Significance
RNA provides a link between variation at the DNA and phenotypic levels. We measured the abundances of RNA products of protein-coding genes and novel transcribed regions in a population of wild-derived inbred strains of Drosophila melanogaster whose genome sequences are also available. We exploited this unique resource to characterize the genetic basis of transcriptome diversity. We found high complexity of the genetic control of gene expression, including widespread sexual dimorphism, highly modularized expression patterns with involvement of novel RNA transcripts, and frequent epistatic interactions among expression quantitative trait loci (QTLs) which often give rise to variance expression QTLs. This study highlights the importance and general applicability of integrating expression phenotypes to understand the genetic architecture of complex quantitative phenotypes.
Keywords: genome-wide association, novel transcribed regions, mean eQTLs, variance eQTLs, epistasis
Abstract
Understanding how DNA sequence variation is translated into variation for complex phenotypes has remained elusive but is essential for predicting adaptive evolution, for selecting agriculturally important animals and crops, and for personalized medicine. Gene expression may provide a link between variation in DNA sequence and organismal phenotypes, and its abundance can be measured efficiently and accurately. Here we quantified genome-wide variation in gene expression in the sequenced inbred lines of the Drosophila melanogaster Genetic Reference Panel (DGRP), increasing the annotated Drosophila transcriptome by 11%, including thousands of novel transcribed regions (NTRs). We found that 42% of the Drosophila transcriptome is genetically variable in males and females, including the NTRs, and is organized into modules of genetically correlated transcripts. We found that NTRs often were negatively correlated with the expression of protein-coding genes, which we exploited to annotate NTRs functionally. We identified regulatory variants for the mean and variance of gene expression, which have largely independent genetic control. Expression quantitative trait loci (eQTLs) for the mean, but not for the variance, of gene expression were concentrated near genes. Notably, the variance eQTLs often interacted epistatically with local variants in these genes to regulate gene expression. This comprehensive characterization of population-scale diversity of transcriptomes and its genetic basis in the DGRP is critically important for a systems understanding of quantitative trait variation.
Genetic variation for quantitative traits is a universal property of evolving populations. Elucidating the general principles that underlie the genotype–phenotype map is critical for understanding natural selection and evolution, improving the efficacy of animal and plant breeding, and identifying targets for treating human diseases. Numerous quantitative trait loci (QTLs) have been identified in linkage and association mapping populations by scanning polymorphic markers across the genome. However, QTLs rarely map to genes or causal genetic variants and typically account for only a small fraction of total genetic variation (1, 2), so interpreting the functional roles of QTLs and dissecting the genetic architecture of quantitative traits is particularly challenging.
By extension of the central dogma of molecular biology, it is generally accepted that a QTL generates phenotypic variation by introducing variation in protein sequence and/or the abundance of gene products (3). Variation in the abundance of gene products constitutes an important class of quantitative traits and can be measured with great precision and high throughput. This ability provides the opportunity to identify expression QTLs (eQTLs) that control variation in global mRNA levels. Furthermore, although the relative importance of structural and regulatory variation remains debatable, mounting evidence has indicated that regulatory variation could be a significant source of phenotypic variation. In particular, there is increasing evidence that QTLs associated with organismal phenotypes are more likely to be eQTLs than are other variants with similar allele frequencies in the genome (4).
Genetic studies of global gene expression in a wide range of organisms including yeast (5), animals (6–8), and plants (9, 10) have found that a large fraction of gene-expression traits is heritable. Although both local (cis) and distal (trans) eQTLs have been detected, in most cases eQTLs near genes tend to be more common and have larger effects. Conventionally, individuals within each genotype class of an eQTL share the same mean of expression, which differs among individuals of different genotypes (we call these “mean eQTLs” or simply “eQTLs” throughout this study). More recently, another class of QTLs for which there is a difference in the variance of phenotypes between individuals with different genotypes has been identified for both gene-expression (11–13) and organismal phenotypes (14–16). These variance QTLs are of interest because differences in the variance of gene expression among different genotypes at a focal locus can be induced by epistasis between the focal locus and one or more interacting loci (14), thereby providing a simple approach for identifying QTLs participating in genetic interactions (12). A third class of QTLs comprises those that affect variation among genetically identical individuals, which could be attributed to variable plasticity of traits in response to microenvironmental variation or to fluctuation in developmental processes, both of which are stochastic in nature (17–20). Although both affect the degree of variation in quantitative traits, the QTLs affecting between-individual variance and within-individual variance (or variance among genetically identical individuals) are conceptually different and thus use distinct mapping models.
The Drosophila melanogaster Genetic Reference Panel (DGRP) consists of 205 inbred lines with whole-genome sequences (21, 22). The DGRP harbors molecular variation for more than four million loci (approximately one every 50 bp) and exhibits quantitative genetic variation for many organismal phenotypes (22), facilitating genome-wide association (GWA) mapping in a scenario in which nearly all variants are known. Recent GWA studies in the DGRP indicate that the inheritance of most organismal quantitative traits in Drosophila is complex, involving many genes with small additive effects as well as epistatic interactions (21, 23–25).
A small-scale study of 40 DGRP lines previously revealed substantial quantitative genetic variation in gene expression in the DGRP (6). The genetically variable transcripts cluster into modules of highly correlated expression traits associated with distinct biological processes (6). More recently, an eQTL mapping analysis in this subset of DGRP lines has identified cis-eQTLs within 10 kb of more than 2,000 genes (26).
As QTL mapping studies in the DGRP accumulate information about the genetic basis of many organismal traits, a comprehensive characterization of the diversity of transcriptomes and its genetic basis in the entire DGRP becomes critically important. In the present study, we identify unannotated transcriptional units in the Drosophila genome using RNA sequencing (RNA-seq) and quantify gene expression using genome-tiling microarrays. We then comprehensively characterize the genetic diversity of gene expression in the DGRP. Finally, we identify eQTLs that control the mean and variance of global gene expression and show that the latter frequently can be explained by interactions with cis-eQTLs.
Results
Identification of Novel Transcribed Regions.
Recent efforts to characterize genome-wide transcription in human cells found that approximately three quarters of the human genome is transcribed into primary transcripts and that more than 60% of the genomic bases represent processed mature RNA transcripts (27). Pervasive transcription appears to be a common feature for eukaryotic genomes (28). Approximately 75% of the D. melanogaster genome is transcribed at least temporarily during development, and thousands of novel transcribed regions (NTRs) have been identified, the majority of which do not appear to code for proteins (29). With the exception of a small number of long noncoding RNAs (ncRNAs), whose regulatory roles in mammals are well established (30), the functional implications of pervasive transcription and ncRNAs remain to be resolved.
The DGRP provides a platform to study the molecular quantitative genetics and functions of RNAs by associating them with genetic determinants of gene expression and the expression of other RNAs. To identify unannotated NTRs, we sequenced polyadenylation [poly(A)]-positive RNAs of adult flies pooled from 192 DGRP lines using 100-bp paired-end sequencing for females and males separately. Approximately 100 M cDNA fragments were sequenced in both sexes (SI Appendix, Table S1 and Dataset S1). We aligned the sequence reads to the annotated transcriptome and reference genome and used the resulting overlapping alignments to assemble transcript models. Approximately 4.5 and 6.7% of mapped reads in males and females, respectively, do not overlap with any annotated exons and may represent unannotated transcriptional units (SI Appendix, Table S1 and Dataset S1). We merged overlapping transcript models in females and males and compared them with the FlyBase (Release 5.49) annotation to identify NTRs. We found 1,669 transcripts derived from 1,628 intronic regions and 2,192 transcripts derived from 1,876 intergenic regions, representing a total of 3.6 M unannotated bases in processed RNAs—an ∼11% addition to the existing annotations. In addition, a total of 2,807 previously unreported alternatively spliced isoforms were found for 2,049 annotated genes. We characterized NTRs for the size of processed transcripts they produce, nucleotide composition, sequence conservation, and propensity to harbor polymorphic DNA variants. NTRs do not differ qualitatively from annotated ncRNAs. Compared with protein-coding genes, both NTRs and annotated ncRNAs have shorter transcripts, lower guanine-cytosine (GC) content, weaker sequence conservation, and slightly higher density of DNA variants (SI Appendix, Fig. S1).
We estimated the expression of annotated genes and NTRs in the pooled samples of females and males. Not surprisingly, NTRs generally are expressed at a much lower level than annotated genes (Fig. 1); more highly and ubiquitously expressed genes were more likely to be detected by previous annotation efforts. We reasoned that spurious NTRs identified in RNA-seq would not be genetically variable in subsequent quantitative genetic analyses using an independent expression platform. Therefore, we did not filter NTRs by their expression level, a practice commonly used to eliminate erroneous transcript reconstruction in RNA-seq.
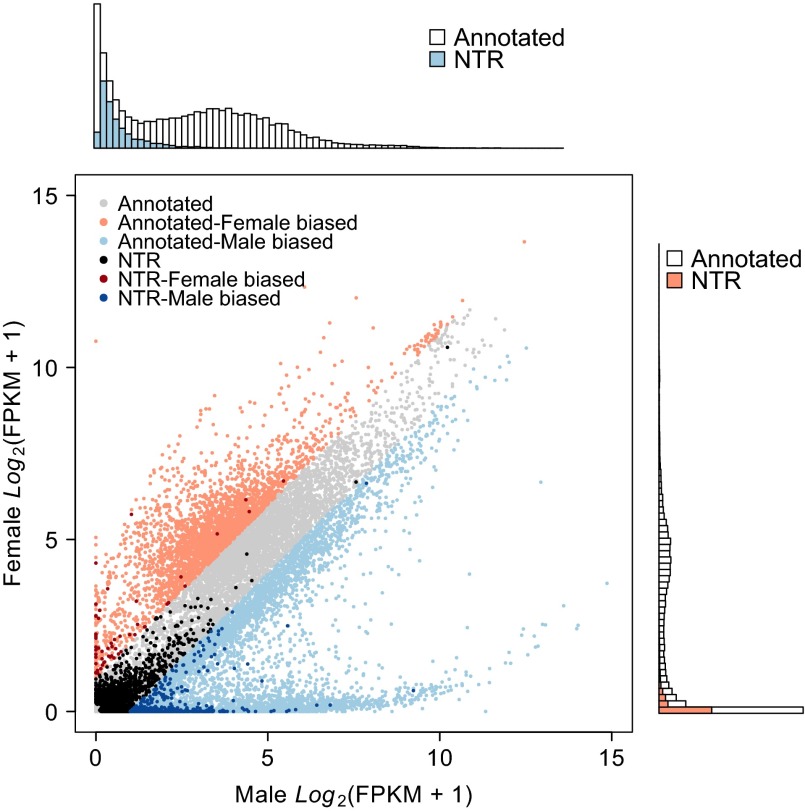
Fig. 1.
RNA-seq in the DGRP reveals many NTRs. The scatter plot compares gene expression of annotated genes and NTRs in females and males. Genes with expression differences of twofold or more between the sexes are considered to have sex-biased expression. The histograms depict the distribution of gene expression in females (Right) and males (Upper), with colored bars (orange, female; blue, male) showing the distributions for NTRs.
Transcriptome Diversity in the DGRP.
We used Affymetrix Drosophila 2.0 genome-tiling arrays to measure the expression of annotated genes and NTRs in 185 DGRP lines, with two biological replicates for each sex. We estimated the overall expression of genes by median polish of background-corrected and quantile-normalized probe expression. Only probes that uniquely and entirely map to constitutive exons and do not contain common (nonreference allele frequency >0.05) variants were used. A small fraction of annotated genes (n = 1,217; 8%) and NTRs (n = 113; 3%) could not be interrogated after the removal of nonconstitutive or variant-overlapping probes.
We used a linear mixed model to test for the effect of sex (sexual dimorphism) and to partition the variance in gene expression into three sources: between-line (genetic) variance, variance in sex-by-line interaction (genetic variance in sexual dimorphism), and within-line (environmental) variance. As expected, given that sexual dimorphism is common for D. melanogaster gene-expression traits (6, 31, 32), the great majority of genes (16,445/18,140, 90.6%) showed significant mean differences [false-discovery rate (FDR) <0.05] between females and males, including NTRs, of which 80.9% (2,743/3,391) show sex-biased expression (SI Appendix, Table S2 and Dataset S1). Gene-set enrichment analysis (GSEA) revealed that genes with female-biased expression were enriched for several biological processes primarily associated with DNA replication, DNA repair, and the cell cycle, whereas genes with male-biased expression were enriched for genes involved in reproduction (SI Appendix, Fig. S2 and Table S3 and Dataset S1). Furthermore, genes with female-biased expression are highly enriched for ovary-specific genes, and genes with male-biased expression are highly enriched for testis-specific genes (SI Appendix, Fig. S3). A substantial fraction of genes (2,388/18,140; 13.2%), of which 106/3,391 (3.1%) were NTRs, show significant (FDR <0.05) sex-by-line interaction, indicating that the degree of sexual dimorphism as a quantitative trait is genetically variable for these genes (SI Appendix, Table S2 and Dataset S1). The lower proportion of NTRs showing sexual dimorphism and sex-by-line interaction is likely a result of their low expression and thus smaller effects. Because of the widespread sexual dimorphism and sex-by-line interaction, we performed all subsequent analyses in females and males separately.
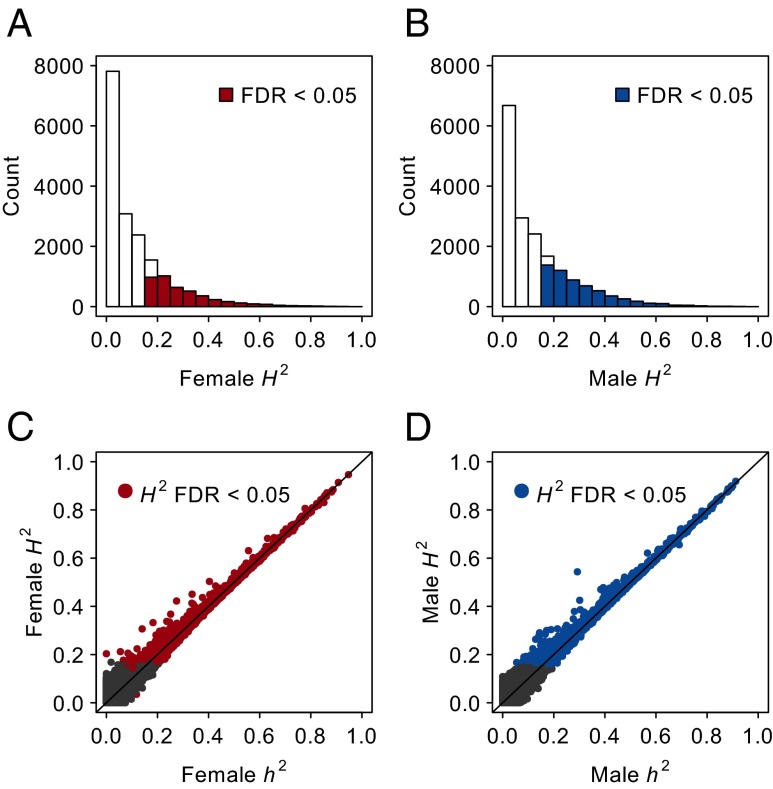
We next asked to what extent variation in gene expression is heritable. We tested the significance of the among-line variance component and estimated the broad-sense heritability (H2) for each gene-expression trait as the proportion of total variance explained by between-line differences. Among the 18,140 annotated genes and NTRs, a total of 7,626 unique genes showed significant (FDR <0.05) genetic variability in expression in either sex (SI Appendix, Table S4 and Dataset S1). Among these genetically variable transcripts, including NTRs, 4,308 had a significant genetic component in females (1,812 occurred only in females), 5,814 had a significant genetic component in males (3,318 occurred only in males), and 2,496 had a significant genetic component in both sexes (Fig. 2 A and B, SI Appendix, Tables S4 and S5 and Dataset S1). Remarkably, 231 NTRs are genetically variable in females (120 occur only in females), and 430 NTRs are genetically variable in males (319 occur only in males); 111 NTRs are genetically variable in both sexes (SI Appendix, Tables S4 and S5 and Dataset S1). Estimates of H2 for genes with heritable variation in expression range from 0.034 to 0.946 in both sexes (Fig. 2).
Fig. 2.
Genetic variation of gene expression. (A and B) Distribution of H2 for gene-expression traits. (A) Females. (B) Males. (C and D) Relationship between h2 and H2 for gene-expression traits. (C) Females. (D) Males. Genetically variable genes (FDR <0.05) are color-coded as indicated.
Given the availability of complete genome sequences, we can compute the genetic covariance among the DGRP lines, which measures the genetic similarity between pairs of lines assuming an infinitesimal model. This method allows us to estimate the proportion of phenotypic variance in gene expression explained by the additive genetic variance (or narrow-sense heritability, h2) using a mixed-effects model (33, 34). Interestingly, h2 of the great majority of genes captures most of the total genetic variance (Fig. 2 C and D, SI Appendix, Table S5 and Dataset S1). Although large differences between h2 and H2 indicate a large contribution of nonadditive gene action (i.e., dominance and/or epistasis), the opposite is not necessarily true (25). Epistatic gene action can lead to largely additive variance if the minor allele frequencies (MAF) of interacting loci are low (25, 35).
Among the 185 DGRP lines, 99 were infected with the endosymbiotic bacterium Wolbachia pipientis (22). We tested the effect of Wolbachia infection on gene expression, conditional on five polymorphic major inversions and the first 10 principal components (PCs) of common variants. Because lines are nested within the Wolbachia infection, it is not possible to separate the Wolbachia effect from between-line variation. Nevertheless, by accounting for major inversions and top genotypic PCs, we aim to test for the effect of Wolbachia independent of genetic differentiation between the lines. Overall, Wolbachia infection has only minor effects on gene expression, and the effects are female specific (SI Appendix, Fig. S4). In particular, genes that are down-regulated in lines positive for Wolbachia are largely ovary specific (SI Appendix, Fig. S4).
Many large chromosomal inversions are polymorphic in the DGRP, some of which occur at relatively high frequency. We tested the effects of each of the five major segregating inversions on the expression of genetically variable transcripts. For each inversion, we grouped lines segregating for the inversion into a third genotype class in addition to the two inbred genotypes, noting that frequencies of inversions within these lines may vary. At an FDR < 0.05, there are 125 (20), 9 (13), 35 (26), 17 (32), and 21 (39) genes in females (males) whose expression is affected by Inversion (2L)t [In(2L)t], Inversion (2R) from Nova Scotia [In(2R)NS], Inversion (3R) of Payne [In(3R)P], Inversion (3R) of Kodani [In(3R)K], and Inversion (3R) from Missouri [In(3R)Mo], respectively (SI Appendix, Table S6 and Dataset S1). We also tested whether inversions preferentially affect the expression of genes within the inverted regions. Such a local effect could indicate the accumulation of cis-regulatory mutations after the inversions arose in the population. Interestingly, In(2L)t and In(3R)Mo preferentially affect genes within the boundaries of their respective inversions in both sexes, but other inversions do not appear to do so (SI Appendix, Fig. S5).
Modules of Genetically Correlated Transcripts.
We have shown previously, using 40 DGRP lines, that genetically variable transcripts are not independent but instead cluster into a smaller number of genetically correlated coexpression modules whose members often contribute to the same biological processes (6). To expand the investigation to the entire DGRP, we first estimated the genetic component of expression for genetically variable transcripts after adjusting for Wolbachia infection status. Because inversions affect the expression of only a small number of genes (SI Appendix, Fig. S5) and are genuine genetic effects, we did not adjust for their effects in our analysis of correlated gene expression.
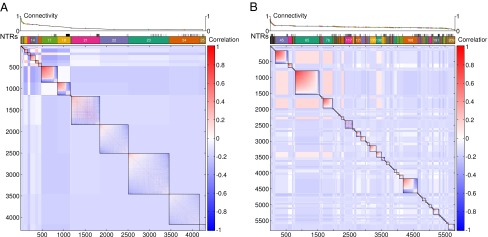
We used Modulated Modularity Clustering (MMC) (6, 36) to identify clusters of genetically correlated genes. This algorithm derives modules such that the absolute value of the genetic correlation among transcripts is maximized within modules and minimized between modules. We found a few large modules of high connectivity in both sexes (Fig. 3, SI Appendix, Table S7 and Dataset S1). These modules are not merely statistical constructs but are frequently enriched for genes within the same gene ontology (GO) terms (SI Appendix, Fig. S6 and Tables S7–S9 and Dataset S1), indicating that genes with genetically correlated transcripts tend to fall within the same biological pathways. Indeed, the genetic correlation in the expression of genes belonging to the same GO pathways is significantly higher than that between genes in different GO pathways (SI Appendix, Figs. S7 and S8). Therefore, functions of computationally predicted genes within these modules can be inferred with functional annotations of other genes in the module using the principle of “guilt by association” (37, 38).
Fig. 3.
Genetically correlated modules of gene-expression traits shown as heat maps from MMC analyses. Genetically variable transcripts are ordered based on their cluster membership and connectivity, which decreases from the top left corner to the bottom right corner of the heat maps. The correlation between transcripts within and between modules is depicted by the color-scale bars. The modules are indicated by the colored rectangles above the heat maps, and NTRs are denoted by short vertical bars. The average connectivity within each module is given at the top of the plots. (A) Females. (B) Males.
The remaining transcripts are organized into either large modules with low connectivity (especially in females; Fig. 3A) or smaller modules with relatively high connectivity (especially in males; Fig. 3B). The choice between a few large modules with low connectivity versus many small modules with high connectivity is affected by both the specific genetic correlation structure and the object function in the MMC clustering algorithm (36). We focused our biological inference on relatively large modules with high connectivity, which are less affected by stochastic noise in estimates of genetic correlation. Consistent with the small effect of Wolbachia on gene-expression traits, the overall patterns of genetic correlation before and after adjusting for Wolbachia are largely similar (SI Appendix, Fig. S9); therefore we performed subsequent inferences based on the clustering after adjusting for Wolbachia infection.
Remarkably, expression of NTRs is negatively correlated in general with the expression of protein-coding genes from the same expression modules, especially in males, suggesting that NTRs may act as negative regulators for the expression of protein-coding genes (Fig. 3 and SI Appendix, Fig. S10). The mechanism by which NTRs regulate gene expression is unclear. Most NTRs, regardless of the strength of their association with protein-coding genes, are distant from the protein-coding genes with which they are associated (SI Appendix, Fig. S11), suggesting that NTRs function in trans. Among the 5,733 pairs of NTRs and protein-coding genes whose genetic correlation exceeds 0.25 in females and the 11,519 such pairs in males, only 6 and 26, respectively, had very weak homology, and all were shorter than 30 bp, suggesting that NTRs do not function through base-pairing with mRNAs.
The genetic correlation in expression between NTRs and annotated genes allows us to infer putative functions of NTRs by coexpression. We used GSEA to associate (FDR <0.05) 105 of 231 genetically variable NTRs in females and 208 of 430 genetically variable NTRs in males with at least one GO or Kyoto Encyclopedia of Genes and Genomes pathway (SI Appendix, Table S10 and Dataset S1). The majority of these associations are negative. Several pathways, such as mitotic spindle organization, unfolded protein binding, and mitosis in females and translation initiation factor activity, protein binding, and ubiquitin-protein ligase activity in males, appear to recruit a large number of NTRs (SI Appendix, Fig. S12).
QTLs Associated with Mean Transcript Abundance.
To characterize the genetic architecture of quantitative variation in gene expression, we performed GWA analyses to map mean eQTLs that regulate mean expression for all genetically variable genes. We fitted linear mixed models to adjust for Wolbachia, inversions, and 10 significant PCs of the genotypes and estimated line means for each genetically variable transcript using best linear unbiased prediction (BLUP). The significance of association between each of the 1,913,487 individual common variants (MAF ≥0.05) and mean of gene-expression traits was evaluated by single-marker regression of the BLUP line means on marker genotypes. The empirical FDR for each gene expression trait was estimated by dividing the expected number of associations under the null hypothesis (n = 100 permutations) at variable P-value thresholds by the observed number of associations at the same P-value thresholds.
As expected, fewer significant eQTLs are detected as increasingly stringent FDR thresholds are applied (Table 1). By arbitrarily defining eQTLs as variants within ± 1 kb of the genes they influence as cis-eQTLs, more than 50% of genes with eQTLs have at least one cis-eQTL at FDR <0.05. More trans-eQTLs are detected at more lenient FDR thresholds, but the increase in the number of cis-eQTLs is relatively small (Table 1). This result is consistent with the observation that cis-eQTLs are more strongly associated with variation in gene expression (SI Appendix, Fig. S13). At an empirical FDR <0.20, 941 genetically variable gene-expression traits in females and 1,339 genetically variable gene-expression traits in males have at least one cis- and/or trans-eQTL; of these, 31 are NTRs in females and 114 are NTRs males (SI Appendix, Tables S11 and S12 and Dataset S1). Interestingly, the proportion of genes with cis-eQTLs is substantially larger for males than for females (Table 1). The association between DNA variants and gene expression is much stronger around transcription start sites (TSS) and transcription end sites (TES) (SI Appendix, Fig. S13), where regulatory elements for transcription and RNA stability are concentrated. This observation is consistent with the distribution of cis-eQTLs previously found in Drosophila and other organisms (26, 39–41).
Table 1.
Number of genes with at least one significant eQTL at different FDR thresholds
| Sex | FDR threshold (cis + trans)* | |||
| 0.05 | 0.10 | 0.15 | 0.20 | |
| Female | 503 (263 + 240) | 671 (287 + 384) | 807 (297 + 510) | 941 (308 + 633) |
| Male | 837 (533 + 304) | 1,029 (568 + 461) | 1,189 (594 + 595) | 1,339 (608 + 731) |
Number of genes with at least one cis-eQTL (within 1 kb of genes) and number of genes with only trans-eQTLs.
We compared eQTLs mapped in females and males and asked whether the genetic control of gene expression by individual eQTLs is preserved in the two sexes. As is consistent with the widespread prevalence of sexual dimorphism and sex-by-line interaction in gene expression, only 185 genes have at least one common eQTL in both sexes (SI Appendix, Fig. S14). The remaining genes either contain sex-specific eQTLs or do not vary genetically in the other sex (SI Appendix, Fig. S14).
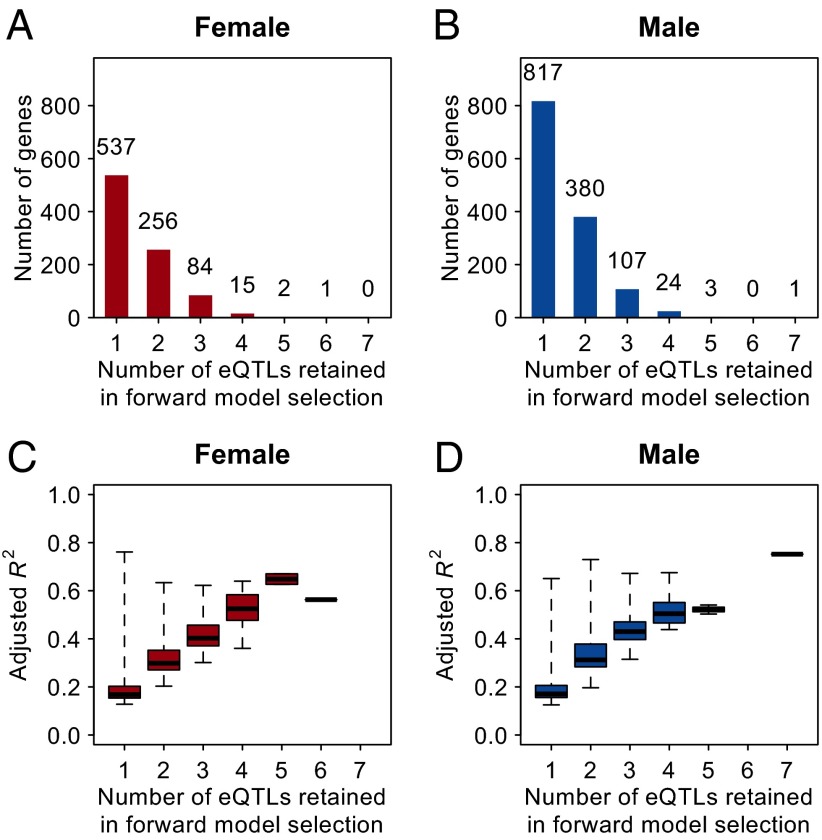
To assess the fraction of total genetic variance explained by mapped eQTLs, we first identified eQTLs for each expression trait that are largely independent. To do so, we performed forward model selection to add eQTLs successively to an additive genetic model for each genetically variable gene-expression trait, requiring that the conditional P value of each added eQTL be smaller than 10−5. The number of eQTLs selected by the forward selection ranged from one to seven, with the majority of gene-expression traits having one or two independent eQTLs (Fig. 4 A and B). For most genes, the selected eQTLs explained a substantial fraction of genetic variance (Figs. 4 C and D).
Fig. 4.
Variance in gene expression explained by independent eQTLs. (A and B) Distributions of the numbers of eQTLs retained in forward model selection. (A) Females. (B) Males. (C and D) Genetic variance explained by detected eQTLs (as measured by adjusted R2) versus the number of selected eQTLs. (C) Females. (D) Males.
Finally, we performed gene-based tests to search for groups of low frequency (MAF <0.05) variants within 1 kb of gene boundaries that collectively affect local gene expression. We used permutation to estimate the empirical FDR. At an FDR <0.20, 626 genes in females and 1,153 genes in males are significantly associated with cis low-frequency variants (SI Appendix, Tables S13 and S14 and Dataset S1). Remarkably, 216 of these genes in females and 408 of these genes in males also contain common eQTLs in cis, accounting for more than 75% of all genes with a common cis-eQTL. This result suggests that mapping eQTLs with common frequencies also captures effects induced by rare variants collectively.
QTLs Associated with Variance of Expression.
To search for variance eQTLs (veQTLs) for which lines carrying different alleles differ in their variance of expression among lines carrying the same allele, we performed a genome-wide scan for each gene-expression trait using Levene’s test (42) for homogeneity of variance between two groups. At an FDR <0.20, 925 genes in females and 412 genes in males contained at least one veQTL (Table 2); among these genes, 47 genes in females and no genes in males were NTRs (SI Appendix, Tables S4, S15, and S16 and Dataset S1). The great majority of these genes are trans-veQTLs (Table 2) and, correspondingly, the strength of association between veQTLs and variance among lines within the same genotype class showed only weak concentration around TSS and TES (SI Appendix, Fig. S15).
Table 2.
Number of genes with at least one significant veQTL at different FDR thresholds
| Sex | FDR threshold (cis + trans)* | |||
| 0.05 | 0.10 | 0.15 | 0.20 | |
| Female | 319 (6 + 313) | 544 (8 + 536) | 743 (9 + 734) | 925 (9 + 916) |
| Male | 162 (3 + 159) | 247 (6 + 241) | 353 (7 + 436) | 412 (7 + 405) |
Number of genes with at least one cis-veQTL (within 1 kb of genes) and number of genes with only trans-veQTLs.
To obtain veQTLs that are independent of each other, we successively selected veQTLs from those that met the initial FDR thresholds. For each gene with more than one significant veQTL, we started with the most significant veQTL and scaled the variance of gene expression within the major and minor allele classes to unit variance while preserving their means. We then tested the next veQTL in the P value-ranked list of veQTLs using the scaled phenotype and continued this process until no veQTL could be added with a P value smaller than 10−5. Similar to the mean eQTL analysis, this forward selection procedure also led to few veQTLs that independently controlled the variance of gene expression (SI Appendix, Fig. S16). Consistent with the observation that veQTLs were concentrated only weakly around genes (SI Appendix, Fig. S15), few genes with veQTLs contained cis-veQTLs (Table 2) after forward selection, in sharp contrast to eQTLs (Table 1).
Of the 941 genes in females and 1,339 genes in males whose expression was controlled by at least one eQTL, 248 genes in females and 107 genes in males also had veQTLs. In total, 1,618 genes in females and 1,644 genes in males had at least one eQTL or veQTL, i.e., were at least partially under the control of regulatory DNA variants. We could not assess whether genes with eQTLs are more likely to have veQTLs because the magnitude of variation between lines affects the power to detect both veQTLs and eQTLs. We further asked whether there were variants that control both the mean and the variance in expression of the same genes. Sixteen of the 1,432 eQTL gene pairs retained in forward model selection in females and six of the 2,029 eQTL gene pairs retained in forward model selection in males were significantly associated with the same genes as veQTLs. Of these mean eQTLs that also were veQTLs, one in females and none in males were in cis (within <1 kb of a gene), and the remaining were in trans. On the other hand, 24 of the 1,170 veQTL pairs in females and 15 of the 484 veQTL pairs in males also were significantly associated with the same genes as eQTLs, and four in females and four in males were in cis. Moreover, only 37 of the 1,170 veQTLs in females and 28 of 484 veQTLs in males showed significant association with the mean expression of any genes, suggesting that the variance-controlling effects of veQTLs generally were not caused by their effects on changing the mean level of expression of other genes. Taken together, these results suggest that the genetic architectures for mean and variance of gene expression are largely independent.
veQTLs Are Involved in Epistatic Interactions with cis-eQTLs.
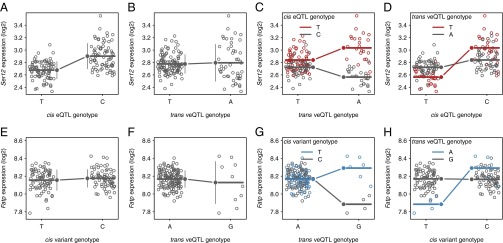
Because veQTLs can be emergent effects of underlying epistatic interactions for mean expression, we looked for variants that interact with veQTLs to affect gene expression epistatically. Because of the large number of possible epistatic pairs genome wide, we limited the search to interactions between veQTLs and variants that are in cis (within 1 kb) to the genes affected by the veQTLs. At an empirical FDR <0.20, the great majority of veQTLs (727/925 for females and 348/412 for males) for genes interacted with at least one cis variant (SI Appendix, Fig. S17). Moreover, among the 248 genes in females and 107 genes in males that had both eQTLs and veQTLs, 86 and 41, respectively, had detectable interactions between the cis-eQTLs and the veQTLs. For example, the expression of the serine protease 12 (Ser12) gene in females was associated with a cis-eQTL (Fig. 5A) and a trans-veQTL (Fig. 5B), which interacted epistatically to change the mean of expression for individuals carrying the same allelic combinations (Fig. 5C). The effect of the cis-eQTL for Ser12 therefore depended on the genotype of the trans-veQTL (Fig. 5D), which nevertheless was detected by ignoring the veQTL genotype in this specific case. However, many more cis variants have veQTL-dependent effects that could not be detected by single-marker regression (Fig. 5 E–H), highlighting the complexity and importance of context-dependent effects in the genetic architecture of gene expression.
Fig. 5.
veQTLs are involved in epistatic interactions with cis variants. (A–D) Scatter plots of Ser12 (2L:2250431.0.2251275) expression in females versus eQTL or veQTL genotypes. (A) The effect of a cis-eQTL (2L_2251218_SNP) on the mean but not on the variance of expression in individuals carrying the same genotypes. (B) The effect of a trans-veQTL (2L_11857529_SNP) on the variance but on not the mean of expression in individuals carrying the same genotypes. (C) The effect of the trans-veQTL on the mean expression is dependent on the cis-eQTL genotype. (D) The effect of the cis-eQTL on the mean expression is dependent on the trans-veQTL genotype. (E and F) Scatter plots of Fatp (2L:10510672.0.10517218) expression in males versus eQTL or veQTL genotypes. (E) A cis variant (2L_10510716_SNP) has no effect on the mean or variance of expression. (F) The effect of a trans-veQTL (3L_17881605_SNP) on the variance but not on the mean of expression. (G) The effect of the trans-veQTL on the mean expression is dependent on the cis variant genotype. (H) The effect of the cis variant on the mean expression is dependent on the trans-veQTL genotype.
Discussion
We have performed a comprehensive population-scale genetic characterization of the D. melanogaster transcriptome in a genetic reference population of sequenced, inbred, wild-derived lines. Similar to a previous study based on a subset of DGRP lines, we find that there is pervasive sexual dimorphism in mean gene expression and that a substantial fraction of the transcriptome is genetically variable (6, 26). In contrast to the previous studies, which used Affymetrix 3′ IVT microarrays, this analysis used genome-tiling microarrays. With an average resolution of 38 bp and more than 98% of exons in the fly transcriptome exceeding this size, this array provides sufficient resolution to detect the majority of genes. However, we found lower levels of genetic variance, higher within-line variation, and correspondingly lower average heritabilities than observed previously. Nevertheless, this decrease in precision was offset by our ability to assess the considerable contribution of NTRs to genetic variation in gene expression.
The abundances of genetically variable genes are not independent but covary and form highly connected gene-expression modules in a wide range of organisms (6, 43–45); this covariance may be the basis for pleiotropic trans-eQTLs and could be used to infer causal structure among gene expression traits and between expression and phenotype (45). In Arabidopsis, for example, the enzyme AOP2 was identified by linking eQTL and metabolite QTLs; modifying the expression level of AOP2 causally affected both enzyme and metabolite levels in the glucosinolate biosynthesis pathway (46).
The highly genetically correlated transcriptome sets the stage for annotating genes for which there is no functional information by using the guilt-by-association principle, which is particularly useful for NTRs that have not been annotated previously. Several hundred of these NTRs were genetically variable and tend to correlate negatively with protein-coding genes. We functionally annotated many of the previously unknown NTRs based on their genetic correlations with gene expression of known genes. Despite their weak conservation and low expression levels, many NTRs may have biological functions based on their association with genes of known functions. Further characterization of these NTRs and their mechanism(s) of regulation of transcription is an exciting area for future investigation.
We performed GWA analyses to identify eQTLs for mean gene expression as well as for variance of expression in the DGRP. In both cases we used a stringent forward model selection procedure to avoid over-fitting QTLs. These analyses revealed that the genetic basis of transcriptional regulation is sex specific and largely independent for the mean and variance. Most transcripts had single eQTLs or veQTLs (a consequence of the model selection criteria), although 40% of mean expression traits had more than one eQTL, and 15–23% of variance expression traits had more than one veQTL. Males had relatively more eQTLs and fewer veQTLs than females. At an FDR <0.05, most eQTLs are in cis to the gene whose expression they regulate and typically map near TSS and TES, as shown previously in D. melanogaster and other species (26, 39–41). The numbers of trans-eQTLs increase as the FDR threshold is lowered. In contrast, the majority of veQTLs are in trans to the gene for which they regulate variance in expression, and the fraction of cis-veQTLs remains low as the FDR threshold is lowered.
eQTLs in humans are enriched in cis regulatory elements such as DNase I hypersensitive sites, chromatin marks, and transcription factor-binding sites (47). In contrast, little is known about the regulatory nature of veQTLs. It has been postulated that veQTLs might reflect underlying genetic (epistatic) or genotype-by-environment interactions (12). Here, we demonstrated that trans-veQTLs frequently interact epistatically with cis-variants to modulate gene-expression levels (Fig. 5 and SI Appendix, Fig. S17). However, these interacting cis-variants are not the same as those affecting mean gene expression. The exact mechanisms are likely gene specific and remain to be studied.
The influences of sex and genetic interactions on gene expression fall into the broad framework of context-dependent effects, which provide the basis for dynamic gene-expression programs during development and in response to different physical and social environments. Indeed, a substantial fraction of the Drosophila transcriptome is plastic and sensitive to changing environments (48). However, the genetic basis of such plasticity is yet to be determined. The present study provides a baseline for further studies that investigate transcriptome diversity under various conditions.
In summary, the genetic architecture of Drosophila gene expression is complex and sex specific, with pervasive genetic correlation between gene-expression traits presumably caused in part by pleiotropy and loci affecting both mean and variance in expression, the latter being frequently attributable to epistatic interactions (Fig. 6). Epistatic interactions also have been implicated in the genetic architecture of complex traits (23, 25). These complexities need to be incorporated into systems genetics models seeking to predict organismal level phenotypes for quantitative traits from gene-expression data (3). Further, our estimates of gene expression were from tiling arrays, which have a narrow dynamic range relative to digital gene-expression estimates from RNA sequencing, and were from entire flies at a single age and environmental condition. Further work is needed to assess to what extent these features of the genetic architecture of gene expression are robust or plastic in different tissues, developmental stages, and social and physical environments.
Fig. 6.
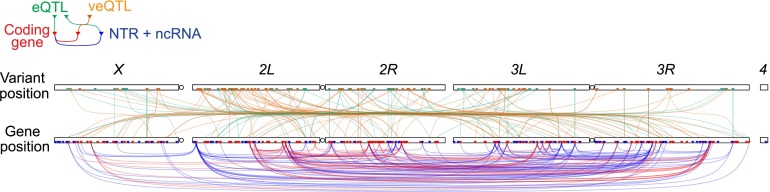
Architecture of genetic variation and genetic correlation in gene expression. The relationships between eQTL–gene, veQTL–gene, and gene–gene pairs are shown. Physical locations of DNA variants (chromosomes on top) and genes (chromosomes on bottom) are indicated by triangles; green, brown, red, and blue triangles denote eQTL, veQTL, protein-coding genes, and NTR or ncRNA, respectively. Green lines connect eQTLs and their associated genes; brown lines connect veQTLs and their associated genes; red lines connect genes whose expression correlate at r > 0.75; and blue lines connect genes whose expression correlate at r < −0.5.
Methods
Drosophila Lines.
We used inbred lines of the D. melanogaster DGRP. These lines were established by 20 generations of full sibling inbreeding from isofemale lines established from gravid females collected at a farmer’s market in Raleigh, NC. Complete genome sequences of the DGRP lines have been obtained using the Illumina platform. SNPs, indels, and other complex non-SNP variants have been genotyped using an integrated genotyping method (22).
Fly Husbandry and Collection.
All lines were reared under standard culture conditions (cornmeal-molasses-agar medium, 25 °C, 60–75% relative humidity, and a 12-h light/dark cycle) at equal larval densities. For each line, we collected two replicates per sex for analysis of gene expression, consisting of 25 female flies or 40 male flies per replicate (∼25 mg each), for a total of 768 samples. Because it was not possible to collect all replicates from all lines simultaneously, we used a strict randomized experimental design for sample collection. We collected mated 3- to 5-d-old flies between 1:00 and 3:00 PM. We transferred the flies into empty culture vials, froze them over ice supplemented with liquid nitrogen, and sexed the frozen flies. The samples were transferred to 2.0-mL nuclease-free microcentrifuge tubes (Ambion) and stored at −80 °C until processing.
RNA Extraction.
The flies were homogenized with 1 mL of QIAzol lysis reagent (Qiagen) and two 0.25-in ceramic beads (MP Biomedical) using the TissueLyser (Qiagen) adjusted to a frequency of 15 Hz for 1 min. Total RNA was extracted using the miRNeasy 96 kit (Qiagen) with on-column DNase I digestion and following the spin technology protocol as outlined in the manufacturer’s manual. The RNA was eluted with 45 μL RNase-free water. The eluted samples contain total RNA including miRNAs and other small RNAs (≥18 nucleotides). Total RNA was quantified using a NanoDrop 8000 spectrophotometer (Thermo Scientific), and the concentrations of the RNA samples were adjusted to 1 μg/μL for preparation of biotin-labeled double-stranded cDNA.
RNA-Seq Annotation of DGRP Lines.
We pooled 200 ng total RNA from each of 192 DGRP lines, separately for males and females. Poly(A)+ RNA-seq libraries were prepared from each pool according to the Illumina TruSeq mRNA-seq protocol, multiplexed, and sequenced by 100-bp paired ends in one lane of the HiSEq 2000 platform. Approximately 100-M fragments were sequenced for each of the male and female libraries. Sequence reads were mapped to the transcriptome (FlyBase annotation r5.49) and genome (FlyBase r5.49) using TopHat (49), allowing a maximum edit distance of 6 bp (nondefault options: –read-mismatches 6–read-gap-length 6–read-edit-dist 6–read-realign-edit-dist 0–mate-inner-dist -20–mate-std-dev 80–min-intron-length 20–max-intron-length 25000–solexa1.3-quals–max-multihits 20–library-type fr-unstranded–segment-mismatches 2–segment-length 25–min-segment-intron 20–max-segment-intron 25000–no-coverage-search–GTF flybase-r5.49.gtf). Gene models were assembled for male and female separately from the cDNA alignments using Cufflinks (50, 51) with the guide of the reference annotation (nondefault options: –multiread-correct–GTF-guide flybase-r5.49.gtf -M flybase-r5.46(ribosomal and mitochondrial RNAs).gtf -b flybase-r5.49.fa–library-type fr-unstranded -N–total-hits-norm–max-bundle-frags 1000000–min-isoform-fraction 0.1–premrna-fraction 0.05–max-intron-length 25000–small-anchor-fraction 0.08–min-frags-per-transfrag 10–overhang-tolerance 8–max-bundle-length 400000–min-intron-length 20–trim-3-avgcov-thresh 10–trim-3-dropoff-frac 0.1–max-multiread-fraction 0.50–overlap-radius 50–3-overhang-tolerance 200–intron-overhang-tolerance 50). The transcript assemblies from males and females were merged and compared with the reference annotation to identify transcripts in previously unannotated intronic and intergenic NTRs.
Preparation of Whole-Transcript Double-Stranded cDNA.
For each of the two replicates for each line and each sex, first-strand cDNA was prepared from 7 μg of total RNA (1 μg/μL) with 1 μL of random primers (3 μg/μL) (Invitrogen) and incubation at 70 °C for 5 min, 25 °C for 5 min, and 4 °C for 10 min. We added 5× first-strand buffer (4 μL) (Invitrogen), 0.1 M DTT (2 μL) (Invitrogen), 10 mM dNTP+dUTP (1 μL) (Promega), RNase Inhibitor (1 μL) (Invitrogen), and SuperScript II (4 μL) (Invitrogen) and incubated the reactions in a thermal cycler (with a heated lid) using the following program: 25 °C for 10 min; 42 °C for 90 min; 70 °C for 10 min; and 4 °C for 10 min. Second-strand cDNA was synthesized by adding 17.5 mM MgCl2 (8 μL) (Sigma), 10 mM dNTP+dUTP (1 μL) (Promega), DNA Polymerase I (1.2 μL) (Promega), RNase H (0.5 μL) (Promega), and RNase-free water (9.3 μL) (Ambion) to the first-strand cDNA reactions. The reactions were incubated in a thermal cycler at 16 °C for 2 h (without a heated lid) followed by 75 °C for 10 min (with a heated lid) and 4 °C for 10 min. Double-stranded cDNA was purified using the QIAquick 96 PCR kit (Qiagen) by following the manufacturer’s protocol except that buffer PN (Qiagen) was used instead of buffer PM (Qiagen). The cDNA was eluted with 45 μL of RNase-free water and was quantified using a NanoDrop 8000 spectrophotometer (Thermo Scientific).
Fragmentation and Biotin Labeling of Double-Stranded cDNA.
The double-stranded cDNA (7.5 μg) was fragmented with 4.8 μL 10× fragmentation buffer (Affymetrix), 1.5 μL Uracil-DNA glycosylase (10 U/μL) (Affymetrix), 2.25 μL apurinic/apyrimidinic endonuclease 1 (100 U/μL) (Affymetrix), and RNase-free water (up to 48 μL) (Affymetrix) using a thermal cycler (with a heated lid) and the following program: 37 °C for 1 h, 93 °C for 2 min, and 4 °C for10 min. The fragmented dsDNA (45 μL) was biotin-labeled by incubation with 12 μL of 5× terminal deoxynucleotidyl transferase (TdT) buffer (Affymetrix), 2 μL of 30 U/μL TdT (Affymetrix), and 1 μL of 5 mM DNA-labeling reagent (Affymetrix) in a thermal cycler (with a heated lid) using the following protocol: 37 °C for 1 h, 70 °C for 10 min, and 4 °C for10 min. Hybridization mixture (164 μL) was added to 7 μg of fragmented and labeled double-stranded cDNA for hybridization to Drosophila 2.0R Tiling Arrays (Affymetrix). We randomized RNA extraction, labeling, and hybridization across all samples.
Quality Control.
We visualized the spatial distribution of probe intensities using the R package Starr to identify technical artifacts on the arrays (e.g., salt rings from reagents). We also considered arrays to be outliers if the mean expression of probes on the array was ± 2 SD of the sample mean from all arrays in the study or if the variance of probe expression was ± 2 SD from the sample mean variance of arrays in the study. We rehybridized samples from all arrays with visible spatial artifacts and all outlier arrays to new arrays, using the same labeled samples used for the original arrays. Of the 192 lines that initially were hybridized to Affymetrix arrays, we retained 185 lines that have sequence data for analysis. Finally, within each sex, we removed replicates that contained excessive numbers of genes that were ± 2 SD from the sample mean. A total of three replicate arrays (two female replicates and one male replicate) were removed.
Preprocessing of Tiling Array Data.
Raw intensities of tiling arrays were extracted from the .CEL files using the R package AffyTiling and were subjected to background correction on a per-array basis using functions modified from the gcrma (version 2.30.0) package to work with tiling arrays. Briefly, nonspecific binding affinities were calculated using 33,886 background probes on each array with varying degrees of GC content. The affinity information then was used to adjust for background hybridization for all D. melanogaster genomic probes on each array through a model-based approach (52). We mapped probes to the reference genome using the Burrows–Wheeler aligner (53) and removed probes that perfectly matched multiple genomic locations. Probes that fell entirely within nonoverlapping constitutive exons as defined by the FlyBase annotation (5.49) and NTRs discovered in the RNA-seq annotation were retained. We further removed probes that overlapped with common (nonreference allele frequency >0.05) variants in the DGRP Freeze 2.0 data (22). Background-corrected intensities for the remaining 499,817 probes were quantile normalized (54) within each sex across arrays using the limma (version 3.14.1) package. Expression for each gene was summarized using median polish.
Quantitative Genetics of Gene Expression.
For each gene-expression trait, we fitted a linear mixed model to partition variation in gene expression into the fixed effects of sex (S, sexual dimorphism in gene expression) and random effects of line (L, genetic variance) and the sex-by-line (SL, genetic variation in the magnitude of sex dimorphism) interaction. The significance of sex effect was tested using a likelihood ratio test comparing the full model and a reduced model without the sex effect. The models were fitted using the lme4 package (version 0.999999-0) in R by maximum likelihood. The significance of the sex-by-line variance was tested using an F test comparing the variance for the SL term and error variance. To estimate H2 for each gene-expression trait in females and males separately, we fitted a linear mixed model with L as a random effect and estimated H2 as /(), where and are the between- and within-line variance components, respectively. h2 was estimated using a mixed linear model with L as a random effect and the covariance matrix determined by the genetic covariance among lines (22), using the rrBLUP package (version 4.0) in R. The effect of Wolbachia and inversions were tested by a likelihood ratio test comparing the full model including Wolbachia infection status, inversion genotypes for In(2L)t, In(2R)NS, In(3R)P, In(3R)K, and In(3R)Mo, and the first 10 PCs of the genotype matrix as fixed effects and L as a random effect, with a reduced nested model without the tested term. PCs were obtained using the EIGENSTRAT software (55) on linkage disequilibrium-pruned genotypes and excluding regions harboring the inversions.
GSEA.
We performed GSEA on the list of genes ranked by their sex effect using a previously described procedure (56). We transformed t statistics to a signed correlation score , where n is the number of lines and s(t) indicates the sign of the t statistic. An empirical FDR was determined by permuting the sex label within each line 1,000 times and estimating the expected number of gene sets passing a certain threshold under the null hypothesis. Because the sex effect is large, unbalanced permutation can bias the estimated sex effect substantially. We removed one line from the dataset to ensure that balanced permutation (the same number of females and males) can be performed properly. A similar GSEA was performed to annotate NTRs in which the GSEA operated on the ranked list of annotated genes based on their correlation with the NTR.
Mapping eQTL for Mean Transcript Abundance.
eQTLs for mean gene expression were mapped using linear regression implemented in PLINK (57), separately for males and females. The BLUP line means were first estimated using a mixed model adjusting for Wolbachia, inversions, and PCs and then were regressed on marker genotypes to obtain a P value for each pair of markers and transcripts. To estimate the empirical FDR, we permuted line labels 100 times, retaining the correlation structure among the genes, and performed the same single-marker regressions for the permuted phenotypes. The FDR was estimated by dividing the average number of significant markers meeting a certain threshold in the 100 permutations by the number of significant markers in the observed dataset. To arrive at a model with independent associations, forward model selection was performed on significant markers. In each step, a marker with the smallest type III F test P value was added to the model until no marker could be added with a P < 10−5. Gene-based association tests were performed using the sequence kernel association test (SKAT) (58) implemented in the SKAT (version 0.95) package in R. The empirical FDR was determined using the same permuted dataset and a procedure similar to that described above for the marker-based tests.
Mapping veQTL and Epistasis.
For each gene, veQTLs were mapped by testing for equal variance among the lines carrying the two alleles for each marker using Levene’s test. Empirical FDR was estimated by permutation as described above. To select for markers that independently control variance of gene expression, a forward selection procedure was performed on significant veQTLs. In each step, a marker with the smallest Levene’s test P value was retained; then the variance within each genotype class was scaled to unit variance while preserving the phenotypic mean. This process was repeated with the remaining markers until no marker could be added with a P value smaller than 10−5. To identify cis variants that interact epistatically with veQTLs, the model y = μ + Mv + Mc + Mv:Mc + e was fitted to each gene, where y is the adjusted gene expression, μ is an intercept, Mv, Mc, and Mv:Mc are the effects of the veQTL, cis variant, and their interaction, respectively, and e is residual. This model was fitted for all pairs of veQTLs and all cis (within 1 kb) variants of the gene. The significance of the interaction term was evaluated using an F test. The empirical FDR was calculated by permuting the gene expression and veQTL genotype together (thus a veQTL is still a veQTL after permutation) 100 times and dividing the observed number of significant hits by the expected number of significant hits at variable thresholds.
Availability of Supporting Data.
The pooled RNA sequences from 192 DGRP lines have been deposited in Gene Expression Omnibus (GEO accession no. GSE67505). All tiling array CEL files used in this study have been deposited at ArrayExpress (accession no. E-MTAB-3216).
Supplementary Material
Acknowledgments
We thank Gunjan Arya, Julien Ayroles, Terry Campbell, Kultaran Chohan, Charlene Couch, Kyle Craver, Laura Duncan, Alden Hearn, George Khan, Faye Lawrence, Lenovia McCoy, Tatiana Morozova, Beth Ruedi, Yazmin Serrano-Negron, Shilpa Swarup, Crystal Tabor, Lavanya Turlapati, Allison Weber, Akihiko Yamamoto, and Shanshan Zhou for technical assistance and collecting samples for gene-expression analysis. This work was supported by NIH Grants R01 GM45146 (to T.F.C.M., R.R.H.A., and E.A.S.) and R01 AA016560, R01 GM076083, and R01 GM59469 (to T.F.C.M. and R.R.H.A.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the Gene Expression Omnibus data bank (accession no. GSE67505) and in the ArrayExpress database (accession no. E-MTAB-3216).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1519159112/-/DCSupplemental.
References
- 1.Flint J, Mackay TFC. Genetic architecture of quantitative traits in mice, flies, and humans. Genome Res. 2009;19(5):723–733. doi: 10.1101/gr.086660.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Manolio TA, et al. Finding the missing heritability of complex diseases. Nature. 2009;461(7265):747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mackay TFC, Stone EA, Ayroles JF. The genetics of quantitative traits: Challenges and prospects. Nat Rev Genet. 2009;10(8):565–577. doi: 10.1038/nrg2612. [DOI] [PubMed] [Google Scholar]
- 4.Nicolae DL, et al. Trait-associated SNPs are more likely to be eQTLs: Annotation to enhance discovery from GWAS. PLoS Genet. 2010;6(4):e1000888. doi: 10.1371/journal.pgen.1000888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brem RB, Yvert G, Clinton R, Kruglyak L. Genetic dissection of transcriptional regulation in budding yeast. Science. 2002;296(5568):752–755. doi: 10.1126/science.1069516. [DOI] [PubMed] [Google Scholar]
- 6.Ayroles JF, et al. Systems genetics of complex traits in Drosophila melanogaster. Nat Genet. 2009;41(3):299–307. doi: 10.1038/ng.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schadt EE, et al. Genetics of gene expression surveyed in maize, mouse and man. Nature. 2003;422(6929):297–302. doi: 10.1038/nature01434. [DOI] [PubMed] [Google Scholar]
- 8.Cheung VG, et al. Natural variation in human gene expression assessed in lymphoblastoid cells. Nat Genet. 2003;33(3):422–425. doi: 10.1038/ng1094. [DOI] [PubMed] [Google Scholar]
- 9.West MAL, et al. Global eQTL mapping reveals the complex genetic architecture of transcript-level variation in Arabidopsis. Genetics. 2007;175(3):1441–1450. doi: 10.1534/genetics.106.064972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang X, Cal AJ, Borevitz JO. Genetic architecture of regulatory variation in Arabidopsis thaliana. Genome Res. 2011;21(5):725–733. doi: 10.1101/gr.115337.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hulse AM, Cai JJ. Genetic variants contribute to gene expression variability in humans. Genetics. 2013;193(1):95–108. doi: 10.1534/genetics.112.146779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brown AA, et al. 2014. Genetic interactions affecting human gene expression identified by variance association mapping. Elife 2014(3):e01381.
- 13.Nelson RM, Pettersson ME, Li X, Carlborg Ö. Variance heterogeneity in Saccharomyces cerevisiae expression data: Trans-regulation and epistasis. PLoS One. 2013;8(11):e79507. doi: 10.1371/journal.pone.0079507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rönnegård L, Valdar W. Detecting major genetic loci controlling phenotypic variability in experimental crosses. Genetics. 2011;188(2):435–447. doi: 10.1534/genetics.111.127068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shen X, Pettersson M, Rönnegård L, Carlborg Ö. Inheritance beyond plain heritability: Variance-controlling genes in Arabidopsis thaliana. PLoS Genet. 2012;8(8):e1002839. doi: 10.1371/journal.pgen.1002839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang J, et al. FTO genotype is associated with phenotypic variability of body mass index. Nature. 2012;490(7419):267–272. doi: 10.1038/nature11401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hall MC, Dworkin I, Ungerer MC, Purugganan M. Genetics of microenvironmental canalization in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2007;104(34):13717–13722. doi: 10.1073/pnas.0701936104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ansel J, et al. Cell-to-cell stochastic variation in gene expression is a complex genetic trait. PLoS Genet. 2008;4(4):e1000049. doi: 10.1371/journal.pgen.1000049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jimenez-Gomez JM, Corwin JA, Joseph B, Maloof JN, Kliebenstein DJ. Genomic analysis of QTLs and genes altering natural variation in stochastic noise. PLoS Genet. 2011;7(9):e1002295. doi: 10.1371/journal.pgen.1002295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morgante F, Sørensen P, Sorensen DA, Maltecca C, Mackay TFC. Genetic architecture of micro-environmental plasticity in Drosophila melanogaster. Sci Rep. 2015;5:9785. doi: 10.1038/srep09785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mackay TFC, et al. The Drosophila melanogaster Genetic Reference Panel. Nature. 2012;482(7384):173–178. doi: 10.1038/nature10811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang W, et al. Natural variation in genome architecture among 205 Drosophila melanogaster Genetic Reference Panel lines. Genome Res. 2014;24(7):1193–1208. doi: 10.1101/gr.171546.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang W, et al. Epistasis dominates the genetic architecture of Drosophila quantitative traits. Proc Natl Acad Sci USA. 2012;109(39):15553–15559. doi: 10.1073/pnas.1213423109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Swarup S, Huang W, Mackay TFC, Anholt RRH. Analysis of natural variation reveals neurogenetic networks for Drosophila olfactory behavior. Proc Natl Acad Sci USA. 2013;110(3):1017–1022. doi: 10.1073/pnas.1220168110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mackay TFC. Epistasis and quantitative traits: Using model organisms to study gene-gene interactions. Nat Rev Genet. 2014;15(1):22–33. doi: 10.1038/nrg3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Massouras A, et al. Genomic variation and its impact on gene expression in Drosophila melanogaster. PLoS Genet. 2012;8(11):e1003055. doi: 10.1371/journal.pgen.1003055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Djebali S, et al. Landscape of transcription in human cells. Nature. 2012;489(7414):101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dinger ME, Amaral PP, Mercer TR, Mattick JS. Pervasive transcription of the eukaryotic genome: Functional indices and conceptual implications. Brief Funct Genomics Proteomics. 2009;8(6):407–423. doi: 10.1093/bfgp/elp038. [DOI] [PubMed] [Google Scholar]
- 29.Graveley BR, et al. The developmental transcriptome of Drosophila melanogaster. Nature. 2011;471(7339):473–479. doi: 10.1038/nature09715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee JT. Epigenetic regulation by long noncoding RNAs. Science. 2012;338(6113):1435–1439. doi: 10.1126/science.1231776. [DOI] [PubMed] [Google Scholar]
- 31.Ranz JM, Castillo-Davis CI, Meiklejohn CD, Hartl DL. Sex-dependent gene expression and evolution of the Drosophila transcriptome. Science. 2003;300(5626):1742–1745. doi: 10.1126/science.1085881. [DOI] [PubMed] [Google Scholar]
- 32.Parisi M, et al. A survey of ovary-, testis-, and soma-biased gene expression in Drosophila melanogaster adults. Genome Biol. 2004;5(6):R40. doi: 10.1186/gb-2004-5-6-r40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang J, et al. Common SNPs explain a large proportion of the heritability for human height. Nat Genet. 2010;42(7):565–569. doi: 10.1038/ng.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ober U, et al. Using whole-genome sequence data to predict quantitative trait phenotypes in Drosophila melanogaster. PLoS Genet. 2012;8(5):e1002685. doi: 10.1371/journal.pgen.1002685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hill WG, Goddard ME, Visscher PM. Data and theory point to mainly additive genetic variance for complex traits. PLoS Genet. 2008;4(2):e1000008. doi: 10.1371/journal.pgen.1000008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stone EA, Ayroles JF. Modulated modularity clustering as an exploratory tool for functional genomic inference. PLoS Genet. 2009;5(5):e1000479. doi: 10.1371/journal.pgen.1000479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ayroles JF, Laflamme BA, Stone EA, Wolfner MF, Mackay TF. Functional genome annotation of Drosophila seminal fluid proteins using transcriptional genetic networks. Genet Res. 2011;93(6):387–395. doi: 10.1017/S0016672311000346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81(1):145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ronald J, Brem RB, Whittle J, Kruglyak L. Local regulatory variation in Saccharomyces cerevisiae. PLoS Genet. 2005;1(2):e25. doi: 10.1371/journal.pgen.0010025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stranger BE, et al. Population genomics of human gene expression. Nat Genet. 2007;39(10):1217–1224. doi: 10.1038/ng2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Veyrieras JB, et al. High-resolution mapping of expression-QTLs yields insight into human gene regulation. PLoS Genet. 2008;4(10):e1000214. doi: 10.1371/journal.pgen.1000214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Levene H. Robust tests for equality of variances. In: Olkin I, Ghurye SG, Hoeffding W, Madow WG, Mann HB, editors. Contributions to Probability and Statistics, Essays in Honor of Harold Hotelling. Stanford Univ Press; Palo Alto, CA: 1960. pp. 278–292. [Google Scholar]
- 43.Stuart JM, Segal E, Koller D, Kim SK. A gene-coexpression network for global discovery of conserved genetic modules. Science. 2003;302(5643):249–255. doi: 10.1126/science.1087447. [DOI] [PubMed] [Google Scholar]
- 44.Kliebenstein DJ, et al. Identification of QTLs controlling gene expression networks defined a priori. BMC Bioinformatics. 2006;7:308. doi: 10.1186/1471-2105-7-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Emilsson V, et al. Genetics of gene expression and its effect on disease. Nature. 2008;452(7186):423–428. doi: 10.1038/nature06758. [DOI] [PubMed] [Google Scholar]
- 46.Wentzell AM, et al. Linking metabolic QTLs with network and cis-eQTLs controlling biosynthetic pathways. PLoS Genet. 2007;3(9):1687–1701. doi: 10.1371/journal.pgen.0030162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brown CD, Mangravite LM, Engelhardt BE. Integrative modeling of eQTLs and cis-regulatory elements suggests mechanisms underlying cell type specificity of eQTLs. PLoS Genet. 2013;9(8):e1003649. doi: 10.1371/journal.pgen.1003649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou S, Campbell TG, Stone EA, Mackay TFC, Anholt RRH. Phenotypic plasticity of the Drosophila transcriptome. PLoS Genet. 2012;8(3):e1002593. doi: 10.1371/journal.pgen.1002593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Trapnell C, Pachter L, Salzberg SL. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28(5):511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roberts A, Pimentel H, Trapnell C, Pachter L. Identification of novel transcripts in annotated genomes using RNA-Seq. Bioinformatics. 2011;27(17):2325–2329. doi: 10.1093/bioinformatics/btr355. [DOI] [PubMed] [Google Scholar]
- 52.Wu Z, Irizarry RA, Gentleman R, Martinez-Murillo F, Spencer F. A model-based background adjustment for oligonucleotide expression arrays. J Am Stat Assoc. 2004;99(468):909–917. [Google Scholar]
- 53.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bolstad BM, Irizarry RA, Åstrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19(2):185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 55. Price AL, et al. (2006) Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 38(8):904–909. [DOI] [PubMed]
- 56.Subramanian A, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Purcell S, et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu MC, et al. Rare-variant association testing for sequencing data with the sequence kernel association test. Am J Hum Genet. 2011;89(1):82–93. doi: 10.1016/j.ajhg.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.