Significance
Maintenance of glucose concentrations within a homeostatic range is essential for preserving the function of glucose-sensitive tissues. Perturbations in the mechanisms that control this homeostasis give rise to a continuum of glucopathologies associated with aberrant carbohydrate metabolism. Here we show Steroid Receptor Coactivator 2 (SRC-2) to be an integral coregulator that couples gene output with energetic demand by stabilizing and amplifying transcriptional complexes. This study highlights the collective importance of transcriptional coregulators for coordination of gene expression events and may provide insight for understanding components of polygenic diseases such as type 2 diabetes mellitus.
Keywords: Steroid Receptor Coactivator 2, SRC-2, glucose homeostasis, glucokinase, polygenic disease
Abstract
Despite extensive efforts to understand the monogenic contributions to perturbed glucose homeostasis, the complexity of genetic events that fractionally contribute to the spectrum of this pathology remain poorly understood. Proper maintenance of glucose homeostasis is the central feature of a constellation of comorbidities that define the metabolic syndrome. The ability of the liver to balance carbohydrate uptake and release during the feeding-to-fasting transition is essential to the regulation of peripheral glucose availability. The liver coordinates the expression of gene programs that control glucose absorption, storage, and secretion. Herein, we demonstrate that Steroid Receptor Coactivator 2 (SRC-2) orchestrates a hierarchy of nutritionally responsive transcriptional complexes to precisely modulate plasma glucose availability. Using DNA pull-down technology coupled with mass spectrometry, we have identified SRC-2 as an indispensable integrator of transcriptional complexes that control the rate-limiting steps of hepatic glucose release and accretion. Collectively, these findings position SRC-2 as a major regulator of polygenic inputs to metabolic gene regulation and perhaps identify a previously unappreciated model that helps to explain the clinical spectrum of glucose dysregulation.
Glucose homeostasis is a tightly regulated biological process requiring intricate communication between multiple tissues of the body. Central to this process are the essential functions of the liver that protect the organism during the feeding-to-fasting transition. The feeding-responsive regulation of glucose homeostasis by the liver illustrates how humoral metabolic cues control the delicate balance of glucose in the body. Insulin produced by the endocrine pancreas is released prandially and drives the simultaneous uptake, catabolism, and storage of excess glucose. Conversely, glucagon is released during conditions of fasting in response to low blood sugar, which acts primarily on the liver to initiate glycogenolysis and gluconeogenesis. Together, both of these pathways function to release glucose for use by obligate glucose-using tissues such as the brain and red blood cells.
At the molecular level, transcriptional activation and repression are key cellular events in the feeding-to-fasting transition that fine tunes cellular programmatic shifts between glycolysis and gluconeogenesis. The transcriptional activation of glucose storage and utilization programs and the repression of glucose-producing programs during feeding are ultimately the result of signaling cascades that instruct the actions of transcriptional machinery. A similar process occurs during fasting whereby glucose-producing genes are actively transcribed and glucose-using and storage programs are repressed. This fine balance of activation and repression is often perturbed in diseases of glucose metabolism such as type 2 diabetes mellitus (T2DM), insulin-resistant fatty liver, and various glycogenopathies (1). To enhance our understanding of the molecular pathogenesis of these diseases, it is necessary to define the transcriptional machinery that is responsive to metabolic signals.
Published studies from our laboratory, as well as others, have established Steroid Receptor Coactivator 2 (SRC-2) as a powerful pleiotropic metabolic modulator (2). In adipose tissue, SRC-2 is a key regulator of lipolytic (white fat) and thermogenic gene programs (brown fat) (3). In muscle, SRC-2 facilitates mitochondrial function (skeletal) (4) and promotes fatty acid utilization (cardiac) (5, 6). In the liver, SRC-2 was identified as an essential coactivator for brain and muscle Arnt-like 1 (BMAL1) for synchronizing peripheral and central circadian functions (7). Additional hepatic roles for SRC-2 have highlighted its importance for maintaining fasting glycemia through coactivation of retinoic acid receptor-related orphan receptor (RORα) (8).
Recently published work from our laboratory showing that coregulators form hormonally responsive transcriptional complexes suggests that platform coactivators such as SRC-2 may differentially influence transcriptional complexes in response to metabolic cues (9, 10). The mapping of protein complexes that provide the foundation for the coregulator “complexome” further supports this notion (9). The dynamic interaction of multiple protein networks is collectively referred to as the “interactome,” and it is acutely responsive to cellular signaling cues (10). Despite these conceptual advances, the way that energetic signals (i.e., fasting and feeding) alter the transcriptional complexome and expression of reparative metabolic gene programs remains largely undefined.
We propose that dynamic coregulator complex composition is a fundamental mechanism for the transcriptional switch from glycolysis to gluconeogenesis. To interrogate this hypothesis, we have developed a DNA pull-down technique coupled with mass spectrometry (MS) to evaluate the impact of metabolic fluctuations on coregulator complex formation on two opposing rate-limiting genes regulating glucose metabolism [i.e., Glucokinase (Gck) and Glucose-6-phosphatase (G6pc)]. Congenic ablation or acute knockdown of SRC-2 leads to a marked reduction in Gck and G6pc gene expression. Consistent with these findings, we show that SRC-2KO mice are hypoglycemic during the fasted state, yet surprisingly display postprandial hyperglycemia. Using our newly developed DNA pull-down system, we identified metabolically sensitive, SRC-2–dependent coregulator complexes on the Gck and G6pc gene promoters. Validation of these complex components identified an interaction of SRC-2 with the transcription factor (TF) C/EBPα on the Gck promoter during the fed state. Perturbing this interaction resulted in alterations in glucose homeostasis. From a broader perspective, our findings implicate SRC-2 as a pleiotropic factor that coordinates polygenic inputs for fine-tuning metabolic gene transcription through establishment of promoter-specific coregulator complexes. The technical advance highlighted in this study combined with our findings on the activating and repressive coregulator complexes that are sensitive to the actions of SRC-2 provide insight into how whole animal energetics coordinately direct metabolic transcriptional events. Conceptually, these findings establish SRC-2 as a model for polygenic disease by identifying a previously unappreciated mechanism by which metabolic cues integrate a hierarchy of transcriptional inputs that precisely govern glucose availability.
Materials and Methods
Complete details of materials and methods are described in SI Materials and Methods.
Animal Experiments.
All vertebrate animal experiments were performed in accordance with the Animal Care Research Committee at Baylor College of Medicine.
ChIP-Quantitative PCR (qPCR).
Liver tissue was isolated and flash frozen from WT or SRC-2 knockout mice as indicated. Chromatin was isolated using the SimpleChIP Enzymatic Chromatin IP Kit (Cell Signaling) and performed per the manufacturer’s suggestion. Different antibodies were used at 2 μg per reaction. qPCR was performed with gene-specific primers and SyberGreen technology (Applied Biosystems) with normalization to total input DNA. ChIP-qPCR primer sequences are listed in Table S1.
Table S1.
Gene-specific qPCR primer sequences and ChIP-qPCR primer sequences
| Gene | Forward and reverse primer sequences |
| qPCR primer sequences | |
| Cebpa | F, AAACAACGCAACGTGGAGA |
| R, GCGGTCATTGTCACTGGTC | |
| SRC-2 | F, GATGGACCTTCACCCTTG |
| R, GCTCCCTGTTGAGTCCTTGTE | |
| Gck | F, GGAACAACATCGTGGGACTT |
| R, ATTGCCACCACATCCATCTC | |
| G6pc | F, TCTGTCCCGGATCTACCTTG |
| R, GAAAGTTTCAGCCACAGCAA | |
| Gpi | F, CCCTCTTTATAATCGCCTCCA |
| R, GAAACCACTCCTTTGCTGTCTC | |
| Pfkfb1 | F, AGCCTTTGGATGAGGAATTG |
| R, GTGTGCCCACATCGAAGAT | |
| Pfkl | F, GGAACTCAAGGGGAACTCG |
| R, AGCACAGTAGACCCCCACAT | |
| Fbp1 | F, TATACCCCGCCAACAAGAAA |
| R, AAGCTATGGGGTTGCACTCA | |
| Fbp2 | F, GAAGAGAATAAAGAGGCGGTGA |
| R, AGGGTCAAAGCAAACCACAT | |
| Aldob | F, CACACAGCTTCTGATACCTTGG |
| R, AATCGGTGAGCCATGATGA | |
| Tpi | F, AAACCAAGGTCATCGCAGA |
| R, CCCGGAGCTTCTCGTGTA | |
| Gapdh | F, CAATGAATACGGCTACAGCAAC |
| R, TTACTCCTTGGAGGCCATGT | |
| Pgk1 | F, GTATTTTCTTTCCTGCCTTTGG |
| R, CGCTAACACCAAATGGAGATG | |
| Pgm1 | F, TTTGCATTTGAAGAAGCTATCG |
| R, ACGCCATCTTTGTCCAGAAC | |
| Eno1 | F, GAGGCGCTTAGTGCTGCT |
| R, CTGCAAAGCAAGGAAAGAGG | |
| Pklr | F, CATTGTGCTGACAAAGACTGG |
| R, TGGGCAGAACGAGTCACA | |
| Pck1 | F, GCAGCATGGGGTGTTTGTA |
| R, TGATGATCTTGCCCTTGTGT | |
| Pcx | F, GGGACTCCTTTGGACACAGA |
| R, GCCCCTTCCCAGTACTCACT | |
| Glut1 | F, ATGGATCCCAGCAGCAAG |
| R, CCAGTGTTATAGCCGAACTGC | |
| Glut2 | F, TCTTCACGGCTGTCTCTGTG |
| R, AATCATCCCGGTTAGGAACA | |
| Glut4 | F, GACGGACACTCCATCTGTTG |
| R, GCCACGATGGAGACATAGC | |
| ChIP-qPCR primer sequences | |
| Gck | F, GAGGATAAGCAGGGGTCGTT |
| R, TGCCTGTCTTTTGCAACACT | |
| G6pc | F, CCCTGAACATGTTTGCATCA |
| R, GTAGGTCAATCCAGCCCTGA | |
| UTR10 | F, TACACATGAGGCCAGGATCA |
| R, CTCCTGGAACATGCCTAACC |
DNA Pull-Down Assays.
DNA pull-downs were performed as previously described (10). Briefly, 60 μL of Dynabeads M280 streptavidin (Life Technologies) beads were pelleted with standard magnetic racks (Life Technologies), and 4 μg of biotinylated Gck or G6pc DNA were bound to the Dynabeads in 150 μL d-PBS by rotation for ∼1 h at 4 °C. Biotinylated Gck and G6pc DNAs were synthesized by PCR using 5′-biotinylated primers (as described in SI Materials and Methods) and mouse genomic DNA subcloned into either pCR2-Topo or pCR4-Topo. Bead-immobilized DNAs were then washed, and the final PBS removed. Nuclear extract (NE) was thawed on ice and clarified by centrifugation at 4 °C for 10 min. We added NE (1 mg) to resuspend beads, and reactions were incubated with rotation at 4 °C for 1.5 h. Protein complexes were cross-linked with 1% formaldehyde for 10 min at room temperature (RT) and quenched with 1× glycine for 5 min at RT. Beads were washed using ice-cold buffers [two washes in 100 mM NaCl, 0.5 mM EDTA, 20 mM Tris-HCl (pH 8.0), 0.5% Nonidet P-40 (10), followed by one wash in d-PBS]. After PBS removal, beads were resuspended in 20–30 μL 5× SDS sample buffer (Thermo Fisher). After boiling, protein samples were loaded on 4–15% (wt/vol) Mini-PROTEAN TGX Precast gels (Bio-Rad) for immunoblotting or on NuPage gels (8% Bis–Tris in Mops buffer; Invitrogen) for preparative separation-gel slice excision for MS.
Protein Identification by MS.
For MS analysis, the entire DNA pull-down reaction was scaled up twofold and was separated on 1D SDS/PAGE. After staining proteins in SDS/PAGE gels with Coomassie blue, gel lanes were sliced into different bands and in-gel digested overnight at 37 °C with trypsin. After digestion, peptides were extracted twice in 200 μL of acetonitrile with resuspension in 20 μL of 2% (wt/vol) formic acid before a second extraction, dried in a Savant SpeedVac, and dissolved in a 5% (wt/vol) methanol/0.1% formic acid solution. The samples were loaded through a 2 cm C18 trap followed by 1 h 0–30% (wt/vol) acetonitrile gradients on a 10 cm C18 column (packed in-house with Reprosil-Pur Basic C18 3 µm beads; Dr. Maisch GmbH) and measured online with the Thermo Orbitrap Velos or QExactive instruments (Thermo Scientific). The raw data were searched with Proteome Discoverer 1.3 Mascot engine against a human RefSeq database using lenient 1%/5% restricted/relaxed peptide-specific matches FDR (false discovery rate), and the data were further grouped into gene products that were assigned homology and identification quality groups using an in-house developed algorithm. All protein gene products chosen for follow-up in this study were required to have at least one identification where a spectral match passing <1% FDR and >20 ion score or <5% FDR and >30 ion score thresholds was present. The amount of each gene product was estimated with a label-free intensity-based absolute quantification (iBAQ) approach (as the sum of peptide areas normalized to the theoretical tryptic peptide potential) (11) and reported as a fraction of total protein iBAQ amount per experiment (in 10−5 units for visual comprehension). All MS datasets described in this study are available online at epicome.org/index.php/msprojects/src2metabolismresource.
SI Materials and Methods
Animal Experiments.
The generation of the SRC-2KO, SRC-2flox/flox, and SRC-2LKO (liver-specific knockout) mice has been described previously (7, 31). Congenic SRC-2WT and SRC-2KO male littermate mice (ages 10–20 wk) were used for all studies. All mice were entrained under strict temperature control, ad libitum food access (normal chow, 2920X Teklad Global), and water under 12 h light/dark conditions. All experiments were performed in accordance with the Animal Care Research Committee at Baylor College of Medicine. For fasting experiments, mice were fasted overnight with free access to water only. The following morning mice were fed with normal chow for 6 h, after which blood glucose was measured using a hand-held glucometer (One Touch Ultra, Lifescan). Liver glycogen was measured 6 h after refeeding following the manufacturer’s instructions (Biovision).
Primary Hepatocytes.
Mouse primary hepatocytes were isolated from SRC-2WT and SRC-2KO male littermate mice, pooled, and plated in triplicate as previously described (7). Rat primary hepatocytes were isolated exactly as described (3, 32). Plated hepatocytes were treated as indicated with either insulin (100 nM) or forskolin (10 nM) for 3 h, unless otherwise noted. For hormone treatment studies of primary hepatocytes, targeted acute knockdown of SRC-2 was accomplished using siRNA (40 nM) directed against SRC-2 (l-020159–00-0010, GE Dharmacon) for 2 d. siRNA knockdown of SRC-2 (3 nM) or C/EBPα (3 nM) in primary hepatocytes was performed using Silencer Select siRNAs (Life Technologies SRC-2, s70425 and s70426 and C/EBPα, s63853 and s63854). All hepatocytes were isolated for RNA in triplicate using the RNeasy Mini Kit (Qiagen).
qPCR.
Total mRNA was isolated from the liver of SRC-2WT and SRC-2KO mice with RNeasy Kit (Qiagen). Reverse transcription was carried out using SuperScript III (Life Technologies) per the manufacturer’s instructions. For gene expression analyses, qPCR was performed using the Taqman system with sequence-specific primers and the LNA probes from the Universal Probe Library (Roche). All data were analyzed with 18S as the endogenous control. All qPCR primer sequences are listed in Table S1.
ChIP-Seq Data Analysis.
Galaxy/Cistrome’s SeqPos motif tool (v1.0.0) (cistrome.dfci.harvard.edu/ap/root) was used on the entire published SRC-2 ChIP-SEq (7) interval set (20,599 intervals) for de novo (MDscan algorithm) and reference-based (Transfac, JASPAR, pbm, and hPDI databases) sequence motif analyses, and their implementation of the CEAS tools for enrichment analyses on chromosomes and genomic annotations. Database for Annotation, Visualization and Integrated Discovery Bioinformatics Resources v6.7 (david.ncifcrf.gov/) was used on protein coding genes to identify enriched biological themes. SRC-2 cistromic binding sites were related to mouse reference gene annotations (ftp://ftp.ncbi.nlm.nih.gov/genomes/MapView/Mus_musculus/sequence/BUILD.37.1) and formed into a relational database system for easy querying and data sharing. For generation of images of SRC-2 binding sites on glycolytic or gluconeogenic gene promoters, the UCSC genome browser (https://genome.ucsc.edu/cgi-bin/hgGateway) was used.
Luciferase Assays.
For transient-transfection assays, HeLa cells were plated overnight and transfected with expression plasmids for SRC-2 (pSG5-HA-SRC-2), C/EBPα (pSPORT-C/EBPα), pGL3-Basic-Gck (–1465/+124), pGL3-Basic-G6pc (–1227/+57), or Renilla (pRL-TK) plasmids containing the luciferase reporter using Lipofectamine 2000 (Life Technologies). Cell lysates were prepared 72 h posttransfection, and luciferase activities were measured with a luciferase assay kit (Promega) and normalized Renilla.
NE Preparation.
NE was made from whole liver following modifications to the standard protocol (33). Protein concentrations were determined by Bradford assays (Bio-Rad), and aliquots were snap-frozen in liquid N2 and stored at –80 °C until use.
Biotinylated Gck and G6pc Promoters.
A 1,129 bp doubly biotinylated fragment from the mouse G6pc gene promoter was made by PCR using pCR2-Topo-G6pc as a template and forward 5′-biotin-CGCCAAGCTATTTAGGTG and reverse 5′-biotin-ATCTGCAGAATTCGCCCTT primers biotinylated at their 5′ ends. pCR2-Topo-G6pc was constructed by cloning a 1,129 bp PCR fragment made with Taq DNA polymerase (Denville) and NIH 3T3 mouse genomic DNA (NEB). Primers used for this PCR (without 5′-biotin moieties) are forward 5′-TCTCATGTGCATTGGTGGCT and reverse 5′-GCTGGAGGACAGCCATTTCT. A 1,017 bp doubly biotinylated fragment from the mouse Gck gene promoter was made by PCR using pCR4-Topo-Gck as a template and forward 5′-biotin-AGTCCTGCAGGTTTAAACGA and reverse 5′-biotin-ACTCACTATAGGGCGAATTG biotinylated at their 5′ ends. pCR4-Topo-Gck was constructed by cloning a 1,017 bp PCR fragment made with Taq DNA polymerase (Denville) and NIH 3T3 mouse genomic DNA (NEB). Primers used for this PCR (without 5′-biotin moieties) are forward 5′-ATCTGAGGATAAGCAGGGGTC and reverse 5′-TGGGAAAGTGATCAATCGTG. To remove any free biotinylated primers after PCR, reactions were passed through PCR Kleen Spin Columns (Bio-Rad).
Metabolomic Profiling.
The metabolomics analyses of all samples were executed using the protocol described previously (7). In brief, the raw data (LC-MS output) were normalized using internal standards. Before normalization, the median coefficient of variation of each internal standard was measured to select the appropriate standard for normalization to confirm consistency.
13C Glucose Flux.
Nutrients labeled with 13C were purchased from Cambridge Isotope Laboratories. Primary hepatocytes isolated from either wild-type (SRC-2flox/flox) or SRC-2LKO mice were plated in six-well plates in William’s E media, followed by overnight starvation and then addition of 11 mM d-[U-13C6]glucose supplemented with RPMI[–]glucose, 10% (wt/vol) dialyzed FBS, and 1% Penicillin Streptomycin. Culture medium was collected, cells were washed with PBS, and equal numbers of cells were snap-frozen with liquid nitrogen. Cells were scraped into a 0.5-mL mixture of 1:1 water/methanol, sonicated for 1 min (two 30-s pulses), and then mixed with 450 μL ice-cold chloroform. The resulting homogenate was then mixed with 150 μL ice-cold water and vortexed again for 2 min. The homogenate was incubated at –20 °C for 20 min and centrifuged at 4 °C for 10 min to partition the aqueous and organic layers. The aqueous and organic layers were combined and dried at 37 °C for 45 min in an automatic Environmental Speed Vac system (Thermo Fisher Scientific). The extract was reconstituted in a 500-μL solution of ice-cold methanol/water (1:1) and filtered through a 3-kDa molecular filter (Amicon Ultracel 3-kDa Membrane) at 4 °C for 90 min to remove proteins. The filtrate was dried at 37 °C for 45 min in a speed vacuum and stored at –80 °C until MS analysis. Before MS analysis, the dried extract was resuspended in a 50 μL solution of methanol/water (1:1) containing 0.1% formic acid and then analyzed using multiple reaction monitoring. Ten microliters were injected and analyzed using a 6490 QQQ triple quadrupole mass spectrometer (Agilent Technologies) coupled to a 1290 Series HPLC system via selected reaction monitoring. Metabolites were targeted in both positive and negative ion modes: The electrospray source ionization voltage was +4,000 V in positive ion mode and –3,500 V in negative ion mode. Approximately 9–12 data points were acquired per detected metabolite. For 13C-labeled experiments, selected reaction monitoring was performed for expected 13C incorporation in various forms for targeted LC-MS/MS. Mass isotopomer distribution was calculated as fractional incorporation = (13C/13C+12C) × 100 and corrected for natural abundance.
MS Profiling.
For data quality control, the proteomic profile data were quantile-normalized by the Bioconductor lumi package (34). Differences in gene expression between different genotypes, feeding state, and/or promoter were inferred using the Bioconductor limma package (35) (P < 0.05) and imposing a fold change exceeding 1.5×, using the R statistical system. Heat maps were generated using the R statistical system. All MS datasets and tables from this study are available online at epicome.org/index.php/msprojects/src2metabolismresource under msProjects/SRC-2 Metabolism Resource.
Immunoblotting.
Immunoblot analyses were performed as described previously (10). Briefly, proteins separated by SDS/PAGE were transferred to nitrocellulose membranes, blocked in tris-buffered saline + Tween-20 supplemented with 5% (wt/vol) BSA, and incubated overnight with primary antibody. Blots were incubated with an appropriate secondary antibody coupled to horseradish peroxidase, reacted with ECL reagents per the manufacturer’s (Thermo) suggestion, and detected on X-ray film by autoradiography. The antibodies used are as follows: SRC-2 (Bethyl, A300-346A), C/EBPα (Santa Cruz, sc-61), SRC-1 (Cell Signaling Technology, 128E7), SRC-3 (Cell Signaling Technology, 5E11), PHB2 (Cell Signaling Technology, E1Z5A), NF-YA (Santa Cruz, sc-10779), E4BP4 (Santa Cruz, sc-28203), LSD1 (Cell Signaling Technology, C69G12), HNF-4α (Santa Cruz, sc-8987), TAF5 (Bethyl, A303-686), ERRα (Abcam, ab16363), HDAC1 (Bethyl, A300-713A), RBP1 (Upstate, 50623), BMAL1 (Abcam, ab93806), and INO80 (Bethyl, A303-371A).
Statistical Analyses.
The raw data (LC-MS output) were separately normalized in each method using the internal standard with the lowest coefficient of variation for that specific method. G-6-P/F-6-P, pyruvate, and glucose levels were normalized using l-Tryptophan. All data are represented as the mean ± SEM. Standard statistical comparison of different groups was performed using two-tailed unpaired Student’s t test with differences of P ≤ 0.05 considered statistically significant.
Results
SRC-2 Dynamically Regulates Glycemia via Opposing Transcriptional Programs.
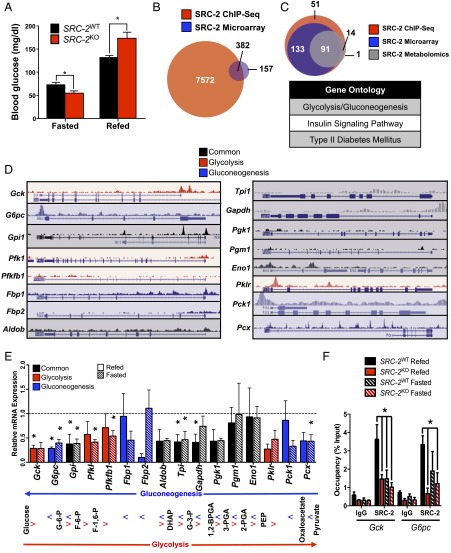
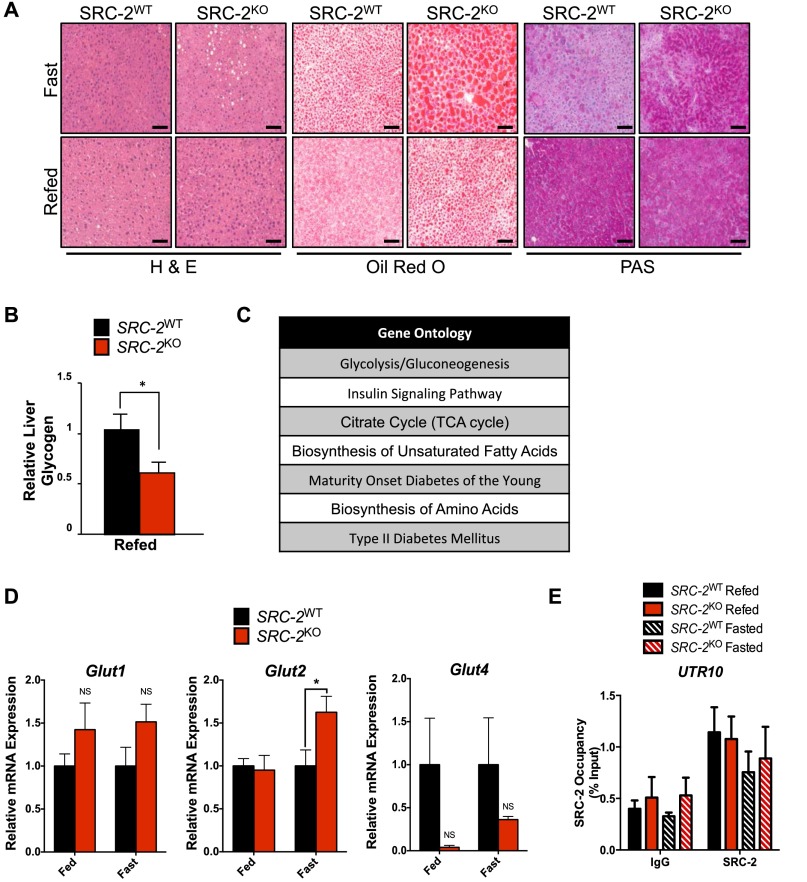
Previous characterization of SRC-2KO mice revealed fasting hypoglycemia accompanied with increased liver glycogen stores (8). These phenotypes result from the aberrant expression of G6pc in the absence of SRC-2, which coactivates RORα on the G6pc promoter. In addition to fasting hypoglycemia, we found that SRC-2KO mice surprisingly displayed postprandial hyperglycemia (Fig. 1A). In line with these data, fasted and refed SRC-2KO mice also show disturbances in glycogen storage/utilization as well as lipid deposition, consistent with aberrant glucose metabolism in the feeding-to-fasting transition (Fig. S1 A and B). These findings point to an overarching role for SRC-2 in the dynamic regulation of glucose homeostasis. Supporting this notion, integration of SRC-2 liver cistromic and transcriptomic data revealed glycolysis/gluconeogenesis as the top ontology of the candidate genes controlled by SRC-2 (Fig. 1B and Fig. S1C). Additionally, integration of metabolomic signatures from livers of SRC-2WT and SRC-2KO mice further specifies glycolysis/gluconeogenesis as the top candidate pathway controlled by SRC-2 (Fig. 1C). Our laboratory has demonstrated by ChIP-Seq that SRC-2 has a preference for the proximal promoter of its target genes (7). Analysis of the hepatic SRC-2 cistrome identified SRC-2 binding sites in the promoter regions of multiple glycolytic and gluconeogenic genes, suggesting SRC-2 may coordinately regulate gene programs that control glycemia (Fig. 1D).
Fig. 1.
SRC-2 dynamically regulates glycemia via opposing transcriptional programs. (A) Blood glucose (mg/dL) measurements of SRC-2WT and SRC-2KO mice (n = 5 each) after either an overnight fast or an overnight fast followed by a 6 h refeed. (B) Venn diagram representation of integrated hepatic SRC-2 ChIP-Seq binding sites with liver microarray data from SRC-2KO mice. (C) Venn diagram overlap of Kyoto Encyclopedia of Genes and Genomes (KEGG) gene ontologies from liver SRC-2 ChIP-Seq (red), SRC-2KO microarray (blue), and metabolomics data (gray). The top KEGG gene ontologies from the integration of these three ontological sets are listed in the accompanying table. (D) University of California, Santa Cruz (UCSC) genome browser representations of SRC-2 binding sites as determined by ChIP-Seq (7) on common (black) or unique glycolytic (red) or gluconeogenic (blue) gene promoters in wild-type mouse liver. (E) qPCR analysis of common (black) or unique glycolytic (red) or gluconeogenic (blue) genes in livers isolated from SRC-2WT and SRC-2KO mice (n = 5 each) after either a 24 h fast or a 24 h fast followed by a 3 h refeed. Data are represented as relative to wild-type gene expression for each feeding condition, which was set to 1. Accompanying the qPCR data is an outline of glycolysis and gluconeogenesis indicating where each gene product functions in those processes. (F) ChIP-qPCR analysis of SRC-2 occupancy on the gene promoters of Gck and G6pc in SRC-2WT and SRC-2KO mice (n = 3 each) after either a 24 h fast or a 24 h fast followed by a 3 h refeed. ChIP data are represented as a percent of the input, whereas all other data are graphed as the mean ± SEM. *P < 0.05.
Fig. S1.
SRC-2 regulates glycemia through transcriptional mechanisms. (A) Representative H&E, Oil Red O, and periodic acid–Schiff (PAS) of SRC-2WT and SRC-2KO fasted for 24 h or 24 h fasting followed by a 3 h refeed. (B) Quantitative measurement of liver glycogen levels in SRC-2WT and SRC-2KO mice (n = 5 each) that were fasted overnight and refed for 6 h. (C) Top KEGG gene ontologies of shared genes between the liver SRC-2 ChIP-Seq and SRC-2KO microarray. (D) qPCR analysis of Glut1, Glut2, and Glut4 in livers isolated from SRC-2WT and SRC-2KO mice (n = 5 each) after either a 24 h fast or a 24 h fast followed by a 3 h refeed. (E) ChIP-qPCR analysis of SRC-2 occupancy on UTR10 in SRC-2WT and SRC-2KO mice (n = 3 each) after either a 24 h fast or a 24 h fast followed by a 3 h refeed. ChIP data are represented as percent of the input, whereas all other data are graphed as the mean ± SEM. *P < 0.05.
To determine the enzymes in the glucose metabolism pathways that may be responsible for the glycemic phenotypes of the SRC-2KO mice, we performed a qPCR analysis for genes of the glycolytic and gluconeogenic programs in SRC-2WT and SRC-2KO mice that were fasted for 24 h or fasted and then refed for 3 h. This qPCR screen revealed significant decreases in the expression of multiple glycolytic and gluconeogenic target genes (Fig. 1E). We also tested the effects of SRC-2 ablation on genes encoding the liver-specific glucose transporters (i.e., Glut1, Glut2, Glut4), but the mild alterations in these genes are unlikely to fully explain the glycemic phenotypes of the SRC-2KO mice (Fig. S1D). Ablation of SRC-2, however, resulted in a significant reduction of Gck expression during the fed state, whereas the expression of G6pc was decreased under both fasted and refed conditions. These changes are consistent with the effects of SRC-2 on fasting versus postprandial glycemia (Fig. 1E). Additionally, we found that Gpi and Tpi expression were decreased in the absence of SRC-2, however neither of these gene products represent rate-limiting steps of glycolysis or gluconeogenesis such as those controlled by Gck and G6pc. Therefore, we sought to explore the mechanism(s) by which SRC-2 controlled the expression of these two opposing rate-limiting stages of hepatic glucose entry and release.
To confirm the presence of SRC-2 on the Gck and G6pc promoters as suggested by the SRC-2 ChIP-Seq (Fig. 1D), we performed ChIP-qPCR using chromatin isolated from livers of SRC-2WT and SRC-2KO mice that were either fasted or fasted and then refed. We found that SRC-2 localized to the proximal promoter region of Gck in the refed state in SRC-2WT mice, and this recruitment is attenuated during fasting (Fig. 1F). These data verified the specificity of our SRC-2 antibody as we observed minimal recruitment of SRC-2 in the SRC-2KO livers (Fig. 1F). Somewhat unexpectedly, we found SRC-2 recruitment to the G6pc promoter to be highest during the refed state with no significant differences in SRC-2 recruitment in fasted versus refed conditions (Fig. 1F). These data suggest that SRC-2 may function differently in the regulation of G6pc and Gck such that feeding signals may recruit SRC-2 to the Gck promoter, whereas the recruitment and dismissal of SRC-2 to the G6pc promoter may be a part of a poised complex. Importantly, no recruitment of SRC-2 to the UTR10 gene region was observed, which is consistent with our SRC-2 ChIP-Seq data (Fig. S1E) (5).
SRC-2 Directly Controls the Cell-Autonomous Expression of Gck and G6pc.
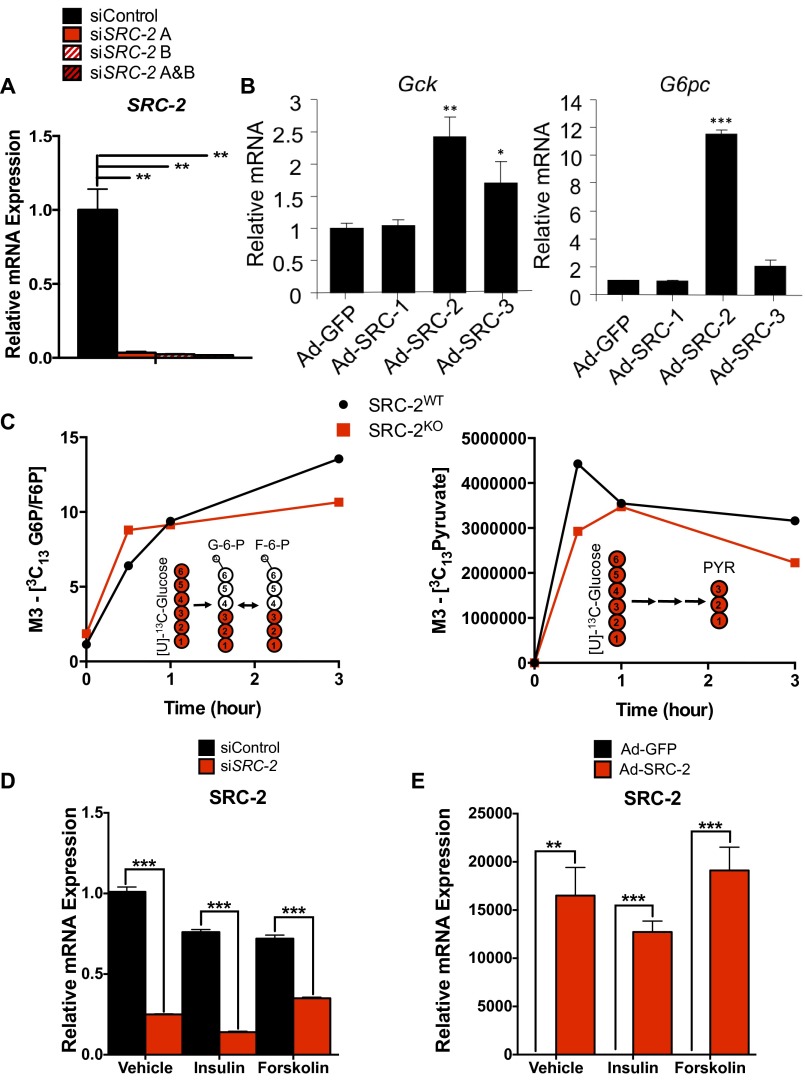
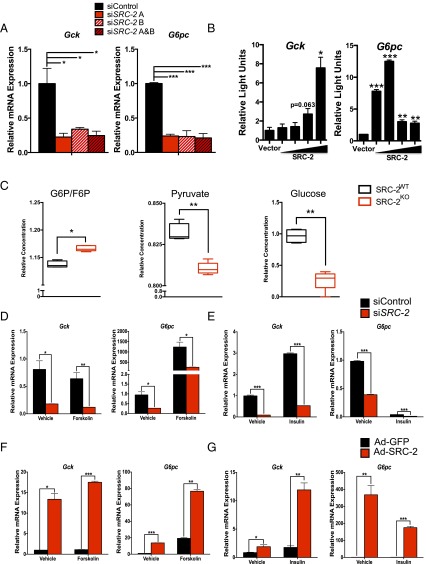
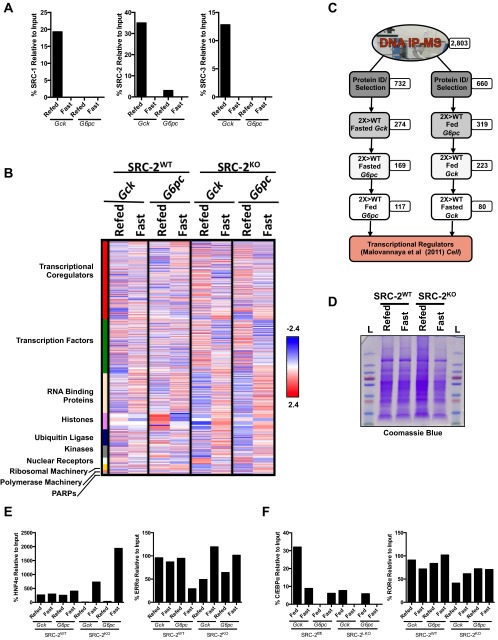
To assess the cell-autonomous activity of SRC-2 on the promoters of Gck and G6pc, we knocked down SRC-2 in primary hepatocytes using two unique siRNAs, which shows greater than 90% reduction in SRC-2 mRNA (Fig. S2A). Following acute knockdown of SRC-2, Gck and G6pc expression were significantly decreased as measured by qPCR (Fig. 2A). To confirm SRC-2 activity on the Gck and G6pc promoters, we transfected HeLa cells with either a Gck– or G6pc–promoter–luciferase reporter construct in the presence or absence of HA-SRC-2. These data demonstrate that SRC-2 overexpression potentiates luciferase expression driven by both Gck and G6pc promoters, although the amount of HA–SRC-2 needed for maximal activation differs on these two promoters (Fig. 2B). Adenoviral overexpression of SRC-1, SRC-2, or SRC-3 revealed that SRC-2 confers the greatest increased expression of both Gck and G6pc relative to the other SRCs (Fig. S2B).
Fig. S2.
SRC-2 regulates the expression of Gck and G6pc. (A) qPCR analysis of SRC-2 gene expression in primary hepatocytes transfected with individual or combined siRNAs against SRC-2. (B) qPCR analysis of Gck and G6pc expression in wild-type primary hepatocytes infected with adenovirus expressing GFP, SRC-1, SRC-2, or SRC-3. (C) Glycolytic flux of [U]-13C–labeled glucose in primary hepatocytes isolated from SRC-2WT or SRC-2KO mice. Data are represented as M3-13C–labeled G6pc and pyruvate, respectively (M indicates nominal mass). (D) qPCR analysis of SRC-2 expression in wild-type primary hepatocytes transfected with control siRNA or siSRC-2 in the presence or absence of insulin or forskolin. (E) qPCR analysis of SRC-2 expression in wild-type primary hepatocytes infected with adenovirus expressing GFP or SRC-2 in the presence or absence of insulin or forskolin. Data are graphed as the mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001.
Fig. 2.
SRC-2 directly controls the expression of Gck and G6pc. (A) qPCR analysis of Gck and G6pc gene expression in primary hepatocytes transfected with individual or combined siRNAs against SRC-2. (B) Transactivation assays in HeLa cells transfected with Gck or G6pc luciferase reporter constructs along with increasing amounts of SRC-2 expression vector. (C) Metabolomics analysis of glucose-6-phosphate/fructose-6-phosphate (G6P/F6P), pyruvate, and glucose in liver tissue from SRC-2WT and SRC-2KO mice. (D) qPCR analysis of Gck and G6pc expression in wild-type primary hepatocytes transfected with control siRNA or siSRC-2 in the presence or absence of forskolin. (E) qPCR analysis of Gck and G6pc expression in wild-type primary hepatocytes transfected with control siRNA or siSRC-2 in the presence or absence of insulin. (F) qPCR analysis of Gck and G6pc expression in wild-type primary hepatocytes infected with adenovirus expressing GFP or SRC-2 in the presence or absence of forskolin. (G) qPCR analysis of Gck and G6pc expression in wild-type primary hepatocytes infected with adenovirus expressing GFP or SRC-2 in the presence or absence of insulin. Data are graphed as the mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001.
Further supporting our phenotypic data, quantitative metabolomic analysis of primary hepatocytes from SRC-2WT or SRC-2KO mice revealed a significant increase in glucose-6-phosphate/fructose-6-phosphate (G6P/F6P), consistent with the observed decrease of G6pc gene expression (Fig. 2C and Fig. S2C). Likewise, loss of SRC-2 also led to a significant reduction in pyruvate due to low glycolytic flux (Fig. 2C and Fig. S2C), indicating the importance of SRC-2 for maintaining Gck expression to preserve the flow of glucose carbons. In line with the impaired expression of G6pc and Gck, we found that SRC-2 ablation markedly reduced the levels of glucose produced from primary hepatocytes devoid of SRC-2 (Fig. 2C). Taken together, these data suggest that loss of SRC-2 is sufficient to perturb glucose entry due to impaired expression of Gck, although we cannot rule out the contribution of G6pc expression on glucose release that could partially account for the observed reduction in glucose levels in SRC-2KO primary hepatocytes.
To determine the cell-autonomous acute effects of SRC-2 on Gck or G6pc expression, SRC-2 was either knocked down (siRNA) or overexpressed (adenoviral infection) in primary hepatocytes treated with the fasting mimetic forskolin or feeding mimetic insulin. Primary hepatocytes treated with siRNA targeting SRC-2, which has previously demonstrated an 80% reduction in SRC-2 protein expression (12), showed decreased mRNA expression of SRC-2, Gck, and G6pc (Fig. 2 D and E and Fig. S2D). As expected, forskolin treatment increased the expression of G6pc but failed to impact Gck expression (Fig. 2D). Knockdown of SRC-2 in hepatocytes treated with forskolin resulted in decreased expression of both Gck and G6pc relative to control (Fig. 2D). Hepatocytes treated with insulin showed increased expression of Gck and decreased expression of G6pc relative to vehicle controls (Fig. 2E). However, knockdown of SRC-2 decreased the expression of both Gck and G6pc upon insulin treatment (Fig. 2E).
Conversely, we used adenoviral overexpression of SRC-2 in primary hepatocytes to test its effect on the expression of Gck and G6pc. Ad–SRC-2 primary hepatocytes increased SRC-2, Gck, and G6pc in vehicle-treated hepatocytes relative to the Ad-GFP hepatocytes (Fig. 2 F and G and Fig. S2E). Treatment of Ad–SRC-2–expressing hepatocytes with forskolin resulted in increased expression of G6pc (Fig. 2F). This treatment, however, failed to overcome the increased expression of Gck with overexpression of SRC-2 (Fig. 2F). These data are highly consistent with the effect of siRNA knockdown of SRC-2 whereby treatment with forskolin fails to suppress Gck expression. Treatment of Ad–SRC-2–expressing hepatocytes with insulin, however, resulted in increased expression of Gck relative to Ad-GFP insulin-treated hepatocytes (Fig. 2G). Similarly, insulin treatment of Ad–SRC-2 hepatocytes resulted in decreased expression of G6pc relative to the vehicle treatment, but the expression was still significantly increased relative to the Ad-GFP insulin-treated hepatocytes (Fig. 2G). Taken together, these studies confirm that SRC-2 localizes to, and transcriptionally coactivates, the Gck and G6pc promoters to regulate their dynamic expression in response to hormonal cues that recapitulate the feeding-to-fasting transition.
SRC-2 Recruits Distinct Coregulator Complexes to Regulate Gck and G6pc Expression.
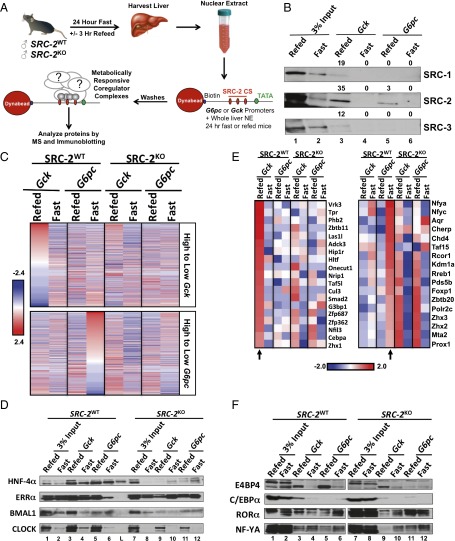
Published work from our laboratory has demonstrated that the SRCs participate in hormonally sensitive coregulator complexes on the promoter regions of an endogenous ERα target gene (10). We sought to determine if differential coregulator complex recruitment plays a mechanistic role in the dynamic regulation of Gck and G6pc by SRC-2. To that end, we performed DNA pull-down assays using endogenous Gck and G6pc promoter fragments that were biotinylated, bound to streptavidin magnetic beads, and incubated with liver NE isolated from SRC-2WT and SRC-2KO mice that were fasted for 24 h or fasted followed by a 3 h refeed (Fig. 3A). Immunoblotting and quantification confirmed the predominant recruitment of SRC-2 to the Gck and G6pc promoters (Fig. 3B and Fig. S3A). These data are congruent with the effects of SRC-2 to potentiate Gck and G6pc-luciferase activity whereby lower levels of SRC-2 recruitment to the G6pc promoter are required to coactivate gene expression compared with that of Gck (Fig. 2B). Using unbiased MS, we identified protein complexes that are recruited to the Gck or G6pc promoters in an SRC-2–dependent and/or metabolically sensitive manner (fasting versus refed) (Fig. 3C and table 1, hosted at epicome.org/index.php/msprojects/src2metabolismresource). Stratification of MS-identified proteins enriched on the Gck promoter during the fed state were dismissed in the fasted state (Fig. 3C, Upper). Conversely, proteins highly recruited to the G6pc promoter in the fasted state were subsequently reduced during the postprandial state (Fig. 3C, Lower). In general, proteins ontologically defined as TFs or transcriptional coregulators were among the most highly recruited and dynamic with respect to the metabolic state, further emphasizing the importance of these protein classes for controlling metabolic gene transcription (Fig. S3B and table 2, hosted at epicome.org/index.php/msprojects/src2metabolismresource). We also identified other regulatory classes of proteins that contain important enzymatic functions such as ADP ribosylases, kinases, and ubiquitin ligases that likely provide signaling inputs to these transcriptional complexes via posttranslational modifications (Fig. S3B) (13).
Fig. 3.
SRC-2 interacts with distinct coregulator complexes to regulate Gck and G6pc expression. (A) Schematic of the DNA pull-down assay system. NE was isolated from livers of SRC-2WT or SRC-2KO male mice that were either fasted for 24 h or fasted for 24 h followed by a 3 h refeed. Biotinylated promoter fragments of Gck or G6pc genes were bound to magnetic Dynabeads and incubated with liver NE. Bound proteins were identified by unbiased MS. SRC-2 ChIP-Seq (CS) denotes cistromic locations of SRC-2 from our liver ChIP-Seq. (B) Immunoblot validation of SRC-1, SRC-2, and SRC-3 binding to Gck or G6pc promoters from fasted or refed wild-type liver NE. (C) Heat map representation of thresholded MS analysis of Gck and G6pc DNA pull-downs from liver NE from fasted or refed SRC-2WT or SRC-2KO mice. Proteins were stratified by those with the greatest input-normalized fraction of total (iFOT) signal on either the Gck promoter during the refed condition (Top) or the G6pc promoter in the fasted state (Bottom). (D) Immunoblot validation of candidate TFs identified from C. Experimental lanes are numbered; L denotes the protein marker. (E) Heat map representation of selected MS analysis of Gck and G6pc DNA pull-downs from liver NE from fasted or refed SRC-2WT or SRC-2KO mice. Proteins listed are enriched for binding to the Gck promoter in the refed state (Left) or the G6pc promoter in the fasted state (Right). Arrows indicate lanes used to determine enrichment. (F) Immunoblot validation of candidate complex components identified from stratification of Gck or G6pc promoter MS data in E. Lanes between pull-downs from SRC-2WT or SRC-2KO liver NE were cropped out of the same image to allow for consistent data representation, as this lane contained a protein marker. Heat maps use official gene names, and immunoblot data use common protein names.
Fig. S3.
SRC-2 influences TF–coregulator complexes to regulate Gck and G6pc expression. (A) Quantification of the DNA pull-down and immunoblot (IB) of SRC-1, 2, or 3 on either Gck or G6pc promoters from refed or fasted wild-type nuclear extract. (B) Heat map representation of thresholded MS analysis of Gck and G6pc DNA pull-downs from liver nuclear extract from fasted or refed SRC-2WT or SRC-2KO mice. Identified proteins are functionally grouped based on ontological function. (C) Flow diagram illustrating the stratification of proteins recruited to the Gck promoter during the refed condition or the G6pc promoter during the fasted condition. (D) Coomasie blue stained gel of nuclear extract from SRC-2WT and SRC-2KO fasted and refed mouse liver. (E) Quantification of the DNA pull-down and IB of HNF4α and ERRα on either Gck or G6pc promoters from refed or fasted SRC-2WT and SRC-2KO nuclear extract. (F) Quantification of the DNA pull-down and IB of C/EBPα and RORα on either Gck or G6pc promoters from refed or fasted SRC-2WT and SRC-2KO nuclear extract.
Consistent with the classical definition of a transcriptional coactivator as an amplifier and stabilizer of coregulator complexes (2, 9, 10, 14, 15), loss of SRC-2 strikingly reduced the recruitment of metabolically responsive proteins to both Gck and G6pc promoters (Fig. 3C). These findings highlight the collective importance of SRC-2 for establishing functional, biochemically stable TF–coregulator complexes on gene promoters that permit optimal transcriptional output in response to metabolic demand.
We confirmed the differential recruitment of top candidate TFs to both the Gck and G6pc promoters (i.e., HNF4α, BMAL1, CLOCK, and ERRα) via DNA pull-down followed by immunoblotting (Fig. 3D and Fig. S3E). Coomassie blue staining was used to confirm loading of NE inputs (Fig. S3D). Although these pleiotropic hepatic TFs demonstrated dependency on SRC-2 for stable recruitment to both Gck and G6pc promoters, the fact that they lack preferential metabolic recruitment to either promoter failed to offer a plausible mechanism for the metabolic-sensitive regulation of these two rate-limiting genes by SRC-2 (Fig. 3D). To identify candidate TFs that were recruited to either the Gck or G6pc promoters in response to fasting or refeeding, we stratified the DNA pull-down MS data to highlight proteins that preferentially localize to the Gck promoter in the refed state or the G6pc promoter in the fasted state (Fig. 3E, Fig. S3C, and table 3, hosted at epicome.org/index.php/msprojects/src2metabolismresource). Using immunoblot analysis, we confirmed that C/EBPα and E4BP4 were selectively recruited to the Gck promoter during the refed state (Fig. 3F and Fig. S3F). Consistent with published data, we found that RORα (8) was selectively recruited to G6pc during the fasted state, yet this effect was blunted in the absence of SRC-2 (Fig. 3F and Fig. S3F). Our immunoblot analysis also identified that NF-YA was preferentially recruited during the fasted state, although its recruitment was less dependent on SRC-2 (Fig. 3F).
SRC-2 Selectively Coactivates C/EBPα to Promote Gck Expression.
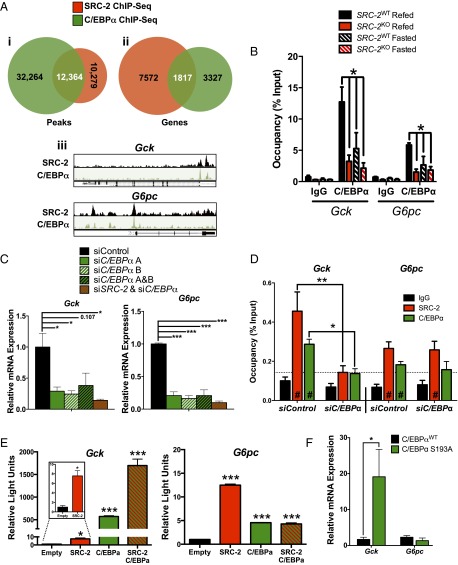
As our laboratory has previously elucidated the mechanism by which SRC-2 coactivates RORα to facilitate G6pc expression during fasting (8), stratification of our MS data identified C/EBPα as a predominant TF by which SRC-2 may coactivate Gck gene transcription in the postprandial state (Fig. 3 E and F and Fig. S3F). C/EBPα is of particular interest as a putative SRC-2 interacting partner for coactivating Gck expression as the C/EBP motif was among the top predicted TF binding sites from our SRC-2 ChIP-Seq (7). Integration of hepatic SRC-2 (7) and C/EBPα (16) ChIP-Seq datasets revealed a robust cistromic overlap (∼55% of SRC-2 peaks shared with C/EBPα) (Fig. 4 A, i). These overlapping binding sites corresponded to over 1,800 genes on which SRC-2 and C/EBPα share cistromic occupancy (Fig. 4 A, ii). Integration of published hepatic C/EBPα and SRC-2 cistromic data revealed identical binding sites for these two factors on the Gck promoter, providing further evidence that SRC-2 may cooperate with C/EBPα to drive Gck transcription (Fig. 4 A, iii).
Fig. 4.
SRC-2 coactivates C/EBPα function for selective regulation of Gck expression. (A, i) Venn diagram representation of integrated hepatic SRC-2 and C/EBPα ChIP-Seq binding sites in wild-type mice. (A, ii) Venn diagram representation of integrated hepatic SRC-2 and C/EBPα ChIP-Seq overlapping genes called from binding sites in wild-type mice. (A, iii) UCSC genome browser representations of overlapping SRC-2 and C/EBPα binding sites as determined by ChIP-Seq on the Gck and G6pc promoters. (B) Occupancy of C/EBPα on the Gck and G6pc promoter as determined by ChIP-qPCR. (C) qPCR analysis of Gck and G6pc expression in wild-type primary hepatocytes treated with siSRC-2, siC/EBPα, or both. (D) ChIP-qPCR analysis of SRC-2 and C/EBPα occupancy on the Gck and G6pc promoters in wild-type primary hepatocytes transfected with control siRNA or siC/EBPα. (E) Transactivation assays in HeLa cells transfected with Gck or G6pc luciferase reporter constructs along with overexpression of SRC-2, C/EBPα, or both. (F) qPCR analysis of Gck and G6pc expression in livers from mice harboring a congenic mutation in C/EBPα at serine 193 to alanine (S193A). ChIP data are represented as percent of input, whereas all other data are graphed as the mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001. #, significance over IgG.
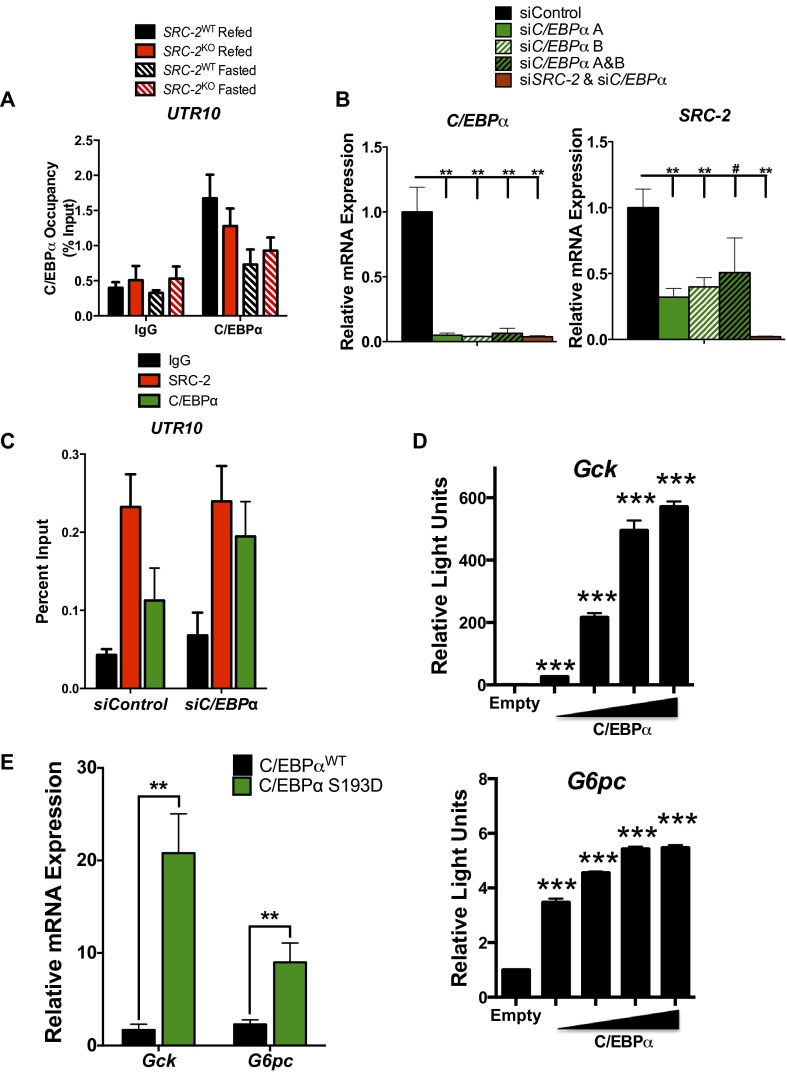
To confirm the occupancy of C/EBPα on the Gck promoter, we performed ChIP-qPCR on liver chromatin prepared from SRC-2WT and SRC-2KO mice that were fasted or refed. We found that C/EBPα is enriched on the Gck promoter during the refed state, but not UTR10, and its occupancy is largely dependent upon SRC-2 (Fig. 4B and Fig. S4A). C/EBPα also was recruited to the G6pc promoter during refeeding but to a lesser extent than to the Gck promoter, suggesting that C/EBPα may bind these promoters with different affinities or perhaps is tethered to the G6pc promoter rather than direct binding (Fig. 4B). To determine the function of C/EBPα in mediating transcription of Gck or G6pc, we knocked down C/EBPα expression by siRNA in primary hepatocytes. Our results demonstrated that knockdown of C/EBPα with individual siRNAs or in combination results in significantly lower expression of Cebpa, Gck, and G6pc (Fig. 4C and Fig. S4B) (12, 17). As expected, knockdown of SRC-2 along with C/EBPα provided no additional inhibition of Gck or G6pc as it is likely C/EBPα is required for recruitment to either of these target gene promoters (Fig. 4C and Fig. S4B). To test whether C/EBPα is required for recruitment of SRC-2 to the Gck promoter, we knocked down C/EBPα with two unique siRNAs in primary hepatocytes and performed ChIP-qPCR with primers designed around the overlapping SRC-2 and C/EBPα ChIP-Seq peaks on the promoters of Gck and G6pc (Fig. 4 A, iii). We found that loss of C/EBPα selectively decreased the recruitment of SRC-2 to the Gck promoter, but not the G6pc promoter or the UTR10 gene region (Fig. 4D and Fig. S4C). To further assess the ability of SRC-2 to preferentially coactivate C/EBPα on the Gck promoter, we performed luciferase assays with either Gck or G6pc promoters in the presence of SRC-2 and/or C/EBPα. We determined that SRC-2 synergistically coactivates C/EBPα on the Gck promoter but not the G6pc promoter (Fig. 4E). Additionally, titration of increasing amounts of C/EBPα into the luciferase assay was sufficient to dose-dependently potentiate luciferase expression from the Gck promoter while only marginally impacting G6pc promoter activity (Fig. S4D).
Fig. S4.
SRC-2 synergizes with C/EBPα to regulate Gck expression. (A) ChIP-qPCR analysis of C/EBPα occupancy on the UTR10 region in SRC-2WT and SRC-2KO mice (n = 3 each) after either a 24 h fast or a 24 h fast followed by a 3 h refeed. (B) qPCR analysis of C/EBPα and SRC-2 expression in wild-type primary hepatocytes treated with siSRC-2, siC/EBPα, or both. (C) ChIP-qPCR analysis of SRC-2 and C/EBPα occupancy on UTR10 in wild-type primary hepatocytes transfected with control siRNA or siC/EBPα. (D) Transactivation assays in HeLa cells transfected with Gck or G6pc luciferase reporter constructs along with overexpression of C/EBPα. (E) qPCR analysis of Gck and G6pc expression in livers from mice harboring congenic mutation in C/EBPα at serine 193 to aspartic acid (S193D). ChIP data are represented as percent of input, whereas all other data are graphed as the mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001.
The transcriptional activity of C/EBPα is influenced by phosphorylation that alters its interaction with coregulators (18). Insulin activation leads to PP2A-induced dephosphorylation of S193 on C/EBPα (19). Consistent with these findings, loss of the regulatory domain containing this serine residue was sufficient to increase Gck expression but failed to impact the expression of G6pc (18, 20). Supporting these data, mice harboring a congenic loss of function mutation (S193A) in C/EBPα displayed a marked increase in Gck expression but showed no effect on G6pc expression (Fig. 4F) (18). Conversely, both Gck and G6pc expression are elevated in mice carrying a constitutively active mutation (S193D) in C/EBPα (Fig. S4E). Taken together, these findings substantiate the importance of phosphorylation of C/EBPα at S193 for controlling the recruitment of coregulators like SRC-2 to specify target gene transcription.
Discussion
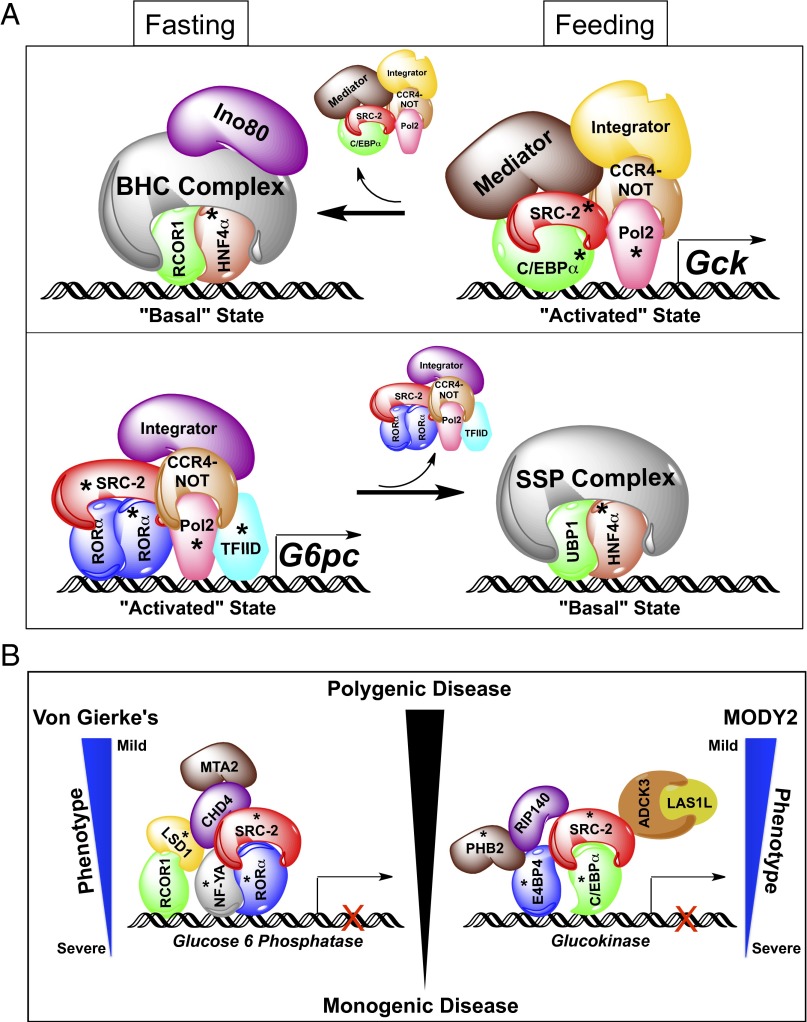
Through the use of DNA pull-down technology coupled with the unbiased power of MS, we have elucidated two overarching roles for SRC-2 in the metabolic regulation of rate-limiting genes for maintenance of glucose homeostasis. First, our data demonstrate that SRC-2 dictates the composition of TF–coregulator complexes on the promoters of Gck and G6pc in the fasted and refed states. Second, SRC-2 is essential for establishing a functional threshold of transcriptional machinery required for meeting metabolic demand. Stratification, integration, and selective validation of candidate complex components lead to the concept of a metabolic switch whereby rate-limiting glycemic genes are transformed from a “basal” to an “activated” state in response to energetic cues (Fig. 5A).
Fig. 5.
Proposed model for transcriptional regulation of glucose homeostasis by SRC-2. (A) Schematic representation of SRC-2 regulation of the opposing enzymatic functions of Gck and G6pc in regulation of glucose homeostasis. (B) Schematic summary of protein complex components enriched on the Gck or G6pc promoters as identified by MS in the fasting versus feeding states. Asterisks indicate complex components analyzed by immunoblot. BHC, BRAF–histone deacetylase complex; SSP, stage selector protein complex.
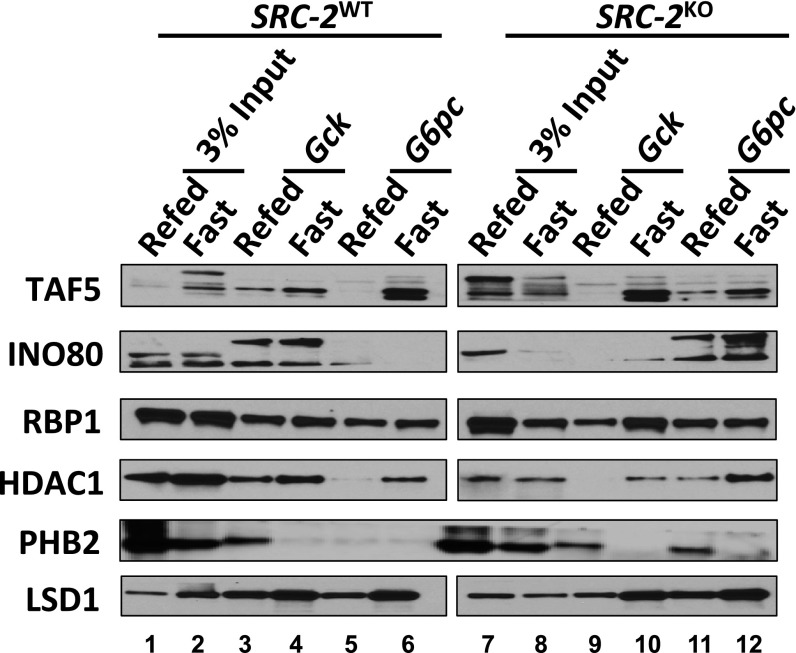
Although our MS data clearly highlight the existence of a fasting–feeding switch for counterregulation of genes that control glucose homeostasis, we chose candidate TFs for our model that were most robustly responsive to these metabolic conditions (Fig. 3 D and F and table 4, hosted at epicome.org/index.php/msprojects/src2metabolismresource). During postprandial conditions, we identified an activated state complex on the Gck promoter that synergizes the activities of C/EBPα and SRC-2 to stabilize recruitment of the Mediator and Integrator complexes along with POL2 and components of the CCR4–NOT complex to drive Gck expression (Fig. 5 A, Top, Fig. S5, and table 4, hosted at epicome.org/index.php/msprojects/src2metabolismresource). In the fasted state, these components are dismissed, leading to the recruitment of members of the repressive BRAF–histone deacetylase (BHC) complex that maintains the gene in a basal state (Fig. 5 A, Top, Fig. S5, and table 4, hosted at epicome.org/index.php/msprojects/src2metabolismresource). Supporting our published data, during the fasted state RORα and SRC-2 function in concert with components of the Integrator complex, CCR4–NOT complex, POL2, and TFIID on the G6pc promoter to amplify its expression (8) (Fig. 5 A, Bottom, Fig. S5, and table 4, hosted at epicome.org/index.php/msprojects/src2metabolismresource). In the postprandial state, this activating machinery is replaced by members of the stage selector protein (SSP) complex that facilitates repression of G6pc transcription. These findings establish the importance of SRC-2 for dynamic TF–coregulator complex recruitment to modulate metabolic gene transcription.
Fig. S5.
TF–coregulator complex components influenced by SRC-2 and immunoblot analysis of TAF5, INO80, RBP1, HDAC1, PHB2, and LSD1 binding to Gck or G6pc promoters from fasted or refed wild-type liver nuclear extract. Experimental lanes are numbered. Lanes between pull-downs from SRC-2WT or SRC-2KO liver nuclear extract were cropped out of the same image to allow for consistent data representation, as this lane contained a protein marker.
Our findings raise the intriguing possibility that SRC-2 may serve as a model to explain a part of the spectrum of polygenic diseases such as T2DM (Fig. 5B). Genetic ablation of SRC-2 represents a monogenic insult with polygenic consequences on the expression of nearly two-thirds of programmatic genes essential for glycolysis and gluconeogenesis (Fig. 1E). By definition, the milieu of transcriptional machinery recruited to either Gck in the fed state or G6pc during the fasted state represents the byproducts of polygenic inputs. Our findings suggest that a primary role of SRC-2 action is to stabilize metabolically responsive transcriptional complexes, thus establishing the set point for optimal gene output. As such, the repercussions of impaired SRC-2 function are amplified by perturbing complex formation on target gene promoters, resulting in blunted gene transcription in response to metabolic duress. Based on this logic, one might predict that mutations that diminish the function of a platform coregulator such as SRC-2 may explain a portion of the polygenic inputs to disorders of glucose regulation (Fig. 1E). In fact, these findings provide the most convincing evidence to date of our contention that coregulators like SRC-2 can serve as an entrée to understand a subset of polygenic diseases (21). It is not surprising that monogenic diseases give rise to the most severe phenotype, as is the case of G6pc for von Gierke’s disease (22, 23) or Gck for maturity onset diabetes of the young (MODY2) (24, 25) where pathogenic mutations often lead to complete loss of gene function. However, our findings suggest that disturbances in coregulator function not only perturb TF binding but also dampen the recruitment of secondary and tertiary coregulators that fractionally contribute to gene output. Loss of SRC-2 function leads to impaired Gck and G6pc expression, resulting in aberrant glucose homeostasis reminiscent of von Gierke’s and MODY2 phenotypes (22–25).
To date, numerous genome-wide association studies and whole exome sequencing efforts have been insufficient to clarify definitive genetic contributions to polygenic diseases like T2DM (26–28). These shortcomings are understandable as these approaches use a phenotype-centric strategy wherein participants are selected that display a certain predefined definition of phenotype/disease (29, 30). In the case of T2DM, the disease spectrum ranges from simple hyperglycemia to a constellation of accompanying comorbidities defined as the metabolic syndrome (i.e., hypertension, dyslipidemia, abdominal obesity, insulin resistance/glucose intolerance, proinflammatory, and prothrombotic states). Given such heterogeneity in the clinical definition of T2DM, discovery of definitive genetic underpinnings that strongly correlate with the disease have remained elusive. The wealth of genetic sequencing information accumulated by these investigations provides a unique opportunity to evaluate whether small nucleotide polymorphisms or pathogenic mutations in genes like SRC-2 correlate with patient phenotypes. Based on our findings of the expansive polygenic effects of SRC-2 disruption, the future integration of various sequencing datasets with matched transcriptomic analyses may permit a better understanding of how perturbations in pleiotropic factors like SRC-2 contribute to the phenotypic spectrum of diseases like T2DM.
Acknowledgments
We thank Rainer Lanz for lessons on relational database management. We thank Dr. Pradip Saha from the Mouse Metabolic Core Facility at Baylor College of Medicine and Cancer Prevention Research Institute of Texas (CPRIT) Core Facility Support Award RP120092 “Proteomic and Metabolomic Core Facility” for technical support. This research was supported by NIH Grants F31CA171350 (to E.S.) and P01 DK59820 and 3U19DK062434-10W1 (to B.W.O.) and Diabetes Research Center Grant NIDDK-P30 DK079638. Partial support for this work was provided by NIH Grant GM033976 to Anthony R. Means as well as a grant to Baylor College of Medicine from the Howard Hughes Medical Institutes through the Med Into Grad Initiative, Medical Scientist Training Program National Institute of General Medical Sciences (NIGMS) T32 GM7330 and DK56338 to the Texas Medical Center Digestive Diseases Center, and Susan G. Komen Research Grant PDF14300468 (to S.D.). Additionally, research support was provided by the Welch Foundation (Q1521) and from the Center for the Advancement of Science in Space Integrated OMICs Award (to C.C.D., B.Y., and B.W.O.). The authors acknowledge the joint participation by Adrienne Helis Malvin Medical Research Foundation through its direct engagement in the continuous active conduct of medical research in conjunction with Baylor College of Medicine and the “CaMKK2 Inhibitors for Therapeutic Treatment of Hepatic Cancer” program.
Footnotes
The authors declare no conflict of interest.
Data deposition: The datasets reported in this paper have been deposited at epicome.org/index.php/msprojects/src2metabolismresource.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1519073112/-/DCSupplemental.
References
- 1.Knowler WC, et al. Reduction in the incidence of type 2 diabetes with lifestyle intervention or metformin. N Engl J Med. 2002;346(6):393–403. doi: 10.1056/NEJMoa012512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stashi E, York B, O’Malley BW. Steroid receptor coactivators: Servants and masters for control of systems metabolism. Trends Endocrinol Metab. 2014;25(7):337–347. doi: 10.1016/j.tem.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Picard F, et al. SRC-1 and TIF2 control energy balance between white and brown adipose tissues. Cell. 2002;111(7):931–941. doi: 10.1016/s0092-8674(02)01169-8. [DOI] [PubMed] [Google Scholar]
- 4.Duteil D, et al. The transcriptional coregulators TIF2 and SRC-1 regulate energy homeostasis by modulating mitochondrial respiration in skeletal muscles. Cell Metab. 2010;12(5):496–508. doi: 10.1016/j.cmet.2010.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reineke EL, et al. Steroid receptor coactivator-2 is a dual regulator of cardiac transcription factor function. J Biol Chem. 2014;289(25):17721–17731. doi: 10.1074/jbc.M113.539908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reineke EL, et al. SRC-2 coactivator deficiency decreases functional reserve in response to pressure overload of mouse heart. PLoS One. 2012;7(12):e53395. doi: 10.1371/journal.pone.0053395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stashi E, et al. SRC-2 is an essential coactivator for orchestrating metabolism and circadian rhythm. Cell Reports. 2014;6(4):633–645. doi: 10.1016/j.celrep.2014.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chopra AR, et al. Absence of the SRC-2 coactivator results in a glycogenopathy resembling Von Gierke’s disease. Science. 2008;322(5906):1395–1399. doi: 10.1126/science.1164847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Malovannaya A, et al. Analysis of the human endogenous coregulator complexome. Cell. 2011;145(5):787–799. doi: 10.1016/j.cell.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Foulds CE, et al. Proteomic analysis of coregulators bound to ERalpha on DNA and nucleosomes reveals coregulator dynamics. Mol Cell. 2013;51(2):185–199. doi: 10.1016/j.molcel.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schwanhausser B, et al. Global quantification of mammalian gene expression control. Nature. 2011;473(7347):337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 12.Dasgupta S, et al. Coactivator SRC-2-dependent metabolic reprogramming mediates prostate cancer survival and metastasis. J Clin Invest. 2015;125(3):1174–1188. doi: 10.1172/JCI76029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mann M, Jensen ON. Proteomic analysis of post-translational modifications. Nat Biotechnol. 2003;21(3):255–261. doi: 10.1038/nbt0303-255. [DOI] [PubMed] [Google Scholar]
- 14.York B, et al. Reprogramming the posttranslational code of SRC-3 confers a switch in mammalian systems biology. Proc Natl Acad Sci USA. 2010;107(24):11122–11127. doi: 10.1073/pnas.1005262107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheskis BJ, et al. Hierarchical affinities and a bipartite interaction model for estrogen receptor isoforms and full-length steroid receptor coactivator (SRC/p160) family members. J Biol Chem. 2003;278(15):13271–13277. doi: 10.1074/jbc.M211031200. [DOI] [PubMed] [Google Scholar]
- 16.Schmidt D, et al. Five-vertebrate ChIP-seq reveals the evolutionary dynamics of transcription factor binding. Science. 2010;328(5981):1036–1040. doi: 10.1126/science.1186176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Safdar H, et al. Modulation of mouse coagulation gene transcription following acute in vivo delivery of synthetic small interfering RNAs targeting HNF4alpha and C/EBPalpha. PLoS One. 2012;7(6):e38104. doi: 10.1371/journal.pone.0038104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jin J, et al. Cooperation of C/EBP family proteins and chromatin remodeling proteins is essential for termination of liver regeneration. Hepatology. 2015;61(1):315–325. doi: 10.1002/hep.27295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang GL, Iakova P, Wilde M, Awad S, Timchenko NA. Liver tumors escape negative control of proliferation via PI3K/Akt-mediated block of C/EBP alpha growth inhibitory activity. Genes Dev. 2004;18(8):912–925. doi: 10.1101/gad.1183304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pedersen TA, et al. Distinct C/EBPalpha motifs regulate lipogenic and gluconeogenic gene expression in vivo. EMBO J. 2007;26(4):1081–1093. doi: 10.1038/sj.emboj.7601563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lonard DM, Kumar R, O’Malley BW. Minireview: The SRC family of coactivators: An entree to understanding a subset of polygenic diseases? Mol Endocrinol. 2010;24(2):279–285. doi: 10.1210/me.2009-0276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chou JY. The molecular basis of type 1 glycogen storage diseases. Curr Mol Med. 2001;1(1):25–44. doi: 10.2174/1566524013364112. [DOI] [PubMed] [Google Scholar]
- 23.Yang Chou J, Mansfield BC. Molecular genetics of type 1 glycogen storage diseases. Trends Endocrinol Metab. 1999;10(3):104–113. doi: 10.1016/s1043-2760(98)00123-4. [DOI] [PubMed] [Google Scholar]
- 24.Martin D, et al. Long-term follow-up of oral glucose tolerance test-derived glucose tolerance and insulin secretion and insulin sensitivity indexes in subjects with glucokinase mutations (MODY2) Diabetes Care. 2008;31(7):1321–1323. doi: 10.2337/dc07-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garcia-Herrero CM, et al. Functional analysis of human glucokinase gene mutations causing MODY2: Exploring the regulatory mechanisms of glucokinase activity. Diabetologia. 2007;50(2):325–333. doi: 10.1007/s00125-006-0542-7. [DOI] [PubMed] [Google Scholar]
- 26.Bonnefond A, Froguel P, Vaxillaire M. The emerging genetics of type 2 diabetes. Trends Mol Med. 2010;16(9):407–416. doi: 10.1016/j.molmed.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 27.Groop L, Pociot F. Genetics of diabetes—Are we missing the genes or the disease? Mol Cell Endocrinol. 2014;382(1):726–739. doi: 10.1016/j.mce.2013.04.002. [DOI] [PubMed] [Google Scholar]
- 28.de Bruin C, Dauber A. Insights from exome sequencing for endocrine disorders. Nat Rev Endocrinol. 2015;11(8):455–464. doi: 10.1038/nrendo.2015.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McGuire AL, Cho MK, McGuire SE, Caulfield T. Medicine. The future of personal genomics. Science. 2007;317(5845):1687. doi: 10.1126/science.1147475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McGuire SE, McGuire AL. Don’t throw the baby out with the bathwater: Enabling a bottom-up approach in genome-wide association studies. Genome Res. 2008;18(11):1683–1685. doi: 10.1101/gr.083584.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chopra AR, et al. Cellular energy depletion resets whole-body energy by promoting coactivator-mediated dietary fuel absorption. Cell Metab. 2011;13(1):35–43. doi: 10.1016/j.cmet.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tackett BC, et al. P2Y2 purinergic receptor activation is essential for efficient hepatocyte proliferation in response to partial hepatectomy. Am J Physiol Gastrointest Liver Physiol. 2014;307(11):G1073–G1087. doi: 10.1152/ajpgi.00092.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11(5):1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Du P, Kibbe WA, Lin SM. lumi: A pipeline for processing Illumina microarray. Bioinformatics. 2008;24(13):1547–1548. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- 35.Smyth GK. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:Article3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]