Significance
Cyclodextrins are cyclic oligosaccharides used in the pharmaceutical industry to improve drug delivery. Here, we show that cyclodextrins improve the antifungal activity of the anabaenolysins A and B lipopeptides and that are both produced by the same cyanobacteria. This study identifies the putative biosynthetic gene cluster involved in the synthesis of these unique cyanobacterial lipopeptide anabaenolysins.
Keywords: natural products, bioactive compounds, hydroxyamino fatty acid, NRPS, PKS
Abstract
Cyclodextrins are cyclic oligosaccharides widely used in the pharmaceutical industry to improve drug delivery and to increase the solubility of hydrophobic compounds. Anabaenolysins are lipopeptides produced by cyanobacteria with potent lytic activity in cholesterol-containing membranes. Here, we identified the 23- to 24-kb gene clusters responsible for the production of the lipopeptide anabaenolysin. The hybrid nonribosomal peptide synthetase and polyketide synthase biosynthetic gene cluster is encoded in the genomes of three anabaenolysin-producing strains of Anabaena. We detected previously unidentified strains producing known anabaenolysins A and B and discovered the production of new variants of anabaenolysins C and D. Bioassays demonstrated that anabaenolysins have weak antifungal activity against Candida albicans. Surprisingly, addition of the hydrophilic fraction of the whole-cell extracts increased the antifungal activity of the hydrophobic anabaenolysins. The fraction contained compounds identified by NMR as α-, β-, and γ-cyclodextrins, which undergo acetylation. Cyclodextrins have been used for decades to improve the solubility and bioavailability of many drugs including antifungal compounds. This study shows a natural example of cyclodextrins improving the solubility and efficacy of an antifungal compound in an ancient lineage of photosynthetic bacteria.
Cyclodextrins are small cyclic oligosaccharides composed of six (α), seven (β), or eight (γ) glucose units (1). Cyclodextrins were discovered in 1891 from Bacillus amylobacter and had their chemical structure determined by X-ray crystallography in the 1940s (2). Strains producing higher amounts of cyclodextrins were obtained in the 1970s, thereby enabling pharmaceutical applications (3). Cyclodextrins are capable of forming inclusion complexes with different compounds, thereby improving their solubility, reactivity, and stability, a property that has been widely exploited in the pharmaceutical industry (4–6). Cyclodextrins are used in the pharmaceutical industry to improve drug delivery and the encapsulation of antibiotics for the prevention and treatment of infections (1, 4, 7).
Cyanobacteria are photosynthetic bacteria capable of producing a broad range of bioactive compounds (8, 9). More than 600 peptides or peptidic compounds have been detected in different genera of cyanobacteria (8). Lipopeptides are commonly produced by cyanobacteria and present a broad range of bioactivity, including different types of cytotoxicity, growth, and enzyme inhibition of various organisms (fungi, bacteria, plants, protozoa, microalgae, and viruses) (9). A large number of lipopeptides have been described from cyanobacteria, such as barbamide (10), jamaicamide (11), curacin A (12), lyngbyatoxin (13) hassallidin (14), and puwainaphycins (15). Many lipopeptides are synthesized by large multifunctional enzymes known as polyketide synthases (PKSs) and nonribosomal peptide synthetases (NRPSs) (10–13). The lipid structure of lipopeptides is synthesized in a variety of ways. PKS modules directly synthesize the lipophilic part of the curacin A (12), whereas the activation of the fatty acid chain incorporated in jamaicamide involves an acyl-acyl carrier protein synthetase (JamA) (10). In the hassallidin gene cluster, produced by Anabaena sp., the involvement of an acyl-protein synthetase and ligase (HasG), an acyl carrier protein (HasH), and a 3-oxoacyl (acyl-carrier-protein) reductase (HasL) are predicted to be involved in the fatty acid synthesis and incorporation in this nonribosomal glycolipopeptide (14), whereas the synthesis of the hybrid puwainaphycins in Cylindrospermum alatosporum involves a fatty-acyl ligase (FAAL) (15).
Anabaenolysins are lipopeptides produced by species of Anabaena isolated from the Baltic Sea (16). Our previous study found them to consist of two glycines, a 2-(3-amino-5-oxytetrahydrofuran-2-yl)-2-hydroxyacetic acid and a long (C16–C19) unsaturated α-hydroxy-β-amino carboxylic acid (16). Anabaena sp. XPORK15F produces 10 variants of anabaenolysins, which differ in the length of the methylene units and the degree of unsaturation of the hydroxyamino fatty acid. Anabaenolysins may account for up to 0.1% of the dry weight of Anabaena strains (16). Anabaenolysins are able to permeabilize mammalian cells in a cholesterol-dependent manner and show more hemolytic potency than do the plant saponin digitonin and the bacterial cyclic peptide surfactin (17). In this study, we report the discovery of the anabaenolysin gene cluster and demonstrate that anabaenolysins exhibit antifungal activity. Furthermore, the ananbaenolysins work in synergy with the cyclodextrins produced by the same Anabaena strains, resulting in an increased antifungal activity.
Results
Synergistic Antifungal Activity of Cyclodextins and Anabaenolysins.
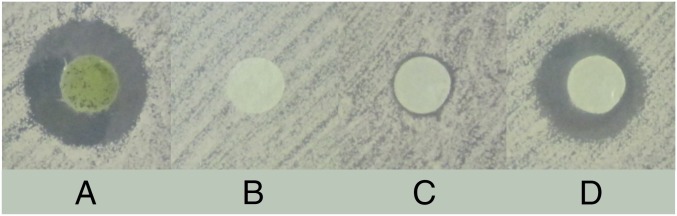
We screened 151 cyanobacterial isolates from diverse sources and various genera to detect new producers of the lipopeptide anabaenolysin (SI Appendix, Table S1). We used high-performance liquid chromatography (HPLC) to screen the methanol extracts from the strains, and detection was based on the characteristic UV spectrum of the anabaenolysins resulting from three conjugated double bonds in the hydroxyamino fatty acid (16). We then tested strains containing the characteristic UV spectra against bacteria (data not shown) and fungi to verify the activity of the cell extracts. Extracts containing anabaenolysins exhibited antifungal activity against Candida albicans and Aspergillus spp. (SI Appendix, Table S2). However, bioassays of isolated anabaenolysin B against C. albicans showed a much smaller zone of inhibition (Fig. 1C). Combining isolated anabaenolysin B with a fraction containing hydrophilic compounds from the same Anabaena sp. XPORK1D strain restored the stronger antifungal activity (Fig. 1D). Efforts to identify the synergistic compound led to the detection of α-, β-, and γ-cyclodextrins (Fig. 2).
Fig. 1.
Disk diffusion assay showing synergistic antifungal activity against C. albicans HAMBI 261 of anabaenolysin B and cyclodextrin. Whole-cell methanol extract from Anabaena sp. XPORK1D (A); a fraction containing cyclodextrins (B); anabaenolysin B (C); and a fraction containing cyclodextrins and anabaenolysin B combined (D).
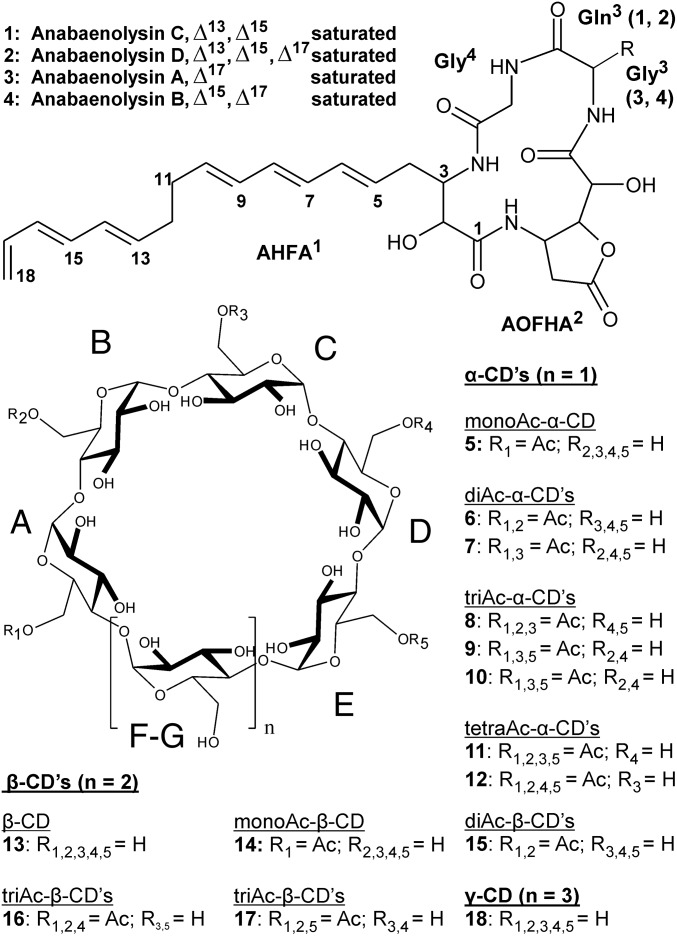
Fig. 2.
Structures of anabaenolysins A–D (compounds 1–4) and α-, β-, and γ-cyclodextrins (compounds 5–18). The newly discovered anabaenolysins C and D differ mainly in the glutamine in position 3 from anabaenolysins A and B, which have a glycine molecule in position 3. Ac, acetyl; AHFA, (5E,7E,9E)-3-amino-2-hydroxyoctadeca-5,7,9,17-tetraenoic acid; AOFHA, (3-amino-5-oxotetrahydrofuran-2-yl)(hydroxy)acetic acid; CD, cyclodextrin.
Anabaenolysins.
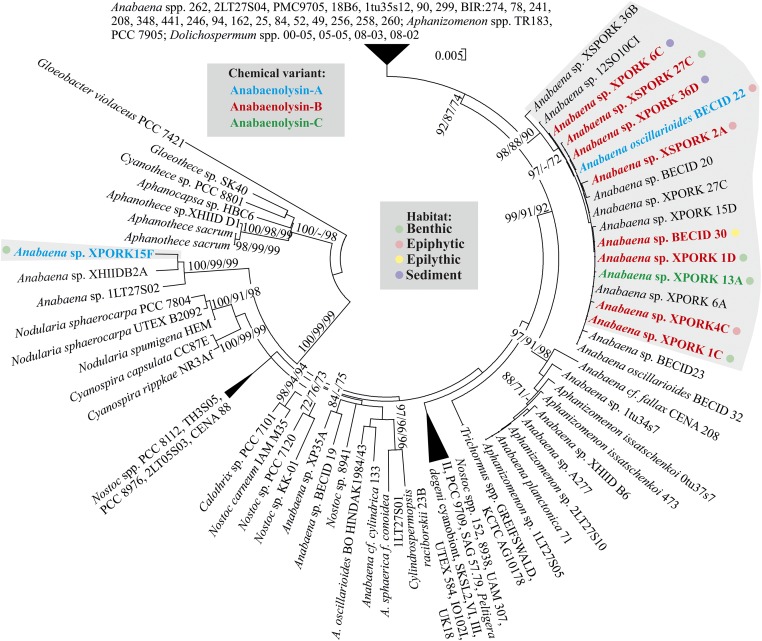
We detected anabaenolysins A–D (Fig. 2) in 11 strains of the genus Anabaena isolated from the brackish water of the Baltic Sea (Fig. 3 and SI Appendix, Table S3). Each strain produced roughly similar amounts of only one major anabaenolysin variant. These 11 cyanobacterial strains were isolated from benthic, epiphytic, epilithic, and sediments environments (Fig. 3). The cells contained almost all of the anabaenolysins and very little of the lipopeptides were present in the culture medium (SI Appendix, Fig. S1). A phylogenetic tree based on the 16S rRNA gene indicates that most of the anabaenolysin producers are grouped with other Anabaena spp. strains isolated from the Baltic Sea (Fig. 3). However, Anabaena sp. XPORK15F is more distantly related to the other anabaenolysins producers (Fig. 3). Comparison of the 16S rRNA gene sequences shows low identity of Anabaena sp. XPORK15F and other anabaenolysin producers (94%).
Fig. 3.
Phylogenetic tree of cyanobacteria based on 16S rRNA gene sequences. The phylogenetic tree was constructed by using neighbor-joining, maximum parsimony, and maximum likelihood methods using 1,000 bootstraps. Strains producing anabaenolysins are marked according to the main variant produced: blue (anabaenolysin A), red (anabaenolysin B), and green (anabaenolysin C).
Anabaenolysins A, B, and C were the major variants present (Figs. 2 and 3), but we also detected minor variants (present in quantities of a small percentage of the main variant) of anabaenolysins in these Anabaena strains (data not shown). We determined the two previously unidentified variants of anabaenolysins C and D from Anabaena sp. XPORK13A (Fig. 2). Abl-C (1.94 mg) was purified from 2.4 g of dried biomass resulting to minimum of 0.8‰ of Abl-C in the Anabaena sp. XPORK13A dry cell mass. NMR and amino acid analysis indicated the presence of a glutamine instead of glycine in position 3 (SI Appendix, Figs. S2 and S3 and Table S4). Anabaenolysin D lacks a double bond in the α-hydroxy-β-amino fatty acid (AHFA1) (Fig. 2). The presence of single bond is shown as smaller mass (by two units) and in the NMR signals δC 27.9 ppm and δH 1.26 ppm for 17-CH2, and δC 13.6 ppm and δH 0.85(t) ppm for 18-CH3, from anabaenolysin D, which demonstrates that the last double bond is saturated.
Cyclodextrins.
We selected Anabaena sp. XSPORK2A for the isolation of the synergistic compound. A sharp peak containing different variants of cyclodextrins was isolated by high-performance liquid chromatography (HPLC) and analyzed by HPLC–ion-trap mass spectrometry (ITMS), ultra-performance liquid chromatography–quadrupole time of flight mass spectrometry (UPLC-QTOF), and nuclear magnetic resonance (NMR) (Fig. 2 and SI Appendix, Figs. S4 and S5 and Table S5). NMR showed that the cyclodextrin fraction contained mainly di- and triAc-α-cyclodextrins as well as mono- and diAc-β-cyclodextrins. The results showed the presence of glucose units, acetyl groups in 6-OH groups, and the absence of free anomeric carbon and proton signals (SI Appendix, Fig. S6 and Table S6). Also, 1H and 13C signals were typical for cyclodextrin but different from linear glucooligosaccharide signals. Accurate masses of sodiated di- and triAc-α-cyclodextrin ions m/z 1079.3291 (∆ 1.7 ppm) and m/z 1121.3383 (∆ 0.4 ppm) provided elemental compositions of C40H64O32Na and C42H66O33Na, respectively, which confirms the cyclodextrin structures. We detected cyclodextrins in all anabaenolysin-producing strains (Table 1). The cyclodextrins had a diverse chemical structures with mono-, di-, and triacetylated-α- as well as non-, mono-, and diacetylated-β-cyclodextrins, formed by six and seven hexoses and decorated with acetyl (Ac) groups (Fig. 2 and Table 1). Most of the strains also produced tetra-Ac-α-cyclodextrins, tri-Ac-β-cyclodextrins, and nonacetylated γ-cyclodextrins. Acetyl groups were always bound to the 6-OH of the glucose unit.
Table 1.
Relative (%) abundance of the cyclodextrins (CDs) identified in the anabaenolysin-producing strains
| α-CDs | β-CDs | ||||||||||
| Strain | monoAc-α-CD | diAc-α-CD | triAc-α-CD | tetraAc-α-CD | SUM | β-CD | monoAc-β-CD | diAc-β-CD | triAc-β-CD | SUM | γ-CD |
| XPORK1C | <1 | 31 | 54 | 1 | 86 | 3 | 4 | 6 | ND | 13 | <1 |
| XPORK1D | <1 | 37 | 47 | 1 | 86 | 3 | 4 | 6 | ND | 14 | <1 |
| XSPORK2A | <1 | 18 | 45 | 5 | 68 | 7 | 12 | 11 | <1 | 30 | 2 |
| XPORK4C | 2 | 56 | 15 | ND | 73 | 4 | 12 | 11 | ND | 27 | <1 |
| XPORK6C | 2 | 42 | 42 | <1 | 87 | 4 | 5 | 4 | <1 | 13 | <1 |
| XPORK13A | 1 | 49 | 41 | <1 | 92 | 2 | 3 | 3 | <1 | 7 | <1 |
| XPORK15F | 4 | 46 | 24 | <1 | 75 | 2 | 5 | 17 | 1 | 25 | <1 |
| XSPORK27C | <1 | 57 | 35 | <1 | 92 | <1 | 2 | 5 | <1 | 8 | <1 |
| XPORK36D | <1 | 28 | 52 | 4 | 84 | 4 | 4 | 8 | <1 | 16 | <1 |
| BECID22 | ND | 18 | 54 | 3 | 76 | <1 | 6 | 17 | <1 | 24 | <1 |
| BECID30 | <1 | 31 | 52 | <1 | 84 | 1 | 6 | 8 | <1 | 16 | ND |
The relative abundance of the main cyclodextrin produced is in bold. Relative abundances were calculated from the peak areas of the extracted sodiated cyclodextrin ion chromatograms. ND, not detected.
Biosynthetic Genes of Anabaenolysins and Cyclodextrins.
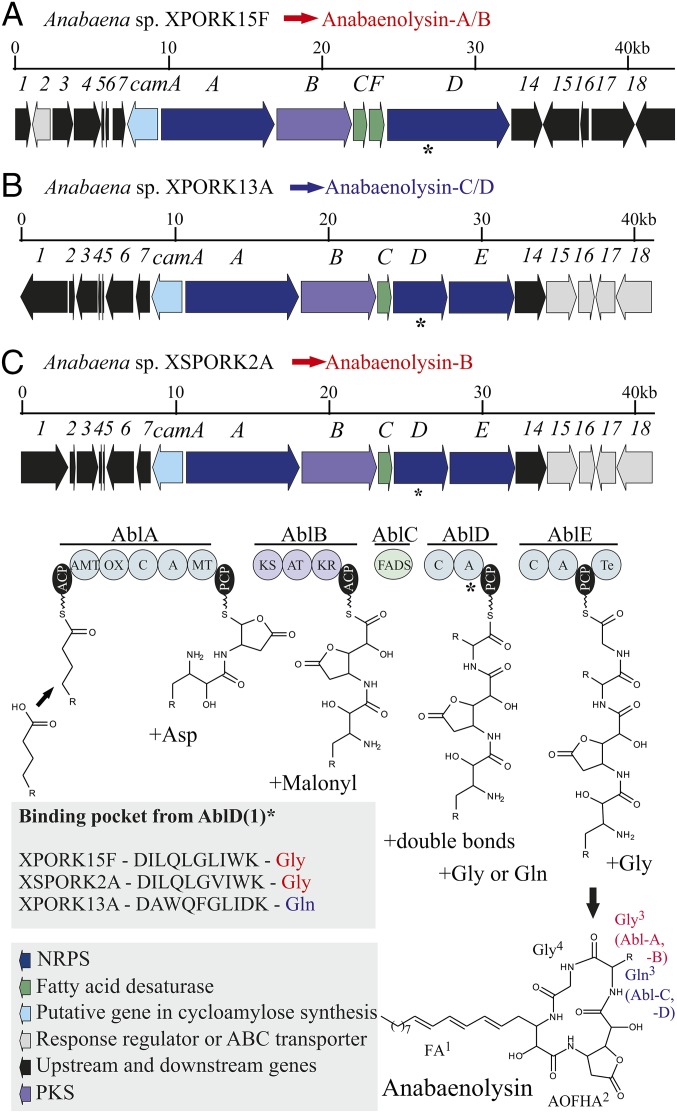
Genome mining identified a putative 23-kb anabaenolysin gene cluster (abl) in the partial genome of Anabaena sp. XSPORK2A (Fig. 4). The putative abl gene cluster encodes hybrid NRPS-PKS enzymes and a fatty acid desaturase (Fig. 4). AblA is a 278-kDa NRPS protein predicted to be involved in the incorporation of the 3-amino-2-hydroxyoctadecanoic acid, a hydroxyl group, an asparagine, and a methyl group into the growing anabaenolysin peptide (Fig. 4 and SI Appendix, Table S7). AblB is a 171-kDa PKS and is likely to incorporate a malonyl-CoA precursor unit and another hydroxyl group. AblC is a 35-kDa delta fatty acid desaturase (FADS) predicted to be responsible for the formation of double bonds in the 3-amino-2-hydroxyoctadecanoic acid. AblD and AblE are, respectively, 124-kDa and 157-kDa NRPS proteins that incorporate l-glycine into the peptide intermediate (SI Appendix, Table S8), and a thioesterase domain present in the latter catalyzes the cyclization and release of the compound. CamA encodes a 75-kDa protein containing two S-layer homology domains and a glycoside hydrolase domain with an unclear role in anabaenolysin biosynthesis. However, this enzyme may play a role in the formation of cyclodextrins. The anabaenolysin gene cluster is surrounded upstream by proteins unlikely to be involved in anabaenolysin biosynthesis (e.g., peptidoglycan binding protein, transposase, lysin, and transmembrane sensor-like protein) and downstream by DNA repair and response regulator proteins (SI Appendix, Table S8).
Fig. 4.
Putative anabaenolysin gene clusters from Anabaena spp. XPORK15F (A), XPORK13A (B), and XSPORK2A (C). The main variant of anabaenolysin (A or B in red, C and D in blue) produced per strain is indicated in the figure. The main difference between the variants is the glycine (A/B) or glutamine (C/D) in position 3. The signature of the binding pockets from the adenylation domain of AblD(1) appears in the figure, and an asterisk (*) denotes the AblD(1) region in the gene cluster.
The abl gene cluster was found in the draft genomes of two additional anabaenolysin producers, Anabaena sp. XPORK13A and Anabaena sp. XPORK15F (Fig. 4). The only genetic region with lower similarity is the adenylation domain of AblD, which encodes the NRPS that incorporates the amino acid in position 3 of the anabaenolysin gene clusters (SI Appendix, Table S9). XSPORK2A produces anabaenolysin B with glycine in position 3, whereas XPORK13A produces a variant with glutamine in this position (Fig. 4). The downstream response regulator proteins from both strains are similar, but the upstream proteins vary, and the XPORK13A strain encodes more proteins with unknown function (SI Appendix, Table S8).
Anabaena spp. XPORK13A and XSPORK2A strains are more closely related to one another than to the XPORK15F strain based on the 16S rRNA gene phylogenetic tree (Fig. 3). The 24.3-kb anabaenolysin gene cluster of Anabaena sp. XPORK15F encodes a second 35-kDa fatty acid desaturase (AblF). This strain contains the 276-kDa NRPS protein AblD, which corresponds to both AblD and AblE proteins in the XSPORK2A and XPORK13A anabaenolysin gene clusters (Fig. 4). Comparison of the entire anabaenolysin gene cluster showed less similarity to the sequences of XPORK15F than to those of the XSPORK2A strain (62% of coverage and 79% of identity). Nevertheless, these two strains synthesize the same chemical variant of anabaenolysin, anabaenolysin B (Figs. 2 and 3). Anabaena sp. XPORK15F also synthesizes anabaenolysin A, the main chemical variant produced by this strain, which lacks a saturated bond in the fatty acid chain characteristic of anabaenolysin B (16). Proteins upstream and downstream of the anabaenolysin gene cluster differ in both XPORK15F and XSPORK2A strains (SI Appendix, Table S9).
A phylogenetic tree based on the sequences of the condensation domains present in the anabaenolysin gene cluster indicates that the first condensation domain of AblA is grouped with domains containing a monooxygenase and/or aminotransferase domains prior to them, whereas the condensation domains of AblD and AblE are grouped with other LCL condensation domains (SI Appendix, Fig. S8). How the 3-amino-2-hydroxyoctadecanoic acid is activated and incorporated into anabaenolysin remains unclear. However, we detected FAAL encoded at distant locations in the three Anabaena spp. genomes (data not shown). The adenylation domain in the AblA is predicted to activate asparagine (NRPSpredictor2) or aspartic acid (PKS/NRPS Analysis) (SI Appendix, Table S7). The AOFHA moiety, which is the 2-(3-amino-5-oxotetrahydrofuran-2-yl)-2-hydroxyacetic acid (Fig. 2) in the anabaenolysin structure, is predicted to be derived from an aspartic acid. The methyltransferase-like domain embedded in the adenylation domain might be involved in the formation of AOFHA. However, the exact biosynthetic mechanism underpinning the biosynthesis of the AOFHA is unclear and remains to be elucidated (Fig. 4).
The CamA has a domain predicted to act as an α-amylase. A phylogenetic tree based on diverse α-amylases shows that CamA sequences are more closely related to sequences predicted to be 4-α-glucanotransferases and amylo-α-1,6-glucosidases (SI Appendix, Fig. S8). We found similar amino acid sequences in the genomes of Anabaena sp. 90 (71–84% identity and 100% coverage) and Anabaena sp. 1tu33S10 (70–82% identity and 100% coverage) (data not shown). However, methylated and disparately derivatized cyclodextrins were present only in Anabaena sp. 90 cell extracts (data not included); no cyclodextrin glucanotransferase (CGTase) sequences were present in the three partial genomes obtained from Anabaena sp. XSPORK2A, XPORK13A, or XPORK15F.
Discussion
Synergistic Antifungal Activity.
Cyanobacteria are known to produce lipopeptides with antifungal activity, such as nostofungicidine (18), laxaphycins (19–21), lobocyclamides (22), and hassallidins (14, 23, 24). In this study, we show that anabaenolysins have a weak antifungal activity (Fig. 1). Isolated anabaenolysins are known to permeabilize mammalian cells, which contain cholesterol (17), and it could also make the ergosterol containing fungal cells permeable. The structure of these two sterols differs only by two double bonds and a methyl group.
We found that all anabaenolysin producers also produce cyclodextrins, which increased the antifungal activity of anabaenolysins (Fig. 1D). Previously, lipopeptides, such as laxaphycins (19–21) and lobocyclamides (22), showed synergistic antifungal activity. Portoamides A and B have also shown synergistic allelophatic activity inhibiting the green microalgae Chlorella vulgaris (25). However, the combination of two chemically different variants of the same compound improved their antifungal or allelophatic activity. In the present study, the two unrelated compounds worked in synergy to produce a potent antifungal activity. Previous studies have shown that structurally unrelated compounds produced by Streptomyces spp., such as the β-lactam antibiotic cephamycin C and clavulanic acid, an inhibitor of β-lactamases, functioned synergistically (26).
Cyclodextrins are known to improve the solubility of compounds because of the hydrophobic and hydrophilic portions of their cone-shaped structures (1). The hydrophobic portion of a compound penetrates the hydrophobic inner cavity of a cyclodextrin molecule, resulting in an inclusion complex that increases hydrophilicity/water solubility. This feature makes them an important tool in drug delivery (1, 4, 7). β-cyclodextrins are also used to extract cholesterol from the plasma cell membrane (27) and, in a similar way, could act on the ergosterol present in the fungal membrane. Most of the cyclodextrins produced by Anabaena (73–92%), are α-cyclodextrins, which are capable of forming inclusion complexes with hydrocarbon chains similar to those present in anabaenolysins (2). A small percentage of the cyclodextrins produced by Anabaena (7–30%), are β-cyclodextrins, which are capable of forming inclusion complexes with membrane sterols. Anabaena-producing α-cyclodextrins may form inclusion complexes with anabaenolysins to increase their solubility so that the concentration of anabaenolysins against the target increases. β-cyclodextrins produced by the same Anabaena strains could target fungal ergosterol-containing membranes by extracting ergosterol from the membrane, thereby changing the native function of the membrane. Commercially available amphotericin B and cyclodextrins used together showed an increase of 60% in antifungal activity compared with amphotericin B alone (7). Another example of improved efficacy in the treatment of oral candidosis in patients with AIDS comes from the use of commercially available itraconazole in complex with cyclodextrins (28). Cyclodextrins produced by the cyanobacterium Tolypothrix byssoidea blocked the antifungal activity of the tolytophycins produced by this cyanobacterium (29). However, in this study we showed that Anabaena spp. from the Baltic Sea produce both cyclodextrins and anabaenolysins and that anabaenolysin antifungal activity improves in the presence of cyclodextrins.
Benthic Cyanobacteria Producing Anabaenolysins and Cyclodextrins.
Cyanobacteria isolated from terrestrial and marine, brackish, and freshwater environments produce lipopeptides (9, 30–32). In the present study, anabaenolysins were detected in benthic cyanobacteria isolated from the Baltic Sea (Fig. 3). We used HPLC to analyze 151 methanol extracts from 14 different genera of cyanobacteria (SI Appendix, Table S1). However, only Anabaena species produced anabaenolysins and cyclodextrins (Fig. 3 and Table 1). Anabaenolysins A and B were first described as products of Anabaena spp. XPORK15F and XSPORK27C (16). We detected previously unidentified variants of anabaenolysins C and D produced by Anabaena sp. XPORK13A. Clarifying whether Anabaena strains exclusively produce anabaenolysins and determining the geographical distribution of anabaenolysin producers will require further analysis with a broader range of cyanobacterial genera and a larger selection of habitats.
The rRNA gene phylogeny groups most anabaenolysin producers together with nonproducers (Fig. 3). However, Anabaena sp. XPORK15F is located in a different part of the phylogenetic tree (Fig. 3). Previous studies indicate that benthic cyanobacteria are genetically more diverse than planktonic ones (33, 34).
Anabaenolysin producers were found to contain differently acetylated variants of α- and β-cyclodextrins as well as nonacetylated β- and γ-cyclodextrins. Anabaena sp. 90 can produce methylated and disparately derivatized cyclodextrins, which have been described as products of T. byssoidea (29). Putative genes involved in the synthesis of cyclodextrins were reported from Nostoc sp. PCC 9229 (35). In this study, bioinformatic analysis aided the discovery of a new cyclodextrin producer known as planktonic Anabaena sp. 90. However, our results indicate that Anabaena strains present in the benthic habitat of the Baltic Sea are the only known producers of anabaenolysins.
Potential Biosynthetic Gene Cluster of Anabaenolysin and Cyclodextrins.
Many of the cyanobacterial bioactive natural products reported to date are synthesized by a hybrid of NRPS and PKS pathways (36). We discovered in the partial genomes of Anabaena spp. XSPORK2A, XPORK13A, and XPORK15F a putative anabaenolysin gene cluster (Fig. 4). The anabaenolysin gene cluster consists of a hybrid NRPS/PKS, one or two fatty acid desaturase(s), and a hypothetical protein (Fig. 4). The incorporation of the fatty acid chain into the anabaenolysin is unclear. The phylogenetic tree groups the condensation domain of AblA with other condensation domains that come after aminotransferases and/or monooxygenases. Previous studies indicate that condensation domains are involved in the incorporation a β-hydroxyl fatty acid into lipopeptides (37–39). Surfactin, lichenysin, fengycin, and arthrofactin are examples of lipopeptides that are produced by using a starter condensation domain (37). Fatty acids, activated by FAAL enzyme, can be incorporated into nonribosomal peptide synthesis (40). The lipidation of daptomycin involves the DptE FAAL enzyme and the DptF acyl carrier protein (40). The anabaenolysin gene cluster contains an acyl carrier protein in the AblA (Fig. 4). Anabaena sp. XSPORK2A has three FAALs, and XPORK13A and XPORK15F have two FAALs encoded elsewhere in its genome. However, a full understanding of anabaenolysin lipidation will require further analysis. The adenylation domain of AblA is predicted to incorporate an aspartic acid (SI Appendix, Table S7). The methyltransferase-like activity embedded in this domain could result in the formation of AOFHA. AOFHA formation may occur in a manner homologous to Ahp (3-amino-6-hydroxy-2-piperodone) formation in anabaenopeptilides, where it is predicted to form from glutamine, thus involving an unusual methyltransferase-like domain (41). This methyltransferase-like domain is putatively involved in the heterocyclization of anabaenolysins and anabaenopeptilides. Future biochemical characterization will improve our understanding of the function of these enzymes.
The presence of a gene encoding the putative α-amylase CamA in the anabaenolysin gene cluster suggests that Anabaena may coordinate the production of anabaenolysins and cyclodextrins. Phylogenetic analysis shows CamA sequences to be more closely related to 4-α-glucanotransferases, which produce cycloamyloses (e.g., ref. 42). The action of 4-α-glucanotransferases synthesizes larger cycloamylases, whereas cyclodextrin glucanotransferases (CGTases) synthesize cyclodextrins (5, 6, 41). Phylogenetic studies based on the α-amylase (glycoside hydrolase 13 family) help to predict the function of enzymes among this family, but may be unreliable for a small number of proteins (43). Recombinant 4-α-glucanotransferases from Synechocystis sp. PCC 6803 could convert sucrose and amylopeptin in cycloamyloses (42, 44). Cyclodextrins are known to act synergistically to improve the potency of a range of antifungal compounds (e.g., ref. 7). In this study, we show that living organisms use the synergistic application of cyclodextrins and lipopeptides to produce an antifungal effect. Interestingly, we found an α-amylase–encoded gene, which may be involved in cyclodextrin synthesis, upstream of the putative anabaenolysin biosynthetic gene cluster. The presence of enzymes involved in the synthesis of anabaenolysins and cyclodextrins in the same gene cluster indicates that they may be cosynthesized and act together. Lipopeptides with a unique chemical structure, such as anabaenolysins, can be used topically to treat fungal infections. Our knowledge of anabaenolysin biosynthetic genes improves the potential for further modifications in the chemical structure or for the heterologous expression of this peptide.
Materials and Methods
Details of cyanobacterial strains (SI Appendix, Table S1) and their cultivation as well as DNA isolation, 16S rRNA gene sequencing, and phylogenetic tree construction are presented in SI Appendix, SI Materials and Methods (primers in SI Appendix, Table S10). Accession numbers of 16S rRNA genes obtained in this study are as follows: KP715718 (Anabaena sp. XPORK1C), KP715719 (Anabaena sp. XPORK1D), KP715720 (Anabaena sp. XPORK4C), KP715721 (Anabaena sp. BECID30), and KP715722 (Anabaena sp. XPORK13A). Genome sequences of Anabaena spp. XSPORK2A, XPORK13A and XPORK15F were obtained and the biosynthetic gene clusters annotated as detailed in SI Appendix, SI Materials and Methods. The anabaenolysin gene clusters were deposited into the GenBank (NCBI) with the following accession numbers: Anabaena sp. XPORK15F (KP761740), Anabaena sp. XPORK13A (KP761741), and Anabaena sp. XSPORK2A (KP761742). Detection of anabaenolysins and cyclodextrins was based on UV spectrum and mass spectrometry (SI Appendix, SI Materials and Methods). The disk diffusion bioassay was used to detect the antifungal activity (SI Appendix, SI Materials and Methods). Previously unidentified anabaenolysins (C and D) were identified based on amino acid analysis, mass spectrometry, and NMR. Mass spectrometry and NMR solved the structures of cyclodextrins.
Supplementary Material
Acknowledgments
We thank M. Sc. Lyudmila Saari for maintaining and handling the cyanobacteria strains, T.K.S. thanks Prof. Mervin Bibb and Dr. Govind Chandra for all their teaching in analyzing bacterial genomes. This work was supported by Academy of Finland Grants 1258827, 1273798 (to K.S.) and 1259505 (to D.P.F.). T.K.S. received partial funding from the Helsinki Graduate Program in Biotechnology and Molecular Biology, now the Integrative Life Science Doctoral Program; the São Paulo Research Foundation Grant 2009/13455-0; Centre for International Mobility Grant TM-09-6506; and the Finnish Cultural Foundation. E.R. received funding from “Futuro in Ricerca” Grant RBFR126B8I_003.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. KP715718 (Anabaena sp. XPORK1C), KP715719 (Anabaena sp. XPORK1D), KP715720 (Anabaena sp. XPORK4C), KP715721 (Anabaena sp. BECID30), and KP715722 (Anabaena sp. XPORK13A)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1510432112/-/DCSupplemental.
References
- 1.Nardello-Rataj V, Leclercq L. Encapsulation of biocides by cyclodextrins: Toward synergistic effects against pathogens. Beilstein J Org Chem. 2014;10:2603–2622. doi: 10.3762/bjoc.10.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Del Valle EM. Cyclodextrins and their uses: A review. Process Biochem. 2004;39:1033–1046. [Google Scholar]
- 3.Cal K, Centkowska K. Use of cyclodextrins in topical formulations: Practical aspects. Eur J Pharm Biopharm. 2008;68(3):467–478. doi: 10.1016/j.ejpb.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 4.Szejtli J. Introduction and general overview of cyclodextrin chemistry. Chem Rev. 1998;98(5):1743–1754. doi: 10.1021/cr970022c. [DOI] [PubMed] [Google Scholar]
- 5.Sträter N, et al. Structural basis of the synthesis of large cycloamyloses by amylomaltase. Biologia (Bratisl) 2002;11:93–99. [Google Scholar]
- 6.Qi Q, Zimmermann W. Cyclodextrin glucanotransferase: From gene to applications. Appl Microbiol Biotechnol. 2005;66(5):475–485. doi: 10.1007/s00253-004-1781-5. [DOI] [PubMed] [Google Scholar]
- 7.Ruiz HK, et al. New amphotericin B-gamma cyclodextrin formulation for topical use with synergistic activity against diverse fungal species and Leishmania spp. Int J Pharm. 2014;473(1-2):148–157. doi: 10.1016/j.ijpharm.2014.07.004. [DOI] [PubMed] [Google Scholar]
- 8.Welker M, von Döhren H. Cyanobacterial peptides - nature’s own combinatorial biosynthesis. FEMS Microbiol Rev. 2006;30(4):530–563. doi: 10.1111/j.1574-6976.2006.00022.x. [DOI] [PubMed] [Google Scholar]
- 9.Burja AM, Banaigs B, Abou-Mansour E, Burgess JG, Wright PC. Marine cyanobacteria – a prolific source of natural products. Tetrahedron. 2001;57:9347–9377. [Google Scholar]
- 10.Chang Z, et al. The barbamide biosynthetic gene cluster: A novel marine cyanobacterial system of mixed polyketide synthase (PKS)-non-ribosomal peptide synthetase (NRPS) origin involving an unusual trichloroleucyl starter unit. Gene. 2002;296(1-2):235–247. doi: 10.1016/s0378-1119(02)00860-0. [DOI] [PubMed] [Google Scholar]
- 11.Edwards DJ, et al. Structure and biosynthesis of the jamaicamides, new mixed polyketide-peptide neurotoxins from the marine cyanobacterium Lyngbya majuscula. Chem Biol. 2004;11(6):817–833. doi: 10.1016/j.chembiol.2004.03.030. [DOI] [PubMed] [Google Scholar]
- 12.Chang Z, et al. Biosynthetic pathway and gene cluster analysis of curacin A, an antitubulin natural product from the tropical marine cyanobacterium Lyngbya majuscula. J Nat Prod. 2004;67(8):1356–1367. doi: 10.1021/np0499261. [DOI] [PubMed] [Google Scholar]
- 13.Edwards DJ, Gerwick WH. Lyngbyatoxin biosynthesis: Sequence of biosynthetic gene cluster and identification of a novel aromatic prenyltransferase. J Am Chem Soc. 2004;126(37):11432–11433. doi: 10.1021/ja047876g. [DOI] [PubMed] [Google Scholar]
- 14.Vestola J, et al. Hassallidins, antifungal glycolipopeptides, are widespread among cyanobacteria and are the end-product of a nonribosomal pathway. Proc Natl Acad Sci USA. 2014;111(18):E1909–E1917. doi: 10.1073/pnas.1320913111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mareš J, Hájek J, Urajová P, Kopecký J, Hrouzek P. A hybrid non-ribosomal peptide/polyketide synthetase containing fatty-acyl ligase (FAAL) synthesizes the β-amino fatty acid lipopeptides puwainaphycins in the Cyanobacterium Cylindrospermum alatosporum. PLoS One. 2014;9(11):e111904. doi: 10.1371/journal.pone.0111904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jokela J, et al. Anabaenolysins, novel cytolytic lipopeptides from benthic Anabaena cyanobacteria. PLoS One. 2012;7(7):e41222. doi: 10.1371/journal.pone.0041222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oftedal L, et al. The lipopeptide toxins anabaenolysin A and B target biological membranes in a cholesterol-dependent manner. Biochim Biophys Acta. 2012;1818(12):3000–3009. doi: 10.1016/j.bbamem.2012.07.015. [DOI] [PubMed] [Google Scholar]
- 18.Kajiyama S, Kanzaki H, Kawazu K, Kobayashi A. Nostofungicidine, an antifungal lipopeptide from the field-grown terrestrial blue-green alga Nostoc commune. Tetrahedron Lett. 1998;39:3737–3740. [Google Scholar]
- 19.Frankmölle WP, et al. Antifungal cyclic peptides from the terrestrial blue-green alga Anabaena laxa. I. Isolation and biological properties. J Antibiot (Tokyo) 1992a;45(9):1451–1457. doi: 10.7164/antibiotics.45.1451. [DOI] [PubMed] [Google Scholar]
- 20.Frankmölle WP, Knübel G, Moore RE, Patterson GM. Antifungal cyclic peptides from the terrestrial blue-green alga Anabaena laxa. II. Structures of laxaphycins A, B, D and E. J Antibiot (Tokyo) 1992b;45(9):1458–1466. doi: 10.7164/antibiotics.45.1458. [DOI] [PubMed] [Google Scholar]
- 21.Bonnard I, et al. Total structure and inhibition of tumor cell proliferation of laxaphycins. J Med Chem. 2007;50(6):1266–1279. doi: 10.1021/jm061307x. [DOI] [PubMed] [Google Scholar]
- 22.MacMillan JB, Ernst-Russell MA, de Ropp JS, Molinski TF. Lobocyclamides A-C, lipopeptides from a cryptic cyanobacterial mat containing Lyngbya confervoides. J Org Chem. 2002;67(23):8210–8215. doi: 10.1021/jo0261909. [DOI] [PubMed] [Google Scholar]
- 23.Neuhof T, et al. Hassallidin A, a glycosylated lipopeptide with antifungal activity from the cyanobacterium Hassalia sp. J Nat Prod. 2005;68(5):695–700. doi: 10.1021/np049671r. [DOI] [PubMed] [Google Scholar]
- 24.Neuhof T, Schmieder P, Seibold M, Preussel K, von Döhren H. Hassallidin B--second antifungal member of the hassallidin family. Bioorg Med Chem Lett. 2006;16(16):4220–4222. doi: 10.1016/j.bmcl.2006.05.094. [DOI] [PubMed] [Google Scholar]
- 25.Leão PN, et al. Synergistic allelochemicals from a freshwater cyanobacterium. Proc Natl Acad Sci USA. 2010;107(25):11183–11188. doi: 10.1073/pnas.0914343107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Challis GL, Hopwood DA. Synergy and contingency as driving forces for the evolution of multiple secondary metabolite production by Streptomyces species. Proc Natl Acad Sci USA. 2003;100(Suppl 2):14555–14561. doi: 10.1073/pnas.1934677100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.López CA, de Vries AH, Marrink SJ. Molecular mechanism of cyclodextrin mediated cholesterol extraction. PLOS Comput Biol. 2011;7(3):e1002020. doi: 10.1371/journal.pcbi.1002020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cartledge JD, Midgely J, Gazzard BG. Itraconazole solution: Higher serum drug concentrations and better clinical response rates than the capsule formulation in acquired immunodeficiency syndrome patients with candidosis. J Clin Pathol. 1997;50(6):477–480. doi: 10.1136/jcp.50.6.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Entzeroth M, Moore RE, Niemczura WP, Patterson GML. O-Acetyl-O- butyryl-O- carbamoyl-O, O -dimethyl-α-cyclodextrins from the Cyanophyte Tolypothrix byssoidea. J Org Chem. 1986;51:5307–5310. [Google Scholar]
- 30.Stratmann K, Burgoyne DL, Moore RE, Patterson GML. Hapalosin, a cyanobacterial cyclic depsipeptide with multidrug-resistance reversing activity. J Org Chem. 1994;59:7219–7226. [Google Scholar]
- 31.Murakami M, et al. Aeruginosins 98-A and B, trypsin inhibitors from the blue-green alga Microcystis aeruginosa (NIES-98) Tetrahedron Lett. 1995;36:2785–2788. [Google Scholar]
- 32.Fujii K, et al. Comparative study of toxic and non-toxic cyanobacterial products: A novel glycoside, suomilide, from non-toxic Nodularia spumigena HKVV. Tetrahedron Lett. 1997;38:5529–5532. [Google Scholar]
- 33.Lyra C, Laamanen M, Lehtimäki JM, Surakka A, Sivonen K. Benthic cyanobacteria of the genus Nodularia are non-toxic, without gas vacuoles, able to glide and genetically more diverse than planktonic Nodularia. Int J Syst Evol Microbiol. 2005;55(Pt 2):555–568. doi: 10.1099/ijs.0.63288-0. [DOI] [PubMed] [Google Scholar]
- 34.Halinen K, et al. Genetic diversity in strains of the genus Anabaena isolated from planktonic and benthic habitats of the Gulf of Finland (Baltic Sea) FEMS Microbiol Ecol. 2008;64(2):199–208. doi: 10.1111/j.1574-6941.2008.00461.x. [DOI] [PubMed] [Google Scholar]
- 35.Wouters J, Bergman B, Janson S. Cloning and expression of a putative cyclodextrin glucosyltransferase from the symbiotically competent cyanobacterium Nostoc sp. PCC 9229. FEMS Microbiol Lett. 2003;219(2):181–185. doi: 10.1016/S0378-1097(02)01204-1. [DOI] [PubMed] [Google Scholar]
- 36.Méjean A, Ploux O. A genomic view of secondary metabolite production in cyanobacteria. In: Chauvat F, Cassier-Chauvat C, editors. Advances in Botanical Research: Genomics of Cyanobacteria. Vol 65. Academic; San Diego: 2013. pp. 189–234. [Google Scholar]
- 37.Rausch C, Hoof I, Weber T, Wohlleben W, Huson DH. Phylogenetic analysis of condensation domains in NRPS sheds light on their functional evolution. BMC Evol Biol. 2007;7:78. doi: 10.1186/1471-2148-7-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kraas FI, Helmetag V, Wittmann M, Strieker M, Marahiel MA. Functional dissection of surfactin synthetase initiation module reveals insights into the mechanism of lipoinitiation. Chem Biol. 2010;17(8):872–880. doi: 10.1016/j.chembiol.2010.06.015. [DOI] [PubMed] [Google Scholar]
- 39.Qian CD, et al. Identification and functional analysis of gene cluster involvement in biosynthesis of the cyclic lipopeptide antibiotic pelgipeptin produced by Paenibacillus elgii. BMC Microbiol. 2012;12:197. doi: 10.1186/1471-2180-12-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wittmann M, Linne U, Pohlmann V, Marahiel MA. Role of DptE and DptF in the lipidation reaction of daptomycin. FEBS J. 2008;275(21):5343–5354. doi: 10.1111/j.1742-4658.2008.06664.x. [DOI] [PubMed] [Google Scholar]
- 41.Rouhiainen L, et al. Genes encoding synthetases of cyclic depsipeptides, anabaenopeptilides, in Anabaena strain 90. Mol Microbiol. 2000;37(1):156–167. doi: 10.1046/j.1365-2958.2000.01982.x. [DOI] [PubMed] [Google Scholar]
- 42.Lee BH, Oh DK, Yoo SH. Characterization of 4-alpha-glucanotransferase from Synechocystis sp. PCC 6803 and its application to various corn starches. N Biotechnol. 2009;26(1-2):29–36. doi: 10.1016/j.nbt.2009.06.981. [DOI] [PubMed] [Google Scholar]
- 43.Stam MR, Danchin EG, Rancurel C, Coutinho PM, Henrissat B. Dividing the large glycoside hydrolase family 13 into subfamilies: Towards improved functional annotations of alpha-amylase-related proteins. Protein Eng Des Sel. 2006;19(12):555–562. doi: 10.1093/protein/gzl044. [DOI] [PubMed] [Google Scholar]
- 44.Kim JH, et al. One-pot synthesis of cycloamyloses from sucrose by dual enzyme treatment: Combined reaction of amylosucrase and 4-α-glucanotransferase. J Agric Food Chem. 2011;59(9):5044–5051. doi: 10.1021/jf2002238. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.