Significance
The human proton channel hHv1 is responsible for the extrusion protons during a series of important physiological events with clinical relevance. Yet, the mechanistic details underlying voltage-dependent gating, ion selectivity, and cooperativity between homodimers remain to be determined. We carried out a structural characterization of hHv1 in a lipid bilayer using spectroscopic methods and revealed a strongly focused electric field across its putative selectivity filter. In combination with computational methods, we showed that the recently determined crystal structure of CiVSD-WT is the best structural template for dimeric hHv1, and constructed a full-length model of dimeric hHv1 with spectroscopic constraints. We proposed an explicit mechanism for hHv1 gating with functional predictions according to the one-click model recently established in CiVSD.
Keywords: proton channel, voltage sensing domain, EPR, MD simulation
Abstract
The voltage-gated proton channel Hv1 plays a critical role in the fast proton translocation that underlies a wide range of physiological functions, including the phagocytic respiratory burst, sperm motility, apoptosis, and metastatic cancer. Both voltage activation and proton conduction are carried out by a voltage-sensing domain (VSD) with strong similarity to canonical VSDs in voltage-dependent cation channels and enzymes. We set out to determine the structural properties of membrane-reconstituted human proton channel (hHv1) in its resting conformation using electron paramagnetic resonance spectroscopy together with biochemical and computational methods. We evaluated existing structural templates and generated a spectroscopically constrained model of the hHv1 dimer based on the Ci-VSD structure at resting state. Mapped accessibility data revealed deep water penetration through hHv1, suggesting a highly focused electric field, comprising two turns of helix along the fourth transmembrane segment. This region likely contains the H+ selectivity filter and the conduction pore. Our 3D model offers plausible explanations for existing electrophysiological and biochemical data, offering an explicit mechanism for voltage activation based on a one-click sliding helix conformational rearrangement.
Voltage-gated proton channels (hHv1) represent a remarkable evolutionary adaptation of canonical voltage sensing domain (VSD). In hHv1, both voltage-sensing and ion (H+) conduction are carried out by a single domain, and undergo a global conformational rearrangement (1, 2). In humans, hHv1 is widely expressed and required for a variety of physiological processes (3), including optimal reactive oxygen species production by NADPH oxidase (4–6), B-cell proliferation and differentiation (7), and regulation of human sperm motility (8). hHv1 folds as an antiparallel four-helix bundle with its fourth transmembrane segment (S4) containing three putative gating charges. Just as in the VSDs from ion channels and enzymes, the positively charged S4 moves outwardly in response to a depolarization (9). Beyond sensing the transmembrane voltage, S4 reorientation in hHv1 participates in the formation of a proton-selective permeation pathway responsible for the generation of the proton currents that underlie its multiple physiological functions (10, 11).
Despite high sequence similarity and a common structural blueprint, VSDs from ion channels and enzymes display a wide range of electrophysiological properties; these include large variations in effective gating charge (z = ∼0.9 to ∼3.6 e−) and midpoints of activation (V1/2 = ∼−150 to ∼+150 mV). Existing VSD crystal structures (12–16) have provided key data on the arrangement and orientation of the transmembrane helices S1–S4, while at the same time revealed a considerable structural heterogeneity—particularly, the number and positions of both the gating charges and their compensating countercharges in relation to the hydrophobic “plug” or “gasket” electrically separating the intra- and extracellular spaces (16, 17).
Electrophysiological studies on the human proton channel (hHv1) have identified several functionally critical residues (18, 19) and suggested a list of potential charge–charge interactions (20–22) for different functional states. A number of hHv1 molecular models (19, 20, 23, 24) have been proposed using VSD structures from KvAP and the Kv1.2 chimera as templates. However, no overall consensus exists so far, in part due to the structural heterogeneity among voltage sensors. A recent 3.45-Å resolution crystal structure of a chimeric construct of the mouse proton channel mHv-cc, containing parts of mHv, CiVSD (VSD of voltage sensitive phosphatase from Ciona intestinalis, CiVSP), and a fragment of the GCN4 coiled coil (25), has provided the first glimpses of a proton channel structure, including its close agreement with a consensus VSD scaffold and the relative positions of functionally critical residues. However, due to the unorthodox nature of the chimeric construct crystallization (it forms a trimer in the lattice), details on the individual role of the gating charges, plus the mechanisms of proton permeation and cooperative gating, remain to be established (25).
Ultimately, a complete understanding of the mechanistic principles that define hHv1 voltage activation and proton conduction requires the determination of its conformational states in an environment that is as close as possible to native conditions. Here, we have carried out a systematic EPR spectroscopic analysis of membrane-embedded hHv1, together with biochemical and computational approaches to generate a structural model that accounts for most of the existing functional data (19, 20). hHv1 showed significant solvent penetration along S4, suggesting a highly focused electric field in the region near the first two gating charges, R1 and R2, an observation that explicitly narrows the potential location of the selectivity filter and the pore of the proton channel. Evaluation of existing VSD structures supports our hypothesis that the CiVSD-WT structure (16) represents the best structural template for 3D models that are optimally compatible with existing hHv1 experimental data, including its dimer interface. This model not only provides mechanistic insights to interpret the chimeric construction of recent mHv-cc model, but also explicitly suggests a path toward defining hHv1 open state.
Results
Biochemical Analyses of Human Proton Channel.
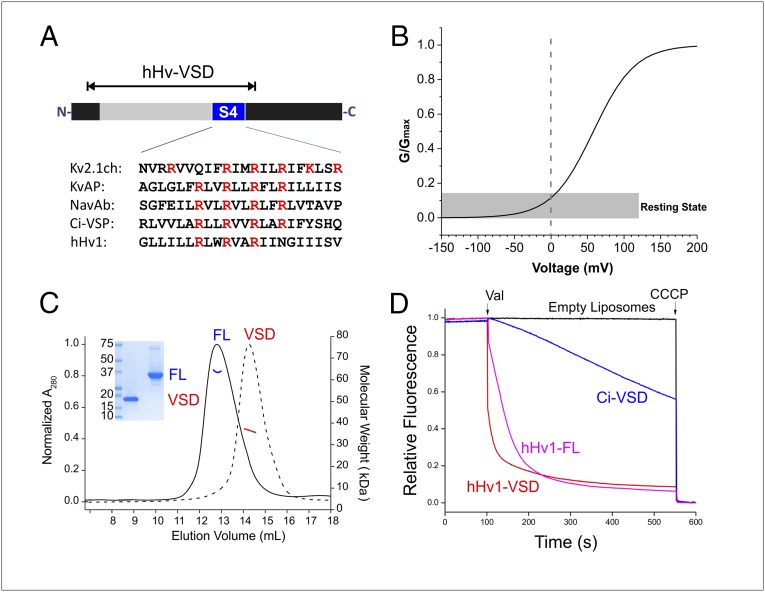
Full length-hHv1 (hHv1-FL; including residues 1–273) and a VSD only construct (hHv1-VSD; residues 75–223; Fig. 1A) can be expressed in Escherichia coli at moderate yields (0.7 mg/L) at the expected molecular weights, as confirmed by electrospray ionization (ESI)-TOF mass spectrometry. Both hHv1-FL and hHv1-VSD are homogenous and stable after solubilization in Fos-choline 14 and Fos-choline 12 (Fig. 1C). Purified hHv1 displays high helical contents in solution (helicity 75% for hHv1-FL, 82% for hHv1-VSD; SI Appendix, Fig. S1A), which indicate proper secondary structure, consistent with the expected four-helical bundle scaffold. According to its voltage dependence (Fig. 1B; V1/2 = 58 mV and z = 0.90) (19), hHv1 populates its resting or “down” state at 0 mV. After reconstitution into 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC):1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1′-rac-glycerol) (POPG) at molar ratio 3:1 (26–28), both the hHv1-FL and hHv1-VSD constructs efficiently catalyze proton fluxes determined by an established fluorescence-based assay (29) (Fig. 1D), which is consistent with previous electrophysiological data showing that hHv1-VSD alone serves as the pore and selectivity filter for proton conduction (10). In comparison, reconstituted Ci-VSD conducts protons at a much slower rate (Fig. 1D), whereas KvAP-VSD can only sustain a marginal level of proton conduction, as shown previously (29).
Fig. 1.
Biochemical preparation of hHv1 protein. (A) Schematic presentation of hHv1 constructs. Voltage-sensing domain (gray) and the fourth transmembrane segment (blue) were highlighted on full-length construct. The hHv1-VSD construct, which covered the entire VSD, was used as a template for cysteine scanning and EPR studies. (B) Simulated G–V curve of hHv1 with V1/2 = 58 mV and z = 0.90 (19). At 0 mV, hHv1 populates at the resting state. (C) Characterization of hHv1 full-length (FL) and hHv1-VSD (VSD) by SDS/PAGE gel, SEC (FL, solid black line; VSD, dotted black line), and MALS (FL, blue solid line; VSD, red solid line) methods. Expected molecular mass is 34 kDa for hHv1-FL and 19 kDa for hHv1-VSD. Both hHv1-FL and hHv1-VSD are dimers in the buffer of 20 mM Tris (pH 8.0), 150 mM NaCl, and 4 mM Fos-choline 12 at studied concentration (∼40 μM). (D) ACMA-based flux assay for reconstituted liposome. The truncated hHv1-VSD (red) conducts the proton as well as the full-length hHv1-FL (magenta). CiVSD (blue) has a small leakage of proton current.
Dynamics and Helical Packing in a Lipid Environment.
We used site-directed spin labeling (SDSL) in combination with EPR spectroscopy to probe the local environment of hHv1-VSD in POPC:POPG liposomes. The single native cysteine in the construct (C107) was mutated into serine to generate a cysteineless hHv1-VSD. A total of 149 single cysteine mutants, covering the entire VSD scaffold in hHv1-VSD background, were expressed, purified, spin-labeled, and reconstituted in POPC:POPG liposomes. Continuous-wave EPR spectra were acquired for individual samples (SI Appendix, Figs. S2 and S3), and spectra were analyzed using established methods (30). We focused on two types of EPR spectra parameters to deduce structural information: first, nitroxide motional freedom was estimated from the inverse of the central line width of the first derivative absorption spectra (ΔHo−1) (31). Second, solvent accessibility was evaluated by collisional relaxation methods (32), where nonpolar oxygen (ΠO2) serves as a contrast agent to evaluate membrane exposure, and polar Ni(II) ethylenediaminediacetic acid (NiEdda) reports on the extent of aqueous exposure.
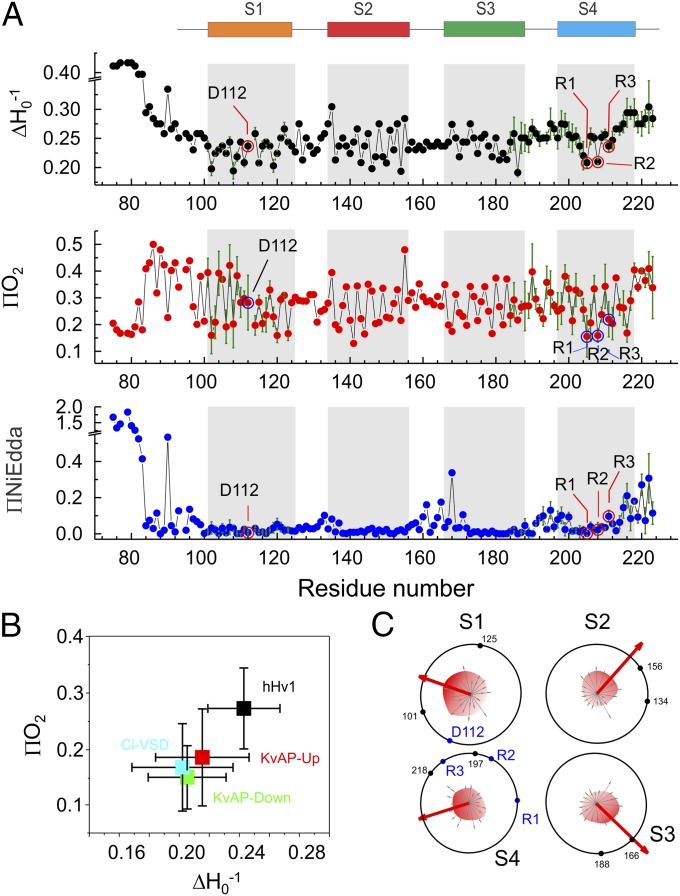
Fig. 2A shows the EPR environmental parameter profile for hHv1-VSD, highlighting the distinct dynamic features of hHv1. Indeed, among all VSDs studied by EPR spectroscopy (16, 26–28), hHv1 is by far the most dynamic one (Fig. 2B). Both the N- and C-terminal regions extending outside of four-helical bundle are significantly more mobile than the transmembrane regions. We find clear periodicity in the accessibility data along the transmembrane regions, where the three putative gating charges (R205: R1; R208: R2; and R211: R3) appear at local minima for ΔHo−1 (red circles in Fig. 2A, Top), consistent with an expected tertiary contact. The ΠO2 profile (Fig. 2A, Middle) similarly reflects the dynamic nature of hHV1 in membranes: high in lipid-exposed residues but low in residues with tertiary contacts. Two-dimensional projections of ΠO2 for the four individual helices clearly points to the overall direction of tertiary contacts (Fig. 2C). Within this projection, functionally critical residues such as the three putative gating arginines and D112 (18) appear naturally inside the bundle (Fig. 2C), fully consistent with the notion that these charged residues are protected away from the hydrophobic environment of the lipid bilayer.
Fig. 2.
Environmental parameters of hHv1 in lipids. (A) Mobility (∆Ho−1), oxygen accessibility (ΠO2), and NiEdda accessibility (ΠNi) of hHv1-VSD in POPC:POPG proteoliposome. Repeats for selected positions (n = 3–5) in S1 and S4 are shown. Error bars, SD. The putative transmembrane segments (S1, S2, S3, and S4) are shown on the top and indicated in gray in the plot. The critical residues D112, R205 (R1), R208 (R2), and R211 (R3) are indicated with circles. (B) Average mobility and oxygen accessibility of four transmembrane helices of four VSD systems hHv1-VSD (black), CiVSD (cyan), KvAP-Up (red), and KvAP-Down (green), which are studied by EPR methods, are plotted with SD. hHv1-VSD is the most dynamic VSD system in lipids by far. (C) A 2D wheel presentation of oxygen accessibility of four transmembrane helices. The vectorial sums (red arrow) of oxygen accessibility are significantly larger than values of individual residues, and preferably point to the lipid-exposing environment away from the core of the four-helix bundle. The functionally critical residues (D112, R1, R2, and R3) are on the opposite side of the lipid environment and protected by side-chain interactions. Boundaries of each transmembrane segment are shown by black dot with residue numbers for reference.
Finally, NiEdda accessibility between transmembrane regions (low ΠNiEdda) and the exposed loops (high ΠNiEdda) unambiguously define the boundaries of the four individual helices (S1–S4; gray regions of Fig. 2A, Bottom). Together with the amino acid sequence, the ΠNiEdda profile is able to explicitly define the boundary of the transmembrane helices and the position of the residues relative to the lipid bilayer as in homologous systems KvAP-VSD and CiVSD (SI Appendix, Fig. S4) (12, 16, 28). On the basis of this dataset, the gating charges (R1, R2, and R3) appear located toward the inner leaflet of the bilayer, reminiscent of the location of the gating charges in CiVSD at resting state, but in strong contrast to KvAP-VSD. More importantly, we observe deep water penetration at both ends of the S4 segments. The present data highlights the existence of deep water-filled crevasses in hHv1 and suggests that only a very narrow region of the sensor (about two turns of helix, between residues 201 and 209) remains fully inaccessible to NiEdda in the resting (down) conformation. This region includes the top two gating charges, R1 and R2, leaving R3 at least partially exposed to intracellular crevasse and thus, accessible to interactions with water.
A Spectroscopically Constrained Model of the hHv1 VSD.
We have taken advantage of the present EPR data set to evaluate the 3D structure of resting (down) hHv1 in a lipid bilayer. We have carried out this exercise independent of the recently reported X-ray structure of the mHv1/CiVSD/GNC4 chimera. Due to the common overall architecture of VSDs, structural models for hHv1 have been constructed based on existing structures from either KvAP or Kv1.2 VSDs (3, 6, 20, 24). We point out that given local sequence similarity, the recently determined VSD from Ci-VSP (CiVSD) represents a more logical and accurate structural template for hHv1 (16).
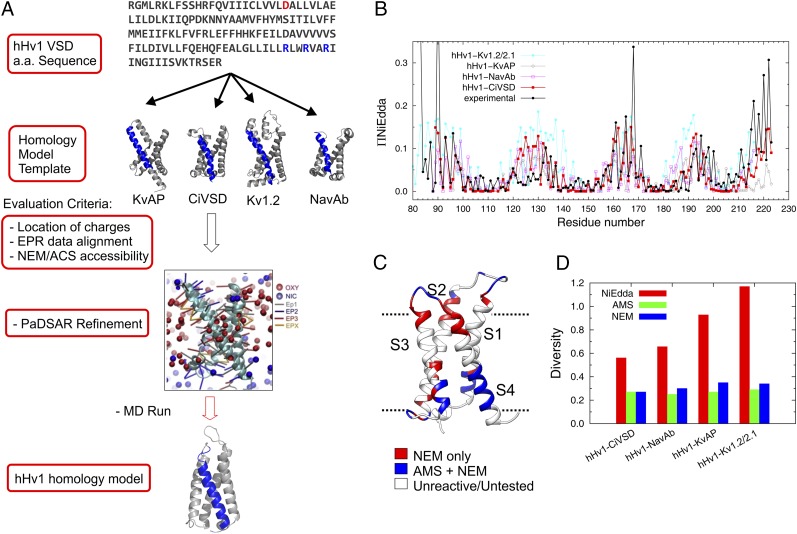
We carried out an unbiased evaluation of all available VSD structures as a way to generate more appropriate models of hHv1 at resting state. To this end, we constructed four structural models based on pairwise amino acid sequence alignments of hHv1-VSD with sequences from four existing VSD structures: KvAP (12), Kv1.2_ch (13), NavAb (14), and CiVSD-WT (16) (Fig. 3A and SI Appendix, Fig. S5; see Experimental Procedures for details). Based on each of the four preliminary models in explicit lipid environments, we calculated the predicted accessibility for each residue using molecular dynamic simulation methods and applied Kullback–Leibler divergence (33) to compare the predicted patterns with both EPR-based NiEdda accessibility (current data; Fig. 3B) and NEM/AMS chemical reactivity accessibility (Fig. 3C) (34). Models based on CiVSD-WT as a template display the best agreement with both NEM/AMS reactivity and ΠNiEdda. It is worth noting that the ΠNiEdda profile is a more complete and quantitative dataset than that derived from NEM/AMS reactivity, and thus more sensitive to structural models with larger differences in Kullback–Leibler divergences (Fig. 3D). Not surprisingly, CiVSD-WT is indeed most similar to hHv1 not only in sequence homology (SI Appendix, Fig. S5, Bottom), but also in their voltage dependences and apparent slopes of activation (1, 35). Our initial CiVSD-WT–based structural model was further refined by EPR constraints using a pseudoatom-driven solvent accessibility refinement method PaDSAR (26, 36) that uses experimental solvent accessibilities as soft constraints. The resulting model was equilibrated in explicit lipid POPC for 400 ns and proved to be stable as a representation of the resting state of hHv1-VSD in lipid bilayer.
Fig. 3.
Structural modeling for hHv1-VSD at resting state. (A) Strategy of structural modeling by MD simulation. Starting from the sequence alignments, four structural models of hHv1-VSD were built on the templates of crystal structures of KvAP, CiVSD-WT, Kv1.2_chimera, and NavAb respectively. Each of the structural models was equilibrated with explicit lipid POPC and stabilized for 300 ns by MD simulation. The four MD stable models were evaluated with experimental data primarily with ΠNiEdda and NEM/AMS accessibility data. The structural model correlating best with experimental accessibilities was subjected to an additional MD simulation with constraints from present EPR spectroscopic data using an established computational method, PaDSAR (36). The constrained model was equilibrated in explicit lipid POPC for 400 ns, which serves our working model for the resting state of hHv1-VSD. (B) Predicted solvent accessibility from four structural models and the experimental NiEdda accessibility. (C) NEM/AMS accessibility data were mapped onto hHv1-VSD structural model from CiVSD-WT. (D) The correlation between predicted accessibilities and experimental accessibilities for each of the four hHv1-VSD structural models was evaluated by Kullback–Leibler divergence. The structural model from CiVSD-WT with lowest divergence was chosen to further construct the structural model for hHv1-VSD.
Mapped Environmental Parameters: Solvent Penetration on S4.
We then mapped the EPR environmental data onto our equilibrated model of hHv1-VSD to evaluate its basic structural features. The mapped nitroxide probe mobility shows the highly dynamic features of hHv1-VSD (Fig. 4C). Among the four transmembrane segments, S4 appears to be the most dynamic one (Fig. 4C, Left). The most restricted regions (in blue) are located around the middle of hHv1-VSD and scattered on S1, S2, and S3. The oxygen accessibility profile (ΠO2) confirms that the sites with low accessibilities are packed in the interior of the four-helix bundle (Fig. 4D), suggesting the agreement between the present hHv1 model and experimental data.
Fig. 4.
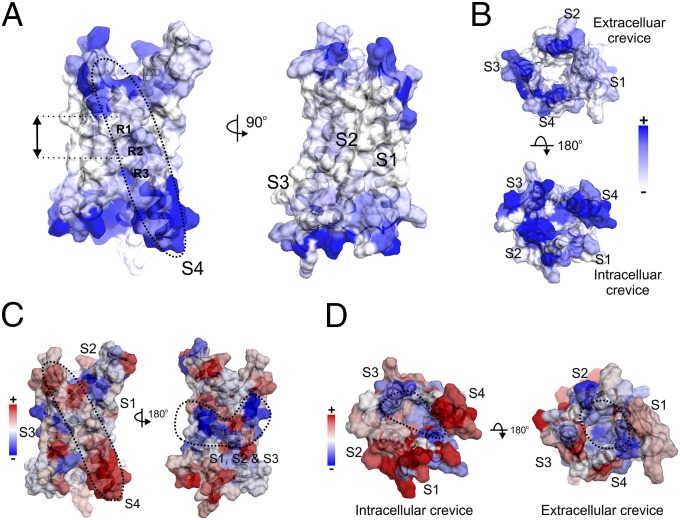
Mapping the EPR environmental parameters on the hHv1-VSD model. (A) ΠNiEdda data were mapped onto hHv1-VSD structural model. Apparent ΠNiEdda were observed along S4 segment (Left, dotted circle) and two turns of helix around the first two gating charges R1 and R2 (Left, black arrow), together with all other transmembrane regions in S1, S2, and S3 (Right), have minimum ΠNiEdda. (B) Both extracellular and intracellular crevices have significant ΠNiEdda. There is apparent solvent penetration through the intracellular crevice, but not obvious in the extracellular crevice. The scale bar is the same for both A and B. (C) The mapped mobility shows the S4 segment is most mobile (Left, dotted circle) among all transmembrane segments. There is a cluster of motion-restriction residues in the middle of transmembrane segments S1, S2, and S3 (Right, dotted circle); they are in the same vertical position around R1 and R2 with minimum ΠNiEdda. (D) The mapped oxygen accessibilities shows the regions with lowest oxygen accessibility are located roughly inside the four-helix bundle away from the lipid environment.
The key feature in the present model becomes evident from the mapped solvent accessibility (ΠNiEdda). As seen from the ΠNiEdda profile (Fig. 2A), we find extensive solvent penetration from both extracellular and intracellular sides of hHv1, particularly along S4 (Fig. 4A, Left). The observed low-oxygen accessibilities around both intracellular and extracellular crevices (Fig. 4D) were consistent with this solvent penetration, because oxygen contents are lower in solvent than in lipid environment. At the same time, there is minimal solvent penetration on the central regions of S1 and S2 (Fig. 4A, Right). Based on the present data and 3D model, the water penetration from both sides of the membrane appears to ebb around R1 and R2 in hHv1, leaving a very narrow region of putatively low dielectric environment and, thus, locally focusing the electric field surrounding the gating charges.
The hHv1 Dimer Interface.
In solution, hHv1-VSD is a dimer at ∼40 µM as shown by the multiple-angle light-scattering (MALS; Fig. 1C). To evaluate the strength of the dimer interface, we labeled hHv1-VSD-193C with fluorescein maleimide and monitored its size-exclusion elution profile by fluorescence at low concentrations. At ∼40 µM, the elution profile of fluorescein-labeled hHv1-VSD-193 is the same as native hHv1-VSD dimer. However, serial dilutions to 200 nM show clear concentration dependence of dimer dissociation, evident from elution volume shifts toward lower molecular weights (SI Appendix, Fig. S6A). Protein concentration estimates from the predicted extinction coefficient (A280) at the peak of elution profiles and the dilution factors, we calculated an apparent Kd of 3.2 µM for hHv1-VSD dimerization in solution (SI Appendix, Fig. S6A).
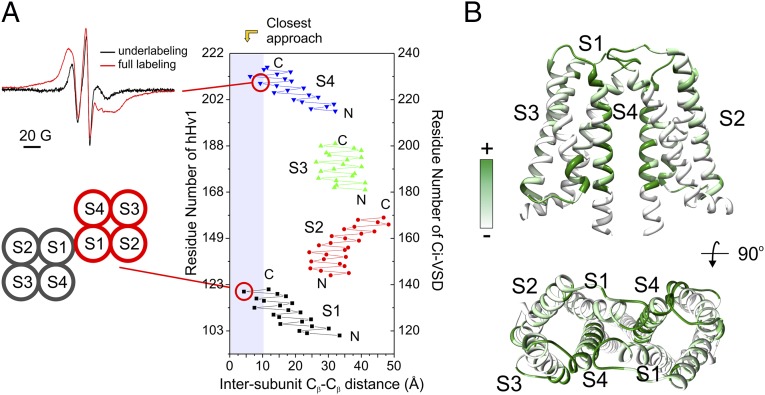
As a result of the saturation spin-labeling of hHv1-VSD for EPR studies, we observed strong spin–spin coupling at the C-terminal end of S4 (residue 206; Fig. 5A), which unambiguously shows that hHv1-VSD is oligomeric under the present experimental conditions. Because there is only one spin label attached on each hHv1 monomer, position 206 has to be close to each other at the oligomer interface. As expected, mixing spin-labeled hHv1-VSD-206 with unlabeled protein at a 1:3 molar ratio before reconstitution sharply diminished the dipolar coupling (Fig. 5A). Using the dipolar coupling as readout for dimer formation, we reconstituted the spin-labeled hHv1-VSD-206 at different starting concentrations, ranging from 360 nM to 360 µM. Consistent with dilution experiments in detergent solution, the apparent Kd in proteoliposome was shown to be 3.4 µM from fits of the concentration dependence of dipolar coupling (SI Appendix, Fig. S6B), which strongly suggests that the dimerization of hHv1-VSD in proteoliposomes is a direct reflection of its state in solution before the reconstitution and confirms that at a 1:1,000 protein:lipid reconstitution ratio (that of all our EPR spectroscopy and proton flux measurements), hHv1-VSD is a dimer in the lipid bilayer. The observed dipolar coupling is also consistent with FRET data that hHv1-VSD is an oligomer when reconstituted at a 1:1,000 protein:lipid molar ratio in a variety of lipid compositions (SI Appendix, Fig. S1B).
Fig. 5.
The hHv1-VSD dimer interface agrees with CiVSD-WT dimer structure. (A) (Upper Left) Strong dipolar coupling from spin-labeled hHv1-VSD-206C. (Lower Left) Mutant 121C at top of S1 can form spontaneous disulfide linkage in native hHv1 (38). (Right) Cβ distances between hHv1 monomer according to the CiVSD-WT dimer (PDB ID code 4G80). Both residues 121 and 206 are among the closest residues (<10 Å). (B) hHv1-VSD dimer model built by aligning two monomer structural models with CiVSD-WT dimer crystal structure. The cross-linking data from mHv was mapped onto the dimer model (40), and regions with high cross-linking propensities, including S4 and top of S1, are in close proximity in the dimer model.
It is worth noting, however, that the C-terminal truncated hHv1-VSD has been shown by single-molecule fluorescence bleaching methods (10) to be monomeric when expressed in Xenopus oocytes. We suggest a simple explanation to the apparent discrepancy with the present set of results. Stoichiometry counting requires a minimal level of expression to easily resolve single spots on the cell surface. It is likely that these low expression levels in oocytes fail to reach the concentration threshold for dimer formation, which is in the low-micron range for hHv1-VSD in solution as shown in Fig. 4A. For comparison purposes, we reconstituted fluorescein-labeled hHv1-VSD-193C protein at a protein-to-lipid ratio of ∼1:1 million and applied the single-molecule bleaching methods to determine the stoichiometry. We were able to obtain evenly distributed hHv1-VSD spots (SI Appendix, Fig. S1C, Top), where hHv1-VSD was predominantly monomeric (70% single-step bleaching; SI Appendix, Fig. S1C). However, at the 2D densities required to reach an equivalent low-micron concentration, it is practically impossible to separate single molecules, and thus we were unable to properly carry out the photobleaching experiment. These results are further evidence showing that the oligomeric state of hHv1-VSD is highly dependent on its density in the membrane.
The C-terminal coiled coil of hHv1 has been crystallized as a dimer with a covalent disulfide bond between 249C (37). We set out to test whether this covalent linkage is relevant in the formation of the native hHv1-FL dimer. Mild oxidizing conditions (5 mM CuSO4 for 10 min at room temperature) led to the formation of >50% hHv1-FL covalent dimer as shown in nonreducing SDS/PAGE (SI Appendix, Fig. S16C). Mutating the two hHv1-FL native cysteines (107C and 249C) individually into serine had differential effects on dimer formation: the C107S mutant is predominantly (>90%) dimer, whereas the C249S mutant shows only trace amounts (<5%), confirming that 249C is responsible for the covalent linkage of hHv1-FL. Higher-order oligomers can only be seen in the present of both cysteines in hHv1-FL (SI Appendix, Fig. S1C).
An hHv1 Dimer Model in the Resting Conformation.
Extensive cross-linking results have proposed two different dimer interfaces for hHv1-VSD through either S1 (38) or S4 (39). CiVSD-WT, the closest structural template of hHv1, was crystallized as a homodimer in the asymmetric unit (16) and analysis of Cβ distances between CiVSD-WT monomers shows that the dimer interface is formed by both S1 and S4, with the closest proximity at the bottom of S4 and top of S1. Indeed, the mutant 121C at the extracellular loop of S1 can form a spontaneous disulfide link (38) and shows shortest Cβ–Cβ distance in the equivalent position of the CiVSD dimer (Fig. 5A). At the same time, the observed dipolar coupling at position 206 in hHv1 (Fig. 5A) unambiguously places the bottom of S4 at the dimer interface. These data together strongly suggest that the dimer interface of hHv1-VSD is homologous to CiVSD-WT. Consequently, we aligned two structural models of hHv1-VSD monomers with the CiVSD-WT dimer as a template to generate the hHv1-VSD dimer model (Fig. 5B). Mapping all of the available cross-linking data (40) onto this dimer model shows that most of the residues with high cross-linking propensity are precisely located at the dimer interface predicted from CiVSD-WT.
Finally, to generate a dimer model of hHv1, we combined the present structural model of hHv1-VSD dimer (residues 89–226) and the crystal structure of hHv1 C-terminal dimer (residues 226–266) (37) by simple fusion. This full-length hHv1 dimer model was equilibrated for 300 ns by molecular dynamics (MD) simulation in a POPC lipid bilayer, applying secondary structure constraints to ensure the continuity of the α-helix (41) from S4 to the end of C-terminal coiled coil (40) (SI Appendix, Fig. S7A).
Discussion
Based on the present spectroscopic and biochemical data, we evaluated existing VSD structures to generate a structural model of full-length hHv1, using as templates the structures of CiVSD-WT (16) and isolated hHv1 C-terminal coiled coils (37). Based on the voltage dependence of hHv1 (1), both our dimer model and the trimeric mHv-cc structure (25) likely represent the resting state of hHv1 at 0 mV.
Mechanistic Interpretation.
One of the striking features from the present EPR data, compared with equivalent data from KvAP (26, 28, 42), NaChBac (43), and CiVSD (16, 27), is the deep solvent penetration along S4 (Fig. 4A). This water penetration is consistent with the large opening of the intracellular crevice in the mHv-cc trimer structure (25) and would lead, presumably, to a sharply focused electric field around the R1 and R2 and, indeed, a focused electric field has been observed, to different degrees, in several VSDs (44–46) and appears to be a key property of VSD at the resting state.
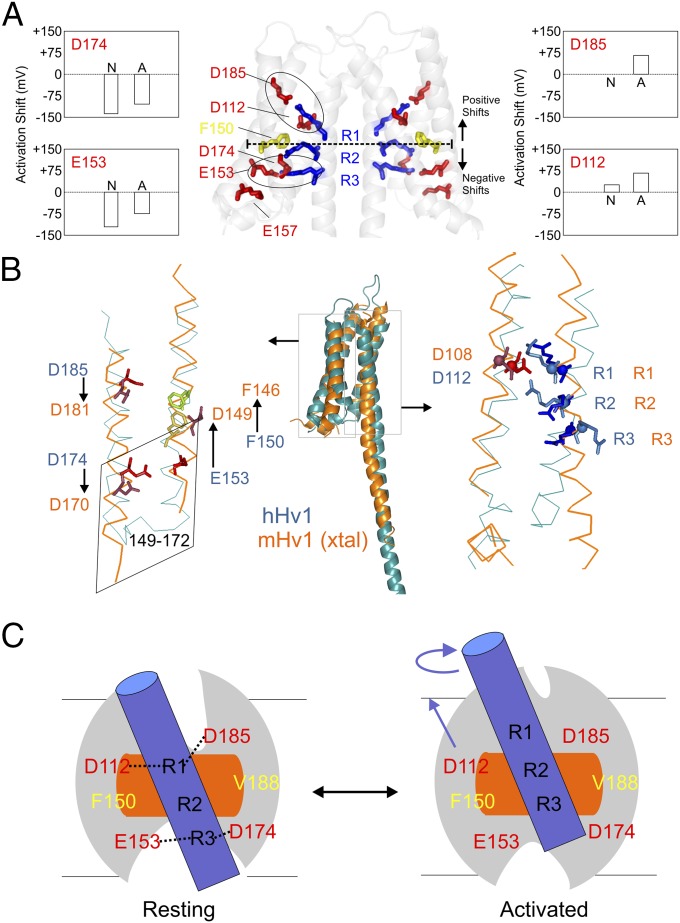
Interestingly, a highly conserved phenylalanine (F150 in hHv1) lies between R1 and R2 (Fig. 6A) where it is solvent excluded (Fig. 4A) and motionally restricted (Fig. 4C). It is thus likely that F150 serves as a barrier for proton permeation in its nonconductive resting state. As such, F150 might be putatively involved in the formation of the gating charge transfer center (17) or hydrophobic gasket (16) that separates intracellular and extracellular continuity with the electric field in canonical VSDs. If, as expected, the hHv1 gating mechanism is similar to that in existing VSDs (where S1, S2, and S3 remain relatively steady as the scaffold), the occluded region at rest should become permeable and selective to proton upon activation. Therefore, the present data points to a narrow solvent-occluded region that includes D112 (S1), F150 (S2), V178 (S3), R205 (S4), and R208 (S4) (Fig. 6 A and B) as the likely site for both activation gate and selectivity filter during proton conduction. In fact, D112 has previously been suggested to be a key component of the selectivity filter in hHv1 (18). Thus, the molecular basis of proton permeation is likely defined by relatively small side-chain reorientations in residues lining this narrow region. However, a definitive demonstration of this hypothesis will likely require further structural data at significantly higher resolutions than those presently available.
Fig. 6.
Model comparison and mechanistic implications. (A) Functionally critical residues in the hHv1-VSD dimer model. D174 and E153 are located below F150, the putative hydrophobic gasket, and D185 and D112 are located above. Neutralization of D174 and E153 causes left-shift in the G–V curve, whereas neutralization of D185 and D112 causes right-shift. The two electrostatic clusters D185-R1-D112 and E153-R3-D174 are highlighted with black circles. (B) Structural model of hHv1 was aligned with crystal structure of mHv-chimeric by the Cα of S1 + S2 + S3a. The residues on S1 (D112) and S4 (R1, R2, and R3) agreed very well (Right) with obvious differences of the residues on S2 and S3 between two models. To match mHv1-chimeric structure, the residues on S2 (D153 and F150) need to upshift one turn of helix, whereas the residues on S3 (D174 and D185) need to downshift one turn of helix. The chimeric region from CiVSD on mHv-chimera spanned from residues 149 to 172 (corresponding to residue 153–176 in hHv1), which are highlighted in the rhomboidal box. The structures of the identical sequence in CiVSD and mHv-chimera are distinctly different. (C) Proposed gating model of hHv1 according to the one-click gating mechanism in CiVSD. At resting-state hHv1-VSD in lipid bilayer, solvent penetrates from both intracellular and extracellular crevices leaving a focused electric field around R1 and R2 region at the level of hydrophobic gasket. hHv1 scaffold is shown by the gray oval; S4 by blue cylinder; hydrophobic gasket by orange rectangle. The two salt-bridge clusters (D112-R1-D183 and E153-R3-D174) are connected by black dotted lines. Up activation: the S4 of hHv1 rotates and moves three residues up.
The present structural model of hHv1 at rest explicitly suggests two electrostatic clusters, D112-R1-D185 and E153-R3-D174, as the basis of the energetic stabilization of the resting state. The predicted role of these clusters is in good agreement with existing electrophysiological data from charge mutants: neutralization of either D112 or D185 above the hydrophobic gasket dramatically right-shift the I–V curve (19), whereas neutralization of either D174 or E153 below hydrophobic gasket left-shifts the I–V curve (19) (Fig. 6A). In our model, K157 does not appear to be within electrostatic range of other charged residues, a prediction that seems to be supported by a lack of a functional effect for the K157A neutralization (19). Furthermore, given that both clusters are exposed to water, we suggest they might also play a role in hHv1 sensitivity to pH because the protonation state of these charges will have a profound effect on the stability of the electrostatic interactions in the cluster.
Model Comparison.
We have shown that the position of the gating charges in CiVSD-WT (toward the inner leaflet of the bilayer) is, by far, the lowest among VSDs with explicit crystal structures (16). However, previous hHv1 structural models have been based on structural templates from voltage-gated ion channels (19, 23, 24), all of which populate the activated (up) state at 0 mV, according to their voltage dependences. In comparison with the present structural model, early hHv1 homology models tend to place the three gating charges toward the extracellular leaflet of the bilayer; thus, they would (in principle) be structurally closer to the activated state of the hHv1.
A comparison between the present structural model and the mHv-cc structure was carried out by aligning the backbone Cα of S1 + S2 + S3a (Fig. 6B, S5) (16). We find excellent agreement regarding the position of the critical residues on S1 (D112) and S4 (R1, R2, R3) between our model and mHv-cc. In fact, mHv-cc arginines are in positional agreement only with CiVSD-WT (down conformation) and, as such, are located further toward the inner leaflet of the bilayer than in other existing VSD structures (16). However, though there is overall agreement in the location of S1 and S4, there are clear differences between the residues on S2 and S3 between trimeric mHv-cc and our hHv1 model (Fig. 6B). To correct this discrepancy, S2 residues in our model (D153 and F150) need to be shifted upwardly by one turn of helix, whereas the residues on S3 (D174 and D185) require a downward shift of one helical turn (Fig. 6B).
These apparent discrepancies can be explained as follows. First, though appropriate overall, it is possible that our choice of CiVSD as structural model for hHv1 might not be able to reproduce the conformation of S2 and S3 in hHv1. Because there is a large structural heterogeneity in the relative position of the gating charges among existing VSD structures (16), it is reasonable to expect some variation in the relative positions of S1, S2, or S3, the least-conserved regions of VSDs. A second option is that given the unorthodox nature of the chimeric construct used to solve the structure of mHv-cc, it is possible that the sequence incorporated from Ci-VSP in the region of S2–S3 (164–188) locally perturbs hHv1, disrupting some of the native interactions in the region of S2 and S3 and changing its register.
A recent set of functional data, probing the pairwise interaction between two residues by thermocycle analysis (20), has suggested a list of interacting residue pairs in the proton channel. Unfortunately, most of the predictions are inconsistent with both the present structural model and mHv-cc structure (SI Appendix, Table S1). Thermocycle analysis relies on the functional perturbation by mutations, which, by definition are evaluated in the activated state of the proton channel (because it relies on ion conduction as the functional readout). The distances between most of the suggested interaction pairs in the activated state appear too far in the resting state, and there is no simple movement of S4 to satisfy those interactions. As expected, these large discrepancies indicate that the activated state of hHv1 is very different from the resting state, and there might be movements from other segments as well during the gating process (47). Another possibility is that the observed interaction may only exist in the presence of a particular mutation but is not apparent in the native state. For instance, mutation R205A in hHv1 barely shifts its voltage dependence (19), whereas R205C in CiHv1 left shifts V1/2 more than 50 mV (20).
Structural Basis of Cooperative Gating.
As a functional dimer, hHv1 shows strong cooperativity during activation gating, involving at least two conformational changes (10, 48, 49); in view of this, and considering the present data, we suggest that cooperativity in hHv1 dimer derives from three independent interactions: the presence of a covalent linkage at C249, interactions within the C-terminal coiled coil (37), and the ability of the VSDs to dimerize through the S1/S4 interface. It has been shown that the C-terminal cytoplasmic domain is essential for hHv1’s cooperativity (10); however, whether the cooperative is directly affected by coiled-coil interaction or by the VSD interface, which is enhanced by the coiled-coil interaction, remains an open question. In the absence of its C-terminal coiled coils, hHv1-VSD dimerizes in solution with an apparent Kd ∼3 µM, and we expect this interaction to be enhanced in the full-length construct. Our finding that redox potential modulates the disulfide linkage through 249C (SI Appendix, Fig. S6C) argues in favor of the hypothesis that hHv1 cooperativity is sensitive to the redox potential of the intracellular environment (40), because this would affect the stability of the C-terminal coiled coil.
Using a similar strategy as that described here, Okamura et al. (41) recently proposed a full-length dimer hHv1 model by combining the VSD from crystal structure of mHv-cc with the existing structure of the isolated C-terminal coiled-coil domain. Not surprisingly, both hHv1-FL models have essentially the same topology: a long coiled-coil extending out from S4 in a membrane-embedded VSD dimer. However, the similarity between structural models ends at the VSD dimer interface. Based on cross-linking data alone (34), S4 dominates as the predominant contact between VSDs in Okamura’s model. However, the dimer interface in our model involves both S1 and S4, and agrees well with cross-linking data from different sources (34, 38). Still, the existence of additional cross-linking regions at the bottom of S3 (Fig. 5B) appears to be inconsistent with both Okamura’s and the present model, likely as the result of the highly dynamic nature of hHv1 in the membrane (Fig. 2B) and the fact that copper phenanthroline is known to promote cys–cys linkage even when the sulfhydryl groups are up to 15 Å apart (50).
A Voltage-Dependent Gating Mechanism Based on the One-Click Model of CiVSD.
At present, CiVSD is the only VSD system where the structures of both the resting (down) and activated (up) conformations are known. Based on these structures, a one-click sliding helix/helical screw motion gating mechanism was proposed in which gating charges are stabilized by sequential sets of countercharges as they translocate through a hydrophobic gasket (16). Here, we presented quantitative evidence suggesting that CiVSD-WT is a reasonable structural model for dimeric hHv1 in its resting state. The voltage dependence for hHv1 (V1/2 = 58 mV and z = 0.90) (19) is similar to CiVSD (V1/2 = 62 mV and z = 1.10) (35), so it is tempting to suggest a mechanistic model where hHv1 gates in a similar way as CiVSD. In this model (Fig. 6C), the S4 segment moves upward to the activated state by a one-click (three residues within an α-helix) rotation/translation movement, whereas transmembrane segments S1, S2, and S3 remain as a relatively static scaffold. Kulleperuma et al. (24) have shown that histidine mutant at R3 position is accessible to internally applied Zn2+ in the open state, which is highly consistent with minimum movement of S4 in the present model. Furthermore, this S4 movement rearranges the electrostatic clusters present in the resting state: the D112-R1-D185 triad becomes D112-R2-D185 in the activated state, and R3 will move out of the interaction range with E153 and D174. This movement brings R1 much closer to E119, allowing the formation of a R1–E119 pair, which we hypothesize stabilizes the activated state. These predictions finds experimental support, because disruption of R1-E119 interaction by neutralization (E119A) right-shifts hHv1 V1/2 more than 20 mV (6), suggesting that additional energy is needed to stabilize hHv1 in the activated state. Because the hydrophobic gasket is formed by side-chain contributions from S1 + S2 + S3, the voltage-driven upward movement of S4 will likely move R1 across the gasket, positioning R3 at or very close to it. However, the proposed one-click model alone cannot account for the approximately three transferred gating charges per VSD in hHv1 (51). The presence of additional contributions, such as field refocusing, and additional S4 movements remains to be illustrated. At the moment, the complete mechanism of gating, the nature of the conduction pathway, and the molecular underpinnings of proton selectivity in hHv1 are beyond the resolution of existing data, but we surmise that they are intimately coupled to the predicted changes in the electrostatic clusters and arginine positions as they move through the hydrophobic gasket.
Experimental Procedures
Molecular Biology, Protein Expression, and Purification.
The native genes of hHv1 full-length and hHv1-VSD (residues 75–223) were individually codon optimized toward E. coli K12. The synthetic genes were cloned and inserted into pET15b vector with an N-terminal 6XHis tag. The plasmids were transformed into fresh E. coli BL21 (DE3) competent cells, and grown overnight at 37 °C for 12∼16 h. The saturated overnight cultures were diluted at 1:100 ratio into LB medium (Invitrogen) with 0.2% glucose and grown at 37 °C for ∼2 h, and the expression was induced at OD600 ∼1.0 with 0.6 mM isopropyl-β-d-thiogalactopyranoside for ∼2–4 h. The cells were harvested by centrifugation and the pellet was resuspended in Tris buffer [20 mM Tris (pH 8.0), 150 mM NaCl] in 1 mM phenylmethanesulfonyl fluoride (PMSF), 0.1 mg/mL DNase, 5 mM MgCl2, 0.1 µg/mL pepstatin, 1 µg/mL aprotinin, and 1 µg/mL leupeptin. The mixture was homogenized and spun down at 100,000 × g for 35 min. The membrane pellet was resuspended in Tris buffer and solubilized with 2 mM Fos-choline 14 in 1 mM PMSF and 10 mM imidazole at room temperature for 1 h. The homogenate was spun down at 100,000 × g for 35 min, and the supernatant was loaded on to cobalt resin (2 mL resin per liter of cell culture). The resin was washed with 20 times resin volume of Tris buffer in 1 mM Fos-choline 14 and 5 mM imidazole. The hHv1 protein was eluted with Tris buffer in presence of 1 mM Fos-choline 14 and 300 mM imidazole. For the purification of single cysteine mutants, all washing buffers contained an additional 0.5 mM Tris(2-carboxyethyl)phosphine hydrochloride and 0.5 mM Tris(2-cyanoethyl)phosphine. The eluate from the cobalt column was further purified by size-exclusion chromatography (SEC) using a Superdex 200 HR 10/30 column (GE Healthcare) with the buffer 20 mM Tris (pH 8.0), 150 mM NaCl, 4 mM Fos-choline 12.
The endogenous cysteines (107C and 249C) on hHv1-FL were mutated to serine to generate the cysteineless template, which was used to generate single cysteine mutant for probing methods with fluorescence and EPR spectroscopy. To probe which cysteine is responsible for the covalent linkage between hHv1-FL dimer, hHv1-FL native and two mutants (C107S and C249S) were purified under reducing condition in 5 mM DTT. Right after removing the DTT by SEC using a Superdex 200 column in buffer of 20 mM Tris (pH 8.0), 150 mM NaCl, and 4 mM Fos-choline 12, each protein was split into three samples with addition of 10 mM DTT, 5 mM CuSO4, and equal volume of Tris buffer. After incubation at room temperature for 10 min, all samples were added with SDS loading without reducing reagent and run on SDS/PAGE.
Protein Characterization and Reconstitution.
The purity of hHv1 proteins was checked by SDS/PAGE; their secondary structures in detergent were determined by circular dichroism (CD) spectroscopy. The protein concentration was adjusted to ∼10 μM, and the spectra were measured on an AVIV circular dichroism spectrophotometer model 202 (AVIV Instrument Inc.) between 190 and 260 nm at a 1-nm interval in a 1-mm path-length strain-free cylindrical cuvette (Hellma). Differential absorbance was converted into molar ellipticity [θ]. The percent helicity of hHv1 proteins were calculated from θ222 following standard methods.
The molecular mass and homogeneity of hHv1–detergent complexes were determined by MALS. An AKTA FPLC system equipped with a Superdex 200 HR 10/30 size-exclusion column was coupled to a MALS system with a flow cell. The light-scattering was detected with a HELEOS system (Wyatt Technology Corp.) equipped with a 60-mW GaAs laser at 658 nm and 18 detectors at angles from 22.5° to 147.0°. Refractive index was determined from Optilab T-rEX (Wyatt Technology Corp.). UV absorbance was measured at 280 nm by the detector from an AKTA system (GE Healthcare) and converted to analog between 0–1 V into a HELEOS system. Data were acquired and analyzed by ASTRA software (Wyatt Technology Corp.). Even though self-correlation between the intensity of light-scattering at discrete angles is sufficient to determine the molecular mass without standard, a standard BSA sample was used to ensure correct settings and data analysis. Then, the molecular weight of the protein–detergent complex and the protein content of the complex were analyzed for the peak of interest by the system template Protein Conjugate of ASTRA software.
Prior to reconstitution, lipids were added with 10 mM DM and rotated at room temperature for 30 min. The concentrated hHv1 (∼5 mg/mL) samples were incubated with the softened lipids for 30 min. The mixture was dialyzed against the 20-mM Tris (pH 8.0), 150-mM NaCl overnight (up to 5 d). The oligomerization behavior of hHv1-VSD after reconstitution was characterized by a standard fluorescence-based assay. In brief, hHv1-VSD 223C was purified and individually labeled with either fluorescein 5-maleimide (excitationmax = 494 nm; emissionmax = 518 nm) or tetramethylrhodamine 5-maleimide (excitationmax = 544 nm; emissionmax = 572 nm) fluorescence dye. The individually labeled hHv1-VSD 223C was mixed at donor:acceptor = 1:1 ratio, and reconstituted into liposomes at protein:lipid = 1:2,000 (molar ratio). The fluorescence emission was recorded from 500 to 650 nm, and excited at 494 nm in a PTI fluorimeter (PTI Technology). The relative FRET intensity in the 570– to 580-nm range indicates the closeness between donor and acceptor, as the relative aggregation level of hHv1-VSD.
Single-Molecule Fluorescence Bleaching.
Single cysteine mutant 193C proteins were prepared in both hHv1-VSD and hHv1-FL background. The single cysteine mutants were labeled with BODIPY fluorescein maleimide (Invitrogen). Fluorescence-labeled hHv1 proteins were reconstituted into POPC:POPG (3:1) liposomes at molar ratios of ∼1:10 million. The protein concentrations prior to reconstitution were varied at 1 nM and 10 µM. For example, for the fluorescence-labeled hHv1-VSD-193C, one sample was prepared by mixing 1 ng protein at the concentration of 1 nM (0.019 ng/µL) with 5 mg softened liposome. The other sample was prepared by mixing 200 ng protein at the concentration of 10 µM (190 ng/µL) with 5 mg softened liposome and diluted 200 times with liposomes after reconstitution. The reconstituted proteoliposomes were dialyzed over 20 mM Tris (pH 8.0), 150 mM NaCl for 48 h, and extruded 20 times with a 0.1-µm filter before bleaching experiments.
Glass coverslips (Fisher) were cleaned by immersion for 60 min in piranha solution (70% H2SO4/30% H2O2), then copiously rinsed in deionized water and stored under water for up to 2 wk. Immediately before use, a coverslip would be dried under a nitrogen stream and a chamber made from cured Sylgard 184 elastomer (Dow Corning) placed on the coverslip. Each chamber had up to four wells so that multiple liposome preparations could be tried on the same coverslip. Liposome suspensions were placed into the wells for 15 s before being extensively rinsed with buffer to remove excess liposomes. Coverslips were imaged using an Olympus IX71 inverted fluorescence microscope and a 60× 1.45 N.A. oil-immersion total internal reflection fluorescence objective lens (Olympus). Excitation was performed using a 50-mW 473-nm DPSS laser (Shanghai Dream Lasers). Typical illumination intensity was ∼200 W/cm2. Fluorescence was captured with an Evolve 128 EMCCD camera (Photometrics); frame rates used ranged from 20 to 50 Hz.
ACMA-Based Proton Flux Assay in Liposome.
The proton conduction of purified hHv1 protein was assayed by an established fluorescence-based flux assay in the proteoliposomes. The proteoliposome were prepared with high K+ inside and diluted into low K+ buffer. The addition of K+-specific carrier valinomycin leads to outflux of K+ and generates inward electrochemical potential to drive H+ into liposomes through proton channels. The resulting pH gradient across membrane leads to accumulation of protonated fluorescence dye 9-amino-6-chloro-2-methoxyacridine (ACMA) and the quenching of fluorescence signal. The time-based fluorescence change was monitored by a PTI fluorimeter (PTI Technology) with excitation at 410 nm and emission at 490 nm.
EPR Spectroscopy.
A total of 149 single cysteine mutants covering the residues 75–223 on the scaffold of hHv1-VSD were purified and labeled using 1:20 molar excess spin label (1-oxy-2,2,5,5,-tetramethylpyrrolidin-3-yl)methanethiosulfonate at room temperature for 20 min. Excess spin label was removed with a PD10 column (GE Healthcare). The spin-labeled hHv1-VSD was reconstituted into POPC/POPG 3:1 liposomes at the molar ratio protein:lipid = 1:2,000. For the underlabeling experiments, the spin label (1-oxy-2,2,5,5,-tetramethylpyrrolidin-3-yl)methanethiosulfonate was premixed with a dimagnet label at molar ratio of 1:4 before adding to the single cysteine mutant hHv1-VSD 206C.
Continuous-wave EPR spectroscopic measurements were carried on a Bruker EMX X-band spectrometer equipped with a dielectric resonator and a gas-permeable TPX plastic capillary at room temperature. Spectra were recorded at 2.0-mW incident power, 100-kHz modulation frequency, 1.0 Gaussian amplitude modulation, 20.48-ms time constant, 40.96-ms conversion time, and 120 Gaussian scan width. The motion of the spin label at a specific site was quantified by anisotropy of spectra shape with an empirical parameter as the inverse of peak-to-peak width of the central (mI = 0) resonance, which was conventionally termed as mobility (ΔHo−1) (31). The mobility will be low for the region involving tertiary or quaternary contacts, which can constrain the motion of spin label, but high for the loop region where spin label has more freedom of motion. The accessibility (Π) of spin label to the paramagnetic relaxing reagent, which can present predominantly in lipid (O2) or water (NiEdda), can be measured from power-saturation methods (52). In brief, P1/2, the power when amplitude is reduced to half of the value as if no saturation, can be obtained by fitting the peak-to-peak amplitude of the central line of EPR spectra as a function of increasing incident microwave power with the equation: . A is the peak-to-peak amplitude of the central line of EPR spectra, P is the incident power used in the experiment, P1/2 is the incident power, ε is the saturation factor in the range of 1–1.5. The accessibilities are calculated by normalizing the P1/2 differences between conditions with and without paramagnetic relaxing reagents with following equation: . ΔP1/2 is the P1/2 difference, the ΔHo−1 is the inverse central line width, P1/2 (DPPH) is the P1/2 value of standard sample DPPH (2,2-diphenyl-1-picrylhydrazyl), ΔHo−1(DPPH) is the central line width of DPPH spectra. The measurements for each spin-labeled mutant can be done after equilibration with air (21% O2) or N2 (0% O2) for oxygen accessibility (ΠO2), or with 25 mM NiEdda + N2 or N2 (0 mM NiEdda) for NiEdda accessibility (ΠNi). High accessibility of the spin probe to O2 (ΠO2) is suggestive of a lipid-exposed environment within membrane, whereas high NiEdda accessibility is a reflection of the probe being exposed to aqueous solvent outside membrane.
Structural Modeling by Molecular Dynamic Simulation Methods.
Sequences of the VSDs of Kv1.2–2.1 chimera, KvAP, NavAb, Ci-VSD-WT, and hHv1 were aligned using the VMD plug-in MultiSeq. (53, 54), which is analogous to previous alignment (24). The average distance tree of the above sequences was generated by Jalview (55). The crystal structures of the VSD of Kv1.2–2.1 (2R9R), KvAP (1ORS), NavAb (3RVY), and CiVSD (WT, 4G80) were used as templates to build four individual structural models following standard procedure and using the I-TASSER server (56).
Each of the four models was embedded in an explicit POPC lipid bilayer, solvated by 150 mM NaCl solutions and first equilibrated in the isothermal–isobaric ensemble at 300 K and 1 bar, with a time step of 2 fs for 10 ns using molecular dynamics simulations with NAMD (57). The CHARMM36 parameter set (58, 59) was used for protein and lipid, and a TIP3P model for water (60). The four models were further equilibrated for 300 ns with ANTON (61). The temperature, pressure, and time step were set to the same as those used in the initial 10-ns MD simulations.
Model Comparison.
The four models after 300-ns equilibration were used to evaluate their agreement with experimental data. A spin label (ONP0) pseudoatom, representing the nitroxide spin label, was covalently attached to the alpha carbon atom of each residue (62, 63). Sufficient solvent OXY and NIC pseudoatoms (36), representing O2 and NiEdda, were introduced into the system and distributed into the membrane and water compartments, respectively. The interactions between pseudoatoms and water; pseudoatoms and ions; and OXY and lipids were turned off. In addition, pseudospins see neither pseudosolvents nor other pseudospins.
The solvent accessibility simulations were carried out with NAMD for 5 ns using the same simulation protocol for equilibration of the structural model systems. The distribution of OXY along the normal axis of the membrane, based on the oxygen accessibility of nitroxide radical-containing lipids, was restrained using the grid forces in NAMD. Solvation numbers of the spin-label pseudoatoms were measured by calculating the average number of OXY or NIC within 6 Å of each pseudospin during the last 2 ns of simulations. In comparison with the O2 and NiEdda accessibility derived from EPR spectra, all solvation numbers were scaled with a constant scaling factor such that they have the smallest rmsd with respect to the experimental data. The Kullback–Leibler divergence (33) was used to measure the difference between the profiles of solvation number and solvent accessibility. The solvation number for each pseudospin was also used to predict the 4-acetamido-4′-maleimidylstilbene-2,2′-disulfonic acid (AMS) and N-ethylmaleimide (NEM) accessibility patterns (34). A pseudospin was treated to be AMS or NEM accessible when the solvation number of this pseudospin larger than a threshold. Different threshold values were chosen for the AMS and NEM accessibility profiles to make the model predicted accessibility have the largest similarity with the experimental ones.
Structural Refinement with Experimental Constraints.
The O2 and NiEdda accessibility data from EPR experiments were used as structural restraints to further refine the hHv1-VSD structural model based on CiVSD-WT with the PaDSAR approach (26, 36). Three types of spin-label pseudoatoms (ONP1, ONP2, and ONP3) and three types of environmental pseudoatoms (PROT, OXY, NIC) were introduced into the system. ONP1, ONP2, and ONP3 represent nitroxide spin labels with both low O2 and NiEdda accessibility values (buried within the protein itself), with low O2 but high NiEdda accessibility values (exposed to the water solution) and with high O2 but low NiEdda accessibility values (exposed to the lipid bilayer), respectively. PROT, OXY, and NIC represent protein residues O2 and NiEdda, respectively. The different types of van der Waals interactions between the spin-label pseudoatoms and the environmental pseudoatoms provided the restraint driving forces during the course of simulations. The equilibrated hHv1-VSD was used to construct a starting configuration for the restrained molecular dynamics simulation. ONP1, ONP, ONP3, and PROT were covalently attached to the alpha atom of the assigned residues. The protein with patched pseudoatoms was then embedded in a box of OXY with a slab of NIC on each side mimicking lipids and water, respectively. The restrained simulations were run in an NVT ensemble (300 K) with NAMD, and the secondary structure of the four transmembrane helices were restrained during the course of simulations. The resulting model was further equilibrated in an explicit POPC membrane for 400 ns.
The refined structural model of hHv1-CiVSD was used to build a dimer of hHv1 with the C-terminal coiled-coil domain (with residues 89–266), consulting the crystal structures of the Ci-VSD dimer (WT, 4G80) and the hHv1 C-terminal dimer (37). The linker between the S4 helix and the coiled-coil domain (with residues 217–228) was rebuilt as an α-helix, considering the heptad repeat of coiled-coil structures (40). The dimer model was first equilibrated for 5 ns in vacuum and then equilibrated for 300 ns in an explicit POPC lipid bilayer at 300 K using NAMD. The secondary structure restraint and the symmetry restraint were imposed during the simulation.
Supplementary Material
Acknowledgments
We thank James Hinshaw, Kevin Song, and Tobin Sosnick for experimental help on mass spectrometry and circular dichroism measurements, and members of the E.P., F.B., and Roux laboratories for helpful discussions and invaluable experimental advice. This work was supported in part by NIH Grants R01-GM57846 and U54 GM74946. The computational resources were supported in part by NIH Grants 1S10OD018495-01 and P41GM103712-S1.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 13430.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1515043112/-/DCSupplemental.
References
- 1.Ramsey IS, Moran MM, Chong JA, Clapham DE. A voltage-gated proton-selective channel lacking the pore domain. Nature. 2006;440(7088):1213–1216. doi: 10.1038/nature04700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sasaki M, Takagi M, Okamura Y. A voltage sensor-domain protein is a voltage-gated proton channel. Science. 2006;312(5773):589–592. doi: 10.1126/science.1122352. [DOI] [PubMed] [Google Scholar]
- 3.DeCoursey TE. Voltage-gated proton channels: Molecular biology, physiology, and pathophysiology of the H(V) family. Physiol Rev. 2013;93(2):599–652. doi: 10.1152/physrev.00011.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Henderson LM, Chappell JB, Jones OT. The superoxide-generating NADPH oxidase of human neutrophils is electrogenic and associated with an H+ channel. Biochem J. 1987;246(2):325–329. doi: 10.1042/bj2460325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DeCoursey TE, Morgan D, Cherny VV. The voltage dependence of NADPH oxidase reveals why phagocytes need proton channels. Nature. 2003;422(6931):531–534. doi: 10.1038/nature01523. [DOI] [PubMed] [Google Scholar]
- 6.Ramsey IS, Ruchti E, Kaczmarek JS, Clapham DE. Hv1 proton channels are required for high-level NADPH oxidase-dependent superoxide production during the phagocyte respiratory burst. Proc Natl Acad Sci USA. 2009;106(18):7642–7647. doi: 10.1073/pnas.0902761106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Capasso M, et al. HVCN1 modulates BCR signal strength via regulation of BCR-dependent generation of reactive oxygen species. Nat Immunol. 2010;11(3):265–272. doi: 10.1038/ni.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lishko PV, Botchkina IL, Fedorenko A, Kirichok Y. Acid extrusion from human spermatozoa is mediated by flagellar voltage-gated proton channel. Cell. 2010;140(3):327–337. doi: 10.1016/j.cell.2009.12.053. [DOI] [PubMed] [Google Scholar]
- 9.Gonzalez C, Koch HP, Drum BM, Larsson HP. Strong cooperativity between subunits in voltage-gated proton channels. Nat Struct Mol Biol. 2010;17(1):51–56. doi: 10.1038/nsmb.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tombola F, Ulbrich MH, Isacoff EY. The voltage-gated proton channel Hv1 has two pores, each controlled by one voltage sensor. Neuron. 2008;58(4):546–556. doi: 10.1016/j.neuron.2008.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koch HP, et al. Multimeric nature of voltage-gated proton channels. Proc Natl Acad Sci USA. 2008;105(26):9111–9116. doi: 10.1073/pnas.0801553105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang Y, et al. X-ray structure of a voltage-dependent K+ channel. Nature. 2003;423(6935):33–41. doi: 10.1038/nature01580. [DOI] [PubMed] [Google Scholar]
- 13.Long SB, Tao X, Campbell EB, MacKinnon R. Atomic structure of a voltage-dependent K+ channel in a lipid membrane-like environment. Nature. 2007;450(7168):376–382. doi: 10.1038/nature06265. [DOI] [PubMed] [Google Scholar]
- 14.Payandeh J, Gamal El-Din TM, Scheuer T, Zheng N, Catterall WA. Crystal structure of a voltage-gated sodium channel in two potentially inactivated states. Nature. 2012;486(7401):135–139. doi: 10.1038/nature11077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang X, et al. Crystal structure of an orthologue of the NaChBac voltage-gated sodium channel. Nature. 2012;486(7401):130–134. doi: 10.1038/nature11054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Q, et al. Structural mechanism of voltage-dependent gating in an isolated voltage-sensing domain. Nat Struct Mol Biol. 2014;21(3):244–252. doi: 10.1038/nsmb.2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tao X, Lee A, Limapichat W, Dougherty DA, MacKinnon R. A gating charge transfer center in voltage sensors. Science. 2010;328(5974):67–73. doi: 10.1126/science.1185954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Musset B, et al. Aspartate112 is the selectivity filter of the human voltage-gated proton channel. Nature. 2011;480(7376):273–277. doi: 10.1038/nature10557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ramsey IS, et al. An aqueous H+ permeation pathway in the voltage-gated proton channel Hv1. Nat Struct Mol Biol. 2010;17(7):869–875. doi: 10.1038/nsmb.1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chamberlin A, et al. Hydrophobic plug functions as a gate in voltage-gated proton channels. Proc Natl Acad Sci USA. 2014;111(2):E273–E282. doi: 10.1073/pnas.1318018111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berger TK, Isacoff EY. The pore of the voltage-gated proton channel. Neuron. 2011;72(6):991–1000. doi: 10.1016/j.neuron.2011.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hong L, Pathak MM, Kim IH, Ta D, Tombola F. Voltage-sensing domain of voltage-gated proton channel Hv1 shares mechanism of block with pore domains. Neuron. 2013;77(2):274–287. doi: 10.1016/j.neuron.2012.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wood ML, et al. Water wires in atomistic models of the Hv1 proton channel. Biochim Biophys Acta. 2012;1818(2):286–293. doi: 10.1016/j.bbamem.2011.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kulleperuma K, et al. Construction and validation of a homology model of the human voltage-gated proton channel hHV1. J Gen Physiol. 2013;141(4):445–465. doi: 10.1085/jgp.201210856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takeshita K, et al. X-ray crystal structure of voltage-gated proton channel. Nat Struct Mol Biol. 2014;21(4):352–357. doi: 10.1038/nsmb.2783. [DOI] [PubMed] [Google Scholar]
- 26.Li Q, Wanderling S, Sompornpisut P, Perozo E. Structural basis of lipid-driven conformational transitions in the KvAP voltage-sensing domain. Nat Struct Mol Biol. 2014;21(2):160–166. doi: 10.1038/nsmb.2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Q, Jogini V, Wanderling S, Cortes DM, Perozo E. Expression, purification, and reconstitution of the voltage-sensing domain from Ci-VSP. Biochemistry. 2012;51(41):8132–8142. doi: 10.1021/bi300980q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chakrapani S, Cuello LG, Cortes DM, Perozo E. Structural dynamics of an isolated voltage-sensor domain in a lipid bilayer. Structure. 2008;16(3):398–409. doi: 10.1016/j.str.2007.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee SY, Letts JA, MacKinnon R. Functional reconstitution of purified human Hv1 H+ channels. J Mol Biol. 2009;387(5):1055–1060. doi: 10.1016/j.jmb.2009.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perozo E, Cortes DM, Cuello LG. Three-dimensional architecture and gating mechanism of a K+ channel studied by EPR spectroscopy. Nat Struct Biol. 1998;5(6):459–469. doi: 10.1038/nsb0698-459. [DOI] [PubMed] [Google Scholar]
- 31.Mchaourab HS, Lietzow MA, Hideg K, Hubbell WL. Motion of spin-labeled side chains in T4 lysozyme. Correlation with protein structure and dynamics. Biochemistry. 1996;35(24):7692–7704. doi: 10.1021/bi960482k. [DOI] [PubMed] [Google Scholar]
- 32.Altenbach C, Flitsch SL, Khorana HG, Hubbell WL. Structural studies on transmembrane proteins. 2. Spin labeling of bacteriorhodopsin mutants at unique cysteines. Biochemistry. 1989;28(19):7806–7812. doi: 10.1021/bi00445a042. [DOI] [PubMed] [Google Scholar]
- 33.Kullback S, Leibler RA. On information and sufficiency. Ann Math Stat. 1951;22(1):142–143. [Google Scholar]
- 34.Kurokawa T, Okamura Y. Mapping of sites facing aqueous environment of voltage-gated proton channel at resting state: A study with PEGylation protection. Biochim Biophys Acta. 2014;1838(1 Pt B):382–387. doi: 10.1016/j.bbamem.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 35.Murata Y, Iwasaki H, Sasaki M, Inaba K, Okamura Y. Phosphoinositide phosphatase activity coupled to an intrinsic voltage sensor. Nature. 2005;435(7046):1239–1243. doi: 10.1038/nature03650. [DOI] [PubMed] [Google Scholar]
- 36.Sompornpisut P, Roux B, Perozo E. Structural refinement of membrane proteins by restrained molecular dynamics and solvent accessibility data. Biophys J. 2008;95(11):5349–5361. doi: 10.1529/biophysj.108.142984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li SJ, et al. The role and structure of the carboxyl-terminal domain of the human voltage-gated proton channel Hv1. J Biol Chem. 2010;285(16):12047–12054. doi: 10.1074/jbc.M109.040360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee SY, Letts JA, Mackinnon R. Dimeric subunit stoichiometry of the human voltage-dependent proton channel Hv1. Proc Natl Acad Sci USA. 2008;105(22):7692–7695. doi: 10.1073/pnas.0803277105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fujiwara Y, Takeshita K, Nakagawa A, Okamura Y. Structural characteristics of the redox-sensing coiled coil in the voltage-gated H+ channel. J Biol Chem. 2013;288(25):17968–17975. doi: 10.1074/jbc.M113.459024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fujiwara Y, Kurokawa T, Okamura Y. Long α helices projecting from the membrane as the dimer interface in the voltage-gated H(+) channel. J Gen Physiol. 2014;143(3):377–386. doi: 10.1085/jgp.201311082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Okamura Y, Fujiwara Y, Sakata S. Gating mechanisms of voltage-gated proton channels. Annu Rev Biochem. 2015;84:685–709. doi: 10.1146/annurev-biochem-060614-034307. [DOI] [PubMed] [Google Scholar]
- 42.Cuello LG, Cortes DM, Perozo E. Molecular architecture of the KvAP voltage-dependent K+ channel in a lipid bilayer. Science. 2004;306(5695):491–495. doi: 10.1126/science.1101373. [DOI] [PubMed] [Google Scholar]
- 43.Chakrapani S, Sompornpisut P, Intharathep P, Roux B, Perozo E. The activated state of a sodium channel voltage sensor in a membrane environment. Proc Natl Acad Sci USA. 2010;107(12):5435–5440. doi: 10.1073/pnas.0914109107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang N, Horn R. Evidence for voltage-dependent S4 movement in sodium channels. Neuron. 1995;15(1):213–218. doi: 10.1016/0896-6273(95)90078-0. [DOI] [PubMed] [Google Scholar]
- 45.Starace DM, Bezanilla F. A proton pore in a potassium channel voltage sensor reveals a focused electric field. Nature. 2004;427(6974):548–553. doi: 10.1038/nature02270. [DOI] [PubMed] [Google Scholar]
- 46.Asamoah OK, Wuskell JP, Loew LM, Bezanilla F. A fluorometric approach to local electric field measurements in a voltage-gated ion channel. Neuron. 2003;37(1):85–97. doi: 10.1016/s0896-6273(02)01126-1. [DOI] [PubMed] [Google Scholar]
- 47.Mony L, Berger TK, Isacoff EY. A specialized molecular motion opens the Hv1 voltage-gated proton channel. Nat Struct Mol Biol. 2015;22(4):283–290. doi: 10.1038/nsmb.2978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Qiu F, Rebolledo S, Gonzalez C, Larsson HP. Subunit interactions during cooperative opening of voltage-gated proton channels. Neuron. 2013;77(2):288–298. doi: 10.1016/j.neuron.2012.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tombola F, Ulbrich MH, Kohout SC, Isacoff EY. The opening of the two pores of the Hv1 voltage-gated proton channel is tuned by cooperativity. Nat Struct Mol Biol. 2010;17(1):44–50. doi: 10.1038/nsmb.1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dmitriev O, Jones PC, Jiang W, Fillingame RH. Structure of the membrane domain of subunit b of the Escherichia coli F0F1 ATP synthase. J Biol Chem. 1999;274(22):15598–15604. doi: 10.1074/jbc.274.22.15598. [DOI] [PubMed] [Google Scholar]
- 51.Musset B, et al. Detailed comparison of expressed and native voltage-gated proton channel currents. J Physiol. 2008;586(10):2477–2486. doi: 10.1113/jphysiol.2007.149427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Altenbach C, Greenhalgh DA, Khorana HG, Hubbell WL. A collision gradient method to determine the immersion depth of nitroxides in lipid bilayers: Application to spin-labeled mutants of bacteriorhodopsin. Proc Natl Acad Sci USA. 1994;91(5):1667–1671. doi: 10.1073/pnas.91.5.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roberts E, Eargle J, Wright D, Luthey-Schulten Z. MultiSeq: Unifying sequence and structure data for evolutionary analysis. BMC Bioinformatics. 2006;7:382. doi: 10.1186/1471-2105-7-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Humphrey W, Dalke A, Schulten K. 1996. VMD: Visual molecular dynamics. J Mol Graph 14(1):33–38, 27–28. [DOI] [PubMed]
- 55.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25(9):1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Roy A, Kucukural A, Zhang Y. I-TASSER: A unified platform for automated protein structure and function prediction. Nat Protoc. 2010;5(4):725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Phillips JC, et al. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26(16):1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Best RB, et al. Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone φ, ψ and side-chain χ(1) and χ(2) dihedral angles. J Chem Theory Comput. 2012;8(9):3257–3273. doi: 10.1021/ct300400x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Klauda JB, et al. Update of the CHARMM all-atom additive force field for lipids: Validation on six lipid types. J Phys Chem B. 2010;114(23):7830–7843. doi: 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79(2):926–935. [Google Scholar]
- 61.Shaw DE, et al. Anton, a special-purpose machine for molecular dynamics simulation. Commun ACM. 2008;51(7):91–97. [Google Scholar]
- 62.Roux B, Islam SM. Restrained-ensemble molecular dynamics simulations based on distance histograms from double electron-electron resonance spectroscopy. J Phys Chem B. 2013;117(17):4733–4739. doi: 10.1021/jp3110369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Islam SM, Stein RA, McHaourab HS, Roux B. Structural refinement from restrained-ensemble simulations based on EPR/DEER data: Application to T4 lysozyme. J Phys Chem B. 2013;117(17):4740–4754. doi: 10.1021/jp311723a. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.