Abstract
The serotonin 1A receptor (5-HT1A), a critical regulator of the brain serotonergic tone, is implicated in major depressive disorder (MDD) where it is often found to be dys-regulated. However, the extent to which stress and antidepressant treatment impact 5-HT1A expression in adults remains unclear. To address this issue, we subjected adult male BALB/c mice to unpredictable chronic mild stress (UCMS) to induce a depression-like phenotype that was reversed by chronic treatment with the antidepressant imipramine. In prefrontal cortex (PFC) and midbrain tissue, UCMS increased 5-HT1A RNA and protein levels, changes that are expected to decrease the brain serotonergic activity. The stress-induced increase in 5-HT1A expression was paralleled by a specific increase in DNA methylation of the conserved -681 CpG promoter site, located within a Sp1-like element. We show that the -681 CpG site is recognized and repressed by Sp4, the predominant neuronal Sp1-like factor and that Sp4-induced repression is attenuated by DNA methylation, despite a stress-induced increase in PFC Sp4 levels. These results indicate that adult life stress induces DNA methylation of the conserved promoter site, antagonizing Sp4 repression to increase 5-HT1A expression. Chronic imipramine treatment fully reversed the UCMS-induced increase in methylation of the -681 CpG site in the PFC but not midbrain of stressed animals and also increased 5-HT1A expression in the PFC of control animals. Incomplete reversal by imipramine of stress-induced changes in 5-HT1A methylation and expression indicates a persistence of stress vulnerability, and that sustained reversal of behavioral impairments may require additional pathways.
Keywords: serotonin, UCMS, raphe, DNA methylation, Sp4, antidepressant, 5-HT1A receptor, epigenetic, plasticity, gene expression
Introduction
Major Depressive Disorder (MDD) affects 1 in 6 individuals during their lifetime and is influenced both by genetic and environmental factors (Northoff, 2013; Vialou et al., 2013). A decrease in brain serotonergic activity is thought to underlie MDD (Asberg and Traskman, 1981; Jans et al., 2007; Mann, 1999) and altered expression of serotonergic genes has often been reported in individuals suffering from MDD. For example, reductions in 5-HT transporter (5-HTT) sites and 5-HTT-labelled axons and in 5-HT1A receptor levels have been observed in postmortem prefrontal cortex (PFC) following suicide (Arango et al., 1995; Austin et al., 2002) and MDD (Lopez-Figueroa et al., 2004), suggesting that cortical 5-HT neurotransmission is altered. Conversely, elevated levels of 5-HT1A autoreceptor levels have been reported in post-mortem MDD/suicide midbrain samples (Boldrini et al., 2008; Stockmeier et al., 1998), which could mediate reduced serotonergic activity (Albert and Lemonde, 2004). Genetic polymorphisms that alter transcription of serotonin genes including the 5-HT1A receptor gene have been associated with MDD in meta-analysis studies (Kishi et al., 2013), and this association is strengthened in individuals subjected to early life stress (Karg et al., 2011; Zhang et al., 2009), highlighting that depression arises from the interplay of genetic and environmental factors.
Epigenetic modifications such as histone modification or DNA methylation can induce long lasting changes in gene expression that could explain the persistent and relapsing nature of depression and the need for chronic drug treatment (Tsankova et al., 2007; Vialou et al., 2013). Social defeat was shown to decrease hippocampal expression of brain-derived neurotrophic factor (BDNF) by increasing the repressive histone methylation of its promoters (Tsankova et al., 2006). Similarly, chronic mild stress was associated with dynamic changes in histone methylation/acetylation and GDNF gene expression in the striatum (Uchida et al., 2011). In both paradigms, chronic imipramine treatment normalized gene expression. Persistent changes in DNA methylation have been mainly associated with early life stress, which affects lifelong behavioral phenotype through modulation of gene expression. For example, pups from mothers providing intensive maternal care develop significantly lower stress responses in adulthood. This difference is associated with decreased DNA methylation of an NGFIA-site in the glucocorticoid receptor (GR) promoter, leading to increased hippocampal expression of the receptor and greater negative feedback on the hypothalamus-pituitary-adrenal (HPA) axis (Weaver et al., 2004).
Given the accumulating evidence of maladaptive changes in 5-HT1A expression in human depression and following stress (Jovanovic et al., 2011; Parsey et al., 2010) and the critical role of this receptor in the regulation of serotonergic neurotransmission (Albert et al., 2011), we investigated the impact of unpredictable chronic mild stress (UCMS) and chronic imipramine treatment on 5-HT1A expression in pre- and post-synaptic brain regions. Adult animals subjected to UCMS and chronic imipramine displayed region specific changes in 5-HT1A expression and 5-HT1A promoter methylation which were only partially reversed by chronic imipramine. Our results indicate that DNA methylation is a dynamic and tissue-specific event that could play an important role in the persistent and relapsing nature of depression.
Materials and methods
Animals, UCMS paradigm and behavior tests
Six-week old male BALB/c mice (Centre d’Élevage Janvier, France) were housed for 2 weeks in groups of 15 under a 12/12 light/dark cycle (lights on at 9:00 pm), 21±2°C, with food and water ad libitum before initiating the UCMS protocol. The stress protocol used was described previously (Willner et al., 1992), adapted for mice by our laboratory (Ducottet and Belzung, 2004; Santarelli et al., 2003). Stressed mice were maintained in isolation cages (8.5 × 22 cm); non-stressed mice were in groups of 5 in standard cages (21 × 38 cm), with a shelter and some tubes and did not undergo UCMS. Nociceptive stressors, restraint and food/water deprivation were excluded. The stressors included: removal of bedding, wet bedding, repeated changes of bedding, tilting of cages 45°, replacing bedding with 2-cm water, exposure to predator sounds (15 min), social stress 1 (transfer to foreign mouse cage), social stress 2 (transfer to foreign mouse cage; return to home cage that was occupied by foreign mouse), lights on during the dark phase, lights off during the light phase, a succession of light and dark periods (30 min), switching light/dark cycle. UCMS mice were first subjected to a 2-week drug-free stress procedure and then treated for 7 weeks with 10 ml/kg 0.9% NaCl (vehicle) or 20mg/kg/day imipramine (Sigma) intraperitonally at 1:00 pm each day, except after behavioral testing.
Coat condition was evaluated for head, neck, dorsal coat, ventral coat, tail, front and hind paws and genital areas. Each area was scored (0=good condition; 1= bad condition (disordered, piloerection) and summed for each area as described (Ducottet et al., 2003; Santarelli et al., 2003). Body weight was assessed at the end of the UCMS procedure.
In the nest test, a piece of Nestlet (PLEXX, Netherlands) (5×5cm, 3g) was placed in the cage (8 am) and nest-building scored 24 h later (Deacon, 2006), with 1=square untouched; 2=partially torn (50–90% intact); 3=shredded without nest site; 4 =flat nest and 5=perfect nest (Nollet et al., 2013). Two mice died between this test and the following scoring.
Home cage activity was measured for 2 hr between 12:30–4:30 pm using an actograph with a score of 1 applied for each quadrant crossed. Four mice escaped and were excluded from analysis.
Sacrifice and tissue collection
Transcardiac perfusions were performed in half of the mice from each experimental group. Mice were anesthetized with pentobarbital (injected intraperitoneally at a dose of 40 mg/kg body weight in a volume of 10 ml/kg of body weight), followed by transcardiac perfusion with 80 ml of 0.9% NaCl followed by 180 ml of 4% paraformaldehyde (PFA) in 0.1 M PBS (pH 7.4). Brains were dissected out, post-fixed in PFA for 2 hours and then cryoprotected in 20% sucrose solution at 4°C. Microdissections were performed on the other half of the mice. Brains were rapidly removed from CO2-killed mice and placed in ice-cold slurry of 0.9% NaCl. Rostro-caudal sections (2 mm) were quickly obtained on a brain tissue blocker. Samples were stored in RNAlater (Qiagen) at −80°C.
DNA methylation analysis
Levels of DNA methylation and RNA were quantified in the same tissue samples (whole PFC or midbrain). Genomic DNA was extracted from brain tissues using QIAamp DNA mini-kit (Qiagen) and digested with HindIII overnight. The DNA was denatured (99°C, 5 min), incubated in 0.3M NaOH (39°C, 5 min) and treated with bisulfite (4 M sodium bisulfite, 6 μM hydroquinone, 0.3M guanidine HCl) at 55°C for 16 hr. Bisulfite-treated genomic DNA was purified using the Wizard kit (Promega), desulfonated in 0.3 M NaOH (37°C, 15 min), desulfonation terminated by addition of NH4OAc, and co-precipitated with 20 μg linear acrylamide in EtOH overnight at −20°C. Amplification of the 5-HT1A proximal promoter was performed by nested PCR using the following primers: (fwd: TAGTAGATTAGAGTAGATATATGAAG; rev: TATCCCGAAAAACTCCA; nfwd: TGAGTGTGGTGTGGATT; nrev: CCAAAACTAAACATATCCAT). Cycling conditions were: 95°C, 1 min; 35 cycles at 95°C, 30 sec; 45°C, 90 sec; 72°C, 60 sec; 72°C, 5 min; 4°C. PCR products were cloned in pGEM-T-easy (Promega) and 12–18 minipreps from 2–3 independent PCR reactions/ligations were randomly selected and sequenced to quantify DNA methylation in each sample.
RNA extraction, reverse transcription and real-time PCR
Frozen brain tissue was homogenized in TRI-reagent (Ambion), extracted with chloroform and RNA was co-precipitated with 20 μg linear acrylamide in isopropanol and treated with TURBO DNase (Ambion) for (37°C, 30 min). cDNA synthesis was carried out with M-MLV reverse transcriptase (NEB), using 2 μg RNA and 250 ng of random oligonucleotides (Invitrogen) or a control reaction without reverse transcriptase. Real-time PCR quantification was performed using TaqMan Gene expression Assay kits (Applied Biosystems) for mouse 5-HT1A (Mm00434106_s1), GAPDH (Mm99999915_s1), Sp4 (Mm00599719_m1), NR2A (Mm00433802_m1) and TPH (Mm00557717_m1) in a 20-μl reaction on a Rotor Gene 3000 (Corbett Research) at: 95°C, 10 min; 40 cycles of 95°C, 15 s; 60°C, 60 s. Relative quantification was performed using the ΔΔCt method (Livak and Schmittgen, 2001), using GAPDH as a reference gene.
Immunofluorescence
Coronal brain slices (30 μm) from brain tissue were obtained using the following coordinates (Franklin and Paxinos, 2007): Dorsal Raphe Nucleus (DRN) (Bregma −4.2 to −4.96 mm) and PFC (Bregma +1.3 to +1.90 mm) including infralimbic and prelimbic cortex (PFC) which express high levels of 5-HT1A receptors and have an important role in stress-induced regulation of the serotonergic system (Amat et al., 2005). The sections were stained as described (Czesak et al., 2012), incubated in target retrieval solution (DakoCytomation, Mississauga, Ontario)(95°C, 30 min), washed 3x in PBS, blocked 1 hr in PBS with 1% BSA, 10% goat serum, 0.1% Triton X-100; and incubated overnight at 4°C with 1:50 rabbit anti-5-HT1A with or without 1:100 sheep anti-TPH (Chemicon). Sections were washed 3 times in PBS followed by 2-hr incubation with an Alexa Fluor 488/Fluor 594 donkey anti-rabbit IgG and/or Alexa Fluor 594 donkey anti-sheep IgG (Molecular Probes) in blocking solution. Images were acquired with the Axiovision imaging software on a Zeiss Axio Observer D1 microscope. Quantification of 5-HT1A, TPH or 5-HT1A/TPH positive cells was performed blindly using sections from three independent animals. The rabbit 5-HT1A antibody used in immunohistochemistry experiments has been characterized and validated in our previous study (Czesak et al., 2012).
Cell lines, plasmids, and luciferase assays
SKN-SH and SN48 cells were maintained in DMEM (Wisent) supplemented with 10% FBS, while Neuro2A cells were cultured in DMEM/F12 supplemented with 10%FBS at 37°C in 5% CO2. Full-length cDNA clones (Open Biosystems) for human Sp1 (MHS1011-9199783) and Sp4 (MHS4426-98361223) were amplified by PCR and the open reading frame cloned in the SmaI/HindIII sites of the pTriEx4 vector (Novagen) in frame with the N-terminal His and S-tags. The pGL3P/-681CpG reporter construct was obtained by cloning the conserved double stranded 25-bp sequence (AGGGAGAGGAGGGCGGGGACCCAGA) in the KpnI/XhoI sites of pGL3P. To study the impact of methylation of this site, double-stranded methylated or unmethylated oligonucleotides containing the 25-bp Sp1-like sequence were ligated overnight at 16 °C into the KpnI/XhoI sites of pGL3P using a 3:1 insert vector ratio. Prior to transfection, an aliquot of each ligation was resolved on a 1% agarose gel to ensure that ligation efficiencies were similar between samples.
For luciferase assays, 3×105 cells were plated in 6-well plates, allowed to attach overnight and transfected with 1 μg each of reporter, pTriEx4-Sp1, pTriEx4-Sp4 or empty vector and CMV-β-galactosidase using calcium phosphate or Lipofectamine 2000 (Invitrogen). Cells were incubated 36 to 48h post-transfection and lyzed with reporter lysis buffer. Luciferase activity was measured on a Victor V3 apparatus (Perkin Elmer) and normalized to β-galactosidase activity to correct for transient transfection efficiency.
EMSA
Sense and antisense oligonucleotides of the unmethylated -681mCpG -681 CpG site (5′-GAGAGGAGGGCGGGGACCCAGA) were generated with 5′GG overhangs, annealed and [32P]-dCTP-labelled with Klenow fragment DNA polymerase. Bacterially expressed and purified recombinant Sp4 protein (10ug/reaction) was pre-incubated with or without competition in a reaction mixture containing DNA binding buffer (20 mM HEPES, 0.2 mM EDTA, 0.2 mM EGTA, 100 mM KCl, 5% glycerol, 2 mM DTT, pH 7.9), poly-dIdC (1 μg) and probe (100,000 cpm/reaction) for 30 minutes at room temperature. Binding of Sp4 was competed by cold Sp1 consensus (5′-TTGCTTACGGGGCGGGGCGTCAATC) as well as cold methylated or unmethylated CpG -681 CpG site. Protein-DNA complexes were resolved on a non-denaturing 5% acrylamide gel at 4°C, which was then dried and exposed at −80°C with intensifying screens.
Chromatin Immunoprecipitation
Tissue from PFC and hippocampus was harvested from 12-wk old C57Bl6 mice and placed and minced in ice-cold PBS with protease inhibitors and then cross-linked in 1.67% formaldehyde in PBS for 20 min at 22°C. Crosslinking was halted with glycine for 5 min and samples were pelleted at 1000g and washed twice and resuspended in 1 mL PBS (plus protease inhibitors) per 25 mg tissue. The tissue was homogenized in a pre-chilled 7 mL glass dounce homogenizer, with 25 strokes of a large clearance pestle, followed by 25 strokes with a small clearance pestle. Cell suspensions were pelleted at 1000g, 4°C and resuspended in 100 ul/25mg tissue sample in ChIP buffer (50 mM Tris HCl, pH 7.4, with 150 mM NaCl, 1 mM EDTA, and 1% TRITON X-100) with protease inhibitors. Samples were incubated on ice for 10 min, then sonicated 14 × 30 sec. Fragment size was assessed on an agarose gel prior to immunoprecipitation. Cleared lysates (1500μg) were incubated overnight with Sp4-V20 (Santa Cruz Biotech) (1:2000) antibody at 4°C with rotation. The following day, 60 μL protein agarose A beads was added and tubes were incubated with rotation 2 h at 4°C. Beads were washed once with lysis buffer, four times in LiCl buffer (100 mM Tris-HCl, pH 7.5, 500 mM LiCl, 1% Nonidet P-40, and 1% deoxycholate) and once in TE before eluting in 1% SDS, 0.1M NaHCO3 at 65°C for 1 h. Samples were de-crosslinked with 0.2M NaCl overnight at 65°C. DNA was extracted with phenol/chloroform and precipitated with 2 vol of ethanol and 20μg linear acrylamide overnight at −20°C. DNA was analyzed by PCR with primers designed to amplify a 276-bp region (−870/−595 bp) of the putative Sp4 site. Primer sequences were: fwd 5′-CCTCTAGTTTAATAGGTAAGAGGC; rev 5′-CTTCTCACCTCTCTTTCTTTAGTC. Samples (15uL) of each elution and inputs (1:100) were used in a 25-uL reaction mixture containing 1X Thermopol buffer, 5mM MgSO4, 0.2uM forward and reverse primers, 200 uM dNTPs and 2.5U Taq DNA polymerase (NEB). Cycling parameters were as follows: 94°C × 1 min, and 40 cycles of 94°C × 20 s, 59°C × 30 s, 72°C × 45 s, with a final extension of 72°C × 10 min.
Statistical Analysis
Statistical analyses were performed using Statistica Software. Behavioral and physical test results were analysed by ANOVA with LSD post-hoc test or Kruskal-Wallis test followed by a Mann-Whitney U-test when required. Methylation, mRNA and protein expression data were analyzed using a one-way ANOVA followed by Tukey’s post-hoc test, while luciferase assays were analyzed using an unpaired t-test.
Results
Impact of UCMS and chronic imipramine on behavior of BALB/c mice
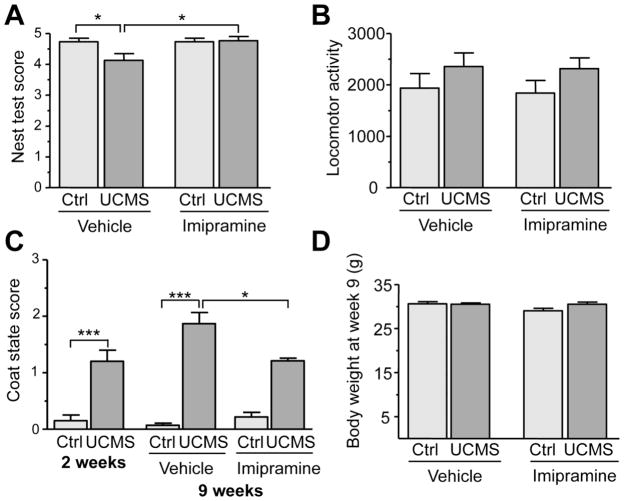
At the end of the UCMS procedure, self-oriented behaviors were assessed with the nest test and by determining the coat state of the animals, two tests indicative of a depressed phenotype (Nollet et al., 2013). Nest quality showed significant differences among groups in the Kruskal-Wallis test (H(3,58)=9.250386 p=0.026) (Fig. 1A) and was significantly reduced in the stressed/vehicle group compared to the non-stressed/vehicle group (p < 0.05). Treatment with imipramine significantly reversed this effect (p < 0.05 when compared to the stressed vehicle group). The Kruskal-Wallis test also revealed significant differences in the coat state among groups (H(3,56)=41.94482 p<0.0001). UCMS + vehicle induced a significant deterioration of coat state after 2 weeks that persisted at 9 weeks (Fig. 1C) compared to non-stressed vehicle group (p<0.000001). Treatment with imipramine, initiated after 2 weeks, significantly reversed the deterioration of coat state (stressed-vehicle group vs stressed-imipramine group, p< 0.05). Neither UCMS nor imipramine induced significant change in overall locomotor activity (Kruskal-Wallis test (H(3,52)=4.235444 p=0.237) (Fig. 1B). The body weight of the animals was also monitored and similarly to locomotor activity, no effect of UCMS or treatment was observed among the four groups (H(3,56)=6.774429 p=0.079) (Fig. 1D). In summary, the UCMS regimen led to a decrease in self-oriented behaviors (nest building and grooming), effects that were significantly reversed by chronic imipramine treatment, validating its usefulness as an animal model of depression and antidepressant response.
Figure 1. Behavioural phenotype of the BALB/c mice following UCMS and imipramine treatment.
Self-oriented behaviours, locomotor activity and weight of BALB/c mice of control mice (n=14–15) and stressed mice (n=11–15) treated with saline or imipramine (20mg/kg/day) at the end of the 9 week of the UCMS procedure. (A) The nest building ability (self-oriented behaviour) of control and stressed mice was scored after 24h using the Deacon scale. A score of 0 represents a complete lack of nest building while a score of 5 was given for a perfect nest. Data represents mean ±SEM. *p<0.05 Kruskal-Wallis H test. (B) Home cage locomotor activity of the animals was measured using an actograph for a 2 hour period. Data presented as mean ±SEM. No significant changes were observed. (C) The overall coat state of the animals was measured before initiating the imipramine treatment (after 2 weeks of UCMS) and at the end of the UCMS procedure for vehicle and imipramine treated mice (after the 9 week paradigm). A score of 0 was given to region where the coat was in a good state while a score of 1 was given for each region with poor condition. A total of 7 areas were scored for each animal. Data represents mean ±SEM. *p<0.05, ***p<0.01, Kruskal-Wallis H test. (D) Body weight of control and stressed mice treated with vehicle or imipramine at the end of the 9 week paradigm. Data represents mean ±SEM.
Chronic stress and imipramine treatment alter 5-HT1A receptor expression
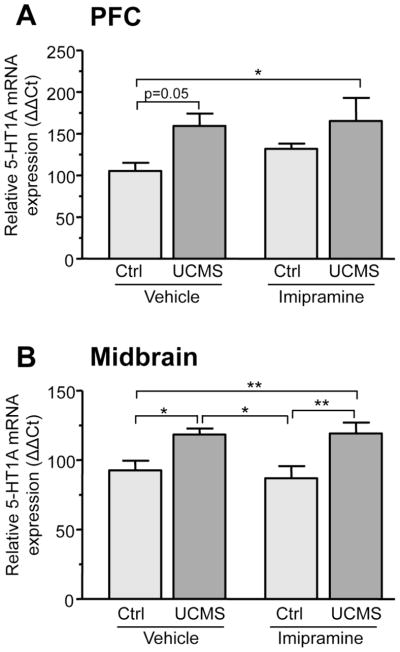
In order to determine the impact of stress and antidepressant treatment on 5-HT1A expression, we measured 5-HT1A mRNA and protein levels in pre- and post-synaptic brain compartments (midbrain and PFC). The PFC was chosen as a post-synaptic site of interest since several imaging and post-mortem studies indicate alterations in 5-HT1A receptor levels in this region in MDD and suicide (Arango et al., 1995; Parsey et al., 2010). Significant changes in 5-HT1A mRNA levels between groups occurred in both the PFC (F(3,31)=3.401 p=0.031) and the midbrain (F(3,20)=5.224 p=0.01). Compared to control animals, UCMS increased 5-HT1A mRNA levels in the PFC by 50% (Fig. 2A). Unexpectedly, imipramine treatment also showed a trend for a 30% increase in 5-HT1A mRNA levels in the PFC of control animals. Stressed animals treated with imipramine displayed higher 5-HT1A RNA levels compared to control, an effect that could be due to stress, imipramine or a combination of both. In the midbrain, UCMS also led to a 30% increase in 5-HT1A mRNA levels (Fig. 2B). In contrast to the PFC, imipramine treatment alone had no effect on 5-HT1A mRNA levels in midbrain. Furthermore, stressed animals treated with imipramine displayed 5-HT1A mRNA levels comparable to untreated stressed animals, indicating that imipramine did not reverse the increase in midbrain 5-HT1A RNA induced by stress. RNA levels of another midbrain serotonergic marker, tryptophan hydroxylase (TPH), were also measured and were unaffected by stress or imipramine (data not shown). Therefore, the changes in midbrain 5-HT1A levels are specifically induced by stress and do not reflect broad changes in serotonergic gene expression.
Figure 2. Altered 5-HT1A mRNA levels in the prefrontal cortex and midbrain of mice subjected UCMS and chronic imipramine.
Relative 5-HT1A mRNA expression in the PFC (A) and midbrain (B) of control and stressed animals treated with vehicle or imipramine. Relative 5-HT1A mRNA levels were calculated using the ΔΔCt method, with GAPDH serving as the reference gene. (A) Control animals treated with vehicle or imipramine (20mg/kg/day) (n=9) and animals subjected to UCMS treated with vehicle or imipramine (20mg/kg/day) (n=7). (B) Control animals treated with vehicle or imipramine (20mg/kg/day) (n=6) and animals subjected to UCMS treated with vehicle (n=5) or imipramine (20mg/kg/day) (n=4). **p<0.01, *p<0.05 One-way ANOVA Tukey’s posthoc analysis.
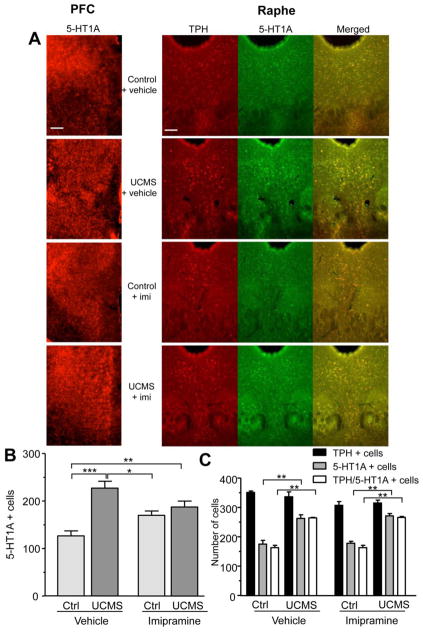
To determine whether the observed changes in 5-HT1A RNA resulted in changes in 5-HT1A protein levels, we examined 5-HT1A immunoreactivity in the medial PFC and dorsal raphe nucleus. Following stress, the number of 5-HT1A positive cells was increased in the prelimbic and infralimbic cortex by 80% (Fig. 3A and 3B). Imipramine treatment also increased the number of 5-HT1A labelled cells in both control and stressed animals, by 30% and 50% respectively. Therefore, in agreement with changes in 5-HT1A mRNA, both stress and chronic imipramine appear to induce the expression of the 5-HT1A in this cortical region. Similar to the mPFC, UCMS led to a 50% increase in the number of 5-HT1A-reactive cells in the dorsal raphe nucleus (Fig. 3A and 3C). In control animals, 5-HT1A immunoreactivity was detected in 50% of TPH+ neurons compared to almost 80% colocalization in stressed mice. This demonstrates that the increase in 5-HT1A was localized to serotonergic neurons and represented higher levels of 5-HT1A autoreceptors (Fig. 3C). Imipramine treatment did not alter control or stress-induced raphe 5-HT1A autoreceptor levels compared to untreated animals. Neither stress nor imipramine treatment affected the number of TPH+ neurons (Fig. 3A and 3C), confirming that the observed changes are specific to the 5-HT1A receptor and do not reflect changes in the overall number of serotonin neurons. In both the medial PFC and the raphe, changes in receptor levels paralleled changes observed in 5-HT1A mRNA levels (Fig. 2).
Figure 3. Altered 5-HT1A receptor levels in the medial prefrontal cortex and the raphe of mice subjected to UCMS and chronic imipramine.
(A) Immunohistochemistry staining of 5-HT1A receptors in the infralimbic cortex (left panel) and TPH (red) and 5-HT1A receptors (green) in the dorsal raphe nucleus (right panel) of control and stressed (UCMS) animals treated with vehicle or imipramine (imi, 20mg/kg/day). Representative images taken at 10x magnification are shown. Yellow represents colocalization between TPH and 5-HT1A signal in the DRN. Scale bar represents 100 microns. (B) Quantification of the number of 5-HT1A positive cells in the infralimbic cortex of control (C) and stressed (UCMS) animals treated with vehicle or imipramine. 3 sections were counted per group. Data represents mean ±SEM. ***p<0.001, **p<0.01, *p<0.05 One-way ANOVA Tukey’s posthoc analysis. (C) Quantification of the number TPH, 5-HT1A positive and TPH/5-HT1A positive cells in the DRN of control and stressed (UCMS) animals treated with vehicle or imipramine. The number of TPH positive cells (serotonergic neurons) is unaffected by stress or imipramine treatment, while the number of 5-HT1A positive cells and 5HT1A/TPH positive cells is increased following UCMS. Data presented as mean ±SEM. **p<0.01 by one-way ANOVA with Tukey’s post-hoc analysis vs. respective control animals (vehicle or imipramine-treated).
Altered DNA methylation at a conserved 5-HT1A promoter site following UCMS and imipramine treatment
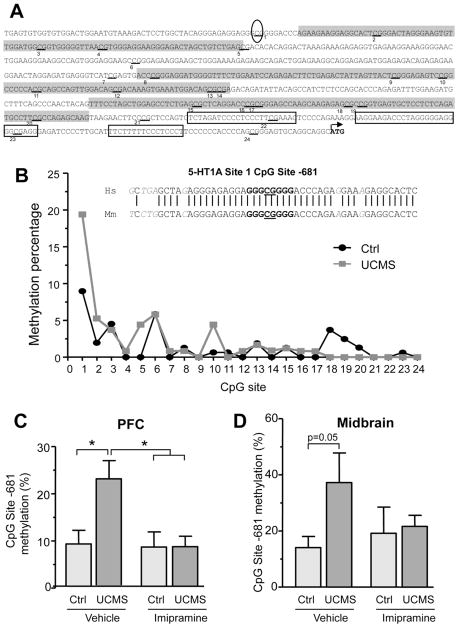
We hypothesized that stress and imipramine might alter the epigenetic status of the 5-HT1A promoter to enhance 5-HT1A receptor expression. The mouse 5-HT1A proximal promoter region extending to −750bp (relative to the ATG) contains a total 24 CpG sites and 3 predicted CpG islands, making it a potential target for DNA methylation (Fig. 4A). Therefore, we analyzed the DNA methylation profile of the 5-HT1A promoter in the PFC and the midbrain of control and stressed animals treated with vehicle or imipramine. In both brain regions, methylation of the proximal region immediately upstream of the coding sequence (from −1 bp to −350 bp) was almost undetectable while the methylation of the distal region of the promoter was more pronounced and ranged from 2 to 8% on average (Fig 4B). The CpG site located 681 bp upstream of the start codon not only displayed the highest level of basal DNA methylation but its methylation was significantly increased in the PFC (F(3,30)=4.383 p=0.012) and midbrain (F(3,28)= 3.023 p=0.05) of stressed animals (Fig. 4C and 4D). Overall DNA methylation of the 5-HT1A proximal promoter was not affected by stress or imipramine treatment (data not shown), indicating that the increase in DNA methylation occurred specifically at the -681 CpG site. Despite the limited conservation of the upstream 5-HT1A regulatory regions, the region containing this CpG site lies within a predicted Sp1 consensus binding site that is highly conserved between human and mouse (Fig. 4B, inset), suggesting a functional role for this site in the regulation of 5-HT1A expression. Importantly, stress-induced methylation of the -681 CpG site paralleled the increase in 5-HT1A RNA and protein observed in both midbrain and PFC. Chronic imipramine treatment, which reversed the depressive phenotype of stressed animals, reduced DNA methylation at this site to control levels in the PFC of UCMS mice (p<0.05) (Fig. 4C), while only a partial decrease in DNA methylation was observed in the midbrain (Fig. 4D). Notably, all midbrains samples from stressed animals (treated with imipramine or not) displayed methylation at the -681 CpG site, in sharp contrast to the control group where the majority of animals displayed limited or no methylation at this site. In summary, stress induced specific DNA methylation of the conserved 5-HT1A promoter -681 CpG site, an effect that was attenuated by chronic imipramine treatment, suggesting that epigenetic mechanisms may contribute to dys-regulation of 5-HT1A levels.
Figure 4. Differential DNA methylation of a conserved 5-HT1A promoter CpG site in the mouse PFC and midbrain following UCMS and imipramine treatment.
(A) Sequence and functional elements of the mouse 5-HT1A proximal promoter region (−724bp to +3bp relative to the translational start site). The three putative CpG islands are highlighted in grey and the 24 CpG sites are underlined and numbered. The previously characterized MAZ/Sp1 binding sites are boxed and the location of -681 CpG site is circled. (B) Methylation profile of the 24 CpG sites present in the mouse 5-HT1A proximal promoter (relative to the ATG) in control mice (n=9) and animal subjected to UCMS for 9 weeks (n=7). The data are presented as the mean methylation at each of the CpG sites; the SEM was within ±10% of the mean (not shown for clarity). Inset shows the alignment of CpG site-681 homologous sequences of the human (Hs, −691 to −654bp, top) and mouse (Mm, −699 to −662bp, bottom) 5-HT1A promoter. The predicted Sp1 consensus site is shown in bold. Methylation percentage at the -681bp CpG site in the prefrontal cortex (C) or the midbrain (D), of control animals treated with vehicle or imipramine (20mg/kg/day) (n=7–9) and animals subjected to UCMS treated with vehicle or imipramine (20mg/kg/day) (n=6–7). Data represents mean ±SEM. * p<0.05 One-way ANOVA, Tukey’s posthoc analysis.
Sp4 acts as a transcriptional repressor at the -681 CpG site
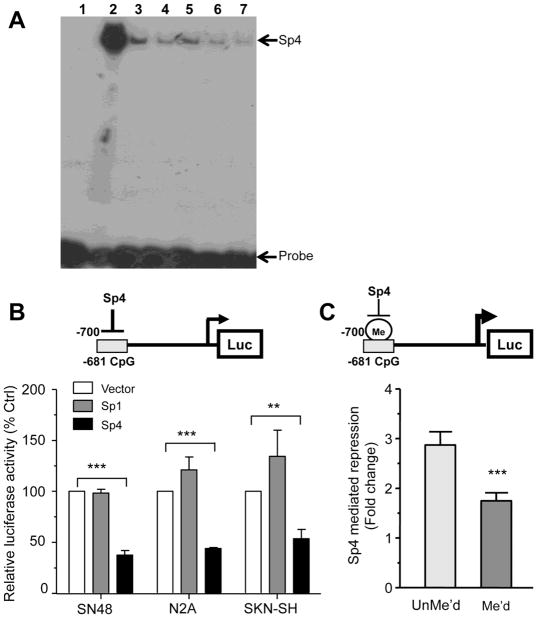
The -681 CpG site is within a putative Sp1-like DNA element and because its methylation correlates with higher expression levels of the 5-HT1A receptor in vivo, we hypothesized that this site may regulate 5-HT1A transcription. Since Sp4 is the predominant Sp1 homologue expressed in neurons (Mao et al., 2007), we first examined whether Sp4 can bind to this site using EMSA. Purified Sp4 protein bound specifically to the 5-HT1A conserved Sp1 element and was competed by excess unlabelled probe or Sp1 consensus site (Fig. 5A). However, the binding of Sp4 to the conserved element was equally competed by methylated and unmethylated oligonucleotides containing the conserved CpG element (Fig. 5A, lanes 3–6). Thus, although Sp4 binds specifically to the -681 CpG site, methylation does not impede Sp4 binding in vitro.
Figure 5. Sp4 repressor activity is decreased following methylation of 5-HT1A -681 CpG Site.
(A) Sp4 binds the Sp1-like element at CpG site -681 in vitro in EMSA assays. Bacterially-expressed Sp4 was incubated with a radio-labelled 5-HT1A CpG site -681 probe in the presence of 5- or 10-fold excess of methylated or unmethylated CpG site -681 double-stranded primers as well as Sp1 consensus oligonucleotides, and reactions were resolved on a 5% acrylamide gel. Band at the top of the gel represents Sp4/DNA complex while the free probe is at the bottom. Lane 1: CpG -681 probe alone; lane 2: CpG -681 probe + rSp4; lanes 3 & 4: CpG -681 probe+ rSp4 + 5x (3) or 10x (4) cold unmethylated CpG -681 competitor; lanes 5 & 6: CpG -681 probe+ rSp4 + 5x (5) or 10x (6) cold methylated CpG -681 competitor; lane 7: CpG -681 probe+ rSp4 + 10x cold Sp1 consensus. (B) Sp4 repressor activity at the -681 CpG element (model above). SN48, Neuro2A or SKN-SH cells were co-transfected with 1 ug of the pGL3P/-681CpG construct, Sp1, Sp4 or empty pTRIEX4 expression vector and pCMV β-gal. After 36h to 48h cells were lyzed and both luciferase and β-galactosidase activities were measured. Luciferase activity was normalized to β-galactosidase activity to correct for transfection efficiency. Data are presented as mean ±SEM of three independent experiments. ***p<0.001, **p<0.01, unpaired t-test. (C) Sp4 repressor activity is impaired by DNA methylation of 5-HT1A -684 CpG site. 25 bp oligomers containing methylated or unmethylated versions of the conserved promoter site were ligated in the KpnI/XhoI sites of pGL3P and transfected in Neuro2A cells along with the Sp4 or pTRIEX4 vector and pCMV β-gal. After 36h to 48h cells were lyzed and both luciferase and β-gal activities were measured. Luciferase activity was normalized to β-galactosidase activity to correct for transient transfection efficiency. Data presented as the ratio of pTRIEX4/pTRIEX4-Sp4 activities and is the mean ±SEM of four independent experiments. ***p<0.001 unpaired t-test.
In order to address the function of this element, the activity of both Sp1 and Sp4 on 5-HT1A transcription at the -681 bp CpG site was compared using a reporter construct containing the 25-bp conserved region upstream of the SV40 promoter (pGL3P CpG site -681). Sp4 repressed transcriptional activity of this reporter construct by 50-70% of control (pTRIEX4) in N2A, SN48, and SKN-SH cells (Fig. 5B), while Sp1 had no significant effect in any of these cell lines. These results demonstrate that Sp4 displays a strong repressor activity at the -681 CpG site. Consistent with previous studies showing that the 5-HT1A proximal promoter contains multiple MAZ/Sp1 sites that positively regulate the gene transcription (Parks and Shenk, 1996), Sp1 was able to stimulate the activity of a mouse 5-HT1A promoter-luciferase reporter (data not shown). To determine the effect of methylation at this site on Sp4 activity, we designed a reporter assay where methylated or unmethylated versions of the conserved -681 CpG site were cloned in pGL3P and co-transfected in N2A cells with Sp4 or vector plasmid (Fig. 5C). Sp4 repressed the unmethylated reporter construct by about 3-fold in N2A cells. Upon methylation of the -681 CpG site, Sp4 repression was attenuated to less than 2-fold. Therefore, methylation of the conserved CpG element reduced Sp4 repressor activity by 40% (Fig. 5C), while it had no effect on basal promoter activity. This demonstrates that specific methylation of the conserved -681 CpG site impairs Sp4 repressor activity, leading to increased 5-HT1A transcription.
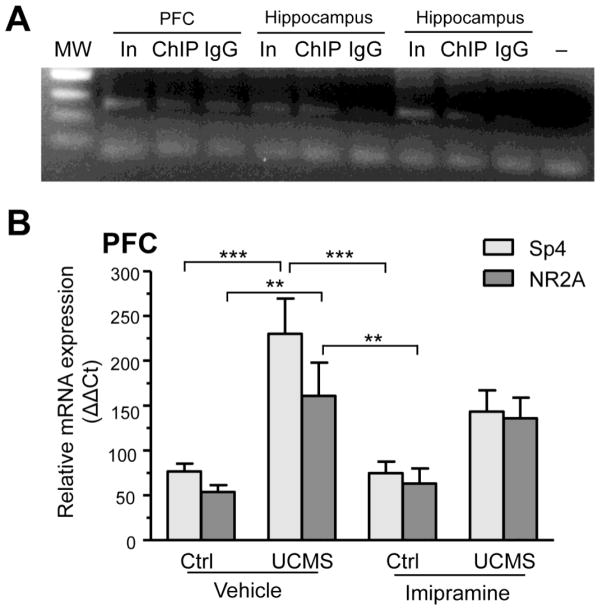
To determine whether Sp4 specifically binds the 5-HT1A promoter in vivo, we conducted Sp4 chromatin immunoprecipitation in brain extracts from PFC and hippocampus and amplified a 276-bp 5-HT1A promoter fragment containing the CpG-681 site by PCR. The 5-HT1A CpG-681 site was consistently enriched in Sp4 immunoprecipitates from hippocampus (compared to IgG control), however, we were unable to detect this enrichment in the PFC because of higher background levels (Fig. 6A). The preferential interaction of Sp4 with the 5-HT1A promoter in hippocampus may reflect the higher abundance of 5-HT1A receptors and Sp4 in this brain region compared to the PFC. Nonetheless, these data indicate that Sp4 binds to the 5-HT1A promoter at the CpG site -681 in vivo.
Figure 6. Sp4 is recruited to the 5-HT1A promoter in vivo and upregulated by stress but not imipramine.
(A) Binding of endogenous Sp4 to 5-HT1A promoter in vivo. Chromatin immunoprecipitation was done on tissue extracts of PFC and hippocampus using anti-Sp4 (ChIP) or control antibody (IgG). Following DNA purification, a 276-bp portion of the 5-HT1A promoter containing the CpG -681 site was amplified by PCR and resolved on a 2% agarose gel. Diluted input (In) was used as positive control while IgG or no template (−) served as a negative controls. Sp4 binding to the 5-HT1A promoter was detected in hippocampal samples but not PFC. A representative gel of three independent experiment is shown here. (B) Relative Sp4 and NR2A mRNA expression in the PFC of control and stressed animals treated with vehicle or imipramine. Relative mRNA levels were calculated using the ΔΔCt method, with GAPDH serving as the reference gene. Control animals treated with vehicle or imipramine (20mg/kg/day) (n=9) and animals subjected to UCMS treated with vehicle or imipramine (20mg/kg/day) (n=7). ***p<0.001, **p<0.01 one-way ANOVA Tukey’s posthoc analysis.
Finally, we addressed if impairment of Sp4 repression at the 5-HT1A promoter following UCMS involves a decrease in Sp4 expression by measuring Sp4 mRNA levels in the PFC and midbrain of control and UCMS mice. UCMS + vehicle led to a 3-fold increase in Sp4 levels in the PFC (Fig. 6B), a change expected to enhance repression of the 5-HT1A promoter. Furthermore, the mRNA level of NR2A (Grin2a), a neuronal gene positively regulated by Sp4 (Priya et al., 2013), was also increased by stress. A trend for an increase in both Sp4 and Grin2A RNA was also detected in the PFC of UCMS mice treated with imipramine, indicating that stress-induced changes were not fully reversed by imipramine. No significant changes in Sp4 or NR2A mRNA levels were observed in the midbrain of these animals (data not shown). Despite adaptive changes in Sp4 levels following UCMS, specific DNA methylation at the CpG-681 site attenuates Sp4-mediated repression of the 5-HT1A promoter leading to increased 5-HT1A expression.
Discussion
Impact of stress and antidepressants on 5-HT1A expression in adults
The unpredictable chronic mild stress (UCMS) is an animal model intended to mimic the persistent environmental stressors that lead to human depression (Ibarguen-Vargas et al., 2008; Yalcin et al., 2008). In accordance with previous studies, our results show that UCMS induced a deterioration of self-oriented behaviours indicative of a depressed phenotype, an effect that was reversed by chronic imipramine treatment. We used this model to address whether stress and/or antidepressants can impact 5-HT1A receptor expression in adults and present evidence of region-specific and plastic changes in 5-HT1A expression.
Importantly, we find that chronic adult stress consistently increases 5-HT1A RNA and receptor levels in both the medial PFC and the midbrain. Similarly, human depressed subjects display increased 5-HT1A receptor levels in the raphe (Boldrini et al., 2008; Stockmeier et al., 1998) and PFC compared to control subjects (Arango et al., 1995; Hesselgrave and Parsey, 2013). However, opposite findings have also been reported (Savitz et al., 2009), highlighting the need for animal models to better understand these changes. Studies in rodents revealed that the type of stress (e.g., escapable vs. inescapable; severe vs. mild; acute vs. chronic) plays an important role in determining changes in 5-HT1A receptor levels. In the chronic social defeat model, 5-HT1A RNA down-regulation has been reported in PFC (Kieran et al., 2010); on the other hand, acute restraint stress increased both prelimbic PFC and raphe 5-HT1A receptors (Fogaca et al., 2014), while others found no change (Steciuk et al., 2000). These differences could reflect the severity and duration of stress (Szewczyk et al., 2014), suggesting that changes in 5-HT1A levels may constitute an adaptive response to stress and/or alterations in 5-HT neurotransmission. Reduced activation of the mPFC region projecting to the raphe is implicated in the actions of stress, leading to depression-related behaviours in rodents (Amat et al., 2005; Warden et al., 2012). Conversely, stimulation of the mPFC in rodents and humans has an antidepressant effect (Covington et al., 2010; Nahas et al., 2010). Our findings show that following UCMS, depressed animals have higher 5-HT1A levels in the mPFC, a change expected to lower neuronal activity (Beique et al., 2004; Hajos et al., 2003) and have a pro-depression effect. Similarly, an increase in raphe 5-HT1A autoreceptor levels is expected to decrease serotonergic activity throughout the brain, consistent with lower 5-HT levels in depressed individuals (Asberg and Traskman, 1981; Mann, 1999).
In contrast to stress, the effect of chronic imipramine treatment on 5-HT1A expression was more complex. While imipramine had no effect in midbrain/DRN, in the PFC it increased 5-HT1A levels to almost the same extent as stress. Although stress and imipramine both increase 5-HT1A expression, they may up-regulate different 5-HT1A populations (interneurons vs. pyramidal cells) to oppositely regulate mPFC network excitability (Puig, Gulledge 2011 Mol Neurobiol) and behaviour. A stress-mediated increase in 5-HT1A expression in pyramidal neurons could be compensated by an imipramine-driven increase in 5-HT1A receptors on GABAergic interneurons leading to disinhibition of pyramidal neuronal firing (Hajos et al., 1999). In fact, chronic depression is associated with reduced PFC 5-HT1A receptor levels (Savitz et al., 2009; Stockmeier et al., 2009) that may reflect inability to compensate for severe stress. Also, increased 5-HT1A levels in mPFC interneurons could compensate for the stress-induced increase in 5-HT1A autoreceptors that persists following imipramine treatment. Further work is required to identify the cell types in which these stress-induced dynamic changes take place.
Epigenetic mechanisms mediate the effect of stress on 5-HT1A expression
Epigenetic modifications can induce long-lasting changes in gene expression and mediate the emergence of persistent behavioral phenotypes (Tsankova et al., 2007). Several studies have documented changes in histone modifications and gene expression in adult animals subjected to stress (Tsankova et al., 2006; Uchida et al., 2011); however, to date, alterations in DNA methylation have most often been documented following early life stress. Our data demonstrate that in stress-sensitive BALB/c mice, increased 5-HT1A levels following chronic stress are paralleled by increased DNA methylation at a unique conserved 5-HT1A promoter CpG site. The close association of 5-HT1A promoter methylation, mRNA and protein levels in the midbrain and PFC of UCMS mice suggests that the methylation of -681CpG site is the major driver of stress-induced 5-HT1A transcription in adult midbrain and PFC in vivo.
Following chronic imipramine treatment, stress-driven methylation of the CpG-681 was decreased in the PFC, demonstrating that in adult PFC changes in DNA methylation are dynamically regulated. In contrast, chronic imipramine only slightly decreased CpG-681 methylation levels in the midbrain, with a majority of animals still displayed stress-induced methylation after treatment. The maintenance of increased DNA methylation at this site may represent a persistent epigenetic mark that promotes increased 5-HT1A autoreceptor transcription and sustains vulnerability to depression. Interestingly, our findings are reminiscent of the chronic defeat stress model in which chronic imipramine reversed BDNF expression and the behavioural phenotype, despite the fact that repressive histone modification at the BDNF promoter remained unchanged (Tsankova et al., 2006). Previous studies have shown that early life stress can induce life-long changes in gene expression and behavior that is dependent on specific DNA methylation of a single regulatory CpG site, as shown for the GR and FKBP5 genes (Klengel et al., 2013; McGowan et al., 2009; Weaver et al., 2004). We find that chronic stress and antidepressant treatment in adult mice also converge on a specific conserved CpG site to alter 5-HT1A receptor expression, rather than impacting the global DNA methylation of the 5-HT1A promoter. Therefore, stress in adulthood can also induce long-term plasticity by altering DNA methylation and gene expression to affect behavior.
CpG -681 site methylation attenuates Sp4-mediated repression of the 5-HT1A promoter
Promoter methylation of CpG islands is generally associated with gene repression, but stress induced methylation of the 5-HT1A CpG-681 site correlated with increased 5-HT1A expression. We identified Sp4 as a transcriptional repressor of 5-HT1A gene transcription at this site, linking increased DNA methylation with increase expression. Sp4, the predominant neuronal Sp1 isoform, is known to function as a repressor or enhancer, depending on the promoter context (Chu et al., 2011; Priya et al., 2013; Ramos et al., 2009). Our data demonstrate that Sp4 specifically binds to the CpG-681 site to repress transcription and that methylation of this conserved site is sufficient to attenuate Sp4 repressor activity. Our in vitro assays show that this effect is indirect and probably involve the specific recruitment of methyl binding proteins such as MeCP2 that may compete with Sp4 to bind to the methylated -681 CpG site, to prevent repression or directly enhance transcription (Chahrour et al., 2008). Interestingly, both Sp4 and NR2A levels (a transcriptional target of Sp4) were significantly increased in PFC by stress. The increase in Sp4 may represent a compensatory response aimed at normalizing 5-HT1A expression levels and enhancing stress coping. However, increased DNA methylation at the CpG-681 site prevents effective repression by Sp4 despite the increased levels of the transcription factor following stress. Taken together, our data indicate that neuronal 5-HT1A expression is primarily regulated at the -681 CpG site by Sp4-mediated repression, and that DNA methylation at this site leads to increased expression by disrupting Sp4 function.
How DNA methylation is specifically targeted to the -681 CpG site remains unclear, but may involve Sp4 binding to the site and recruitment of DNMT3b to direct site-specific DNA methylation (Hervouet et al., 2009). DNMT3b is induced in the PFC of MDD individuals (Poulter et al., 2008) and could also mediate a genome-wide increase DNA methylation at Sp4 promoter sites. Interestingly, sequence variants in the Sp4 gene have been implicated in MDD or recurrent early onset MDD in genome-wide association studies (Shi et al., 2011; Shyn et al., 2011), suggesting that dysregulation of Sp4 or its target genes (such as 5-HT1A) could be involved in the etiology of MDD. Therefore, stress-induced DNA methylation of Sp4 target sites or altered levels of this transcription factor may play an important role in depression, especially given the role of this transcription factor in the regulation of 5-HT1A levels.
Highlights.
Chronic stress in adult mice induced depression that was reversed by imipramine.
Stress increased 5-HT1A RNA and protein in PFC and raphe.
Stress increased methylation of a 5-HT1A Sp1-like site that prevented Sp4 repression.
Stress-induced 5-HT1A methylation/expression was partly reversed by imipramine.
Adult stress induces reversible plasticity of methylation and gene expression.
Acknowledgments
This research was supported by grants from Canadian Institutes of Health Research (CIHR MOP-36437 and MOP-115098 to PRA), AstraZeneca/CIHR/Rx&D/Canadian Psychiatric Research Foundation Neurobiology Program (PRA). Equipment used in these studies was supported by Heart and Stroke Foundation Canadian Partnership for Stroke Recovery (PRA). Fellowship support was from CIHR Frederick Banting and Charles Best Canada Graduate Scholarship and Ontario Graduate Scholarship and Queen Elizabeth II Graduate Scholarships in Science and Technology (to AM); National Science and Engineering Research Council Scholarship and Undergraduate Research Opportunity Program (to JS).
Footnotes
The authors declare no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Albert PR, et al. Transcriptional dysregulation of 5-HT1A autoreceptors in mental illness. Mol Brain. 2011;4:21. doi: 10.1186/1756-6606-4-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert PR, Lemonde S. 5-HT1A Receptors, Gene Repression, and Depression: Guilt by Association. Neuroscientist. 2004;10:575–93. doi: 10.1177/1073858404267382. [DOI] [PubMed] [Google Scholar]
- Amat J, et al. Medial prefrontal cortex determines how stressor controllability affects behavior and dorsal raphe nucleus. Nat Neurosci. 2005;8:365–71. doi: 10.1038/nn1399. [DOI] [PubMed] [Google Scholar]
- Arango V, et al. Localized alterations in pre- and postsynaptic serotonin binding sites in the ventrolateral prefrontal cortex of suicide victims. Brain Res. 1995;688:121–33. doi: 10.1016/0006-8993(95)00523-s. [DOI] [PubMed] [Google Scholar]
- Asberg M, Traskman L. Studies of CSF 5-HIAA in depression and suicidal behaviour. Adv Exp Med Biol. 1981;133:739–52. doi: 10.1007/978-1-4684-3860-4_41. [DOI] [PubMed] [Google Scholar]
- Austin MC, et al. Localized decrease in serotonin transporter-immunoreactive axons in the prefrontal cortex of depressed subjects committing suicide. Neuroscience. 2002;114:807–15. doi: 10.1016/s0306-4522(02)00289-0. [DOI] [PubMed] [Google Scholar]
- Beique JC, et al. Serotonergic regulation of membrane potential in developing rat prefrontal cortex: coordinated expression of 5-hydroxytryptamine (5-HT)1A, 5-HT2A, and 5-HT7 receptors. J Neurosci. 2004;24:4807–17. doi: 10.1523/JNEUROSCI.5113-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldrini M, et al. Serotonin-1A autoreceptor binding in the dorsal raphe nucleus of depressed suicides. J Psychiatr Res. 2008;42:433–42. doi: 10.1016/j.jpsychires.2007.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chahrour M, et al. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science. 2008;320:1224–9. doi: 10.1126/science.1153252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu C, et al. Transcription factors Sp1 and Sp4 regulate TRPV1 gene expression in rat sensory neurons. Mol Pain. 2011;7:44. doi: 10.1186/1744-8069-7-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covington HE, 3rd, et al. Antidepressant effect of optogenetic stimulation of the medial prefrontal cortex. J Neurosci. 2010;30:16082–90. doi: 10.1523/JNEUROSCI.1731-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czesak M, et al. Increased serotonin-1A (5-HT1A) autoreceptor expression and reduced raphe serotonin levels in Deformed Epidermal Autoregulatory Factor-1 (Deaf-1) gene knock-out mice. Journal of Biological Chemistry. 2012;287:6615–27. doi: 10.1074/jbc.M111.293027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deacon RM. Assessing nest building in mice. Nat Protoc. 2006;1:1117–9. doi: 10.1038/nprot.2006.170. [DOI] [PubMed] [Google Scholar]
- Ducottet C, Belzung C. Behaviour in the elevated plus-maze predicts coping after subchronic mild stress in mice. Physiology and Behavior. 2004;81:417–26. doi: 10.1016/j.physbeh.2004.01.013. [DOI] [PubMed] [Google Scholar]
- Ducottet C, et al. Effects of the selective nonpeptide corticotropin-releasing factor receptor 1 antagonist antalarmin in the chronic mild stress model of depression in mice. Progress in Neuro-Psychopharmacology and Biological Psychiatry. 2003;27:625–31. doi: 10.1016/S0278-5846(03)00051-4. [DOI] [PubMed] [Google Scholar]
- Fogaca MV, et al. Effects of intra-prelimbic prefrontal cortex injection of cannabidiol on anxiety-like behavior: involvement of 5HT1A receptors and previous stressful experience. Eur Neuropsychopharmacol. 2014;24:410–9. doi: 10.1016/j.euroneuro.2013.10.012. [DOI] [PubMed] [Google Scholar]
- Franklin K, Paxinos G. The Mouse Brain in Stereotaxic Coordinates with CDROM. Academic Press; New York, NY, USA: 2007. [Google Scholar]
- Hajos M, et al. In vivo inhibition of neuronal activity in the rat ventromedial prefrontal cortex by midbrain-raphe nuclei: role of 5-HT1A receptors. Neuropharmacology. 2003;45:72–81. doi: 10.1016/s0028-3908(03)00139-4. [DOI] [PubMed] [Google Scholar]
- Hajos M, et al. Role of the medial prefrontal cortex in 5-HT1A receptor-induced inhibition of 5-HT neuronal activity in the rat. Br J Pharmacol. 1999;126:1741–50. doi: 10.1038/sj.bjp.0702510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hervouet E, et al. Dnmt3/transcription factor interactions as crucial players in targeted DNA methylation. Epigenetics. 2009;4:487–99. doi: 10.4161/epi.4.7.9883. [DOI] [PubMed] [Google Scholar]
- Hesselgrave N, Parsey RV. Imaging the serotonin 1A receptor using [11C]WAY100635 in healthy controls and major depression. Philos Trans R Soc Lond B Biol Sci. 2013;368:20120004. doi: 10.1098/rstb.2012.0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibarguen-Vargas Y, et al. Multifaceted strain-specific effects in a mouse model of depression and of antidepressant reversal. Psychoneuroendocrinology. 2008;33:1357–68. doi: 10.1016/j.psyneuen.2008.07.010. [DOI] [PubMed] [Google Scholar]
- Jans LA, et al. Serotonergic vulnerability and depression: assumptions, experimental evidence and implications. Mol Psychiatry. 2007;12:522–43. doi: 10.1038/sj.mp.4001920. [DOI] [PubMed] [Google Scholar]
- Jovanovic H, et al. Chronic stress is linked to 5-HT(1A) receptor changes and functional disintegration of the limbic networks. Neuroimage. 2011;55:1178–1188. doi: 10.1016/j.neuroimage.2010.12.060. [DOI] [PubMed] [Google Scholar]
- Karg K, et al. The Serotonin Transporter Promoter Variant (5-HTTLPR), Stress, and Depression Meta-analysis Revisited: Evidence of Genetic Moderation. Arch Gen Psychiatry. 2011;68:444–54. doi: 10.1001/archgenpsychiatry.2010.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieran N, et al. Chronic social defeat downregulates the 5-HT1A receptor but not Freud-1 or NUDR in the rat prefrontal cortex. Neurosci Lett. 2010;469:380–4. doi: 10.1016/j.neulet.2009.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishi T, et al. The serotonin 1A receptor gene confer susceptibility to mood disorders: results from an extended meta-analysis of patients with major depression and bipolar disorder. European Archives of Psychiatry and Clinical Neuroscience. 2013;263:105–18. doi: 10.1007/s00406-012-0337-4. [DOI] [PubMed] [Google Scholar]
- Klengel T, et al. Allele-specific FKBP5 DNA demethylation mediates gene-childhood trauma interactions. Nat Neurosci. 2013;16:33–41. doi: 10.1038/nn.3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lopez-Figueroa AL, et al. Serotonin 5-HT1A, 5-HT1B, and 5-HT2A receptor mRNA expression in subjects with major depression, bipolar disorder, and schizophrenia. Biol Psychiatry. 2004;55:225–33. doi: 10.1016/j.biopsych.2003.09.017. [DOI] [PubMed] [Google Scholar]
- Mann JJ. Role of the serotonergic system in the pathogenesis of major depression and suicidal behavior. Neuropsychopharmacology. 1999;21:99S–105S. doi: 10.1016/S0893-133X(99)00040-8. [DOI] [PubMed] [Google Scholar]
- Mao X, et al. Glutamate receptor activation evokes calpain-mediated degradation of Sp3 and Sp4, the prominent Sp-family transcription factors in neurons. Journal of Neurochemistry. 2007;100:1300–14. doi: 10.1111/j.1471-4159.2006.04297.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGowan PO, et al. Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. Nat Neurosci. 2009;12:342–8. doi: 10.1038/nn.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahas Z, et al. Bilateral epidural prefrontal cortical stimulation for treatment-resistant depression. Biological Psychiatry. 2010;67:101–9. doi: 10.1016/j.biopsych.2009.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nollet M, et al. Models of depression: unpredictable chronic mild stress in mice. Curr Protoc Pharmacol. 2013;Chapter 5(Unit 5):65. doi: 10.1002/0471141755.ph0565s61. [DOI] [PubMed] [Google Scholar]
- Northoff G. Gene, brains, and environment-genetic neuroimaging of depression. Current Opinion in Neurobiology. 2013;23:133–42. doi: 10.1016/j.conb.2012.08.004. [DOI] [PubMed] [Google Scholar]
- Parks CL, Shenk T. The serotonin 1a receptor gene contains a TATA-less promoter that responds to MAZ and Sp1. Journal of Biological Chemistry. 1996;271:4417–30. doi: 10.1074/jbc.271.8.4417. [DOI] [PubMed] [Google Scholar]
- Parsey RV, et al. Higher serotonin 1A binding in a second major depression cohort: modeling and reference region considerations. Biol Psychiatry. 2010;68:170–8. doi: 10.1016/j.biopsych.2010.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulter MO, et al. GABAA receptor promoter hypermethylation in suicide brain: implications for the involvement of epigenetic processes. Biol Psychiatry. 2008;64:645–52. doi: 10.1016/j.biopsych.2008.05.028. [DOI] [PubMed] [Google Scholar]
- Priya A, et al. Specificity protein 4 functionally regulates the transcription of NMDA receptor subunits GluN1, GluN2A, and GluN2B. Biochim Biophys Acta. 2013;1833:2745–56. doi: 10.1016/j.bbamcr.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos B, et al. Sp4-dependent repression of neurotrophin-3 limits dendritic branching. Mol Cell Neurosci. 2009;42:152–9. doi: 10.1016/j.mcn.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santarelli L, et al. Requirement of hippocampal neurogenesis for the behavioral effects of antidepressants. Science. 2003;301:805–9. doi: 10.1126/science.1083328. [DOI] [PubMed] [Google Scholar]
- Savitz J, et al. 5-HT(1A) receptor function in major depressive disorder. Prog Neurobiol. 2009;88:17–31. doi: 10.1016/j.pneurobio.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, et al. Genome-wide association study of recurrent early-onset major depressive disorder. Mol Psychiatry. 2011;16:193–201. doi: 10.1038/mp.2009.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyn SI, et al. Novel loci for major depression identified by genome-wide association study of Sequenced Treatment Alternatives to Relieve Depression and meta-analysis of three studies. Mol Psychiatry. 2011;16:202–15. doi: 10.1038/mp.2009.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steciuk M, et al. Acute stress does not alter 5-HT1A receptor density. Progress in Neuro-Psychopharmacology and Biological Psychiatry. 2000;24:155–61. doi: 10.1016/s0278-5846(99)00078-0. [DOI] [PubMed] [Google Scholar]
- Stockmeier CA, et al. Antagonist but not agonist labeling of serotonin-1A receptors is decreased in major depressive disorder. J Psychiatr Res. 2009;43:887–94. doi: 10.1016/j.jpsychires.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockmeier CA, et al. Increase in serotonin-1A autoreceptors in the midbrain of suicide victims with major depression-postmortem evidence for decreased serotonin activity. J Neurosci. 1998;18:7394–401. doi: 10.1523/JNEUROSCI.18-18-07394.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szewczyk B, et al. Stress-induced alterations in 5-HT1A receptor transcriptional modulators NUDR and Freud-1. Int J Neuropsychopharmacol. 2014:1–13. doi: 10.1017/S146114571400100X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsankova N, et al. Epigenetic regulation in psychiatric disorders. Nat Rev Neurosci. 2007;8:355–67. doi: 10.1038/nrn2132. [DOI] [PubMed] [Google Scholar]
- Tsankova NM, et al. Sustained hippocampal chromatin regulation in a mouse model of depression and antidepressant action. Nat Neurosci. 2006;9:519–25. doi: 10.1038/nn1659. [DOI] [PubMed] [Google Scholar]
- Uchida S, et al. Epigenetic status of Gdnf in the ventral striatum determines susceptibility and adaptation to daily stressful events. Neuron. 2011;69:359–72. doi: 10.1016/j.neuron.2010.12.023. [DOI] [PubMed] [Google Scholar]
- Vialou V, et al. Epigenetic mechanisms of depression and antidepressant action. Annual Review of Pharmacology and Toxicology. 2013;53:59–87. doi: 10.1146/annurev-pharmtox-010611-134540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warden MR, et al. A prefrontal cortex-brainstem neuronal projection that controls response to behavioural challenge. Nature. 2012;492:428–32. doi: 10.1038/nature11617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver IC, et al. Epigenetic programming by maternal behavior. Nat Neurosci. 2004;7:847–54. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- Willner P, et al. Chronic mild stress-induced anhedonia: a realistic animal model of depression. Neuroscience and Biobehavioral Reviews. 1992;16:525–34. doi: 10.1016/s0149-7634(05)80194-0. [DOI] [PubMed] [Google Scholar]
- Yalcin I, et al. Mouse strain differences in the unpredictable chronic mild stress: a four-antidepressant survey. Behav Brain Res. 2008;193:140–3. doi: 10.1016/j.bbr.2008.04.021. [DOI] [PubMed] [Google Scholar]
- Zhang KR, et al. The combined effects of the 5-HTTLPR and 5-HTR1A genes modulates the relationship between negative life events and major depressive disorder in a Chinese population. Journal of Affective Disorders. 2009;114:224–231. doi: 10.1016/j.jad.2008.07.012. [DOI] [PubMed] [Google Scholar]