Abstract
Immunological protection against microbial pathogens is dependent on robust generation of functionally diverse T lymphocyte subsets. Upon microbial infection, naïve CD4+ or CD8+ T lymphocytes can give rise to effector- and memory-fated progeny that together mediate a potent immune response. Recent advances in single-cell immunological and genomic profiling technologies have helped elucidate early and late diversification mechanisms that enable the generation of heterogeneity from single T lymphocytes. We discuss these findings here and argue that one such mechanism, asymmetric cell division, creates an early divergence in T lymphocyte fates by giving rise to daughter cells with a propensity towards the terminally differentiated effector or self-renewing memory lineages, with cell-intrinsic and -extrinsic cues from the microenvironment driving the final maturation steps.
Keywords: adaptive immunity, asymmetric division, T lymphocyte fate determination, T lymphocyte differentiation, single-cell analyses
Diversification of T lymphocyte fates in the immune response to infection
Heterogeneity in T lymphocyte responses to microbial infection is essential for establishing protective immunity. Upon activation, naïve antigen-specific T cells differentiate into two distinct classes of cellular progeny: terminal effector cells that mediate acute protection and self-renewing memory cells that provide long-term protective immunity. It is now well appreciated that substantial heterogeneity in effector and memory subsets can arise from a single activated T lymphocyte. However, a fundamental question in the field remains unanswered: how does this cellular diversity arise from a single cell?
Various models incorporating the influence of TCR signal-strength, environmental cues, and transcriptional changes on T cell differentiation have been proposed and discussed extensively by others (reviewed by Kaech and Cui [1] and discussed by Ahmed, et al. [2]). One possibility is that the progeny of an activated T lymphocyte differentiate along a linear pathway, with cells transiting through an equipotent effector phase before a subset of these cells later diverges to form the memory lymphocyte pool [3, 4]. This model can be envisioned as one of late divergence, such that diversification into distinct T cell fates does not occur until later stages of the immune response. The signal-strength model proposes that the initial strength of TCR and cytokine signals received during priming dictate whether the progeny of an activated naïve T cell will adopt a terminal effector or memory cell fate. The decreasing-potential model proposes that memory-fated T cells arise as a result of fewer cumulative encounters with antigen and cytokines during the immune response, and are therefore less differentiated than effector-fated T cells. A fourth model is an early divergent model of T lymphocyte diversification whereby a propensity towards terminal effector or self-renewing memory fates can be conferred as early as the first cell division owing to asymmetric division [5], with continued maturation toward the final cell fates controlled by a combination of cell-intrinsic and -extrinsic signals. During an asymmetric cell division, important fate determinants and cellular components, including protein, RNA, and organelles, are unequally inherited by the two nascent daughter cells, thus enabling them to adopt distinct fates [6]. Importantly, it should be emphasized that these models are not mutually exclusive, and data supporting one model do not necessarily rule out other models.
Differentiation into CD4+ and CD8+ T cell effector and memory fates is driven, in part, by the expression and activities of several transcription factors, as demonstrated by prior investigation of the transcriptional networks and molecular mechanisms that control T lymphocyte fate specification [7–11]. While the traditional microarray approaches used by these studies have provided fundamental insights into the molecular basis and timing in gene expression changes underlying the functional differences in effector and memory CD4+ and CD8+ T cell subsets, it should be noted that these analyses were performed on bulk cell populations. As such, conclusions from these studies have been restricted to the cell population level, and cell-to-cell differences in gene expression may have been hidden as a result of averaged gene expression from seemingly homogeneous populations of cells. Recent technological advances in high-throughput single-cell gene expression profiling, such as microfluidics-based qRT-PCR analyses and RNA sequencing, have been combined with computational analyses to analyze the in vivo transcriptional changes in thousands of single cells in an array of diverse biological systems [12–15]. Systematic modeling of temporal changes in single-cell transcription pattern dynamics has uncovered substantial heterogeneity within a number of diverse cell populations, including immune cells [16, 17], murine embryonic tissue [14], human colon tumors [13] and primary glioblastomas [18]. Moreover, cell-intrinsic fate determinants critical in driving the formation of cellular diversity have been identified [14, 19]. For instance, high expression of Id2 and Sox2 have been found to indicate early fate commitment into the outer and inner cell lineages, respectively, during mouse embryogenesis [14], thus highlighting the importance of dissecting gene expression heterogeneity at the single-cell level.
Tracking individual lymphocytes as they progress through the early stages of the immune response has been difficult due to biological and technical constraints, such as the inability to sample adequate endogenous antigen-experienced cell numbers due to low precursor frequencies of cells specific for a particular antigen (on the order of 10 to 100) [20, 21]. Recent advances in magnetic bead-based strategies have enabled the enrichment of antigen-specific T cells at early phases of the immune response, during which these cells are virtually undetectable [20]. Combining the approaches described above has recently made it possible to analyze transcriptional changes in individual T lymphocytes early after microbial infection [16], thereby providing some initial insights into two fundamental questions: how is T cell diversification achieved and when does this divergence in fates occur?
Here, we explore these questions as we discuss recent studies aimed at interrogating the pathways by which single activated T cells differentiate towards effector- and memory-fated lineages. We highlight how asymmetric division is exploited by T lymphocytes to yield robust immune responses and draw attention to several gaps in our current understanding of how asymmetric division may shape T lymphocyte diversification. A detailed understanding of how and when T lymphocyte fate specification occurs may have far-reaching implications in the design of vaccination and therapeutic approaches to enhance long-term protective immunity against infectious agents.
Generating T lymphocyte diversity from a single cell
It is well established that heterogeneity in CD8+ and CD4+ T cell responses is required for robust immunity [22]. For the purposes of this review, we will focus on terminal effector CD8+ T cells, long-lived central memory (TCM) and effector memory (TEM) CD8+ T cells (see Glossary), CD4+ T helper type 1 (TH1) cells, and CD4+ follicular helper T (TFH) cells. Pioneering in vivo cell tracing studies provided the first experimental evidence to support the idea that heterogeneous cellular progeny can be derived from a single activated naïve T cell. Terminal effector (KLRG1hiIL-7Rlo), TEM (CD44hiCD62Llo), and TCM (CD44hiCD62Lhi) CD8+ T lymphocyte subsets were shown to arise from a single T cell receptor (TCR) transgenic OT-1 CD8+ T cell adoptively transferred into a congenic recipient infected with Listeria monocytogenes expressing ovalbumin (Lm-OVA) [23]. The development of ‘DNA-barcode’ technologies, in which DNA sequences (barcodes) are retrovirally introduced into thymocytes, has permitted the generation of naïve T cells harboring genetic tags [24]. This strategy has allowed a single barcode-labeled naïve T cell and its progeny to be traced following in vivo infection to better understand the developmental histories of individual cells [24, 25]. Applications of limiting dilution strategies have shown that pathogen-induced environmental cues influence the differentiation path of single activated CD8+ T cells responding to Lm-OVA or vesicular stomatitis virus infection [26] and that diversity derived from single CD4+ T lymphocytes can also be achieved in response to several attenuated Lm strains [27]. In the latter study, single naïve CD4+ T lymphocytes were capable of producing each of the TH1, TFH, and germinal center TFH effector subsets; however, the ratios of these subsets within the generated effector pool were found to be influenced by the peptide:MHCII dwell times specific to unique TCRs [27].
The aforementioned studies have undoubtedly illustrated the capacity of a single lymphocyte to give rise to differentially fated cellular progeny and have highlighted the influence of TCR avidity, pathogen-specific differences, and the cytokine milieu on the generation of cellular diversity. However, single-cell transfer experiments using limiting dilution approaches should be interpreted with caution as the adoptive transfer of a single cell is dependent on Poisson probabilities [26] and the assumption that ~20% of transferred cells will survive in recipient mice [27]; thus, it is a challenge to guarantee the transfer of an individual cell. Nonetheless, a key question remains unanswered – how do the progeny of a single activated T cell differentiate into effector and memory cells? While the analysis of antigen-specific T cell populations during the early (days 6–8 post-infection) and late (days 30 and later post-infection) phases of the immune response in these studies [23, 24, 26, 27] has informed us that single activated naïve T lymphocytes can give rise to distinct subsets during primary and secondary infections, they do not reveal how or when terminal effector and memory subsets are formed.
Early acquisition of disparate transcriptional programs
In recent years, improvements in T cell enrichment techniques along with advances in single-cell genomics approaches and computational modeling analyses have made it possible to begin to dissect how and when single T lymphocytes differentiate into terminal effector and memory subsets. We recently investigated how an activated naïve CD8+ T lymphocyte differentiates in vivo into one of three fates: terminal effector, TCM, and TEM cell [16]. Single-cell gene expression analyses of activated CD8+ T cells at sequential timepoints following Lm-OVA infection suggested that an early divergent model may be the most likely pathway that underlies T lymphocyte fate specification, at least in this experimental system [16] (Figure 1A; Key Figure). Unsupervised clustering analysis revealed that single CD8+ T cells exhibit marked molecular heterogeneity during early phases of the immune response (at the first division and at day 3 post-infection), owing to acquisition of distinct transcriptional programs conferring disparate propensities towards the terminal effector versus memory cell fates. Our findings raised the possibility that T lymphocyte fates may already begin to be specified prior to the onset of phenotypic differences in cell surface marker expression, such as KLRG1 and IL-7R, that are observed only at later stages of infection [28].
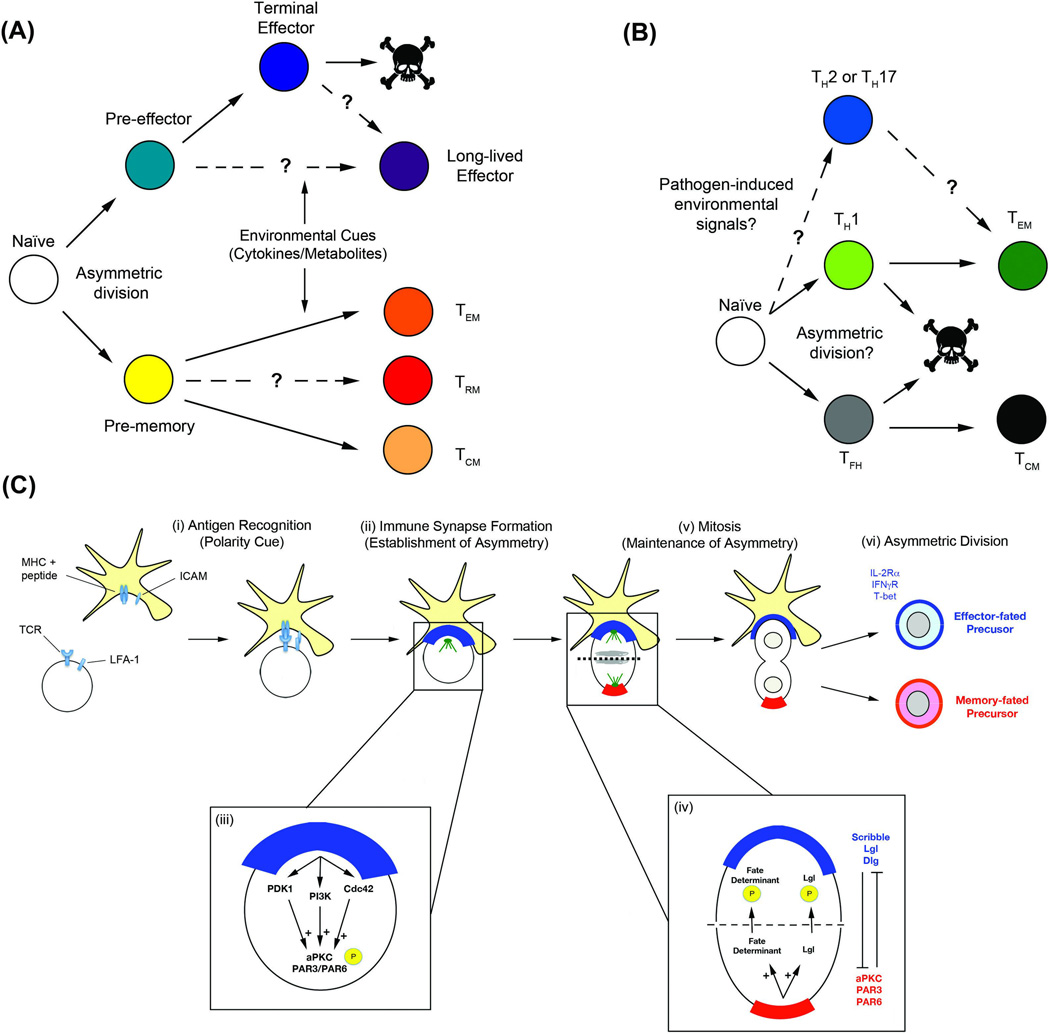
Figure 1.
T lymphocyte differentiation into diverse fates. (A) Hypothetical model of CD8+ T lymphocyte fate specification. Asymmetric division of an activated naïve cell (white) gives rise to pre-effector (teal) and pre-memory (yellow) daughter cells, while environmental cues, such as cytokines and metabolites, may differentially affect progression into the terminal effector (blue) or TEM (orange) and TCM (light orange) cell fates, respectively. Dotted lines indicate the proposed ontogeny of long-lived effector (dark blue) and TRM (red) cells. (B) Hypothetical model of CD4+ T lymphocyte fate specification. Differentiation into TH1 effector (light green) and TFH (gray) cells diverges early during an immune response to intracellular pathogens, as a possible result of asymmetric division. A subset of TH1 effector cells survives to form the TEM pool (dark green), while a subset of TFH cells survive to form TCM cells (black). It remains to be determined whether infections by extracellular pathogens induce divergent pathways of differentiation into TH2 or TH17 effector cells (blue) and TFH/TCM cells. (C) Potential mechanisms regulating establishment and maintenance of asymmetric T lymphocyte division. (i) An antigen-presenting cell provides an initial polarity cue (MHC/peptide and ICAM) to a T lymphocyte through interaction with the TCR and LFA-1 [5, 57], leading to (ii) formation of an immune synapse and establishment of asymmetry. (iii) Critical components of the immune synapse, including PDK1 [88], PI3K [89], and Cdc42 [90], which have been previously shown to be activators of aPKC in adipocytes and Drosophila and C. elegans [91–93], may function similarly in T lymphocytes to transmit the initial polarity cue to aPKC. (iv) Activated aPKC, bound with PAR3 and PAR6 [58], can then phosphorylate, and thereby exclude, important fate determinants that enter the distal pole (red) of the T lymphocyte, thus (v) maintaining asymmetry through the completion of division. In Drosophila and C. elegans, the Scribble and PAR complexes have an antagonistic relationship (i.e. aPKC phosphorylates Lgl to induce polarization, whereas Lgl represses aPKC activity), which serves to restrict aPKC activity to one pole of a dividing cell and coordinate the maintenance of asymmetry [94]. It remains to be determined whether a similar antagonistic mechanism functions in T lymphocytes to facilitate (vi) the generation of effector-fated daughter cells, which express IL-2Ra, IFNγR, and T-bet (blue), and memory-fated daughter cells via asymmetric division. Abbreviations: TEM, effector memory cell; TCM, central memory cell; TRM, tissue resident memory cell; TH, helper T cell; TFH, follicular helper T cell; TCR, T cell receptor; LFA, lymphocyte function-associated antigen; aPKC, atypical protein kinase C; PDK1, phosphoinositide-dependent kinase-1; PI3K, phosphoinositide-3 kinase; Cdc42, cell division control protein 42; PAR, partitioning defective; Lgl, lethal giant larvae; IL-2Rα, interleukin-2 receptor α; IFNγR, interferon-γ receptor.
To test this possibility, we applied a Hidden Markov Model (HMM) approach to capture dynamic changes in gene expression patterns of single activated CD8+ T lymphocytes derived at multiple intermediate time points (first division, days 3 and 5 post-infection) between the naïve ‘state’ and a differentiated ‘fate’ (terminal effector, TCM, or TEM cell). Coordinated changes in the transcriptional profiles of antigen-experienced CD8+ T cells were associated with the progression towards disparate transitional states and eventual fates. Most notably, high and low expression of Il2ra were predictive of cells that had adopted a pre-terminal effector versus pre-memory path of differentiation, respectively. In support of this observation, CD62LloIL-2Rαhi CD8+ T cells within the population of cells that had undergone their first division in vivo exhibited enhanced production of IFNγ and Granzyme B upon ex vivo stimulation. Conversely, first division CD62LhiIL-2Rαlo cells appeared to represent pre-memory cells, as they survived long-term following adoptive transfer into infection-matched recipients and mounted a robust proliferative response to rechallenge. The observation of asymmetric IL-2Rα segregation in CD8+ T lymphocytes undergoing their first division in vivo suggested that asymmetric division might play a role in mediating this early divergence in transcriptional programming.
Asymmetric division regulates early T lymphocyte fate diversification
Our recent work characterizing CD8+ T lymphocytes deficient in either isoform of the evolutionary conserved polarity protein, atypical protein kinase C (aPKC), has provided some of the first evidence suggesting that asymmetric division is an important first step in specifying T lymphocyte fates [29]. The aPKC isoforms - PKCζ and PKλ/ι - have been identified as important regulators of asymmetry in Drosophila and C. elegans (Box 1) and function during the first division of activated naïve CD8+ T lymphocytes to mediate asymmetric segregation of effector fate-associated factors, including IL-2Rα, IFNγR, and T-bet. Loss of either aPKC isoform increased the symmetric distribution of these effector fate-associated molecules, resulting in a striking reduction in the molecular heterogeneity exhibited by individual cells that had undergone their first division and increased the proportion of effector-fated precursor cells. Although asymmetric division was only partially reduced, the subsequent alterations to the initial balance of effector-versus memory-fated precursor cells increased differentiation towards the effector T lymphocyte fates at the expense of memory T cell formation [29]. While complete ablation of asymmetric division would be necessary to definitively test the full extent of its role in cell fate specification, the currently available data suggest that asymmetric division, by virtue of excluding important effector fate-associated factors from one of the two nascent daughter cells, enables the simultaneous generation of effector- and memory-fated precursor cells.
Box 1. Asymmetric cell division in Drosophila and C. elegans.
Asymmetric division has been well studied in Drosophila neuroblasts [95] and C. elegans embryos, which undergo one or more rounds of asymmetric division to yield self-renewing and terminally differentiating cell lineages [96]. Intrinsic and extrinsic polarity and spatial cues regulate the initiation and advancement into and through an asymmetric division [97]. The process is often guided by a pair of evolutionarily conserved polarity complexes, including the Scribble complex, which contains Scribble, Discs large (Dlg), and Lethal giant larvae (Lgl), and the partition defective (Par) complex, which contains Par3, Par6, and atypical protein kinase C (aPKC) [98–100]. In Drosophila and C. elegans model systems, these polarity proteins play fundamental roles in both establishing and maintaining asymmetry following an initial symmetry breaking event [101].
In a dividing Drosophila neuroblast, aPKC phosphorylates Lgl [99, 102], which serves to inactivate Lgl and restrict its localization to one pole of the dividing cell [103]. Conversely, Lgl can bind to aPKC, repressing its activity and restricting aPKC to the opposite pole [104, 105]. Thus, an antagonistic relationship between the Scribble and Par complexes serves to establish and maintain an axis of polarity within a dividing neuroblast following an initial extrinsic polarity cue [6]. In addition to Lgl, aPKC can phosphorylate Numb [106], which is an inhibitor of Notch signaling [107], and Miranda [98], which binds to the fate determinants, Prospero and Brain tumor (Brat) [108], causing these proteins to segregate into the daughter cell destined for terminal differentiation. In this manner, aPKC functions to direct protein localization during division and regulate the balance between self-renewal and terminal differentiation [6, 94].
Activating proteins upstream of the Par polarity complex in Drosophila may exert a threshold effect that regulates asymmetric division [109] similar to that induced by TCR signal strength in T lymphocytes [57]. The cell cycle kinase, Cdc2, can activate Aurora A [110], which in turn can activate aPKC [111]. Furthermore, the levels of Cdc2 within a dividing Drosophila neuroblast determine whether or not a cell divides and whether or not the division is asymmetric [109]. While it remains unclear whether intrinsic or extrinsic cues are responsible for setting the levels of Cdc2, these types of threshold effects may be important for dictating cell fate by balancing the frequencies of asymmetric versus symmetric divisions. Future studies that utilize lessons in polarity learned from Drosophila and C. elegans model systems will be critical for understanding the biology of asymmetric division by T lymphocytes.
Asymmetric division has been observed in CD4+ T cells responding in vivo to Leishmania infection [5], raising the possibility that asymmetric division may also regulate an early divergence in CD4+ T lymphocyte fates. During an immune response to LCMV or Lm, TH1 effector cells, identified by high expression of IL-2Rα and T-bet, and TFH cells, identified by high expression of CXCR5 and Bcl6, were detected as early as the second division [30, 31], suggesting that divergence of these two cell fates occurs early during the immune response. Moreover, the adoptive transfer of CXCR5loT-bethi TH1 cells or CXCR5hiT-betlo TFH cells into recipient mice at 7 days post-infection showed that a subset of TH1 effector cells can survive to become TEM cells, while a subset of CXCR5hi cells forms the TCM pool [31] (Figure 1B). T-bet can be asymmetrically degraded during the first division by activated naïve CD4+ T cells as a result of unequal distribution of the proteasome degradation machinery [32]. Because pseudo-substrate inhibition of PKCζ has been shown to reduce asymmetric division in CD4+ T lymphocytes [32], it is intriguing to speculate that asymmetric division may play a functionally important role in CD4+ T cell fate specification, similar to that observed in CD8+ T cells [29]. In this way, asymmetric division might also specify heterogeneous CD4+ T cell fates at the first division and potentiate divergent pathways of differentiation into TH1/TEM or TFH/TCM cell fates; this hypothesis, however, remains to be addressed experimentally. Moreover, whether other CD4+ TH subsets follow similar divergent pathways of differentiation as TH1 and TFH cells remains to be investigated (see Outstanding Questions Box). There is evidence to suggest that the various effector TH subsets can exhibit reversible plasticity between fates [33], indicating that effector TH cells may be derived from the same precursor cells. Furthermore, TFH cells have been shown to upregulate the lineage-defining transcription factors of other TH subsets, depending on the type of infection [34–36]. It is therefore possible that the inflammatory environment produced in the presence of a particular pathogen could dictate the production of the appropriate effector TH subset, whereas asymmetric division by activated naïve CD4+ T lymphocytes may serve as a mechanism to preserve TCM and TFH cell differentiation regardless of the type of infection. Alternatively, CD4+ T cells can modulate their differentiation programs through repeated encounters with antigen-presenting cells after the first division [37], which raises the possibility that CD4+ T lymphocytes could undergo successive rounds of asymmetric division to produce a full spectrum of TH subsets responding to any given infection.
Outstanding Questions Box.
Can a multi-faceted asymmetry of cell fate-determining mechanisms in the progeny of initial responding T lymphocytes be identified early in the immune response?
How are the distinct gene expression patterns that are found in effector- and memory-fated first daughter T cells regulated by RNA- and epigenetic-based mechanisms?
Can a disparity in metabolic function be detected amongst differentially fated T lymphocytes early in the immune response?
What molecules are responsible for transmitting the initial polarity cue downstream to activate aPKC during the first division by activated T lymphocytes?
Do naïve CD4+ T lymphocytes make use of asymmetric division to differentiate into TH subsets (TH17, TH9, TH2, regulatory T cells, etc.) in response to different pathogens?
Is asymmetric division by CD8+ T lymphocytes compromised in the absence of CD4+ T cells?
Can surface markers be found earlier in the immune response that better predict and represent functional heterogeneity?
What is the function of evolutionarily conserved polarity proteins in regulating asymmetric division and fate specification of CD8+ T lymphocytes?
Additional lymphocyte subsets, including non-circulating tissue resident memory T cells [38, 39] and long-lived effector CD8+ T lymphocytes [29, 40, 41], have also been identified after acute infection; however, the ontogeny of these subsets remains less defined (Figure 1A). The observation that memory CD8+ T lymphocytes can undergo asymmetric division upon rechallenge [42] raises the possibility that additional asymmetric divisions during the primary immune response may mediate further divergence of early responding T cells into these additional effector and memory lineages. However, aPKC does not appear to be involved in CD8+ T cell fate determination after the first division, based on the observation that wild-type and aPKC-deficient pre-memory cells, when adoptively transferred in equal numbers into infection-matched recipients, were capable of responding robustly to rechallenge [29]. If additional rounds of asymmetric division were to occur, therefore, it is possible that other members of the polarity network could be involved. Studies of these polarity proteins, however, have not yet addressed their role in asymmetric T lymphocyte division or have been limited to previously activated T lymphocyte populations [43–53]. Moreover, loss of Scribble does not appear to affect CD8+ T cell responses to influenza virus [53]. Characterization of immune responses in which polarity proteins are absent prior to the first division would yield insights into how T lymphocytes regulate asymmetric division, how polarity network proteins impact the generation of functional immune responses, and whether they are required for further asymmetric divisions after the first cell division. Investigating the precise role of polarity proteins in T lymphocytes may also enhance our understanding of these proteins in regulating cell fate decisions in other immune cells (Box 2)
Box 2. Polarity protein regulation of asymmetric division in hematopoietic cell lineages.
T lymphocytes are not the only cells of the hematopoietic system that have been shown to undergo asymmetric division. Hematopoietic stem cells (HSCs) undergo symmetric and asymmetric divisions to balance self-renewal and differentiation [112, 113]. Thymocytes have long been proposed to undergo asymmetric division on the basis of cell-kinetic and morphological studies [114, 115]. B lymphocytes have also been observed to undergo asymmetric divisions during germinal center reactions [116] and can asymmetrically distribute antigen following activation [117]. However, a functional role of asymmetric division, regulated by the evolutionarily conserved polarity complexes, in determining cell fate choices in these hematopoietic lineages has been elusive.
In HSCs, an RNA interference screen identified Pard6a (Par6) and Prkcz (PKCζ) as important players in retaining HSC self-renewal capabilities [118], while deficiency of Lgl1, a phosphorylation target of aPKC in Drosophila [99, 102], increased the number of HSCs with a capacity for self-renewal [119]. However, studies of PKCζ and PKλ/ι in HSCs have shown that aPKC may be dispensable for HSC function and differentiation [120]. In thymocytes, knockdown of Scribble blocks T cell development at the double negative 3 (DN3) stage [121], but Dlg1 and PKCζ knockout mice display normal numbers and percentages of peripheral T cells [45, 122] and Scribble-deficient, Dlg1-deficient, or Lgl1-deficient fetal liver cells generate normal percentages of CD3+ T cells in the blood 6–10 weeks after transfer into recipient mice [53]. In B cells, deletion of Scribble, Dlg1, and Lgl1 did not appear to have an effect on B cell development or B cell antibody production in vitro or in vivo, following immunization with influenza antigen [53].
Notably, because the aforementioned studies did not investigate an impact on asymmetric division in the absence of the studied polarity proteins, it remains unclear whether asymmetric division has a functional impact on cell fate determination in these cell lineages. The discovery of an additional related polarity protein, Par3-like, that acts independently of the normal polarity complexes to regulate mammary stem cell maintenance [123, 124] suggests that regulation of asymmetric division in mammalian cells may be more complex than previously appreciated. Moreover, while often expressed as single genes in lower organisms, vertebrate species often express multiple isoforms and multiple splice variants of a given polarity protein [123, 125], further complicating the interpretations of single knockout studies in mammalian systems, as compensatory mechanisms could be at play. Future studies that correlate alterations in asymmetric division with readouts of cell fate determination will be necessary to fully understand the role of asymmetric division and polarity proteins in hematopoietic-derived cell lineages.
Our work has also provided insight into the cell biology underlying asymmetric division by T lymphocytes. Contrary to findings from Drosophila neuroblasts and C. elegans embryos (Box 1), symmetry breaking and establishment of asymmetry in activated naïve CD4+ and CD8+ T lymphocytes does not appear to involve the Scribble or PAR polarity complexes, as Scribble is polarized late following activation [54] and loss of PKCζ or PKCλ/ιdid not affect immune synapse formation [29]. Instead, two important functions of the immune synapse are required: TCR engagement and integrin-binding interactions. Adoptive transfer of naïve CD8+ T cells into RAG-deficient mice [5], which led to TCR-independent homeostatic expansion [55], or into ICAM1-deficient mice, in which dendritic cells exhibit impaired binding to T cells [56], followed by microbial infection [5] resulted in greatly reduced asymmetric division. Moreover, King et al. have demonstrated that low affinity TCR interactions were unable to induce asymmetric division [57], which suggests that a T lymphocyte must overcome a certain threshold to undergo an asymmetric division. This threshold effect may be related to T lymphocyte conjugation time with dendritic cells, as sustained contact with an antigen-presenting cell is thought to be required for asymmetric division [58] and low affinity TCR interactions decreased conjugation times with dendritic cells [57]. As noted above, the maintenance of asymmetry through the completion of mitosis requires aPKC [29], suggesting an important role for the polarity complexes in regulating this latter stage of asymmetric division. Together, these findings suggest a model of asymmetric division whereby an antigen-presenting cell provides an initial extrinsic polarity cue to establish a polarized immune synapse and orient the plane of asymmetry, while polarity complex proteins provide further intrinsic cues to maintain this asymmetry through the completion of mitosis (Figure 1C). Although many proteins involved in immune synapse formation have been identified, additional studies will be required to fully elucidate their impact in controlling asymmetric T cell division and T lymphocyte fate.
Extrinsic signals contribute to late diversification of single-cell derived progeny
While asymmetric segregation of fate-determining factors in mitotic T cells can direct the divergence of daughter cells towards different lineages early in the immune response, cell-extrinsic signals derived from the microenvironment may help shape the development of single-cell derived progenies into terminal effector and long-lived memory subsets at the population level during later stages of a primary infection. In two recent studies that combined single-cell lineage tracing methodologies (DNA-barcode labeling [25] or cell tracking by the detection of combinations of congenic markers [4]) with mathematical modeling approaches, the progenies of individual activated naïve T lymphocytes responding to microbial infection in vivo were found to differ in their size and expression of KLRG1, CD27, and CD62L. These data suggested that individual naïve T cells may contribute differentially to the terminal effector and self-renewing memory T cell subsets, and that the functional heterogeneity observed at the population level during the immune response may be the result of averaging stochastic behaviors of individual cells, such that multiple activated naïve cells may be required to yield a uniform reproducible immune response. Mathematical modeling of the differentiation path of activated CD8+ T lymphocytes were suggestive of a linear model of differentiation, i.e. naïve→TCM → TEM→effector cells [4], rather than an early divergent model. One limitation of these studies, however, was that analysis of the proliferative activities [4] or T cell family size [25] of single-cell derived progenies were performed only at late time points after infection (days 5–8 post-infection) using a limited set of phenotypic markers to distinguish between putative pre-effector and pre-memory cells. Thus, these limitations may have precluded the ability of these studies to detect an early divergence in T cell fates.
The use of selected phenotypic markers in the preceding studies highlights a current challenge for the field of linking phenotypic diversity with functional heterogeneity. Phenotypic heterogeneity in T cell subsets is typically distinguished by differences in the expression of cell surface molecules, such as CD27, KLRG1, and IL-7R, during the expansion phase of the immune response [4, 28, 59, 60]. For instance, KLRG1 and IL-7R are used to distinguish terminal effector (KLRG1hiIL-7Rlo) and putative memory precursor (KLRG1loIL-7Rhi) T lymphocyte subsets that are presumed to be destined for death or survival after pathogen clearance, respectively [28, 60]. While these markers can be useful, they are not always predictive of cell death and survival [28]. At the single-cell transcriptional level, individual CD8+ T cells with high IL-7R protein expression at day 7 post-infection were found to comprise a mixture of terminal effector and memory cells [16]. Thus, similarity in the expression of phenotypic markers, particularly at later stages of the immune response, does not necessarily equate with functional similarity. These findings underscore the importance of identifying alternative molecular and phenotypic markers early in the immune response that are more predictive of distinct cell fates.
Understanding diversification using single-cell technologies
A comprehensive understanding of the ontogeny of effector and memory T lymphocytes responding to infection will require a temporal analysis that combines in vivo single-cell lineage tracing technologies with high-throughput transcriptomic, epigenetic, proteomic, and metabolomics approaches (Figure 2). Resolving the precise differentiation path of individual T lymphocytes into different effector and memory cell fates will necessitate the tracking and sampling of molecules (DNA, RNA, protein) within the same cell of single-cell progenies over time and at different anatomical locations. While this is not yet technically feasible, in vivo lineage tracking methods [24] and emerging single-cell technologies (Box 3) are available to interrogate individual activated T cells at sequential time points in the immune response. MARS-Seq and Drop-Seq approaches have recently been applied to analyze heterogeneity in the hematopoietic system and mouse retinal tissue composition [61, 62], respectively, using dissociation of lymphoid and non-lymphoid tissues followed by single-cell RNA sequencing and analysis in parallel. Because these approaches employ multiple levels of barcoding (molecular and cellular) prior to RNA sequencing and subsequent computational analyses, the cell of origin for a single transcript can be delineated. Thus, cellular heterogeneity can be dissected at single-cell resolution in an unbiased way by integrating the transcriptional networks of non-preselected cell types. These deconstructive [61] methods, coupled with model systems of infectious disease and the analysis of different anatomical locations, could be applied to create a time-series snapshot of how individual precursor T lymphocytes differentiate into effector, memory, and tissue resident memory T lymphocytes, and may provide additional information about their respective migratory patterns.
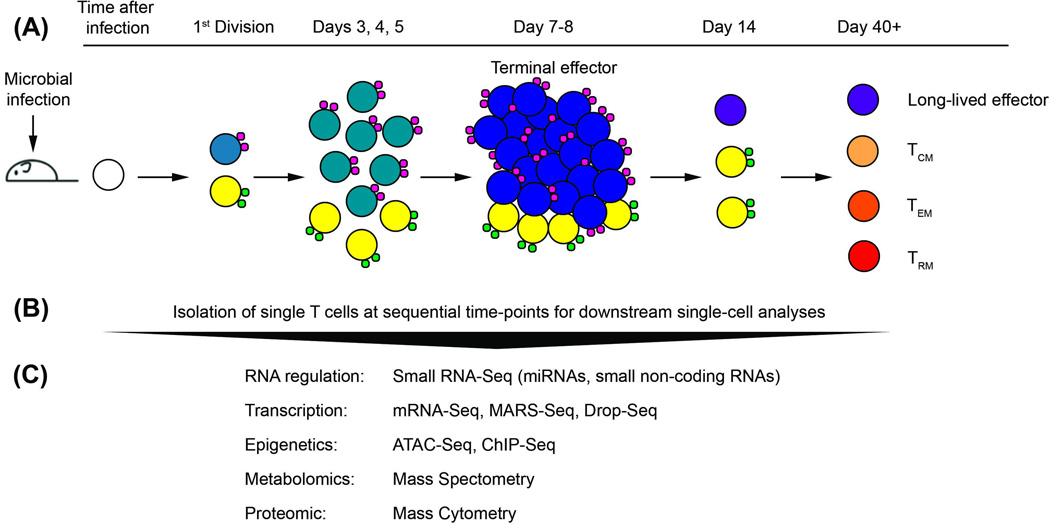
Figure 2.
Possible approach to study T lymphocyte differentiation combining in vivo fate mapping with single-cell technologies. Microbial infection triggers the adaptive immune response. T lymphocytes are activated and undergo multiple rounds of cell division for clonal expansion and differentiation into effector and memory subsets. (A) Asymmetric division at the first cell division of an activated naïve T cell (white) results in unequal inheritance of a hypothetical fate-associated molecule (small green or pink circles) into one of the two daughter cells (pre-memory cells (yellow); pre-effector cells (teal)). Fusion of the fate-determining molecule to a fluorescent protein could enable the tracking of progenies throughout the immune response, if expression of the fate-determining molecule is maintained and detectable in the progenies of only one daughter cell. (B) Single cells sorted on the basis of fusion fluorescent protein expression at early and late times post-infection, together with cell surface markers that distinguish terminal effector cells (dark blue) at days 7–8 post-infection and memory cell subsets at day 30 post-infection and beyond (long-lived effector (purple), TCM (light orange), TEM (orange), TRM (red)) by flow cytometry. (C) Single-cell technologies enable the interrogation of individual responding T lymphocytes at multiple levels of regulation: RNA, gene expression, epigenetic, cellular metabolism, and protein expression. Combinatorial analyses of these approaches with computational modeling create a global overview of when and how T cell fates are acquired during an immune response. Abbreviations: TCM, central memory cell; TEM, effector memory cell; TRM, tissue resident memory cell; miRNA, microRNA; MARS-Seq, Massively Parallel Single-Cell RNA-Seq; ATAC-Seq, Assay for Transposase-Accessible Chromatin with high-throughput sequencing; ChIP, chromatin immunoprecipitation; Seq, sequencing.
Box 3. Emerging single-cell technologies.
As a growing number of complex biological systems have been found to consist of coordinated effects of individual behaviors of single cells [12–19], new technologies are being developed to assess the transcriptomic, epigenetic, and metabolic composition of individual cells in order to predict subsequent behavior and cell fate choices. Massively parallel single-cell RNA-sequencing (MARS-seq) is an automated high-throughput platform that combines molecular barcoding with single-cell RNA sequencing, which allows for the assessment of molecular heterogeneity without making a priori assumptions based on surface markers or previously defined cell types [61]. Similarly, newly developed Drop-seq technologies encapsulate single cells into nanoliter-sized aqueous droplets with different DNA-barcoded material for high-throughput sequencing of thousands of cells in parallel, helping to reveal population structure, gene regulatory linkages, and rare sub-populations within any given system [62, 126]. Regarding epigenetic regulation, assays for transposase-accessible chromatin with high-throughput sequencing (ATAC-Seq) have now reached single-cell resolution, allowing accessible regions of chromatin to be probed in individual cells. Single-cell ATAC-Seq has been applied to human and mouse cells, revealing regulatory elements that might be responsible for the cell-to-cell variability in gene expression and heterogeneity observed within a given population [127, 128]. Additionally, mass cytometry techniques have been developed that combine time-of-flight mass spectrometry with metal-labeling technology for multiparameter protein expression analyses at the single-cell level, allowing for extensive investigation of cellular heterogeneity and developmental processes [129]. Finally, single-cell mass spectrometry has been developed and used to investigate metabolic and small-molecule networks that underlie commitment to neuronal, epidermal, and hindgut fates in single cells of developing Xenopus embryos [75]. Applications of these emerging technologies during T lymphocyte differentiation will prove invaluable in revealing mechanistic insights controlling lymphocyte diversification and fate decisions.
The finding that distinct transcriptional signatures predictive of an effector- or memory-fated lineage can be identified in single cells early after infection [16] suggests that disparate fate propensities may be imprinted into the two daughter cells at the first cell division. In recent years, there has been considerable interest in understanding the epigenetic landscape of memory T lymphocyte transcription patterning (reviewed by Weng et al. [63] and Youngblood et al. [64]) and the role of regulatory noncoding RNAs in CD4+ (reviewed by Pagani et al. [65]) and CD8+ T cell differentiation [66]. Recent profiling of the dynamics of chromatin modifications during different stages of hematopoietic differentiation has identified chromatin patterns that are lineage-specific [67]. Whether terminal effector and memory T lymphocytes display lineage-specific patterns of epigenetic imprinting and whether an asymmetry in the epigenetic landscape of T lymphocytes can be detected early during an immune response remain to be determined. Future exploration into the temporal dynamics of transcription networks and gene expression regulatory mechanisms will be needed to yield mechanistic insights underlying differential gene expression [16], cell proliferation [68], homing [69], and functional diversity [70] that, collectively, underpin the differentiation of T lymphocytes into distinct fates.
In addition to potential transcriptional regulatory ‘programs’, metabolic changes have recently been implicated in driving differentiation into effector and memory T lymphocyte fates [22, 71, 72]. Genes that encode for molecules of the glycolysis, glutaminolysis, and lipid biosynthesis pathways are highly expressed in effector T cells but downregulated in memory T cells [72, 73], and these distinct metabolic programs have been shown to sustain the respective functions of terminal effector and memory cells [71, 72]. As fate-associated molecules that are asymmetrically segregated in dividing T cells begin to be uncovered, can an early asymmetry in cellular metabolic processes be identified at the single-cell level? Measuring metabolites in single T lymphocytes early in the immune response using high-throughput microfluidic cytometry, single-cell microarrays, and single-cell mass spectrometry [74, 75] will be key to understanding whether disparate metabolic processes drive T cell fate formation or are solely a product of differentiation into the effector or memory lineages. This knowledge will be important for the rational design of metabolically targeted immunotherapies that enhance immune responses.
Further study of how asymmetric division contributes to T lymphocyte diversity will require an experimental system with the capacity to trace multigenerational lineages in vivo at the single-cell level for downstream transcriptomic, epigenetic, and proteomic analyses on the cellular progenies. Applications of fluorescent protein technologies have allowed the progeny of Drosophila germline stem cells and neuroblasts to be traced following an asymmetric division [76–78]. To effectively study the role of asymmetric division in the immune response, an ideal system would employ a genetically encoded fluorescent reporter to follow the expression of a fate-determining gene or protein that is asymmetrically segregated in dividing T cells, maintained during subsequent divisions, and is detectable in the progenies of only one daughter cell (Figure 2A). However, considerable overlap in gene expression between naïve and memory cells [63] and rapid upregulation of effector-associated molecules in activated naïve CD8+ T cells, such as Granzyme B and T-bet [80], even prior to their first division will make fluorescent labeling specifically in early effector- or memory-fated daughter cells challenging to implement and interpret. Nonetheless, future studies investigating the role of asymmetric division in the immune response will benefit from the application of sophisticated microscopy tools that can visually track cell division events and potential asymmetric signaling molecules in dividing T cells after infection. Such technologies include intravital microscopy, dynamic in situ cytometry (DISC) [79], intravital dynamics-immunosignal correlative microscopy [80], which have been used to analyze single T cell dynamic behavior in situ, and high-throughput two-photon microscopy [81] for in vivo imaging of T cells at different anatomical locations. Future studies combining the aforementioned techniques should allow for an in-depth analysis of the role of asymmetric division in shaping heterogeneous T lymphocyte responses.
Concluding remarks
The findings reviewed here have suggested that early and late diversification mechanisms [3–5, 16, 25, 29, 32, 57, 58, 69, 82, 83] likely cooperate to shape heterogeneous adaptive immune responses to infection, ultimately resulting in the formation of terminally differentiated effector and self-renewing memory T lymphocytes that are required for durable immunity (Figure 1). However, several gaps in our understanding remain. If asymmetric division is completely ablated, can memory cells still form? Additionally, while asymmetric division appears to result in daughter cells with distinct fate predispositions, to what extent does asymmetric division modulate other aspect of T lymphocyte biology? First division daughter cells can inherit differential amounts of homing receptors, CD62L and LFA-1 [5, 16], but how this affects the subsequent function of effector- and memory-fated precursors is unknown. Similarly, beyond early specification of the effector and memory fates, does asymmetric division function to specify other fates, including tissue resident memory cells and “virtual” memory cells, which may arise via non-antigenic stimulation [84–87]? Whether a T lymphocyte is capable of undergoing multiple rounds of asymmetric division to potentiate additional heterogeneity within an adaptive immune response remains to be determined. While these questions require further exploration, understanding the process of T lymphocyte differentiation warrants continued application of single-cell technologies to analyze differences in gene expression and associated regulatory mechanisms at early and late times after microbial infection (Figure 2). Such studies will be crucial to identifying novel T cell fate-associated markers and will yield new insights into how heterogeneous T lymphocyte fates are specified during the complex orchestration of an immune response.
Trends Box.
Recent studies using limiting dilution strategies or novel cell tracing techniques have demonstrated that a single naïve CD4+ or CD8+ T lymphocyte can generate a heterogeneous adaptive immune response.
Application of emerging single-cell technologies has revealed an early divergence of terminal effector and self-renewing memory lymphocyte fates that can only be observed at the single-cell level.
Impaired asymmetric division at the initiation of the adaptive immune response reduces early molecular heterogeneity of CD8+ T lymphocytes, thus altering the balance of effector- and memory-fated precursor cells and reducing differentiation into the memory T lymphocyte fates.
Single-cell approaches will be essential to improving our understanding of T lymphocyte fate determination and advancing vaccination and therapeutic approaches that enhance long-term protective immune responses.
Acknowledgements
We thank members of the Chang lab for helpful comments and suggestions. This work was supported by US National Institutes of Health (DK080949, OD008469, and AI095277 to J.T.C).
Glossary Box
- Atypical protein kinase C (aPKC)
An evolutionarily conserved polarity protein that regulates asymmetric division
- Hidden Markov Model (HMM)
A statistical Markov model that ascertains hidden states that remain unobserved during biological processes
- Long-lived effector T lymphocyte
Subset of effector T cells that survives into the memory phase of the adaptive immune response, exerts potent protective responses, but exhibits poor proliferative capacities; ontogeny is less defined
- Partitioning defective (PAR) polarity complex
Important polarity complex known to regulate asymmetric division; members include PAR3, PAR6, and aPKC
- Proximal and distal daughter cells
Upon activation, a T lymphocyte forms an immune synapse with an antigen-presenting cell (APC); the T lymphocyte maintains this contact as the cell divides. The nascent daughter cell that was in close proximity to the APC has been termed the ‘proximal’ daughter cell. whereas the nascent daughter cell away from the APC and distal to the immune synapse has been called the ‘distal’ daughter cell [5]. It has been hypothesized that proximal daughter cells have a predisposition towards the terminal effector fate. whereas distal daughter cells have a predisposition towards the memory fates
- Scribble polarity complex
Antagonist of the PAR polarity complex to maintain asymmetry within a cell: members include Scribble, Discs large (Dlg), and Lethal giant larvae (Lgl)
- TCM
central memory T lymphocyte Long-lived memory subset defined by high expression of CD62L and CCR7; TCM cells patrol lymphoid organs and undergo robust proliferation upon antigen rechallenge
- TEM, effector memory T lymphocyte
Long-lived memory subset defined by low expression of CD62L and CCR7; TEM cells exert immediate effector function upon antigen rechallenge
- Terminal effector T lymphocyte
Mediates cytotoxic activities to provide acute protection against microbial infection; characterized by high rates of proliferation early during the immune response; followed by rapid apoptosis after pathogen clearance
- TRMtissue resident memory T lymphocyte
T lymphocyte subset defined by high expression of αE integrin CD103 and CD69 for both CD4+ and CD8+ TRM cells; TRM cells reside within peripheral tissues to mediate host protective responses (reviewed by Park et al [130]); ontogeny is less defined
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kaech SM, Cui W. Transcriptional control of effector and memory CD8+ T cell differentiation. Nature reviews. Immunology. 2012;12:749–761. doi: 10.1038/nri3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahmed R, et al. The precursors of memory: models and controversies. Nature reviews. Immunology. 2009;9:662–668. doi: 10.1038/nri2619. [DOI] [PubMed] [Google Scholar]
- 3.Ahmed R, Gray D. Immunological memory and protective immunity: understanding their relation. Science. 1996;272:54–60. doi: 10.1126/science.272.5258.54. [DOI] [PubMed] [Google Scholar]
- 4.Buchholz VR, et al. Disparate individual fates compose robust CD8+ T cell immunity. Science. 2013;340:630–635. doi: 10.1126/science.1235454. [DOI] [PubMed] [Google Scholar]
- 5.Chang JT, et al. Asymmetric T lymphocyte division in the initiation of adaptive immune responses. Science. 2007;315:1687–1691. doi: 10.1126/science.1139393. [DOI] [PubMed] [Google Scholar]
- 6.Knoblich JA. Mechanisms of asymmetric stem cell division. Cell. 2008;132:583–597. doi: 10.1016/j.cell.2008.02.007. [DOI] [PubMed] [Google Scholar]
- 7.Best JA, et al. Transcriptional insights into the CD8(+) T cell response to infection and memory T cell formation. Nat Immunol. 2013;14:404–412. doi: 10.1038/ni.2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Holmes S, et al. Memory T cells have gene expression patterns intermediate between naive and effector. Proc Natl Acad Sci U S A. 2005;102:5519–5523. doi: 10.1073/pnas.0501437102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaech SM, et al. Molecular and functional profiling of memory CD8 T cell differentiation. Cell. 2002;111:837–851. doi: 10.1016/s0092-8674(02)01139-x. [DOI] [PubMed] [Google Scholar]
- 10.Liu K, et al. Augmentation in expression of activation-induced genes differentiates memory from naive CD4+ T cells and is a molecular mechanism for enhanced cellular response of memory CD4+ T cells. J Immunol. 2001;166:7335–7344. doi: 10.4049/jimmunol.166.12.7335. [DOI] [PubMed] [Google Scholar]
- 11.Willinger T, et al. Molecular signatures distinguish human central memory from effector memory CD8 T cell subsets. J Immunol. 2005;175:5895–5903. doi: 10.4049/jimmunol.175.9.5895. [DOI] [PubMed] [Google Scholar]
- 12.Buganim Y, et al. Single-cell expression analyses during cellular reprogramming reveal an early stochastic and a late hierarchic phase. Cell. 2012;150:1209–1222. doi: 10.1016/j.cell.2012.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dalerba P, et al. Single-cell dissection of transcriptional heterogeneity in human colon tumors. Nature biotechnology. 2011;29:1120–1127. doi: 10.1038/nbt.2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo G, et al. Resolution of cell fate decisions revealed by single-cell gene expression analysis from zygote to blastocyst. Dev Cell. 2010;18:675–685. doi: 10.1016/j.devcel.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 15.Lu R, et al. Tracking single hematopoietic stem cells in vivo using high-throughput sequencing in conjunction with viral genetic barcoding. Nature biotechnology. 2011;29:928–933. doi: 10.1038/nbt.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arsenio J, et al. Early specification of CD8+ T lymphocyte fates during adaptive immunity revealed by single-cell gene-expression analyses. Nat Immunol. 2014;15:365–372. doi: 10.1038/ni.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shalek AK, et al. Single-cell RNA-seq reveals dynamic paracrine control of cellular variation. Nature. 2014;510:363–369. doi: 10.1038/nature13437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Patel AP, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344:1396–1401. doi: 10.1126/science.1254257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Buettner F, et al. Computational analysis of cell-to-cell heterogeneity in single-cell RNA-sequencing data reveals hidden subpopulations of cells. Nature biotechnology. 2015;33:155–160. doi: 10.1038/nbt.3102. [DOI] [PubMed] [Google Scholar]
- 20.Moon JJ, et al. Naive CD4(+) T cell frequency varies for different epitopes and predicts repertoire diversity and response magnitude. Immunity. 2007;27:203–213. doi: 10.1016/j.immuni.2007.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Obar JJ, et al. Endogenous naive CD8+ T cell precursor frequency regulates primary and memory responses to infection. Immunity. 2008;28:859–869. doi: 10.1016/j.immuni.2008.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chang JT, et al. Molecular regulation of effector and memory T cell differentiation. Nat Immunol. 2014;15:1104–1115. doi: 10.1038/ni.3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stemberger C, et al. A single naive CD8+ T cell precursor can develop into diverse effector and memory subsets. Immunity. 2007;27:985–997. doi: 10.1016/j.immuni.2007.10.012. [DOI] [PubMed] [Google Scholar]
- 24.Gerlach C, et al. One naive T cell, multiple fates in CD8+ T cell differentiation. J Exp Med. 2010;207:1235–1246. doi: 10.1084/jem.20091175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gerlach C, et al. Heterogeneous differentiation patterns of individual CD8+ T cells. Science. 2013;340:635–639. doi: 10.1126/science.1235487. [DOI] [PubMed] [Google Scholar]
- 26.Plumlee CR, et al. Environmental cues dictate the fate of individual CD8+ T cells responding to infection. Immunity. 2013;39:347–356. doi: 10.1016/j.immuni.2013.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tubo NJ, et al. Single naive CD4+ T cells from a diverse repertoire produce different effector cell types during infection. Cell. 2013;153:785–796. doi: 10.1016/j.cell.2013.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Joshi NS, et al. Inflammation directs memory precursor and short-lived effector CD8(+) T cell fates via the graded expression of T-bet transcription factor. Immunity. 2007;27:281–295. doi: 10.1016/j.immuni.2007.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Metz PJ, et al. Regulation of asymmetric division and CD8+ T lymphocyte fate specification by protein kinase Czeta and protein kinase Clambda/iota. J Immunol. 2015;194:2249–2259. doi: 10.4049/jimmunol.1401652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Choi YS, et al. ICOS receptor instructs T follicular helper cell versus effector cell differentiation via induction of the transcriptional repressor Bcl6. Immunity. 2011;34:932–946. doi: 10.1016/j.immuni.2011.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pepper M, et al. Opposing signals from the Bcl6 transcription factor and the interleukin-2 receptor generate T helper 1 central and effector memory cells. Immunity. 2011;35:583–595. doi: 10.1016/j.immuni.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chang JT, et al. Asymmetric proteasome segregation as a mechanism for unequal partitioning of the transcription factor T-bet during T lymphocyte division. Immunity. 2011;34:492–504. doi: 10.1016/j.immuni.2011.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O’Shea JJ, Paul WE. Mechanisms underlying lineage commitment and plasticity of helper CD4+ T cells. Science. 2010;327:1098–1102. doi: 10.1126/science.1178334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Glatman Zaretsky A, et al. T follicular helper cells differentiate from Th2 cells in response to helminth antigens. J Exp Med. 2009;206:991–999. doi: 10.1084/jem.20090303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johnston RJ, et al. Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science. 2009;325:1006–1010. doi: 10.1126/science.1175870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmitt N, et al. The cytokine TGF-beta co-opts signaling via STAT3-STAT4 to promote the differentiation of human TFH cells. Nat Immunol. 2014;15:856–865. doi: 10.1038/ni.2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Celli S, et al. CD4 T cells integrate signals delivered during successive DC encounters in vivo. J Exp Med. 2005;202:1271–1278. doi: 10.1084/jem.20051018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gebhardt T, et al. Memory T cells in nonlymphoid tissue that provide enhanced local immunity during infection with herpes simplex virus. Nat Immunol. 2009;10:524–530. doi: 10.1038/ni.1718. [DOI] [PubMed] [Google Scholar]
- 39.Masopust D, et al. Dynamic T cell migration program provides resident memory within intestinal epithelium. J Exp Med. 2010;207:553–564. doi: 10.1084/jem.20090858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hikono H, et al. Activation phenotype, rather than central- or effector-memory phenotype, predicts the recall efficacy of memory CD8+ T cells. J Exp Med. 2007;204:1625–1636. doi: 10.1084/jem.20070322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Olson JA, et al. Effector-like CD8(+) T cells in the memory population mediate potent protective immunity. Immunity. 2013;38:1250–1260. doi: 10.1016/j.immuni.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ciocca ML, et al. Cutting edge: Asymmetric memory T cell division in response to rechallenge. J Immunol. 2012;188:4145–4148. doi: 10.4049/jimmunol.1200176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Crocetti J, et al. Selective phosphorylation of the Dlg1AB variant is critical for TCR-induced p38 activation and induction of proinflammatory cytokines in CD8+ T cells. J Immunol. 2014;193:2651–2660. doi: 10.4049/jimmunol.1401196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gmyrek GB, et al. Polarity gene discs large homolog 1 regulates the generation of memory T cells. Eur J Immunol. 2013;43:1185–1194. doi: 10.1002/eji.201142362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Humphries LA, et al. Characterization of in vivo Dlg1 deletion on T cell development and function. PLoS One. 2012;7:e45276. doi: 10.1371/journal.pone.0045276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martin P, et al. Control of T helper 2 cell function and allergic airway inflammation by PKCzeta. Proc Natl Acad Sci U S A. 2005;102:9866–9871. doi: 10.1073/pnas.0501202102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Real E, et al. Cutting edge: Atypical PKCs regulate T lymphocyte polarity and scanning behavior. J Immunol. 2007;179:5649–5652. doi: 10.4049/jimmunol.179.9.5649. [DOI] [PubMed] [Google Scholar]
- 48.Round JL, et al. Dlgh1 coordinates actin polymerization, synaptic T cell receptor and lipid raft aggregation, and effector function in T cells. J Exp Med. 2005;201:419–430. doi: 10.1084/jem.20041428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Silva O, et al. Discs Large Homolog 1 Splice Variants Regulate p38 -Dependent and -Independent Effector Functions in CD8+ T Cells. PLoS One. 2015;10:e0133353. doi: 10.1371/journal.pone.0133353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xavier R, et al. Discs large (Dlg1) complexes in lymphocyte activation. J Cell Biol. 2004;166:173–178. doi: 10.1083/jcb.200309044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang JQ, et al. Loss of PKC lambda/iota impairs Th2 establishment and allergic airway inflammation in vivo. Proc Natl Acad Sci U S A. 2009;106:1099–1104. doi: 10.1073/pnas.0805907106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zanin-Zhorov A, et al. Scaffold protein Disc large homolog 1 is required for T-cell receptor-induced activation of regulatory T-cell function. Proc Natl Acad Sci U S A. 2012;109:1625–1630. doi: 10.1073/pnas.1110120109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hawkins ED, et al. Regulation of asymmetric cell division and polarity by Scribble is not required for humoral immunity. Nat Commun. 2013;4:1801. doi: 10.1038/ncomms2796. [DOI] [PubMed] [Google Scholar]
- 54.Yeh JH, et al. Regulation of a late phase of T cell polarity and effector functions by Crtam. Cell. 2008;132:846–859. doi: 10.1016/j.cell.2008.01.013. [DOI] [PubMed] [Google Scholar]
- 55.Goldrath AW, Bevan MJ. Low-affinity ligands for the TCR drive proliferation of mature CD8+ T cells in lymphopenic hosts. Immunity. 1999;11:183–190. doi: 10.1016/s1074-7613(00)80093-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dustin ML, Springer TA. T-cell receptor cross-linking transiently stimulates adhesiveness through LFA-1. Nature. 1989;341:619–624. doi: 10.1038/341619a0. [DOI] [PubMed] [Google Scholar]
- 57.King CG, et al. T cell affinity regulates asymmetric division, effector cell differentiation, and tissue pathology. Immunity. 2012;37:709–720. doi: 10.1016/j.immuni.2012.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Oliaro J, et al. Asymmetric cell division of T cells upon antigen presentation uses multiple conserved mechanisms. J Immunol. 2010;185:367–375. doi: 10.4049/jimmunol.0903627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Marshall HD, et al. Differential expression of Ly6C and T-bet distinguish effector and memory Th1 CD4(+) cell properties during viral infection. Immunity. 2011;35:633–646. doi: 10.1016/j.immuni.2011.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sarkar S, et al. Functional and genomic profiling of effector CD8 T cell subsets with distinct memory fates. J Exp Med. 2008;205:625–640. doi: 10.1084/jem.20071641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jaitin DA, et al. Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science. 2014;343:776–779. doi: 10.1126/science.1247651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Macosko EZ, et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Weng NP, et al. The molecular basis of the memory T cell response: differential gene expression and its epigenetic regulation. Nature reviews. Immunology. 2012;12:306–315. doi: 10.1038/nri3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Youngblood B, et al. T-cell memory differentiation: insights from transcriptional signatures and epigenetics. Immunology. 2013;139:277–284. doi: 10.1111/imm.12074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pagani M, et al. Role of microRNAs and long-non-coding RNAs in CD4(+) T-cell differentiation. Immunol Rev. 2013;253:82–96. doi: 10.1111/imr.12055. [DOI] [PubMed] [Google Scholar]
- 66.Pang KC, et al. Genome-wide identification of long noncoding RNAs in CD8+ T cells. J Immunol. 2009;182:7738–7748. doi: 10.4049/jimmunol.0900603. [DOI] [PubMed] [Google Scholar]
- 67.Lara-Astiaso D, et al. Immunogenetics. Chromatin state dynamics during blood formation. Science. 2014;345:943–949. doi: 10.1126/science.1256271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kinjyo I, et al. Real-time tracking of cell cycle progression during CD8+ effector and memory T-cell differentiation. Nat Commun. 2015;6:6301. doi: 10.1038/ncomms7301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jung YW, et al. Differential localization of effector and memory CD8 T cell subsets in lymphoid organs during acute viral infection. J Immunol. 2010;185:5315–5325. doi: 10.4049/jimmunol.1001948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jameson SC, Masopust D. Diversity in T cell memory: an embarrassment of riches. Immunity. 2009;31:859–871. doi: 10.1016/j.immuni.2009.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.van der Windt GJ, Pearce EL. Metabolic switching and fuel choice during T-cell differentiation and memory development. Immunol Rev. 2012;249:27–42. doi: 10.1111/j.1600-065X.2012.01150.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang R, Green DR. Metabolic reprogramming and metabolic dependency in T cells. Immunol Rev. 2012;249:14–26. doi: 10.1111/j.1600-065X.2012.01155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kidani Y, et al. Sterol regulatory element-binding proteins are essential for the metabolic programming of effector T cells and adaptive immunity. Nat Immunol. 2013;14:489–499. doi: 10.1038/ni.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dittrich P, Ibanez AJ. Analysis of metabolites in single cells-what is the best micro-platform? Electrophoresis. 2015 doi: 10.1002/elps.201500045. [DOI] [PubMed] [Google Scholar]
- 75.Onjiko RM, et al. Single-cell mass spectrometry reveals small molecules that affect cell fates in the 16-cell embryo. Proc Natl Acad Sci U S A. 2015;112:6545–6550. doi: 10.1073/pnas.1423682112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Conduit PT, Raff JW. Cnn dynamics drive centrosome size asymmetry to ensure daughter centriole retention in Drosophila neuroblasts. Curr Biol. 2010;20:2187–2192. doi: 10.1016/j.cub.2010.11.055. [DOI] [PubMed] [Google Scholar]
- 77.Januschke J, et al. Drosophila neuroblasts retain the daughter centrosome. Nat Commun. 2011;2:243. doi: 10.1038/ncomms1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yamashita YM, et al. Asymmetric inheritance of mother versus daughter centrosome in stem cell division. Science. 2007;315:518–521. doi: 10.1126/science.1134910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Moreau HD, et al. Dynamic in situ cytometry uncovers T cell receptor signaling during immunological synapses and kinapses in vivo. Immunity. 2012;37:351–363. doi: 10.1016/j.immuni.2012.05.014. [DOI] [PubMed] [Google Scholar]
- 80.Chodaczek G, et al. Body-barrier surveillance by epidermal gammadelta TCRs. Nat Immunol. 2012;13:272–282. doi: 10.1038/ni.2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pozzi P, et al. High-throughput spatial light modulation two-photon microscopy for fast functional imaging. Neurophotonics. 2015;2:015005. doi: 10.1117/1.NPh.2.1.015005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Haring JS, et al. Inflaming the CD8+ T cell response. Immunity. 2006;25:19–29. doi: 10.1016/j.immuni.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 83.Plumlee CR, et al. Early Effector CD8 T Cells Display Plasticity in Populating the Short-Lived Effector and Memory-Precursor Pools Following Bacterial or Viral Infection. Scientific reports. 2015;5:12264. doi: 10.1038/srep12264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Martinet V, et al. Type I interferons regulate eomesodermin expression and the development of unconventional memory CD8(+) T cells. Nat Commun. 2015;6:7089. doi: 10.1038/ncomms8089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chiu BC, et al. Cutting edge: Central memory CD8 T cells in aged mice are virtual memory cells. J Immunol. 2013;191:5793–5796. doi: 10.4049/jimmunol.1302509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lee JY, et al. Virtual memory CD8 T cells display unique functional properties. Proc Natl Acad Sci U S A. 2013;110:13498–13503. doi: 10.1073/pnas.1307572110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Akue AD, et al. Derivation and maintenance of virtual memory CD8 T cells. J Immunol. 2012;188:2516–2523. doi: 10.4049/jimmunol.1102213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Park SG, et al. The kinase PDK1 integrates T cell antigen receptor and CD28 coreceptor signaling to induce NF-kappaB and activate T cells. Nat Immunol. 2009;10:158–166. doi: 10.1038/ni.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fruman DA, Bismuth G. Fine tuning the immune response with PI3K. Immunol Rev. 2009;228:253–272. doi: 10.1111/j.1600-065X.2008.00750.x. [DOI] [PubMed] [Google Scholar]
- 90.Tskvitaria-Fuller I, et al. Specific patterns of Cdc42 activity are related to distinct elements of T cell polarization. J Immunol. 2006;177:1708–1720. doi: 10.4049/jimmunol.177.3.1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Atwood SX, et al. Cdc42 acts downstream of Bazooka to regulate neuroblast polarity through Par-6 aPKC. J Cell Sci. 2007;120:3200–3206. doi: 10.1242/jcs.014902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bandyopadhyay G, et al. PKC-zeta mediates insulin effects on glucose transport in cultured preadipocyte-derived human adipocytes. J Clin Endocrinol Metab. 2002;87:716–723. doi: 10.1210/jcem.87.2.8252. [DOI] [PubMed] [Google Scholar]
- 93.Uberall F, et al. Evidence that atypical protein kinase C-lambda and atypical protein kinase C-zeta participate in Ras-mediated reorganization of the F-actin cytoskeleton. J Cell Biol. 1999;144:413–425. doi: 10.1083/jcb.144.3.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Prehoda KE. Polarization of Drosophila neuroblasts during asymmetric division. Cold Spring Harb Perspect Biol. 2009;1:a001388. doi: 10.1101/cshperspect.a001388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yamashita YM, et al. Polarity in stem cell division: asymmetric stem cell division in tissue homeostasis. Cold Spring Harb Perspect Biol. 2010;2:a001313. doi: 10.1101/cshperspect.a001313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Doe CQ, Bowerman B. Asymmetric cell division: fly neuroblast meets worm zygote. Curr Opin Cell Biol. 2001;13:68–75. doi: 10.1016/s0955-0674(00)00176-9. [DOI] [PubMed] [Google Scholar]
- 97.Williams SE, Fuchs E. Oriented divisions, fate decisions. Curr Opin Cell Biol. 2013;25:749–758. doi: 10.1016/j.ceb.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Atwood SX, Prehoda KE. aPKC phosphorylates Miranda to polarize fate determinants during neuroblast asymmetric cell division. Curr Biol. 2009;19:723–729. doi: 10.1016/j.cub.2009.03.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Betschinger J, et al. The Par complex directs asymmetric cell division by phosphorylating the cytoskeletal protein Lgl. Nature. 2003;422:326–330. doi: 10.1038/nature01486. [DOI] [PubMed] [Google Scholar]
- 100.Suzuki A, Ohno S. The PAR-aPKC system: lessons in polarity. J Cell Sci. 2006;119:979–987. doi: 10.1242/jcs.02898. [DOI] [PubMed] [Google Scholar]
- 101.Ohno S. Intercellular junctions and cellular polarity: the PAR-aPKC complex, a conserved core cassette playing fundamental roles in cell polarity. Curr Opin Cell Biol. 2001;13:641–648. doi: 10.1016/s0955-0674(00)00264-7. [DOI] [PubMed] [Google Scholar]
- 102.Plant PJ, et al. A polarity complex of mPar-6 and atypical PKC binds, phosphorylates and regulates mammalian Lgl. Nat Cell Biol. 2003;5:301–308. doi: 10.1038/ncb948. [DOI] [PubMed] [Google Scholar]
- 103.Betschinger J, et al. Phosphorylation-induced autoinhibition regulates the cytoskeletal protein Lethal (2) giant larvae. Curr Biol. 2005;15:276–282. doi: 10.1016/j.cub.2005.01.012. [DOI] [PubMed] [Google Scholar]
- 104.Yamanaka T, et al. Mammalian Lgl forms a protein complex with PAR-6 and aPKC independently of PAR-3 to regulate epithelial cell polarity. Curr Biol. 2003;13:734–743. doi: 10.1016/s0960-9822(03)00244-6. [DOI] [PubMed] [Google Scholar]
- 105.Lee CY, et al. Lgl, Pins and aPKC regulate neuroblast self-renewal versus differentiation. Nature. 2006;439:594–598. doi: 10.1038/nature04299. [DOI] [PubMed] [Google Scholar]
- 106.Smith CA, et al. aPKC-mediated phosphorylation regulates asymmetric membrane localization of the cell fate determinant Numb. The EMBO journal. 2007;26:468–480. doi: 10.1038/sj.emboj.7601495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Guo M, et al. Control of daughter cell fates during asymmetric division: interaction of Numb and Notch. Neuron. 1996;17:27–41. doi: 10.1016/s0896-6273(00)80278-0. [DOI] [PubMed] [Google Scholar]
- 108.Fuerstenberg S, et al. Identification of Miranda protein domains regulating asymmetric cortical localization, cargo binding, and cortical release. Molecular and cellular neurosciences. 1998;12:325–339. doi: 10.1006/mcne.1998.0724. [DOI] [PubMed] [Google Scholar]
- 109.Tio M, et al. cdc2 links the Drosophila cell cycle and asymmetric division machineries. Nature. 2001;409:1063–1067. doi: 10.1038/35059124. [DOI] [PubMed] [Google Scholar]
- 110.Hutterer A, et al. Mitotic activation of the kinase Aurora-A requires its binding partner Bora. Dev Cell. 2006;11:147–157. doi: 10.1016/j.devcel.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 111.Wirtz-Peitz F, et al. Linking cell cycle to asymmetric division: Aurora-A phosphorylates the Par complex to regulate Numb localization. Cell. 2008;135:161–173. doi: 10.1016/j.cell.2008.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ting SB, et al. Asymmetric segregation and self-renewal of hematopoietic stem and progenitor cells with endocytic Ap2a2. Blood. 2012;119:2510–2522. doi: 10.1182/blood-2011-11-393272. [DOI] [PubMed] [Google Scholar]
- 113.Wu M, et al. Imaging hematopoietic precursor division in real time. Cell Stem Cell. 2007;1:541–554. doi: 10.1016/j.stem.2007.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Metcalf D, Wiadrowski M. Autoradiographic analysis of lymphocyte proliferation in the thymus and in thymic lymphoma tissue. Cancer Res. 1966;26:483–491. [PubMed] [Google Scholar]
- 115.Sugimoto M, Yasuda T. Asymmetric (differential) cell division of thymic lymphocytes by means of cytoplasmic polarization: possible biological meanings. Thymus. 1983;5:297–310. [PubMed] [Google Scholar]
- 116.Barnett BE, et al. Asymmetric B cell division in the germinal center reaction. Science. 2012;335:342–344. doi: 10.1126/science.1213495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Thaunat O, et al. Asymmetric segregation of polarized antigen on B cell division shapes presentation capacity. Science. 2012;335:475–479. doi: 10.1126/science.1214100. [DOI] [PubMed] [Google Scholar]
- 118.Hope KJ, et al. An RNAi screen identifies Msi2 and Prox1 as having opposite roles in the regulation of hematopoietic stem cell activity. Cell Stem Cell. 2010;7:101–113. doi: 10.1016/j.stem.2010.06.007. [DOI] [PubMed] [Google Scholar]
- 119.Heidel FH, et al. The cell fate determinant Llgl1 influences HSC fitness and prognosis in AML. J Exp Med. 2013;210:15–22. doi: 10.1084/jem.20120596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Sengupta A, et al. Atypical protein kinase C (aPKCzeta and aPKClambda) is dispensable for mammalian hematopoietic stem cell activity and blood formation. Proc Natl Acad Sci U S A. 2011;108:9957–9962. doi: 10.1073/pnas.1103132108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Pike KA, et al. Immature T-cell clustering and efficient differentiation require the polarity protein Scribble. Proc Natl Acad Sci U S A. 2011;108:1116–1121. doi: 10.1073/pnas.1018224108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Leitges M, et al. Targeted disruption of the zetaPKC gene results in the impairment of the NF-kappaB pathway. Mol Cell. 2001;8:771–780. doi: 10.1016/s1097-2765(01)00361-6. [DOI] [PubMed] [Google Scholar]
- 123.Gao L, et al. Multiple splice variants of Par3 and of a novel related gene, Par3L, produce proteins with different binding properties. Gene. 2002;294:99–107. doi: 10.1016/s0378-1119(02)00681-9. [DOI] [PubMed] [Google Scholar]
- 124.Huo Y, Macara IG. The Par3-like polarity protein Par3L is essential for mammary stem cell maintenance. Nat Cell Biol. 2014;16:529–537. doi: 10.1038/ncb2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Goldstein B, Macara IG. The PAR proteins: fundamental players in animal cell polarization. Dev Cell. 2007;13:609–622. doi: 10.1016/j.devcel.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Klein AM, et al. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell. 2015;161:1187–1201. doi: 10.1016/j.cell.2015.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Buenrostro JD, et al. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature. 2015;523:486–490. doi: 10.1038/nature14590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Cusanovich DA, et al. Epigenetics. Multiplex single-cell profiling of chromatin accessibility by combinatorial cellular indexing. Science. 2015;348:910–914. doi: 10.1126/science.aab1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Bendall SC, et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science. 2011;332:687–696. doi: 10.1126/science.1198704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Park CO, Kupper TS. The emerging role of resident memory T cells in protective immunity and inflammatory disease. Nature medicine. 2015;21:688–697. doi: 10.1038/nm.3883. [DOI] [PMC free article] [PubMed] [Google Scholar]