Significance
Holocentric chromosomes are characterized by kinetochore activity along each sister chromatid. Although the kinetochore structure seems to be well conserved, as in monocentric organisms, the organization of holocentromeres is still elusive, and no centromeric repeat has been found associated with centromeric histone H3 variant-positive centromeric nucleosomes for any holocentric organism studied hitherto. We demonstrate that holocentrics of the sedge (Cyperaceae) Rhynchospora pubera possess different classes of centromere-specific repeats. Holocentromeres are composed of multiple centromeric units interspersing the gene-containing chromatin, and, as a functional adaption, a cell-cycle–dependent shuffling of centromeric units results in the formation of functional (poly)centromeres during cell division. The genome-wide distribution of centromeric repeat arrays interspersing the euchromatin provides a previously unidentified type of centromere organization.
Keywords: centromere, satellite DNA, holokinetic, chromosome, evolution
Abstract
Holocentric chromosomes lack a primary constriction, in contrast to monocentrics. They form kinetochores distributed along almost the entire poleward surface of the chromatids, to which spindle fibers attach. No centromere-specific DNA sequence has been found for any holocentric organism studied so far. It was proposed that centromeric repeats, typical for many monocentric species, could not occur in holocentrics, most likely because of differences in the centromere organization. Here we show that the holokinetic centromeres of the Cyperaceae Rhynchospora pubera are highly enriched by a centromeric histone H3 variant-interacting centromere-specific satellite family designated “Tyba” and by centromeric retrotransposons (i.e., CRRh) occurring as genome-wide interspersed arrays. Centromeric arrays vary in length from 3 to 16 kb and are intermingled with gene-coding sequences and transposable elements. We show that holocentromeres of metaphase chromosomes are composed of multiple centromeric units rather than possessing a diffuse organization, thus favoring the polycentric model. A cell-cycle–dependent shuffling of multiple centromeric units results in the formation of functional (poly)centromeres during mitosis. The genome-wide distribution of centromeric repeat arrays interspersing the euchromatin provides a previously unidentified type of centromeric chromatin organization among eukaryotes. Thus, different types of holocentromeres exist in different species, namely with and without centromeric repetitive sequences.
The centromere is the chromosome region where the microtubules attach to the chromatids to enable their movement to the daughter cells during mitosis and meiosis. This region is often enriched in repetitive DNA families, with satellite DNAs (satDNAs) and transposable elements, such as Ty3/gypsy-like retrotransposons being the most frequent ones in plants. However, there is little sequence conservation among species (1, 2), and centromere-specific sequences are neither sufficient nor required for the centromere identity (3). Instead, the assembly site for the kinetochore complex of most active centromeres is epigenetically determined by the chromosomal location of the centromeric histone H3 variant CENH3, also known as “CENP-A” (4). Nevertheless, some evolutionary preferences seem to exist, and long-established centromeres are frequently formed on long arrays of satDNAs and/or transposable elements (1).
In certain independent eukaryotic lineages holocentric (also called “holokinetic”) chromosomes occur. These holocentrics lack a primary constriction, and they form kinetochores distributed along almost the entire poleward surface of the chromatids, to which the spindle fibers attach (5, 6). The best-analyzed organisms possessing holocentric chromosomes are the nematode Caenorhabditis elegans (7, 8) and some species of the plant genus Luzula (9–11). In both cases, a longitudinal CENH3-positive centromere structure was observed during mitosis. In the rush Luzula (Juncaceae), the longitudinal centromere forms a groove (here referred as the “centromere groove”) in each sister chromatid along almost the whole metaphase chromosome except for the most terminal regions (9–11). Recently, a similar centromere organization was found in the sedge species Rhynchospora pubera (12). The absence of CENH3 and the centromeric protein C (CENP-C) in some lineages of holocentric insects (13) challenges the general notion of a conserved molecular composition of centromeres in mono- and holokinetic chromosome species. Furthermore, no centromere-specific repeat has been identified thus far for any species possessing holocentric chromosomes (14–17). For instance, Heckmann, et al. (15) have characterized the high-copy fraction of the Luzula elegans genome in detail, and none of the identified repeats showed colocalization with the holokinetic centromere. Even in C. elegans, where robust studies [Chip-on chip (ChIP-chip) and ChIP sequencing (ChIP-seq)] were performed, no centromere-specific repeats were identified (14, 16).
ChIP-chip experiments suggested that nearly half of the C. elegans genome may be associated with CENH3 domains, although only 4% of them form the holocentromeres. Therefore, CENH3 nucleosomes may assemble at random positions (14). However, a recent study based on native CENH3 ChIP sequencing proposed that the holocentromeres in C. elegans consist of about 700 individual centromeric units distributed along the length of the chromosomes. Each of these units is formed by only one CENH3-containing nucleosome, where microtubules attach during cell division. These centromeric sites coincide with transcription factor hotspots which are occupied by many transcription factors, without having high binding affinity for any of them (16). The observation that any sequence of C. elegans can be propagated as an extrachromosomal array suggests that no specialized sequences are required for the segregation of holocentric chromosomes (18). On the other hand, that DNA arrays did not segregate with the same fidelity as normal chromosomes indicates that these extrachromosomal arrays lack certain features that promote mitotic stability of wild-type chromosomes (19).
To test the assumption that all holocentromeres are devoid of centromere-specific repeats, we analyzed R. pubera (12, 20–22). We report here on the first (to our knowledge) conserved centromere-specific repeats of a holocentric species and propose a model of chromosome organization for species with centromeric repeats distributed throughout the entire genome.
Results
Satellite Repeat Tyba Shows a Holocentromere-Specific Localization.
To identify putative centromeric repeats in the genome of the holocentric plant R. pubera, with diploid chromosome number 2n = 10 and a genome size of the unreplicated reduced chromosome complement (1C) = 1.61 Gbp, we performed high-throughput shotgun sequencing. A randomly sampled proportion (8.89 million) of generated paired-end reads then was subjected to bioinformatic analysis, implemented within the clustering-based repeat identification pipeline (23, 24). This analysis resulted in thousands of clusters, or groups of reads, with overlapping sequences, each representing a single repeated element or part of it. After repeat classification within major clusters, the global repeat composition of the genome was determined by taking into account the sizes (number of reads) of individual clusters, which are proportional to the genomic abundance of the corresponding repeats.
The R. pubera genome was found to be relatively poor in repeated sequences, with highly and moderately repetitive elements represented by clusters with genome proportions of at least 0.01%, collectively making up 41.16% of the genome. The majority of these sequences were classified into the major classes of repetitive DNA. DNA transposons (8.81% of the genome), with more than half represented by miniature inverted-repeat transposable elements (MITEs), accounted for the most frequent repetitive elements, followed by Ty1/copia (8.68%) and Ty3/gypsy (5.14%) LTR retrotransposons. Other classes of repetitive DNA, such as non-LTR retrotransposons, pararetroviruses, hAT DNA transposons, and helitrons were found in much lower genomic proportions (SI Appendix, Table S1).
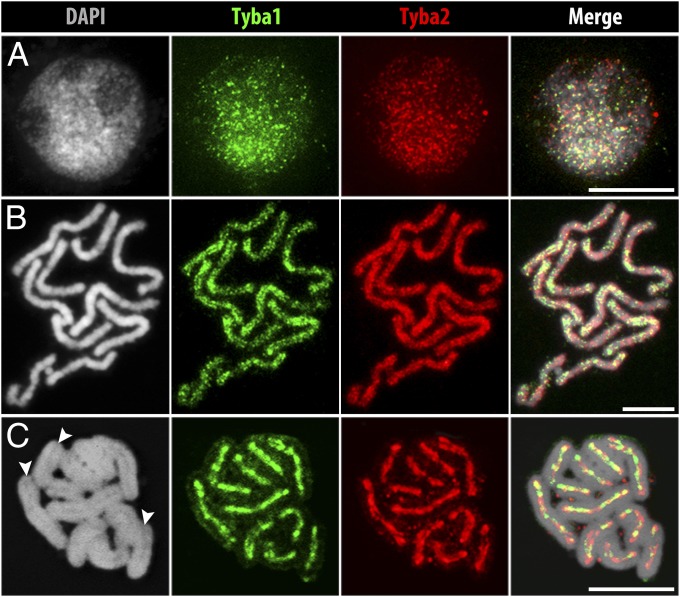
All satDNA reads were grouped into two highly abundant clusters, representing 2.24% and 1.36% of the genome. Because of the high similarity of reads (∼70%) present in both clusters, they were considered as two subfamilies of the same satDNA repeat, named “Tyba” (meaning “abundance” in Tupi-Guarani, a language spoken by many Brazilian native tribes), and were designated “Tyba1” and “Tyba2.” The consensus monomer length of Tyba1 and -2 is 172 bp (SI Appendix, Fig. S2C), which is in the range of monomer lengths of many other known centromeric satDNAs (1, 25, 26). After FISH with Tyba1- and Tyba2-specific probes, interphase nuclei showed dispersed dot-like signals (Fig. 1A), contrasting with the typical strong clustered distribution of satDNA in interphase nuclei of other species. With the onset of chromosome condensation, Tyba signals associate and form dot-like lines along the sister chromatids (Fig. 1B and SI Appendix Fig. S1A). At metaphase, all chromosomes showed line-like FISH signals colocalizing with the longitudinal DAPI-negative centromere groove (Fig. 1C and Movie S1). The hybridization intensity of Tyba1 and -2 probes varied along the chromatids, but only minor labeling was found outside the groove. Both the repetitive nature and the chromosomal distribution of Tyba1 and -2 indicate that this satDNA family is centromere specific in R. pubera. In contrast, other high-copy sequences, such as MITE DNA transposons, showed dispersed labeling throughout the chromosomes (SI Appendix, Fig. S1B).
Fig. 1.
FISH localization of Tyba1 and -2 in R. pubera. (A) Hybridization signals of both Tyba subfamilies in an interphase nucleus show a genome-wide dot-like labeling. (B) Prometaphase chromosomes show a line-like but dispersed labeling on the poleward surface of each chromatid and (C) a distinct labeling along the centromere groove of both sister chromatids during metaphase. Arrowheads in C indicate grooves. (Scale bars: 5 μm.)
Because no other high-copy satellite repeat other than Tyba was found in R. pubera, and no heterochromatic domains are visible throughout the cell cycle, we asked whether this genome is organized on a large scale into euchromatin- and heterochromatin-enriched subregions. Therefore, we applied antibodies against typical euchromatin- and heterochromatin-associated histone methylation marks (H3K4me3 and H3K9me2, respectively). In agreement with other holocentric species (15, 27) the euchromatin and heterochromatic domains in R. pubera were found dispersed along the entire chromosomes (SI Appendix, Fig. S1 C and D). Thus, on a large scale, euchromatin and heterochromatic domains are interspersed.
Tyba satDNA Interacts with CENH3-Containing Nucleosomes.
The centromere specificity of Tyba repeats was tested by colocalization analysis and ChIP with a R. pubera-specific CENH3 antibody. First, the pollen mother cell transcriptome of R. pubera was determined, and assembled RNA-sequencing (RNA-seq) reads were used to identify CENH3. RT-PCR revealed, in addition to the 680-bp-long CENH3-like sequence (RpCENH3_1, GenBank accession no. KR029618), a second CENH3-like variant (RpCENH3_2, GenBank accession no. KR029619) of 708 bp, characterized by an insertion in the C-terminal tail after the stop codon (SI Appendix, Fig. S2A). Both are expressed mainly in anthers and roots but less in leaf tissue, most likely because of the higher number of dividing cells in anthers and roots (SI Appendix, Fig. S2D). Alignment of the R. pubera CENH3 candidates with CENH3s of other plant species supported the correct identification (SI Appendix, Fig. S2B). Phylogenetic analysis grouped both RpCENH3s as a sister branch of Juncaceae and other monocot CENH3s (SI Appendix, Fig. S2E) (transcriptome assemblies, alignments, and trees are available through the iPlant Data Store and can be accessed via iPlant Discovery Environment or at https://de.iplantcollaborative.org/dl/d/8258A143-C5F5-4DF1-84F2-88C94BE8EA8F/R_pubera_holocentromeres_data.rar).
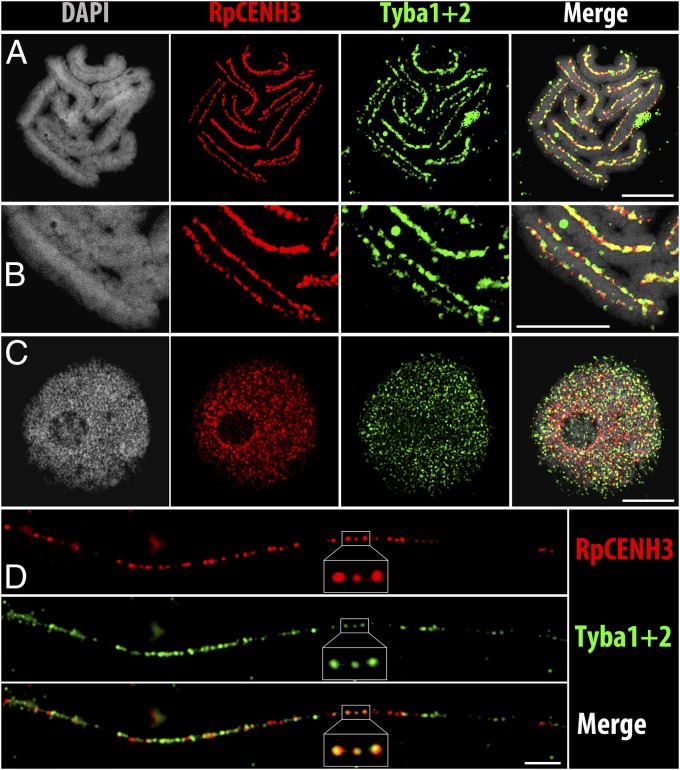
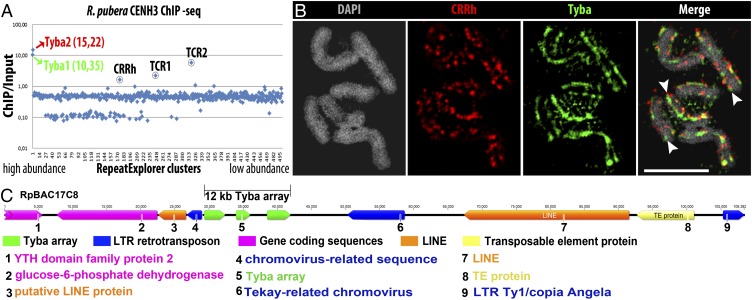
Next, an antibody (anti-RpCENH3) designed to recognize both CENH3 variants of R. pubera was generated and used for immunostaining. A specific labeling of the centromere groove was found at metaphase (SI Appendix, Fig. S3A and Movie S2). In interphase nuclei, a dispersed distribution of CENH3 foci was found, whereas prophase chromosomes displayed line-like signal arrangements (SI Appendix, Fig. S3 B and C). Colocalization experiments using the CENH3-specific antibody and a Tyba-specific FISH probe revealed a coincidence of both signal patterns along the holocentromeres of metaphase chromosomes (Fig. 2 A and B and Movie S3) and in interphase nuclei (Fig. 2C). Detailed analyses using super-resolution microscopy at a lateral resolution of ∼120 nm quantified an overlap of 57.8% of CENH3 signals with Tyba signals and a 69.1% overlap of Tyba with CENH3 in interphase (n = 4). Within the holocentromeres of metaphase chromosomes (n = 8) these values amounted to 53.6% and 76.0%, respectively. Double-labeling experiments on extended chromatin fibers confirmed the segmental overlap of the two signal types (Fig. 2D). Hence, as in other species e.g., Arabidopsis, maize, rice, and humans (28–31), not all centromeric repeat copies interact with CENH3-containing chromatin, and vice versa. Finally, the association of the satDNA with functional centromeres was analyzed by CENH3-ChIP. Quantitative PCR showed a higher than fourfold enrichment of Tyba1 and -2 in the RpCENH3-ChIP-DNA compared to the input DNA (SI Appendix, Fig. S2F). Analysis of ChIP-seq reads demonstrated that Tyba1 and -2 showed the highest level of association in the immunoprecipitated fraction, with 10- and 15-fold enrichment, respectively (Fig. 3A). Thus, both members of the Tyba satDNA interact with CENH3-containing nucleosomes.
Fig. 2.
Colocalization of RpCENH3 and Tyba1 and -2 on R. pubera holocentromeres. (A) Metaphase. (B) Enlarged metaphase chromosome. (C) Interphase nucleus. (D) Extended chromatin fiber. A–C are superresolution microscopy (SIM) images. (Scale bars: 5 µm.)
Fig. 3.
Characterization of CENH3-interacting sequences. (A) CENH3 ChIP-seq reads mapped against the main RepeatExplorer clusters of the R. pubera genome. Colored circles and names indicate the main centromeric sequences in R. pubera CENH3 ChIP-seq. (B) SIM image showing FISH with CRRh (CL175) and Tyba1+2 on metaphase chromosomes. Arrowheads indicate the longitudinal centromere grooves. (Scale bar: 5 µm.) (C) Annotation of an R. pubera BAC (RpBAC17C8) containing a centromere unit showing a Tyba array of ∼12 kb divided into three subarrays inserted in the protein domains region of a chromovirus-related sequence. Additional transposable elements and single-copy coding sequences were found in close neighborhood.
To validate the tandem-repeat organization and to check whether Tyba is present also in related species, we performed PCR and Southern hybridization using DraI-digested genomic DNA of different species of Rhynchospora and four other Cyperaceae genera. PCR using Tyba1-specific primers resulted in amplification products in all tested species. However, Southern hybridizations using Tyba1 from Rhynchospora tenuis exhibited only tandem repeat-typical ladder-like patterns in all Rhynchospora species (SI Appendix, Fig. S3D), indicating a conserved abundance of this repeat within this genus. Indeed, immuno-FISH using R. pubera Tyba probes and antibodies against CENH3 confirmed a colocalized holocentric signal distribution in R. tenuis and R. ciliata (SI Appendix, Fig. S3 E and F) similar to that in R. pubera. Additionally, as reported for other centromeric sequences (32), Tyba1 and -2 are transcriptionally active in all tissues of R. pubera analyzed by RT-PCR (SI Appendix, Fig. S3G) and RNA-seq search.
Although Tyba is clearly the most abundant centromeric repeat found in R. pubera, detailed analysis of ChIP-seq data revealed that CENH3-containing chromatin is also associated with at least three additional families of repetitive sequences: the Ty3/gypsy LTR retrotransposon CRRh and two different Tyba-containing repeats, hereafter referred to as “TCR1” and “TCR2” (Fig. 3A). The CRRh retrotransposon family, representing about 0.2% of the genome, was found to be heterogeneous, including two autonomous elements (CRRh-1 and CRRh-2) and three nonautonomous elements (noaCRRh-1, noaCRRh-2, and noaCRRh-3). Each autonomous element possessed a single ORF of 1,473 and 1,463 codons, respectively. The putative polyprotein sequences were relatively divergent (59.5% identity), but they both contained all the domains necessary for replication and integration (Gag, protease, reverse transcriptase, RNase H, and integrase) (SI Appendix, Fig. S4A). In contrast, the nonautonomous elements did not encode any of the protein domains found in the autonomous elements, but each of them possessed an ORF of unknown function (SI Appendix, Fig. S4A). Although the internal sequences of noaCRRh-1 and -2 had no significant similarity to either of the autonomous elements, their LTR sequences shared 73.3 and 72.7% similarity, respectively, with the LTR sequence of CRRh-1, suggesting that they are derivates of CRRh-1. On the other hand, noaCRRh-3 seemed to have originated from CRRh-2, because these elements shared significant similarities at both LTR termini and in a short region in the 5′ UTR. Phylogenetic analysis based on reverse transcriptase domain sequences revealed that the CRRh elements belong to the group B of the centromeric retrotransposon of maize (CRM) clade of chromoviruses (SI Appendix, Fig. S4B). Similar to other CRM chromoviruses of this group, both CRRh elements lacked the putative targeting domain at the C terminus of integrase (SI Appendix, Fig. S4E) (2). In-depth analysis of ChIP enrichment showed that only CRRh-1, noaCRRh-1, noaCRRh-2, and noaCRRh-3 elements are associated with CenH3-containing chromatin, but CRRh-2 is not (SI Appendix, Fig. S4C). Inspection of sequences at insertion sites further revealed that all four ChIP-enriched elements are frequently integrated into Tyba arrays (SI Appendix, Fig. S4D). The observed number of CRRh insertions into Tyba was 13- to 21-fold higher than expected (P value < 2.2e-6), implying preferential CRRh insertion into Tyba regions. This observation was confirmed experimentally by FISH, which detected CRRh-specific signals intermingled with Tyba signals within the longitudinal centromere groove (Fig. 3B), although the CRRh signals were less abundant. Thus, the holocentromeres of R. pubera are enriched in different types of repetitive sequences, with the satDNA Tyba being the main sequence contributing to the formation of active centromeric units, and CRRh showing only moderate enrichment.
Unlike CRRh, whose association with Tyba is clearly a result of its integration preferences, Tyba is a constituent of the repetitive unit in both TCR1 and TCR2. The sequence of the repetitive element TCR1 could be reconstructed in part, because the ∼5-kb-long fragments represent around 0.14% of the genome. One end of the reconstructed consensus sequence likely represents the real boundary of the repetitive unit, because the analysis of this end in reads from different genomic loci revealed a high diversity in the flanking sequences, as is typical for insertion sites. The other end possessed a sequence with high similarity to Tyba. Because our attempts to bridge the Tyba region were not successful, the other boundary of the TCR1 repeat remained unknown. Nevertheless, the presence of putative insertion sites at the Tyba-lacking terminus strongly suggests that TCR1 is a transposable element. In contrast, the repetitive element TCR2 (0.02% of the genome) lacked any sign of insertion sites, and both ends of the in silico reconstructed fragment possessed sequences with high similarity to Tyba. PCR with primers designed from the Tyba-unrelated part of the repeat and directed outwards from the repetitive unit amplified three major fragments about 2.5, 5.0, and 7.5 kb in length, suggesting that TCR2 is a tandem repeat. Sequencing of four randomly selected clones revealed that the monomer of TCR2 is 2,551 bp long and possess nine monomers of Tyba.
Centromeric Sequences Are Composed of High-Order Tyba Tandem Repeats Interspersing the Gene-Containing Chromatin.
The higher-order organization of centromeric DNA was analyzed using seven Tyba-containing genomic BACs of R. pubera. The identified Tyba arrays were found to be rather small, varying from 3 to 16 kb. As typical for centromeric satDNAs (25, 33), we found the Tyba arrays forming high-order repeat (HOR) structures. Pentamers (830–870 bp) were the most frequent HORs in all BACs analyzed, whereas dimers (∼344 bp) were found only occasionally in two BACs (8P1 and 23M1) (SI Appendix, Table S2). Tyba arrays occurred as a continuous array in five of the seven BACs; in BACs 17C8 and 23H8 the array was divided into subarrays (Fig. 3C and SI Appendix, Fig. S3H). The regions between these subarrays did not show similarity to any known sequence.
A close proximity between Tyba and putative centromeric retroelements was found in three BACs (17C8, 9H8, and 8P1). To confirm these elements as centromeric retroelements, their reverse transcriptase-domain sequences were analyzed phylogenetically. Only the element found in BAC 8P1 grouped within other centromeric retroelements, relatively close to the CRRh clade. The elements found in BACs 17C8 and 9H8 represent most likely a chromovirus of the Tekay clade and a pararetrovirus, respectively (SI Appendix, Fig. S3H). Interestingly, in BACs 17C8 and 9H8 Tyba arrays were found very close to the protein domain regions of these elements, but in BAC 8P1 the CRRh-like element was present ∼20 kb upstream of the Tyba array (Fig. 3C and SI Appendix, Fig. S3H). Thus, although we frequently detected CRRh elements integrated into Tyba arrays (SI Appendix, Fig. S4D), we did not observe this integration in the sequenced BACs.
In addition to a long Tyba array, BAC 23M1 showed a short Tyba-like cluster (∼700 bp) inserted into an LTR retrotransposon-like sequence (SI Appendix, Fig. S3H). In addition, centromeric sequences were generally found to be flanked by gene-coding sequences and other transposable elements (Fig. 3C and SI Appendix, Fig. S3H). A transcriptional activity of these genes is likely, because BLAST comparison of these coding sequences found high similarity between them and the pollen mother cell transcriptome.
Discussion
Origin and Genome-Wide Spreading of a Holocentromeric Satellite.
How could a centromeric satellite repeat evolve along a chromosome in thousands of interspersed arrays? The chromosomes of R. pubera differ in their organization from all other holocentric species studied so far because no other repeat-based centromeres have been reported to date. SatDNA evolves according to the principles of concerted evolution. Within a genome, mutations are homogenized among repeats by the mechanisms of nonreciprocal sequence transfer, such as unequal crossover, gene conversion, rolling circle replication, and transposition-related mechanisms (34). Although the centromere traditionally was treated as a region of suppressed recombination, unequal crossing-over, and gene conversion have been identified as the most widespread mechanism involved in satDNA dynamics (35–37). In addition, segmental duplication has been proposed as a mechanism for massive amplification of satDNA arrays (38, 39).
However, the origin of novel satellite repeats remains elusive. Tandem repeats with homology to parts of retrotransposons have been identified in several plants, e.g., potato (40). Here, we found that the centromeric retrotransposon CRRh is very frequently (>40%) integrated into Tyba satellite arrays, although so far there is no evidence for the presence of Tyba inside CRRh. Thus, CRRh most likely did not contribute to the origin and spreading of Tyba. Further, because CRRh integrates into Tyba at different positions, the targeting is most likely sequence independent. Furthermore, the finding that Tyba is a constituent of the repetitive unit in the transposable element TCR1 might explain the origin and spreading of Tyba throughout the genome. Additionally, a preferential insertion/stabilization of Tyba sequences into centromeric chromatin followed by positive selection might have been facilitated by CENH3 itself, thereby contributing to Tyba accumulation along the centromere groove in Rhynchospora. Alternatively, Tyba at a certain level might define the preferential sites for CENH3 accumulation.
A Cell-Cycle–Dependent Dynamic Shuffling of Multiple Centromeric Units Results in the Formation of Holocentromeres During Metaphase.
Mitotic chromosomes of R. pubera exhibit a line-like centromere organization comprising a high number of centromeric units composed of consecutive CENH3 nucleosomes and enriched in centromeric tandem repeats and retroelements. In contrast to monocentric chromosomes, the prerequisite for this holokinetic centromere organization is the intermingling of coding and noncoding regions and the genome-wide interspersion of euchromatic and heterochromatic domains at large scale. Indeed, no distinguishable large-scale patterns of euchromatin- and heterochromatin-typical epigenetic marks were found along mitotic chromosomes of R. pubera. Furthermore, analysis of R. pubera BACs revealed a close proximity of the centromeric Tyba satDNA with gene-coding sequences and different classes of repetitive DNA, such as class I (Maximus/SIRE clade of copia-like and Ogre/Tat, and Athila clades of gypsy-like) and II (hAT-like, MITE-like, and MuDR-like) transposable elements. Many transposable elements are randomly dispersed in the genomes, but others may appear concentrated in specific chromosomal regions, such as the centromeric retrotransposon CRRh, which belongs to the chromoviral clade CRM (2, 41). Our finding that the CENH3-interacting Tyba satellite and CRRh are inserted into transcriptionally active gene-containing chromatin corroborates the assumption that the centromere organization type influences the organization of the genome at the global chromosomal level.
Our results show that the CENH3/repeat-containing centromeric units are the basic components of the holocentric centromere organization in R. pubera supporting the classical polycentric model of holocentricity (42). In support, a study of elongated polycentric chromosomes in Pisum sativum, representing a potential evolutionary intermediate between monocentric and holocentric chromosomes, demonstrated that all functional centromere domains are tightly associated with clusters of 13 distinct satDNA families and with one centromeric retrotransposon (CR) family (43).
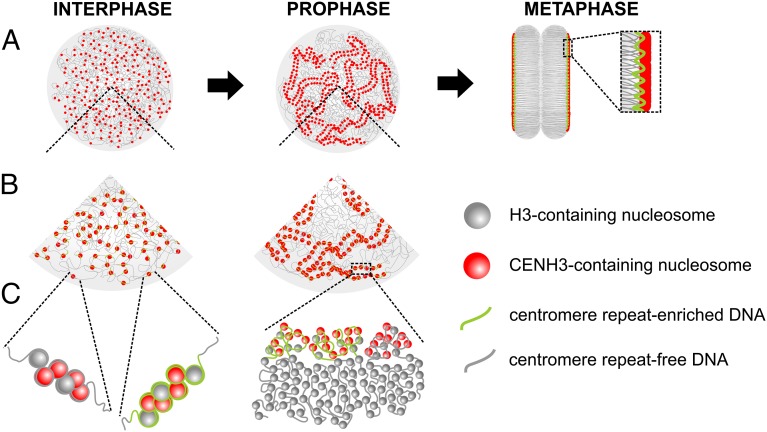
To explain the observed dynamic distribution of centromeric units during the cell cycle, we propose a model in which, during interphase, holocentromeres dissociate into individual CENH3/centromere repeat-containing units. Then, in prophase and metaphase they reassociate and form the holocentromeres along the centromere groove (Fig. 4). A similar dot-like CENH3 distribution in interphase nuclei was shown for Luzula species (9, 10) and C. elegans (8), indicating that holocentromeres are composed of hundreds of individual centromeric units. Because no centromere-specific repeats were found in these species (15, 16), the dynamic cell cycle-dependent shuffling of centromeric units occurs independently of centromeric repeats. It is tempting to speculate that the dissociation of holocentromeres during interphase is required to ensure the transcription of genes located close to the centromeric units.
Fig. 4.
Model of cell-cycle–dependent changes in the holocentromere chromatin organization of R. pubera. During interphase, holocentromeres dissociate into individual CENH3/centromere repeat-containing units. In prophase and metaphase they reassociate and form holocentromeres along the sister chromatids. Most of the CENH3-containing nucleosomes associate with centromere repeat-enriched sequences. Some CENH3-containing nucleosomes associate with centromere repeat-free sequences. Increasing levels of resolution are shown in A, B, and C.
In parallel with the process of chromosome condensation toward metaphase, centromeric units join to form a line-like “polycentromere” within the longitudinal centromere groove to ensure faithful segregation of chromosomes. The mechanism behind this dynamic shuffling of centromeric units remains unknown. A reversible cohesive association of centromeric units might lead to the progressive shuffling of individual centromeric units, finally resulting in the formation of a line-like kinetochore composed of multiple units.
How Long Are the Centromeric Arrays of R. pubera?
Because about 4% of the genome of this species is composed of centromeric repeats, each of the five chromosomes should harbor a sum of multiple centromeric arrays comprising about 13 Mbp of centromeric DNA, based on an estimated genome size of 1,614 Mb per 1C. In our analysis, the length of Tyba arrays varied from 3 to 16 kb, although we cannot exclude the existence of smaller and/or larger arrays. Assuming 10–15 kb as an average size and that Tyba arrays serve as preferential sites for CENH3-recruitment, each chromosome could harbor between 800 and 1,300 centromeric subunits. Considering that the R. pubera genome is about 16-fold larger than that of C. elegans (44), which was considered to harbor 707 centromeric units (16), a correlation of centromeric units per million base pairs in these genomes will give a slightly higher abundance of CENH3-hotspots in C. elegans (∼7.2 centromeric units per million base pairs) than in R. pubera (2.5–4 Tyba arrays per million base pairs). Despite the small size of single centromeric DNA arrays in Rhynchospora compared to other species, the total amount of potential centromeric DNA per chromosome is among the largest reported for any species so far.
Conclusions
Although the mechanism behind the spreading of Tyba along the centromeric chromatin remains elusive, a preferential integration to CENH3-positive chromatin followed by positive selection might have occurred. On the other hand, the alternative option in which Tyba satellite repeat-rich regions may work as preferred sites for the deposition of centromeric nucleosomes and thus serve as potential kinetochore attachment sites in R. pubera cannot be excluded. Finally, the genome-wide distribution of centromeric repeat arrays interspersing the euchromatin observed in this species provides a previously unidentified variant of centromere organization. Thus, it is evident that different types of holocentromeres, namely centromeres with and without specific repetitive sequences and with or without CENH3/CENP-C, exist in different species. Further studies of species with holocentric chromosomes will broaden our mainly monocentric chromosome-biased knowledge about centromere organization and may help elucidate the centromere plasticity among eukaryotes.
Materials and Methods
Detailed materials and methods are described in SI Appendix, Materials and Methods. Briefly, high-copy repeats were identified by graph-based clustering (23) of genomic Illumina reads of R. pubera. A Rhynchospora CENH3-specific antibody was generated and used for indirect immunostaining, ChIP, ChIP-qPCR, and ChIP-seq. Centromeric repeats were characterized by FISH and sequence analysis.
Supplementary Material
Acknowledgments
We thank Wayt Thomas for kindly identifying the plant material; Manuela Knauft for pulsed-field electrophoresis analysis; Karin Lipfert for art work; and Jan Vrána, Radka Tušková, and Eva Jahnová for flow sorting, BAC library construction, and filter preparation. The Brazilian Federal Agency for the Support and Evaluation of Graduate Education within the Ministry of Education of Brazil (CAPES) provided a Special Visiting Researcher Grant and project funding (to A.H.) and scholarships (to A.M. and T.R.). We thank the Fundação de Amparo à Ciência e Tecnologia do Estado de Pernambuco (FACEPE) (AMD-0025-2.00-14) for visiting research grant (to A.M.). The Brazilian National Council of Technological and Scientific Development (CNPq) provided financial support for A.P.-H. The construction of the BAC library was funded by Ministry of Education, Youth and Sports of the Czech Republic National Program of Sustainability I Grant Award LO1204. The Czech Science Foundation and the Czech Academy of Sciences provided financial support for J.M. (GBP501/12/G090 and RVO:60077344).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Assembled BAC sequences are available through the iPlant Data Store and can be accessed via iPlant Discovery Environment or at https://de.iplantcollaborative.org/dl/d/8258A143-C5F5-4DF1-84F2-88C94BE8EA8F/R_pubera_holocentromeres_data.rar. Original ChIP-sequencing sample data, original Illumina sequencing data for the genomic DNA and BACs, and RNA-sequencing data have been deposited at the Sequence Read Archive (www.ebi.ac.uk/ena/) [accession nos. PRJEB9647 (ChIP-sequencing sample data), PRJEB9643 (Illumina sequencing data for genomic DNA), PRJEB9649 (Illumina sequencing data for BACs), and PRJEB9645 (RNA-sequencing data)]. CENH3 cDNA sequences variants were deposited in the GenBank database [accession nos. KR029618 (RpCENH3_1) and KR029619 (RpCENH3_2)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1512255112/-/DCSupplemental.
References
- 1.Plohl M, Meštrović N, Mravinac B. Centromere identity from the DNA point of view. Chromosoma. 2014;123(4):313–325. doi: 10.1007/s00412-014-0462-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Neumann P, et al. Plant centromeric retrotransposons: A structural and cytogenetic perspective. Mob DNA. 2011;2(4):1–16. doi: 10.1186/1759-8753-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marshall OJ, Chueh AC, Wong LH, Choo KH. Neocentromeres: New insights into centromere structure, disease development, and karyotype evolution. Am J Hum Genet. 2008;82(2):261–282. doi: 10.1016/j.ajhg.2007.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Earnshaw WC, et al. Esperanto for histones: CENP-A, not CenH3, is the centromeric histone H3 variant. Chromosome Res. 2013;21(2):101–106. doi: 10.1007/s10577-013-9347-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heckmann S, Houben A. 2013. Holokinetic centromeres. Plant Centromere Biology, eds Jiang J. Birchler JA (Wiley-Blackwell, Oxford, UK), pp 83–94.
- 6.Guerra M, et al. Neocentrics and holokinetics (holocentrics): Chromosomes out of the centromeric rules. Cytogenet Genome Res. 2010;129(1-3):82–96. doi: 10.1159/000314289. [DOI] [PubMed] [Google Scholar]
- 7.Moore LL, Morrison M, Roth MB. HCP-1, a protein involved in chromosome segregation, is localized to the centromere of mitotic chromosomes in Caenorhabditis elegans. J Cell Biol. 1999;147(3):471–480. doi: 10.1083/jcb.147.3.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buchwitz BJ, Ahmad K, Moore LL, Roth MB, Henikoff S. A histone-H3-like protein in C. elegans. Nature. 1999;401(6753):547–548. doi: 10.1038/44062. [DOI] [PubMed] [Google Scholar]
- 9.Nagaki K, Kashihara K, Murata M. Visualization of diffuse centromeres with centromere-specific histone H3 in the holocentric plant Luzula nivea. Plant Cell. 2005;17(7):1886–1893. doi: 10.1105/tpc.105.032961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heckmann S, et al. Holocentric chromosomes of Luzula elegans are characterized by a longitudinal centromere groove, chromosome bending, and a terminal nucleolus organizer region. Cytogenet Genome Res. 2011;134(3):220–228. doi: 10.1159/000327713. [DOI] [PubMed] [Google Scholar]
- 11.Wanner G, Schroeder-Reiter E, Ma W, Houben A, Schubert V. The ultrastructure of mono- and holocentric plant centromeres: An immunological investigation by structured illumination microscopy and scanning electron microscopy. Chromosoma. June 6, 2015 doi: 10.1007/s00412-015-0521-1. [DOI] [PubMed] [Google Scholar]
- 12.Cabral G, Marques A, Schubert V, Pedrosa-Harand A, Schlögelhofer P. Chiasmatic and achiasmatic inverted meiosis of plants with holocentric chromosomes. Nat Commun. 2014;5:5070. doi: 10.1038/ncomms6070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chmátal L, et al. Centromere strength provides the cell biological basis for meiotic drive and karyotype evolution in mice. Curr Biol. 2014;24(19):2295–2300. doi: 10.1016/j.cub.2014.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gassmann R, et al. An inverse relationship to germline transcription defines centromeric chromatin in C. elegans. Nature. 2012;484(7395):534–537. doi: 10.1038/nature10973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heckmann S, et al. The holocentric species Luzula elegans shows interplay between centromere and large-scale genome organization. Plant J. 2013;73(4):555–565. doi: 10.1111/tpj.12054. [DOI] [PubMed] [Google Scholar]
- 16.Steiner FA, Henikoff S. Holocentromeres are dispersed point centromeres localized at transcription factor hotspots. eLife. 2014;3:e02025. doi: 10.7554/eLife.02025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Subirana JA, Messeguer X. A satellite explosion in the genome of holocentric nematodes. PLoS One. 2013;8(4):e62221. doi: 10.1371/journal.pone.0062221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mello CC, Kramer JM, Stinchcomb D, Ambros V. Efficient gene transfer in C. elegans: Extrachromosomal maintenance and integration of transforming sequences. EMBO J. 1991;10(12):3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Riddle DL, Blumenthal T, Meyer BJ, Priess JR. 1997. Introduction to C. elegans. C. elegans II, eds Riddle DL, Blumenthal T, Meyer BJ, Priess JR (Cold Spring Harbor Lab Press, Cold Spring Harborn, NY), 2nd Ed.
- 20.Luceno M, Vanzela ALL, Guerra M. Cytotaxonomic studies in Brazilian Rhynchospora (Cyperaceae), a genus exhibiting holocentric chromosomes. Canadian Journal of Botany. 1998;76(3):440–449. [Google Scholar]
- 21.Vanzela ALL, Cuadrado A, Jouve N, Luceño M, Guerra M. Multiple locations of the rDNA sites in holocentric chromosomes of Rhynchospora (Cyperaceae) Chromosome Res. 1998;6(5):345–349. doi: 10.1023/a:1009279912631. [DOI] [PubMed] [Google Scholar]
- 22.Vanzela ALL, Guerra M, Luceno M. Rhynchospora tenuis Link (Cyperaceae), a species with the lowest number of holocentric chromosomes. Cytobios. 1996;88(355):219–228. [Google Scholar]
- 23.Novák P, Neumann P, Macas J. Graph-based clustering and characterization of repetitive sequences in next-generation sequencing data. BMC Bioinformatics. 2010;11:378. doi: 10.1186/1471-2105-11-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Novák P, Neumann P, Pech J, Steinhaisl J, Macas J. RepeatExplorer: A Galaxy-based web server for genome-wide characterization of eukaryotic repetitive elements from next-generation sequence reads. Bioinformatics. 2013;29(6):792–793. doi: 10.1093/bioinformatics/btt054. [DOI] [PubMed] [Google Scholar]
- 25.Plohl M, Luchetti A, Mestrović N, Mantovani B. Satellite DNAs between selfishness and functionality: Structure, genomics and evolution of tandem repeats in centromeric (hetero)chromatin. Gene. 2008;409(1-2):72–82. doi: 10.1016/j.gene.2007.11.013. [DOI] [PubMed] [Google Scholar]
- 26.Melters DP, et al. Comparative analysis of tandem repeats from hundreds of species reveals unique insights into centromere evolution. Genome Biol. 2013;14(1):R10. doi: 10.1186/gb-2013-14-1-r10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kelly WG, et al. X-chromosome silencing in the germline of C. elegans. Development. 2002;129(2):479–492. doi: 10.1242/dev.129.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nagaki K, et al. Chromatin immunoprecipitation reveals that the 180-bp satellite repeat is the key functional DNA element of Arabidopsis thaliana centromeres. Genetics. 2003;163(3):1221–1225. doi: 10.1093/genetics/163.3.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nagaki K, et al. Sequencing of a rice centromere uncovers active genes. Nat Genet. 2004;36(2):138–145. doi: 10.1038/ng1289. [DOI] [PubMed] [Google Scholar]
- 30.Zhong CX, et al. Centromeric retroelements and satellites interact with maize kinetochore protein CENH3. Plant Cell. 2002;14(11):2825–2836. doi: 10.1105/tpc.006106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vafa O, Sullivan KF. Chromatin containing CENP-A and alpha-satellite DNA is a major component of the inner kinetochore plate. Curr Biol. 1997;7(11):897–900. doi: 10.1016/s0960-9822(06)00381-2. [DOI] [PubMed] [Google Scholar]
- 32.Hall LE, Mitchell SE, O’Neill RJ. Pericentric and centromeric transcription: A perfect balance required. Chromosome Res. 2012;20(5):535–546. doi: 10.1007/s10577-012-9297-9. [DOI] [PubMed] [Google Scholar]
- 33.Iwata A, et al. Identification and characterization of functional centromeres of the common bean. Plant J. 2013;76(1):47–60. doi: 10.1111/tpj.12269. [DOI] [PubMed] [Google Scholar]
- 34.Dover GA. Molecular drive in multigene families - How biological novelties arise, spread and are assimilated. Trends Genet. 1986;2(6):159–165. [Google Scholar]
- 35.Mahtani MM, Willard HF. Physical and genetic mapping of the human X chromosome centromere: Repression of recombination. Genome Res. 1998;8(2):100–110. doi: 10.1101/gr.8.2.100. [DOI] [PubMed] [Google Scholar]
- 36.Smith GP. Evolution of repeated DNA sequences by unequal crossover. Science. 1976;191(4227):528–535. doi: 10.1126/science.1251186. [DOI] [PubMed] [Google Scholar]
- 37.Talbert PB, Henikoff S. Centromeres convert but don’t cross. PLoS Biol. 2010;8(3):e1000326. doi: 10.1371/journal.pbio.1000326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Horvath JE, et al. Punctuated duplication seeding events during the evolution of human chromosome 2p11. Genome Res. 2005;15(7):914–927. doi: 10.1101/gr.3916405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ma J, Jackson SA. Retrotransposon accumulation and satellite amplification mediated by segmental duplication facilitate centromere expansion in rice. Genome Res. 2006;16(2):251–259. doi: 10.1101/gr.4583106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tek AL, Song J, Macas J, Jiang J. Sobo, a recently amplified satellite repeat of potato, and its implications for the origin of tandemly repeated sequences. Genetics. 2005;170(3):1231–1238. doi: 10.1534/genetics.105.041087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gorinsek B, Gubensek F, Kordis D. Evolutionary genomics of chromoviruses in eukaryotes. Mol Biol Evol. 2004;21(5):781–798. doi: 10.1093/molbev/msh057. [DOI] [PubMed] [Google Scholar]
- 42.Schrader F. The role of the kinetochore in the chromosomal evolution of the heteroptera and homoptera. Evolution. 1947;1:134–142. [Google Scholar]
- 43.Neumann P, et al. Stretching the rules: Monocentric chromosomes with multiple centromere domains. PLoS Genet. 2012;8(6):e1002777. doi: 10.1371/journal.pgen.1002777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.C. elegans Sequencing Consortium Genome sequence of the nematode C. elegans: A platform for investigating biology. Science. 1998;282(5396):2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.