Significance
Paper mulberry, a common East Asian tree used for paper making, is propagated across the Pacific for making barkcloth, a practical and symbolic component of Austronesian material culture. Using chloroplast DNA sequences, we demonstrate a tight genealogical link between its populations in South China and North Taiwan, and South Taiwan and Remote Oceania by way of Sulawesi and New Guinea, presenting the first study, to our knowledge, of a commensal plant species transported to Polynesia whose phylogeographic structure concurs with expectations of the “out of Taiwan” hypothesis of Austronesian expansion. As a commensal plant likely transported across the full range of Austronesian expansion from South China to East Polynesia, paper mulberry may also be the most widely transported fiber crop in human prehistory.
Keywords: Broussonetia papyrifera, commensal approach, DNA of herbarium specimens, out of Taiwan hypothesis, Voyaging Corridor Triple I
Abstract
The peopling of Remote Oceanic islands by Austronesian speakers is a fascinating and yet contentious part of human prehistory. Linguistic, archaeological, and genetic studies have shown the complex nature of the process in which different components that helped to shape Lapita culture in Near Oceania each have their own unique history. Important evidence points to Taiwan as an Austronesian ancestral homeland with a more distant origin in South China, whereas alternative models favor South China to North Vietnam or a Southeast Asian origin. We test these propositions by studying phylogeography of paper mulberry, a common East Asian tree species introduced and clonally propagated since prehistoric times across the Pacific for making barkcloth, a practical and symbolic component of Austronesian cultures. Using the hypervariable chloroplast ndhF-rpl32 sequences of 604 samples collected from East Asia, Southeast Asia, and Oceanic islands (including 19 historical herbarium specimens from Near and Remote Oceania), 48 haplotypes are detected and haplotype cp-17 is predominant in both Near and Remote Oceania. Because cp-17 has an unambiguous Taiwanese origin and cp-17–carrying Oceanic paper mulberries are clonally propagated, our data concur with expectations of Taiwan as the Austronesian homeland, providing circumstantial support for the “out of Taiwan” hypothesis. Our data also provide insights into the dispersal of paper mulberry from South China “into North Taiwan,” the “out of South China–Indochina” expansion to New Guinea, and the geographic origins of post-European introductions of paper mulberry into Oceania.
The peopling of Remote Oceania by Austronesian speakers (hereafter Austronesians) concludes the last stage of Neolithic human expansion (1–3). Understanding from where, when, and how ancestral Austronesians bridged the unprecedentedly broad water gaps of the Pacific is a fascinating and yet contentious subject in anthropology (1–8). Linguistic, archaeological, and genetic studies have demonstrated the complex nature of the process, where different components that helped to shape Lapita culture in Near Oceania each have their own unique history (1–3). Important evidence points to Taiwan as an Austronesian ancestral homeland with a more distant origin in South China (S China) (3, 4, 9–12), whereas alternative models suggest S China to North Vietnam (N Vietnam) (7) or a Southeast Asian (SE Asian) origin based mainly on human genetic data (5). The complexity of the subject is further manifested by models theorizing how different spheres of interaction with Near Oceanic indigenous populations during Austronesian migrations have contributed to the origin of Lapita culture (1–3), ranging from the “Express Train” model, assuming fast migrations from S China/Taiwan to Polynesia with limited interaction (4), to the models of “Slow Boat” (5) or “Voyaging Corridor Triple I,” in which “Intrusion” of slower Austronesian migrations plus the “Integration” with indigenous Near Oceanic cultures had resulted in the “Innovation” of the Lapita cultural complex (2, 13).
Human migration entails complex skills of organization and cultural adaptations of migrants or colonizing groups (1, 3). Successful colonization to resource-poor islands in Remote Oceania involved conscious transport of a number of plant and animal species critical for both the physical survival of the settlers and their cultural transmission (14). In the process of Austronesian expansion into Oceania, a number of animals (e.g., chicken, pigs, rats, and dogs) and plant species (e.g., bananas, breadfruit, taro, yam, paper mulberry, etc.), either domesticated or managed, were introduced over time from different source regions (3, 8, 15). Although each of these species has been shown to have a different history (8), all these “commensal” species were totally dependent upon humans for dispersal across major water gaps (6, 8, 16). The continued presence of these species as living populations far outside their native ranges represents legacies of the highly skilled seafaring and navigational abilities of the Austronesian voyagers.
Given their close association to and dependence on humans for their dispersal, phylogeographic analyses of these commensal species provide unique insights into the complexities of Austronesian expansion and migrations (6, 8, 17). This “commensal approach,” first used to investigate the transport of the Pacific rat Rattus exulans (6), has also been applied to other intentionally transported animals such as pigs, chickens, and the tree snail Partula hyalina, as well as to organisms transported accidentally, such as the moth skink Lipinia noctua and the bacterial pathogen Helicobacter pylori (see refs. 2, 8 for recent reviews).
Ancestors of Polynesian settlers transported and introduced a suite of ∼70 useful plant species into the Pacific, but not all of these reached the most isolated islands (15). Most of the commensal plants, however, appear to have geographic origins on the Sahul Plate rather than being introduced from the Sunda Plate or East Asia (16). For example, Polynesian breadfruit (Artocarpus altilis) appears to have arisen over generations of vegetative propagation and selection from Artocarpus camansi that is found wild in New Guinea (18). Kava (Piper methysticum), cultivated for its sedative and anesthetic properties, is distributed entirely to Oceania, from New Guinea to Hawaii (16). On the other hand, ti (Cordyline fruticosa), also a multifunctional plant in Oceania, has no apparent “native” distribution of its own, although its high morphological diversity in New Guinea suggests its origin there (19). Other plants have a different history, such as sweet potato, which is of South American origin and was first introduced into Oceania in pre-Columbian times and secondarily transported across the Pacific by Portuguese and Spanish voyagers via historically documented routes from the Caribbean and Mexico (17).
Of all commensal species introduced to Remote Oceania as part of the “transported landscapes” (1), paper mulberry (Broussonetia papyrifera; also called Wauke in Hawaii) is the only species that has a temperate to subtropical East Asian origin (15, 20, 21). As a wind-pollinated, dioecious tree species with globose syncarps of orange–red juicy drupes dispersed by birds and small mammals, paper mulberry is common in China, Taiwan, and Indochina, growing and often thriving in disturbed habitats (15, 20, 21). Because of its long fiber and ease of preparation, paper mulberry contributed to the invention of papermaking in China in A.D. 105 and continues as a prime source for high-quality paper (20, 21). In A.D. 610, this hardy tree species was introduced to Japan for papermaking (21). Subsequently it was also introduced to Europe and the United States (21). Paper mulberry was introduced to the Philippines for reforestation and fiber production in A.D. 1935 (22). In these introduced ranges, paper mulberry often becomes naturalized and invasive (20–22). In Oceania, linguistic evidence suggests strongly an ancient introduction of paper mulberry (15, 20); its propagation and importance across Remote Oceanic islands were well documented in Captain James Cook’s first voyage as the main material for making barkcloth (15, 20).
Barkcloth, generally known as tapa (or kapa in Hawaii), is a nonwoven fabric used by prehistoric Austronesians (15, 21). Since the 19th century, daily uses of barkcloth have declined and were replaced by introduced woven textiles; however, tapa remains culturally important for ritual and ceremony in several Pacific islands such as Tonga, Fiji, Samoa, and the SE Asian island of Sulawesi (23). The symbolic status of barkcloth is also seen in recent revivals of traditional tapa making in several Austronesian cultures such as Taiwan (24) and Hawaii (25). To make tapa, the inner bark is peeled off and the bark pieces are expanded by pounding (20, 21, 23). Many pieces of the bark are assembled and felted together via additional poundings to create large textiles (23). The earliest stone beaters, a distinctive tool used for pounding bark fiber, were excavated in S China from a Late Paleolithic site at Guangxi dating back to ∼8,000 y B.P. (26) and from coastal Neolithic sites in the Pearl River Delta dating back to 7,000 y B.P. (27), providing the earliest known archaeological evidence for barkcloth making. Stone beaters dated to slightly later periods have also been excavated in Taiwan (24), Indochina, and SE Asia, suggesting the diffusion of barkcloth culture to these regions (24, 27). These archaeological findings suggest that barkcloth making was invented by Neolithic Austric-speaking peoples in S China long before Han-Chinese influences, which eventually replaced proto-Austronesian language as well as culture (27).
In some regions (e.g., Philippines and Solomon Islands), tapa is made of other species of the mulberry family (Moraceae) such as breadfruit and/or wild fig (Ficus spp.); however, paper mulberry remains the primary source of raw material to produce the softest and finest cloth (20, 23). Before its eradication and extinction from many Pacific islands due to the decline of tapa culture, paper mulberry was widely grown across Pacific islands inhabited by Austronesians (15, 20). Both the literature (15, 20) and our own observations (28–30) indicate that extant paper mulberry populations in Oceania are only found in cultivation or as feral populations in abandoned gardens as on Rapa Nui (Easter Island), with naturalization only known from the Solomon Islands (20). For tapa making, its stems are cut and harvested before flowering, and as a majority of Polynesian-introduced crops (16), paper mulberry is propagated clonally by cuttings or root shoots (15, 20), reducing the possibility of fruiting and dispersal via seeds. The clonal nature of the Oceanic paper mulberry has been shown by the lack of genetic variability in nuclear internal transcribed spacer (ITS) DNA sequences (31). With a few exceptions (30), some authors suggest that only male trees of paper mulberry were introduced to Remote Oceania in prehistoric time (15, 20). Furthermore, because paper mulberry has no close relative in Near and Remote Oceania (20), the absence of sexual reproduction precludes the possibility of introgression and warrants paper mulberry as an ideal commensal species to track Austronesian migrations (6, 30).
To increase our understanding of the prehistoric Austronesian expansion and migrations, we tracked geographic origins of Oceanic paper mulberry, the only Polynesian commensal plant likely originating in East Asia, using DNA sequence variation of the maternally inherited (32) and hypervariable (SI Text) chloroplast ndhF-rpl32 intergenic spacer (33). We sampled broadly in East Asia (Taiwan, S China, and Japan) and SE Asia (Indochina, the Philippines, and Sulawesi) as well as Oceanic islands where traditional tapa making is still practiced. Historical herbarium collections (A.D. 1899–1964) of Oceania were also sampled to strengthen inferences regarding geographic origins of Oceanic paper mulberry. The employment of ndhF-rpl32 sequences and expanded sampling greatly increased phylogeographic resolution not attainable in a recent study (31) using nuclear ITS sequences (also see SI Text and Fig. S1) and intersimple sequence repeat (ISSR) markers with much smaller sampling.
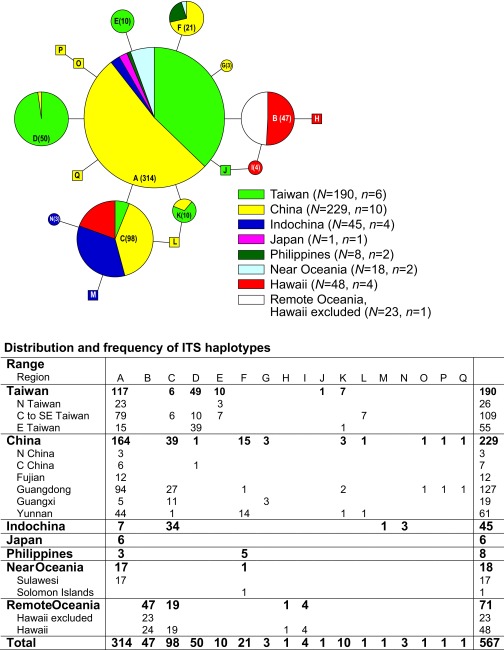
Fig. S1.
ITS haplotype network (n = 17, A–Q) and haplotype distribution and frequency. The haplotype network was reconstructed using TCS (34), with alignment gaps treated as missing data. The sizes of the circles and pie charts are proportional to the frequency of the haplotype (shown in parentheses). Squares denote unique haplotypes (haplotype found only in one individual).
Results and Discussion
DNA Sequence Variation in ndhF-rpl32.
We successfully sequenced ndhF-rpl32 spacer in 604 samples, including 19 historical herbarium collections of Oceania (Fig. 1 and Dataset S1). The DNA matrix includes 1,233 aligned positions of which 54 sites are variable and 39 sites are parsimoniously informative. Variation in the ndhF-rpl32 conforms to expectations of neutrality both by Tajima’s criterion (D = −1.33422, P > 0.10) and by Fu and Li’s criteria (D* = −1.94131, 0.10 > P > 0.05; F* = −1.99663, 0.10 > P > 0.05). With alignment gaps treated as missing, the 604 ndhF-rpl32 DNA sequences were classified into 48 haplotypes, including 31 shared and 17 unique haplotypes (Table 1). The high level of sequence variation in ndhF-rpl32 provided sufficient polymorphism to track the origin of Oceanic paper mulberry. Based on statistic parsimony implemented by the program TCS (34), a single, fully resolved haplotype network was reconstructed (Fig. 1), with haplotype cp-20 (numbers artificially designated) inferred as the most likely ancestral haplotype (outgroup weight of 0.16).
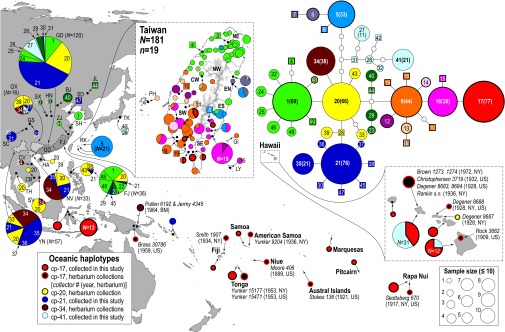
Fig. 1.
Distribution of ndhF-rpl32 haplotypes and haplotype network. The sizes of the circles and pie charts are proportional to the number of individuals sampled (N) for both distribution maps and haplotype network. In the haplotype network, numbered and colored circles represent shared haplotypes and squares denote unique haplotypes. Thicker lined circles and squares (cp-1 to cp-19) are haplotypes found in Taiwan. The distributions of haplotypes in Taiwan (and its offshore islands) and the Hawaii islands are shown in enlarged maps. The map of Taiwan is demarcated by the seven major climatic regions of the island (36), with areas above 2,000 m shown in gray. Detailed distributions of haplotypes in Fujian (FJ) and Guangdong (GD) are shown in Fig. S2. Voucher information for herbarium collections of Oceania is shown as collector number (year; Herbarium). BJ, Beijing; CB, Cambodia; CV, central Vietnam; FJ, Fujian; GD, Guangdong; GI, Green Island; GS, Gansu; GX, Guangxi; HA, Hainan; HN, Henan; JL, Jilin; LY, Lanyu; NV, northern Vietnam; PH, Penghu; RK, Ryukyus; SC, Sichuan; SD, Shandong; SH, Shanghai; SV, southern Vietnam; SX, Shanxi; TH, Thailand; TK, Tokyo; YN, Yunnan; ZJ, Zhejiang.
Table 1.
Distribution and frequency of ndhF-rpl32 haplotypes
| Shared haplotypes | Region-specific haplotypes [haplotype (N)] | Unique haplotypes | N | n | ||||||||||||
| Range and region | 1 | 5 | 12 | 16 | 17 | 20 | 21 | 28 | 29 | 34 | 35 | 39 | ||||
| Taiwan | 43 | 12 | 8 | 39 | 8 | N = 62, n = 5 | 2, 4, 7, 8, 10, 11, 15, 18, 19 | 181 | 19 | |||||||
| N Taiwan | 35 | 1 | 2, 4 | 38 | 4 | |||||||||||
| C to SE Taiwan | 4 | 7 | 17 | 7 | 3 (2), 9 (44), 13 (5), 14 (4) | 10, 11, 15, 18, 19 | 95 | 13 | ||||||||
| E Taiwan | 4 | 11 | 1 | 22 | 1 | 6 (7) | 7, 8 | 48 | 8 | |||||||
| China | 26 | 44 | 66 | 1 | 6 | 18 | 18 | 5 | N = 57, n = 12 | 26, 30, 31, 38, 44, 45, 47 | 248 | 27 | ||||
| N China | 1 | 40 (6) | 44, 47 | 9 | 4 | |||||||||||
| C China | 4 | 4 | 38 | 9 | 3 | |||||||||||
| Fujian (FJ) | 9 | 5 | 1 | 1 | 22 (5), 23 (3), 46 (7), 48 (4) | 45 | 36 | 9 | ||||||||
| Guangdong (GD) | 13 | 27 | 52 | 1 | 5 | 24 (5), 25 (4), 27 (11) | 26, 30, 31 | 121 | 11 | |||||||
| Guangxi (GX) | 4 | 2 | 3 | 5 | 32 (2) | 16 | 5 | |||||||||
| Yunnan (YN) | 8 | 6 | 15 | 18 | 33 (5), 36 (3), 37 (2) | 57 | 7 | |||||||||
| Indochina | 19 | 7 | 18 | 3 | 1 | 48 | 5 | |||||||||
| N Vietnam (NV) | 7 | 7 | 16 | 3 | 1 | 34 | 5 | |||||||||
| S Vietnam (SV) | 8 | 2 | 10 | 2 | ||||||||||||
| Cambodia (CB) | 1 | 1 | 1 | |||||||||||||
| Thailand (TH) | 3 | 3 | 1 | |||||||||||||
| Japan | 21 | 2 | 42 | 24 | 3 | |||||||||||
| Tokyo (TK) | 2 | 42 | 3 | 2 | ||||||||||||
| Ryukyus (RK) | 21 | 21 | 1 | |||||||||||||
| Philippines | 2 | 1 | N = 5, n = 1; 43 (5) | 8 | 3 | |||||||||||
| Near Oceania | 17 | 2 | 2 | 21 | 3 | |||||||||||
| Sulawesi | 16 | 16 | 1 | |||||||||||||
| New Guinea | 1 | 2 | 3 | 2 | ||||||||||||
| Solomon Islands | 2 | 2 | 1 | |||||||||||||
| Remote Oceania | 52 | 1 | N = 21, n = 1 | 74 | 3 | |||||||||||
| Fiji | 2 | 2 | 1 | |||||||||||||
| Tonga | 5 | 5 | 1 | |||||||||||||
| Samoa | 2 | 2 | 1 | |||||||||||||
| Niue | 1 | 1 | 1 | |||||||||||||
| Hawaii | 34 | 1 | 41 (21) | 56 | 3 | |||||||||||
| Austral Islands | 1 | 1 | 1 | |||||||||||||
| Marquesas | 1 | 1 | 1 | |||||||||||||
| Pitcairn | 1 | 1 | 1 | |||||||||||||
| Rapa Nui | 5 | 5 | 1 | |||||||||||||
| Total | 69 | 33 | 8 | 39 | 77 | 66 | 76 | 3 | 6 | 38 | 21 | 6 | N = 145, n = 19 | n = 17 | 604 | 48 |
N, sample size; n, number of haplotypes. Definition of regions (see Fig. 1 for abbreviations): N Taiwan, NW and NE; C to SE Taiwan, CW, PH, SW, SE, GI, and LY; E Taiwan, EN and ES; N China, JL, BJ, SD, SX, HN, and GS; C China, SH, SC, and ZJ; Guangdong (GD), GD and HA; N Vietnam, NV and CV.
Native Range Genetic Structure of Paper Mulberry.
Across the wide sampling range of paper mulberry, East Asia (China and Taiwan) and Indochina (Vietnam, Cambodia, and Thailand) harbor the inferred ancestral haplotype (cp-20) and a majority of DNA sequence variation of ndhF-rpl32 (Table 1 and Fig. 1), conforming to the expectation as the native range of a species (35). China has the highest number of haplotypes (n = 27) and haplotype diversity (Hd = 0.873 ± 0.013), with 20 haplotypes exclusive to this vast region (Table 1). Five haplotypes were detected in Indochina (Hd = 0.691). Surprisingly, Taiwan, regardless of its relatively small size (Fig. 1), harbors the highest nucleotide diversity (π = 0.00268 vs. 0.00235 in China) and the second highest number of haplotypes (n = 19) and haplotype diversity (Hd = 0.831), with 16 haplotypes exclusive to the island and its offshore islets (Table 1). The 19 haplotypes in Taiwan, however, were clustered in three separate and geographically largely nonoverlapping groups (Fig. 1). The “green” haplotype group (cp-1 to cp-4) is predominant in N Taiwan (NW and NE in Fig. 1); cp-1 was elsewhere also found in central and S China (Fig. S2). The “blue” haplotype group (cp-5 to cp-8) was found primarily in E Taiwan (EN and ES in Fig. 1), with cp-5 further distributed in the southern Ryukyu islands (Iriomote, Ishigaki, and Miyako), Japan. The “red” haplotype group (cp-9 to cp-19) was distributed almost entirely in central to southern Taiwan [central west (CW), SW, and SE], except for cp-17, which is also found widely in Near and Remote Oceanic islands (Fig. 1). Although most of Taiwan is characterized by a monsoonal (summer rain) climate (CW, SW, SE, EN, and ES), the north (NE and NW) is generally wet throughout the year (36).
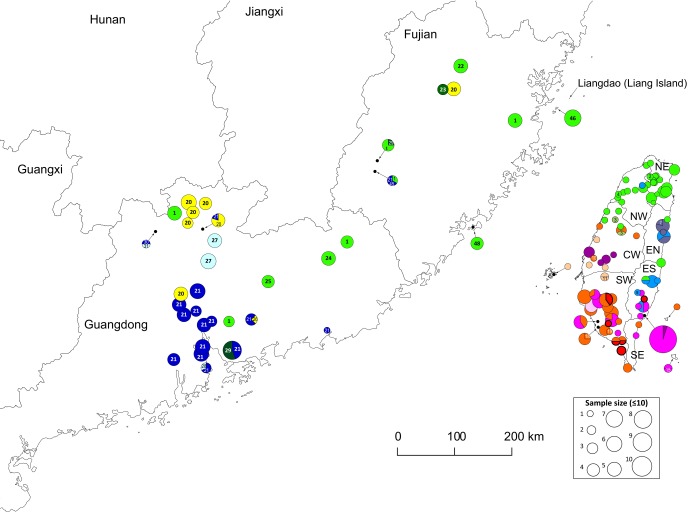
Fig. S2.
Distribution of ndhF-rpl32 haplotypes in Fujian, Guangdong, and Taiwan.
Despite being a common, wind-pollinated, and animal-dispersed weedy tree species, distributions of haplotypes in its native range were highly geographically structured, especially in Taiwan (Fig. 1). The marked phylogeographic pattern is also evident in the analysis of molecular variance (AMOVA; Table S1) and pairwise FST (Table S2). Specifically, values of pairwise FST among populations of northern (NW and NE), eastern (EN and ES), and southern (CW, SW, and SE) Taiwan are highly significant (Table S2), suggesting marked population differentiation. Because paper mulberry is dioecious and its chloroplast is maternally inherited (32), the significant genetic structure, especially the high population differentiation, implies a limited seed dispersal (35), suggesting that its occurrence outside its native range is most likely human-mediated.
Table S1.
AMOVA for the native range (China, Taiwan, and Indochina) paper mulberry
| Source of variance | df | Sum of squares | Variance components | Percentage of variance | Fixation index | P |
| Among ranges | 2 | 18.607 | 0.01339 | 2.79 | ФCT, 0.0279 | 0.19832 |
| Among regions within ranges | 7 | 34.283 | 0.11388 | 23.74 | ФST, 0.26530 | <0.0001 |
| Within ranges | 467 | 164.596 | 0.35245 | 73.47 | ФSC, 0.24420 | <0.0001 |
| Total | 476 | 217.486 | 0.47973 |
The four regions of Indochina were treated as one single population in the analyses.
Table S2.
Pairwise FST calculated based on pairwise distance (upper diagonal) and haplotype frequencies (lower diagonal)
| Region | N Taiwan | W Taiwan | E Taiwan | N China | C China | Fujian | Guangdong | Guangxi | Yunnan | Indochina |
| N Taiwan | — | 0.80959*** | 0.44257*** | 0.73919*** | 0.55452*** | 0.14762*** | 0.30455*** | 0.52805*** | 0.55524*** | 0.53893*** |
| W Taiwan | 0.46845*** | — | 0.3754*** | 0.74312*** | 0.74720*** | 0.73355*** | 0.68872*** | 0.72062*** | 0.73701*** | 0.74871*** |
| E Taiwan | 0.50296*** | 0.19512*** | — | 0.31749*** | 0.27957*** | 0.36066*** | 0.36088*** | 0.28547*** | 0.39206*** | 0.3753*** |
| N China | 0.73866*** | 0.31028*** | 0.32665*** | — | 0.42975*** | 0.40434*** | 0.32094*** | 0.34519*** | 0.45533*** | 0.47943*** |
| C China | 0.45296*** | 0.26991*** | 0.27117*** | 0.34253** | — | 0.18514** | −0.00149n.s. | 0.11174* | 0.10482* | 0.18938* |
| Fujian | 0.34416*** | 0.19441*** | 0.19256*** | 0.24378*** | 0.11084* | — | 0.17926*** | 0.25183*** | 0.35661*** | 0.28661*** |
| Guangdong | 0.42306*** | 0.25144*** | 0.25704*** | 0.27431*** | 0.06335 | 0.13905*** | — | 0.07677* | 0.10564*** | 0.09801*** |
| Guangxi | 0.60098*** | 0.22426*** | 0.23409*** | 0.27127*** | 0.20175** | 0.12064** | 0.12358*** | — | 0.14543*** | 0.10008** |
| Yunnan | 0.48804*** | 0.22902*** | 0.23650*** | 0.26874*** | 0.21259** | 0.14918*** | 0.16291*** | 0.0994** | — | 0.1055*** |
| Indochina | 0.55889*** | 0.2801*** | 0.29300*** | 0.33565*** | 0.27042*** | 0.17663*** | 0.15027*** | 0.06531* | 0.07779*** | — |
The four regions of Indochina were treated as one single population in the analyses. *P < 0.05; **P < 0.005; ***P < 0.001.
Haplotypes in Tokyo and Los Baños (Philippines).
Two haplotypes, cp-28 and cp-42, were detected in three samples from Tokyo. Although cp-28 is also found in S China, cp-42 is a derived haplotype from cp-28 currently known only in Tokyo (Fig. 1). Of the eight samples from Los Baños sequenced, three haplotypes (cp-20, -21, and -43) were detected. Although the former two haplotypes are both abundant in China and Indochina, cp-43 is derived from cp-20 and currently only known from this region. Because introductions of paper mulberry to Japan (21) and the Philippines (22) are both well documented, the ndhF-rpl32 dataset suggests that S China is the most probable source of introduction for the Tokyo paper mulberry, whereas S China and Indochina are both probable sources for the population of Los Baños.
Haplotype Variation in Near and Remote Oceania.
In Near and Remote Oceania, five generally unrelated haplotypes were present in the 95 samples (Table 1 and Fig. 1), suggesting multiple introductions to the vast region. In the Near Oceanic samples, cp-17, -21, and -34 were found. Cp-17 was the most frequent (17/21, or 81%) haplotype in the region, present in all samples of Sulawesi (n = 16) and one of New Guinea. Two samples from the Solomon Islands derived from modern introduction (20) carried cp-21, a common and widespread haplotype in S China–Indochina. Two herbarium accessions (Pullen 6192 and Jermy 4345) from a highland village of New Guinea carried cp-34, which is also dominant in S China–Indochina (Fig. 1). In Remote Oceania, cp-17 was also present in a majority of samples (52/74, or 70%). Together, the 69 cp-17–carrying Oceanic samples also included 16 herbarium accessions from New Guinea (n = 1), Fiji (n = 1), Tonga (n = 2), American Samoa (n = 1), Niue (n = 1), Austral Islands (n = 1), Rapa Nui (n = 1), and Hawaii (n = 8) dated from A.D. 1899–1959 (Fig. 1). Elsewhere, cp-17 was only known from eight plants restricted to S Taiwan. Because cp-17 is a tip haplotype derived from within the red haplotype group endemic to Taiwan, cp-17 has an unambiguous Taiwanese origin. The second most frequent haplotype in Remote Oceania was cp-41, carried by 21 (37.5%) of the Hawaiian samples. Because cp-41 is a tip haplotype derived from cp-28 carried by two samples of Guangdong and Tokyo, these two areas are likely sources of the Hawaiian introduction. Cp-20, the inferred ancestral and geographically most widespread haplotype in China–Indochina, was present in one herbarium specimen (Degener 8687) from Molokai, Hawaii.
Paper Mulberry “into” Taiwan.
With high genetic variation and the presence of 17 endemic haplotypes, paper mulberry should have been growing naturally in Taiwan for a long time. However, for plants that have been used by humans for millennia, the nature of their distribution in a given region needs to be interpreted with great caution (20). In Taiwan, because paper mulberry grows abundantly in secondary forests and disturbed habitats, Matthews (20) questioned its native status and attributed its weediness to naturalization following introduction from S China by Austronesian ancestors. Interestingly, in a pollen core drilled in Wuku, Taipei (NE in Fig. 1), pollen identified to Moraceae appeared and peaked abruptly at ∼5,000 y B.P. (37). Because contemporary airborne pollen flora of N Taiwan is dominated by paper mulberry (38), this 5,000-y-old moraceous pollen record is most likely attributable to paper mulberry. This likely pollen record of paper mulberry also coincided with the earliest records of stone beaters in N Taiwan at 5,500–4,500 y B.P. (27), suggesting that paper mulberry in N Taiwan might have been associated with early Austronesian settlers.
Of the 19 haplotypes found in Taiwan, the NW and NE are dominated by cp-1 of the green haplotype group, which is elsewhere found mainly in Fujian and central China (Table 1 and Fig. S2). During the Pleistocene glacial periods, the lowering of global sea levels exposed the continental shelf of the present-day Taiwan Strait, freely allowing migrations of flora and fauna between Taiwan and Fujian (39). Therefore, paper mulberry carrying haplotype cp-1 could have naturally dispersed to Taiwan from Fujian via the Taiwan Strait land bridge during the Pleistocene glacial periods. However, although Fujian is Taiwan’s closest neighbor, phytogeographic studies indicate that the lowland tropical flora of Taiwan generally has its geographical origin in Indochina and/or southernmost China with rare influences from Fujian (39). Considering the coincidental occurrences of stone beaters and moraceous pollen at ∼5,000 y B.P. that considerably postdated the end of the last ice age at ∼18,000 y B.P., the cp-1–carrying paper mulberry in N Taiwan might not be native but may represent the genetic trace left from the initial Austronesian “out of China” expansion from Fujian as part of their transported landscape. Indeed, this scenario could also explain the relatively low sequence variation of the green haplotype group in Taiwan compared with the native red and blue haplotype groups that were likely present in Taiwan for a much longer time. Additionally this conjecture is compatible with inferences drawn from a recently excavated ∼8,000-y-old male skeleton of Austronesian ancestry from the small islet Liangdao (Fig. S2) off the coast of Fujian (10). Based on analyses of mitochondrial genome sequences, Ko and coworkers (14) not only revealed a close relationship between Liangdao Man and extant Formosan aboriginal groups but inferred further a rapid southward population expansion after their initial settlement in N Taiwan, ∼6,000 y ago. If cp-1–carrying paper mulberry was a commensal species transported from Fujian to N Taiwan along with the initial Austronesian expansion out of S China, its further southward transportation would have been unnecessary because native paper mulberry carrying haplotypes of the red and blue groups in central, southern, and eastern Taiwan would provide an abundant source of materials for tapa making, explaining why cp-1 was codistributed with red and blue haplotypes only in central but not southern Taiwan (Fig. 1).
Paper Mulberry “Out” of Taiwan.
Contrary to cp-1 in the north, the presence of 11 endemic haplotypes of the red haplotype group in the central and southern parts of the island firmly attests to the native status of paper mulberry in Taiwan (Fig. 1). Given this, the predominance and wide distribution of cp-17 in Oceania, from Sulawesi all of the way to Rapa Nui, and its disjunct geographic origin in Taiwan (Fig. 1) strongly support a tight link between the two regions. Because Oceanic samples carrying cp-17 were all collected from asexually propagated plantations and private/abandoned gardens for tapa making (28–30), the dominance of cp-17 also complies with the expectation of its clonal nature in Near and Remote Oceania (20, 31). Taken together, our dataset strongly supports a human-mediated dispersal of the Oceanic paper mulberry and its origin in Taiwan, a scenario highly congruent with the “out of Taiwan” hypothesis of Austronesian expansion (3, 4, 11). Additionally, the distribution of cp-17 mainly in SW and SE Taiwan (Fig. 1) also concurs with recent human genetic studies indicating that only the southern tribes of the Formosan aborigines had been involved in the initial out of Taiwan expansion (11). Given this, it is expected that cp-17 should be detected in the Philippines, as the archipelago is the first stepping stone of the out of Taiwan expansion. However, except for recent introductions (22), there is no historical botanical record of paper mulberry in the Philippines (20). Because paper mulberry generally survives only under cultivation in an equatorial climate (20), Matthews (20) surmised that its absence could have been related to the early introduction of hand-woven fabrics to SE Asia, resulting in the diminishing of paper mulberry cultivation in the region and eventual extinction in areas such as the Philippines. Alternatively the absence of cp-17 from the Philippines actually consolidates the endemic status of cp-17 to Taiwan, further strengthening the Taiwanese origin of Oceanic paper mulberry.
Origins of New Guinea Paper Mulberry.
In New Guinea, barkcloth is made by some Papua groups using various tree species including paper mulberry, which is also planted as a source for cordage (23). In addition to cp-17, haplotype cp-34 was also detected in two herbarium specimens collected half a century ago from a remote village (Fig. 1), concurring with the report of multiple cultivars of paper mulberry in the island (20). However, the marked genetic divide between haplotypes in Taiwan and continental Asia suggests that the cp-34 haplotype in New Guinea could have been transported instead from Indochina, where cp-34 is common (Fig. 1). Therefore, the haplotype diversity of paper mulberry planted in mountain villages of New Guinea not only testify for an intrusion and integration descending from the out of Taiwan expansion foreseen by the Voyaging Corridor Triple I model (2, 13) but might also signify the genetic trace of the “out of S China–Indochina” expansion that converged in coastal New Guinea (7), a scenario also highly congruent with recent findings in human genetics (12) and archaeology (40).
Possible Historic Introductions.
Historically, although Hawaii was known for the finest kapa and Wauke was widely grown, its barkcloth production and paper mulberry cultivation had ceased by the end of the 19th century (25). The presence of cp-17 that is common in both contemporary and herbarium collections across the archipelago attests to the introduction of paper mulberry to Hawaii by Austronesian voyagers. However, the history of Hawaiian paper mulberry is complicated by contemporary samples carrying cp-41, which is derived from cp-28 known only from Guangdong and Tokyo. In addition, the finding of cp-20, the most widespread and probable ancestral haplotype, in a 1928 herbarium specimen collected from Molokai, suggests a third introduction from mainland Asia. Coincidentally the detection of three unrelated haplotypes in Hawaiian Wauke samples is consistent with the study by Meilleur et al. (25), where three cultivars were identified by isozyme variability. These authors attributed the existence of multiple Wauke cultivars in Hawaii to post-European introductions (25). In the late 19th century, waves of Chinese and Japanese farmers were recruited to Hawaii as contract laborers to work in sugarcane plantations. As a plant used for medicine and as a source for making paper, cordage, and clothing in both China and Japan, paper mulberry could have been introduced to Hawaii along with other food crops or seeds of familiar vegetables to increase the limited and repetitious diet in the early immigrant community (41), although there is no specific record for its introduction.
Conclusion
Our chloroplast sequence data of paper mulberry demonstrate a tight genealogical link between populations of S China and N Taiwan, and S Taiwan and Remote Oceania by way of Sulawesi and New Guinea, presenting the first study, to our knowledge, of a commensal plant species transported to Polynesia at some point in prehistory whose phylogeographic pattern concurs with expectations of the out of Taiwan hypothesis of Austronesian expansion. Additionally the haplotype diversity in New Guinea further suggests both influences of the out of Taiwan and the out of S China–Indochina expansion. Our data also detect probable recent introductions of paper mulberry to Hawaii, indicating that ndhF-rpl32 is an ideal marker for tracking origins of this weedy species worldwide. The holistic picture of Austronesian expansion and migrations revealed by the phylogeography of Pacific paper mulberry is in close agreement with the archaeological, linguistic, and human genetic data, suggesting that further studies using genomic data of this common East Asian tree are promising in unveiling further insights into prehistorical and historical human movements in the Pacific.
Materials and Methods
Sampling.
Our sampling scheme was designed to detect the extent of genetic variation in the native range and track geographic origins of Oceanic paper mulberry. Fieldwork was conducted in Taiwan, S China, Vietnam, the Philippines, Sulawesi, Hawaii, Fiji, Tonga, Samoa, Rapa Nui, and Marquesas Islands by the authors, with additional samples provided by third parties from Japan, Thailand, Cambodia, the Solomon Islands, and Pitcairn. Except for the naturalized populations in the Solomon Islands, Oceanic paper mulberry is only known from plantations, private gardens, or abandoned feral populations (15, 20). Seelenfreund et al. (30) and Chang (28, 29) provide details regarding the ecology and ethnobotanical aspects of the samples from Remote Oceanic islands and Sulawesi. In the field, leaves were desiccated and preserved using silica gel. Although paper mulberry is common in China (20, 21), considering our main objective, our sampling efforts were concentrated in the southern provinces (Fujian, Guangdong, Guangxi, and Yunnan) where stone beaters have been excavated (26, 27) and ancestral Austronesian peoples lived (3). We also successfully sequenced 19 historical (A.D. 1899–1964) herbarium collections from New Guinea, Fiji, Tonga, American Samoa, Hawaii, Niue, Austral Islands, and Rapa Nui (Fig. 1).
DNA Sequences and Data Analyses.
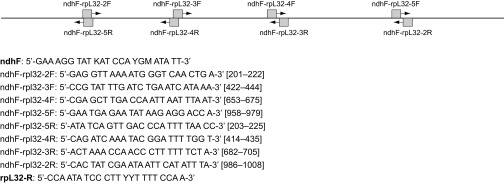
Genomic DNA was extracted using the cetyltrimethylammonium bromide (CTAB) protocol (31). For herbarium samples, genomic DNA was extracted using the protocol of CTAB + Wizard DNA Clean-Up System (42). SI Text details the process of DNA marker selection; chloroplast ndhF-rpl32 intergenic spacer was chosen for its high variability (33) and ease of PCR amplification. Primer sequences and PCR conditions followed Shaw et al. (33), with herbarium materials amplified using internal primers designed for short (ca. 250 bp) PCR amplification (Fig. S3). DNA sequences were aligned using MUSCLE implemented in the software MEGA5 (43) with minor adjustments. Parameters of DNA sequence variation, including haplotype diversity (Hd), nucleotide diversity (π), and θ estimated by segregation sites (S), were calculated using DnaSP v5 (44). DNA haplotype network was reconstructed using TCS with alignment gaps treated as missing (34). Population structure was examined by calculating pairwise FST and performing an AMOVA using Arlequin version 3.5 (45).
Fig. S3.
Schematic figure of primer locations and primer sequences designed for amplifying herbarium collections. Primers in bold are adopted from Shaw et al. (33) for amplifying the entire region of ndhF-rpl32 of fresh collected materials.
SI Text
Selection of Molecular Markers.
We began this project from expanding and sequencing nuclear ITS sequences (31), one of the fastest evolving DNA regions in flowering plants. We obtained 567 ITS sequences (Fig. S1) from samples collected from China, Taiwan, Vietnam, Thailand, Japan, the Philippines, Sulawesi, Solomon Islands, and Pacific islands (Fiji, Tonga, Samoa, Marquesas, Pitcairn, and Rapa Nui). Among the 17 haplotypes (haplotype A–Q) detected in the dataset, four (B, C, H, and I) grouped in two clades were present in the Pacific islands (Fig. S1). However, the ITS haplotypes present in the Pacific samples were either also present in a wide geographic range (i.e., haplotype C) or derived from a widespread haplotype (haplotype A), providing inconclusive information regarding the precise origin of the Pacific paper mulberry. Additionally we also failed to amplify and acquire any quality ITS DNA sequences in our historical herbarium samples.
Because paper mulberry is a wind-pollinated and dioecious tree species propagated clonally in the Pacific, chloroplast markers that are maternally inherited in B. papyrifera (32) are better suited for tracking its transportation. To search for suitable chloroplast markers, we screened DNA sequence variation of 10 samples [Taiwan (n = 4), China (n = 4), Sulawesi (n = 1), Hawaii (n = 1)] in 12 noncoding chloroplast DNA sequence regions (atpI-atpH, ndhF-rpl32, rpl16 intron, rps16 intron, psaI-accD, psbD-trnT, psbJ-petA, rpoB-trnC, trnC-ycf6, trnD-trnT, trnL-trnF, and trnS-trnG) that were reported to be hypervariable and suitable for phylogeographic studies (33). To effectively and efficiently obtain quality data, we seek markers that (i) are highly variable and easy for PCR amplification and DNA sequencing, (ii) can detect native range phylogeographic structure, and (iii) can track the origins of the Pacific paper mulberry. We choose the ndhF-rpl32 intergenic spacer (ndhF-rpl32) in the small single-copy region, as this region fitted all three criteria. This region is also among the best choices for low-level molecular investigation (33).
Other Supporting Information Files
Supplementary Material
Acknowledgments
We thank Q.-E. Yang, W.-B. Xu, J. Regalado, H. Nguyen, I. Kailey (Province Museum of Central Sulawesi), and H.-C. Kuo for field assistance and the following people for providing plant materials: Y.-C. Chiang and C.-I. Peng (China), K. Nakamura (Japan), H. Won (Cambodia), T. Phuttai (Thailand), T.-Y.A. Yang (Solomon Islands), C. Walter (Pitcairn), and B. Kam (Bishop Museum), P.v. Dyke (Amy BH Greenwell Ethnobotanical Garden), M. DeMotta and R. Matthews (National Tropical Botanical Garden, NTBG), K. Aiona (NTBG Kahanu Garden), J. Paman (Maui Nui Garden), B. Mulloy, and D. Tanahy (Hawaii). We are also grateful to P. Matthews, S.-H. Chen, and Y.-C. Liu for helpful discussions; two reviewers for their constructive review; and J. Soto-Aguilar and V. Torres for analysis of parts of the Polynesian samples. We thank the Easter Island National Park for a collecting permit (to A.S.); the herbaria of the Natural Historical Museum, New York Botanic Garden, Missouri Botanical Garden, and Smithsonian Institute for permission to sample herbarium collections; and B. Torke, V. Funk, and S.-H. Liu for sampling. This work was supported by National Science Council, Taiwan Grant NSC-102-2621-B-002-007 (to K.-F.C.), Fondo Nacional de Ciencia y Tecnología (FONDECYT), Chile Grants 1080633 and 1120175 (to A.S.), and funding from the Office of World Austronesian Studies, Bureau of International Cultural and Educational Affairs, Ministry of Education, Taiwan (to C.-S.C., H.-L.L., and D.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. KP728381–KP728458).
See Commentary on page 13432.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1503205112/-/DCSupplemental.
References
- 1.Kirch PV. Peopling of the Pacific: A holistic anthropological perspective. Annu Rev Anthropol. 2010;39(1):131–148. [Google Scholar]
- 2.Addison DJ, Matisoo-Smith E. Rethinking Polynesians origins: A West-Polynesia Triple-I Model. Archaeol Ocean. 2010;45(1):1–12. [Google Scholar]
- 3.Bellwood P, Chambers G, Ross M, Hung H-C. Are ‘cultures’ inherited? Multidisciplinary perspectives on the origins and migrations of Austronesian-speaking peoples prior to 1000 BC. In: Roberts BW, Vander Linden M, editors. Investigating Archaeological Cultures. Springer; New York: 2011. pp. 321–354. [Google Scholar]
- 4.Diamond JM. Express train to Polynesia. Nature. 1988;336(6197):307–308. [Google Scholar]
- 5.Oppenheimer SJ, Richards M. Polynesian origins. Slow boat to Melanesia? Nature. 2001;410(6825):166–167. doi: 10.1038/35065520. [DOI] [PubMed] [Google Scholar]
- 6.Matisoo-Smith E, Robins JH. Origins and dispersals of Pacific peoples: Evidence from mtDNA phylogenies of the Pacific rat. Proc Natl Acad Sci USA. 2004;101(24):9167–9172. doi: 10.1073/pnas.0403120101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tsang C-H. Once again on the Austronesian origin and dispersal. Journal of Austronesian Studies. 2012;3(1):87–119. [Google Scholar]
- 8.Storey AA, et al. DNA and Pacific commensal models: Applications, construction, limitations, and future prospects. J Island Coast Archaeol. 2013;8(1):37–65. [Google Scholar]
- 9.Gray RD, Drummond AJ, Greenhill SJ. Language phylogenies reveal expansion pulses and pauses in Pacific settlement. Science. 2009;323(5913):479–483. doi: 10.1126/science.1166858. [DOI] [PubMed] [Google Scholar]
- 10.Ko AM-S, et al. Early Austronesians: Into and out of Taiwan. Am J Hum Genet. 2014;94(3):426–436. doi: 10.1016/j.ajhg.2014.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mirabal S, Cadenas AM, Garcia-Bertrand R, Herrera RJ. Ascertaining the role of Taiwan as a source for the Austronesian expansion. Am J Phys Anthropol. 2013;150(4):551–564. doi: 10.1002/ajpa.22226. [DOI] [PubMed] [Google Scholar]
- 12.Lipson M, et al. Reconstructing Austronesian population history in Island Southeast Asia. Nat Commun. 2014;5:4689. doi: 10.1038/ncomms5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Green RC. Lapita and the cultural model for intrusion, integration and innovation. In: Anderson A, Murray T, editors. Australian Archaeologist: Collected Papers in Honour of Jim Allen. Coombs Academic Publishing, Australian National University; Canberra, Australia: 2000. pp. 372–392. [Google Scholar]
- 14.Bourdieu P. Cultural reproduction and social reproduction. In: Karabel J, Halsey AH, editors. Power and Ideology in Education. Oxford Univ Press; New York: 1977. pp. 487–511. [Google Scholar]
- 15.Whistler WA. Plants of the Canoe People. National Tropical Botanical Garden; Kaua'i, HI: 2009. [Google Scholar]
- 16.Lebot V. La domestication des plantes en Océanie et les contraintes de la voie asexuée. J Soc Ocean. 2002;114–115(1):45–61. [Google Scholar]
- 17.Roullier C, Benoit L, McKey DB, Lebot V. Historical collections reveal patterns of diffusion of sweet potato in Oceania obscured by modern plant movements and recombination. Proc Natl Acad Sci USA. 2013;110(6):2205–2210. doi: 10.1073/pnas.1211049110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zerega NJC, Ragone D, Motley TJ. Complex origins of breadfruit (Artocarpus altilis, Moraceae): Implications for human migrations in Oceania. Am J Bot. 2004;91(5):760–766. doi: 10.3732/ajb.91.5.760. [DOI] [PubMed] [Google Scholar]
- 19.Hinkle AE. Population structure of Pacific Cordyline fruticosa (Laxmanniaceae) with implications for human settlement of Polynesia. Am J Bot. 2007;94(5):828–839. doi: 10.3732/ajb.94.5.828. [DOI] [PubMed] [Google Scholar]
- 20.Matthews PJ. Ethnobotany, and the origins of Broussonetia papyrifera in Polynesia: An essay on tapa prehistory. In: Davidson JM, Irwin G, Leach BF, Pawley A, Brown D, editors. Oceanic Culture History: Essays in Honour of Roger Green. New Zealand Journal of Archaeology Special Publication; Dunedin, New Zealand: 1996. pp. 117–132. [Google Scholar]
- 21.Barker C. Plate 432. Broussonetia papyrifera. Curtis’s Bot Magazine. 2002;19(1):8–18. [Google Scholar]
- 22.Florece LM, Coladilla JO. Spatial distribution and dominance of paper mulberry (Broussonetia papyrifera) in the vicinities of Mt. Makiling, Philippines. Journal of Environmental Science and Management. 2006;9(2):54–65. [Google Scholar]
- 23.Howard MC, editor. Bark-Cloth in Southeast Asia. White Lotus Co., Ltd; Bangkok, Thailand: 2006. [Google Scholar]
- 24.Chang C-S, editor. Felting Bark to Make Cloth—Catalogue of the Tapa Collections of the National Museum of Prehistory, Taiwan. National Museum of Prehistory, Taiwan; Taitung: 2011. [Google Scholar]
- 25.Meilleur BA, Maigret MB, Manshardt R. Hala and Wauke in Hawai’i. Bishop Museum Press; Honolulu, HI: 1997. [Google Scholar]
- 26.Li D, et al. The oldest bark cloth beater in southern China (Dingmo, Bubing basin, Guangxi) Quat Int. 2014;354(1):184–189. [Google Scholar]
- 27.Cameron J. Trans-oceanic transfer of bark-cloth technology from South China-Southeast Asia to Mesoamerica? In: O'Connor S, Clark G, Leach F, editors. Islands of Inquiry: Colonisation, Seafaring and the Archaeology of Maritime Landscapes. ANU E Press; Canberra, Australia: 2008. pp. 203–210. [Google Scholar]
- 28.Chang C-S. Barkcloth and paper mulberry. Forestry Research Newsletter. 2012;20(2):62–66. [Google Scholar]
- 29.Chang C-S. Searching the Fuya: The field report of bark cloth, Central Sulawesi, Indonesia. Journal of Austronesian Studies. 2013;4(1):75–94. [Google Scholar]
- 30.Seelenfreund D, et al. Paper mulberry (Broussonetia papyrifera) as a commensal model for human mobility in Oceania: Anthropological, botanical and genetic considerations. NZ J Bot. 2010;48(3-4):231–247. [Google Scholar]
- 31.González-Lorca J, et al. Ancient and modern introduction of Broussonetia papyrifera ([L.] Vent.; Moraceae) into the Pacific: Genetic, geographical and historical evidence. NZ J Bot. 2015;53(2):75–89. [Google Scholar]
- 32. Zhang Q, Liu Y, Sodmergen (2003) Examination of the cytoplasmic DNA in male reproductive cells to determine the potential for cytoplasmic inheritance in 295 angiosperm species. Plant Cell Physiol 44(9):941–951. [DOI] [PubMed]
- 33.Shaw J, et al. Chloroplast DNA sequence utility for the lowest phylogenetic and phylogeographic inferences in angiosperms: The tortoise and the hare IV. Am J Bot. 2014;101(11):1987–2004. doi: 10.3732/ajb.1400398. [DOI] [PubMed] [Google Scholar]
- 34.Clement M, Posada D, Crandall KA. TCS: A computer program to estimate gene genealogies. Mol Ecol. 2000;9(10):1657–1659. doi: 10.1046/j.1365-294x.2000.01020.x. [DOI] [PubMed] [Google Scholar]
- 35.Templeton AR. Nested clade analyses of phylogeographic data: Testing hypotheses about gene flow and population history. Mol Ecol. 1998;7(4):381–397. doi: 10.1046/j.1365-294x.1998.00308.x. [DOI] [PubMed] [Google Scholar]
- 36.Su H-J. Studies on the climate and vegetation types of natural forests in Taiwan (III): A scheme of geographical climatic regions. Quarterly Jounal of Chinese Forestry. 1985;18(3):33–44. [Google Scholar]
- 37.Tseng M-H, Liew P-M. A preliminary probe on sporo-pollen assemblages and paleoenvironments of the recent 20 ka in Taipei Basin. In: Chen C-H, editor. Central Geological Survey Special Publication No 11. Ministry of Economic Affairs, Republic of China; Taipei: 1999. pp. 160–179. [Google Scholar]
- 38.Yang Y-L, Huang T-C, Chen S-H. Diurnal variations of airborne pollen and spores in Taipei City, Taiwan. Taiwania. 2003;43(3):168–179. [Google Scholar]
- 39.Huang S-F. Historical biogeography of the flora of Taiwan. Journal of the National Taiwan Museum. 2011;64(3):33–63. [Google Scholar]
- 40.Gaffney D, et al. Earliest pottery on New Guinea mainland reveals Austronesian influences in highland environments 3000 years ago. PLoS One. 2015;10(9):e0134497. doi: 10.1371/journal.pone.0134497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Odo F, Sinoto K. A Pictorial History of the Japanese in Hawai’i, 1885–1924. Bishop Museum Press; Honolulu, HI: 1985. [Google Scholar]
- 42.Särkinen T, Staats M, Richardson JE, Cowan RS, Bakker FT. How to open the treasure chest? Optimising DNA extraction from herbarium specimens. PLoS One. 2012;7(8):e43808. doi: 10.1371/journal.pone.0043808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tamura K, et al. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28(10):2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Librado P, Rozas J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25(11):1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 45.Excoffier L, Lischer HEL. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour. 2010;10(3):564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.