Significance
Dogs were the first domesticated species, but the precise timing and location of domestication are hotly debated. Using genomic data from 5,392 dogs, including a global set of 549 village dogs, we find strong evidence that dogs were domesticated in Central Asia, perhaps near present-day Nepal and Mongolia. Dogs in nearby regions (e.g., East Asia, India, and Southwest Asia) contain high levels of genetic diversity due to their proximity to Central Asia and large population sizes. Indigenous dog populations in the Neotropics and South Pacific have been largely replaced by European dogs, whereas those in Africa show varying degrees of European vs. indigenous African ancestry.
Keywords: admixture, domestication, linkage disequilibrium, introgression, haplotype diversity
Abstract
Dogs were the first domesticated species, originating at least 15,000 y ago from Eurasian gray wolves. Dogs today consist primarily of two specialized groups—a diverse set of nearly 400 pure breeds and a far more populous group of free-ranging animals adapted to a human commensal lifestyle (village dogs). Village dogs are more genetically diverse and geographically widespread than purebred dogs making them vital for unraveling dog population history. Using a semicustom 185,805-marker genotyping array, we conducted a large-scale survey of autosomal, mitochondrial, and Y chromosome diversity in 4,676 purebred dogs from 161 breeds and 549 village dogs from 38 countries. Geographic structure shows both isolation and gene flow have shaped genetic diversity in village dog populations. Some populations (notably those in the Neotropics and the South Pacific) are almost completely derived from European stock, whereas others are clearly admixed between indigenous and European dogs. Importantly, many populations—including those of Vietnam, India, and Egypt—show minimal evidence of European admixture. These populations exhibit a clear gradient of short-range linkage disequilibrium consistent with a Central Asian domestication origin.
The domestic dog, Canis lupus familiaris, is found living with and around humans throughout the globe. Selective breeding of dogs has been practiced for thousands of years, but the majority of modern breeds are less than 200 y old and of European ancestry (1, 2). Most dogs in the world are not purebred or even mixed-breed dogs, but rather belong to free-breeding human-commensal populations (“village dogs”) (1, 3, 4). The history and lineage of most modern breeds is well established (5, 6), but the genetic relationships among village dog populations and between village dogs and breeds is less understood.
Global surveys of mitochondrial and Y chromosome diversity in dogs have concluded that domestication occurred in southern China less than 16,500 yBP (7–10). In contrast, the earliest archeological evidence for dog-like canids occurs in Europe and Siberia, and Mt haplotypes found in ancient and modern gray wolves appear to be consistent with an origin of dogs from European wolves (11). These conflicting observations could be due to demographic processes after domestication (bottlenecks, migration, and admixture), altering patterns of genetic diversity or simply a consequence of a sparse archeological record in East Asia during this period. Archeologists and geneticists agree that dogs evolved from Eurasian gray wolves at least 15,000 yBP (2), but precise determination of the domestication origin(s) is elusive.
Whereas the Y and Mt chromosomes are just two inherited loci, autosomal markers offer a vastly richer picture of the patterning of genetic variation genome-wide and better resolution for demographic inference. Efforts to identify the basis of phenotypic diversity and genetic diseases in dogs have yielded large genomic datasets of purebred dogs readily available for demographic inference (6, 12, 13). Genomic comparisons of purebred dogs and wolves show Middle Eastern wolves have more haplotype sharing with dogs than other wolf populations (6), but this is likely due to dog-wolf introgression in the Middle East (14) rather than an indication of Middle Eastern origins.
Inference of early population history using purebred dogs is hampered by the confounding effects of artificial selection and bottlenecks and by the relative dearth of breeds without European ancestry. Genetic analyses have identified fewer than 20 “basal” breeds that have remained isolated enough from modern admixture to retain genetic signatures reflecting their geographic origins, and most of these lineages were severely depleted by genetic bottlenecks in the modern era (1, 2, 6). Whereas patterns of linkage disequilibrium (LD) in people can be used to trace human origins to Africa (15), similar analyses in purebred dogs show LD patterns dominated by breed-specific bottlenecks without any spatial trends to suggest a domestication origin, even in basal breeds (12, 16–18).
Because village dogs are geographically widespread and genetically diverse, they can be highly informative of dog population history if recent admixture with foreign dogs is minimal (4, 19, 20). Bottlenecks and artificial selection have drastically skewed genetic diversity within breeds, but the larger effective population size () of village dogs make them a better reflection of the genetic structure present in dogs before the modern era (21). Village dog populations that are relatively free of admixture should show genetic signatures reflecting the origins and movement of early dogs [and humans (22)], including the spread of pastoralism into Europe, the Bantu expansion in Africa, the peopling of the Americas, the settlement of the Pacific, and, most recently, European colonialism throughout the Americas and elsewhere.
Village dogs and local breeds represent an important but underused resource for disentangling the complicated evolutionary history of dogs. To this end, we genotyped a diverse panel of 549 village dogs from 38 countries and 4,676 purebred dogs from 161 breeds on a semicustom Illumina CanineHD array consisting of 185,805 markers, including 582 and 336 Mt and Y markers, respectively (13). We combined this with existing Mt and array data (6, 8, 11, 13, 23–27) to amass the largest canine diversity panel assembled to date, allowing efficient comparison of Y, Mt, and autosomal loci to evaluate the forces patterning genetic variation in diverse dog populations.
Results
Breed Dogs Represent a Fraction of Global Dog Diversity.
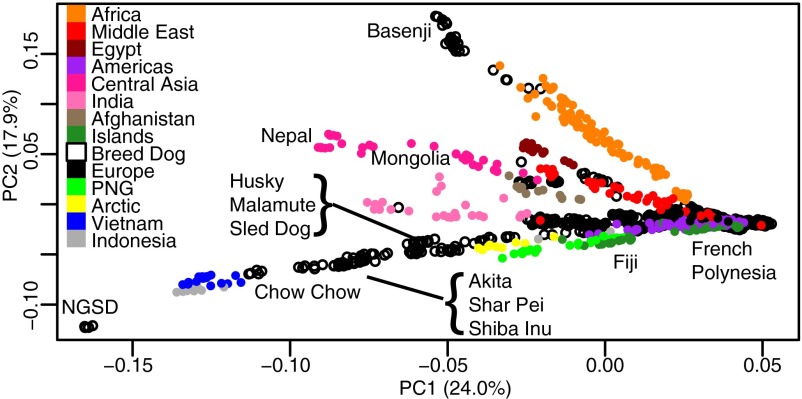
Although breed dogs are phenotypically diverse, they contain only a fraction of dog genetic diversity (4). Principal component analysis (PCA) of village dogs clusters European dogs tightly, whereas others are dispersed according to geography (Fig. 1 and SI Appendix, Fig. S4). Although some of this diversity is reflected in basal dog breeds that have been projected onto the PC plot (e.g., East Asia), much of the diversity, particularly in Central Asia, India, and the Middle East, is absent from the breeds studied. PCA indicates a clear divergence between East Asia (Vietnam and Island Southeast Asia), Central Asia (Mongolia and Nepal), India, the Middle East (Egypt, Lebanon, Qatar, Turkey, and Afghanistan), and sub-Saharan Africa. These groupings are consistent with regional definitions based on human cultural history (28).
Fig. 1.
PCA of village dogs, with breed dogs projected onto the PC space (Methods). Village dogs are colored according to origin; projected breed dogs are black circles.
Our array contained 360 Mt and 206 Y markers passing quality control, enabling detection of 373 and 72 haplotypes, respectively (Methods). Although only 15.0% of the dogs (15.6% of the males) were village dogs, 36.6% of the Mt and 38.9% of the Y haplotypes were only found in village dogs (SI Appendix, Table S3). In contrast, only 40.6% of the Mt and 18.1% of the Y haplotypes were exclusive to breed dogs, despite being 85% of the cohort. Most breeds have Y haplotypes from a single haplogroup, HG1-3, and Mt haplotypes from haplogroups A1 and B1, whereas village dogs are much more variable (SI Appendix, Figs. S5–S10 and S13). Mt and Y haplotype heterozygosity ( and ) is also higher in village dogs (Table 1 and SI Appendix, Tables S4 and S5). East Asia contains the largest number of Mt haplogroups, but fewer Y haplogroups than Africa, India, Central Asia, or Southwest Asia.
Table 1.
Mt and Y haplotype diversity in village dogs
| Population | Mt | Y | ||
| N | H | N | H | |
| Africa | 91 | 0.96 | 64 | 0.93 |
| America | 171 | 0.94 | 99 | 0.86 |
| Arctic | 11 | 0.82 | 6 | 0.87 |
| Oceania | 105 | 0.93 | 47 | 0.94 |
| Central Asia | 26 | 0.95 | 17 | 0.84 |
| East Asia | 118 | 0.99 | 24 | 0.92 |
| Europe | 18 | 0.96 | 14 | 0.93 |
| India | 35 | 0.97 | 27 | 0.89 |
| Southwest Asia | 80 | 0.96 | 46 | 0.94 |
Sample size (N) and haplotype heterozygosity (H) by region.
Village Dog Populations Differ in Their Level of Admixture with European Dogs.
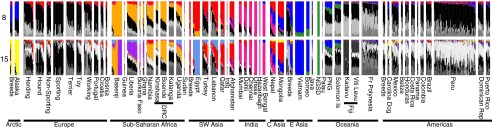
A striking pattern in the data is the varying levels of European admixture across village dog populations (Fig. 2 and SI Appendix, Fig. S11). European admixture is also evident in several basal breeds (SI Appendix, Fig. S14). Dogs in the Neotropics and South Pacific are almost exclusively descended from European stock, even though large dog populations lived in these regions before European contact (22, 29) (SI Appendix, Fig. S12 and Table S6).
Fig. 2.
ADMIXTURE analysis at K = 8 and 15 for unrelated village and breed dogs. European ancestry components are in grayscale ancestry. Vertical lines are individual dogs with one dog per breed (except for Basenjis, NGSDs, and Carolina Dogs).
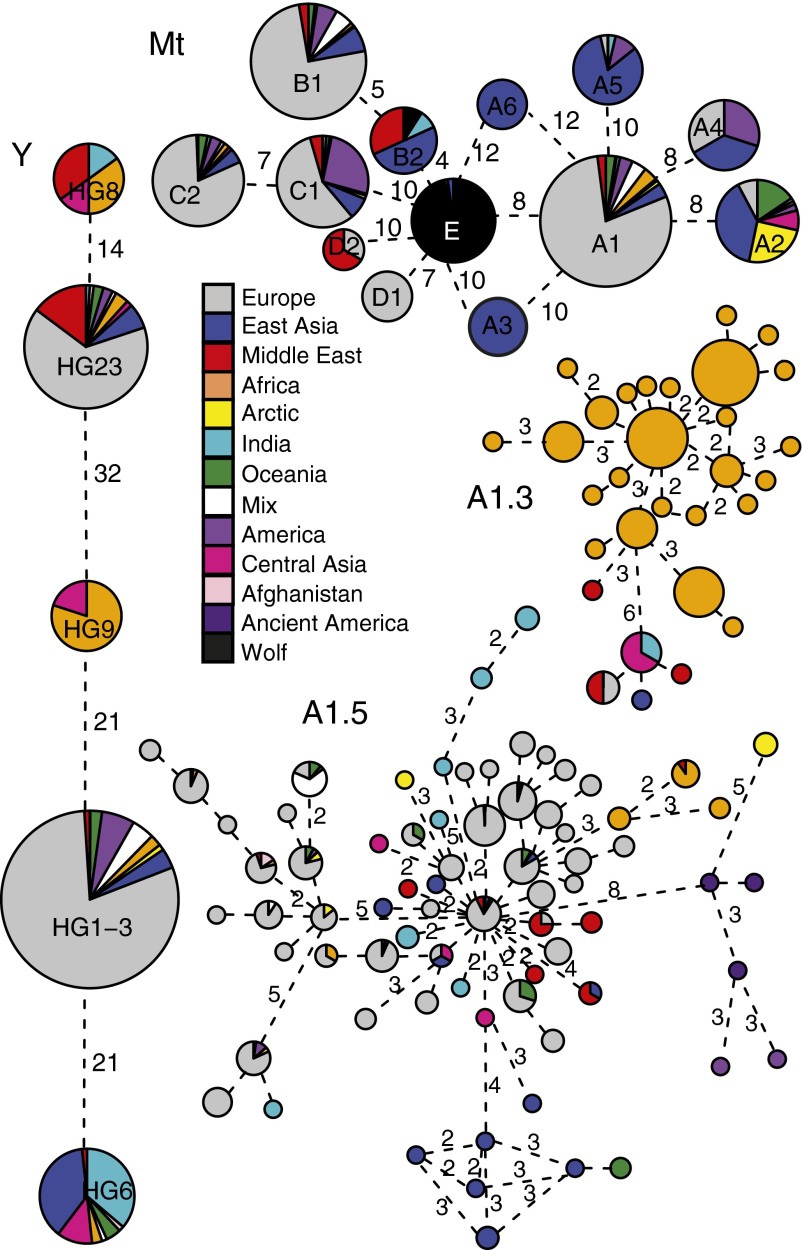
Nevertheless, a subcluster in Mt haplogroup A1.5 contains only haplotypes from ancient and modern American dogs (Fig. 3 and SI Appendix, Fig. S7). We also see indigenous Mt haplotypes segregating in Carolina dogs and Xoloitzcuintlis, but no unique Y haplotypes indicative of indigenous ancestry were found in American dogs outside of the Arctic. Alaskan village dogs and Arctic breeds form a cluster based on nuclear markers, whereas other American dogs show little, if any, non-European ancestry, except for Carolina dogs, which contain between 10% and 35% pre-Columbian ancestry based on clustering with East Asia (SI Appendix, Fig. S12).
Fig. 3.
Minimum spanning networks of Y and Mt haplogroups. Haplotypes within A1.3 and A1.5 Mt haplogroups also shown. Within each network, circle size is proportional to haplotype/group frequency and line length is determined by number of mutations separating haplotypes (indicated by numerals when greater than one mutation).
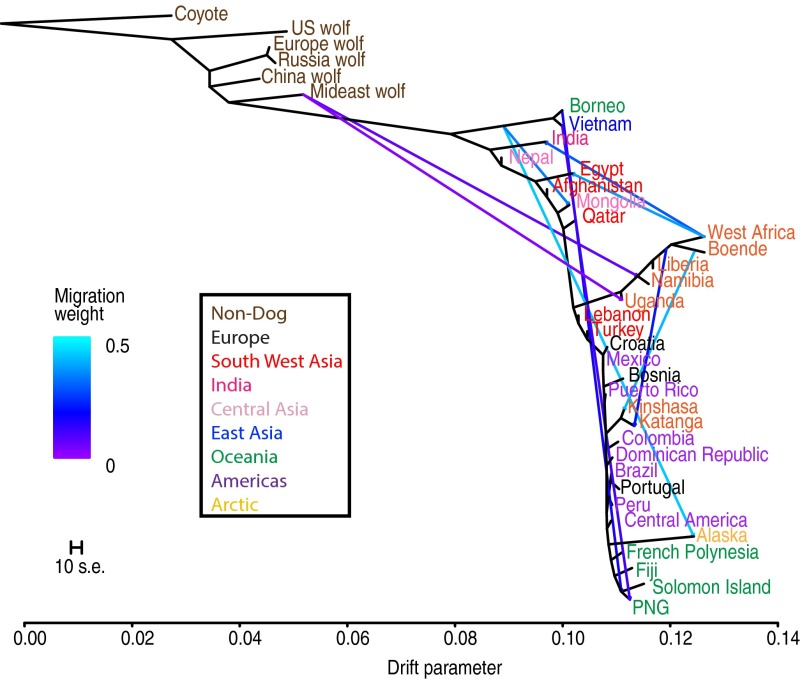
Fijian and French Polynesian dogs also retain little indigenous ancestry (Fig. 4) and contain only universal Y and Mt haplotypes. Nevertheless, these dogs are clearly genetically distinct ( 0.01–0.04 between island groups), with additional genetic structure within both countries (SI Appendix, Table S8), suggesting genetic drift has occurred on these islands since the introduction of European dogs. In contrast, Papua New Guinea and Solomon Island dogs retain a mix of European and indigenous ancestry (68–78% and 78–86% European ancestry, respectively; SI Appendix, Table S6), whereas Borneo dogs have no detectable European ancestry and are only moderately differentiated from Vietnamese dogs ().
Fig. 4.
Maximum likelihood tree of village dogs and select wolves with 10 migration edges. Edge color shows the proportion of source population ancestry found in the sink population. Populations are colored by region (outgroups are brown).
African dogs have indigenous ancestry components and European admixture. African ancestry components reflect geography, with Basenjis containing two components reflecting Benin and Democratic Republic of Congo (DRC) imports, whereas European admixture is strongest in Katanga and Kinshasa (86–88% and 74–75%, respectively). African dogs exhibit high Y and Mt haplotype diversity (Table 1), and haplotypes are geographically structured, including a high frequency Y haplogroup found only in Africa and Mongolia, and a cluster of closely related uniquely African Mt haplotypes (Fig. 3), demonstrating significant diversification of dogs within Africa (SI Appendix, Table S9).
In Eurasia, regional ancestry components predominate, with appreciable European ancestry occurring mainly in regions proximate to Europe (e.g., Turkey and Lebanon), consistent with gene flow between neighboring populations (SI Appendix, Fig. S17 and Table S10). Fine-scale population structure is also evident; for example, in Namibia one sample came from north of the Red Line veterinary cordon and clearly clusters with European rather than Namibian dogs, consistent with a previous study (4). In Lebanon and Egypt, dogs near Beruit and Cairo have more European ancestry than dogs from elsewhere. Similar patterns in Papua New Guinea (Port Moresby), Nepal (Kathmandu), and India (Mumbai) suggest that foreign dogs are more likely to be brought to (or survive in) urban areas than in more remote regions. Although some regions (e.g., Peru) were extensively sampled in remote areas, in other regions, sampling was largely limited to urban areas (e.g., Java and Turkey) or certain isolated populations (e.g., Roatán Island, Honduras), so caution is warranted in interpreting results from these areas.
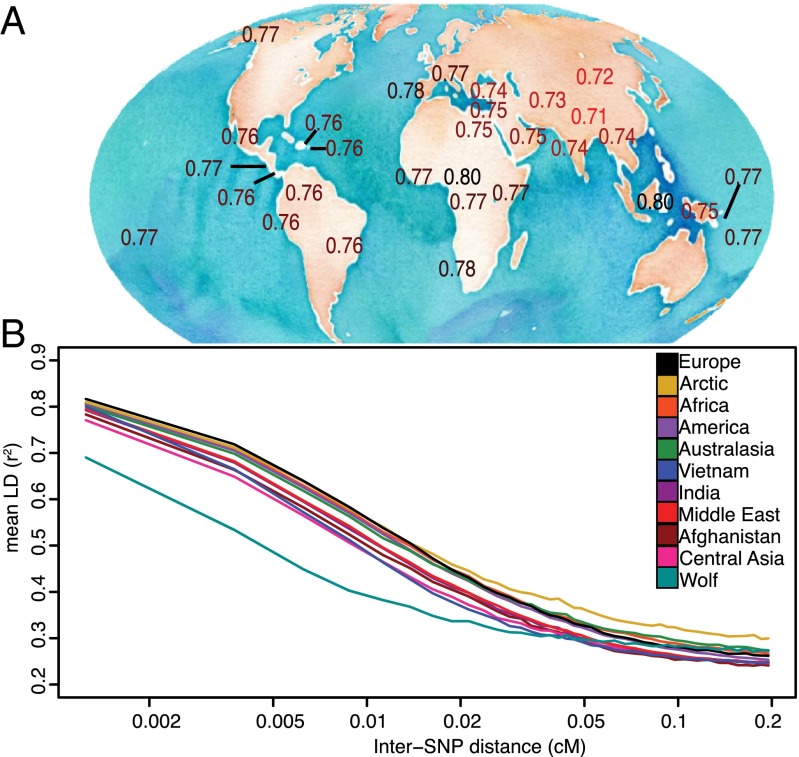
Global Patterns of LD Indicate a Central Asian Origin for Dogs.
The average rate of decay of LD between nearby autosomal markers is strongly influenced by population history. Four specific patterns are (i) populations with large show faster decay rates (30), (ii) populations with recent admixture show greater LD than unadmixed populations, particularly at large inter-SNP distances (31), (iii) bottlenecked populations show elevated LD (32), and (iv) expanding populations have a greater slope of LD decay versus stable or contracting populations as long-range LD is mainly determined by recent recombination events but short-range LD is affected by more ancient events (33).
As expected, village dog populations show elevated short-range LD compared with gray wolves, reflecting a bottleneck during domestication (Fig. 5 and SI Appendix). The rate of LD decay is greater in dogs than wolves, reflecting larger postdomestication for dogs vs. wolves and leading to similarly low LD in dogs and wolves at large inter-SNP distances ( 0.05 cM). Across village dog populations, LD is lowest in Asia. Specifically, LD is lowest in Afghanistan and Central Asia at short inter-SNP distances ( 0.0005 cM) and lowest in Vietnam at intermediate distances (0.01–0.05 cM), with rates increasing in other populations depending on their isolation and distance from Asia. These patterns of LD decay strongly suggest a Central Asian origin for domestic dogs with a subsequent population expansion (larger contemporary ) in East Asia and elsewhere. These patterns are consistent if physical, rather than genetic, inter-SNP distance is measured, or if different subsets of dogs are used for each population (SI Appendix, Fig. S18).
Fig. 5.
LD decay for village dog populations worldwide. (A) Mean LD for village dog populations at short inter-SNP distances (<0.005 cM). (B) LD decay curves for select regions calculated from N = 6 dogs per region (100 replicates).
Discussion
This study represents the largest-ever survey of worldwide canine genetic diversity using nuclear, Y, and Mt markers. We confirm high diversity and low LD in village compared with purebred dogs (4, 12, 19) and show how village dog populations improve inference of dog evolutionary history. This increased geographic and genetic resolution reveals the effects of bottlenecks and admixture in extant populations, as well as evidence for an origin of dogs in Central Asia.
Like previous studies, we find high levels of Mt and Y haplotype diversity in East Asia (8–10, 29, 34), but we also find high levels of Mt and Y diversity in India and Southwest Asia, respectively (Table 1). Whereas previous studies have used the high levels of uni-parentally inherited haplotype diversity as evidence for an East Asian, specifically Southern Chinese, origin for dogs, genome-wide LD patterns among populations suggests a different process. Namely, domestication occurred in Central Asia where early dogs carrying nearly the full complement of Mt and Y haplotypes spread to nearby Asian regions, including Afghanistan, India, and Vietnam. The substantial in these regions, particularly East Asia, allowed these haplogroups to survive and diversify to a greater extent than in Central Asia. Higher in East vs. Central Asia is supported both by census estimates (35) and by the more negative slope of the LD decay curve in East vs. Central Asia (Fig. 5), because recent population history has a greater impact on long-range vs. short-range LD (30, 33).
Gray wolves were clearly present in Central Asia during the Mesolithic, and both wolves and human hunter-gatherers were exploiting large mammals during this time (36). Increasing human population density, blade and hunting technology, and/or climate change during the Late Paleolithic in Central Asia (28) may have altered prey densities and made scavenging crucial to the survival of some wolf populations. Adaptations to scavenging such as tameness, small body size, and a decreased age of reproduction would reduce hunting efficiency further, eventually leading to obligate scavenging (37). Whether these earliest dogs were simply human-commensal scavengers or they played some role as companions or hunters that hastened their spread is uncertain, but clearly adaptation to conditions outside this initial domestication origin [e.g., efficient starch digestion (38) and aseasonal breeding (39, 40)] has also been important in dog evolution.
Although SNP array data are poorly suited for estimating the timing of ancient population events, it does shed light on the conflicting estimates of dog origins in previous genetic studies. Because there is incomplete lineage sorting between dogs and wolves, estimates based on Mt or Y haplotype diversity are sensitive to assumptions regarding the number of founder haplotypes in early dogs (8, 41). Nuclear datasets offer better resolution for parameterizing demographic models, but two such studies have yielded widely varying results [14 vs. 32 kya (14, 42)]. Our LD data support a relatively strong domestication bottleneck in dogs followed by substantial population expansion, particularly in East Asia. An ancient origin with a weak domestication bottleneck and small current in Asian village dogs is also consistent with the allele frequency data in ref. 42, but a more recent, stronger domestication bottleneck, and large current could be consistent with both allele frequency data and LD decay rates, bringing the inferred timing of dog origins more in line with archeological estimates.
Central Asia has been considered a likely domestication origin for dogs by some archeologists (43), but it has been poorly represented in previous genetic studies of dog origins. The pattern of reduced short-range LD in populations near Central Asia is most parsimoniously explained by an origin of dogs somewhere in this region, but we cannot rule out the possibility that dogs were domesticated elsewhere and subsequently, either through migration or a separate domestication event, arrived and diversified in Central Asia. For example, European dog populations have undergone extensive turnover over the last 15,000 y (44), erasing the genomic signatures of early European population history. Although it is difficult to explain the clear gradient of short-range LD out of Central Asia if dogs were domesticated from a far-flung region, studies of extant dogs cannot exclude the possibility of earlier domestication events that subsequently died out, or were overwhelmed by more modern populations.
Further analysis of diverse dogs throughout Central Asia and surrounding regions is crucial for precisely resolving the origin and history of early dogs. Refining the timing of dog domestication could yield substantial insights into the process by which dogs became domesticated and the wolf and human population(s) involved. Ancient DNA analysis will surely contribute to our understanding of early dog populations, but where ancient specimens are unavailable, village dogs are often the best proxy we have to ancient populations. Many indigenous populations have already succumbed to swamping gene flow from foreign dogs, so further work characterizing remaining indigenous populations genetically, morphologically, and behaviorally, is vital for building an improved understanding of dog evolutionary history.
Methods
Sample Collection.
The majority of samples used in this study come from blood stored in the Cornell Veterinary Biobank collected in accordance with Cornell animal care protocols 2005-0151 and 2011-0061. These samples include 4,676 purebred dogs from 161 breeds, 167 mixed breed dogs, and 549 village dogs from 38 different countries (SI Appendix, Table S11). Blood was stored in EDTA, and DNA was extracted by salt precipitation.
Genotyping.
Samples were genotyped on a semicustom Illumina SNP array containing 173,662 SNPs from the CanineHD array (13) and 12,143 markers identified using whole genome sequencing (45). A total of 166,171 markers remained after filtering markers with % missing data, discordant genotypes between technical replicates, or extreme divergence from Hardy–Weinberg expectations (HWEs; observed heterozygosity vs. HWE ratio <0.25 or >1.0). Genotype and geographical data have been deposited in Dryad (datadryad.org, doi:10.5061/dryad.v9t5h).
PCA.
PCA of unrelated village dogs was run using the smartpca program distributed in the Eigenstrat v5.0.1 software package (46). Village dogs were used to define the PCA space, and Basenjis, Carolina Dogs, New Guinea Singing Dogs (NGSDs), and one dog each from other breeds were projected onto it.
Haplotype Analysis.
The array included 582 Mt markers, of which 367 were polymorphic and passed quality control filtering. Additionally, seven markers that introduced multiple cycles in the haplotype network suggesting genotyping error were removed. We added 431 additional dogs with published complete Mt sequences (8, 11, 23–27) based on their genotypes at the marker positions. Haplotypes were named according to published convention (8), with some published haplotypes mapping to multiple haplotypes in this study due to the markers we used outside the control region. These haplotypes were split and are indicated with a letter. Conversely, some of the sequenced haplotypes are identical across all 360 marker positions on the array and are included as a single combined haplotype (e.g., C1_2).
The array included 336 Y chromosome markers, of which 207 were polymorphic and passed quality control filtering. One of these was removed because it introduced several cycles in the haplotype network. Haplotypes were named to correspond with Ding et al. (10), subdividing haplotypes to account for our enriched marker set.
Haplotype networks were constructed in R v3.1.0 using the Ape and Pegas packages (47–49). The distance matrix was calculated based on the count of differences, and then a minimum spanning forest was calculated. Networks were visualized in R using the igraph package (50). We defined haplogroups as groups of haplotypes at least 2 SDs further apart than the average distance between haplotypes, as measured by number of differences at the array marker positions.
Regional haplotype diversity (H) was computed with regions defined by geography and PCA. To control for sample size differences, we subsampled () dogs 100 times within regions and counted the number of observed haplotypes.
LD Decay.
LD is a reflection of , with LD at proximate SNPs reflecting historic and LD at distant SNPs reflecting in more recent times (30, 33). To ensure estimates of LD were not biased by particular individuals or by the choice of sample size, we used the PLINK 1.0.7 (51) --genome command to remove related ( outliers) and the --het command to remove inbred () individuals. We then performed two parallel analyses, one retaining 6 individuals per population and one retaining 20, randomly selecting the individuals 100 times to compute means and SEs. LD was calculated using the command --maf 0.3 --r2 --ld-window 999 --ld-window-r2 0 --ld-window-kb 200, and averaging within bins based on inter-SNP distance was performed using a C script (12).
Admixture.
Ancestry of individual dogs was determined using ADMIXTURE software (52). For a global view of village dog ancestry, we included all unrelated individuals from NGSDs, Basenjis, Carolina dogs, and village dogs, and a single individual from select dog breeds. For each K, 10 replicates were run using a different random seed. The replicate with the lowest cross-validation score for each K is reported.
To estimate historical relationships between populations we used TreeMix (53). We built admixture trees for dog breeds with gray wolves as the root, and for village dogs with coyotes as the root. For the village dog tree, we combined our data with previously published wolf and coyote Affymetrix v2 data (13). Only SNPs genotyped on both arrays and passing quality control were included (36,358 total). Trees were calculated using a range of numbers of migration events (m); we report the trees where further migration edges do not appreciably improve the fit. With the same populations, we calculated pairwise values using a custom C script.
We formally tested for admixture (indigenous vs. European) for African and Pacific Island dogs by computing f3 statistics using the Admixtools package (54) for each population with Europe as one source population and Basenji, Vietnam, or Borneo as the other. Populations were the same as for the village dog TreeMix analysis, and wolves were the outgroup for testing the bounds of the admixture percentage.
For American dogs, a suitable unadmixed source population was not available, so we used a PCA-based approach (55) to identify the extent of European ancestry in individual breed and village dogs. Village dogs from Europe, Alaska, and Vietnam were used to define the PCA space, and then dogs from the Americas were projected onto it. The same approach was used to investigate indigenous versus European ancestry proportions from East Asian breeds using village dogs from Europe, Borneo, Vietnam, Mongolia, and Vietnam to define the principal components.
Supplementary Material
Acknowledgments
We thank the countless dog owners and enthusiasts who facilitated sample collecting, including Carol Beuchat, Laura Colín, Jon Curby, Gautum Das, Ricardo de Matos, Baird Fleming, George Hicks, Gary S. Johnson, Warren Johnson, Janice Koler-Matznick, Leonard Kuwale, Kris Kvam, Judith Liggio, Stephanie Little Wolf, Gaby Matshimba, Mark Neff, Casey Quimby, Sue Ann Sandusky, Myrna Shiboleth, Jo Thompson, Steve Wooten, Asociación de Amigos por los Animales de Sosúa-Judy’s Pet Lodge, Animal Care in Egypt, Animals Fiji, Beirut for the Ethical Treatment of Animals, Liberia Animal Welfare and Conservation Society, Mongolian Bankhar Dog Project, Gump South Pacific Research Station, and the Qatar Animal Welfare Society. We thank Joy Li (Cornell Veterinary Biobank) and the Cornell University Genomics Core Facility for technical help. We thank A. G. Clark, S. Gravel, S. M. Myles, and R. K. Wayne for critical input. Funding for this project came from National Science Foundation Grant 0516310, National Geographic Society Expedition Council Grants EC0492-11 and 1P-14, Zoetis Animal Health, Cornell University Center for Advanced Technology, Cornell University, and dozens of PetriDish donors, including Sandra Coliver, Buck Farmer, Richard Gardner, Kathryn Sikkink, and Elaine and Chris McLeod.
Footnotes
Conflict of interest statement: A.R.B. and R.H.B. are cofounders and officers of Embark Veterinary, Inc., a canine genetics testing company.
This article is a PNAS Direct Submission.
Data deposition: Genotype and geographical data have been deposited in Dryad, datadryad.org (doi:10.5061/dryad.v9t5h).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1516215112/-/DCSupplemental.
References
- 1.Parker HG, et al. Genetic structure of the purebred domestic dog. Science. 2004;304(5674):1160–1164. doi: 10.1126/science.1097406. [DOI] [PubMed] [Google Scholar]
- 2.Larson G, et al. Rethinking dog domestication by integrating genetics, archeology, and biogeography. Proc Natl Acad Sci USA. 2012;109(23):8878–8883. doi: 10.1073/pnas.1203005109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coppinger R, Coppinger L. Dogs: A Startling New Understanding of Canine Origin, Behavior & Evolution. Simon and Schuster; New York: 2001. [Google Scholar]
- 4.Boyko AR, et al. Complex population structure in African village dogs and its implications for inferring dog domestication history. Proc Natl Acad Sci USA. 2009;106(33):13903–13908. doi: 10.1073/pnas.0902129106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Club AK. The Complete Dog Book. 20th Ed Ballantine Books; New York: 2006. [Google Scholar]
- 6.Vonholdt BM, et al. Genome-wide SNP and haplotype analyses reveal a rich history underlying dog domestication. Nature. 2010;464(7290):898–902. doi: 10.1038/nature08837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Savolainen P. Zhang YP, Luo J, Lundeberg J, Leitner T (2002) Genetic evidence for an East Asian origin of domestic dogs. Science. 298(5598):1610–1613. doi: 10.1126/science.1073906. [DOI] [PubMed] [Google Scholar]
- 8.Pang JF, et al. mtDNA data indicate a single origin for dogs south of Yangtze River, less than 16,300 years ago, from numerous wolves. Mol Biol Evol. 2009;26(12):2849–2864. doi: 10.1093/molbev/msp195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown SK, et al. Phylogenetic distinctiveness of Middle Eastern and Southeast Asian village dog Y chromosomes illuminates dog origins. PLoS One. 2011;6(12):e28496. doi: 10.1371/journal.pone.0028496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ding ZL, et al. Origins of domestic dog in southern East Asia is supported by analysis of Y-chromosome DNA. Heredity (Edinb) 2012;108(5):507–514. doi: 10.1038/hdy.2011.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thalmann O, et al. Complete mitochondrial genomes of ancient canids suggest a European origin of domestic dogs. Science. 2013;342(6160):871–874. doi: 10.1126/science.1243650. [DOI] [PubMed] [Google Scholar]
- 12.Boyko AR, et al. A simple genetic architecture underlies morphological variation in dogs. PLoS Biol. 2010;8(8):e1000451. doi: 10.1371/journal.pbio.1000451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vaysse A, et al. LUPA Consortium Identification of genomic regions associated with phenotypic variation between dog breeds using selection mapping. PLoS Genet. 2011;7(10):e1002316. doi: 10.1371/journal.pgen.1002316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Freedman AH, et al. Genome sequencing highlights the dynamic early history of dogs. PLoS Genet. 2014;10(1):e1004016. doi: 10.1371/journal.pgen.1004016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reich DE, et al. Linkage disequilibrium in the human genome. Nature. 2001;411(6834):199–204. doi: 10.1038/35075590. [DOI] [PubMed] [Google Scholar]
- 16.Lindblad-Toh K, et al. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature. 2005;438(7069):803–819. doi: 10.1038/nature04338. [DOI] [PubMed] [Google Scholar]
- 17.Gray MM, et al. Linkage disequilibrium and demographic history of wild and domestic canids. Genetics. 2009;181(4):1493–1505. doi: 10.1534/genetics.108.098830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Axelsson E, Webster MT, Ratnakumar A, Ponting CP, Lindblad-Toh K. LUPA Consortium Death of PRDM9 coincides with stabilization of the recombination landscape in the dog genome. Genome Res. 2012;22(1):51–63. doi: 10.1101/gr.124123.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Irion DN, Schaffer AL, Grant S, Wilton AN, Pedersen NC. Genetic variation analysis of the Bali street dog using microsatellites. BMC Genet. 2005;6:6. doi: 10.1186/1471-2156-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pedersen N, Liu H, Theilen G, Sacks B. The effects of dog breed development on genetic diversity and the relative influences of performance and conformation breeding. J Anim Breed Genet. 2013;130(3):236–248. doi: 10.1111/jbg.12017. [DOI] [PubMed] [Google Scholar]
- 21.Boyko AR. The domestic dog: Man’s best friend in the genomic era. Genome Biol. 2011;12(2):216. doi: 10.1186/gb-2011-12-2-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Witt KE, et al. DNA analysis of ancient dogs of the Americas: Identifying possible founding haplotypes and reconstructing population histories. J Hum Evol. 2015;79:105–118. doi: 10.1016/j.jhevol.2014.10.012. [DOI] [PubMed] [Google Scholar]
- 23.Kim KS, Lee SE, Jeong HW, Ha JH. The complete nucleotide sequence of the domestic dog (Canis familiaris) mitochondrial genome. Mol Phylogenet Evol. 1998;10(2):210–220. doi: 10.1006/mpev.1998.0513. [DOI] [PubMed] [Google Scholar]
- 24.Björnerfeldt S, Webster MT, Vilà C. Relaxation of selective constraint on dog mitochondrial DNA following domestication. Genome Res. 2006;16(8):990–994. doi: 10.1101/gr.5117706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baranowska I, et al. Sensory ataxic neuropathy in golden retriever dogs is caused by a deletion in the mitochondrial tRNATyr gene. PLoS Genet. 2009;5(5):e1000499. doi: 10.1371/journal.pgen.1000499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Webb KM, Allard MW. Mitochondrial genome DNA analysis of the domestic dog: Identifying informative SNPs outside of the control region. J Forensic Sci. 2009;54(2):275–288. doi: 10.1111/j.1556-4029.2008.00952.x. [DOI] [PubMed] [Google Scholar]
- 27.Matsumura S, Inoshima Y, Ishiguro N. Reconstructing the colonization history of lost wolf lineages by the analysis of the mitochondrial genome. Mol Phylogenet Evol. 2014;80:105–112. doi: 10.1016/j.ympev.2014.08.004. [DOI] [PubMed] [Google Scholar]
- 28. Unesco (1992) History of Civilizations of Central Asia (UNESCO Publishing, Paris), Vol 1.
- 29.Sacks BN, et al. Y chromosome analysis of dingoes and southeast asian village dogs suggests a neolithic continental expansion from Southeast Asia followed by multiple Austronesian dispersals. Mol Biol Evol. 2013;30(5):1103–1118. doi: 10.1093/molbev/mst027. [DOI] [PubMed] [Google Scholar]
- 30.Hill WG. Estimation of effective population size from data on linkage disequilibrium. Genet Res. 1981;38(03):209–216. [Google Scholar]
- 31.Pritchard JK, Rosenberg NA. Use of unlinked genetic markers to detect population stratification in association studies. Am J Hum Genet. 1999;65(1):220–228. doi: 10.1086/302449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Slatkin M. Linkage disequilibrium--understanding the evolutionary past and mapping the medical future. Nat Rev Genet. 2008;9(6):477–485. doi: 10.1038/nrg2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hayes BJ, Visscher PM, McPartlan HC, Goddard ME. Novel multilocus measure of linkage disequilibrium to estimate past effective population size. Genome Res. 2003;13(4):635–643. doi: 10.1101/gr.387103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ardalan A, et al. Comprehensive study of mtDNA among Southwest Asian dogs contradicts independent domestication of wolf, but implies dog-wolf hybridization. Ecol Evol. 2011;1(3):373–385. doi: 10.1002/ece3.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dong W, et al. 2014. The dog-human-wildlife interface: Assessing the scope of the problem. Free-Ranging Dogs and Wildlife Conservation, ed Gompper ME (Oxford Univ Press, Oxford, UK), pp 9–45.
- 36.Dong W, et al. Late Pleistocene mammalian fauna from Wulanmulan Paleolithic Site, Nei Mongol, China. Quat Int. 2014;347(9):139–147. [Google Scholar]
- 37.Coppinger R, Feinstein M. How Dogs Work. Univ of Chicago Press; Chicago: 2015. [Google Scholar]
- 38.Axelsson E, et al. The genomic signature of dog domestication reveals adaptation to a starch-rich diet. Nature. 2013;495(7441):360–364. doi: 10.1038/nature11837. [DOI] [PubMed] [Google Scholar]
- 39.Driscoll CA, Macdonald DW, O’Brien SJ. From wild animals to domestic pets, an evolutionary view of domestication. Proc Natl Acad Sci USA. 2009;106(Suppl 1):9971–9978. doi: 10.1073/pnas.0901586106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lord K, Feinstein M, Smith B, Coppinger R. Variation in reproductive traits of members of the genus Canis with special attention to the domestic dog (Canis familiaris) Behav Processes. 2013;92:131–142. doi: 10.1016/j.beproc.2012.10.009. [DOI] [PubMed] [Google Scholar]
- 41.Vilà C, Seddon J, Ellegren H. Genes of domestic mammals augmented by backcrossing with wild ancestors. Trends Genet. 2005;21(4):214–218. doi: 10.1016/j.tig.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 42.Wang GD, et al. The genomics of selection in dogs and the parallel evolution between dogs and humans. Nat Commun. 2013;4:1860. doi: 10.1038/ncomms2814. [DOI] [PubMed] [Google Scholar]
- 43.Derr M. How the Dog Became the Dog: From Wolves to Our Best Friends. Overlook Books; New York: 2013. [Google Scholar]
- 44.Malmström H, et al. Barking up the wrong tree: Modern northern European dogs fail to explain their origin. BMC Evol Biol. 2008;8:71. doi: 10.1186/1471-2148-8-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Auton A, et al. Genetic recombination is targeted towards gene promoter regions in dogs. PLoS Genet. 2013;9(12):e1003984. doi: 10.1371/journal.pgen.1003984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Price AL, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38(8):904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 47.Paradis E, Claude J, Strimmer K. APE: Analyses of phylogenetics and evolution in r language. Bioinformatics. 2004;20(2):289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- 48.Paradis E. pegas: An R package for population genetics with an integrated-modular approach. Bioinformatics. 2010;26(3):419–420. doi: 10.1093/bioinformatics/btp696. [DOI] [PubMed] [Google Scholar]
- 49.R Core Team . R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna: 2014. [Google Scholar]
- 50.Csardi G, Nepusz T. 2006. The igraph software package for complex network research, InterJournal, Complex Systems 1695. Available at igraph.org.
- 51.Purcell S, et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19(9):1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pickrell JK, Pritchard JK. Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genet. 2012;8(11):e1002967. doi: 10.1371/journal.pgen.1002967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Patterson N, et al. Ancient admixture in human history. Genetics. 2012;192(3):1065–1093. doi: 10.1534/genetics.112.145037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bryc K, et al. Genome-wide patterns of population structure and admixture in West Africans and African Americans. Proc Natl Acad Sci USA. 2010;107(2):786–791. doi: 10.1073/pnas.0909559107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.