Abstract
Oligodendrocyte:oligodendrocyte (O:O) gap junction (GJ) coupling is a widespread and essential feature of the CNS, and is mediated by connexin47 (Cx47) and Cx32. Loss of function mutations affecting Cx47 results in a severe leukodystrophy, Pelizeus-Merzbacher-like disease (also known as Hypomyelinating Leukodystrophy 2), which can be reproduced in mice lacking both Cx47 and Cx32. Here we report the gene expression profile of the cerebellum – an affected brain region – in mice lacking both Cx47 and Cx32. Of the 43,174 mRNA probes examined, we find decreased expression of 23 probes (corresponding to 23 genes) and increased expression of 545 probes (corresponding to 348 genes). Many of the genes with reduced expression map to oligodendrocytes, and two of them (Fa2h and Ugt8a) are involved in the synthesis of myelin lipids. Many of the genes with increased expression map to microglia and lymphocytes, and to leukotriene/prostaglandin synthesis and chemokine/cytokine pathways. In accord, immunostaining showed activated microglia and astrocytes, as well as T- and B-cells in the cerebella of mutant mice. Thus, in addition to the loss of GJ coupling, there is a prominent immune response in mice lacking both Cx47 and Cx32.
Introduction
Gap junctions (GJs) are intercellular channels between apposed cell membranes. They permit the electrical communication between cells as well as the diffusion of ions and small molecules typically less than 1000 Da (Bruzzone, et al. 1996). In vertebrates, GJs are comprised of connexins (Cxs) - a family of integral membrane proteins that are named according to their predicted molecular mass (Willecke, et al. 2002). In humans, mutations in GJB1, the gene that encodes Cx32, cause X-linked Charcot–Marie–Tooth disease (CMT1X), the second most common kind of inherited demyelinating neuropathy (Kleopa and Scherer 2006). Many CMT1X patients also have slowed central conduction, and subsets of patients develop overt CNS manifestations including spasticity, hyperreflexia, ataxia, and acute reversible encephalopathy with white matter abnormalities on MRI (Abrams and Scherer 2011). Recessive mutations in GJC2, the gene that encodes Cx47, cause Pelizaeus–Merzbacher-like disease (PMLD; also known as hypomyelinating leukodystrophy 2), a severe leukodystrophy with childhood onset, characterized by nystagmus, progressive spasticity, and ataxia (Bugiani, et al. 2006, Uhlenberg, et al. 2004). Cx32 is expressed by oligodendrocytes and Schwann cells (Scherer, et al. 1995), and Cx47 is expressed by oligodendrocytes (Menichella, et al. 2003, Menichella, et al. 2006, Odermatt, et al. 2003), so that the demyelination that is seen in CMT1X and PMLD is thought to be caused by cell autonomous effects of GJB1 and GJC1 mutations, respectively. In addition, both Cx32 and Cx47 GJs are also reduced in and around chronic lesions in multiple sclerosis and animal models of multiple sclerosis (Kleopas, et al. 2013, Markoullis, et al. 2012a, Markoullis, et al. 2012b, Masaki 2013), raising the possibility that the loss of these connexins contributes to clinical disability in acquired demyelinating diseases.
How the loss of oligodendrocytes Cxs lead to demyelination has been investigated in rodents. Mice that lack both Cx32 and Cx47 are a model of PMLD as they exhibit a progressive movement disorder and dysmyelination (Menichella, et al. 2003, Menichella, et al. 2006, Odermatt, et al. 2003). Previous electron microscopic studies provided anatomical evidence that oligodendrocytes were GJ coupled only to astrocytes (O:A coupling) but not to themselves (O:O coupling) (Massa and Mugnaini 1982, Massa and Mugnaini 1985, Rash, et al. 2001). However, recent electrophysiological studies using dye transfer in acute brain slices in the corpus callosum in mice lacking Cx32 and/or Cx47 demonstrated extensive O:O coupling mediated by Cx47:Cx47 and Cx32:Cx32 homotypic GJs (Maglione, et al. 2010, Wasseff and Scherer 2011). O:O coupling is found in other white matter tracts (Wasseff and Scherer, submitted), and is thus likely to be typical.
To determine how the loss of oligodendrocytes GJs coupling leads to the pathology of these disorders, we used microarrays and pathway analysis to compare the steady state mRNA levels of brains from Cx32//Cx47-double-null (Gjb1−/Y//Gjc2−/−) mice versus wild type cerebella. The observed changes in Gjb1−/Y//Gjc2−/− mice would be predicted to reduce the synthesis of myelin-related lipids. We also found evidence of immune activation in Gjb1−/Y//Gjc2−/− mice-higher mRNA levels for key enzymes required for leukotriene synthesis from arachidonic acid, as well as for chemokines, interleukins, complement components, regulators of natural killer (NK) cells, B-cells and T-cells. Immunostaining showed lymphocytic infiltration, as well as activated microglia and astrocytes. Our results suggest that oligodendrocytes connexins/coupling is required for normal CNS lipid/myelin metabolism, and is associated with a substantial immune response.
Methods
Microarray RNA analyses and qRT-PCR
We generated Gjb1−/Y//Gjc2−/− and Gjb1+/Y//Gjc2+/+ mice from our colony of Gjb1-null (Nelles, et al. 1996) and Gjc2-null mice (Odermatt, et al. 2003), which have been maintained on a C57BL/6 background for more than 10 generations. These mice develop the full phenotype about the fourth postnatal week (Menichella, et al. 2003, Odermatt, et al. 2003). P29 Gjb1−/Y//Gjc2−/− mice (n=4) and their Gjb1+/Y //Gjc2+/+ littermates (n=4) were euthanized, and their cerebella were dissected, immediately placed in Trizol reagent (Invitrogen), homogenized for 30–60 sec using Omni motor tissue homogenizer, snap frozen in liquid nitrogen. Upon thawing, total RNA was extracted from Trizol reagent according to the manufacturer's instructions. The purity and concentration of the RNA of each sample was determined prior to labeling and hybridization using the Agilent 2100 Bioanalyzer, and dual color expression analysis was conducted using whole Mouse Genome Microarray Kit (Agilent Technologies) using 43,174 probes to analyze the expression of mRNAs. In this array, more than one different probe may be used for different parts of the same gene, and in some instances identical probes are replicated for quality control purposes.
mRNAs that were expressed significantly different levels in the Gjb1−/Y//Gjc2−/− cerebella were analyzed with the Cell Type-Specific Expression Analysis tool (CSEA; described in (Xu, et al. 2014) to identify the candidate cell population of the transcript. Results were further analyzed based cell specific gene profile reported previously to see which genes are specific to microglia (Hickman, et al. 2013) and other CNS cells using RNA sequencing (Zhang, et al. 2014). For pathway analysis, we used DAVID Bioinformatics Resources 6.7 (Huang da, et al. 2009) to examine gene-disease association, functionally related genes and pathway mapping using Kyoto Encyclopedia of Genes and Genomes (KEGG), and we complemented these analyses with using the Panther classification system; mRNA lists were uploaded, and Mus musculus was selected, and subsequently the functional classification was viewed as pie charts.
Gene expression was quantified in quadruplicate (from the same samples used for the microarray analysis) by qRT-PCR using the mouse TaqMan Assay Kits (Applied Biosystems by Life Technologies, Foster City, CA, USA). The reverse transcription reaction was carried out with High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Quantitative PCR was run on a QuantStudio 12K Flex Real-Time PCR system (Applied Biosystems) and the reaction mixtures were incubated at 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. The threshold cycle (Ct) values were calculated with QuanStudio Software version 1.2.2 (Applied Biosystems). The housekeeping gene Gapdh, and Actb were included in the analysis as controls, and water was included as a negative control. Fold change in the gene was calculated by the equation 2-ΔΔCt, the expression was normalized by the housekeeping gene Gapdh, using DataAssist software version 1.2.2.
Immunohistochemistry
P22 Gjb1−/Y//Gjc2−/− mice (n=3) and their Gjb1+/Y//Gjc2+/+ littermates (n=3) were perfused with PBS followed by 4% paraformaldehyde in PBS, the cerebra and cerebella were dissected and fixed for another hour, infiltrated overnight in 10% sucrose in PBS at 4°C, then embedded in OCT. Cryostat sections (10 µm thick) were thaw-mounted on Super Frost Plus glass slides (Fisher Scientific, Pittsburgh, PA) and stored at −20°C. A spleen was dissected and processed in a similar way and used as a control through the immunostaining procedures. Tissue sections were permeabilized by immersion in −20°C acetone for 10 min, incubated for 1 h in blocking solution (0.1% Triton X-100, 5% fish skin gelatin in PBS), incubated overnight at 4°C with one of the following antibodies: a rat monoclonal antibody against Ly6c (1:200 dilution; Sigma), a rabbit antisera against CD3 (1:200 dilution; Santa Cruz ), CD72 (1:200 dilution; Sigma), glial fibrillary acid protein (GFAP; 1:200 dilution ; Sigma), or Iba1 (1:500 dilution; Wako), washed several times in PBS, incubated with rhodamine-conjugated donkey anti-mouse, anti-rat, or anti-rabbit antisera (1:200 dilution; Jackson ImmunoResearch Laboratories), washed in PBS, mounted with Vectashield with DAPI (Vector laboratories), and examined by epifluorescence with appropriate optical filters (Leica DMR).
Results
Altered levels of mRNAs expressed by oligodendrocytes
We compared the expression of 43,174 mRNA probes in individual cerebella from P29 Gjb1−/Y//Gjc2−/− mice (n=4) to their Gjb1+/Y//Gjc2+/+ littermates (n=4) using RNA microarrays, because myelinated axons are prominently affected in the cerebellar white matter of Gjb1−/Y//Gjc2−/− mice (Menichella, et al. 2003). A total of 545 probes had significantly different levels (1.5 fold change or more with less than 10% false discovery rate; Supplemental File 1) - 522 probes (corresponding to 348 different mRNAs) were expressed at higher levels in Gjb1−/Y//Gjc2−/− mice, and 23 probes (corresponding to 23 different mRNAs) were expressed at lower levels. To identify the cellular origin of these mRNAs (Fig. 1), we used the Cell Type-Specific Expression Analysis tool (CSEA; http://genetics.wustl.edu/jdlab/csea-tool-2/), which utilizes the expression of an EGFP-L10a ribosomal transgene in specific cell populations specified by different Bacterial Artificial Chromosomes (BACs). The polysomes are immunoaffinity purified, and their mRNAs are identified; this is called translating ribosome affinity purification (TRAP); (Dougherty, et al. 2012, Doyle, et al. 2008, Xu, et al. 2014). For the discussion, we will assume that the cellular origins of these mRNAs is not altered by the leukodystrophy, but this remains to be determined.
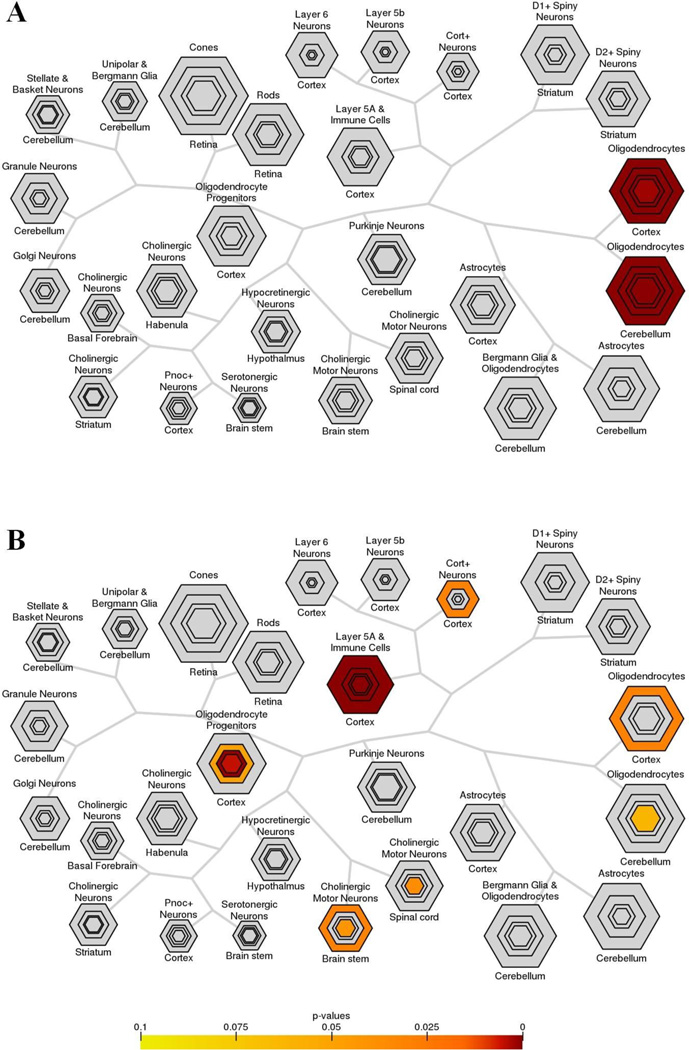
Figure 1. Cerebellar mRNAs analyzed by Cell Type-Specific Expression Analysis (CSEA).
The mRNAs showing lower (A) or higher (B) levels of expression in Gjb1−/Y//Gjc2−/− cerebella were mapped by CSEA (Xu, et al. 2014), which displays the data as hexagrams – their size reflects the number of specific, enriched transcripts at different stringency thresholds, and their color reflects the Fisher’s exact p values. In (A), note that all mRNAs are enriched only in oligodendrocytes at every p value. In (B), note that at any p value, most of the mRNAs map to immune cells (or layer 5a cortical neurons), while at p <0.05, some mRNA map to cortical oligodendrocytes, cerebellar oligodendrocytes, oligodendrocyte progenitors, cholinergic motor neurons in brain stem and spinal cord, and Cort+ cortical interneurons/immune cells.
Figure 1A illustrates the topography of the cell-specific expression of the mRNAs with reduced expression in Gjb1−/Y//Gjc2−/− cerebella. Of the 23 genes whose mRNA levels were reduced (Table 1), 15 mRNAs (Itgb4, Fa2h, Hapln2, Ugt8a, Nkx2–9, Plin3, Trf, Serpinb1a, Anln, Pla2g4a, Dock5, Smtnl2, Klk6, Pkd2l1, and Acy3) correspond to genes that map to cerebellar oligodendrocytes by CSEA at p<0.05. Except for Nkx2–9 and Klk6, these mRNAs also map to cortical oligodendrocytes at p<0.05; none of remaining 10 genes map any other cell type at any given p value. Pathway analysis using the DAVID Bioinformatics Resources tool indicates that these 15 genes are related to signaling pathways and/or metabolic pathways associated with sphingolipid, amino acid, glycerophospholipid, and arachidonic acid metabolism, as well as integrin interactions.
Table 1.
mRNAs expressed at lower levels in Gjb1−/Y//Gjc2−/− cerebella.
The table shows the genes in Gjb1−/Y//Gjc2−/− mice ranked by fold change (FC) in the levels of detected mRNA, and shows the false discovery rate (FDR), adjusted p value for each mRNA, and their expression, mostly according to the CSEA tool based on the work (Xu, et al. 2014) and RNA sequencing (Zhang, et al. 2014). N/A indicates that the mRNA was not reported to map to oligodendrocytes (OL) by the CSEA tool. OL: oligodendrocytes; AS: astrocytes; OPCs: oligodendrocytes precursor cells.
| name | FC | FDR | P | CSEA | Expression; function |
|---|---|---|---|---|---|
| Plin3 | −1.6 | 9.9 | 0.01 | OL | All CNS cells but more enriched in myelinating OL; mannose 6-phosphate receptor-binding protein |
| Birc7 | −1.6 | 4.7 | 0.007 | N/A | AS; inhibitor of apoptosis |
| Fa2h | −1.6 | 5.7 | 0.008 | OL | Myelinating OL and newly formed OL; required for formation of 2-hydroxy fatty acids |
| Dock5 | −1.6 | 4.7 | 0.007 | OL | All CNS cells but more enriched in myelinating OL; small G protein activator |
| Trim16 | −1.7 | 3.7 | 0.005 | N/A | Endothelial cells; tripartite motif family of proteins of yet to be determined exact function |
| Smcr8 | −1.7 | 3.7 | 0.03 | N/A | All CNS cells but more enriched in microglia; Smith- Magenis syndrome chromosomal region |
| Ugt8a | −1.8 | 8.2 | 0.02 | OL | Myelinating OL, newly formed OL, and OPCs; required for synthesis of galactocerebrosides |
| Trf | −1.8 | 3.7 | 0.007 | OL | More enriched in myelinating OL, not expressed in AS or Neurons; iron transport protein |
| Serpinb1a | −1.8 | 5.7 | 0.01 | OL | All CNS cells but more enriched in myelinating OL; serine (or cysteine) proteinase inhibitor |
| Pla2g4a | −1.8 | 1.2 | 0.003 | OL | All CNS cells but more enriched in newly formed OL and myelinating OL; formation of arachidonic acid |
| Pkd2l1 | −1.8 | 0.6 | 0.002 | OL | Myelinating OL and newly formed OL; polycystin protein family involved in cell-cell/matrix interactions |
| Ssxb10 | −1.9 | 3.7 | 0.006 | N/A | newly formed OL; cancer/testis antigen |
| Zfp672 | −1.9 | 1.8 | 0.004 | N/A | CNS, but not restricted to one cell type; zinc finger protein |
| Ssxb1 | −1.9 | 4.7 | 0.009 | N/A | not specific to the CNS; cancer/testis antigen |
| Acy3 | −2.1 | 0.0 | 0.001 | OL | All CNS cells but more enriched in myelinating OL, newly formed OL and endothelial cells; aspartoacylase (aminoacylase) |
| 2610507I0 | −2.3 | 1.0 | 0.006 | N/A | All CNS cells but more enriched in AS; unclassified gene |
| 1Rik | |||||
| Nkx2–9 | −2.5 | 0 | 0.0001 | N/A | Myelinating OL; transcription factor |
| Smtnl2 | −2.6 | 0 | 0.001 | OL | All CNS cells but more enriched in myelinating OL; functionally uncharacterized protein |
| Itgb4 | −2.7 | 0 | 0.001 | OL | Myelinating OL and newly formed OL; integrin subunit |
| Slc5a11 | −2.9 | 0 | 0.002 | N/A | All CNS cells but more enriched in myelinating OL; actin-binding protein sodium glucose co-transporter |
| Anln | −3.7 | 0 | 0.002 | OL | All CNS cells but more enriched in myelinating OL and newly formed OL; actin-binding protein |
| Hapln2 | −3.9 | 0 | 0.001 | OL | Myelinating OL and newly formed OL; hyaluronan- associated matrix in the CNS |
| Klk6 | −5.5 | 0 | 0.001 | N/A | Myelinating OL and newly formed OL; serine protease |
Using Cnp- and Olig2-BACs to define RNAs from oligodendrocytes, 18/348 mRNAs with increased expression in Gjb1−/Y//Gjc2−/− (Table 2) map to cerebellar oligodendrocytes (Fig. 1B) at p<0.05 (Pdlim2, Sh3bp2, Opalin, Prima1, Ppp1r16b, Bfsp2, Csf1, Ada, 2210011C24Rik, Serinc5, Tgfbi, AA986860, Tnfaip6, Hebp1, Gng8, Ctsc, Cd9, Unc93b1). The same mRNAs, plus LCP1, also map to cortical oligodendrocytes. In addition, 15/348 mRNAs with increased levels map to oligodendrocytes progenitors (as defined by expression of Pdgfar-BAC) at p<0.05 (Rab32, Fam111a, Tmem176a, Opalin, Npas1, Gpr17, Bfsp2, Cdk1, Cdca3, Pbk, Dct, Serinc5, 9630013A20Rik, Top2A, and Cdca5), three of which (Bfsp2, Serinc5, and Opalin) map to both oligodendrocytes and their progenitors. Pathway analysis using the DAVID Bioinformatics Resources tool (Table 3) indicates that these genes are related to signaling pathways associated with NK-cells mediated cytotoxicity, cytokine-cytokine receptors interactions, and chemokine signaling pathways. Thus, many genes that expressed by oligodendrocytes have either lower or higher mRNA levels in Gjb1−/Y//Gjc2−/− cerebella. That some of these mRNAs (e.g. Fa2h and Ugt8a) are involved in the synthesis of myelin-related lipids suggests that they may be down-regulated by disrupted myelination, but many other myelin-related genes (e.g., Plp1, Mag) are not similarly affected.
Table 2.
mRNAs expressed at higher levels in Gjb1−/Y//Gjc2−/− cerebella map to oligodendrocytes or oligodendrocytes precursor cells.
The table shows the genes in Gjb1−/Y//Gjc2−/− mice ranked by fold change (FC) in the levels of detected mRNA, and shows the false discovery rate (FDR), adjusted p value for each mRNA, and their expression, mostly based on the work of (Xu, et al. 2014) and (Zhang, et al. 2014). N/A indicates that mRNA was not mapped to cells by (Zhang, et al. 2014). OL: oligodendrocytes; AS: astrocytes; OPCs: oligodendrocytes precursor cells
| Name | FC | FDR | P | CSEA | expression(Zhang, et al. 2014); function |
|---|---|---|---|---|---|
| Bfsp2 | 3.1 | 0 | 0.06 | OL, OPCs | All CNS cells but more enriched in newly formed OL, myelinating OL and OPCs; cytoskeletal component |
| 2210011C24Rik | 2.7 | 0 | 0.003 | OL | N/A; unclassified gene |
| Fam46a | 2.3 | 0 | 0.002 | OL | CNS, but not restricted to one cell type; uncharacterized function |
| Ctsc | 2.2 | 0.2 | 0.04 | OL | All CNS cells but not enriched in newly formed OL or myelinating OL; dipeptidyl aminopeptidase |
| Tmem176a | 2.1 | 0 | 0.01 | OPCs | CNS, but not restricted to one cell type; dendritic cells/carcinoma antigen |
| Pbk | 1.9 | 0 | 0.02 | OPCs | All CNS cells but more enriched in OPCs; lymphokine- activated killer T cells originated PDZ-binding kinase |
| Pdlim2 | 1.9 | 1.2 | 0.1 | OL | All CNS cells but more enriched in myelinating OL and newly formed OL; STAT-interacting protein |
| Cdk1 | 1.9 | 0 | 0.006 | OPCs | All CNS cells but more enriched in endothelial cells; cyclin- dependent kinase |
| Npas1 | 1.9 | 0.3 | 0.03 | OPCs | All CNS cells but more enriched in Neurons, OPCs, and newly formed OL; transcription factor |
| Rab32 | 1.9 | 1 | 0.09 | OPCs | All CNS cells but more enriched in microglia; controls trafficking to lysosomes |
| AA986860 | 1.8 | 0.6 | 0.05 | OL | All CNS cells but more enriched in myelinating OL; uncharacterized function |
| Ada | 1.8 | 0 | 0.01 | OL | CNS, but not restricted to one cell type; adenosine deaminase, present in high levels in lymphocytes |
| Sh3gl3 | 1.8 | 0 | 0.002 | OL | All CNS cells but more enriched in myelinating OL and newly formed OL; implicated in endocytosis |
| 9630013A20Rik | 1.8 | 0 | 0.006 | OPCs | Myelinating OL, new formed OL and OPCs; unclassified gene |
| Fam111a | 1.8 | 0 | 0.01 | OPCs | All CNS cells but more enriched in endothelial cells; governs parathyroid hormone production & calcium homeostasis, |
| Prima1 | 1.7 | 1.2 | 0.1 | OL | All CNS cells but more enriched in myelinating OL and newly formed OL; membrane anchor of acetylcholinesterase in the brain |
| Dct | 1.7 | 0 | 0.006 | OPCs | Newly formed OL, myelinating OL and OPCs; tyrosine- related protein |
| Lcp1 | 1.7 | 0 | 0.003 | OL | All CNS cells but more enriched in microglia; actin-binding protein |
| Prickle1 | 1.6 | 0 | 0.009 | OL | All CNS cells but more enriched in newly formed OL and myelinating OL; nuclear receptor linked to myoclonic epilepsy |
| Cd9 | 1.6 | 0 | 0.004 | OL | All CNS cells but more enriched in newly formed OL; leukocyte surface glycoprotein |
| Top2A | 1.6 | 0.2 | 0.01 | OPCs | CNS, but not restricted to one cell type; DNA topoisomerase |
| Hebp1 | 1.6 | 6.6 | 0.4 | OL | All CNS cells but more enriched in microglia; promotes chemotaxis in monocytes and dendritic cells |
| Cdca5 | 1.6 | 1.8 | 0.05 | OPCs | CNS, but not restricted to one cell type ; cell cycle-associated protein |
| TnfAIP6 | 1.6 | 0.3 | 0.017 | OL | CNS, but not restricted to one cell type; hyaluronan-binding protein |
| Tgfbi | 1.6 | 0 | 0.006 | OL | All CNS cells but more enriched in microglia; inhibit cell adhesion |
| Gpr17 | 1.6 | 0 | 0.006 | OPCs | Myelinating OL, newly formed OL, and OPCs, also in neurons; leukotriene receptor |
| Gng8 | 1.6 | 1.8 | 0.07 | OL | All CNS cells but more enriched in myelinating OL and newly formed OL; G protein involved in transmembrane signaling |
| Cdca3 | 1.6 | 1.2 | 0.05 | OPCs | All CNS cells but more enriched in OPCs; cell cycle- associated protein |
| Opalin | 1.5 | 2.6 | 0.1 | OL, OPCs | Myelinating OL, newly formed OL; myelin paranodal protein |
| Serinc5 | 1.5 | 0.7 | 0.02 | OL, OPCs | All CNS cells but more enriched in newly formed OL, OPCs and myelinating OL; incorporation of serine into phosphatidylserine and sphingolipids |
| Ppp1r16b | 1.5 | 1.8 | 0.07 | OL | All CNS cells but more enriched in newly formed OL, myelinating OL and OPCs; protein phosphatase regulatory subunit |
Table 3.
mRNAs that were higher in Gjb1−/Y//Gjc2−/− cerebella that map to immune cells.
The table shows the genes in Gjb1−/Y//Gjc2−/− mice ranked by fold change (FC) in the levels of detected mRNA, and shows the false discovery rate (FDR), adjusted p value for each mRNA, and their expression, that map to layer 5a cortical neurons and/or immune cells by the CSEA tool, and is known to map to immune cells based on previous reports, mostly based on the work of (Beutner, et al. 2013) and (Hickman, et al. 2013), and maps to immune cells based on DAVID, and is also known to be involved in immune biological processes based on the Panther classification tool. N/A indicates that the mRNA was not reported to map to immune cells by DAVID or Panther.
| name | FC | FDR | p | DAVID (expression) |
enriched in immune cells/associated immune process |
|---|---|---|---|---|---|
| Tyrobp | 5.0 | 0 | 0.0002 | brain, mast cells | N/A |
| Ly86 | 4.2 | 00 | 0.0006 | B-cells | lymphocyte antigen 86 |
| Fcrls | 3.9 | 0 | 0.0006 | diencephalon | Fc receptor-like S, scavenger receptor; lymphocytes activation, B-cell meditated immunity |
| Slc11a1 | 3.7 | 0 | 0.0006 | pre B-cells | also known as natural resistance-associated macrophage protein 1. |
| Fcer1g | 3.6 | 0 | 0.002 | mast cells | Fc receptor, IgE, high affinity I, gamma polypeptide |
| Fcgr2b | 3.6 | 0 | 0.001 | macrophages, mast cells |
Fc receptor, IgG, low affinity IIb; lymphocyte activation, B-cell meditated immunity |
| Ctss | 3.6 | 0 | 0.001 | brain | cathepsin S; antigen processing and presentation via MHC class II proteolysis |
| Blnk | 3.4 | 0 | 0.001 | lymphoid | B-cell linker |
| Irf8 | 3.4 | 0 | 0.001 | spleen, bone marrow |
interferon regulator factor 8; response to interferon gamma |
| C1qc | 3.0 | 0 | 0.001 | macrophages | complement component 1, q subcomponent, C chain. |
| C1qa | 3.0 | 0 | 0.005 | macrophages | complement component 1, q subcomponent, alpha polypeptide |
| Cd68 | 3.0 | 0 | 0.002 | macrophages | macrophages, monocytes marker (Holness and Simmons 1993) |
| Fcgr3 | 2.9 | 0 | 0.001 | hematopoietic stem cells |
Fc receptor, IgG, low affinity III; lymphocytes activation; B-cell meditated immunity |
| Ptpn6 | 2.9 | 0 | 0.0002 | mast cells, | N/A |
| Cd14 | 2.8 | 0 | 0.001 | macrophages | macrophages, monocytes, dendritic cells |
| C1qb | 2.8 | 0 | 0.002 | striatum, macrophages |
complement component 1, q subcomponent, beta polypeptide; complement activation |
| Laptm5 | 2.8 | 0 | 0.001 | mast cells | N/A |
| Fyb | 2.4 | 0 | 0.001 | macrophages, T- cells |
N/A |
| Lyz2 | 2.3 | 0 | 0.001 | bone marrow macrophages |
N/A |
| Cyba | 2.2 | 0 | 0.001 | macrophages | N/A |
| Inpp5d | 2.1 | 0 | 0.003 | macrophages, T- cells |
N/A |
| Tnfaip8l2 | 2.0 | 0 | 0.006 | spinal cord | tumor necrosis factor alpha-induced protein 8- like protein 2 |
| Csf1r | 2.0 | 0 | 0.003 | macrophages | N/A |
| Emr1 | 1.9 | 0 | 0.02 | brain | macrophages activation |
| Rnase4 | 1.8 | 8.2 | 0.6 | N/A | microglia |
| Fermt3 | 1.7 | 0 | 0.003 | hematopoietic stem cell |
N/A |
| Ptpn18 | 1.7 | 1.2 | 0.08 | mast cells | N/A |
| P2ry13 | 1.5 | 2.6 | 0.09 | hippocampus, hypothalamus |
microglia |
| Fes | 1.5 | 0 | 0.008 | mast cells, spleen |
N/A |
Altered levels of mRNAs related to the immune system
In addition to the mRNAs that are enriched in oligodendrocytes, 52 mRNAs with significantly (p<0.05) increased expression in P29 Gjb1−/Y//Gjc2−/− cerebella mapped to genes that are enriched in immune (lymphoid) cells and/or layer 5a cortical neurons (Doyle, et al. 2008), as defined by expression with Etv1_tm88-BAC (Fig. 1B). Because we isolated mRNA from the cerebellum, these mRNAs are more likely to be derived from immune cells, and according to the DAVID and the Panther classification system, 22/52 of these genes are immune-related (Table 3). In addition to the immune-related genes that were identified in this manner, we found many other mRNAs with increased expression that are likely to be expressed by immune cells, including Cd52, Cd86, Cd48, Cd33, Cd68, Cd14, Cd84, Cd9, Cd37, and Cd109 (Table 4). We also found many mRNAs that encode for chemokines that are up-regulated Gjb1−/Y//Gjc2−/− cerebella - Ccl2, Ccl3, Ccl4, Ccl6, Ccl9, Ccl10, Ccl12, and one mRNA for a chemokine receptor (Cx3cr1). Further analysis using both the David bioinformatics tool and the Panther classification system revealed that many genes with increased mRNA levels would be predicted to be involved in NK-, B- and T-cell activation, inflammation mediated by chemokines and cytokines, and many metabolic processes (Fig. 2).
Table 4.
Chemokines and cluster of differentiation mRNAs found in Gjb1−/Y//Gjc2−/− cerebella.
The table shows the chemokines and CD genes in Gjb1−/Y//Gjc2−/− mice ranked by fold change (FC) based on the levels of detected mRNA, and shows the false discovery rate (FDR), adjusted p value for each mRNA, and the immune cells known to express them.
| name | FC | FDR | p | expression by cell type |
|---|---|---|---|---|
| Ccl6 | 13 | 0 | 0.0002 | macrophages and neutrophils (Orlofsky, et al. 1991) |
| Ccl3 | 10. | 0 | 0.0002 | microglia (Williams, et al. 2014) |
| Ccl4 | 7.3 | 0 | 0.001 | microglia (Williams, et al. 2014) |
| Ccl9 | 3.7 | 0 | 0.002 | macrophages (Williams, et al. 2014) |
| Cxcl10 | 2.2 | 0 | 0.03 | monocytes, chemokine for monocytes/macrophages and T-cells (Williams, et al. 2014) |
| Cx3cr1 | 2.2 | 0 | 0.003 | microglia (Williams, et al. 2014) |
| Ccl2 | 1.9 | 0 | 0.01 | secreted by monocytes/ macrophages, chemokine for macrophages and T-cells (Orlofsky, et al. 1991); Williams et al., 2014) |
| Cxcl12 | 1.9 | 0 | 0.02 | microglia (Williams, et al. 2014) |
| CD52 | 6.5 | 0 | 0.0002 | mature lymphocytes, monocytes (Buggins, et al. 2002, Domagala and Kurpisz 2001) |
| CD84 | 3.5 | 0 | 0.0006 | memory B-cells (Tangye, et al. 2002) |
| CD68 | 3.0 | 0 | 0.002 | macrophages, monocytes (Holness and Simmons 1993) |
| CD14 | 2.8 | 0 | 0.001 | macrophages, monocytes, dendritic cells (Simmons, et al. 1989) |
| CD72 | 2.5 | 0 | 0.0009 | B- and T-cells (Van de Velde, et al. 1991) |
| CD53 | 2.5 | 0 | 0.002 | leukocyte surface glycoproteins (Horejsi and Vlcek 1991) |
| CD109 | 2.2 | 0 | 0.002 | activated T-cells (Sutherland, et al. 1991) |
| CD86 | 2.1 | 0 | 0.005 | antigen presenting cells, costimulating/activating T- cells (Chen, et al. 1994) |
| CD48 | 2.0 | 0 | 0.009 | B- and T-cells (Yokoyama, et al. 1991) |
| CD37 | 1.9 | 0 | 0.003 | leukocyte surface glycoprotein (Horejsi and Vlcek 1991) |
| CD9 | 1.6 | 0 | 0.004 | leukocyte surface glycoprotein (Horejsi and Vlcek 1991) |
| CD33 | 1.6 | 0.2 | 0.01 | myeloid lineage, lymphoid cells (Garnache-Ottou, et al. 2005, Hernandez-Caselles, et al. 2006, Perez-Oliva, et al. 2011) |
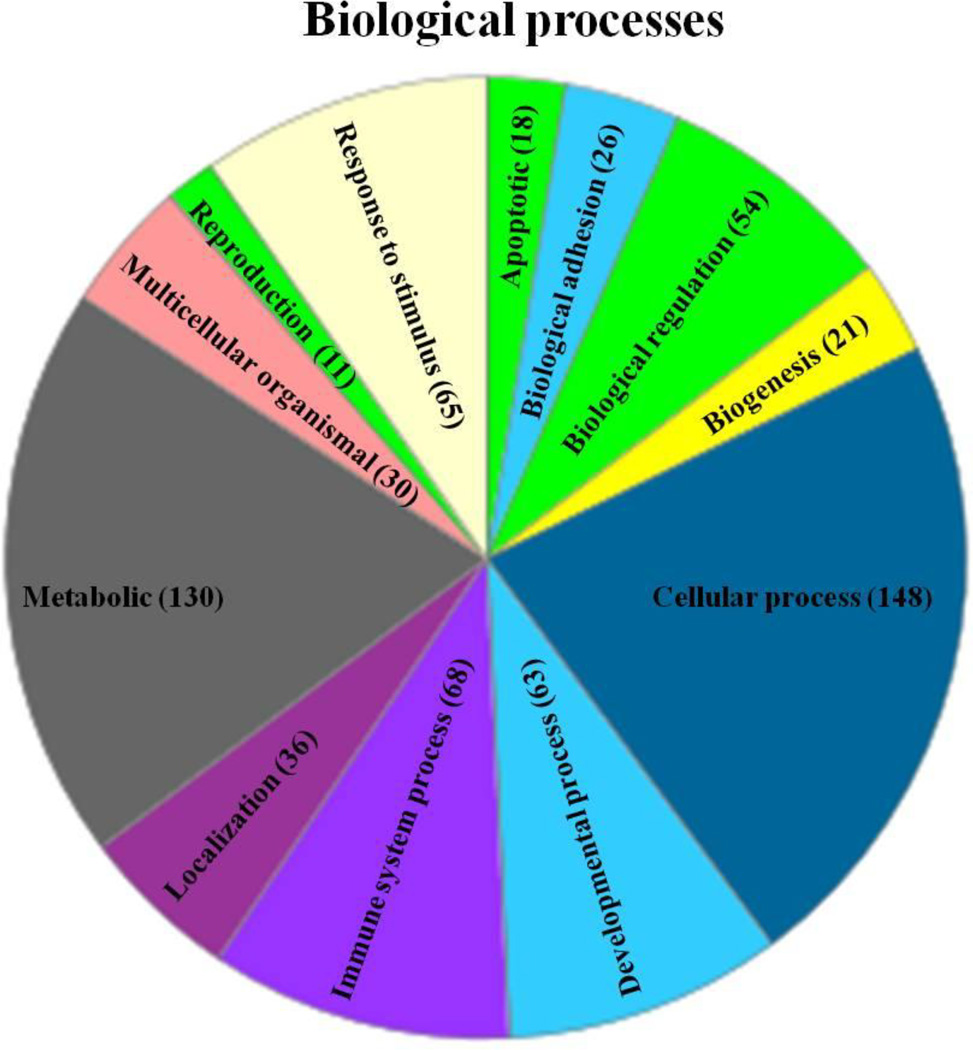
Figure 2. CNS metabolism and immune responses are altered in Gjb1−/Y//Gjc2−/− cerebella.
This is a pie chart generated using the Panther classification system, and shows the biological processes in which the increased mRNA are involved, with the number of the increased mRNA in each process (in parenthesis). The chart shows that mRNAs with increased expression in Gjb1−/Y//Gjc2−/− cerebella are predicted to be involved in the CNS immune and metabolic processes.
Quantitative RT-PCR (qRT-PCR)
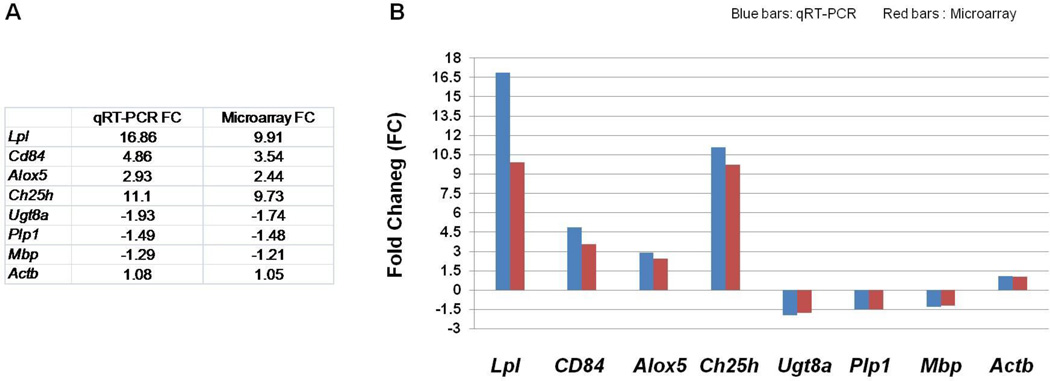
To corroborate our findings, we performed qRT-PCR on selected genes using the same batches of RNA that were used for the microarrays. With the exception of the gene with the highest change in expression (Lpl), the fold change measured by qRT-PCR was similar to that measure by microarrays for all 8 genes (Fig. 3).
Figure 3. qRT-PCR.
The table (A) and a graph (B) that shows qRT-PCR analysis of 8 genes, using the same RNA samples from the 4 P29 Gjb1−/Y//Gjc2−/− mice and 4 littermate controls (Gjb1+/Y//Gjc2+/−) that were used for the microarray analysis. Data were normalized for the housekeeping gene Gapdh. With the exception of Lpl, the fold change (FC) of the mRNA levels qRT-PCR (blue bars in B) were similar to those measured by microarrays (red bars in B).
Altered microglia in Gjb1−/Y//Gjc2−/− cerebella
To determine whether microglia and/or macrophages contribute to the changes in the immune-related mRNAs that we observed, we compared our results to those of Beutner et al. (Beutner, et al. 2013) and Hickman et al. (Hickman, et al. 2013). According to this analysis, 15 of the up-regulated genes are microglial-specific - Hexb, Rnase4, Gpr34, Cx3cr1, Olfml3, P2ry13, Trem2, Ccl4, Aif1, Ccl3, Adora3, Parvg, Ccl12, Gpr84, Asb10, and 2 (Cd68 and Cd14) are expressed by microglia and macrophages (Hickman, et al. 2013). Except for Ccl3, Ccl4, Ccl12 (which are involved in cytokine-cytokine interactions and chemokine signaling), David bioinformatics analysis tool did not indicate that any of the other 12 genes are involved in lipid/myelin metabolism or NK-, B-, or T-cell signaling/pathway activation.
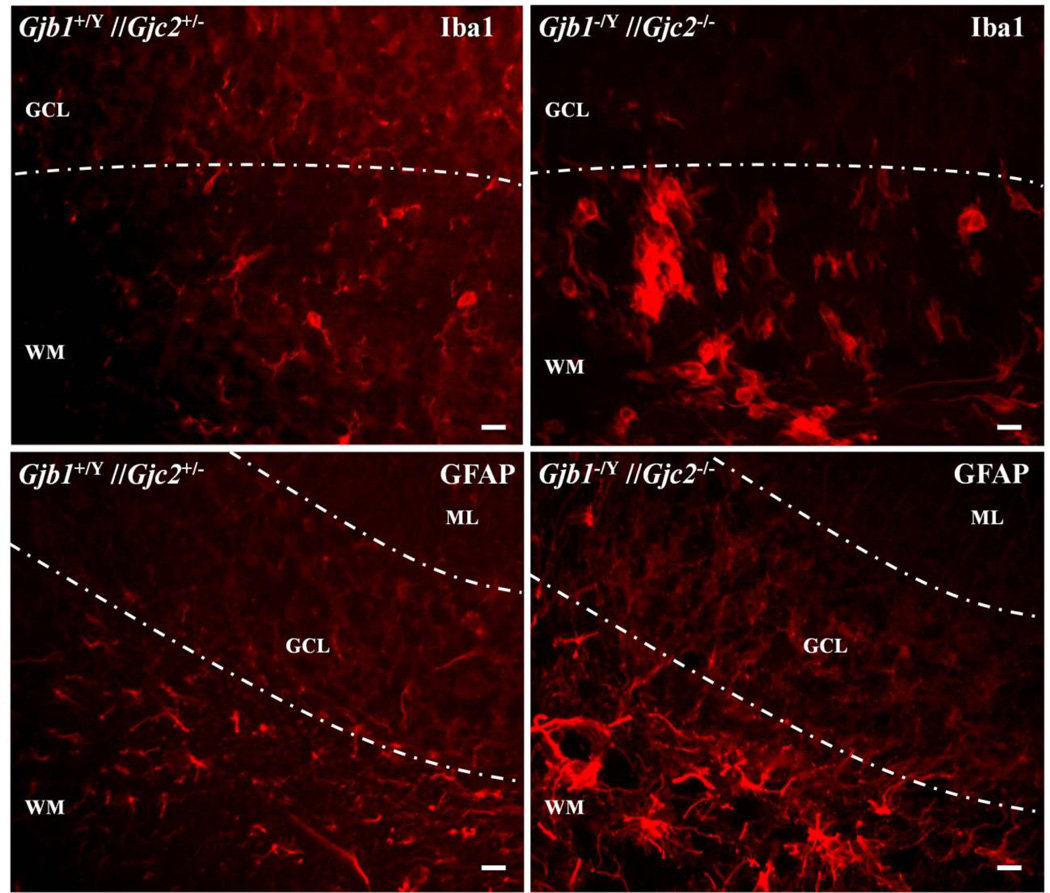
To visualize microglia, we immunostained sections of the cerebellum (and the attached pons) from P29 Gjb1−/Y//Gjc2−/− mice (n=3) and their littermate Gjb1+/Y//Gjc2+/− controls (n=3) for Iba1 (Ito, et al. 1998), which labels microglia and monocytes/macrophages (Imai, et al. 1996), and Ly6c, which is expressed by macrophages and endothelial cells (Jutila, et al. 1988). Iba1 staining was strongly increased in white matter within Gjb1−/Y//Gjc2−/− cerebella and the pons, compared to their littermate controls (Fig. 4); the increased staining appeared to correspond to larger microglia. We did not detect a difference in Ly6 staining (results not shown). Insofar as hypertrophied microglia are a histological proxy for their activation, these findings indicate that microglia are activated in Gjb1−/Y//Gjc2−/− cerebella and pons, as was previously reported in mice models with combined Gjb1/Cx32 and Gjc2/Cx47 mutations (Schiza, et al. 2015, Tress, et al. 2011, Tress, et al. 2012).
Figure 4. Microglial and astrocytic responses in Gjb1−/Y//Gjc2−/− cerebella.
These are digital images of sections from the cerebella from P29 Gjb1−/Y//Gjc2−/− mice and their littermate controls (Gjb1+/Y//Gjc2+/−), immunostained for Iba1 or GFAP. The molecular layer (ML), granular cell layer (GCL), and white matter (WM) are labeled. The upper panels show activated microglia in the WM of a Gjb1−/Y//Gjc2−/− cerebellum but not a littermate control. The lower panels show increased GFAP staining in the WM of a Gjb1−/Y//Gjc2−/− cerebellum compared to the control. Scale bars: 10 µm.
Altered astrocytes in Gjb1−/Y//Gjc2−/− cerebella
Some of the mRNAs with significantly increased expression map to genes that are astrocyte-enriched; Gfap, Gjb, Prodh, and Cybrd1 (Zhang, et al. 2014), so we also immunostained for GFAP to examine the astrocytes in the cerebella of the Gjb1−/Y//Gjc2−/− mice. As shown in Figure 4, there was increased GFAP staining in white matter tracts, compared to the control mice, indicating that astrocytes are activated as was previously reported in mice models with combined Gjb1/Cx32 and Gjc2/Cx47 mutations (Schiza, et al. 2015, Tress, et al. 2011, Tress, et al. 2012).
B-cells and T-cells infiltrate the cerebella of the Gjb1−/Y//Gjc2−/−mice
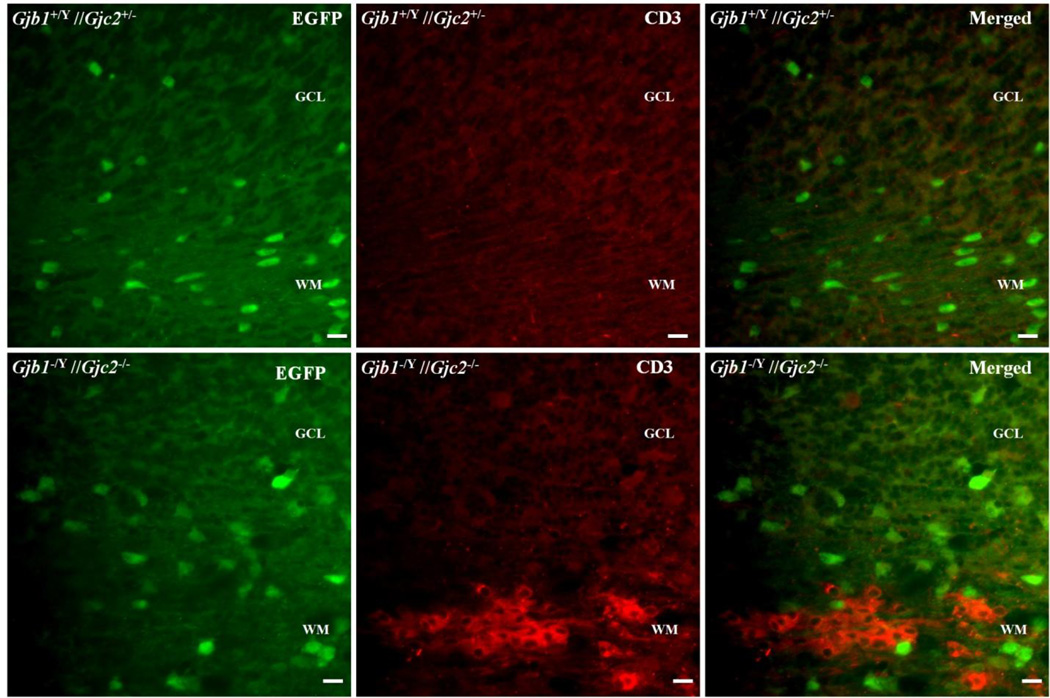
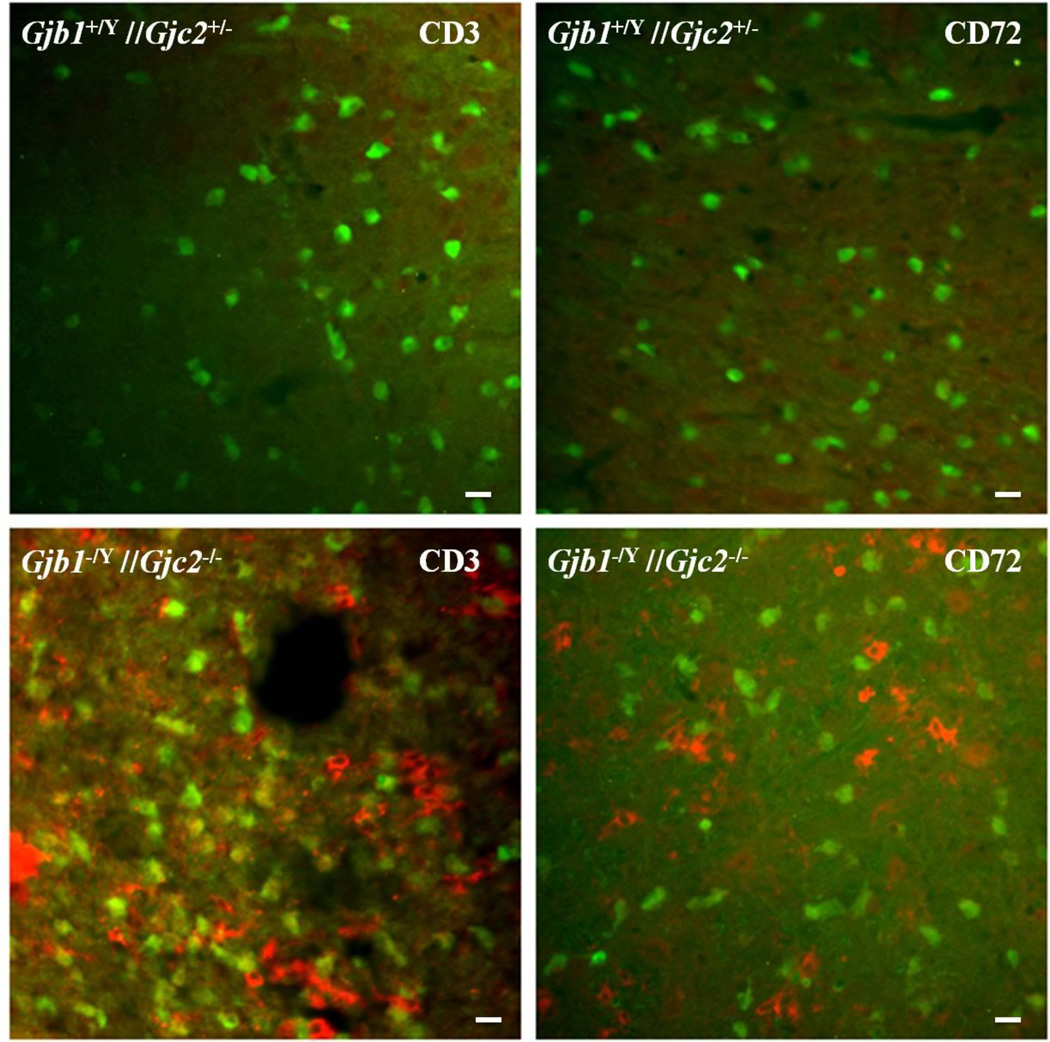
Some of the inflammatory chemokines that are upregulated in Gjb1−/Y //Gjc2−/− cerebella (Williams, et al. 2014) can theoretically attract lymphocytes. To determine whether this occurs, we immunostained cerebellar sections from P29 Gjb1−/Y//Gjc2−/− mice (n=3) and their littermate (Gjb1+/Y //Gjc2+/− ) controls (n=3) for CD3, a marker for T-cells and Cd72, a (Chetty and Gatter 1994) marker for B-cells (Kumanogoh, et al. 2000, Parnes and Pan 2000, Van de Velde, et al. 1991); we did not find an antibody for NK cells that worked for us. We found many CD3-positive cells within and around the white matter tracts of the cerebella (Fig. 5) of Gjb1−/Y//Gjc2−/− mice compared to their littermate controls. Double staining for GFP showed that the CD3-positive cells were distinct from oligodendrocytes. We also found clusters of CD72-positive cells in white matter tracts of the cerebella (Fig. 6); these were not as numerous as the CD3-positive clusters. We also found more clusters of CD3- and CD72-positive cells in the pons (Fig. 7). These findings confirm that B- and T-cell infiltrate the white matter tracts that are known to undergo demyelinating in Gjb1−/Y//Gjc2−/− mice (Menichella, et al. 2003).
Figure 5. T-cells in of Gjb1−/Y//Gjc2−/− cerebella.
These are digital images of sections from the cerebella from a P29 Gjb1−/Y//Gjc2−/− mouse and a littermate control (Gjb1+/Y//Gjc2+/−), immunostained for CD3, a T-cell marker. Because the Egfp gene was “knocked into” the Gjc2- null allele, oligodendrocytes are EGFP-positive in both genotypes. The granular cell layer (GCL), and white matter (WM) are labeled. Note CD3-positive cells in the WM of a Gjb1−/Y//Gjc2−/− cerebellum (lower panels) but not a control cerebellum. Scale bars: 10 µm.
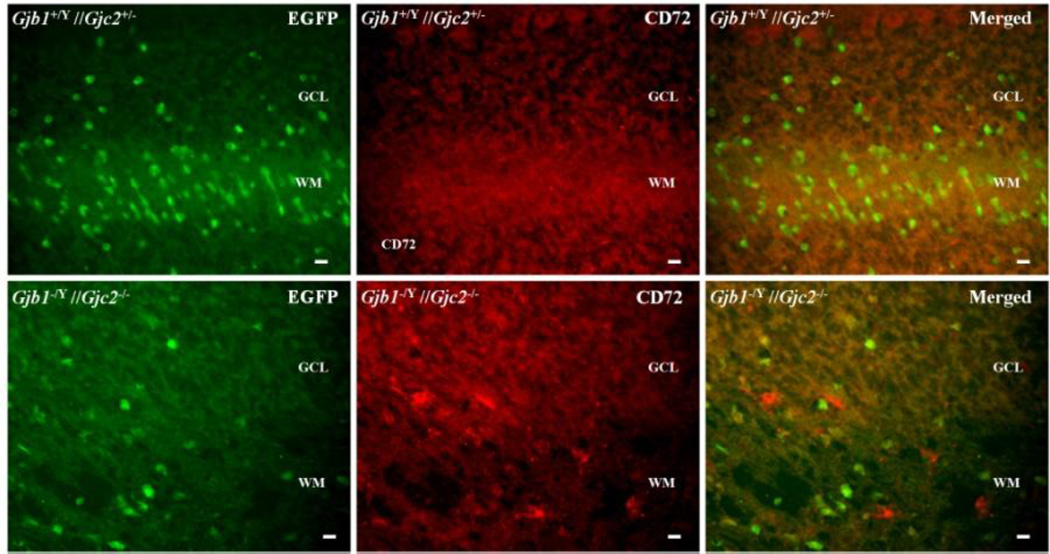
Figure 6. B-cells in Gjb1−/Y//Gjc2−/− cerebella.
These are digital images of sections from cerebella from a P29 Gjb1−/Y//Gjc2−/− mouse and a littermate control (Gjb1+/Y//Gjc2+/−), immunostained with a CD72 antibody, a B-cell marker. Because the Egfp gene was “knocked into” the Gjc2- null allele, oligodendrocytes are EGFP-positive in both genotypes. The granular cell layer (GCL), and white matter (WM) are labeled. Unlike the littermate control, the Gjb1−/Y//Gjc2−/− had scattered CD72-positive cells . Scale bars: 10 µm.
Figure 7. T-cells and B-cells in the pons in Gjb1−/Y//Gjc2−/−.
These are merged digital images of sections from the pons of a P29 Gjb1−/Y//Gjc2−/− mouse and a littermate control (Gjb1+/Y//Gjc2+/−), immunostained for CD3 and CD72, T- and B-cell markers, respectively. Because the Egfp gene was “knocked into” the Gjc2- null allele, oligodendrocytes are EGFP-positive in both genotypes, T- and B-cells are present in the pons of Gjb1−/Y//Gjc2−/−(lower panels) but not in control mice (upper panels). Scale bars: 10 µm.
Discussion
This is the first comprehensive examination of changes in mRNA expression in Gjb1−/Y//Gjc2−/−mice. We find reduced mRNA levels of genes involved in myelin synthesis, and increased mRNAs levels of genes involved in breaking down lipids, releasing arachidonic acid, and creating cellular immune responses. Microglia are activated and B- and T-cells infiltrate affected white matter tracts.
The role of glial GJ coupling
The traditional views regarding the physiological roles of glial GJ coupling are centered around the spatial buffering of K+ released during neural activity (Berger, et al. 1991, Chvatal, et al. 1999, Frankenhaeuser and Hodgkin 1956, Kamasawa, et al. 2005, Menichella, et al. 2006, Orkand, et al. 1966, Wallraff, et al. 2006). More recently, glial GJ coupling has been implicated in the transfer of glucose and/or lactate to generate energy in order to sustain the neural activities, as both glucose and lactate can permeate O:O and O:A GJs (Rouach, et al. 2008)(Funfschilling, et al. 2012, Lee, et al. 2012, Rinholm, et al. 2011, Rinholm and Bergersen 2012). Glucose is also required for fatty acid/lipid synthesis, the generation of ribose-5-phosphate that is used in the synthesis of nucleotides and nucleic acids, and the erythrose-4-phosphate that is used in the synthesis of aromatic amino acids (Janson and Tischler 2012, Murray 2012) . Both O:A and O:O GJ coupling are abrogated in Gjb1−/Y //Gjc2 −/− mice (Maglione, et al. 2010, Wasseff and Scherer 2011), so all of these functions are potentially affected. Gjb1−/Y //Gjc2 −/− mice (Maglione, et al. 2010, Wasseff and Scherer 2011) have a more severe dysmyelination than is seen in Gja1−/−//Gjb6−/− mice, which lack O:A but not O:O GJ coupling (Lutz, et al. 2009). This discrepancy implies that the severe leukodystrophy in PMLD likely results from disrupted O:O GJ coupling in white matter tracts, and that Cx47 is the main connexin that mediates O:O coupling in humans.
mRNAs with decreased expression - related to myelin
We found reduced levels of 23 mRNAs, 2 of which encode enzymes that have essential roles in myelin lipid metabolism. Ugt8a encodes UDP galactosyltransferase, an enzyme that is essential for synthesis of galactosylceramide (GalCer), the major myelin lipid (Morell 1977). Fa2h encodes fatty acid 2-hydroxylase, which is the enzyme essential for synthesis of 2-hydroxy fatty acids (Eckhardt, et al. 2005). Recessive mutations in Ugt8a and Fa2h cause leukodystrophies (Edvardson, et al. 2008, Potter, et al. 2011), so that the reduced expression of Ugt8a and Fa2h could contribute to the demyelination and the phenotype observed in Gjb1−/Y//Gjc2−/− mice. The isolated decrease of Ugt8a and Fa2h mRNA is unexpected because their transcriptional profiles usually follow those of other myelin-related mRNAs (Bujalka, et al. 2013, Emery, et al. 2009, Srinivasan, et al. 2012).
Increased expression of mRNAs related to lipid metabolism
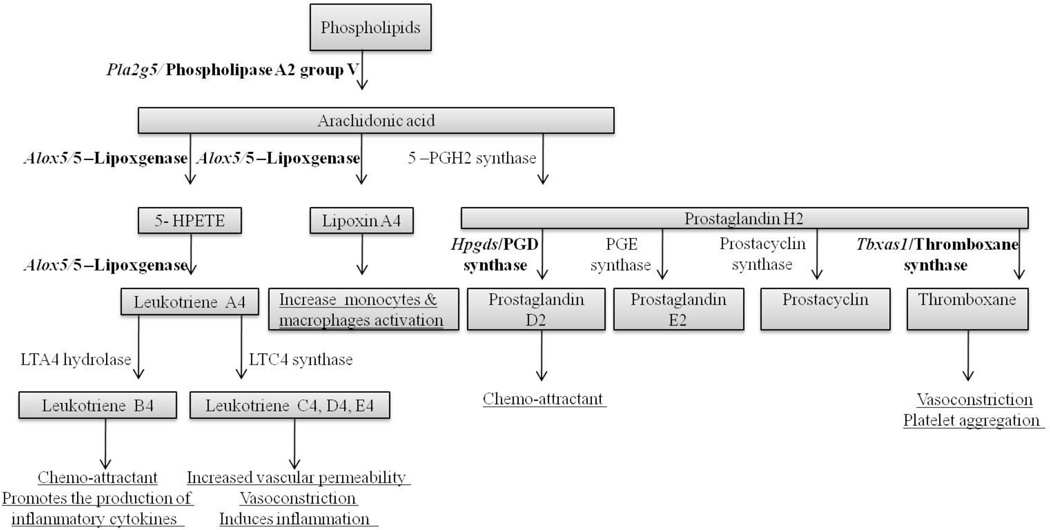
Some of the mRNAs with increased expression encode enzymes/proteins involved in forming pro-inflammatory molecules through ecosanoid metabolism (Fig. 8). The mRNA level of Alox5 was increased 2.5-fold; Alox5 encodes arachidonate 5-lipoxygenase, the key enzyme involved in the biosynthesis of leukotrienes from fatty acids, and the only lipoxygenase that can catalyze the formation of leukotrienes (Back, et al. 2014, Ford-Hutchinson, et al. 1994, Janson and Tischler 2012, Siegel, et al. 2006). This enzyme is also required for lipoxinA4 formation, which activates monocytes and macrophages (Ford-Hutchinson, et al. 1994, Murray 2012). The mRNA level of Hpdgs, which encodes prostaglandin D synthase was increased 1.8-fold; this catalyzes the formation of prostaglandin D2, which is mainly produced by oligodendrocytes in the normal CNS (Urade, et al. 1993) but by activated microglia in twitcher mice, which are a genetically authentic model of Krabbe disease (Mohri, et al. 2006a). Prostaglandin D2 is a chemo-attractant (Hirai, et al. 2001), provides neuroprotection (Taniike, et al. 2002), and may mediate demyelinating in twitcher mice (Mohri, et al. 2006b). The mRNA level of Tbxas1, which encodes thromboxane A synthase 1, was increased 3.3-fold; this catalyzes the formation of thromboxane A – a powerful inducer of vasoconstriction and platelet aggregation (Ford-Hutchinson, et al. 1994, Murray 2012). The mRNA levels of two phospholipases were increased - Pla2g5 (1.6-fold) and Plcg2 (1.5-fold). Pla2g5 encodes phospholipase A2 group V, the enzyme that catalyzes the hydrolysis of membrane phospholipids to generate lysophospholipids and free fatty acids, including arachidonic acid (Balsinde and Dennis 1997). It also induces leuokotrines (eicosanods) biosynthesis in neighboring inflammatory cells (Wijewickrama, et al. 2006). Plcg2 encodes the transmembrane signaling enzyme phospholipase C gamma 2 (Hernandez, et al. 1994), which catalyzes the conversion of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate to 1D–myo-inositol 1,4,5-trisphosphate (IP3) and diacylglycerol (DAG) (Berridge 1987, Berridge 2005) – both important secondary messengers that transmit signals from surface receptors. DAG is well known as a secondary messenger of protein kinase C, which mediates the activities of many receptors (Berridge 2005, Nishizuka 1995), it is also a precursor for arachidonic acid through the action of phospholipase A2 (Murray 2012) or triacylglycerol through the action of diglyceride acyltransferase (Bishop and Hajra 1984). These findings indicate that the loss of O:O and/or O:A GJ coupling shifts fatty acids metabolism in the CNS toward the biosynthesis of proinflammatory molecules such as prostaglandin D2, and the secondary messengers such as DAG involved in lymphocytes activation and signaling pathways.
Figure 8. Increased mRNAs are involved in leukotriene synthesis in Gjb1−/Y//Gjc2−/− mice.
This is a chart that shows the arachidonic acid metabolism, with the enzymes/proteins that would be predicted to increase in Gjb1−/Y//Gjc2−/− mice as a result of the increase in their mRNAs (bold). Pla2g5, encodes for phospholipase A2, group V, which catalyzes the release of arachidonic acid from cell membrane phospholipids. Arachidonic acid is then metabolized to form prostaglandins, prostacyclin, and thromboxane synthase. Alox5 encodes arachidonate 5-lipoxgenase (also known as 5-lipoxgenase), which catalyzes the metabolism of arachidonic acid to form leukotrienes and also activates lipoxin 4 to increase monocytes and macrophages activation. Tbxas1, which encodes thromboxan A synthase 1, was increased 3.3-fold; this catalyzes the formation of thromboxane A. Hpdgs encodes prostaglandin D synthase, which catalyzes the formation of prostaglandin D2. The predicted effects of these increased leukotrienes are underlined -production of proinflammatory molecules, change in vascular tone and chemo-attraction.
Increased mRNA expression was also found in other genes that encode enzymes that play important role in lipid metabolism. Lpl (9.9-fold) encodes lipoprotein lipase, the key enzyme required for the breakdown of the lipoproteins (Mead, et al. 2002, Merkel, et al. 2002). Several lipoprotein genes had higher mRNA levels - Apoc1 (3.2-fold), Apoc2 (3.1-fold), Apoc4 (2.5-fold), and Apoe (1.6-fold) - encoding apolipoproteins CI (that interferes with cellular fatty acid uptake; (Shachter 2001), CII (which is a coenzyme for and activates lipoprotein lipase; (Musliner, et al. 1977, Stocks and Galton 1980), CIV (which leads to cellular triglycerides accumulation(Kotite, et al. 2003)(Kim, et al. 2008), and E (which is required for cholesterol transportation and cellular uptake in a redistribution (Mahley 1988, Zlokovic 2013). Ch25h (9.7-fold) encodes cholesterol 25-hydroxylase, which metabolizes cholesterol into 25-hydroxy cholesterol, which suppresses endogenous cellular cholesterol synthesis (Diczfalusy, et al. 2009, Lagace, et al. 1997, Lund, et al. 1998). Cholesterol is a major myelin lipid (Morell 1977), and disabling cholesterol synthesis in oligodendrocytes results in deficient myelination (Saher, et al. 2005).
Immune responses in Gjb1−/Y//Gjc2−/− brains
We found B- and T-cells by immunohistochemistry, which matched the predictions of the CSEA tool, David, and Panther. Of the many genes with increased expression, 18 genes, including some with the most pronounced increases, such Clc6 (13-fold), Clc3 (10-fold), and Clc4 (7.3-fold), are involved in recruiting immune cells (Williams, et al. 2014), and 10 more genes are related to chemokine signaling pathways; some are listed in Table 4. Inflammatory cytokines also up-regulate the level of Cxcl12 mRNA, which is widely expressed in the CNS. Cxcl12 enhances T cell responses via co-stimulation of T-cell receptors (Smith, et al. 2013), and recruits leukocytes in experimental autoimmune encephalomyelitis (EAE) and multiple sclerosis (Williams, et al. 2014).
Mouse models demonstrate that infiltration of B- and T-cells is not an inevitable consequence of demyelination. Genetically killing oligodendrocytes in mice causes demyelination and reactive microglia, but does not result in B- or T-cell infiltration (Ghosh, et al. 2011, Gritsch, et al. 2014, Locatelli, et al. 2012, Oluich, et al. 2012, Traka, et al. 2010). Infiltrating lymphocytes are not seen in mice lacking PLP (Tatar, et al. 2010), a model of PMD, Arsa-null mice, which are a genetically authentic model of metachromatic leukodystrophy (Gieselmann 2003), or cuprizone-induced demyelination (Hiremath, et al. 1998). T-cells but not B-cells were found in mice in which oligodendrocytes lack Pex5 (a model of adrenoleukodystrophy that has inflammation but not demyelination), although increased levels of some chemokines/cytokines were also found in affected brains (Kassmann, et al. 2007). Similarly, few T-cells and no B-cells were found in mice lacking 2’,3’-phosphodiesterase (Wieser, et al. 2013), a myelin-related protein. T-cell infiltration has been reported in twitcher mouse caused by a mutation in Galc gene (Ohno, et al. 1993, Taniike, et al. 1997). Overexpression of PLP in mice results in T-cell infiltration, which contributes to the inflammation (Bradl, et al. 2005, Ip, et al. 2006); whether this is also the case in people who have extra copies of the PLP1 gene remains to be shown. In humans, T-cells are a prominent feature of demyelinating CNS lesions in patients with adrenoleukodystrophy, but not of other leukodystrophies (Eichler and Van Haren 2007), and we are not aware of an autopsied case of PMLD. If T-cells and B-cells mediated cellular inflammation were a prominent feature of PMLD, it seems appropriate to consider immune-modulating therapies, as PMLD is a devastating disease for which no treatments are currently known.
Many of the mRNAs with elevated levels in Gjb1−/Y//Gjc2−/− cerebella are also increased in multiple sclerosis and multiple sclerosis animal models. These include Tnfrsf1b (1.6-fold), which is a multiple sclerosis susceptibility locus (De Jager, et al. 2009, Tseveleki, et al. 2010), as well as C1qc and C1qa (both 3-fold), which encode complement components that have been identified in multiple sclerosis lesions (Tseveleki, et al. 2010). Tlr2 (3.4-fold) encodes toll-like receptor 2, which is expressed by oligodendrocytes and observed in MS lesions, where it is thought to mediate hyaluronan’s inhibition of oligodendrocyte precursor cells maturation (Sloane, et al. 2010). Cst7 (30-fold) encodes cystatin F (also known as leukocystatin), is up-regulated in microglia during acute demyelination (Banik 1992, Ma, et al. 2007, Ma, et al. 2011). Alox5 (2.5-fold) is increased in multiple sclerosis and multiple sclerosis models (Whitney, et al. 2001), and deleting Alox5 in a mouse model attenuated the neuroinflammation and axonal damage (Yoshikawa, et al. 2011). The molecular targets of several multiple sclerosis medications have increased mRNA levels in Gjb1−/Y//Gjc2−/− cerebella. S1pr3 (1.5-fold) encodes sphingosine 1-phosphate receptor, which is targeted by fingolimod; Ada (1.9-fold) encodes adenosine deaminase, which is targeted by cladribine, and Cd52 (6.5-fold) encodes the CD52 antigen targeted by Alemtuzumab.
In summary, our results show that in addition to the previously described demyelination, the loss of O:O and O:A GJ coupling results in extensive changes in gene expression and an immune response. The genes with reduced mRNA expression mostly map to oligodendrocytes, and include genes that encode key enzymes required for myelin lipids. The genes with increased expression are implicated in diverse responses and likely originate from different cell types. Many map to the immune system, and we show directly that T- and B-cells infiltrate the CNS. These findings raise questions about how lymphocytes are recruited to the CNS in acquired demyelinating diseases, and whether lymphocytes contribute to the pathogenesis of PMLD.
Conversely, one wonders whether the loss of Cx32 and Cx47 GJs in and around chronic demyelinating lesions in multiple sclerosis contributes to clinical disability (Kleopas, et al. 2013, Markoullis, et al. 2012a, Markoullis, et al. 2012b, Masaki 2013).
Supplementary Material
Highlights.
CNS lymphocytes activation with the loss of oligodendrocytes gap junctions (GJs).
Oligodendrocytes GJs are required for normal CNS lipid and myelin metabolism.
CNS oligodendrocytes GJs loss alters the CNS immune status without external triggers.
Immune-modulating drugs might be useful in leukodystrophies caused by GJs mutations.
Acknowledgments
This work was supported by the NIH (NS055284) and the National Multiple Sclerosis Society (to S.S.S.). We thank Jonathan Schug, Ph.D., and Olga Smirnova from the Functional Genomics Core at the Institute of Diabetes, Obesity and Metabolism at the Perelman School of Medicine at the University of Pennsylvania for the RNA microarray analysis. We thank Kathakali Addya, Ph.D., from the Molecular Profiling Facility at the Perelman School of Medicine at the University of Pennsylvania for the qRT-PCR.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abrams CK, Scherer SS. Gap junctions in inherited human disorders of the central nervous system. Biochim. Biophys. Acta. 2011;18:2030–2047. doi: 10.1016/j.bbamem.2011.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Back M, Powell WS, Dahlen SE, Drazen JM, Evans JF, Serhan CN, Shimizu T, Yokomizo T, Rovati GE. International Union of Basic and Clinical Pharmacology. Update on Leukotriene, Lipoxin and Oxoeicosanoid Receptors: IUPHAR Review "X". Br. J. Pharmacol. 2014 doi: 10.1111/bph.12665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balsinde J, Dennis EA. Function and inhibition of intracellular calcium-independent phospholipase A2. J. Biol. Chem. 1997;272:16069–16072. doi: 10.1074/jbc.272.26.16069. [DOI] [PubMed] [Google Scholar]
- Banik NL. Pathogenesis of myelin breakdown in demyelinating diseases: role of proteolytic enzymes. Crit. Rev. Neurobiol. 1992;6:257–271. [PubMed] [Google Scholar]
- Berger T, Schnitzer J, Kettenmann H. Developmental changes in the membrane current pattern, K+ buffer capacity, and morphology of glial cells in the corpus callosum slice. J. Neurosci. 1991;11:3008–3024. doi: 10.1523/JNEUROSCI.11-10-03008.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berridge MJ. Unlocking the secrets of cell signaling. Annu. Rev. Physiol. 2005;67:1–21. doi: 10.1146/annurev.physiol.67.040103.152647. [DOI] [PubMed] [Google Scholar]
- Berridge MJ. Inositol trisphosphate and diacylglycerol: two interacting second messengers. Annu. Rev. Biochem. 1987;56:159–193. doi: 10.1146/annurev.bi.56.070187.001111. [DOI] [PubMed] [Google Scholar]
- Beutner C, Linnartz-Gerlach B, Schmidt SV, Beyer M, Mallmann MR, Staratschek-Jox A, Schultze JL, Neumann H. Unique transcriptome signature of mouse microglia. Glia. 2013;61:1429–1442. doi: 10.1002/glia.22524. [DOI] [PubMed] [Google Scholar]
- Bishop JE, Hajra AK. Biosynthesis of triglyceride and other fatty acyl esters by developing rat brain. J. Neurochem. 1984;43:1046–1051. doi: 10.1111/j.1471-4159.1984.tb12842.x. [DOI] [PubMed] [Google Scholar]
- Bradl M, Bauer J, Flugel A, Wekerle H, Lassmann H. Complementary contribution of CD4 and CD8 T lymphocytes to T-cell infiltration of the intact and the degenerative spinal cord. Am. J. Pathol. 2005;166:1441–1450. doi: 10.1016/S0002-9440(10)62361-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruzzone R, White TW, Paul DL. Connections with connexins: the molecular basis of direct intercellular signaling. Eur. J. Biochem. 1996;238:1–27. doi: 10.1111/j.1432-1033.1996.0001q.x. [DOI] [PubMed] [Google Scholar]
- Buggins AG, Mufti GJ, Salisbury J, Codd J, Westwood N, Arno M, Fishlock K, Pagliuca A, Devereux S. Peripheral blood but not tissue dendritic cells express CD52 and are depleted by treatment with alemtuzumab. Blood. 2002;100:1715–1720. [PubMed] [Google Scholar]
- Bugiani M, Al Shahwan S, Lamantea E, Bizzi A, Bakhsh E, Moroni I, Balestrini MR, Uziel G, Zeviani M. GJA12 mutations in children with recessive hypomyelinating leukoencephalopathy. Neurology. 2006;67:273–279. doi: 10.1212/01.wnl.0000223832.66286.e4. [DOI] [PubMed] [Google Scholar]
- Bujalka H, Koenning M, Jackson S, Perreau VM, Pope B, Hay CM, Mitew S, Hill AF, Lu QR, Wegner M, Srinivasan R, Svaren J, Willingham M, Barres BA, Emery B. MYRF is a membrane-associated transcription factor that autoproteolytically cleaves to directly activate myelin genes. PLoS Biol. 2013;11:e1001625. doi: 10.1371/journal.pbio.1001625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Gault A, Shen L, Nabavi N. Molecular cloning and expression of early T cell costimulatory molecule-1 and its characterization as B7-2 molecule. J. Immunol. 1994;152:4929–4936. [PubMed] [Google Scholar]
- Chetty R, Gatter K. CD3: structure, function, and role of immunostaining in clinical practice. J. Pathol. 1994;173:303–307. doi: 10.1002/path.1711730404. [DOI] [PubMed] [Google Scholar]
- Chvatal A, Anderova M, Ziak D, Sykova E. Glial depolarization evokes a larger potassium accumulation around oligodendrocytes than around astrocytes in gray matter of rat spinal cord slices. J. Neurosci. Res. 1999;56:493–505. doi: 10.1002/(SICI)1097-4547(19990601)56:5<493::AID-JNR5>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- De Jager PL, Jia X, Wang J, de Bakker PI, Ottoboni L, Aggarwal NT, Piccio L, Raychaudhuri S, Tran D, Aubin C, Briskin R, Romano S International MS Genetics Consortium. Baranzini SE, McCauley JL, Pericak-Vance MA, Haines JL, Gibson RA, Naeglin Y, Uitdehaag B, Matthews PM, Kappos L, Polman C, McArdle WL, Strachan DP, Evans D, Cross AH, Daly MJ, Compston A, Sawcer SJ, Weiner HL, Hauser SL, Hafler DA, Oksenberg JR. Meta-analysis of genome scans and replication identify CD6, IRF8 and TNFRSF1A as new multiple sclerosis susceptibility loci. Nat. Genet. 2009;41:776–782. doi: 10.1038/ng.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diczfalusy U, Olofsson KE, Carlsson AM, Gong M, Golenbock DT, Rooyackers O, Flaring U, Bjorkbacka H. Marked upregulation of cholesterol 25-hydroxylase expression by lipopolysaccharide. J. Lipid Res. 2009;50:2258–2264. doi: 10.1194/jlr.M900107-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domagala A, Kurpisz M. CD52 antigen--a review. Med. Sci. Monit. 2001;7:325–331. [PubMed] [Google Scholar]
- Dougherty JD, Fomchenko EI, Akuffo AA, Schmidt E, Helmy KY, Bazzoli E, Brennan CW, Holland EC, Milosevic A. Candidate pathways for promoting differentiation or quiescence of oligodendrocyte progenitor-like cells in glioma. Cancer Res. 2012;72:4856–4868. doi: 10.1158/0008-5472.CAN-11-2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JP, Dougherty JD, Heiman M, Schmidt EF, Stevens TR, Ma G, Bupp S, Shrestha P, Shah RD, Doughty ML, Gong S, Greengard P, Heintz N. Application of a translational profiling approach for the comparative analysis of CNS cell types. Cell. 2008;135:749–762. doi: 10.1016/j.cell.2008.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckhardt M, Yaghootfam A, Fewou SN, Zoller I, Gieselmann V. A mammalian fatty acid hydroxylase responsible for the formation of alpha-hydroxylated galactosylceramide in myelin. Biochem. J. 2005;388:245–254. doi: 10.1042/BJ20041451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edvardson S, Hama H, Shaag A, Gomori JM, Berger I, Soffer D, Korman SH, Taustein I, Saada A, Elpeleg O. Mutations in the fatty acid 2-hydroxylase gene are associated with leukodystrophy with spastic paraparesis and dystonia. Am. J. Hum. Genet. 2008;83:643–648. doi: 10.1016/j.ajhg.2008.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichler F, Van Haren K. Immune response in leukodystrophies. Pediatr. Neurol. 2007;37:235–244. doi: 10.1016/j.pediatrneurol.2007.06.011. [DOI] [PubMed] [Google Scholar]
- Emery B, Agalliu D, Cahoy JD, Watkins TA, Dugas JC, Mulinyawe SB, Ibrahim A, Ligon KL, Rowitch DH, Barres BA. Myelin gene regulatory factor is a critical transcriptional regulator required for CNS myelination. Cell. 2009;138:172–185. doi: 10.1016/j.cell.2009.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford-Hutchinson AW, Gresser M, Young RN. 5-Lipoxygenase. Annu. Rev. Biochem. 1994;63:383–417. doi: 10.1146/annurev.bi.63.070194.002123. [DOI] [PubMed] [Google Scholar]
- Frankenhaeuser B, Hodgkin AL. The after-effects of impulses in the giant nerve fibres of Loligo. J. Physiol. 1956;131:341–376. doi: 10.1113/jphysiol.1956.sp005467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funfschilling U, Jockusch WJ, Sivakumar N, Mobius W, Corthals K, Li S, Quintes S, Kim Y, Schaap IA, Rhee JS, Nave KA, Saher G. Critical time window of neuronal cholesterol synthesis during neurite outgrowth. J. Neurosci. 2012;32:7632–7645. doi: 10.1523/JNEUROSCI.1352-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnache-Ottou F, Chaperot L, Biichle S, Ferrand C, Remy-Martin JP, Deconinck E, de Tailly PD, Bulabois B, Poulet J, Kuhlein E, Jacob MC, Salaun V, Arock M, Drenou B, Schillinger F, Seilles E, Tiberghien P, Bensa JC, Plumas J, Saas P. Expression of the myeloid-associated marker CD33 is not an exclusive factor for leukemic plasmacytoid dendritic cells. Blood. 2005;105:1256–1264. doi: 10.1182/blood-2004-06-2416. [DOI] [PubMed] [Google Scholar]
- Ghosh A, Manrique-Hoyos N, Voigt A, Schulz JB, Kreutzfeldt M, Merkler D, Simons M. Targeted ablation of oligodendrocytes triggers axonal damage. PLoS One. 2011;6:e22735. doi: 10.1371/journal.pone.0022735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gieselmann V. Metachromatic leukodystrophy: recent research developments. J. Child Neurol. 2003;18:591–594. doi: 10.1177/08830738030180090301. [DOI] [PubMed] [Google Scholar]
- Gritsch S, Lu J, Thilemann S, Wortge S, Mobius W, Bruttger J, Karram K, Ruhwedel T, Blanfeld M, Vardeh D, Waisman A, Nave KA, Kuner R. Oligodendrocyte ablation triggers central pain independently of innate or adaptive immune responses in mice. Nat. Commun. 2014;5:5472. doi: 10.1038/ncomms6472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez D, Egan SE, Yulug IG, Fisher EM. Mapping the gene that encodes phosphatidylinositol-specific phospholipase C-gamma 2 in the human and the mouse. Genomics. 1994;23:504–507. doi: 10.1006/geno.1994.1533. [DOI] [PubMed] [Google Scholar]
- Hernandez-Caselles T, Martinez-Esparza M, Perez-Oliva AB, Quintanilla-Cecconi AM, Garcia-Alonso A, Alvarez-Lopez DM, Garcia-Penarrubia P. A study of CD33 (SIGLEC-3) antigen expression and function on activated human T and NK cells: two isoforms of CD33 are generated by alternative splicing. J. Leukoc. Biol. 2006;79:46–58. doi: 10.1189/jlb.0205096. [DOI] [PubMed] [Google Scholar]
- Hickman SE, Kingery ND, Ohsumi TK, Borowsky ML, Wang LC, Means TK, El Khoury J. The microglial sensome revealed by direct RNA sequencing. Nat. Neurosci. 2013;16:1896–1905. doi: 10.1038/nn.3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai H, Tanaka K, Yoshie O, Ogawa K, Kenmotsu K, Takamori Y, Ichimasa M, Sugamura K, Nakamura M, Takano S, Nagata K. Prostaglandin D2 selectively induces chemotaxis in T helper type 2 cells, eosinophils, and basophils via seven-transmembrane receptor CRTH2. J. Exp. Med. 2001;193:255–261. doi: 10.1084/jem.193.2.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiremath MM, Saito Y, Knapp GW, Ting JP, Suzuki K, Matsushima GK. Microglial/macrophage accumulation during cuprizone-induced demyelination in C57BL/6 mice. J. Neuroimmunol. 1998;92:38–49. doi: 10.1016/s0165-5728(98)00168-4. [DOI] [PubMed] [Google Scholar]
- Holness CL, Simmons DL. Molecular cloning of CD68, a human macrophage marker related to lysosomal glycoproteins. Blood. 1993;81:1607–1613. [PubMed] [Google Scholar]
- Horejsi V, Vlcek C. Novel structurally distinct family of leucocyte surface glycoproteins including CD9, CD37, CD53 and CD63. FEBS Lett. 1991;288:1–4. doi: 10.1016/0014-5793(91)80988-f. [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Imai Y, Ibata I, Ito D, Ohsawa K, Kohsaka S. A novel gene iba1 in the major histocompatibility complex class III region encoding an EF hand protein expressed in a monocytic lineage. Biochem. Biophys. Res. Commun. 1996;224:855–862. doi: 10.1006/bbrc.1996.1112. [DOI] [PubMed] [Google Scholar]
- Ip CW, Kroner A, Bendszus M, Leder C, Kobsar I, Fischer S, Wiendl H, Nave KA, Martini R. Immune cells contribute to myelin degeneration and axonopathic changes in mice overexpressing proteolipid protein in oligodendrocytes. J. Neurosci. 2006;26:8206–8216. doi: 10.1523/JNEUROSCI.1921-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito D, Imai Y, Ohsawa K, Nakajima K, Fukuuchi Y, Kohsaka S. Microglia-specific localisation of a novel calcium binding protein, Iba1. Brain Res. Mol. Brain Res. 1998;57:1–9. doi: 10.1016/s0169-328x(98)00040-0. [DOI] [PubMed] [Google Scholar]
- Janson LW, Tischler M. The big picture: medical biochemistry. New York: McGraw-Hill Medical; 2012. [Google Scholar]
- Jutila MA, Kroese FG, Jutila KL, Stall AM, Fiering S, Herzenberg LA, Berg EL, Butcher EC. Ly-6C is a monocyte/macrophage and endothelial cell differentiation antigen regulated by interferon-gamma. Eur. J. Immunol. 1988;18:1819–1826. doi: 10.1002/eji.1830181125. [DOI] [PubMed] [Google Scholar]
- Kamasawa N, Sik A, Morita M, Yasumura T, Davidson KG, Nagy JI, Rash JE. Connexin-47 and connexin-32 in gap junctions of oligodendrocyte somata, myelin sheaths, paranodal loops and Schmidt-Lanterman incisures: implications for ionic homeostasis and potassium siphoning. Neuroscience. 2005;136:65–86. doi: 10.1016/j.neuroscience.2005.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassmann CM, Lappe-Siefke C, Baes M, Brugger B, Mildner A, Werner HB, Natt O, Michaelis T, Prinz M, Frahm J, Nave KA. Axonal loss and neuroinflammation caused by peroxisome-deficient oligodendrocytes. Nat. Genet. 2007;39:969–976. doi: 10.1038/ng2070. [DOI] [PubMed] [Google Scholar]
- Kim E, Li K, Lieu C, Tong S, Kawai S, Fukutomi T, Zhou Y, Wands J, Li J. Expression of apolipoprotein C-IV is regulated by Ku antigen/peroxisome proliferator-activated receptor gamma complex and correlates with liver steatosis. J. Hepatol. 2008;49:787–798. doi: 10.1016/j.jhep.2008.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleopa KA, Scherer SS. Molecular genetics of X-linked Charcot-Marie-Tooth disease. Neuromolecular Med. 2006;8:107–122. doi: 10.1385/nmm:8:1-2:107. [DOI] [PubMed] [Google Scholar]
- Kleopas K, Irene S, Kyriaki M. Connexin pathology in chronic multiple sclerosis and experimental autoimmune encephalomyelitis. Clin. Exp. Neuroimmunol. 2013;4:45–58. [Google Scholar]
- Kotite L, Zhang LH, Yu Z, Burlingame AL, Havel RJ. Human apoC-IV: isolation, characterization, and immunochemical quantification in plasma and plasma lipoproteins. J. Lipid Res. 2003;44:1387–1394. doi: 10.1194/jlr.M300087-JLR200. [DOI] [PubMed] [Google Scholar]
- Kumanogoh A, Watanabe C, Lee I, Wang X, Shi W, Araki H, Hirata H, Iwahori K, Uchida J, Yasui T, Matsumoto M, Yoshida K, Yakura H, Pan C, Parnes JR, Kikutani H. Identification of CD72 as a lymphocyte receptor for the class IV semaphorin CD100: a novel mechanism for regulating B cell signaling. Immunity. 2000;13:621–631. doi: 10.1016/s1074-7613(00)00062-5. [DOI] [PubMed] [Google Scholar]
- Lagace TA, Byers DM, Cook HW, Ridgway ND. Altered regulation of cholesterol and cholesteryl ester synthesis in Chinese-hamster ovary cells overexpressing the oxysterol-binding protein is dependent on the pleckstrin homology domain. Biochem. J. 1997;326(Pt 1):205–213. doi: 10.1042/bj3260205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Morrison BM, Li Y, Lengacher S, Farah MH, Hoffman PN, Liu Y, Tsingalia A, Jin L, Zhang PW, Pellerin L, Magistretti PJ, Rothstein JD. Oligodendroglia metabolically support axons and contribute to neurodegeneration. Nature. 2012;487:443–448. doi: 10.1038/nature11314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locatelli G, Wortge S, Buch T, Ingold B, Frommer F, Sobottka B, Kruger M, Karram K, Buhlmann C, Bechmann I, Heppner FL, Waisman A, Becher B. Primary oligodendrocyte death does not elicit anti-CNS immunity. Nat. Neurosci. 2012;15:543–550. doi: 10.1038/nn.3062. [DOI] [PubMed] [Google Scholar]
- Lund EG, Kerr TA, Sakai J, Li WP, Russell DW. cDNA cloning of mouse and human cholesterol 25-hydroxylases, polytopic membrane proteins that synthesize a potent oxysterol regulator of lipid metabolism. J. Biol. Chem. 1998;273:34316–34327. doi: 10.1074/jbc.273.51.34316. [DOI] [PubMed] [Google Scholar]
- Lutz SE, Zhao Y, Gulinello M, Lee SC, Raine CS, Brosnan CF. Deletion of astrocyte connexins 43 and 30 leads to a dysmyelinating phenotype and hippocampal CA1 vacuolation. J. Neurosci. 2009;29:7743–7752. doi: 10.1523/JNEUROSCI.0341-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Tanaka KF, Shimizu T, Bernard CC, Kakita A, Takahashi H, Pfeiffer SE, Ikenaka K. Microglial cystatin F expression is a sensitive indicator for ongoing demyelination with concurrent remyelination. J. Neurosci. Res. 2011;89:639–649. doi: 10.1002/jnr.22567. [DOI] [PubMed] [Google Scholar]
- Ma J, Tanaka KF, Yamada G, Ikenaka K. Induced expression of cathepsins and cystatin C in a murine model of demyelination. Neurochem. Res. 2007;32:311–320. doi: 10.1007/s11064-006-9183-y. [DOI] [PubMed] [Google Scholar]
- Maglione M, Tress O, Haas B, Karram K, Trotter J, Willecke K, Kettenmann H. Oligodendrocytes in mouse corpus callosum are coupled via gap junction channels formed by connexin47 and connexin32. Glia. 2010;58:1104–1117. doi: 10.1002/glia.20991. [DOI] [PubMed] [Google Scholar]
- Mahley RW. Apolipoprotein E: cholesterol transport protein with expanding role in cell biology. Science. 1988;240:622–630. doi: 10.1126/science.3283935. [DOI] [PubMed] [Google Scholar]
- Markoullis K, Sargiannidou I, Gardner C, Hadjisavvas A, Reynolds R, Kleopa KA. Disruption of oligodendrocyte gap junctions in experimental autoimmune encephalomyelitis. Glia. 2012a;60:1053–1066. doi: 10.1002/glia.22334. [DOI] [PubMed] [Google Scholar]
- Markoullis K, Sargiannidou I, Schiza N, Hadjisavvas A, Roncaroli F, Reynolds R, Kleopa KA. Gap junction pathology in multiple sclerosis lesions and normal-appearing white matter. Acta Neuropathol. 2012b;123:873–886. doi: 10.1007/s00401-012-0978-4. [DOI] [PubMed] [Google Scholar]
- Masaki K. Connexin pathology in acute multiple sclerosis, Balo's dieases, and neuromyelitis optica. Clin Exp Neuroimmunol. 2013;4:36–44. [Google Scholar]
- Massa PT, Mugnaini E. Cell-cell junctional interactions and characteristic plasma membrane features of cultured rat glial cells. Neuroscience. 1985;14:695–709. doi: 10.1016/0306-4522(85)90320-3. [DOI] [PubMed] [Google Scholar]
- Massa PT, Mugnaini E. Cell junctions and intramembrane particles of astrocytes and oligodendrocytes: a freeze-fracture study. Neuroscience. 1982;7:523–538. doi: 10.1016/0306-4522(82)90285-8. [DOI] [PubMed] [Google Scholar]
- Mead JR, Irvine SA, Ramji DP. Lipoprotein lipase: structure, function, regulation, and role in disease. J. Mol. Med. (Berl) 2002;80:753–769. doi: 10.1007/s00109-002-0384-9. [DOI] [PubMed] [Google Scholar]
- Menichella DM, Majdan M, Awatramani R, Goodenough DA, Sirkowski E, Scherer SS, Paul DL. Genetic and physiological evidence that oligodendrocyte gap junctions contribute to spatial buffering of potassium released during neuronal activity. J. Neurosci. 2006;26:10984–10991. doi: 10.1523/JNEUROSCI.0304-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menichella DM, Goodenough DA, Sirkowski E, Scherer SS, Paul DL. Connexins are critical for normal myelination in the CNS. J. Neurosci. 2003;23:5963–5973. doi: 10.1523/JNEUROSCI.23-13-05963.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkel M, Eckel RH, Goldberg IJ. Lipoprotein lipase: genetics, lipid uptake, and regulation. J. Lipid Res. 2002;43:1997–2006. doi: 10.1194/jlr.r200015-jlr200. [DOI] [PubMed] [Google Scholar]
- Mohri I, Taniike M, Okazaki I, Kagitani-Shimono K, Aritake K, Kanekiyo T, Yagi T, Takikita S, Kim HS, Urade Y, Suzuki K. Lipocalin-type prostaglandin D synthase is up-regulated in oligodendrocytes in lysosomal storage diseases and binds gangliosides. J. Neurochem. 2006a;97:641–651. doi: 10.1111/j.1471-4159.2006.03753.x. [DOI] [PubMed] [Google Scholar]
- Mohri I, Taniike M, Taniguchi H, Kanekiyo T, Aritake K, Inui T, Fukumoto N, Eguchi N, Kushi A, Sasai H, Kanaoka Y, Ozono K, Narumiya S, Suzuki K, Urade Y. Prostaglandin D2-mediated microglia/astrocyte interaction enhances astrogliosis and demyelination in twitcher. J. Neurosci. 2006b;26:4383–4393. doi: 10.1523/JNEUROSCI.4531-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morell P. Myelin. New York: Plenum Press; 1977. [Google Scholar]
- Murray RK. Harper's illustrated biochemistry. New York: McGraw-Hill Medical; 2012. [Google Scholar]
- Musliner TA, Church EC, Herbert PN, Kingston MJ, Shulman RS. Lipoprotein lipase cofactor activity of a carboxyl-terminal peptide of apolipoprotein C-II. Proc. Natl. Acad. Sci. U. S. A. 1977;74:5358–5362. doi: 10.1073/pnas.74.12.5358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelles E, Butzler C, Jung D, Temme A, Gabriel HD, Dahl U, Traub O, Stumpel F, Jungermann K, Zielasek J, Toyka KV, Dermietzel R, Willecke K. Defective propagation of signals generated by sympathetic nerve stimulation in the liver of connexin32-deficient mice. Proc. Natl. Acad. Sci. U. S. A. 1996;93:9565–9570. doi: 10.1073/pnas.93.18.9565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizuka Y. Protein kinase C and lipid signaling for sustained cellular responses. FASEB J. 1995;9:484–496. [PubMed] [Google Scholar]
- Odermatt B, Wellershaus K, Wallraff A, Seifert G, Degen J, Euwens C, Fuss B, Bussow H, Schilling K, Steinhauser C, Willecke K. Connexin 47 (Cx47)-deficient mice with enhanced green fluorescent protein reporter gene reveal predominant oligodendrocytic expression of Cx47 and display vacuolized myelin in the CNS. J. Neurosci. 2003;23:4549–4559. doi: 10.1523/JNEUROSCI.23-11-04549.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno M, Komiyama A, Martin PM, Suzuki K. MHC class II antigen expression and T-cell infiltration in the demyelinating CNS and PNS of the twitcher mouse. Brain Res. 1993;625:186–196. doi: 10.1016/0006-8993(93)91058-z. [DOI] [PubMed] [Google Scholar]
- Oluich LJ, Stratton JA, Xing YL, Ng SW, Cate HS, Sah P, Windels F, Kilpatrick TJ, Merson TD. Targeted ablation of oligodendrocytes induces axonal pathology independent of overt demyelination. J. Neurosci. 2012;32:8317–8330. doi: 10.1523/JNEUROSCI.1053-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orkand RK, Nicholls JG, Kuffler SW. Effect of nerve impulses on the membrane potential of glial cells in the central nervous system of amphibia. J. Neurophysiol. 1966;29:788–806. doi: 10.1152/jn.1966.29.4.788. [DOI] [PubMed] [Google Scholar]
- Orlofsky A, Berger MS, Prystowsky MB. Novel expression pattern of a new member of the MIP-1 family of cytokine-like genes. Cell Regul. 1991;2:403–412. doi: 10.1091/mbc.2.5.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parnes JR, Pan C. CD72, a negative regulator of B-cell responsiveness. Immunol. Rev. 2000;176:75–85. doi: 10.1034/j.1600-065x.2000.00608.x. [DOI] [PubMed] [Google Scholar]
- Perez-Oliva AB, Martinez-Esparza M, Vicente-Fernandez JJ, Corral-San Miguel R, Garcia-Penarrubia P, Hernandez-Caselles T. Epitope mapping, expression and post-translational modifications of two isoforms of CD33 (CD33M and CD33m) on lymphoid and myeloid human cells. Glycobiology. 2011;21:757–770. doi: 10.1093/glycob/cwq220. [DOI] [PubMed] [Google Scholar]
- Potter KA, Kern MJ, Fullbright G, Bielawski J, Scherer SS, Yum SW, Li JJ, Cheng H, Han X, Venkata JK, Khan PA, Rohrer B, Hama H. Central nervous system dysfunction in a mouse model of FA2H deficiency. Glia. 2011;59:1009–1021. doi: 10.1002/glia.21172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rash JE, Yasumura T, Dudek FE, Nagy JI. Cell-specific expression of connexins and evidence of restricted gap junctional coupling between glial cells and between neurons. J. Neurosci. 2001;21:1983–2000. doi: 10.1523/JNEUROSCI.21-06-01983.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinholm JE, Bergersen LH. Neuroscience: The wrap that feeds neurons. Nature. 2012;487:435–436. doi: 10.1038/487435a. [DOI] [PubMed] [Google Scholar]
- Rinholm JE, Hamilton NB, Kessaris N, Richardson WD, Bergersen LH, Attwell D. Regulation of oligodendrocyte development and myelination by glucose and lactate. J. Neurosci. 2011;31:538–548. doi: 10.1523/JNEUROSCI.3516-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouach N, Koulakoff A, Abudara V, Willecke K, Giaume C. Astroglial metabolic networks sustain hippocampal synaptic transmission. Science. 2008;322:1551–1555. doi: 10.1126/science.1164022. [DOI] [PubMed] [Google Scholar]
- Saher G, Brugger B, Lappe-Siefke C, Mobius W, Tozawa R, Wehr MC, Wieland F, Ishibashi S, Nave KA. High cholesterol level is essential for myelin membrane growth. Nat. Neurosci. 2005;8:468–475. doi: 10.1038/nn1426. [DOI] [PubMed] [Google Scholar]
- Scherer SS, Deschenes SM, Xu YT, Grinspan JB, Fischbeck KH, Paul DL. Connexin32 is a myelin-related protein in the PNS and CNS. J. Neurosci. 1995;15:8281–8294. doi: 10.1523/JNEUROSCI.15-12-08281.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiza N, Sargiannidou I, Kagiava A, Karaiskos C, Nearchou M, Kleopa KA. Transgenic replacement of Cx32 in gap junction-deficient oligodendrocytes rescues the phenotype of a hypomyelinating leukodystrophy model. Hum. Mol. Genet. 2015;24:2049–2064. doi: 10.1093/hmg/ddu725. [DOI] [PubMed] [Google Scholar]
- Shachter NS. Apolipoproteins C-I and C-III as important modulators of lipoprotein metabolism. Curr. Opin. Lipidol. 2001;12:297–304. doi: 10.1097/00041433-200106000-00009. [DOI] [PubMed] [Google Scholar]
- Siegel GJ, Albers RW, Brady S, Price DL. Basic neurochemistry: molecular, cellular and medical aspects. San Diego: Elsevier Academic, Burlington, MA; 2006. [Google Scholar]
- Simmons DL, Tan S, Tenen DG, Nicholson-Weller A, Seed B. Monocyte antigen CD14 is a phospholipid anchored membrane protein. Blood. 1989;73:284–289. [PubMed] [Google Scholar]
- Sloane JA, Batt C, Ma Y, Harris ZM, Trapp B, Vartanian T. Hyaluronan blocks oligodendrocyte progenitor maturation and remyelination through TLR2. Proc. Natl. Acad. Sci. U. S. A. 2010;107:11555–11560. doi: 10.1073/pnas.1006496107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith X, Schneider H, Kohler K, Liu H, Lu Y, Rudd CE. The chemokine CXCL12 generates costimulatory signals in T cells to enhance phosphorylation and clustering of the adaptor protein SLP-76. Sci. Signal. 2013;6:ra65. doi: 10.1126/scisignal.2004018. [DOI] [PubMed] [Google Scholar]
- Srinivasan R, Sun G, Keles S, Jones EA, Jang SW, Krueger C, Moran JJ, Svaren J. Genome-wide analysis of EGR2/SOX10 binding in myelinating peripheral nerve. Nucleic Acids Res. 2012;40:6449–6460. doi: 10.1093/nar/gks313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stocks J, Galton DJ. Activation of the phospholipase A1 activity of lipoprotein lipase by apoprotein C-II. Lipids. 1980;15:186–190. doi: 10.1007/BF02540967. [DOI] [PubMed] [Google Scholar]
- Sutherland DR, Yeo E, Ryan A, Mills GB, Bailey D, Baker MA. Identification of a cell-surface antigen associated with activated T lymphoblasts and activated platelets. Blood. 1991;77:84–93. [PubMed] [Google Scholar]
- Tangye SG, van de Weerdt BC, Avery DT, Hodgkin PD. CD84 is up-regulated on a major population of human memory B cells and recruits the SH2 domain containing proteins SAP and EAT-2. Eur. J. Immunol. 2002;32:1640–1649. doi: 10.1002/1521-4141(200206)32:6<1640::AID-IMMU1640>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- Taniike M, Mohri I, Eguchi N, Beuckmann CT, Suzuki K, Urade Y. Perineuronal oligodendrocytes protect against neuronal apoptosis through the production of lipocalin-type prostaglandin D synthase in a genetic demyelinating model. J. Neurosci. 2002;22:4885–4896. doi: 10.1523/JNEUROSCI.22-12-04885.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniike M, Marcus JR, Popko B, Suzuki K. Expression of major histocompatibility complex class I antigens in the demyelinating twitcher CNS and PNS. J. Neurosci. Res. 1997;47:539–546. [PubMed] [Google Scholar]
- Tatar CL, Appikatla S, Bessert DA, Paintlia AS, Singh I, Skoff RP. Increased Plp1 gene expression leads to massive microglial cell activation and inflammation throughout the brain. ASN Neuro. 2010;2:e00043. doi: 10.1042/AN20100016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traka M, Arasi K, Avila RL, Podojil JR, Christakos A, Miller SD, Soliven B, Popko B. A genetic mouse model of adult-onset, pervasive central nervous system demyelination with robust remyelination. Brain. 2010;133:3017–3029. doi: 10.1093/brain/awq247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tress O, Maglione M, May D, Pivneva T, Richter N, Seyfarth J, Binder S, Zlomuzica A, Seifert G, Theis M, Dere E, Kettenmann H, Willecke K. Panglial gap junctional communication is essential for maintenance of myelin in the CNS. J. Neurosci. 2012;32:7499–7518. doi: 10.1523/JNEUROSCI.0392-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tress O, Maglione M, Zlomuzica A, May D, Dicke N, Degen J, Dere E, Kettenmann H, Hartmann D, Willecke K. Pathologic and phenotypic alterations in a mouse expressing a connexin47 missense mutation that causes Pelizaeus-Merzbacher-like disease in humans. PLoS Genet. 2011;7:e1002146. doi: 10.1371/journal.pgen.1002146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseveleki V, Rubio R, Vamvakas SS, White J, Taoufik E, Petit E, Quackenbush J, Probert L. Comparative gene expression analysis in mouse models for multiple sclerosis, Alzheimer's disease and stroke for identifying commonly regulated and disease-specific gene changes. Genomics. 2010;96:82–91. doi: 10.1016/j.ygeno.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlenberg B, Schuelke M, Ruschendorf F, Ruf N, Kaindl AM, Henneke M, Thiele H, Stoltenburg-Didinger G, Aksu F, Topaloglu H, Nurnberg P, Hubner C, Weschke B, Gartner J. Mutations in the gene encoding gap junction protein alpha 12 (connexin 46.6) cause Pelizaeus-Merzbacher-like disease. Am. J. Hum. Genet. 2004;75:251–260. doi: 10.1086/422763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urade Y, Kitahama K, Ohishi H, Kaneko T, Mizuno N, Hayaishi O. Dominant expression of mRNA for prostaglandin D synthase in leptomeninges, choroid plexus, and oligodendrocytes of the adult rat brain. Proc. Natl. Acad. Sci. U. S. A. 1993;90:9070–9074. doi: 10.1073/pnas.90.19.9070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Velde H, von Hoegen I, Luo W, Parnes JR, Thielemans K. The B-cell surface protein CD72/Lyb-2 is the ligand for CD5. Nature. 1991;351:662–665. doi: 10.1038/351662a0. [DOI] [PubMed] [Google Scholar]
- Wallraff A, Kohling R, Heinemann U, Theis M, Willecke K, Steinhauser C. The impact of astrocytic gap junctional coupling on potassium buffering in the hippocampus. J. Neurosci. 2006;26:5438–5447. doi: 10.1523/JNEUROSCI.0037-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasseff SK, Scherer SS. Cx32 and Cx47 mediate oligodendrocyte:astrocyte and oligodendrocyte:oligodendrocyte gap junction coupling. Neurobiol. Dis. 2011;42:506–513. doi: 10.1016/j.nbd.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney LW, Ludwin SK, McFarland HF, Biddison WE. Microarray analysis of gene expression in multiple sclerosis and EAE identifies 5-lipoxygenase as a component of inflammatory lesions. J. Neuroimmunol. 2001;121:40–48. doi: 10.1016/s0165-5728(01)00438-6. [DOI] [PubMed] [Google Scholar]
- Wieser GL, Gerwig UC, Adamcio B, Barrette B, Nave KA, Ehrenreich H, Goebbels S. Neuroinflammation in white matter tracts of Cnp1 mutant mice amplified by a minor brain injury. Glia. 2013;61:869–880. doi: 10.1002/glia.22480. [DOI] [PubMed] [Google Scholar]
- Wijewickrama GT, Kim JH, Kim YJ, Abraham A, Oh Y, Ananthanarayanan B, Kwatia M, Ackerman SJ, Cho W. Systematic evaluation of transcellular activities of secretory phospholipases A2. High activity of group V phospholipases A2 to induce eicosanoid biosynthesis in neighboring inflammatory cells. J. Biol. Chem. 2006;281:10935–10944. doi: 10.1074/jbc.M512657200. [DOI] [PubMed] [Google Scholar]
- Willecke K, Eiberger J, Degen J, Eckardt D, Romualdi A, Guldenagel M, Deutsch U, Sohl G. Structural and functional diversity of connexin genes in the mouse and human genome. Biol. Chem. 2002;383:725–737. doi: 10.1515/BC.2002.076. [DOI] [PubMed] [Google Scholar]
- Williams JL, Holman DW, Klein RS. Chemokines in the balance: maintenance of homeostasis and protection at CNS barriers. Front. Cell. Neurosci. 2014;8:154. doi: 10.3389/fncel.2014.00154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Wells AB, O'Brien DR, Nehorai A, Dougherty JD. Cell type-specific expression analysis to identify putative cellular mechanisms for neurogenetic disorders. J. Neurosci. 2014;34:1420–1431. doi: 10.1523/JNEUROSCI.4488-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama S, Staunton D, Fisher R, Amiot M, Fortin JJ, Thorley-Lawson DA. Expression of the Blast-1 activation/adhesion molecule and its identification as CD48. J. Immunol. 1991;146:2192–2200. [PubMed] [Google Scholar]
- Yoshikawa K, Palumbo S, Toscano CD, Bosetti F. Inhibition of 5-lipoxygenase activity in mice during cuprizone-induced demyelination attenuates neuroinflammation, motor dysfunction and axonal damage. Prostaglandins Leukot. Essent. Fatty Acids. 2011;85:43–52. doi: 10.1016/j.plefa.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR, O'Keeffe S, Phatnani HP, Guarnieri P, Caneda C, Ruderisch N, Deng S, Liddelow SA, Zhang C, Daneman R, Maniatis T, Barres BA, Wu JQ. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J. Neurosci. 2014;34:11929–11947. doi: 10.1523/JNEUROSCI.1860-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlokovic BV. Cerebrovascular effects of apolipoprotein E: implications for Alzheimer disease. JAMA Neurol. 2013;70:440–444. doi: 10.1001/jamaneurol.2013.2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.