Significance
Aedes albopictus is a highly adaptive species that thrives worldwide in tropical and temperate zones. From its origin in Asia, it has established itself on every continent except Antarctica. This expansion, coupled with its ability to vector the epidemic human diseases dengue and Chikungunya fevers, make it a significant global public health threat. A complete genome sequence and transcriptome data were obtained for the Ae. albopictus Foshan strain, a colony derived from mosquitoes from its historical origin. The large genome (1,967 Mb) comprises an abundance of repetitive DNA classes and expansions of the numbers of gene family members involved in insecticide resistance, diapause, sex determination, immunity, and olfaction. This large genome repertory and plasticity may contribute to its success as an invasive species.
Keywords: mosquito genome, transposons, flavivirus, diapause, insecticide resistance
Abstract
The Asian tiger mosquito, Aedes albopictus, is a highly successful invasive species that transmits a number of human viral diseases, including dengue and Chikungunya fevers. This species has a large genome with significant population-based size variation. The complete genome sequence was determined for the Foshan strain, an established laboratory colony derived from wild mosquitoes from southeastern China, a region within the historical range of the origin of the species. The genome comprises 1,967 Mb, the largest mosquito genome sequenced to date, and its size results principally from an abundance of repetitive DNA classes. In addition, expansions of the numbers of members in gene families involved in insecticide-resistance mechanisms, diapause, sex determination, immunity, and olfaction also contribute to the larger size. Portions of integrated flavivirus-like genomes support a shared evolutionary history of association of these viruses with their vector. The large genome repertory may contribute to the adaptability and success of Ae. albopictus as an invasive species.
The Asian tiger mosquito, Aedes albopictus, is an aggressive daytime-biting insect that is an increasing public health threat throughout the world (1). Its impact on human health results from its rapid and aggressive spread from its native home range, along with its ecological adaptability in different traits, including feeding behavior, diapause, and vector competence (2). This species is indigenous to East Asia and islands of the western Pacific and Indian Ocean, but has spread in the past 40 y to every continent except Antarctica (1). This widespread geographic distribution includes tropical and temperate zones, which is unusual for mosquitoes. Ae. albopictus is a competent vector for at least 26 arboviruses, and is important in the transmission of those that cause dengue and Chikungunya fevers (2, 3). It also is implicated as a vector of filarial nematodes of veterinary and zoonotic significance (4, 5). Although this species is considered a less efficient dengue vector than Aedes aegypti (2), it is the sole vector of recent outbreaks in southern China, Hawaii, and Gabon, and the first local (autochthonous) transmission in Europe (1, 2). Ae. albopictus vector competence for viruses is dynamic (3): for example, recent Chikungunya fever outbreaks in Réunion (Island), Mauritius, Madagascar, and Mayotte (2005–2007); Central Africa (2006–2007); and Italy (2007) were caused by viruses carrying at least one mutation that improved their transmission efficiency by the mosquito, making it the primary vector (2). The genome sequence of Ae. albopictus provides the basis for probing and understanding the mechanisms underlying its fast expansion and the development of strategies for controlling it and the pathogens it transmits.
Results and Discussion
Genome Properties and Evolution.
Genome sequencing and assembly.
A total of 23 libraries with insert sizes ranging from 170 bp to 20 kb in length were sequenced to yield a total of 689.59 Gb after filtering low-quality reads. An assembly generated a total of 1,967 Mb of sequence with scaffold and contig N50s of 195.54 kb and 17.28 kb, respectively, and an estimated whole-genome coverage of ∼350 fold (SI Appendix, Figs. S1.1–S1.4 and Tables S1.1–S1.6).
Genome size variation.
The Ae. albopictus genome is the largest of any mosquito species sequenced to date, which vary from 174 Mb for Anopheles darlingi to 540 Mb and 1,376 Mb for Culex quinquefasciatus and Ae. aegypti, respectively (6–9). Genome size variation also is observed among different populations of Ae. albopictus (10). An analysis of 47 geographic isolates from 18 countries showed a 2.5-fold variation in haploid genome weights (i.e., C-value) ranging from 0.62 pg in a population from Koh Samui (Thailand) to 1.66 pg in those recovered from Houston, TX (11). Inter- and intraspecific variation in genome size among mosquitoes appears to be caused mainly by changes in the amounts and organization of repetitive DNA. Increases in abundance of all classes of repetitive DNA sequences are correlated linearly with total genome size (12) (SI Appendix, Fig. S1.5 and Table S1.7).
Gene predictions.
A total of 17,539 protein-encoding gene models were predicted de novo and supported by evidence-based searches using reference gene sets from mosquito (Ae. aegypti, An. gambiae, and Cx. quinquefasciatus) and fruit fly (Drosophila melanogaster) genome annotations, and this number is larger than those species with the exception of Cx. quinquefasciatus (SI Appendix, Table S1.9). These predictions were supported by RNA sequencing (RNA-seq)-based transcriptome data from multiple developmental stages (SI Appendix, Table S1.8). Approximately 93.6% of the predicted proteins matched entries in the SWISS-PROT, InterPro, or TrEMBL databases (SI Appendix, Tables S1.10–S1.12). Noncoding RNAs discovered in RNA-seq analyses include as many as 57 previously undescribed miRNAs putatively unique to Ae. albopictus. (SI Appendix, Tables S1.13 and S4.4).
Evolution.
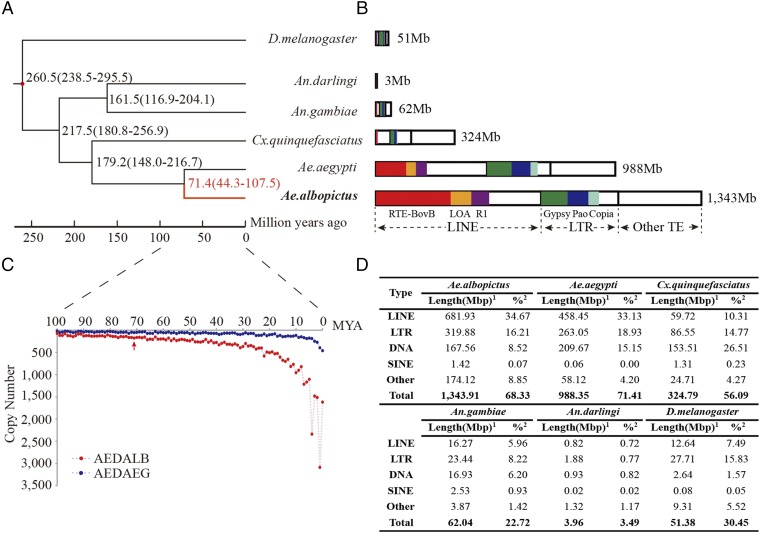
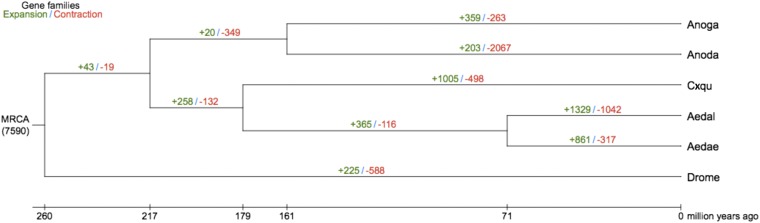
Phylogenetic relationships based on 2,096 single-copy orthologous genes from five mosquito and one fruit fly species are consistent with previous reports that place Ae. albopictus within the Culicinae (www.fossilrecord.net/) and estimate a divergence time of ∼71 Mya from Ae. aegypti (Fig. 1), longer than a previous estimate of ∼60 Mya (13). Mosquito/fruit fly divergence is estimated to have occurred ∼260 Mya. A total of 86 expansion (773 genes) and 26 contraction (108 genes) Ae. albopictus gene families were identified, and function enrichment of the former was determined (Fig. 2 and SI Appendix, Fig. S1.6 and Tables S1.14 and S1.15). Furthermore, 239 of the 2,096 orthologs (∼11%) show evidence of positive selection in Ae. albopictus with 32 Gene Ontology (GO) classes exhibiting significant enrichment (P < 0.05; SI Appendix, Table S1.16)
Fig. 1.
Comparative analyses of the Ae. albopictus genome. (A) Phylogeny and divergence time estimation by molecular clock analysis. The mosquitoes An. gambiae, An. darlingi, Cx. quinquefasciatus, Ae. albopictus, and Ae. aegypti are estimated to have last shared a common ancestor with the fruit fly D. melanogaster ∼260 Mya. The anopheline and culicine branches are estimated to have diverged 217 Mya (SE = 180.8–256.9 Mya). Ae. albopictus and Ae. aegypti are estimated to have last shared a common ancestor 71.4 Mya (SE = 44.3–107.5 Mya). (B) Comparisons of repeat content among the six species. Families of repetitive elements are represented by colors: green, Gypsy; blue, Pao, light blue, Copia, red, RTE-BovB; orange, LOA; purple, R1; white, other. The horizontal extent of each bar represents the relative length of repeat within each family. Numbers to the right represent the total length of repeats in the respective genomes. (C) Copy number of TE insertions relative to the estimated time of insertion. Shown here are type I LINE (LINE/I) transposons. AEDALB (red), Ae. albopictus; AEDAEG (blue), Ae. aegypti. (D) Statistics of TE representation in the genomes of six species. 1, Length in base pairs; 2, percentage of genome represented. DNA, DNA transposon; SINE, short interspersed nuclear element.
Fig. 2.
Expansion and contraction of gene families among mosquito species. The numbers designate the number of gene families that have expanded (green) and contracted (red) since the split from the last common ancestor. The most recent common ancestor (MRCA) has 7,590 gene families. Aedae, Ae. aegypti; Aedal, Ae. albopictus; Anoda, An. darlingi; Anoga, An. gambiae; Cxqu, Cx. quinquefasciatus; Drome, D. melanogaster.
Properties of Specific Gene Categories.
Repetitive DNA.
The Ae. albopictus genome harbors all major groups of transposable elements (TEs) (Fig. 1). Repetitive sequences represent ∼68% of the genome, the most of all sequenced mosquito species (9). This high repeat content is consistent with the large genome size, and the total length of these DNAs is ∼40% more than that of Ae. aegypti, a member of the same subgenus, Stegomyia, and the only other mosquito with a sequenced genome larger than 1 Gb (5) (SI Appendix, Table S2). Non-LTR (long terminal repeats) retrotransposons or long interspersed nuclear elements (LINEs) showed the highest genome abundance in both species (Fig. 1). The LINE family, RTE-Bov, represents ∼15.7% (308 Mb) of the entire Ae. albopictus genome. Interestingly, a single Ae. albopictus LINE element, Duo (SI Appendix, Fig. S2.1), and its Ae. aegypti homolog, TF000022, occupy ∼4.1% (∼82 Mb) and ∼3.17% (∼44 Mb) of their respective genomes. The shared element and its abundance support the conclusion that it was present in the ancestral lineage of the two species. In contrast, >20% of the Ae. albopictus genome is occupied by interspersed repeats that have no similarity (i.e., e-value ≤ 1e-5) to Ae. aegypti sequences, and this provides support for the hypothesis that there was a rapid expansion of repeat DNA after divergence of the species.
The relative times of insertion of LINE and LTR retrotransposons were estimated by comparing sequence similarities among the best matching TE pairs within clusters (SI Appendix, Fig. S2.2). This analysis determined that the highest number of insertions in Ae. albopictus occurred within the last 10 My. Similar recent activity maxima were not observed or were at lower levels in Ae. aegypti TEs of the same clade. Thus, recent transposition of LTR and LINE retrotransposons contributes to the expansion of the Ae. albopictus genome.
Varied deletion rates also drive genome size differences (14, 15). Deletion rate analysis using the “dead-on-arrival” (i.e., neutrally evolving) non-LTR retrotransposon sequences from Ae. albopictus, Ae. aegypti, and Cx. quinquefasciatus reveals that there are more deletions than insertions, a result consistent with what is seen in other similarly analyzed organisms (15) (Table 1). Ae. albopictus has a slightly lower DNA loss rate than Ae. aegypti and Cx. quinquefasciatus, and this also may contribute to its large genome size.
Table 1.
DNA deletion rates in mosquitoes
| Species | No. alignments* | Insertions | Deletions | Substitutions | Length, bp | Loss rate† | |
| Deleted | Inserted | ||||||
| Ae. albopictus | 15,188 | 22,200 | 39,626 | 795,858 | 239,230 | 75,052 | 0.206290569 |
| Ae. aegypti | 34,812 | 39,891 | 87,866 | 1,592,084 | 544,096 | 106,804 | 0.274666412 |
| C. quinquefasciatus | 1,057 | 643 | 1,538 | 15,222 | 5,570 | 1,679 | 0.25561687 |
Number of alignments analyzed.
DNA of base pairs lost per substitution.
Flavivirus-like sequences in the Ae. albopictus genome.
Sequences with similarity to flaviruses are detected in the genome of Ae. albopictus (16–18). Integrations from nonretroviral RNA viruses are referred to as nonretroviral integrated RNA viruses (NIRVs) (19, 20); the first integrations from flaviviruses in the Ae. albopictus genome were referred to as Cell Silent Agent (16). NIRV representation in the genomes of the Ae. albopictus Foshan strain, Ae. aegypti (Assembly AaegL3), An. gambiae (AgamP4 assembly), and Cx. quinquefasciatus (CpipJ2 assembly) were queried bioinformatically by using 261 sequences of previously characterized NIRVs along with the complete or portions of the genomes of representative insect-specific flaviviruses (ISFs), mosquito-borne viruses (MBVs), and tick-borne viruses (TBVs) and flaviviruses with no known vector (NBVs; Dataset S2). No matches were returned for An. gambiae or Cx. quinquefasciatus, whereas thousands with e-values <10−4 were detected in Ae. albopictus and Ae. aegypti (Datasets S3 and S4). Ae. albopictus has more variability than Ae. aegypti among viral types, including those with similarities to dengue viruses, and integrations were longer in length. Analyses and functional annotation of the sequences corresponding to basic local alignment search tool (BLAST) hits in Ae. albopictus revealed 24 sequences spanning partial or complete flaviviral ORFs, primarily NS1 and NS5, across 10 scaffolds (Dataset S5). NIRVs were embedded in regions rich with LTR retrotransposon sequences, primarily Ty1-copia and Ty3-gypsy (21, 22). No nucleotide repeats (direct or inverted) were observed at integration sites, supporting the conclusion that flaviviral integrations were derived from ectopic recombination with retrotransposons rather than being catalyzed by classical transposition activity (22, 23).
The larger number of NIRVs identified in the Foshan strain with respect to previous reports may result from the fact that past characterizations were based on gene-amplification analyses with flavivirus-specific primers (16, 18, 24). Alternatively, the larger number may indicate that these are ancestral integrations and that migration out of its native range results in integration loss associated with founder effects. The presence of a variable number of integrations across geographic populations also may contribute to the observed variation in genome size of Ae. albopictus populations (10). The current variability in the NIRV integration sites and sequences support the conclusion that different regions of different length of the flavivirus genome can integrate. NIRVs phylogenetic relationship with respect to previously characterized NIRVs, ISFs and MBVs, TBVs, and NBVs indicate that flaviviral integrations may occur in germ-line cells and may be inherited in Ae. albopictus as mosquitoes of the Foshan strain have been excluded from contact with wild-caught mosquitoes and viruses for more than 30 y (SI Appendix, Figs. S3.1–S3.4). At the same time, these data support the hypothesis that integrations of flaviviral sequences may be an ongoing regional process because NIRVs reported recently for Ae. albopictus collected in northern Italy (17) formed a separate cluster from any of the identified NIRVs (SI Appendix, Fig. S3.1), and sequences with an intact ORF and a high level of identity to circulating viruses were detected along with sequences harboring extensive rearrangements. Whether these sequences affect the replication and/or dissemination of mosquito-infecting arboviruses and contribute to vector competence is unknown.
Diapause-related genes.
Diapause in insects is characterized by no or low growth, low metabolic activity, and an increased ability to survive environmental temperature and humidity extremes, and may be specific to one developmental stage. In mosquitoes, diapause may occur at the embryonic, larval, or adult stage (25), and it is observed in Ae. albopictus at low frequencies in populations derived from subtropical habitats (26). The Foshan strain was not tested for a diapause response. However, our analysis addresses regions of the genome that have been demonstrated to be expressed differentially as part of the diapause program in temperate, fully diapause-capable populations based on extensive previously published RNA-seq studies (27–30). A total of 71 genes with a putative diapause function were annotated based on these previous studies (Dataset S9). Of these, 14 are duplicated in the Ae. albopictus genome, including several with known diapause-related functions such as lipid metabolism, elongation of long-chain hydrocarbons, and hormone signaling.
Approximately 211 Ae. albopictus genes in expansion families are represented in transcriptomes of mosquitoes in diapause and nondiapause conditions in preadult and adult stages (27–30) (Dataset S10). Of these, 140 (66%) are expressed differentially during at least one of the life-cycle stages examined under diapause vs. nondiapause conditions, which is a greater percentage than the overall proportion of differentially expressed gene models in the transcriptome as a whole (P = 0.022; SI Appendix, Table S4.3). Furthermore, 96 of the 140 differentially expressed genes represent superfamilies of stress response, lipid metabolism, gene expression regulation, serine protease-related, and other genes (SI Appendix, Table S4.1). The proportion of genes within each category that show contrasting patterns of diapause-associated differential transcript accumulation (e.g., higher under diapause conditions at the preadult stage, no differential accumulation or lower under diapause conditions at the adult stage) was determined across preadult vs. adult stages of the life cycle. The superfamily categories of lipid metabolism (P = 0.043), gene expression regulation (P = 0.012), serine protease-related (P = 0.003), and stress response (P = 0.043) were enriched significantly for genes with contrasting patterns of diapause-associated differential transcript accumulation across the life cycle relative to the transcriptome database as a whole. These results are consistent with the hypothesis that gene family expansion can give rise to flexible gene expression across the life cycle and thereby contribute to the tolerance of environmental heterogeneity. Lipid metabolism also was implicated previously as an important transcriptional component of the diapause program in Ae. albopictus, and gene expression regulation is implicated as an important component of diapause-based extensive differential transcript accumulation (>5,000 genes represented) under diapause and nondiapause conditions (27–30). The role of contrasting differential transcript accumulation for serine protease genes across the life cycle remains unclear.
Detoxification (cytochrome-oxidase P450, carboxyl/cholinesterase, glutathione S-transferases) and ABC transporter gene families.
The Ae. albopictus genome contains 186 full-length cytochrome-oxidase P450 (CYP) genes, compared with 168, 104, and 87 in Ae. aegypti, An. gambiae, and D. melanogaster, respectively (Table 2, SI Appendix, Figs. S5.1–S5.6 and Table S5.3, and Datasets S12 and S13), and 196 are reported in Cx. quinquefasciatus (31). Approximately 24 CYP pseudogenes also were found. Approximately 20% of the Ae. albopictus genes are clustered on three scaffolds (scaffolds 64, 501, and 4011). All orthologs of the CYP9J family, the main pyrethroid metabolizers in Ae. aegypti (32), were identified in Ae. albopictus.
Table 2.
Numbers of genes belonging to different xenobiotic and resistance gene families
| Gene type | D. melanogaster | An. gambiae | Ae. aegypti | Cx. quinquefasciatus | Ae. albopictus* |
| Cytochrome P450s† | 87 | 104 | 168 | 196 | 186 (210) |
| Glutathione-S-transferases†‡ | 37 | 28 | 26 | 35 | 32 (37) |
| CCEs† | 34 | 46 | 59 | 71 | 64 (71) |
| ABC transporters§ | 56 | 52 | 58 | — | 71 |
The total number of genes including pseudogenes for Ae. albopictus is shown in parentheses.
Numbers derived from this study, Strode et al., 2008 (41), Yan et al., 2012 (31), VectorBase (92), and FlyBase (93).
Cytosolic glutathione-S-transferases only.
Numbers derived from Dermauw and Van Leeuwen (94) and the present study.
Most insects have one or two genes in the CYP4G family (33), and we identified three each in Ae. albopictus—AalbCYP042, AalbCYP052, and AalbCYP125—and Ae. aegypti (SI Appendix, Figs. S5.2 and S5.3). Cyp4g1 in D. melanogaster is the most highly expressed of all fruit fly CYP genes, and encodes an insect-specific P450 oxidative decarboxylase with a role in cuticular hydrocarbon biosynthesis (33). Phylogenetic analysis shows that two Ae. albopictus CYP4Gs (042 and 052) cluster together and do not cluster on a 1:1 basis with the Ae. aegypti CYP4Gs, supporting the conclusion that gene duplication likely occurred after divergence of the two lineages. The abundant expression of AalbCYP042 and AalbCYP052 during egg formation and diapause (27) is consistent with a potential role in promoting mosquito survival during unfavorable environmental conditions and may contribute to the invasion success of the species. An ortholog of D. melanogaster Cyp4g15 in the wild silk moth Antheraea yamamai (CYP4G25) is also expressed highly during diapause of the pharate first-instar larvae (34).
Sixty-four full-length carboxyl/cholinesterase (CCE) genes were identified in Ae. albopictus, a number similar to that found in Ae. aegypti and Cx. quinquefasciatus, but higher than the numbers found in D. melanogaster and An. gambiae (Table 2, SI Appendix, Fig. S5.7 and Table S5.6, and Datasets S14 and S15). A CCE gene, CCEae3A, implicated in temephos resistance in Ae. aegypti (35) and Ae. albopictus (36), is present in Ae. albopictus as two tandemly duplicated genes (AalBCCE013 and AalbCCE014). Acetylcholinesterases are major insecticide targets, and, in contrast to other insects that only have one or two such genes, three (AalbCCE031 and AalbCCE100, orthologs of Ace1, and AalbCCE101, an ortholog of Ace2) are annotated in the Ae. albopictus genome. Furthermore, a notable expansion to 18 and 9 genes was found for the subfamily of cricklet co-orthologs and juvenile hormone esterases, respectively (37, 38). In D. melanogaster the cricklet gene is located at a locus essential for mediating the response of adult tissues to juvenile hormone (37, 38) and allelic variants in the gene contribute to altitudinal variation in development time (39). Among the mosquito species, Ae. albopictus had the highest number of cricklet co-orthologs, with five cases in which Ae. albopictus has two or three copies compared with one in Ae. aegypti (SI Appendix, Fig. S5.7). The numbers of glutactins in Ae. albopictus and Ae. aegypti are nearly double those found in An. gambiae and D. melanogaster. The function of glutactins is not well understood, but a role in the formation of the eggshell matrix was proposed (7, 40).
Ae. albopictus has 32 full-length cytosolic glutathione S-transferase (GST) genes (Table 2, SI Appendix, Fig. S5.8 and Table S5.9, and Datasets S16 and S17), more than Ae. aegypti and An. gambiae, but fewer than D. melanogaster and Cx. quinquefasciatus. This expansion results mainly from the higher number of delta- and epsilon-class GSTs, the majority of which are associated with insecticide resistance (41). Finally, we annotated 71 putative ABC genes in Ae. albopictus, more copies than are found in Ae. aegypti, D. melanogaster and most other insect species (Table 2, SI Appendix, Figs. S5.9–S5.16 and Table S5.12, and Datasets S18 and S19). Orthologs of ABC proteins conserved widely in metazoans were identified, with five cases of duplicated ABCC transporter genes, a family known for its role in multidrug resistance in humans. Similarly, six duplications of Ae. aegypti ABCG genes are found in Ae. albopictus. Human ABCG transporters are involved in lipid transport, and the duplication in Ae. albopictus of genes encoding these proteins may be related to the complex regulation of increased lipid content in diapausing vs. nondiapausing pharate larvae. These combined findings provide genomic support for the potential of a robust response of Ae. albopictus to environmental stresses and insecticides.
Odorant-binding and odorant receptor proteins.
A total of 86 odorant-binding proteins (OBPs) and 158 odorant receptor (OR) genes are predicted in the Ae. albopictus genome (Table 3 and SI Appendix, Table S6.1). All the OBPs are members of the pheromone-binding protein (PBP)/GOBP family, 47 of which are PBPs with putative functions associated with communication (42–45). Orthologs of 156 of the OR genes could be found in Ae. aegypti. Comparisons of the Ae. albopictus repertoire with An. gambiae, Ae. aegypti, Cx. quinquefasciatus, and D. melanogaster confirmed previous reports of smaller numbers of genes encoding OBPs and ORs in the fruit fly than the mosquito species (46–56) (Table 3 and SI Appendix, Fig. S6.2). Both Ae. albopictus and Ae. aegypti have more of both classes of genes than An. gambiae and Cx. quinquefasciatus, and 43 OBPs and two OR putative novel genes (i.e., no orthologs identified in other species) contribute to these differences (SI Appendix, Table S6.1). Most of the putative OBP genes encode a predicted N-terminal signal peptide, a feature characteristic of their respective proteins (52, 56, 57), and had molecular weights ranging from 14 to 41 kDa. Conserved domain database (CDD) predictions showed that they belong to the PBP/GOBP family, and amino acid alignments confirmed the conservation of six characteristic cysteines (SI Appendix, Fig. S6.3). The putative OR genes encode seven transmembrane domain proteins characteristic of this family.
Table 3.
Numbers of annotated genes encoding OBP and OR
| Protein | Ae. albopictus | Ae. aegypti | An. gambiae | Cx. quinquefasciatus | D. melanogaster |
| OBP | 86 | 64 | 58 | 50 | 51 |
| OR | 158 | 110 | 80 | 88 | 61 |
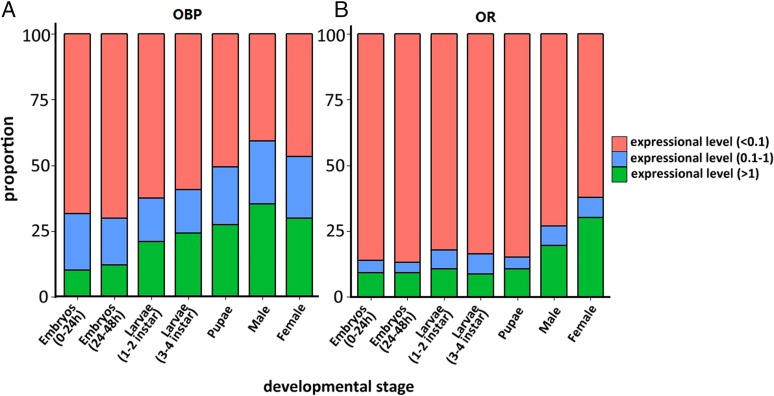
The expression profiles of OBPs and ORs in Ae. albopictus and other mosquitoes in which data are available show an increasing complexity in the number of transcriptionally active genes as the insects progress through development (47, 56, 57) (Fig. 3 and SI Appendix, Fig. S6.4 and Table S6.2). Furthermore, although their mRNAs are present at relatively low abundance, these genes exhibit distinct temporal- and tissue-specific expression. The increasing transcriptional activity may contribute to the ability of this group of insects to navigate increasingly complex environments as they transition from food location in aqueous larval habitats to the mate-detection, host-seeking, feeding-preference and oviposition site-identification abilities of the adults.
Fig. 3.
Proportion of OBP and OR gene transcripts in each expression level through development. The proportion of (A) OBP and (B) OR genes, containing nonexpressed genes, in each expressional level of each stage was calculated. The red, blue, and green columns refer to high (>1), medium (0.1–1) and low (<0.1) RPKM levels, respectively.
Sex-biased gene expression.
Sex-biased gene expression is responsible for the extensive phenotypic and behavioral differences exhibited by male and female mosquitoes (58). RNA-seq analysis of separate samples derived from Ae. albopictus adult females and males identified 8,559 and 4,140 genes, respectively, with sex-biased expression profiles, and the total represents ∼50% of all annotated genes in the genome (Dataset S24). A total of 246 and 268 genes in females and males, respectively, exhibited sex-specific expression (Datasets S25 and S26). Genes with sex-biased expression are enriched significantly (P < 0.01) in GO terms for 26 biological process, 11 cellular compartment, and 34 molecular function categories, with the highest representations in RNA metabolic processes, nucleus, and ion binding (Datasets S27–S30). Further studies are needed to link many of these genes with specific roles in sex-specific biology.
Sex-determination genes.
Aedes mosquitoes, including Ae. albopictus, have a homomorphic sex-determining chromosome with a small male-specific region called the M-locus containing DNA functioning as a dominant male-determining factor (M factor) (59, 60). Importantly, an Ae. aegypti gene, Nix, encoding the phenotypic properties of the M factor (59), has an ortholog, KP765684, in Ae. albopictus. Orthologs of other genes likely involved in the sex-determination pathway also were found. Several orthologs of transformer2 (tra2) and those of doublesex and fruitless, the terminal regulatory genes in the sex-determination pathway, were identified (SI Appendix, Table S7.4). No ortholog of transformer was found, most likely because it evolves rapidly resulting in sequence divergence (61–63).
Immune-related genes.
Comparative analysis with curated sets of immune-related genes (64, 65) identified 554, 476, 536, 400, and 345 immunity genes (includes candidate pseudogenes) in Ae. albopictus, Ae. aegypti, An. gambiae, Cx. quinquefasciatus, and D. melanogaster, respectively (SI Appendix, Table S8.1). Expansions of several gene families (SPZ, BGBP, SRRP, GALE, TOLL, SCR, TOLLPATH, SOD, APHAG, PPO, and CLIP) account for the large Ae. albopictus immunity-related gene repertory. Analyses of the transcriptome data reveal increased abundances of the immune-related gene products in the postembryonic insects (SI Appendix, Fig. S8.6). A total of 468 immune-related transcripts [representing ∼88% (486 of 554) of the total predicted genes] were found in adult mosquitoes, including 166 related to immune recognition, 106 involved in gene modulation, 100 in signal transduction, and 96 in effector molecule (SI Appendix, Fig. S8.7). The top three most abundant transcripts represent effector AMPs, recognition LRRs, and modulation CLIPs (56, 53, and 51, respectively).
Summary and Conclusions
It was known previously that Ae. albopictus had a large genome, and this is confirmed in the present report. This large size is evident in the greater number of repetitive DNA elements, expansion in all protein-encoding gene categories examined, and the amount of the genome represented by insertions of DNA copies of RNA viruses. The genome size may account in part for why this mosquito is successful as an invasive species. The large repertory of noncoding and coding DNA may provide the genetic substrates from which adaptation emerges following selection in novel environments. A draft sequence was published recently of an Ae. albopictus strain, Fellini, that recently invaded Italy (66). Detailed molecular comparisons of that strain with the ancestral one presented here are expected to highlight aspects of genome evolution as this species adapts to more temperate zones.
Materials and Methods
Mosquitoes.
The Foshan strain of Ae. albopictus was obtained from the Center for Disease Control and Prevention of Guangdong Province, China, where it has been in culture since 1981. Mosquitoes were reared at 28 °C and 70–80% relative humidity with 14/10 h light/dark cycles. Larvae were reared in pans and fed on finely ground fish food mixed at a 1:1 ratio with yeast powder. Adults were kept in 30-cm3 cages and allowed access to a cotton wick soaked in 0.2 g/ml sucrose as a carbohydrate source. Adult females were allowed to feed on anesthetized mice 3–4 d after eclosion.
DNA Sequencing.
Approximately 1.414 μg of genomic DNA isolated from a single Ae. albopictus pupa of a ninth-generation isofemale line was subjected to whole-genome amplification (67) to produce 243.2 μg of DNA. Amplified DNA was used to construct paired-end short-insert (170,500 and 800 bp in length) and mate-paired long-insert (2 kb, 5 kb, 10 kb, and 20 kb) genomic libraries, and these were sequenced by using the HisEq.2000 platform.
Data Quality Control and Assembly.
The raw sequence data were filtered before assembly by removing duplicated reads caused by gene amplification and reads contaminated by adapters, trimming continuous low-quality bases on 5′ ends according to quality graphs, and filtering reads with a significant excess of “N” and low-quality bases. The assembler SOAPdenovo (version 2.04) (68), SSPACE (version 2.0) (69), and Gapcloser (version 1.10) (68) were used for genome assembly. Overlapped pair-end reads from the 170 insert-size libraries were connected first to yield long sequences. A 97-bp sequence from the connected long reads was used next to construct contigs. All usable reads from different insert-size libraries then were realigned to the contigs by using SSPACE. The resulting linking information was used to produce the final scaffold construction, and this was followed by gap-filling of the scaffolds. The sequences of Wolbachia pipientis were aligned to the assembly, and the scaffolds matching them were removed to avoid the contamination.
Accuracy of Genome Assembly.
The quality of the draft genome was evaluated by assessing the sequencing depth and coverage by using available mRNA and fosmid sequences. All useable sequence reads were realigned to the draft genome by using SOAP2 (70).
Transcriptome Sequencing (RNA-Seq).
Transcriptomes were derived from libraries comprising mRNA from seven developmental stages of the Foshan strain: mixed-sex samples of 100 embryos at 0–24 h post deposition (hpd), 100 embryos at 24–48 hpd, a combined pool of 8 first-and second-instar larvae, a combined pool of five third- and fourth-instar larvae, five pupae of all stages, and five each of adult males and sugar-fed adult females. TRIzol reagent (Invitrogen) and RNase-free DNase I were used to extract and treat total RNA. Polyadenylated (i.e., polyA+) mRNA was enriched by using oligo-dT beads, fragmented, and primed randomly during the first-strand synthesis by reverse transcription. Second-strand cDNA was synthesized by using RNase H and DNA polymerase I to create double-stranded fragments. The ds cDNA was applied to 200-bp paired-end RNA-seq libraries per Illumina protocols and sequenced with 90 bp at each end on the Illumina HiSEq 2000 platform. The cDNA library was normalized by the duplex-specific nuclease method (71) followed by cluster generation on the Illumina HiSEq.2000 platform. Transcript reads were mapped by TopHat and analyzed subsequently with custom Perl scripts. Gene expression levels were calculated as reads per kilobase of exon model per million mapped reads (RPKM) (72). Genes expressed differentially between two samples were detected by using a method based on a Poisson distribution, and samples were normalized for differences in the RNA output size, sequencing depth, and gene length. Genes identified in at least one experiment with a minimum twofold difference (RPKM) in two experiments and an false discovery rate of <0.001 were defined as differentially expressed. Enrichment analysis was performed by using Enrich Pipeline (73).
Gene Annotation.
De novo gene prediction by using RNA-seq data and Ae. aegypti, D. melanogaster, An. gambiae, and Cx. quinquefasciatus protein sequences aligned to the Ae. albopictus genome with TBLASTN (74) was performed to produce homology-based predictions. Putatively homologous genome sequences were aligned with the matching proteins by using GeneWise (75) to define gene models. Augustus (76) and Genscan (77) were used with appropriate parameters for de novo prediction of coding genes. Homology-based and de novo-derived gene sets were merged to form a comprehensive and nonredundant reference gene set using GLEAN (sourceforge.net/projects/glean-gene). The transcriptome reads from the seven different samples were mapped to the genome assembly by using TopHat (78) to give RNA-seq-based predictions. TopHat mapping results were combined, and Cufflinks (79) was applied to predict transcript structures. A total of 1,000 intact genes also were selected from the homology-based prediction to pass a fifth-order Markov model, then to predict the ORFs of RNA transcripts based on the hidden Markov model. Finally, the RNA transcripts were integrated with the GLEAN gene set to form the final nonredundant gene set.
Manual annotation of putative diapause-related genes was performed by using Web Apollo (80) to integrate the original GLEAN/Cuff annotations on the scaffolds with Maker annotations (81) based on a comprehensive diapause transcriptome (27–30). Annotated genes included those involved in chromatin remodeling, lipid metabolism, hormonal regulation, circadian rhythms, and other functions. Final annotations were based on the presence of a start codon, stop codon, canonical splice sites, and extended 5′ or 3′ UTRs that were supported by Maker or exonerate (82) alignment of contigs from the transcriptome.
Gene Functional Annotation.
Ae. albopictus protein sequences were aligned by using InterPro (83), Swiss-Prot (84), Kyoto Encyclopedia of Genes and Genomes (KEGG) (85), and TrEMBL (84) to infer their biological functions or their molecular pathways. GO descriptions of gene products were retrieved from InterPro. The symbol of each gene was assigned based on the best match derived from the alignments with Swiss-Prot databases by using BLASTP. Motifs and domains were annotated by InterPro by searching publicly available databases, including Pfam, PRINTS, PANTHER, PROSITE, ProDom, and SMART. Genes also were mapped to KEGG pathway maps by searching KEGG databases and finding the best hit for each gene.
Gene Family Clustering.
The TreeFam methodology (86) was used to define gene families using data from five mosquito species (Ae. albopictus, An. gambiae, Ae. aegypti, Cx. quinquefasciatus, and An. darlingi) as references, and the fruit fly D. melanogaster was used as the outgroup. BLASTP was used to find all homologous relationships among protein sequences of the six species with e-values <1e-10, and Solar (in-house software, version 0.9.6) was used to conjoin high-scoring segment pairs between each pair of protein homologs. Protein sequence similarity was assessed with bit-score, and protein encoding genes clustered into gene families by a hierarchical clustering algorithm (an implementation included in the Treefam pipeline, version 0.5.0) with an algorithm analogous to average-linkage clustering with the parameters set to be “-w 5 -s 0.33 -m 100000”.
Phylogenetic Tree Construction and Divergence Time Estimate.
A total of 2,096 single-copy gene families defined as orthologous genes according to the Treefam pipelines chosen in this analysis were assigned to a coding sequence (CDS) based on the alignment results. All CDSs and the 4d sites (fourfold degenerate synonymous sites) were extracted from each alignment and concatenated to one super gene for the six species. PhyMLv3.0 (parameters: -m HKY85, other default) was used to construct a phylogenetic tree for the six species. The chain length was set to 100,000 (1 sample/100 generations), and the first 1,000 samples were burned in. The transition/transversion ratio was estimated as a free parameter. Divergence time was estimated by using the program MCMCTREE (version 4), which was part of the PAML package. “JC69” models in MCMCTREE program were used in our calculations.
Expansion and Contraction of Gene Families.
Computational Analysis of gene Family Evolution (version 2.1) (87) was used to detect gene family expansion and contraction in Ae. albopictus, An. gambiae, Ae. aegypti, Cx. quinquefasciatus, An. darlingi, and D. melanogaster with the parameters “P-value threshold 0.05, number of random 10,000, and search for the λ value.” Gene families with P values <0.05 were analyzed manually.
Detection of Positively Selected Genes.
BLASTP and TreeFam methodologies were used to define orthologs among Ae. albopictus, An. gambiae, Ae. aegypti, Cx. quinquefasciatus, An. darlingi, and D. melanogaster. The coding sequences of the orthologs were aligned by using Prank software (88) (code.google.com/p/prank-msa/) with default parameters. The genes were filtered even if the alignment rate of the gene was less than 80% in only one species. Ka (nonsynonymous substitution rates) and Ks (synonymous substitution rates) were calculated for the aligned orthologs by using KaKs calculator software (89) (version 1.2, parameter “-m YN”) with default parameters.
DNA Loss Analysis in Mosquito Genomes.
The DNA loss rates for neutrally evolved DNA sequences in mosquito genomes were estimated by using a previously described method (90). In brief, the consensus sequences of autonomous non-LTR retrotransposons in the focal mosquito genomes were collected. The consensus sequences for Ae. aegypti and Cx. quinquefasciatus were downloaded from TEfam (tefam.biochem.vt.edu/tefam/index.php). The consensus sequences for Ae. albopictus were generated in the present study by using RepeatScout (91). Second, the consensus sequences were trimmed to keep only the protein-coding regions. Third, the consensus sequences after trimming were used as a repeat library to mask their corresponding genomic sequences by RepeatMasker (www.repeatmasker.org/) to generate pairwise alignment files. We used the obtained alignments to eliminate all non-LTR sequences with nonrandom distributions of substitutions across codon positions (χ2 test, P < 0.05) to avoid counting substitutions that occurred along master element lineages. Finally, for each remaining non-LTR element copy, the numbers of insertions, deletions, and substitutions relative to the consensus sequence were obtained based on the RepeatMasker-generated alignment, and the sums of these values for every individual element copy were used to represent the total amounts of DNA gained and lost through small indels (≤30 bp) in the focal mosquito genome (base pairs deleted minus base pairs inserted/substitution).
Analyses of Specific Gene Features, Gene Families, and Developmentally Regulated Gene Expression.
The specific materials and methods used in the discovery and analysis of TEs, and integrated flavivirus-like sequences, are described in the SI Appendix. Discovery and analysis of gene family members involved in insecticide resistance, diapause, sex determination, immunity, and olfaction are also described in the SI Appendix.
Supplementary Material
Acknowledgments
This work was supported by National Natural Science Foundation of China Grants U0832004, 81371845, and 81420108024 (to X.-G.C.); Research Team Program of Natural Science Foundation of Guangdong Grant 2014A030312016 (to X.-G.C.); Scientific and Technological Program of Guangdong Grant 2013B051000052 (to X.-G.C.); International Cooperation Program of Guangzhou Grant 2013J4500016 (to X.-G.C.); National Institute of Allergy and Infectious Diseases Grants AI083202 (to X.-G.C.), D43TW009527 (to G.Y.), and R37AI029746 (to A.A.J.); Marie Curie International Outgoing Fellowship PIOF-GA-2011-303312 (to R.M.W.); and the Leading Scholar Program of Guangdong (G.Y.). W.D. is a postdoctoral fellow of the Fund for Scientific Research Flanders.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the GenBank database (accession nos. SRA245721 and SRA215477), National Center for Biotechnology Information (NCBI; ID code JXUM00000000 [genome assembly]), and NCBI Transcriptome Shotgun Assembly database, www.ncbi.nlm.nih.gov/genbank/TSA.html (ID code GCLM00000000).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1516410112/-/DCSupplemental.
References
- 1.Bonizzoni M, Gasperi G, Chen X, James AA. The invasive mosquito species Aedes albopictus: Current knowledge and future perspectives. Trends Parasitol. 2013;29(9):460–468. doi: 10.1016/j.pt.2013.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paupy C, Delatte H, Bagny L, Corbel V, Fontenille D. Aedes albopictus, an arbovirus vector: From the darkness to the light. Microbes Infect. 2009;11(14-15):1177–1185. doi: 10.1016/j.micinf.2009.05.005. [DOI] [PubMed] [Google Scholar]
- 3.Bonizzoni M, et al. Complex modulation of the Aedes aegypti transcriptome in response to dengue virus infection. PLoS One. 2012;7(11):e50512. doi: 10.1371/journal.pone.0050512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cancrini G, et al. Aedes albopictus is a natural vector of Dirofilaria immitis in Italy. Vet Parasitol. 2003;118(3-4):195–202. doi: 10.1016/j.vetpar.2003.10.011. [DOI] [PubMed] [Google Scholar]
- 5.Pietrobelli M. Importance of Aedes albopictus in veterinary medicine. Parassitologia. 2008;50(1-2):113–115. [PubMed] [Google Scholar]
- 6.Nene V, et al. Genome sequence of Aedes aegypti, a major arbovirus vector. Science. 2007;316(5832):1718–1723. doi: 10.1126/science.1138878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arensburger P, et al. Sequencing of Culex quinquefasciatus establishes a platform for mosquito comparative genomics. Science. 2010;330(6000):86–88. doi: 10.1126/science.1191864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marinotti O, et al. The genome of Anopheles darlingi, the main neotropical malaria vector. Nucleic Acids Res. 2013;41(15):7387–7400. doi: 10.1093/nar/gkt484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Neafsey DE, et al. Mosquito genomics. Highly evolvable malaria vectors: The genomes of 16 Anopheles mosquitoes. Science. 2015;347(6217):1258522. doi: 10.1126/science.1258522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Severson DW, Behura SK. Mosquito genomics: Progress and challenges. Annu Rev Entomol. 2012;57:143–166. doi: 10.1146/annurev-ento-120710-100651. [DOI] [PubMed] [Google Scholar]
- 11.Rai KS, Black WC., 4th Mosquito genomes: Structure, organization, and evolution. Adv Genet. 1999;41:1–33. doi: 10.1016/s0065-2660(08)60149-2. [DOI] [PubMed] [Google Scholar]
- 12.Black WC, 4th, Rai KS. Genome evolution in mosquitoes: Intraspecific and interspecific variation in repetitive DNA amounts and organization. Genet Res. 1988;51(3):185–196. doi: 10.1017/s0016672300024289. [DOI] [PubMed] [Google Scholar]
- 13.Reidenbach KR, et al. Phylogenetic analysis and temporal diversification of mosquitoes (Diptera: Culicidae) based on nuclear genes and morphology. BMC EvolBiol. 2009;9:298. doi: 10.1186/1471-2148-9-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Petrov DA. Mutational equilibrium model of genome size evolution. Theor Popul Biol. 2002;61(4):531–544. doi: 10.1006/tpbi.2002.1605. [DOI] [PubMed] [Google Scholar]
- 15.Sun C, et al. LTR retrotransposons contribute to genomic gigantism in plethodontid salamanders. Genome Biol Evol. 2012;4(2):168–183. doi: 10.1093/gbe/evr139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crochu S, et al. Sequences of flavivirus-related RNA viruses persist in DNA form integrated in the genome of Aedes spp. mosquitoes. J Gen Virol. 2004;85(pt 7):1971–1980. doi: 10.1099/vir.0.79850-0. [DOI] [PubMed] [Google Scholar]
- 17.Rizzo F, et al. Molecular characterization of flaviviruses from field-collected mosquitoes in northwestern Italy, 2011-2012. Parasit Vectors. 2014;7:395. doi: 10.1186/1756-3305-7-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roiz D, Vázquez A, Seco MP, Tenorio A, Rizzoli A. Detection of novel insect flavivirus sequences integrated in Aedes albopictus (Diptera: Culicidae) in Northern Italy. Virol J. 2009;6:93. doi: 10.1186/1743-422X-6-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tromas N, Zwart MP, Forment J, Elena SF. Shrinkage of genome size in a plant RNA virus upon transfer of an essential viral gene into the host genome. Genome BiolEvol. 2014;6(3):538–550. doi: 10.1093/gbe/evu036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ballinger MJ, Bruenn JA, Taylor DJ. Phylogeny, integration and expression of sigma virus-like genes in Drosophila. Mol Phylogenet Evol. 2012;65(1):251–258. doi: 10.1016/j.ympev.2012.06.008. [DOI] [PubMed] [Google Scholar]
- 21.Goic B, et al. RNA-mediated interference and reverse transcription control the persistence of RNA viruses in the insect model Drosophila. Nat Immunol. 2013;14(4):396–403. doi: 10.1038/ni.2542. [DOI] [PubMed] [Google Scholar]
- 22.Geuking MB, et al. Recombination of retrotransposon and exogenous RNA virus results in nonretroviral cDNA integration. Science. 2009;323(5912):393–396. doi: 10.1126/science.1167375. [DOI] [PubMed] [Google Scholar]
- 23.Barrón MG, Fiston-Lavier AS, Petrov DA, González J. Population genomics of transposable elements in Drosophila. Annu Rev Genet. 2014;48:561–581. doi: 10.1146/annurev-genet-120213-092359. [DOI] [PubMed] [Google Scholar]
- 24.Vázquez A, et al. Novel flaviviruses detected in different species of mosquitoes in Spain. Vector Borne Zoonotic Dis. 2012;12(3):223–229. doi: 10.1089/vbz.2011.0687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Denlinger DL, Armbruster PA. Mosquito diapause. Annu Rev Entomol. 2014;59:73–93. doi: 10.1146/annurev-ento-011613-162023. [DOI] [PubMed] [Google Scholar]
- 26.Lounibos LP, Escher RL, Nishimura N. Retention and adaptiveness of photoperiodic EGG diapause in Florida populations of invasive Aedes albopictus. J Am Mosq Control Assoc. 2011;27(4):433–436. doi: 10.2987/11-6164.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang X, Poelchau MF, Armbruster PA. Global transcriptional dynamics of diapause induction in non-blood-fed and blood-fed Aedes albopictus. PLoS Negl Trop Dis. 2015;9(4):e0003724. doi: 10.1371/journal.pntd.0003724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poelchau MF, Reynolds JA, Denlinger DL, Elsik CG, Armbruster PA. Transcriptome sequencing as a platform to elucidate molecular components of the diapause response in the Asian tiger mosquito, Aedes albopictus. Physiol Entomol. 2013;38(2):173–181. doi: 10.1111/phen.12016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poelchau MF, Reynolds JA, Elsik CG, Denlinger DL, Armbruster PA. RNA-Seq reveals early distinctions and late convergence of gene expression between diapause and quiescence in the Asian tiger mosquito, Aedes albopictus. J Exp Biol. 2013;216(pt 21):4082–4090. doi: 10.1242/jeb.089508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Poelchau MF, Reynolds JA, Elsik CG, Denlinger DL, Armbruster PA. 2013. Deep sequencing reveals complex mechanisms of diapause preparation in the invasive mosquito, Aedes albopictus. Proc Biol Sci 280(1759), 20130143.
- 31.Yan L, et al. Transcriptomic and phylogenetic analysis of Culex pipiens quinquefasciatus for three detoxification gene families. BMC Genomics. 2012;13:609. doi: 10.1186/1471-2164-13-609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stevenson BJ, Pignatelli P, Nikou D, Paine MJ. 2012. Pinpointing P450s associated with pyrethroid metabolism in the dengue vector, Aedes aegypti: Developing new tools to combat insecticide resistance. PLoS Negl Trop Dis 6(3):e1595. [DOI] [PMC free article] [PubMed]
- 33.Qiu Y, et al. An insect-specific P450 oxidative decarbonylase for cuticular hydrocarbon biosynthesis. Proc Natl Acad Sci USA. 2012;109(37):14858–14863. doi: 10.1073/pnas.1208650109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang P, Tanaka H, Kuwano E, Suzuki K. A novel cytochrome P450 gene (CYP4G25) of the silkmoth Antheraea yamamai: Cloning and expression pattern in pharate first instar larvae in relation to diapause. J Insect Physiol. 2008;54(3):636–643. doi: 10.1016/j.jinsphys.2008.01.001. [DOI] [PubMed] [Google Scholar]
- 35.Poupardin R, Srisukontarat W, Yunta C, Ranson H. Identification of carboxylesterase genes implicated in temephos resistance in the dengue vector Aedes aegypti. PLoS Negl Trop Dis. 2014;8(3):e2743. doi: 10.1371/journal.pntd.0002743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grigoraki L, et al. Transcriptome profiling and genetic study reveal amplified carboxylesterase genes implicated in temephos resistance, in the Asian Tiger Mosquito Aedes albopictus. PLoS Negl Trop Dis. 2015;9(5):e0003771. doi: 10.1371/journal.pntd.0003771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Campbell PM, et al. Identification of a juvenile hormone esterase gene by matching its peptide mass fingerprint with a sequence from the Drosophila genome project. Insect Biochem Mol Biol. 2001;31(6-7):513–520. doi: 10.1016/s0965-1748(01)00035-2. [DOI] [PubMed] [Google Scholar]
- 38.Shirras AD, Bownes M. Cricklet: A locus regulating a number of adult functions of Drosophila melanogaster. Proc Natl Acad Sci USA. 1989;86(12):4559–4563. doi: 10.1073/pnas.86.12.4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mensch J, et al. Stage-specific effects of candidate heterochronic genes on variation in developmental time along an altitudinal cline of Drosophila melanogaster. PLoS One. 2010;5(6):e11229. doi: 10.1371/journal.pone.0011229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fakhouri M, et al. Minor proteins and enzymes of the Drosophila eggshell matrix. Dev Biol. 2006;293(1):127–141. doi: 10.1016/j.ydbio.2006.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Strode C, et al. Genomic analysis of detoxification genes in the mosquito Aedes aegypti. Insect Biochem Mol Biol. 2008;38(1):113–123. doi: 10.1016/j.ibmb.2007.09.007. [DOI] [PubMed] [Google Scholar]
- 42.Xu YL, et al. Large-scale identification of odorant-binding proteins and chemosensory proteins from expressed sequence tags in insects. BMC Genomics. 2009;10:632. doi: 10.1186/1471-2164-10-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu W, Cornel AJ, Leal WS. Odorant-binding proteins of the malaria mosquito Anopheles funestus sensustricto. PLoS One. 2010;5(10):e15403. doi: 10.1371/journal.pone.0015403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mao Y, et al. Crystal and solution structures of an odorant-binding protein from the southern house mosquito complexed with an oviposition pheromone. Proc Natl Acad Sci USA. 2010;107(44):19102–19107. doi: 10.1073/pnas.1012274107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vogt RG, Große-Wilde E, Zhou JJ. The Lepidoptera Odorant Binding Protein gene family: Gene gain and loss within the GOBP/PBP complex of moths and butterflies. Insect Biochem Mol Biol. 2015;62:142–153. doi: 10.1016/j.ibmb.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 46.Bohbot JD, et al. Conservation of indole responsive odorant receptors in mosquitoes reveals an ancient olfactory trait. Chem Senses. 2011;36(2):149–160. doi: 10.1093/chemse/bjq105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bohbot J, et al. Molecular characterization of the Aedes aegypti odorant receptor gene family. Insect Mol Biol. 2007;16(5):525–537. doi: 10.1111/j.1365-2583.2007.00748.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hill CA, et al. G protein-coupled receptors in Anopheles gambiae. Science. 2002;298(5591):176–178. doi: 10.1126/science.1076196. [DOI] [PubMed] [Google Scholar]
- 49.Fox AN, Pitts RJ, Zwiebel LJ. A cluster of candidate odorant receptors from the malaria vector mosquito, Anopheles gambiae. Chem Senses. 2002;27(5):453–459. doi: 10.1093/chemse/27.5.453. [DOI] [PubMed] [Google Scholar]
- 50.Graham LA, Davies PL. The odorant-binding proteins of Drosophila melanogaster: Annotation and characterization of a divergent gene family. Gene. 2002;292(1-2):43–55. doi: 10.1016/s0378-1119(02)00672-8. [DOI] [PubMed] [Google Scholar]
- 51.Kent LB, Walden KK, Robertson HM. The Gr family of candidate gustatory and olfactory receptors in the yellow-fever mosquito Aedes aegypti. Chem Senses. 2008;33(1):79–93. doi: 10.1093/chemse/bjm067. [DOI] [PubMed] [Google Scholar]
- 52.Pelletier J, Leal WS. Genome analysis and expression patterns of odorant-binding proteins from the Southern House mosquito Culex pipiens quinquefasciatus. PLoS One. 2009;4(7):e6237. doi: 10.1371/journal.pone.0006237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pelletier J, Hughes DT, Luetje CW, Leal WS. An odorant receptor from the southern house mosquito Culex pipiens quinquefasciatus sensitive to oviposition attractants. PLoS One. 2010;5(4):e10090. doi: 10.1371/journal.pone.0010090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu PX, Zwiebel LJ, Smith DP. Identification of a distinct family of genes encoding atypical odorant-binding proteins in the malaria vector mosquito, Anopheles gambiae. Insect Mol Biol. 2003;12(6):549–560. doi: 10.1046/j.1365-2583.2003.00440.x. [DOI] [PubMed] [Google Scholar]
- 55.Xia Y, Zwiebel LJ. Identification and characterization of an odorant receptor from the West Nile virus mosquito, Culex quinquefasciatus. Insect Biochem Mol Biol. 2006;36(3):169–176. doi: 10.1016/j.ibmb.2005.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou JJ, He XL, Pickett JA, Field LM. Identification of odorant-binding proteins of the yellow fever mosquito Aedes aegypti: Genome annotation and comparative analyses. Insect MolBiol. 2008;17(2):147–163. doi: 10.1111/j.1365-2583.2007.00789.x. [DOI] [PubMed] [Google Scholar]
- 57.Deng Y, et al. Molecular and functional characterization of odorant-binding protein genes in an invasive vector mosquito, Aedes albopictus. PLoS One. 2013;8(7):e68836. doi: 10.1371/journal.pone.0068836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stamboliyska R, Parsch J. Dissecting gene expression in mosquito. BMC Genomics. 2011;12(1):297. doi: 10.1186/1471-2164-12-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hall AB, et al. SEX DETERMINATION. A male-determining factor in the mosquito Aedes aegypti. Science. 2015;348(6240):1268–1270. doi: 10.1126/science.aaa2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McClelland K, Bowles J, Koopman P. Male sex determination: Insights into molecular mechanisms. Asian J Androl. 2012;14(1):164–171. doi: 10.1038/aja.2011.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Verhulst EC, van de Zande L, Beukeboom LW. Insect sex determination: It all evolves around transformer. Curr Opin Genet Dev. 2010;20(4):376–383. doi: 10.1016/j.gde.2010.05.001. [DOI] [PubMed] [Google Scholar]
- 62.Geuverink E, Beukeboom LW. Phylogenetic distribution and evolutionary dynamics of the sex determination genes doublesex and transformer in insects. Sex Dev. 2014;8(1-3):38–49. doi: 10.1159/000357056. [DOI] [PubMed] [Google Scholar]
- 63.Verhulst EC, Beukeboom LW, van de Zande L. Maternal control of haplodiploid sex determination in the wasp Nasonia. Science. 2010;328(5978):620–623. doi: 10.1126/science.1185805. [DOI] [PubMed] [Google Scholar]
- 64.Waterhouse RM, et al. Evolutionary dynamics of immune-related genes and pathways in disease-vector mosquitoes. Science. 2007;316(5832):1738–1743. doi: 10.1126/science.1139862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bartholomay LC, et al. Pathogenomics of Culex quinquefasciatus and meta-analysis of infection responses to diverse pathogens. Science. 2010;330(6000):88–90. doi: 10.1126/science.1193162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dritsou V, et al. 2015. A draft genome sequence of an invasive mosquito: an Italian Aedes albopictus. Pathog Glob Health Sep 14:2047773215Y0000000031.
- 67.Spits C, et al. Whole-genome multiple displacement amplification from single cells. Nat Protoc. 2006;1(4):1965–1970. doi: 10.1038/nprot.2006.326. [DOI] [PubMed] [Google Scholar]
- 68.Luo R, et al. SOAPdenovo2: An empirically improved memory-efficient short-read de novo assembler. Gigascience. 2012;1(1):18. doi: 10.1186/2047-217X-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Boetzer M, Henkel CV, Jansen HJ, Butler D, Pirovano W. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics. 2011;27(4):578–579. doi: 10.1093/bioinformatics/btq683. [DOI] [PubMed] [Google Scholar]
- 70.Li R, et al. SOAP2: An improved ultrafast tool for short read alignment. Bioinformatics. 2009;25(15):1966–1967. doi: 10.1093/bioinformatics/btp336. [DOI] [PubMed] [Google Scholar]
- 71.Zhulidov PA, et al. Simple cDNA normalization using kamchatka crab duplex-specific nuclease. Nucleic Acids Res. 2004;32(3):e37. doi: 10.1093/nar/gnh031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5(7):621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 73.Chen Y, Liu M, Yan G, Lu H, Yang P. One-pipeline approach achieving glycoprotein identification and obtaining intact glycopeptide information by tandem mass spectrometry. Mol Biosyst. 2010;6(12):2417–2422. doi: 10.1039/c0mb00024h. [DOI] [PubMed] [Google Scholar]
- 74.Gertz EM, Yu YK, Agarwala R, Schäffer AA, Altschul SF. Composition-based statistics and translated nucleotide searches: Improving the TBLASTN module of BLAST. BMC Biol. 2006;4:41. doi: 10.1186/1741-7007-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Birney E, Durbin R. Using GeneWise in the Drosophila annotation experiment. Genome Res. 2000;10(4):547–548. doi: 10.1101/gr.10.4.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Stanke M, Waack S. Gene prediction with a hidden Markov model and a new intron submodel. Bioinformatics. 2003;19(suppl 2):ii215–ii225. doi: 10.1093/bioinformatics/btg1080. [DOI] [PubMed] [Google Scholar]
- 77.Salamov AA, Solovyev VV. Ab initio gene finding in Drosophila genomic DNA. Genome Res. 2000;10(4):516–522. doi: 10.1101/gr.10.4.516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Trapnell C, Pachter L, Salzberg SL. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Roberts A, Pimentel H, Trapnell C, Pachter L. Identification of novel transcripts in annotated genomes using RNA-Seq. Bioinformatics. 2011;27(17):2325–2329. doi: 10.1093/bioinformatics/btr355. [DOI] [PubMed] [Google Scholar]
- 80.Lee E, et al. Web Apollo: A Web-based genomic annotation editing platform. Genome Biol. 2013;14(8):R93. doi: 10.1186/gb-2013-14-8-r93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cantarel BL, et al. MAKER: An easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res. 2008;18(1):188–196. doi: 10.1101/gr.6743907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Slater GS, Birney E. Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics. 2005;6:31. doi: 10.1186/1471-2105-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mulder N, Apweiler R. InterPro and InterProScan: Tools for protein sequence classification and comparison. Methods Mol Biol. 2007;396:59–70. doi: 10.1007/978-1-59745-515-2_5. [DOI] [PubMed] [Google Scholar]
- 84.Bairoch A, Apweiler R. The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res. 2000;28(1):45–48. doi: 10.1093/nar/28.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Li H, et al. TreeFam: A curated database of phylogenetic trees of animal gene families. Nucleic Acids Res. 2006;34(database issue):D572–D580. doi: 10.1093/nar/gkj118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.De Bie T, Cristianini N, Demuth JP, Hahn MW. CAFE: A computational tool for the study of gene family evolution. Bioinformatics. 2006;22(10):1269–1271. doi: 10.1093/bioinformatics/btl097. [DOI] [PubMed] [Google Scholar]
- 88.Wheeler TJ, Kececioglu JD. Multiple alignment by aligning alignments. Bioinformatics. 2007;23(13):i559–i568. doi: 10.1093/bioinformatics/btm226. [DOI] [PubMed] [Google Scholar]
- 89.Zhang Z, et al. KaKs_Calculator: Calculating Ka and Ks through model selection and model averaging. Genomics Proteomics Bioinformatics. 2006;4(4):259–263. doi: 10.1016/S1672-0229(07)60007-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sun C, LópezArriaza JR, Mueller RL. Slow DNA loss in the gigantic genomes of salamanders. Genome Biol Evol. 2012;4(12):1340–1348. doi: 10.1093/gbe/evs103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Price AL, Jones NC, Pevzner PA. De novo identification of repeat families in large genomes. Bioinformatics. 2005;21(suppl 1):i351–i358. doi: 10.1093/bioinformatics/bti1018. [DOI] [PubMed] [Google Scholar]
- 92.Megy K, et al. VectorBase Consortium VectorBase: Improvements to a bioinformatics resource for invertebrate vector genomics. Nucleic Acids Res. 2012;40(database issue):D729–D734. doi: 10.1093/nar/gkr1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.St Pierre SE, Ponting L, Stefancsik R, McQuilton P. FlyBase Consortium FlyBase 102--advanced approaches to interrogating FlyBase. Nucleic Acids Res. 2014;42(database issue):D780–D788. doi: 10.1093/nar/gkt1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Dermauw W, Van Leeuwen T. The ABC gene family in arthropods: Comparative genomics and role in insecticide transport and resistance. Insect Biochem Mol Biol. 2014;45:89–110. doi: 10.1016/j.ibmb.2013.11.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.