Significance
The number of hematopoietic stem cell clones contributing to T-cell development is restricted at entry of and during further development inside the thymus. However, despite this severe restriction, a fully diverse T-cell receptor repertoire can be generated, indicating that hematological and immunological clonality are independently regulated.
Keywords: thymus, T-cell receptor, hematopoietic stem cell, T-cell development, T lymphocyte
Abstract
The fate and numbers of hematopoietic stem cells (HSC) and their progeny that seed the thymus constitute a fundamental question with important clinical implications. HSC transplantation is often complicated by limited T-cell reconstitution, especially when HSC from umbilical cord blood are used. Attempts to improve immune reconstitution have until now been unsuccessful, underscoring the need for better insight into thymic reconstitution. Here we made use of the NOD-SCID-IL-2Rγ−/− xenograft model and lentiviral cellular barcoding of human HSCs to study T-cell development in the thymus at a clonal level. Barcoded HSCs showed robust (>80% human chimerism) and reproducible myeloid and lymphoid engraftment, with T cells arising 12 wk after transplantation. A very limited number of HSC clones (<10) repopulated the xenografted thymus, with further restriction of the number of clones during subsequent development. Nevertheless, T-cell receptor rearrangements were polyclonal and showed a diverse repertoire, demonstrating that a multitude of T-lymphocyte clones can develop from a single HSC clone. Our data imply that intrathymic clonal fitness is important during T-cell development. As a consequence, immune incompetence after HSC transplantation is not related to the transplantation of limited numbers of HSC but to intrathymic events.
Hematopoietic stem cell transplantation (HSCT) has become common clinical practice in the treatment of leukemia, lymphoma, and certain inherited immune and metabolic disorders. After transplantation there is an immediate need for de novo production of granulocytes, erythrocytes, and platelets, referred to as “hematopoietic reconstitution,” later followed by recovery of lymphocyte numbers, termed “immune reconstitution.” Although often successful, HSCT is associated with a number of complications arising from poor immune reconstitution, which presents one of the most important causes of morbidity after HSCT (1, 2). Poor myeloid reconstitution is directly linked to low numbers of HSCs in the transplant, but the reasons for poor immune and, in particular, T-lymphocyte reconstitution are much less clear.
A study on the application of antithymocyte globulin in cord blood transplantation showed that antithymocyte globulin administration in pretransplantation conditioning results in decreased T-cell reconstitution and survival (3), indicating the need for a better understanding of de novo T-cell development after HSCT. As T cells develop in the thymus, in contrast to all other blood lineages that develop in the bone marrow (BM), HSC-derived progenitors need to seed the thymus. Earlier work in mice has indicated that relatively few progenitors seed the thymus (4, 5), yet their numbers and the subsequent fate of the progeny derived from an HSC clone has remained elusive. The nature of the thymus-seeding cell has been subject of much debate, certainly in humans. CD34+CD38−CD7+ cells (6, 7), CD34+CD38−CD10−CD62L+ cells (8), and others have been proposed as thymus-seeding cells in the human situation. In contrast to mice, the earliest human thymocytes show a much broader lineage developmental potential, including low levels of erythroid and megakaryocytic potential (9), and may therefore be much closer related to HSCs or the proposed myeloid lymphoid precursors.
In this study, we aimed to address the functional relationship between HSCs and developing T cells by using DNA barcoding. This method allows marking of each stem cell and its progeny by a unique, neutral DNA sequence that can be retrieved by next-generation sequencing. Here we used replication-deficient lentiviruses to transfer a DNA barcode and the GFP into phenotypically defined human HSCs. These marked HSCs were transplanted, using an optimized protocol (10), into NOD/SCID/Il-2Rγ−/− (NSG) mice (11), which allowed us to determine the dynamics of clonal reconstitution in the thymus in great detail. We used umbilical cord blood (UCB) as source of human stem cells, because UCB is frequently used as a readily available HSC source with less stringent HLA (human leukocyte antigen)-matching requirements (12). However, T-cell immune reconstitution, a general problem in allogeneic HSCT, is more problematic in UCB. Therefore, UCB provides an appropriate HSC source to address questions of lineage tracing and clonality in the blood and immune system. As we demonstrate here, only a fraction of HSC clones in UCB contributes to the T-cell pool, yet a fully diverse T-cell receptor (TCR) repertoire can be generated in the thymus.
Results
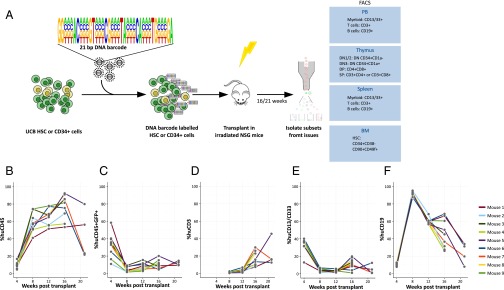
We set out to better understand the functional relationship between HSCs in the BM and developing T cells in the thymus. Cellular barcoding allows this question to be addressed in a quantitative sense. To do so, human HSC were isolated from UCB and were transduced with a lentiviral DNA barcoding vector that carried 485 individual barcodes (13, 14) (Fig. 1A). We aimed to transduce <20% of the target cells to limit the occurrence of multiple lentiviral insertions per cell. A well-defined library with restricted complexity was used to allow full quantification yet also to contain enough diversity to individually track HSC, given the number of transduced HSCs per mouse. In three independent experiments, a total of 17 mice were transplanted with either low (26,000; n = 4) or high dose (150,000; n = 4) of CD34+ cells or ∼1,000 highly purified HSCs (n = 9), defined as CD34+CD38−CD45RA−CD90+CD49f+ (15).
Fig. 1.
Barcoding human HSC and xenotransplantation. (A) Human HSC (CD34+CD38−CD45RA−CD90+CD49f+) or CD34+ cells were isolated and transduced with the PTGZ barcode library carrying 21 variable bases in the barcode with a total complexity of 485 different barcodes. The transduced cells were transplanted in to 5- to 6-wk-old sublethally irradiated NSG mice. The mice were bled monthly and myeloid cells, T cells, and B cells were isolated from PB. At 16 or 21 wk, the experiment was terminated and, in addition to PB, cells isolated from thymus, spleen, and BM were sorted as indicated and the barcode content of the samples was analyzed. From the thymus, DN, DP, and SP cells were isolated using the indicated markers. Chimerism in the PB of nine mice transplanted with 1,000 purified human HSCs as determined by human CD45 expression (B) and GFP marking of human cells (C). Development of CD3+ T cells (D), CD13+/CD33+ myeloid cells (E), and CD19+ B cells (F) within the human CD45+ population were followed in time after transplantation of the transduced cells.
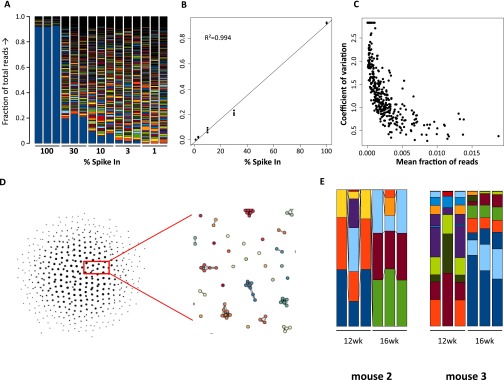
To ensure that viral DNA barcoding methodology allows quantification of marked clones, calibration experiments were performed. The mixtures of DNA barcodes resulting from the spike-in experiments demonstrate a decrease of the sample from the 100% down to the 1% spike-in sample (Fig. S1A). Linear regression analysis showed a Pearson correlation coefficient R2 = 0.994 (Fig. S1B). In a separate experiment, nine repeat measurements of the polyclonally transduced samples were analyzed for reproducibility. Read fractions below 0.5% of the sample threshold were progressively more difficult to quantify reliably as demonstrated by the increase in coefficient of variation at lower fractions of reads (Fig. S1C). Deep sequencing is prone to generate sequence misreads at low frequency, and because our analysis is dependent on the ability to assign and distinguish individual barcodes, we compared all identified DNA barcodes against each other and determined their dissimilarity. We considered barcodes with up to two dissimilar bases to originate from the same original barcode (Fig. S1D). Repeated measurements on CD19+ cells isolated from peripheral blood (PB) (Fig. S1E) also demonstrated that the method results in limited variation between samples. Thus, the applied barcoding strategy can reliably quantify progeny of individual HSC clones in this setting.
Fig. S1.
Spike-in experiments to calibrate the DNA barcoding method. A plasmid sample was spiked in at equimolar ratio and diluted into a bulk barcoded sample. The bargraph shows the measured fraction of deep-sequencing reads in each sample, with the lowest blue bar in the graph indicating the spiked-in plasmid (A). Triplicate analysis of the fraction of reads made up by the spiked-in barcode shows the least-squares fitted line and the Pearson correlation coefficient. (B). In an additional experiment, nine repeated measures of the same sample were performed and the coefficient of variation was determined and plotted against the mean fraction of reads, showing that the precision of quantification increases with the amount of reads (C). A higher number of barcodes than the library complexity was retrieved from the analysis of these samples. To correct for these sequencing errors, the number of dissimilar bases between all barcodes was determine and a directed acyclic graph was built for all barcodes that had less than three bases difference. Using this graph and an empirically determined threshold of two bases differences between barcodes, the reads of barcodes with <three bases difference were combined. All barcoding data in the report was processed using this procedure (D). Samples of PB CD19+ B cells were analyzed in triplicate to determine the variation of the method between samples (E). Fraction of total reads is indicated on the y axis.
Transplantation of human HSC from UCB resulted in a robust reconstitution (median 81%, range 53–92% at 16 wk) (Fig. 1B) in all nine mice. GFP marking was initially around 20% and stabilized at 16 wk at median 8.9% (range 4.5–20%) (Fig. 1C), consistent with the intended low transduction rate, which limits the occurrence of multiple integrations per cell. After 12 wk, CD3+ T cells were detectable for the first time (Fig. 1D), which coincides with a concomitant reduction in the percentage of CD19+ B cells (Fig. 1F), which form the majority of repopulating human cells.
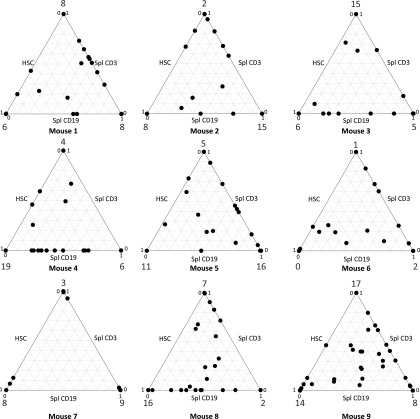
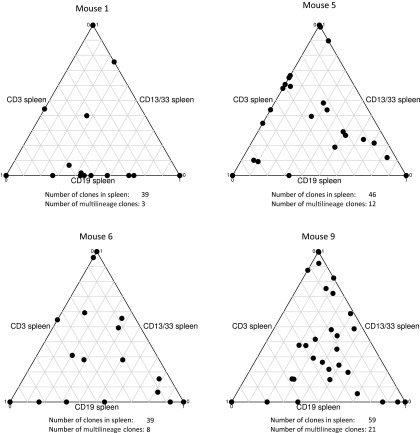
Analysis of the barcodes present during repopulation demonstrated that a limited repertoire of hematopoietic clones was responsible for the generation of myeloid cells, B cells, and T cells. We detected a median number of 70 clones (range 51–88) per mouse in the PB, rising to a median of 83 (range 57–107) when BM and thymus samples with different barcodes were included. Because the barcodes in purified HSCs are inherited by the daughter cells, we could also investigate the contribution of the marked clones in HSC to the B- and T-cell lineage. We therefore analyzed the extent to which a clone retrieved from the human HSC isolated from the NSG mice was also detected in CD19+ B cells or CD3+ T cells in the spleen. The ternary plots (Fig. 2 and Fig. S2) showed that a considerable fraction of clones was shown to be present in two or three of these three cell types, demonstrating contribution to sorted HSC as well as the adaptive lymphoid lineage. This finding confirms that true HSCs with multilineage differentiation capacity were marked by lentiviral barcoding. The existence of myeloid, lymphoid, or mixed progenitors is in line with previous reports in mice (16) and human cells in the NSG xenograft model (17). However, none of these studies specifically addressed the thymus or T-cell reconstitution.
Fig. 2.
Clonal contribution to HSC and adaptive immune cells. Multilineage engraftment is demonstrated by contribution of barcoded clones to HSC, splenic CD3+ T cells and CD19+ B cells in nine NSG mice that were transplanted with purified barcoded human HSCs. The barcode content of these compartments was determined by comparing the normalized contribution of clones. The relative contributions to the total human HSC (BM) and T- and B-lymphoid lineages (spleen) in each mouse is shown as a ternary plot. Each point represents the contribution of a clone to HSC, CD19+ B-cell, and CD3+ T-cell lineages. The extent of contribution is depicted by the location of the point along the three axes, with clones contributing equally to all lineages being closer to the center of the triangle. In most mice, but especially in mouse 8 and mouse 9, true multilineage contribution of HSC clones can be observed. The numbers at the points of the triangle indicate the number of detected clones for which contribution to only one lineage was detected.
Fig. S2.
Barcoded HSC result in multlineage engraftment in NSG mice. NSG mice were transplanted with barcoded HSC and after 21 wk, barcode content in CD19+ B cells, CD3+ T cells, and CD13+/CD33+ myeloid cells was determined. Ternary plots show the contribution of a clone to each lineage. Multilineage contribution is defined as contribution of a clone to each of these three lineages. The total number of clones retrieved from these spleen samples is indicated, as is the number of these clones that contribute to all three lineages.
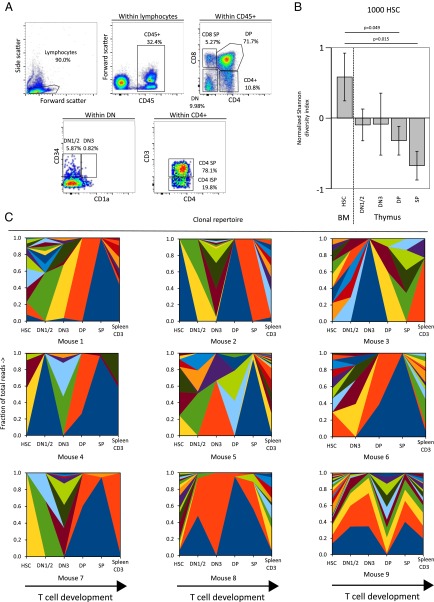
To study thymic repopulation, various stages of human T-cell development, ranging from early double-negative (DN) (CD4−CD8−) cells to double-positive (DP) (CD4+CD8+) and single-positive (SP) mature cells (CD4+ or CD8+) were isolated from the thymus, as described (DN1/2: CD3−CD4−CD8−CD34+CD1a−; DN3: CD3−CD4−CD8−CD34+CD1a+; DP: CD4+CD8+; SP: CD3+ CD8+ or CD3+ CD4+) (Fig. 3A) (18). During differentiation from polyclonal HSC to mature CD3+ T cells, a dramatic clonal skewing was seen in the form of reduction of the number of clones contributing more than 1% of the thymic populations. We used the Shannon diversity index, a measure that can be used to describe biological diversity (19), to capture the frequency and abundance of barcoded clones. Using this parameter, the HSC (Fig. 3B) and high-dose CD34+ (Fig. S3 A and C) experiments show high diversity in BM but much lower in the thymus, with clear clonal restriction at DN1/2 and subsequently a further restriction at the DN3 and DP stages. The reconstitution with the 26,000 CD34+ cell dose containing lower numbers of true HSCs led to lower diversity in BM, but still only one clone dominates at the thymic SP or DP stage (Fig. S3 B and D). Analysis of the DN3, DP, and SP compartments indicated a further reduction in the number of hematopoietic clones (Fig. 3B and Fig. S3A), but the numbers of isolated cells actually increase going from DN1/2 through DN3 to DP, which demonstrates that clonal restriction is not a result of smaller thymic subpopulations investigated.
Fig. 3.
Clonal restriction during T-cell development. Thymus was homogenized and cells were sorted into their developmental stages (DN1/2, DN3, DP, SP) (A). The number of clones contributing >1% of the sample in nine mice transplanted with barcoded HSCs or CD34+ cells were counted and the Shannon diversity index calculated. The normalized Shannon diversity was compared between HSC in the BM and the developmental stages in the thymus. Mean normalized Shannon diversity indices for nine mice transplanted with 1,000 HSC are shown with error bars indicating SEM (B). Wilcoxon P values are shown in B. (C) Clonal repertoire, as determined by barcode analysis of sorted population in the thymus in stacked area graphs (as in A), with colors identifying the individual clones. Note that the colors are chosen for the purpose of display, which means that the same color depicts different clones in different mice.
Fig. S3.
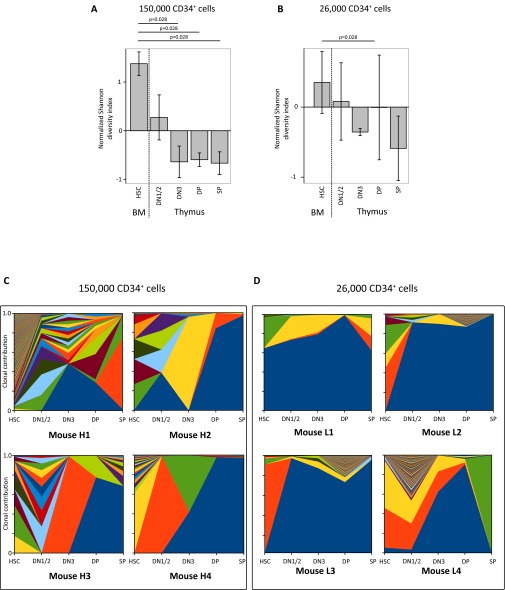
Clonal restriction during T-cell development after CD34+ cell transplantation. Similar to the data in Fig. 2, the normalized Shannon diversity was compared between the developmental stages in the thymus of four mice transplanted with 150,000 CD34+ cells (A) or four mice transplanted with 26,000 CD34+ cells (B). Kruskal–Wallis P values are shown in A and B. (C and D) The clonal contribution in the thymic subsets of mice transplanted with 150,000 or 26,000 barcoded human CD34+ cells is shown in stacked area graphs using colors to mark the individual barcoded clones in mice receiving a high (150,000 cells, C) or low (26,000 cells, D) dose of barcoded CD34+ cells.
To investigate whether the restriction of clones is a result of a limitation in the number of transplanted cells, we performed similar experiments with 150,000 or 26,000 CD34+ cells per mouse (containing ∼625 and ∼108 HSC, respectively) (15). Whereas 150,000 CD34+ cells are capable of generating a repopulation pattern in the thymus similar to that observed after transplantation of 1,000 purified HSC (Fig. S3C), transplantation of 26,000 CD34+ cells per mouse shows more limited clonal diversity, with most compartments consisting of a few clones (Fig. S3D). Nevertheless, further clonal restriction during T-cell development was observed despite the transplantation of a sixfold higher number of CD34+ cells, or when a 10-fold higher number of phenotypically defined HSC was transplanted, suggesting that thymic clonality is primarily determined by intrathymic events rather than based on cell dose. Hence, clinically there is merit in transplanting a higher dose of CD34+ cells to obtain a more diverse repertoire, yet a plateau is readily reached.
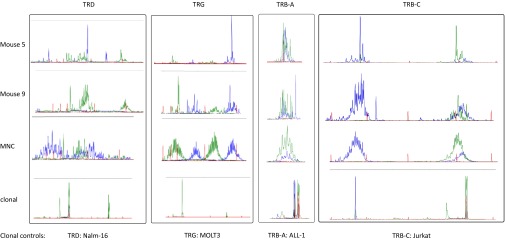
Having observed the limited number of hematological clones in the thymus, we asked the question whether this restricted number of cells would negatively influence the TCR repertoire. At DP and SP stages, a fully polyclonal pattern of TCRβ (TRB) rearrangements is visible, with even further diversity in the splenic CD3+ T cells (Fig. 4). From these data, we conclude that a fully diverse TCR repertoire can be generated from progeny with a high level of clonal restriction. It needs to be noted that the polyclonality in the spleen can be interpreted as the result of ongoing thymic output, whereas the thymic analysis is more of a “snapshot.” Nevertheless, even at this snapshot the progeny of HSC clones is very small, whereas thymic DP and SP cells have a fully diverse TCR repertoire, which is further expanded in the spleen by the accumulation of cells egressing from the thymus. Because only a portion of the cells in the thymus and spleen were lentivirally transduced, the untransduced cells might contribute to this diverse repertoire; we therefore also sorted GFP+-transduced cells from the spleen to confirm that the TCR repertoire of the transduced cells was indeed polyclonal. This analysis demonstrated diverse repertoires in TCRδ (TRD), TCRγ (TRG), and TRB, thereby confirming that the limited clonal repertoire of transduced cells does lead to the observed TRB repertoire (Fig. S4). Our data show strong intrathymic competition between the progeny of stem cell clones. Therefore, clonality at the HSC level (hematological clonality) and a fully diverse TCR repertoire (immunological clonality) are independently regulated.
Fig. 4.
Separation of hematological and immunological clonality. TRB repertoires in DP and SP thymic subsets, and sorted CD3 splenocytes of mice transplanted with human HSC were determined. Electropherograms of two collections of primers amplifying the TRB locus (green and blue) show the TRB diversity for a representative mouse. The positive control shows peripheral blood mononuclear cells (PBMC) of a healthy individual.
Fig. S4.
Hematological and immunological clonality in splenocytes. Rearrangements of TRD, TRG, and TRB of sorted transduced human splenic CD3 cells are shown using two collections of primers to amplifying the respective loci. PBMC of a healthy individual was used a positive control and clonal rearrangements in Nalm-16 (TRD), MOLT3 (TRG), ALL-1 (TRB-A), and Jurkat (TRB-C) are shown for comparison.
Discussion
The relationship between hematopoietic stem cells and T cells in quantitative terms previously was largely unexplored. Previous studies in mice showed that a limited number of HSC can be marked, transplanted, and identified this way. Initially, such studies were performed using the unique marks that virus insertion form when they integrate into the host genome (20, 21). From these studies, several models for behavior of the hematopoietic system were postulated and mathematically modeled (22). DNA barcoding technology allowed a much more refined insight into the composition of the blood compartments and allowed the study of aging HSC (13) and distribution of HSC clones through the different locations in the body (14), in addition to the clonal dynamics of repopulation of the BM subset hierarchy after transplantation (23, 24). The existence of myeloid, lymphoid, or mixed progenitors, which had previously been observed in mice (16), was demonstrated with human cells in the NSG xenograft model (17), and although these seminal studies in the xenograft system demonstrated the dynamics of lymphoid development, they did not address T-cell development nor thymic function.
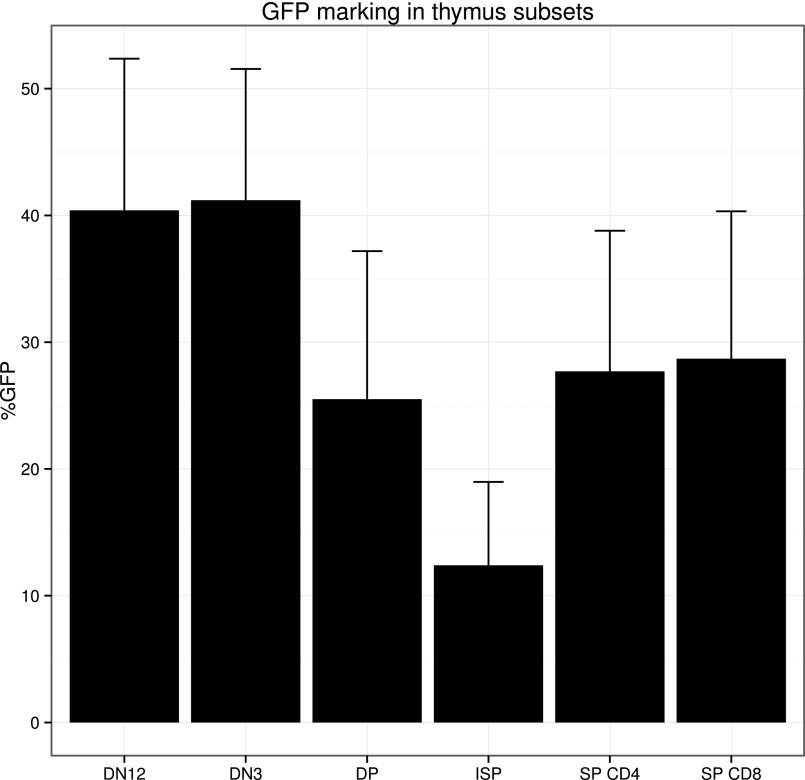
Using lentiviral cellular barcoding of purified human HSCs or CD34+ progenitor cells, we here show at least 10% of the HSC clones are GFP-marked in the thymus (Fig. S5) and show considerable barcode-defined clonal restriction. How this restriction in clonal repertoire in the thymus occurs is not yet understood. It is conceivable that competition of DN cells for thymic niches (specific location in the thymus that need to be reached for further development), fitness of the individual clone for progression in development, or stochastic events drive competition between clones.
Fig. S5.
GFP marking in the thymus. The percentage of GFP marking in the different thymocyte subsets after transplantation of barcoded HSC is displayed as mean + SEM (n = 9).
Intrathymic events that regulate TCR repertoire and the integrity and quality of thymic epithelial cells are likely an important factor. As a consequence, efforts to improve thymic reconstitution using cytokines [IL7 (25), SCF, KFG (26, 27), FLT3L (28)] or hormones [growth hormone (29), thyroid-stimulating hormone (30), and ablation of sex hormones (31)] are only likely to be effective if they selectively act on the thymic microenvironment or mimic signals given by the thymic epithelial cells to developing thymocytes. Transplantation of committed T-cell progenitors together with HSCT, as shown before with mouse hematopoietic cells cultured ex vivo on OP9-DL1 expressing stromal support—which provide the cells with the Notch signals required to direct T-cell development (32)—as well as diminishing the influence of male sex hormones known to deregulate intrathymic Notch ligand DLL4 (31) may work well to improve thymic function and support diverse TCR repertoire formation. There is one interesting physiological situation in which hematopoietic reconstitution can be stimulated: namely, by exposure to noninherited maternal antigens of the HLA-locus (33). This phenomenon is now widely recognized in UCB transplantations and indicates that effective T-cell responses can be mounted with beneficial (e.g., graft vs. leukemia) effects (34, 35). Although such exposure would theoretically lead to increased thymic output, the effect on TCR repertoire is expected to be limited; hence efforts directed at increasing overall thymic output are warranted.
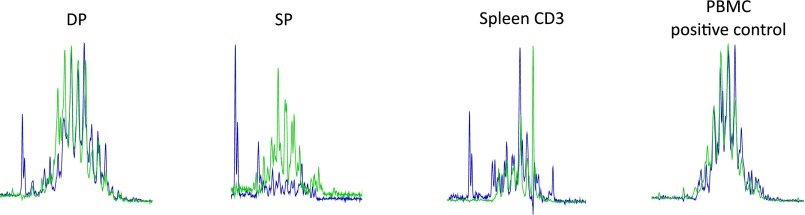
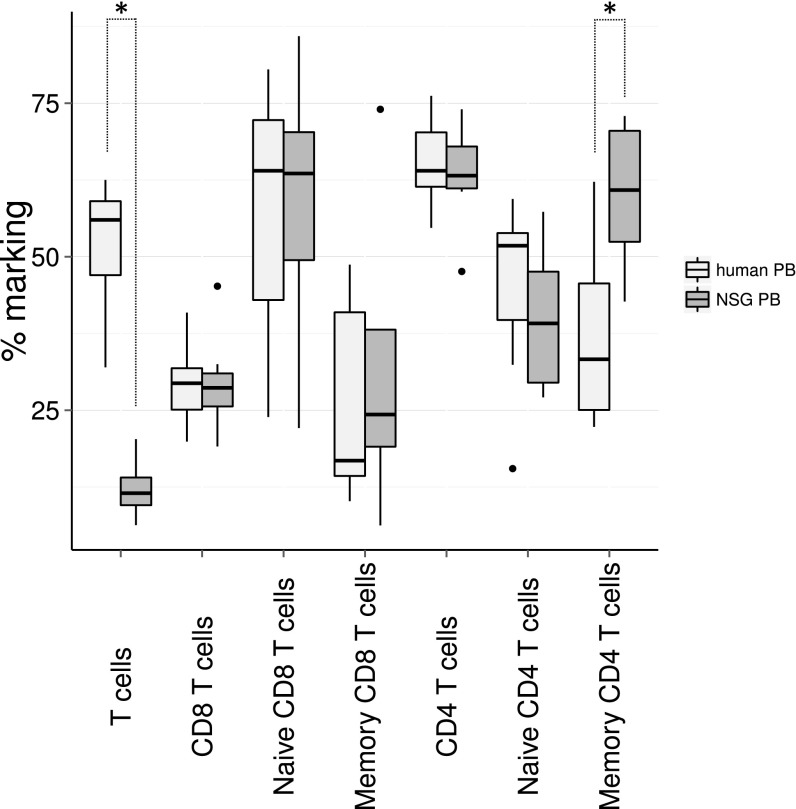
Xenotransplantation of human cells in mice is a valuable approximation of the normal development of human cells. In NSG mice, the human cells develop into mature functional T cells that are responsive to immunization (10, 36) and the thymus shows highly similar phenotype to normal human thymi (37). Compared with human control samples (Fig. S6), xenotransplanted NSG mice show lower CD3+ cell counts (Wilcoxon test, P = 0.0015), but within the CD3+ cells, the percentage of CD4 and CD8 T cells is comparable. Furthermore, human and xenografted CD8 T cells show similar distribution of CD45RA+ naive cells and CD45RO+ memory cells, but CD4 CD45RO+ memory T cells are present in a significantly higher proportion in the transplanted NSG mice (P = 0.009), which might point to ongoing homeostatic proliferation in the CD4 compartment. Analysis of the interaction between murine thymic stroma and human T cells showed that T cells are capable of migrating to the site where they are expected to reside, corresponding to their developmental stage in mouse thymus in response to murine Ccl25, Cxcl12, and Ccl21, all chemokines that attract T cells to the thymus (38). Several papers have addressed the TCR repertoire in NSG mice in the presence or absence of transgenic human HLA-A2. First, Shultz et al. (39) demonstrated a functional Epstein–Barr virus infection in xenografted NSG mice and compared the responses of xenografted NSG or NSG with transgenic expression of HLA-A2. No differences were observed in the frequency of naive, central memory and effector-memory CD8 T cells in the spleen or for Granzyme A and B or Perforin expression. The only demonstrable difference between NSG and HLA-A2-expressing NSG is seen in response to HLA-A2 restricted BMLF and LMP1 proteins. Second, the issue of interaction of the xenografted cells with transgenic human HLA-A2 was elegantly addressed by Halkias et al. (40), where HLA-A2 transgenic NSG were compared with normal NSG after transplantation of HLA-A2+ or HLA-A2− human UCB cells. No differences were observed in repopulation and T cells in the spleen. Thus, the reported findings on T-cell development appear to be relevant for allogeneic human HSCT transplantation. However, being a xenograft model, some aspects of the results should be interpreted with caution. Important T-cell subsets, such as regulatory T cells (40) and PLZF1+ innate T cells (41), which are both critical components of immune competence, do not develop or function properly in NSG mice. This likely relates to the fact that MHC restriction occurs mostly on mouse epithelial cells and less frequently on human APC or via T–T interactions. Thus, clinically significant differences in the TCR repertoires of T cells selected on mouse thymus versus T cells that are selected on human thymus exist. Nevertheless, in experiments where CD34+ cells were transplanted into NSG mice, a survey of the IgH locus and TCR showed a comparable combinatorial diversity (42), although the screening method used in this study (multiplexed PCR of V-J rearrangements) might not be sensitive enough to detect small differences in TCR repertoire between human and xenografted T-cell samples.
Fig. S6.
Comparison of T-cell subsets in human PB and xenografted NSG mice. PB of NSG mice (n = 8) transplanted with lentivirally transduced UCB CD34+ cells at 20 wk posttransplantation and human PB control samples (n = 7, female, ages 20–52 y) were analyzed for the expression of CD45RO and CD45RA in the CD4+ and CD8+ T cells by flow cytometry. Wilcoxon P values smaller than 0.05 are indicated with an asterisk.
Recent work by Rodewald and colleagues proposed cell competition as a mechanism underlying selection for cellular fitness during T-cell development (43). This hypothesis explains the further clonal restriction at DN3, DP, and SP stages that we observe in many mice. Apparently there is selection for progeny of certain HSCs clones while others are selected against. Although this could be merely stochastic (neutral competition), the possibility that clonal fitness (e.g., metabolic activity, quiescence, other factors) plays a role is not unlikely given the existence of thymic stem cell clones with differential gene-expression programs (43). We therefore propose that the progeny from different HSC clones have inherently different capacity to pass through various developmental checkpoints in the thymus. Alternatively, in the experiments where barcoded CD34+ cells were transplanted, we cannot exclude that in some cases progenitors rather than bona fide HSCs would be the ancestors of the developing thymocytes. These progenitors are likely to have reduced fitness as well. In the experiments where phenotypically defined HSCs were transplanted, a role for such progenitors seems much less likely because of the purity of the transplanted cells. This notion is supported by the fact that in human gene-therapy settings, a considerable fraction of T-cell may have arisen from progenitors rather than from HSCs (44).
As a consequence, immune incompetence after HSCT is not related to the transplantation of limited numbers of HSC but to intrathymic events. Although transplantation of more HSC may improve hematological reconstitution, our data suggests it will do little to improve immunological reconstitution. Thymus seeding is quickly saturated when the transplanted cell dose increases; therefore, efforts aimed at improving function of the thymic epithelium (45) rather than increasing the number of transplanted cells, most likely will be most successful in improving immunological immune reconstitution after stem cell transplantation.
Materials and Methods
CD34+ and HSC Isolations from UCB.
Human CD34+ cells were isolated from UCB samples. Mononuclear cells of UCB were isolated by Ficoll [Leiden University Medical Center (LUMC) apothecary] density centrifugation, washed, and stored in liquid nitrogen until further use. Single UCB units (HSCT) or 5 to 10 combined UCB units (CD34+ transplantations) were selected, thawed, and selected for CD34+ progenitors using the CD34 Microbead Kit (Miltenyi Biotec) according to the manufacturer’s protocol. Purity of the CD34 selection was verified by flowcytometry and exceeded 95% purity. HSCs were isolated by staining with anti-human CD38 (HIT2) PECy7, CD90 (ebio5e10) APC, CD49f (ebioG0H3) PerCPEfluor710 (all eBioscience), CD34 (8G12) PE, CD45 (HI30) V450, CD45RA (L48) FITC (BD), and subsequent sorting of CD34+CD38−CD45RA−CD90+CD49f+ cells using a BD Aria II SORP cell sorter (Beckton-Dickinson). The cells were subsequently cultured in Stemspan (Stem Cell Technologies), in the presence of 10 ng/mL stem cell factor (a gift from Amgen, Thousand Oakes, CA), 20 ng/mL recombinant human thrombopoietin (R&D Systems), 20 ng/mL recombinant mouse insulin-like growth factor 2 (R&D Systems), 10 ng/mL recombinant human fibroblast growth factor 1 (Peprotech), and penicillin/streptomycin (Lonza) on retronectin-coated plates (Takara). The cells were incubated for 24 h at 37 °C and 5% (vol/vol) CO2.
Virus Production and Transduction Procedure.
Lentiviral vector pTGZ is derivative of pGIPZ (Thermo, Openbiosystems) where the Puromycin-IRES-MCS site locus (BsrGI-MluI) was replaced first by BsrGI-ClaI-BamHI-MluI adapter, then a barcode linker was integrated via BsrGI-BamHI sites between tGFP and the WPRE element, as described previously (13, 14). The barcode sequence, named B322, was as follows: AGGNNNACNNNGTNNNCGNNNTANNNCANNNTGNNNGAC. Viral particles were generated in 293T cells by transfection of pEnv VSVG, pMD2 GagPol, pREV and the barcoded vector backbone pTGZ-B322. Titers were determined on isolated CD34+ cells to achieve a transduction efficiency of 20% to minimize the occurrence of multiple virus integrations per cell, while maintaining a reasonably high marking rate. UCB CD34+ cells were cultured as described above at 6,500 cells/cm2 density. After 24 h, the cells were spinoculated with virus supernatant at a multiplicity of infection = 1, after which the supernatant was replaced with fresh Stemspan medium with cytokines. The cells were kept for 16 h at 37 °C and 5% (vol/vol) CO2 and then prepared for transplantation. Because lentiviral vectors cause pseudotransduction (protein expression from the preintegration complex), assessment of the transduction rate at the moment of transplantation is not accurate. Instead, a fraction of transduced cells was kept in culture for an additional week until the nonintegrated preintegration complexes were diluted out and the transduction rate could be determined by flow cytometry.
Animals and Transplantation.
NSG mice (NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ, NSG) were obtained from Charles River Laboratories and bred in individually ventilated cages under specific pathogen-free conditions. Mice (aged 5–6 wk) were sublethally irradiated with 1.91 Gy X-rays using orthovoltage irradiation and transplanted 24 h after irradiation via tail vein injection with the indicated cell numbers. Mice were fed Diet Gel recovery (Clear H2O) and kept on antibiotics [560 µg/L polymixin B (Bupha), 700 µg/L ciprofloxacin (Bayer), 800 µg/L amphotheracin B (Bristol-Myers Squibb)] in autoclaved acidified drinking water until white blood cell counts were recovered at 4 wk posttransplant. Blood samples were obtained at 4, 8, 12, 16, and 21 wk by lateral tail vein incision. At 16 or 21 wk, mice were killed by O2/CO2 inhalation and thymus, BM, PB, and spleen were isolated. BM cell suspensions were made by crushing the bones in IMDM (Lonza) and passing the remaining cells through a 70-µm filter. Spleen and thymus were homogenized by passing them through a 70-µm filter.
Flowcytometry.
Erythrocytes in the PB samples were lysed using an isotonic NH4Cl buffer (LUMC apothecary), after which the cells were stained with the indicated antibodies in PBS/0.2% BSA /0.1% sodium azide buffer for 30 min on ice. Cells were subsequently sorted using a FACS Aria II SORP (BD).
Spleen and PB cells were stained using anti-human CD45 (HI30) v450, CD19 (SJ25C1) APCCy7, CD3 (SK7) PE, CD4 (RPA-T4) V500, CD8 (SK1) PECy7, CD13 (WM15) APC, and CD33 (WM53) APC (all BD). PB cells were sorted into T cells (huCD45+ CD3+) B cells (huCD45+ CD19+) and myeloid cells (huCD45+ CD13+/CD33+) and splenocytes into B-cell (huCD45+CD19+) and T-cell (huCD45+CD3+) populations. BM cells were stained using anti-human CD45 (HI30) V450, CD34 (8G12) PE, (BD), CD38 (HIT2) PECy7, CD90 (ebio5e10) APC, and CD49f(ebioG0H3) PerCPEf710 (eBioscience). HSC (huCD45+CD34+CD38−CD90+CD49f+) were sorted. Thymus cells were stained using human CD45 (HI30) V450, CD1a (HI149) APC, CD34 (8G12) PE, CD3e (UCHT1) PECy5, CD4 (RPA-T4) APCCy7, and CD8 (SK1) PECy7 (all BD), after which DN1/2 (huCD45+CD3−CD34+CD1a−) and DN3 (huCD45+CD3−CD34+CD1a+), DP (huCD45+CD3+CD4+CD8+), and SP (huCD45+CD3+CD4+ and huCD45+CD3+CD8+) were sorted.
Spike-in Experiment.
Plasmid DNA with a known barcode was mixed with DNA from polyclonally transduced population of cells at 0%, 1%, 3%, 10%, 30%, and 100% based on the estimation that one virus was inserted per genome (3.3 × 109 bp). The size of the plasmid was 10,201 bp and carried one DNA barcode. The samples were then processed for barcode determination as described below.
DNA Preparation and Deep Sequencing.
DNA from the sorted populations was extracted using Sigma GenElute columns (Sigma-Aldrich), according to the manufacturer’s instructions, with the addition of 5-µg herring sperm DNA as carrier material. The DNA barcode was amplified using LWGFP_pTGZ (CACATGCACTTCAAGAGCGCCAT) and LWPRE_pTGZ (TGAAAGCCATACGGGAAGCA) primers (Sigma-Aldrich), at 0.6 µM, using GoTaq polymerase (Promega) and a temperature profile of 95 °C for 5 min, followed by 35 cycles of 95 °C for 30 s, 60° for 30 s, and 72 °C for 30s, with a final extension of product at 72 °C for 10 min. The 379-bp resulting product was cleaned using Qiagen PCR clean-up columns according to the manufacturer’s instructions (Qiagen). The cleaned products were then multiplexed by adding an Iontorrent adapter with sample multiplexing barcodes to one side CCATCTCATCCCTGCGTGTCTCCGACTCAG [eight base multiplexing barcode] GCAGATGCCGGTGAAGAATAATGTAC and a common IonTorrent adapter CCTCTCTATGGGCAGTCGGTGATAGTCAATCTTTCACAAATTTTGTA to the other side (95 °C for 5 min, followed by 35 cycles of 95 °C for 30 s, 60° for 30 s, and 72 °C for 30s). The resulting 208-bp amplicons were separated on a 2% (wt/vol) agarose gel, the products were excised and cleaned using Qiagen gel extraction columns, mixed in equimolar amounts, and submitted to the LUMC sequencing core facility (Leiden Genome Technology Center), where library concentration was determined using a High Sensitivity DNA kit on an Agilent Bioanalyzer (Agilent). The library was amplified using emulsion PCR using the Ion PGM Template OT2 200 kit (Agilent) and subsequently sequenced using the Ion PGM Sequencing 200 Kit v2 (Agilent) and 314 or 316 chips, according to the manufacturer’s instructions.
Sequence Analysis.
FASTQ files were retrieved from the sequencer and underwent overall quality control using FASTQC (Simon Andrews, Babraham Institute, Cambridge, United Kingdom; available at www.bioinformatics.babraham.ac.uk/projects/fastqc/), after which the sequences were separated by sample multiplexing barcode using a custom BioPerl script (www.bioperl.org/wiki/Main_Page) that uses regular expressions to determine primer and sample barcode sequences. From the resulting FASTA files, the DNA barcodes were extracted using a custom BioPerl script that determined barcode location based on a regular expression matching the surrounding virus sequences and the invariable doublets within the DNA barcode. Samples with >100 associated reads were considered for analysis. These barcodes were clustered using R (R-3.0.0, www.r-project.org) by calculating dissimilarity between all barcodes for all samples of an animal. Read numbers for barcodes with dissimilarity of less than 2 nt were clustered and added together, thereby correcting for sequencing errors (46). Simulation of random sets of 485 barcodes showed that the frequency of 1- to 2-nt distances would occur in fewer than 1/1,000 reads; therefore, a dissimilarity threshold of 2 nt seems appropriate to eliminate false barcode reads in sequencing results caused by PCR and sequencing errors. By applying this strategy to the sequenced library pool, this resulted in a set of barcodes that agreed with the expected size of our DNA barcode library (485 barcodes by sequencing with noise removal, compared with the 500 single bacterial clones that were selected for the preparation of the vector library). The contribution of each clone to a particular tissue was normalized by dividing the reads for each clustered clone by the total number of reads per sample. The resulting fraction of contribution was then displayed.
TCR Analysis.
Rearrangements of TRB, TRG, and TRD loci were analyzed by means of BIOMED-2 multiplex PCR assays (47) and visualized via GeneScan analysis. All assays were performed using the proper clonal, polyclonal and negative controls, and according to the published BIOMED-2 multiplex PCR protocol.
Statistics.
To describe the population diversity in the thymus, we calculated the Shannon diversity index using the cell number isolated from a tissue (HSC or thymus DN1/2, DN3, DP, and SP, or spleen CD3 cells) multiplied by the fraction of each clone as determined by barcode sequencing (shown in Fig. 3A) for all reads making up more than 1% of the reads recovered in these tissues. The Shannon indices obtained in this way were then normalized per animal to allow comparison between animals. Differences in Shannon diversity between subsets were analyzed using a Wilcoxon rank sum test, considering P < 0.05 significant.
Study Approval.
Experimental procedures were approved by the Ethical Committee on Animal Experiments of the Leiden University Medical Center. The UCB samples used in this study were collected at the Diakonessenhuis hospital, Leiden, after informed consent was given by the parents. PB control samples were obtained with informed consent under guidelines issued by the Medical Ethics Committee of the Leiden University Medical Center (Leiden, The Netherlands).
Acknowledgments
We thank Dr. Ramon Arens for critically reading the manuscript and Jolanda de Roo for providing materials. This work was supported by Grant FES0908 from the Dutch government (to the Netherlands Institute for Regenerative Medicine); and a Japanese Society of Hematology/European Hematology Association fellowship (to M.H.B.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1519118112/-/DCSupplemental.
References
- 1.Szabolcs P, Cairo MS. Unrelated umbilical cord blood transplantation and immune reconstitution. Semin Hematol. 2010;47(1):22–36. doi: 10.1053/j.seminhematol.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Talvensaari K, et al. A broad T-cell repertoire diversity and an efficient thymic function indicate a favorable long-term immune reconstitution after cord blood stem cell transplantation. Blood. 2002;99(4):1458–1464. doi: 10.1182/blood.v99.4.1458. [DOI] [PubMed] [Google Scholar]
- 3.Admiraal R, et al. Association between anti-thymocyte globulin exposure and CD4+ immune reconstitution in paediatric haemopoietic cell transplantation: A multicentre, retrospective pharmacodynamic cohort analysis. Lancet Haematol. 2015;2(5):e194–e203. doi: 10.1016/S2352-3026(15)00045-9. [DOI] [PubMed] [Google Scholar]
- 4.Wilkinson B, Owen JJ, Jenkinson EJ. Factors regulating stem cell recruitment to the fetal thymus. J Immunol. 1999;162(7):3873–3881. [PubMed] [Google Scholar]
- 5.Champion S, Imhof BA, Savagner P, Thiery JP. The embryonic thymus produces chemotactic peptides involved in the homing of hemopoietic precursors. Cell. 1986;44(5):781–790. doi: 10.1016/0092-8674(86)90844-5. [DOI] [PubMed] [Google Scholar]
- 6.Hao QL, et al. Identification of a novel, human multilymphoid progenitor in cord blood. Blood. 2001;97(12):3683–3690. doi: 10.1182/blood.v97.12.3683. [DOI] [PubMed] [Google Scholar]
- 7.Haddad R, et al. Molecular characterization of early human T/NK and B-lymphoid progenitor cells in umbilical cord blood. Blood. 2004;104(13):3918–3926. doi: 10.1182/blood-2004-05-1845. [DOI] [PubMed] [Google Scholar]
- 8.Kohn LA, et al. Lymphoid priming in human bone marrow begins before expression of CD10 with upregulation of L-selectin. Nat Immunol. 2012;13(10):963–971. doi: 10.1038/ni.2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weerkamp F, et al. Human thymus contains multipotent progenitors with T/B lymphoid, myeloid, and erythroid lineage potential. Blood. 2006;107(8):3131–3137. doi: 10.1182/blood-2005-08-3412. [DOI] [PubMed] [Google Scholar]
- 10.Wiekmeijer A-S, et al. Sustained engraftment of cryopreserved human bone marrow CD34(+) cells in young adult NSG mice. Biores Open Access. 2014;3(3):110–116. doi: 10.1089/biores.2014.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shultz LD, et al. Human lymphoid and myeloid cell development in NOD/LtSz-scid IL2R gamma null mice engrafted with mobilized human hemopoietic stem cells. J Immunol. 2005;174(10):6477–6489. doi: 10.4049/jimmunol.174.10.6477. [DOI] [PubMed] [Google Scholar]
- 12.Eapen M, et al. Children’s Oncology Group; Center for International Blood and Marrow Transplant Research Outcomes after HLA-matched sibling transplantation or chemotherapy in children with acute lymphoblastic leukemia in a second remission after an isolated central nervous system relapse: A collaborative study of the Children’s Oncology Group and the Center for International Blood and Marrow Transplant Research. Leukemia. 2008;22(2):281–286. doi: 10.1038/sj.leu.2405037. [DOI] [PubMed] [Google Scholar]
- 13.Gerrits A, et al. Cellular barcoding tool for clonal analysis in the hematopoietic system. Blood. 2010;115(13):2610–2618. doi: 10.1182/blood-2009-06-229757. [DOI] [PubMed] [Google Scholar]
- 14.Verovskaya E, et al. Asymmetry in skeletal distribution of mouse hematopoietic stem cell clones and their equilibration by mobilizing cytokines. J Exp Med. 2014;211(3):487–497. doi: 10.1084/jem.20131804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Notta F, et al. Isolation of single human hematopoietic stem cells capable of long-term multilineage engraftment. Science. 2011;333(6039):218–221. doi: 10.1126/science.1201219. [DOI] [PubMed] [Google Scholar]
- 16.Benz C, et al. Hematopoietic stem cell subtypes expand differentially during development and display distinct lymphopoietic programs. Cell Stem Cell. 2012;10(3):273–283. doi: 10.1016/j.stem.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 17.Cheung AMS, et al. Analysis of the clonal growth and differentiation dynamics of primitive barcoded human cord blood cells in NSG mice. Blood. 2013;122(18):3129–3137. doi: 10.1182/blood-2013-06-508432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dik WA, et al. New insights on human T cell development by quantitative T cell receptor gene rearrangement studies and gene expression profiling. J Exp Med. 2005;201(11):1715–1723. doi: 10.1084/jem.20042524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hill MO. Diversity and evenness: A unifying notation and its consequences. Ecology. 1973;54(2):427–432. [Google Scholar]
- 20.Schmidt M, et al. Polyclonal long-term repopulating stem cell clones in a primate model. Blood. 2002;100(8):2737–2743. doi: 10.1182/blood-2002-02-0407. [DOI] [PubMed] [Google Scholar]
- 21.Kuramoto K, et al. The impact of low-dose busulfan on clonal dynamics in nonhuman primates. Blood. 2004;104(5):1273–1280. doi: 10.1182/blood-2003-08-2935. [DOI] [PubMed] [Google Scholar]
- 22.Roeder I, et al. Characterization and quantification of clonal heterogeneity among hematopoietic stem cells: A model-based approach. Blood. 2008;112(13):4874–4883. doi: 10.1182/blood-2008-05-155374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu R, Neff NF, Quake SR, Weissman IL. Tracking single hematopoietic stem cells in vivo using high-throughput sequencing in conjunction with viral genetic barcoding. Nat Biotechnol. 2011;29(10):928–933. doi: 10.1038/nbt.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grosselin J, et al. Arrayed lentiviral barcoding for quantification analysis of hematopoietic dynamics. Stem Cells. 2013;31(10):2162–2171. doi: 10.1002/stem.1383. [DOI] [PubMed] [Google Scholar]
- 25.Morrissey PJ, et al. Administration of IL-7 to mice with cyclophosphamide-induced lymphopenia accelerates lymphocyte repopulation. J Immunol. 1991;146(5):1547–1552. [PubMed] [Google Scholar]
- 26.Min D, et al. Sustained thymopoiesis and improvement in functional immunity induced by exogenous KGF administration in murine models of aging. Blood. 2007;109(6):2529–2537. doi: 10.1182/blood-2006-08-043794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wils E-J, et al. Keratinocyte growth factor and stem cell factor to improve thymopoiesis after autologous CD34+ cell transplantation in rhesus macaques. Biol Blood Marrow Transplant. 2012;18(1):55–65. doi: 10.1016/j.bbmt.2011.09.010. [DOI] [PubMed] [Google Scholar]
- 28.Fry TJ, et al. Flt3 ligand enhances thymic-dependent and thymic-independent immune reconstitution. Blood. 2004;104(9):2794–2800. doi: 10.1182/blood-2003-11-3789. [DOI] [PubMed] [Google Scholar]
- 29.Welniak LA, Sun R, Murphy WJ. The role of growth hormone in T-cell development and reconstitution. J Leukoc Biol. 2002;71(3):381–387. [PubMed] [Google Scholar]
- 30.van der Weerd K, et al. Thyrotropin acts as a T-cell developmental factor in mice and humans. Thyroid. 2014;24(6):1051–1061. doi: 10.1089/thy.2013.0396. [DOI] [PubMed] [Google Scholar]
- 31.Velardi E, et al. Sex steroid blockade enhances thymopoiesis by modulating Notch signaling. J Exp Med. 2014;211(12):2341–2349. doi: 10.1084/jem.20131289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zakrzewski JL, et al. Adoptive transfer of T-cell precursors enhances T-cell reconstitution after allogeneic hematopoietic stem cell transplantation. Nat Med. 2006;12(9):1039–1047. doi: 10.1038/nm1463. [DOI] [PubMed] [Google Scholar]
- 33.van Rood JJ, Roelen DL, Claas FHJ. The effect of noninherited maternal antigens in allogeneic transplantation. Semin Hematol. 2005;42(2):104–111. doi: 10.1053/j.seminhematol.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 34.van Rood JJ, et al. Effect of tolerance to noninherited maternal antigens on the occurrence of graft-versus-host disease after bone marrow transplantation from a parent or an HLA-haploidentical sibling. Blood. 2002;99(5):1572–1577. doi: 10.1182/blood.v99.5.1572. [DOI] [PubMed] [Google Scholar]
- 35.van Rood JJ, et al. Reexposure of cord blood to noninherited maternal HLA antigens improves transplant outcome in hematological malignancies. Proc Natl Acad Sci USA. 2009;106(47):19952–19957. doi: 10.1073/pnas.0910310106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ishikawa F, et al. Development of functional human blood and immune systems in NOD/SCID/IL2 receptor gamma chain(null) mice. Blood. 2005;106(5):1565–1573. doi: 10.1182/blood-2005-02-0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weerkamp F, et al. Age-related changes in the cellular composition of the thymus in children. J Allergy Clin Immunol. 2005;115(4):834–840. doi: 10.1016/j.jaci.2004.10.031. [DOI] [PubMed] [Google Scholar]
- 38.Halkias J, et al. Opposing chemokine gradients control human thymocyte migration in situ. J Clin Invest. 2013;123(5):2131–2142. doi: 10.1172/JCI67175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shultz LD, et al. Generation of functional human T-cell subsets with HLA-restricted immune responses in HLA class I expressing NOD/SCID/IL2r gamma(null) humanized mice. Proc Natl Acad Sci USA. 2010;107(29):13022–13027. doi: 10.1073/pnas.1000475107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Halkias J, et al. Conserved and divergent aspects of human T-cell development and migration in humanized mice. Immunol Cell Biol. 2015;93(8):716–726. doi: 10.1038/icb.2015.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee YJ, et al. Generation of PLZF+ CD4+ T cells via MHC class II-dependent thymocyte-thymocyte interaction is a physiological process in humans. J Exp Med. 2010;207(1):237–246. doi: 10.1084/jem.20091519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marodon G, et al. High diversity of the immune repertoire in humanized NOD.SCID.gamma c−/− mice. Eur J Immunol. 2009;39(8):2136–2145. doi: 10.1002/eji.200939480. [DOI] [PubMed] [Google Scholar]
- 43.Martins VC, et al. Cell competition is a tumour suppressor mechanism in the thymus. Nature. 2014;509(7501):465–470. doi: 10.1038/nature13317. [DOI] [PubMed] [Google Scholar]
- 44.Fischer A, Hacein-Bey-Abina S, Cavazzana-Calvo M. 20 years of gene therapy for SCID. Nat Immunol. 2010;11(6):457–460. doi: 10.1038/ni0610-457. [DOI] [PubMed] [Google Scholar]
- 45.Tuckett AZ, et al. Image-guided intrathymic injection of multipotent stem cells supports lifelong T-cell immunity and facilitates targeted immunotherapy. Blood. 2014;123(18):2797–2805. doi: 10.1182/blood-2013-10-535401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Csárdi G, Nepusz T. The igraph software package for complex network research. InterJournal Complex Systems. 2006;1695 . Available at igraph.org. [Google Scholar]
- 47.van Dongen JJM, et al. Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T-cell receptor gene recombinations in suspect lymphoproliferations: report of the BIOMED-2 concerted action BMH4-CT98-3936. Leukemia. 2003;17(12):2257–2317. doi: 10.1038/sj.leu.2403202. [DOI] [PubMed] [Google Scholar]