Significance
Five noncoding small RNAs (sRNAs) called the Qrr1-5 sRNAs act at the heart of the Vibrio harveyi quorum-sensing cascade. The Qrr sRNAs posttranscriptionally regulate 20 mRNA targets. Here, we use a method we call RSort-Seq that is based on unbiased high-throughput screening to define the critical bases in Qrr4 that specify its function. The power of our study comes from using the screening results to pinpoint particular nucleotides for follow-up biological analyses that define function. Using this approach, we discover how Qrr4 differentially regulates two of its targets, luxO and luxR. We also show how this strategy can be used to identify intramolecular suppressor mutations. This approach can be applied to any sRNA and any mRNA target.
Keywords: quorum sensing, regulatory sRNA, Qrr, RSort-Seq, regulation
Abstract
Five homologous noncoding small RNAs (sRNAs), called the Qrr1-5 sRNAs, function in the Vibrio harveyi quorum-sensing cascade to drive its operation. Qrr1-5 use four different regulatory mechanisms to control the expression of ∼20 mRNA targets. Little is known about the roles individual nucleotides play in mRNA target selection, in determining regulatory mechanism, or in defining Qrr potency and dynamics of target regulation. To identify the nucleotides vital for Qrr function, we developed a method we call RSort-Seq that combines saturating mutagenesis, fluorescence-activated cell sorting, high-throughput sequencing, and mutual information theory to explore the role that every nucleotide in Qrr4 plays in regulation of two mRNA targets, luxR and luxO. Companion biochemical assays allowed us to assign specific regulatory functions/underlying molecular mechanisms to each important base. This strategy yielded a regional map of nucleotides in Qrr4 vital for stability, Hfq interaction, stem-loop formation, and base pairing to both luxR and luxO, to luxR only, and to luxO only. In terms of nucleotides critical for sRNA function, the RSort-Seq analysis provided strikingly different results from those predicted by commonly used regulatory RNA-folding algorithms. This approach is applicable to any RNA–RNA interaction, including sRNAs in other bacteria and regulatory RNAs in higher organisms.
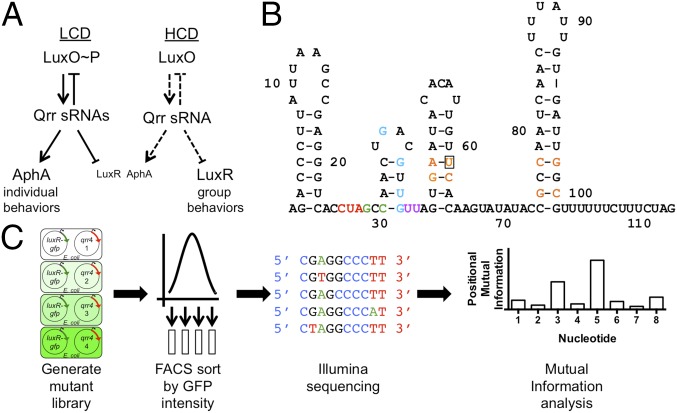
Quorum sensing is a bacterial cell–cell communication process that involves the production, release, and population-wide detection of extracellular signaling molecules called autoinducers (AIs) (1, 2). Quorum sensing controls collective behaviors including bioluminescence, virulence factor production, and biofilm formation (3–5). Vibrio harveyi produces three AIs that are decoded by three cognate receptors (6–12). At low cell density (LCD), the receptors act as kinases and shuttle phosphate to the response regulator LuxO (Fig. 1A) (13, 14). LuxO∼P activates transcription of genes encoding five homologous noncoding small RNAs (sRNAs) called the quorum-regulatory RNAs 1–5, or Qrr1-5 (15, 16). The Qrr sRNAs, in conjunction with the chaperone Hfq, activate translation of the LCD master regulator called AphA and destabilize the mRNA encoding LuxR, the high-cell density (HCD) master regulator (16, 17). AphA controls genes that underpin individual behaviors (17, 18). At HCD, AI binding converts the receptors to phosphatases, resulting in dephosphorylation and inactivation of LuxO, and cessation of qrr expression (13, 15). This event terminates Qrr activation of AphA translation, and luxR mRNA is stabilized, leading to LuxR production. LuxR controls the expression of genes that underlie collective processes (17–20). The Qrr sRNAs feed back to repress luxO, and they repress or activate 17 additional mRNA targets (21).
Fig. 1.
Qrr sRNAs play a central role in V. harveyi quorum sensing. (A) Key features of Qrr sRNA regulation of quorum sensing. (B) Predicted structure of Qrr4. Bases important for repression of luxO (red), repression of luxR (blue), repression of both luxO and luxR (green), Hfq binding (purple), and stem-loop formation (orange) as revealed by our analysis. Nucleotide U61, enclosed by a box, plays an additional role in Qrr4 function. (C) Flowchart summarizing the RSort-Seq procedure.
The Qrr sRNAs control mRNA translation by at least four regulatory mechanisms, which are dictated by the Qrr-target mRNA base-pairing patterns (22). All five Qrr sRNAs are predicted to fold into similar secondary structures possessing four stem-loops (e.g., Qrr4 in Fig. 1B) (16). The sequences of the Qrr sRNAs are up to 90% identical, and conservation is especially high within the predicted base-pairing regions (16). Thus, the elements driving specificity in mRNA target selection must reside in the 5′-UTRs of the target mRNAs and in nonconserved bases in the Qrr sRNAs that reside outside of the predicted base-pairing regions. Our goal was to define the contributions each individual Qrr nucleotide makes to function to learn the determinants that govern Qrr sRNA’s discriminatory power.
Traditional approaches to discovering what roles particular nucleotides in sRNAs play in binding to target mRNAs have relied on predicting sRNA–mRNA pairing with algorithms, engineering of mutations in one or a few nucleotides in the sRNA, assaying for a defect, and subsequently exploring restoration of function by generating compensatory mutations in the partner mRNA target (23–26). Such strategies have provided insight into sRNA regulatory mechanisms; however, they are labor intensive so only a few nucleotides can be reasonably analyzed at a time. Furthermore, the findings are restricted to functions that can be predicted algorithmically, e.g., base-pairing and stem-loop formation. Additionally, the role one sRNA nucleotide plays in regulation of one mRNA target (e.g., base pairing) might not be identical to the role it plays in regulating another mRNA target. Furthermore, often a single nucleotide in an sRNA can play multiple regulatory roles, for example in base pairing and in Hfq binding, complicating these kinds of analyses.
Inspired by the development of the Sort-Seq method that enabled highly parallel quantitative interrogation of protein–DNA interactions and deep-sequencing methods developed to study mutations in bacterial sRNAs and 5′-UTRs of target mRNAs (27–30), we crafted an unbiased, quantitative, high-throughput strategy to systematically define which nucleotides in an sRNA are critical for its function (Fig. 1C). We call our method “RSort-Seq.” Our goal was to rapidly pinpoint vital nucleotides so that follow-up mechanistic analyses could be restricted to only a few, and very particular, nucleotides. Here, we coupled saturating mutagenesis with fluorescence-activated cell sorting (FACS) and Illumina sequencing to define every alteration in qrr4 that disrupted repression of luxR, of luxO, or of both luxR and luxO. We examined the contribution of each nucleotide to regulation of each target by using mutual information (MI) theory to compute the information footprint of the single mutations on the distribution of fluorescence intensities (27, 28). We used a set of in vivo and in vitro assays to define how the identified bases specifically contribute to Qrr function. Our analysis identified the nucleotides in Qrr4 required for stability, Hfq binding, base pairing to one or both mRNA targets, and stem-loop formation, enabling us to understand how Qrr sRNAs select specific mRNA targets for regulation.
Results
RSort-Seq Analysis Identifies Nucleotides in Qrr4 Critical for Regulation of luxR and luxO.
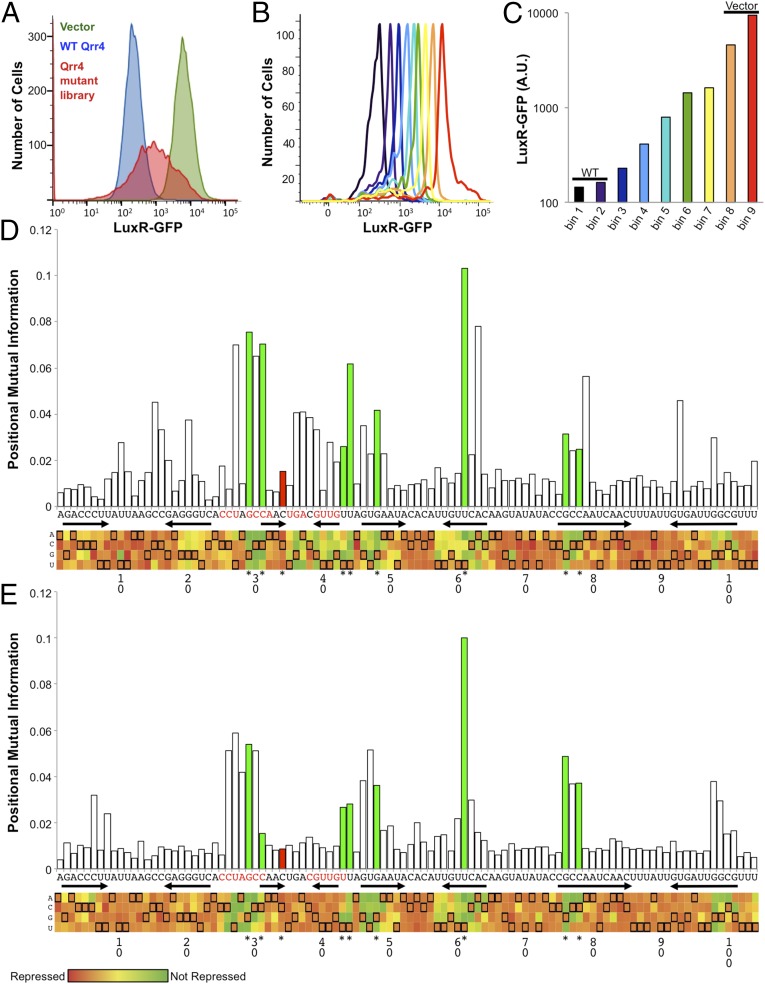
To understand the contributions of individual Qrr4 bases to mRNA target regulation and to specifying target preferences, we designed a screen to probe the importance of every nucleotide of Qrr4 in posttranscriptional repression of two targets, luxR and luxO. We explain the strategy, which we call RSort-Seq (28), using luxR as the target; the identical procedure was used for luxO. We introduced inducible, plasmid-borne luxR-gfp (21, 22) into recombinant Escherichia coli. Upon induction, the cells produced high levels of GFP, as measured by flow cytometry (Fig. 2A, green). Subsequent introduction and induction of a plasmid carrying qrr4 caused a 20-fold decrease in GFP production, showing that wild-type (WT) Qrr4 represses luxR in our heterologous expression system (Fig. 2A, blue). LuxR-GFP exhibited a single narrow peak in the presence and absence of Qrr4 production, showing that LuxR-GFP production is uniform across the population.
Fig. 2.
RSort-Seq provides a regional footprint of bases important for Qrr4 function. (A) Flow cytometry traces of E. coli cells expressing luxR-gfp alone (green), in combination with WT Qrr4 (blue), and in the presence of a Qrr4 mutant library (red). (B) Flow cytometry traces of sorted bins of E. coli carrying the Qrr4 mutant library (resorted red peak in A). (C) Average fluorescence from luxR-gfp in each bin containing subsets of Qrr4 mutants. (D) MI footprint of Qrr4 with respect to repression of luxR-gfp. (Top) Positional MI between fluorescence bin and base identity in bits for each base position. Each bar represents a base in Qrr4 from 5′ to 3′ going Left to Right. Green bars represent retested base positions with high MI, where mutations impair regulation. The red bar represents a control base position that possesses low MI, where mutations do not impair regulation. (Bottom) The heat map shows the average fluorescence of each mutation in Qrr4. Each column represents a base in Qrr4 from 5′ to 3′ going Left to Right. Each square represents a potential mutation. The WT sequence is shown by the orange squares with black outlines. The color of each square represents the effect of that Qrr4 mutation on average fluorescence from decreased fluorescence (red) to little effect (orange) to increased fluorescence (green). Arrows denote the locations of predicted stem-loops. The red nucleotides show the location of the predicted base-pairing region. Asterisks denote bases that were retested to verify the screen results. (E) As in D, positional MI footprint and heat map for Qrr4 with respect to regulation of luxO.
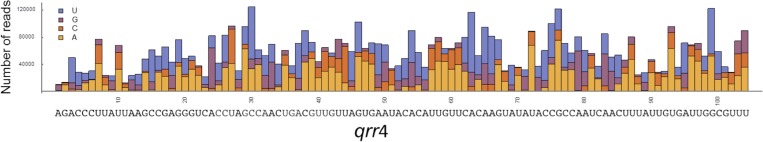
We next used saturating mutagenesis to generate a library of ∼100,000 qrr4 mutants, primarily containing single point mutations (Table S1). The method provides full coverage with all three possible substitutions at each position in qrr4 (Fig. S1). We introduced the mutant qrr4 library into the E. coli strain carrying luxR-gfp, grew this culture to stationary phase, and measured gfp expression. Following mutagenesis, the average luxR-gfp expression was reduced sevenfold and the range of GFP levels produced expanded 50-fold (Fig. 2A, red). We interpret this to mean that the different mutant Qrr4 sRNAs repress luxR-gfp expression to different extents. The culture containing the library was sorted using FACS, and clones carrying different Qrr4 alleles were partitioned into nine bins of ∼150,000 clones/bin based on repression activity (Fig. 2B). Thus, each bin contained Qrr4 mutants displaying a similar level of repression of luxR-gfp (Fig. 2C). WT Qrr4 repression of luxR-gfp was captured by bins 1 and 2. No repression (e.g., empty vector control) was approximately captured by bins 8 and 9 (Fig. 2C). All of the qrr4 genes in each bin were sequenced simultaneously using Illumina next-generation sequencing.
Fig. S1.
Total number of reads for each single substitution qrr4 mutant identified by RSort-Seq (related to Fig. 2). The number of reads is shown as a function of each position. All three possible substitutions were obtained at each position in qrr4 as assessed by RSort-Seq with luxR as the target. Although not uniform, all single mutations were represented by at least many hundreds of reads.
To screen for nucleotides that are crucial for Qrr4 regulatory functions, we quantified the effects of single mutations on repression levels of luxR-gfp and we show the results using a positional MI footprint (Fig. 2D) (27, 28, 31, 32). This information footprint was obtained by computing, for each position in the Qrr4 sequence, the MI between the identity of the nucleotide at that position and the repression level of luxR-gfp. Importantly, MI is the most general measure of the statistical dependence between mutation and repression level, which may take the form of mutation-dependent mean repression, mutation-dependent variance in repression, or more complex dependences (33, 34). Intuitively, if a nucleotide at position i in the sRNA plays no role in mRNA target regulation, the bin distribution of FACS-sorted cells carrying a mutation at position i will be the same as the WT distribution. Thus, one cannot gain information about the nucleotide identity at position i by observing the level of repression: the MI is zero. By contrast, if a mutation at position i substantially alters repression levels, the distribution of FACS-sorted cells carrying the mutation will differ from the WT distribution, possibly in a complex fashion, but always yielding a nonzero MI. Specifically, the value of the MI measures how well one can predict the identity of the nucleotide by observing the fluorescence-bin distribution of mutated cells, providing us with a score to assess the importance of the nucleotide in the Qrr4 sequence.
MI encapsulates many aspects of statistical dependence in one number. In the present context, the statistical dependence measured by the MI is well captured by the mean repression level of luxR-gfp. Positions with a high MI score (Fig. 2D) are those for which repression is substantially impaired on average. To visualize and interpret the effect of every possible nucleotide change in Qrr4, we represented the mean repression levels of luxR-gfp as a heat map (Fig. 2D). For every position in the sequence, the three colored squares show the population-averaged repression level for the three possible single mutations of Qrr4 at that position. Thus, the ith column of the heat map depicts the “mutation profile” of position i. A black frame singles out the repression level for the WT Qrr4 sequence, color-coded by the same orange hue at every WT sequence position. Low average fluorescence, indicated by red squares, pinpoints mutations predicted to have little to no effect on Qrr4 repression of luxR-gfp (e.g., Qrr4-C34G). High average fluorescence, denoted by green squares, shows nucleotides that, when altered, are predicted to severely impair Qrr4 repression of luxR-gfp (e.g., Qrr4-U44A). Colors on the spectrum spanning red to green (i.e., orange and yellow) indicate intermediate defects in repression. We note that, at some positions, each of the three possible nucleotide substitutions results in a different level of repression (e.g., Qrr4-C78) and presumably a correspondingly different severity in phenotype.
We performed exactly this analysis using luxO-gfp as the mRNA target (Fig. 2E). Several nucleotide positions exhibit loss of repression for both targets. These nucleotides are located throughout the length of Qrr4 and thus include nucleotides in the proposed base-pairing region (e.g., Qrr4-G29) as well as outside of that region (e.g., Qrr4-U61). Most of the nucleotides that exhibit different MI with respect to luxR and luxO regulation lie within the previously proposed base-pairing region (e.g., Qrr4-U41) (Fig. 2 D and E). In summary, the MI analysis allowed us to rapidly identify interesting Qrr4 nucleotides for further study.
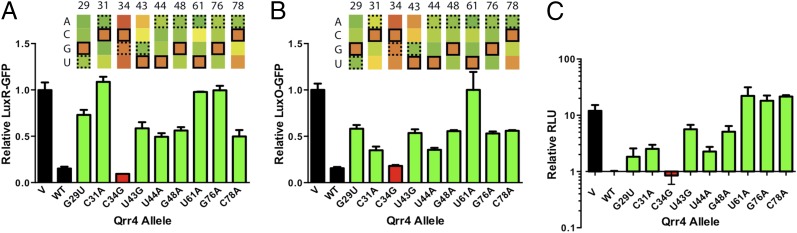
Validating the Predictive Power of MI in E. coli and V. harveyi.
To confirm that the MI scores track with the ability/inability of the Qrr4 mutants to repress luxR, luxO, or both mRNA targets, we constructed representative Qrr4 mutants and measured regulation of luxR-gfp and luxO-gfp in E. coli (Fig. 3 A and B). First, we focused on mutations that the MI analysis predicted affect both luxR and luxO regulation. We selected positions spanning the entirety of Qrr4: in the predicted base-pairing region (Qrr4-G29U, Qrr4-C31A, Qrr4-U43G, and Qrr4-U44A), SL2 (Qrr4-C31A), SL3 (Qrr4-G48A and Qrr4-U61A), and SL4 (Qrr4-G76A and Qrr4-G78A) (Figs. 2 D and E and 3 A and B, green). Qrr4-C34G, which has low MI for both targets, served as a control for a mutant with WT function (Figs. 2 D and E and 3 A and B, red).
Fig. 3.
RSort-Seq predicts mutations that impair Qrr4 function in E. coli and in V. harveyi. (A) Individual mutations in Qrr4 predicted to have defects examined for repression of luxR-gfp in E. coli. Black bars show fluorescence levels from luxR-gfp in the presence of a vector control (V) and WT Qrr4 (WT). Green bars denote mutations that impair repression. The red bar shows a control base that displays WT repression. The heat map presents results from the RSort-Seq analysis for reference, as in Fig. 2D. Error bars are SDs of at least three trials. (B) As in A for luxO-gfp. (C) The effects of Qrr4 mutations on light production in V. harveyi at OD600 = 0.1 are shown. Relative light units (RLU) are counts per minute per milliliter per OD600. Error bars are SDs of at least three trials.
WT Qrr4 repressed luxR and luxO 10-fold and 6-fold, respectively, under our conditions (Fig. 3 A and B, black). Mutations at positions with high MI for both targets (Fig. 3 A and B, green) dramatically impaired Qrr4 repression in both cases. By contrast, Qrr4-C34G (Fig. 3 A and B, red), the control mutant exhibiting low MI, behaved similarly to WT. In most instances, the higher the MI, the stronger the Qrr4 defect in target repression. For example, compare regulation of luxR-gfp and luxO-gfp by Qrr4-C31A, which had a significantly higher MI score for luxR than for luxO, and would thus be predicted to be more defective in regulation of luxR than luxO. Consistent with this prediction, the Qrr4-C31A mutation eliminated Qrr4 repression of luxR-gfp but only modestly impaired Qrr4 repression of luxO-gfp.
We next investigated whether the representative Qrr4 mutations affect quorum sensing in vivo in V. harveyi. We introduced our qrr4 mutants into a V. harveyi Δqrr1-5 strain and used bioluminescence, the canonical quorum-sensing behavior, as the readout (16). At OD600 = 0.1, (i.e., LCD), the Δqrr1-5 V. harveyi strain was 13-fold brighter than the strain expressing WT qrr4 from a plasmid (Fig. 3C). Introduction of Qrr4-C34G, which had low MI, produced the WT phenotype. By contrast, all of the Qrr4 mutants that had high MI had impaired quorum-sensing–dependent regulation of light production. Mutations residing in SL3 (Qrr4-U61A) and SL4 (Qrr4-G76A and Qrr4-C78A) produced the strongest phenotypes (Fig. 3C).
We note a major difference between our findings in V. harveyi and those in recombinant E. coli with respect to the severity of the Qrr4 defects. The MI analysis and the E. coli experiments suggest that substitutions in the Qrr4 base-pairing region produce effects similar to mutations in SL3 and SL4. However, in V. harveyi, SL mutations were more severe than base-pairing mutations. We hypothesize that this discrepancy occurs because, in vivo, mutations in the Qrr4 base-pairing region likely affect only a subset of Qrr targets, even if those targets are affected severely. In vivo, multiple mRNA targets vie for attention of Qrr4; loss of regulation of one target due to mutation could result in a surplus of Qrr4 available for regulation of another target, thus modulating the effect of a mutation (22). By contrast, mutations in SL3 and SL4 impair Qrr4 regulation of all targets, and thus produce a stronger overall quorum-sensing defect in vivo. In recombinant E. coli, we examined one mRNA target in isolation, thus enabling us to exactly quantify the effect of a given Qrr4 mutation on a given target. In V. harveyi, by contrast, we quantified the effect of a given Qrr4 mutation on the entire quorum-sensing process.
Definition of the Mechanisms Underpinning the Qrr4 Defects.
Our next goal was to define the mechanisms by which individual mutations disrupted Qrr4 function. We used assays for decreased Qrr4 stability, loss of Qrr4 binding to Hfq, defective Qrr4 stem-loop formation, and altered Qrr4-mRNA target base pairing.
Reduced Qrr4 levels.
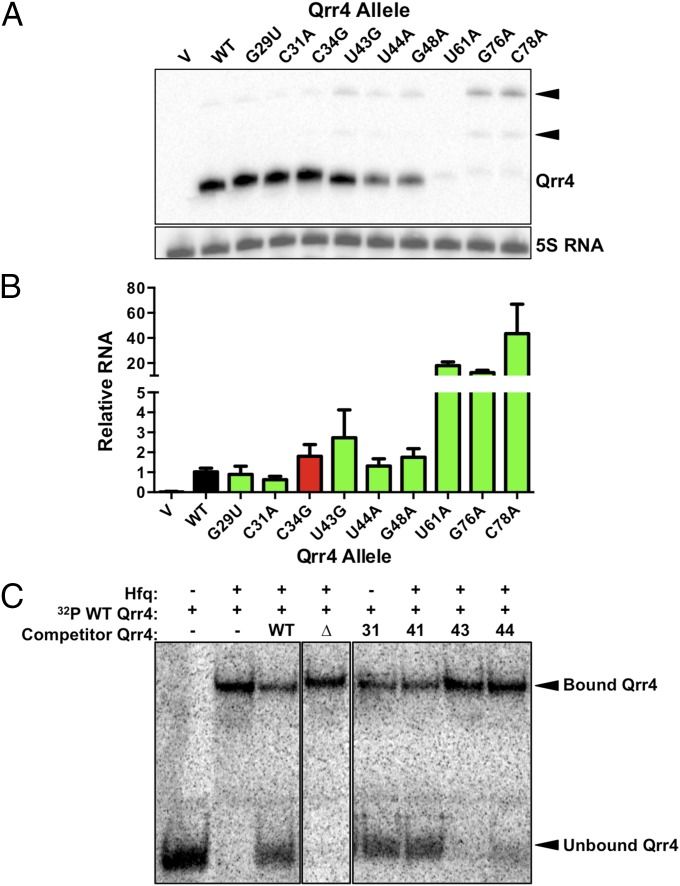
First, we measured the levels of all of our Qrr4 mutants to identify mutations that altered Qrr4 production and/or stability. We isolated the WT and the mutant Qrr4 RNAs from E. coli (in the absence of mRNA targets) and assessed Qrr4 levels by Northern blot (Fig. 4A). Qrr4 was expressed from the identical inducible promoter in all cases, so differences in Qrr4 levels could be attributed to altered stability or postinitiation changes in transcription caused by the mutation. The control Qrr4-C34G mutation and the Qrr4-G29U, Qrr4-C31A, and Qrr4-U43G mutations in the putative Qrr4-target mRNA base-pairing region did not cause any decrease in Qrr4 levels compared with WT, whereas Qrr4-U44A is present at modestly reduced levels (Fig. 4A). The most drastic effects on Qrr4 levels occurred in Qrr4 variants with mutations in SL3 and SL4. With respect to SL3, Qrr4-G48A showed a modest decrease, whereas Qrr4-U61A was nearly undetectable by Northern blot (Fig. 4A). Both of the SL4 Qrr4-G76A and Qrr4-C78A mutants exhibited strikingly lower amounts of RNA than WT at the expected size. Larger RNA species were present on the blot in both cases, showing that terminator readthrough occurs, which is consistent with SL4 acting as the terminator. We conclude that Qrr4-U44A, Qrr4-G48A, Qrr4-U61A, Qrr4-G76A, and Qrr4-C78A are defective because decreased levels of these mutant Qrr4 RNAs are present to regulate target mRNAs.
Fig. 4.
Qrr4 mutations can decrease the levels of Qrr4 and its ability to bind Hfq. (A) Representative Northern blot showing WT and mutant Qrr4 levels in E. coli. The WT size band is denoted Qrr4. Larger RNA species are denoted by black triangles. 5S RNA is the loading control. (B) Real-time PCR analysis of WT and mutant Qrr4 levels in V. harveyi at OD600 = 0.1. The black bar shows the relative amount of WT Qrr4. Green bars show relative amounts of Qrr4 mutants with impaired repression. The red bar denotes the level of a control Qrr4 mutant that displays WT repression. (C) Representative native gel showing unbound Qrr4 and Qrr4 bound to Hfq. The Δ represents Qrr4Δ68-115, a non-Hfq-binding control; 31 represents Qrr4-C31A; 41 represents Qrr4-U41A; 43 represents Qrr4-U43G; and 44 represents Qrr4-U44A.
We also measured the levels of the above Qrr4 alleles in V. harveyi at LCD. As in E. coli, none of the Qrr4 mutants with alterations in the putative base-pairing region or in SL2 exhibited significantly altered Qrr4 levels (Fig. 4B). Surprisingly however, Qrr4-U61A, Qrr4-G76A, and Qrr4-C78A, which had the most dramatically reduced levels in E. coli, were present at significantly higher levels than WT Qrr4 in V. harveyi (Fig. 4B). We hypothesize that this result is a consequence of the loss of feedback repression on luxO that occurs in V. harveyi but not in recombinant E. coli (35). Specifically, Qrr4-U61A, Qrr4-G76A, and Qrr4-C78A are impaired for repression of luxO (Fig. 3B). The absence of the Qrr–|LuxO negative-feedback loop enables LuxO to drive increased production of Qrr4-U61A, Qrr4-G76A, and Qrr4-C78A (Fig. 1, Left). A second hypothesis is that the increased levels in V. harveyi are a result of loss of target regulation. Base pairing with certain targets results in degradation of the Qrr sRNA (22); thus, loss of regulation could cause increased levels of Qrr4. Even with the increased levels, Qrr4-U61A, Qrr4-G76A, and Qrr4-C78A remained incapable of properly controlling light production (Fig. 3C). This result suggests that their lowered stability in E. coli is not the sole cause of their inability to properly regulate target mRNAs. We analyze the mechanisms underlying these defects in a later section. This result highlights the power of studying sRNAs both endogenously in V. harveyi and exogenously in recombinant E. coli: the loss of stability could not be revealed in V. harveyi and the loss of feedback could not be revealed in E. coli.
Loss of Hfq binding.
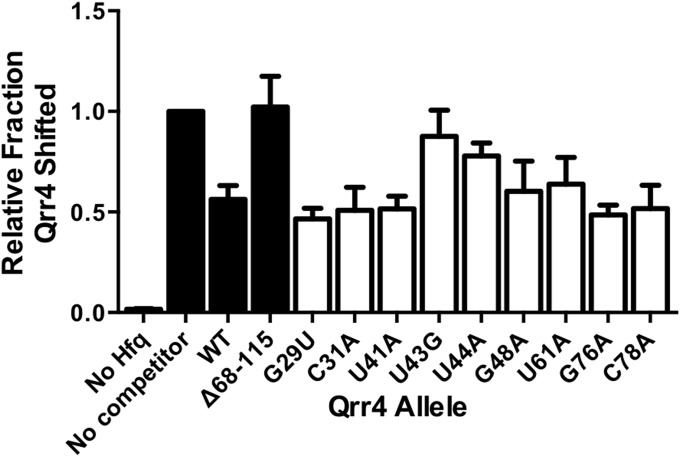
Binding to the chaperone Hfq is critical for bacterial sRNA-mediated target mRNA regulation (36, 37), and some of the defects in our set of Qrr4 mutants could reflect a loss of Hfq binding. We used an in vitro competition assay with purified Hfq protein to test this possibility. Briefly, radioactively labeled WT Qrr4 was bound to Hfq, and subsequently, the complex was competed with unlabeled WT or mutant Qrr4. The outcome was assessed by gel shift (Fig. 4C and Fig. S2). We used WT Qrr4 at a concentration that produced 50% competition against itself, and thus we could assay Qrr4 mutants that had improved or defective Hfq binding. To validate our assay, we constructed a Qrr4 truncation (Δ68-115) that removes SL4 and associated bases proposed to be crucial for Hfq binding (21). Indeed, when Qrr4-Δ68-115 was added, the truncated Qrr4 did not compete. Qrr4-U43G and Qrr4-U44A were also defective in competition for Hfq binding. This was not, however, the case for of all of the Qrr4 mutants analyzed here. None of the other mutations significantly affected the competition (Fig. S2). Loss of Hfq binding by Qrr4-U43G and Qrr4-U44A could account for the defects in Qrr4-mediated regulation of both luxO and luxR. Indeed, Qrr4-U43G and Qrr4-U44A had high MI for both luxR and luxO, and these nucleotides reside immediately upstream of a stem-loop, SL3 (Figs. 1B and 2 D and E), which is consistent with what is known about Hfq binding preferences (37). Qrr4-U43 and Qrr4-U44 are conserved between all five of the Qrr4 sRNAs, as well as in Qrr sRNAs of other Vibrio species, fitting with their essential roles in overall Qrr4 function (16, 38).
Fig. S2.
Quantitation of data from native gels shown as fraction Hfq shifted relative to the sample lacking cold competitor Qrr4 (related to Fig. 4). Black bars depict control samples; white bars depict Qrr4 mutants with impaired repression. In all panels, error bars are SDs of at least three trials.
Defective stem-loop formation.
A third mechanism that could impair Qrr4 regulation of both luxR-gfp and luxO-gfp would be mutations that interfere with proper Qrr secondary-structure formation. Formation of SL1 has previously been shown to be crucial for Qrr stability (21). However, our RSort-Seq did not identify single mutations in SL1 with high MI. We reason that multiple mutations in SL1 may be required to significantly impair its stability. We are currently investigating this hypothesis.
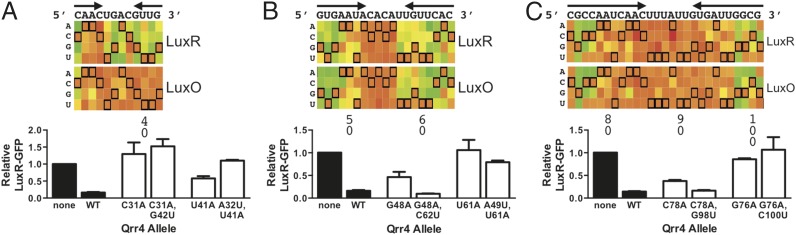
With respect to SL2, the profile shows that some, but not all, bases predicted by RNAfold (rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi) (16) to be involved in SL2 formation are crucial for repression (Fig. 5A). The SL2 region overlaps with the base-pairing region (Figs. 1B and 2 D and E), and thus substitutions in SL2 have the potential to affect both base pairing and Qrr4 secondary structure. If SL2 can be disabled by a single mutation on either side of the stem, one would predict a mirror image in the heat map for the two sides of the stem: the effect of disrupting a base on one side of the stem would match the effect of disrupting the partner base on the other side of the stem. This is not the case for SL2 (Fig. 5A). With respect to regulation of LuxR, mutating bases on the 3′ side of the stem of SL2 impairs repression, whereas mutating bases on the 5′ side of the stem has little effect on LuxR regulation. With respect to regulation of LuxO, except for mutation of Qrr4-C31, base alterations in the stem of SL2 do not impair repression. The mutation profiles align closely with those of nucleotides predicted to be important for base pairing, as discussed below (Fig. 6 B and C). To provide proof that mutations in the 3′ side of SL2 that affect regulation of luxR are not due to roles in stem-loop formation, we analyzed single and double mutants in which we disabled SL2 base pairing, introduced compensating mutations to reestablish SL2, and tested whether function was restored (implicating SL formation as the original defect) or not (implicating base pairing as the original defect). For example, Qrr4-C31 is predicted by RNAfold to form an intramolecular base pair with Qrr4-G42 (Fig. 1B). Qrr4-C31A disrupted repression of luxR, but repression was not restored by the compensatory mutation Qrr4-G42U (Fig. 5A). This result suggests that loss of SL2 formation is not the cause of the Qrr4-C31A defect. Likewise, regulation was not restored for the Qrr4-U41A mutant through the compensating SL2 mutation Qrr4-A32U (Fig. 5A). These cases indicate that the ability to form SL2 is not essential for regulation of luxR-gfp. Rather, these two mutations impair Qrr4 function by disrupting base pairing with the target mRNA, as we experimentally demonstrate below (Fig. 6 B and C).
Fig. 5.
Qrr4 mutations can block stem-loop formation. (Top) Regions of heat maps depicting the results from the RSort-Seq analysis for (A) SL2, (B) SL3, and (C) SL4. Refer to Fig. 2 for explanations of nomenclature and colors. (Bottom) Examination of stem-loop formation mutant phenotypes. Black bars denote fluorescence from luxR-gfp in the presence of the empty vector and WT Qrr4. White bars denote single and double mutants of Qrr4. Error bars are SDs of at least three trials.
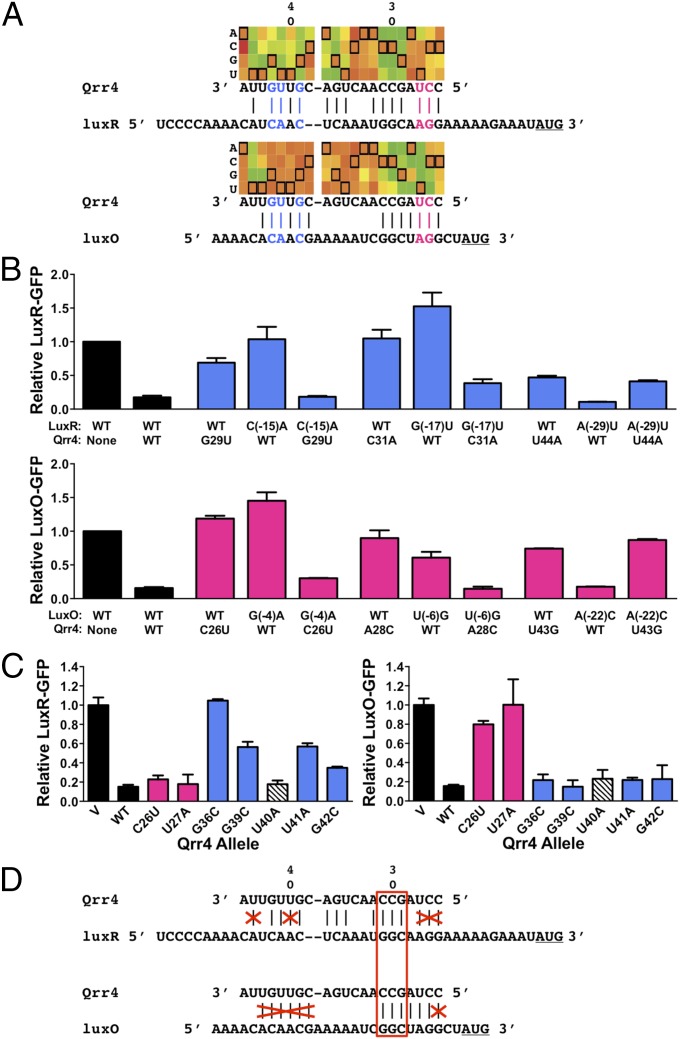
Fig. 6.
Qrr4 mutations can block base pairing. (A) Region of heat map showing results of RSort-Seq analysis for regulation of luxR and luxO in the putative base-pairing region, the corresponding Qrr4 sequence, and the aligned target mRNA 5′-UTR sequence. See Fig. 2 for explanations of nomenclature and colors. RSort-Seq predicts Qrr4 bases used to regulate only luxR (blue) or only luxO (pink). (B) WT and mutant Qrr4 regulation of luxR-gfp (Upper, blue) or luxO-gfp (Lower, pink) in E. coli. (C) Comparison of the ability of WT and Qrr4 mutants to repress luxR and luxO. Blue bars show nucleotides essential for luxR regulation, pink bars show nucleotides essential for luxO regulation, and the white hatched bar shows that Qrr4-U40 is not required for regulation of luxR or luxO. (D) Model for predicted and actual critical base pairing between Qrr4 and luxR and Qrr4 and luxO. Bases crossed out in red represent nucleotides that have the potential to base pair but that are not important for regulation of the given target. The red box shows the only 3 nucleotides required for base pairing to and regulation of both targets. In all panels, error bars are SDs of at least three trials.
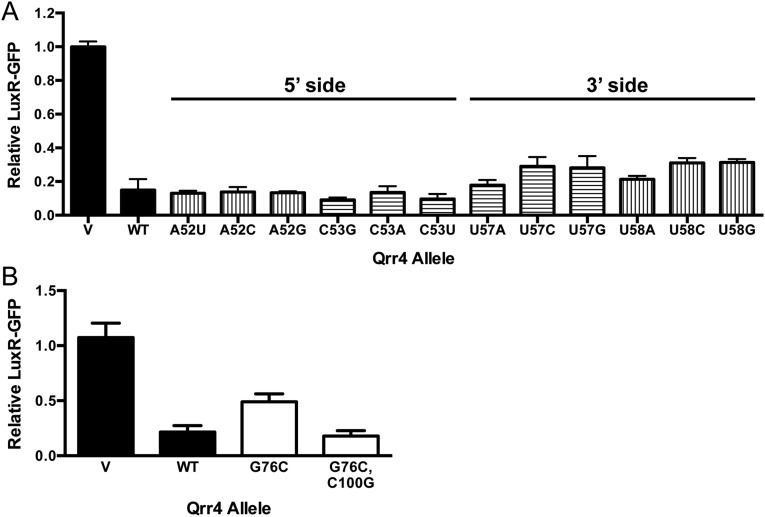
A surprising mutation profile was obtained for SL3 (Fig. 5B). Similar to SL2, a mirror image in the mutation profile is not present as expected for a stem-loop. However, unlike SL2, the pattern of the mutation profile is the same for Qrr4 regulation of LuxR and LuxO. This finding suggests that SL3 serves a non–target-specific regulatory role that does not involve stem formation. Some SL3 bases are indeed required for stem-loop formation. For example, Qrr4-G48A eliminated Qrr4 repression of luxR, and the defect could be rescued through the complementary mutation Qrr4-C62U that restored stem-loop formation (Fig. 5B). However, this pattern did not exist for all of SL3. Specifically, Qrr4-A49, Qrr4-A50, Qrr4-U51, Qrr4-A52, and Qrr4-C53, which are bases on the 5′ side of SL3, impair repression less than Qrr4-U61, Qrr4-U60, Qrr4-G59, Qrr4-U58, and Qrr4-U57 on the 3′ side of SL3. Consistent with this result, Qrr4-U61A was severely impaired for luxR regulation; however, additionally mutating Qrr4-A49U, to make a double mutant in which the stem-loop should be reestablished, did not restore WT regulation (Fig. 5B). This result suggests that bases on the 3′ side of SL3 play a role in Qrr4 function that is independent of stem-loop formation. To further explore this notion, we examined additional bases at the top of the stem (Qrr4-A52 and Qrr4-U58; Fig. S3A, bars with vertical lines) and at the bottom of the loop (Qrr4-C53 and Qrr4-U57; Fig. S3A, bars with horizontal lines) by mutating each base to every other nucleotide. Indeed, mutating bases on the 3′ side of the stem-loop (Qrr4-U57 and Qrr4-U58) resulted in more severe defects in luxR repression than did changing bases on the 5′ side of the stem-loop (Qrr4-A52 and Qrr4-C53) (Fig. S3A). These results provide proof that an additional role, beyond hydrogen bonding in the stem, exists for the bases on the 3′ side of the stem of SL3. We return to this point below.
Fig. S3.
Features of stem-loop formation (related to Fig. 5). (A) Fluorescence production from luxR-gfp in the presence of Qrr4 mutants at the top of the stem (vertical stripes) and the bottom of the loop (horizontal stripes) of SL3. Black bars show fluorescence production in the presence of a vector or WT Qrr4. (B) Fluorescence production from luxR-gfp in the presence of Qrr4 mutants in SL4. Vector and WT Qrr4 are denoted by black bars. White bars show mutations in SL4 that disrupt, and subsequently restore, stem-loop formation. In both panels, error bars represent SDs of at least three trials.
The two arms of SL4 exhibited symmetric mutation profiles for regulation of both LuxR and LuxO (Fig. 5C), and the profile suggests that mutating 3 nucleotides at the base of the stem impairs repression much more than mutating bases in the upper region of the stem. We tested two of the three bases at the bottom of the stem to assess if the defect in repression could be attributed to loss of stem-loop formation. Indeed, the Qrr4-C78A mutation disrupted repression of luxR, but regulation was regained in the double Qrr4-C78A, G98U mutant, which restored stem-loop formation (Fig. 5C). This was not the case for Qrr4-G76A. Here, constructing the double mutant (Qrr4-G76A, C100U) to reestablish stem-loop formation was not sufficient to restore regulation of luxR (Fig. 5C). However, we reasoned that changing a G/C base pair to an A/U base pair in SL4 simply made an intrinsically less stable secondary structure. To test this possibility, we constructed Qrr4-G76C (Fig. S3B) to disrupt base pairing in SL4 and we made the double-mutant Qrr4-G76C, C100G to reestablish base pairing. In so doing, we replaced a G/C pair with a C/G pair. We note that the possibility exists that this alteration changes the steric packing in the stem-loop; however, the binding energy within the base pair is maintained. The Qrr4-G76C mutation disrupted repression of luxR-gfp, and the Qrr4-G76C, C100G mutant restored the WT phenotype. We conclude that Qrr4-G76 is involved in stem-loop formation and that a G/C base pair in this position is essential for proper SL4 formation.
Defective base pairing.
A final possible mechanism by which Qrr4 repression could be impaired is through disruption of base pairing to the target mRNA. Not surprisingly, there exist several nucleotides in the predicted base-pairing region with high MI scores, corresponding to severely impaired repression (Figs. 2 D and E and 6A). To test whether these bases have high MI scores due to roles in base pairing, we made partner compensatory mutations in the 5′-UTRs of the mRNA targets. For example, the Qrr4-G29U and Qrr4-C31A mutants were disrupted for regulation of luxR (Fig. 6B), and repression was restored when the mutations were partnered with luxR-C(-15)A and luxR-G(-17)U, respectively. Importantly, WT Qrr4 did not repress luxR-C(-15)A or luxR-G(-17)U. We obtained the identical results for Qrr4-C26U and Qrr4-A28C when paired with luxO-G(-4)A and luxO-U(-6)G, respectively (Fig. 6B).
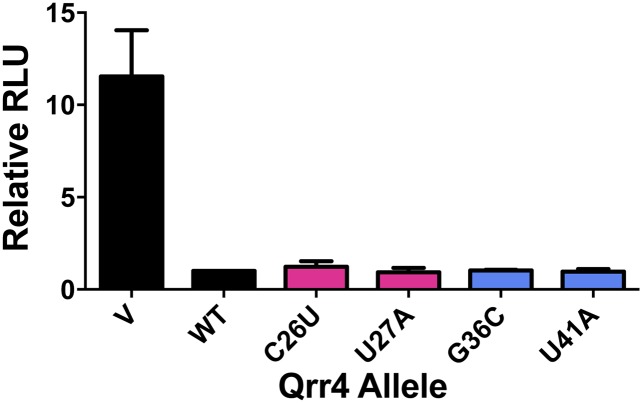
By contrast, the Qrr4-U44A and Qrr4-U43G defects could not be compensated by the corresponding mutations in the luxR and luxO 5′-UTRs luxR-A(-29)U and luxO-A(-22)C, respectively. luxR-A(-29)U and luxO-A(-22)C are fully repressed by WT Qrr4, further suggesting that Qrr4-U44 and Qrr4-U43 (and luxR-A29 and luxO-A22) do not mediate interactions between Qrr4 and the target mRNAs. Thus, Qrr4-U43 and Qrr4-U44 are critical for Qrr4 to function in regulation; however, this is not due to roles in base pairing (Fig. 6B). As discussed above, these two bases are essential because of their roles in Hfq binding (Fig. 4C). We conclude that, of the nucleotides with high MI in the putative base-pairing region, Qrr4-G29 and Qrr4-C31 are critical for Qrr4 paring to luxR, and Qrr4-C26 and Qrr4-A28 are essential for pairing to luxO.
Our RSort-Seq analysis showed that multiple bases in the putative base-pairing region of Qrr4 are not crucial for regulation even though RNAfold absolutely predicts them to base pair with luxR mRNA (Fig. 6A, Top, pink) and with luxO mRNA (Fig. 6A, Bottom, blue). Indeed, in the present context, RNA-folding algorithms predict that 15 nucleotides in Qrr4 are available to pair with the luxR mRNA and 13 Qrr4 nucleotides can pair with luxO mRNA. Ten of these nucleotides are common between luxR and luxO mRNA. According to these programs, only 5 nucleotides are unique to the putative luxR–Qrr4 interaction and 3 nucleotides are unique to the putative luxO–Qrr4 interaction (Fig. 6A). The high-throughput screening paired with MI analysis provides a strikingly different view. Only a small subset of these bases had high MI, suggesting that the base-pairing pattern predicted by RNAfold is much more extensive than what is used by bacteria for regulation (Fig. 6A). Support for this idea comes from our analysis (Fig. 6C): the Qrr4-G36C, Qrr4-G39C, Qrr4-U41A, and Qrr4-G42C mutations affected only luxR regulation, whereas the Qrr4-C26U and Qrr4-U27A mutations affected only luxO regulation (Fig. 6C). Notably, mutation of Qrr4-U40, which is predicted by RNAfold to base pair with both targets, did not disrupt regulation of either target (Fig. 6C, white hatched), which matches its MI mutation profile (Fig. 2). Visualizing all of the mutation profiles in a heat map revealed the minimal base-pairing region and, thus, the nucleotides that are not used for target regulation (Fig. 6D, red crosses). In so doing, our analysis reduced the number of bases in Qrr4 that are used by both luxR and luxO during regulation to only 3 nucleotides (Fig. 6D, red box). Companion analysis using the V. harveyi light production assay showed that Qrr mutations that affect only luxR (Qrr4-G36C or Qrr4-U41A) or only luxO (Qrr4-C26U or Qrr4-U27A) in E. coli did not result in altered regulation of light production (Fig. S4), implying that simultaneous Qrr regulation of both luxR and luxO is not strictly necessary for proper quorum sensing.
Fig. S4.
Disrupting Qrr4 regulation of LuxR or LuxO alone does not impair bioluminescence in V. harveyi (related to Fig. 6). Light production in V. harveyi following expression of WT or the designated Qrr4 mutants. Blue denotes nucleotides essential for luxR regulation, and pink denotes nucleotides essential for luxO regulation. In all panels, error bars are SDs of at least three trials.
Suppressor Analysis Reveals a Structural Role for Qrr4-U61.
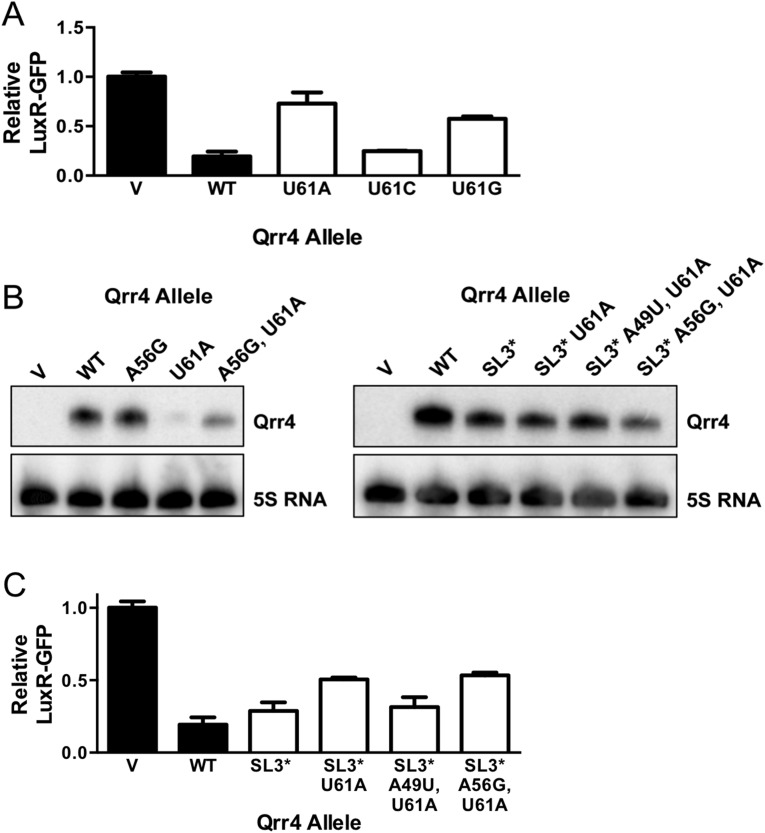
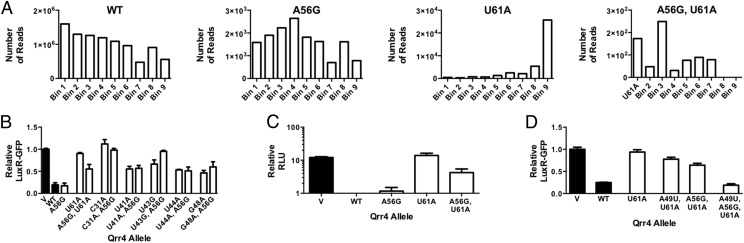
To understand the function of Qrr4 mutants that are defective in target mRNA regulation but that cannot be explained by the above four mechanisms, we leveraged our large mutant pool to probe for double mutants possessing disabling mutations as single mutants but that had regained WT repression of luxR-gfp and luxO-gfp when combined with a second mutation. Our rationale was that we had, in effect, carried out a suppressor analysis using the Qrr4 library if there existed second mutations that overcame original defects. We did not perform our Qrr4 mutagenesis with the aim of gaining saturating coverage in double mutants so we could not carry out this analysis for every disabling mutant. As a proof-of-principle for this line of thinking, we focused our attention exclusively on Qrr4-U61A in SL3. As described above, the disabling Qrr4-U61A mutation could not be restored to the WT luxR repression level by reestablishing stem-loop formation (Fig. 5B). Furthermore, different mutations at position 61 elicited distinct levels of impairment of luxR-gfp regulation (Fig. S5A). We reasoned that the Qrr4-U61A mutation could perturb an additional Qrr4 function beyond stem-loop formation and that a suppressor mutation could provide insight into this role. We found that the double mutant, Qrr4-A56G, U61A exhibited a distribution similar to WT following FACS (Fig. 7A). We reconstructed Qrr4-A56G, U61A and expressed it in E. coli to test its ability to regulate luxR-gfp (Fig. 7B). Qrr4-A56G displayed WT luxR-gfp repression. As shown above, Qrr4-U61A was defective. The double mutant, Qrr4-A56G, U61A, functioned more effectively than the single Qrr4-U61A mutant. Introduction of Qrr4-A56G likewise partially rescued the light production defect associated with the Qrr4-U61A mutation in V. harveyi (Fig. 7C). The Qrr4 double A56G, U61A mutant also showed increased stability on a Northern blot relative to Qrr4-U61A (Fig. S5B).
Fig. S5.
The function of Qrr4-U61 (related to Fig. 7). (A) Fluorescence production from luxR-gfp in the presence of Qrr4 carrying all possible mutations of Qrr4-U61. Black bars show mean fluorescence levels in the presence of a vector control or WT Qrr4. White bars show mean fluorescence levels in the presence of the different Qrr4–U61 mutants. Error bars represent SDs of at least three trials. (B) Representative Northern blot analysis of Qrr4 mutants from E. coli. 5S RNA was used as the loading control. (C) As in A with the designated Qrr4 mutants containing an inverted SL3 (denoted SL3*). Mutant number corresponds to the location of the mutation in WT Qrr4. In all panels, error bars represent SDs of at least three trials.
Fig. 7.
Double-mutant analysis reveals a suppressor of the Qrr4-U61A defect. (A) Number of reads per bin from the original RSort-Seq analysis for WT Qrr4, Qrr4-A56G, Qrr4-U61A, and Qrr4-A56G, U61A. (B) Fluorescence levels from luxR-gfp in the presence of Qrr4 variants. Black bars show luxR-gfp in the presence of a vector control or WT Qrr4. White bars depict mutant Qrr4 repression of luxR-gfp with and without the secondary mutation Qrr4-A56G. (C) The effect of mutation of Qrr4-A56G on the ability of WT Qrr4 and Qrr4-U61A to regulate light production in V. harveyi at OD600 = 0.1. (D) As in B with the designated Qrr4 mutants.
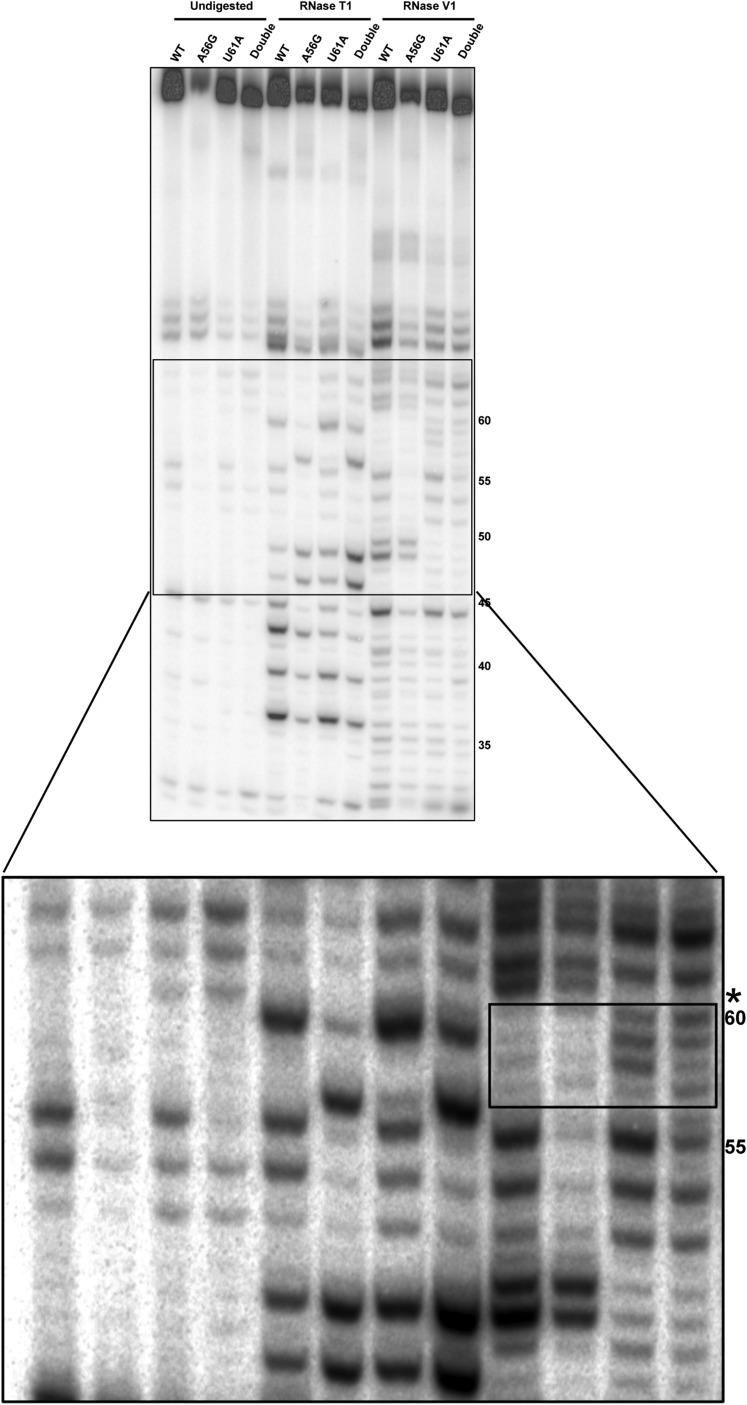
We were next interested to probe how the Qrr4-A56G mutation suppressed the original Qrr4-U61A defect. First, we assayed whether suppression by Qrr4-A56G is unique to Qrr4-U61A. Qrr4-A56G could not rescue defects associated with Qrr4-C31A, Qrr4-U41A, Qrr4-U43G, Qrr4-U44A, or Qrr4-G48A (Fig. 7B). This result suggests that Qrr4-A56G is not a generic suppressor. Second, we probed whether suppression by Qrr4-A56G is additive with the partial suppression conferred by the Qrr4-A49U mutation in the Qrr4-U61A mutant [as a reminder, the Qrr4-A49U change reestablishes SL3 formation in the context of the Qrr4-U61A mutation (Fig. 5B)]. Indeed, the triple mutant Qrr4-A49U, A56G, U61A functions as WT (Fig. 7D). Thus, the partial suppression conferred by Qrr4-A56G onto the Qrr4-U61A mutant occurs by a mechanism that does not rely on base pairing with Qrr4-U61. Finally, we tested whether the ability of Qrr4-A56G to rescue the Qrr4-U61A defect depended on the orientation of SL3. Introduction of the U61A alteration into Qrr4 containing an inverted SL3 (denoted SL3*) decreased the severity of the Qrr4-U61A regulatory defect and, likewise, partially rescued its stability defect (Fig. S5 B and C). Furthermore, WT luxR-gfp repression was fully rescued by restoring stem-loop formation (Qrr4-SL3*-A49U, U61A) but not through introduction of the original suppressor mutation, Qrr4-A56G into the SL3 inverted mutant (Qrr4-SL3*-A56G, U61A; Fig. S5C). Together, these results suggest that Qrr4-U61A plays a function in addition to its role in stem-loop formation, and this role requires that the nucleotide be located on the 3′ side of SL3.
To garner insight into how Qrr4-U61 controls Qrr4 activity and how Qrr4-A56G suppresses the Qrr4-U61 defect, we used RNase digestion and compared WT Qrr4, Qrr4-A56G, Qrr4-U61A, and Qrr4-A56G, U61A with a focus on SL3 (Fig. S6). With respect to WT Qrr4, digestion with RNase V1 shows that U48, A49, U61, C62, and C64 make base pairs so they are in the stem of SL3. From this, we can assume that G46, U47, and A63 also form base pairs, but as RNase V1 typically cuts preferentially on one side of dsRNA, these partner bases do not produce digestion products. With respect to the mutants, Qrr4-U61A and Qrr4-A56G, U61A both lack a dsRNA signature for U61 (Fig. S6, asterisk), showing that the Qrr4-U61A mutation indeed disrupts base pairing in the stem. Furthermore, four additional bands (Fig. S6, box in expanded view), at U57, U58, G59, and U60, are present in Qrr4-U61A and Qrr4-A56G, U61A digested with RNase V1 that are not present in WT Qrr4. Together, the data suggest that Qrr4-U61A disrupts base pairing in SL3, which likely underpins its decreased stability. There are modest differences between the digestion patterns of Qrr4-U61A and Qrr4-A56G, U61A that could explain the suppression in the double mutant. In the Qrr4-A56G, U61A double mutant, RNase V1 digestion at U57 is enhanced, whereas digestion at U58 is decreased compared with the Qrr4-U61A single mutant (Fig. S6, compare lanes 11 and 12). We hypothesize that Qrr4-A56G, U61A adopts an alternative secondary structure(s) that partially restores stability and Qrr4 regulatory activity. The triple Qrr4 Qrr4-A49U, A56G, U61A mutant, as mentioned, is fully restored for base pairing, for function, and for stability.
Fig. S6.
Structure probing reveals changes in base pairing in Qrr4-U61A. Structure probing of SL3 of WT Qrr4, Qrr4-A56G, Qrr4-U61A, and Qrr4-A56G, U61A. The solid box denotes the enlarged region. The asterisk shows the loss of a band at Qrr4-U61 in Qrr4-U61A and Qrr4-A56G, U61A. The box in the expanded view shows bands representing alternative base pairs in Qrr4-U61A and Qrr4-A56G, U61A.
Discussion
Cell density-dependent gene expression needs to be precisely regulated to ensure that bacteria make accurate transitions into and out of quorum-sensing mode. In vibrios, the Qrr sRNAs mediate these transitions. Presumably, sRNAs are an appropriate biological tool to use for this function because they are highly dynamic due to their small size, rapid synthesis, and fast turnover rates (25). Even with these features, it remains mysterious how one sRNA distinguishes between multiple mRNA targets and how multiple sRNAs work simultaneously to control gene expression dynamics. In the present context, the five homologous V. harveyi Qrr sRNAs control at least 20 mRNA targets. This arrangement makes the Qrr sRNAs and their partner targets ideal to probe how subtle variations in sRNA sequence can drive potency and specificity in target regulation, and in turn, how these differences contribute to proper quorum-sensing regulation.
RSort-Seq Analysis Allows Unbiased Assessment of sRNA–mRNA Interactions.
Past analyses to define nucleotides that contribute to sRNA function relied on computational algorithms to identify important bases, typically followed by directed mutagenesis in a laborious nucleotide-by-nucleotide manner to test function. To accelerate this process, we developed a general procedure we call RSort-Seq, and we used it to map the residues that are essential for Qrr4 to control two mRNA targets, luxR and luxO. An analogous method has recently been successfully deployed to assess the importance of individual bases in the sRNAs DsrA and RyhB with respect to regulation of their targets (30) and to analyze the 5′-UTR of csgD (29). These previous studies focused on identifying nucleotides driving sRNA stability and stem-loop formation. The authors used computational assessment to vet the results. The power of our procedure stems from a set of biological assays that verify the initial computational predictions and, furthermore, reveal the molecular mechanism underlying the functions of crucial bases. Thus, our method culminates in a highly accurate regional map of the key nucleotides involved in sRNA stability, Hfq binding, stem-loop formation, partner mRNA binding specificity, and intramolecular suppression. By modifying the conditions, it could be possible to reveal additional features of sRNA-mediated regulation of mRNA targets. In our experiments, both the target gfp reporter and the sRNA regulator were expressed at high levels. The goal was to achieve the maximum possible dynamic range between no regulation (empty vector) and full repression by WT Qrr4. A consequence of this strategy could be that, due to the high level production of Qrr4, mutations that confer modest effects would likely not be identified. For example, this aspect of our strategy could explain the lack of single mutations with high MI located in SL1, a structural motif that we know is essential for Qrr4 stability. In principle, performing the RSort-Seq procedure at lower Qrr4 sRNA levels or at lower target reporter levels could uncover additional single substitutions important for function.
Qrr4 Target Discrimination Region and Target Selection.
Distinct base-pairing patterns exist among the Qrr sRNAs and their partner target mRNAs, and these patterns are sufficient to define the mechanisms of regulation (22). These pairing patterns drive Qrr regulatory potency and dynamics of target mRNA regulation as well as determine the ability of different mRNA targets to compete for the attention of the Qrr sRNAs (22). However, these parameters alone cannot fully explain mRNA target selection and regulation because Qrr sRNAs with identical predicted base-pairing regions have the capacity to differentially regulate target mRNAs. Deciphering which bases encode this feature and how they function is the topic of the present work.
RNAfold predicts that Qrr4 target selection relies on a stretch of 38 nucleotides that span SL1 and SL2 and form base pairs with the target mRNAs (38). RSort-Seq revealed, and our biological analyses verified, the fact that many fewer bases than predicted are critical for Qrr4 base pairing with target mRNAs. Previously, critical nucleotides could not be distinguished from nonessential bases within the pairing region because the vital nucleotides are not identical for all mRNA targets. Furthermore, it is difficult to predict the importance of individual nucleotides residing within a contiguous stretch of bases available for pairing. For example, Qrr4-U40 (Fig. 6C), which is flanked by one 5′ base and two 3′ bases within the base-pairing region, is not important for regulation of luxR; however, the adjacent base, Qrr4-U41, is absolutely required. These issues, which have plagued sRNA–mRNA studies, highlight the power of unbiased screening to define the critical bases within the possible base-pairing region. We are currently extending our analyses to other Qrr4 mRNA targets to generate a complete map of the critical bases used for pairing to each mRNA target. We are also examining the other Qrr sRNAs to define whether the crucial bases are identical or different among the Qrr sRNAs.
Our analysis suggests that identifying nucleotides that are critical for base pairing may be key to understanding differential regulation by sRNAs. RNAfold predicts that 10 nucleotides in Qrr4 could be used to regulate both luxR and luxO. However, RSort-Seq shows that few, but very particular, bases within the larger base-pairing region play the essential roles that mediate the sRNA–mRNA interaction (Fig. 6D). Somewhat surprisingly, our analysis indicates that few single-nucleotide changes have significant effects on regulation under the conditions we tested. Thus, the potential exists for individual bases in the larger base-pairing region to be used to differentiate between targets. Furthermore, we hypothesize that bases that are predicted to base pair but are not crucial for functional regulation could play supporting roles in fine-tuning Qrr–mRNA interactions in the context of other noncrucial nucleotides. Although such nucleotides cannot be revealed by our analysis, we are examining a double-mutant library in the base-pairing region to define the nature of such defects. This analysis should determine whether Qrr4 exploits all possible 38 nucleotides available for base pairing in different combinations to elicit its large diversity of quorum-sensing regulatory effects or if in fact much simpler, condensed pairing schemes suffice.
RSort-Seq Reveals Information About Stem-Loop Formation.
Formation of the proper secondary structure is vital to sRNA function; stem-loops aid in increasing stability of the sRNA, allow the sRNA to interact with Hfq, and ensure proper termination of sRNA transcription (21). Computational algorithms predict stem-loop formation based on base pairing and resulting free-energy estimates. Here, we show that RSort-Seq can hone these predictions, revealing those bases that are critical to stem-loop formation. RSort-Seq also revealed that SL1 formation is indifferent to single mutations, SL2 formation is irrelevant for function, and a unique structural motif exists within SL3.
Even though our library contained only a small percentage of double mutants, RSort-Seq nonetheless uncovered the suppressor Qrr4-A56G of the Qrr4-U61A defect. Our analysis of this suppressor serves as a proof-of-principle for the importance of extending the RSort-Seq technology to sRNAs harboring multiple mutations. Our initial analyses of the Qrr4-A56G, U61A mutant shows that the phenotype depends on the orientation of the stem on which the mutations reside. We hypothesize that A56G suppresses Qrr4-U61A by forming a higher-order intramolecular structure or possibly by enabling an interaction with a cofactor. We are currently analyzing these possibilities.
Conclusions
To understand how the Qrr sRNAs properly regulate quorum sensing, we must define their individual and combined contributions to target mRNA regulation. Toward that goal, here we have probed all individual bases in Qrr4 to generate a map of nucleotides vital to its function with respect to two mRNA targets. We contrast our strategy to previous ones that have been used with similar goals in mind. Commonly used RNA-folding algorithms, as they should, predict all possible base pairs between an sRNA and its mRNA target and all base pairs available for forming the stems of stem-loops. Our analysis underscores that, at least in our example, the majority of these pairs are dispensable for sRNA function. Of the nucleotides that are required for regulation, our RSort-Seq strategy further identifies those that are target specific and those that are universally used. Most importantly, the RSort-Seq analysis constrains the number of mutants that need to be considered for follow-up biochemical analysis to define mechanism. By expanding this line of thinking to the other Qrr sRNAs and to additional mRNA targets, we will next define the unique role each Qrr plays in quorum-sensing gene regulation and, in so doing, derive clues to their combined function.
In summary, combining high-throughput biological assessment of nucleotide essentiality for target regulation with MI theory enables a comprehensive understanding of the precise intermolecular and intramolecular interactions required for sRNA function. This strategy can be used for any set of sRNA–mRNA pairs.
Materials and Methods
Plasmids and Strains.
See Tables S2 and S3 for lists of plasmids and primers, respectively. Information on growth conditions and construction of constructs is provided in SI Experimental Procedures.
RSort-Seq.
See SI Experimental Procedures for detailed methods. Briefly, a random mutant library of qrr4 was constructed using GeneMorph II EZClone Domain Mutagenesis (Agilent Technologies). A majority of the qrr4 mutants had single substitutions (Table S1 shows the distribution of single, double, triple, etc., mutations obtained). Although each possible mutation is not represented uniformly in the final analysis (Fig. S1), they are all present at high numbers, alleviating concerns of nonhomogenous representation. The library was coexpressed with pLF128 (luxR-gfp) or pLF129 (luxO-gfp) and sorted based on repression strength using a FACSAria (BD Biosciences). The reporters and the sRNA were expressed to near-maximal levels to enable us to observe the largest possible range of regulation. Mutant qrr4 clones from each bin were amplified by PCR using the primers in Table S3. This method introduced Illumina priming sites and barcode sequences. Our approach amplified only the qrr4 gene, and trimmed the flanking sequence. The clones were subsequently sequenced by Illumina sequencing. In the RSort-Seq experiment with pLF128, we obtained 11 million total sequences (∼1.3 million per bin), and with pLF129, we obtained 13 million sequences (∼1.6 million per bin). MI was used to assess the phenotype of all qrr4 mutants.
Assessment of Mutants.
Select mutations determined by RSort-Seq to be important for Qrr4 function were reconstructed and assayed in both E. coli and V. harveyi. Qrr4 defects were assessed by secondary mutation analysis, Northern blot, real-time PCR, in vitro Hfq binding, and structure probing to determine the basis of the phenotype. See SI Experimental Procedures for detailed information.
SI Experimental Procedures
Growth Conditions.
Escherichia coli strains were grown in LB medium at 37 °C with aeration. Vibrio harveyi strains were grown in Luria-marine medium at 30 °C with aeration. Antibiotics (Sigma) were supplied at the following concentrations: tetracycline (Tet), 5 μg/mL; chloramphenicol (Cm), 10 μg/mL; ampicillin (Amp), 100 μg/mL; spectinomycin (Spec), 60 μg/mL.
Plasmids and Strains.
E. coli hfq was amplified with primers STR947 and STR948, and subsequently cloned into the NcoI and XhoI sites in pET28b, using standard cloning procedures (39). This strategy resulted in pSTR1351, which contains Hfq with a C-terminal 6-His tag driven by the T7 promoter. Mutations in qrr4 were engineered using site-directed mutagenesis of plasmid LF127, LF128, or LF129 using PfuUltra polymerase (QuikChange II; Agilent Technologies) (21, 22). Plasmids containing qrr4 mutants were transformed into electrocompetent BW-R1 E. coli cells (40) harboring either LF128 or LF129 (40). qrr4 mutations for use in V. harveyi were amplified to introduce NheI and BamHI sites and subcloned into the XbaI and BamHI sites in pLAFR2 (41). Following digestion with the above enzymes (New England Biolabs), inserts were ligated into pLAFR2 using T4 ligase (New England Biolabs). Plasmids were conjugated into V. harveyi KT282 (Δqrr1-5) (16). See Table S2 for a list of plasmids and Table S3 for a list of primers used in this study.
Library Construction.
GeneMorph II EZClone Domain Mutagenesis (Agilent Technologies) was used to introduce random mutations into qrr4 in pLF127 using primers STR912 and STR913. Conditions suggested by the manufacturer to obtain high and medium mutation frequencies were used, resulting a majority of qrr4 mutants possessing single substitutions and a minority of mutants harboring two or more substitutions (Table S3). The library was transformed into XL10-Gold ultracompetent E. coli cells (Agilent) in three separate transformations, and the entirety of the transformations was spread on LB-Cm plates. Following overnight incubation at 37 °C, a total of ∼82,000 colonies was collected from the plates and pooled. Approximately 20 individual colonies were randomly selected, and the qrr4 region was sequenced to confirm the mutagenesis. No mutations were detected outside of the mutated qrr4 region in these clones (data not shown). The pooled cells were resuspended in 8 mL of LB and divided into eight 1-mL aliquots, and plasmids were prepared from each suspension (Zymo Research). The plasmid preparations were pooled. The plasmid library was subsequently transformed by electroporation into the reporter strain BW-R1 carrying pLF128 (luxR-gfp) or pLF129 (luxO-gfp). The entire transformation reaction was plated on LB-Spec, Cm, Amp plates. Approximately 60,000–80,000 colonies were grown overnight at 37 °C. Transformants were collected, pooled, and suspended in 5 mL of LB-Spec, Cm, Amp with 20% (vol/vol) glycerol. One-milliliter aliquots were flash frozen with liquid N2 and stored at −80 °C.
FACS and Preparation for Illumina Sequencing.
A 1-mL aliquot of the qrr4 mutant library was thawed on ice and used to inoculate 4 mL of fresh LB-Spec, Cm, Amp with 0.5 mM isopropyl β-d-1-thiogalactopyranoside (IPTG). The culture was divided into two aliquots that were grown with and without 15 ng/mL anhydrotetracycline (aTc) (Clontech). All cultures were incubated with shaking at 30 °C for 5 h, 15 min. Ten 50-μL aliquots of each culture were removed and diluted into 10 FACS tubes containing 500 μL of sterile PBS. Samples were sorted on a FACSAria Special Order Research Product running FACSDiva software (BD Biosciences). Each of the 10 aliquots was sorted into nine activity bins based on FITC intensity. The resulting bins from each aliquot were pooled such that each bin contained 150,000–200,000 events in 100 μL of PBS. Mean GFP of each postsort activity bin was assessed (10,000 events) as well as the presort induced and uninduced samples. Cells from each activity bin were pelleted by centrifugation, suspended in 10 μL of sterile water, and boiled for 10 min. Cell debris was pelleted by centrifugation, and the supernatant was removed to a fresh tube and brought up to 20 μL in sterile water.
The qrr4 genes in each activity bin were amplified by PCR (iProof High-Fidelity DNA Polymerase; Bio-Rad) using forward (either STR1012, STR1114, STR1115, or STR1116) and reverse primers that encoded Illumina adapters and priming sites (STR1013, STR1034-STR1044, STR1118-1129) (Table S2). Offset primers were used to generate sufficient diversity in the resulting sequences to enable productive Illumina sequencing reads. Additionally, the reverse primer for each bin had a unique barcode (Table S2). PCR was carried out for 29 rounds. Products were analyzed by gel electrophoresis, purified (QIAquick PCR purification kit; Qiagen), and DNA was quantified. Equal amounts of DNA from each reaction (bin) were pooled and diluted to 2.5 ng/μL before submission to the Princeton University Sequencing Core facility for next generation sequencing by Illumina HiSeq. RSort-Seq experiments with pLF128 yielded a total of 11 million sequences with ∼1.3 million per bin. Experiments with pLF129 yielded a total of 13 million sequences with ∼1.6 million per bin.
Mutual Information Analysis.
To probe for residues that are crucial for Qrr4 regulation, we quantified the effect of single mutations at a given position on the distribution of fluorescence intensities. Denoting the ith nucleotide in the WT sequence as , we estimated the probability for a bacterium with the specific single mutation m ≠ ni at position i to be sorted into fluorescence bin b. As the best estimate for this probability, we used the empirical frequency:
where is the number of sequences with mutation m at position i found in bin b, and is the total number of sequences with mutation m at position i. This treatment defines three conditional probability distributions for each position, corresponding to the three possible single mutations. The bin distribution of the WT bacterium determines a fourth probability distribution, corresponding to Positions of interest in the Qrr4 sequence are those for which the bin distribution for some or all of the single mutations differs markedly from the WT distribution. Such a difference indicates a strong statistical dependence between bin and residue identity at position . We quantified this statistical dependence via the mutual information between and :
where we have taken a flat prior for the nucleotide probability, i.e., . Choosing a flat prior for corrects for biases in the mutational sampling of the sequence space.
E. coli GFP Reporter Assay.
The E. coli GFP reporter assay was modified from ref. 21. Briefly, E. coli strains were grown overnight, diluted into fresh LB medium containing 0.5 mM IPTG to induce pZE12G-LuxR-GFP or pZE12G-LuxO-GFP, and 30 ng/mL aTc was added to induce Qrr4 production from pZA31-Qrr4. Following overnight growth in 96-well plates (Fisher Scientific), GFP fluorescence and OD600 were measured using an EnVision Multilabel Plate Reader (PerkinElmer). Average fluorescence/OD600 was calculated and normalized to that of the uninduced samples (grown without aTc). Changes in fluorescence following induction were compared with that of the vector control strain. At least three trials were performed.
Bioluminescence Quorum-Sensing Assay.
V. harveyi bioluminescence was measured using a previously described assay (11). V. harveyi KT282 (16) harboring plasmids carrying the designated mutant qrr4 genes was grown overnight and diluted to OD600 = 0.005. Following 2 h growth, OD600 was measured on a DU 800 Spectrophotometer (Beckman Coulter) and bioluminescence was measured on a Tri-Carb 2810 TR (PerkinElmer) every 30 min to generate a light production curve. Relative light units (RLU) are counts per minute per milliliter per OD600. GraphPad Prism was used to designate the best fit of the curve and to determine the light production at OD600 = 0.1. The average light production, relative to that of WT Qrr4 at OD600 = 0.1, over at least three trials is shown.
Northern Blot Analysis.
E. coli strains were grown overnight, and Qrr4 production was induced as above. Cultures were subsequently incubated with RNAprotect (Qiagen) for 5 min. RNA was extracted using TRIzol, as previously published (17). DNA was eliminated using a DNase treatment and removal kit (Life Technologies). RNA was subjected to Northern blot analysis, as previously described (42).
Quantitative Real-Time PCR.
V. harveyi KT282 carrying plasmids containing mutant qrr4 genes was grown overnight, diluted to OD600 = 0.005, and subsequently grown to OD600 = 0.1. Cells were pelleted, and RNA was extracted using TRIzol (Life Technologies) (17). A DNase treatment and removal kit (Life Technologies) was used to eliminate DNA. Equal amounts of total RNA were used to make cDNA using SuperScript III Reverse Transcriptase (Life Technologies). Real-time PCR analysis was performed using the ABI Prism 7900HT Sequence Detection System (ABI) or QuantStudio6 Real-Time PCR system (Life Technologies) with PerfeCTa SYBR Green FastMix (Quanta Biosciences).
Hfq Binding Assay.
Hfq was purified from pSTR1351 using a previously established protocol (43). Qrr4 RNA was produced in vitro using the MEGAshortscript T7 Transcription Kit (Life Technologies). WT Qrr4 was synthesized with and without [32P]UTP incorporation. Resulting RNA was purified using Illustra Microspin G-25 columns (GE Healthcare Life Sciences). Following denaturation and reannealing, 32P-labeled WT Qrr4 RNA was incubated with purified Hfq protein at room temperature at a molar ratio of 1:5. Twice the amount of unlabeled WT Qrr4 RNA or mutant Qrr4 RNA was used as competitor. Truncated Qrr4 RNA, lacking bases 67–115, was used as the nonbinding control. Following incubation with competitor RNA, all samples were subjected to native gel electrophoresis. Gels were dried (model 583 gel dryer; Bio-Rad), exposed overnight to a phosphor screen (GE Healthcare Life Sciences), and visualized using a Typhoon scanner (GE Healthcare Life Sciences).
Qrr4 Structure Probing.
RNA structure probing was performed as described previously (44). Briefly, RNA was produced in vitro and labeled at the 5′ end using [32P]ATP (45). Resulting RNA was cleaned and extracted. Following incubation with structure buffer and yeast RNA, labeled RNA was digested with RNase T1 (Ambion; diluted 1:100) for 3 min or RNase V1 (Ambion; diluted 1:50) for 1 min at 37 °C. Digestion was terminated by the addition of 1× sequencing buffer (Ambion). T1 ladder was generated by incubating denatured labeled WT Qrr4 with RNase T1 (diluted 1:10) for 5 min at 37 °C in sequencing buffer. OH ladder was made by incubating labeled WT Qrr4 in alkaline sequencing buffer for 5 min at 95 °C. All samples were denatured at 95 °C for 1 min before loading onto a 10% (vol/vol) polyacrylamide/7 M urea sequencing gel. The gel was dried and visualized as above.
Supplementary Material
Acknowledgments
We thank members of the laboratories of B.L.B. and N.S.W. for helpful discussion, and Kathrin Fröhlich and Kai Papenfort for help with structure probing. We thank Curtis G. Callan for his gracious help in the design of the screen and the MI analysis. This work was supported by the Howard Hughes Medical Institute, NIH Grant 5R01GM065859, and National Science Foundation Grant MCB-0343821 (to B.L.B.). Work in the laboratory of N.S.W. was supported by Defense Advanced Research Projects Agency Biochronicity Program Grant D12AP00025 and the W. M. Keck Foundation. S.T.R. was supported by NIH Fellowship F32AI085922.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1518958112/-/DCSupplemental.
References
- 1.Rutherford ST, Bassler BL. Bacterial quorum sensing: Its role in virulence and possibilities for its control. Cold Spring Harb Perspect Med. 2012;2(11):a012427. doi: 10.1101/cshperspect.a012427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ng WL, Bassler BL. Bacterial quorum-sensing network architectures. Annu Rev Genet. 2009;43:197–222. doi: 10.1146/annurev-genet-102108-134304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davies DG, et al. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science. 1998;280(5361):295–298. doi: 10.1126/science.280.5361.295. [DOI] [PubMed] [Google Scholar]
- 4.Engebrecht J, Silverman M. Identification of genes and gene products necessary for bacterial bioluminescence. Proc Natl Acad Sci USA. 1984;81(13):4154–4158. doi: 10.1073/pnas.81.13.4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Kievit TR. Quorum sensing in Pseudomonas aeruginosa biofilms. Environ Microbiol. 2009;11(2):279–288. doi: 10.1111/j.1462-2920.2008.01792.x. [DOI] [PubMed] [Google Scholar]
- 6.Cao JG, Meighen EA. Purification and structural identification of an autoinducer for the luminescence system of Vibrio harveyi. J Biol Chem. 1989;264(36):21670–21676. [PubMed] [Google Scholar]
- 7.Schauder S, Shokat K, Surette MG, Bassler BL. The LuxS family of bacterial autoinducers: Biosynthesis of a novel quorum-sensing signal molecule. Mol Microbiol. 2001;41(2):463–476. doi: 10.1046/j.1365-2958.2001.02532.x. [DOI] [PubMed] [Google Scholar]
- 8.Henke JM, Bassler BL. Three parallel quorum-sensing systems regulate gene expression in Vibrio harveyi. J Bacteriol. 2004;186(20):6902–6914. doi: 10.1128/JB.186.20.6902-6914.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen X, et al. Structural identification of a bacterial quorum-sensing signal containing boron. Nature. 2002;415(6871):545–549. doi: 10.1038/415545a. [DOI] [PubMed] [Google Scholar]
- 10.Higgins DA, et al. The major Vibrio cholerae autoinducer and its role in virulence factor production. Nature. 2007;450(7171):883–886. doi: 10.1038/nature06284. [DOI] [PubMed] [Google Scholar]
- 11.Bassler BL, Wright M, Showalter RE, Silverman MR. Intercellular signalling in Vibrio harveyi: Sequence and function of genes regulating expression of luminescence. Mol Microbiol. 1993;9(4):773–786. doi: 10.1111/j.1365-2958.1993.tb01737.x. [DOI] [PubMed] [Google Scholar]
- 12.Bassler BL, Wright M, Silverman MR. Multiple signalling systems controlling expression of luminescence in Vibrio harveyi: Sequence and function of genes encoding a second sensory pathway. Mol Microbiol. 1994;13(2):273–286. doi: 10.1111/j.1365-2958.1994.tb00422.x. [DOI] [PubMed] [Google Scholar]
- 13.Freeman JA, Bassler BL. A genetic analysis of the function of LuxO, a two-component response regulator involved in quorum sensing in Vibrio harveyi. Mol Microbiol. 1999;31(2):665–677. doi: 10.1046/j.1365-2958.1999.01208.x. [DOI] [PubMed] [Google Scholar]
- 14.Lilley BN, Bassler BL. Regulation of quorum sensing in Vibrio harveyi by LuxO and sigma-54. Mol Microbiol. 2000;36(4):940–954. doi: 10.1046/j.1365-2958.2000.01913.x. [DOI] [PubMed] [Google Scholar]
- 15.Lenz DH, et al. The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell. 2004;118(1):69–82. doi: 10.1016/j.cell.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 16.Tu KC, Bassler BL. Multiple small RNAs act additively to integrate sensory information and control quorum sensing in Vibrio harveyi. Genes Dev. 2007;21(2):221–233. doi: 10.1101/gad.1502407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rutherford ST, van Kessel JC, Shao Y, Bassler BL. AphA and LuxR/HapR reciprocally control quorum sensing in vibrios. Genes Dev. 2011;25(4):397–408. doi: 10.1101/gad.2015011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Kessel JC, Rutherford ST, Shao Y, Utria AF, Bassler BL. Individual and combined roles of the master regulators AphA and LuxR in control of the Vibrio harveyi quorum-sensing regulon. J Bacteriol. 2013;195(3):436–443. doi: 10.1128/JB.01998-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Waters CM, Bassler BL. The Vibrio harveyi quorum-sensing system uses shared regulatory components to discriminate between multiple autoinducers. Genes Dev. 2006;20(19):2754–2767. doi: 10.1101/gad.1466506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henke JM, Bassler BL. Quorum sensing regulates type III secretion in Vibrio harveyi and Vibrio parahaemolyticus. J Bacteriol. 2004;186(12):3794–3805. doi: 10.1128/JB.186.12.3794-3805.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shao Y, Feng L, Rutherford ST, Papenfort K, Bassler BL. Functional determinants of the quorum-sensing non-coding RNAs and their roles in target regulation. EMBO J. 2013;32(15):2158–2171. doi: 10.1038/emboj.2013.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feng L, et al. A qrr noncoding RNA deploys four different regulatory mechanisms to optimize quorum-sensing dynamics. Cell. 2015;160(1-2):228–240. doi: 10.1016/j.cell.2014.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vogel J, Wagner EG. Target identification of small noncoding RNAs in bacteria. Curr Opin Microbiol. 2007;10(3):262–270. doi: 10.1016/j.mib.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 24.Li W, Ying X, Lu Q, Chen L. Predicting sRNAs and their targets in bacteria. Genomics Proteomics Bioinformatics. 2012;10(5):276–284. doi: 10.1016/j.gpb.2012.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Storz G, Vogel J, Wassarman KM. Regulation by small RNAs in bacteria: Expanding frontiers. Mol Cell. 2011;43(6):880–891. doi: 10.1016/j.molcel.2011.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Waters LS, Storz G. Regulatory RNAs in bacteria. Cell. 2009;136(4):615–628. doi: 10.1016/j.cell.2009.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kinney JB, Tkacik G, Callan CG., Jr Precise physical models of protein–DNA interaction from high-throughput data. Proc Natl Acad Sci USA. 2007;104(2):501–506. doi: 10.1073/pnas.0609908104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kinney JB, Murugan A, Callan CG, Jr, Cox EC. Using deep sequencing to characterize the biophysical mechanism of a transcriptional regulatory sequence. Proc Natl Acad Sci USA. 2010;107(20):9158–9163. doi: 10.1073/pnas.1004290107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holmqvist E, Reimegård J, Wagner EG. Massive functional mapping of a 5′-UTR by saturation mutagenesis, phenotypic sorting and deep sequencing. Nucleic Acids Res. 2013;41(12):e122. doi: 10.1093/nar/gkt267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peterman N, Lavi-Itzkovitz A, Levine E. Large-scale mapping of sequence-function relations in small regulatory RNAs reveals plasticity and modularity. Nucleic Acids Res. 2014;42(19):12177–12188. doi: 10.1093/nar/gku863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shannon CE. Communication in the presence of noise. P Ire. 1949;37(1):10–21. [Google Scholar]
- 32.Cover TM, Thomas JA. Elements of Information Theory. 2nd Ed Wiley-Interscience; Hoboken, NJ: 2006. [Google Scholar]
- 33.Tkacik G, Callan CG, Jr, Bialek W. Information capacity of genetic regulatory elements. Phys Rev E Stat Nonlin Soft Matter Phys. 2008;78(1 Pt 1):011910. doi: 10.1103/PhysRevE.78.011910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tkacik G, Callan CG, Jr, Bialek W. Information flow and optimization in transcriptional regulation. Proc Natl Acad Sci USA. 2008;105(34):12265–12270. doi: 10.1073/pnas.0806077105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tu KC, Long T, Svenningsen SL, Wingreen NS, Bassler BL. Negative feedback loops involving small regulatory RNAs precisely control the Vibrio harveyi quorum-sensing response. Mol Cell. 2010;37(4):567–579. doi: 10.1016/j.molcel.2010.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brennan RG, Link TM. Hfq structure, function and ligand binding. Curr Opin Microbiol. 2007;10(2):125–133. doi: 10.1016/j.mib.2007.03.015. [DOI] [PubMed] [Google Scholar]
- 37.Vogel J, Luisi BF. Hfq and its constellation of RNA. Nat Rev Microbiol. 2011;9(8):578–589. doi: 10.1038/nrmicro2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shao Y, Bassler BL. Quorum-sensing non-coding small RNAs use unique pairing regions to differentially control mRNA targets. Mol Microbiol. 2012;83(3):599–611. doi: 10.1111/j.1365-2958.2011.07959.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1989. [Google Scholar]
- 40.Levine E, Zhang Z, Kuhlman T, Hwa T. Quantitative characteristics of gene regulation by small RNA. PLoS Biol. 2007;5(9):e229. doi: 10.1371/journal.pbio.0050229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Friedman AM, Long SR, Brown SE, Buikema WJ, Ausubel FM. Construction of a broad host range cosmid cloning vector and its use in the genetic analysis of Rhizobium mutants. Gene. 1982;18(3):289–296. doi: 10.1016/0378-1119(82)90167-6. [DOI] [PubMed] [Google Scholar]
- 42.Urban JH, Vogel J. Translational control and target recognition by Escherichia coli small RNAs in vivo. Nucleic Acids Res. 2007;35(3):1018–1037. doi: 10.1093/nar/gkl1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mikulecky PJ, et al. Escherichia coli Hfq has distinct interaction surfaces for DsrA, rpoS and poly(A) RNAs. Nat Struct Mol Biol. 2004;11(12):1206–1214. doi: 10.1038/nsmb858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Papenfort K, Förstner KU, Cong JP, Sharma CM, Bassler BL. Differential RNA-seq of Vibrio cholerae identifies the VqmR small RNA as a regulator of biofilm formation. Proc Natl Acad Sci USA. 2015;112(7):E766–E775. doi: 10.1073/pnas.1500203112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fröhlich KS, Papenfort K, Fekete A, Vogel J. A small RNA activates CFA synthase by isoform-specific mRNA stabilization. EMBO J. 2013;32(22):2963–2979. doi: 10.1038/emboj.2013.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.