchanged in the multi-miR–injected group [arginine vasopressin (AVP), t (10) = 0.19, P = 0.85); oxytocin (OT), t (10) = 0.00027, P = 0.99); corticotropin releasing factor (CRF), t (10) = 0.48, P = 0.64); integrin alpha M (CD11b) t (10) = 0.49, P = 0.64); glial fibrillary acidic protein (GFAP), t (10) = 0.50, P = 0.62)] (Fig. 3A). Stress-regulatory genes in peripheral sites of the HPA axis were also unchanged in the multi-miR–injected group [pituitary corticotropin releasing factor receptor 1 (Crfr1), treatment (F1,24 = 0.0006, P = 0.98) by sex (F1,24 = 4.57, P = 0.043) with no interaction (F1,24 = 0.13, P = 0.72); pituitary proopiomelanocortin (POMC), treatment (F1,26 = 0.0017, P = 0.97) by sex (F1,26 = 2.73, P = 0.11) with no interaction (F1,26 = 0.060, P = 0.81); adrenal melanocortin receptor 2 (Mc2r), treatment (F1,26 = 0.098, P = 0.76) by sex (F1,26 = 33.53, P < 0.0001) with interaction (F1,26 = 5.43, P = 0.023) driven by an effect of treatment in males [t (14) = 2.46, P = 0.028], but not in females [t (13) = 1.47, P = 0.16]; adrenal corticosteroid 11β-dehydrogenase (11βHSD-1), treatment (F1,27 = 0.013, P = 0.91) by sex (F1,27 = 3.69, P = 0.065) with no interaction (F1,27 = 0.78, P = 0.38)] (Fig. 3 B–E).
Fig. 3.
No differences in adult expression of stress-regulatory genes in the HPA axis. (A) Markers of major neuronal and nonneuronal cell populations in the PVN were assessed by qRT-PCR and were unchanged in the multi-miR–injected group: Avp, arginine vasopressin; Ot, oxytocin; Crf, corticotropin releasing factor; Cd11b, integrin alpha M; Gfap, glial fibrillary acidic protein. n = 6 mice per group. (B–E) Stress-axis related gene expression in the pituitary and adrenal glands was not significantly different between PBS and multi-miR–injected animals. No effect was observed for pituitary Crfr1, pituitary Pomc, adrenal Mc2r, or adrenal 11βhsd-1. Data presented as mean ± SEM, n = 6–9 mice per group. *P < 0.05.
Reduction of Stored Maternal mRNA by Sperm miRs.
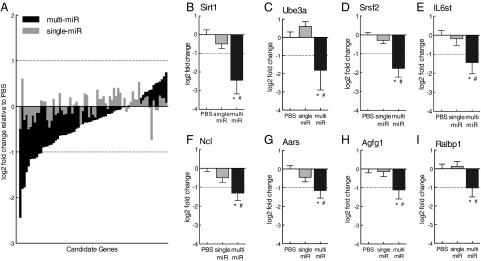
To investigate the potential function of sperm miRs postfertilization, we directly assessed sperm miR targeting of stored maternal mRNAs, using single-cell amplification technology to examine gene expression levels in miR-injected zygotes 24 h postinjection (Fig. 1A). Candidate genes included in the analysis (n = 75) were selected as predicted targets of one or more of the nine miRs cross-referenced against maternal mRNAs reported in the mouse late-stage oocyte and/or single-cell zygote (30–32), (Table S4). We found that the multi-miR injection reduced the expression of many more stored maternal mRNA than the single miR injection (χ2 = 11.53, P < 0.005), (Fig. 4A). Further, we detected a significant two- to fourfold decrease in expression of eight genes: Sirt1 (F2,31 = 8.54, P = 0.0011), Ube3a (F2,31 = 4.00, P = 0.029), Srsf2 (F2,31 = 12.39, P < 0.0001), IL6st (F2,31 = 3.80, P = 0.033), Ncl (F2,31 = 6.37, P = 0.0048), Aars (F2,31 = 4.52, P = 0.019), Agfg1 (F2,31 = 3.47, P = 0.044), and Ralbp1 (F2,31 = 3.39, P = 0.047) in the multi-miR–injected group (Fig. 4 B–I). The down-regulation of these eight genes was not maintained into the mature brain, as their expression in the adult PVN was unaffected (Fig. S1).
Fig. 4.
Single-cell amplification studies confirmed sperm miR targeting of stored maternal mRNA. (A) Expression of many more maternal mRNA candidate genes in zygotes was reduced by multi-miR injection than by the single-miR injection. P < 0.005. (B–I) Expression of 8 of the 75 candidate genes—Sirt1, Ube3a, Srsf2, IL6st, Ncl, Aars, Agfg1, and Ralbp1—was significantly reduced by the multi-miR injection, with expression levels measuring log2 fold change > |1| (dotted lines) relative to PBS controls. Expression of these genes was unaffected in single-miR injection controls. Data are shown relative to PBS-injected zygotes and presented as mean ± SEM, n = 10–13 zygotes per group. *P < 0.05 multi-miR vs. PBS, P < 0.05 multi-miR vs. single miR.
Discussion
Posttranslational histone modifications, DNA methylation patterns, and populations of small noncoding RNAs in sperm have been implicated in the transgenerational transmission of paternal experience, with changes in these epigenetic marks observed following male exposure to such diverse stimuli as stress, malnutrition, and drugs of abuse (11–21). In particular, the role of sperm RNA as a mechanistic link between paternal experience and its consequences on offspring behavior and physiology has been emphasized by recent studies that characterize offspring phenotypes following in vitro fertilization and/or the experimental manipulation of total sperm RNA content (12, 20). In our model of paternal stress, a reduced HPA axis response in offspring was associated with the increased expression of nine miRs (miR-29c, miR-30a, miR-30c, miR-32, miR-193-5p, miR-204, miR-375, miR-5323p, and miR-698) in paternal sperm following chronic stress exposure (11). In the current study, to confirm causality of these miR changes in offspring neurodevelopmental programming, the nine miRs (multi-miR injection) were microinjected into single-cell zygotes. Two components were critical to control for in these experiments to ensure outcomes were clearly interpreted: (i) the injection process itself, and (ii) the increased overall miR concentration, which could overwhelm and inhibit the endogenous activity of the zygote RNA-induced silencing complex (RISC) (33). Therefore, control injection conditions included a PBS injection to control for the zygote injection process itself, and a single-miR injection (randomly selected from one of the nine used in the composite) at the same overall concentration to control for miR concentration effects.
Remarkably, and important for our understanding of paternal RNA involvement in postfertilization development, the multi-miR injection produced a phenotype nearly identical to that of our paternal stress model, where both male and female mice from the multi-miR injections mounted a significantly blunted corticosterone response to an acute restraint stress as adults. A striking overlap was observed in the magnitude of effect on corticosterone production and its rate of rise and of recovery between multi-miR–injected animals and our previously reported paternal stress offspring (as shown in the schematic in Fig. 1F). The corticosterone response curve elicited by an acute stressor is multifaceted, with aspects of the curve (e.g., baseline, maximal rise, and rates of rise and recovery) regulated by specific brain regions, including the PVN, thalamus, and hippocampus (34). Thus, parallels in the shape of the corticosterone curves between this study and our paternal stress model emphasize similarities in programming mechanisms elicited by paternal sperm miRs. The single-miR injection did not affect the HPA axis corticosterone levels, suggesting that the specific and combinatorial activity of these sperm miRs alters stress axis responsivity. Certainly, as we did not examine each miR independently or in all possible combinations to determine the minimal complement necessary to produce the effect, future studies will need to examine the contribution of each of the nine miRs. Additionally, although expected sex differences in corticosterone levels were observed, sex did not interact to impact miR programming of stress responsivity. Further, similar to what we reported following paternal stress exposure, the expression of offspring stress-regulatory genes in the pituitary (CRF receptor-1 and proopiomelanocortin) and adrenal glands (melanocortin receptor-2 and 11βHSD-1) were unaffected in the zygote multi-miR–injected mice as adults. However, the interaction of zygote miR injection with sex on adrenal Mc2r expression suggests that miR reprogramming of the HPA axis may involve a convergent sex difference, whereas the endpoint effect on the HPA is ultimately the same (35).
In examination of the long-term changes in neurodevelopmental programming in the multi-miR–injected mice, we completed RNA-seq analysis on micropunches from the adult PVN, the hypothalamic regulator of the HPA stress axis. GSEA of the PVN transcriptome revealed a robust enrichment of extracellular matrix and collagen formation gene sets in control animals, reflecting a decreased expression of these genes in the PVN of the multi-miR–injected group. Functional annotation clustering of differentially expressed genes using DAVID tools confirmed this outcome, where affected genes primarily associated with GO terms for the extracellular matrix and collagen organization. In fact, 17 unique collagen genes were down-regulated in the multi-miR–injected group PVN compared with controls. These findings may reflect deficits in cerebral circulation and/or blood–brain barrier permeability in the PVN, effects that would have significant impact on neuroendocrine function (36, 37). Furthermore, the PVN is one of the most highly vascularized regions in the brain, suggesting that it may be particularly susceptible to developmental changes in circulation or permeability (38). This heightened plasticity of stress homeostatic mechanisms to transgenerational reprogramming could confer a selective advantage or disadvantage depending on environmental conditions.
Additionally, in examination of RNA-seq results, we noted that the majority of differentially expressed genes (288 of 298) in the PVN exhibited a dramatic decrease in expression, similar to what we reported previously in our paternal stress model and supporting the idea that an increase of these specific nine miRs in the zygote in either model (delivered through sperm in response to stress or experimentally by microinjection) can elicit long-term genetic reprogramming (11). As the overall focus of these repressed genes is in the extracellular matrix and collagen organization, this mouse model suggests that hypothalamic reprogramming governs neuroendocrine function in control of the HPA axis. Gross PVN development was not impacted by the zygote injection, as markers of the primary and dominant cell populations within the PVN were examined and did not differ between control and multi-miR–injected groups. Instead, this robust suppression of the transcriptome may suggest regulation by an upstream epigenetic mechanism, such as a histone repressive mark. The programming of such a change at the level of the brain is likely a complex and multifaceted process, whereby miRs initiate a cascade of molecular events at fertilization that, through many steps, impact regulatory mechanisms such as chromatin modifications or DNA methylation to eventually alter neurodevelopment and related behaviors or physiology (9). As we would predict, we found that the expression of the nine specific sperm miRs was unchanged in the adult PVN. Ultimately, stepwise investigation of the complex cascade by which a germ cell mark, such as sperm miRs, can impart changes in the developing offspring is critical to our understanding of the transgenerational transmission of paternal experiences, and must begin with an investigation of its activity immediately following fertilization in the single-cell zygote.
During normal embryogenesis, stores of maternal mRNA initially present in the zygote are selectively degraded postfertilization, and miR function within the zygote RISC complex is essential to this developmental process (39, 40). As this regulatory role has previously only been attributed to maternally derived miRs present in the oocyte, and as a postfertilization function of paternally derived miRs has not been established, we examined the ability of these nine paternally derived sperm miRs to impact stores of maternal mRNA. To do so, injected zygotes were cultured to the one- to two-cell stage, and the mRNA within each individual zygote was then amplified to examine differences in expression. The specific mRNAs examined were selected as being both present in published lists of the transcriptome in mouse late-stage oocytes and/or single-cell zygotes and predicted targets of one or more of the nine sperm miRs in the miRWalk database (30–32). Impressively, the pattern of candidate maternal mRNAs expression in single-cell zygotes was overall reduced in the multi-miR–injected group, supporting our hypothesized function that sperm miRs selectively target maternal mRNA leading to the posttranscriptional silencing of expression. Further, eight of the candidate mRNA—Sirt1, Ube3a, Srsf2, IL6st, Ncl, Aars, Agfg1, and Ralbp1—were dramatically affected by the multi-miR zygote injection, with expression levels reduced to between a half and a quarter of control levels. Importantly, the known developmental functions of the targeted genes, including the notable involvement of Ube3a and Sirt1 in chromatin remodeling and neurodevelopmental disorders (41, 42), support the hypothesis that sperm miRs may initiate lasting, programmatic changes that ultimately impact neurodevelopment and stress reactivity of the developing embryo. Again, as the single-miR injection did not affect the expression of stored maternal mRNA, the reduction in mRNA levels appears related to the specific composite activity of the nine miRs and not simply to changes in overall miR concentration. Future studies will need to determine how the targeted reduction in stored maternal mRNAs by paternal sperm miRs can induce reprogramming of the hypothalamus, but these findings certainly suggest an important influence of the paternal epigenome on early zygote development, a process previously considered maternally driven.
Together, our findings demonstrate a definitive involvement of the sperm miRs previously identified in our paternal stress model in the recapitulation of offspring HPA stress axis phenotype and hypothalamic reprogramming (11). Identification of a novel postfertilization function of these miRs in targeting stored maternal mRNAs offers exciting insight into the role of paternal RNA transmission and epigenetic marks. These studies emphasize a stepwise mechanistic approach toward a greater understanding of the transgenerational transmission of paternal lifetime experiences and offer valuable insight into the novel factors influencing offspring disease risk and resilience.
Materials and Methods
Microinjection.
C57/Bl6:129S6/SvEvTac hybrid mouse zygotes were randomly assigned for microinjection of nine miRs together (1 ng/μL final concentration of 0.11 ng/μL each miRIDIAN mimic miR-29c, miR-30a, miR-30c, miR-32, miR-193-5p, miR-204, miR-375, miR-532-3p, miR-698 in 1× DPBS) (Table S5) or to one of two control conditions: 1× DPBS only, to control for the injection process itself, or a single miR (1 ng/μL final concentration of miR-193-5p in 1× DPBS) to control for the total increased concentration of miRs within the zygote while minimizing nonspecific targeting effects that would be produced by alternate groups of miRs. Injected zygotes were transferred into recipient CD-1 females (Charles River) or cultured overnight in potassium-supplemented simplex optimised medium (KSOM, EMD Millipore) and frozen at −80 °C. Experimental protocols were approved by the University of Pennsylvania Institutional Animal Care and Use Committee and conducted in accordance with the NIH Guide for the Care and Use of Laboratory Animals (43).
HPA Axis Assessment.
Naïve 13–14 wk male and female mice were restrained in flat bottom animal restrainers (Braintree Scientific) for 15 min beginning at time 0 min and were immediately returned to their home cage at the conclusion of restraint. As previously described, a single tail snip was made at time 0 min, removing <1 mm tissue from the distal tip. A total of 10 μL tail blood was collected by micropipette at 0, 15, 30, and 120 min into EDTA-treated tubes, first blotting the blood clot and then gently milking the tail to stimulate blood flow (11). Corticosterone levels were determined by radioimmunoassay (MP Biomedicals) in 3 μL plasma. All testing was completed 2–5 h after lights on.
RNA-Seq.
Illumina single-end cDNA libraries of adult male PVN mRNA were prepared from 250 ng total RNA using the TruSeq Stranded mRNA Kit with poly-A enrichment (RS-122-2101) according to the manufacturer’s protocol. Six biological replicates per group (12 total cDNA libraries) were multiplexed and sequenced on two identical HiSeq2000 lanes (Illumina).
Single-Cell Amplification.
Criteria for selection of candidate genes for analysis in individual zygotes were as follows: present in late-stage MII mouse oocytes3a, in single-cell mouse zygotes3b, and/or homologously in human and mouse mature oocytes4 (two or more of three lists) (30, 31) and predicted target of one or more of nine miRs in both miRWalk and miRanda algorithms (32). qRT-PCR was conducted with the two-step single-cell protocol with EvaGreen Supermix on the BioMark HD System using DELTAgene assays (Table S4) according to the manufacturer (Fluidigm).
Statistics.
Corticosterone levels were analyzed by two-way ANOVA with time as a repeated measure. Corticosterone AUC, body weight and length, and adrenal and pituitary gene expression were analyzed by two-way ANOVAs. Litter statistics were analyzed by one-way ANOVA and PVN gene expression was analyzed by two-tailed t tests. Significance was set at P < 0.05. When appropriate, a post hoc Fisher’s least significant difference or Student’s t test was used to explore main effects. Differential gene expression in single-cell analysis was calculated with the Fluidigm Singular toolset 2.0 using P < 0.05. The expression pattern of all candidate genes was compared using a χ2 test. GSEA (v2.2.0, Broad Institute) of c2_CP gene sets from the Molecular Signature Database (MsigDB v5.0, Broad Institute) was run using 1,000 gene_set permutations (26, 27). Differential gene expression analysis of RNA-seq data was conducted by a modified Fisher’s exact test with EdgeR (44). Statistical criteria for significance were an adjusted P < 0.05, FDR < 0.05, and log2 fold change >|0.585| (1.5 linear fold change). Functional annotation clustering of all GO annotations was performed with DAVID v6.7, with cluster enrichment score of 1.3, equal to an α of 0.05, considered significant enrichment (28, 29). FDR correction for multiple comparisons was applied to GO annotation enrichment P values.
SI Materials and Methods
Microinjection.
F1 hybrid fertilized mouse zygotes were harvested following superovulation of naïve C57/Bl6 females (The Jackson Laboratories) by administration of 5 IU pregnant mare serum gonadotropin (Sigma-Aldrich) and 5 IU human chorionic gonadotropin (HCG, Sigma-Aldrich) and mating with 129S6/SvEvTac males (Taconic). F1 hybrid fertilized zygotes from donor females were harvested 14–16 h after HCG treatment and randomly assigned for microinjection, with zygotes from each donor being divided between the three treatment groups. Microinjection into the male pronucleus of each single-cell zygote was performed with a Cell Tram Air controller (Eppendorf) using gravity to generate natural flow. Each injection was considered successful upon observation of significant swelling of the pronucleus (∼10 pL per zygote). As mature sperm cells contain 10–20 fg of total RNA (45), of which 0.4% (or 0.04 fg) is miR (46), the amount of miRs injected into zygotes (∼0.01 fg) did not surpass approximate physiological levels, although precise measurement of each miR concentration in single sperm following stress is not currently methodically feasible.
Following injection, zygotes for offspring studies were transferred into recipient CD-1 females (Charles River). Recipient females were mated with vasectomized males and identified by copulation plug on the day of zygote transfer. Each recipient female was implanted with 10–13 zygotes in a single-injection group, sourced from multiple donor females. Dams were individually housed with ad libitum access to food and water on a 12:12 light–dark cycle. Offspring were weaned at postnatal day 28 into same-sex group housing (three to four per cage).
Mouse Tissue Dissection.
Male and female mice were anesthetized with isoflurane and decapitated 5–6 wk after HPA axis assessment. Whole brains were immediately frozen on dry ice, preserved at −80 °C, and cryosectioned. Whole pituitary and left adrenal glands were flash frozen in liquid nitrogen. PVN tissue for each animal was micropunched from two successive 300-µm brain slices (−0.50 to −0.80 and −0.80 to −1.10 relative to bregma) using a hollow 1.0-mm needle (Ted Pella). Tissue was treated with TRIzol reagent (Invitrogen) and bath sonicated on ice with 30 s on/off cycles for 2.5 min (brain) or 7 min (peripheral). Total brain RNA was isolated by miRNeasy kit (Qiagen) and total pituitary and adrenal RNA was isolated by ethanol precipitation. RNA concentration was quantified using a Nanodrop 2000 (Thermo Scientific).
RNA-Seq.
For analysis via the Illumina TruSeq Stranded mRNA kit and HiSEq. 2000 lanes, cDNA fragments were ligated to indexed adapter sequences (Illumina: AR002, AR004–AR007, AR012–AR016, AR018, and AR019) and enriched through 15 PCR cycles (10 s at 98 °C, 30 s at 60 °C, and 30 s at 72 °C). cDNA libraries (n = 6 mice per group, 12 total cDNA libraries) were multiplexed in a single pool and sequenced on 2 HiSeq 2000 lanes (Illumina) at the University of Pennsylvania Next Generation Sequencing Core, obtaining an average of 31,450,856 total 91-nt reads per sample. Quality control was performed with a Bioanalyzer 2100 (Agilent Technologies) and Qubit 2.0 fluorometer (Life Technologies). RNA-seq data were aligned with Bowtie 2.2.3 (47) and processed with the RUM pipeline (48).
Quantitative RT-PCR.
cDNA was synthesized for pituitary gland, adrenal gland, and PVN mRNA or miR expression analyses using the High-Capacity cDNA Reverse Transcriptase Kit or the TaqMan MicroRNA Reverse Transcription Kit, respectively. mRNA and miR levels were quantified by qRT-PCR using TaqMan Gene Expression or Small RNA Assays (Applied Biosystems): in pituitary, CRFr1 and POMC; in adrenal, Mc2r and 11βHSD-1; in female PVN, miR-29c, miR-30a, miR-30c, miR-32, miR-193-5p, miR-204, miR-375, miR-532-3p, and miR-698 and cell population markers AVP, OT, CRF, CD11b, and GFAP (Table S5). Samples (n = 6–9 mice per group) were run in technical triplicate and calculated Ct values were normalized to endogenous controls (pituitary, Gapdh; adrenal, Actb; PVN miRs, U6 and sno202; and PVN mRNA, Gapdh). Relative expression analysis performed by comparative Ct method, normalizing to PBS controls and excluding samples with Ct > 35 (49).
Single-Cell Amplification.
cDNA for qRT-PCR on the BioMark HD System (Fluidgim) was synthesized from total cell lysate of cultured two-cell zygotes using the SuperScript VILO cDNA Synthesis Kit (Invitrogen) with SUPERase-In (Ambion). Specific target amplification was performed over 20 PCR cycles (5 s at 96 °C, 4 min at 60 °C). Amplified cDNA was diluted 1:5 in suspension buffer (Teknova) and loaded into a 96.96 dynamic array IFC chip (Fluidigm). Fluidigm real-time PCR analysis software calculated cycle threshold (Ct) values. Four genes with undetermined expression in >10% of samples were excluded from further analysis.
Statistics.
For analyses of variance (ANOVAs), normality was examined and a Bartlett’s test for equal variance was performed. Data met the assumptions of the statistical tests used. Data greater than 2 SDs above each group mean were removed from analyses: for AUC (and therefore removed from all corticosterone measures), one PBS male, one single-miR male, one multi-miR male, and one single-miR female; and for Mc2r, one multi-miR female. In single-cell analysis with Singular 2.0, the identify outliers program removed one multi-miR–injected sample from analysis.
Supplementary Material
Acknowledgments
The authors thank D. Beiting for statistical consultation; N. Li and C. Lengner for assistance with Fluidigm assays; the Functional Genomics Core (FGC) director, J. Schug, for his assistance; the University of Pennsylvania Molecular Profiling Core; and the University of Pennsylvania Diabetes Research Center for use of the FGC (P30-DK19525). This work was supported by NIH Grants MH073030, MH091258, MH087597, MH099910, MH104184, and MH108286.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1508347112/-/DCSupplemental.
References
- 1.Toth M. Mechanisms of non-genetic inheritance and psychiatric disorders. Neuropsychopharmacology. 2015;40(1):129–140. doi: 10.1038/npp.2014.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yehuda R, et al. Influences of maternal and paternal PTSD on epigenetic regulation of the glucocorticoid receptor gene in Holocaust survivor offspring. Am J Psychiatry. 2014;171(8):872–880. doi: 10.1176/appi.ajp.2014.13121571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Van den Bergh BRH, Van Calster B, Smits T, Van Huffel S, Lagae L. Antenatal maternal anxiety is related to HPA-axis dysregulation and self-reported depressive symptoms in adolescence: A prospective study on the fetal origins of depressed mood. Neuropsychopharmacology. 2008;33(3):536–545. doi: 10.1038/sj.npp.1301450. [DOI] [PubMed] [Google Scholar]
- 4.Dietz DM, et al. Paternal transmission of stress-induced pathologies. Biol Psychiatry. 2011;70(5):408–414. doi: 10.1016/j.biopsych.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morgan CP, Bale TL. Early prenatal stress epigenetically programs dysmasculinization in second-generation offspring via the paternal lineage. J Neurosci. 2011;31(33):11748–11755. doi: 10.1523/JNEUROSCI.1887-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harris A, Seckl J. Glucocorticoids, prenatal stress and the programming of disease. Horm Behav. 2011;59(3):279–289. doi: 10.1016/j.yhbeh.2010.06.007. [DOI] [PubMed] [Google Scholar]
- 7.Moisiadis VG, Matthews SG. Glucocorticoids and fetal programming part 2: Mechanisms. Nat Rev Endocrinol. 2014;10(7):403–411. doi: 10.1038/nrendo.2014.74. [DOI] [PubMed] [Google Scholar]
- 8.Bale TL. Lifetime stress experience: Transgenerational epigenetics and germ cell programming. Dialogues Clin Neurosci. 2014;16(3):297–305. doi: 10.31887/DCNS.2014.16.3/tbale. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rodgers AB, Bale TL. Germ cell origins of posttraumatic stress disorder risk: The transgenerational impact of parental stress experience. Biol Psychiatry. 2015;78(5):307–314. doi: 10.1016/j.biopsych.2015.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Babenko O, Kovalchuk I, Metz GAS. Stress-induced perinatal and transgenerational epigenetic programming of brain development and mental health. Neurosci Biobehav Rev. 2015;48:70–91. doi: 10.1016/j.neubiorev.2014.11.013. [DOI] [PubMed] [Google Scholar]
- 11.Rodgers AB, Morgan CP, Bronson SL, Revello S, Bale TL. Paternal stress exposure alters sperm microRNA content and reprograms offspring HPA stress axis regulation. J Neurosci. 2013;33(21):9003–9012. doi: 10.1523/JNEUROSCI.0914-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dias BG, Ressler KJ. Parental olfactory experience influences behavior and neural structure in subsequent generations. Nat Neurosci. 2014;17(1):89–96. doi: 10.1038/nn.3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vassoler FM, White SL, Schmidt HD, Sadri-Vakili G, Pierce RC. Epigenetic inheritance of a cocaine-resistance phenotype. Nat Neurosci. 2013;16(1):42–47. doi: 10.1038/nn.3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lambrot R, et al. Low paternal dietary folate alters the mouse sperm epigenome and is associated with negative pregnancy outcomes. Nat Commun. 2013;4:2889. doi: 10.1038/ncomms3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guerrero-Bosagna C, Weeks S, Skinner MK. Identification of genomic features in environmentally induced epigenetic transgenerational inherited sperm epimutations. PLoS One. 2014;9(6):e100194. doi: 10.1371/journal.pone.0100194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Milekic MH, et al. Age-related sperm DNA methylation changes are transmitted to offspring and associated with abnormal behavior and dysregulated gene expression. Mol Psychiatry. 2015;20(8):995–1001. doi: 10.1038/mp.2014.84. [DOI] [PubMed] [Google Scholar]
- 17.Radford EJ, et al. In utero effects. In utero undernourishment perturbs the adult sperm methylome and intergenerational metabolism. Science. 2014;345(6198):1255903. doi: 10.1126/science.1255903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fullston T, et al. Paternal obesity initiates metabolic disturbances in two generations of mice with incomplete penetrance to the F2 generation and alters the transcriptional profile of testis and sperm microRNA content. FASEB J. 2013;27(10):4226–4243. doi: 10.1096/fj.12-224048. [DOI] [PubMed] [Google Scholar]
- 19.Bohacek J, et al. Pathological brain plasticity and cognition in the offspring of males subjected to postnatal traumatic stress. Mol Psychiatry. 2015;20(5):621–631. doi: 10.1038/mp.2014.80. [DOI] [PubMed] [Google Scholar]
- 20.Gapp K, et al. Implication of sperm RNAs in transgenerational inheritance of the effects of early trauma in mice. Nat Neurosci. 2014;17(5):667–669. doi: 10.1038/nn.3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Petropoulos S, Matthews SG, Szyf M. Adult glucocorticoid exposure leads to transcriptional and DNA methylation changes in nuclear steroid receptors in the hippocampus and kidney of mouse male offspring. Biol Reprod. 2014;90(2):43. doi: 10.1095/biolreprod.113.115899. [DOI] [PubMed] [Google Scholar]
- 22.Gannon JR, Emery BR, Jenkins TG, Carrell DT. The sperm epigenome: Implications for the embryo. Adv Exp Med Biol. 2014;791:53–66. doi: 10.1007/978-1-4614-7783-9_4. [DOI] [PubMed] [Google Scholar]
- 23.Belleannée C, Calvo É, Caballero J, Sullivan R. Epididymosomes convey different repertoires of microRNAs throughout the bovine epididymis. Biol Reprod. 2013;89(2):30. doi: 10.1095/biolreprod.113.110486. [DOI] [PubMed] [Google Scholar]
- 24.Păunescu TG, et al. High-resolution helium ion microscopy of epididymal epithelial cells and their interaction with spermatozoa. Mol Hum Reprod. 2014;20(10):929–937. doi: 10.1093/molehr/gau052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rassoulzadegan M, et al. RNA-mediated non-mendelian inheritance of an epigenetic change in the mouse. Nature. 2006;441(7092):469–474. doi: 10.1038/nature04674. [DOI] [PubMed] [Google Scholar]
- 26.Subramanian A, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mootha VK, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34(3):267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 28.Huang W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37(1):1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 30.Potireddy S, Vassena R, Patel BG, Latham KE. Analysis of polysomal mRNA populations of mouse oocytes and zygotes: Dynamic changes in maternal mRNA utilization and function. Dev Biol. 2006;298(1):155–166. doi: 10.1016/j.ydbio.2006.06.024. [DOI] [PubMed] [Google Scholar]
- 31.Stanton JL, Green DP. A set of 840 mouse oocyte genes with well-matched human homologues. Mol Hum Reprod. 2001;7(6):521–543. doi: 10.1093/molehr/7.6.521. [DOI] [PubMed] [Google Scholar]
- 32.Dweep H, Sticht C, Pandey P, Gretz N. miRWalk--database: Prediction of possible miRNA binding sites by “walking” the genes of three genomes. J Biomed Inform. 2011;44(5):839–847. doi: 10.1016/j.jbi.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 33.Grimm D, et al. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441(7092):537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- 34.Myers B, McKlveen JM, Herman JP. Glucocorticoid actions on synapses, circuits, and behavior: Implications for the energetics of stress. Front Neuroendocrinol. 2014;35(2):180–196. doi: 10.1016/j.yfrne.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McCarthy MM, Arnold AP, Ball GF, Blaustein JD, De Vries GJ. Sex differences in the brain: The not so inconvenient truth. J Neurosci. 2012;32(7):2241–2247. doi: 10.1523/JNEUROSCI.5372-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goldstein JM, Handa RJ, Tobet SA. Disruption of fetal hormonal programming (prenatal stress) implicates shared risk for sex differences in depression and cardiovascular disease. Front Neuroendocrinol. 2014;35(1):140–158. doi: 10.1016/j.yfrne.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Quaegebeur A, Lange C, Carmeliet P. The neurovascular link in health and disease: Molecular mechanisms and therapeutic implications. Neuron. 2011;71(3):406–424. doi: 10.1016/j.neuron.2011.07.013. [DOI] [PubMed] [Google Scholar]
- 38.Frahm KA, Schow MJ, Tobet SA. The vasculature within the paraventricular nucleus of the hypothalamus in mice varies as a function of development, subnuclear location, and GABA signaling. Horm Metab Res. 2012;44(8):619–624. doi: 10.1055/s-0032-1304624. [DOI] [PubMed] [Google Scholar]
- 39.Pauli A, Rinn JL, Schier AF. Non-coding RNAs as regulators of embryogenesis. Nat Rev Genet. 2011;12(2):136–149. doi: 10.1038/nrg2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lykke-Andersen K, et al. Maternal Argonaute 2 is essential for early mouse development at the maternal-zygotic transition. Mol Biol Cell. 2008;19(10):4383–4392. doi: 10.1091/mbc.E08-02-0219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Glessner JT, et al. Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature. 2009;459(7246):569–573. doi: 10.1038/nature07953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Herskovits AZ, Guarente L. SIRT1 in neurodevelopment and brain senescence. Neuron. 2014;81(3):471–483. doi: 10.1016/j.neuron.2014.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Committee on Care and Use of Laboratory Animals (1996) Guide for the Care and Use of Laboratory Animals (Natl Inst Health, Bethesda), DHHS Publ No (NIH) 85-23.
- 44.Robinson MD, McCarthy DJ, Smyth GK. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kiani J, Rassoulzadegan M. A load of small RNAs in the sperm: How many bits of hereditary information? Cell Res. 2013;23(1):18–19. doi: 10.1038/cr.2012.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kawano M, Kawaji H, Grandjean V, Kiani J, Rassoulzadegan M. Novel small noncoding RNAs in mouse spermatozoa, zygotes and early embryos. PLoS One. 2012;7(9):e44542. doi: 10.1371/journal.pone.0044542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grant GR, et al. Comparative analysis of RNA-Seq alignment algorithms and the RNA-Seq unified mapper (RUM) Bioinformatics. 2011;27(18):2518–2528. doi: 10.1093/bioinformatics/btr427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3(6):1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.