Significance
Modulation of actin filament architecture underlies a plethora of cellular processes including cell shape, division, adhesion, and motility. In heart muscle cells actin-containing thin filaments form highly organized structures with precisely regulated lengths. This precision is required for efficient interaction with myosin-containing filaments and provides the basis for contraction. The mechanism whereby heart muscle cells regulate thin filament assembly and its consequences for cardiac physiology are largely unknown. We discovered that Leiomodin 2 (Lmod2) elongates thin filaments to a proper length. Mice lacking Lmod2 have abnormally short thin filaments, experience severe contractile dysfunction and ventricular chamber enlargement consistent with dilated cardiomyopathy, and die at age ∼3 wk. Therefore, Lmod2 and proper thin filament lengths are essential for heart function.
Keywords: actin-thin filaments, cardiomyopathy, cytoskeletal dynamics
Abstract
Leiomodin 2 (Lmod2) is an actin-binding protein that has been implicated in the regulation of striated muscle thin filament assembly; its physiological function has yet to be studied. We found that knockout of Lmod2 in mice results in abnormally short thin filaments in the heart. We also discovered that Lmod2 functions to elongate thin filaments by promoting actin assembly and dynamics at thin filament pointed ends. Lmod2-KO mice die as juveniles with hearts displaying contractile dysfunction and ventricular chamber enlargement consistent with dilated cardiomyopathy. Lmod2-null cardiomyocytes produce less contractile force than wild type when plated on micropillar arrays. Introduction of GFP-Lmod2 via adeno-associated viral transduction elongates thin filaments and rescues structural and functional defects observed in Lmod2-KO mice, extending their lifespan to adulthood. Thus, to our knowledge, Lmod2 is the first identified mammalian protein that functions to elongate actin filaments in the heart; it is essential for cardiac thin filaments to reach a mature length and is required for efficient contractile force and proper heart function during development.
Striated muscle cells contain arrays of protein filaments assembled into contractile units that are nearly crystalline in structure. Efficient contraction at the molecular level is predicated upon accurate overlap of actin-containing thin and myosin-containing thick filaments. Therefore, proper control of filament assembly is absolutely critical.
In striated muscle it is currently thought that the thin-filament pointed end capping protein tropomodulin (Tmod) is the predominant regulator of thin filament length, with Tmod1 being the sole isoform expressed in cardiomyocytes (1). Extensive in vitro work has revealed that Tmod1 uses two actin- and two tropomyosin-binding sites to associate with the end of the thin filament and to prevent addition or loss of actin monomers, thereby controlling length of the thin filament (2–7). Tmod1 is essential for life; Tmod1-KO mice are embryonic lethal because of cardiac defects (8–11).
Identification of additional but structurally different members of the Tmod family of proteins, the leiomodins (Lmods), raises the possibility that thin filament lengths are not regulated solely by Tmod at thin filament pointed ends (12). Although there are three Lmod genes (Lmod1–3), Lmod2 and 3 are expressed in striated muscle with Lmod2 being the predominant isoform in cardiac muscle and Lmod3 the predominant isoform in skeletal muscle (12–16). The Lmods share ∼40% sequence identity at the protein level with the Tmods but do not contain a recognizable second tropomyosin-binding domain and have an additional C-terminal extension that includes a proline-rich region and an actin-binding Wiskott–Aldrich syndrome protein homology 2 (WH2) domain (12, 17). Lmod2 has been proposed to be the long-sought muscle actin filament nucleator because it robustly nucleates actin filament formation in vitro (because of its three actin-binding sites) and is reportedly required for proper sarcomere assembly in cultured cardiomyocytes (17). Like Tmod1, Lmod2 assembly at the pointed end of the thin filament requires association with tropomyosin; however unlike Tmod1, Lmod2 assembly also is dependent on contractility and the availability of polymerizable actin (18). Although part of the Tmod family of proteins, Lmod2 does not demonstrate actin filament-capping activity, and its overexpression displaces Tmod1; it is not known if this displacement is a direct or indirect effect (13). Nevertheless, Lmod2 overexpression results in the elongation of thin filaments in cells in culture (13). Limited data regarding the function of Lmod2 suggest it could play an important role in sarcomeric actin assembly, but the physiological function of Lmod2 has yet to be studied.
Here we show that Lmod2 functions as an actin filament elongation factor in the heart. Our search for the mechanism by which Lmod2 functions revealed that Lmod2 promotes actin assembly and dynamics at the pointed end of the thin filament, is not necessary for myofibrillogenesis, but is required for thin filaments to attain a mature length. Our results also indicate that Lmod2 is essential for normal heart function and suggest that dysregulation of the thin filament length is causative for dilated cardiomyopathy (DCM).
Results
Lmod2−/− Mice Die ∼3 Wk After Birth.
To decipher the in vivo function of Lmod2, we generated Lmod2-KO mice (Fig. S1). Lmod2-KO mice are born in the expected Mendelian ratios of 1:2:1 (23% Lmod2+/+, 54% Lmod2+/−, and 22% Lmod2−/−) and die 15–33 d after birth with a median survival of 20 d (Fig. 1A). Mice heterozygous for Lmod2 have normal life spans and present with no discernable phenotype. Genotyping with primers located within exon 2 confirmed loss of the Lmod2 gene (Fig. 1B). Immunoblot analysis revealed complete loss of Lmod2 protein in the left ventricle (LV) of Lmod2-KO mice (Fig. 1C).
Fig. S1.
Strategy used by Regeneron Pharmaceuticals to knock out the Lmod2 gene in mice. Lines represent introns, and boxes indicate exons; filled boxes designate the coding sequence. Homologous recombination results in the replacement of the complete coding sequence of Lmod2 with a cassette containing the LacZ gene from Escherichia coli. The locations of primers used in genotyping are specified above their corresponding exons (Lmod2 GP and LacZ GP).
Fig. 1.
Lmod2-KO mice die before weaning with no detectable Lmod2 protein in the heart. Lmod2 expression is restricted to striated muscle in the mouse embryo. (A) Survival curve of Lmod2+/+ (WT, black line) and Lmod2−/− (KO, gray line) mice. The KO curve is significantly different from WT, P < 0.0001, log-rank test. (B) Genotyping with Lmod2 and LacZ cassette-specific primers. WT mice produce a 231-bp Lmod2 band, KO mice produce a 684-bp LacZ cassette band, and heterogeneous (HET) mice produce both bands. (C) Immunoblots of LV lysate from P1 Lmod2 WT and KO mice. Lysate was probed with anti-Lmod2 and anti-GAPDH antibodies. (D) β-Gal staining of Lmod2+/− embryos at E8.5–E10.5. Arrows denote the heart, arrowheads the pharyngeal arches, and asterisks the somites. (Scale bars: 0.5 mm.)
Lmod2 Expression Is Restricted to Striated Muscle.
To determine the spatial and temporal expression pattern of Lmod2, embryos at various stages of development were stained for β-gal activity. Lmod2 expression is restricted to striated muscle and is first detected in the heart at embryonic day 8.5 (E8.5) and in the somites at E9.5 (Fig. 1D, arrows and asterisks respectively); Lmod2 continues to be expressed in these tissues throughout development (Fig. 1D). Slight staining also was detected in the pharyngeal arches, which will form the muscles of the head and neck (Fig. 1D, arrowheads).
Lmod2-KO Hearts Have Shorter Thin Filaments as Early as E12.5.
Because Lmod2 has been implicated in the regulation of thin filament assembly in vitro, we analyzed thin filaments in Lmod2-KO hearts using deconvolution microscopy. To measure thin filament lengths, cryosections of relaxed whole hearts (E12.5) or stretched LV tissue (postnatal days 6 and 15; P6, P15) were stained with fluorescently conjugated phalloidin to label filamentous actin. Thin filament length then was determined accurately using Distributed Deconvolution (DDecon) software (19). An example of the staining and intensity profiles used to measure thin filament length is illustrated in Fig. 2A. Strikingly, from as early as E12.5, the Lmod2-KO hearts have shorter thin filaments than WT hearts (with up to ∼15% reduction at P15) (Fig. 2B). Thin filaments are significantly longer in the hearts of neonatal than fetal WT mice, but their length does not change over this same time period in the KO mice (Fig. 2B). As an internal control, the lengths of myosin filaments did not differ in KO and WT mice (WT: 1.76 ± 0.025 μm; KO: 1.73 ± 0.027 μm; mean ± SEM; n = 3; P = 0.42). Thin filament lengths were unchanged in two muscles of the leg, the extensor digitorum longus (EDL) and soleus (Fig. 2C). Overall, these results suggest there is an alteration in actin-myosin cross-bridge stoichiometry in the hearts of KO mice.
Fig. 2.
Lmod2-KO mice have shorter cardiac thin filaments. (A, Upper) Representative image of F-actin stain from WT and Lmod2-KO stretched LV tissue at P6; pink lines denote a gap in F-actin staining across the M line (center of sarcomere). B, barbed end; P, pointed end. (Scale bar: 1 μm.) (Lower) An example of the intensity profiles used by the DDecon analysis program to determine thin filament length accurately. (B) Thin filament (TF) lengths in the LV of WT (black bars) and Lmod2-KO (white bars) mice at various developmental time points; n = 3 or 4. (C) Thin filament lengths in the EDL and soleus muscles of WT (black bars) and Lmod2-KO (white bars) mice at P15; n = 2. (D) Thin filament lengths from neonatal mouse cardiomyocytes (NMCM) in culture isolated from WT (black bars) and Lmod2-KO (white bars) hearts followed by transduction with GFP or GFP-Lmod2 Adv at a multiplicity of infection (MOI) of 2; n = ∼110 total measurements from 8–12 cells per culture, three cultures. All values are mean ± SEM.
Lmod2-KO Neonatal Cardiomyocytes Have Shorter Thin Filaments: Rescue with GFP-Lmod2.
Primary cultures of neonatal cardiomyocytes were used to take advantage of their flexibility and amenability to functional manipulations and the ability to analyze them live by microscopy and to avoid the confounding influences of other tissues. Identical to observations in the cardiac tissue of Lmod2-KO mice, neonatal cardiomyocytes from P1–P2 KO mice that were cultured for 5–6 d have shorter thin filaments than cells obtained from WT mice (Fig. 2D). Thus, cardiomyocytes in culture strikingly recapitulate the thin filament alterations present in Lmod2-KO hearts in vivo. Additionally, reduction in thin filament length is caused specifically by the loss of Lmod2, because adenoviral (Adv)-mediated transduction of GFP-Lmod2 restores thin filament length in KO cells to that found in WT cells (Fig. 2D and Fig. S2 A and B demonstrate that GFP-Lmod2 expresses at ∼40% of endogenous Lmod2 levels in the rescue experiment and assembles at the pointed end of the thin filament).
Fig. S2.
Determination of the relative protein levels of GFP-Lmod2 that rescue the thin filament length deficit in Lmod2-KO cardiomyocytes in culture and localization of GFP-Lmod2. (A, Left) Immunoblot analysis of Lmod2 protein levels in WT and Lmod2-KO neonatal cardiomyocytes transduced with 2 MOI of GFP or GFP-Lmod2 Adv. Note: Endogenous Lmod2 runs between 70–100 kDa and GFP-Lmod2 between 100–130 kDa. (Right) Mean relative Lmod2 protein expression in two cultures ± SEM. (B) Rat cardiomyocytes plated on flexible culture plates (Flexcell Inc.), transduced with GFP-Lmod2 and subjected to ∼20% equibiaxial stretch before fixation. Costaining for α-actinin marks the Z-disk. (Scale bar: 2 μm.)
Lmod2-KO Mice Present with DCM.
The Lmod2-KO mice are indistinguishable from their WT littermates and have no overt signs of distress until just before death. Interestingly, histological analysis of Lmod2-KO hearts at P15, a time point when they begin to die, revealed enlarged ventricular lumens and thin ventricular walls, consistent with DCM, with no observable increase in fibrosis (Fig. 3A). Transthoracic M-mode echocardiography at the level of the papillary muscle confirmed the histology results: (i) thickness of the LV walls at end diastole is reduced significantly in KO hearts compared with WT hearts (Fig. 3B and Table S1), and (ii) the internal diameter of the LV is significantly larger in the KO heart than in the WT heart (Fig. 3C). Accordingly, the ratio of wall thickness to chamber diameter is decreased in the KO heart, as is consistent with eccentric remodeling (Table S1). Eccentric remodeling is often associated with elongation of cardiomyocytes. Indeed, at P19 isolated Lmod2-KO cardiomyocytes are 37% longer on average than WT but have no change in width (Fig. S3). The cellular elongation is likely caused by the addition of new sarcomeres and not by the elongation of existing sarcomeres, because the isolated KO cells have reduced sarcomere lengths (WT: 1.83 ± 0.01 μm; KO: 1.74 ± 0.01 μm; mean ± SEM; n = 36–50, P < 0.0001, Student's t test). Finally, the ejection fraction is reduced by nearly 60% in the KO mice, indicating that systolic performance is severely compromised after the loss of Lmod2 (Fig. 3D).
Fig. 3.
Lmod2-KO hearts display large ventricular lumens, thin ventricular walls, and reduced systolic performance. (A) Longitudinal (Upper) and transverse (Lower) sections of P15 paraffin-embedded hearts stained with Masson's Trichrome. RV, right ventricle. (Scale bar: 0.5 mm.) (B–D) Echocardiography analysis of WT (black bars) and Lmod2-KO (white bars) hearts at P2, P6, and P15. (B) LV posterior wall in diastole. (C) LV end diastolic diameter. (D) Ejection fraction (EF). Data are shown as mean ± SEM; n = 6 or 7.
Table S1.
Echocardiographic analysis of WT and Lmod2-KO mice at days P2, P6, and P15
| Age | Genotype | EF, % | LVAW d, mm | LVAW s, mm | LVPW d, mm | LVPW s, mm | LVID d, mm | LVID s, mm | Eccentricity, WT/LVDD | HR, bpm |
| P2 n = 7 | WT | 74.59 ± 1.18 | 0.42 ± 0.03 | 0.61 ± 0.05 | 0.36 ± 0.01 | 0.53 ± 0.02 | 1.44 ± 0.07 | 0.87 ± 0.05 | 0.55 ± 0.04 | 492 ± 7 |
| KO | 76.08 ± 1.86 | 0.46 ± 0.03 | 0.64 ± 0.04 | 0.36 ± 0.02 | 0.54 ± 0.01 | 1.51 ± 0.07 | 0.88 ± 0.05 | 0.54 ± 0.03 | 479 ± 14 | |
| P6 n = 8–11 | WT | 68.54 ± 1.48 | 0.40 ± 0.01 | 0.64 ± 0.02 | 0.43 ± 0.02 | 0.64 ± 0.04 | 1.97 ± 0.04 | 1.26 ± 0.04 | 0.43 ± 0.02 | 524 ± 14 |
| KO | 54.15 ± 4.81** | 0.36 ± 0.02* | 0.51 ± 0.03** | 0.33 ± 0.01** | 0.50 ± 0.04* | 2.02 ± 0.11 | 1.50 ± 0.14 | 0.35 ± 0.20* | 531 ± 8 | |
| P15 n = 6–7 | WT | 75.80 ± 2.50 | 0.63 ± 0.04 | 0.98 ± 0.07 | 0.64 ± 0.02 | 0.99 ± 0.06 | 2.17 ± 0.12 | 1.24 ± 0.17 | 0.60 ± 0.06 | 628 ± 9 |
| KO | 30.55 ± 5.58**** | 0.45 ± 0.01*** | 0.60 ± 0.03*** | 0.41 ± 0.03**** | 0.52 ± 0.06*** | 3.40 ± 0.16**** | 2.94 ± 0.23**** | 0.26 ± 0.02*** | 620 ± 21 |
Measurements include ejection fraction (EF), LV anterior wall thickness (LVAW), LV posterior wall thickness (LVPW), LV internal diameter (LVID), eccentricity (wall thickness/LV end diastolic diameter), and heart rate (HR). bpm, beats per minute; d, diastole, s, systole. Measurements are means ± SEM. Bolded text indicates values that are statistically significantly different between WT and KO (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001). n = 6–11.
Fig. S3.
Cardiomyocytes are significantly longer in Lmod2-KO mice. Length and width measurements of cardiomyocytes isolated from P19 WT (black bars) and KO (white bars) mice. Mean ± SEM; n = 3; *P < 0.05, Student's t test.
Onset of DCM Is Rapid in Lmod2-KO Mice.
We next set out to analyze the progression of cardiac dysfunction in Lmod2-KO mice. Echocardiography performed on mice just after birth (P2) revealed no difference in wall thickness, chamber diameter, or systolic performance (Fig. 3 B–D and Table S1). Interestingly, echocardiography at P6 was markedly variable; the data revealed thinner LV walls and reduced ejection fraction on average, with no significant change in chamber dimension in KO hearts (Fig. 3 B–D and Table S1). P6 likely represents a developmental stage in KO mice just before the pathological remodeling of cardiac dilation.
Although echocardiography revealed that Lmod2-KO hearts are dilated by P15, most of the common indicators of cardiac failure are not present at P15. There is no significant change in heart weight or heart weight-to-body weight ratio (Table S2), and there is a significant alteration in the expression of only two of six molecular markers of heart failure [B-type natriuretic peptide (BNP) and sarcoplasmic reticulum CA2+ ATPase (SERCA2a); Fig. S4A]. Because the age at death varied over a 2-wk period, we analyzed KO hearts at a later time point. Mice at P19 (an age by which ∼50% of the KOs have died) have a significant increase in heart weight and in the heart weight-to-body weight ratio (Table S2), and the expression of all the molecular markers of heart failure we analyzed is altered significantly (Fig. S4B). Thus, Lmod2-KO mice develop an unusually rapid-onset DCM, displaying typical markers of cardiomyopathy only very late in the progression of the pathology.
Table S2.
Lmod2-KO mice have significantly larger hearts at P19
| Age | Genotype | HW, mg | LW, mg | BW, g | TL, mm | HW/BW. mg/g | HW/TL, mg/mm | LW/BW, mg/g | LW/TL, mg/mm |
| P15 n = 6–11 | WT | 50.4 ± 1.6 | 84.8 ± 2.4 | 8.15 ± 0.22 | 10.48 ± 0.23 | 6.21 ± 0.17 | 4.87 ± 0.12 | 10.43 ± 0.23 | 8.20 ± 0.36 |
| KO | 52.2 ± 3.4 | 81.4 ± 2.7 | 8.43 ± 0.27 | 10.21 ± 0.12 | 6.40 ± 0.52 | 5.14 ± 0.34 | 9.65 ± 0.13 | 7.71 ± 0.21 | |
| P19 n = 7–8 | WT | 56.4 ± 4.4 | 86.9 ± 3.6 | 8.45 ± 0.64 | 11.31 ± 0.22 | 6.73 ± 0.28 | 4.95 ± 0.28 | 10.47 ± 0.36 | 7.68 ± 0.25 |
| KO | 76.2 ± 4.4** | 83.7 ± 5.6 | 8.33 ± 0.64 | 11.37 ± 0.23 | 9.26 ± 0.39*** | 6.70 ± 0.35** | 10.10 ± 0.20 | 7.32 ± 0.34 |
BW, body weight; HW, heart weight; LW, lung weight; TL, tibia length. Measurements are means ± SEM. Bolded text indicates values that are statistically significantly different between WT and KO (**P < 0.01, ***P < 0.001). n = 6–11.
Fig. S4.
Expression of four of six molecular markers of heart failure is not altered significantly at P15, but all markers are significantly changed at P19. RT-qPCR analysis of gene expression in the LV of WT and Lmod2-KO mice at P15 (A) and P19 (B). Data are shown as mean ± SEM; n = 5–11.
The Ultrastructure of Lmod2-KO Hearts Displays Multiple Pathological Hallmarks of Myopathy Right Before Death.
Lmod2-KO hearts were analyzed at the ultrastructural level to determine if there are any additional changes in addition to shorter thin filaments. Although Lmod2-KO hearts have a detectable functional deficit as assessed by echocardiography at P6 (see above), analysis by EM revealed remarkably unperturbed structure (Fig. 3 B–D and Fig. S5A), except for the presence of broader Z-discs in the KO hearts (WT: 99 ± 23 nm; KO: 136 ± 30 nm at sarcomere lengths of 1.3–1.4 μm; n = 36–50, P < 0.0001). At P1, WT and KO hearts show no difference in Z-disk width and appear structurally identical to each other. Late-stage (P20) Lmod2-KO hearts display general myofibril disarray and hallmarks of DCM (Fig. S5B). Specific changes include (i) myofibril misalignment; (ii) broad Z-discs; (iii) T-tubule and sarcoplasmic reticulum dilation; (iv) mislocalization and increased convolutions of intercalated discs; and (v) mitochondrial abnormalities including swelling and loss of cristae with intact outer double membranes. In addition, mitochondria are less abundant in KO than in WT hearts (WT: 0.40 ± 0.04 mitochondria/μm2; KO: 0.28 ± 0.05 mitochondria/μm2; mean ± SEM, n = 10 images, P < 0.0001), indicating possible dysfunction of mitochondrial biogenesis.
Fig. S5.
The hearts of Lmod2-KO mice present with few detectable defects early in neonatal development but exhibit myofibril disarray and common features of DCM ∼2 wk later by EM. (A) Electron micrographs of cardiac LV tissue from P6 WT and Lmod2-KO mice. (Scale bar: 1 μm.) (B) LV micrographs of P20 WT (a, c, e, and g) and KO (b, d, f, and h) mice. (a and c) WT myocardium has compact filaments, with sarcomeres in register and mitochondria with intact cristae. (b and d) Lmod2-KO mouse hearts present with decreased mitochondrial (M) number, mitochondrial swelling with disorganized cristae, and dilated sarcoplasmic reticulum (SR). (e and f) Lmod2-KO intercalated discs have a significantly higher degree of convolutions compared with WT (arrows). (g and h) The sarcoplasmic reticulum (SR) is dilated in Lmod2-KO mice, and the mitochondria show degradation. (Scale bars: 2 μm in a and b; 1 μm in c–h.) (C) Skeletal muscle of the KO mice does not display any detectable defects. Electron micrographs of EDL muscle at P16. Z, Z-disk. (Scale bar: 1 μm.)
The ability of Lmod2-KO mice to suckle and maintain normal mobility for 2–3 wk of life suggests that there are no significant skeletal muscle defects. Ultrastructural analysis of the EDL muscle of the leg revealed no obvious differences between KO and WT mice (Fig. S5C), as is consistent with the mice dying of cardiac dysfunction.
Lmod3 and Tmod1 Protein Levels Are Not Altered in Lmod2-KO Hearts.
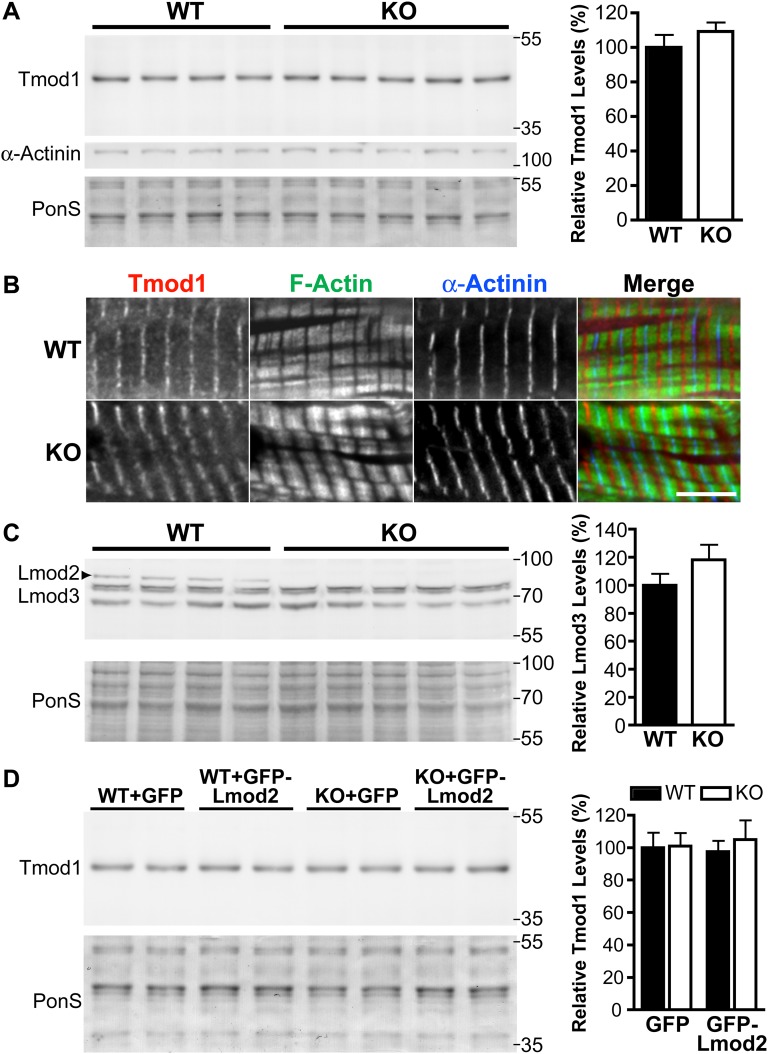
Because Lmod2 appears to have an antagonistic relationship with Tmod1 in cultured myocytes (13), we analyzed the effect that the loss of Lmod2 has on Tmod1. At a time point when cardiac dysfunction was first observed (P6–P9) Tmod1 levels are unchanged in the LV of the KO mice (Fig. S6A). Immunofluorescence staining revealed that Tmod1 remains localized at the pointed end of the thin filament in the KO mice but often displays a slightly broader distribution (Fig. S6B). We also found that protein levels of Lmod3, the other Lmod family member expressed in the heart, are not altered significantly upon ablation of Lmod2 (Fig. S6C).
Fig. S6.
Tmod1 and Lmod3 protein levels and Tmod1 localization are not significantly altered in Lmod2-KO mice, and the introduction of GFP-Lmod2 AAV does not alter Tmod1 protein levels. (A, Left) Immunoblot analysis of Tmod1 of protein levels in WT and Lmod2-KO mice at P9. (Right) Mean relative Tmod1 protein expression in four or five mice ± SEM. (B) Immunofluorescence staining of Tmod1 in the LV of WT and Lmod2-KO mice at P6. Staining for α-actinin marks the Z-disk. (Scale bar: 5 μm.) (C, Left) Immunoblot analysis of Lmod3 protein levels in WT and Lmod2-KO mice at P9. (Right) Mean relative Lmod3 protein expression in four or five mice ± SEM. Note: The Lmod3 antibody appears to cross-react with Lmod2 (top bands that disappear in the KO). (D) Immunoblot analysis of Tmod1 in the LV of WT and Lmod2-KO mice injected with GFP or GFP-Lmod2 AAV. (Left) Representative blot of two animals in each group. (Right) Mean relative Tmod1 protein expression in four to six animals ± SEM.
Lmod2-KO Cardiomyocytes Have Decreased Contractile Force.
The contractile force of single isolated cardiomyocytes was assessed to determine if the contractile dysfunction observed in the Lmod2-KO heart extends to the cellular level. Because of their biocompatibility, poly-dimethyl siloxane (PDMS) micropillar arrays can be used to measure nonmuscle cellular traction force (20, 21). We modified this technique to measure the contractile force of cardiomyocytes. WT and Lmod2-KO neonatal cardiomyocytes were plated on micropillar arrays (Fig. S7A). The cardiomyocytes spread over and attached to the pillars. After 4–5 d in culture, contractile force was determined based on the concept of beam-bending theory, in which force is related to the lateral deflection of the pillars (Fig. S7B). Lmod2-KO cardiomyocytes have an ∼20% reduction in contractile force compared with WT cardiomyocytes (Fig. 4A). Transduction of Adv GFP-Lmod2 is able to restore contractile force in the KO cells to that observed in WT cells (Fig. 4B). Thus, Lmod2-KO mice display contractile dysfunction at both the intact organ and cellular levels.
Fig. S7.
Micropillar array fabrication. (A) Schematic of micropillar fabrication process. (B) Lateral deflection of each pillar (δ) is measured and then multiplied by the pillar’s spring constant (k = 3πED4/64L3) to obtain a force value.
Fig. 4.
Lmod2-KO cardiomyocytes have reduced contractile force. (A) Contractile force of WT (black bar) and Lmod2-KO (white bar) neonatal cardiomyocytes plated on micropillar arrays. Data are shown as mean ± SEM; n = ∼70 cells from four cultures. (B) Contractile force of WT (black bar) and KO (white bar) neonatal cardiomyocytes transduced with GFP, and KO neonatal cardiomyocytes transduced with GFP-Lmod2 (gray vertically striped bar). n = 30–70 cells from two or three cultures.
Introduction of GFP-Lmod2 Elongates Thin Filaments in the Heart and Rescues Lmod2-KO Mice.
To confirm that the pathophysiology of the Lmod2-KO hearts is the primary cause of death and to determine the necessity of Lmod2 in fetal development, we reintroduced Lmod2 into Lmod2-KO mice. GFP-Lmod2 adeno-associated virus (rAAV2/9) was injected into the pericardial cavity of neonatal mice at day P4 (before detection of cardiac dysfunction). Analysis of these mice at P17–P19 (a time when ∼50% of Lmod2-KO mice die) revealed that GFP-Lmod2 is expressed in the LV at ∼30–40% of endogenous Lmod2 levels [Fig. 5A; note: This level of Lmod2 overexpression does not alter Tmod1 protein levels (Fig. S6D)]. Remarkably, thin filament length is restored to nearly normal in KO mice injected with GFP-Lmod2-AAV (Fig. 5B). The expression of molecular markers of heart failure [atrial natriuretic peptide (ANF) and BNP] is also rescued to normal levels in KO mice (Fig. 5C). Furthermore, echocardiography of GFP-Lmod2 AAV-injected mice revealed improved cardiac morphology and function as opposed to the enlarged ventricular chambers and systolic dysfunction evident in KO mice injected with GFP-AAV (Fig. 5D). Wall thickness is not restored upon the introduction of GFP-Lmod2 into the KO hearts. Finally, to determine the longevity of rescued Lmod2-KO mice, we allowed three mice injected with GFP-Lmod2 to live beyond the standard point of collection; one mouse died at P32, and two mice lived to adulthood. As a reference, without injection, only one mouse (of 48) lived past day 29; it died at day 33 (Fig. 1A).
Fig. 5.
Introduction of GFP-Lmod2 AAV rescues Lmod2-KO mice structurally and functionally. (A) Immunoblot analysis of Lmod2 protein levels in the LV of WT and KO mice injected with GFP or GFP-Lmod2 AAV. (Left) Representative blots of two animals in each group. Note: Endogenous Lmod2 runs between 70–100 kDa and GFP-Lmod2 between 100–130 kDa. (Right) Mean relative Lmod2 protein expression ± SEM; n = 4–6 animals. (B) LV thin filament lengths. (C) RT-qPCR of molecular markers of heart failure. Note: Because of a high degree of variation, ANF was not statistically significantly up-regulated in the KO mice injected with GFP-AAV. (D) Echocardiographic analysis of injected mice. Wall thickness, LV posterior wall in diastole; LV diameter, LV end diastolic diameter; EF, ejection fraction. All analyses are of P17–P19 mice. Data are shown as mean ± SEM; n = 5–6.
Lmod2 Promotes Actin Dynamics and Assembly at the Thin Filament Pointed End.
To probe for the mechanism by which Lmod2 is able to promote elongation of thin filaments, we analyzed the assembly of actin after the expression of Lmod2 in isolated cardiomyocytes. To mark the location of newly incorporated actin effectively, Rhodamine-labeled actin (Rho-actin) was microinjected into cardiomyocytes, and after 1-h incubation the cells were relaxed and fixed. Microinjected actin assembles similarly at both the pointed and barbed ends of thin filaments in GFP-expressing cells, as determined by costaining for α-actinin (Fig. 6 A and A′). However, analysis of the ratio of the intensity of Rho actin fluorescence at the pointed vs. barbed end of thin filaments in cells expressing GFP-Lmod2 indicated a significant increase in actin incorporation at the pointed end compared with that observed in cells expressing GFP alone (pointed end/barbed end incorporation = 1.55 ± 0.11 vs. 1.25 ± 0.07, respectively; mean ± SEM, n = 13–32 cells, P < 0.05). These results indicate that Lmod2 enhances the assembly of actin at the thin filament pointed end.
Fig. 6.
Lmod2 enhances actin incorporation and dynamics at thin filament pointed ends. (A) Microinjection of Rho-actin in GFP (Upper) and GFP-Lmod2 overexpressing (OE) (Lower) neonatal rat cardiomyocytes. Staining for α-actinin marks the Z-disk where the barbed ends of the actin filament are located (pink arrows); pointed ends are denoted by blue arrowheads. Note: GFP-Lmod2 often localizes to the pointed end and along the length of the thin filament but is excluded from the Z-disk; the non–pointed-end localization is likely of low affinity and/or nonspecific (see ref. 13). (Scale bar: 1 μm.) (A′) Plot profile of Rho-actin in cells transduced with GFP (pink) and GFP-Lmod2 (orange). (B–D) FRAP of GFP-cardiac actin in rat cardiomyocytes transduced with mCherry or mCherry-Lmod2. (B) Representative images of rat cardiomyocytes before and after photo bleaching. Barbed (pink arrow) and pointed (blue arrowheads) ends of the actin filaments are marked. (Scale bar: 1 μm.) (C) Mean relative recovery following photobleaching over time ± SEM. (D) Mean slow and fast mobile fractions ± SEM; n = 9–15. *P < 0.05, ****P < 0.0001.
To determine how Lmod2 affects actin dynamics, fluorescence recovery after photobleaching (FRAP) was used. Cardiomyocytes were cotransfected with GFP-actin and either mCherry or mCherry-Lmod2. GFP-actin was photobleached in an area comprising multiple sarcomeres, and fluorescence recovery was followed independently at the pointed and barbed ends of the thin filament (Fig. 6 B and C). The mean slow mobile fraction and half-time of recovery for actin at the pointed end is significantly larger in myocytes expressing mCherry-Lmod2 than in those expressing mCherry alone (Fig. 6D and Table S3). The total mobile fraction of actin at the pointed end increased by nearly 50% after Lmod2 overexpression. The mean slow mobile fraction for actin at the barbed end also was significantly increased upon Lmod2 expression, although the extent of increase was slight. Therefore, excess Lmod2 greatly promotes actin turnover at thin filament pointed ends (and possibly somewhat at the barbed end), thereby producing a more dynamic thin filament.
Table S3.
Summary of FRAP data
| Transduction | n | M(fast) | t1/2(fast) (S) | M(slow) | t1/2(slow) (S) | M(fast)+M(slow) |
| Cherry (P) | 9 | 0.172 ± 0.015 | 9.60 ± 0.97 | 0.231 ± 0.020 | 182.9 ± 32.3 | 0.404 ± 0.035 |
| Cherry-Lmod2 (P) | 15 | 0.115 ± 0.008 | 16.0 ± 1.7 | 0.467 ± 0.023**** | 278.3 ± 32.0* | 0.581 ± 0.032**** |
| Cherry (B) | 9 | 0.152 ± 0.015 | 9.51 ± 0.88 | 0.188 ± 0.016 | 164.6 ± 29.5 | 0.341 ± 0.031 |
| Cherry-Lmod2 (B) | 15 | 0.113 ± 0.009 | 13.3 ± 0.93 | 0.256 ± 0.019* | 221.4 ± 19.4 | 0.369 ± 0.028 |
Recovery data were best fit using nonlinear regression curves with a two-exponential association equation {R = M(fast) × [1− exp(−k(fast) × t)] + M(slow) × [1 − exp(−k(slow) × t)]}. R is the relative recovery of fluorescence at time t. Mean mobile fraction (M), and half-time of recovery (t1/2) are indicated for both slow and fast components of recovery ± SEM. The total mobile fraction is the sum of M(fast) and M(slow). Bolded text indicates values that are statistically significantly different between Cherry and Cherry-Lmod2 (*P < 0.05, **P < 0.01, ****P < 0.0001). n = 9–15. P refers to thin-filament pointed ends; B refers to thin-filament barbed ends.
Discussion
Actin is the most abundant protein in most cell types, and regulation of actin filament architecture is critical for proper cellular function. Striated muscle cells display one of the most extreme examples of actin filament organization found in nature, with thin filaments assembling to remarkably uniform lengths. In the present study we discovered the function of Lmod2 in the context of the heart. Lmod2 is essential for cardiac thin filaments to reach a mature length. Moreover, our data suggest that short filaments are detrimental to the heart, resulting in a unique, rapid-onset DCM.
Fetal expression of Lmod2 suggests that it regulates actin filament assembly early in development. However, microscopic analysis during fetal development and early after birth revealed no detectable changes in thin filament organization or abundance within Lmod2-KO hearts. Consistent with this finding, isolated neonatal KO cardiomyocytes are able to reassemble their thin filaments as well as WT myocytes. Thus, our results reveal that Lmod2 is not essential for myofibrillogenesis and is not the initial nucleator of actin filament assembly in the heart in vivo, as proposed previously by others (17).
Lmod2 is clearly necessary for thin filaments to reach mature lengths in the heart, because we observed a reduction in thin filament length in the KO hearts, as well as in isolated KO neonatal cardiomyocytes, as early as E12.5. Because the lengths of myosin filaments are unchanged in the KO hearts, these results suggest the potential for an alteration in thin–thick filament overlap and thus in the number of force-generating cross-bridges in the hearts of KO mice. With extensive morphological and functional analysis, no other abnormalities were observed until ∼6 d after birth; this result strongly suggests that defects in thin filament length are the primary mechanism of disease progression in Lmod2-KO mice.
Both the loss of Lmod2 in vivo (resulting in shorter filaments, as reported in this study) and overexpression of Lmod2 in neonatal cardiomyocytes [resulting in longer filaments (13)] indicate that Lmod2 functions to regulate thin filament length by promoting the elongation of the filament. To our knowledge, this is the first mammalian actin filament elongation factor identified in vivo. To decipher the mechanism of how Lmod2 functions, we analyzed actin filament assembly/turnover following overexpression of Lmod2 in neonatal cardiomyocytes by two independent methods. First, we microinjected the cardiomyocytes with Rho-actin to assess the location of newly incorporated actin. Consistent with previously published data, Rho-actin assembles to a greater degree at the pointed end than at the barbed end of the thin filament in control cells (22). Lmod2 overexpression exacerbates this difference, indicating that Lmod2 promotes actin incorporation at the pointed end of the filament. Second, we analyzed the dynamics of actin assembly using FRAP. This approach revealed two distinct mobile fractions of GFP-actin. The fast mobile fraction may represent a highly dynamic population of actin, perhaps filaments not associated with actin-binding proteins (e.g., Lmod2 or Tmod1), and the slow mobile fraction may represent a more stable population of actin. In this regard, two populations of actin filaments have been reported previously, and dynamic actin has been associated with active contractility (23, 24). We also discovered that Lmod2 increases the turnover of actin at the pointed end within the more stable population of filaments as evidenced by an increase in the slow mobile fraction of actin following GFP-Lmod2 expression, indicating that Lmod2 promotes a more dynamic thin filament. Together these data are consistent with a model in which excess Lmod2 binds the pointed end of the filament, preventing the association (and subsequent capping activity) of Tmod1. Because Lmod2 itself is unable to cap the filament (13), it likely allows or even promotes, actin incorporation (elongation) at the end of the filament. Our results do not exclude the possibility that actin nucleation by Lmod2 [as demonstrated in vitro (17)] contributes to the mechanism of elongation.
Lmod2-KO mice present with cardiac abnormalities consistent with DCM. The onset of DCM is rapid, and the DCM seems to progress without evidence of intervening hypertrophic cardiomyopathy. To our knowledge, the rapid disease progression of Lmod2-KO mice is unique compared with other mouse models of DCM that are caused by defects in integral sarcomeric proteins [i.e., in most cases the KO/mutation is embryonic lethal, or the mice live to adulthood (e.g., see refs. 25–31)].
Introduction of GFP-Lmod2 via AAV transduction at P4 remarkably rescues most of the structural and contractile defects present in Lmod2-KO mice, including deficit in the thin filament length. These results indicate that phenotypes observed in Lmod2-KO mice are specific to the loss of Lmod2 in the heart and to our knowledge are the first demonstration that abnormally short thin filaments can be lengthened experimentally in hearts in vivo. The data also indicate that there is a window in development that extends from just after birth at least until weaning in which Lmod2 and mature thin filament length are essential for proper cardiac function.
Interestingly, two additional observations about thin filament regulation via Lmod2 were made in this study. First, relatively low levels of GFP-Lmod2 (∼40% of endogenous levels in the LV) are able to rescue thin filaments to normal lengths in the Lmod2-KO hearts. However, expression of >10× GFP-Lmod2 in cultured cardiomyocytes (13) is needed to elongate thin filaments beyond normal lengths. Second, we noted that thin filaments in hearts of Lmod2-KO mice assemble to lengths that are ∼85% of the lengths in WT hearts. Thus, there seems to be a “core” filament that does not require Lmod2 for assembly. The lengths of filaments that assemble in the absence of Lmod2 may not all be uniform, because Tmod1, which localizes to the end of filament, often displays a broader distribution in KO than in WT hearts. Our analysis of WT mice also reveal that thin filaments in the heart elongate throughout development, because they are significantly longer at P6 and P15 than at E12.5. The developmental elongation observed in WT hearts is Lmod2-dependent, because thin filament lengths remain unchanged in Lmod2-KO hearts over this same time period.
Our discovery that loss of Lmod2 results in cardiomyocytes with abnormally short thin filaments provides critical in vivo support for a model in which Lmod2 and Tmod1 have antagonistic roles in thin filament assembly, with Lmod2 functioning as a thin filament elongation factor and Tmod1 as a capper/stabilizer. Also consistent with this model is that the phenotypes described in Tmod1 overexpression transgenic (TOT) mice are similar to the phenotypes displayed by Lmod2-KO mice described in this study. TOT mice appear to have shorter thin filaments and to develop DCM; 50% of the mice die 14–21 d after birth (8, 32). Therefore, an increase in Tmod1 levels (i.e., more capping/stability) mimics an absence of Lmod2 (i.e., loss of elongation), supporting the antagonistic roles of these molecules.
The myofibrillar disarray resulting from loss of Lmod2 in the mouse heart is also similar to that described following the loss or mutation of Lmod3 in skeletal muscle of humans, mice, Xenopus, and zebrafish (14–16). Like Lmod2, Lmod3 has been implicated in the regulation of thin filament length (15). Lmod3 is expressed in the heart, but its ablation results in only slight cardiac dysfunction (16). Thus, Lmod2 appears to be critical for proper heart function, whereas Lmod3 is critical for skeletal muscle function. Although protein levels of Lmod3 are not up-regulated upon knockout of Lmod2, it is possible that Lmod3 could partially compensate for the loss of Lmod2, delaying the onset of cardiac dysfunction observed in the Lmod2-KO mice.
Although not homologous by sequence, Lmod2 appears to be functionally similar to the Drosophila protein, sarcomere length short (SALS). SALS also associates with thin filaments, and its levels correlate with thin filament length (33). However, SALS only has two identified actin-binding sites (WH2 domains) and does not appear to nucleate actin filaments in vitro. In fact, SALS inhibits polymerization from the pointed end of the actin filament in vitro, but Lmod2 does not (13, 33).
The inception of disease during neonatal development in the Lmod2-KO mice may be linked to a period in which the heart is subjected to increased afterload as a result of increased vascular resistance following birth. The increased afterload might give rise to a longer end-diastolic sarcomere length with reduced force caused by shorter thin filament lengths. Consistent with this prediction, the first change in cardiac performance we detected in Lmod2-KO neonates is a loss of systolic function (i.e., reduced ejection fraction) from as early as P6. This functional deficit is likely caused by intracellular mechanisms (e.g., thin filament and sarcomere structure), because contractile force is reduced in neonatal cardiomyocytes isolated from Lmod2-KO hearts as measured using previously undescribed micropillar arrays. Cardiac dysfunction is likely the cause of death in the Lmod2-KO mice because the analysis of EDL muscle by EM indicated no detectable defects in skeletal muscle. Supporting this conclusion, thin filament lengths also are unaltered in skeletal muscles in the KO mice. Although to date Lmod2 has not been linked to human disease, our results may have important implications for human health: A recent study revealed that mutations in Lmod3 that result in shorter thin filaments cause nemaline myopathy in humans (15).
Materials and Methods
Embryos heterozygous for Lmod2 were obtained from the knockout mouse project (KOMP) repository at the University of California, Davis (https://www.komp.org). These mice were generated by Regeneron Pharmaceuticals Inc. on a C57BL/6NTac strain. All animal procedures were approved by The Institutional Animal Care and Use Committee at the University of Arizona.
Further information regarding the Lmod2-KO mice, genotyping, immunoblotting, whole-mount detection of β-gal activity, microscopy, cell isolation and transfection/transduction, Adv and AAV generation, histology, echocardiography, quantitative RT-PCR (RT-qPCR), EM, myocyte contractility on micropillar arrays, microinjection, FRAP, and statistical analyses can be found in SI Materials and Methods.
SI Materials and Methods
Generation of Lmod2-KO Mice.
All animal procedures were approved by The Institutional Animal Care and Use Committee at the University of Arizona. The entire Lmod2 gene, starting at the translational start site and ending at the translational stop site, was replaced with a lacZ reporter cassette encoding β-gal (Fig. S1). The embryos were re-derived in the Genetically Engineered Transgenic Core at the University of Arizona, and a colony was established by crossing to a C57BL/6J strain (Jackson Laboratory stock no. 000664).
Genotyping.
Genomic DNA was isolated by digesting tail tips in 100 μL digestion buffer (50 mM KCl, 1.5 mM MgCl2, 0.1% gelatin, 0.45% Nonidet P-40, 0.45% Tween-20, 10 mM Tris⋅HCl, pH 8.3) plus ∼6 U proteinase K at 60 °C for 20 min and then at 100 °C for 3 min. PCR was performed with primers to exon 2 of Lmod2 (forward: 5′-GAGGAGGTGTGTACAGAAGATGAGGAAGAGTC; reverse: 5′- GGAGTTCCTCTGTGTTCTTCCACTGTTG, which amplified a 231-bp product) and within the LacZ cassette (forward: 5′-CTTGGAGAAACAGTGAGGAAGCTAGGACAG and reverse: 5′-CCTGCCATAAAGAAACTGTTACCCGTAGGT, which amplified a 684-bp product). PCR conditions were as follows: 95 °C for 2 min, then 95 °C for 30 s/66 °C for 30 s/72 °C for 45 s repeated 35 times, then 72 °C for 5 min.
Whole-Mount Detection of β-Gal Activity.
Embryos resulting from an Lmod2+/+ × Lmod2+/− breeding were placed in ice-cold PBS. The embryos were fixed in β-gal fixative solution [0.2% glutaraldehyde, 1.5% (wt/vol) paraformaldehyde, 5 mM EGTA, 2 mM MgCl2, 100 mM sodium phosphate buffer, pH 8.0] for 20 min (E8.5 embryos), 30 min (E9.5 embryos), or 40 min (E10.5 embryos) on ice followed by three washes of 15–30 min in β-gal wash buffer (2 mM MgCl2, 0.01% sodium deoxycholate, 0.02% Nonidet P-40, 100 mM sodium phosphate buffer, pH 8.0) at room temperature. The embryos then were incubated in β-gal staining solution [1 mg/mL X-Gal, 5 mM K4Fe(CN)6, 5 mM K3Fe(CN)6] diluted in β-gal wash buffer] overnight at room temperature followed by three 30-min washes in PBS.
Immunofluorescence Microscopy.
A section of LV free wall was stretched (to resolve thin filament pointed ends effectively), fixed overnight in 4% (wt/vol) paraformaldehyde/PBS, washed extensively in PBS, embedded in Tissue-Tek O.C.T. compound (Sakura Finetek), and frozen immediately in 2-methlybutane cooled in liquid N2. Then 5-μm cryosections were cut and mounted onto number 1.5 coverslips. Cryosections and neonatal cardiomyocytes were permeabilized in 0.2% Triton X-100/PBS for 20 min at room temperature, blocked with 2% (wt/vol) BSA plus 1% normal donkey serum/PBS for 1 h at room temperature, and incubated overnight at 4 °C with rabbit polyclonal anti-Tmod1 (2 μg/mL), mouse monoclonal anti–α-actinin (1:200) (EA-53; Sigma), or anti-myosin heavy chain (F59; Developmental Studies Hybridoma Bank) antibodies. Sections then were washed with PBS for 20 min and incubated with secondary antibodies/PBS for 1.5 h. Secondary antibodies (Invitrogen) included Alexa Fluor 488-conjugated goat anti-mouse IgG (1:1,000), Alexa Fluor 488-conjugated donkey anti-rabbit IgG (1:1,000), and Alexa Fluor 350-conjugated goat anti-mouse IgG (1:200). Texas Red- or Alexa Fluor 488-conjugated phalloidin (1:50) was used to stain F-actin. Sections were washed with PBS for 20 min and then were mounted onto slides with Aqua Poly/Mount (Polysciences Inc.). Images were captured using a Deltavision RT system (Applied Precision) with a 100× NA 1.3 objective and a CCD camera (CoolSNAP HQ; Photometrics). Images were deconvolved using SoftWoRx software and processed using Photoshop CS (Adobe). Thin filament and sarcomere lengths were measured using the DDecon plugin for Image J (19). Reported changes in thin filament length were confirmed to be independent of sarcomere length.
Immunoblotting.
A section of LV free wall (10–30 mg) was placed in a microcentrifuge tube containing 300 μL of lysis buffer [150 mM NaCl, 1.5 mM MgCl2, 1 mM EGTA, 10 mM sodium pyrophosphate, 10 mM sodium fluoride, 0.1 mM sodium deoxycholate, 1% Triton X-100, 1% SDS, 10% (vol/vol) glycerol, 25 mM Hepes, pH 7.4, plus protease inhibitors] and 200 mg of stainless steel beads (0.9–2.0 mm diameter blend; Next Advance Inc.) and was homogenized in a Bullet Blender (BBX24; Next Advance Inc.) at speed 10 for 4 min at 4 °C. Samples were spun down for 15 min at 16,000 × g at 4 °C. Protein concentration was normalized by BCA assay (Thermo Scientific). Samples were prepared in Laemmli sample buffer at 100 °C for 7 min, resolved on a 10% SDS polyacrylamide gel, and transferred to a nitrocellulose membrane (0.2 μm; Amersham Protran GE Healthcare). Total lane density of transferred proteins stained with Ponceau S was used to control for loading/transfer differences. The membrane was blocked with 2% (wt/vol) BSA/PBS for 1.5 h at 37 °C, washed five times (5 min each washing) in wash buffer (150 mM NaCl, 1 mM EDTA, 0.2% Triton X-100, 10 mM sodium phosphate buffer, pH 7.4), and incubated with primary antibodies in antibody dilution buffer (150 mM NaCl, 1 mM EDTA, 0.2% Triton X-100, 40 mg/mL BSA, 10 mM sodium phosphate buffer, pH 7.4) overnight at 4 °C. Primary antibodies included rabbit polyclonal anti-leiomodin 2 (0.1 μg/mL) (E13; Santa Cruz Biotechnology), rabbit polyclonal anti-LMOD3 (0.16 μg/mL) (14948-1-AP; Proteintech), rabbit polyclonal anti-Tmod1 (0.2 μg/mL), rabbit polyclonal anti-GFP (1:3,000) (AB290; Abcam), and mouse monoclonal anti-GAPDH (0.9 μg/mL) (clone 6C5; Life Technologies). Following five 5-min washes the membrane was incubated for 1.5 h at room temperature with HRP-conjugated anti-rabbit IgG (1:25,000) or anti-mouse IgG (1:15,000) (Jackson ImmunoResearch) diluted in antibody dilution buffer, followed by SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific). Blots were imaged and analyzed using a G-Box Chemi-XR5 chemiluminescent imaging system with GeneTools software (Syngene).
Cell Isolation, Culture, and Transduction.
Cardiomyocytes were isolated from P3 or younger rats or mice as described (34). Briefly, for mouse cardiomyocyte cultures, hearts from three to six mice were isolated and separated based on genotype (WT or Lmod2 KO), and two parallel cultures were completed. The hearts were digested in pancreatin and collagenase five times for 15 min each, with shaking at 230 rpm at 37 °C. Isolated cells were plated on 35-mm tissue-culture dishes containing 12-mm round glass coverslips thin-coated with Matrigel (1:100) (BD Biosciences) at ∼4.5 × 105 cells per dish and were maintained in DMEM with 1 g/L glucose (Gibco) with the addition of 10% (vol/vol) FBS (HyClone) and 1% penicillin/streptomycin (Cellgro). Mouse cardiomyocytes in rescue experiments were transduced 2–3 d after plating with 2 MOI of Adv. Rat cardiomyocytes used for microinjection experiments were transduced 2–3 d after plating with 5–10 MOI of Adv. Cardiomyocytes were isolated from P19 mice as previously described (35).
Adv and AAV Generation.
A replication-deficient Adv vector expressing GFP-Lmod2 or GFP alone was constructed using the AdEasy Adenoviral Vector System (Agilent Technologies). Briefly, GFP-Lmod2 cDNA was subcloned into a pShuttle-CMV plasmid and linearized before transformation of BJ5183 cells containing the pAdEasy-1 vector. After homologous recombination, the purified pAdEasy-1 vector containing GFP-Lmod2 or GFP alone was transfected into HEK293 cells for Adv propagation. The Adv then was purified by CsCl gradient centrifugation, and viral titer was determined by a dilution assay in HEK293 cells grown in 96-well plates. At a MOI of 5–10, >95% of cardiomyocytes were transfected, as determined by GFP-positive cells.
AAV was generated by the insertion of GFP-Lmod2 or GFP into pAAV-MCS (Agilent Technologies). Virus was generated both onsite and by the Vector Development/Proteomics Laboratory (Jody Martin) at Loyola University Medical Center, Maywood, IL. Briefly, pAAV-GFP-Lmod2 or pAAV-GFP plasmids were cotransfected with AAV helper and AAV-RC2/9 at a 1:1:1 molar ratio. Plasmid DNA used in the transfection was purified using a QIAGEN plasmid Midi Kit. Fifteen 15-cm plates of AAV 293T cells at 60% confluence were transfected as one batch. The ratio of polyethylenimine (Polysciences) to DNA was 4:1 (vol:wt). After 72 h, cells were collected and lysed by three freeze-thaw cycles. The lysate then was treated with benzonase (250 U) (Sigma) at 37 °C for 1 h. The virus was purified from the benzonase-treated cell crude lysates over an iodixanol density gradient (OptiPrep; Greiner Bio-One Inc.). Finally, virus was concentrated, exchanged into modified PBS (2.5 mM KCl, 1 mM MgCl2, and 0.0001% Tween20) using centrifugal concentrators (50 k molecular weight cutoff) (Millipore), and stored at 4 °C. Genome-containing particles were determined by a dot blot assay. Vector genomes (2.5 × 1011) were injected into the pericardial cavity of P4–5 mice.
Histology.
Hearts were excised, washed in PBS, fixed in 10% neutral buffered formalin (Sigma) overnight, dehydrated, and embedded in paraffin. Longitudinal or transverse sections (8 μm) were stained using a Trichrome Stain (Masson) Kit (Sigma) and mounted with Permount (Sigma). Bright-field images were captured on a Zeiss Axio Imager M1with a 5× NA 0.16 objective and a Zeiss AxioCam MRc camera.
Echocardiography.
Conscious mice were placed in dorsal recumbence on a heated (37 °C) platform for echocardiography. Transthoracic echocardiographic images were obtained with a Vevo 700 or Vevo 2100 High Resolution Imaging System (Visual-Sonics) using the model 707B or MS-550D transducer arrays. Images were collected and stored as a digital cine loop for off-line calculations. Standard imaging planes and functional calculations were obtained according to American Society of Echocardiography guidelines. M-mode images at the level of the papillary muscles were used to determine LV wall thicknesses, chamber dimensions, and ejection fraction.
Cardiomyocyte Contractility on Micropillar Arrays.
A two-step molding process was used to prepare micropillar arrays (see Fig. S7A). First, a master mold was fabricated using conventional photolithography (36). SU-8 2015 negative epoxy photoresist (Micro Chem) was spin coated onto a silicon wafer followed by UV exposure, development, and cleaning. Second, a negative duplicate of the master mold was fabricated using PDMS. To facilitate the subsequent micropillar release, the PDMS mold was silanized with (tridecafluoro-1,1,2,2-tetrahydrooctyl)-1-trichlorosilane (United Chemical) for 2 h in a vacuum chamber (37–39). A 1:10 ratio (curing agent:base) of PDMS with a stiffness of 2.15 MPa was used to prepare the micropillar arrays (40). For the PDMS used in all steps, the solution was degassed for 3 min in a vacuum chamber and heat-cured on a hot plate for 2 h at 75 °C. The fabricated micropillars have an average height of 15.3 µm and an average diameter of 4.6 µm. To promote cell attachment, the micropillar arrays were exposed to atmospheric plasma (PDC-001; Harrick Plasma) for 10 min (40) and were coated with a thin layer of Matrigel (diluted 1:20 in DMEM; BD Biosciences) for 1 h at 37 °C and 5% CO2. The deflection of micropillars was quantified using ImageJ (NIH) from phase-contrast videos captured on a Zeiss Axiovert 135 with a 32× NA 0.4 objective and a 1.6× optovar using a FinePix digital camera (640 × 480 pixels at 30 frames/s) (FinePix S9000; Fujifilm). For each cell, a minimum of three pillars with the highest deflection were measured. Based on the concept of beam-bending theory, contractile force is related to the lateral deflection of the pillars by F = (3πED4/64L3)δ, where F, E, D, L, and δ are the exerted force, Young’s modulus, diameter, height, and deflection of the pillar, respectively (Fig. S7B).
RT-qPCR.
LV free wall was homogenized in RLT lysis buffer (Qiagen) as outlined in the immunoblot protocol above, except that RNase-free microcentrifuge tubes and beads were used. Total RNA was extracted using the RNeasy Fibrous Tissue Mini Kit, including the completion of an on-column DNase digestion step (Qiagen). cDNA was synthesized from 500 ng of total RNA using the Maxima cDNA Kit (Thermo Scientific). Five microliters of template cDNA (diluted 1:25) were used in a PCR with Maxima SYBR Green qPCR master mix (Thermo Scientific) on a Rotor-Gene thermocycler (Qiagen). To determine relative gene expression, the 2^-∆∆CT method was used. Each sample was normalized to ornithine decarboxylase (ODC) (∆CT) and then was compared with the mean ∆CT of a control group (WT or WT+GFP) (∆∆CT). The mean 2^-∆∆CT for each group then was plotted (Mean Relative Expression). Primers sequences were obtained from the qPrimerDepot (NIH) and were tested to ensure efficiencies of amplification were nearly equal. Primers included: ANF (forward: 5-GGGGGTAGGATTGACAGGAT and reverse: 5-AGGGCTTAGGATCTTTTGCG; 130-bp product); BNP (forward: 5′-ACAAGATAGACCGGATCGGA and reverse: 5′-ACCCAGGCAGAGTCAGAAAC; 110-bp product); α-MHC (forward: 5′-GCGCATTGAGTTCAAGAAGA and reverse: 5′-CTTCATCCATGGCCAATTCT; 103-bp product); β-MHC (forward: 5′- GTGGCTCCGAGAAAGGAAG and reverse: 5′- GAGCCTTGGATTCTCAAACG; 98-bp product), ODC (forward: 5′-ACATCCAAAGGCAAAGTTGG and reverse: 5′-AGCCTGCTGGTTTTGAGTGT; 102-bp product); SERCA2a (forward: 5′-AATATGAGCCTGAAATGGGC and reverse: 5′-TCAGCAGGAACTTTGTCACC; 124-bp product); and skeletal actin (forward: 5′-CTCTCCTCAGGACGACAATC and reverse: 5′-TTTTCCATTTCCTTTCCACA; 177-bp product).
Transmission EM.
Sections of LV free wall (0.5–1 mm2) were dissected in 30 mM KCl (to arrest the muscle in diastole), fixed in 2.5% (vol/vol) glutaraldehyde/0.1 M Pipes, and washed in 0.1 M Pipes. Samples were postfixed in 1% osmium tetroxide/0.1 M Pipes followed by 2% (wt/vol) uranyl acetate. The tissue was dehydrated in increasing amounts of ethanol and embedded in Spurr’s resin. Sixty-nanometer sections were cut onto uncoated copper mesh grids and stained with 2% (wt/vol) lead acetate. Samples were analyzed on a Technai G2 Spirit BioTWIN electron microscope, and images were collected with a 4 × 4 digital camera (Optronics). Z-disk width was measured using ImageJ at the widest point of the Z-disk. Mitochondrial number was calculated in Photoshop (Adobe) by laying a 0.8–1.9 μm2 grid on top of the images and counting the number of times mitochondria passed through the gridline (41).
Microinjection.
We adapted an assay developed by Littlefield et al. (22).Two to three days after transduction, neonatal rat cardiomyocytes in culture were microinjected with rhodamine-labeled G-actin (Cytoskeleton) resuspended to 1 mg/mL in 5 mM Tris (pH 8.0), 10 μM MgCl2, 0.2 mM ATP, and 1 mM DTT using an Eppendorf Transjector. After a 30 min to 1 h incubation (i.e., a short time interval to allow actin incorporation only at the ends of the thin filaments), the cells were incubated in relaxing buffer [150 mM KCl, 5 mM MgCl2, 10 mM 3-(N-morpholino)propanesulfonic acid (pH 7.4), 1 mM EGTA, and 4 mM ATP] for 15 min and fixed with 2% (wt/vol) paraformaldehyde in relaxing buffer for 15 min.
FRAP.
A Leica SP5-II confocal microscope with a 63× NA 1.4 objective and a 488-nm argon laser was used for FRAP experiments. Cells were plated on glass-bottomed dishes (MatTek) and were maintained at 37 °C and ∼5% CO2 for the duration of the experiment. Three prebleach images were recorded, followed by photobleaching for ∼2 s at 80% total laser power. To monitor recovery after photobleaching, images were captured in successive intervals of 1 s (for a duration of 30 s), 5 s (for a duration of 150 s), and 10 s (for a duration of 600 s). Analysis of recovery was completed as described previously with minor modifications (42). In brief, images were imported into ImageJ (NIH), and mean intensities from bleached, nonbleached, and background regions were recorded. After background subtraction and correction for photobleaching caused by image acquisition, intensities were normalized so that prebleach intensity was set to 1 and postbleach intensity was set to 0. Mobile fractions and half-times were determined from nonlinear regression curves best fit using a two-exponential association equation {R = M(fast) × [1− exp(−k1 × t)] + M(slow) × [1 − exp(−k2 × t)]} with Prism 4 software (GraphPad Software, Inc.). R is the relative recovery at time t, and the total mobile fraction is the combination of M(fast) and M(slow). Half times are 0.69/k1 and 0.69/k2. Recovery was determined independently at two or three barbed and pointed thin filament ends per cell.
Statistics.
All statistical analyses were completed using GraphPad Prism (GraphPad Software). Student's t tests were used to compare two groups (Figs. 3 and 4A, Figs. S3 and S4, and Tables S1 and S2). For comparisons of multiple groups one-way (Fig. 4B) or two-way (Figs. 2, 5, and 6 and Table S3) ANOVAs with Tukey's post hoc test were used. All values are mean ± SEM; P < 0.05 was considered significant. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Acknowledgments
We thank Elisa Namdarian for assistance with filament length measurements, David O’Neil Lyons for genotyping, Anthony Day (Arizona Health Sciences Center Imaging Core) for processing samples for EM, Jody Martin and the Vector Development/Proteomics Laboratory at Loyola University Medical Center for adeno-associated virus generation, Marcus DiMarco for preparing myocyte cultures, Eleni Constantopoulos for assistance with echocardiography, Velia Fowler and David Gokhin for generously sharing DDecon software, and Stefanie Novak for insightful discussions. This work was supported by Walter and Vinnie Hinz Memorial; Frank and Alex Frazer; Stephen Michael Schneider Family University of Arizona Sarver Heart Center Research Grants; NIH Grants T32HL007249 (to C.T.P.), R01HL108625 and R01HL123078 (to C.C.G.), DP2OD007161 (to P.K.W.), and R01HL062881 (to H.L.G.); a donation from Linda and Jim Lee (C.C.G.); and a Sarnoff Cardiovascular Research Foundation Fellowship (to Z.H.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1508273112/-/DCSupplemental.
References
- 1.Gokhin DS, Fowler VM. Tropomodulin capping of actin filaments in striated muscle development and physiology. J Biomed Biotechnol. 2011;2011:103069. doi: 10.1155/2011/103069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fowler VM, Sussmann MA, Miller PG, Flucher BE, Daniels MP. Tropomodulin is associated with the free (pointed) ends of the thin filaments in rat skeletal muscle. J Cell Biol. 1993;120(2):411–420. doi: 10.1083/jcb.120.2.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weber A, Pennise CR, Babcock GG, Fowler VM. Tropomodulin caps the pointed ends of actin filaments. J Cell Biol. 1994;127(6 Pt 1):1627–1635. doi: 10.1083/jcb.127.6.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gregorio CC, Weber A, Bondad M, Pennise CR, Fowler VM. Requirement of pointed-end capping by tropomodulin to maintain actin filament length in embryonic chick cardiac myocytes. Nature. 1995;377(6544):83–86. doi: 10.1038/377083a0. [DOI] [PubMed] [Google Scholar]
- 5.Gregorio CC, Fowler VM. Mechanisms of thin filament assembly in embryonic chick cardiac myocytes: Tropomodulin requires tropomyosin for assembly. J Cell Biol. 1995;129(3):683–695. doi: 10.1083/jcb.129.3.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sussman MA, et al. Altered expression of tropomodulin in cardiomyocytes disrupts the sarcomeric structure of myofibrils. Circ Res. 1998;82(1):94–105. doi: 10.1161/01.res.82.1.94. [DOI] [PubMed] [Google Scholar]
- 7.Kostyukova AS, Choy A, Rapp BA. Tropomodulin binds two tropomyosins: A novel model for actin filament capping. Biochemistry. 2006;45(39):12068–12075. doi: 10.1021/bi060899i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sussman MA, et al. Myofibril degeneration caused by tropomodulin overexpression leads to dilated cardiomyopathy in juvenile mice. J Clin Invest. 1998;101(1):51–61. doi: 10.1172/JCI1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chu X, et al. E-Tmod capping of actin filaments at the slow-growing end is required to establish mouse embryonic circulation. Am J Physiol Heart Circ Physiol. 2003;284(5):H1827–H1838. doi: 10.1152/ajpheart.00947.2002. [DOI] [PubMed] [Google Scholar]
- 10.Fritz-Six KL, et al. Aberrant myofibril assembly in tropomodulin1 null mice leads to aborted heart development and embryonic lethality. J Cell Biol. 2003;163(5):1033–1044. doi: 10.1083/jcb.200308164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McKeown CR, Nowak RB, Moyer J, Sussman MA, Fowler VM. Tropomodulin1 is required in the heart but not the yolk sac for mouse embryonic development. Circ Res. 2008;103(11):1241–1248. doi: 10.1161/CIRCRESAHA.108.178749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Conley CA, Fritz-Six KL, Almenar-Queralt A, Fowler VM. Leiomodins: Larger members of the tropomodulin (Tmod) gene family. Genomics. 2001;73(2):127–139. doi: 10.1006/geno.2000.6501. [DOI] [PubMed] [Google Scholar]
- 13.Tsukada T, et al. Leiomodin-2 is an antagonist of tropomodulin-1 at the pointed end of the thin filaments in cardiac muscle. J Cell Sci. 2010;123(Pt 18):3136–3145. doi: 10.1242/jcs.071837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nworu CU, Kraft R, Schnurr DC, Gregorio CC, Krieg PA. Leiomodin 3 and tropomodulin 4 have overlapping functions during skeletal myofibrillogenesis. J Cell Sci. 2015;128(2):239–250. doi: 10.1242/jcs.152702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yuen M, et al. Leiomodin-3 dysfunction results in thin filament disorganization and nemaline myopathy. J Clin Invest. 2014;124(11):4693–4708. doi: 10.1172/JCI75199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cenik BK, et al. Severe myopathy in mice lacking the MEF2/SRF-dependent gene leiomodin-3. J Clin Invest. 2015;125(4):1569–1578. doi: 10.1172/JCI80115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chereau D, et al. Leiomodin is an actin filament nucleator in muscle cells. Science. 2008;320(5873):239–243. doi: 10.1126/science.1155313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Skwarek-Maruszewska A, et al. Different localizations and cellular behaviors of leiomodin and tropomodulin in mature cardiomyocyte sarcomeres. Mol Biol Cell. 2010;21(19):3352–3361. doi: 10.1091/mbc.E10-02-0109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Littlefield R, Fowler VM. Measurement of thin filament lengths by distributed deconvolution analysis of fluorescence images. Biophys J. 2002;82(5):2548–2564. doi: 10.1016/S0006-3495(02)75598-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schoen I, Hu W, Klotzsch E, Vogel V. Probing cellular traction forces by micropillar arrays: Contribution of substrate warping to pillar deflection. Nano Lett. 2010;10(5):1823–1830. doi: 10.1021/nl100533c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sniadecki NJ, Chen CS. Microfabricated silicone elastomeric post arrays for measuring traction forces of adherent cells. Methods Cell Biol. 2007;83:313–328. doi: 10.1016/S0091-679X(07)83013-5. [DOI] [PubMed] [Google Scholar]
- 22.Littlefield R, Almenar-Queralt A, Fowler VM. Actin dynamics at pointed ends regulates thin filament length in striated muscle. Nat Cell Biol. 2001;3(6):544–551. doi: 10.1038/35078517. [DOI] [PubMed] [Google Scholar]
- 23.Skwarek-Maruszewska A, Hotulainen P, Mattila PK, Lappalainen P. Contractility-dependent actin dynamics in cardiomyocyte sarcomeres. J Cell Sci. 2009;122(Pt 12):2119–2126. doi: 10.1242/jcs.046805. [DOI] [PubMed] [Google Scholar]
- 24.Ehler E, Fowler VM, Perriard JC. Myofibrillogenesis in the developing chicken heart: Role of actin isoforms and of the pointed end actin capping protein tropomodulin during thin filament assembly. Dev Dyn. 2004;229(4):745–755. doi: 10.1002/dvdy.10482. [DOI] [PubMed] [Google Scholar]
- 25.Cheng H, et al. Loss of enigma homolog protein results in dilated cardiomyopathy. Circ Res. 2010;107(3):348–356. doi: 10.1161/CIRCRESAHA.110.218735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arber S, et al. MLP-deficient mice exhibit a disruption of cardiac cytoarchitectural organization, dilated cardiomyopathy, and heart failure. Cell. 1997;88(3):393–403. doi: 10.1016/s0092-8674(00)81878-4. [DOI] [PubMed] [Google Scholar]
- 27.Zheng M, et al. Cardiac-specific ablation of Cypher leads to a severe form of dilated cardiomyopathy with premature death. Hum Mol Genet. 2009;18(4):701–713. doi: 10.1093/hmg/ddn400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gramlich M, et al. Stress-induced dilated cardiomyopathy in a knock-in mouse model mimicking human titin-based disease. J Mol Cell Cardiol. 2009;47(3):352–358. doi: 10.1016/j.yjmcc.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Purevjav E, et al. Nebulette mutations are associated with dilated cardiomyopathy and endocardial fibroelastosis. J Am Coll Cardiol. 2010;56(18):1493–1502. doi: 10.1016/j.jacc.2010.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chu G, Haghighi K, Kranias EG. From mouse to man: Understanding heart failure through genetically altered mouse models. J Card Fail. 2002;8(6) Suppl:S432–S449. doi: 10.1054/jcaf.2002.129284. [DOI] [PubMed] [Google Scholar]
- 31.Nonaka M, Morimoto S. Experimental models of inherited cardiomyopathy and its therapeutics. World J Cardiol. 2014;6(12):1245–1251. doi: 10.4330/wjc.v6.i12.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sussman MA, et al. Pathogenesis of dilated cardiomyopathy: Molecular, structural, and population analyses in tropomodulin-overexpressing transgenic mice. Am J Pathol. 1999;155(6):2101–2113. doi: 10.1016/S0002-9440(10)65528-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bai J, Hartwig JH, Perrimon N. SALS, a WH2-domain-containing protein, promotes sarcomeric actin filament elongation from pointed ends during Drosophila muscle growth. Dev Cell. 2007;13(6):828–842. doi: 10.1016/j.devcel.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 34.Brand NJ, Lara-Pezzi E, Rosenthal N, Barton PJ. Analysis of cardiac myocyte biology in transgenic mice: A protocol for preparation of neonatal mouse cardiac myocyte cultures. Methods Mol Biol. 2010;633:113–124. doi: 10.1007/978-1-59745-019-5_9. [DOI] [PubMed] [Google Scholar]
- 35.Granzier HL, et al. Deleting titin’s I-band/A-band junction reveals critical roles for titin in biomechanical sensing and cardiac function. Proc Natl Acad Sci USA. 2014;111(40):14589–14594. doi: 10.1073/pnas.1411493111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xia Y, Whitesides GM. Soft lithography. Annu Rev Mater Sci. 1998;28(1):153–184. [Google Scholar]
- 37.Ting LH, Feghhi S, Han SJ, Rodriguez ML, Sniadecki NJ. Effect of silanization film thickness in soft lithography of nanoscale features. J Nanotechnol Eng Med. 2011;2(4):041006. [Google Scholar]
- 38.Tan JL, et al. Cells lying on a bed of microneedles: An approach to isolate mechanical force. Proc Natl Acad Sci USA. 2003;100(4):1484–1489. doi: 10.1073/pnas.0235407100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gates BD, et al. New approaches to nanofabrication: Molding, printing, and other techniques. Chem Rev. 2005;105(4):1171–1196. doi: 10.1021/cr030076o. [DOI] [PubMed] [Google Scholar]
- 40.Nam KH, et al. Probing mechanoregulation of neuronal differentiation by plasma lithography patterned elastomeric substrates. Sci Rep. 2014;4:6965. doi: 10.1038/srep06965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Medeiros DM. Assessing mitochondria biogenesis. Methods. 2008;46(4):288–294. doi: 10.1016/j.ymeth.2008.09.026. [DOI] [PubMed] [Google Scholar]
- 42.Wang J, et al. Dynamics of Z-band based proteins in developing skeletal muscle cells. Cell Motil Cytoskeleton. 2005;61(1):34–48. doi: 10.1002/cm.20063. [DOI] [PMC free article] [PubMed] [Google Scholar]