Significance
Blooms of phytoplankton can shift the ecosystem state of low-nutrient ocean regions between net heterotrophic and autotrophic conditions, increasing carbon sequestration and driving carbon export to the deep sea. Little is known about the traits that govern the timing and magnitude of these bloom events. We used metatranscriptomics to assess phytoplankton functional group-specific metabolic shifts experimentally during simulated blooms in the North Pacific Subtropical Gyre. The results indicate blooms form when phytoplankton are released from limitation by resources (nutrients, vitamins, and trace metals) and that the mechanistic basis for the success of one functional group over another may be driven by how efficiently the transcriptome is modulated following a nutrient pulse.
Keywords: metatranscriptomics, phytoplankton, ecological traits, biogeochemistry, blooms
Abstract
A diverse microbial assemblage in the ocean is responsible for nearly half of global primary production. It has been hypothesized and experimentally demonstrated that nutrient loading can stimulate blooms of large eukaryotic phytoplankton in oligotrophic systems. Although central to balancing biogeochemical models, knowledge of the metabolic traits that govern the dynamics of these bloom-forming phytoplankton is limited. We used eukaryotic metatranscriptomic techniques to identify the metabolic basis of functional group-specific traits that may drive the shift between net heterotrophy and autotrophy in the oligotrophic ocean. Replicated blooms were simulated by deep seawater (DSW) addition to mimic nutrient loading in the North Pacific Subtropical Gyre, and the transcriptional responses of phytoplankton functional groups were assayed. Responses of the diatom, haptophyte, and dinoflagellate functional groups in simulated blooms were unique, with diatoms and haptophytes significantly (95% confidence) shifting their quantitative metabolic fingerprint from the in situ condition, whereas dinoflagellates showed little response. Significantly differentially abundant genes identified the importance of colimitation by nutrients, metals, and vitamins in eukaryotic phytoplankton metabolism and bloom formation in this system. The variable transcript allocation ratio, used to quantify transcript reallocation following DSW amendment, differed for diatoms and haptophytes, reflecting the long-standing paradigm of phytoplankton r- and K-type growth strategies. Although the underlying metabolic potential of the large eukaryotic phytoplankton was consistently present, the lack of a bloom during the study period suggests a crucial dependence on physical and biogeochemical forcing, which are susceptible to alteration with changing climate.
It has been postulated that the net oxygen state of oligotrophic systems is controlled by irregular blooms of large eukaryotic phytoplankton (1), which have been shown to respond to nutrient input in open ocean ecosystems (2). Nutrient loading shifts the community away from a tightly regenerating microbial loop, based on small phytoplankton (typically bacterioplankton), toward a community with a higher proportion of larger phytoplankton cells (typically eukaryotic phytoplankton) and, consequently, more carbon export (2). Data from Station ALOHA in the oligotrophic North Pacific Subtropical Gyre (NPSG) during the summer period have identified increased export of organic carbon, diatom-associated biogenic silica (BSi), and, to a lesser extent, particulate inorganic carbon (PIC) associated with calcification (3). These increases are potentially a result of nutrient-driven community shifts. Blooms of eukaryotic phytoplankton in oligotrophic environments are often dominated by the diatom functional group (4) and may determine the balance between net heterotrophic and net autotrophic conditions (3). Such blooms, however, are thought to be undersampled and may often go undetected in satellite ocean color records (5). Consequently, the drivers of bloom formation and concomitant carbon export remain poorly understood, particularly in critical oligotrophic systems.
Marine phytoplankton accounts for roughly half of global primary production (6). Although central to balancing global biogeochemical models of gross primary production (7), knowledge of the biogeochemical drivers that govern the dynamics of these bloom-forming organisms in oligotrophic systems is limited. Nutrient environments are integral to the structuring of phytoplankton communities (8–10) and initiating blooms. Originally thought to be a stable low-fluctuating habitat, long-term monitoring at Station ALOHA has demonstrated that within the constraints of an oligotrophic ecosystem, the nutrient regime can be quite dynamic, alternating in controlling factors over many time scales (11). These oscillations may be driven by biological [e.g., bloom of N2-fixing cyanobacteria, which draw down phosphorus or iron while injecting new nitrogen into the system (12)], physical [e.g., nutrient supply from transient eddies (13)], or anthropogenic [e.g., atmospheric deposition of nutrients (14)] forcing. Regardless of the source, these nutrient-loading events could act to stimulate blooms in the large eukaryotic phytoplankton community. Historically, the distributions of key eukaryotic phytoplankton function groups have been tracked relative to nutrient stoichiometries to examine how nutrients influence the physiological ecology of different functional groups like diatoms and haptophytes (4, 15). Although valuable, it is still difficult to relate stoichiometries to the dynamics of bloom formation and, in particular, to patterns in key groups like diatoms without specific measures of functional group physiology.
Molecular-level tools that can track transcripts, proteins, or even metabolites and biochemicals in a taxon-specific way are increasingly being used in cultures and field populations to track metabolic capacity and physiological responses (16–23). Molecular assessment of physiology for eukaryotic populations is most tractable in coastal systems with high biomass (17, 18); thus, in oligotrophic ocean regions, molecular studies of physiology have typically been limited to the numerically abundant members of the microbial community: picoplankton (cyanobacteria, heterotrophic bacteria, and small picoeukaryotes). Groundbreaking studies have demonstrated the responsiveness of the dominant picoplankton community to pulses of deep seawater (DSW) (24) and dissolved organic matter (DOM) (25), as well as the diel synchrony of the photosynthetic and heterotrophic communities (20), yet were limited in their coverage of the rare, large eukaryotic fraction due to both the low biomass of their standing stock and the lack of reference sequences. A recent world ocean survey has highlighted the diversity encompassed within the eukaryotic planktonic community (26) and suggested that the structuring of these communities may be driven by physical and chemical oceanographic features (27). However, again, the responsiveness of these keystone eukaryotic phytoplankton communities to changing environmental conditions remains poorly described and understood.
New resources, including a new marine microbial eukaryote sequencing initiative (28), in combination with the bioinformatic pipeline used here are making possible the study of oligotrophic eukaryotic phytoplankton with unprecedented mechanistic resolution. Here, we examined potential biogeochemical controls on phytoplankton bloom formation in the NPSG and the metabolic traits that govern the distribution and responses of unique functional groups.
Results and Discussion
We leveraged increases in reference sequence availability (28) to identify the expressed metabolic capabilities of different phytoplankton functional groups. Total RNA from the eukaryotic community (>5 μm) of the surface mixed layer at Station ALOHA was sequenced (60 million 100-bp, poly-A–selected, paired-end reads) from three samples collected during August–September 2012 (S1–S3). To investigate the underlying functional group-specific metabolism associated with blooms, three blooms were simulated via on-deck incubations. Although there are many potential nutrient inputs or bloom drivers (e.g., biologically fixed nitrogen), these blooms were simulated through the addition of DSW to mimic nutrient loading by upwelling. These incubation experiments were modeled after a study by McAndrew et al. (2) and were performed in conjunction with each of the in situ samples, amending surface water with a 10% (vol/vol) mixture of water from below the nutricline (700 m). Sequence reads from the in situ samples (S1–S3) and the bloom simulations (E1–E3) were mapped to (i) nonsymbiotic microalgal genomes and (ii) a custom database composed of all publicly available transcriptomes as of March 17, 2014 from the Marine Microbial Eukaryotic Transcriptome Project (MMETSP) (28).
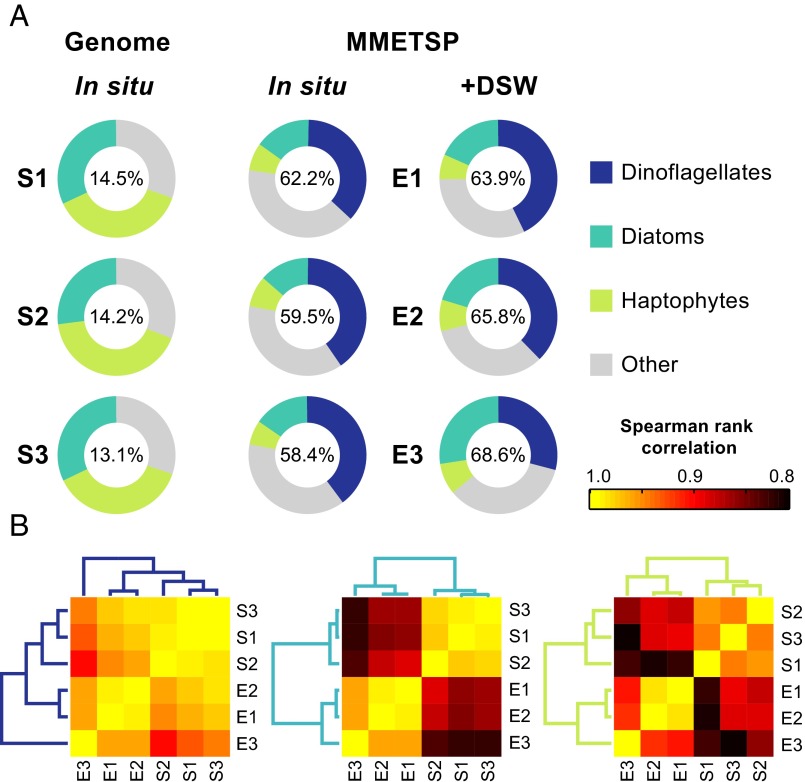
The MMETSP provides a 50-fold expansion of molecular data across the eukaryotic tree of life, which both better reflects the broad diversity within the protists and adds higher definition coverage for well-studied groups, such as diatoms and dinoflagellates (28). Our leveraging of this database for the pipeline developed herein enabled unprecedented identification of taxonomic composition (58–62% of reads identified) compared with mapping the same dataset to genomes (12–14% of reads identified), which have been used in previous studies (16). Due to the taxonomic bias of the nonsymbiotic algal genomes, the majority of reads from the in situ eukaryote community mRNA samples were annotated as diatoms or haptophytes. When mapped to the custom MMETSP database, however, dinoflagellates dominated, constituting 36–40% of the mapped reads, with the diatoms and haptophytes being the next most highly represented functional groups (Fig. 1A). The dominance of dinoflagellates in the 5-μm size fraction, confirmed in historic surface pigment analyses at Station ALOHA (29), stands in contrast to previous eukaryotic metatranscriptome studies in the oligotrophic ocean, where they accounted for <5% of reads (16). Although clearly present and important to the community, dinoflagellate read abundance here and in sediment trap data collected during the same cruise series (30) may be magnified by their large transcript pool (31, 32).
Fig. 1.
Taxonomic distribution in mRNA mapped reads consistent across time but altered by DSW addition. Sequences collected during the summer of 2012 at Station ALOHA (S1: August 6, S2: August 24, S3: September 2) and corresponding DSW incubation experiments (E1–E3) were mapped to two custom databases: (i) nonsymbiotic microalgal genomes and (ii) all freely available transcriptomes from the MMETSP as of March 17, 2014. (A) Taxonomic affiliation of reads across the three most abundant functional groups—dinoflagellates, diatoms, and haptophytes—mapped to both the genome and MMETSP databases for S1–S3. The corresponding DSW addition incubations E1–E3 were only mapped to the MMETSP database. The percentage of total reads mapped is indicated inside each of the circles. (B) Spearman rank correlation for species composition shifts within each of the three functional groups across S1–S3 and E1–E3.
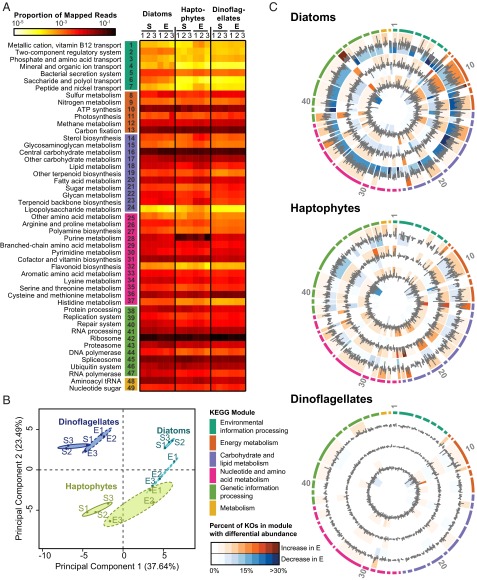
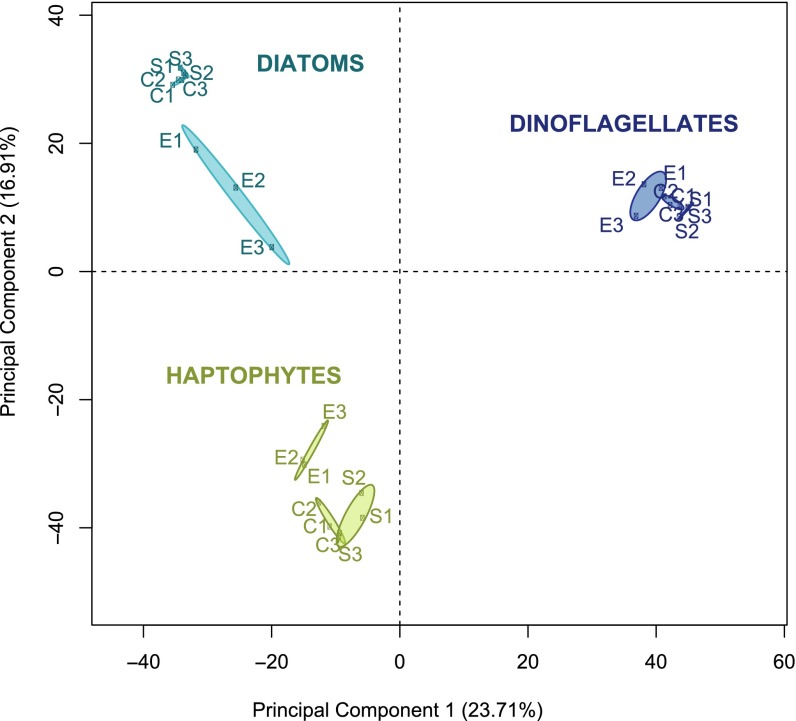
The species composition of the functional groups, reflected in rank abundance (Fig. 1B), was highly conserved across all three in situ samples (S1–S3), underscoring the stability of phytoplankton populations in this well-studied oligotrophic system. Between 18.1% and 20.7% of reads mapped to the MMETSP database were annotated with Kyoto Encyclopedia of Genes and Genomes (KEGG) orthology, elucidating differences in the mRNA distribution between functional groups at the pathway level. Looking at the module level, the general distribution of transcripts in proportion to the total was assessed using quantitative metabolic fingerprint(ing) (QMF) (17). Diatoms have a larger proportion of mRNA in the transport-related system (e.g., metallic cation, vitamin B12, phosphate, amino acid) compared with haptophytes and dinoflagellates (Fig. 2A). Purine metabolism was consistently a large component of haptophyte QMF (5.6–27%), an order of magnitude higher than diatoms and dinoflagellates (1–2%). Purine nucleotides may represent a source of dissolved organic nitrogen accessible to haptophytes, because haptophytes have been found to grow on purines as their sole nitrogen source (33). As the precursor for nucleic acid biosynthesis, purine uptake in the ocean has also been attributed to nucleotide salvage (34). These functional group differences observed at the module level are underscored by principal component analysis (PCA), with the QMF for each functional group differing with 95% confidence (Fig. 2B), and are suggestive of metabolic partitioning between functional groups. Within a functional group, the QMF was stable across time (Fig. 2A), in contrast to the variability observed in coastal systems over similar time scales (17, 18). This stability likely reflects the unique physiological attributes of oligotrophic phytoplankton as well as the comparatively static geochemical environment.
Fig. 2.
QMF and patterns of differential expression across KEGG orthology following DSW addition underscore functional group traits. (A) Relative metabolic partitioning of the mRNA pool across the three in situ samples (S1–S3) and corresponding DSW incubation experiments (E1–E3) was assessed using QMF. The summed proportion of mapped reads falling into each of the KEGG modules is depicted as a heat map. (B) PCA of the QMF signals for each of the functional groups across S1–S3 and E1–E3; 95% confidence ellipses are indicated for each of the sample types by functional group. (C) Log fold change and significance of differential expression between DSW amendments and in situ samples for KEGG orthologs are visualized with Circos (54) for the diatoms, haptophytes, and dinoflagellates. Outermost ring colors indicate the KEGG supermodule, with individual wedges of the pie corresponding to KEGG modules as numbered in A. Concentric circles indicate expression of the three replicated DSW addition experiments compared with in situ samples: E3 (outer), E2 (middle), and E1 (inner). The log fold change of individual KEGG orthologs is depicted as a bar plot bounded −3 to 3. The background color of individual KEGG modules identifies the percentage of genes within the module that were significantly (twofold change, post-p > 0.95) increased (orange) or decreased (blue) in abundance, where darker colors indicate that a higher percentage of genes within that module were significantly different.
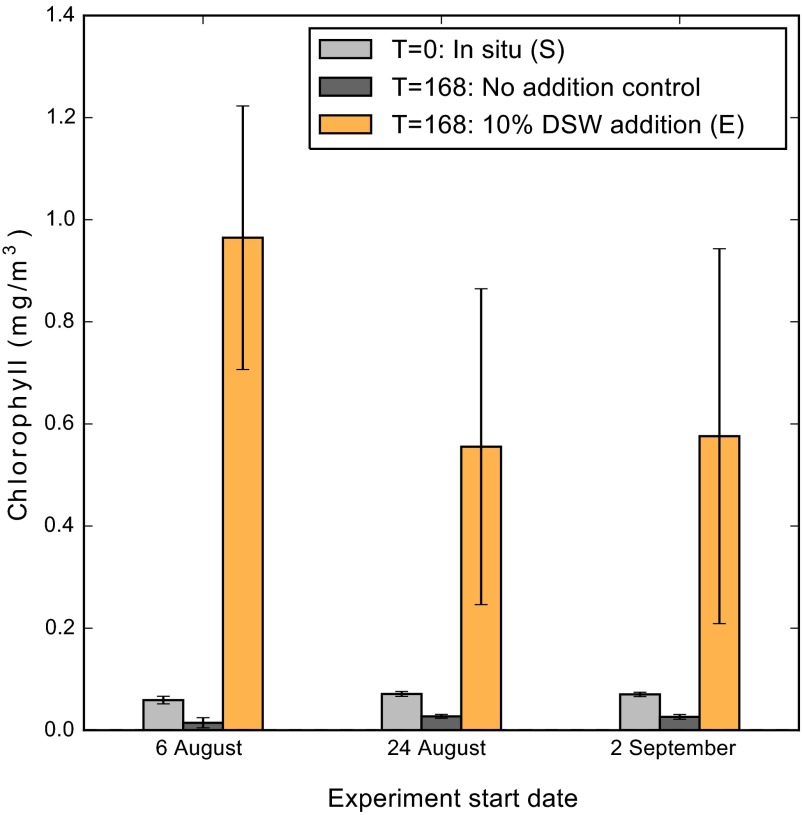
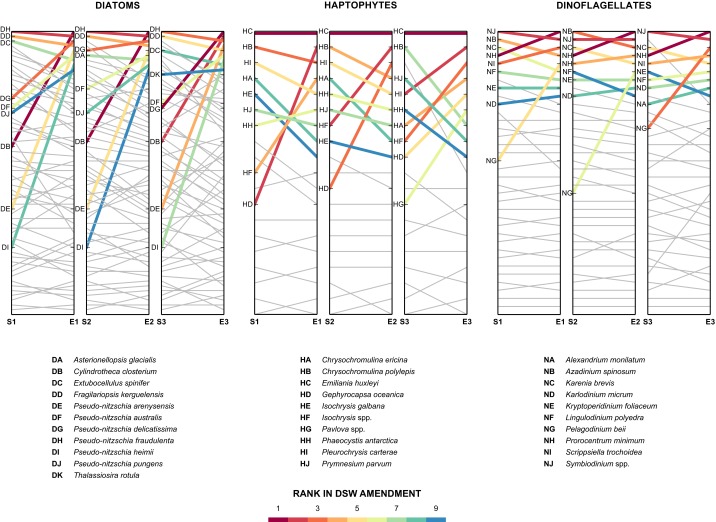
The static nature of population structure and functional group QMF was altered by replicated nutrient-rich DSW addition experiments, which led to a 7- to 17-fold increase in chlorophyll a, increases not observed in the control treatment (Fig. S1). This increase was consistent with previous studies, which also noted increases in diatoms (2). Diatom-associated mRNA reads increased in each of the DSW experiments (Fig. 1A). Although species designations can be influenced by the composition of the database used for read mapping, apparent taxonomic shifts occurred at the species level for diatoms and haptophytes (Fig. S2). Shifts in taxonomic composition were consistent with DSW addition for diatoms and haptophytes, with rank abundance clustering for all experiments (Fig. 1B). Taxonomic shifts in diatoms were driven by an increase in the rank abundance of certain species, which were pennate forms in all cases (Fig. S2), including genera known to be present in the NPSG, such as Pseudo-nitzschia (35), and common in oligotrophic nutrient amendment incubation studies (16, 36). Although species shifts also occurred within the haptophtyes, Emiliania huxleyi was always the most dominant taxon (Fig. S2). The shifts in diatom dominance compared with the consistent dominance of E. huxleyi may reflect differences in evolutionary strategies, with metabolic diversity spread across many species in the diatoms and a single species complex with a pangenome, E. huxleyi, in the haptophytes (37). The QMF of DSW addition was significantly different from the QMF of the in situ community for both diatoms and haptophytes, but not for dinoflagellates (Fig. 2B). Following DSW addition, the QMF for both diatoms and haptophytes was characterized by increased expression of modules associated with growth, such as carbon fixation (Fig. 2A). These shifts are not the result of changes in species composition, because the patterns of expression from individual species tracked the summed community (Fig. S3). The lack of change in the QMF of dinoflagellates (Fig. 2B) likely reflects their range of life strategies (32) and minimal transcriptional regulation of gene expression as observed in culture studies (31).
Fig. S1.
Chlorophyll a of replicated experiments for in situ samples (S), a no-addition control, and a 10% DSW amendment (E) are shown. Incubation samples were harvested after 168 h. T, postinoculation time in hours.
Fig. S2.
Rank abundance shifts in the species composition of diatoms, haptophytes, and dinoflagellates for the three experiments. The relative shift in rank abundance for each species is depicted for each incubation experiment (E1–E3) following DSW addition. The nine most abundant taxa following DSW addition are highlighted for each of the functional groups. Although the species that recruited the reads are denoted here, the recruitment is highly driven by the composition of the database and does not necessarily indicate the actual species present, but rather the closest species present in the database.
Fig. S3.
Comparison of the QMF between the whole functional group and representative taxa. The proportion of reads falling into each of the modules depicted in Fig. 2 is plotted for S1–S3 and E1–E3, comparing the summed functional group signal and the signal of a representative taxon. The color of the marker indicates the sample; solid and dashed lines mark the 1:1 and 1:2 lines, respectively.
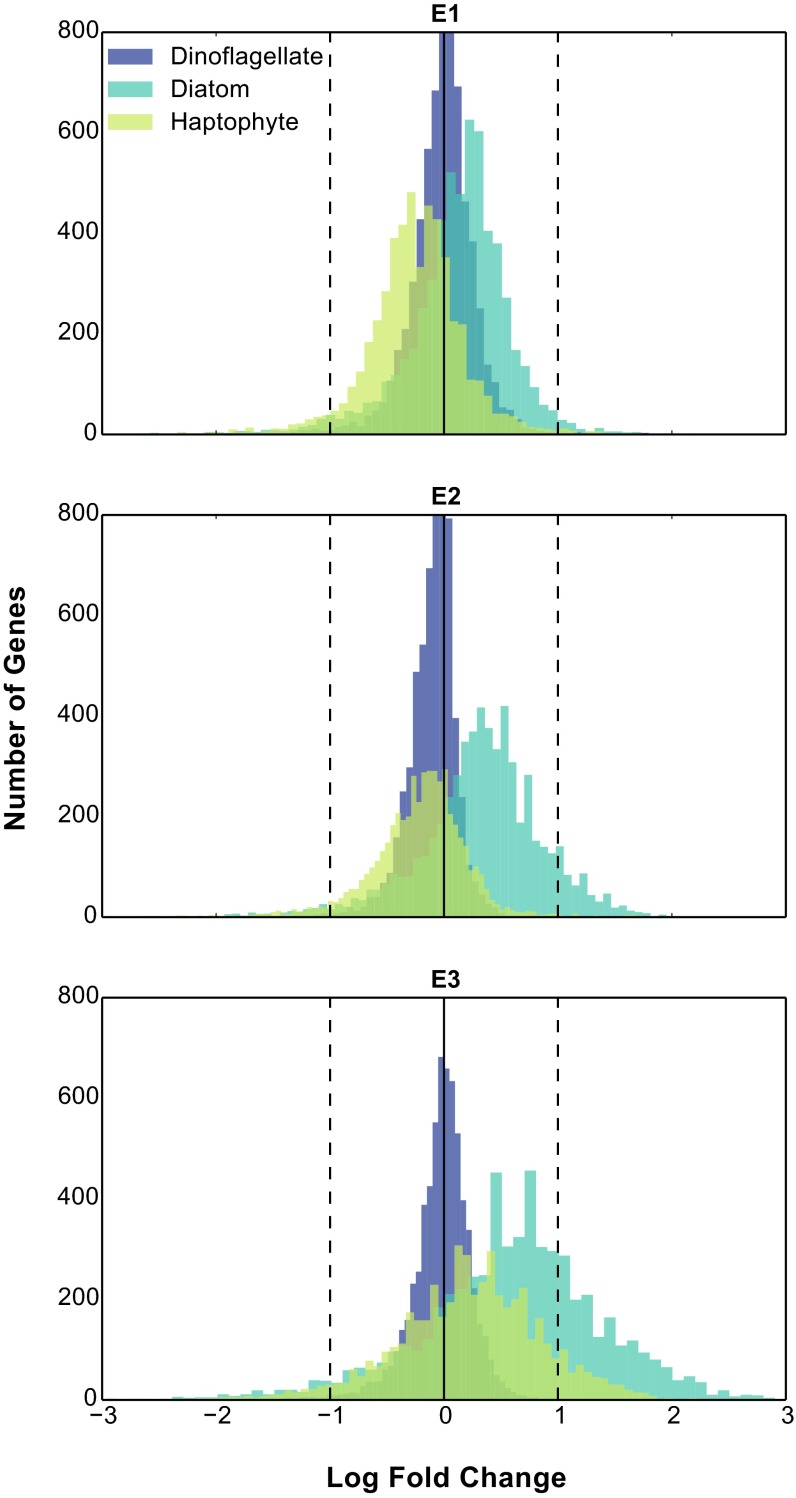
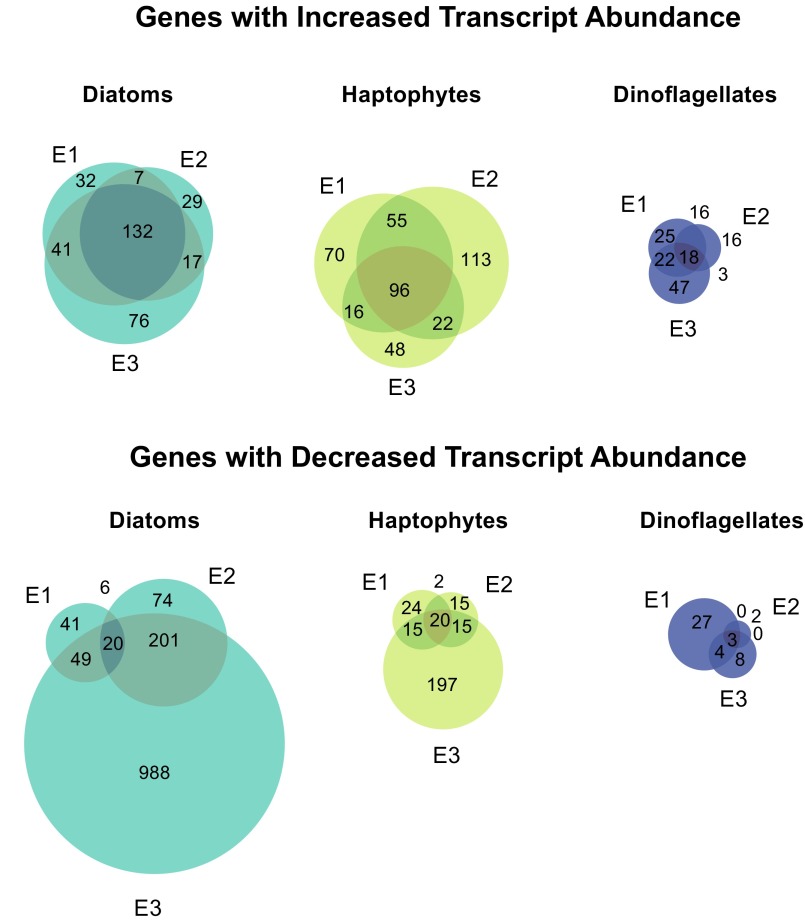
Variability within the QMF modules was resolved by statistically assessing the changes in abundance of individual genes for each functional group using a Bayesian approach (38) (Fig. 2C and Fig. S4). The statistical significance [twofold change, posterior probability (post-p) > 0.95] of differential abundance was examined for 4,038 KEGG orthologs common to diatoms, haptophytes, and dinoflagellates. As with the QMF (Fig. 2A), dinoflagellates demonstrated little to no significant changes in gene expression (Fig. 2C). The suites of transcripts with significantly increased transcript abundance following DSW were highly conserved for both diatoms and haptophytes across the three replicate experiments (40% of 334 and 19% of 490 genes in common, respectively) but differed between functional groups (41 of the total 824 significantly increased transcript genes in common) (Fig. S5). Of the genes with increased transcript abundance, many of those genes conserved across all three experiments for diatoms and haptophytes were associated with growth [e.g., ATP synthesis (10), photosynthesis (11), carbon fixation (13)] (Fig. 2C and Fig. S5). Additionally, following DSW addition, diatoms had signals indicative of the incorporation of both nitrogen (increasing abundance of glutamate and glutamine synthase) and iron (switching from NADPH to ferredoxin sulfate reductase). These changes in transcriptional patterns indicate that both diatoms and haptophytes increase fundamental metabolic processes required for photosynthetic growth in response to DSW.
Fig. S4.
Distribution of log fold change following DSW addition. Histograms of the number of genes falling within each of the log fold change bins for diatoms, haptophytes, and dinoflagellates are shown. The solid line indicates no fold change; dashed lines indicate twofold change, both up and down.
Fig. S5.
Weighted Venn diagrams of genes with significantly different abundances following DSW addition by functional group. The uniqueness of KEGG orthologs with increased or decreased abundances as determined by ASC (twofold change, post-p > 0.95) across experiments was assessed for diatoms, haptophytes, and dinoflagellates.
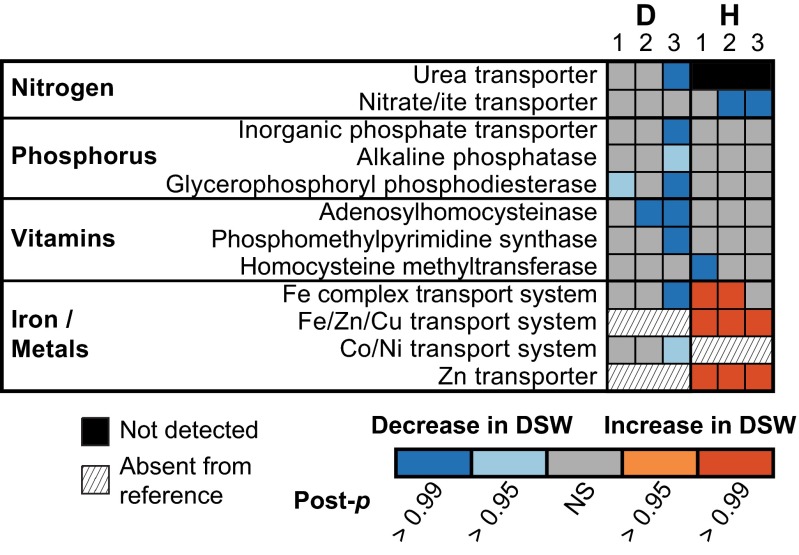
For diatoms and haptophytes, the consistency in genes with increased transcript abundance stands in contrast to the patterns of genes with significantly decreased transcript abundance following DSW addition (Fig. S5). Diatoms were typified by significant decreases in transcript abundance of many genes following DSW, consisting of a large portion (1,389 genes) of their metabolism compared with haptophytes (490 genes) (Fig. 2C). Genes with decreased transcript abundance were variable across the three experiments (1.5% and 6.9% similar for diatoms and haptophytes, respectively) (Fig. S5) and imply a tailoring of basal metabolism to the change in biogeochemical environment from DSW amendment. Across the KEGG modules, specific genes known to be markers of nutrient limitation significantly decreased in abundance following DSW addition (Fig. 3), potentially signifying a limitation in the in situ population that was alleviated following resupply. Such genes or their protein products are frequently used as proxies to identify limitation in the field (39). In E1 and E2, there was a decrease in diatom-associated glycerophosphoryl phosphodiesterase (22) and adenosylhomocysteinase (40), indicative of phosphorus and vitamin limitation, respectively (Fig. 3). Silica transporters, although not in KEGG, were surveyed and not found to have significant shifts in abundance. E3 was characterized by a decrease in abundance of many genes indicative of limitation in the diatoms, including but not limited to a urea transporter (41), a phosphate transporter (22), and metal transporters (Fig. 3). This finding is suggestive of diatom colimitation in E3, similar to patterns of colimitation recently observed in picocyanobacteria in the Pacific Ocean (39). There was a decrease in transcript abundance for haptophyte-associated nitrate and nitrite transporters in E2 and E3 and for homocysteine methyltransferase in E1 (Fig. 3). This decrease in transcript abundance may be indicative of nitrogen and vitamin limitation based on the pattern in other organisms (40, 41), but the regulation of these targets in haptophytes is poorly understood. Metal transport proteins were significantly increased for haptophytes in E1–E3 and indicated metabolic strategies following the addition of resources that differ from diatoms. In contrast to the diatoms, no markers of phosphorus limitation, such as the phosphate transporter or alkaline phosphatase in E. huxleyi (42–44), were significantly decreased for haptophytes, consistent with their known tolerance for phosphorus limitation (45). These data evince that diatoms and haptophytes are not under the same biogeochemical controls in situ and use disparate strategies following DSW addition to capitalize on newly available resources. Being able to identify and characterize multiple markers of limitation in a genera-specific manner for these eukaryotes is of central importance to the modeling of aperiodic blooms of these groups in oligotrophic systems.
Fig. 3.
Shifts in transcript abundance of genes responsive to biogeochemical forcing. The significance of changes in abundance (twofold change, post-p > 0.95 or post-p > 0.99) for genes known to be associated with nitrogen, phosphorus, vitamin, iron, or other trace metal metabolism for diatoms (D) or haptophytes (H) is indicated as blue (decrease) or orange (increase). Genes present within the reference transcriptome but not detected in the field are marked in black, and genes absent from the reference are hatched. KEGG identifications are as follows: urea transporter (K11959), nitrite/ate transporter (K02575), phosphate transporter (K08176), glycerophosphoryl diester phosphodiesterase (K01126), adenosylhomocysteinase (K01251), phosphomethylpyrimidine synthase (K03147), 5-methyltetrahydrofolate–homocysteine methyltransferase (K00548), iron complex transport system (K02013), iron/zinc/copper transport system (K11706), cobalt/nickel transport system (K02006), and zinc transporter (K14715).
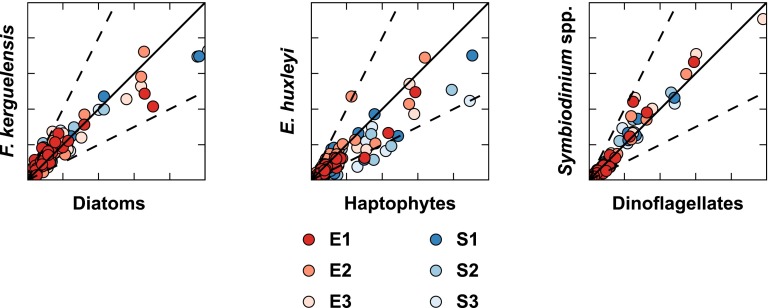
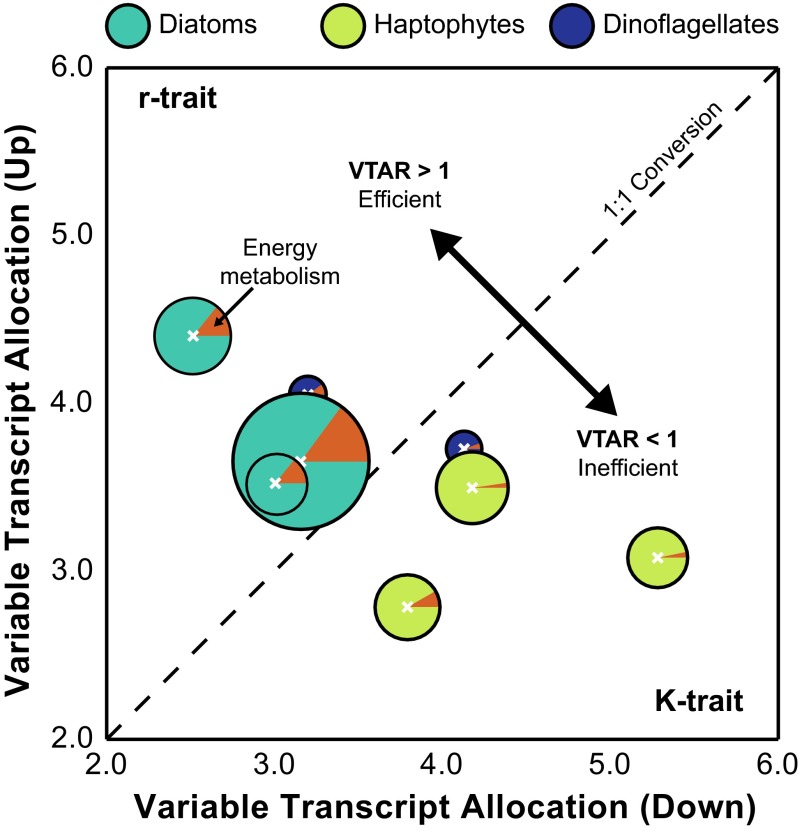
The above analyses focus primarily on the gene content and transcriptional patterns; however, the underlying ecoevolutionary metabolic traits for a functional group may be better described by considering the shift in the total transcript pool, as reported in tags per million (TPM). Using the statistically resolved patterns of increases and decreases in transcript abundance, a variable transcript allocation ratio (VTAR) was calculated to model functional group transcriptional responsiveness to DSW amendment (Fig. 4). VTAR > 1 indicates an efficient reallocation of the transcriptional potential from genes with decreased abundance to genes with increased abundance following DSW amendment, whereas VTAR < 1 indicates an inefficient reallocation. The dinoflagellates had variable transcript allocation (VTA) scores due to their small pool of differentially abundant genes; however, the VTA scores consistently resulted in disparate patterns for the diatom and the haptophytes (Fig. 4). In every experiment, the diatoms fell above the 1:1 line, with VTAR > 1 (1.15–1.75), whereas haptophytes fell below, with VTAR < 1 (0.58–0.83) (Fig. 4). The relative efficiency of reallocation, here defined by VTAR, reflects differences in the metabolic traits of these functional groups and aligns with preexisting ecological traits as defined by Margalef (8). Diatoms are r-selected with high maximum uptake rates that enhance their competition under high or fluctuating nutrients (e.g., a DSW upwelling event). This trait is reflected in the significant decrease in transcript abundance of many genes across a broad metabolic range coupled with the targeted increase of a subset of genes largely falling within the energy metabolism super module (Fig. 4), a pattern that was also observed with gene-focused analyses using trimmed mean of M value (TMM) normalization (16, 46) (Fig. S6). Haptophytes are K-selected, possessing a low half-saturation constant that enhances growth under low-nutrient conditions, but are unable to capitalize on nutrient pulses like r-selected competitors. Again, this ecological trait is reflected in changes in the haptophyte transcript pool. Although numerically fewer of their genes significantly decreased in abundance with DSW addition, the total TPM represented by those genes with decreased abundance exceeded the total TPM of those genes induced, defined by VTAR < 1 (Fig. 4). This finding is also reinforced by the gene-specific analysis (Fig. S6). It has been speculated that the mechanistic basis for the r- and K-tradeoff dichotomy in the phytoplankton lies in the disparate investment in growth or resource acquisition machinery (47). The large portion of the transcript pool that increased (VTAR > 1) for diatoms shows an ability to capitalize on newly available resources, with 14.2–14.9% of the increased TPM in the KEGG energy metabolism supermodule (Fig. 4). Haptophytes (VTAR < 1), by contrast, do not efficiently reallocate the transcript pool, with only 5.5–10.2% of the increased TPM in energy metabolism (Fig. 4). In short, haptophytes do not appear to modulate their transcript pool to capitalize on growth processes as efficiently as diatoms (Fig. 4).
Fig. 4.
Variable transcript allocation ratio (VTAR, Eq. 5) differentiates functional group strategies. The variable transcript allocation (VTA) score of the genes with significantly increased (VTAUp, Eq. 3) or decreased (VTADn, Eq. 4) abundance in the DSW amendment relative to the in situ sample is plotted for diatoms, haptophytes, and dinoflagellates for E1–E3. The size of the pie indicates the total number of genes with significantly different transcript abundances between the in situ and DSW-amended treatments. The proportion of increased TPM in DSW-amended treatments within the energy metabolism supergroup is illustrated as a pie slice in orange.
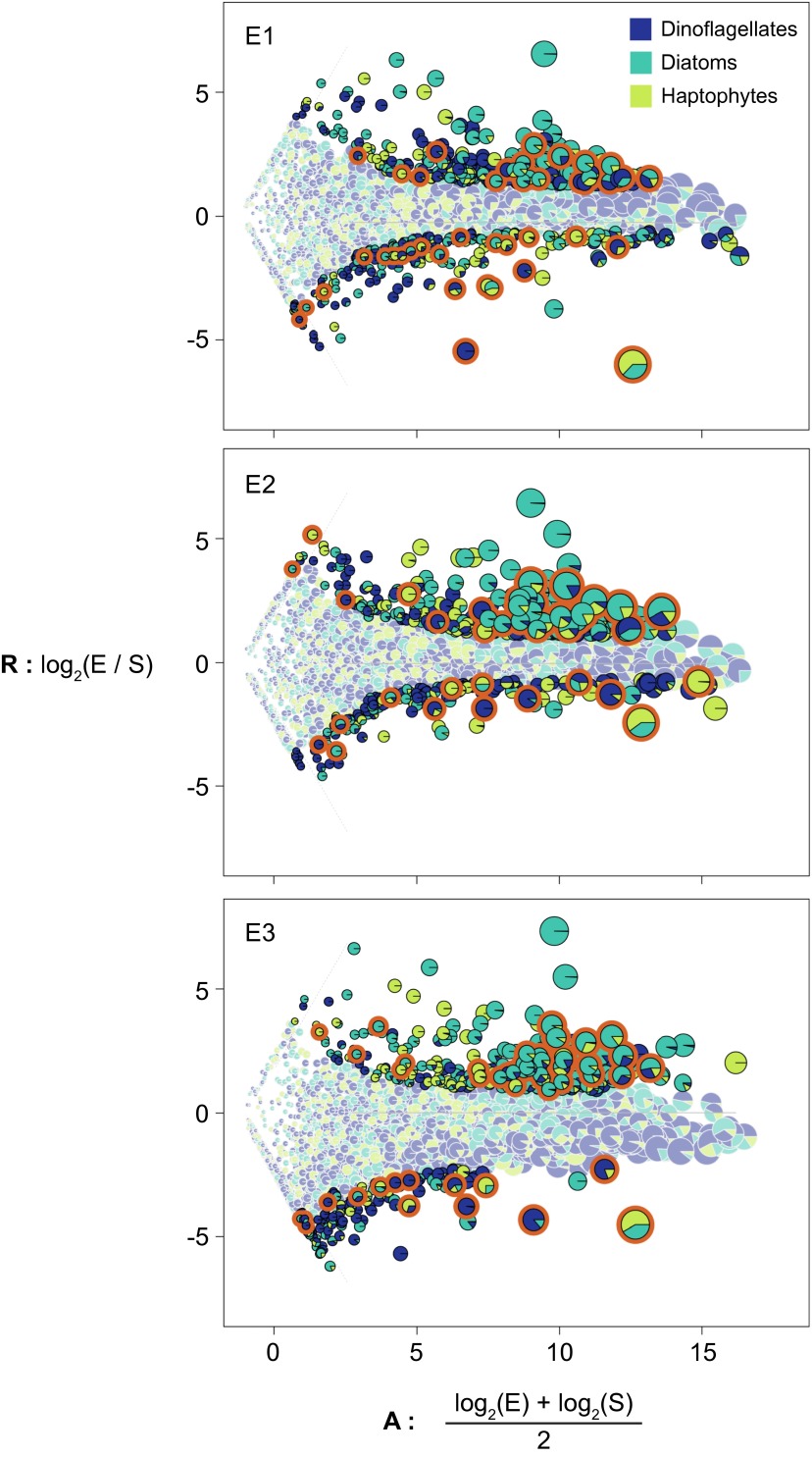
Fig. S6.
Microbial assemblage normalized transcript analysis (MANTA) ratio-averaged plots for global shifts in expression of KEGG orthologs. Fold change ratio (R) and average read count (A) are plotted for read counts in the in situ (S) and DSW amendment (E) samples across the three sample pairs (S1:E1, S2:E2, and S3:E3). The trimmed mean of fold change values is noted as a gray solid line; orthologs unique to one library are separated by gray dashed lines. Pies indicate the taxonomic distribution of orthologous reads across the three functional groups. KEGG orthologs that were significantly differentially expressed (DE) (adjusted P > 0.05) are outlined in black and highlighted in brighter colors, and those KEGG orthologs not significantly DE are outlined in gray. DE KEGG orthologs that fall in the energy metabolism KEGG module are outlined in orange.
These functional group-specific molecular and metabolic mechanisms underpin the aperiodic eukaryotic phytoplankton blooms in the oligotrophic ocean. Whereas both diatoms and haptophytes, including calcifying groups, likely contribute to a shift toward a net autotrophic condition when there is a nutrient pulse, the ecosystem function of oligotrophic systems may ultimately hinge on the unique trait of the diatoms to turn over their scavenger metabolism more efficiently to one of enhanced production. This finding is consistent with the dominance of diatom-associated BSi export relative to PIC export during summer in the NPSG (3). Unlike the preceding 13 years of study at Station ALOHA (3), enhanced production and export characteristic of bloom events were not observed during the summer of 2012, which exhibited a period of sustained net heterotrophy. We demonstrated through simulated blooms that the metabolic capacity for enhanced production is inherent in the large eukaryotic phytoplankton regardless of water mass, suggesting that the lack of bloom in 2012 was variably due to deficiency in macronutrients, vitamins, and metals. Because the conditions observed during the summer of 2012 may be increasingly encountered in a future ocean (48), modeling the molecular traits and tradeoffs of these populations will help to predict ecosystem state and metabolic balance of the ocean.
Methods
Sample Collection.
Seawater for the in situ eukaryote community mRNA analysis was collected at Station ALOHA (22°45′ N, 158°00′ W) from a depth of 25 m at 1400 h (local time) on three occasions during August and September 2012 as part of the HOE–DYLAN research expedition aboard the R/V Kilo Moana. Hydrocasts for sampling were performed using a conductivity-temperature-depth rosette water sampler equipped with 24 Scripps 12-L sampling bottles. Water was collected in acid-washed 20-L carboys, and ∼60 L of seawater was prescreened through 200-μm mesh and then filtered onto polycarbonate filters (5.0-μm pore size, 47 mm; Whatman) by way of a peristaltic pump. This size fraction was targeted to sample large eukaryotic phytoplankton, which are known to form blooms and contribute to export flux in the NPSG. Filters were changed every 20 min or when the flow rate decreased. Filters were placed in cryovials and stored in liquid nitrogen until mRNA extraction. The total length of filtration time did not exceed 3 h. Nutrient delivery in the ocean is highly variable; here, we chose to model a single nutrient pulse. The incubation experiments were performed with two treatments: a DSW treatment, which included 10% 0.2-μm filtered seawater collected from 700 m added to whole seawater collected at 25 m and a control treatment with no addition (Table S1). Triplicate 20-L carboys of each treatment were incubated at 30% surface light levels using on-deck incubators for 7 d and processed as described above on the final day at 1400 h (local time). Nutrient concentrations for phosphate [PO4], nitrate and nitrite [NO2+NO3], and silicate [SiO4] (Table S1) were measured by filtering 125 mL of seawater through a 0.2-μm, 47-mm polycarbonate filter, and stored frozen (−20 °C) in acid washed bottles until analysis at the Chesapeake Bay Lab at the University of Maryland according to the facility's protocols. Chlorophyll a was measured on whole-water samples collected onto GF/F filters (25 mm; Whatman) using a 90% acetone extraction and assayed by fluorescence using an AquaFluor Turner TD700 fluorometer/turbidimeter (49). There was little difference between the in situ sample and the control treatment with regard to both total chlorophyll a concentration and transcriptional profile (Figs. S1 and S7). There was no significant difference in QMF between the control treatment at the final time point and the in situ sample taken at the start of the incubation (Fig. S7). To compare nonbloom and bloom scenarios most conservatively, analyses thus focused on the comparison of the in situ community to the DSW-amended samples.
Table S1.
Macronutrient concentrations in control no addition (C), DSW-amended incubations (E), and 700 m water used in DSW amendment incubations
| Treatment | Time postinoculation, h | NO2 + NO3, μM | PO4, μM | Silica, μM |
| C (control no addition)* | 168 | 0.12 ± 0.03 | 0.12 ± 0.02 | 1.91 ± 0.2 |
| E (+10% DSW)* | 168 | 1.9 ± 0.93 | 0.23 ± 0.05 | 8.46 ± 3.11 |
| DSW (700 m water)* | N/A | 37.5 ± 1.68 | 3.14 ± 0.03 | 83.4 ± 9.33 |
N/A, not applicable.
Nutrient data averaged for E1 and E2; nutrients were not assayed on E3.
Fig. S7.
PCA of the QMF signals across in situ, control no addition, and DSW amended samples. PCA of the QMF signals for each of the functional groups across in situ (S1–S3), control no addition (C1–C3), and DSW amendment (E1–E3); 95% confidence ellipses are indicated for each of the sample types by functional group.
RNA Extraction and Sequencing.
RNA was extracted from individual filters with an RNeasy Mini Kit (Qiagen), following a modified version of the yeast protocol. Briefly, lysis buffer and RNA-clean zirconia/silica beads were added to the filter, and samples were vortexed for 1 min, placed on ice for 30 s, and then vortexed again for 1 min. Samples were then processed following the yeast protocol. The resulting RNA was eluted in water and then treated for possible DNA contamination using a TURBO DNA-free Kit (Ambion) following the rigorous DNase protocol. RNA from individual filters was then pooled by sample, using the RNA Cleanup Protocol (Qiagen) from the RNeasy Mini Kit. The resulting RNA sample thus represented ∼60 L of total seawater for the in situ sample. Filters were pooled across like triplicate bottles by treatment, totaling 60 L from each of the incubation treatments. The total RNA sample was then enriched for eukaryotic mRNA through a poly-A pull-down procedure. The resulting enriched mRNA sample then went through library preparation with an Illumina TruSeq mRNA Prep Kit. Libraries were sequenced with an Illumina HiSeq2000 at the Columbia Genome Center. Each sample was sequenced to produce a targeted 60 million 100-bp, paired-end reads. These environmental sequence data are deposited in the Sequence Read Archive through the National Center for Biotechnology Information under accession no. SRP056385. Raw sequence data quality was visualized using FastQC and then cleaned and trimmed using Trimmomatic version 0.27 (paired-end mode, 6-bp-wide sliding window for quality below 20, minimum length of 25 bp).
Genome Database Creation and Mapping.
Environmentally relevant algal genomes (Aureococcus anophagefferens CCMP 1984 v1.0, E. huxleyi CCMP 1516 v1.0, Fragilariopsis cylindrus CCMP 1102 v1.0, Micromonas pusilla RCC 299 v3.0, Ostreococcus lucimarinus CCMP 2514 v2.0, Ostreococcus tauri RCC 141 v2.0, Phaeodactylum tricornutum CCMP 632 v2.0, Pseudo-nitzschia multiseries CLN-47 v1.0, and Thalassiosira pseudonana CCMP 1335 v3.0) were collected from the Joint Genome Institute database (genome.jgi.doe.gov/) and concatenated. Trimmed, paired-end reads from each of the samples were mapped to this concatenated genome library using the Burrows–Wheeler Aligner (50) (BWA-mem, parameters: −k 10 −aM) and then counted using the HTSeq 0.6.1 package (51).
MMETSP Database Creation and Mapping.
Transcriptome sequences and annotations generated through the MMETSP that were made public as of March 17, 2014 were collected, representing 401 transcriptomes across 209 species or cultured isolates. Transcriptomes from like species (regardless of strain or condition) and cultured isolates were pooled and clustered using CD-HIT-EST (98% identity, word size of 9). The resulting clustered set of transcripts was considered to be the representative transcriptome for the species or cultured isolate. Transcriptomes were annotated with KEGG orthology annotations using the bidirectional best hit method through the KEGG Automatic Annotation Server (52). It is worth noting that the KEGG module names are human-defined and some genes are artificially grouped in the context of phytoplankton metabolism. For example, the “methane metabolism” module includes a suite of genes related to carbon, and not specifically methane, metabolism. The 209 transcriptomes created in this manner were concatenated to form a comprehensive species-level transcriptome database from the MMETSP library. Due to the large size of the resulting MMETSP database, trimmed reads were mapped to the MMETSP using the Burrows–Wheeler Aligner (50) (BWA-mem, parameters: −k 10 −aM) and then counted using the HTSeq 0.6.1 package (51).
Differential Expression Analysis.
Counts obtained from HTSeq were pooled for like KEGG orthologs across all species in a functional group. The QMF was assessed by normalizing global patterns of expression at the module level to the total mapped reads, an approach similar to those approaches used in several metagenomic and metatranscriptomic studies focused on prokaryotes (20, 24, 53). PCA and confidence ellipses of the QMF signals by functional group and sample type (in situ, control no addition, and DSW addition) were calculated and visualized using the FactorMineR package in R (Fig. 2 and Fig. S7). No significant difference was seen between the in situ and additional control samples (Fig. S7). For each functional group, the pooled KEGG counts from the in situ samples (S1–S3) were compared with those samples from the corresponding DSW amendment (E1–E3) using analysis of sequence counts (ASC), an empirical Bayes method (38). Genes were considered to be differentially abundant between treatments if for a fold change of 2.0, the post-p was greater than 0.95 (22). Patterns of differential abundance were visualized using Circos (54). Global shifts in the expression of genes independent of functional group were assessed with TMM normalization using the Microbial Assemblage Normalized Transcript Analysis package (MANTA, version 1.12.0) (16).
Variable Transcript Allocation Modeling.
VTA following DSW amendment was calculated for each functional group. Although there was a normal distribution of log fold change across all functional groups, the means were offset for the diatoms and the haptophytes (Fig. S4). From the set of all genes (G), the genes that had statistically significant increased transcript abundance (Eq. 1) and decreased transcript abundance (Eq. 2) as identified with ASC (twofold change, post-p > 0.95) (38) in DSW-amended treatments (E) relative to the in situ samples (S) were considered. A VTA score was calculated for the set of genes with both increased (Eq. 3) and decreased (Eq. 4) abundance, taking the ratio of the summed transcripts per million (T) for the in situ samples (S) and experimental DSW-amended treatments (E) of every gene (u or d) within the set of significantly differentially abundant genes (U or D). VTA scores were calculated so as always to be greater than 1; thus, the VTADn sums the reciprocal of the ratio summed in VTAUp (Eqs. 3 and 4). VTADn is the magnitude of the decreased transcriptional response following DSW addition, whereas VTAUp is the magnitude of the increased transcriptional response following DSW addition. VTAUp and VTADn, as ratios, focus on the total transcript pool shifted between S and E rather than on the number of genes with differential abundance. As such, we can directly compare these two ratios with VTAR (Eq. 5) and assess the metabolic trait of reallocation efficiency. If the VTAR was >1, there was a larger transcript pool (TPM) in genes that were increased than in genes that were decreased, indicating an efficient reallocation of the transcript pool. By contrast, if the VTAR was <1, less TPM was increased than was decreased. This model defines the total metabolic responsiveness as a trait that can be compared between the functional groups:
| [1] |
| [2] |
| [3] |
| [4] |
| [5] |
Acknowledgments
We thank the captain and crew of the R/V Kilo Moana and the participants in the HOE–DYLAN cruise series for their help. We also thank Bethany Jenkins for useful discussion. The MMETSP samples used in this study were sequenced, assembled, and annotated with the Assembly by Short Sequences (ABySS) pipeline at the National Center for Genome Resources. This research was supported by funds from the Center for Microbial Oceanography: Research and Education (C-MORE, National Science Foundation Grant EF04-24599) and by grants from the Simons Foundation (to D.M.K. and S.T.D.), and is a contribution of the Simons Collaboration on Ocean Processes and Ecology (SCOPE). Support was also provided by the Gordon and Betty Moore Foundation through Grant 2637 to the National Center for Genome Resources for the MMETSP and Grant 3794 (to D.M.K.). H.A. was supported by the Department of Defense through the National Defense Science & Engineering Graduate Fellowship Program.
Footnotes
The authors declare no conflict of interest.
Data deposition: Hydrographic data associated with sample collection reported in this paper have been deposited in the Center for Microbial Oceanography: Research and Education (C-MORE) data site. Raw sequence data reported in this paper have been deposited in the National Center for Biotechnology Information Sequence Read Archive (accession no. SRP056385).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1518165112/-/DCSupplemental.
References
- 1.Karl DM, Laws EA, Morris P, Williams PJL, Emerson S. Global carbon cycle: Metabolic balance of the open sea. Nature. 2003;426(6962):32. doi: 10.1038/426032a. [DOI] [PubMed] [Google Scholar]
- 2.McAndrew P, et al. Metabolic response of oligotrophic plankton communities to deep water nutrient enrichment. Mar Ecol Prog Ser. 2007;332:63–75. [Google Scholar]
- 3.Karl DM, Church MJ, Dore JE, Letelier RM, Mahaffey C. Predictable and efficient carbon sequestration in the North Pacific Ocean supported by symbiotic nitrogen fixation. Proc Natl Acad Sci USA. 2012;109(6):1842–1849. doi: 10.1073/pnas.1120312109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Villareal TA, Brown CG, Brzezinski MA, Krause JW, Wilson C. Summer diatom blooms in the North Pacific subtropical gyre: 2008-2009. PLoS One. 2012;7(4):e33109. doi: 10.1371/journal.pone.0033109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Villareal TA, Adornato L, Wilson C, Schoenbaechler CA. Summer blooms of diatom-diazotroph assemblages and surface chlorophyll in the North Pacific Gyre: A disconnect. J Geophys Res. 2011;116(C3):C03001. [Google Scholar]
- 6.Field CB, Behrenfeld MJ, Randerson JT, Falkowski P. Primary production of the biosphere: Integrating terrestrial and oceanic components. Science. 1998;281(5374):237–240. doi: 10.1126/science.281.5374.237. [DOI] [PubMed] [Google Scholar]
- 7.López-Urrutia A, San Martin E, Harris RP, Irigoien X. Scaling the metabolic balance of the oceans. Proc Natl Acad Sci USA. 2006;103(23):8739–8744. doi: 10.1073/pnas.0601137103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Margalef R. Life-forms of phytoplankton as survival alternatives in an unstable environment. Oceanol Acta. 1978;1(4):493–509. [Google Scholar]
- 9.Tilman D. Resource competition between plankton algae: An experimental and theoretical approach. Ecology. 1977;58(2):338–348. [Google Scholar]
- 10.Cavender-Bares KK, Karl DM, Chisholm SW. Nutrient gradients in the western North Atlantic Ocean: Relationship to microbial community structure and comparison to patterns in the Pacific Ocean. Deep Sea Res Part I Oceanogr Res Pap. 2001;48(11):2373–2395. [Google Scholar]
- 11.Karl DM, et al. Ecological nitrogen-to-phosphorus stoichiometry at station ALOHA. Deep Sea Res Part II Top Stud Oceanogr. 2001;48(8-9):1529–1566. [Google Scholar]
- 12.Karl DM. Nutrient dynamics in the deep blue sea. Trends Microbiol. 2002;10(9):410–418. doi: 10.1016/s0966-842x(02)02430-7. [DOI] [PubMed] [Google Scholar]
- 13.Benitez-Nelson CR, et al. Mesoscale eddies drive increased silica export in the subtropical Pacific Ocean. Science. 2007;316(5827):1017–1021. doi: 10.1126/science.1136221. [DOI] [PubMed] [Google Scholar]
- 14.Kim I-N, et al. Chemical oceanography. Increasing anthropogenic nitrogen in the North Pacific Ocean. Science. 2014;346(6213):1102–1106. doi: 10.1126/science.1258396. [DOI] [PubMed] [Google Scholar]
- 15.Cortés MYM, Bollmann J, Thierstein HHR. Coccolithophore ecology at the HOT station ALOHA, Hawaii. Deep Sea Res Part II Top Stud Oceanogr. 2001;48(8-9):1957–1981. [Google Scholar]
- 16.Marchetti A, et al. Comparative metatranscriptomics identifies molecular bases for the physiological responses of phytoplankton to varying iron availability. Proc Natl Acad Sci USA. 2012;109(6):E317–E325. doi: 10.1073/pnas.1118408109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alexander H, Jenkins BD, Rynearson TA, Dyhrman ST. Metatranscriptome analyses indicate resource partitioning between diatoms in the field. Proc Natl Acad Sci USA. 2015;112(17):E2182–E2190. doi: 10.1073/pnas.1421993112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dupont CL, et al. Genomes and gene expression across light and productivity gradients in eastern subtropical Pacific microbial communities. ISME J. 2015;9(5):1076–1092. doi: 10.1038/ismej.2014.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pearson GA, et al. Metatranscriptomes reveal functional variation in diatom communities from the Antarctic Peninsula. ISME J. 2015;9(10):2275–2289. doi: 10.1038/ismej.2015.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ottesen EA, et al. Multispecies diel transcriptional oscillations in open ocean heterotrophic bacterial assemblages. Science. 2014;345(6193):207–212. doi: 10.1126/science.1252476. [DOI] [PubMed] [Google Scholar]
- 21.Hennon GMM, et al. Diatom acclimation to elevated CO2 via cAMP signalling and coordinated gene expression. Nat Clim Change. 2015;5(8):761–765. [Google Scholar]
- 22.Dyhrman ST, et al. The transcriptome and proteome of the diatom Thalassiosira pseudonana reveal a diverse phosphorus stress response. PLoS One. 2012;7(3):e33768. doi: 10.1371/journal.pone.0033768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Amin SA, et al. Interaction and signalling between a cosmopolitan phytoplankton and associated bacteria. Nature. 2015;522(7554):98–101. doi: 10.1038/nature14488. [DOI] [PubMed] [Google Scholar]
- 24.Shi Y, McCarren J, DeLong EF. Transcriptional responses of surface water marine microbial assemblages to deep-sea water amendment. Environ Microbiol. 2012;14(1):191–206. doi: 10.1111/j.1462-2920.2011.02598.x. [DOI] [PubMed] [Google Scholar]
- 25.McCarren J, et al. Microbial community transcriptomes reveal microbes and metabolic pathways associated with dissolved organic matter turnover in the sea. Proc Natl Acad Sci USA. 2010;107(38):16420–16427. doi: 10.1073/pnas.1010732107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Vargas C, et al. Tara Oceans Coordinators Eukaryotic plankton diversity in the sunlit ocean. Science. 2015;348(6237):1261605. doi: 10.1126/science.1261605. [DOI] [PubMed] [Google Scholar]
- 27.Villar E, et al. Tara Oceans Coordinators Environmental characteristics of Agulhas rings affect interocean plankton transport. Science. 2015;348(6237):1261447. doi: 10.1126/science.1261447. [DOI] [PubMed] [Google Scholar]
- 28.Keeling PJ, et al. The Marine Microbial Eukaryote Transcriptome Sequencing Project (MMETSP): Illuminating the functional diversity of eukaryotic life in the oceans through transcriptome sequencing. PLoS Biol. 2014;12(6):e1001889. doi: 10.1371/journal.pbio.1001889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Letelier RM, et al. Temporal variability of phytoplankton community structure based on pigment analysis. Limnol Oceanogr. 1993;38(7):1420–1437. [Google Scholar]
- 30.Fontanez KM, Eppley JM, Samo TJ, Karl DM, DeLong EF. Microbial community structure and function on sinking particles in the North Pacific Subtropical Gyre. Front Microbiol. 2015;6:469. doi: 10.3389/fmicb.2015.00469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moustafa A, et al. Transcriptome profiling of a toxic dinoflagellate reveals a gene-rich protist and a potential impact on gene expression due to bacterial presence. PLoS One. 2010;5(3):e9688. doi: 10.1371/journal.pone.0009688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hackett JD, Anderson DM, Erdner DL, Bhattacharya D. Dinoflagellates: A remarkable evolutionary experiment. Am J Bot. 2004;91(10):1523–1534. doi: 10.3732/ajb.91.10.1523. [DOI] [PubMed] [Google Scholar]
- 33.Palenik BP, Henson SE. The use of amides and other organic nitrogen sources by the phytoplankton Emiliania huxleyi. Limnol Oceanogr. 1997;42(7):1544–1551. [Google Scholar]
- 34.Winn CD, Karl DM. Laboratory calibrations of the [h]adenine technique for measuring rates of RNA and DNA synthesis in marine microorganisms. Appl Environ Microbiol. 1984;47(4):835–842. doi: 10.1128/aem.47.4.835-842.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Silver MW, et al. Toxic diatoms and domoic acid in natural and iron enriched waters of the oceanic Pacific. Proc Natl Acad Sci USA. 2010;107(48):20762–20767. doi: 10.1073/pnas.1006968107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marchetti A, et al. Iron and silicic acid effects on phytoplankton productivity, diversity, and chemical composition in the central equatorial Pacific Ocean. Limnol Oceanogr. 2005;55(1):11–29. [Google Scholar]
- 37.Read BA, et al. Emiliania huxleyi Annotation Consortium Pan genome of the phytoplankton Emiliania underpins its global distribution. Nature. 2013;499(7457):209–213. doi: 10.1038/nature12221. [DOI] [PubMed] [Google Scholar]
- 38.Wu Z, et al. Empirical bayes analysis of sequencing-based transcriptional profiling without replicates. BMC Bioinformatics. 2010;11(1):564. doi: 10.1186/1471-2105-11-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saito MA, et al. Multiple nutrient stresses at intersecting Pacific Ocean biomes detected by protein biomarkers. Science. 2014;345(6201):1173–1177. doi: 10.1126/science.1256450. [DOI] [PubMed] [Google Scholar]
- 40.Bertrand EM, et al. Influence of cobalamin scarcity on diatom molecular physiology and identification of a cobalamin acquisition protein. Proc Natl Acad Sci USA. 2012;109(26):E1762–E1771. doi: 10.1073/pnas.1201731109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bender SJ, Durkin CA, Berthiaume CT, Morales RL, Armbrust EV. 2014. Transcriptional responses of three model diatoms to nitrate limitation of growth. Front Mar Sci 1:3.
- 42.Dyhrman ST, et al. Long serial analysis of gene expression for gene discovery and transcriptome profiling in the widespread marine coccolithophore Emiliania huxleyi. Appl Environ Microbiol. 2006;72(1):252–260. doi: 10.1128/AEM.72.1.252-260.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dyhrman ST, Palenik B. Characterization of ectoenzyme activity and phosphate-regulated proteins in the coccolithophorid Emiliania huxleyi. J Plankton Res. 2003;25(10):1215–1225. [Google Scholar]
- 44.Xu Y, Wahlund TM, Feng L, Shaked Y, Morel FMM. A novel alkaline phosphatase in the cocolithophore Emiliania huxleyi (Prymnesiophyceae) and its regulation by phosphorus. J Phycol. 2006;42(4):835–844. [Google Scholar]
- 45.Lessard EJ, Merico A, Tyrrell T. Nitrate:phosphate ratios and Emiliania huxleyi blooms. Limnol Oceanogr. 2005;50(3):1020–1024. [Google Scholar]
- 46.Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11(3):R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Litchman E, Klausmeier CA. Trait-based community ecology of phytoplankton. Annu Rev Ecol Evol Syst. 2008;39(1):615–639. [Google Scholar]
- 48.Doney SC, et al. Climate change impacts on marine ecosystems. Annu Rev Mar Sci. 2012;4(1):11–37. doi: 10.1146/annurev-marine-041911-111611. [DOI] [PubMed] [Google Scholar]
- 49.Parsons TR, Yoshiaki M, Lalli CM. A Manual of Chemical & Biological Methods for Seawater Analysis. Pergamon; Oxford: 1984. [Google Scholar]
- 50.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26(5):589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31(2):166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M. KAAS: An automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007;35(Web Server issue):W182–W185. doi: 10.1093/nar/gkm321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shi Y, Tyson GW, Eppley JM, DeLong EF. Integrated metatranscriptomic and metagenomic analyses of stratified microbial assemblages in the open ocean. ISME J. 2011;5(6):999–1013. doi: 10.1038/ismej.2010.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Krzywinski M, et al. Circos: An information aesthetic for comparative genomics. Genome Res. 2009;19(9):1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]