Significance
MerR-family regulators act on suboptimal promoters to control the transcriptions of genes that help bacteria defend against a diverse set of metals and drugs. How they modulate RNA polymerase (RNAP) activity to control transcription initiation remains unclear, however. Here we show that CueR—a Cu+-responsive MerR-family metalloregulator—biases the kinetic sampling of RNAP binding events that lead to two noninterconverting states: a dead-end complex to repress or an open complex to activate transcription, constituting a branched pathway distinct from the linear pathway prevalent for transcription initiation at optimal promoters. This mechanistic insight contributes new fundamental knowledge to bacterial transcription regulation, and may help develop antibiotics that target this regulation mechanism to compromise bacterial defenses.
Keywords: single-molecule FRET, protein–DNA interaction dynamics, MerR-family regulators, metal-responsive transcription regulation
Abstract
Metalloregulators respond to metal ions to regulate transcription of metal homeostasis genes. MerR-family metalloregulators act on σ70-dependent suboptimal promoters and operate via a unique DNA distortion mechanism in which both the apo and holo forms of the regulators bind tightly to their operator sequence, distorting DNA structure and leading to transcription repression or activation, respectively. It remains unclear how these metalloregulator−DNA interactions are coupled dynamically to RNA polymerase (RNAP) interactions with DNA for transcription regulation. Using single-molecule FRET, we study how the copper efflux regulator (CueR)—a Cu+-responsive MerR-family metalloregulator—modulates RNAP interactions with CueR’s cognate suboptimal promoter PcopA, and how RNAP affects CueR−PcopA interactions. We find that RNAP can form two noninterconverting complexes at PcopA in the absence of nucleotides: a dead-end complex and an open complex, constituting a branched interaction pathway that is distinct from the linear pathway prevalent for transcription initiation at optimal promoters. Capitalizing on this branched pathway, CueR operates via a “biased sampling” instead of “dynamic equilibrium shifting” mechanism in regulating transcription initiation; it modulates RNAP’s binding–unbinding kinetics, without allowing interconversions between the dead-end and open complexes. Instead, the apo-repressor form reinforces the dominance of the dead-end complex to repress transcription, and the holo-activator form shifts the interactions toward the open complex to activate transcription. RNAP, in turn, locks CueR binding at PcopA into its specific binding mode, likely helping amplify the differences between apo- and holo-CueR in imposing DNA structural changes. Therefore, RNAP and CueR work synergistically in regulating transcription.
Maintaining cellular metal homeostasis is essential for bacteria, which often dwell in environments with high concentrations of metals. Some of these metals are purely toxic to bacteria, such as Cd and Hg. Many others are required for cellular function, such as Zn and Cu, but can be toxic in excess. Cells have thus developed many ways to regulate intracellular metal concentrations (1–9). Metal-responsive transcriptional regulation is one of them, where metalloregulators respond to intracellular metal ions and regulate transcription of metal efflux, uptake, or other metal homeostasis genes (4–9).
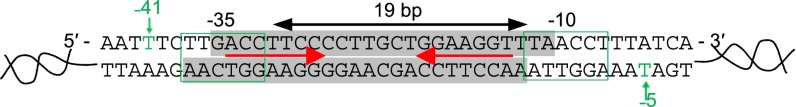
In Gram-negative bacteria, MerR-family metalloregulators act on σ70-dependent suboptimal promoters to repress or activate transcription of metal resistance genes (7, 8). These suboptimal promoters have elongated spacing, 19–20 bp (Fig. 1), compared with the optimal 17 ± 1 bp, between the −35 and −10 elements. This elongated spacing causes a misalignment of these two recognition elements, impairing proper interactions with the RNA polymerase (RNAP) and leading to a weak basal level of transcription. Functioning as stable homodimers, MerR-family metalloregulators bind tightly to specific dyad-symmetric sequences within the spacer region in both unmetallated apo and metallated holo forms. The apoprotein acts as a repressor; its binding to DNA bends and distorts the promoter region, further impairing the ability of RNAP to productively engage with the −35 and −10 elements as required for open complex formation. The holoprotein has a different conformation, and its binding distorts the DNA by bending and unwinding the binding site. This distortion shortens the distance between the −35 and −10 elements and better aligns them for RNAP binding, thereby facilitating open complex formation and activating transcription. This DNA distortion mechanism for repressing and activating transcription (7, 10–14), so far unique to MerR-family metalloregulators, is confirmed by the protein crystal structures in complex with DNA (14–17), which show the bending and unwinding of the DNA in the repressed and activated states.
Fig. 1.
The σ70-dependent suboptimal promoter of the E. coli copA gene regulated by the MerR-family metalloregulator CueR. Red arrows, the dyad-symmetric sequence recognized by CueR; gray shades, CueR footprint (18). The RNAP footprint spans from approximately −55 to +29 (10) (SI Appendix, Fig. S1 A and B). Our DNA construct PcopACy3@−5 spans from position −69 to +41; PcopACy3@−41 spans from position −69 to +61.
The DNA distortion model has provided much insight into how metalloregulator-imposed DNA structural changes may affect RNAP−DNA interactions, especially for transcription activation. Little is known, however, on how metalloregulator−DNA interactions are coupled dynamically to RNAP−DNA interactions for transcription initiation. For example, how do the holo-metalloregulators facilitate the formation of the RNAP–promoter open complex, so as to activate transcription? How do the apo-metalloregulators repress transcription initiation? Does RNAP affect metalloregulator−DNA interactions and, if so, how? Answering these questions will help elucidate the unique regulation mechanism of MerR-family regulators and contribute fundamental knowledge to our understanding of transcription initiation, the most regulated phase of transcription (19).
Single-molecule FRET (smFRET) has proven to be powerful for dissecting the mechanisms of transcription initiation and elongation by RNAP at optimal promoters (20, 21). Here we report a smFRET study of how CueR—a Cu+-responsive MerR-family metalloregulator—modulates RNAP interactions with CueR’s cognate suboptimal promoter and how RNAP in turn affects CueR-promoter interactions for transcription regulation. In Escherichia coli, CueR regulates the transcription of copper-resistance genes, including CopA, an inner membrane copper ATPase, and CueO, a periplasmic multicopper oxidase (18, 22), which both help to remove copper from the cell. We find that even in the absence of nucleotides, RNAP can form two complexes at the suboptimal promoter of the copA gene: a dead-end closed-like complex and an open complex, constituting a branched mechanistic pathway for transcription initiation, unlike the linear pathway prevalent for RNAP interactions with optimal promoters. Capitalizing on this branched pathway, CueR biases RNAP’s binding–unbinding kinetics in forming these two complexes: in its apo-repressor form, CueR reinforces the dominance of the dead-end complex to repress transcription; in its holo-activator form, it shifts the interactions toward the open complex to activate transcription. In either case, no interconversions between the dead-end and open complexes are observed. RNAP in turn locks CueR binding at the promoter into the specific binding mode, likely helping amplify the differences between apo- and holo-CueR in imposing DNA structural changes. Therefore, RNAP and CueR work synergistically at the promoter in regulating transcription.
Results and Analysis
RNAP Forms Two, Noninterconverting, Complexes with the Suboptimal Promoter PcopA.
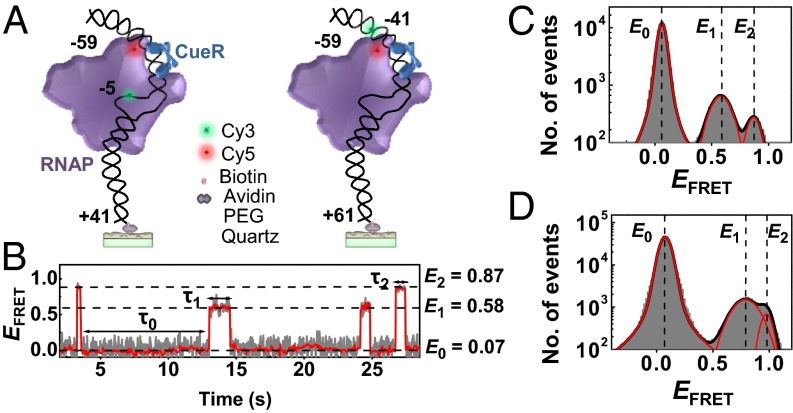
We used smFRET to study RNAP interactions with a surface-immobilized 100- or 120-bp double-strand DNA that spans the entire footprint of RNAP at the suboptimal promoter of the copA gene, containing the −10 and −35 elements as well as the dyad-symmetric sequence recognized by CueR (Fig. 2A, Left; SI Appendix, Fig. S1 A and B). We refer to this DNA as PcopA. We labeled E. coli RNAP holoenzyme with the FRET acceptor Cy5 at R596C of the σ70 factor (i.e., RNAPCy5), which was shown to not interfere with RNAP function (23, 24). We labeled PcopA with the FRET donor Cy3 at its −5 T base on the template strand (i.e., PcopACy3@−5; Fig. 1). Based on an RNAP−DNA complex structure (SI Appendix, Fig. S1C) (25), this label position is ∼6 nm from the acceptor on RNAPCy5; it is also within the transcription bubble region, so the FRET should be sensitive to RNAP–promoter open complex formation. In vitro transcription assay further showed that both RNAPCy5 and PcopACy3@−5 are functional (SI Appendix, Fig. S1 E and F).
Fig. 2.
SmFRET of RNAPCy5−PcopACy3 interactions. (A) Scheme for surface immobilization and labeling of PcopACy3@−5 (Left) or PcopACy3@−41 (Right) for interacting with σ70-containing RNAPCy5 holoenzyme (and CueR) in solution. Scheme generated based on a structure model (SI Appendix, Fig. S1 C and D). (B) Single-molecule EFRET trajectory of an immobilized PcopACy3@−5 interacting with RNAPCy5 (2 nM). The dwell times on the E0, E1, and E2 states are τ0, τ1, and τ2, respectively. Gray line, original data; red line, after nonlinear filtering. (C) Histogram of EFRET trajectories of RNAPCy5−PcopACy3@−5 interactions at 2 nM RNAPCy5. Red lines are resolved peaks centered at EFRET ∼0.07, ∼0.58, and ∼0.87, with percentage areas of 92.1 ± 2.4, 6.2 ± 0.6, and 1.7 ± 0.5%, respectively. (D) Same as C, but with PcopACy3@−41. Three peaks centered at EFRET ∼0.07, ∼0.78, and ∼0.95, with percentage areas of 92.4 ± 1.9, 5.9 ± 1.4, and 1.7 ± 0.2%, respectively.
Analysis of the FRET efficiency (EFRET) vs. time trajectory for a single immobilized PcopACy3@−5 interacting with 2 nM RNAPCy5 in solution reveals three distinct EFRET states at E0 ∼0.07, E1 ∼0.58, and E2 ∼0.87 (Fig. 2 B and C). The E0 state corresponds to free PcopACy3@−5 DNA, as verified by control experiments in the absence of RNAPCy5. The observation of two additional states at higher EFRET indicates that RNAP can form two distinct complexes with PcopA in the absence of nucleotides. These two complexes differ in stability, reflected by the ∼3.6:1 ratio of their peak areas in the EFRET histogram with the E1 complex being the major complex (Fig. 2C).
To confirm that the E1 and E2 states are not just from different label orientations in its microenvironment, we moved the Cy3 label on PcopA to −41 (i.e., PcopACy3@−41; Fig. 1 and Fig. 2A, Right), which is also a functional transcription template (SI Appendix, Fig. S1F). Again, besides the free PcopA state at E0 ∼0.07, two distinct higher EFRET states were observed at ∼0.78 and ∼0.95 (Fig. 2D). The relative stabilities of the two states are ∼3.5:1, within error to that when using PcopACy3@−5, corroborating that these two states indeed reflect two RNAP−PcopA complexes with different structural configurations. Moreover, with increasing RNAPCy5 concentration, their populations expectedly increase relative to the free PcopA (SI Appendix, Fig. S2B). The extracted dissociation constant for the major RNAP−PcopA complex E1 is K1D ∼30 ± 3 nM, and that for the minor complex E2 is K2D ∼105 ± 26 nM (SI Appendix, Table S1).
As controls, we studied a Cy3-labeled DNA that does not contain the promoter sequence. Expectedly, only nonspecific complexes with RNAPCy5 were observed; they exhibit a broad distribution of EFRET and have an overall dissociation constant of 1.1 ± 0.4 μM (SI Appendix, Fig. S3), consistent with reported RNAP interactions with nonspecific DNA (KD ∼10−3 to 10−6 M, depending on the salt concentrations) (26). Moreover, with Cy5-labeled σ70-factor and without the RNAP core enzyme, no complex was observed with PcopA, consistent with σ70 not binding to DNA alone (27).
In the EFRET vs. time trajectories (Fig. 2B), frequent E0 ↔ E1 and E0 ↔ E2 state transitions were observed, indicating that RNAP can bind and unbind reversibly to PcopA to form either of the two complexes. Surprisingly, almost no direct E1 ↔ E2 transitions were observed (i.e., merely one over ∼100 min during which 1,481 E0 ↔ E1 and E0 ↔ E2 transitions occurred), indicating that the two RNAP–PcopA complexes, once formed, essentially do not interconvert (rate is ∼10−4 s−1).
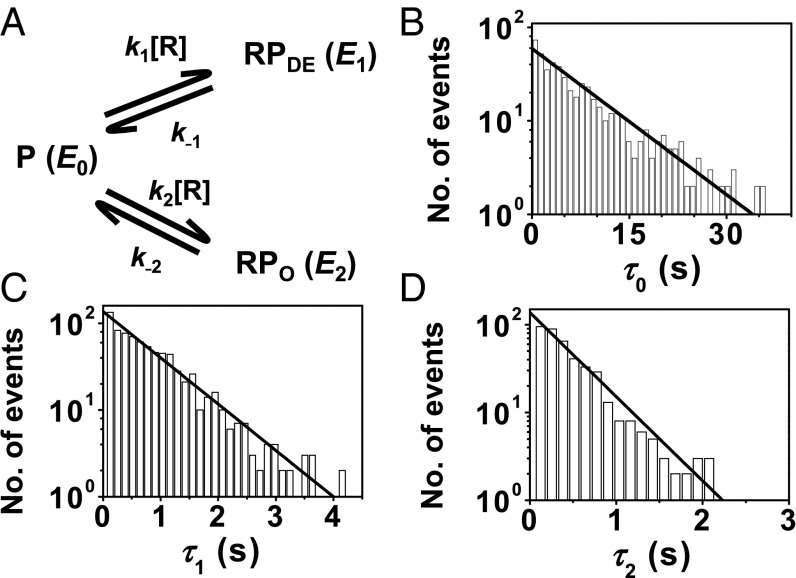
The distributions of the microscopic dwell time on the three states τ0, τ1, and τ2 all follow single-exponential decays, indicating that the reversible binding–unbinding of RNAP to PcopA all follow single-step kinetics, and each of the three states contains just one dominant species (Fig. 3 B–D). Analyzing these distributions gives the rate constants of the associated kinetic steps in a minimal three-state kinetic mechanism (Fig. 3A), with the binding rate constants on the order of 106 M−1⋅s−1, and unbinding rate constants on the order of 100 s−1 (Table 1). The higher stability of the major E1 complex results from both its larger binding rate constant (k1) and its smaller unbinding rate constant (k−1), compared with those (i.e., k2 and k−2) of the minor E2 complex.
Fig. 3.
RNAP−PcopA interaction kinetics and mechanism. (A) Minimal kinetic mechanism. RNAP (R) binds reversibly to PcopA (P, E0 state) to form the dead-end (RPDE, E1 state) and open (RPO, E2 state) complexes that do not interconvert. k’s, kinetic constants; [R], RNAP concentration. (B) Distribution of τ0 for RNAPCy5−PcopACy3@−5 interactions at 2 nM [RNAPCy5]. Solid line is a fit with Nγexp(−γτ); γ = (k1 + k2)[R]; N is a scaling factor. Fitted parameters are in Table 1. k1/k2 = (ratio of observed E0 → E1 and E0 → E2 transitions). (C) Same as B but for τ1. Solid line is a fit with Nk−1exp(−k−1τ). (D) Same as B but for τ2. Solid line is a fit with Nk−2exp(−k−2τ).
Table 1.
Kinetic and thermodynamic parameters for RNAP−PcopA interactions in the absence or presence of 200 nM apo-CueR or holo-CueR
| Processes | Kinetic parameters | RNAP | RNAP + holo-CueR | RNAP + apo-CueR |
| Dead-end complex (RPDE, E1) | ||||
| Binding | k1 (×106 M−1⋅s−1) | 34.9 ± 4.7 | 29.3 ± 3.1 | 42.4 ± 6.1 |
| Unbinding | k−1, s−1 | 1.23 ± 0.03 | 2.03 ± 0.15 | 1.04 ± 0.04 |
| Dissociation constant | K1D, nM | 35 ± 5 | 69 ± 9 | 24 ± 4 |
| Open complex (RPO, E2) | ||||
| Binding | k2 (×106 M−1⋅s−1) | 17.5 ± 2.3 | 44.9 ± 4.8 | 23.2 ± 3.4 |
| Unbinding | k−2, s−1 | 2.21 ± 0.06 | 1.80 ± 0.05 | 2.47 ± 0.12 |
| Dissociation constant | K2D, nM | 126 ± 17 | 40 ± 4 | 106 ± 16 |
k’s and K’s in the presence holo- or apo-CueR are apparent rate constants and dissociation constants of the respective RNAP–PcopA complexes.
Holo-CueR, the Activator, Shifts RNAP−PcopA Interactions Toward the Original Minor Complex.
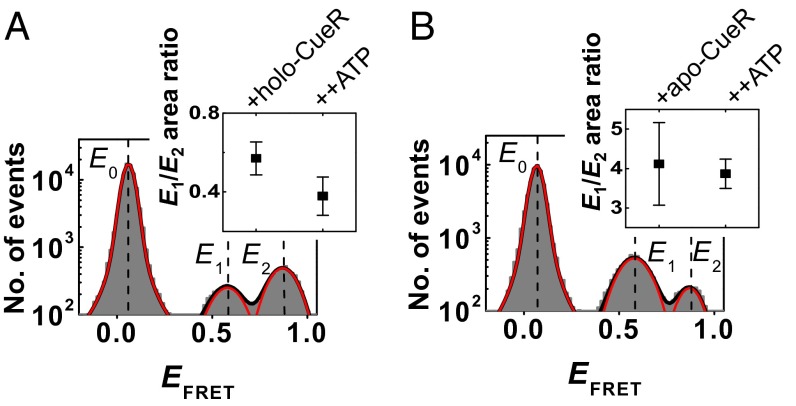
We next examined how holo-CueR, the transcription activator, would affect RNAPCy5−PcopACy3@−5 interactions. In the presence of 200 nM holo-CueR, a concentration significantly higher than the KD (∼120 nM) for CueR binding to PcopA (28), the same three EFRET states are present at E0 ∼0.07, E1 ∼0.58, and E2 ∼0.87 (Fig. 4A), corresponding to the free PcopA and the two RNAP−PcopA complexes. However, the relative populations of the two complexes have shifted significantly: the E1:E2 state population ratio is now ∼0.57:1 (Fig. 4A), compared with ∼3.6:1 in the absence of holo-CueR (Fig. 2C). And this shift comes more from the stabilization of the E2 complex and less from the destabilization of the E1 complex, as reflected by their populations relative to the free PcopA and their apparent dissociation constants (Table 1). Therefore, the original minor RNAPCy5–PcopACy3@−5 complex at E2 ∼0.87 becomes the dominant complex upon interaction with the activator holo-CueR. The same shift toward the minor complex induced by holo-CueR is also observed for the alternatively labeled RNAPCy5-PcopACy3@−41 interactions, and this shift is proportional to the concentration of holo-CueR (SI Appendix, Fig. S4).
Fig. 4.
RNAPCy5−PcopACy3@−5 interactions in the presence of CueR. (A) EFRET histogram with 2 nM [RNAPCy5] in the presence of 200 nM unlabeled holo-CueR. Red lines are resolved peaks at EFRET∼0.07, ∼0.58, and ∼0.87, with percentage areas of 92.0 ± 2.8, 2.9 ± 0.3, and 5.1 ± 0.6%, respectively. (Inset) The corresponding E1/E2 area ratio and that with further adding 1 mM ATP. (B) Same as A, but in the presence of 200 nM unlabeled apo-CueR. The three peaks at EFRET ∼0.07, ∼0.58, and ∼0.87 have percentage areas of 90.8 ± 4.6, 7.4 ± 0.7, and 1.8 ± 0.8%, respectively. (Inset) The corresponding E1/E2 area ratio and that with further adding 1 mM ATP.
In addition to holo-CueR, we further added 1 mM ATP, the initiating nucleotide, which was shown to stabilize open complex formation at millimolar concentrations (29, 30). For both RNAPCy5−PcopACy3@−5 and RNAPCy5−PcopACy3@−41 interactions, a further (although slight) shift toward the E2 complex is clear: the E1:E2 complex population ratio drops to ∼0.4:1 (Fig. 4A, Inset). Further addition of the next nucleotide, GTP, did not cause any discernible changes (SI Appendix, Fig. S5C). As a control, we added ATP in the absence of holo-CueR; no discernible change was observed in the RNAP−PcopA interactions (SI Appendix, Fig. S6).
Apo-CueR, the Repressor, Reinforces the Original Major RNAP−PcopA Complex.
In the presence of 200 nM apo-CueR, the transcription repressor, the same three EFRET states are present in RNAPCy5−PcopACy3@−5 interactions: the free PcopA (E0) and the two complexes (E1 and E2; Fig. 4B). The dominance of the E1 complex is slightly reinforced (E1:E2 population ratio is ∼4.1:1 compared with the original ∼3.6:1), in contrast with the ability of holo-CueR to flip their relative stabilities. The same reinforcement by apo-CueR is observed for the alternatively labeled RNAPCy5−PcopACy3@−41 interactions (SI Appendix, Fig. S7B). This reinforcement results from the further stabilization of the E1 complex, whereas the stability of the E2 complex is only increased slightly, as reflected by their populations relative to the free PcopA (SI Appendix, Fig. S7 D and E). Adding the initiating nucleotide ATP did not cause any further changes (Fig. 4B, Inset).
Assignments of Two RNAP–PcopA Complexes: CueR Shifts RNAP–PcopA Interactions Between Open and Dead-End Complexes.
We can assign the major RNAP–PcopA complex (E1) in the absence of CueR as a dead-end closed-like complex (i.e., RPDE) and the minor complex (E2) as an open complex (i.e., RPO). The major complex should be a dead end because it cannot convert to the open complex. As such, holo-CueR activates transcription by shifting the RNAP–PcopA interactions toward the open complex, whereas apo-CueR represses transcription by reinforcing this dead-end closed-like complex. These assignments are based on the following rationales. (i) PcopA is a suboptimal, weakly repressed promoter. Therefore, in the absence of a regulator, the formed complexes should be dominated by an inactive closed-like complex, but should still contain some fraction that is capable of progressing to the open complex, allowing for leakage transcription detectable by in vitro transcription assay (SI Appendix, Fig. S1E) (13, 28). (ii) Holo-CueR, the activator, is expected to enhance those active complexes that can successfully transition to productive open complexes during transcription initiation. Consistently, holo-CueR shifts the RNAP–PcopA interactions toward the original minor complex (which is on the active transcription initiation pathway), rendering it dominant (Fig. 4A). (iii) Apo-CueR, the repressor, is expected to secure the inactive closed-like complex for transcription repression. Consistently, apo-CueR reinforces the original major RNAP-PcopA complex (Fig. 4B). Previous studies of MerR-family metalloregulators also showed the apo-repressor–DNA complex to be dominated by the closed complex (11, 13). (iv) ATP, the initiating nucleotide, is expected to stabilize the open complex while not affecting much the closed-like complex. Consistently, ATP further shifts RNAP–PcopA interactions toward the original minor complex in the presence of the activator holo-CueR (Fig. 4A, Inset) while causing no discernible changes in the presence of the repressor apo-CueR (Fig. 4B, Inset).
Several evidences further support that the DNA strands are likely to be separated in the open complex E2. (i) Past DNA footprinting showed that in the presence of holo-activator, the transcription bubble is clearly formed at these suboptimal promoters (10, 13). (ii) For PcopACy3@−5, the FRET donor Cy3 is located on the template strand within the region for forming the transcription bubble. The structure of an open RNAP–DNA complex shows that this part of template strand is closer to the Cy5 labeling position on RNAP than the nontemplate strand after bubble formation (SI Appendix, Fig. S1C) (25, 31), suggesting that bubble formation would bring Cy3 closer to the Cy5; this would lead to a higher EFRET value for the open complex than that of the inactive closed-like complex, which is consistent with our assignment—for RNAPCy3−PcopACy3@−5 interactions, the open complex E2 ∼0.87, higher than the dead-end complex E1 ∼0.58 (Fig. 2C). (iii) The initiating nucleotide ATP, besides further stabilizing the open complex, does not change the EFRET value of the open complex (SI Appendix, Fig. S5A), supporting that the bubble is already present and no further strand separation is needed for ATP binding.
It is worth noting that for RNAPCy5–PcopACy3@−41 interactions, the open complex (E2 ∼0.95) has a higher EFRET value than the dead-end complex (E1 ∼0.78), indicating that the formation of the open complex moves the upstream DNA segment (i.e., around position −41) closer toward RNAP. Therefore, the DNA structural changes associated with open complex formation at PcopA involve not only those at the transcription bubble but also those at upstream locations, which may be related to the upstream DNA wrapping around RNAP (13, 25, 32, 33).
CueR Changes RNAP Binding–Unbinding Kinetics in Forming Open or Dead-End Complex Without Allowing for Interconversion.
To determine the kinetic basis of the population shifts between the open and dead-end RNAP-PcopA complexes caused by holo- and apo-CueR, we extracted kinetic rate constants from the distributions of the microscopic dwell times τ0, τ1, and τ2 using the minimal model in Fig. 3A (Table 1). Regarding the dead-end complex, holo-CueR does not change RNAP’s binding rate constant (k1) significantly, but increases its unbinding (k−1) by ∼65%, resulting in its weakening. Regarding the open complex, holo-CueR significantly increases RNAP’s binding rate constant (k2) by ∼157%, while slightly decreasing its unbinding (k−2) by ∼19%, leading to its strengthening. Overall, holo-CueR modulates RNAP−PcopA interactions by accelerating RNAP’s unbinding from the dead-end complex and more significantly by increasing RNAP’s binding to form the open complex, leading to transcription activation.
In contrast, apo-CueR slightly increases the binding rate constant (k1) of the dead-end complex by ∼21% and slightly decreases the unbinding (k−1) by ∼15%, leading to a strengthening of the dead-end complex. For the open complex, apo-CueR increases slightly both its binding (k2) and its unbinding (k−2) rate constants by ∼33% and ∼12%, respectively, resulting in its slight strengthening. Overall, apo-CueR modulates RNAP–PcopA binding–unbinding kinetics, leading to a net reinforcement of the dead-end complex and thus transcription repression.
Regardless of whether holo- or apo-CueR was present, essentially no interconversion between the dead-end and open complexes of RNAP-PcopA was observed (i.e., two interconversions in ∼240 min during which 5,180 binding–unbinding events occurred, corresponding to a rate of ∼10−5 s−1). Therefore, apo-CueR and holo-CueR bias RNAP’s kinetic sampling of the dead-end or open complexes, giving rise to transcription repression or activation.
RNAP Locks CueR−PcopA Interactions into the Specific Binding Mode.
To probe how RNAP would affect CueR−DNA interactions, we used smFRET to examine CueRCy5−PcopACy3@−41 in the presence of unlabeled RNAP. We have previously shown that singly labeled CueR (at C129; SI Appendix, section 1b) is functional, and we characterized CueR interactions with the PcopA operator site (28). We found that CueR, in both its apo and holo forms, can bind to the operator site with two different binding modes: one in which CueR recognizes the specific sequence and distorts the DNA structure, and the other in which CueR likely interacts with the DNA in a nonspecific fashion (SI Appendix, Fig. S11). Furthermore, once forming the specific binding mode at the recognition site, both apo- and holo-CueR can undergo an assisted dissociation or direct substitution process, where a CueR molecule from the surrounding solution helps carry away or replaces the incumbent CueR on DNA, respectively.
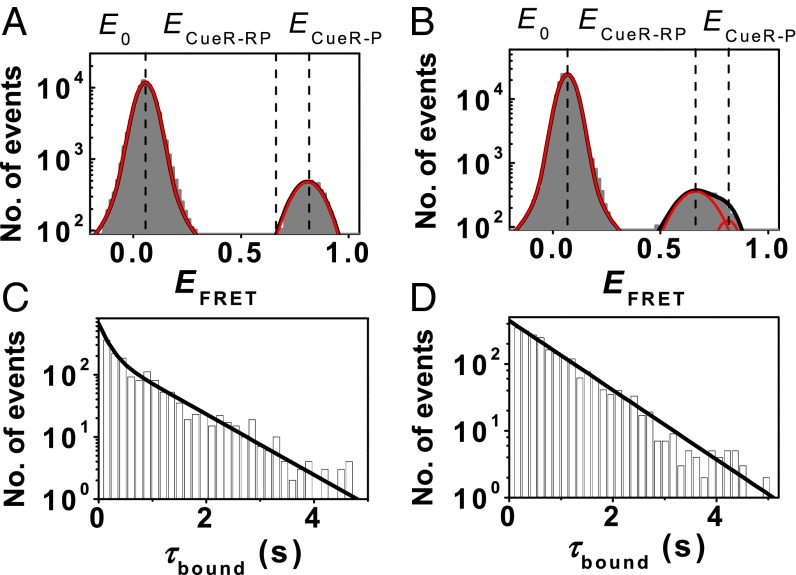
Fig. 5A shows the single-molecule EFRET histogram of holo-CueRCy5−PcopACy3@−41 interactions. The peak at E0 ∼0.07 is the free PcopA state. The peak at ECueR-P ∼0.81 is a holo-CueRCy5–PcopACy3@−41 complex, in which the homodimeric CueR binds in an orientation such that its Cy5 label on one monomer is closer to the Cy3 label on PcopA than in the other orientation, whose EFRET value is small (∼0.18; SI Appendix, Fig. S12A), and the corresponding peak is buried by the large E0 peak. The two different binding orientations of CueRCy5 on PcopACy3@−41 results from that the single Cy5 label on one monomer breaks the symmetry of the homodimeric CueR, as we have shown (28). The presence of the two different CueR binding modes in each orientation cannot be resolved by their EFRET, and instead, is reflected by the distribution of the dwell time τbound on the CueR-bound ECueR-P state: it is a double-exponential distribution (Fig. 5C) (28). The assisted dissociation or direct substitution process of CueR on PcopA is reflected by that 〈τbound〉−1, the apparent single-molecule rate for CueR−PcopA complex dissociation, increases with the CueR concentration in the solution; and these two processes only occur in the specific CueR−PcopA binding mode, as we have shown (28).
Fig. 5.
CueRCy5−PcopACy3@−41 interactions in the absence and presence of RNAP. (A) EFRET histogram of holo-CueRCy5−PcopACy3@−41 interactions at 2 nM [holo-CueRCy5]. Two resolved peaks centered at E0 ∼0.07 and ECueR-P ∼0.81. (B) Same as A, but in the presence of 100 nM unlabeled RNAP. Three resolved peaks centered at E0 ∼0.07, ECueR-RP ∼0.68, and ECueR-P ∼0.81. (C) Distribution of τbound for holo-CueRCy5−PcopACy3@−41 interactions at 2 nM holo-CueRCy5. Solid line is a fit with a sum of two exponentials. (D) Distribution of τbound for holo-CueRCy5−PcopACy3@−41 at 2 nM holo-CueRCy5 and in the presence of 100 nM unlabeled RNAP. Solid line is a single exponential fit.
With the addition of 100 nM RNAP (a concentration comparable to the KD’s for RNAP binding to PcopA; Table 1), a new FRET state, ECueR-RP, appears in the EFRET histogram at ∼0.68, whereas the ECueR-P state of the holo-CueRCy5−PcopACy3@−41 complex diminishes (Fig. 5B). The same behavior was observed for apo-CueRCy5 interactions with PcopACy3@−41 in the presence of RNAP (SI Appendix, Fig. S8). Furthermore, the increase in the ECueR-RP peak area and the decrease in the ECueR-P peak area both scale with increasing RNAP concentration (SI Appendix, Fig. S9). In contrast, core RNAP (i.e., without the σ70 factor), which can only interact with DNA nonspecifically (KD ∼10−4 to 10−6 M, depending on the salt concentrations) (26, 34), did not cause any discernible perturbation (SI Appendix, Fig. S10), suggesting that core RNAP does not bind significantly to the CueR−PcopA complex. Together, these results show that RNAP can directly interact with the CueR−PcopA complex at the promoter region, forming a ternary complex in which the CueR–PcopA interaction structure is perturbed compared with that in the absence of RNAP. The observation of the ternary complex here is consistent with previous studies of RNAP-DNA-MerR interactions (10, 35, 36).
Interestingly, with the addition of RNAP, the distribution of the dwell time τbound on the holo-CueR bound state now follows a single-exponential decay (Fig. 5D), suggesting that one of the two CueR binding modes vanished. However, 〈τbound〉−1 still increases with increasing [holo-CueR] (SI Appendix, Fig. S12B), indicating that the assisted dissociation or direct substitution process still occur to CueR bound on PcopA while interacting with RNAP. Because the assisted dissociation and direct substitution only occur to a CueR−DNA complex in the specific binding mode wherein CueR recognizes the specific sequence and distorts DNA structure, the remaining binding mode of CueR on PcopA must be the specific binding mode. Assuming various rate constants for CueR−DNA interactions and using the previously determined CueR−DNA interaction mechanism (28), we simulated the distributions of τbound. These simulations further support the notion that the distribution of τbound converts from a double-exponential decay into a single-exponential decay because only the specific binding mode remains (SI Appendix, section 9c and Fig. S13). Together, these results show that RNAP locks CueR−PcopA interactions into the specific binding mode at the recognition site.
Discussion
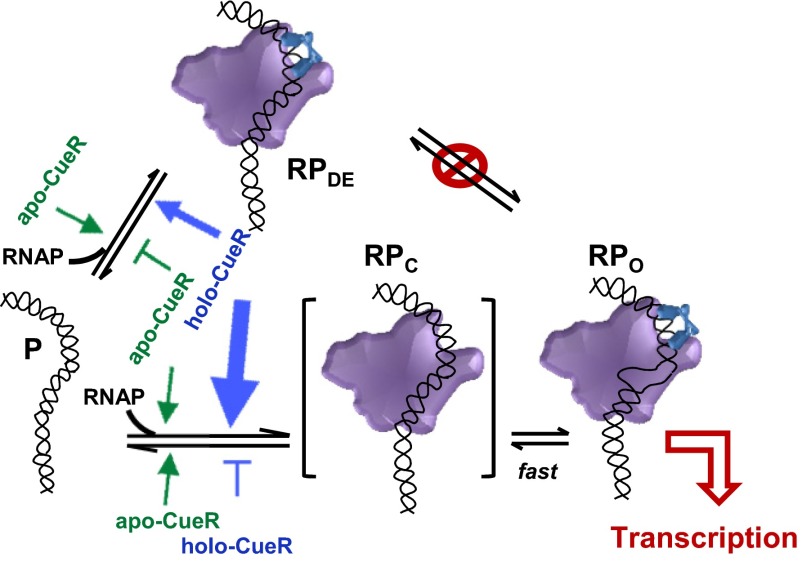
Using smFRET measurements we have quantified the dynamic interactions among E. coli RNAP, the suboptimal promoter PcopA, and the MerR-family metalloregulator CueR. Without CueR, RNAP can bind to PcopA reversibly to form two distinct, noninterconverting RNAP−PcopA complexes (Fig. 6). The major RNAP−PcopA complex can be assigned as a dead-end closed-like complex (RPDE); its dominance can account for the weakly repressed nature of the promoter PcopA. The minor complex can be assigned as an open complex (RPO); its presence can account for the leakage transcription observable at this promoter. And for both complexes, their formation and dissociation kinetics follow effective single-step kinetics without detectable kinetic intermediates within our time resolution (30 ms).
Fig. 6.
The biased sampling mechanism for CueR modulating RNAP–PcopA interactions for transcription regulation. P is the free promoter. RPC is a transient closed complex upon initial RNAP binding to PcopA. RPDE is the dead-end complex. RPO is the open complex. The pointed and flat-end arrows (green and blue), respectively, denote the enhancing and inhibiting kinetic effects of holo-CueR and apo-CueR on RNAP binding–unbinding on PcopA; the widths and lengths of these arrows both represent the strength of enhancement or inhibition effects.
The observed dead-end complex RPDE is a terminally unproductive complex (i.e., off pathway toward active transcription; Fig. 6) because it cannot convert to the open complex. On the pathway toward the open complex (RPO), during which the transcription bubble forms, there should be an earlier closed complex (RPC) upon initial RNAP binding to PcopA (Fig. 6). RPC here should be transient within our time resolution, because there is no detectable kinetic intermediate in forming the open-complex RPO. The formation of the dead-end complex (RPDE) should not go through this RPC, which would otherwise provide an interconversion pathway to the open complex that is inconsistent with our observations.
Holo–CueR, the transcription activator, predominantly accelerates RNAP’s unbinding from RPDE and even more significantly accelerates its binding to form RPO, thus shifting the RNAP−PcopA interactions toward RPO and leading to transcription activation (Fig. 6). In contrast, apo-CueR, the transcription repressor, slightly increases both the binding and unbinding for forming RPO, but more significantly, it increases RNAP’s binding and decreases its unbinding for forming RPDE, overall reinforcing the dominance of RPDE and leading to transcription repression (Fig. 6). Moreover, neither holo- nor apo-CueR enables interconversions between RPDE and RPO. Therefore, CueR regulates transcription via a biased sampling mechanism, in which the RNAP’s binding and unbinding kinetics to the promoter are tuned in forming the dead-end or open complex, depending on CueR’s metallation state, rather than a dynamic equilibrium shift mechanism, in which the interconversion rates between the dead-end and open complexes are altered to shift the equilibrium toward one or the other.
At the molecular level, CueR likely achieves the biasing by distorting the DNA structure at the promoter. The bending and unwinding of DNA imposed by the holo-CueR (10, 11, 13–17) are likely the structural basis for it to bias RNAP’s kinetic sampling toward the open complex that we discovered here. Apo-CueR mainly bends the DNA at the recognition site and the structural changes differ from that imposed by holo-CueR (14); this different structural distortion could be the basis for apo-CueR’s biasing RNAP toward sampling more of the dead-end complex.
From the perspective of CueR−PcopA interactions, we found that RNAP can lock CueR into its specific binding mode at the recognition site, suppressing its nonspecific binding mode, which likely reflects a synergistic effect in forming the ternary RNAP−PcopA−CueR open or dead-end complex. Consistently, RNAP’s overall affinity to PcopA is increased in the presence of holo- or apo-CueR, whereas CueR’s overall affinity to PcopA is also increased in the presence of RNAP (SI Appendix, Fig. S14).
The presence of the dead-end, off-pathway complex RPDE here constitutes a distinct “branched” RNAP−PcopA interaction pathway, compared with the linear pathway prevalent for RNAP interactions with optimal promoters. For optimal promoters, RNAP binding generally results in an initial closed complex, which then quickly isomerizes to the open complex which is competent for transcription initiation (19, 30, 33). This branched pathway of RNAP–PcopA interactions is also distinct from the earlier branched pathway for transcription initiation at λPRAL and T7A1 promoters, where the branching occurs after forming the closed complex (37, 38). Exploiting this branched interaction pathway, CueR biases the kinetic sampling of RNAP between the off-pathway RPDE and the RPO, leading to transcription repression or activation. This biased sampling also builds on the unique feature that the apo-repressor and holo-activator forms of MerR-family metalloregulators both bind to the recognition sequences tightly; their tight bindings allow them to exert DNA structural distortions, which may differ depending on CueR’s metallation state, thus leading to different biasing. Because MerR-family regulators encompass not only many metal-sensing regulators but also drug- and redox-sensing ones (1), the operation mechanism of CueR elucidated here should contribute a broad understanding to this mechanistically unique but functionally diverse family of transcription regulators, and may help the development of antibiotics that target this regulation process to compromise bacteria’s defense.
Materials and Methods
Materials and methods are described in SI Appendix, section 1, and include the design, labeling, and preparation of DNA constructs; expression, purification, and labeling of RNAP and CueR; in vitro transcription assay; and single-molecule FRET imaging and data analysis.
Supplementary Material
Acknowledgments
We thank R. H. Ebright for providing the plasmid and purification protocol of σ70; C. He and P. R. Chen for providing the plasmid and purification protocol of CueR; and C. Kinsland for access to the protein production facility. This research was supported by NIH Grants GM109993, GM106420, and AI117295, and Army Research Office Grant W911NF1510268. D.J.M. was supported by NIH Chemistry and Biology Interface Trainee Grant 5T32GM008500.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1515231112/-/DCSupplemental.
References
- 1.Hobman JL, Wilkie J, Brown NL. A design for life: Prokaryotic metal-binding MerR family regulators. Biometals. 2005;18(4):429–436. doi: 10.1007/s10534-005-3717-7. [DOI] [PubMed] [Google Scholar]
- 2.Waldron KJ, Rutherford JC, Ford D, Robinson NJ. Metalloproteins and metal sensing. Nature. 2009;460(7257):823–830. doi: 10.1038/nature08300. [DOI] [PubMed] [Google Scholar]
- 3.Moore CM, Helmann JD. Metal ion homeostasis in Bacillus subtilis. Curr Opin Microbiol. 2005;8(2):188–195. doi: 10.1016/j.mib.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 4.Rensing C, Grass G. Escherichia coli mechanisms of copper homeostasis in a changing environment. FEMS Microbiol Rev. 2003;27(2-3):197–213. doi: 10.1016/S0168-6445(03)00049-4. [DOI] [PubMed] [Google Scholar]
- 5.Ma Z, Jacobsen FE, Giedroc DP. Coordination chemistry of bacterial metal transport and sensing. Chem Rev. 2009;109(10):4644–4681. doi: 10.1021/cr900077w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dosanjh NS, Michel SLJ. Microbial nickel metalloregulation: NikRs for nickel ions. Curr Opin Chem Biol. 2006;10(2):123–130. doi: 10.1016/j.cbpa.2006.02.011. [DOI] [PubMed] [Google Scholar]
- 7.Summers AO. Untwist and shout: A heavy metal-responsive transcriptional regulator. J Bacteriol. 1992;174(10):3097–3101. doi: 10.1128/jb.174.10.3097-3101.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O’Halloran TV. Transition metals in control of gene expression. Science. 1993;261(5122):715–725. doi: 10.1126/science.8342038. [DOI] [PubMed] [Google Scholar]
- 9.Hobman JL, Crossman LC. Bacterial antimicrobial metal ion resistance. J Med Microbiol. 2015;64(Pt 5):471–497. doi: 10.1099/jmm.0.023036-0. [DOI] [PubMed] [Google Scholar]
- 10.O’Halloran TV, Frantz B, Shin MK, Ralston DM, Wright JG. The MerR heavy metal receptor mediates positive activation in a topologically novel transcription complex. Cell. 1989;56(1):119–129. doi: 10.1016/0092-8674(89)90990-2. [DOI] [PubMed] [Google Scholar]
- 11.Frantz B, O’Halloran TV. DNA distortion accompanies transcriptional activation by the metal-responsive gene-regulatory protein MerR. Biochemistry. 1990;29(20):4747–4751. doi: 10.1021/bi00472a001. [DOI] [PubMed] [Google Scholar]
- 12.Ansari AZ, Bradner JE, O’Halloran TV. DNA-bend modulation in a repressor-to-activator switching mechanism. Nature. 1995;374(6520):371–375. doi: 10.1038/374370a0. [DOI] [PubMed] [Google Scholar]
- 13.Outten CE, Outten FW, O’Halloran TV. DNA distortion mechanism for transcriptional activation by ZntR, a Zn(II)-responsive MerR homologue in Escherichia coli. J Biol Chem. 1999;274(53):37517–37524. doi: 10.1074/jbc.274.53.37517. [DOI] [PubMed] [Google Scholar]
- 14.Philips SJ, et al. Allosteric transcriptional regulation via changes in the overall topology of the core promoter. Science. 2015;349(6250):877–881. doi: 10.1126/science.aaa9809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heldwein EE, Brennan RG. Crystal structure of the transcription activator BmrR bound to DNA and a drug. Nature. 2001;409(6818):378–382. doi: 10.1038/35053138. [DOI] [PubMed] [Google Scholar]
- 16.Newberry KJ, Brennan RG. The structural mechanism for transcription activation by MerR family member multidrug transporter activation, N terminus. J Biol Chem. 2004;279(19):20356–20362. doi: 10.1074/jbc.M400960200. [DOI] [PubMed] [Google Scholar]
- 17.Newberry KJ, et al. Structures of BmrR-drug complexes reveal a rigid multidrug binding pocket and transcription activation through tyrosine expulsion. J Biol Chem. 2008;283(39):26795–26804. doi: 10.1074/jbc.M804191200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Outten FW, Outten CE, Hale J, O’Halloran TV. Transcriptional activation of an Escherichia coli copper efflux regulon by the chromosomal MerR homologue, cueR. J Biol Chem. 2000;275(40):31024–31029. doi: 10.1074/jbc.M006508200. [DOI] [PubMed] [Google Scholar]
- 19.Saecker RM, Record MT, Jr, Dehaseth PL. Mechanism of bacterial transcription initiation: RNA polymerase–promoter binding, isomerization to initiation-competent open complexes, and initiation of RNA synthesis. J Mol Biol. 2011;412(5):754–771. doi: 10.1016/j.jmb.2011.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kapanidis AN, et al. Initial transcription by RNA polymerase proceeds through a DNA-scrunching mechanism. Science. 2006;314(5802):1144–1147. doi: 10.1126/science.1131399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robb NC, et al. The transcription bubble of the RNA polymerase-promoter open complex exhibits conformational heterogeneity and millisecond-scale dynamics: Implications for transcription start-site selection. J Mol Biol. 2013;425(5):875–885. doi: 10.1016/j.jmb.2012.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stoyanov JV, Hobman JL, Brown NL. CueR (YbbI) of Escherichia coli is a MerR family regulator controlling expression of the copper exporter CopA. Mol Microbiol. 2001;39(2):502–511. doi: 10.1046/j.1365-2958.2001.02264.x. [DOI] [PubMed] [Google Scholar]
- 23.Caslake LF, Ashraf SI, Summers AO. Mutations in the alpha and sigma-70 subunits of RNA polymerase affect expression of the mer operon. J Bacteriol. 1997;179(5):1787–1795. doi: 10.1128/jb.179.5.1787-1795.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mukhopadhyay J, et al. Fluorescence resonance energy transfer (FRET) in analysis of transcription-complex structure and function. Methods Enzymol. 2003;371:144–159. doi: 10.1016/S0076-6879(03)71010-6. [DOI] [PubMed] [Google Scholar]
- 25.Hudson BP, et al. Three-dimensional EM structure of an intact activator-dependent transcription initiation complex. Proc Natl Acad Sci USA. 2009;106(47):19830–19835. doi: 10.1073/pnas.0908782106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.deHaseth PL, Lohman TM, Burgess RR, Record MT., Jr Nonspecific interactions of Escherichia coli RNA polymerase with native and denatured DNA: Differences in the binding behavior of core and holoenzyme. Biochemistry. 1978;17(9):1612–1622. doi: 10.1021/bi00602a006. [DOI] [PubMed] [Google Scholar]
- 27.Feklístov A, Sharon BD, Darst SA, Gross CA. Bacterial sigma factors: A historical, structural, and genomic perspective. Annu Rev Microbiol. 2014;68:357–376. doi: 10.1146/annurev-micro-092412-155737. [DOI] [PubMed] [Google Scholar]
- 28.Joshi CP, et al. Direct substitution and assisted dissociation pathways for turning off transcription by a MerR-family metalloregulator. Proc Natl Acad Sci USA. 2012;109(38):15121–15126. doi: 10.1073/pnas.1208508109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gaal T, Bartlett MS, Ross W, Turnbough CL, Jr, Gourse RL. Transcription regulation by initiating NTP concentration: rRNA synthesis in bacteria. Science. 1997;278(5346):2092–2097. doi: 10.1126/science.278.5346.2092. [DOI] [PubMed] [Google Scholar]
- 30.deHaseth PL, Zupancic ML, Record MT., Jr RNA polymerase-promoter interactions: The comings and goings of RNA polymerase. J Bacteriol. 1998;180(12):3019–3025. doi: 10.1128/jb.180.12.3019-3025.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y, et al. Structural basis of transcription initiation. Science. 2012;338(6110):1076–1080. doi: 10.1126/science.1227786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Coulombe B, Burton ZF. DNA bending and wrapping around RNA polymerase: A “revolutionary” model describing transcriptional mechanisms. Microbiol Mol Biol Rev. 1999;63(2):457–478. doi: 10.1128/mmbr.63.2.457-478.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dangkulwanich M, Ishibashi T, Bintu L, Bustamante C. Molecular mechanisms of transcription through single-molecule experiments. Chem Rev. 2014;114(6):3203–3223. doi: 10.1021/cr400730x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hansen UM, McClure WR. Role of the sigma subunit of Escherichia coli RNA polymerase in initiation. I. Characterization of core enzyme open complexes. J Biol Chem. 1980;255(20):9556–9563. [PubMed] [Google Scholar]
- 35.Heltzel A, Lee IW, Totis PA, Summers AO. Activator-dependent preinduction binding of sigma-70 RNA polymerase at the metal-regulated mer promoter. Biochemistry. 1990;29(41):9572–9584. doi: 10.1021/bi00493a011. [DOI] [PubMed] [Google Scholar]
- 36.Kulkarni RD, Summers AO. MerR cross-links to the α, β, and σ 70 subunits of RNA polymerase in the preinitiation complex at the merTPCAD promoter. Biochemistry. 1999;38(11):3362–3368. doi: 10.1021/bi982814m. [DOI] [PubMed] [Google Scholar]
- 37.Susa M, Sen R, Shimamoto N. Generality of the branched pathway in transcription initiation by Escherichia coli RNA polymerase. J Biol Chem. 2002;277(18):15407–15412. doi: 10.1074/jbc.M112481200. [DOI] [PubMed] [Google Scholar]
- 38.Susa M, Kubori T, Shimamoto N. A pathway branching in transcription initiation in Escherichia coli. Mol Microbiol. 2006;59(6):1807–1817. doi: 10.1111/j.1365-2958.2006.05058.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.