Significance
In obligate bacterial predators such as Bdellovibrio bacteriovorus the cell-replication cycle is tightly coupled to the predatory act. Using a novel growth system, in which the content of emptied prey cells is manipulated, we show that B. bacteriovorus uses two spatially and temporally separated prey-derived cues to regulate the steps from prey recognition to predatory cell growth through differential gene expression. A previously unidentified cell-cycle checkpoint is uncovered, during which prey quality, as defined by nutrient availability, may be assessed.
Keywords: microbial ecology, bacterial physiology, bacterial cell cycle, predatory bacteria, Bdellovibrio bacteriovorus
Abstract
Predators feed on prey to acquire the nutrients necessary to sustain their survival, growth, and replication. In Bdellovibrio bacteriovorus, an obligate predator of Gram-negative bacteria, cell growth and replication are tied to a shift from a motile, free-living phase of search and attack to a sessile, intracellular phase of growth and replication during which a single prey cell is consumed. Engagement and sustenance of growth are achieved through the sensing of two unidentified prey-derived cues. We developed a novel ex vivo cultivation system for B. bacteriovorus composed of prey ghost cells that are recognized and invaded by the predator. By manipulating their content, we demonstrated that an early cue is located in the prey envelope and a late cue is found within the prey soluble fraction. These spatially and temporally separated cues elicit discrete and combinatory regulatory effects on gene transcription. Together, they delimit a poorly characterized transitory phase between the attack phase and the growth phase, during which the bdelloplast (the invaded prey cell) is constructed. This transitory phase constitutes a checkpoint in which the late cue presumably acts as a determinant of the prey’s nutritional value before the predator commits. These regulatory adaptations to a unique bacterial lifestyle have not been reported previously.
Predators obtain their nutrients by killing prey organisms. This trophic mode is common in microorganisms such as protozoa that feed on bacteria and phages that use bacterial cells for replication. Predation impacts on food webs have been established, and its effect on parameters such as bacterial prey size, distribution, or palatability has been investigated (1–3). However, predation between bacteria, the most abundant cellular living entities on Earth, is poorly understood. The known diversity of predatory bacteria is low (4), and few studies have addressed predation dynamics in natural or man-made environments (5–7). An important aspect of predatory interactions is prey choice. Prey choice can have important consequences for the predator, because it defines how much energy is spent in obtaining prey (8, 9). Thus, the predator first may discriminate between prey and nonprey and then determine the nutritional value of the prey (10). Whether such concerns exist in the bacterial realm is not known. In other words, do predatory bacteria, in which nutrient acquisition and cell reproduction are tightly coupled, modulate their interactions with their bacterial prey?
BALOs (Bdellovibrio and similar organisms) constitute an exclusive group of obligate predatory Gram-negative bacteria that prey solely on other Gram-negative bacteria (11, 12). As such, they are considered as potential biocontrol agents and living antibiotics (13, 14). Predation by BALOs is either periplasmic or epibiotic, with both strategies being carried out by closely related predators (15). BALOs are well distributed in nature, being found in both terrestrial and aquatic ecosystems and possibly in animal (including human) intestines (11, 16, 17). Because obligate predators cannot replicate in the absence of adequate prey, the structure of BALO populations is under environmental selection (6, 7, 18), whereas natural selection and arms race may shape predator–prey interactions (19, 20). Indeed, BALO isolates possess different prey spectra that are adapted to the prey available in their surroundings (18, 21, 22). A central goal of BALO research is to understand the mechanisms by which a prey cell is recognized and how this event sets the predator’s cell cycle toward cell growth and cell multiplication. BALOs thus constitute a unique system for studying ecological effects on bacterial development and differentiation.
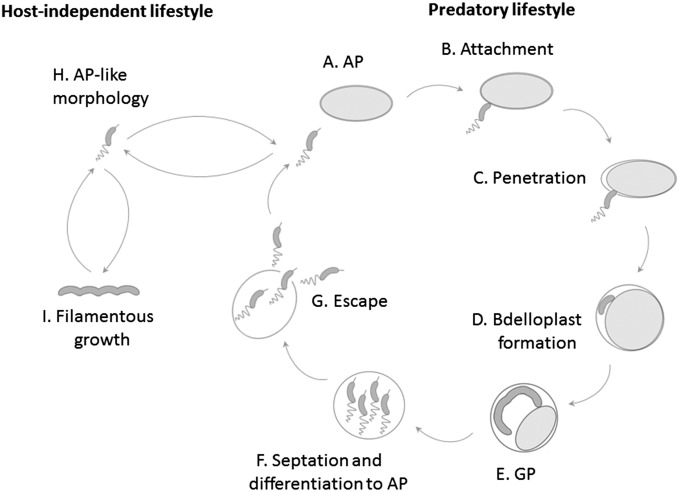
Bdellovibrio bacteriovorus, a periplasmic obligate predator, has a biphasic and dimorphic cell cycle during which a short, fast-swimming, and nonreplicative attack-phase (AP) cell searches for prey, attaches to the prey’s envelope, and perforates the envelope to penetrate and settle within the prey periplasm, forming a bdelloplast (12). Numerous functions, such as modifications of the prey’s cell wall (23–27), shedding of the predator’s flagellum (28), and killing of the prey cell (29), generate the conditions required for entering the growth phase (GP). During the GP, the prey is consumed, and the predatory cell grows as a multinucleoid filament that finally splits into progeny AP cells that leave the now depleted bdelloplast (30, 31). Thus, B. bacteriovorus' cell cycle includes spatial and temporal separation of a motile nonreplicative phase and a sessile growth and replication phase (Fig. S1). Although manipulation of WT B. bacteriovorus’ growth conditions is complicated by its total dependence on prey cells, earlier works have shown that predation is triggered by two distinct but still unidentified prey-derived cues. One cue presumably elicits the predatory process, promoting prey penetration, bdelloplast formation, and prey consumption (32, 33). A second soluble cue promotes DNA synthesis and is required for sustained cellular growth (33–35).
Fig. S1.
The cell cycle of B. bacteriovorus: WT B. bacteriovorus maintains an obligate predatory biphasic lifecycle. Nonreplicating fast-swimming vibrioid AP cells carrying a predatory essential type IVa pilus at the nonflagellated pole roam the environment in search for prey (A). Encounter with the prey cell is followed by stable attachment (B) and then by type IVa pilus-dependent invasion into the prey periplasm (C). During entry and settlement the bdelloplast is formed (D). Within the bdelloplast, the predator enters the GP, consumes the prey and grows as a multinucleoid filament (E). Upon nutrient depletion, the filament divides to progeny AP cells (F) that puncture and escape from the bdelloplast, ready to initiate another predation cycle (G). HI mutants are not restricted to the predatory lifestyle and whilst remaining predatory, they can grow saprophytically in the presence of prey-extract (type I HI mutants) or axenically in a rich medium (type II HI mutants), retaining a dimorphic cell cycle (H and I).
These cues can be by-passed by mutations occurring in host-independent (HI) mutants (Fig. S1). Mutations in gene bd0108 yield type I HI mutants that grow in the absence of prey in a rich medium supplemented with prey extract, i.e., they require the second cue (32, 36, 37). Type II HI mutants eliminate the requirement for prey extract; they are generated by a mutation in rhlB or in pcnB, genes that encode for RNA degradosome subunits (37). Although useful in research, HI mutants exhibit deregulated transitions between phases, as demonstrated by alterations in gene and protein expression (38, 39). Several efforts were made in the past to elicit growth of WT B. bacteriovorus with prey extract. These attempts met with varying degrees of success and could not be replicated (40, 41).
Here, we developed a novel ex vivo system for growing WT B. bacteriovorus independently of prey. This system is based on the use of intact cell envelopes (ghost cells) of Escherichia coli devoid of cytoplasm. These ghosts reliably mimic the prey’s envelope but do not promote growth unless supplemented with prey extract. With this system, growth control in B. bacteriovorus was characterized, revealing a transitory phase including a checkpoint during which the prey’s nutritional value is assessed.
Results
Production and Characterization of Prey Envelopes.
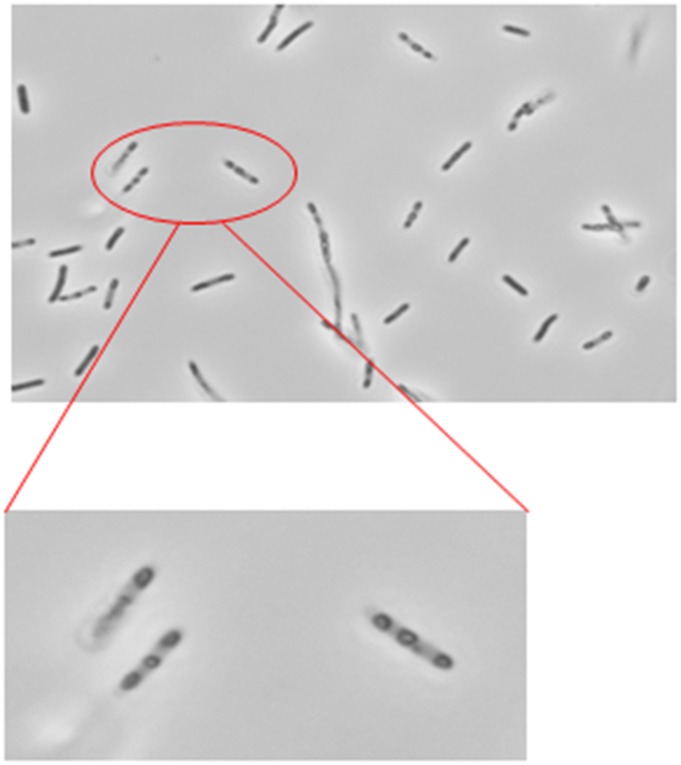
Plasmid pRK-kPR-cI-E-SNA bearing the bacteriophage PhiX174 lysis E gene was introduced into normal E. coli S17-1 cells to produce E. coli ghosts (42). Although these E. coli cells are preyed upon by B. bacteriovorus 109J, their ghosts were not recognized as prey when added to a fresh culture of AP B. bacteriovorus 109J. The predators transiently adhered to the ghost cells but were incapable of anchoring themselves permanently as occurs with live prey cells. Careful observation of ghost preparations by light microscopy revealed large, pore-like alterations of the envelopes that probably were generated by the expression of the PhiX174 gene E during their production (Fig. S2). Conceivably, these alterations prevented attachment, suggesting that prey cell envelopes with extensive structural damage are not recognized by the predator.
Fig. S2.
Phase-contrast microscopy of E. coli S17-1 ghosts produced by induction of the lysis E gene. Pore-like structures conceivably prevented recognition by B. bacteriovorus cells.
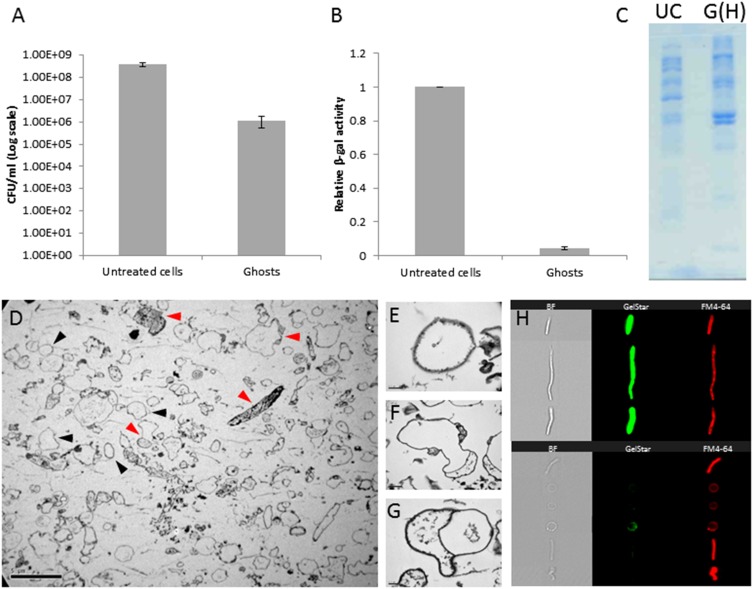
In contrast, functional prey envelopes were obtained by osmotic shock, based on Eisenbach and Adler’s procedure (43) for the production of integral E. coli envelopes, with modifications. The resulting empty cells (hereafter referred to as “ghosts”) were heterogeneous in size, shape, and light refraction when observed under phase-contrast microscopy, the last parameter probably reflecting different levels of peptidoglycan. Ghost suspensions were evaluated using several parameters. (i) Viability, as estimated by plate counts on LB plates, was reduced by two to three orders of magnitude compared with untreated cells (Fig. S3A), suggesting that the great majority of treated cells had died. (ii) β-Galactosidase activity, an indicator of cytoplasmic enzymatic activities, decreased by 95.7 ± 0.9%, indicating an almost complete loss of cytosolic content (Fig. S3B). The loss of cytosolic content was confirmed again by (iii) cryo-transmission electron microscopy (TEM) showing swollen ghosts harboring residual or no cytoplasm and confirming that the production of envelopes yielded whole, integral, cell-like empty structures (Fig. S3 D–G), and by (iv) SDS/PAGE, which markedly distinguished between ghosts’ and viable cells’ protein composition, the former presumably enriched with membrane proteins (Fig. S3C). Finally, (v) image cytometry, a technology combining flow cytometry and imaging, concomitantly counting and photographing cells and revealing their size, shape, and fluorescence (ImageStreamX; Amnis), showed that 81.1 ± 5.4% of the cells did not contain detectable levels of nucleic acids (Fig. S3H).
Fig. S3.
Characterization of E. coli AT984 ghosts produced via osmotic shock compared with untreated cells. (A) Viable counts are reduced by two to three orders of magnitude during ghost preparation, while retaining a similar number of cells, as determined with a Neubauer chamber. Values are averages of three independent experiments. Error bars indicate SD. (B) β-Galactosidase activity was used as a proxy to determine the loss of cytoplasmic content. In the ghost treatment, activity decreased by 95.7 ± 0.9%. Values are averages of three independent experiments. Error bars indicate SD. (C) SDS/PAGE protein profile of untreated cells (UC) and ghosts containing Hepes buffer [G(H)] demonstrates a large shift in protein display upon osmotic shock and loss of the cytoplasm. Both lanes were loaded with equal amounts of protein (20 µL of 2.7 mg/mL protein samples). (D) The diversity in ghost shape and content, as captured by TEM. Black arrowheads mark representative ghosts devoid of cytosol, and red arrowheads mark representative ghosts retaining various partial cytosolic contents. (Scale bar, 5 µm.) (E–G) Images of individual ghosts, showing complete (E) and partial (F and G) losses of the cytosol. (Scale bar, 0.2 µm.) (H, Right) Image cytometry (ImageStreamX) visualization of individual elongated E. coli cells before osmotic shock (Upper) and of ghosts (Lower) stained with the nucleic acid dye GelStar and the membrane stain FM4-64. (Left) Bright-field imaging (BF). Compared with intact cells, ghosts exhibit a severe loss of nucleic content (81.1 ± 5.4% of the ghost population had no detectible levels of nucleic acids) and showed a reduction in light refraction).
Prey Ghosts Are Recognized and Penetrated by WT Predators.
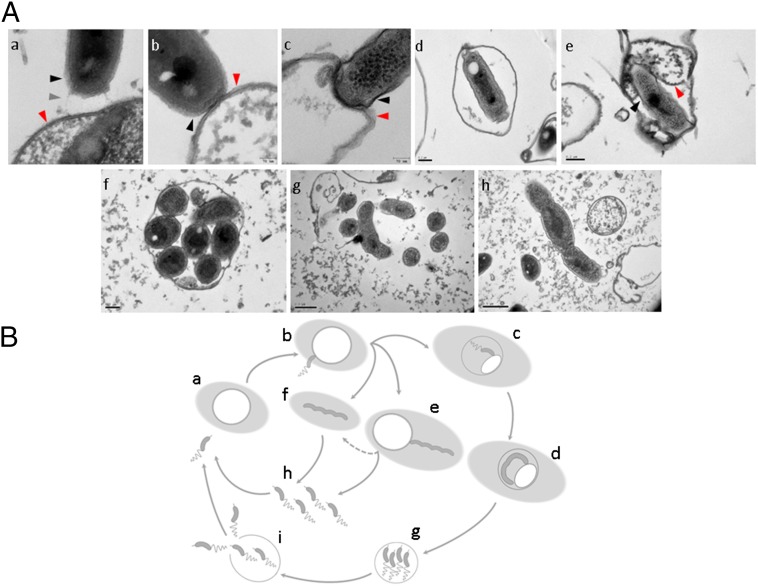
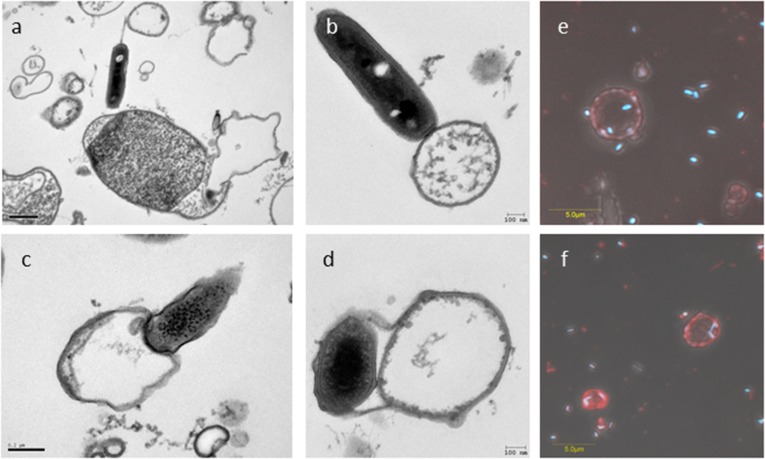
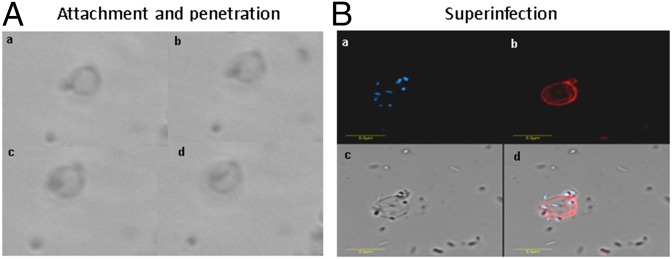
When prey ghosts suspended in buffer were mixed with fresh B. bacteriovorus 109J AP cells, rapid attachment occurred, possibly mediated by pilus-like structures, followed by stable binding (Fig. 1 A and B and Fig. S4 A–D). Within minutes, AP cells penetrated the envelopes, confining themselves to the empty vesicles, unable to escape the ghosts or enter GP (Fig. 2A and Movie S1). It is noteworthy that penetrating AP cells did not lose their flagella and remained motile inside the ghosts, as described earlier (44), suggesting that penetration itself is not sufficient for flagellar shedding. Superinfection (penetration by more than one predator) was also observed (Fig. 2B and Fig. S4 E and F), perhaps because of the high multiplicity of infection (MOI) used in this experiment (a 5:1 predator:ghost ratio), which leads to multiple infections in live prey (23, 31). The predator appears to maintain contact with the ghost’s cytoplasmic membrane during penetration as well as during growth (see contact points in Fig. 1 A, c and e and Fig. S4D), as previously observed with live prey cells (45). Upon prolonged incubation, neither escape from the prey ghosts nor cellular growth (cell elongation) of the predatory cell were observed (Fig. 3 A and C and Fig. S5), demonstrating that ghosts did not retain enough cytoplasm to support growth and replication of the B. bacteriovorus cells and the ensuing exit of attack cells from the depleted prey.
Fig. 1.
A full ex vivo growth cycle. (A) B. bacteriovorus 109J AP cells were incubated with ghost cells for 1 h (a–e) or with ghost cells and prey extract for 16 h (f–h) before processing for TEM. (a) A B. bacteriovorus AP cell interacting with the ghost envelope by means of two pili protruding from the invasive pole (one pilus is indicated by a gray arrowhead). (b) An AP cell attaching to a ghost cell. (c and d) An AP cell penetrating into and settling in a cytoplasm-depleted ghost cell. (e) A predatory cell residing inside a ghost cell with cytosol remains and interacting with its cytosolic membrane. (f and g) Filamentous growth of B. bacteriovorus within a ghost cell and outside a ghost cell in the presence of prey extract. (h) An ex vivo-grown filamentous B. bacteriovorus cell in the process of septation and differentiation into AP progeny cells. In a–c and e, capturing predatory synapses; black arrowheads mark the predator's cell envelope; red arrowheads mark the ghost cell envelope in a–c and the cytoplasmic membrane in e. A broader view of images a–c is presented in Fig. S4 A–C. (B) The ex vivo cultivation system supplies the requirements of the predatory life cycle. B. bacteriovorus AP cells can be cultivated ex vivo only if cytosol-depleted E. coli ghosts are complemented with prey extract (shaded light-gray background) (a). The AP cell stably attaches to the ghost cell (b) and subsequently either invades the ghost (c) and replicates intracellularly (d) or carries out an epibiotic growth cycle (e). Unattached GP cells are probably a consequence of ghost disintegration (fragmented arrow), because an initial interaction with the ghost cell is compulsory for growth (f). Growth is terminated by septation and differentiation to progeny AP cells (g). In the case of intracellular growth (h), progeny AP cells must lyse and evade the ghost cell (h).
Fig. S4.
Early interaction between B. bacteriovorus 109J and ghost cells. (A–D) B. bacteriovorus AP cells captured by TEM interacting with the ghost envelope by means of two pili protruding from the invasive pole (A), attaching to a small-sized empty ghost cell (B), penetrating into a cytoplasm-depleted ghost cell (C), and (residing inside a ghost cell and interacting with its cytosolic membrane (D). (E and F) Two confocal microscopy composite images of AP cells penetrating ghost cells under a high MOI (5:1 predator:ghost ratio). DAPI-stained AP cells are shown in blue; FM4-64-stained membranes of empty envelopes are shown in red. A closer view of the predation synapses in A–C is presented in Fig. 1 A, a–c.
Fig. 2.
Recognition of prey ghosts by B. bacteriovorus. (A) Tracking of a B. bacteriovorus 109J AP cell using phase-contrast microscopy. The predator is seen (a) attaching to and (b) penetrating a ghost cell (c) without damaging it, and (d) settling within the empty ghost cell space. (B) Confocal microscopy rendering of numerous AP cells penetrating a ghost cell under high multiplicity of infection (5:1 predator:ghost ratio). (a) DAPI stain. (b) FM4-64 membrane stain. (c) Bright-field imaging. (d) Composite of a–c.
Fig. 3.
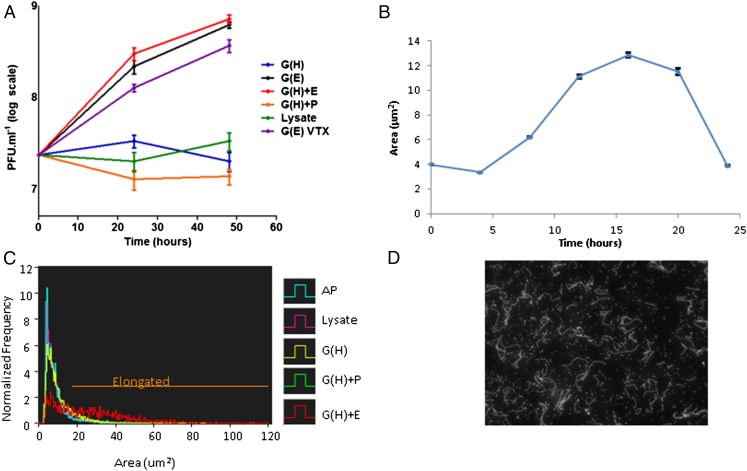
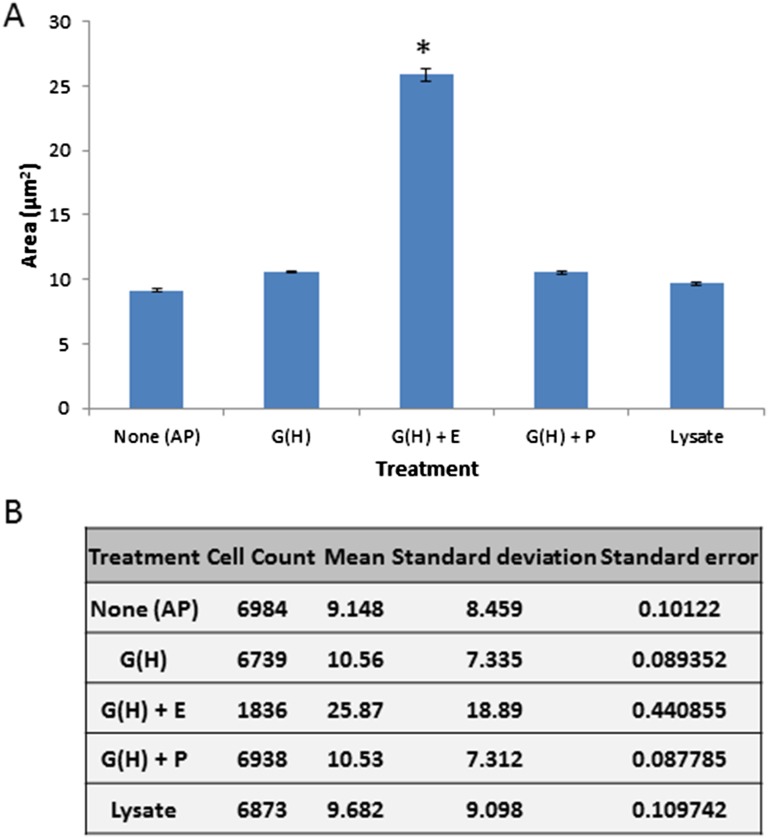
Characterization of ex vivo growth of B. bacteriovorus on ghost cells. (A) Viable counts of B. bacteriovorus 109J 24 and 48 h after inoculation of ∼2 × 107 pfu/mL AP cells with ghost cells containing Hepes buffer [G(H)], ghosts containing prey extract [G(E)], ghosts mixed with prey extract [G(H)+E], ghosts mixed with PYE medium [G(H)+P], prey lysate (Lysate), or with ghost cells after vigorous vortexing [G(E) VTX]. Values are averages of three independent experiments, each performed in triplicate. Error bars indicate SD. (B) Growth and septation of B. bacteriovorus 109J (∼2 × 107 pfu/mL) incubated with ghost cells and prey extract, measured as changes in average cell area (in square micrometers) of a representative batch monitored by FISH coupled with image cytometry. Filaments reach maximal area (in square micrometers) 16 h after inoculation. Error bars indicate SE. The number of images analyzed per time point were 0 h, 7,261; 4 h, 7,774; 8 h, 7,633; 12 h, 7,269; 16 h, 8,143; 20 h, 7,047; and 24 h, 6,775. (C) Area (in square micrometers) of AP cells taken from a predation culture (AP) or after 16 h of incubation in various conditions (abbreviations are as in A). The area (in square micrometers) was monitored as in B. Sixty percent of the predatory cells elongate (values below the “Elongated” line threshold) only in the presence of ghost cells and prey extract. The number of images analyzed per treatment were AP, 6,984; lysate, 6,873; G(H), 6,739; G(H)+P, 6,938; and G(H)+E, 1,836. The population incubated with G(H)+E elongated significantly and increased its average cell area (one-way ANOVA: df = 4, F = 1,328.8, P = 0; Tukey’s HSD post hoc test: P < 10−4), compared with the other treatments in which the cell area distribution was similar to that of untreated AP cells (for more information see Fig. S4). (D) DAPI stain of filamentous ex vivo growth of B. bacteriovorus cultured for 16 h with ghost cells and prey extract. Filaments are composed of 20 ± 5.7 daughter cells (distinct nucleoids) per filament. Growth is not confined to the ghost interior.
Fig. S5.
Increase in B. bacteriovorus’ cell area requires both ghost cells and prey extract. (A) Approximately 2 × 107 pfu/mL AP cells were harvested from a predation culture (AP) and after further incubation for 16 h (the time when filamentous GP cells reach their maximal cell area ex vivo; Fig. 3B) with ghost cells containing Hepes buffer [G(H)], ghosts mixed with prey extract [G(H)+E], ghosts mixed with PYE medium [G(H)+P], or with prey lysate (Lysate). Cell area (in square micrometers) was monitored by FISH coupled with image cytometry. Incubation with G(H)+E resulted in a significant increase in average cell area (one-way ANOVA: df = 4, F = 1,328.8, P = 0; Tukey’s HSD post hoc test: *P < 10−4) compared with the other treatments, in which the average cell area was similar to that of untreated AP cells. Error bars indicate SE. (B) Statistical data of the five populations.
Predator Growth Requires Prey Envelopes and Prey Extract.
Prey ghosts supplemented with debris-free prey extract were necessary to promote B. bacteriovorus 109J growth and to sustain its full life cycle (Figs. 1 and 3 A and C and Fig. S5). Mixing both components together (exogenous application) or loading envelopes with prey extract during their production were similarly effective (Fig. 3A). About 60% of the B. bacteriovorus cells elongated, exhibiting the characteristic filamentous growth of the GP as polynucleoid filaments (Fig. 3 C and D), typically attaining maximal length 16 h postinoculation (Fig. 3B) as determined by image cytometry. The average population cell area (25.87 ± 18.89 μm2) was significantly higher than the average area attained in the other treatments [one-way ANOVA: df = 4, F = 1,328.8, P = 0; Tukey’s honestly significant difference (HSD) post hoc test: P < 10−4] (Fig. S5). The attainment of maximal length was followed soon after by septation, yielding 20 ± 5.7 AP daughter cells per GP cell (Fig. 3D). Filamentous growth was observed inside the prey envelopes but also epibiotically, with the predator externally attached to the ghosts. Extended filaments also could be observed unattached to ghosts (Fig. 1 A, g and h). Because growth was not achieved in the absence of ghosts, we postulate that these filaments were originally bound to ghosts that later disintegrated. No growth was observed when B. bacteriovorus was incubated with (i) prey extract alone, (ii) prey lysate alone, which comprises prey cell debris in addition to the soluble components of the cell, or (iii) peptone yeast extract (PYE), a rich medium, even when supplemented with prey envelopes (Fig. 3A). Furthermore, a predator mutated in pcnB, a mutation that enables type I HI mutants to become completely axenic (37), still required the presence of ghosts for growth (see below and Fig. 5).
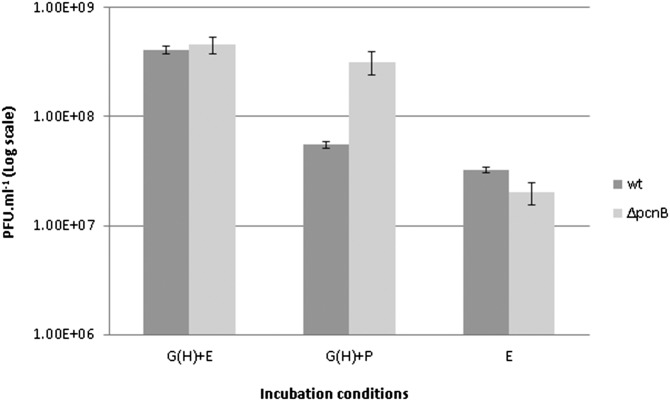
Fig. 5.
pcnB deletion mutant bypasses the requirement for prey extract. Approximately 2 × 107 pfu/mL of B. bacteriovorus HD100 AP cells and of B. bacteriovorus HD100 ∆pcnB AP cells were incubated separately for 24 h with ghost cells and prey extract [G(H)+E], with ghosts and PYE medium [G(H)+P], or with prey extract alone (E). The number of progeny was determined by viable counts. In contrast to the WT strain that was unable to proliferate, the ∆pcnB mutant replicated when cultured with ghost cells and PYE medium. Values are averages of three independent experiments, each performed in triplicate. Error bars indicate SE.
These results strongly suggest that a cell envelope-associated cue(s) is required for growth and is uniquely provided by the ghosts, whereas a second cue(s) is included in the soluble fraction of the prey extract. Evidently these two cues are spatially and temporally separated: A membrane-associated component is sensed first, priming the cell and enabling it to exploit a second soluble cue that promotes growth initiation, i.e., phase transition (henceforth the two terms “growth initiation” and “phase transition” are used interchangeably). Growth induction by prey ghosts was not a specific phenomenon of the strain used in these experiments; type strain HD100 also was able to grow ex vivo when presented with E. coli ghosts and prey extract (see Fig. 5). Ghosts were found to be quite stable and sturdy: when kept at 4 °C, their growth-promoting activity was retained for several weeks. Furthermore, vigorous mixing did not hinder their activity (Fig. 3A).
The ex Vivo System Supports the Growth of WT Predators.
Type I HI mutants arise at low frequency (10−7) and can grow saprophytically in the presence of prey extract (32, 37). To confirm that prey extract-dependent ex vivo growth did not result from the selection of HI mutants but was still seen in WT cells, DNA was purified from AP cells and from ex vivo-cultivated GP cells. bd0108, a marker gene of the HI phenotype (46), was subjected to deep sequencing. We posited that if ex vivo cultivation selects for host independence, then mutated alleles of bd0108 should dominate. To assure that the observed bd0108 genotype was stable after ex vivo growth, AP cells originating from ex vivo cultures were incubated further with live prey. DNA was extracted from the following generation of AP cells, and the bd0108 locus was sequenced. Of 3,180, 11,332, and 1,3465 reads obtained from AP cells, ex vivo GP cells, and AP cells from progeny samples, respectively, 98.48, 99.06, and 99.05 matched the WT allele of bd0108 (Table S1), suggesting that the WT genotype was conserved during and following ex vivo growth.
Table S1.
Allele typing of the bd0108 gene in B. bacteriovorus cell populations during and following ex vivo cultivation
| Population | No. of reads | No. of WT bd0108 reads | Percentage of WT bd0108 reads |
| AP cells | 3,229 | 3,180 | 98.48 |
| Ex vivo GP cells | 13,594 | 11,332 | 99.06 |
| Postpredation ex vivo progeny AP cells | 11,440 | 13,465 | 99.05 |
Genomic DNA was purified from B. bacteriovorus AP cells, from GP cells after 16 h ex vivo cultivation with ghosts and prey extract, and from AP cell progeny derived from ex vivo cultivation after a full growth cycle on live E. coli prey. The bd0108 gene was amplified and sequenced by high-throughput sequencing. Approximately 99% of the reads in the three populations were of the WT bd0108 allele. Mutations in bd0108 are hallmarks of HI variants; therefore this gene is used as a marker for host dependence. The high conservation of the WT bd0108 allele indicates that ex vivo conditions are not enriching for HI mutants.
Genuine Cell-Cycle–Regulated Gene Expression Occurs During ex Vivo Growth.
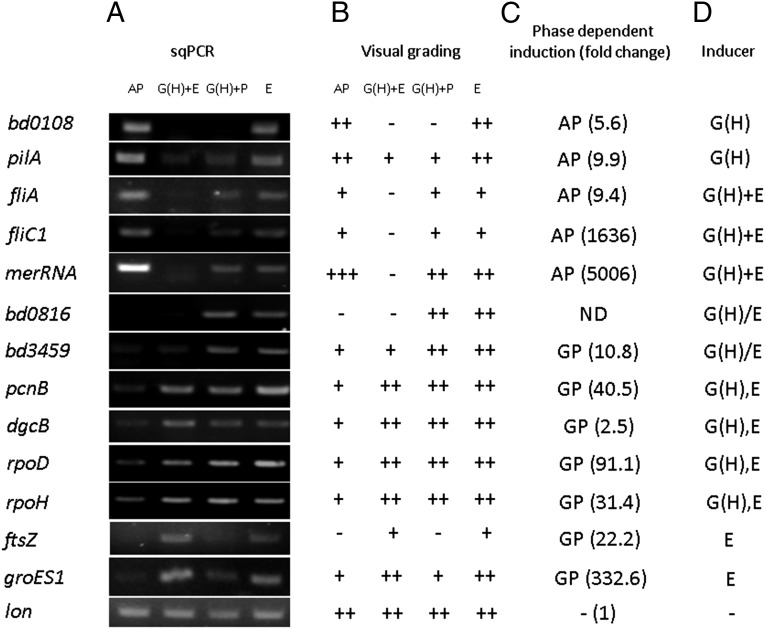
Gene expression of AP and GP B. bacteriovorus HD100 was recently shown to be almost completely exclusive (47). This cell-cycle–based transcriptional separation was exploited to characterize the ex vivo system further by measuring the expression of chosen AP and GP marker genes of B. bacteriovorus HD100 in ex vivo cultures by semiquantitative PCR. All the AP-specific genes, i.e., bd0108, pilA (bd1290), fliA (bd3318), fliC1 (bd0604), and merRNA (47), as well as the GP-specific genes pcnB (bd3464), dgcB (bd0742), rpoD (bd0242), rpoH (bd3314), groES1 (bd0097), and ftsZ (bd3189) [except the evenly expressed control gene lon (bd3749)] exhibited the expected phase-dependent expression: AP-expressed genes were down-regulated when incubated with ghosts and prey extracts, and GP-expressed genes were up-regulated [Fig. 4, AP and G(H)+E lanes]. This result confirmed that the phenotypes and physiologies observed in the ex vivo system were consistent with their predicted gene expression.
Fig. 4.
Prey cues control gene expression in AP B. bacteriovorus HD100. (A) Total RNA was purified from fresh AP cells before (AP, lane 1) and after 6 h of incubation with ghost cells and prey extract [G(H)+E, lane 2], with ghosts and PYE-rich medium [G(H)+P, lane 3], or with prey extract alone (E, lane 4). Transcript levels of selected genes were assessed by semiquantitative RT-PCR. (B) Visual grading of the band intensity of the transcription profile shown in A: −, no band; +, low intensity; ++, medium intensity; +++, high intensity. (C) Phase-dependent induction of the genes shown in A obtained by transcriptomic analysis of AP and GP B. bacteriovorus growing with E. coli prey (46). (D) Combinatorial effects of ghost cells and prey extract on B. bacteriovorus gene expression. Specific effect: G(H), ghosts (bd0108, pilA); E, prey extract (ftsZ, groES1). Additive effect: G(H)+E (merRNA). Synergistic effect: G(H)+E (fliA, filC1). Nonadditive effect: G(H),E (pcnB, dgcB, rpoD, and rpoH). Antagonistic effect: G/PE (bd0816, bd3459). No effect: - (lon).
Spatially and Temporally Separated Cues Affect the Transition from AP to GP.
The modularity of the ex vivo system enabled us to explore the GP-inducing cues further and to analyze the transcriptional effect elicited by each, separately and together, using the selected marker genes. In addition to the growth-promoting conditions (ghosts and prey extract), predatory AP cells were incubated with either ghosts and PYE [G(H)+P, in Fig. 4; and thereby were provided with only the first cue and nutrients] or with prey extract (E, in Fig. 4; which provided only the second cue). Down-regulation of AP genes was nonuniform: bd0108 and pilA were specifically suppressed by ghost cells in any given treatment [G(H)+PYE and G(H)+E, in Fig. 4], the former being completely silenced. merRNA was also down-regulated by the presence of ghost cells [G(H)+PYE, in Fig. 4] and was further silenced in the presence of both cues together [G(H)+E, in Fig. 4]. fliA and fliC1 were not affected by the first cue alone [G(H)+PYE, in Fig. 4] and similarly required both cues [G(H)+E, in Fig. 4] to be silenced. Similarly, expression of the GP genes groES1 and ftsZ was selectively induced by the prey extract (which provided the second cue), whereas expression of the GP genes pcnB, dgcB, rpoD, and rpoH was brought about by either cue or by both. Surprisingly, the genes bd0816 and bd3459, which are involved in peptidoglycan restructuring (48), a function essential for the transformation of the prey cell into a bdelloplast, were up-regulated only when either cue was provided but were repressed in the presence of both. Genes affected by both cues presented additive (merRNA), synergistic (fliA and fliC1), nonadditive (pcnB, dgcD, rpoD, and rpoH), or antagonistic (bd0816 and bd3459) responses (Fig. 4). Apparently, not all genes are affected by the cues, because lon was evenly expressed under all conditions, regardless of the cues present, constituting (to our knowledge) the first bona fide constitutive gene in B. bacteriovorus (Fig. 4). Finally, earlier work had shown that deletion of pcnB, encoding for poly(A) polymerase, alleviates growth dependency on prey extract in type II HI mutants (37). We hypothesized that a ∆pcnB mutant in an otherwise WT background would create a strain still dependent on the presence of ghosts but independent of prey extract. Indeed, a ∆pcnB strain proved capable of growth and replication when cultured with ghosts and prey extract or with ghosts and PYE; however, no growth was observed when the culture was supplemented with prey extract only, demonstrating that PcnB represses the transduction of the soluble cue present in the prey extract that is required to initiate growth (Fig. 5). This result provides genetic evidence that, although prey extract alone is nutritionally adequate to support B. bacteriovorus growth, it is not sufficient to promote ex vivo growth of WT cells.
Discussion
In this study we present a novel ex vivo system in which WT B. bacteriovorus grows in a medium supplemented with intact prey cell envelopes and prey extract. We show that the system successfully mimics predation on live prey, because the ex vivo GP is morphologically, genetically, and physiologically indistinguishable from the genuine GP. Based on our data, we define a stepwise sensory-based AP-to-GP phase transition that defines an intermediate uncharacterized transitory phase. This transitory phase exhibits a third, distinct gene-expression profile, making B. bacteriovorus’ cell cycle triphasic, and constitutes a previously unidentified checkpoint that we posit is set to determine the availability of prey-derived nutrients before the predator cell is committed to growth.
Previous studies suggested that prey extracts might elicit prey-independent growth of WT B. bacteriovorus (40, 41). However, we were unable to replicate these results. Follow-up studies found that although prey extracts sustained extracellular filamentous growth and DNA replication, they could not promote the differentiation of the nonreproductive AP cell to the GP (32–35). This result suggested that two separate prey-derived cues, one in the soluble prey extract fraction and the other hitherto undefined, were necessary for the AP-to-GP transition to occur (33). We therefore hypothesized that (i) the undefined cue is associated with the prey cell envelope and is absent from the prey extract (this cue therefore is detected during the interaction between the predator and the prey cell) and (ii) because both cues are required for phase transition, they act differentially upon the AP cell. The cues and their effects cannot be differentiated in live prey, but they can be put in evidence using the ex vivo cultivation system for WT B. bacteriovorus.
Upon encounter of BALO with prey, nonspecific, transient attachment is followed by stable binding, suggesting that the latter is coupled to recognition. It has been proposed that components of the outer membrane (components of the prey envelope) are recognized by the predator (49, 50), bringing about the initial stages of the interaction. Here, prey envelopes altered by the expression of the pore-forming lysis E gene encoded by bacteriophage PhiX174 (40, 51) did not enable stable attachment, and bacterial cell lysates containing debris of the cell envelope did not promote growth, indicating that the integrity of the prey cell envelope is an important determinant of recognition and/or attachment. In contrast, B. bacteriovorus stably attached to and penetrated intact prey envelopes (Figs. 1 A, a–e and 2A and Fig. S4 A–D) that enabled ex vivo growth when supplemented with prey extract. This finding locates the initial elicitor(s) for recognition to an intact prey envelope, spatially and temporally separated from the second soluble component(s) that controls growth initiation and also sustains it (34). Thus the first interaction between the predator and the prey requires cell–cell contact; the requirement for cell–cell contact is abrogated in type I HI mutants defective in bd0108, an AP-expressed gene that is turned off rapidly with attachment and penetration (32, 37), effectively relieving the need for the prey envelope.
It was proposed recently that Bd0108 controls the extrusion/retraction of a type IVa pilus in B. bacteriovorus (52) that is essential for predation and prey entry but is not involved in prey attachment (53–55). Recognition thus includes signaling and stable attachment, and both processes are sustained by prey ghosts. The pilus is anchored to the prey inner membrane before prey penetration and during the GP (54), suggesting that sensing and transduction of the first cue is continuous during growth rather than relevant only to prey penetration. The interaction with the inner membrane is maintained with ghosts during ex vivo growth (Fig. 1 A, e and Fig. S4D). Inactivation of the type IVa pilin-coding gene pilA in HI mutants results in apiliated cells (53). Therefore, the interaction with the ghost inner membrane probably is mediated similarly by type IVa pili (Fig. 1 A, a).
AP cells incubated with ghost preparations and nutrients but without prey extract were not altered in their morphology but showed changes in gene expression: bd0108 was silenced, and merRNA [a highly expressed putative cyclic di-GMP riboswitch (47)] and pilA were down-regulated, whereas genes involved in bdelloplast formation through the remodeling of the prey peptidoglycan were up-regulated, i.e., the specific peptidocarboxylase-coding genes bd0816 and bd3459 (48). In parallel, some GP genes (pcnB, dgcB, rpoD, and rpoH) were induced, priming the cell for growth. Importantly, ftsZ, encoding for a major septation protein that probably is required toward the end of the GP (39), was not induced under these conditions. Moreover, the core of AP genes, as determined by fliC1 and fliA [the latter encoding for the central AP regulator σ28 (47)], was not affected and remained expressed, extending the span of the σ28 regulon beyond the AP to the early stages of predation, when flagellar motility is evidently still functional (Movie S1). These conditions, which are not attainable when using live prey, reveal a transitory phase during which the predatory cell still appears morphologically as an AP cell but actually expresses a different gene set and is capable of shaping and probably consuming prey but remains nonreplicative.
Strikingly, a different gene-expression profile was obtained in AP cells incubated with prey extract without ghosts: bd0108 and pilA were not silenced, and groES1 and ftsZ, both strongly enhanced in the GP (40, 56), were specifically induced. The other GP marker genes (pcnB, dgcB, rpoD, and rpoH) as well as the bdelloplast formation genes (bd0816 and bd3459) and merRNA responded similarly as when incubated with the ghost preparation. Thus, the complement of GP genes (47) is composed of subsets that respond differentially to the prey cues: GP-initiation genes are elicited at the prey-sensing and penetration stage upon detection of the cell envelope-borne cue, whereas core GP genes are up-regulated by the soluble prey-derived cues. Finally, the presence of both cues, i.e., prey ghosts and prey extracts, led to the repression of genes that are differentially controlled during the AP and during prey penetration, i.e., the central AP gene regulator coding gene fliA and consequently the flagellar marker gene fliC1, merRNA, the expression of which decreased below detection level (47), and the bdelloplast niche construction genes (48). Thus, the combined sensing of the two cues has a general effect on gene expression and is required to bring about a full, genuine GP, the B. bacteriovorus equivalent of the exponential growth phase.
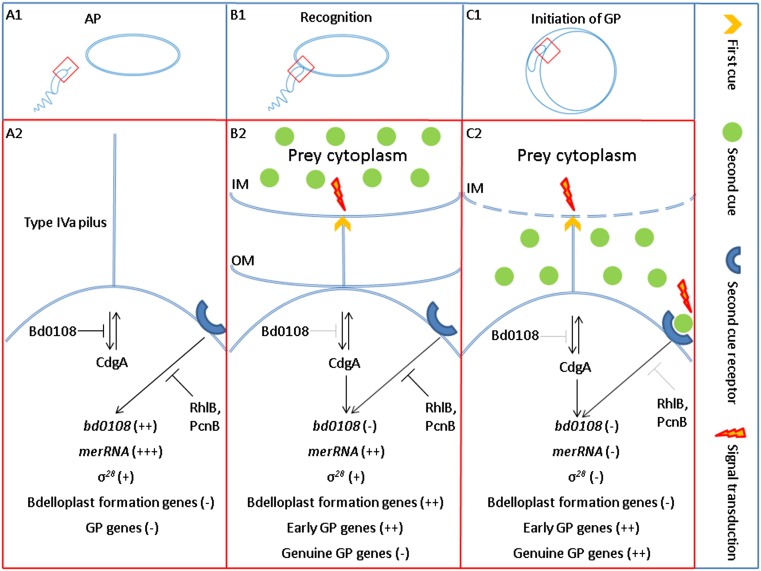
To interpret these data and integrate them with current knowledge, we propose the following model for B. bacteriovorus’ growth control (Fig. 6): Being an obligate predator, B. bacteriovorus must reach and exploit a spatially and temporally discontinuous food source (prey cells). To do so, the bacterium must (i) distinguish between prey vs. nonprey cells, because only the prey cell represents a potential source of nutrition and (ii) assess availability of nutrients, because only when nutrients are available does commitment to the GP become fruitful. We thus propose that this scheme constitutes a control on the quality of the interaction, i.e., prey nutritional value. During the AP, prey sensing is down-regulated: Early transduction of the first, envelope-borne cue via the type IVa pilus (52) is inhibited by Bd0108 (Fig. 6A2). The second, prey extract-derived cue is transduced by a late signaling pathway through an unknown receptor. This pathway is repressed by the RNA degradosome subunits RhlB or PcnB (Fig. 6A2). The AP cell expresses a default transcriptional profile, the AP program, during which merRNA is massively transcribed and bd0108 and the σ28 regulon are expressed, but bdelloplast-formation genes and GP genes are silent. When an AP cell stably binds to a prey cell through a still unknown mechanism, concomitant sensing of the first cue, located in the cytoplasmic membrane (54), alleviates Bd0108 inhibition, altering the extrusion/retraction state of the type IVa pilus found at the nonflagellated, invasive pole with which Bd0108 is associated (52), leading to signal transduction. This process may be mediated, at least in part, through CdgA, a cyclic di-GMP effector required for efficient penetration of the prey cell (57) that interacts with the pilus’ regulatory protein complex (55). The cyclic di-GMP machinery is essential for both prey entry and progeny escape at the end of the predator’s replicative cycle (57), indicating that this second messenger is involved in transducing the first cue. The second, late signal transduction remains inactive (Fig. 6B2). Early sensing then shifts the transcriptional profile from an AP program to a recognition program (the transition phase), reflected by complete bd0108 silencing and partial merRNA repression, possibly releasing cyclic di-GMP (47); bdelloplast-construction genes (e.g., peptidoglycan- shaping genes) and early GP genes are induced, enabling penetration and bdelloplast formation as well as the expression of growth-related functions. However, the σ28 regulon remains active (Fig. 6B2). At this stage, the predatory cell does not commit to entering the GP, and, although morphologically it still resembles an AP cell, it is in an intermediary state and, in fact, is stopped at a checkpoint (Fig. 4 and Movie S1). This transitory phase can be maintained for 1 h, the time that elapses between prey entry and initiation of growth (34), during which the bdelloplast’s cytoplasmic membrane becomes completely permeable to small molecules (58). Relief from this point and progress into the GP requires that the second, soluble cue, probably originating from the prey cytosol (35), be released into the periplasm of the bdelloplast, where the predator can sense it. Thus, consequent to spatial separation, the sensing of the first, envelope-borne cue is temporally separated from the sensing of the second, cytosolic cue, bringing about successive waves of change in gene expression. While the predator remains attached to the cytoplasmic membrane, both cues are sensed concomitantly, resulting in shutting off the σ28 regulon and the bdelloplast-formation genes, in total silencing of merRNA, and in the up-regulation of core GP genes, thus yielding a full-fledged GP cell (Fig. 6C2).
Fig. 6.
Model for the stepwise phase transition in B. bacteriovorus. For simplicity, only the prey cell is portrayed with outer and cytoplasmic membranes. A2, B2, and C2 are magnifications of the predatory pole enclosed by red rectangles in A1, A2, and A3, respectively. (A1) A nonreplicative B. bacteriovorus AP cell searches for a prey. (A2) During AP there is no prey sensing. Early signal transduction, mediated by the type IVa pilus and assembled at the nonflagellated pole, is inhibited by Bd0108. Late signal transduction, introduced by an unknown receptor, is repressed by the RNA degradosome subunits RhlB or PcnB. The AP cell expresses a default transcriptional profile, the AP program, in which merRNA is massively expressed, bd0108 and the σ28 regulon are up-regulated, and bdelloplast-formation genes and GP genes are silenced. (B1) The AP cell stably attaches to and recognizes a prey cell. (B2) Recognition, but not attachment, is mediated by a change in the extrusion/retraction state of the type IVa pilus (shown here as a retraction), achieved after sensing of the first cue located in the cytoplasmic membrane. As a result, the inhibition by Bd0108 is alleviated, setting off signal transduction, possibly in part through CdgA, a cyclic di-GMP effector required for efficient penetration to the prey cell. CdgA interacts with the pilus’ regulatory protein complex (55). Early sensing induces the recognition program: bd0108 is completely silenced, and merRNA levels decline, but the σ28 regulon remains active, and the bdelloplast-formation genes and the early GP genes are induced, enabling penetration and bdelloplast formation as well as the production of growth-related functions. (C1) Inside the bdelloplast periplasm the predator remains attached to the cytoplasmic membrane, consumes the prey, and initiates growth. (C2) Perforation of the cytoplasmic membrane (58, 61, 62) leads to the release of the second cue from the cytosol. Sensing of the two cues promotes a second, timely alteration of the transcriptional profile to the GP program in which all active AP genes (merRNA and the σ28 regulon) as well as the bdelloplast-formation genes are down-regulated and genuine GP genes are induced to enable de novo DNA replication and growth.
The growth-promoting activity induced by the second cue is concentration dependent (35) but does not contribute directly to nutrition (34, 35). We propose that its occurrence may signal that enough prey-derived nutrients are present in the intracellular medium to support growth, thereby acting as a marker of prey quality and enabling growth initiation. Unlike free-living bacteria that adjust their growth rates according to nutrient concentrations, BALOs meet discrete food packets that are sufficient or insufficient to sustain their growth and replication. Thus, the predator may need to assess the quantity rather than the concentration of the available nutrients. Nutrient concentration may affect BALO's growth decisions otherwise: it had been previously shown that under oligotrophic conditions, the bdelloplasts of Bdellovibrio strain W and of marine BALOs enter a nonreplicative dormant state, and that the predator is resuscitated upon addition of nutrients to the external medium. Hence, nutrient availability serves as a proxy for the presence of enough prey for the resuscitated predatory cell upon its release from the bdelloplast (45, 59). It is noteworthy that the two schemes may not be mutually exclusive.
We posit that the stepwise growth control illustrated here decouples predation from growth. This scheme restricts the investment made by the AP cell under uncertain growth-enabling conditions to a limited and possibly reversible expression profile, while conserving an AP-like status, uncommitted to growth, until the environment provides the adequate second cue for growth. In effect, this scheme represents a prudent betting strategy. In contrast to the successful recognition of the carefully produced prey ghosts, in nature B. bacteriovorus, an effective predator of biofilms (56, 60), encounters prey remains from which the first cue is hypothesized to be easily lost, as observed with prey lysate and perforated envelopes (Results).
Interestingly, small E. coli cells are preyed upon without supporting B. bacteriovorus’ growth and replication (31). In soil and water, where B. bacteriovorus is found, a very large fraction of the bacterial community consists of small cells, <0.5 µm in width (58, 61, 62), not larger than B. bacteriovorus AP cells themselves (12). Our model predicts that B. bacteriovorus would prey successfully on such poorly nutritious prey cells, which may be too small to accommodate a full intracellular growth cycle but do have a limited nutritional value.
Prey quality is an important parameter in animals because it affects behaviors and ultimately fitness (63, 64). In bacteria, fitness (generation time), cell growth, and division are tightly coupled to feeding. The ability to separate the different cues and determine their effects through the ex vivo system helped uncover how predatory bacteria assess the internal nutritional status of their prey to inform cell-cycle decisions.
Materials and Methods
Bacterial Strains and Growth Conditions.
B. bacteriovorus strains HD100-S1 and 109J-S1 were grown in two-membered suspension cultures with ∼2 × 109 pfu/mL of E. coli ML35 (ATCC no. 43827) in Hepes buffer (25 mM Hepes, 2 mM CaCl2⋅2H2O, 3 mM MgCl2⋅6H2O, pH 7.4–7.6) supplemented with 50 µg/mL streptomycin at 28 °C and 180 rpm. Fresh AP cells were obtained from overnight cultures and used for ex vivo cultivation experiments. Plaques, used for viable counts and isolation, were achieved by double-layer agar plating: 100 µL of serially diluted predator and 7 × 108 E. coli ML35 cells were resuspended in 3 mL of soft dilute nutrient broth (DNB) agar [0.7 g/L nutrient broth, 2 mM CaCl2⋅2H2O, 3 mM MgCl2⋅6H2O, pH 7.4–7.6 and 0.7% agar) and overlaid on DNB agar (0.7 g/L nutrient broth, 2 mM CaCl2⋅2H2O, 3 mM MgCl2⋅6H2O (pH 7.4–7.6), and 1.5% agar]. E. coli strains were cultured at 37 °C and 180 rpm in LB broth supplemented, when stated, with 100 µg/mL ampicillin, 50 µg/mL diaminopimelic acid (DAP), and 50 µg/mL kanamycin or 20 µg/mL tetracycline. Strains and their origins are listed in Table S2.
Table S2.
Bacterial strains and plasmids
| Bacterial strain or plasmid | Genotype | Reference or source |
| E. coli ML35 | Type strain used as prey model | (29, 74) |
| E. coli S17-1 | TpR,SmR, recA, thi, pro, hsdR-M+RP4: 2-Tc:Mu: Km, Tn7, λpir | (75) |
| E. coli S17-1:: pSSK10:∆pcnB | Used as a donor for conjugating pSSK10:∆pcnB into B. bacteriovorus HD100-S1 | This study |
| E. coli S17-1:: pRK-kPR-cI-E-SNA | Used for production of ghosts by over expression of the PhiX174 lysis E gene | This study |
| E. coli AT984 | λ−, e14-, dapE6, relA1, spoT1, thiE1; DAP auxotroph | (76, 77) |
| E. coli AT984:: pBADsulA-FLAG | Used for production of intact envelopes by osmotic shock | This study |
| B. bacteriovorus 109J-S1 | Streptomycin-resistant derivative of the type strain 109J | Laboratory collection |
| B. bacteriovorus HD100-S1 | Streptomycin-resistant derivative of the type strain HD100 | (37) |
| B. bacteriovorus HD100-S1 ∆pcnB | pcnB mutant | This study |
| pSSK10 | oriR6K, mobRP4, cat, sacB, kmR; Suicide vector used for gene knockouts | (65) |
| pRK-kPR-cI-E-SNA | Used for production of ghosts by temperature-based induction of the PhiX174 lysis E gene and the Staphylococcal nuclease A gene. | (42) |
| pBADsulA-FLAG | pBAD24 harboring sulA CDS, encoding for the division inhibitor SulA, under the ara promoter, | (78) |
Plasmids and Primers.
Plasmids used in this study are listed in Table S2. Oligonucleotides used in this study are listed in Table S3.
Table S3.
Oligonucleotides used in this study
| Name | Sequence (5′-3′) | Description | Reference or source |
| BDE525 | GATCCCTCGTCTTACCGC | B. bacteriovorus 16S rRNA probe (5′ Cy3) | (69) |
| F_bd0108 | CCTCTTGGATATCATTGCTCCC | bd0108 454 sequencing | (46) |
| R_bd0108 | TGTGTAAGGCGAGTGTCAGG | bd0108 454 sequencing | (46) |
| F_sq_bd0108 | GCTTCTCCTTTGCGGGAACAG | spPCR primer | This study |
| R_sq_bd0108 | AGTCGCCCAAAGCCGATGTC | spPCR primer | This study |
| F_sq_pilA | GGTTTCTCGCTTGTAGAGT | spPCR primer | This study |
| R_sq_pilA | AGCTTTCTCTGACGTGTACA | spPCR primer | This study |
| F_sq_fliA | GAGAATATGCTCCGCTAA | spPCR primer | This study |
| R_sq_fliA | CGATGACACCTGCAGAA | spPCR primer | This study |
| F_sq_fliC1 | CAACAAACACCGCATCTAT | spPCR primer | This study |
| R_sq_fliC1 | TCCTTTGAAGTTCTCAGAAAT | spPCR primer | This study |
| F_sq_merRNA | GGAGGTTCCTAGGGTTATA | spPCR primer | This study |
| R_sq_merRNA | TTCCTCAGGTGACTCATGA | spPCR primer | This study |
| F_sq_bd0816 | AAAGCTGGGCGTCGACTATC | spPCR primer | This study |
| R_sq_bd0816 | GCGGTTCAGTTCAGATATCAG | spPCR primer | This study |
| F_sq_bd3459 | CAACTGCCAACGGTTCTTACG | spPCR primer | This study |
| R_sq_bd3459 | TTCCGCCAACCACGCCAAC | spPCR primer | This study |
| F_sq_pcnB | GGATTCATCCGAAAGATT | spPCR primer | This study |
| R_sq_pcnB | GACGGAAAGTTGCAACTT | spPCR primer | This study |
| F_sq_dgcB | ATGGCTCACAACGATGAC | spPCR primer | This study |
| R_sq_dgcB | GCCGATGACGATATCACT | spPCR primer | This study |
| F_sq_rpoD | AAGAGCCTGTGAAGGTTCT | spPCR primer | This study |
| R_sq_rpoD | GATGGAGCGACAATTTCA | spPCR primer | This study |
| F_sq_rpoH | CTCCAAAGCGCTGACTT | spPCR primer | This study |
| R_sq_rpoH | CAGGGTCTTTGCTTTCAA | spPCR primer | This study |
| F_sq_ftsZ | TCGTTGCCAATACAGACA | spPCR primer | This study |
| R_sq_ftsZ | ACGATCTCATTGTAGGATTC | spPCR primer | This study |
| F_sq_groES1 | GCCCACTTCATGACAGAA | spPCR primer | This study |
| R_sq_groES1 | CTTTATCGCCAACTTTAACT | spPCR primer | This study |
| F_sq_lon | CTTTGCAATCGGAACAGT | spPCR primer | This study |
| R_sq_lon | AACGATGTTGGTAGGATCTT | spPCR primer | This study |
| F_UP_KO_pcnB | CCTTTTTAACCCATCACATATACCTGCCGTTTATACTCCGGTCTGAAGCTC | Primer | This study |
| R_UP_KO_pcnB | CAAAATTCAAACCTGAATAACA | Primer | This study |
| F_DN_KO_pcnB | CGGTCTTTTGTTATTCAGGTTTGAATTTTGGTCCGCTAAGGGTCTCAG | Primer | This study |
| R_DN_KO_pcnB | CGTTTGGATTGCAACTGGTCTATTTTCCTCTAGCGCGAACATCGAACAA | Primer | This study |
| F_Val_KO_pcnB | ATTCTGAGAATTTCAAGTTTC | Primer | This study |
| R_Val_KO_pcnB | TATTATGAAAACATGGAAGG | Primer | This study |
Prey Extracts.
E. coli ML35 was cultured overnight in LB broth at 37 °C and 180 rpm. The culture was resuspended in Hepes buffer, concentrated 50-fold, and lysed by sonication. The soluble fraction of the crude lysate was centrifuged twice (20,000 × g for 30 min at 4 °C) and filtered through 0.2-µm syringe filters (Whatman). Protein concentration was determined by the Bradford protein assay (Quick Start Bradford ×1 protein dye reagent; Bio-Rad), and the debris-free extract was divided to aliquots, flash-frozen in liquid nitrogen, and kept at −20 °C until use.
pcnB Deletion Mutant Strain.
The pcnB deletion procedure is based on the protocol described in refs. 65 and 66. The suicide vector pSSK10:∆pcnB, harboring the pcnB knockout cassette, i.e., pcnB CDS flanking regions (each 1-kb long) ligated to each other, was constructed following the restriction-free cloning procedure for multicomponents assembly (66). The vector was introduced into B. bacteriovorus HD100-S1 by conjugation in which an E. coli S17-1 transformant, cultivated with 50 µg/mL kanamycin, was used as a donor. Exconjugants were inoculated into a predation culture (Hepes buffer and 2 × 109 cfu/mL E. coli ML35), and recombinants were selected for with 50 µg/mL kanamycin. After three subsequent rounds of predation without kanamycin, leading to plasmid excision, 5% sucrose was added as a substrate for the vector-encoded levansucrase (SacB) to select against remaining recombinants. Predator cells were isolated on semisolid double-agar plates and screened, first for plasmid excision via loss of kanamycin resistance, and next for pcnB deletion by PCR.
Production of E. coli Ghosts by Overexpression of the PhiX174 Lysis E Gene.
The production of E. coli ghosts was based on the method described in ref. 42 with minor adjustments. Briefly, E. coli S17-1 (ATCC no. 47055) transformed with pRK-kPR-cI-E-SNA (courteously provided by Ki Hong Kim, Department of Aquatic Life Medicine, Pukyong National University, Busan, Korea) were cultivated to midlog phase (OD600 = 0.4) in LB broth supplemented with 20 µg/mL tetracycline at 30 °C and 180 rpm. Then, 1 mM MgCl2 and 10 mM CaCl2 were added, and the temperature was shifted to 42 °C, resulting in the expression of the lysis E gene and the Staphylococcal nuclease A gene. After 4 h incubation bacteria were washed with double-distilled water and resuspended in Hepes buffer. The survival ratio was 10−3–10−4, and the resultant ghosts presented porous envelopes (Fig. S2).
Production of E. coli Ghosts by Osmotic Shock.
The protocol for the production of E. coli ghosts was based on the method described in ref. 43, with modifications. E. coli AT984, a DAP auxotroph (Coli Genetic Stock Center no. 4558), transformed with pBAD24sulA-FLAG (courteously provided by Eliora Z. Ron, Department of Molecular Microbiology and Biotechnology, Tel Aviv University, Tel Aviv, Israel and MIGAL Galil Research Center, Kiriat Shmone, Israel), was cultured in LB broth amended with 50 µg/mL DAP and 100 µg/mL ampicillin at 37 °C and 180 rpm to OD600 = 0.2. Then 0.2% arabinose was added to induce sulA for 40 min, resulting in cell elongation by approximately fourfold. The culture was washed twice to remove DAP (10,000 × g for 5 min), resuspended in LB broth with 0.2% arabinose and 100 µg/mL ampicillin, and incubated for 20 min at 37 °C, 180 rpm, to ensure complete consumption of residual DAP. Then it was diluted 1:4 with Penassay solution (17.5 g/L antibiotic medium III with 20% sucrose, 9 mM MgSO4, 0.2% arabinose, and 103 U/mL penicillin G) and incubated for 120 min at 37 °C. During that period, bacterial envelopes were partially lysed and swelled but did not erupt because of the high hypertonicity of the solution. Then, the culture was centrifuged (10,000 × g for 5 min), and the pellet was resuspended in a hypotonic solution (Hepes buffer). This treatment resulted in the opening and immediate resealing of the envelopes, accompanied by the release of the bacterial cell content. For encapsulation of cell extract, Hepes buffer was amended with prey extract (3 mg/mL total protein) before resuspension. The culture was incubated for an additional 15 min with 20 µg/mL DNase I (Sigma-Aldrich) and then was centrifuged gently (1,000 × g for 5 min at 4 °C) to precipitate unaffected cells. The supernatant was collected, centrifuged (10,000 × g for 15 min at 4 °C), and resuspended in Hepes buffer. Particle counting was carried out using a Neubauer chamber and by viable counts on LB plates amended with 100 µg/mL ampicillin and 50 µg/mL DAP.
β-Galactosidase Assay.
The β-galactosidase assay, used to assess the average loss of cytosolic content in ghosts compared with untreated bacteria, was performed as described in ref. 67.
Protein Electrophoresis.
SDS/PAGE was performed to assess the altered protein content in ghosts compared with untreated bacteria, as described in ref. 67.
Ex Vivo Experiments.
AP B. bacteriovorus cells (∼2 × 107 pfu/mL) were incubated at 28 °C and 180 rpm with E. coli AT984 ghosts (∼2 × 109 particles/mL) without additives or (i) encapsulated prey extract (3 mg/mL total protein), (ii) supplemented with exogenous prey extract, or (iii) supplemented with PYE (10 g/L Bacto peptone, 3 g/L yeast extract, 2 mM CaCl2·2H2O, and 3 mM MgCl2·6H2O, pH 7.4–7.6). AP cells also were incubated with crude prey lysate (3 mg/mL soluble protein) or prey extract.
FISH.
The FISH procedure was adapted from ref. 68 to avoid aggregation of the ghosts. Ex vivo cultures were mixed with three volumes of 4% paraformaldehyde dissolved in PBS (140 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4∙7 H2O, and 1.8 mM KH2PO4, pH 7.2) and were incubated overnight (up to 16 h) at 4 °C. The fixed sample was pelleted and washed twice with equal volume of cold PBS (3,500 × g for 10 min) and then resuspended in one fourth of the volume with cold PBS. Five microliters of the fixed sample and 5 ng/µL of the labeled oligonucleotide probe BDE525 (69) were added to 50 µL of prewarmed (46 °C) hybridization buffer (0.9 M NaCl, 20 mM Tris/HCl, 0.1% SDS, pH 7.2). The solution was incubated for 2 h at 46 °C and then was pelleted (6,000 × g for 2 min) and resuspended in 100 µL hybridization buffer. The sample was incubated further for 20 min at 48 °C and then was pelleted (6,000 × g for 2 min), resuspended in 100 µL ice-cold PBS (pH 8.4), and kept on ice in the dark until use.
Imaging Flow Cytometry.
For ghost analysis, E. coli AT984 envelopes were stained with the membrane dye FM4-64 (Life Technologies) and the nucleic acid stain GelStar (Lonza). For ex vivo growth analysis, a streptomycin-resistant derivative of B. bacteriovorus 109J (ATCC no. 43826), termed 109J-S1, was hybridized with a Cy3-labeled probe BDE525 (as described above). The cells were visualized using imaging flow cytometry (ImageStreamX flow cytometer; Amnis Corporation). Approximately 104 cells were collected from each sample, and data were analyzed using image analysis software (IDEAS 6.0; Amnis Corporation). Images were compensated for fluorescent dye overlaps by using single-stain controls. Cells were gated for focused cells using the Gradient RMS feature (70). Beads and debris were separated using the area and side-scatter features. E. coli envelopes were gated based on their membrane integrity, low maximum pixel intensity (the highest-intensity pixel in each cell) and low pixel intensity per cell area of GelStar. Ghosts’ cell area in square micrometers was calculated from the bright-field image. B. bacteriovorus 109J-S1 growth was evaluated by the increase in the quantified area of the Cy3 staining in square micrometers.
Semiquantitative PCR.
A streptomycin-resistant derivative of B. bacteriovorus HD100 (ATCC no. 15356), termed HD100-S1 (37), was incubated for 6 h with prey ghosts and prey extract (3:1 predator:ghost ratio), prey ghosts and PYE, or prey extract only. Predators interacting with ghosts were separated from noninteracting AP cells via filtration and collection of 0.45-µm filters (Sartorius AG). RNA was stabilized using RNAlater (Life Technologies) and purified with the MasterPure kit (EpicentreBio). DNA was digested by the TURBO DNA-free kit (Life Technologies). Reverse transcription was carried out from equal amounts of RNA using random primers (Promega) and the ImProm-II reverse-transcription system (Promega). Semiquantitative PCR was performed using Taq Plus Master Mix (Lamda Biotech).
Bright-Field and Epi-Fluorescence Microscopy.
Microscopic observations and image acquisitions were performed at a magnification of 1,000× with a BX51 phase-contrast and epi-fluorescence microscope (Olympus) equipped with a Digital Sight cooled monochrome CCD camera (Nikon) and NIS-elements, edition 3.0, software (Nikon). Cells stained with DAPI (Sigma-Aldrich) were used to quantify the average number of daughter cells per ex vivo-cultivated GP cell by counting concatenated DAPI-stained (Sigma-Aldrich) genomes in GP cells after ex vivo cultivation for 16 h (Fig. 3D).
Confocal Microscopy.
AP B. bacteriovorus 109J-S1 cells were mixed with E. coli AT984 ghosts under a high MOI (5:1 predator:ghost ratio). The culture was stained with DAPI and FM4-64 and after 5-min incubation the culture was visualized by confocal microscopy, performed with an IX-81 laser-scanning confocal microscope (FV 500; Olympus) equipped with a 405-nm diode laser and 488-nm argon-ion laser with a PlanApo60× 1.4 NA oil immersion objective. To observe DAPI fluorescence, a 405-nm laser for excitation and BA430-460 nm barrier filter were used. For visualization of FM4-64–dyed cells, a 488-nm excitation line and BA610IF emission filter were used. Transmitted light images were obtained using Nomarski differential interference contrast microscopy.
TEM.
Fresh cultures of B. bacteriovorus 109J-S1 incubated with (i) E. coli AT984 ghosts for 1 h and (ii) ghosts and prey extract for 16 h were concentrated by centrifugation (10,000 × g for 5 min 4 at °C). Pellets were mounted on an aluminum disk with a depth of 100 µm (Engineering Office M. Wohlwend GmbH) and covered with a flat disk. The sandwiched samples were frozen in a HPM010 high-pressure freezing machine (Bal-Tec) and subsequently were freeze-substituted in an AFS2 freeze substitution device (Leica Microsystems) in anhydrous acetone containing 2% glutaraldehyde and 0.2% tannic acid osmium tetroxide for 3 d at −90 °C. They then were warmed to −30 °C over 24 h. Samples were washed three times with acetone, incubated for 1 h at room temperature with 2% osmium tetroxide, washed three times with acetone, and infiltrated for 5–7 d at room temperature in a series of increasing concentrations of Epon (Agar Scientific) in acetone. After polymerization at 60 °C, 60- to 80-nm sections were stained with uranyl acetate and lead citrate and were examined in a Tecnai T12 transmission electron microscope (FEI) operating at 120 kV, using an Erlangshen ES500W CCD camera (Gatan).
High-Throughput Sequencing and Sequence Analysis.
bd0108 gene sequences (read length 405 bp) were obtained by 454 sequencing (Mr. DNA). All reads were cleaned using MOTHUR v1.29 (71). First, Fasta and quality data were extracted from the raw SFF file. Sequences were grouped according to barcode and primer, allowing one mismatch to the barcode and two mismatches to the primer. Denoising was achieved by the AmpliconNoise algorithm (72), removing both 454 sequencing errors and PCR single-base errors. Next, sequences were trimmed to remove barcode and primer sequences, all sequences with homopolymers (i.e., AAAA) longer than 8 bp, and all sequences shorter than 300 bp. All sequences were aligned and filtered so that they all overlapped perfectly with no overhang or no-data base pairs, resulting in a 300-bp alignment. To reduce sequencing errors further, preclustering of the sequences was performed based on the algorithm of Huse et al. (73). Pairwise distances were calculated between all DNA reads, which subsequently were clustered into operational taxonomic units (OTUs) at the unique level, meaning that only sequences which were 100% identical were considered to be the same OTU.
Supplementary Material
Acknowledgments
This research was support by Israel Science Foundation Grants 1583/12 and 1290/08 and German-Israeli Foundation for Scientific Research Grant I-1217-342.13/2012.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1515749112/-/DCSupplemental.
References
- 1.Jezbera J, Hornák K, Simek K. Prey selectivity of bacterivorous protists in different size fractions of reservoir water amended with nutrients. Environ Microbiol. 2006;8(8):1330–1339. doi: 10.1111/j.1462-2920.2006.01026.x. [DOI] [PubMed] [Google Scholar]
- 2.Jousset A, et al. Predators promote defence of rhizosphere bacterial populations by selective feeding on non-toxic cheaters. ISME J. 2009;3(6):666–674. doi: 10.1038/ismej.2009.26. [DOI] [PubMed] [Google Scholar]
- 3.Jürgens K. Predation on bacteria and bacterial resistance mechanisms: Comparative aspects among different predator groups in aquatic systems. In: Jurkevitch E, editor. Predatory Prokaryotes. Springer; Berlin: 2006. pp. 57–92. [Google Scholar]
- 4.Pasternak Z, et al. By their genes ye shall know them: Genomic signatures of predatory bacteria. ISME J. 2013;7(4):756–769. doi: 10.1038/ismej.2012.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kandel PP, Pasternak Z, van Rijn J, Nahum O, Jurkevitch E. Abundance, diversity and seasonal dynamics of predatory bacteria in aquaculture zero discharge systems. FEMS Microbiol Ecol. 2014;89(1):149–161. doi: 10.1111/1574-6941.12342. [DOI] [PubMed] [Google Scholar]
- 6.Chauhan A, Cherrier J, Williams HN. Impact of sideways and bottom-up control factors on bacterial community succession over a tidal cycle. Proc Natl Acad Sci USA. 2009;106(11):4301–4306. doi: 10.1073/pnas.0809671106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen H, Athar R, Zheng G, Williams HN. Prey bacteria shape the community structure of their predators. ISME J. 2011;5(8):1314–1322. doi: 10.1038/ismej.2011.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scantlebury DM, et al. Mammalian energetics. Flexible energetics of cheetah hunting strategies provide resistance against kleptoparasitism. Science. 2014;346(6205):79–81. doi: 10.1126/science.1256424. [DOI] [PubMed] [Google Scholar]
- 9.Williams TM, et al. Mammalian energetics. Instantaneous energetics of puma kills reveal advantage of felid sneak attacks. Science. 2014;346(6205):81–85. doi: 10.1126/science.1254885. [DOI] [PubMed] [Google Scholar]
- 10.Mayntz D, Raubenheimer D, Salomon M, Toft S, Simpson SJ. Nutrient-specific foraging in invertebrate predators. Science. 2005;307(5706):111–113. doi: 10.1126/science.1105493. [DOI] [PubMed] [Google Scholar]
- 11.Snyder AR, Williams HN, Baer ML, Walker KE, Stine OC. 16S rDNA sequence analysis of environmental Bdellovibrio-and-like organisms (BALO) reveals extensive diversity. Int J Syst Evol Microbiol. 2002;52(Pt 6):2089–2094. doi: 10.1099/00207713-52-6-2089. [DOI] [PubMed] [Google Scholar]
- 12.Stolp H, Starr MP. Bdellovibrio bacteriovorus gen. et sp. n., a predatory, ectoparasitic, and bacteriolytic microorganism. Antonie van Leeuwenhoek. 1963;29:217–248. doi: 10.1007/BF02046064. [DOI] [PubMed] [Google Scholar]
- 13.Sockett RE, Lambert C. Bdellovibrio as therapeutic agents: A predatory renaissance? Nat Rev Microbiol. 2004;2(8):669–675. doi: 10.1038/nrmicro959. [DOI] [PubMed] [Google Scholar]
- 14.Dwidar M, Monnappa AK, Mitchell RJ. The dual probiotic and antibiotic nature of Bdellovibrio bacteriovorus. BMB Rep. 2012;45(2):71–78. doi: 10.5483/BMBRep.2012.45.2.71. [DOI] [PubMed] [Google Scholar]
- 15.Chanyi RM, Ward C, Pechey A, Koval SF. To invade or not to invade: Two approaches to a prokaryotic predatory life cycle. Can J Microbiol. 2013;59(4):273–279. doi: 10.1139/cjm-2013-0041. [DOI] [PubMed] [Google Scholar]
- 16.Schwudke D, Strauch E, Krueger M, Appel B. Taxonomic studies of predatory bdellovibrios based on 16S rRNA analysis, ribotyping and the hit locus and characterization of isolates from the gut of animals. Syst Appl Microbiol. 2001;24(3):385–394. doi: 10.1078/0723-2020-00042. [DOI] [PubMed] [Google Scholar]
- 17.Iebba V, et al. Higher prevalence and abundance of Bdellovibrio bacteriovorus in the human gut of healthy subjects. PLoS One. 2013;8(4):e61608. doi: 10.1371/journal.pone.0061608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jurkevitch E, Minz D, Ramati B, Barel G. Prey range characterization, ribotyping, and diversity of soil and rhizosphere Bdellovibrio spp. isolated on phytopathogenic bacteria. Appl Environ Microbiol. 2000;66(6):2365–2371. doi: 10.1128/aem.66.6.2365-2371.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gallet R, et al. Predation and disturbance interact to shape prey species diversity. Am Nat. 2007;170(1):143–154. doi: 10.1086/518567. [DOI] [PubMed] [Google Scholar]
- 20.Gallet R, Tully T, Evans ME. Ecological conditions affect evolutionary trajectory in a predator-prey system. Evolution. 2009;63(3):641–651. doi: 10.1111/j.1558-5646.2008.00559.x. [DOI] [PubMed] [Google Scholar]
- 21.Rice TD, Williams HN, Turng BF. Susceptibility of bacteria in estuarine environments to autochthonous bdellovibrios. Microb Ecol. 1998;35(3):256–264. doi: 10.1007/s002489900081. [DOI] [PubMed] [Google Scholar]
- 22.Davidov Y, Friedjung A, Jurkevitch E. Structure analysis of a soil community of predatory bacteria using culture-dependent and culture-independent methods reveals a hitherto undetected diversity of Bdellovibrio-and-like organisms. Environ Microbiol. 2006;8(9):1667–1673. doi: 10.1111/j.1462-2920.2006.01052.x. [DOI] [PubMed] [Google Scholar]
- 23.Thomashow MF, Rittenberg SC. Intraperiplasmic growth of Bdellovibrio bacteriovorus 109J: Attachment of long-chain fatty acids to escherichia coli peptidoglycan. J Bacteriol. 1978;135(3):1015–1023. doi: 10.1128/jb.135.3.1015-1023.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thomashow MF, Rittenberg SC. Intraperiplasmic growth of Bdellovibrio bacteriovorus 109J: N-deacetylation of Escherichia coli peptidoglycan amino sugars. J Bacteriol. 1978;135(3):1008–1014. doi: 10.1128/jb.135.3.1008-1014.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thomashow MF, Rittenberg SC. Intraperiplasmic growth of Bdellovibrio bacteriovorus 109J: Solubilization of Escherichia coli peptidoglycan. J Bacteriol. 1978;135(3):998–1007. doi: 10.1128/jb.135.3.998-1007.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cover WH, Rittenberg SC. Change in the surface hydrophobicity of substrate cells during bdelloplast formation by Bdellovibrio bacteriovorus 109J. J Bacteriol. 1984;157(2):391–397. doi: 10.1128/jb.157.2.391-397.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ruby EG, Rittenberg SC. Attachment of diaminopimelic acid to bdelloplast peptidoglycan during intraperiplasmic growth of Bdellovibrio bacteriovorus 109J. J Bacteriol. 1984;158(2):597–602. doi: 10.1128/jb.158.2.597-602.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shilo M. Morphological and physiological aspects of the interaction of Bdellovibrio with host bacteria. Curr Top Microbiol Immunol. 1969;50:174–204. doi: 10.1007/978-3-642-46169-9_6. [DOI] [PubMed] [Google Scholar]
- 29.Rittenberg SC, Shilo M. Early host damage in the infection cycle of Bdellovibrio bacteriovorus. J Bacteriol. 1970;102(1):149–160. doi: 10.1128/jb.102.1.149-160.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kessel M, Shilo M. Relationship of Bdellovibrio elongation and fission to host cell size. J Bacteriol. 1976;128(1):477–480. doi: 10.1128/jb.128.1.477-480.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fenton AK, Kanna M, Woods RD, Aizawa SI, Sockett RE. Shadowing the actions of a predator: Backlit fluorescent microscopy reveals synchronous nonbinary septation of predatory Bdellovibrio inside prey and exit through discrete bdelloplast pores. J Bacteriol. 2010;192(24):6329–6335. doi: 10.1128/JB.00914-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thomashow MF, Cotter TW. Bdellovibrio host dependence: The search for signal molecules and genes that regulate the intraperiplasmic growth cycle. J Bacteriol. 1992;174(18):5767–5771. doi: 10.1128/jb.174.18.5767-5771.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gray KM, Ruby EG. Intercellular signalling in the Bdellovibrio developmental cycle. In: Dworkin M, editor. Microbial Cell-Cell Interactions. American Society for Microbiology; Washington, DC: 1991. pp. 333–336. [Google Scholar]
- 34.Ruby EG, Rittenberg SC. Differentiation after premature release of intraperiplasmically growing Bdellovibrio bacteriovorous. J Bacteriol. 1983;154(1):32–40. doi: 10.1128/jb.154.1.32-40.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gray KM, Ruby EG. Prey-derived signals regulating duration of the developmental growth phase of Bdellovibrio bacteriovorus. J Bacteriol. 1990;172(7):4002–4007. doi: 10.1128/jb.172.7.4002-4007.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cotter TW, Thomashow MF. Identification of a Bdellovibrio bacteriovorus genetic locus, hit, associated with the host-independent phenotype. J Bacteriol. 1992;174(19):6018–6024. doi: 10.1128/jb.174.19.6018-6024.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roschanski N, Klages S, Reinhardt R, Linscheid M, Strauch E. Identification of genes essential for prey-independent growth of Bdellovibrio bacteriovorus HD100. J Bacteriol. 2011;193(7):1745–1756. doi: 10.1128/JB.01343-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lambert C, Chang CY, Capeness MJ, Sockett RE. The first bite--profiling the predatosome in the bacterial pathogen Bdellovibrio. PLoS One. 2010;5(1):e8599. doi: 10.1371/journal.pone.0008599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dori-Bachash M, Dassa B, Pietrokovski S, Jurkevitch E. Proteome-based comparative analyses of growth stages reveal new cell cycle-dependent functions in the predatory bacterium Bdellovibrio bacteriovorus. Appl Environ Microbiol. 2008;74(23):7152–7162. doi: 10.1128/AEM.01736-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reiner AM, Shilo M. Host-independent growth of Bdellovibrio bacteriovorus in microbial extracts. J Gen Microbiol. 1969;59:401–410. [Google Scholar]
- 41.Horowitz AT, Kessel M, Shilo M. Growth cycle of predacious Bdellovibrios in a host-free extract system and some properties of the host extract. J Bacteriol. 1974;117(1):270–282. doi: 10.1128/jb.117.1.270-282.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kwon SR, et al. Generation of Vibrio anguillarum ghost by coexpression of PhiX 174 lysis E gene and staphylococcal nuclease A gene. Mol Biotechnol. 2009;42(2):154–159. doi: 10.1007/s12033-009-9147-y. [DOI] [PubMed] [Google Scholar]
- 43.Eisenbach M, Adler J. Bacterial cell envelopes with functional flagella. J Biol Chem. 1981;256(16):8807–8814. [PubMed] [Google Scholar]
- 44.Lambert C, et al. Characterizing the flagellar filament and the role of motility in bacterial prey-penetration by Bdellovibrio bacteriovorus. Mol Microbiol. 2006;60(2):274–286. doi: 10.1111/j.1365-2958.2006.05081.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sánchez-Amat A, Torrella F. Formation of stable bdelloplasts as a starvation-survival strategy of marine bdellovibrios. Appl Environ Microbiol. 1990;56(9):2717–2725. doi: 10.1128/aem.56.9.2717-2725.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wurtzel O, Dori-Bachash M, Pietrokovski S, Jurkevitch E, Sorek R. Mutation detection with next-generation resequencing through a mediator genome. PLoS One. 2010;5(12):e15628. doi: 10.1371/journal.pone.0015628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Karunker I, Rotem O, Dori-Bachash M, Jurkevitch E, Sorek R. A global transcriptional switch between the attack and growth forms of Bdellovibrio bacteriovorus. PLoS One. 2013;8(4):e61850. doi: 10.1371/journal.pone.0061850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lerner TR, et al. Specialized peptidoglycan hydrolases sculpt the intra-bacterial niche of predatory Bdellovibrio and increase population fitness. PLoS Pathog. 2012;8(2):e1002524. doi: 10.1371/journal.ppat.1002524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Varon M, Shilo M. Attachment of Bdellovibrio bacteriovorus to cell wall mutants of Salmonella spp. and Escherichia coli. J Bacteriol. 1969;97(2):977–979. doi: 10.1128/jb.97.2.977-979.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schelling M, Conti S. Host receptor sites involved in the attachment of Bdellovibrio bacteriovorus and Bdellovibrio stolpii. FEMS Microbiol Lett. 1986;36(2-3):319–323. [Google Scholar]
- 51.Witte A, Wanner G, Sulzner M, Lubitz W. Dynamics of PhiX174 protein E-mediated lysis of Escherichia coli. Arch Microbiol. 1992;157(4):381–388. doi: 10.1007/BF00248685. [DOI] [PubMed] [Google Scholar]
- 52.Capeness MJ, et al. Activity of Bdellovibrio hit locus proteins, Bd0108 and Bd0109, links Type IVa pilus extrusion/retraction status to prey-independent growth signalling. PLoS One. 2013;8(11):e79759. doi: 10.1371/journal.pone.0079759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Evans KJ, Lambert C, Sockett RE. Predation by Bdellovibrio bacteriovorus HD100 requires type IV pili. J Bacteriol. 2007;189(13):4850–4859. doi: 10.1128/JB.01942-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mahmoud KK, Koval SF. Characterization of type IV pili in the life cycle of the predator bacterium Bdellovibrio. Microbiology. 2010;156(Pt 4):1040–1051. doi: 10.1099/mic.0.036137-0. [DOI] [PubMed] [Google Scholar]
- 55.Milner DS, et al. Ras GTPase-like protein MglA, a controller of bacterial social-motility in Myxobacteria, has evolved to control bacterial predation by Bdellovibrio. PLoS Genet. 2014;10(4):e1004253. doi: 10.1371/journal.pgen.1004253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kadouri D, O’Toole GA. Susceptibility of biofilms to Bdellovibrio bacteriovorus attack. Appl Environ Microbiol. 2005;71(7):4044–4051. doi: 10.1128/AEM.71.7.4044-4051.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hobley L, et al. Discrete cyclic di-GMP-dependent control of bacterial predation versus axenic growth in Bdellovibrio bacteriovorus. PLoS Pathog. 2012;8(2):e1002493. doi: 10.1371/journal.ppat.1002493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jürgens K, Pernthaler J, Schalla S, Amann R. Morphological and compositional changes in a planktonic bacterial community in response to enhanced protozoan grazing. Appl Environ Microbiol. 1999;65(3):1241–1250. doi: 10.1128/aem.65.3.1241-1250.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tudor JJ, Conti SF. Characterization of bdellocysts of Bdellovibrio sp. J Bacteriol. 1977;131(1):314–322. doi: 10.1128/jb.131.1.314-322.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dashiff A, Kadouri DE. Predation of oral pathogens by Bdellovibrio bacteriovorus 109J. Mol Oral Microbiol. 2011;26(1):19–34. doi: 10.1111/j.2041-1014.2010.00592.x. [DOI] [PubMed] [Google Scholar]
- 61.Christensen H, Hansen M, Sorensen J. Counting and size classification of active soil bacteria by fluorescence in situ hybridization with an rRNA oligonucleotide probe. Appl Environ Microbiol. 1999;65(4):1753–1761. doi: 10.1128/aem.65.4.1753-1761.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Portillo MC, Leff JW, Lauber CL, Fierer N. Cell size distributions of soil bacterial and archaeal taxa. Appl Environ Microbiol. 2013;79(24):7610–7617. doi: 10.1128/AEM.02710-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Toft S, Wise DH. Growth, development, and survival of a generalist predator fed single- and mixed-species diets of different quality. Oecologia. 1999;119(2):191–197. doi: 10.1007/s004420050776. [DOI] [PubMed] [Google Scholar]
- 64.Toft S, Wise DH. Behavioral and ecophysiological responses of a generalist predator to single- and mixed- species diets of different quality. Oecologia. 1999;119(2):198–207. doi: 10.1007/s004420050777. [DOI] [PubMed] [Google Scholar]
- 65.Steyert SR, Pineiro SA. Development of a novel genetic system to create markerless deletion mutants of Bdellovibrio bacteriovorus. Appl Environ Microbiol. 2007;73(15):4717–4724. doi: 10.1128/AEM.00640-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Peleg Y, Unger T. Application of the Restriction-Free (RF) cloning for multicomponents assembly. Methods Mol Biol. 2014;1116:73–87. doi: 10.1007/978-1-62703-764-8_6. [DOI] [PubMed] [Google Scholar]
- 67.Sambrook J, Russell DW. Molecular Cloning. A Laboratory Manual. Cold Spring Harbor Lab Press; Cold Spring Harbour, NY: 2001. [Google Scholar]
- 68.Wallner G, Amann R, Beisker W. Optimizing fluorescent in situ hybridization with rRNA-targeted oligonucleotide probes for flow cytometric identification of microorganisms. Cytometry. 1993;14(2):136–143. doi: 10.1002/cyto.990140205. [DOI] [PubMed] [Google Scholar]
- 69.Mahmoud KK, McNeely D, Elwood C, Koval SF. Design and performance of a 16S rRNA-targeted oligonucleotide probe for detection of members of the genus Bdellovibrio by fluorescence in situ hybridization. Appl Environ Microbiol. 2007;73(22):7488–7493. doi: 10.1128/AEM.01112-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.George TC, et al. Quantitative measurement of nuclear translocation events using similarity analysis of multispectral cellular images obtained in flow. J Immunol Methods. 2006;311(1-2):117–129. doi: 10.1016/j.jim.2006.01.018. [DOI] [PubMed] [Google Scholar]
- 71.Schloss PD, et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75(23):7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Quince C, Lanzen A, Davenport RJ, Turnbaugh PJ. Removing noise from pyrosequenced amplicons. BMC Bioinformatics. 2011;12:38. doi: 10.1186/1471-2105-12-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Huse SM, Welch DM, Morrison HG, Sogin ML. Ironing out the wrinkles in the rare biosphere through improved OTU clustering. Environ Microbiol. 2010;12(7):1889–1898. doi: 10.1111/j.1462-2920.2010.02193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hespell RB. Intraperiplasmic growth of Bdellovibrio bacteriovorus on heat-treated Escherichia coli. J Bacteriol. 1978;133(3):1156–1162. doi: 10.1128/jb.133.3.1156-1162.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Simon R, Priefer U, Pühler A. A broad host range mobilization system for in vivo genetic engineering: Transposon mutagenesis in gram negative bacteria. Nat Biotechnol. 1983;1(9):784–791. [Google Scholar]
- 76.Bukhari AI, Taylor AL. Genetic analysis of diaminopimelic acid- and lysine-requiring mutants of Escherichia coli. J Bacteriol. 1971;105(3):844–854. doi: 10.1128/jb.105.3.844-854.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Richaud F, Richaud C, Ratet P, Patte JC. Chromosomal location and nucleotide sequence of the Escherichia coli dapA gene. J Bacteriol. 1986;166(1):297–300. doi: 10.1128/jb.166.1.297-300.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mizrahi I, Dagan M, Biran D, Ron EZ. Potential use of toxic thermolabile proteins to study protein quality control systems. Appl Environ Microbiol. 2007;73(18):5951–5953. doi: 10.1128/AEM.01067-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.