Abstract
Fibroblasts from patients with Type I bipolar disorder (BPD) and their unaffected siblings were obtained from an Old Order Amish pedigree with a high incidence of BPD and reprogrammed to induced pluripotent stem cells (iPSCs). Established iPSCs were subsequently differentiated into neuroprogenitors (NPs) and then to neurons. Transcriptomic microarray analysis was conducted on RNA samples from iPSCs, NPs and neurons matured in culture for either 2 weeks (termed early neurons, E) or 4 weeks (termed late neurons, L). Global RNA profiling indicated that BPD and control iPSCs differentiated into NPs and neurons at a similar rate, enabling studies of differentially expressed genes in neurons from controls and BPD cases. Significant disease-associated differences in gene expression were observed only in L neurons. Specifically, 328 genes were differentially expressed between BPD and control L neurons including GAD1, glutamate decarboxylase 1 (2.5 fold) and SCN4B, the voltage gated type IV sodium channel beta subunit (-14.6 fold). Quantitative RT-PCR confirmed the up-regulation of GAD1 in BPD compared to control L neurons. Gene Ontology, GeneGo and Ingenuity Pathway Analysis of differentially regulated genes in L neurons suggest that alterations in RNA biosynthesis and metabolism, protein trafficking as well as receptor signaling pathways may play an important role in the pathophysiology of BPD.
Introduction
Bipolar disorder (BPD) is a highly heritable mood disorder characterized in many patients by dramatic and unpredictable mood swings between depression and mania or hypomania. Nearly 50% of bipolar patients attempt suicide and half of them succeed [1]. Available therapeutic agents effectively treat symptoms in only a subset of patients, accounting for the high morbidity and mortality associated with BPD [2,3]. Recent research efforts have focused on identifying the underlying genes and genetic factors for BPD with the goal of using these molecular genetic insights to design therapeutics with greater efficacy [4,5].
Studies on genetic or population isolates offer a powerful approach to identifying genetic variants accounting for disease heritability due to their relatively reduced genetic, environmental and phenotypic variability. The Old Order Amish of Lancaster County represent a genetic isolate of European ancestry where a number of disease causing genetic mutations have been identified including several Mendelian disorders [6,7]. We have recently reported an in-depth genetic analysis of a large, multigenerational Amish pedigree with BPD [8]. Although the small sample size may have hindered the identification of risk variants or a common etiologic pathway, a set of credible candidate genes were identified for further examination in other Amish pedigrees or in large-scale population-based studies. In addition to genetic studies, gene expression studies on postmortem brain tissue from BPD patients and controls have been conducted as an orthogonal approach to elucidate mechanisms underlying BPD [9–15]. A major caveat of post mortem brain studies, however, is the difficulty in differentiating disease etiology-associated changes (e.g. gene expression) from those caused by post-mortem artifact, life-long illness or prior drug treatment. Recent advances in human induced pluripotent stem cell (iPSC) technology, on the other hand, have the potential to address some of the shortcomings of postmortem studies. For example, iPSC-derived neurons from neurodegenerative diseases with Mendelian inheritance such as familial Amyotrophic Lateral Sclerosis (ALS) and Parkinson’s disease recapitulate key pathological mechanisms associated with the disorders [16–18]. Interestingly, even in a complex psychiatric disorder such as schizophrenia, iPSC-derived neurons have been shown to display morphological and gene expression changes that may be relevant to underlying disease biology [19,20].
Recent studies have applied iPSC technology to the study of BPD. Chen et al [21] generated iPSCs from 3 unrelated patients with Type 1 BPD and saw differential gene regulation in neurons derived from these cell lines when compared to neurons from lines derived from unaffected, unrelated controls. Madison et al [22] generated iPSCs from 2 affected and 2 unaffected individuals in a pedigree enriched for BPD. Comparison of gene expression profiles between BPD and control NPs and BPD and control neurons identified differentially regulated genes. To gain further insight into BPD, we chose to generate iPSCs and their derivatives from individuals within a 5 generation Old Order Amish pedigree with high prevalence of BPD [8]. We reasoned that transcriptomic analysis of iPSC-derived neurons from affected and unaffected first-degree relatives might not only offer a better approach to gain insights into the molecular changes of BPD by reducing both genetic and non- genetic variability, but moreover, might leverage the existing genetic data from this pedigree. The iPSC lines from 4 affected and 4 unaffected relatives were differentiated into neuroprogenitors (NPs) and neurons in vitro. Using microarray analysis, we confirmed stages of differentiation of the iPSCs to neurons by characteristic gene expression patterns, and observed a subset of differentially expressed genes (DEGs) between BPD subjects and their healthy siblings only in L neurons. Hierarchical clustering and pathway analyses revealed differences in genes involved in RNA biosynthesis, protein trafficking and receptor signaling pathways.
Methods
Diagnostic Guidelines and Reliability
When the Amish Study, which includes the cases reported here, began in 1976 it established a project Psychiatric Board of five clinicians charged with diagnosis of patient case materials using strict research diagnostic criteria (both RDC and DSM III–IV) blind to existing medical “treatment opinions” of practitioners and unaware of family relatedness. Each BPD case was re-evaluated blindly at least twice over the decades resulting in consensus diagnoses. Follow-up of BPD patients and their unaffected relatives for genetic research, including those used for the present study, was carried out yearly and has spanned 2–3 decades [23]. A second diagnostic procedure tracked course-of-illness by both the Board and a project Seasonality Panel comprised of three psychiatrists, who coded episodes that extended beyond 20–30 years for the patients in this study. Medical records documented the extent to which lithium was an effective mood stabilizing medication for the BPD patients used to generate iPSC lines. Data for 87 individuals from four large Amish families, the largest being core Pedigree 110, location of the present research subjects, has been described [24]. The project Board and Panel were in agreement regarding medication compliance and the effectiveness of lithium. Several independent sources provided evidence about breakthrough episodes (due to poor compliance) and subsequent recovery with lithium as the mood stabilizing drug.
Patient Population
A multi-generational Old Order Amish pedigree with high prevalence of Type I BPD was chosen for the current study [7,25,26]. Fig 1 shows the pedigree and associated demographics. Fibroblast cell lines derived from four BPD patients (ABP03, 05, 07 and 11) and four unaffected siblings (ABP04, 06, 08 and 12), were obtained from the NIGMS Human Genetic Cell Repository at Coriell Institute for Medical Research (all samples in this biorepository have fulfilled the required informed consent guidelines detailed at https://catalog.coriell.org/1/NIGMS/About/Submitting-Samples). The 4 BPD patients all received high rating for classic presentation of the disease as determined by special coding of the project Board. Their ages at onset of BPD (Fig 1) are reflective of the range for onset age in the total BPD Amish sample. The unaffected control samples had a similar age range. All controls were well beyond the age of risk for BPD in these families.
Fig 1. Selection of fibroblasts from the NIGMH repository for generation of iPSCs.
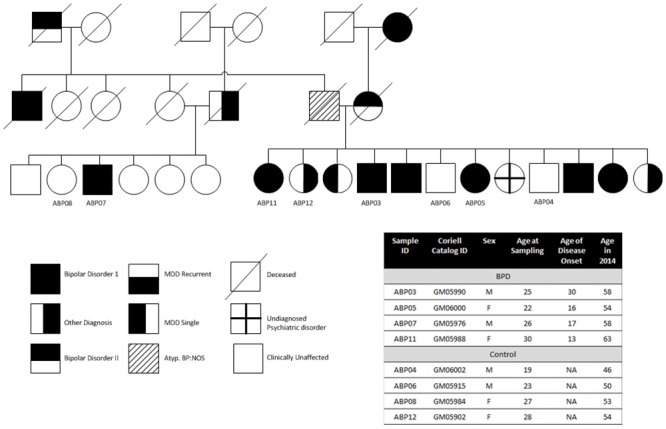
Fibroblasts from patients diagnosed with Type I BPD and their sibling controls of Old Order Amish pedigree were obtained from the NIGMS Human Genetic Cell Repository at Coriell Institute for Medical Research for reprogramming into iPSCs using the Sendai virus method. The pedigree has been updated according to DSM-IV. Circles and squares represent females and males, respectively. MDD = Major Depressive Disorder; Atyp. BP:NOS = Atypical Bipolar (not otherwise specified). Other diagnosis for the patient from which ABP12 was derived was post-partum depression.
Generation of human iPSCs
Fibroblasts (passages 3–8) were reprogrammed using CytoTune®-iPS kit (Life Technologies) following methods described previously [27]. Three iPSC lines with characteristic morphology were generated for each patient fibroblast cell line. Cytogenetic analysis (Cell Line Genetics) on 20 G-banded metaphase cells was performed from each cell line and all 20 cells demonstrated normal karyotype. No abnormal cells were detected. We randomly chose one clone from each patient for subsequent studies.
Selected iPSCs were expanded and cultured on irradiated mouse embryonic fibroblasts (MEF) (GlobalStem) in ES media, which contains DMEM/F12 (1:1) (HyClone) with 20% Knockout serum replacement (Invitrogen), 1X penicillin/Streptomycin (10U/ml/10μg/ml, Hyclone), 1X GlutaMax (Invitrogen), 1X Non-essential amino acids (Hyclone), 55 μM 2-mercaptoethanol (Gibco) and 10ng/ml human bFGF (R&D Systems) with daily media replacement.
Differentiation of iPSC into neurons
Before initial differentiation, iPSCs were transferred to plates coated with Matrigel™ (BD Bioscience) following the manufacturer’s instruction and maintained in mTeSR1 media (StemCell Technologies) until MEF feeder cells were eliminated. Neural Induction Media (NIM) was prepared to contain DMEM/F12 (1:1) with 1X N2 supplements (Invitrogen), 1X penicillin/Streptomycin (10U/ml/10μg/ml, Hyclone), 1X GlutaMax (Invitrogen), 1X Non-essential amino acids (Hyclone). Neural Differentiation Media (NDM) was prepared by adding 1X N2 supplements, 1X B27 without vitamin A (Invitrogen), 1X penicillin/Streptomycin, 1X GlutaMax, 1X Non-essential amino acids (Hyclone) to Neurobasal media (Life Technologies).
Human iPSCs were differentiated using published protocols [28,29] with few modifications to increase differentiation efficiency as illustrated in S1 Fig. Briefly, hiPSCs at between 10 and 25 passages were rinsed with 1X Ca2+/Mg2+ free Dulbecco’s Phosphate-Buffered Saline (HyClone; DPBS) and then treated with Accutase (MP Biomedicals) for 5 ~ 7 minutes at 37°C to make a single cell suspension. Live cells were counted using Vi-Cell (Beckman-Coulter). At day 0 of differentiation, three million cells were added to each well of an AggreWell800 plate (Stem Cell Technologies, Inc.) following manufacturer’s protocol in NIM with 250 ng/ml noggin (R&D Systems) and 10 μM SB431542 (R&D Systems). 50 ~ 75% of the media was replaced daily for 5 days with fresh NIM supplemented with noggin and SB431532. On the fifth day of differentiation, cell clusters were collected from a well of the AggreWell800 plate and re-plated in 6-well plates coated with Matrigel™ in NIM supplemented with noggin and SB431532. Media was replaced every other day for an additional four to five days until neural rosettes composed of NPs were detected in most areas of the wells.
NPs were dissociated from the neural rosettes with Accutase and expanded by culturing on Matrigel-coated plates at >0.1 x 106 cells/cm2 or seeded at 0.33 x 106 cells/ml in Ultra-Low Attachment plates (Corning) for suspension culture in NDM supplemented with 10 ng/ml hLIF (Millipore) and 20 ng/ml bFGF (R&D Systems). After two passages, NPs were dissociated as single-cell suspensions and cryopreserved for future use in 50% NDM, 40% Knockout serum replacement and 10% dimethyl sulfoxide (Sigma).
To further differentiate NPs into neurons, NPs were cultured on plates coated with 0.1% poly-l-ornithine (Sigma) and 10ng/ml of laminin (Invitrogen) in NDM without hLIF and bFGF at 25,000 ~ 50,000 cells/cm2. Media was replaced every 2 ~ 3 days. Differentiation was continued for up to 4 weeks in culture. Cells were harvested and RNA samples were obtained after two weeks to represent an early time point of neuronal differentiation (E) and four weeks to represent a later time point (L) of neuronal differentiation.
Immunocytochemistry
Immunocytological analysis of iPSCs and embryoid bodies was as described previously [30]. Neural rosettes, NPs and neurons were fixed with 4% paraformaldehyde and stained with Pax6 (Covance), Nestin (Covance), MAP2 (AbCam) and β-tubulin III (TUJ1 clone; Covance) antibodies using standard methods.
RNA isolation for gene expression studies
Undifferentiated iPSCs, iPSC-derived NPs or neurons at E and L stage of differentiation were rinsed with 1X DPBS followed by RNA extraction using RNeasy Mini kit (Qiagen) using the manufacturer’s protocol. RNA was quantitated using a Nanodrop Spectrometer ND-1000 (Thermo Scientific).
Reverse transcription and quantitative reverse transcription PCR
Reverse transcription PCR (RT-PCR) analyses for iPSC and embryoid body (EB) characterization were as described previously [30,31], using the same primers, except for PAX6, FLK1 and GATA2 which are listed in Part A of S1 Table. For all quantitative reverse transcription PCR (qRT-PCR) analyses, complementary DNA (cDNA) synthesis was performed using High Capacity cDNA reverse transcriptase kit (Life Technologies), according to the manufacturer’s instructions. Quantitative PCR analysis of reverse transcribed cDNA was conducted in triplicate using the Applied Biosystems ABI 7900 HT system with TaqMan® Assays and TaqMan® Gene Expression Master Mix (Life Technologies, Carlsbad, CA) with primer probes as listed in Part B of S1 Table.
Microarray analysis
Samples for messenger RNA (mRNA) profiling studies were processed by Asuragen, Inc. (Austin, TX, USA) using GeneGhip® Human Genome U133 (Affymetrix, Santa Clara, CA) according to the company's standard operating procedures as described previously in detail [32].
Statistical and bioinformatics data analysis
A summary of the image signal data, detection calls and gene annotations for every gene interrogated on the arrays was generated using the Affymetrix Statistical Algorithm MAS 5.0 (GCOS v1.3) (scaling factor = 1500) and quantile normalization across samples was applied. Log2 transformation and mean-centered standardization was further performed. For genes with multiple probes on the chip, signal intensities were averaged. Each probe ID was converted and collapsed into gene symbols (http://www.genenames.org/). The final 15391 genes that were reliably detected on at least 80% of the arrays with a signal intensity of 64 or greater were used for further analysis.
For Principal Component analysis (PCA), the replicates per group and time points were aggregated by calculating their means and the first two principal components (PCs) were used to visualize the global similarity of the data. Analysis of variance (ANOVA) was applied to determine differentially expressed genes (DEGs) in pair-wise comparisons of the groups (e.g., BPD vs. control for NP, E and L, respectively) and combined for all groups. The ANOVA assessed the main effects of disease status (BPD vs. control) and differentiation stages (NP, E, L) as well as interactions between the two variables. ANOVA was conducted for p-values for each of the three effects (disease status, differentiation stage, and interaction between two variables) along with adjusted p-values for multiple testing using Benjamini-Hochberg-false discovery rate (FDR) correction. Differentially Expressed Genes (DEG) were defined as the probes which had FDR < = 0.2, mean signals > = 250 in at least one group (control or BPD) and absolute fold change > = 1.5 in all chips.
Hierarchical Clustering
The gene sets constituting pathways relevant to neuronal development and differentiation were retrieved from Molecular Signatures Database (MsigDB 3.0, C2 & C5) [33]. Each pathway-specific data set was analyzed by hierarchical average-linkage clustering. The clustering was performed using Gene Cluster 3.0 (http://bonsai.ims.utokyo.ac.jp/~mdehoon/software/cluster/) or using R programs.
The resulting numerical output was used in R package (“gplots”) to generate the associated heat maps and clustering dendrograms.
Enrichment analysis
Enrichment analyses of Gene Ontology (GO), GeneGo, KEGG and Ingenuity Pathway Analysis (IPA) pathways were performed for DEGs using the hypergeometric test and Fisher’s Exact Test. To account for multiple hypothesis testing, FDR correction (FDR = 5%), was applied according to Benjamini-Hochberg.
Results
Characterization of iPSCs
Cytogenetic analysis confirmed that all 8 iPSC lines were karotypically normal. Gene expression of pluripotency markers and inactivation of Sendai-virus particles were confirmed in all iPSC lines by qRT-PCR as described by Park et al. [30] and Chan et al [31] (Panel A of S2 Fig). Protein expression of pluripotency markers was demonstrated by immunocytochemistry (Panel B of S2 Fig). To confirm the capacity of iPS cells to spontaneously differentiate into three germ layers, EBs were generated and maintained in suspension for spontaneous differentiation. Expression of genes specific for three germ layers was demonstrated by RT-PCR (Panel C of S2 Fig).
Differentiation of iPSCs to NPs and neurons
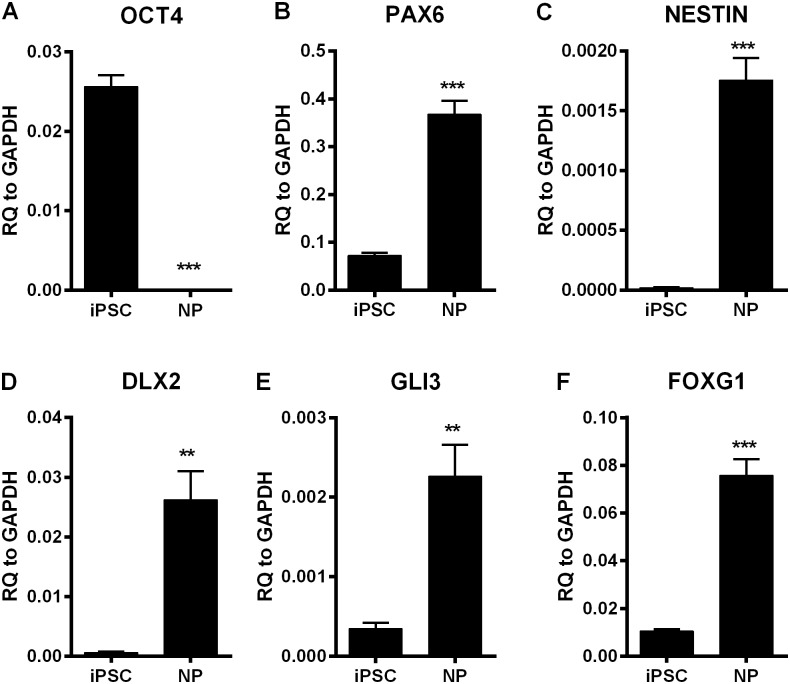
Four BPD patient-derived iPSC clones (ABP03, 05, 07 and 11) and four unaffected sibling-derived (control) iPSC clones (ABP04, 06, 08 and 12), were differentiated into neuronal progenitors (NPs) and neurons as described in Materials and Methods section. S1 Fig shows the key characteristics of the cultures along the stages of neuronal differentiation. L neuronal cultures exhibited characteristic neuronal morphology including axons, dendrites and neurites in addition to positive immunostaining for MAP2 and β-tubulin III (S1 Fig). For microarray studies and further characterization, RNA samples from the four BPD and four controls were collected from NP, E neurons (2 weeks of differentiation) and L neurons (4 weeks of differentiation) as well as undifferentiated iPSCs. Expression of markers of pluripotency and neuronal differentiation were examined using qRT-PCR. The differentiation of iPSCs into NPs resulted in a dramatic decrease in expression of the pluripotency marker, OCT4 (Fig 2A). At the same time, PAX6 and NESTIN expression, early markers of neuroectoderm, increased indicating desired lineage specification (Fig 2B and 2C)[34]. The generation of NPs was further confirmed by increased expression of the dorsoventral neuronal specification markers DLX2, GLI3 and FOXG1 (Fig 2D–2F) [35,36]. Gene expression for endodermal (AFP and SOX17) [37] and mesodermal (T/Brachyury) markers [38] decreased during differentiation (data not shown), consistent with differentiation towards the neuroectodermal lineage.
Fig 2. Quantitative RT-PCR of RNA samples from iPSCs and NPs.
RNA samples were collected from iPSC and NPs and gene expression of markers of developmental stages were evaluated. Changes in pluripotency and neuroprogenitor markers were measured (A-F). Each bar represents mean ± standard error, n = 8. Statistical analysis was performed using unpaired student t-test. *** p < 0.0001, ** p < 0.001.
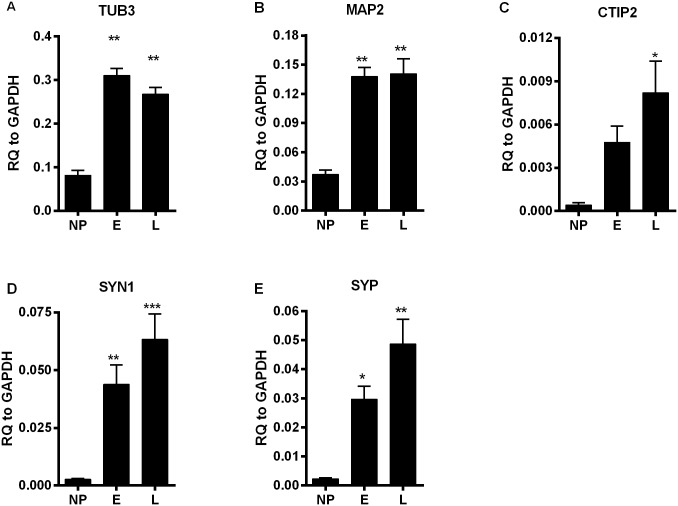
Neuronal differentiation was initiated by growth factor withdrawal from NPs. Successful neuronal commitment was demonstrated in E neurons by an increase in the expression of pan-neuronal markers, tubulin β-III (TUB3) and microtubule associated protein 2 (MAP2) [39,40] (Fig 3A and 3B). Expression of CTIP2, a marker for deep cortical neurons [41], was significantly increased by four weeks of neuronal differentiation (L) (Fig 3C). Expression levels of the synaptic markers, synapsin 1 (SYN1) and synaptophysin (SYP), was positively correlated with the time of neuronal differentiation. (Fig 3D and 3E). Gene expression of several neuronal ion channel proteins also tended to increase as NPs matured into neurons (S3 Fig), although some of the markers did not reach statistical significance. Collectively, the gene expression analyses indicate that E neurons already exhibit key neuronal identifiersbut markers for neuronal function continue to increase their expression level with further differentiation to L neurons.
Fig 3. Quantitative RT-PCR of RNA samples from NPs and neurons.
RNA samples were collected from NPs, E and L neurons and markers of neuronal gene expression were evaluated. Pan-neuronal markers, (A,B), cortical neuronal marker (C), and synaptic markers (D,E) were evaluated. Each bar represents mean ± standard error (n = 8, combining 4 BPD and 4 controls). One-way ANOVA was performed and Bonferroni’s Multiple Comparison post-hoc test was done. * p < 0.01, ** p < 0.001, *** p < 0.0001 compared to NP.
Differentially Regulated Genes (DEGs) in Neuronal Differentiation
RNA samples isolated from NP, E and L neurons of the four BPD and four control donors were subjected to Affymetrix microarrays. RNA samples from four undifferentiated iPSCs (ABP 05, ABP06, ABP11 and ABP12) were also included.
To analyze global gene expression changes associated with different stage of differentiation, Principal Component Analysis (PCA) was performed on the microarray data. As shown in Fig 4A, PCA identified three distinct clusters corresponding to iPSCs, NPs and neurons at either E or L stage with no differences between BPD and controls at any stage of cellular differentiation.
Fig 4. Microarray data analysis.
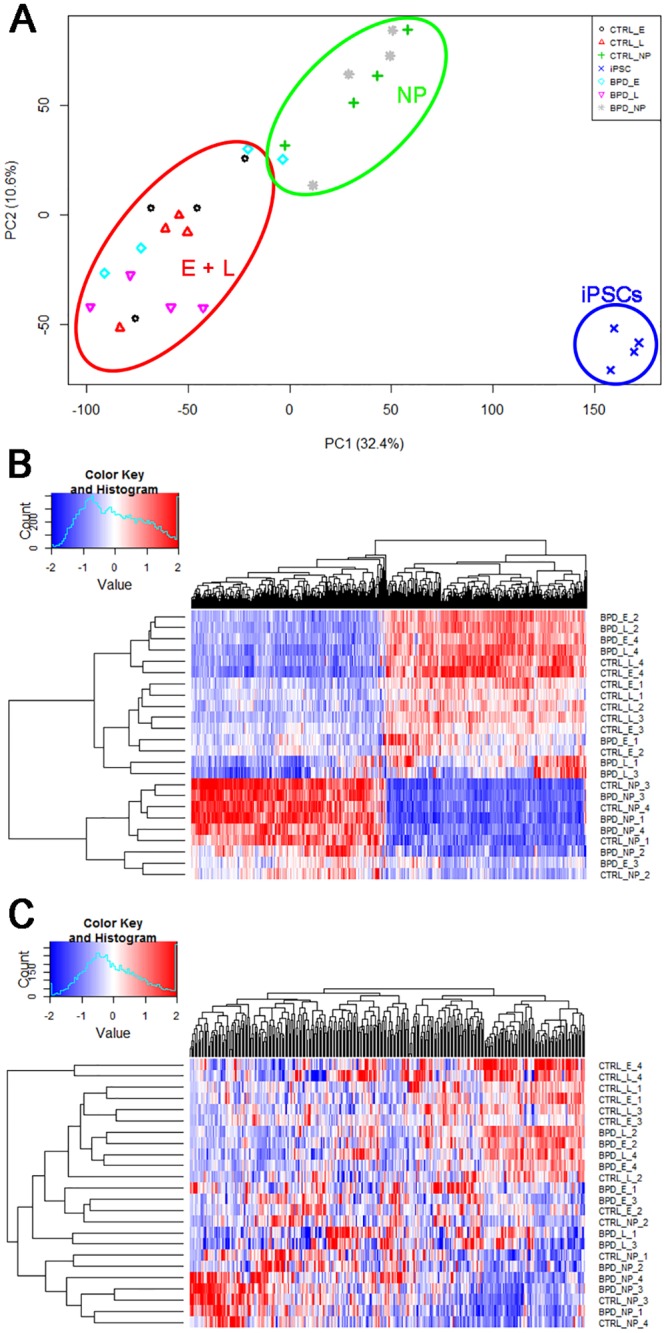
A. Principal Component Analysis (PCA) plot shows three distinct clusters: iPSCs, NPs and E plus L neurons as outlined with blue, green and red lines, respectively. The symbols represent undifferentiated iPSCs (blue x), control NP (green cross; +), control E (black circle; ○), control L (red triangle; Δ) and BPD NP (grey asterisk; *), BPD E (turquoise diamond; ♢) and BPD L (pink inverted triangle; ▽). B. Clustered heat map of differentially expressed genes in both control and BPD neurons demonstrates that the genes are clustered closely for E and L neurons regardless of disease status and separate from NPs and iPSCs. Horizontal axis shows genes used for clustering in the S2 Table and vertical axis shows sample clustering. C. Changes in expression of genes associated with axonal guidance (Ingenuity pathway analysis) shown as a clustered heat map show that the expression of genes associated with axonal guidance was higher in E and L neurons than in NPs. Horizontal axis shows genes used for clustering as listed in S3 Table.
Subsequently, DEGs between NP and E neurons, as well as NP and L neurons, were identified and processed for hierarchical clustering of gene expression, which illustrated close assembly of E and L neuron expression profiles separating from those of iPSCs and NPs (Fig 4B). BPD_E_3 was an apparent outlier. Without technical replicates it is not possible to determine if this variation comes from culture effects, clonal variation or inherent differences between patients. There was no clear segregation of BPD and control cells. This was consistent with the lack of morphological differences between BPD and control cells.
To annotate functions to genes regulated during differentiation, enrichment analysis of DEGs was performed. The GO analysis of DEGs demonstrated that the progressive differentiation from NP to E and L neurons was associated primarily with genes involved in cell cycle regulation and neurogenesis in both BPD and control populations (S4 and S5 Tables). There was a robust alteration in the expression of genes associated with neuronal differentiation and specification as exemplified by the axonal guidance signaling pathway (defined by Ingenuity Pathway Analysis) upon differentiation from iPSC to NP to E and L neurons (Fig 4C). Importantly, the patterns of gene expression were similar in RNAs isolated from BPD and control donors. Collectively, these data demonstrate that our differentiation paradigm was robust and BPD iPSCs differentiated similarly to the control iPSCs.
Differentially Regulated Genes (DEGs) between BPD and control cells
Having confirmed similar pattern and time course of differentiation between BPD and control iPSCs, analysis of DEGs between BPD and controls at each time point was performed. Interestingly, there were no significant differences between BPD and controls at either NP or E stages of differentiation. However, L neurons from BPD cases compared to controls showed differential expressions for 697 probes, which corresponded to 324 genes. Out of the 324 genes, 292 genes showed upregulation in BPDs compared to their sibling controls while only 32 genes were downregulated. The complete list of DEGs between BPD and control L neurons is provided in Table 1.
Table 1. List of differentially expressed genes (DEGs) in control and BPD L neurons.
| Upregulated in BPD neurons | ||||||
| PTPN3 | ZNF669 | APOPT1 | SMCHD1 | DHX57 | COL4A3BP | BDP1 |
| CLTC | SNRPN | SPTAN1 | ATXN3 | ZCCHC6 | ZNF274 | NFYC |
| GSK3B | CSNK1G1 | FUBP3 | UBE2W | KMT2C | MINK1 | KLHL24 |
| C9orf64 | ZNF800 | OCIAD1 | CECR1 | LINC00537 | TRIM66 | AGAP4 |
| TMEM87A | NFX1 | G6PC2 | CPEB4 | MALAT1 | RBM39 | PLD6 |
| KMT2A | CCZ1B | DICER1-AS1 | DBT | CROCCP2 | REEP2 | MGEA5 |
| HCG18 | PDCD6 | SLC22A3 | PAPD4 | PWRN2 | SCFD1 | KIF13A |
| MGC57346 | LETMD1 | BRWD1 | FNBP4 | FCRL2 | GREB1L | LMBRD2 |
| HNRNPD | CRB3 | UBE2H | KANSL1 | ZNF654 | GPATCH2L | SLC23A2 |
| CMBL | RIOK3 | PDIA2 | SLC25A27 | ZXDC | PKN2 | ZBTB11 |
| CLASP2 | CSNK1A1 | FNBP1 | NFATC3 | JAK2 | PIK3C2A | SYMPK |
| DYNC1H1 | SBNO1 | SFTPB | DIP2A | DIO2-AS1 | MTX3 | VPS53 |
| C10orf12 | LOC102546299 | L3MBTL1 | FKSG49 | ZMYM5 | POLR1B | TTC5 |
| MINOS1P1 | HIST1H2BD | NPAS3 | PHF20L1 | NLN | STAG2 | CEP95 |
| ZNF335 | TMEM161B | UBE2D3 | DUSP16 | C9orf84 | RAB21 | CAPN1 |
| TRAF4 | TRAF3IP2-AS1 | HAUS2 | NDFIP2 | TCP11L2 | NKTR | N4BP2L2 |
| PPP2R5C | WDR11 | CCAR1 | CWF19L2 | WDR41 | CYLD | COX17 |
| CTTN | NFASC | TTBK2 | FGFR1 | ZNF160 | MDM2 | MRPS5 |
| SCD5 | FGFR1OP2 | LOC151121 | SLC35E1 | KPNA5 | ANKRD10-IT1 | OSBP |
| CCDC90B | GTF2A2 | TXNL1 | ITGB8 | SPG7 | MYLK3 | ZBTB7A |
| LRCH3 | TRMT13 | FXR1 | PPM1A | TMEM33 | SIK3 | CKMT2-AS1 |
| TRPS1 | ACVR1B | PHF3 | MEG3 | SLC6A15 | ZNF493 | CNTFR-AS1 |
| KLF12 | CBFA2T2 | VPS13C | ABI2 | KIDINS220 | AZI2 | C4orf29 |
| DIP2B | RBM5 | RBM33 | SLC25A36 | AHI1 | ZBTB38 | MACF1 |
| USP34 | LOC100272216 | SUPT20H | TTC37 | ERP44 | ATP6V1H | ZCCHC11 |
| RFX7 | AFF4 | RASAL2 | LRRIQ1 | PHKB | ZFYVE16 | TGDS |
| RAB18 | KIF21A | ZC3H7B | CLEC4F | ANKRD12 | LOC728093 | CAMSAP2 |
| SNRPA1 | SCAF11 | GALNT2 | CYHR1 | FAM169A | NCAPH2 | RBBP6 |
| IFNL1 | ZNF791 | MGC12488 | PDXDC1 | ZNF518A | ELOVL5 | CLOCK |
| STRAP | LINC01007 | NFAT5 | STK32A | PAAF1 | PNISR | KIAA1522 |
| TMEM91 | CNOT4 | GRM5 | CDK13 | SGSM1 | PGF | PCMTD1 |
| ICA1 | DLGAP4 | PTEN | TMF1 | CRYZL1 | FOXK1 | LOC101926996 |
| MYO6 | VPS8 | LPXN | TTC17 | GNG2 | ANKRD13A | PGAP1 |
| PRKAA1 | URGCP | AGFG1 | MZT2B | HELB | AUTS2 | ATP9A |
| KLHDC10 | SEPT2 | ALMS1 | NDST4 | CAPN3 | C11orf80 | GOPC |
| SBF2 | GOSR1 | MORN4 | SART1 | CEP290 | GAD1 | RFPL3S |
| EEA1 | RBM14 | ARHGAP5 | FAM132A | BOD1L1 | ATF7IP | LOC100507311 |
| FCF1 | ZNF577 | GOLGA4 | LOC283045 | KIAA0754 | PIP5K1A | CLINT1 |
| RAB12 | FAM161B | MTMR9 | EPG5 | ZCCHC7 | PPP1R3E | NBR1 |
| FAM208B | UACA | NAALAD2 | PPWD1 | RHOQ | TTLL3 | MED6 |
| CCNT1 | SERPINB6 | FLJ41455 | STK38 | CRIPAK | MON2 | CSAD |
| TMEM67 | PPA2 | ZNF207 | MKLN1 | WDR61 | ||
| Downregulated in BPD neurons | ||||||
| PRCD | C3orf70 | CDKN2C | MVK | RAB22A | TTL | TRAM2-AS1 |
| VPS13B | SLC6A20 | MKI67 | NEK1 | LYRM7 | GPR125 | MYO1D |
| ZADH2 | NOL12 | ALMS1 | TMEM44 | LOC728705 | ACADM | KCTD15 |
| MCOLN1 | PBK | PMEPA1 | GAB1 | SCN4B | ELOVL2 | ASPH |
| C6orf203 | FAM105A | PARD3 | RPTOR | |||
Total DEGs in L neurons between BPD and control (combined up and down regulated DEGs) were annotated by multiple pathway analysis tools. GO analysis performed on all DEGs suggested that the group differences reflect enrichment of genes involved in metabolic processes, including macromolecule and cellular RNA metabolism and biosynthesis pathways. Also identified were pathways involving protein transport including post-Golgi vesicle mediated transport and Golgi to plasma membrane protein transport (Table 2). Many of the pathways found for the total DEGs were seen when up and down regulated DEGs were analyzed separately eg cilium assembly, metabolic processes (Table 2). Although the number of down-regulated genes was small (32 genes), GO analysis revealed cell cycle pathways (Table 2).
Table 2. TOP 10 list of GO pathway analysis of DEGs between BPD and control in L neurons.
| TOTAL | ||||||
| Ranking | GOBPID | Term | Size of GO Term | Matched | P value | FDR |
| 1 | GO:0006892 | post-Golgi vesicle-mediated transport | 75 | 8 | 4.89E-05 | 0.001623 |
| 2 | GO:0042384 | cilium assembly | 78 | 8 | 6.49E-05 | 0.001623 |
| 3 | GO:0044260 | cellular macromolecule metabolic process | 6283 | 139 | 0.000115 | 0.001911 |
| 4 | GO:0006355 | regulation of transcription, DNA-dependent | 2724 | 71 | 0.0002 | 0.002024 |
| 5 | GO:0008152 | metabolic process | 8966 | 183 | 0.000202 | 0.002024 |
| 6 | GO:0051252 | regulation of RNA metabolic process | 2804 | 72 | 0.000281 | 0.002346 |
| 7 | GO:0015031 | protein transport | 1202 | 37 | 0.000525 | 0.00333 |
| 8 | GO:0043001 | Golgi to plasma membrane protein transport | 23 | 4 | 0.000629 | 0.00333 |
| 9 | GO:0032774 | RNA biosynthetic process | 3087 | 76 | 0.000664 | 0.00333 |
| 10 | GO:0010556 | regulation of macromolecule biosynthetic process | 3046 | 75 | 0.000731 | 0.00333 |
| UP-REGULATED | ||||||
| Ranking | GOBPID | Term | Size of GO Term | Matched | P value | FDR |
| 1 | GO:0006355 | regulation of transcription, DNA-dependent | 2724 | 69 | 1.76E-05 | 0.000431 |
| 2 | GO:0006892 | post-Golgi vesicle-mediated transport | 75 | 8 | 2.31E-05 | 0.000431 |
| 3 | GO:0051252 | regulation of RNA metabolic process | 2804 | 70 | 2.44E-05 | 0.000431 |
| 4 | GO:0032774 | RNA biosynthetic process | 3087 | 74 | 5.26E-05 | 0.000652 |
| 5 | GO:0010556 | regulation of macromolecule biosynthetic process | 3046 | 73 | 6.15E-05 | 0.000652 |
| 6 | GO:0051171 | regulation of nitrogen compound metabolic process | 3326 | 77 | 0.000115 | 0.00099 |
| 7 | GO:0031326 | regulation of cellular biosynthetic process | 3171 | 74 | 0.000131 | 0.00099 |
| 8 | GO:0071704 | organic substance metabolic process | 8524 | 160 | 0.000165 | 0.001017 |
| 9 | GO:0090304 | Nucleic acid metabolic process | 4168 | 91 | 0.000173 | 0.001017 |
| 10 | GO:0042384 | cilium assembly | 78 | 7 | 0.000227 | 0.00112 |
| DOWN- REGULATED | ||||||
| Ranking | GOBPID | Term | Size of GO Term | Matched | P value | FDR |
| 1 | GO:0007049 | cell cycle | 1508 | 8 | 0.00265 | 0.008484 |
| 2 | GO:0055117 | regulation of cardiac muscle contraction | 66 | 2 | 0.005583 | 0.008484 |
| 3 | GO:0035383 | thioester metabolic process | 68 | 2 | 0.005916 | 0.008484 |
| 4 | GO:0051283 | negative regulation of sequestering of calcium ion | 75 | 2 | 0.007155 | 0.008484 |
| 5 | GO:0010991 | negative regulation of SMAD protein complex assembly | 5 | 1 | 0.008484 | 0.008484 |
| 6 | GO:0031585 | regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity | 5 | 1 | 0.008484 | 0.008484 |
| 7 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase | 5 | 1 | 0.008484 | 0.008484 |
| 8 | GO:0045329 | carnitine biosynthetic process | 5 | 1 | 0.008484 | 0.008484 |
| 9 | GO:0086067 | AV node cell to bundle of His cell communication | 5 | 1 | 0.008484 | 0.008484 |
| 10 | GO:1901339 | regulation of store-operated calcium channel activity | 5 | 1 | 0.008484 | 0.008484 |
GO pathway analysis was performed on all DEGs identified in L neurons and up- and down-regulated DEGs separately.
GeneGo analysis of total DEGs and upregulated DEGs (S6 Table) revealed potential involvement of protein transport as well as the Wnt signaling pathway. Due to the small sample size together with the finding that <2 matched genes were observed for each GeneGo pathway, conclusions could not be drawn for the down-regulated DEGS using this particular analysis tool (data not shown). From Ingenuity Pathway Analysis (IPA), receptor signaling pathways were enriched between BPD and control L neurons (Table 3). Similar results were obtained when analysis was performed on the up-regulated DEGS only (Table 3). As for GeneGo, an analysis of down-regulated DEGs was not appropriate for IPA. KEGG pathway analysis on DEGs was also performed (S7 Table) and supported the results from the other analyses, identifying receptor-mediated signaling, RNA metabolism, and protein trafficking as major processes potentially dysregulated in BPD. Interestingly, many of the enriched pathways involved GSK3β, which has been implicated as a therapeutic target of lithium [42], the first-line treatment for BPD [43].
Table 3. TOP 10 list of IPA pathway analysis of DEGs between BPD and control in L neurons.
| TOTAL | ||
| RANK | Pathway | Hits Symbol |
| 1 | Integrin Signaling | GSK3B,FNBP1,PIK3C2A,CAPN1,CTTN,ITGB8,MYLK3,PTEN,CAPN3,ARHGAP5,RHOQ |
| 2 | Insulin Receptor Signaling | GSK3B,JAK2,PIK3C2A,GAB1,PTEN,RPTOR,RHOQ |
| 3 | Amyloid Processing | GSK3B,CSNK1A1,CAPN1,CAPN3 |
| 4 | HER-2 Signaling in Breast Cancer | GSK3B,PIK3C2A,MDM2,ITGB8,PARD3 |
| 5 | PI3K/AKT Signaling | GSK3B,JAK2,PPP2R5C,MDM2,ITGB8,GAB1,PTEN |
| 6 | ILK Signaling | GSK3B,FNBP1,PIK3C2A,PPP2R5C,ITGB8,PGF,PTEN,RHOQ |
| 7 | Taurine and Hypotaurine Metabolism | GAD1,CSAD |
| 8 | Role of NFAT in Regulation of the Immune Response | GSK3B,CSNK1G1,CSNK1A1,NFATC3,PIK3C2A,KPNA5,NFAT5,GNG2 |
| 9 | Hypoxia Signaling in the Cardiovascular System | UBE2H,UBE2D3,MDM2,PTEN |
| 10 | IL-4 Signaling | NFATC3,JAK2,PIK3C2A,NFAT5 |
| UP-REGULATED | ||
| RANK | Pathway | Hits Symbol |
| 1 | Integrin Signaling | GSK3B,FNBP1,PIK3C2A,CAPN1,CTTN,ITGB8,MYLK3,PTEN,CAPN3,ARHGAP5,RHOQ |
| 2 | Amyloid Processing | GSK3B,CSNK1A1,CAPN1,CAPN3 |
| 3 | ILK Signaling | GSK3B,FNBP1,PIK3C2A,PPP2R5C,ITGB8,PGF,PTEN,RHOQ |
| 4 | Role of NFAT in Regulation of the Immune Response | GSK3B,CSNK1G1,CSNK1A1,NFATC3,PIK3C2A,KPNA5,NFAT5,GNG2 |
| 5 | Taurine and Hypotaurine Metabolism | GAD1,CSAD |
| 6 | Hypoxia Signaling in the Cardiovascular System | UBE2H,UBE2D3,MDM2,PTEN |
| 7 | IL-4 Signaling | NFATC3,JAK2,PIK3C2A,NFAT5 |
| 8 | IL-17A Signaling in Airway Cells | GSK3B,JAK2,PIK3C2A,PTEN |
| 9 | Melanoma Signaling | PIK3C2A,MDM2,PTEN |
| 10 | PI3K/AKT Signaling | GSK3B,JAK2,PPP2R5C,MDM2,ITGB8,PTEN |
IPA pathway analysis of total DEGs and up-regulated DEGs performed independently shows enrichment of receptor signaling pathways.
Confirmational qRT-PCR
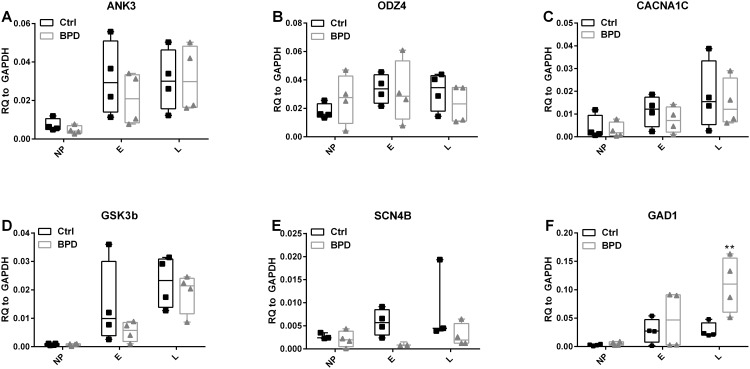
To validate DEGs identified from microarray studies, expression of selected genes was measured by qRT-PCR from an independent aliquot of RNA to that used for the microarray analysis. We first studied the expression of previously reported candidate genes from the literature on genetic studies of BPD: ANK3, ODZ4 and CACNA1C [44–46]. Consistent with the microarray data, qRT-PCR failed to show alterations in the expression of these genes in BPD versus control samples (Fig 5A–5C). We also examined the expression of mRNA encoding GSK3β, a molecule implicated in the mechanism of therapeutic response to lithium [42, 47], which showed small changes (Fold change of BPD_L/CTRL_L = 1.6) in the microarray, but failed to show a significant difference by qRT-PCR (Fig 5D). Voltage gated type IV sodium channel beta subunit, SCN4B, was identified as one of the highly down-regulated genes by the microarray in L neurons of BPD samples (fold change = -14.6). Expression of SCN4B mRNA also showed a decrease by qRT-PCR, but did not reach statistical significance due to high variability between samples (Fig 5E). Glutamate decarboxylase 1 (GAD1) showed 2.5-fold increase in the microarray analysis of L neurons, which was confirmed by qRT-PCR (Fig 5F).
Fig 5. Confirmational quantitative RT-PCR.
RNA samples collected during differentiation at NP, E and L neuron stages for microarray studies were analyzed by quantitative RT-PCR. Genes previously implicated to be involved with BPD from genomic studies were evaluated: ANK3, ODZ4 and CACNA1C were analyzed (A-C) and did not show differences in expression at any stage. GSK3B did not show significant differences in BPD and control at any stage (D). SCN4B showed trends of down regulation both in E and L neurons (E), but did not meet the statistical significance. Expression of GAD1 was upregulated in L of BPD compare to control. The box on the plots represents the 25 and 75 percentiles. The horizontal bar within the box represents the median. Individual data points are represented by triangles (BPD) or squares (Controls). n = 4 except for SCN4B NP and L controls and E BPD where n = 3. Two-way ANOVA was performed and Bonferroni’s Multiple Comparison post-hoc test was done. ** p < 0.01 when compared to control.
Samples from neuronal differentiation of 2 additional BPD and 2 control iPSC lines, were also analyzed by qRT-PCR. These lines were derived from fibroblasts from the same Old Amish population isolate (Coriell Catalog ID: GM05926, GM05934, GM05904, GM05930) using the methods described above. These cases had the same diagnostic reliability, and for the BPD patients, lithium responsiveness. Inclusion of data from these samples did not change the overall expression profiles of candidate genes assessed (data not shown).
Discussion
In this study, we report the successful generation and characterization of iPSC lines from 4 affected and 4 unaffected BPD individuals from an Old Order Amish pedigree and their differentiation into NPs and neurons. We purposely chose subjects with an unequivocal diagnosis of BPD and their healthy unaffected first-degree relatives, the presence or absence of affective disorders being ascertained longitudinally over the years and ratings being carried out blindly and conceded by a psychiatric review board. Consistent with literature, differentiation of the iPSCs into NPs and neurons was accompanied by changes in the expression of signature markers of pluripotency, neuroectoderm and neurons. This is the largest cohort reported to date for iPSC generation from a population isolate with a high prevalence of BPD. By selecting samples from a culturally and genetically isolated Amish population, we anticipated to increase the probability of identifying molecular changes associated with BPD.
One major objective of this study was to conduct global gene expression analysis on iPSC-derived neurons of control and BPDs in order to generate hypotheses related to BPD biology. To ensure that the differentiation paradigm employed in this study was robust, we used the Affymetrix microarray platform to analyze RNA from iPSCs, NPs and E and L neurons. Consistent with signature gene markers analyzed by qPCR, the data subjected to Principal Component Analysis clearly demonstrated three isolated clusters of gene expression corresponding to iPSCs, NPs and neurons, validating the differentiation method we employed. Moreover, the Gene Ontology and Ingenuity Pathway analyses on DEGs associated with progressive differentiation of iPSCs confirmed the expected enrichment of genes and pathways associated with cell cycle regulation, neurogenesis and axon guidance as cells progressed from iPSCs to NPs to E or L neurons. These data are largely consistent with the recent report by Chen et al. [21] who also implemented withdrawal of growth factors to induce commitment of iPSCs to neuronal differentiation and observed enrichment of neuron specific markers in the transcriptome of neurons versus undifferentiated iPSCs.
Critically, we observed that BPD and control iPSCs differentiated similarly to NPs and neurons, thereby paving the way to identify DEGs between controls and BPDs at a given stage of differentiation of iPSCs. This is in contrast to Madison et al [22], who observed a deficit in the proliferative characteristics of NPs as well as in neuronal differentiation ability in their BPD lines. Although we presented one iPSC cell line/donor for 4 donors in the current study, we did confirm the NP differentiation capacity of 2 additional lines from each of the 4 BPD donors and 3 lines from 2 additional affected family members (data not shown). Chen et al [21] did not report a lack of differentiation capacity in their BPD iPSCs.
Analysis of microarray data to identify DEGs between BPD and control samples failed to detect significant changes at NP or E stage neurons. It should be noted that we applied criteria that we believed would minimize false positives in our identification of DEGs (see Materials and Methods). The ability to identify DEGs only in the L neuronal population indicates that the maturity level of neurons may be an important factor to consider for transcriptomic studies of iPSC-derived neurons. This contention is supported by our observations (Fig 3 and S5 Table) that expression of key synaptic markers and proteins involved in the regulation of neuronal function progressively increase in expression from E to L neurons. The timing of neuronal differentiation from iPSCs has been intensively debated in the field with a longer differentiation duration reported to have clearer impact on neuronal function [48].
We conducted qRT-PCR analyses for 2 genes whose expression was highly altered in the microarray analysis. Consistent with microarray data, the expression of GAD1, the gene encoding glutamate acid decarboxylase, a key enzyme for gamma amino butyric acid (GABA) synthesis, showed a significant increase in BPD L neurons. The decrease in the expression of SCN4B (encodes the beta subunit of voltage gated Type IV sodium channel) observed by microarray analysis was also seen as a reduction in L neurons of BPD cases by qRT-PCR, but this finding failed to reach statistical significance. SCN4B is known to regulate neuronal activity and appears to be especially important during brain development [49]. Changes in the expression of both of these genes have been reported in postmortem tissues from schizophrenia [50,51]. Thus the increase in GAD1 concurrently with a decrease in SCN4B indicates that BPD may be associated with an imbalance of excitatory and inhibitory neurotransmission in developing neurons. Interestingly, postmortem data on autopsied hippocampus indicate a decrease in GAD1 mRNA in BPD patients [11]. These data raise the question whether the increase in GAD1 mRNA in developing neurons differentiated from iPS cells is a compensatory response. Of note, Madison et al. [22] also found GAD1 to be differentially regulated between BPD and control CXCR4+ NPs.
We studied by qRT-PCR three other genes, ANK3, CACNA1C and ODZ4 that have been implicated by genetic studies of BPD [44–46]. As with the microarray study, the mRNA for these genes did not show significant differences between BPD and controls in L neurons by qRT-PCR. Similar to our findings at 4 weeks of neural differentiation, Madison et. al. [22] did not detect differences in expression of these genes at 6 weeks using the PsychGene NanoString probe set. Bavamian et al [52] did show decreases in ANK3 gene expression in BPD lines compared to controls at 2 and 8 weeks of neural differentiation but not at 4 or 6 weeks.
The DEGs between BPD and controls identified by microarray analysis were subjected to multiple pathway analysis to understand their potential functional significance. Gene Ontology (GO) indicated that the DEGs reflected alterations in biosynthetic and metabolic pathways for several macromolecules, including RNAs. Defective RNA metabolism contributes to a variety of neurologic diseases, especially motor neuron diseases [53]. Importantly, our findings are consistent with the GO analysis of the microarray study of postmortem brain samples of BPD and controls, where Chen et al [15] reported changes in protein and macromolecule metabolic processes. In addition, these investigators also observed changes in markers of synaptic transmission. Taken together, changes in pre-mRNA processing or RNA editing of neuronal receptors and ion channels that regulate synaptic activity may be important phenotypes associated with BPD.
GO pathway analysis revealed that DEGs between BPD and controls were enriched in protein transport as well as post-Golgi transport pathways. Interestingly, GeneGo analysis also identified clathrin-coated vesicle as well ER-to-Golgi processes. Chen et al [15] identified protein transport in their pathway analyses of DEGs in post-mortem BPD brains. This suggests that disruption of vesicular trafficking and protein transport may play a role in BPD and is supported by other recent genomic and proteomic findings implicating the clathrin interactome in psychiatric disorders including BPD [54].
Another enriched pathway identified by GO analysis of total,up and down regulated DEGs implicated differential regulation in cilium assembly in BPD neurons. Cilium, a microtubule enriched cellular structure, appears to play a key role in developmental signaling cascades, especially associated with hedgehog (Hh) signaling molecules [55]. A report by Breunig et. al. [56] showed that primary cilia are required for Sonic Hedgehog (Shh) pathways and that mutations in ciliary components can lead to phenotypes similar to the phenotypes of Hh/Shh pathway dysregulation, leading to many different pathologies including cognitive deficits and behavioral disorders [55]. A recent report demonstrated that disruption in Shh signaling underlies Ellis-van Creveld dwarfism syndrome, which has a protective effect against development of BPD [57]. This observation, together with our identification of cilium assembly as a pathway modulated in BPD, suggests that disruption in cilial assembly and associated developmental signaling cascade may contribute to BPD pathogenesis.
In multiple pathway analyses, signaling pathways were found in the top 10 rankings. GeneGo analysis of the up-regulated DEGs in BPD L neurons identified the wnt signaling pathway including GSK3β. Ingenuity Pathway Analysis of DEGs showed enrichment of receptor-mediated signaling pathways. One observation was that virtually all of the top pathways identified were associated with GSK3β. This may be due to over-prevalence of GSK3β in IPA pathways; however, GSK3β has been suggested to play a role in the therapeutic effects of lithium [42,47]. Lithium is the treatment of choice for preventing episodes of affective disorder in bipolar subjects and specifically in these Amish families, where good responsiveness and compliance has been documented over decades [24]. In particular, the BPD patients in our study were all good lithium responders based on medication histories recorded longitudinally (see Methods section). Quantitative RT-PCR analysis however failed to show a difference in GSK3β mRNA expression. Since the activity of this enzyme is largely regulated by posttranslational phosphorylation [58,59], future studies of BPD-derived iPS neurons might explore GSK3β protein expression and phosphorylation, as well as the response to lithium. Other components identified in our IPA pathway analysis should also be evaluated.
Chen et al. [21] conducted microarray analysis of 3 BPD and 3 control iPSCs and neurons derived from these iPSCs. Although our GO annotations, derived from DEGs in iPSCs versus neurons, are similar to those reported by Chen et al, we did not see a major overlap in DEGs in BPD neurons compared to control between the two studies. A key reason for this underlying discrepancy may be the fact that Chen et al. conducted microarray analysis on neurons differentiated for 8 weeks, whereas ours were differentiated for 4 weeks. Alternatively the genetic heterogeneity of BPD revealed by GWAS [45,60] and, more recently, WGS [8,44] studies may underlie this lack of overlap. Our samples were derived from an Old Order Amish pedigree, whereas Chen et al. studied iPSCs reprogrammed from Caucasian donors from unrelated patients and controls. Madison et al [22] reported seeing differential gene expression as early as the NP stage between BPD and control population. This discrepancy might be due to their use of an enriched NP population (CXCR4+) or the different analytical platforms employed. RNA-seq has a wider dynamic range and is more sensitive than microarray which may also have contributed to the identification of DEGs at this time point. In their study of iPSC-derived neurons differentiated for 6 weeks, Madison et al. found 44 DEGs between BPD and controls using the 352 custom PsychGene Nanostring probe set. We saw no overlap between their DEGs and the DEGs that we identified between BPD and control samples in our L neuronal population.
In our study, we randomly picked one iPSC line per donor on the basis of recent studies demonstrating that the genetic background of individual donors has greater influence on the transcriptome than line-to-line differences or the reprogramming method used [61]. However, we cannot exclude the significance of line-to-line variability in modeling diseases using iPSCs. Even though the variability between lines might not be as significant as the variability coming from each donor, inclusion of multiple lines might be beneficial to reduce technical errors in future experiments.
In summary, our study represents the first report of characterizing iPS cell lines and iPSC-derived neurons derived from a genetic isolate, the Old Order Amish. Having applied a robust differentiation protocol, we were able to assess molecular mechanisms associated with BPD by microarray analysis on neurons derived from the Amish pedigree. The differentially expressed gene expression data lead us to hypothesize that the alterations in RNA metabolic processes, protein trafficking and receptor-mediated signaling contribute to BPD biology. The 4 BPD and 4 control lines from the Amish pedigree now offers us the opportunity to test these hypotheses in order to gain further insights into the biology associated with this devastating disease.
Supporting Information
IPSCs maintained in feeder-free system were dissociated, allowed to form EBs in AggreWell800 for 5 days, and further differentiated after plating prior to collection of NPs. NPs were expanded further before starting neuronal differentiation by growth factor withdrawal. RNA samples were collected from iPSCs, NPs, and from early (E) and late (L) neurons, defined as 2 and 4 weeks after growth factor withdrawal from NPs, respectively. Representative images of each of the stages of differentiation are shown to the right. iPSCs were stained for Oct4, Neural rosettes for Pax6, Neural progenitors for Pax6 (green), Nestin (red) and nuclei (Hoechst33342), and L neurons for MAP2 (red) and β-Tubulin III (green). Photomicrographs of EBs and E neurons are shown.
(TIF)
Successful reprogramming of fibroblasts into iPSCs has been shown by RT-PCR of pluripotent stem cell markers DNMT3b, hTERT, NANOG, OCT4, REX1 and SOX2. Inactivation of viral particles was also confirmed by RT-PCR using Sendai-virus specific primers set (SEV) (A). The iPS cells were shown to be positive for alkaline phosphatase. Expression of pluripotent protein markers was also shown by immunocytochemistry for OCT4, Nanog, Tra1-60, SSEA-3 and SSEA-4 (white bar = 100μm). Fibroblasts surrounding human iPS colonies serve as internal negative controls for IHC staining. (B). The ability of iPSCs to give rise to three germ layers was confirmed by forming EBs and testing expression of mesodermal (FLK2, GATA2), endodermal (AFP, GATA4) and ectodermal (PAX6, N-CAM) markers by RT-PCR (C).
(TIF)
RNA samples, collected from NPs, E and L neurons, were analyzed for ion channel protein expression. Each bar represents mean ± standard error (n = 4). One-way ANOVA was performed and Bonferroni’s Multiple Comparison post-hoc test was done. * p < 0.05, ** p < 0.01 compared to NP. The changes were not statistically significant in some cases, but the trend for increases in expression was observed.
(TIFF)
Gene expression was measured by quantitative RT-PCR using (A) Custom Primer probes designed by Primer3 with indicated annealing temperatures, or (B) pre-designed TaqMan® probes (Life Technologies).
(DOCX)
The genes that were identified to be differentially expressed in neurons (E and L) from NP are listed in this table. The genes are listed in the order generated by hierarchical clustering in the heat map.
(DOCX)
The table lists genes of axonal guidance defined from IPA, which were used for the generation of the heat map in Fig 4C.
(DOCX)
DEGs from microarray data were analyzed for BPD and control lines independently by GO pathway analysis. Both BPD and control showed enrichment in genes associated with cell proliferation, neurogenesis, and axon guidance.
(DOCX)
DEGs from microarray data were analyzed for BPD and control lines independently by GO pathway analysis. Both BPD and control showed enrichment of in cell proliferation, neurogenesis, and axon guidance.
(DOCX)
GeneGo analysis of total and up- regulated DEGs in L neurons was performed and the top 10 ranked pathways listed.
(DOCX)
KEGG analysis on DEGs in L neurons was performed and top10 ranked pathways listed.
(DOCX)
Acknowledgments
We want to thank Chad Cowan and Laurence Daheron (Harvard Stem Cell Institute) for iPSC generation and discussion for neuronal differentiation, and Cara Ann Ruble (Eli Lilly and Company) for bioinformatics assistance. We want to acknowledge the major contribution of members of the Psychiatric Board: James N. Sussex, Abram M. Hostetter, John J. Schwab, David R. Offord, Jon A. Shaw with special consultant Jean Endicott, and the Seasonality Panel: Gianni Faedda, Richard L. Hauger and Demitri F. Papolos.
Data Availability
Data have been deposited to GEO: GSE74358.
Funding Statement
Eli Lilly and Company fully funded this project. The funder provided support in the form of salaries for authors [KHK, JL, RJSG, JLD, RCS, KMM, SMP], but did not have any additional role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. The specific roles of these authors are articulated in the ‘author contributions’ section.
References
- 1. Cassidy F. Risk factors of attempted suicide in bipolar disorder. Suicide & life-threatening behavior. 2011;41(1):6–11. [DOI] [PubMed] [Google Scholar]
- 2. Hauser M, Galling B, Correll CU. Suicidal ideation and suicide attempts in children and adolescents with bipolar disorder: a systematic review of prevalence and incidence rates, correlates, and targeted interventions. Bipolar disorders. 2013;15(5):507–23. 10.1111/bdi.12094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Post RM, Leverich GS, Kupka RW, Keck PE Jr., McElroy SL, Altshuler LL, et al. Early-onset bipolar disorder and treatment delay are risk factors for poor outcome in adulthood. The Journal of clinical psychiatry. 2010;71(7):864–72. 10.4088/JCP.08m04994yel [DOI] [PubMed] [Google Scholar]
- 4. Craddock N, Sklar P. Genetics of bipolar disorder. Lancet. 2013;381(9878):1654–62. 10.1016/S0140-6736(13)60855-7 [DOI] [PubMed] [Google Scholar]
- 5. Kerner B. Genetics of bipolar disorder. The application of clinical genetics. 2014;7:33–42. 10.2147/TACG.S39297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hostetler JA. Amish society. 4th ed Baltimore: Johns Hopkins University Press; 1993. xi, 435: 322–42. [Google Scholar]
- 7. Egeland JA. An epidemiologic and genetic study of affective disorders among the Old Order Amish In: Papolos DF, Lachman HM, editors. Genetic studies in affective disorders: overview of basic methods, current directions, and critical research issues. New York: J. Wiley; 1994. p. 70–90. [Google Scholar]
- 8. Georgi B, Craig D, Kember RL, Liu W, Lindquist I, Nasser S, et al. Genomic view of bipolar disorder revealed by whole genome sequencing in a genetic isolate. PLoS genetics. 2014;10(3):e1004229 10.1371/journal.pgen.1004229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Altar CA, Vawter MP, Ginsberg SD. Target identification for CNS diseases by transcriptional profiling. Neuropsychopharmacology: official publication of the American College of Neuropsychopharmacology. 2009;34(1):18–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Elashoff M, Higgs BW, Yolken RH, Knable MB, Weis S, Webster MJ, et al. Meta-analysis of 12 genomic studies in bipolar disorder. Journal of molecular neuroscience: MN. 2007;31(3):221–43. [DOI] [PubMed] [Google Scholar]
- 11. Konradi C, Eaton M, MacDonald ML, Walsh J, Benes FM, Heckers S. Molecular evidence for mitochondrial dysfunction in bipolar disorder. Archives of general psychiatry. 2004;61(3):300–8. [DOI] [PubMed] [Google Scholar]
- 12. Ogden CA, Rich ME, Schork NJ, Paulus MP, Geyer MA, Lohr JB, et al. Candidate genes, pathways and mechanisms for bipolar (manic-depressive) and related disorders: an expanded convergent functional genomics approach. Molecular psychiatry. 2004;9(11):1007–29. [DOI] [PubMed] [Google Scholar]
- 13. Ryan MM, Lockstone HE, Huffaker SJ, Wayland MT, Webster MJ, Bahn S. Gene expression analysis of bipolar disorder reveals downregulation of the ubiquitin cycle and alterations in synaptic genes. Molecular psychiatry. 2006;11(10):965–78. [DOI] [PubMed] [Google Scholar]
- 14. Sibille E, Arango V, Galfalvy HC, Pavlidis P, Erraji-Benchekroun L, Ellis SP, et al. Gene expression profiling of depression and suicide in human prefrontal cortex. Neuropsychopharmacology: 2004;29(2):351–61. [DOI] [PubMed] [Google Scholar]
- 15. Chen H, Wang N, Zhao X, Ross CA, O’Shea KS, McInnis MG. Gene expression alterations in bipolar disorder postmortem brains. Bipolar Disord. 2013; 15(2): 177–87. 10.1111/bdi.12039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kiskinis E, Sandoe J, Williams LA, Boulting GL, Moccia R, Wainger BJ, et al. Pathways disrupted in human ALS motor neurons identified through genetic correction of mutant SOD1. Cell stem cell. 2014;14(6):781–95. 10.1016/j.stem.2014.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chung CY, Khurana V, Auluck PK, Tardiff DF, Mazzulli JR, Soldner F, et al. Identification and rescue of alpha-synuclein toxicity in Parkinson patient-derived neurons. Science. 2013;342(6161):983–7. 10.1126/science.1245296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cooper O, Seo H, Andrabi S, Guardia-Laguarta C, Graziotto J, Sundberg M, et al. Pharmacological rescue of mitochondrial deficits in iPSC-derived neural cells from patients with familial Parkinson's disease. Science translational medicine. 2012;4(141):141ra90 10.1126/scitranslmed.3003985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Brennand K, Savas JN, Kim Y, Tran N, Simone A, Hashimoto-Torii K, et al. Phenotypic differences in hiPSC NPCs derived from patients with schizophrenia. Molecular psychiatry. 201; 20(3):361–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Brennand KJ, Simone A, Jou J, Gelboin-Burkhart C, Tran N, Sangar S, et al. Modelling schizophrenia using human induced pluripotent stem cells. Nature. 2011;473(7346):221–5. 10.1038/nature09915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chen HM, DeLong CJ, Bame M, Rajapakse I, Herron TJ, McInnis MG, et al. Transcripts involved in calcium signaling and telencephalic neuronal fate are altered in induced pluripotent stem cells from bipolar disorder patients. Translational psychiatry. 2014;4:e375 10.1038/tp.2014.12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Madison JM, Zhou F, Nigam A, Hussain A, Nehme R, van der Ven K, et al. Characterization of bipoplar disorder patient-specific induced pluripotent stem cells from a family reveals neurodevelopmental and mRNA expression abnormalities. Mol. Psychiatry. 2015; 20(6): 703–17. 10.1038/mp.2015.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Egeland JA, Sussex JN, Endicott J, Hostetter AM, Offord DR, Schwab JJ, et al. The impact of diagnoses on genetic linkage study for bipolar affective disorders among the Amish. Psychiatric Genetics. 1990; 1(2):5–18. [Google Scholar]
- 24. Egeland JA, Kidd JR, Frazer A, Kidd KK, Neuhauser VI. Amish study, V: Lithium-sodium countertransport and catechol O-methytransferase in pedigrees of bipolar probands. Am J Psychiatry. 1984; 141(9): 1049–54. [DOI] [PubMed] [Google Scholar]
- 25. Pauls DL, Morton LA, Egeland JA. Risks of affective illness among first-degree relatives of bipolar I old-order Amish probands. Archives of general psychiatry. 1992;49(9):703–8. [DOI] [PubMed] [Google Scholar]
- 26. Egeland JA, Gerhard DS, Pauls DL, Sussex JN, Kidd KK, Allen CR, et al. Bipolar affective disorders linked to DNA markers on chromosome 11. Nature. 1987;325(6107):783–7. [DOI] [PubMed] [Google Scholar]
- 27. Schlaeger TM, Daheron L, Brickler TR, Entwisle S, Chan K, Cianci A, et al. A comparison of non-integrating reprogramming methods. Nature Biotechnol. 2015; 33(1): 58–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chambers SM, Fasano CA, Papapetrou EP, Tomishima M, Sadelain M, Studer L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nature biotechnology. 2009;27(3):275–80. 10.1038/nbt.1529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pankratz MT, Li XJ, Lavaute TM, Lyons EA, Chen X, Zhang SC. Directed neural differentiation of human embryonic stem cells via an obligated primitive anterior stage. Stem Cells. 2007;25(6):1511–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Park IH, Arora N, Huo H, Maherali N, Ahfeldt T, Shimamura A, et al. Disease-Specific Induced Pluripotent Stem Cells. 2008. Cell; 134(5):877–886. 10.1016/j.cell.2008.07.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chan EM, Ratanasirintrawoot S, Park IH, Manos PD, Loh YH, Huo H, et al. Live cell imaging distinguishes bona fide human iPS cells from partially reprogrammed cells. Nature Biotechnol. 2009; 27(11):1033–7. [DOI] [PubMed] [Google Scholar]
- 32. Dage JL, Colvin EM, Fouillet A, Langron E, Roell WC, Li J, et al. Pharmacological characterisation of ligand- and voltage-gated ion channels expressed in human iPSC-derived forebrain neurons. Psychopharmacology. 2014;231(6):1105–24. 10.1007/s00213-013-3384-2 [DOI] [PubMed] [Google Scholar]
- 33. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(43):15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Conti L, Cattaneo E. Neural stem cell systems: physiological players or in vitro entities? Nature reviews Neuroscience. 2010;11(3):176–87. 10.1038/nrn2761 [DOI] [PubMed] [Google Scholar]
- 35. Rallu M, Machold R, Gaiano N, Corbin JG, McMahon AP, Fishell G. Dorsoventral patterning is established in the telencephalon of mutants lacking both Gli3 and Hedgehog signaling. Development. 2002;129(21):4963–74. [DOI] [PubMed] [Google Scholar]
- 36. Manuel MN, Martynoga B, Molinek MD, Quinn JC, Kroemmer C, Mason JO, et al. The transcription factor Foxg1 regulates telencephalic progenitor proliferation cell autonomously, in part by controlling Pax6 expression levels. Neural development. 2011;6:9 10.1186/1749-8104-6-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kwon GS, Viotti M, Hadjantonakis AK. The endoderm of the mouse embryo arises by dynamic widespread intercalation of embryonic and extraembryonic lineages. Developmental cell. 2008;15(4):509–20. 10.1016/j.devcel.2008.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Beddington RS, Puschel AW, Rashbass P. Use of chimeras to study gene function in mesodermal tissues during gastrulation and early organogenesis. Ciba Foundation symposium. 1992;165:61–74. [DOI] [PubMed] [Google Scholar]
- 39. Katsetos CD, Herman MM, Mork SJ. Class III beta-tubulin in human development and cancer. Cell motility and the cytoskeleton. 2003;55(2):77–96. [DOI] [PubMed] [Google Scholar]
- 40. Maccioni RB, Cambiazo V. Role of microtubule-associated proteins in the control of microtubule assembly. Physiological reviews. 1995;75(4):835–64. [DOI] [PubMed] [Google Scholar]
- 41. Chen B, Wang SS, Hattox AM, Rayburn H, Nelson SB, McConnell SK. The Fezf2-Ctip2 genetic pathway regulates the fate choice of subcortical projection neurons in the developing cerebral cortex. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(32):11382–7. 10.1073/pnas.0804918105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Klein PS, Melton DA. A molecular mechanism for the effect of lithium on development. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(16):8455–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Connolly KR, Thase ME. The clinical management of bipolar disorder: a review of evidence-based guidelines. The primary care companion for CNS disorders. 2011;13(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Fiorentino A, O'Brien NL, Locke DP, McQuillin A, Jarram A, Anjorin A, et al. Analysis of ANK3 and CACNA1C variants identified in bipolar disorder whole genome sequence data. Bipolar disorders. 2014;16(6):583–91. 10.1111/bdi.12203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sklar P, Ripke S, Scott LJ, Andreassen OA, Cichon S, Craddock N, et al. Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nature Genetics. 2011;43(10):977–83. 10.1038/ng.943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ferreira MA, O’Donovan MC, Meng YA, Jones IR, Ruderfer DM, Jones L, et al. Collaborative genome-wide association analysis supports a role for ANK3 and CACNA1C in bipolar disorder. Nat Genet. 2008; 40(9): 1056–8. 10.1038/ng.209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Benedetti F, Serretti A, Pontiggia A, Bernasconi A, Lorenzi C, Colombo C, et al. Long-term response to lithium salts in bipolar illness is influenced by the glycogen synthase kinase 3-beta -50 T/C SNP. Neuroscience letters. 2005;376(1):51–5. [DOI] [PubMed] [Google Scholar]
- 48. Odawara A, Saitoh Y, Alhebshi AH, Gotoh M, Suzuki I. Long-term electrophysiological activity and pharmacological response of a human induced pluripotent stem cell-derived neuron and astrocyte co-culture. Biochemical and biophysical research communications. 2014;443(4):1176–81. 10.1016/j.bbrc.2013.12.142 [DOI] [PubMed] [Google Scholar]
- 49. Patino GA, Isom LL. Electrophysiology and beyond: multiple roles of Na+ channel β subunits in development and disease. Neurosci. Lett. 2010; 486(2): 53–9. 10.1016/j.neulet.2010.06.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Guillozet-Bongaarts AL, Hyde TM, Dalley RA, Hawrylycz MJ, Henry A, Hof PR, et al. Altered gene expression in the dorsolateral prefrontal cortex of individuals with schizophrenia. Mol Psychiatry. 2014; 19(4): 478–85. 10.1038/mp.2013.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Akbarian S, Huang HS. Molecular and cellular mechanisms of altered GAD1/GAD67 expression in schizophrenia and related disorders. Brain Res Rev. 2006; 52(2): 293–304. [DOI] [PubMed] [Google Scholar]
- 52. Bavamian S, Mellios N, Lalonde J, Fass DM, Wang J, Sheridan SD, et al. Dysregulation of miR-34a links neuronal development to genetic risk factors for bipolar disorder. Mol Psychiatry. 2015; 20(5): 573–84. 10.1038/mp.2014.176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ramaswami M, Taylor JP, Parker R. Altered ribostasis: RNA-protein granules in degenerative disorders. Cell. 2013;154(4):727–36. 10.1016/j.cell.2013.07.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Schubert KO, Focking M, Prehn JHM, Cotter DR. Hypothesis review: are clathrin-mediated endocytosis and clathrin-dependent membrane and protein trafficking core pathophysiological processes in schizophrenia and bipolar disorder? Mol. Psychiatry; 2012: 17: 669–681. 10.1038/mp.2011.123 [DOI] [PubMed] [Google Scholar]
- 55. Goetz SC Anderson KV. The primary cilium: a signaling centre during vertebrate development. Nat. Rev. Genetics. 2010; 11: 331–44. 56 10.1038/nrg2774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Breunig JJ, Sarkisian MR, Arellano JI, Morozov YM, Ayoub AE, Sojitra S, et al. Primary cilia regulate hippocampal neurogenesis by mediating sonic hedgehog signaling, Proc. Natl. Acad. Sci. U.S.A. 2008; 105(35):13127–32. 10.1073/pnas.0804558105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ginns EI, Galdzicka M, Elston RC, Song YE, Paul SM, Egeland JA. Disruption of sonic hedgehog signaling in Ellis-van Creveld dwarfism confers protection against bipolar affective disorder. Mol. Psychiatry. 2014; 1–7. [DOI] [PubMed] [Google Scholar]
- 58. Hughes K, Nikolakaki E, Plyte SE, Totty NF, Woodgett JR. Modulation of the glycogen synthase kinase-3 family by tyrosine phosphorylation. The EMBO journal. 1993;12(2):803–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wang QM, Fiol CJ, DePaoli-Roach AA, Roach PJ. Glycogen synthase kinase-3 beta is a dual specificity kinase differentially regulated by tyrosine and serine/threonine phosphorylation. The Journal of biological chemistry. 1994;269(20):14566–74. [PubMed] [Google Scholar]
- 60. Muhleisen TW, Leber M, Schulze TG, Strohmaier J, Degenhardt F, Treutlein J, et al. Genome-wide association study reveals two new risk loci for bipolar disorder. Nature communications. 2014;5:3339 10.1038/ncomms4339 [DOI] [PubMed] [Google Scholar]
- 61. Vitale AM, Matigian NA, Ravishankar S, Bellette B, Wood SA, Wolvetang EJ, et al. Variability in the generation of induced pluripotent stem cells: importance for disease modeling. Stem cells translational medicine. 2012;1(9):641–50. 10.5966/sctm.2012-0043 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
IPSCs maintained in feeder-free system were dissociated, allowed to form EBs in AggreWell800 for 5 days, and further differentiated after plating prior to collection of NPs. NPs were expanded further before starting neuronal differentiation by growth factor withdrawal. RNA samples were collected from iPSCs, NPs, and from early (E) and late (L) neurons, defined as 2 and 4 weeks after growth factor withdrawal from NPs, respectively. Representative images of each of the stages of differentiation are shown to the right. iPSCs were stained for Oct4, Neural rosettes for Pax6, Neural progenitors for Pax6 (green), Nestin (red) and nuclei (Hoechst33342), and L neurons for MAP2 (red) and β-Tubulin III (green). Photomicrographs of EBs and E neurons are shown.
(TIF)
Successful reprogramming of fibroblasts into iPSCs has been shown by RT-PCR of pluripotent stem cell markers DNMT3b, hTERT, NANOG, OCT4, REX1 and SOX2. Inactivation of viral particles was also confirmed by RT-PCR using Sendai-virus specific primers set (SEV) (A). The iPS cells were shown to be positive for alkaline phosphatase. Expression of pluripotent protein markers was also shown by immunocytochemistry for OCT4, Nanog, Tra1-60, SSEA-3 and SSEA-4 (white bar = 100μm). Fibroblasts surrounding human iPS colonies serve as internal negative controls for IHC staining. (B). The ability of iPSCs to give rise to three germ layers was confirmed by forming EBs and testing expression of mesodermal (FLK2, GATA2), endodermal (AFP, GATA4) and ectodermal (PAX6, N-CAM) markers by RT-PCR (C).
(TIF)
RNA samples, collected from NPs, E and L neurons, were analyzed for ion channel protein expression. Each bar represents mean ± standard error (n = 4). One-way ANOVA was performed and Bonferroni’s Multiple Comparison post-hoc test was done. * p < 0.05, ** p < 0.01 compared to NP. The changes were not statistically significant in some cases, but the trend for increases in expression was observed.
(TIFF)
Gene expression was measured by quantitative RT-PCR using (A) Custom Primer probes designed by Primer3 with indicated annealing temperatures, or (B) pre-designed TaqMan® probes (Life Technologies).
(DOCX)
The genes that were identified to be differentially expressed in neurons (E and L) from NP are listed in this table. The genes are listed in the order generated by hierarchical clustering in the heat map.
(DOCX)
The table lists genes of axonal guidance defined from IPA, which were used for the generation of the heat map in Fig 4C.
(DOCX)
DEGs from microarray data were analyzed for BPD and control lines independently by GO pathway analysis. Both BPD and control showed enrichment in genes associated with cell proliferation, neurogenesis, and axon guidance.
(DOCX)
DEGs from microarray data were analyzed for BPD and control lines independently by GO pathway analysis. Both BPD and control showed enrichment of in cell proliferation, neurogenesis, and axon guidance.
(DOCX)
GeneGo analysis of total and up- regulated DEGs in L neurons was performed and the top 10 ranked pathways listed.
(DOCX)
KEGG analysis on DEGs in L neurons was performed and top10 ranked pathways listed.
(DOCX)
Data Availability Statement
Data have been deposited to GEO: GSE74358.