Abstract
We previously elucidated the pleotropic role of solute carrier family A1 member 5 (SLC1A5) as the primary transporter of glutamine (Gln), a modulator of cell growth and oxidative stress in non-small cell lung cancer (NSCLC). The aim of our study was to evaluate SLC1A5 as a potential new therapeutic target and candidate biomarker predictive of survival and response to therapy. SLC1A5 targeting was examined in a panel of NSCLC and human bronchial cell lines by RNA interference and by a small molecular inhibitor, gamma-L-glutamyl-p-nitroanilide (GPNA). The effects of targeting SLC1A5 on cell growth, Gln uptake, ATP level, autophagy and cell death were examined. Inactivation of SLC1A5 genetically or pharmacologically decreased Gln consumption, inhibited cell growth, induced autophagy and apoptosis in a subgroup of NSCLC cell lines that overexpress SLC1A5. Targeting SLC1A5 function decreased tumor growth in NSCLC xenografts. A multivariate Cox proportional hazards analysis indicates that patients with increased SLC1A5 mRNA expression have significantly shorter overall survival (p =0.01, HR =1.24, 95% CI: 1.05–1.46), adjusted for age, gender, smoking history and disease stage. In an immunohistochemistry study on 207 NSCLC patients, SLC1A5 protein expression remained highly significant prognostic value in both univariate (p < 0.0001, HR =1.45, 95% CI: 1.15–1.50) and multivariate analyses (p =0.04, HR =1.22, 95% CI: 1.01–1.31). These results position SLC1A5 as a new candidate prognostic biomarker for selective targeting of Gln-dependent NSCLC.
Keywords: lung cancer, glutamine transporters, biomarker
New therapeutic strategies are desperately needed in lung cancer. Targeting cancer-specific biochemical phenotypes, including those determined metabolically, represents an alternative approach to treating patients with lung cancer, the leading cause of cancer deaths in the USA and worldwide.1 L-Glutamine (Gln) is an essential amino acid for non-small cell lung cancer (NSCLC) growth in vitro and in vivo.2–4 Gln has been shown to sustain tumor growth under hypoxia,5 to mediate K-RAS-driven lung cancer growth6 and to support the autophagy-mediated prosurvival pathway in B-RAFV600-driven lung tumors.7 Nonetheless, the specific contribution of Gln transporters to lung cancer remains largely unknown.
What’s new?
New strategies to overcome lung cancer mortality depend heavily on the discovery of novel therapeutic targets. In non-small cell lung cancer (NSCLC), a possible target is solute linked carrier family 1A, member 5 (SLC1A5), a major glutamine transporter in NSCLC. This study furthers the promise of SLC1A5 by showing that its expression levels in lung cancer cells can predict cell sensitivity to the inhibitor gamma-L-glutamyl-p-nitroanilide (GPNA). In NSCLC cell lines, SLC1A5 inactivation led to glutamine starvation and oxidative stress-mediated autophagy and apoptosis. In NSCLC patients, SLC1A5 expression was associated with poor overall survival.
We recently identified by in-depth shotgun proteomic analysis of stage I NSCLCs several therapeutic targets including the solute linked carrier family 1A, member 5 (SLC1A5).8 We were the first to report that SLC1A5 functions as the primary transporter of Gln in a sodium-dependent manner in NSCLC.9 Inhibition of SLC1A5 attenuates cell growth and mTOR signaling.9,10 Although pharmacological strategies to inhibit Gln metabolism using amino acid analogs such as acivicin and 6-diazo-5-oxo-L-norleucine (DON) have been investigated,3,11 the lack of selectivity of these agents has shifted the efforts to developing agents directed at specific nodes of glutamine metabolism instead.12
Given the emerging role of Gln metabolism in cancer13 and the differential expression of SLC1A5 in NSCLC,9 we tested the hypothesis that elevated SLC1A5 expression is a key prosurvival mechanism that promotes NSCLC progression by increasing tumor cells to transport and utilize Gln available in the microenvironment. We investigated the prognostic value of SLC1A5 expression in NSCLC and examined the therapeutic potential of targeting its Gln transport activity in cell and xenografts by inhibiting SLC1A5-dependent Gln transport.
Material and Methods
Cell cultures
Three human squamous carcinoma cell lines (H226, H520 and HCC15), four human lung adenocarcinoma cell lines (H1819, H1435, H1395 and A549), one large-cell carcinoma cell lines (H460), one carcinoid cell line (H727) and two immortalized epithelial cell line (16HBE and BEAS-2B) were purchased from the American Type Culture Collection. One SCC line HCC2450 was a generous gift from Dr. John D. Minna. All cancer cell lines were maintained in RPMI-1640 or DMEM (Life Technologies, Grand Island, NY) with 10% fetal bovine serum (FBS). 16HBE and BEAS-2B were maintained in Dulbecco’s modified eagle medium (DMEM) with 10% FBS. All cells were cultured in 1 mM penicillin/streptomycin.
Growth culture assays
If not otherwise indicated, glutamine deprivation was carried out by incubating cells for 12 hr in glutamine-free medium. To test the growth inhibitor effects of targeting SLC1A5 in human bronchial epithelial cell lines and in NSCLC cell lines (Supporting Information Table S1), cells were plated in 12-well tissue culture plates at a density of 2 × 104 cells per well. The following day, the cells were rinsed once with serum-free L-glutamine-free medium and replaced with RPMI-1640 with the following variations: supplementation with EGF (25 ng/ml) and 1× growth factors cocktail (Invitrogen, Carlsbad, CA) that includes insulin, selenium and transferrin. Cultures were grown for 6 days in the presence of one dose of gamma-L-glutamyl-p-nitroanilide (GPNA) at 500 μM or at increasing doses as indicated in figure legends or DMSO as a control (0.01%, v:v). Cell growth was monitored by measuring the OD490 nm using the Cell-Titer 96-Aqueous colorimetric assay (Promega, Madison, WI). Relative growth rates were expressed as % growth normalized to the control untreated cells.
Western blotting
Whole protein extracts and Western blot (WB) analysis were performed using standard procedures as described in our previous study,9 with detection using the enhanced chemiluminescence system (Life Technologies, Grand Island, NY). Antibodies against SLC1A5 were used at 1:1,000 dilutions (Millipore, Billerica, MA), SN1 at 1:100 (Epitomics, Burlin-game, CA), LAT1 at 1:1,000 (Cell Signaling Technology, Danvers, MA), Caspase 3 at 1:1,000 (Cell Signaling Technology), Caspase 9 at 1:1,000 (Cell Signaling Technology), LC3 1:1,000 (Medical and Biological Laboratories, Nagoya, Japan), Bcl-2 1:1,000 (Cell Signaling Technology) and Bax 1:1000 (Cell Signaling Technology).
Glutamine consumption assay
The concentration of Gln in culture media was determined as previously described.9 Briefly, cells were plated at 105 cells per well in 24-well culture plates (Costar) and allowed to adhere overnight. Before transport assays, the cells were rinsed twice with warm Na+-free Krebs-Ringer Phosphate Buffer (cholKFtP) in which choline chloride and choline phosphate iso-osmotically replaced the corresponding Na+ salts to remove extracellular Na+ and amino acids. The radiotracer used was L-[3,4Gln-3H] glutamine (Amersham) at 500,000 dpm/μmol of the specific activity (5 μCi/ml). For kinetic studies, the amount of unlabeled glutamine in the transport buffer varied from 400 μmol/l to 6.4 mmol/l. Transport values were obtained either in the absence of extracellular Na+ (diffusion and Na-independent uptake) using cholKRP or in the presence of Na+ (total uptake), using NaKRP buffer to determine the Na-dependent rates, reported in units of picomoles per milligram protein per minute. All transport measurements were carried out at 37°C and were terminated after 3 min by adding ice-cold phosphate-buffered saline (PBS) followed by three rapid washes with an ice-cold PBS. Intracellular glutamine was extracted with 0.2 ml per well of 0.2% SDS in 0.2 N NaOH; after 1 hr at room temperature, 0.1 ml of the lysate was neutralized with 2 N HCl and subjected to liquid scintillation spectrophotometry. The remaining lysate was used for the determination of cellular protein by the Pierce BCA Protein assay. Rates of glutamine transport were calculated from the counts per minute (cpm) per sample, and the specific activity of the uptake mix (in cpm/nmol). The concentration of Gln was normalized to total protein and was expressed as nmol Gln/mg protein. Each data point represents the average ±standard error of the mean (SEM) of at least three separate determinations.
Flow cytometry
Flow cytometry assays and cell cycle analysis were performed using propidium iodide (PI; Sigma-Aldrich, St. Louis, MO) staining as previously described.14 A total of 10,000–20,000 stained nuclei were subjected to flow cytometry analysis. Data were collected on a Becton Dickinson FACSCalibur flow cytometer using CellQuest Pro software (BD Biosciences, San Jose, CA). Cell cycle analysis was done using the ModFit LT software (Verity Software House). The percentage of cells in sub-G1 was considered apoptotic.
Mitochondrial potential measurements
The changes in mitochondrial membrane potential were monitored with 5,5′,6,6′-tetrachloro-1,1′,3,3′-tetraethylbenzimida-zolcarbocyanine iodide, JC-1 (Molecular Probes by Life Technologies, Grand Island, NY). In the cytosol, the monomeric (depolarized state) form of this dye fluoresces green (emission read at 527 nm), whereas within the mitochondrial matrix highly concentrated JC-1 forms aggregates (polarized state) that fluoresce red. After 24 hr of GPNA treatment at 50 μM, cells were loaded with JC-1 (3 μM) at 37°C for 30 min. JC-1 aggregates were detectable in the PI channel (emission at 590), JC-1 monomers were detectable in the FITC channel (emission at 520) and the changes in mitochondrial potential were calculated as the red/green ratio in each condition. For quantitation, florescence signals from cells plated in 96-well black sided clear bottom plates were measured using plate reader at the above wavelengths and average. Changes in mitochondrial potential (Δψm) were measured as the ratio of depolarized/polarized state normalized to total cellular protein.
Immunohistochemistry
Tissue microarrays of 207 lung cancer tissues were prepared from formalin-fixed paraffin-embedded (FFPE) blocks as described previously.15 Archived tissue blocks from consecutive anatomic resections between 1989 and 2004 were retrieved from the files of the Vanderbilt University Medical Center and the Nashville Veterans Administration Medical Center pathology departments following an Institutional Review Board approved protocol at both institution. Immunohistochemistry (IHC) protocol and analysis were carried out as previously described.9 The immunohistochemical expression was assessed using light microscopy (magnification, 20×). Both nuclear and cytoplasmic expressions were quantified using a four-value intensity score (0, 1+, 2+ and 3+) and the percentage (0–100%) of reactivity. We defined the intensity categories as follows: 0, no appreciable staining; 1+, barely detectable staining in epithelial cells compared with the stromal cells; 2+, readily appreciable staining and 3+, dark brown staining of cells. Next, an expression score was obtained by multiplying the intensity and reactivity percent values (theoretical range, 0–300). There were three cores of tissue per case in the TMA. On each core, the score was obtained as described earlier. The average of the three cores was used as the score for this case.
Statistical analysis
Spearman correlation between SLC1A5 expression and culture growth was calculated using Graph Pad Prism (Graph Pad Software, San Diego, CA). All other statistical analysis was done in R environment. The relevance of SLC1A5 protein or mRNA expression to the clinical parameters of cancer patients was compared using Student’s t-test, Wilcoxon two-sample test or Kruskal–Wallis test. The Kaplan–Meier method was used to estimate survival as a function of time, and survival difference was analyzed by the log-rank test. Multivariable analyses were performed using Cox proportional hazards model to identify independent prognostic factors. For publically available microarray data sets, Kaplan–Meier Plotter (http://kmplot.com/analysis/index.php?p=service&cancer=lung) open source software were used to assess the prognostic value of SLC1A5 in NSCLC as described elsewhere.16 All experimental data are presented as the mean±SEM of independent measurements. All treatments within each experiment were performed in triplicate wells and repeated on three independent days. Data comparing two experimental conditions were statistically analyzed by two-tailed Student’s t-test and only results with p <0.05 were considered to be statistically significant: *p <0.05, **p <0.005, ***p <0.0005.
Results
Inhibiting SLC1A5 reduces NSCLC cell growth selectively in cells overexpressing the transporter
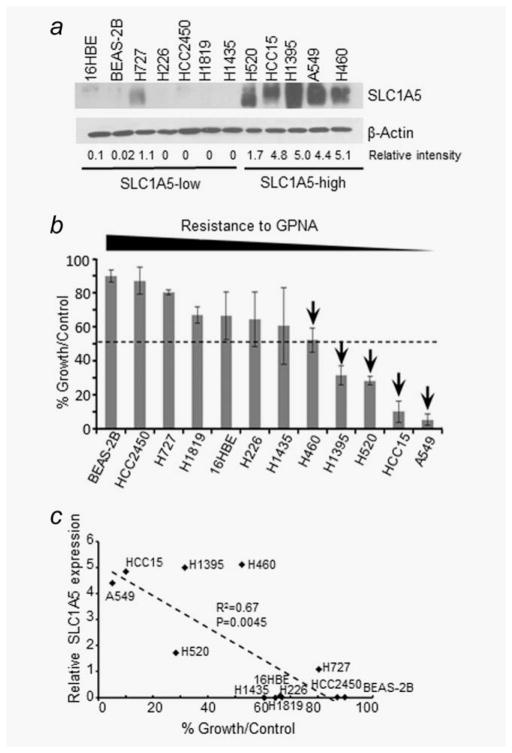
We selected a panel of ten NSCLC cell lines and two human bronchial epithelial cell lines representative of these two distinct subgroups (SLC1A5-high and SLC1A5-low) as a model system for investigating the antitumor effects of inactivating SLC1A5 (Supporting Information Table S1). We cultured the cells that vary in their SLC1A5 expression (Fig. 1a) in the presence of GPNA for 6 days. We designated the cell lines that show 50% or more culture growth inhibition compared to untreated controls as sensitive and the ones with 30% or less inhibition to be resistant (Figs. 1b and 1c). Our results demonstrate that the effect of GPNA on cell growth was dependent on the level of SLC1A5 expression (p =0.0045, R2 =0.67) (Figs. 1b and 1c). GPNA inhibited growth in SLC1A5-high expressing cells in a time- and dose-dependent manner while normal bronchial epithelial cells and low-expressing cancer cells were unaffected (Supporting Information Figs. S1a and S1b). These results indicate that GPNA preferentially inhibits SLC1A5-high expressing NSCLC lines. The selectivity of GPNA was also evident from the lack of response of SLC1A5-low cell lines that express other known Gln/neutral amino acid transporters such as SN1 (SLC38A3) and LAT1 (SLC7A5) that are frequently overexpressed in cancer cells (Supporting Information Fig. S1c).
Figure 1.
SLC1A5 expression predicts response to GPNA in NSCLC. (a) WB analysis of SLC1A5 expression in a panel of non-small cell lung cancer (NSCLC) and human bronchial airway epithelial (HBE) cell lines. A semiquantitative densitometric measurement of the SLC1A5 band intensity normalized to corresponding β-actin using ImageJ software was shown. (b) Sensitivity profile in response to GPNA treatment at 500 μM after 6 days of culture expressed as (% growth/control). All data are the averages ±SD of at least three separate determinations. (c) Spearman correlation showing sensitivity to 500 μM GPNA treatment expressed as relative growth rates shown as % growth normalized to the DMSO-treated cells (control), and protein expression level of SLC1A5 expressed as relative densitometry.
To verify the antiproliferative effect of targeting SLC1A5 in SLC1A5-high expressing lines (H520, A549 and HCC15) we downregulated SLC1A5 expression using siRNA (Supporting Information Fig. S1d). Our results showed that SLC1A5 downregulation (SLC1A5-siRNA) significantly reduced culture growth in H520, A549 and HCC15 compared to a non-specific siRNA control (p <0.005), while 16HBE cells were unaffected (Supporting Information Fig. S1e). In contrast, GPNA treatment led to more cell death on control group than on SLC1A5 KD cells (Supporting Information Fig. S1f), further indicating cancel cells expressing less SLC1A5 are more resistant to GPNA. Together, these results suggest that SLC1A5 expression predicts response to GPNA in NSCLC in vitro.
Antiproliferative effect of targeting SLC1A5 is mediated by inhibition of gln consumption
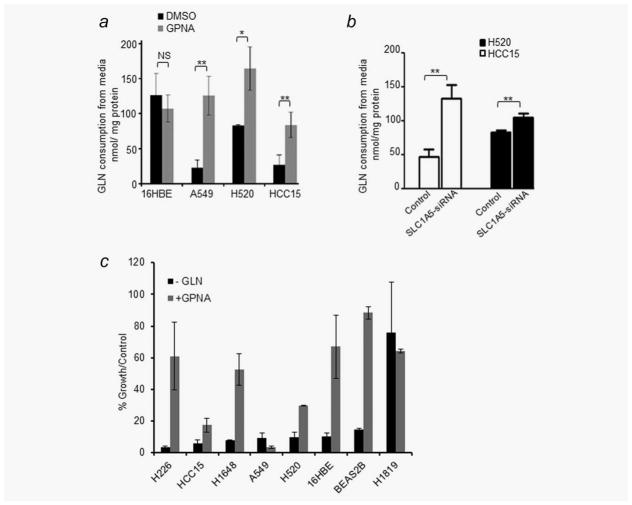
To determine whether the antigrowth effect of targeting SLC1A5 is linked to its Gln transport activity, we measured the level of Gln consumption of cells cultured in growth media treated with GPNA in three SLC1A5-high expressing cell lines (A549, H520 and HCC15) and one SLC1A5-low cell line, 16HBE. GPNA reduced Gln consumption by approximately fivefold in A549, twofold in H520 and threefold in HCC15, while there was no significant change in Gln levels in 16HBE (Fig. 2a). Similarly, siRNA downregulation of SLC1A5 reduced Gln consumption significantly in H520 and HCC15 cells (Fig. 2b). In addition, while Gln deprivation induced a marked reduction in culture growth in seven of the eight cell lines tested, only cell lines overexpressing SLC1A5 (A549, HCC15 and H520) responded to GPNA treatment (Fig. 2c). These results provide a strong biological rationale for targeting SLC1A5 in molecularly defined subset of NSCLC overexpressing the Gln transporter.
Figure 2.
Effect of GPNA on Gln consumption in NSCLC lines. (a) Effect of 500 μM GPNA or DMSO control on glutamine (Gln) consumption in indicated cell lines cultured over 3 days. (b) Gln consumption after 96 hr of silencing HCC15 and H520 with SLC1A5 siRNA or treated with nonspecific siRNA control. (c) Effect of Gln deprivation compared to 500 μM GPNA treatment in six NSCLC and two HBE cell lines that represent SLC1A5-high and SLC1A5-low subgroups after 6 days in culture.
Inhibiting SLC1A5 induces apoptosis via the intrinsic pathway in NSCLC
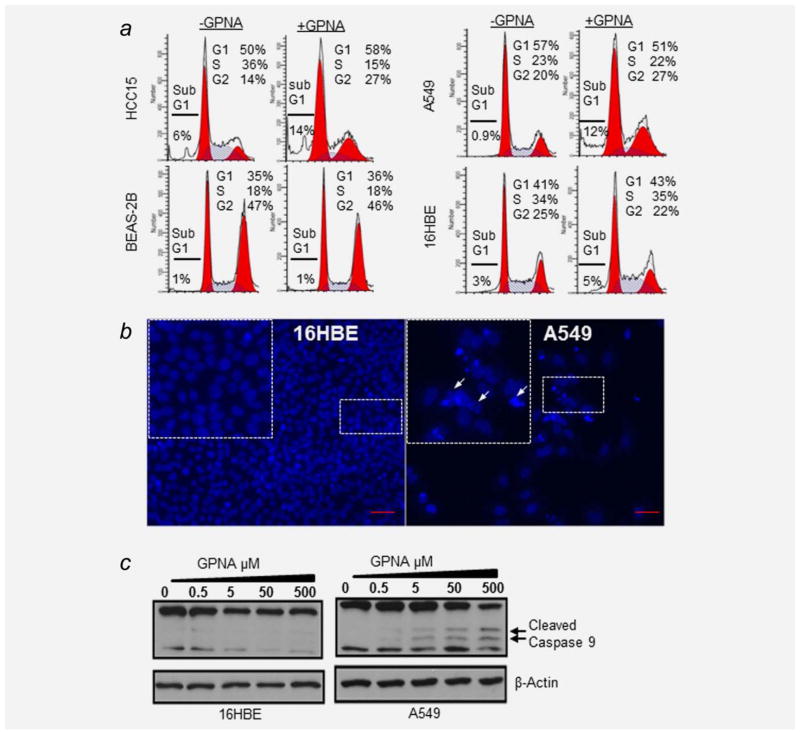
To determine whether the marked reduction in growth caused by GPNA treatment in SLC1A5-high cell lines is attributed to activation of apoptotic cell death, we performed molecular, morphological and cell cycle analyses for apoptotic cell death markers in a panel of six NSCLCs that represent both SLC1A5-high and SLC1A5-low subgroups in the presence of GPNA. Our cell cycle results demonstrated that GPNA treatment caused a marked increase in cell death as evidenced by a threefold increase in the percentage of A549 cells and a 2.3-fold increase of HCC15 cells at the sub-G1 phase (Fig. 3a), while the cell cycle analysis in 16HBE and BEAS2B (HBEs) cells remained unaffected. GPNA treatment also caused DNA condensation and fragmentation in A549 cells but not in 16HBE, consistent with the activation of the late stages of apoptotic cell death (Fig. 3b and Supporting Information Fig. S2a).
Figure 3.
SLC1A5 blockade induces apoptotic cell death. (a) Cell cycle analysis of four cell lines before and after treatment with 500 μM GPNA. GPNA induces cell death in A549 cells and HCC15 (subG1) while has no significant effect on bronchial airway epithelial cell lines. (b) Induction of nuclear condensation and fragmentation by 500 μM GPNA over 4 days shown in A549 but not in 16HBEs. (c) Dose-dependent increase of Caspase 9 cleavage following exposure to GPNA at indicated concentration (arrows point to cleaved forms, 37 and 35 kDa).
To validate these results, we treated A549 cells and 16HBE with increasing concentrations of GPNA and analyzed the levels of intrinsic apoptosis marker-activated Caspase 9 and 317,18 by Western blotting. GPNA treatment induced Caspase 9 cleavage in a dose-dependent manner in A549 (SLC1A5-high expressing cells) but not in 16HBE (SLC1A5-low expressing cells) (Fig. 3c). Consistent with these results, GPNA treatment increased the levels of cleaved Caspase 3, an executioner caspase of the intrinsic pathway in A549 and HCC15 (Supporting Information Fig. S2b). No detectable changes in the expression of antiapoptotic proteins Bcl-2, Bcl-xL, or proapoptotic proteins Bax,19 were observed (Supporting Information Fig. S2b). These results suggest that SLC1A5 blockade by GPNA most likely induces apoptotic cell death via the intrinsic pathway.
SLC1A5-related growth inhibition in NSCLC is mediated by oxidative stress
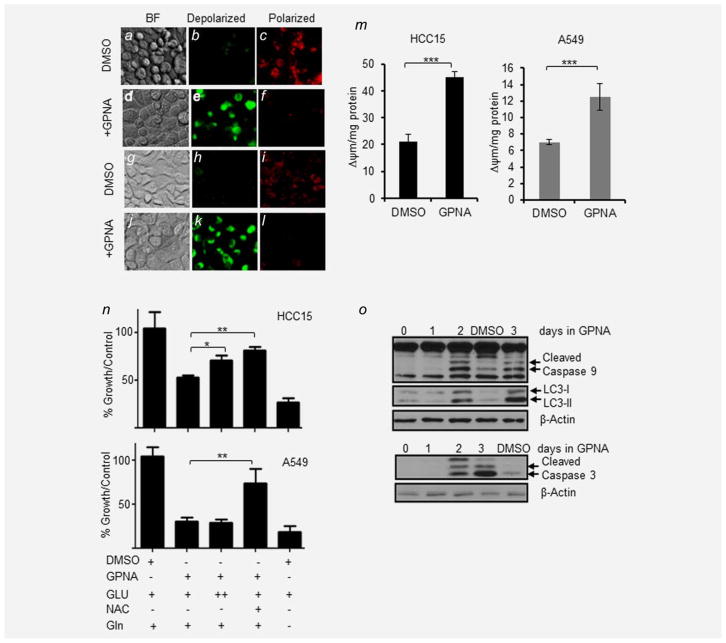
Because oxidative stress induced by mitochondrial disturbances or DNA damage in response to cancer therapeutic agents and hypoxia can trigger apoptosis via the intrinsic pathway,20 we tested the role of oxidative stress in SCL1A5 blockade-induced growth inhibition. We observed significant loss of mitochondrial potential (Δψm) in SLC1A5-high cell lines, HCC15 (Figs. 4af) and A549 (Figs. 4gl), by approximately twofold when treated with GPNA (Fig. 4m). In contrast, GPNA treatment did not significantly alter the mitochondrial potential in SLC1A5-low cell lines H1435 and BEAS-2B (Supporting Information Fig. S2c). Because the antioxidant N-acetylcysteine (NAC)21 was shown to accelerate lung cancer progression in mice by combating oxidative stress damage,22 we postulated that exogenous supplementation of NAC may reduce the antigrowth effects of SLC1A5 blockade in NSCLC cell lines. Our results showed that supplementing the culture medium with 10 mM NAC abrogated the antitumor effect of SLC1A5 blockade by GPNA in both A549 and HCC15 (p =0.0046, 0.034) (Fig. 4n). Supplementing the culture media with tenfold higher concentration of glutamate (Glu) did not have any significant effect on A549 cells but reduced the GPNA antiproliferative effect in HCC15 by 30% (p =0.019). These results suggest that the mechanism of SLC1A5-related growth inhibition in NSCLC is in part mediated by oxidative stress. Upon GPNA treatment, NAC rescues the phenotype. This observation is in support of our previous studies demonstrating a dose-dependent increase in intracellular ROS in response to GPNA.9 Time dependency of the GPNA-induced apoptotic pathway activation was demonstrated in HCC15 and A549 cells for up to 3 days (Fig. 4o). This induction of apoptosis was accompanied by an increase in autophagy as evidenced by the time-dependent increase of LC3-II, an autophagosome marker, and a decrease of both LC3-I and cell size in both cell lines (Fig. 4o and Supporting Information Figs. S2d and S2e). In addition, we observed a significant dose-dependent drop in ATP concentrations in response to GPNA in both HCC15 and A549 (Supporting Information Fig. S2f). On the basis of these molecular, enzymatic and morphological results, we conclude that oxidative stress mediates SLC1A5 prosurvival role in NSCLC cells overexpressing this Gln transporter.
Figure 4.
Targeting SLC1A5 induces autophagy and apoptosis. JC-1 dye was used to measure mitochondrial potential (Δψm) in live HCC15 and A549 cells after 24 hr of 500 μM GPNA or DMSO (0.01%, v/v). (a–c) HCC15 treated with DMSO (0.01%), where (a) is bright field image of HCC15 cells, (b) is a depiction of depolarized mitochondria shown as green fluoresce of JC1 monomers and (c) shows polarized mitochondria as red fluorescence of JC1 aggregates. (d–f) HCC15 treated with GPNA in the same sequence. (g–i) A549 treated with DMSO (0.01%) and (j–l) A549 treated with GPNA. (m) Effect of 500 μM GPNA on mitochondrial potential (Δψm) expressed as the ratio of green/red in HCC15 and A549. (n) Effect of GPNA, glutamate (GLU), glutamine (Gln) and N-acetylcysteine (NAC) on A549 and HCC15 (both high expressing SLC1A5 lines). All data are the averages ±SEM of at least three separate determinations (*p <0.05, **p <0.005). ++, tenfold higher concentration of glutamate was added. (o) Effect of 500 μM GPNA on induction of autophagy (decrease in LC3I and increase of LC3-II) and intrinsic apoptotic markers (Caspase 9 and 3) in a time-dependent manner in HCC15 cells.
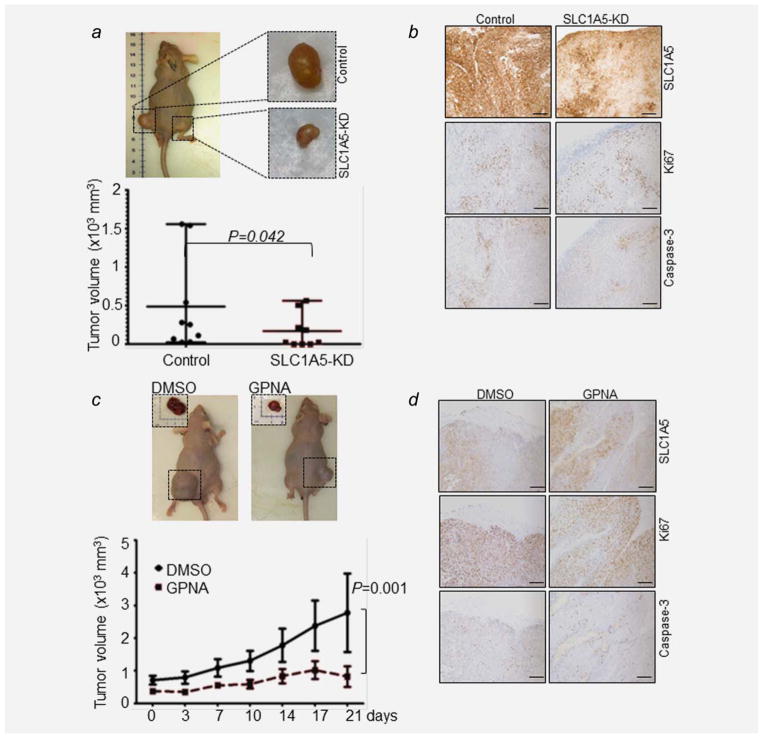
Targeting SLC1A5 attenuates tumor growth in vivo
On the basis of these in vitro data, we sought to determine whether targeting SLC1A5 has an antitumor effect in NSCLC in vivo. To this aim, we generated stable SLC1A5 knockdown (KD) clones using lentiviral shRNA against human SLC1A5 (SLC1A5-KD) and nonspecific shRNA control in A549 cells. We injected stable clones of either SLC1A5-KD or control shRNA subcutaneously into the flank of nude mice. SLC1A5-KD tumor size was significantly smaller than their control shRNA counterparts in all animals tested (p =0.042) (Fig. 5a). IHC analysis revealed that SLC1A5-KD has lower expression level of SLC1A5 protein, decreased in Ki67 and Caspase 3 staining compared to control shRNA (Fig. 5b). Similarly, GPNA treatment significantly reduced the xenograft tumor growth when compared to the DMSO controls (p =0.0014; Fig. 5c). We did not observe a major difference in levels of cleaved Caspase 3 or Ki67 (Fig. 5d) between GPNA- and DMSO-treated tumors. Altogether, these results provide the first in vivo proof-of-concept for targeting SLC1A5 as a therapeutic candidate for NSCLC.
Figure 5.
SLC1A5 blockade attenuates tumor growth in vivo. (a) Effect of inhibition of SLC1A5 expression on tumor volume of isogenic clones of stably transfected A549 cells with either SLC1A5-shRNA (SLC1A5-KD) or nontargeting control shRNA from the same mice (n =9) showing significant growth inhibition in SLC1A5-KD (p =0.042). (b) Representative immunostains for SLC1A5, Ki67 and Caspase 3 of tissue sections of xenografts treated with either SLC1A5-shRNA or nontargeting shRNA. (c) Effect of 500 μM GPNA on xenograft tumor growth over a total of 21 days of treatment. (d) Representative images of immunostaining for SLC1A5, Ki67 and Caspase 3 of tissue sections of xeno-grafts treated with either 500 μM GPNA or DMSO control. Scale bar: 50 μm.
SLC1A5 expression is predictive of poor overall survival in NSCLC
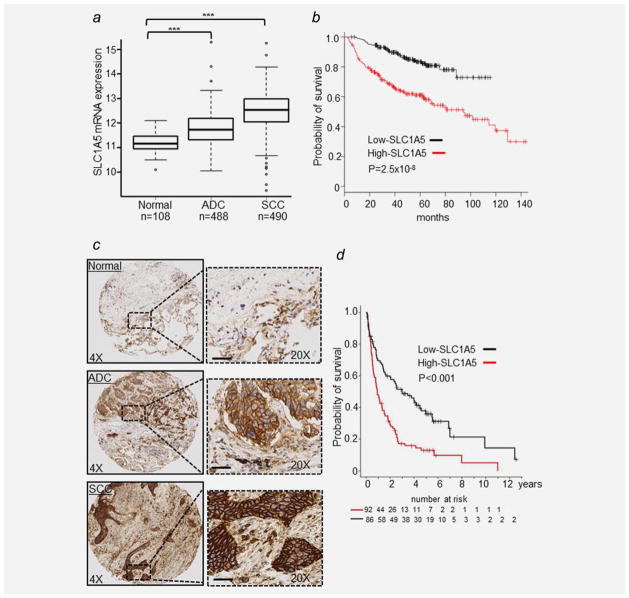
To determine the prevalence of SLC1A5 overexpression in NSCLC, we first tested its mRNA expression in adenocarcinoma (ADC; N = 488), squamous cell carcinoma (SCC; N = 490) and their matched normal lung tissues (N = 108) from The Cancer Genome Atlas (TCGA) database. Our analysis revealed that SLC1A5 is significantly overexpressed in SCC and ADC vs. normal lung tissues (p <0.0001; Fig. 6a), consistent with our previously published proteomics data.8,9 We then used KM plotter, an online survival analysis tool16 to evaluate the prognostic value of SLC1A5 at mRNA level in three independent microarray data sets (GES31210, GES2903 and GES4573) consisting of 411 NSCLCs (Supporting Information Table S2). A multivariate Cox proportional hazards analysis shows that patients with increased SLC1A5 mRNA expression have significantly shorter overall survival (p =0.01, HR =1.24, 95% CI: 1.05–1.46) adjusted for age, gender, smoking history and stage (Supporting Information Table S3 and Fig. 6b).
Figure 6.
SLC1A5 is overexpressed and associated with poor survival in NSCLC. (a) Box plot of SLC1A5 mRNA expression in 488 adenocarcinoma (ADC), 490 squamous cell carcinoma (SCC) and 108 normal lung tissues from the TCGA dataset (***p <0.0001). (b) Kaplan–Meier survival curve of NSCLC from three publically available gene expression data sets (GES31210, GES29013 and GES4573; n =411) using KM plotter online tool16 show that higher mRNA SLC1A5 expression (n =205) is correlated with poor overall survival of patients with NSCLC. Log-rank p values are shown. (c) Representative images of immunostaining for SLC1A5 in normal lung, ADC and SCC. Scale bar: 200 μm. (d) Kaplan–Meier survival curves comparing 178 patients with NSCLC expressing high (red-line) or low SLC1A5 protein (black-line). Log-rank p value is shown.
Furthermore, we assessed protein expression of SCL1A5 using immunohistochemical analysis on 207 NSCLC tissues assembled in tissue microarrays (Supporting Information Table S4) and confirmed that SLC1A5 is differentially expressed along the plasma membrane, with low to negative staining in surrounding stroma of ADC and SCC (Fig. 6c).9 SLC1A5 expression was associated with clinical stage (p <0.001) (Supporting Information Fig. S3a) and with smoking status (p < 0.001) (Supporting Information Fig. S3b). SLC1A5 staining intensity was significantly associated with shorter overall survival in both univariate (p <0.0001, HR =1.45, 95% CI: 1.15–1.50, Fig. 6d) and multivariate analyses (p =0.04, HR =1.22, 95% CI: 1.01–1.31, adjusted for age and stage). Collectively, these results suggest that SLC1A5 expression level is a potential new prognostic biomarker for NSCLC.
Discussion
We report the antitumor effect of a small molecule inhibitor GPNA on SLC1A5-dependent Gln transport in vitro and in vivo in a molecularly defined subset of NSCLCs based on SLC1A5 level of expression. We demonstrated that SLC1A5 antitumor effect is due to apoptosis and is mediated by oxidative stress. We found that high SLC1A5 expression is correlated with poor overall survival in patients with NSCLC in two independent cohorts at the protein (n =207) and gene expression level (n =411). These results demonstrate the potential relevance of SLC1A5 expression as a new candidate companion diagnostic biomarker and a therapeutic target in NSCLC.
To successfully target metabolic pathways involving glutamine metabolism in lung cancer, the dependency of tumor cells from glutamine needs to be established and the mechanisms underlying this dependency understood. This is an area of active investigation.23,24 Here, we identified a subgroup of NSCLCs sensitive to Gln deprivation and determined that its dependency on Gln was largely mediated by SLC1A5 expression. Genetic and pharmacological studies revealed that SLC1A5 is the primary transporter for increasing Gln acquisition to sustain the metabolic demands for the biosynthesis of macromolecules and bioenergetics generation in fast proliferating cells.
To explore the therapeutic potential of targeting SLC1A5 in NSCLC we first determined its expression in NSCLC cell lines using Western blotting and IHC analyses. We then selected a panel of cell lines with low or high expression level for a model system to pursue functional analyses upon SLC1A5 inactivation. Our data revealed that the SLC1A5-high expressing subgroup is specifically sensitive to SLC1A5 inhibition by its selective inhibitor GPNA, while the SLC1A5-low subgroup is resistant. We provide in vitro and in vivo evidence for preclinical targeting of SLC1A5 in NSCLC.
In addition, we demonstrated that blockade of SLC1A5-Gln-mediated activity by GPNA inhibits culture growth and induces apoptotic cell death in the SLC1A5-high subgroup in a selective manner. We elucidated the mechanisms by which GPNA induced cell death. This occurs via the intrinsic apoptotic pathway in a time- and dose-dependent manner. The induction of apoptotic cell death resulted that from SLC1A5 blockade coincided with a measurable increase in an autophagy marker, LC-II, also in a time- and dose-dependent manner. The induction of apoptotic cell death upon SLC1A5 targeting in NSCLC is consistent with findings in hepatocellular carcinoma and in AML.7,25 Induction of apoptosis has also been recently reported in response to blockade of other amino acid transporter including xCT (SLC7A11) in triple negative breast cancer and in SCLC24 and LAT1 (SLC7A5) in human oral epidermoid carcinoma, human osteogenic sarcoma and C6 rat glioma.26
Our results suggest that targeting SLC1A5 in NSCLC cells induces apoptotic cell death by impairing their ability to uptake sufficient Gln and potentially other neutral amino acids (Cys, Ser and Ala) from the extracellular environment. Gln-deprived cells undergo autophagy to sustain their energy supply need for their survival. Extended periods of Gln deprivation eventually lead to a drop in intracellular ATP levels. Decrease in intracellular levels of ATP in turn elicits loss of mitochondrial potential (Δψm) resulting in apoptotic cell death. Recent studies indicate that autophagy plays a critical prosurvival role in preclinical models of lung cancer.7,27,28 For example, BrafV600E-driven lung cancers rely on Gln supplied to the mitochondria by activating autophagy.7 Our results indicate that autophagy is induced in response to SLC1A5 blockade by GPNA most likely as a prosurvival response to intracellular Gln deprivation. Thus, a dual targeting of autophagy and SLC1A5-mediated Gln activity may have additive or synergistic antitumor effects in this molecular subtype of lung cancer.
Our results show that SLC1A5 is differentially expressed at the cell surface of lung cancer cells (Fig. 6c)9 and that tumors harboring high expression of SLC1A5 have a significantly lower survival rate than those of low SLC1A5 expression (Figs. 6b and 6d). Ultimately selecting a population most likely to respond to directed therapy is the goal of personalized therapeutic intervention.13 The identification of bio-markers predictive of response to targeting Gln metabolism is the subject of intensive research24,29–31 with no clinical application as of yet. We believe that SLC1A5 may be an attractive companion diagnostic biomarker and a potential target for well-defined molecular subtypes of NSCLC and deserves further validation.
In summary, this is the first report to elucidate the pro-survival role of SLC1A5 in lung cancer and to establish the transporter as a potential therapeutic target and biomarker of poor prognosis in NSCLC. Ultimately, SLC1A5 may represent a novel companion biomarker of response to therapy targeting a molecularly defined subset of NSCLC.
Supplementary Material
Acknowledgments
Grant sponsor: Lung Cancer Research Foundation (LCRF), National Cancer Institute; Grant numbers: RO1CA102353, UO1CA152662 and P50CA090949
The authors thank Dr. Jamshedur Rahman for his help with handling tissue arrays and Mr. William Alborn for his help with reviewing this manuscript. This work was supported by the Lung Cancer Research Foundation (LCRF) awarded to MH, and RO1CA102353, UO1CA152662 and P50CA090949 to PPM.
Abbreviations
- Δψm
mitochondrial potential
- ADC
adenocarcinoma
- ATP
adenosine 5′-triphosphate
- B-RAF
v-raf murine sarcoma viral oncogene homolog B
- CI
confidence interval
- DMSO
dimethyl sulfoxide
- DON
6-diazo-5-oxo-l-norleucine
- EGF
epidermal growth factor
- Gln
glutamine
- GPNA
gamma-l-glutamyl-p-nitroanilide
- HBE
human bronchial epithelial cell
- HR
hazard ratio
- IHC
immunohistochemistry
- JC-1
first J-aggregate-forming cationic dye
- KD
knockdown
- K-RAS
Kirsten rat sarcoma viral oncogene homolog
- LAT1
large neutral amino acids transporter small subunit 1
- mTOR
mammalian target of rapamycin
- NSCLC
non-small cell lung cancer
- SCC
squamous cell carcinoma
- siRNA
small interfering RNA
- SLC1A5
solute carrier family A1 member 5
- SLC38A3
solute carrier family 38 Member 3
- SLC7A5
solute carrier family 7 member 5
- SN1
system N1 Na+ and H+-coupled glutamine
- TMA
tissue microarray
- WB
Western blot
Footnotes
Conflict of interest: Nothing to report.
Additional Supporting Information may be found in the online version of this article.
References
- 1.Siegel R, DeSantis C, Virgo K, et al. Cancer treatment and survivorship statistics, 2012. CA Cancer J Clin. 2012;62:220–41. doi: 10.3322/caac.21149. [DOI] [PubMed] [Google Scholar]
- 2.Rivera S, Azcon-Bieto J, Lopez-Soriano FJ, et al. Amino acid metabolism in tumour-bearing mice. Biochem J. 1988;249:443–9. doi: 10.1042/bj2490443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huber KR, Mayer EP, Mitchell DF, et al. Cell cycle phase perturbations by 6-diazo-5-oxo-L-norleucine and acivicin in normal and neoplastic human cell lines. Br J Cancer. 1987;55:653–6. doi: 10.1038/bjc.1987.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brower M, Carney DN, Oie HK, et al. Growth of cell lines and clinical specimens of human non-small cell lung cancer in a serum-free defined medium. Cancer Res. 1986;46:798–806. [PubMed] [Google Scholar]
- 5.Metallo CM, Gameiro PA, Bell EL, et al. Reductive glutamine metabolism by idh1 mediates lipo-genesis under hypoxia. Nature. 2012;481:380–4. doi: 10.1038/nature10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gaglio D, Metallo CM, Gameiro PA, et al. Oncogenic K-ras decouples glucose and glutamine metabolism to support cancer cell growth. Mol Syst Biol. 2011;7:523. doi: 10.1038/msb.2011.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Strohecker AM, Guo JY, Karsli-Uzunbas G, et al. Autophagy sustains mitochondrial glutamine metabolism and growth of BrafV600E-driven lung tumors. Cancer Discov. 2013;3:1272–85. doi: 10.1158/2159-8290.CD-13-0397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kikuchi T, Hassanein M, Amann JM, et al. In-depth proteomic analysis of nonsmall cell lung cancer to discover molecular targets and candidate biomarkers. Mol Cell Proteomics. 2012;11:916–32. doi: 10.1074/mcp.M111.015370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hassanein M, Hoeksema MD, Shiota M, et al. slc1a5 mediates glutamine transport required for lung cancer cell growth and survival. Clin Cancer Res. 2013;19:560–70. doi: 10.1158/1078-0432.CCR-12-2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nicklin P, Bergman P, Zhang B, et al. Bidirectional transport of amino acids regulates mTOR and autophagy. Cell. 2009;136:521–34. doi: 10.1016/j.cell.2008.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ovejera AA, Houchens DP, Catane R, et al. Efficacy of 6-diazo-5-oxo-L-norleucine and N-[N-gamma-glutamyl-6-diazo-5-oxo-norleucinyl]-6-diazo-5-oxo-norleucine against experimental tumors in conventional and nude mice. Cancer Res. 1979;39:3220–4. [PubMed] [Google Scholar]
- 12.Ahluwalia GS, Grem JL, Hao Z, et al. Metabolism and action of amino acid analog anti-cancer agents. Pharmacol Ther. 1990;46:243–71. doi: 10.1016/0163-7258(90)90094-i. [DOI] [PubMed] [Google Scholar]
- 13.Hensley CT, Wasti AT, DeBerardinis RJ. Glutamine and cancer: cell biology, physiology, and clinical opportunities. J Clin Invest. 2013;123:3678–84. doi: 10.1172/JCI69600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qian J, Zou Y, Rahman JS, et al. Synergy between phosphatidylinositol 3-kinase/akt pathway and Bcl-xL in the control of apoptosis in adenocarcinoma cells of the lung. Mol Cancer Ther. 2009;8:101–9. doi: 10.1158/1535-7163.MCT-08-0973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Massion PP, Kuo WL, Stokoe D, et al. Genomic copy number analysis of non-small cell lung cancer using array comparative genomic hybridization: implications of the phosphatidylinositol 3-kinase pathway. Cancer Res. 2002;62:3636–40. [PubMed] [Google Scholar]
- 16.Gyorffy B, Surowiak P, Budczies J, et al. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS One. 2013;8:e82241. doi: 10.1371/journal.pone.0082241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reeve JG, Xiong J, Morgan J, et al. Expression of apoptosis-regulatory genes in lung tumour cell lines: relationship to p53 expression and relevance to acquired drug resistance. Br J Cancer. 1996;73:1193–200. doi: 10.1038/bjc.1996.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lorenzo HK, Susin SA. Therapeutic potential of AIF-mediated caspase-independent programmed cell death. Drug Resist Updates. 2007;10:235–55. doi: 10.1016/j.drup.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 19.Guo B, Godzik A, Reed JC. Bcl-G, a novel pro-apoptotic member of the Bcl-2 family. J Biol Chem. 2001;276:2780–5. doi: 10.1074/jbc.M005889200. [DOI] [PubMed] [Google Scholar]
- 20.Oliver L, Vallette FM. The role of caspases in cell death and differentiation. Drug Resist Updates. 2005;8:163–70. doi: 10.1016/j.drup.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 21.Brace JL, Vanderweele DJ, Rudin CM. Svf1 inhibits reactive oxygen species generation and promotes survival under conditions of oxidative stress in Saccharomyces cerevisiae. Yeast. 2005;22:641–52. doi: 10.1002/yea.1235. [DOI] [PubMed] [Google Scholar]
- 22.Sayin VI, Ibrahim MX, Larsson E, et al. Antioxidants accelerate lung cancer progression in mice. Sci Transl Med. 2014;6:221ra15. doi: 10.1126/scitranslmed.3007653. [DOI] [PubMed] [Google Scholar]
- 23.Son J, Lyssiotis CA, Ying H, et al. Glutamine supports pancreatic cancer growth through a KRAS-regulated metabolic pathway. Nature. 2013;496:101–5. doi: 10.1038/nature12040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Timmerman LA, Holton T, Yuneva M, et al. Glutamine sensitivity analysis identifies the xCT anti-porter as a common triple-negative breast tumor therapeutic target. Cancer Cell. 2013;24:450–65. doi: 10.1016/j.ccr.2013.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fuchs BC, Perez JC, Suetterlin JE, et al. Inducible antisense RNA targeting amino acid transporter atb0/asct2 elicits apoptosis in human hepatoma cells. Am J Physiol Gastrointest Liver Physiol. 2004;286:g467–g478. doi: 10.1152/ajpgi.00344.2003. [DOI] [PubMed] [Google Scholar]
- 26.Kim CS, Cho SH, Chun HS, et al. BCH, an inhibitor of system L amino acid transporters, induces apoptosis in cancer cells. Biol Pharm Bull. 2008;31:1096–100. doi: 10.1248/bpb.31.1096. [DOI] [PubMed] [Google Scholar]
- 27.Guo JY, Karsli-Uzunbas G, Mathew R, et al. Autophagy suppresses progression of K-ras-induced lung tumors to oncocytomas and maintains lipid homeostasis. Genes Dev. 2013;27:1447–61. doi: 10.1101/gad.219642.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cardaci S, Rizza S, Filomeni G, et al. Glutamine deprivation enhances antitumor activity of 3-bromopyruvate through the stabilization of monocarboxylate transporter-1. Cancer Res. 2012;72:4526–36. doi: 10.1158/0008-5472.CAN-12-1741. [DOI] [PubMed] [Google Scholar]
- 29.Lieberman BP, Ploessl K, Wang L, et al. PET imaging of glutaminolysis in tumors by 18F-(2S,4R)4-fluoroglutamine. J Nucl Med. 2011;52:1947–55. doi: 10.2967/jnumed.111.093815. [DOI] [PubMed] [Google Scholar]
- 30.Krasikova RN, Kuznetsova OF, Fedorova OS, et al. 4-[18F]fluoroglutamic acid (BAY 85-8050), a new amino acid radiotracer for PET imaging of tumors: synthesis and in vitro characterization. J Med Chem. 2011;54:406–10. doi: 10.1021/jm101068q. [DOI] [PubMed] [Google Scholar]
- 31.Ploessl K, Wang L, Lieberman BP, et al. Comparative evaluation of 18F-labeled glutamic acid and glutamine as tumor metabolic imaging agents. J Nucl Med. 2012;53:1616–24. doi: 10.2967/jnumed.111.101279. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.