Significance
Bacteriophages (phages) are viruses that infect and kill bacteria. Being inanimate, phages rely on diffusion to search for bacterial prey. Here we demonstrate that a phage that adheres weakly to mucus exhibits subdiffusive motion, not normal diffusion, in mucosal surfaces. Supporting theory and experiments revealed that subdiffusive motion increases bacterial encounter rates for phages when bacterial concentration is low. To the best of our knowledge, no other predator has been shown to effectively use a subdiffusive search mechanism. Mucosal surfaces are vulnerable to infection. Mucus-adherent phages reduce bacterial infection of lifelike mucosal surfaces more effectively than nonadherent phages. These findings provide a basis for engineering adherent phages to manipulate mucosal surface microbiomes for protection from infection and other purposes.
Keywords: BAM, virus, mucus, subdiffusion, search strategy
Abstract
Bacteriophages (phages) defend mucosal surfaces against bacterial infections. However, their complex interactions with their bacterial hosts and with the mucus-covered epithelium remain mostly unexplored. Our previous work demonstrated that T4 phage with Hoc proteins exposed on their capsid adhered to mucin glycoproteins and protected mucus-producing tissue culture cells in vitro. On this basis, we proposed our bacteriophage adherence to mucus (BAM) model of immunity. Here, to test this model, we developed a microfluidic device (chip) that emulates a mucosal surface experiencing constant fluid flow and mucin secretion dynamics. Using mucus-producing human cells and Escherichia coli in the chip, we observed similar accumulation and persistence of mucus-adherent T4 phage and nonadherent T4∆hoc phage in the mucus. Nevertheless, T4 phage reduced bacterial colonization of the epithelium >4,000-fold compared with T4∆hoc phage. This suggests that phage adherence to mucus increases encounters with bacterial hosts by some other mechanism. Phages are traditionally thought to be completely dependent on normal diffusion, driven by random Brownian motion, for host contact. We demonstrated that T4 phage particles displayed subdiffusive motion in mucus, whereas T4∆hoc particles displayed normal diffusion. Experiments and modeling indicate that subdiffusive motion increases phage–host encounters when bacterial concentration is low. By concentrating phages in an optimal mucus zone, subdiffusion increases their host encounters and antimicrobial action. Our revised BAM model proposes that the fundamental mechanism of mucosal immunity is subdiffusion resulting from adherence to mucus. These findings suggest intriguing possibilities for engineering phages to manipulate and personalize the mucosal microbiome.
In all animals, mucosal surfaces provide critical immunological services by both protecting against invading bacterial pathogens and supporting large communities of commensal microorganisms (1, 2). Being exposed to the environment, mucosal surfaces are also the infection sites for many important bacterial diseases, including acute diarrhea and cystic fibrosis in humans. This, combined with their accessibility, make mucosal surfaces attractive venues for phage therapy; that is, the use of bacteriophages (phages) to treat and clear bacterial infections (3, 4). Clinical success so far has been erratic (5). The complexities and dynamics of the mucus layer are rarely considered, and the activity of phages therein is mostly unknown. Not surprisingly, phages effective in vitro do not consistently reduce mucosal bacterial host levels in vivo (6, 7). An understanding of the interactions between phages and their bacterial hosts within the relevant physiological environment is critical for consistent success of phage therapy applications.
The multilayered mucus is composed primarily of gel-forming mucin glycoproteins that are continually secreted by the underlying epithelium (8). The mucins self-organize to form a mesh that moves constantly outward from the epithelium, propelled by the ongoing secretion of mucin below (9). As the mucus migrates outward, the mucin concentration decreases and the mesh size (a measure of the distance between neighboring mucin strands) increases. Concurrently, the mucus is degraded by microbial proteases and subjected to shear forces that cause sloughing of the outermost layer (10). Despite this constant outward flux, in a diverse array of animals, phages are enriched on the mucosal surfaces compared with the surrounding milieu (11, 12). Previous in vitro experiments using a T4 phage model (particle size ∼200 nm) demonstrated that this phage adhered to mucus via weak binding between Ig-like domains of the highly antigenic outer capsid protein (Hoc) and the abundant glycans of the mucin glycoproteins (11, 12). Moreover, T4 phage adhering to the mucus layer of tissue culture cells protected the underlying epithelium from bacterial infection. On this basis, we proposed the bacteriophage adherence to mucus (BAM) model, whereby phages adhere to the mucosal surfaces of animals and provide a non-host-derived layer of immunity.
Results
Dynamic Mucus Environment on a Chip.
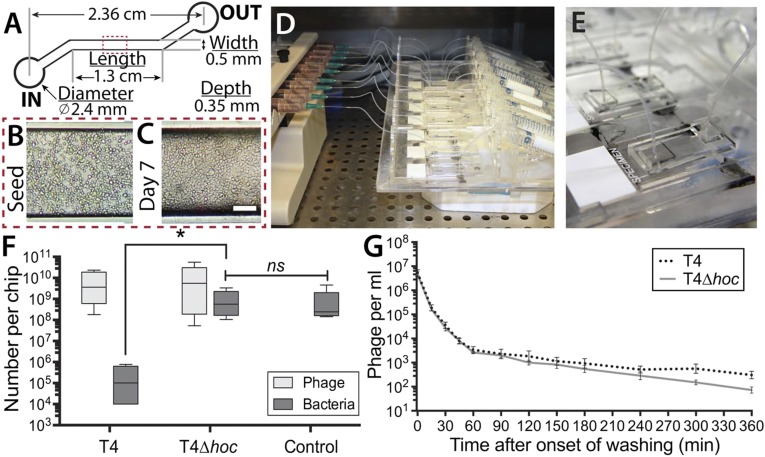
Deciphering the role of phage–bacterial–host interactions in mucosal infections has been hindered by the lack of a reproducible, dynamic, and lifelike experimental mucosal surface. To fill this gap, we developed a microfluidic device (chip) that emulates the microenvironment of a mucus-producing epithelial surface experiencing constant fluid flow across its surface. We used these chips to study the ability of phages to persist in the mucus layer and to protect the epithelium from infection. The chips were made of poly(dimethylsiloxane) and consisted of a single microfluidic channel, with in and out ports, attached to a glass microscope slide (Fig. 1A). Mucus-producing, human lung epithelial cells were seeded into the channel and allowed to attach to the glass surface. Tissue culture media was perfused through up to nine chips simultaneously for 7 d before all experiments. We maintained a lower flow rate (40 μL⋅h–1) for the first 4 d to ensure the establishment of an intact monolayer, and then increased the flow rate to 100 μL⋅h–1 (0.02 dyne⋅cm–2) for the following 3 d and for all experiments to mimic the in vivo fluid flow and shear stresses (13, 14). These conditions consistently provided a confluent epithelial cell layer that exhibited mucus secretion and turnover dynamics (Fig. 1 B–E).
Fig. 1.
Microfluidic devices (chips) simulating a lifelike in vitro mucosal surface. (A) Schematic of chip design. (B) Mucus-producing lung tissue culture cells seeded into the main channel of a chip. (C) Tissue culture cells in the main channel after perfusion for 7 d. (D) Multiplex syringe pump and scaffold perfusing nine chips simultaneously. (E) Close-up of a single chip bonded to a glass microscope slide with microfluidic tubing attached to the in and out ports. (F) Phage therapy experiment. Cell layers were pretreated with T4 or T4∆hoc phage for 12 h, washed for 1 h, and then inoculated with E. coli. Cells with mucus layer were harvested 18 h later, and both the phages and bacteria present were titered (PFU and CFU). (G) Phage detachment. Cell layers were pretreated with T4 or T4∆hoc phage for 12 h and then washed with phage-free media for 6 h, during which time the number of released phages was monitored by titering (PFU).
Interactions Among Bacteria, Phages, and Mucus on a Chip.
We investigated the trilateral interactions among bacteria, phages, and mucosal epithelial cells, using these chip microenvironments. For our model system, we used mucus-producing human lung epithelial cells, T4 phages, and a T4 phage host, Escherichia coli B (E. coli). The phages included a mucus-adherent T4 phage (T4) and a T4 Hoc deletion mutant (T4∆hoc). T4∆hoc does not have the Hoc protein exposed on its capsid and does not adhere to mucus, but is otherwise a normal, infective T4 phage (15). To assay for antibacterial activity of the phages in the mucus layer, we perfused chips with media containing T4 phage (107 mL–1), T4∆hoc phage (107 mL–1), or no phage for 24 h to saturate the mucosal epithelium. Those phage that were not enmeshed within the mucus layer were removed by subsequent perfusion with phage-free media for 1 h. We then inoculated each chip by introducing ∼107 bacteria into the input port, followed immediately by perfusion with phage- and bacteria-free media. Perfusion was continued for 18 h to allow time for both the bacteria and phage to replicate within the mucus layer. We then scraped the mucus-covered epithelium from the glass and titered both the phage and the bacteria present (Fig. 1F). The continual fluid flow during these infection experiments allowed us to selectively observe bacterial and phage replication taking place within the mucus, as both the surrounding fluid and the sloughed mucus layers were continually washed away from the attached cells. Both the T4 and T4∆hoc phage populations actively infected and replicated in the E. coli present. Chips pretreated with T4 phage showed a significant antimicrobial effect, with a >4,000-fold reduction in bacterial abundance compared with the no-phage control; the chips treated with T4∆hoc showed no significant difference from the no-phage control.
Why did only the mucus-adherent T4 phage protect the in vitro epithelial surface from bacterial infection? Our initial BAM model proposed that the enrichment of phages in mucosal surfaces depended on the binding of the Ig-like Hoc proteins exposed on T4 phage capsids to mucin glycans (11). Thus, the difference in antimicrobial effect observed here might be a result of T4 phage accumulating to a higher abundance or persisting longer within the mucus layer. The first possibility was contradicted by the results of the previous experiment (Fig. 1F), with both phages accumulating to comparable abundance in the chips. To test the second, chips were perfused with either T4 or T4∆hoc phage (107 mL–1) for 24 h, after which they were perfused with phage-free media for 6 h while phage detachment was monitored (Fig. 1G). Phage detachment was rapid during the first hour, with comparable numbers of T4 and T4∆hoc phage particles released. By this point, a large portion of the phage particles had already detached, and the subsequent 5 h showed the release of far fewer, with a slightly higher number of T4 phage particles recovered in the perfused media. Because T4 and T4∆hoc phage show the same patterns of accumulation and persistence on a lifelike mucosal surface, irrespective of Hoc-mediated adherence, some other mechanism must account for the observed disparity between their antibacterial effects. More generally, these observations indicate that phage abundance in mucus layers is governed by mucus secretion and turnover dynamics, rather than by the ability of phages to adhere specifically to mucus.
Diffusion of Phages in Mucin Solutions.
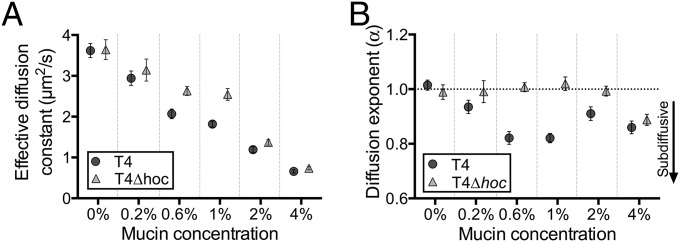
Phages are inert particles dependent on random diffusion driven by Brownian motion to bring about their chance encounters with a bacterial host (16). Because of the key role of diffusion in phage reproductive success, we investigated the diffusive properties of T4 and T4∆hoc phage in mucus. Using high-speed multiple particle tracking, we observed the diffusion of both phage types across a range of physiological mucin concentrations: 0%, 0.2%, 0.6%, 1%, 2%, and 4% (wt/vol) mucin in buffer, with 4% representing the highest concentration typically found in a mucosal layer (17). Fluorescence-labeled phages were mixed with a homogenous mucin solution in a well depression on a microscope slide, and their diffusion was microscopically recorded at a temporal resolution of 43.5 ms. Individual phage particle trajectories were manually tracked, and their effective diffusion constants (μm2/s) were calculated (Fig. 2A) (18).
Fig. 2.
(A) Effective diffusion constants (μm2/s) calculated at 43.5-ms time intervals for T4 and T4∆hoc phage in 0% (buffer), 0.2%, 0.6%, 1%, 2%, and 4% mucin solutions (wt/vol). (B) Diffusion exponents (α) of T4 and T4∆hoc phage in 0% (buffer), 0.2%, 0.6%, 1%, 2%, and 4% mucin solutions (wt/vol). Brownian diffusion α ∼ 1, subdiffusion α < 1.
The effective diffusion constant (K) of both phages was comparable in 0% mucin (buffer). Diffusion decreased with increasing mucin concentration for both phages, with T4 phage being slowed more markedly in 0.6% and 1% mucin than T4∆hoc phage. Specifically, diffusion of T4 phage was 21% less than T4∆hoc phage in 0.6% mucin and 29% less in 1% mucin. At 4% mucin, both phage particle types were effectively “trapped” in the mucin solution. The higher temporal resolution and improved multiple particle tracking methodologies used here yielded lower diffusion constants than previously reported (SI Appendix) (11).
Subdiffusion of Mucus-Adherent Phages.
The diffusive movement of phages can be modeled as a random walk, where the calculated mean square displacement (MSD) of a phage particle represents the area “explored” by the random walker. During normal, Brownian-driven diffusion, a particle’s average position remains unchanged while its MSD increases linearly with time (τ); that is, MSD (τ) = Kτ. However, it is known that within viscoelastic fluids and environments, diffusion can exhibit anomalous characteristics over time, being either enhanced (superdiffusion) or hindered (subdiffusion) (19, 20). Anomalous diffusion is characterized by a nonlinear relationship between a particle’s MSD and time (τ) that can be expressed with the power law exponent alpha (α), where MSD (τ) = Kτα (21, 22). For normal diffusion, α = 1, although superdiffusion is defined by α > 1 and subdiffusion by α < 1.
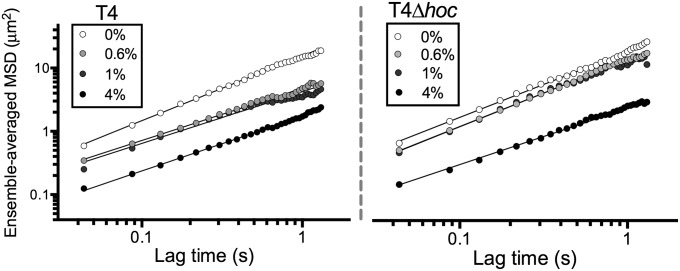
The effective diffusion constant (K) (Fig. 2A) and the diffusion exponent (α) are separate and independent phenomena in the sense that one cannot be taken as indicating the other. To investigate the nature of the observed diffusion of phage particles, we calculated the diffusion exponents (α) from our multiple particle tracking experiments for both phage types at all mucin concentrations (Fig. 2B). For this, we measured the ensemble-averaged MSD (SI Appendix) over time and then derived the values of α from the slope of the line of best fit for each phage–mucin combination (Fig. 3 and SI Appendix, Fig. S1). Both phages displayed normal diffusion (i.e., α ∼ 1) in 0% mucin. As mucin concentrations increased, T4 phage showed a clear subdiffusive signal in 0.6% and 1% mucin solutions (α = 0.82 ± 0.02 and α = 0.82 ± 0.01, respectively), whereas T4∆hoc maintained normal diffusion (α = 1.01 ± 0.01 and α = 1.02 ± 0.02, respectively). At even higher mucin concentrations (4% mucin), both phage particle types showed a comparable subdiffusive signal (α = 0.86 ± 0.02 and α = 0.89 ± 0.02 for T4 and T4∆hoc phage, respectively).
Fig. 3.
Log-log graph of the ensemble-averaged MSD (μm2) of T4 and T4∆hoc phage in 0% (buffer), 0.6%, 1%, and 4% mucin solutions (wt/vol). Solid lines indicate line of best fit from which the diffusion exponent (α) is determined. For full MSD plots, see SI Appendix, Fig. S1.
This experimental system provides a much-sought positive control for subdiffusion research that possesses two previously lacking properties: tunability and mechanism (23, 24). Tunability here is the ability to continuously adjust the mucin concentration, and thereby effect a corresponding change in the degree of subdiffusion (the exponent α). A potential mechanism is provided by the transient binding of the Hoc proteins covering the T4 phage capsids to the ubiquitous mucin glycans.
Theoretical Implications of Phage Subdiffusion in Mucus.
Subdiffusion has been observed in a wide variety of complex biological processes, including the intracellular transport of proteins and nucleic acids. In such situations, one might naively predict the slower diffusion would slow vital processes, as it might take longer for enzymes to “find” substrates or for transcription factors to locate their DNA binding sites. Instead, subdiffusion has been shown to dramatically increase the rate of productive intracellular encounters (23, 25). This is attributed to the search dynamics of subdiffusive particles. These particles remain closer to their initial position during any interval of time, thereby exploring the local area more thoroughly than if moving by normal diffusion (19, 26).
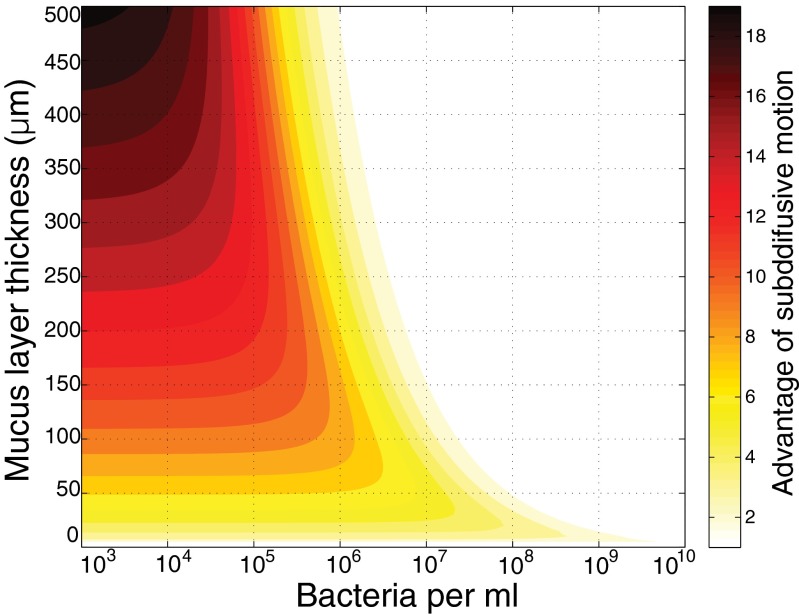
To consider the implications of subdiffusion for phages in mucus, we developed a simple model and compared the theoretical search efficiency for phage particles using normal Brownian motion versus subdiffusive motion (Fig. 4). Comparisons were made over a range of bacterial concentrations and mucus layer thicknesses, two factors that influence the likelihood that any phage particle will encounter a host bacterium before it leaves the mucus. Because increasing the bacterial concentration decreases the area a phage particle must search on average to locate a host, the frequency of phage–bacterial collisions rises with increasing bacterial concentration. The frequency of encounters also depends on the specific search pattern used (27, 28). Phage particles moving by subdiffusion search their local area more thoroughly, independent of bacterial concentration. The other key factor is how long a phage particle remains in the bacteria-enriched mucus before it is sloughed or diffuses out of the mucus layer. The thicker the layer, the longer time, on average, it will remain in the mucus, and thus the more opportunities for collision with a potential host. The interplay of both of these factors with the diffusion exponent α predicts that a subdiffusive phage particle will explore the mucus layer more thoroughly and remain in the subdiffusive zone (Discussion) longer than one moving by normal diffusion (Fig. 4 and SI Appendix). Further, the model predicts greater benefits of subdiffusive motion at lower bacterial concentrations, where phage–bacterial collisions are rare (26, 29). At high bacterial concentrations, where collisions are frequent, more efficient searching by subdiffusion motion is expected to have little effect. Likewise, with thin mucus layers, both subdiffusive and normally diffusive phage particles would be quickly lost from the mucus.
Fig. 4.
Theoretical advantage of a phage–host encounter for a phage particle, using subdiffusive motion compared with Brownian motion across a range of bacterial concentrations and mucus layer thickness. Color scale indicates the ratio of probabilities of bacterial encounter for a subdiffusive phage (α = 0.82) versus a normally diffusing phage (α = 1). The ratio ranges from 19 (black) to 1 (white).
Experimental Verification of Benefit of Subdiffusive Motion.
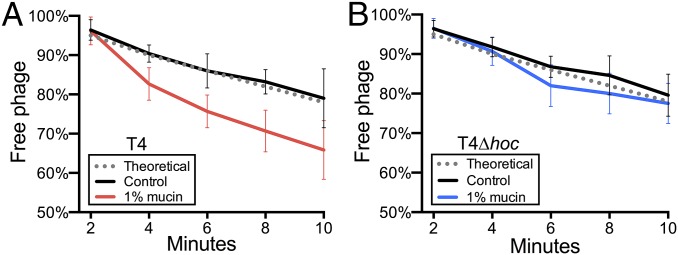
A major challenge in interpreting observations of subdiffusive motion in biological systems is the experimental demonstration of a conferred benefit (23). Here we used the classical adsorption assay (16, 30, 31) to test whether subdiffusion of T4 phage increased their rate of adsorption to bacterial hosts. We compared adsorption of T4 and T4∆hoc phage under conditions where our model predicted a subdiffusive search would be beneficial (i.e., low bacterial densities in 1% mucin). Experimental conditions were selected on the basis of the previously determined adsorption constant of T4 and other T-even phages: 2.4 × 10−9 mL/min (31). Either T4 or T4∆hoc phage was added to control (0%) or 1% mucin solutions containing E. coli to give a final concentration of 2 × 105 phage particles⋅mL–1 and 1 × 107 bacteria⋅mL–1. At these concentrations, ∼22% of phage particles were predicted to adsorb to a bacterial host during a 10-min period (SI Appendix). The suspensions were sampled every 2 min. Chloroform was immediately added to each sample to rapidly destroy all of the bacterial cells and halt phage adsorption. The number of free phage particles in each sample was counted and used to calculate the adsorption constant (k) of T4 and T4∆hoc phage in both 1% mucin and control solutions (32). The adsorption of T4 phage was significantly higher in the 1% mucin solution (k = 4.7 × 10−9 mL/min; n = 6; t = −4.2; P < 0.001) than in the control (k = 2.3 × 10−9 mL/min; Fig. 5A). In contrast, there was no significant difference in the adsorption of the T4∆hoc phage between the 1% mucin (k = 2.6 × 10−9 mL/min; n = 6; t = −1.2; P = 0.22) and the control (k = 2.1 × 10−9 mL/min; Fig. 5B). Because our model predicted that the benefit of a subdiffusive search would decrease with increasing bacterial concentration, we repeated these adsorption assays at a higher bacterial density (7 × 107 mL–1), where ∼80% of phage particles were predicted to adsorb during a 10-min period. Here neither T4 nor T4∆hoc phage showed increased adsorption in 1% mucin (SI Appendix, Fig. S2).
Fig. 5.
Adsorption assay measuring the percentage of free phage remaining during a 10-min period in control (0%) and 1% mucin solutions. Error bars show SD (n = 6). Theoretical values were calculated from the T4 phage adsorption constant (k) = 2.4 × 10−9 mL/min, phage concentration (2 × 105 mL–1), and bacterial concentration (1 × 107 mL–1). (A) T4 phage. (B) T4∆hoc phage.
Discussion
The search for a specific target is a ubiquitous process throughout biology. At the microscopic scale, molecules such as enzymes and repressor proteins perform site-specific searches within a cell; at the macroscopic scale, animals search for food (26, 27). Many predators possess prior knowledge of where prey are located, and can also use their senses to further direct their movement when hunting. For phages, the search for a susceptible bacterial host is effectively “blind.” When limited to random searching, the chance for a successful predator–prey encounter depends largely on the search strategy used (33). In this situation, many motile predators, ranging from microbes to humans, use Lévy flight, a superdiffusive search strategy, to increase their success (28, 34, 35). To the best of our knowledge, no organism has been shown to use a subdiffusive search strategy to the same effect. Phage particles, being inanimate and small (capsid diameters ∼20–200 nm), act as colloidal particles subject to Brownian motion. Our study shows that the mucus-adherent T4 phage uses subdiffusive motion in mucus to increase the frequency of host encounters.
Earlier work had predicted interactions between the T4 phage Hoc protein and mammalian organisms (36, 37). Subsequently, we demonstrated that the Hoc proteins exposed on the T4 capsid bind to the mucin glycoproteins (11). The Hoc-bearing phages were enriched in mucus, reduced the bacterial load, and protected the underlying epithelial cells from infection, effects that we attributed to the putative mucin-binding activity of the Hoc protein (11). However, T4 and T4∆hoc phage particles also carry different capsid charges, as evidenced by their different electrophoretic mobilities (38). We cannot currently rule out the possibility that the observed BAM effects are a result of these charge differences rather than to Hoc binding to the mucins, but our previous results demonstrating high structural homology between the T4 Hoc protein and known glycan-binding proteins support the mucin-binding mechanism (11).
Our previous work using an in vitro model system could not show a statistical difference between T4 and T4∆hoc phage in either their adherence to mucus-producing cells or their antimicrobial effect (11). Here we compared their adherence using a lifelike mucosal surface with fluid flow dynamics and found little difference in either their accumulation or their persistence in the mucosal surface. Therefore, we revise our initial hypothesis, that phage enrichment in mucus occurs via binding interactions, and conclude that the mucus mesh traps both phage types comparably and that phage persistence in mucus is likely governed by mucus secretion and microbial infection dynamics. Regarding their antimicrobial effect, we report a >4,000-fold greater effect for T4 phage than for T4∆hoc phage in a mucosal surface.
On the basis of integration of our previous and current research findings, we have incorporated subdiffusion into our proposed BAM model of immunity. T4 phage with its glycan-binding Hoc proteins exploits the benefits of subdiffusive motion under specific, physiologically relevant mucin concentrations. Subdiffusion of T4 phage particles, but not T4∆hoc particles, was observed in 0.2–1% (wt/vol) mucin. At these concentrations, transient binding to mucins slowed, but did not arrest, T4 phage particles and shifted their motion from normal Brownian motion to subdiffusion. Phage–host encounters assayed in vitro in 1% mucin increased at low, but not high, bacterial concentrations.
As mucin concentration increases (e.g., 4% mucin), the mesh size of the mucin network decreases, likely approaching the capsid diameters of both phage types. As a result, the dense mucus network elicits subdiffusion of both phages, a phenomenon known to occur with particles in viscoelastic fluids such as mucus (39, 40). Subdiffusion at high mucin concentration is accompanied by very restricted diffusivity of both T4 and T4∆hoc phage particles (SI Appendix), which masks the differential effect of transient mucin binding by T4 phage particles. Subdiffusion under these conditions is not predicted to benefit phages.
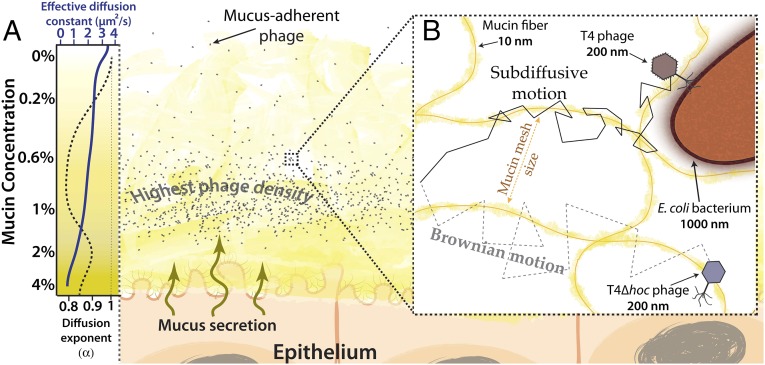
According to our current BAM model, a mucus-adherent phage particle that diffuses into a mucosal surface first encounters the region with the lowest mucin concentration and the most open mucin mesh. If it diffuses closer to the epithelium, mucin concentration increases and its diffusivity decreases. At the critical concentration, diffusion of these particles transitions to subdiffusion. More particles diffuse from the outer region into this subdiffusive zone per unit time than diffuse back out, resulting in their accumulation here (Fig. 6A). Those that diffuse still closer to the epithelium may become temporarily trapped in the more concentrated mucus, but the outward flux of the secreted mucins eventually carries them back into the subdiffusive zone. Within this zone, only the mucus-adherent phage particles transiently bind the mucin network, and this induces the subdiffusive motion that allows them to explore the network more thoroughly. When a resultant phage–host encounter leads to a productive lytic infection, subdiffusion keeps the progeny particles released by host lysis within this zone longer. These two factors contribute to the increase in phage–host encounters that was observed in vitro at low bacterial concentration. However, as the concentration of bacteria increases, so does the chance of random phage–host encounters (31, 41). These encounters become so frequent that they mask the benefit provided by subdiffusive motion that was evident at lower bacterial density.
Fig. 6.
BAM model with subdiffusion. (A) The mucus layer is a dynamic gradient of mucin glycoproteins. Closer to the epithelium, the mucin concentration increases. The corresponding decrease in the mucin mesh size reduces phage diffusion constants. Subdiffusive motion of mucus-adherent phage particles at lower mucin concentrations enriches phage particles within an optimal zone of the mucus layer. (Left) Qualitative representations of the effective diffusion constant (K) (solid blue line) and diffusion exponent (α) (dashed black line) for T4 phage from Fig. 2. (B) Transient binding of mucus-adherent phage to mucin glycoproteins facilitates subdiffusive motion over a range of mucin concentrations present within a mucus layer.
In vivo mucosal surfaces are undoubtedly more complex than our simulated mucus environments. When considering that the in vivo diversity of bacterial host and even greater diversity of phage strains lowers the probability of a successful phage–host encounter, the implications for subdiffusive motion resulting in increased host encounters becomes apparent. In addition, the mucus layer contains a mix of macromolecules including mucin glycoproteins, other diverse proteins, DNA, and polysaccharides, all of which could be expected to affect phage diffusion. Mucosal surfaces, being exposed to the surrounding milieu, experience environmental fluxes that may affect mucus structure and phage particle diffusion. For example, viral production in coral mucus is tightly coupled to temperature and salinity (42, 43). Temperature, salinity, and pH all affect the attachment of Hoc proteins to the T4 capsid (38), and thus have the potential to alter both the subdiffusion of phage particles and the frequency of productive phage–host encounters. Alterations of these physical parameters are known to occur in diseased mucosal surfaces [e.g., the high mucin concentrations (>4% wt/vol) and acidified mucosa characteristic of cystic fibrosis] (40, 44). Although numerous studies have documented the effective use of phages to treat cystic fibrosis-associated bacterial infections (45, 46), how physical and physiological changes in the lung might affect phage diffusion, the frequency of productive phage–host encounters, and bacterial infection dynamics remain to be determined.
Taken together, this suggests that a tightly regulated symbiosis between phages and their metazoan hosts helps metazoans sustain a healthy microbiome and curbs disease at mucosal surfaces. Within this symbiosis, the glycan-binding proteins of some phages have been fine-tuned to provide a balance between the greater speed of normal Brownian diffusion and the more thorough local searching afforded by subdiffusive motion (Fig. 6B).
Subdiffusion of phage particles in mucus has important and previously unrecognized implications for phage therapy. We propose that subdiffusive motion, mediated by mucin binding, underlies the ability of T4 phage, in contrast to T4∆hoc, to reduce bacterial colonization of our mucus-producing epithelial surface on a chip. To date, this protection of an epithelial surface by phage has been demonstrated only for T4 phage, which carries the carbohydrate-binding, Ig-like (Ig-like) domains of its Hoc protein exposed on its capsid (11). Similar protein domains are encoded by ∼25% of the known members of the predominant order of phages (Caudovirales) (47). Given the enormous diversity of phages associated with mucosal surfaces, we expect many will be found to use Ig-like proteins or other carbohydrate-adherent surface proteins to increase their replicative success via subdiffusive motion (48–50). Furthermore, we speculate that their mucin-binding is finely tuned to position the phage particles in the optimal mucus zone where their bacterial hosts reside, further increasing encounter rates.
Phage selection criteria for clinical mucosal phage therapy have been limited to host range, burst size, and growth kinetics—phage characteristics that do not necessarily correlate with in vivo efficacy. An increased understanding of the subdiffusive motion of mucus-adherent phages, combined with physiologically relevant in vitro testing using microfluidic chips, may enable sound prediction and validation of in vivo phage efficacy. Such accurate predictions are essential for the selection or engineering of phages for consistent clinical success. Engineered phages displaying an assortment of BAM domains may allow for unprecedented control to concentrate phages within a mucosal zone that overlaps with bacterial host ranges, leading to personalized therapeutics for mucosal disease.
Materials and Methods
Bacteria Strains, Phage Stocks, Tissue Culture Cell Lines, and Growth Conditions.
E. coli B strain HER 1024 was used for all experiments and was grown in LB (10 g tryptone, 5 g yeast extract, 10 g NaCl, in 1 L dH20) at 37 °C overnight with shaking. The T4 Hoc phage deletion mutant (T4∆hoc) was kindly supplied by Venigalla B. Rao (The Catholic University of America, Washington, D.C.) (15). The human tumorigenic lung epithelial cell line A549 was obtained from the American Type Culture Collection and cultured in F12-K media with 10% FBS and 100 μg·mL–1 penicillin–streptomycin. Tissue culture cells were grown in 50 mL Primaria Tissue Culture Flasks (Becton Dickinson) at 37 °C under 5% CO2.
Microbial Assay of Chips.
T4 phages and E. coli B strain were used at a concentration of 1 × 107 plaque-forming units (PFU) and colony-forming units (CFU) mL–1, respectively, for all chip assays. Before all experiments, chips were perfused with antibiotic- and serum-free medium containing T4 phage for 12 h at a flow rate of 100 μL⋅h–1. For phage-free washes, chips were perfused with antibiotic- and serum-free medium, and effluent from chips was titered. For bacterial infection, chips were inoculated with E. coli followed by perfusion with antibiotic- and serum-free medium for 18 h. The intact mucosal cell layer was then scraped and bacteria and phage titered.
Multiple Particle Tracking.
Five microliters of SYBR Gold-labeled phage suspension (109 PFU mL–1) was added to 45 μL of 0%, 0.2%, 0.6%, 1%, 2%, or 4% (wt/vol) of type II porcine gastric mucin (Sigma) solution in SM buffer supplemented with 1 mM MgSO4 and CaCl2. Solutions were immediately visualized using an Applied Precision OMX structured illumination microscope at 100× (1.40 NA). Movies were captured using Deltavision OMX: 43.5 ms temporal resolution for 100 frames, with >10 analyses per sample. Trajectories were manually tracked in two dimensions using MTrackJ plugin for ImageJ (18), with ∼30 particle trajectories recorded for every analysis. Particle frame number, track ID, x and y coordinates, and time were extracted and analyzed.
Phage Adsorption Assays.
Adsorption assays were performed in LB supplemented with 1 mM MgSO4 and CaCl2 prewarmed to 37 °C, and 1% (wt/vol) type II porcine gastric mucin (Sigma). E. coli B cells (1 × 107 CFU mL–1) were added to adsorption tubes, vortexed, and CFU titered on LB agar plates at 37 °C overnight to ensure accurate bacterial quantification. T4 and T4∆hoc phage (2 × 105 PFU mL–1) were added to respective tubes, immediately vortexed, and sampled without agitation every 2 min continuously for 10 min. Each phage–bacteria sample was added to another tube containing LB media saturated with chloroform, vortexed, and free phage quantified in duplicate by titering.
Supplementary Material
Acknowledgments
We thank Joshua Weitz, Karl Heinz Hoffmann, Gregory Forest, and Scott A. McKinley for helpful discussion and Ryland Young for critical review. This work was supported by the National Institutes of Health (Grant R01GM095384-01 to F.R.), Grants NS047101 and R21AI094534 from the National Institute of General Medical Sciences, and the Gordon and Betty Moore Foundation (Investigator Award 3781). J.J.B. acknowledges funding and support from San Diego State University.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1508355112/-/DCSupplemental.
References
- 1.Johansson MEV, et al. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc Natl Acad Sci USA. 2008;105(39):15064–15069. doi: 10.1073/pnas.0803124105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Linden SK, Sutton P, Karlsson NG, Korolik V, McGuckin MA. Mucins in the mucosal barrier to infection. Mucosal Immunol. 2008;1(3):183–197. doi: 10.1038/mi.2008.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.d’Hérelle F. The Bacteriophage: Its Role in Immunity. Masson et Cie; Paris: 1921. [Google Scholar]
- 4.Brüssow H. Phage therapy: The Escherichia coli experience. Microbiology. 2005;151(Pt 7):2133–2140. doi: 10.1099/mic.0.27849-0. [DOI] [PubMed] [Google Scholar]
- 5.Sulakvelidze A, Alavidze Z, Morris JG., Jr Bacteriophage therapy. Antimicrob Agents Chemother. 2001;45(3):649–659. doi: 10.1128/AAC.45.3.649-659.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chibani-Chennoufi S, et al. In vitro and in vivo bacteriolytic activities of Escherichia coli phages: Implications for phage therapy. Antimicrob Agents Chemother. 2004;48(7):2558–2569. doi: 10.1128/AAC.48.7.2558-2569.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maura D, et al. Intestinal colonization by enteroaggregative Escherichia coli supports long-term bacteriophage replication in mice. Environ Microbiol. 2012;14(8):1844–1854. doi: 10.1111/j.1462-2920.2011.02644.x. [DOI] [PubMed] [Google Scholar]
- 8.Atuma C, Strugala V, Allen A, Holm L. The adherent gastrointestinal mucus gel layer: Thickness and physical state in vivo. Am J Physiol Gastrointest Liver Physiol. 2001;280(5):G922–G929. doi: 10.1152/ajpgi.2001.280.5.G922. [DOI] [PubMed] [Google Scholar]
- 9.Lai SK, et al. Rapid transport of large polymeric nanoparticles in fresh undiluted human mucus. Proc Natl Acad Sci USA. 2007;104(5):1482–1487. doi: 10.1073/pnas.0608611104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johansson MEV, Larsson JMH, Hansson GC. The two mucus layers of colon are organized by the MUC2 mucin, whereas the outer layer is a legislator of host-microbial interactions. Proc Natl Acad Sci USA. 2011;108(Suppl 1):4659–4665. doi: 10.1073/pnas.1006451107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barr JJ, et al. Bacteriophage adhering to mucus provide a non-host-derived immunity. Proc Natl Acad Sci USA. 2013;110(26):10771–10776. doi: 10.1073/pnas.1305923110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barr JJ, Youle M, Rohwer F. Innate and acquired bacteriophage-mediated immunity. Bacteriophage. 2013;3(3):e25857. doi: 10.4161/bact.25857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim HJ, Huh D, Hamilton G, Ingber DE. Human gut-on-a-chip inhabited by microbial flora that experiences intestinal peristalsis-like motions and flow. Lab Chip. 2012;12(12):2165–2174. doi: 10.1039/c2lc40074j. [DOI] [PubMed] [Google Scholar]
- 14.Ishikawa T, Sato T, Mohit G, Imai Y, Yamaguchi T. Transport phenomena of microbial flora in the small intestine with peristalsis. J Theor Biol. 2011;279(1):63–73. doi: 10.1016/j.jtbi.2011.03.026. [DOI] [PubMed] [Google Scholar]
- 15.Fokine A, et al. Structure of the three N-terminal immunoglobulin domains of the highly immunogenic outer capsid protein from a T4-like bacteriophage. J Virol. 2011;85(16):8141–8148. doi: 10.1128/JVI.00847-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schlesinger M. 1932. Adsorption of phages to homologous bacteria. II. Quantitative investigation of adsorption velocity and saturation. Estimation of the particle size of the bacteriophage. Immunitaetsforschung 114:149–160.
- 17.Allen A, Flemström G, Garner A, Kivilaakso E. Gastroduodenal mucosal protection. Physiol Rev. 1993;73(4):823–857. doi: 10.1152/physrev.1993.73.4.823. [DOI] [PubMed] [Google Scholar]
- 18.Meijering E, Dzyubachyk O, Smal I. Methods for cell and particle tracking. Methods Enzymol. 2012;504:183–200. doi: 10.1016/B978-0-12-391857-4.00009-4. [DOI] [PubMed] [Google Scholar]
- 19.Guigas G, Weiss M. Sampling the cell with anomalous diffusion - the discovery of slowness. Biophys J. 2008;94(1):90–94. doi: 10.1529/biophysj.107.117044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saxton MJ. A biological interpretation of transient anomalous subdiffusion. I. Qualitative model. Biophys J. 2007;92(4):1178–1191. doi: 10.1529/biophysj.106.092619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barkai E, Garini Y, Metzler R. Strange kinetics of single molecules in living cells. Phys Today. 2012;65:29. [Google Scholar]
- 22.Ernst D, Köhler J, Weiss M. Probing the type of anomalous diffusion with single-particle tracking. Phys Chem Chem Phys. 2014;16(17):7686–7691. doi: 10.1039/c4cp00292j. [DOI] [PubMed] [Google Scholar]
- 23.Höfling F, Franosch T. Anomalous transport in the crowded world of biological cells. Rep Prog Phys. 2013;76(4):046602. doi: 10.1088/0034-4885/76/4/046602. [DOI] [PubMed] [Google Scholar]
- 24.Saxton MJ. Wanted: A positive control for anomalous subdiffusion. Biophys J. 2012;103(12):2411–2422. doi: 10.1016/j.bpj.2012.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weiss M, Nilsson T. In a mirror dimly: Tracing the movements of molecules in living cells. Trends Cell Biol. 2004;14(5):267–273. doi: 10.1016/j.tcb.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 26.Golding I, Cox EC. Physical nature of bacterial cytoplasm. Phys Rev Lett. 2006;96(9):098102. doi: 10.1103/PhysRevLett.96.098102. [DOI] [PubMed] [Google Scholar]
- 27.Sims DW, et al. Scaling laws of marine predator search behaviour. Nature. 2008;451(7182):1098–1102. doi: 10.1038/nature06518. [DOI] [PubMed] [Google Scholar]
- 28.Viswanathan GM, et al. Lévy flight search patterns of wandering albatrosses. Nature. 1996;381:413–415. doi: 10.1038/nature06199. [DOI] [PubMed] [Google Scholar]
- 29.Halford SE, Marko JF. How do site-specific DNA-binding proteins find their targets? Nucleic Acids Res. 2004;32(10):3040–3052. doi: 10.1093/nar/gkh624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Adams MH. Bacteriophages. Interscience; New York: 1959. [Google Scholar]
- 31.Stent GS. Molecular Biology of Bacterial Viruses. Freeman and Company; San Francisco: 1963. [Google Scholar]
- 32.Hyman P, Abedon ST. In: Practical Methods for Determining Phage Growth Parameters. Clokie MRJ, Kropinski AM, editors. Humana Press; Totowa, NJ: 2009. [DOI] [PubMed] [Google Scholar]
- 33.Humphries NE, Weimerskirch H, Queiroz N, Southall EJ, Sims DW. Foraging success of biological Lévy flights recorded in situ. Proc Natl Acad Sci USA. 2012;109(19):7169–7174. doi: 10.1073/pnas.1121201109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schuster FL, Levandowsky M. Chemosensory responses of Acanthamoeba castellanii: Visual analysis of random movement and responses to chemical signals. J Eukaryot Microbiol. 1996;43(2):150–158. doi: 10.1111/j.1550-7408.1996.tb04496.x. [DOI] [PubMed] [Google Scholar]
- 35.Raichlen DA, et al. Evidence of Levy walk foraging patterns in human hunter-gatherers. Proc Natl Acad Sci USA. 2014;111(2):728–733. doi: 10.1073/pnas.1318616111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dabrowska K, Switała-Jeleń K, Opolski A, Górski A. Possible association between phages, Hoc protein, and the immune system. Arch Virol. 2006;151(2):209–215. doi: 10.1007/s00705-005-0641-7. [DOI] [PubMed] [Google Scholar]
- 37.Dabrowska K, et al. Hoc protein regulates the biological effects of T4 phage in mammals. Arch Microbiol. 2007;187(6):489–498. doi: 10.1007/s00203-007-0216-y. [DOI] [PubMed] [Google Scholar]
- 38.Yamaguchi Y, Yanagida M. Head shell protein hoc alters the surface charge of bacteriophage T4. Composite slab gel electrophoresis of phage T4 and related particles. J Mol Biol. 1980;141(2):175–193. doi: 10.1016/0022-2836(80)90384-8. [DOI] [PubMed] [Google Scholar]
- 39.Lai SK, Wang Y-Y, Wirtz D, Hanes J. Micro- and macrorheology of mucus. Adv Drug Deliv Rev. 2009;61(2):86–100. doi: 10.1016/j.addr.2008.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hill DB, et al. A biophysical basis for mucus solids concentration as a candidate biomarker for airways disease. PLoS One. 2014;9(2):e87681. doi: 10.1371/journal.pone.0087681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wiggins BA, Alexander M. Minimum bacterial density for bacteriophage replication: Implications for significance of bacteriophages in natural ecosystems. Appl Environ Microbiol. 1985;49(1):19–23. doi: 10.1128/aem.49.1.19-23.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nguyen-Kim H, et al. High occurrence of viruses in the mucus layer of scleractinian corals. Environ Microbiol Rep. 2014;6(6):675–682. doi: 10.1111/1758-2229.12185. [DOI] [PubMed] [Google Scholar]
- 43.Nguyen-Kim H, et al. Coral mucus is a hot spot of viral infections. Appl Environ Microbiol. 2015;81(17):5773–5783. doi: 10.1128/AEM.00542-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pezzulo AA, et al. Reduced airway surface pH impairs bacterial killing in the porcine cystic fibrosis lung. Nature. 2012;487(7405):109–113. doi: 10.1038/nature11130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.James CE, et al. Lytic activity by temperate phages of Pseudomonas aeruginosa in long-term cystic fibrosis chronic lung infections. ISME J. 2015;9(6):1391–1398. doi: 10.1038/ismej.2014.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Debarbieux L, et al. Bacteriophages can treat and prevent Pseudomonas aeruginosa lung infections. J Infect Dis. 2010;201(7):1096–1104. doi: 10.1086/651135. [DOI] [PubMed] [Google Scholar]
- 47.Fraser JS, Yu Z, Maxwell KL, Davidson AR. Ig-like domains on bacteriophages: A tale of promiscuity and deceit. J Mol Biol. 2006;359(2):496–507. doi: 10.1016/j.jmb.2006.03.043. [DOI] [PubMed] [Google Scholar]
- 48.Dutilh BE, et al. A highly abundant bacteriophage discovered in the unknown sequences of human faecal metagenomes. Nat Commun. 2014;5:4498. doi: 10.1038/ncomms5498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tariq MA, et al. A metagenomic approach to characterize temperate bacteriophage populations from Cystic Fibrosis and non-Cystic Fibrosis bronchiectasis patients. Front Microbiol. 2015;6:97. doi: 10.3389/fmicb.2015.00097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Edlund A, Santiago-Rodriguez TM, Boehm TK, Pride DT. Bacteriophage and their potential roles in the human oral cavity. J Oral Microbiol. 2015;7:27423. doi: 10.3402/jom.v7.27423. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.