Abstract
Helicobacter pylori colonizes the human stomach and is associated with an increased risk of gastric cancer and peptic ulcer disease. Analysis of H. pylori protein secretion is complicated by the occurrence of bacterial autolysis. In this study, we analyzed the exoproteome of H. pylori at multiple phases of bacterial growth and identified 74 proteins that are selectively released into the extracellular space. These include proteins known to cause alterations in host cells, antigenic proteins, and additional proteins that have not yet been studied in any detail. The composition of the H. pylori exoproteome is dependent on the phase of bacterial growth. For example, the proportional abundance of the vacuolating toxin VacA in culture supernatant is higher during late growth phases than early growth phases, whereas the proportional abundance of many other proteins is higher during early growth phases. We detected marked variation in the subcellular localization of putative secreted proteins within soluble and membrane fractions derived from intact bacteria. By providing a comprehensive view of the H. pylori exoproteome, these results provide new insights into the array of secreted H. pylori proteins that may cause alterations in the gastric environment.
Keywords: mass spectrometry, proteomics, bacterial protein secretion, secretome, exoproteome, VacA, autotransporter, autolysis, outer membrane vesicles, gastric cancer, peptic ulcer disease
Graphical abstract
INTRODUCTION
Helicobacter pylori colonizes the gastric mucosa of humans and is present in about 50% of the population worldwide. A humoral immune response and a gastric mucosal inflammatory response are consistently detectable in H. pylori-infected persons (1, 2), but the bacteria resist clearance by these host defenses (3). Most H. pylori-infected persons remain asymptomatic, but have an increased risk for development of peptic ulcer disease, gastric adenocarcinoma, and gastric lymphoma (4, 5).
H. pylori is localized to the gastric mucus layer overlying gastric epithelial cells and typically does not invade host cells. Therefore, there is considerable interest in identifying proteins released by the bacteria into the extracellular space that can cause alterations in host cells or alterations in the gastric environment (6). The identification of proteins specifically secreted by H. pylori is complicated by the occurrence of bacterial autolysis (resulting in non-selective release of intracellular proteins into the extracellular space) (7–10), the release of membrane vesicles into the extracellular space (11, 12), and a requirement of protein-rich culture medium of undefined composition to support growth of the bacteria to high optical densities.
Three previous studies have utilized proteomic methods to analyze the H. pylori exoproteome (8–10). Two studies used two-dimensional gel electrophoresis methods and evaluated the specificity of H. pylori protein release in comparison to UreB (an abundant cytoplasmic protein that is released into the culture supernatant) (8, 9). One of these studies reported the detection of 33 protein spots when the culture supernatant was analyzed, and identified 26 distinct proteins that were selectively released into the supernatant (8). The second study identified 16 proteins that were selectively released into the supernatant (9). Another study used direct LC-MS/MS methods to analyze the exoproteome of two H. pylori strains (10). Among 130 H. pylori proteins detected in the culture supernatant, 45 were considered to be enriched in the supernatant of one or both strains (based on a >1.5 ratio in the number of unique peptides detected in supernatant compared to the numbers of unique peptides detected in a soluble cell-associated sample). Only four proteins (the vacuolating toxin VacA; “cell binding factor 2” or HP0175; hypothetical protein HP1286; and thioredoxin TrxC or HP1458) were reported to be selectively released into the extracellular space in all three of the previous proteomic studies.
The previous proteomic studies provided valuable insights into the exoproteome of H. pylori, but there are several limitations of the previous studies. For example, two-dimensional gel electrophoresis is dependent on successful resolution of proteins in 2-D gels, is not optimal for identification of low-abundance proteins, and is not well-suited for analyzing large numbers of replicate samples. In addition, the methods used in the previous studies for evaluating the selectivity of protein release were based on measuring gel spot densities and the use of a single protein (UreB) as a reference (8, 9). The use of this protein as a reference may not be optimal, since the findings of one study suggested that UreB is specifically secreted by H. pylori (13). All of the previous studies analyzed protein release at a single time point, and therefore, it was not possible to evaluate growth phase-dependent variations in protein release. Finally, many of the putative secreted proteins identified in the previous studies are orthologs of proteins localized to the cytoplasm, periplasm, or inner membrane in other bacterial species. Thus, the composition of the H. pylori exoproteome is not yet well established, and the mechanisms underlying release of most H. pylori proteins into the extracellular space are not understood.
In this study, we sought to define more clearly the set of H. pylori proteins that are selectively released into the extracellular space. To do this, we undertook a comprehensive analysis of the H. pylori exoproteome, using direct mass spectrometry-based methods to analyze the protein composition of H. pylori broth culture supernatants in comparison to subcellular bacteria fractions derived from intact bacteria. This approach allowed a high level of sensitivity for protein detection, as well as a quantitative means for assessing the selectivity of protein release into the culture supernatant. To facilitate these experiments, we used a culture medium that was optimized to have low protein content (compatible with direct mass spectrometric analysis), while still supporting robust bacterial growth. In total, we identified 74 proteins that are enriched in the culture supernatant compared to a subcellular fraction derived from intact bacteria, thereby indicating selective release of these proteins into the extracellular space. Analysis of the H. pylori exoproteome at multiple phases of bacterial growth allowed us to detect growth phase-dependent differences in the composition of the exoproteome. In addition, we show that there is considerable variation in the subcellular localization of selectively released proteins within soluble and membrane fractions derived from intact bacteria, suggesting that there are multiple mechanisms for the selective release of H. pylori proteins into the extracellular space. By providing a comprehensive view of the H. pylori exoproteome, these results provide new insights into the array of extracellular H. pylori proteins that may cause alterations in the gastric environment.
METHODS
Bacterial strains and culture conditions
All experiments were performed with H. pylori strain 26695, which was originally isolated from a patient with gastritis (14). H. pylori was grown at 37°C in room air supplemented with 5% CO2. The bacteria were routinely passaged on Trypticase soy agar plates containing 5% sheep blood. Liquid cultures were grown in a modified form of sulfite-free Brucella-cholesterol broth, termed “Brucella broth filtrate”. This medium was prepared by passing sulfite-free Brucella broth (15) through a 3 kDa cut-off ultrafiltration membrane (Amicon Ultra-15; EMD Millipore). The resulting filtrate was supplemented with 1X cholesterol (Gibco) (16). The use of this serum-free medium, which had a reduced content of high molecular mass proteins normally present in Brucella broth, facilitated subsequent mass spectrometry analysis.
Bacteria were harvested from blood agar plates after one day of growth, and were inoculated into seed cultures with a starting inoculum of OD600 0.02. Following growth of the seed cultures overnight with shaking at 160 rpm, larger volume cultures (250 ml for most experiments) were inoculated with the seed culture at the same initial density (OD600 0.02). Aliquots (each about 50 ml) were removed at serial time points (12, 24, 30, 36, and 48 hours post-inoculation). The samples were centrifuged at 4500xg at 4°C for 10 minutes, yielding broth culture supernatants and bacterial pellets.
Processing of broth culture supernatants
Supernatants were passed through a 0.22 µm filter to remove any remaining bacteria, and filtered supernatants were concentrated to a final volume of 1 ml using a 10 kDa cut-off centrifugal filter ultrafiltration unit (Amicon Ultra-15; EMD Millipore). Remaining Brucella broth proteins that could potentially interfere with mass spectrometry analysis were removed by buffer exchange with Tris-buffered saline (20 mM Tris, 136 mM NaCl, pH 7.4). Concentrated culture supernatants were centrifuged at 100,000 x g for 2 hours at 4°C to remove outer membrane vesicles or other insoluble components (12). The resulting supernatants were removed and further concentrated to 50 µL by ultrafiltration with a 10 kDa cut-off unit. Protein concentration was quantified by bicinchoninic acid assay (Thermo Scientific).
Bacterial subcellular fractionation
Bacterial subcellular fractions were prepared as described previously (17). Bacterial pellets were washed twice in TNKCM buffer (50 mM Tris [pH 7.4], 100 mM NaCl, 27 mM KCl, 1 mM CaCl2, 0.5 mM MgCl2) and resuspended in resuspension and lysis buffer (50 mM Tris [pH 7.4] and 1 mM MgCl2 with EDTA-free protease inhibitor cocktail [Roche]). Cells were lysed by sonication (5 pulses, 20 seconds on/40 seconds off, 20% amplitude) and lysates were centrifuged for 10 min at 4,500 x g at 4°C. Supernatants (predicted to be enriched in cytoplasmic and periplasmic proteins) were removed and centrifuged at 100,000 x g at 4°C for 2 hours to pellet insoluble proteins. The pellets (predicted to be enriched in membrane proteins) were solubilized in radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris [pH 8.0], 150 mM NaCl, 1 mM EDTA, 1% Nonidet P-40 (U.S. Biologicals), 0.25% sodium deoxycholate) containing protease inhibitor cocktail (Roche). The solubilized pellet contents and supernatants were concentrated, and protein concentration was quantified by bicinchoninic acid assay (Thermo Scientific).
Mass spectronomic analysis of samples
Protein preparations were run about 2 cm into a 10% Bis-Tris NuPAGE gel, stained with colloidal Coomassie blue, and then subjected to in-gel trypsin digestion (18). Samples were then analyzed by either single dimensional LC-MS/MS or multidimensional protein identification technology (MudPIT). Single dimensional LC-MS/MS (1D analysis) was performed using ThermoFisher LTQ equipped with a nano-electrospray source and attached to a Nanoacuity (Waters) HPLC unit with an autosampler. Peptides were resolved via reversed phase separation on a 20 cm by 100 micron column packed emitter tip using an aqueous to organic gradient (2% to 45%). Peptide MS/MS spectra were acquired data-dependently with one full scan MS followed by 5 MS/MS scans. Analysis by 8-step MudPIT was performed essentially as described previously (17) and using same instrumentation. Acidified peptides were loaded onto a 150-µm-inner-diameter (ID) biphasic trapping column comprised of 4-cm strong cation exchange resin (Luna [5-µm particle size]; Phenomenex) followed by 4-cm reverse-phase resin (Jupiter [5-µm particle size, 300-Å pore size]). After offline loading of samples, the trapping column was then attached to a 20-cm-long (Jupiter [3-µm particle size, 300-Å pore size]) 100-µm-ID fused silica analytical column packed into a pulled nanospray tip. Aliquots of ammonium acetate (5 µl) at concentrations of 0, 100, 150, 200, 300, 500, 750, and 1,000 mM were injected by autosampler. After each salt injection, peptides were separated using a 105-min aqueous-to-organic gradient (2% to 35% acetonitrile for all but the last step, which went to 98%). Peptide MS/MS spectra were queried using SEQUEST (full tryptic specificity) against an H. pylori strain 26695 database, to which both common contaminants and reversed versions of the proteins had been appended. Peptide identifications were filtered and collated to proteins using Scaffold 4 (Proteome Systems). Protein identifications required a minimum of 2 unique peptides per protein, and were filtered to a 5% false discovery rate (both peptide and protein).
Analysis of mass spectrometry data
For each sample, we calculated the proportional abundance (% abundance) of each protein detected (number of spectral counts assigned to the protein of interest divided by the total number of assigned spectral counts). To calculate the level of enrichment of an individual protein in the supernatant at a given time point (Esup), we compared its proportional abundance in the supernatant to its proportional abundance in the corresponding subcellular fraction containing cytoplasmic and periplasmic proteins (CP/PP) (Esup= % abundancesup / % abundanceCP/PP). A value of 0.5 was added to raw data values to allow calculation of enrichment values for proteins with zero peptides detected in the CP/PP fraction. To facilitate further analysis, the levels of enrichment were expressed as log2 values (log2 Esup). To analyze the results of 1D mass spectrometric analyses, the data from three independent experiments were merged (19). Esup, Fisher’s exact test, Bonferroni multiple test corrections, and construction of Venn diagrams were performed in R (http://www.R-project.org).
Based on the analysis of 40 ribosomal proteins as controls, we selected criteria for the identification of proteins that were selectively released into the culture supernatant. Specifically, we defined selectively released proteins as those having a log2 Esup value >1.5 (indicating that the proportional abundance in the supernatant was roughly three times greater than the proportional abundance in the CP/PP fraction), with p <0.01 (Fisher’s exact test with Bonferroni correction). The application of these criteria resulted in a protein localization false discovery rate of 5% at the latest time point analyzed (when autolysis is predicted to be maximal), based on analysis of ribosomal proteins. We then applied these same criteria to analyze spectral data for the entire set of proteins identified in the respective preparations.
Cluster analysis
To categorize proteins into groups based on time-dependent patterns of protein enrichment in the supernatant or proportional abundance in the supernatant, we analyzed mass spectrometry data by the Cluster Affinity Search Technique (threshold affinity value = 0.8) using the TM4 Software Suite (20).
Analysis of signal peptides
The presence or absence of signal peptides within proteins of interest was predicted using the program SignalP4.1, using default settings (21).
RESULTS
Identification of H. pylori proteins in broth culture supernatant and subcellular bacterial fractions
H. pylori is typically cultured in complex media containing high concentrations of animal or yeast proteins, which complicates analysis of the extracellular proteome. To facilitate proteomic analysis of the H. pylori exoproteome, we tested the ability of H. pylori to grow in several types of less complex media. One of the simplified media supporting H. pylori growth was Brucella broth filtrate, a serum-free medium containing low molecular mass components of Brucella broth, prepared as described in Methods.
To analyze the release of H. pylori proteins into the extracellular space, H. pylori strain 26695 was cultured in Brucella broth filtrate, and culture aliquots were removed at 12, 24, 30, 36, and 48h time points (corresponding to varying stages of bacterial growth) (Fig. 1A). Culture aliquots were centrifuged to yield a broth culture supernatant fraction and a bacterial pellet. Supernatants were processed as described in Methods to remove residual bacteria, membrane vesicles and other insoluble constituents. Bacterial pellets were sonicated, and the lysates were processed to yield soluble fractions (predicted to be enriched in cytoplasmic and periplasmic proteins) and insoluble fractions (predicted to be enriched in membrane proteins). These subcellular fractions are subsequently designated as cytoplasmic/periplasmic (CP/PP) and membrane fractions. Supernatant, CP/PP and membrane samples collected at each time point were then analyzed by 1D mass spectrometry to identify the H. pylori proteins present in these preparations.
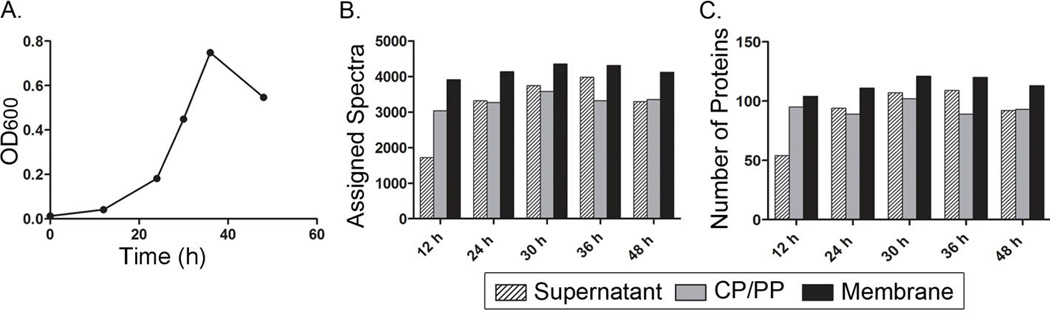
Figure 1. Detection of H. pylori proteins in broth culture supernatant.
Aliquots of an H. pylori broth culture were removed at the indicated time points. The bacteria were pelleted by centrifugation, yielding bacterial pellets and culture supernatants. The bacterial pellets were processed to yield soluble (CP/PP) and insoluble (membrane) fractions. The protein content of the fractions was analyzed by 1D mass spectrometry. (A) Optical density (OD600) of the culture at 12, 24, 30, 36, and 48 hour time points. (B) Total number of spectra assigned to H. pylori proteins, based on analysis of supernatant, CP/PP, and membrane fractions at the indicated time points. (C) Number of H. pylori proteins detected (based on assignment of ≥10 spectra per protein) in fractions at the indicated time points. Data are based on analysis of aliquots taken from a single representative broth culture.
The total numbers of spectra assigned to H. pylori proteins were similar when comparing broth culture supernatants with CP/PP and membrane fractions (Fig. 1B), and the numbers of H. pylori proteins detected in culture supernatants were similar to the numbers detected in the two subcellular fractions (Fig. 1C). There was relatively little variation in the numbers of assigned spectra detected in the supernatants or the numbers of H. pylori proteins detected in the supernatants at different time points. A large number of H. pylori proteins were detected in the broth culture supernatant even at the earliest time points analyzed. The detection of numerous H. pylori proteins in the culture supernatant is consistent with the occurrence of bacterial autolysis.
Relative abundance of selected proteins in culture supernatant compared to a soluble bacterial fraction
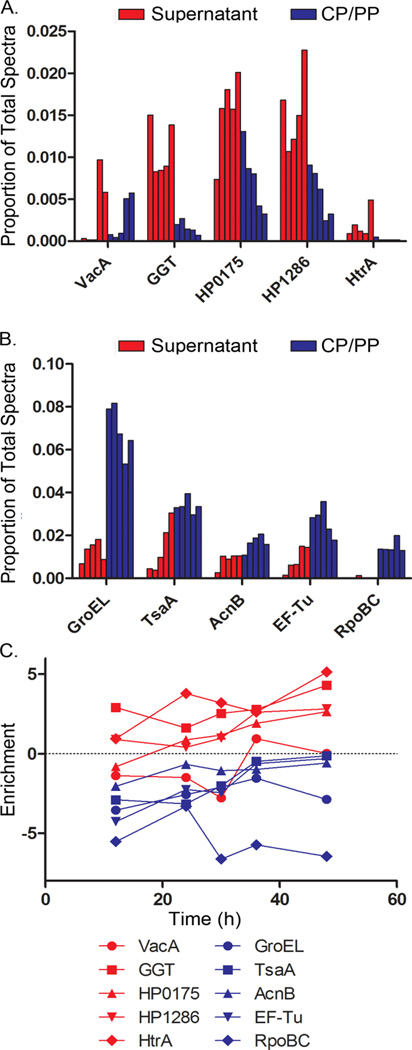
H. pylori broth culture supernatant is predicted to contain proteins non-specifically released during autolysis, as well as proteins specifically secreted by the bacteria. We reasoned that these two groups of proteins might be distributed differently, when comparing relative protein abundance in broth culture supernatant to relative protein abundance in the CP/PP fraction. To test this hypothesis, we did a comparative analysis of five H. pylori proteins previously reported to be present in the culture supernatant and known to be capable of causing alterations in eukaryotic cells (predicted to be specifically secreted) (22–26), as well as five abundant cytoplasmic proteins (predicted to be non-specifically released into the culture supernatant). At each time point, the proportional abundance of the proteins in supernatant and the CP/PP fraction was calculated. The 5 putative secreted proteins [VacA, γ-glutamyl transpeptidase GGT, “cell binding factor 2” (HP0175), hypothetical protein HP1286, and the serine protease HtrA] had a higher proportional abundance in the broth culture supernatant than the CP/PP fraction, while the 5 cytoplasmic proteins (GroEL, TsaA, AcnB, EF-Tu, and RpoBC) had a higher proportional abundance in the CP/PP fraction than in the broth culture supernatant (Fig. 2A and 2B). Levels of enrichment in the supernatant compared to the CP/PP fraction (log2 Esup) were calculated for each protein at each time point (Fig. 2C). The 5 putative secreted proteins generally had positive enrichment values, corresponding to increased relative abundance in the supernatant compared to the CP/PP fraction. Cytoplasmic proteins had negative enrichment values at each time point, corresponding to increased relative abundance in the CP/PP fraction compared to the supernatant. Collectively, these data indicated that the 5 putative secreted proteins are relatively enriched in the supernatant compared to the CP/PP fraction, whereas the 5 cytoplasmic proteins are relatively enriched in the CP/PP fraction compared to the supernatant.
Figure 2. Relative abundance of H. pylori proteins in culture supernatant compared to CP/PP fractions.
Aliquots of an H. pylori broth culture were processed and analyzed as described in Figure 1. (A) Five proteins previously reported to be secreted and known to cause alterations in eukaryotic cells (VacA, GGT, HP0175, HP1286, and HtrA) were selected for analysis. The relative abundance of these proteins in supernatant and CP/PP fractions was assessed by analyzing the proportional abundance of assigned spectral counts (number of spectra assigned to each protein of interest divided by total number of assigned spectra). For each protein, the first five bars (red color) represent the proportional abundance in the supernatant at 12, 24, 30, 36 and 48 h time points (shown in consecutive order), and the second five bars (blue color) represent the proportional abundance in the CP/PP fractions. (B) Five abundant cytoplasmic proteins (GroEL, TsaA, AcnB, EF-Tu, and RpoBC) were selected for analysis, and their relative abundance in the supernatant and CP/PP fractions was calculated and graphed in the same manner. (C) For each of the proteins analyzed in panels A and B, the level of enrichment in the supernatant compared to the CP/PP fraction was assessed at each time point by calculating a log ratio, defined as Esup = log2(proportionsupernatant/proportionCP/PP). Positive log ratio values correspond to enrichment in the supernatant, whereas negative log ratio values indicate enrichment in the CP/PP fraction. Data are based on analysis of aliquots taken from a single representative broth culture.
Identification of proteins enriched in supernatant
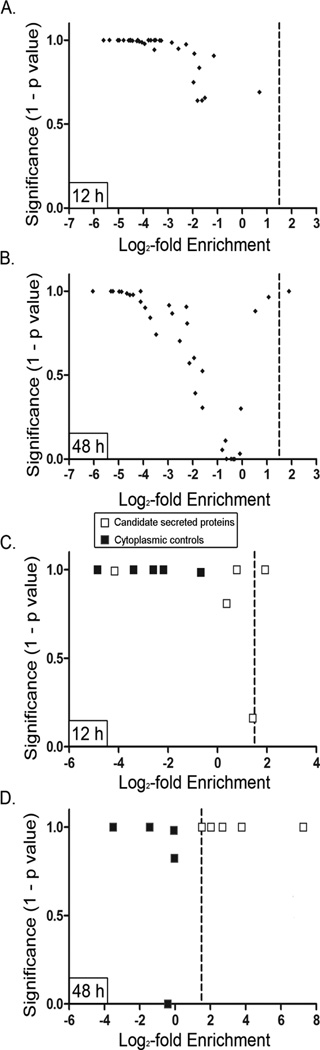
We next undertook a systematic analysis to identify additional proteins that were enriched in the broth culture supernatant compared to the CP/PP fraction. Three independent experiments were performed, in each case sampling 5 distinct phases of bacterial growth, and the merged results of all three experiments were analyzed. To establish criteria for identifying proteins enriched in the supernatant compared to the CP/PP fraction, we analyzed spectral data for 40 proteins previously annotated as ribosomal proteins (Fig. 3). For each protein, we analyzed both the level of enrichment in the supernatant (log2 Esup), as well as the statistical significance of differences in abundance of assigned spectral counts in supernatant compared to the CP/PP fraction (using Fisher’s exact test with Bonferroni correction) (17). As expected, most ribosomal proteins were localized predominantly in the CP/PP fraction at both early (12 h) and late (48 h) time points, with relatively little release of these proteins into the culture supernatant (Fig. 3A and 3B).
Fig. 3. Criteria for identifying H. pylori proteins enriched in culture supernatant.
To identify proteins enriched in culture supernatants compared to CP/PP fractions, we calculated the level of enrichment in the supernatant for each protein (Esup, expressed as log2 values), and also calculated the statistical significance of differences in proportional abundance of spectral counts assigned to each protein when comparing the culture supernatant to the CP/PP fraction (using Fisher’s exact test with the Bonferonni correction). The x axes indicate log2 fold enrichment in the culture supernatant and the y axes indicate statistical significance (1 – p value). We initially analyzed data for 40 proteins annotated as ribosomal proteins at 12 h and 48 h time points (panels A and B, respectively). These ribosomal proteins had low levels of enrichment in the supernatant compared to CP/PP (log2 enrichment values <1). Fisher’s exact test indicated that the differences in distribution of assigned spectral counts (supernatant compared to CP/PP) were highly significant for many of the ribosomal proteins, due to enrichment in the CP/PP fraction. These analyses of ribosomal proteins allowed us to choose appropriate criteria for identifying proteins that were enriched in the supernatant compared to the CP/PP. Specifically, we arbitrarily selected a cutoff of 1.5 for log2 fold-enrichment (vertical lines) and defined a p value of < 0.01 with Bonferonni correction as significant. The use of these criteria resulted in a protein localization false discovery rate of < 5%, based on analysis of ribosomal proteins. We then analyzed 5 putative secreted proteins (VacA, GGT, HP0175, HP1286, and HtrA) and 5 abundant cytoplasmic proteins (GroEL, TsaA, AcnB, EF-Tu, and RpoBC) by the same approach (12 h and 48 time points, panels C and D, respectively). The analysis is based on analysis of merged data from three independent experiments.
Analysis of the relative abundance of ribosomal proteins detected in the supernatant in comparison to the CP/PP fraction facilitated the establishment of criteria to identify proteins that were selectively released into culture supernatant. We defined selectively released proteins as those having a log2 Esup value >1.5 (indicating that the proportional abundance in the supernatant was roughly three times greater than the proportional abundance in the CP/PP fraction), with p <0.01. Application of these criteria to the 40 previously annotated ribosomal proteins resulted in a <5% false discovery rate for selectively released proteins at the latest sampled time point (48 h), when entry of proteins into the supernatant through autolysis is predicted to be maximal.
When these criteria were applied to the 10 proteins analyzed in Figure 2 [five H. pylori proteins previously reported to be present in the culture supernatant and known to be capable of causing alterations in eukaryotic cells (predicted to be specifically secreted), and 5 abundant cytoplasmic proteins], the cytoplasmic control proteins were consistently classified as non-enriched in the culture supernatant. All five putative secreted proteins met the criteria for enrichment in the culture supernatant when data from a late growth phase (48 h) were analyzed, but with the exception of GGT, none of the proteins met the criteria for enrichment in the culture supernatant when an early growth phase (12 h) was analyzed (Fig. 3C and 3D).
We then analyzed the entire data set to identify proteins that were relatively enriched in the broth culture supernatant compared to the CP/PP fraction, using the criteria described above. In total, we identified 74 proteins that were enriched in the culture supernatant at two or more time points (19). Twenty-five of these proteins were significantly enriched in the supernatant at ≥4 time points (Table 1). Among these 25 proteins, the levels of enrichment in the supernatant (log2 Esup) ranged from 1.7 to 6.0. The serine protease HtrA exhibited the highest level of enrichment. Forty-nine proteins were enriched in the supernatant at two or three time points (Table 1). Two of the five putative secreted proteins analyzed in Figure 1 (HtrA and GGT) were selectively enriched in the supernatant at ≥4 time points, whereas two others (VacA and HP1286) were selectively enriched in the supernatant only at late time points. HP0175 (cell binding factor 2) was enriched only at the latest time point. This variation in the kinetics of secretion prompted us to group the 74 selectively released proteins into four categories (constitutive release, early release, late release, and no clear pattern), based on the times when selective release was detected (Table 1).
Table 1.
Proteins identified as enriched in broth supernatant compared to CP/PP fraction
| Name | Accession | Enrichment (all time points)a |
Enrichment (significant time points)b |
Relative abundance in membranec |
|---|---|---|---|---|
| Constitutived | ||||
| serine protease (htrA) | HP1019h | 5.08 | 5.99 | 98.48% |
| protease (pqqE) | HP1012 | 4.68 | 4.68 | 89.39% |
| colicin tolerance-like protein (tolB) | HP1126 | 4.19 | 4.60 | 91.59% |
| conserved hypothetical secreted protein | HP0785 | 3.96 | 3.96 | 37.30% |
| catalase-like protein | HP0485 | 3.74 | 3.74 | 73.21% |
| putative neuraminyllactose-binding hemagglutinin homolog (hpaA) | HP0410h | 3.06 | 3.68 | 81.19% |
| 2’,3’-cyclic-nucleotide 2’-phosphodiesterase (cpdB) | HP0104h | 3.65 | 3.65 | 72.15% |
| membrane-associated lipoprotein (lpp20) | HP1456h | 2.72 | 3.60 | 87.45% |
| processing protease (ymxG) | HP0657 | 3.12 | 3.12 | 76.45% |
| hypothetical protein | HP0408h | 2.73 | 3.07 | 64.10% |
| dipeptide ABC transporter, periplasmic dipeptide- binding protein (dppA) | HP0298 | 3.05 | 3.05 | 42.82% |
| membrane bound endonuclease (nuc) | HP0323 | 3.01 | 3.01 | 79.76% |
| conserved hypothetical secreted protein | HP0235 | 2.74 | 3.00 | 36.32% |
| hypothetical protein | HP0304 | 2.90 | 2.90 | 26.62% |
| conserved hypothetical protein | HP1285h | 2.85 | 2.85 | 67.70% |
| hypothetical protein | HP1173h | 2.71 | 2.71 | 61.34% |
| hypothetical protein | HP1454 | 2.64 | 2.64 | 57.61% |
| gamma-glutamyltranspeptidase (ggt) | HP1118 | 2.43 | 2.43 | 69.40% |
| thiol:disulfide interchange protein (dsbC), putative | HP0377h | 2.39 | 2.39 | 45.95% |
| conserved hypothetical secreted protein | HP0211 | 2.32 | 2.37 | 27.73% |
| hypothetical protein | HP0953 | 2.08 | 2.08 | 24.06% |
| conserved hypothetical secreted protein | HP1098 | 2.06 | 2.06 | 41.83% |
| iron(III) ABC transporter, periplasmic iron-binding protein (ceuE) | HP1562 | 2.01 | 2.01 | 51.38% |
| catalase | HP0875h | 1.77 | 1.98 | 64.17% |
| outer membrane protein | HP1564h | 1.77 | 1.77 | 21.53% |
| Earlye | ||||
| protective surface antigen D15 | HP0655h | 1.89 | 4.74 | 95.85% |
| amino acid ABC transporter, periplasmic binding protein (yckK) | HP0940 | 3.46 | 4.29 | 40.06% |
| flagellar sheath adhesin hpaA | HP0797h | 1.35 | 3.23 | 93.01% |
| outer membrane protein (omp9) | HP0317h | 0.27 | 3.10 | 94.79% |
| hypothetical protein | HP0688 | 1.59 | 3.03 | 14.97% |
| conserved hypothetical protein | HP0507 | 2.19 | 2.89 | 24.74% |
| peptide deformylase (def) | HP0793 | 1.85 | 2.76 | 13.55% |
| purine-nucleoside phosphorylase (deoD) | HP1178 | 1.82 | 2.65 | 11.68% |
| hypothetical protein | HP1265 | 1.67 | 2.61 | 11.92% |
| conserved hypothetical protein | HP0233 | 1.46 | 2.36 | 17.80% |
| phosphoglycerate mutase (pgm) | HP0974 | 1.18 | 2.03 | 12.09% |
| triosephosphate isomerase (tpi) | HP0194 | 1.35 | 1.95 | 8.22% |
| pantoate-beta-alanine ligase (panC) | HP0006 | 1.35 | 1.91 | 4.88% |
| serine hydroxymethyltransferase (glyA) | HP0183 | 1.41 | 1.88 | 28.91% |
| octaprenyl-diphosphate synthase (ispB) | HP0240 | 1.23 | 1.82 | 15.53% |
| hypothetical protein | HP0218 | 1.12 | 1.75 | 20.78% |
| NAD(P)H-flavin oxidoreductase | HP0642 | 1.20 | 1.59 | 6.79% |
| Variedf | ||||
| hypothetical protein | HP1457 | 3.83 | 5.70 | 95.91% |
| outer membrane protein (omp28) | HP1243h | 2.81 | 5.58 | 98.09% |
| outer membrane protein (omp11) | HP0472 | 3.45 | 5.12 | 92.02% |
| outer membrane protein (omp19) | HP0896h | 1.74 | 4.96 | 97.89% |
| conserved hypothetical protein | HP0710h | 3.66 | 4.94 | 97.52% |
| outer membrane protein (omp6) | HP0229h | 2.84 | 4.75 | 95.90% |
| hypothetical protein | HP1455 | 3.99 | 4.59 | 47.31% |
| (3R)-hydroxymyristoyl-(acyl carrier protein) dehydratase (fabZ) | HP1376 | 2.41 | 4.41 | 79.40% |
| CDP-diglyceride hydrolase (cdh) | HP0871 | 2.70 | 3.33 | 40.26% |
| aspartate-semialdehyde dehydrogenase (asd) | HP1189 | 2.10 | 3.30 | 49.80% |
| hypothetical protein | HP0783 | 2.44 | 2.84 | 13.97% |
| outer membrane protein (omp27) | HP1177h | 0.36 | 2.53 | 89.56% |
| hypothetical protein | HP0973h | 1.82 | 2.41 | 47.86% |
| xanthine guanine phosphoribosyl transferase (gpt) | HP0735 | 1.31 | 2.07 | 6.72% |
| arginyl-tRNA synthetase (argS) | HP0319 | 1.49 | 1.93 | 4.37% |
| Lateg | ||||
| ATP-dependent nuclease (addB) | HP0275h | 3.74 | 4.71 | 91.01% |
| outer membrane protein (omp30) | HP1395 | 2.56 | 4.58 | 90.75% |
| methionine amino peptidase (map) | HP1299 | 2.38 | 4.29 | 49.54% |
| membrane fusion protein (mtrC) | HP0606h | 3.81 | 3.62 | 93.81% |
| soluble lytic murein transglycosylase (slt) | HP0645h | 3.10 | 3.15 | 77.55% |
| L-asparaginase II (ansB) | HP0723h | 2.00 | 2.71 | 26.44% |
| hypothetical protein | HP0721 | 1.67 | 2.66 | 46.54% |
| cag pathogenicity island protein (cag24) | HP0545 | 1.18 | 2.54 | 17.02% |
| carbonic anhydrase | HP1186 | 1.99 | 2.36 | 62.21% |
| hypothetical protein | HP0129 | 1.19 | 2.34 | 30.36% |
| conserved hypothetical secreted protein | HP1286 | 1.29 | 2.29 | 22.54% |
| cytochrome c553 | HP1227 | 1.14 | 2.27 | 11.75% |
| iron(III) ABC transporter, periplasmic iron-binding protein (ceuE) | HP1561 | 1.67 | 2.01 | 49.09% |
| hypothetical protein | HP0231h | 1.61 | 2.00 | 54.73% |
| vacuolating cytotoxin | HP0887 | −0.39 | 2.00 | 89.98% |
| hypothetical protein | HP0204 | 1.58 | 1.92 | 24.75% |
| outer membrane protein (omp2) | HP0025h | 1.08 | 1.72 | 83.40% |
Relative abundance in the supernatant compared to the CP/PP fraction (Esup): [Esup=log2(%abundancesupernatant/% abundanceCP/PP)]. Mean Esup values were calculated based on analysis of all five time points.
Average of significant enrichment (Esup) values.
Relative % abundance in the membrane fraction compared to CP/PP fraction: [%abundancemembrane/(%abundancemembrane+%abundanceCP/PP)]. Mean % abundance values were calculated based on analysis of all five time points.
Enriched at 4 or all 5 time points.
Enriched at two of the three earlier time points (12, 24, or 30h).
Enriched at varied time points with no clear pattern.
Enriched at two of the three later time points (30, 36, or 48h).
Previously detected on the bacterial surface by a biotinylation-based method (reference 17).
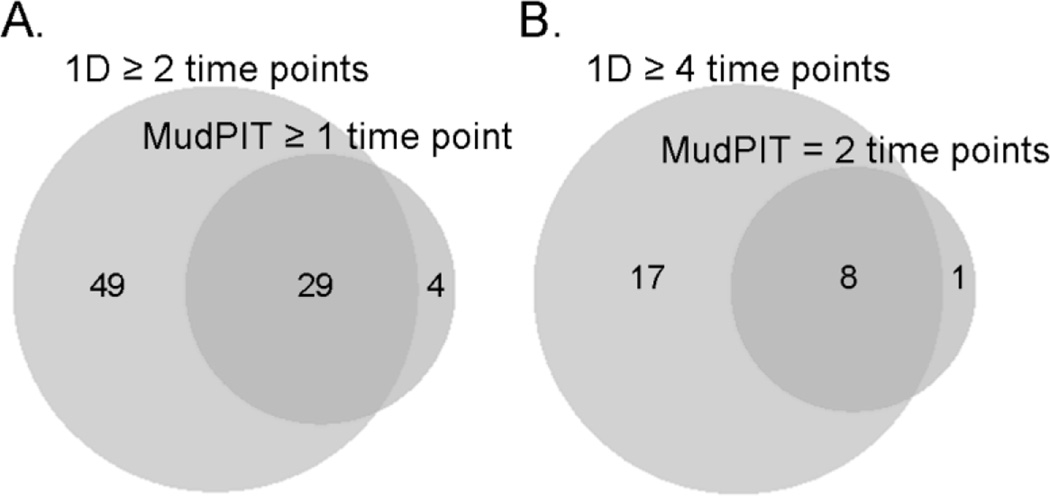
As another approach for identifying proteins enriched in the supernatant compared to the CP/PP fraction, we focused on H. pylori broth culture samples collected at 24h and 36h time points, and analyzed the supernatant, CP/PP, and membrane fractions derived from single representative cultures by multidimensional protein identification technology (MudPIT). Nine proteins were identified as significantly enriched in the supernatant at both 24h and 36h time points (Suppl. Table 1). Three proteins were significantly enriched in the supernatant at 24h only, and 21 proteins at 36h only (Suppl. Table 1). A comparison of the proteins identified as enriched in the 1D experiments to those identified as enriched in the MudPIT experiments indicated that 29 of the 74 proteins identified in the 1D analyses were confirmed in one or both MudPIT experiments. Conversely, 29 of the 33 proteins identified as enriched by MudPIT (at one or both time points) were identified as enriched at two or more time points in the 1D analysis (Fig. 4A). Eight of 9 proteins enriched at both 24 and 36h time points by MudPIT were enriched at ≥4 time points by 1D analysis (Fig. 4B). These MudPIT analyses provided additional evidence for the selective release of specific proteins into the culture supernatant.
Figure 4. Identification of proteins enriched in the culture supernatant, using multiple methods.
(A) Venn diagram analyzing 74 proteins identified as enriched in supernatant at two or more time points based on 1D analysis, compared to 33 proteins identified as enriched in supernatant (24h, 36h, or both time points) by MudPIT. (B) Venn diagram analyzing proteins identified as enriched in supernatant at ≥4 time points by 1D analysis, and identified as enriched in supernatant by MudPIT at both 24 and 36h time points. The 1D data are based on analysis of merged data from three independent experiments, and the MudPIT data are based on analysis of single samples (collected at 24 or 36 h time points).
Kinetics of protein release into culture supernatant
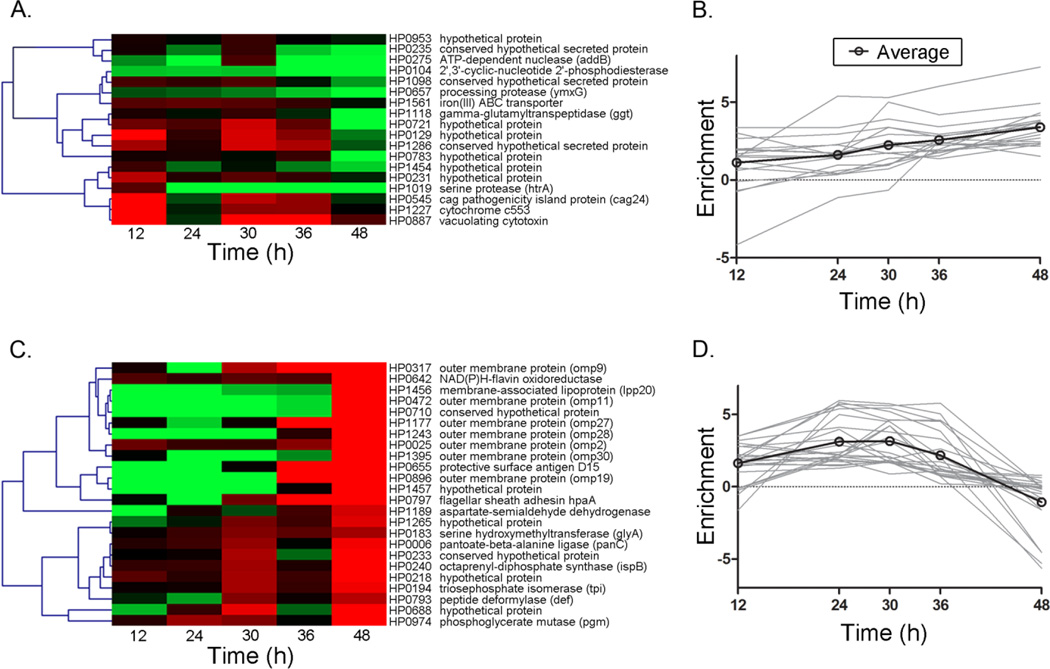
Since some proteins were enriched in the supernatant at only a subset of the time points sampled, this suggested that the composition of the exoproteome might vary depending on the growth phase of the bacteria. To further delineate possible growth phase-dependent patterns of protein release, we analyzed data for the 74 selectively released proteins (enriched in the supernatant at two or more time points, based on 1D analysis) by Cluster Affinity Search Technique (CAST). We analyzed the temporal patterns of protein enrichment in the supernatant (Esup) and the proportional abundance of proteins in the supernatant in two separate analyses. In an analysis of Esup, 42 of the 74 proteins were grouped into two major clusters (Fig. 5A–D), while the remaining proteins were divided among 11 smaller clusters with 6 or fewer proteins per group. One major cluster contained 18 proteins that increased in enrichment over time (Fig. 5A and 5B), and the other cluster contained 24 proteins that decreased in enrichment over time (Fig. 5C and 5D). VacA, GGT, HtrA and HP1286 were grouped together along with 14 other proteins in the cluster exhibiting increased enrichment over time (Fig 5A and 5B). The large cluster demonstrating a decreasing trend line over time (Fig. 5C and 5D) included 7 outer membrane proteins and multiple proteins that are predicted to have an intracellular localization, based on studies of orthologous proteins in other bacterial species.
Figure 5. Growth phase-dependent changes in composition of the H. pylori exoproteome.
Cluster affinity analysis was performed to evaluate growth phase-dependent changes in the enrichment of proteins in the broth culture supernatant compared to the CP/PP fraction. Esup data were analyzed for 74 proteins that were enriched in the supernatant at two or more time points. The proteins were grouped into a total of 13 clusters, based on temporal changes in Esup values over the 5 time points analyzed. The majority of proteins grouped into 2 clusters: one containing 18 proteins that generally increased in enrichment in the supernatant over time as indicated by a shift from red to green (A) and an increasing average trend line (B), and the other containing 24 proteins that generally decreased in enrichment in the supernatant over time, indicated by a shift from green to red (C) and a decreasing average trend line (D). The cluster that increased over time included VacA, GGT, HP1286 and HtrA. The analysis is based on analysis of merged data from three independent experiments.
CAST analysis of the same set of 74 enriched supernatant proteins based on proportional abundance in the supernatant (instead of Esup) yielded 4 main clusters (representing 64 of the 74 proteins) (Suppl. Fig. 1) and several clusters with 4 or fewer proteins per group. The largest cluster contained 20 proteins that exhibited a trend for decreasing proportional abundance over time (Suppl. Fig. 1A–B), and the second largest cluster contained 16 proteins, including VacA, HtrA and HP1286, that increased in proportional abundance over time (Suppl. Fig. 1C–D). Eight of 9 proteins previously annotated as outer membrane proteins (OMPs) grouped into a third cluster containing a total of 13 proteins (Suppl. Fig. 1E–F). One annotated OMP, HP1564, clustered separately from the others. The fourth predominant cluster contained 15 proteins that were constitutively enriched in the supernatant (Suppl. Fig. 1G–H). These analyses provided further evidence for growth phase-dependent variations in the composition of the exoproteome.
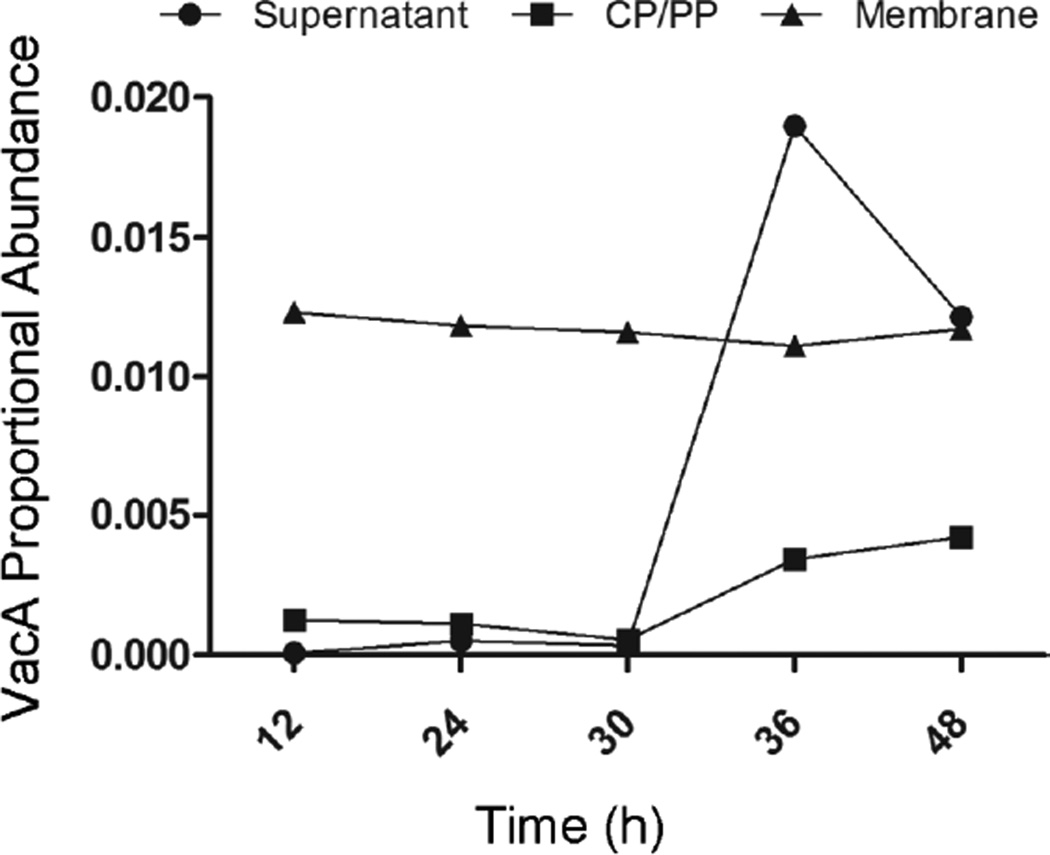
Growth phase-dependent release of VacA into culture supernatant
One of the proteins exhibiting a striking growth phase-dependent variation in proportional abundance in the culture supernatant was VacA. The proportional abundance of VacA in the culture supernatant was low at early time points, but increased markedly at 36h and 48h time points (Fig. 6). The relative abundance of VacA in the membrane and CP/PP fractions remained stable at all time points analyzed (Fig. 6).
Figure 6. Time-dependent increase in relative abundance of VacA in culture supernatant.
Aliquots of an H. pylori broth culture were processed and analyzed as described in Figure 1. The proportional abundance of spectral counts assigned to VacA (number of spectra assigned to VacA divided by total number of assigned spectra) in the supernatant, CP/PP, and membrane fractions was analyzed at each of the indicated time points. The analysis is based on analysis of merged data from three independent experiments.
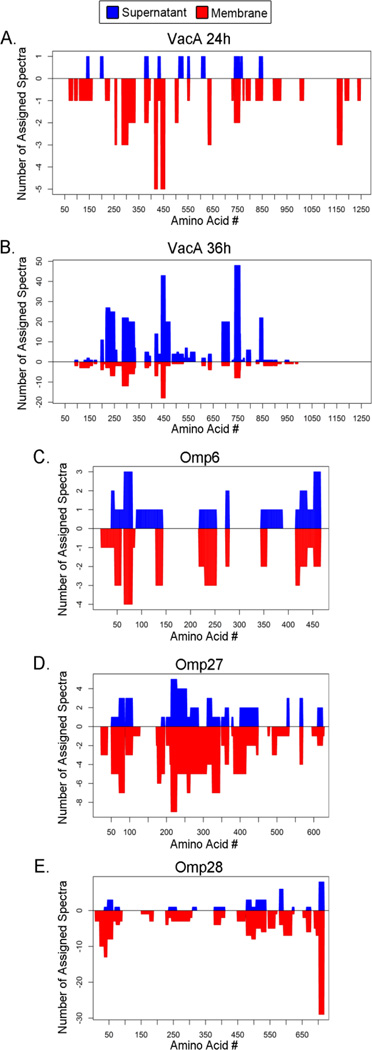
VacA is known to undergo proteolytic processing, resulting in an 88 kDa soluble protein (the active toxin) and a small peptide that are released into the culture supernatant, and a C-terminal β-barrel domain that remains associated with the outer membrane (8, 22, 27, 28). Therefore, we next analyzed growth phase-dependent variation in the abundance of the individual VacA proteolytic products. Specifically, we analyzed spectra assigned to VacA that were detected in the supernatant and membrane fractions, and mapped the distribution of these peptides in relation to the full-length VacA sequence (Fig. 7A and B). At 24h, VacA peptides spanning the full length of the unprocessed protein (1290 amino acids) were detected in the membrane fraction, while VacA peptides detected in the supernatant at this time point only spanned the first 850 amino acids of VacA. At 36h, spectral coverage of VacA in the supernatant was similar to the coverage at 24h, but a substantially higher number of spectral counts were assigned to VacA in the 36h supernatant sample than in the 24h supernatant sample. The VacA spectral coverage differed when comparing membrane fractions from 36h and 24h time points; VacA peptides derived from the C-terminal β-barrel region of VacA were detected in the membrane fraction at 24h but not 36h. Collectively, these data are consistent with release of an 88 kDa soluble VacA protein (the active toxin) into the culture supernatant, and retention of the C-terminal β-barrel domain with the outer membrane (8, 22, 27, 28). These results suggest that the C-terminal membrane-associated region of VacA is degraded at later time points following secretion of the VacA passenger domain.
Figure 7. Analysis of VacA and selected outer membrane proteins in broth culture supernatant and membrane fractions.
H. pylori was grown in broth culture, and the cultures were processed to yield supernatant, CP/PP and membrane fractions. (A, B) Peptides assigned to VacA were analyzed. The graphs depict the distribution of VacA peptides detected in the supernatant and membrane fractions at 24h and 36h. Blue bars indicate results for culture supernatant and red bars indicate results for membrane fractions. The x-axis corresponds to 1290 amino acids that comprise the translated VacA protein. Amino acids 1–33 correspond to a signal sequence, amino acids 34 to about 854 correspond to an 88 kDa secreted VacA protein with toxin activity, and amino acids near the C-terminus are predicted to constitute a β-barrel domain that inserts into the outer membrane. (C–E) Peptides assigned to three outer membrane proteins were analyzed: HopA (Omp6, HP0229), HopQ (Omp27, HP1177) and BabA (Omp28, HP1243). The graphs depict the distribution of assigned peptides detected in the supernatant and membrane fractions at 24h.
Detection of proteins annotated as outer membrane proteins in culture supernatant
Consistent with the results of previous studies (8–10), we detected multiple proteins annotated as outer membrane proteins in the culture supernatant. Outer membrane proteins in the supernatant were detected mainly at intermediate time points (24, 30 and 36h), and were less abundant at 12h or 48h time points. One hypothesis is that the outer membrane proteins are proteolytically processed and the resulting cleavage products are released as soluble proteins, similar to the secretion of the VacA passenger domain. In support of this hypothesis, a recent study analyzed the topology of H. pylori outer membrane proteins on the bacterial surface and noted that many have a topology similar to that of VacA, consistent with an autotransporter mode of secretion (17). To investigate this topic further, we analyzed the peptide sequence distribution maps of several outer membrane proteins that were detected in the culture supernatant, in order to determine if the assigned peptide sequences were derived from the full-length proteins or only limited regions, similar to the pattern observed with VacA. The supernatant peptides assigned to HopA (Omp6; HP0229), HopQ (Omp27; HP1177), and BabA (Omp28; HP1243) spanned the length of the entire proteins at both 24h (Fig. 7C–E) and 36h time points (data not shown). These experimental data suggest that the full-length outer membrane proteins, including the C-terminal regions corresponding to predicted β-barrel domains, are released into the culture supernatant. This pattern is markedly different from the observed secretion of the VacA passenger domain and retention of the VacA β-barrel domain in the outer membrane (Fig. 7).
OMPs could potentially enter the culture supernatant as components of small membrane vesicles or membrane fragments that were not completely removed by ultracentrifugation. To investigate this possibility, we analyzed the protein content of the insoluble material removed from culture supernatant by ultracentrifugation, which is predicted to contain membrane vesicles. The most abundant proteins in these preparations (based on numbers of assigned spectral counts) were outer membrane proteins Omp2, Omp9, and Omp27 (Suppl. Table 2). These were also among the most abundant OMPs detected in the soluble supernatant fraction (Table 1). The similarities between the OMPs detected in the insoluble and soluble supernatant fractions suggest that OMPs may enter the supernatant as membrane vesicles or membrane fragments that are incompletely removed from the culture supernatant by ultracentrifugation.
Distribution of proteins in subcellular bacterial fractions
We hypothesized that among the selectively released proteins identified in these experiments, there might be differences in the subcellular localization of the proteins within intact H. pylori cells, prior to protein release into the culture supernatant. For example, some of the selectively released proteins might be relatively more abundant in the soluble CP/PP bacterial fraction than in the membrane fraction, and others might be relatively more abundant in the membrane fraction than in the CP/PP fraction. Therefore, we investigated the subcellular localization of selectively released proteins in CP/PP and membrane fractions derived from intact bacteria. Specifically, we calculated the relative abundance in the membrane fraction, using the formula [%abundancemembrane/(%abundancemembrane+%abundanceCP/PP)], based on data for all time points.
Among 74 selectively released proteins, 35 had a higher proportional abundance in the membrane fraction than in the soluble CP/PP fraction (>50% relative abundance in membrane fraction), while the remaining 39 had a higher proportional abundance in the CP/PP fraction (<50% relative abundance in membrane fraction) (Table 1). VacA was detected predominantly in the membrane fraction (89.9% relative abundance in membrane fraction). Among the 35 proteins that localized predominantly to the membrane fraction (>50% relative abundance in membrane fraction), 21 were previously localized to the bacterial surface, based on biotinylation experiments (17). Among the 39 proteins with a higher proportional abundance in the CP/PP fraction, only 4 were localized to the bacterial surface in previous biotinylation experiments (17). Many of the proteins detected predominantly in the soluble fraction (<50% relative abundance in membrane fraction) have orthologs that are localized to intracellular sites in other bacterial species. We speculate that the group of proteins localized predominantly to the membrane fraction (>50% relative abundance in membrane fraction) are released into the supernatant by a process requiring localization to the outer membrane, whereas the group of proteins localized predominantly to the soluble fraction are released into the culture supernatant directly from the bacterial cytoplasm or periplasm as soluble proteins without a requirement for membrane localization.
Analysis of signal sequences
The translocation of bacterial proteins across the inner membrane often involves the use of Sec-dependent machinery and cleavage of amino-terminal signal sequences. To evaluate the presence or absence of signal sequences within the 74 proteins that were selectively released into the supernatant, we analyzed the protein sequences using the SignalP 4.1 server. Predicted signal sequences were identified for 34 (46%) of the 74 proteins (Table 2 and 3). Fourteen (56%) of the 25 proteins enriched in the supernatant at 4 or 5 time points were predicted to contain signal sequences. Similarly, 8 (53%) of 15 proteins enriched at varied time points (no clear pattern) were predicted to contain signal sequences, and eleven (65%) of 17 proteins enriched at only late time points were predicted to contain signal sequences. In contrast, only one (6%) of the 17 proteins enriched at exclusively early time points was predicted to contain a signal sequence (p=0.0008 compared to proteins enriched at only late time points).
Table 2.
Selectively released H. pylori supernatant proteins identified in previous studies and confirmed in current study.
| Name | Accession | Methodd | Phase of enrichment |
Reference | Predicted cleavage sitee |
|---|---|---|---|---|---|
| hypothetical protein | HP0129 | 1D | late | b | 21 |
| outer membrane protein (omp6) | HP0229 | 1D | varied | c | - |
| hypothetical protein | HP0231 | 1D | late | a,b | - |
| ATP-dependent nuclease (addB) | HP0275 | both | late | c | 22 |
| dipeptide ABC transporter substrate-binding protein DppA | HP0298 | 1D | constitutive | c | − |
| thiol:disulfide interchange protein (dsbC), putative | HP0377 | both | constitutive | a | 24 |
| putative neuraminyllactose-binding hemagglutinin homolog (hpaA) | HP0410 | both | constitutive | c | 24 |
| outer membrane protein (omp11) | HP0472 | 1D | varied | c | 18 |
| Cag pathogenicity island protein (cag24) | HP0545 | both | late | c | 30 |
| processing protease (ymxG) | HP0657 | both | constitutive | c | 20 |
| conserved hypothetical protein | HP0710 | 1D | varied | c | - |
| hypothetical protein | HP0721 | 1D | late | b | 20 |
| catalase | HP0875 | both | constitutive | c | - |
| vacuolating cytotoxin | HP0887 | both | late | a,b,c | 33 |
| hypothetical protein | HP0953 | both | constitutive | c | 21 |
| hypothetical protein | HP0973 | 1D | varied | b | 28 |
| protease (pqqE) | HP1012 | both | constitutive | c | 28 |
| serine protease (htrA) | HP1019 | both | constitutive | a,c | - |
| conserved hypothetical secreted protein | HP1098 | both | constitutive | a,c | 25 |
| gamma-glutamyltranspeptidase (ggt) | HP1118 | both | constitutive | a,c | 26 |
| hypothetical protein | HP1173 | both | constitutive | a,c | 26 |
| carbonic anhydrase | HP1186 | 1D | late | a,c | 18 |
| conserved hypothetical protein | HP1285 | both | constitutive | c | - |
| conserved hypothetical secreted protein | HP1286 | both | late | a,b,c | 17 |
| outer membrane protein (omp30) | HP1395 | 1D | late | c | 21 |
| hypothetical protein | HP1454 | both | constitutive | a,c | 18 |
| membrane-associated lipoprotein (lpp20) | HP1456 | both | constitutive | c | - |
| hypothetical protein | HP1457 | 1D | varied | c | 28 |
| outer membrane protein | HP1564 | both | constitutive | b | 28 |
Bumann et al. (reference 8)
Kim et al., (reference 9)
Smith et al., (reference 10)
Method indicates the mass spectrometry technique used to identify the protein in the current study. Proteins that were identified in MudPIT and 1D experiments are labeled “both”.
Predicted signal sequence cleavage site. Numbers represent the position of the amino acid N-terminal to the site of predicted cleavage. Dash (−) signifies that no signal sequence was predicted.
Table 3.
Selectively released H. pylori supernatant proteins identified in the current study but not previous studies.
| Name | Accession | Methoda | Phase of enrichment |
Predicted cleavage siteb |
|---|---|---|---|---|
| pantoate-beta-alanine ligase (panC) | HP0006 | 1D | early | - |
| outer membrane protein (omp2) | HP0025 | 1D | late | 20 |
| 2’,3’-cyclic-nucleotide 2’-phosphodiesterase (cpdB) | HP0104 | both | constitutive | - |
| translation elongation factor EF-P (efp) | HP0177 | MudPIT | constitutive | - |
| serine hydroxymethyltransferase (glyA) | HP0183 | 1D | early | - |
| triosephosphate isomerase (tpi) | HP0194 | both | early | - |
| hypothetical protein | HP0204 | both | late | 19 |
| conserved hypothetical secreted protein | HP0211 | 1D | constitutive | - |
| hypothetical protein | HP0218 | 1D | early | - |
| conserved hypothetical protein | HP0233 | 1D | early | - |
| conserved hypothetical secreted protein | HP0235 | both | constitutive | - |
| octaprenyl-diphosphate synthase (ispB) | HP0240 | 1D | early | - |
| hypothetical protein | HP0304 | 1D | constitutive | 19 |
| outer membrane protein (omp9) | HP0317 | 1D | early | 20 |
| arginyl-tRNA synthetase (argS) | HP0319 | both | varied | - |
| membrane bound endonuclease (nuc) | HP0323 | 1D | constitutive | - |
| hypothetical protein | HP0408 | 1D | constitutive | - |
| catalase-like protein | HP0485 | both | constitutive | 24 |
| conserved hypothetical protein | HP0507 | 1D | early | - |
| membrane fusion protein (mtrC) | HP0606 | 1D | late | - |
| adenylate kinase (adk) | HP0618 | MudPIT | constitutive | - |
| NAD(P)H-flavin oxidoreductase | HP0642 | 1D | early | - |
| soluble lytic murein transglycosylase (slt) | HP0645 | 1D | late | - |
| protective surface antigen D15 | HP0655 | 1D | early | - |
| hypothetical protein | HP0688 | 1D | early | - |
| L-asparaginase II (ansB) | HP0723 | both | late | - |
| xanthine guanine phosphoribosyl transferase (gpt) | HP0735 | 1D | varied | - |
| hypothetical protein | HP0783 | 1D | varied | - |
| conserved hypothetical secreted protein | HP0785 | 1D | constitutive | 19 |
| peptide deformylase (def) | HP0793 | 1D | early | - |
| flagellar sheath adhesin hpaA | HP0797 | 1D | early | - |
| CDP-diglyceride hydrolase (cdh) | HP0871 | 1D | varied | 21 |
| outer membrane protein (omp19) | HP0896 | 1D | varied | 19 |
| amino-acid ABC transporter, periplasmic binding protein (yckK) | HP0940 | both | early | - |
| phosphoglycerate mutase (pgm) | HP0974 | 1D | early | - |
| colicin tolerance-like protein (tolB) | HP1126 | both | constitutive | - |
| outer membrane protein (omp27) | HP1177 | 1D | varied | 21 |
| purine-nucleoside phosphorylase (deoD) | HP1178 | 1D | early | - |
| aspartate-semialdehyde dehydrogenase (asd) | HP1189 | 1D | varied | - |
| cytochrome c553 | HP1227 | 1D | late | 19 |
| outer membrane protein (omp28) | HP1243 | 1D | varied | 20 |
| hypothetical protein | HP1265 | 1D | early | - |
| methionine amino- peptidase (map) | HP1299 | 1D | late | - |
| (3R)-hydroxymyristoyl-(acyl carrier protein) dehydratase (fabZ) | HP1376 | 1D | varied | - |
| hypothetical protein | HP1455 | 1D | varied | - |
| iron(III) ABC transporter, periplasmic iron-binding protein (ceuE) | HP1561 | both | late | 31 |
| iron(III) ABC transporter, periplasmic iron-binding protein (ceuE) | HP1562 | both | constitutive | 29 |
Method indicates the mass spectrometry technique used to identify the protein in the current study. Proteins that were identified in MudPIT and 1D experiments are labeled “both”.
Predicted signal sequence cleavage site. Numbers represent the position of the amino acid N-terminal to the site of predicted cleavage. Dash (−) signifies that no signal sequence was predicted.
Of the 74 proteins identified in the current study that are selectively released into the extracellular space, 29 were identified in at least one previous proteomic study (Table 2), and 47 proteins identified in the current study (45 identified by 1D analysis and 2 additional proteins identified by MudPIT) were not identified in any of the previous proteomic studies (Table 3). Conversely, 36 of the proteins identified in the previous studies were not classified as selectively released in the current study (Suppl. Table 3). Of the 29 selectively released proteins that were found in the current study and at least one previous study, 21 (72%) were predicted to contain signal sequences (Table 2). The presence of predicted signal sequences in a large proportion of these proteins is consistent with a model in which these proteins are specifically secreted into the extracellular space through a Sec-dependent process. In contrast, only 5 (12%) of 39 proteins that were reported previously but not confirmed in the current study contained predicted signal sequences (Suppl. Table 3) (p=0.0001). Thirteen (28%) of 46 proteins unique to the current study (not identified in previous proteomic studies) were predicted to have signal sequences (Table 3) (p=0.0003 compared to proteins detected in both the current study and at least one previous study).
Discussion
The results of this study provide a comprehensive view of the H. pylori exoproteome. In total, we identified 74 H. pylori proteins that are selectively released into the extracellular space. By analyzing multiple phases of H. pylori growth, we were able to detect marked differences in the composition of the H. pylori exoproteome at different time points.
Three previous proteomic studies analyzed the H. pylori exoproteome using an assortment of methods (8–10), but only 4 putative secreted proteins were identified by all three of the previous studies. One probable reason for the low level of concordance is related to the use of different growth conditions and multiple H. pylori strains in the individual studies. Two studies used defined media (with compositions similar to RPMI tissue culture medium), and another study used Brain Heart Infusion medium. Cultures were harvested between 20 and 24h in all three studies, but the optical densities of the cultures varied considerably (OD600 values ranging from 0.2 to 0.94). Ultracentrifugation of culture supernatants to remove membrane vesicles was employed in some studies but not others. Another reason for the low level of concordance is related to the use of different analytical methods. Two studies used 2D gel-based methods with UreB as a reference protein, and a third study used a direct analysis of the culture supernatant. Other possible reasons for the low level of concordance include difficulty in detecting low-abundance proteins by 2D gel-based methods, or limitations in the methods for quantitative analysis, with consequent difficulty in distinguishing selectively released proteins from intracellular proteins released during autolysis. The methodology employed in the current study allowed us to directly analyze the composition of the broth culture supernatant without requiring the use of gel-based methods, and the identification of proteins selectively enriched in the supernatant was based on a comparison of the relative abundance of individual proteins in the supernatant to their relative abundance in preparations of soluble proteins derived from intact bacteria.
As one approach for analyzing the 74 H. pylori proteins that were selectively released into the extracellular space, we assessed the localization of these proteins within subcellular fractions derived from intact bacteria. Specifically, we investigated the relative abundance of proteins in the soluble (CP/PP) and membrane fractions, and thereby determined whether individual proteins were localized predominantly to a soluble bacterial fraction (corresponding to cytoplasmic or periplasmic localization) or a membrane-associated bacterial fraction. Many of the selectively released proteins (including several annotated as “conserved hypothetical secreted proteins”) were localized mainly to a soluble bacterial fraction, and we speculate that these proteins are released directly from the cytoplasm or periplasm into the extracellular space (Table 1 and (19)). Other putative secreted proteins, including VacA, were localized mainly to a membrane-associated bacterial fraction. We speculate that these latter proteins may be exported to the surface of the bacteria and remain tethered for some period of time in that location, prior to release into the extracellular space. In support of this hypothesis, many of the proteins localized mainly to a membrane-associated bacterial fraction were previously identified as surface-exposed H. pylori proteins through the use of a biotinylation-based proteomics approach (17) (Table 1).
Some of the selectively released proteins identified in this study met the criteria for enrichment in the supernatant at nearly all phases of bacterial growth, whereas others were considered enriched mainly at early or late growth phases. The mechanisms that govern growth phase-dependent variations in composition of the H. pylori exoproteome are not yet known. One possibility is that growth phase-dependent variations in gene transcription account for the observed patterns. For example, VacA transcription is maximal at late log phase (29), and this could account for the observed increase in relative abundance of VacA in the culture supernatant at later time points. Alternatively, we speculate that there could be growth phase-dependent regulation of specific processes involved in protein secretion or release of proteins from the bacteria, or differences in the stability of proteins after they are released from the bacteria.
There was a strong correlation between the proportional abundance of proteins in the culture supernatant at specific growth phases and the intracellular localization of proteins in intact bacteria. For example, most of the proteins enriched in the supernatant at early growth phases correspond to proteins that are localized mainly to soluble compartments in intact H. pylori. Among 17 proteins enriched in the supernatant at early growth phases, the mean ± standard error relative abundance of these proteins in the membrane fraction was 30 ± 8%; this value was significantly lower than the corresponding mean values for 17 proteins selectively released at later time points and 25 proteins selectively released at >4 time points (54 ± 7% and 60 ± 5% relative abundance in membrane fraction, respectively, p< 0.05) (Table 1). Many of the proteins enriched in the supernatant at early time points are orthologous to proteins known to be localized to the cytoplasm in other bacterial species, and only one of the 17 proteins enriched in the supernatant at early time points contained a predicted signal sequence. Collectively, these results are consistent with a model in which many of the proteins released at early time points enter the extracellular space directly from the cytoplasm. We propose that the process underlying release of proteins at early time points differs from mechanisms underlying release of proteins at later time points.
In addition to manually categorizing proteins into groups based on the time points at which they were enriched in the supernatant (early, late, constitutive, or no clear pattern), we used Cluster Affinity Search Technique (CAST) to classify the 74 selectively released proteins, based on temporal patterns of enrichment or proportional abundance in the supernatant. In an analysis of enrichment in the supernatant, the majority of the proteins were grouped into two major clusters: one that increased in enrichment over time and another cluster that decreased in enrichment over time. Multiple proteins known to cause alterations in host cells (e.g. VacA, GGT, HtrA, and HP1286) were grouped within the former cluster.
Twenty one (72%) of the 29 selectively released proteins that were found in the current study and at least one previous study are predicted to contain signal sequences (Table 2), whereas only 5 (12%) of 39 proteins that were previously reported but not confirmed in the current study are predicted to contain signal sequences (Suppl. Table 3) (p<0.0001). The predicted presence of signal sequences in a large proportion of the former group of proteins is consistent with a model in which these proteins are specifically secreted into the extracellular space through a Sec-dependent pathway. The presence of predicted signal sequence in a very low proportion (12%) of proteins detected in previous studies but not identified in the current study suggests that many of these proteins are not specifically secreted into the extracellular space.
Of the 29 proteins that were identified in the current study as well as one or more previous studies, 15 (52%) were detected at ≥4 of the 5 time points analyzed in the current study, 9 (31%) were detected at late time points, and 4 (14%) were detected at varied time points (no clear pattern). None of the 29 proteins identified in both the current and previous studies were released into the supernatant at only early time points, and conversely, none of the 17 proteins released at early time points were identified in previous studies. Previous studies analyzed supernatant samples at a single time point (either 20 or 24 h), and this likely accounts for why that latter group of proteins were not detected in previous studies.
Collectively, the current study and previous studies provide convincing evidence for the selective release of specific H. pylori proteins into the extracellular space. The secretion of VacA is mediated by a type V (autotransporter) pathway (30, 31), but the mechanisms by which most of the other proteins are secreted remain very poorly understood. H. pylori produces flagella, and the secretion pathway utilized for secretion of flagellar components could potentially mediate the secretion of non-flagellar proteins. Four different type IV secretion pathways have been identified in H. pylori (32), and these could also be responsible for protein secretion. Notably, the H. pylori strain analyzed in the current study has an intact cag type IV secretion pathway (33) and contains genes encoding components of the comB T4SS, but does not produce flagella and lacks two of the T4SSs that are present in a subset of other H. pylori strains. H. pylori is not known to contain type I, type II, type III, or type VI secretion systems. We speculate that some of the selectively released H. pylori proteins identified in this study may be secreted by systems that have not yet been characterized.
Previous secretome studies (8–10) have detected the presence of multiple annotated outer membrane proteins in the culture supernatant, and our results confirm these findings. One hypothesis is that the outer membrane proteins are proteolytically processed and fragments are released as soluble proteins, similar to the secretion of the VacA passenger domain. In the current study, we found that peptides derived from the C-terminal β-barrel region of VacA (secreted by an autotransporter mechanism) were detected in the bacterial membrane fraction at early growth phases (prior to VacA detection in the supernatant) but not later growth phases, and were consistently absent from culture supernatants. In contrast, we found that the peptides detected in the culture supernatant and assigned to outer membrane proteins (OMPs) mapped to the full length of the OMP protein sequences. These data suggest that annotated OMPs are released into the supernatant by a mechanism different from the mechanism of VacA secretion, perhaps involving the release of membrane fragments or very small membrane vesicles that are not completely removed from the supernatant by ultracentrifugation.
One of the annotated outer membrane proteins detected in the current study as well as one previous study (9), HP1564, was localized mainly to the CP/PP fraction instead of the membrane fraction derived from intact bacteria. BLAST analysis indicated that this protein has very little sequence homology to other H. pylori outer membrane proteins. Therefore, we suspect that the original annotation of this protein as an “outer membrane protein” may not be accurate.
Three of the selectively released proteins identified in this study were originally annotated as “conserved hypothetical secreted proteins”, and are now known to belong to a paralogous family of cysteine-rich proteins (34): HcpA (HP0211), HcpC (HP1098), and HcpE (HP0235). HcpA (HP0211) was previously detected in culture supernatant based on its antigenic properties (34), but it was not previously known whether this protein was selectively released into the culture supernatant. HcpA is known to be capable of stimulating cytokine production by splenocytes (35) and has been reported to stimulate differentiation of human myeloid monocytes into macrophages (36). The Hcp family proteins are also of interest because several genes in the family exhibit a very high level of positive selection (37). Only one of the Hcp family proteins, HP1098 (HcpC), was detected in previous secretome studies (8, 10).
In addition to HcpA, several of the other selectively released proteins identified in this study are known to be capable of causing alterations in host cells. These include the vacuolating toxin (VacA), GGT, HtrA, and HP1286 (22–26). Several of the selectively released proteins, including VacA, HcpC (HP1098), and HP0231, are known to elicit an antibody response in H. pylori-infected humans (38–41). Some of these antigenic proteins, including VacA, HpaA paralog (HP0410), and HP0231, have been shown to elicit a protective immune response when administered as vaccine antigens in animal models (42–44).
Many of the selectively released proteins identified in this study are annotated as “hypothetical proteins”, which do not exhibit any relatedness to proteins produced by other bacterial species. Structural studies have been undertaken with several of these hypothetical proteins (6), but their functions remain unknown. Similarly, several selectively released proteins that elicit antibody responses in infected humans or that are required for H. pylori colonization of the stomach have not yet been analyzed at a functional level. In future studies, it will be important to determine the functions of these proteins, and to gain an increased understanding of the mechanisms by which these proteins are secreted.
Supplementary Material
Statement of Significance.
Delineating the composition of the H. pylori exoproteome is necessary for understanding mechanisms of H. pylori protein secretion, and provides insight into the array of H. pylori proteins that may be encountered by host cells in vivo. We demonstrate for the first time that the composition of the H. pylori exoproteome is dependent on the growth phase of the bacteria. The selectively released proteins identified in this study include several that are known to cause alterations in host cells, proteins that elicit a humoral immune response in H. pylori-infected persons, and proteins of unknown function. We propose that the release of these proteins in vivo contributes to a gastric mucosal inflammatory response and alterations in the gastric environment.
Highlights.
H. pylori proteins are selectively released into the extracellular space.
Toxins, antigens, and proteins of unknown function are selectively released.
Composition of the H. pylori exoproteome is dependent on the bacterial growth phase.
Time course analyses provide insight into mechanisms of H. pylori protein secretion.
Acknowledgments
Supported by NIH AI039657, CA116087, and the Department of Veterans Merit Review grant 2I01BX000627. Proteomics experiments were supported by the Vanderbilt Digestive Diseases Research Center (P30DK058404) and Vanderbilt-Ingram Cancer Center (P30 CA068485).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Dooley CP, Cohen H, Fitzgibbons PL, Bauer M, Appleman MD, Perez-Perez GI, et al. Prevalence of Helicobacter pylori infection and histologic gastritis in asymptomatic persons. N Engl J Med. 1989;321(23):1562–1566. doi: 10.1056/NEJM198912073212302. [DOI] [PubMed] [Google Scholar]
- 2.Pérez-Pérez GI, Dworkin BM, Chodos JE, Blaser MJ. Campylobacter pylori antibodies in humans. Ann Intern Med. 1988;109(1):11–17. doi: 10.7326/0003-4819-109-1-11. [DOI] [PubMed] [Google Scholar]
- 3.Algood HM, Cover TL. Helicobacter pylori persistence: an overview of interactions between H. pylori and host immune defenses. Clin Microbiol Rev. 2006;19(4):597–613. doi: 10.1128/CMR.00006-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Suerbaum S, Michetti P. Helicobacter pylori infection. N Engl J Med. 2002;347(15):1175–1186. doi: 10.1056/NEJMra020542. [DOI] [PubMed] [Google Scholar]
- 5.Cover TL, Blaser MJ. Helicobacter pylori in health and disease. Gastroenterology. 2009;136(6):1863–1873. doi: 10.1053/j.gastro.2009.01.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zanotti G, Cendron L. Structural and functional aspects of the Helicobacter pylori secretome. World J Gastroenterol. 2014;20(6):1402–1423. doi: 10.3748/wjg.v20.i6.1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Phadnis SH, Parlow MH, Levy M, Ilver D, Caulkins CM, Connors JB, et al. Surface localization of Helicobacter pylori urease and a heat shock protein homolog requires bacterial autolysis. Infect Immun. 1996;64(3):905–912. doi: 10.1128/iai.64.3.905-912.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bumann D, Aksu S, Wendland M, Janek K, Zimny-Arndt U, Sabarth N, et al. Proteome analysis of secreted proteins of the gastric pathogen Helicobacter pylori . Infect Immun. 2002;70(7):3396–3403. doi: 10.1128/IAI.70.7.3396-3403.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim N, Weeks DL, Shin JM, Scott DR, Young MK, Sachs G. Proteins released by Helicobacter pylori in vitro. J Bacteriol. 2002;184(22):6155–6162. doi: 10.1128/JB.184.22.6155-6162.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith TG, Lim JM, Weinberg MV, Wells L, Hoover TR. Direct analysis of the extracellular proteome from two strains of Helicobacter pylori . Proteomics. 2007;7(13):2240–2245. doi: 10.1002/pmic.200600875. [DOI] [PubMed] [Google Scholar]
- 11.Olofsson A, Vallstrom A, Petzold K, Tegtmeyer N, Schleucher J, Carlsson S, et al. Biochemical and functional characterization of Helicobacter pylori vesicles. Mol Microbiol. 2010;77(6):1539–1555. doi: 10.1111/j.1365-2958.2010.07307.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parker H, Chitcholtan K, Hampton MB, Keenan JI. Uptake of Helicobacter pylori outer membrane vesicles by gastric epithelial cells. Infect Immun. 2010;78(12):5054–5061. doi: 10.1128/IAI.00299-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vanet A, Labigne A. Evidence for specific secretion rather than autolysis in the release of some Helicobacter pylori proteins. Infect Immun. 1998;66(3):1023–1027. doi: 10.1128/iai.66.3.1023-1027.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tomb J-F, White O, Kerlavage AR, Clayton RA, Sutton GG, Fleischmann RD, et al. The complete genome sequence of the gastric pathogen Helicobacter pylori . Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 15.Hawrylik SJ, Wasilko DJ, Haskell SL, Gootz TD, Lee SE. Bisulfite or sulfite inhibits growth of Helicobacter pylori . J Clin Microbiol. 1994;32(3):790–792. doi: 10.1128/jcm.32.3.790-792.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jimenez-Soto LF, Rohrer S, Jain U, Ertl C, Sewald X, Haas R. Effects of cholesterol on Helicobacter pylori growth and virulence properties in vitro. Helicobacter. 2012;17(2):133–139. doi: 10.1111/j.1523-5378.2011.00926.x. [DOI] [PubMed] [Google Scholar]
- 17.Voss BJ, Gaddy JA, McDonald WH, Cover TL. Analysis of surface-exposed outer membrane proteins in Helicobacter pylori . J Bacteriol. 2014;196(13):2455–2471. doi: 10.1128/JB.01768-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Analytical Chemistry. 1996;68(5):850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 19.Snider CA, Voss BJ, McDonald WH, Cover TL. Supporting data for analysis of the Helicobacter pylori exoproteome. Data in Brief. 2015 doi: 10.1016/j.dib.2015.10.008. (submitted) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, et al. TM4: a free, open-source system for microarray data management and analysis. BioTechniques. 2003;34(2):374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 21.Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8(10):785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 22.Cover TL, Blaser MJ. Purification and characterization of the vacuolating toxin from Helicobacter pylori . J Biol Chem. 1992;267(15):10570–10575. [PubMed] [Google Scholar]
- 23.Schmees C, Prinz C, Treptau T, Rad R, Hengst L, Voland P, et al. Inhibition of T-cell proliferation by Helicobacter pylori gamma-glutamyl transpeptidase. Gastroenterology. 2007;132(5):1820–1833. doi: 10.1053/j.gastro.2007.02.031. [DOI] [PubMed] [Google Scholar]
- 24.Li J, Meng FL, He LH, Zhang JZ. Secreted protein HP1286 of Helicobacter pylori strain 26695 induces apoptosis of AGS cells. Biomed Environ Sci. 2012;25(6):614–619. doi: 10.3967/0895-3988.2012.06.002. [DOI] [PubMed] [Google Scholar]
- 25.Basak C, Pathak SK, Bhattacharyya A, Pathak S, Basu J, Kundu M. The secreted peptidyl prolyl cis, trans-isomerase HP0175 of Helicobacter pylori induces apoptosis of gastric epithelial cells in a TLR4- and apoptosis signal-regulating kinase 1-dependent manner. J Immunol. 2005;174(9):5672–5680. doi: 10.4049/jimmunol.174.9.5672. [DOI] [PubMed] [Google Scholar]
- 26.Hoy B, Lower M, Weydig C, Carra G, Tegtmeyer N, Geppert T, et al. Helicobacter pylori HtrA is a new secreted virulence factor that cleaves E-cadherin to disrupt intercellular adhesion. EMBO Rep. 2010;11(10):798–804. doi: 10.1038/embor.2010.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Telford JL, Ghiara P, Dell’Orco M, Comanducci M, Burroni D, Bugnoli M, et al. Gene structure of the Helicobacter pylori cytotoxin and evidence of its key role in gastric disease. J Exp Med. 1994;179(5):1653–1658. doi: 10.1084/jem.179.5.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nguyen VQ, Caprioli RM, Cover TL. Carboxy-terminal proteolytic processing of Helicobacter pylori vacuolating toxin. Infect Immun. 2001;69(1):543–546. doi: 10.1128/IAI.69.1.543-546.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Forsyth MH, Cover TL. Intercellular communication in Helicobacter pylori: luxS is essential for the production of an extracellular signaling molecule. Infect Immun. 2000;68(6):3193–3199. doi: 10.1128/iai.68.6.3193-3199.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fischer W, Buhrdorf R, Gerland E, Haas R. Outer membrane targeting of passenger proteins by the vacuolating cytotoxin autotransporter of Helicobacter pylori . Infect Immun. 2001;69(11):6769–6775. doi: 10.1128/IAI.69.11.6769-6775.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gangwer KA, Mushrush DJ, Stauff DL, Spiller B, McClain MS, Cover TL, et al. Crystal structure of the Helicobacter pylori vacuolating toxin p55 domain. Proc Natl Acad Sci U S A. 2007;104(41):16293–16298. doi: 10.1073/pnas.0707447104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rohrer S, Holsten L, Weiss E, Benghezal M, Fischer W, Haas R. Multiple pathways of plasmid DNA transfer in Helicobacter pylori . PLoS One. 2012;7(9):e45623. doi: 10.1371/journal.pone.0045623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fischer W, Puls J, Buhrdorf R, Gebert B, Odenbreit S, Haas R. Systematic mutagenesis of the Helicobacter pylori cag pathogenicity island: essential genes for CagA translocation in host cells and induction of interleukin-8. Mol Microbiol. 2001;42(5):1337–1348. doi: 10.1046/j.1365-2958.2001.02714.x. [DOI] [PubMed] [Google Scholar]
- 34.Cao P, McClain MS, Forsyth MH, Cover TL. Extracellular release of antigenic proteins by Helicobacter pylori . Infect Immun. 1998;66(6):2984–2986. doi: 10.1128/iai.66.6.2984-2986.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Deml L, Aigner M, Decker J, Eckhardt A, Schutz C, Mittl PR, et al. Characterization of the Helicobacter pylori cysteine-rich protein A as a T-helper cell type 1 polarizing agent. Infect Immun. 2005;73(8):4732–4742. doi: 10.1128/IAI.73.8.4732-4742.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dumrese C, Slomianka L, Ziegler U, Choi SS, Kalia A, Fulurija A, et al. The secreted Helicobacter cysteine-rich protein A causes adherence of human monocytes and differentiation into a macrophage-like phenotype. FEBS Lett. 2009;583(10):1637–1643. doi: 10.1016/j.febslet.2009.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ogura M, Perez JC, Mittl PR, Lee HK, Dailide G, Tan S, et al. Helicobacter pylori evolution: lineage- specific adaptations in homologs of eukaryotic Sel1-like genes. PLoS Comput Biol. 2007;3(8):e151. doi: 10.1371/journal.pcbi.0030151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cover TL, Cao P, Murthy UK, Sipple MS, Blaser MJ. Serum neutralizing antibody response to the vacuolating cytotoxin of Helicobacter pylori . J Clin Invest. 1992;90(3):913–918. doi: 10.1172/JCI115967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mittl PR, Luthy L, Reinhardt C, Joller H. Detection of high titers of antibody against Helicobacter cysteine-rich proteins A, B, C, and E in Helicobacter pylori-infected individuals. Clin Diagn Lab Immunol. 2003;10(4):542–545. doi: 10.1128/CDLI.10.4.542-545.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Haas G, Karaali G, Ebermayer K, Metzger WG, Lamer S, Zimny-Arndt U, et al. Immunoproteomics of Helicobacter pylori infection and relation to gastric disease. Proteomics. 2002;2(3):313–324. doi: 10.1002/1615-9861(200203)2:3<313::aid-prot313>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 41.Bumann D, Holland P, Siejak F, Koesling J, Sabarth N, Lamer S, et al. A comparison of murine and human immunoproteomes of Helicobacter pylori validates the preclinical murine infection model for antigen screening. Infect Immun. 2002;70(11):6494–6498. doi: 10.1128/IAI.70.11.6494-6498.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Del Giudice G, Covacci A, Telford JL, Montecucco C, Rappuoli R. The design of vaccines against Helicobacter pylori and their development. Annual Rev Immunology. 2001;19:523–563. doi: 10.1146/annurev.immunol.19.1.523. [DOI] [PubMed] [Google Scholar]
- 43.Sabarth N, Hurwitz R, Meyer TF, Bumann D. Multiparameter selection of Helicobacter pylori antigens identifies two novel antigens with high protective efficacy. Infect Immun. 2002;70(11):6499–6503. doi: 10.1128/IAI.70.11.6499-6503.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hongying F, Xianbo W, Fang Y, Yang B, Beiguo L. Oral immunization with recombinant Lactobacillus acidophilus expressing the adhesin Hp0410 of Helicobacter pylori induces mucosal and systemic immune responses. Clin Vaccine Immunol. 2014;21(2):126–132. doi: 10.1128/CVI.00434-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.