Abstract
The N-end rule pathway is a proteolytic system in which single N-terminal amino acids of short-lived substrates determine their metabolic half-lives. Substrates of this pathway have been implicated in the pathogenesis of many diseases, including malignancies, neurodegeneration, and cardiovascular disorders. This review provides a comprehensive overview of current knowledge about the mechanism and functions of the N-end rule pathway. Pharmacological strategies for the modulation of target substrate degradation are also reviewed, with emphasis on their in vivo implications. Given the rapid advances in structural and biochemical understanding of the recognition components (N-recognins) of the N-end rule pathway, small-molecule inhibitors and activating ligands of N-recognins emerge as therapeutic agents with novel mechanisms of action.
The N-end Rule Pathway
The ubiquitin (Ub)-proteasome system (UPS) and the autophagy-lysosome system (hereafter referred to as autophagy) are two major regulatory mechanisms of protein catabolism and homeostasis in cells. While the UPS has for decades been considered the principal regulator of irreversible intracellular protein degradation, autophagy has recently emerged as an equally crucial proteolytic pathway implicated in human health and disease [1–4]. Several degradation signals (degrons) of the UPS have been relatively well characterized and generalized. For example, under normal oxygen conditions, the “hydroxy-degron” of hypoxia-inducible factor-1 (HIF-1) is recognized by the von Hippel-Lindau (VHL) E3 ligase, leading to its ubiquitination and proteasomal degradation [5]. When cellular oxygen concentrations reduced, however, the hydroxy-degron is removed, thereby stabilizing HIF-1, which functions as a hypoxia-sensitive transcription factor. Many cyclins and cyclin-dependent kinases (CDKs) contain the Pro-Glu-Ser-Thr (PEST) consensus sequences that function as “phospho-degrons” [6]. These signals, usually generated by specific kinases in various phases of the cell cycle, are recognized by the SCF E3 ligase complex and consequently ubiquitinated and degraded by the UPS. Endoplasmic reticulum (ER)-associated protein degradation (ERAD) system mediates the degradation of misfolded proteins, which are generally characterized by the “hydrophobic degron” and/or specific N-glycans for ubiquitination [7]. In autophagy, the biochemical nature of degron selectivity, spatiotemporal activation of pro-degrons, and the specific interaction between cargoes and adaptor proteins have yet to be identified.
The N-end rule pathway refers to a biological mechanism where N-terminal amino acids determine the half-lives of proteins by serving as an essential component of degradation signals (N-degrons) for UPS-mediated proteolysis [8, 9]. The N-degrons are the first defined degradation signals in eukaryotes [10]. The N-end rule pathway appears to exist in various eukaryotes ranging from mammals to plants and yeasts, and is even present in bacteria, which lack Ub [11]. Global proteomic analysis has revealed that the N-terminal residues (N-proteome) exposed after proteolytic processing in Saccharomyces cerevisiae and Aspergillus niger are indeed mostly “stabilizing” in nature [12, 13]. The pathway's components play roles in various cellular processes, including cardiovascular development, neural tube formation, apoptosis, spermatogenesis, chromosomal stability, oxygen/heme sensing, and muscle protein degradation [14–24]. Various neurodegeneration-associated aggregation-prone proteins, such as tau, α-synuclein, and TDP43, have also been identified as short-lived substrates of the arginylation branch of the N-end rule pathway (Arg/N-end rule pathway) when internal cleavage events expose their N-degrons[25]. Recently, the first mammalian substrate of the acetylation branch of the N-end rule pathway (Ac/N-end rule pathway) was identified [26]. Moreover, the first direct roles of the Arg/N-end rule pathway in autophagy-mediated protein quality control have been recently demonstrated [27]. Thus, the biological insights of the N-end rule pathway in cell life and death as well as in human health and disease are still being clarified.
Since its identification [10], the N-end rule pathway has often been misunderstood as an orphan system without significant roles in cellular processes. However, given the growing numbers of its endogenous substrates and physiological functions identified during the last decade, the importance of this proteolytic system as a key regulator of various critical biological processes is now well appreciated [28, 29]. Herein, we focus on recent efforts to develop small-molecule modulators of the mammalian N-end rule pathway through rational design, high-throughput screening (HTS), and chemical engineering. Of note is a recently identified N-end rule inhibitor, para-chloroamphetamine (PCA), which potently delays the degradation of RGS4 (regulator of G protein signaling 4), a bona fide in vivo N-end rule substrate in the mouse brain [30, 31]. Given that the components in the pathway and their functions are emerging as novel therapeutic targets, we also review recent findings about the biochemical mechanisms and physiological functions of the mammalian N-end rule pathway. Finally, we discuss new approaches to treat human diseases via modulation of the activity of the N-end rule pathway without inhibiting its substrate degradation.
Biochemical machinery of N-end rule-dependent proteolysis
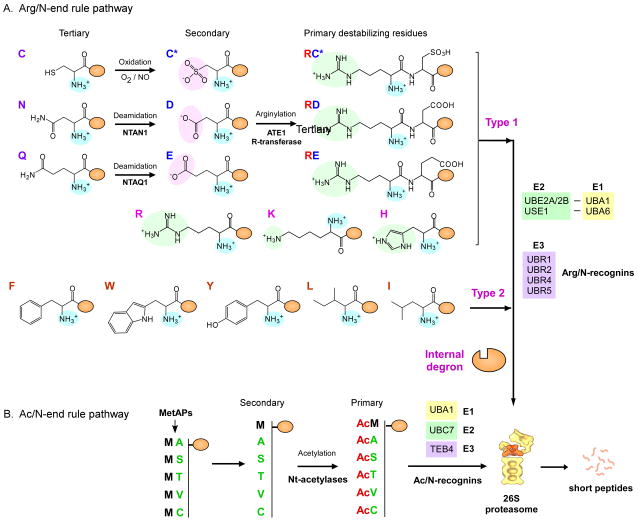
In the N-end rule pathway, a typical N-degron is composed of a “destabilizing” N-terminal amino acid, an internal Lys residue from which a Ub chain grows, and an unstructured N-terminal extension [32–34]. Short-lived substrates that contain N-degrons can be polyubiquitinated by Ub-protein ligase E3 component N-recognin (UBR) proteins, a family of E3 Ub ligases directly recognizing the destabilizing N-terminal residues. The identity of the destabilizing residues defines two branches of the pathway, i.e. the Arg/N-end rule and the Ac/N-end rule branches, in mammals. In the Arg/N-end rule branch, the primary destabilizing N-terminal amino acids include positively charged residues (also known as “type 1” destabilizing residues), such as Arg, Lys, and His, or hydrophobic residues (“type 2”), such as Phe, Trp, Leu, Tyr, and Ile (Figure 1A). Arg/N-degrons can be generated when primary, secondary, or tertiary destabilizing residues, which are normally embedded in the protein inside, are exposed at the N-termini of C-terminal fragments by endoproteolytic cleavage; however, they can also be generated when the substrates undergo structural alteration that exposes the previously sterically sequestered pro-N-degron. Known peptidases that can generate N-degrons include Met-aminopeptidases (MetAPs) that cotranslationally remove N-terminal Met [35, 36], caspases that cleave more than 500 proteins during apoptotic induction [37], the signal peptide peptidase that removes the signal peptides from nascent polypeptides translocating into the ER lumen [27], the mitochondrial processing peptidase (MPP) which cleaves presequences from proteins translocating into the mitochondria [11], calpains that cleave many cellular proteins in a Ca2+ dependent-manner [38], and separase that cleaves the cohesin complex subunit SCC1 [39].
Figure 1.
Mammalian N-end rule pathway. (A) Arginylation branch of the N-end rule pathway (Arg/N-end rule pathway) in mammals. The tertiary destabilizing Cys residues are oxidized by O2 or nitric oxide (NO) into the secondary destabilizing residues Cys sulfinate (Cys-SO2−) or Cys sulfonate (Cys-SO3−). N-terminal Asn and Gln are deamidated into Asp and Glu by NTAN1 and NTAQ1, respectively. All of the secondary destabilizing residues expose negatively charged side chains (pink background). Secondary destabilizing residues, such as oxidized Cys, Asp, and Glu, are substrates of arginylation by ATE1 R-transferases. The positively charged side chains of N-terminal Arg, Lys, and His in type 1 destabilizing residues are shown in green. Type 2 destabilizing residues are hydrophobic residues such as Phe, Trp, Tyr, Leu, and Ile. These destabilizing residues are recognized and polyubiquitinated by Arg/N-recognins. In the Arg/N-end rule pathway, Ub can be activated and transferred by UBA1-UBE2A/2B (canonical) or UBA6-USE1 (noncanonical) cognates as E1–E2 systems. (B) Acetylation branch of the N-end rule pathway (Ac/N-end rule pathway) in mammals. N-terminal acetylation occurs at the newly exposed N-terminal residues, such as Ala, Ser, Thr, Val, and Cys, after the N-terminal Met excision by Met-endopeptidases (MetAPs) or at the retained N-terminal Met residue. These residues are recognized by the mammalian Ac/N-recognin TEB4.
In both arginylation and acetylation branches, N-degrons can be created by a single enzymatic modification or through multistep processes that involve both chemical and enzymatic modifications. For example, in the mammalian Arg/N-end rule pathway, the tertiary destabilizing residues Asn and Gln are deamidated into the secondary destabilizing residues Asp and Glu by NTAN1 and NTAQ1, respectively (Figure 1A). The free α-amine groups of N-terminal Asp and Glu are then conjugated with the carboxylic acid of Arg by ATE1 R-transferases, thus creating the primary destabilizing residue Arg. The tertiary destabilizing residue N-terminal Cys can be converted into an oxidized Cys (C*) via oxidation with oxygen (O2) or nitric oxide (NO). The resulting oxidized Cys residue subsequently undergoes arginylation by ATE1 R-transferases in the same manner as that observed for Asp and Glu N-degrons.
N-degrons in the acetylation branch are generated by N-terminal acetylation, which cotranslationally occurs in approximately 90% of mammalian proteins [40]. The substrates of N-terminal acetylation include the unprocessed N-terminal Met and newly exposed small uncharged N-terminal residues such as Ala, Val, Ser, Thr, or Cys (Figure 1B). These residues are originally present on the penultimate position on the N-termini but are exposed by MetAPs [35, 36]. These secondary destabilizing residues are cotranslationally acetylated by N-terminal acetylases to generate the primary Ac/N-degrons. Although the proteolytic system driven by N-terminal acetylation was originally found in S. cerevisiae, a recent study demonstrated that it operates in mammals as well, in which human TEB4, an ortholog of yeast Doa10 and a 151-kDa integral membrane protein with a RING domain in the ER, is the E3 Ub ligase for Ac/N-degrons [26].
The recognition components of the Arg/N-end rule pathway in mammals, which mediate the formation of polyubiquitin chains on the substrates, include a family of UBR proteins such as UBR1/E3α, UBR2, UBR4/p600, and UBR5/EDD [41–44]. The UBR-type N-recognins are spalogous (with spatial similarity) to each other with the UBR box or N-domain, through which they interact directly with type 1 or type 2 destabilizing residues, respectively [45]. The UBR box is a His/Cys-rich, zinc finger-like motif with a size of approximately 70 residues that mainly utilizes its acidic binding pocket to interact with α-amino groups (-NH3+) and basic side chains of type 1 destabilizing residues [46, 47]. The N-domain of UBR proteins has structural and functional similarities to the Escherichia coli N-recognin ClpS, the only known bacterial N-recognin. A couple of biochemical and computational studies suggest that this domain is essential for binding to type 2 destabilizing residues through a hydrophobic pocket near its surface [48, 49]. Following the recognition of N-degrons, the Arg/N-recognins UBR1 and UBR2 mediate polyubiquitination in cooperation with their cognate E2 Ub transfer enzymes UBE2A and UBE2B (orthologs of yeast Rad6) to form the N-end rule E2-E3 complexes (Figure 1A) [50]. A recent study showed that UBR1 and UBR2 also directly interact with USE1, a specialized E2 enzyme for the non-canonical E1 enzyme UBA6, a unique phenomenon through which different E1 enzymes eventually emerge in the same E3 Ub ligases and target substrates [51].
Given the structural and functional aspects of the Arg/N-end rule pathway, the UBR box and N-domain have been the major targets for the development of small-molecule inhibitors that bear cognate destabilizing residues or their structural mimics. In contrast, little biochemical and structural information is available about the mode of interaction between noncanonical N-recognins, such as UBR4 and UBR5, and their ligands [8].
Physiological implications of the mammalian N-end rule pathway
The physiological functions of the N-end rule pathway have been elucidated through mouse-knockout phenotype analysis as well as human disease-gene mapping and in vivo substrate identification/characterization. The studies have demonstrated that the N-end rule pathway has a strikingly diverse range of developmental and physiological roles, which include the regulation of chromosomal stability [14], spermatogenesis [15], oxygen sensing [16, 19], cardiovascular development [18], muscle wasting [20–23], proteotoxic protein clearance [25], hypertension [26], autophagy [27], neural tube formation [50], bacterial/viral virulence [52, 53], apoptosis [54], and mitophagy [55]. Mutational inactivation of both copies of the UBR1 gene causes Johanson-Blizzard syndrome (JBS), which involves exocrine pancreatic insufficiency and inflammation, physical malformations, and frequent mental retardation [56]. Below we focus on three pathophysiological pathways known to be regulated by components of the mammalian N-end rule pathway. They represent the elaborate cellular machinery controlled by N-end rule biochemistry, which may in turn be regulated by small-molecule antagonists and inhibitors.
Cardiovascular development and hypertension
The ATE1 gene encodes a family of R-transferase isoforms that mediate N-terminal arginylation of pro-N-degrons such as Asp, Glu, and oxidized Cys [57, 58]. ATE1 R-transferases are responsible for all known arginylation activities observed in yeast and mammals, and they provide a central merging point in Arg/N-end rule-related chemical modifications (Figure 1A). Analysis of ATE1−/− mice, along with in vitro HTS for Arg/N-end rule substrates, revealed that RGS4, RGS5 and RGS16 are the substrates of arginylation [18, 19]. These proteins negatively control GPCR signaling through their GTPase activity for Gα subunits of the i, q, and 12 classes (Table 1) [59]. Proteolysis of RGS proteins is also perturbed by either hypoxia or a lack of the N-recognins UBR1 and UBR2, which indicates that additional N-terminal modifications are required for the ubiquitination of RGS proteins. The downstream effectors of the GPCR, including mitogen-activated protein kinase (MAPK), are indeed markedly impaired in ATE1−/− and UBR1−/−UBR2−/− mice [18]. These results established that the Arg/N-end rule pathway, containing the O2-ATE1-UBR1/UBR2 proteolytic circuit, plays a key role in RGS-regulated G protein signaling, including the myocardial hypertrophic response, in the cardiovascular system [19, 60].
Table 1.
Physiological substrates and implications of the mammalian N-end rule pathway
| The mammalian N-end rule pathway | Substrates/Components | Physiological functions |
|---|---|---|
| Arg/N-end rule pathway | Cys2-RGS4 | GPCR signaling pathway Cardiac development |
| Cys2-RGS5 | ||
| Cys2-RGS16 | ||
| Leu2-RGS2 | Hypertension Cardiovascular homeostasis | |
| Arg208-TDP43 | Amyotrophic lateral sclerosis (ALS) Frontotemporal lobar degeneration (FTLD) |
|
| Asp219-TDP43 | ||
| Asp247-TDP43 | ||
| Gln79-Synuclein | Parkinson’s disease (PD) | |
| Asp-Aβ42 | Alzheimer’s disease (AD) | |
| Glu3-Tau | Alzheimer’s disease (AD) | |
| Glu19-Bip1 | Protein quality control through autophagy | |
| PINK1 | Mitochondria quality control through mitophagy | |
| Proapoptotic protein fragments | Apoptosis regulation | |
| myofibrils | Muscle wasting | |
| ATE1−/− | Heart development | |
| Angiogenesis | ||
| Hyperphagia | ||
| Hyperkinesia | ||
| Infertility | ||
| Metabolic defects | ||
| UBR1−/− | Johanson-Blizzard Syndrome(JBS) | |
| UBR1−/−UBR2−/− | Neurogenesis | |
| Cardiogenesis | ||
| UBR4−/− | Regulation of autophagic flux | |
|
| ||
| Ac/N-end rule pathway | Met-Arg-RGS2 | Hypertension Cardiovascular homeostasis |
| Met-Gln-RGS2 | ||
| Met-Leu-RGS2 | ||
In the degradation of Cys-2 bearing RGS proteins, the cleavage of the N-terminal Met by MetAP, which exposes the penultimate residue Cys is a crucial initial step to generate the primary N-degron Arg. It is interesting to note that wild-type RGS2, another R4 subfamily member along with RGS4, RGS5, and RGS16, has a Gln-2 residue but its mutant, which is frequently observed in hypertension patients, has a Leu-2 or Arg-2 residue [61]. These RGS2 proteins are targeted by either the Arg/N-end rule pathway (L-RGS2 as substrate) or the Ac/N-end rule pathway (Ac-ML-RGS2, Ac-MR-RGS2, and Ac-MQ-RGS2 as substrates) (Table 1) [26]. These findings indicate that, despite that they utilize different N-recognins and N-degrons, the two branches of the N-end rule pathway can be functionally complementary to each other to target the same protein in a cell. The communication and possible compensatory activity changes between these branches of the N-end rule pathway during disease progression remain to be further investigated.
Oxidative stress and neurodegenerative diseases
The conjugation of Arg to the pro-N-degrons is a signature reaction of the Arg/N-end rule pathway. The N-terminal Cys residue in RGS4, RGS5, and RGS16 (and possibly other ~500 proteins with the N-terminal Met-Cys repertoire in the human genome [8]) is oxidized to Cys sulfinate (Cys-SO2−) or Cys sulfonate (Cys-SO3−) before its arginylation, suggesting that the Arg/N-end rule pathway may function as an O2 and NO sensor [16, 57]. The relatively higher level of RGS4 observed in the right ventricle of mouse embryos compared with that in the left ventricle may be attributable to its metabolic stabilization under less oxygenated blood [18]. T he N-end rule pathway also functions as a sensor of heme (Fe2+-containing protoporphyrin IX). ATE1 R-transferase is inhibited by hemin (Fe3+-counterpart of heme) through a redox mechanism, involving the formation of a disulfide bridge between its Cys-71 and Cys-72 residues [17]. Given that O2, NO, and carbon monoxide (CO) can bind to heme, it is reasonable to expect that the reciprocal O2 sensing modes of the Arg/N-end rule pathway are implicated in various human diseases. The exact biochemical mechanisms and physiological roles of these processes remain unidentified.
Oxidative stress is also a known risk factor for many neurodegenerative diseases. Reactive oxygen species (ROS) are generated under the conditions of nutrient starvation, aging, mitochondria dysfunction, imbalanced metal ions, impaired antioxidant enzymes, and amyloid deposition [62]. Oxidized proteins are usually efficiently cleared by the UPS; however, under neurodegenerative conditions, they frequently accumulate in the brain [63, 64]. The N-end rule pathway is closely involved in the degradation of neurodegeneration-associated proteins [25]. The aggregates of tau, TDP43, α-synuclein, and amyloid β (Aβ) are etiologically linked with various neurodegenerative diseases (Table 1) [65]. When in the soluble states, these proteotoxic proteins can undergo cleavage to expose C-terminal fragments that bear destabilizing N-terminal residues, which are then subjected to N-end rule pathway-mediated proteolysis [25]. In addition to the N-end rule pathway, tau (primarily in its phosphorylated form) was shown to be targeted for ubiquitination by the E3 Ub ligase CHIP in collaboration with UBCH5B/UBCH5C and HSP70 [66, 67]. Similarly, it has been shown that α-synuclein can be ubiquitinated by a number of Ub ligases, including CHIP [68] and SIAH [69]. These results suggest that a complicated net of proteolytic pathways cooperatively interact with these neurodegeneration-associated proteins, affecting the rates of their aggregation in the neuron.
The intracellular aggregation of TDP43, an RNA/DNA-binding protein, results in a spectrum of related disorders, including amyotrophic lateral sclerosis (ALS) and other neurodegenerative syndromes [70, 71]. The major N-terminal residues of the aggregation-prone C-terminal TDP43 fragments are Arg208, Asp219, and Asp247, all of which are either primary or secondary N-degrons [72, 73]. The levels of these N-degron-bearing fragments are significantly elevated by ATE1 nullification or treatment with N-end rule inhibitors [31], which suggests that the pathological fragment of TDP43 is a substrate for the Arg/N-end rule pathway. Because adult neurons are post-mitotic but must retain synaptic plasticity and self-renewal properties, frequent protein turnover is a central challenge in degradation systems. Thus, the aforementioned observations reflect the critical role of the Arg/N-end rule pathway as a neuroprotective mechanism that regulates the rate at which aggregation-prone fragments and misfolded proteins are destroyed.
Implications in autophagy
Communication between the UPS and autophagy has been suggested by the observation that reduced UPS activity results in autophagy induction [74, 75]. This crosstalk is unlikely to be a simple compensatory mechanism because impaired autophagy decreases UPS flux [76]. Interestingly, recent studies have revealed the unexpected finding that the N-end rule pathway induces proteolysis through autophagy [77]. For example, mouse embryos lacking UBR4 die at approximately embryonic day 9.5 (E.9.5) with pleiotropic abnormalities, including impaired vascular development of the yolk sac. UBR4−/− yolk sac and mouse embryonic fibroblasts show increased formation of microtubule-associated protein 1A/1B-light chain 3 (LC3) puncta associated with enhanced autophagic flux [77]. Autophagic induction by UBR4 inactivation correlates with UBR4 localization to autophagic vacuoles, such as multivesicular bodies (MVBs), and their degradation by lysosomes, which suggests that UBR4 is associated with autophagic cargoes and may regulate their delivery to autophagic vacuoles through the endosome-lysosome pathway (Table 1). Given that UBR4 has the 70-residue UBR box that can bind the N-terminal Arg residue, the function of UBR4 in autophagy may involve an allosteric conformational change induced by N-degrons or their small-molecule mimics that act as activating ligands. In addition, the Arg/N-end rule pathway constitutively degrades C-terminal fragments of PINK1, whose mutations are linked to early-onset familial Parkinson's disease, in normal condition [55]. The accumulation of PINK1 leads to the selective degradation of dysfunctional mitochondria via autophagy, a process called mitophagy.
Another link between the N-end rule pathway and autophagy was unveiled when a set of proteins residing in the ER was found to be N-terminally arginylated by ATE1-encoded R-transferases [27]. These substrates of N-terminal arginylation include major Ca2+-binding molecular chaperones including BiP/GRP78 and oxidoreductases. The N-terminal arginylation of ER proteins is selectively induced under prolonged proteasomal inhibition, which leads to retrotranslocation and cytosolic accumulation of their arginylated species. N-terminally arginylated BiP (R-BiP) in the cytosol is associated with misfolded cytosolic proteins that are initially tagged with Ub but remain unprocessed owing to proteasomal inhibition. R-BiP alone or in association with its cargoes, such as misfolded cytosolic proteins, binds to the autophagic adaptor p62/SQTSM/sequestosome-1 through the interaction between its N-terminal Arg residue and the ZZ domain of p62. This interaction induces an allosteric conformational change in p62, leading to its self-oligomerization. Consequently, R-BiP and p62, along with their cargoes, are co-targeted to the autophagosome for lysosomal degradation (Table 1) [27]. In this autophagic proteolytic pathway, the N-terminal Arg residue of R-BiP acts as a delivery determinant and degron for lysosome targeting. The physiological importance of N-terminal arginylation in protein quality control through autophagy and of the crosstalk between the UPS and autophagy needs to be further investigated.
As illustrated above, the N-end rule pathway actively protects cells from the detrimental effects of accumulated proteotoxic protein fragments through both proteasome-and autophagy-mediated degradation. Excessive levels of oxidized and misfolded proteins inhibit proteasomal degradation, likely by blocking the 20S complex [74]. Therefore, N-end rule-mediated autophagy induction may function as a compensatory neuroprotective mechanism along with direct removal of pathological fragments of proteotoxic proteins through the UPS. In yeasts, the Arg/N-end rule pathway is involved in the quality control mechanism to selectively eliminate misfolded proteins [78, 79]. It remains to be determined whether N-degrons can be generated and eliminated from misfolded proteins during their translational or posttranslational manipulation in mammals.
Chemical modulation of the N-end rule pathway
The physiological functions of the N-end rule pathway have been identified mainly through phenotypic studies of knockout mice. Because the constitutional nullifications of many key pathway components result in embryonic or neonatal lethality, important physiological roles of these components in adulthood remain obscure. Several biocompatible N-end rule inhibitors have been developed to regulate the degradation of target substrates at the experimental level, thus providing a more feasible way in comparison to genetic manipulations. The development of more potent and specific in vivo N-end rule pathway inhibitors is of mounting interest not only for biological study but also for pharmacological applications.
Dipeptide ligands to N-recognins
The concept of employing dipeptides that contain destabilizing N-terminal residues for competitive inhibition was developed almost simultaneously with the identification of the N-end rule pathway in the late 1980's [80–83]. As yeast cells possess a transporter system for amino acid derivatives, including the dipeptides, the ability of dipeptides to inhibit the N-end rule pathway was tested in yeast cells expressing model N-end rule substrates, Ub-X-βgal (X = various residues) [84]. This model substrate exploited the Ub fusion technique in which Ub-X-βgal is cotranslationally cleaved by deubiquitinating enzymes (DUBs) into the Ub reference moiety and X-βgal substrates bearing various N-terminal residues.
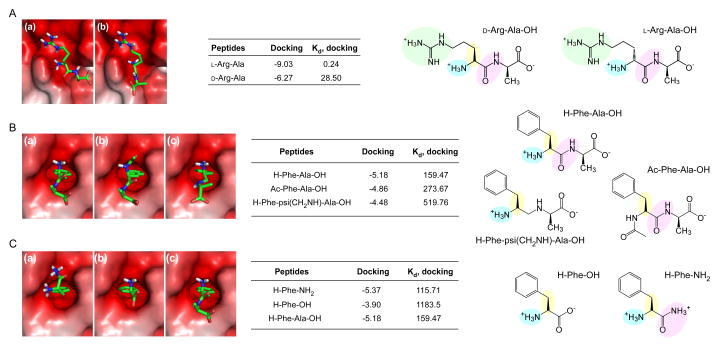
Consistent with the substrate specificity of yeast Ubr1, Arg-Ala and other type-1 dipeptides selectively increased the steady-state levels of X-βgal bearing type-1 residues but not type-2 residues. A similar specificity in N-end rule inhibition was observed with type-2 dipeptides [84]. Notably, the substitution of the N-terminal Arg with its D-form stereoisomers resulted in a dramatic loss in its inhibitory effect (Figure 2A). Even at higher concentrations up to 6 mM, D-Arg-Ala showed virtually no inhibition, indicating that the recognition of UBR proteins and N-degrons is highly stereospecific [85]. The results from yeast cells have been extended and confirmed in in vitro ubiquitination and degradation assays using rabbit reticulocyte lysates [81, 86]. These biochemical studies using dipeptides, together with genetic dissection of yeast Ubr1, revealed that the degradation of N-end rule substrates requires the recognition of type-1 or type-2 destabilizing N-terminal residues by N-recognins. These studies also provide the rationale that the N-end rule pathway can be modulated in vivo with small-molecule inhibitors or activators of N-recognins [84].
Figure 2.
Interaction between N-recognins and N-end rule pathway inhibitors (A) Binding modes calculated through in silico docking analysis of (a) L-Arg-Ala and (b) D-Arg-Ala with the UBR1 box (Protein Data Bank [PDB] code: 3NY3), and binding affinities (kcal/mol) and dissociation constants (μM) of the docked complexes. (B) Binding modes, binding affinities (kcal/mol), and dissociation constants (μM) of (a) Phe-Ala, (b) Ac-Phe-Ala, and (c) Phe-psi(CH2NH)-Ala with the ClpS domain (PDB code: 3DNJ). (C) Same as (B), except that Phe and its derivatives are used for the in silico docking analysis. Positively charged side chains, e.g., guanidino groups in Arg for interaction with the UBR box, are shown in green spheres in the chemical structures. Essential components of small molecules for interaction with UBR proteins, such as L-conformation, protonated α-amine groups, and amide bond characters, are shown in yellow, blue, and pink spheres, respectively.
Various dipeptides found in nature and in the human body are produced from polypeptides and proteins by many different peptidases [87]. Extracellular dipeptides can be imported into cells through their specific transport systems, with efficiencies higher than those of amino acids [88, 89]. In principle, intracellular dipeptides can modulate, positively or negatively, the N-end rule pathway in human cells. Peptide transporter 1 (Ptr1), which is identical to the yeast Ubr1 N-recognin, is essential for peptide transport in S. cerevisiae, along with Ptr2 and Ptr3 [90]. A series of biochemical and genetic studies revealed that imported dipeptides act as ligands to Ubr1 whose binding to the UBR box induces an allosteric conformational change that, in turn, accelerates the ubiquitination and degradation of the homeodomain protein Cup9 [91, 92]. As a consequence of this degradation, which normally suppresses the transcription of Ptr2, the de novo synthesis of Prt2 is induced, mediating the import of extracellular peptides [91, 92]. The discovery of the Ubr1/Ptr1-Cup9-Ptr2 circuit provides the first example of a physiological process controlled by the N-end rule pathway and small peptides with destabilizing residues.
UBR-type N-recognins can mediate ubiquitination of noncanonical N-end rule substrates, i.e., short-lived proteins that do not carry N-degrons but internal degrons (Figure 1A), such as Cup9, Mgt1, histones, and misfolded proteins [93–95]. Internal degron-mediated degradation of Cup9 is accelerated by the binding of type-1 or type-2 short-peptides to the UBR box or N-domain, respectively. As such, dipeptides and their natural and synthetic derivatives (with N-degrons) can act as “activating” ligands for certain functions of N-recognins through opening their autoinhibited internal degron-binding sites. The internal degron-binding site of UBR proteins is not identified yet and could be an interesting target for inhibitor development, for example, to modulate intracellular levels of misfolded proteins.
In addition to their role in importing peptides, dipeptides have been implicated in N-end rule regulation of cell differentiation and neurite outgrowth [96]. These studies have shown that small-molecule ligands of N-recognins can modulate various physiological and pathophysiological processes in vivo. It is expected that endogenous dipeptides and other short peptides may act as modulators of N-recognins in various developmental and pathological processes, especially when cells undergo mass proteolysis such as histone removal during spermatogenesis and muscle wasting during systemic amyotrophism. Indeed, activation of the N-end rule pathway was reported to be implicated in the excessive ubiquitination and degradation of muscle proteins in various catabolic conditions [20, 21]. Moreover, UBR2 levels were significantly upregulated upon tumor-induced muscle loss through the p38β MAPK-C/EBPβ signaling pathway [23]. Interestingly, dipeptides and their derivatives with N-terminal destabilizing residues, such as Lys-Ala, Phe-Ala, Arg-methyl ester (Arg-ME), and Leu-ME, significantly delayed the rates of Ub conjugation and proteasome-mediated degradation of muscle proteins, while Ala-Phe, Ala-Lys, and Ala-ME had little effects [20, 21]. These results suggest that potent N-end rule inhibitors can be of therapeutic benefit to slow systemic muscle wasting in various pathologic conditions such as cancer-associated cachexia and acute diabetes [22, 23].
Heterovalent ligands to N-recognins
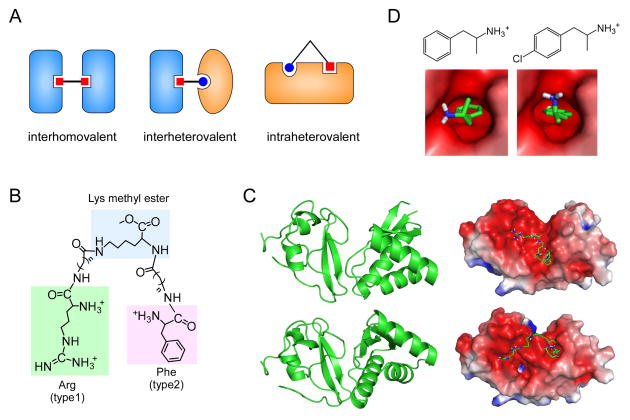
Compared with monomeric binding, multivalent binding provides higher thermodynamic (enhanced binding affinity) and kinetic (reduced dissociation rate) selectivity in protein-protein or protein-ligand interactions. Synthetic multivalent compounds, including interhomovalent, interheterovalent, and intraheterovalent compounds (Figure 3A), have been designed to exploit the cooperative interactions of multivalent ligands to target molecules. Most synthesized multivalent compounds as well as natural substrates are interhomovalent in that they have multiple, identical ligands that interact with the same binding sites on tandem enzymes, for example, those present on the surfaces of viruses, bacteria, or cells [97]. By contrast, interheterovalent compounds utilize non-identical ligands that bind cognate sites on different proteins. For example, rapamycin, a natural compound produced by Streptomyces hygroscopicus, can simultaneously bind to FKBP12 and mTOR, and mTOR binding to the FKBP12-rapamycin complex is ~2,000-fold stronger than that of rapamycin alone (12 nM vs. 26 μM dissociation constant) [98, 99]. Compared with interhomovalent and interheterovalent interactions, intraheterovalent interactions, in which different ligands in a single compound cooperatively bind to a target protein, remain relatively unexploited, mainly because of the lack of such targets in nature.
Figure 3.
Multivalent and monovalent inhibitors of the N-end rule pathway. (A) Types of multivalent ligands bound to their cognate proteins. (B) Structures of heterovalent molecule RF-Cn (where n indicates the length of hydrocarbon chains) tethering the core Lys methyl ester to the N-terminal Arg (type 1 destabilizing residue) and Phe (type 2), which are indicated by green and pink backgrounds, respectively. (C) Two possible structures of the UBR box-N-domain “combined” proteins (left) and their binding modes with the RF-C5 compound (right). Protein structures and docking models were obtained by using the Gramm-X protein-protein docking web server and semi-empirical PM6 method with the Gaussian 09 program [30]. (D) Chemical structures of amphetamine (left) and para-chloroamphetamine (right), and their binding modes with the N-domain (PDB code: 3DNJ) calculated with a ligand-receptor docking computation by using AutoDock version 4.2 with the Lamarckian genetic algorithm [85].
A family of N-recognins were exploited as a target of intraheterovalent interaction. The N-recognins UBR1 and UBR2 have two mutually exclusive binding pockets, the UBR box for type-1 residues (Arg, Lys, and His) and the N-domain for type-2 residues (Phe, Trp, Tyr, Leu, and Ile) (Figure 1A) [58]. Thermodynamically, a heterovalent compound, which carries two different N-terminal destabilizing residues, for example, N-terminal Arg and Phe, simultaneously, is expected to have a synergistically improved binding affinity, compared with a combination of two independent monovalent interactions (Figure 3B). In this heterovalent interaction, the Arg ligand bound to the UBR box will constrain random motions of the unbound Phe ligand, increasing the local Phe concentration in the proximity of the N-domain. This restriction, in turn, will increase the probability that the Phe ligand binds to the N-domain (and vice versa) without affecting the stability of the active site structures.
This conjecture was experimentally tested using RF-C11, a synthetic heterovalent compound that has both an N-terminal Arg (type 1) and an N-terminal Phe (type 2) chemically linked by two C11 hydrocarbon chains (Figure 3B) [30, 100]. Compared with homovalent controls such as RR-C11 (with two N-terminal Arg) and FF-C11 (with two N-terminal Phe) and monovalent dipeptides such as Arg-Ala and Phe-Ala, RF-C11 showed significantly higher inhibitory efficacy for in vitro ubiquitination and degradation of model N-end rule substrates. When the efficacy to inhibit the in vitro degradation of Arg-nsP4 was measured, the IC50 value of RF-C11 was determined to be 16 μM as compared with the homovalent control RR-C11 (67 μM) and the monovalent control Arg-Ala (283 μM) [25]. Similarly, the IC50 value of RF-C11 for Tyr-nsP4 degradation was determined to be 2.7 μM, which was markedly lower than those of FF-C11 (151 μM) and Phe-Ala (21 μM) [30]. RF-C11 also effectively inhibited the degradation of RGS4, a physiological N-end rule substrate, in living cells without noticeable cytotoxicity [30]. In vivo efficacy of RF-C11 was further supported by the finding that RF-C11 delays cell-autonomous hypertrophism in cultured primary cardiomyocytes, identifying a previously unknown function of the N-end rule pathway [100]. The inhibitory activity of RF-C11 was retained even in the absence of bestatin, an endopeptidase inhibitor, consistent with its nonproteinaceous behavior nature.
Thermodynamically, a multivalent inhibitor is most effective when the length of the flexible tether between the ligand is similar to the size of the space between the binding sites because this similarity maximizes the effective concentration near binding sites without creating steric obstruction or decreasing conformational entropy. Jiang et al. [30] determined the optimal linker length of RF-C11 by synthesizing RF-Cn series (where n indicates the linker length between the ligand and the core Lys), along with their homovalent controls RR-Cn and FF-Cn and their negative structural controls GV-Cn bearing the stabilizing amino acids Gly and Val [30, 100]. The results of in vitro degradation assays and in silico docking computational studies for binding energy showed that, compared with other RF-Cn compounds, RF-C5 compounds most efficiently inhibited N-end rule degradation with the lowest binding energy when complexed with UBR proteins (Figure 3C) [30]. In addition, inhibitory efficacy by the heterovalent RF-C5 was much higher than that by a combination of two homovalent compounds, RR-C5 and FF-C5, which further demonstrated the kinetic advantage of multivalent interactions. These prototype heterovalent inhibitors could be further optimized through the structural information of UBR proteins, structure-activity relationship (SAR) studies, and animal models of the N-end rule pathway. In vivo applicable heterovalent inhibitors with optimized linker sizes and relatively more potent N-terminal ligands could prove to be useful not only for probing substrate-UBR protein interactions but also for identifying the novel physiological functions of the Arg/N-end rule pathway.
Amino acid derivatives that act as ligands to N-recognins
The structure of the UBR box suggests that its main recognition groups for type 1 destabilizing residues not only require a free α-amino group and a basic side chain on the N-terminal amino acid residue but also parts of the penultimate N-terminal amino acids [46, 47]. The functions of the N-domain appears to be inhibited by many synthetic monomeric compounds containing essential components for UBR box recognition, such as L-conformation, protonated α-amine groups, and hydrophobic side chains (Figures 2B and 2C). For example, the amino acid derivative leucine methyl ester was reported to inhibit type 2 N-degron (Tyr-βgal) degradation in vivo [84]. More recently, a systemic approach using X-nsP4-based N-end rule model substrates and in vitro biochemical assays identified various Phe-derived monomeric molecules that significantly delay the degradation of both type 1 and type 2 N-end rule model substrates simultaneously [85]. These substrates include simple sympathomimetic amines, such as amphetamine and PCA (Figure 3D) [30, 31]. PCA, a serotonin releaser, has been used as an anti-stimulant drug in the past; however, its use was discontinued when neurotoxicity was identified in several animal studies [101, 102].
Chemical compounds with phenylisopropylamine backbones similar to that of PCA but lacking the chlorine at para-position failed to inhibit N-end rule substrate degradation in vitro [85]. This observation suggests essential pharmacophores of monomeric N-end rule inhibitors for further structure-activity relationship studies. Interestingly, an in silico computational analysis of PCA docking on N-recognins revealed that PCA has strong and specific interactions with both the UBR box and N-domain (binding affinities in kcal/mol: -8.03 (Arg-Ala), -6.01 (amphetamine), and -7.17 (PCA) with the UBR2 box; 5.18 (Phe-Ala), -4.92 (amphetamine), and -5.60 (PCA) with the ClpS box) (unpublished). These results suggest that PCA is a unique small-molecule inhibitor targeting both type 1 and type 2 Arg/N-end rule pathway, raising a possibility that the UBR box and ClpS box descended from a common ancestry bacterial N-recognin. The crystallographic structure of the mammalian N-domain will help verify this hypothesis.
PCA appears to pass through the blood-brain barrier, efficiently inhibiting the degradation of Arg/N-end rule substrates in the mouse brain [31]. PCA treatment stabilized endogenous RGS4 in the frontal cortex and hippocampus, which are known to express high levels of RGS4 mRNA in the brain [103]. Consistent with this observation, the activation of GPCR downstream effectors and target gene expression were also significantly impaired. These effects are similar to the genetic inhibition of the Arg/N-end rule pathway [31, 57]. Therefore, PCA has potential for use in determining the underlying regulatory circuits of the N-end rule pathway and identifying hitherto unknown in vivo N-end rule substrates. In addition, given the recent reports that RGS4 levels were reduced in various pathological states, such as breast cancer metastasis [104, 105], PCA-type compounds may have of a therapeutic value for conditions associated with GPCR hyperactivation.
The scope of PCA-mediated N-end rule inhibition is not limited to the regulation of GPCR signaling. A study using PCA found that the degradation of pathologic C-terminal fragments of TDP43 (Arg-TDP25) is mediated by N-end rule-dependent proteolysis [31]. PCA and its derivatives are expected to show highly efficient systemic delivery, especially to the brain. Therefore, PCA is likely to be a useful tool for understanding the neuroprotective roles of the N-end rule pathway in the brain. Furthermore, UBR-box proteins have been identified to interact directly with the proteasomes [106], which may contribute to the rapid turnover of N-end rule substrates and also suggests that the components of N-end rule-dependent post-translational modifications and ubiquitination are mechanistically coupled with certain proteasome subunits and that the degradation of N-end rule substrates may commence even at the protein translation level.
Recently, the Arg/N-end rule pathway was also determined to be involved in the regulation of non-ER stress responses such as innate immune responses to cytosolic double-stranded DNA and autophagy-mediated quality control of misfolded proteins [27]. Thus, the use of PCA could extend the biological scope of the N-end rule pathway to the pathogenesis of more diverse human diseases. However, because amphetamines also act as ligands to various neurotransmitter receptors [107], more specific N-end rule inhibitors without mediating neurostimulant effects in vivo are desirable.
Inhibitors of the upstream components of the N-end rule pathway
Constitutional deletion of mouse ATE1 is lethal at the embryonic stages due to defective cardiac development and angiogenic remodeling [57]; however, its postnatal deletion using the Cre/loxP-mediated recombination produces strikingly broad phenotypes ranging from growth retardation and defective spermatogenesis to neurological deformities and metabolic/behavioral abnormalities (Table 1) [108]. These outcomes indicate that even a single component of the N-end rule pathway may be involved in a number of regulatory circuits, perhaps due to cooperation with various upstream regulators of the pathway, and that targeting upstream components of the N-end rule pathway could be a promising strategy to treat diseases caused by abnormally enhanced N-end rule activity.
Compared with N-recognins, small-molecule ligands for upstream components of the N-end rule pathway remain relatively unexplored. A recent screening of chemical libraries discovered two inhibitors of ATE1 R-transferases, tannic acid and merbromin, which delayed in vivo degradation of RGS4 [109]. Pharmacological inhibition of R-transferases using these compounds partially reproduced null phenotypes of ATE1-deficient cells and embryos in angiogenesis, actin cytoskeleton, cell leading edge, and cell motility [109]. Despite the observed inhibitory effects, it remains unknown how these compounds inhibit R-transferase activities as the structures of ATE1-encoded R-transferases are yet to be determined.
MetAPs are metalloproteases that cotranslationally generate N-terminal Ala, Val, Ser, Thr, or Cys, which, in turn, can induce N-end rule degradation following N-terminal acetylation or arginylation (Figure 1B). Fumagillin, an antibiotic isolated from a fungus Aspergillus fumigatus fresenius, binds to and irreversibly inactivates MetAP-2 through covalent modification [110]. In mid-1980s, Judah Folkman and his colleagues accidentally found that the proliferation of endothelial cells was inhibited without causing apoptosis when the cells were contaminated with the same fungus. This antiangiogenic compound was later determined to be fumagillin [111]. A series of synthetic fumagillin analogs, such as CKD-732, TNP-470 and PPI-2458, were shown to be more potent than fumagillin and entered clinical trials for the treatment of different types of tumors [110]. Recently, the Arg/N-end rule pathway was reported to have an antiapoptotic function via destabilizing various proapoptotic protein fragments [54]. This is in agreement with other previous findings such as the phenotypes of UBR2−/− mice and JBS patients may originate from excessive apoptosis during developmental stages. Therefore, N-end rule inhibitors in combination with fumagillin may have a therapeutic potential by facilitating apoptosis in tumor cells.
Concluding Remarks
In this review, we describe how single amino acids at the protein N-termini regulate the half-lives of cellular proteins through the UPS or autophagy, and we discuss ongoing efforts to exploit the N-recognins of this proteolytic system as potential therapeutic targets. In addition to N-recognins, N-end rule degradation may be regulated in pathophysiological settings by targeting the upstream components such as caspases, calpains, separases, N-terminal amidases/acetylases, MetAPs, and ATE1 R-transferases. The newly discovered Ac/N-end rule pathway in mammals expands the functional scope of the eukaryotic N-end rule pathway, and the elucidation of the complex enzymatic communications among different N-terminal modifications has recently commenced. Despite these advances, many important questions remain to be answered (Outstanding Questions Box). As additional substrates and functions of the N-end rule pathway are likely to be unveiled in coming years, the pharmacological modulation strategies discussed herein can be used to address these unresolved questions.
Box 1. Outstanding Questions.
Why are certain proteins with N-terminal destabilizing residues extremely short-lived, whereas others are stable? What determines the fate of N-end rule substrates between the arginylation and acetylation branches?
What are the global roles of short peptides (or even single amino acids) with N-terminal destabilizing residue(s)? Are they related with the cellular response mechanism against various stresses such as oxidative stress and proteotoxic stress?
What is the true N-proteome in the cell? What are the primary biological features of the Arg/N-proteome and the Ac/N-proteome? How is its homeostasis regulated, e.g. by a set of endopeptidases?
How can an N-end rule inhibitor achieve its high specificity and selectivity towards certain N-end rule substrates? From a therapeutic point of view, what is currently the most feasible target disease for the N-end rule pathway? Is the neuroprotective mechanism of the Arg/N-end rule pathway impaired in various neurodegenerative diseases?
Various dipeptides bearing simple but distinct N-degrons have been used in numerous studies for biochemical and physiological dissection of this pathway, contributing to our current understanding of the Arg/N-end rule pathway. As a result of their cooperative binding modes at multiple sites on the UBR proteins, heterovalent inhibitors were shown to exhibit more potent inhibitory effects as compared with monovalent dipeptides. RF-C-type heterovalent compounds are also the first non-cytotoxic and cell-permeable inhibitors of N-end rule-mediated proteolysis in mammalian cells. In conjunction with advanced structural and biochemical understanding of N-recognins, the newly identified N-end rule inhibitor PCA has been shown to alter the fates of endogenous N-end rule substrates in the mouse brain. Thus, PCA is not only a tool to dissect the pathway but also has potential pharmacological and biotechnological applications.
Although nearly three decades have passed since the initial discovery of the N-end rule pathway, this proteolytic system continues to provide new biological and physiological insights. Because substrates that newly expose N-degrons exist only briefly prior to rapid degradation, it has been challenging to identify such short-lived protein species and to characterize their functions through standard methodologies. Just as the development of the proteasome inhibitor MG132 in the 1990's fundamentally transformed the methodological approaches of protein metabolism researchers, potent and specific N-end rule inhibitors may have potential for transforming the paradigm of protein metabolism research. More selective targeting of N-end rule components will provide more specific therapeutic windows with increased potency and reduced side effects, while broad-spectrum inhibitors may result in greater therapeutic effects, mainly as anti-cancer agents such as bortezomib and sunitinib [112, 113]. As dipeptides, heterovalent inhibitors, and amphetamines can bind a broad range of N-recognins, more specific inhibitors could be developed by optimizing these pan-N-end rule inhibitors based on the structures of N-recognins. Targeting specific N-recognins along with their upstream modification enzymes, such as fumagillin, may modulate substrate degradation more precisely, providing improved therapeutic benefits.
Potent inhibitors may also enable global screening for unknown N-end rule substrates. Because the N-end rule pathway is relatively insulated from other cellular regulatory systems, its inhibitors may be specific and clinically useful. Finally, inhibitors of the Ac/N-degron TEB4 might be worth developing for their effects on many regulatory proteins, especially relatively long-lived proteins. Recently, many nonproteolytic functions of the N-end rule pathway have been reported, such as the modulation of p62 activity in autophagy. Because the N-end rule pathway and autophagy appear to be closely linked, it would be interesting to investigate how a regulation method in one system affects the degradation flux of the other system.
Trends box.
The N-end rule pathway is a major branch of the Ub-dependent proteolytic system.
The substrates of the N-end rule are involved in various pathophysiological processes.
Inhibitors targeting UBR proteins have been developed via rational design and HTS.
N-end rule inhibitors may have therapeutic potentials in many human diseases.
Acknowledgments
This study was supported by grants from the Disease Oriented Translational Research Program (HI14C0202 to M.J.L.) and the Korea-UK R&D Collaboration Research Program (HI14C2036 to M.J.L.) of the Korea Health Industry Development Institute, and the Basic Science Research Program (2013R1A1A2059793 to J.H.L.) of the National Research Foundation of Korea. YTK was supported by the NIH (HL083365), the Seoul National University (SNU) Nobel Laureates Invitation Program, and SNU Hospital.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Choi AM, et al. Autophagy in human health and disease. N Engl J Med. 2013;368:651–662. doi: 10.1056/NEJMra1205406. [DOI] [PubMed] [Google Scholar]
- 2.Schwartz AL, Ciechanover A. The ubiquitin-proteasome pathway and pathogenesis of human diseases. Annu Rev Med. 1999;50:57–74. doi: 10.1146/annurev.med.50.1.57. [DOI] [PubMed] [Google Scholar]
- 3.Lee MJ, et al. Tau degradation: the ubiquitin-proteasome system versus the autophagy-lysosome system. Prog Neurobiol. 2013;105:49–59. doi: 10.1016/j.pneurobio.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 4.Glickman MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev. 2002;82:373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- 5.Kamura T, et al. Rbx1, a component of the VHL tumor suppressor complex and SCF ubiquitin ligase. Science. 1999;284:657–661. doi: 10.1126/science.284.5414.657. [DOI] [PubMed] [Google Scholar]
- 6.Tyers M, Jorgensen P. Proteolysis and the cell cycle: with this RING I do thee destroy. Curr Opin Genet Dev. 2000;10:54–64. doi: 10.1016/s0959-437x(99)00049-0. [DOI] [PubMed] [Google Scholar]
- 7.Ravid T, Hochstrasser M. Diversity of degradation signals in the ubiquitin-proteasome system. Nat Rev Mol Cell Biol. 2008;9:679–690. doi: 10.1038/nrm2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tasaki T, et al. The N-end rule pathway. Annu Rev Biochem. 2012;81:261–289. doi: 10.1146/annurev-biochem-051710-093308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dougan DA, et al. The N-end rule pathway: from recognition by N-recognins, to destruction by AAA+proteases. Biochim Biophys Acta. 2012;1823:83–91. doi: 10.1016/j.bbamcr.2011.07.002. [DOI] [PubMed] [Google Scholar]
- 10.Bachmair A, et al. In vivo half-life of a protein is a function of its amino-terminal residue. Science. 1986;234:179–186. doi: 10.1126/science.3018930. [DOI] [PubMed] [Google Scholar]
- 11.Sriram SM, et al. The N-end rule pathway: emerging functions and molecular principles of substrate recognition. Nat Rev Mol Cell Biol. 2011;12:735–747. doi: 10.1038/nrm3217. [DOI] [PubMed] [Google Scholar]
- 12.Kim JS, et al. Resin-assisted enrichment of N-terminal peptides for characterizing proteolytic processing. Anal Chem. 2013;85:6826–6832. doi: 10.1021/ac401000q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vogtle FN, et al. Global analysis of the mitochondrial N-proteome identifies a processing peptidase critical for protein stability. Cell. 2009;139:428–439. doi: 10.1016/j.cell.2009.07.045. [DOI] [PubMed] [Google Scholar]
- 14.An JY, et al. UBR2 of the N-end rule pathway is required for chromosome stability via histone ubiquitylation in spermatocytes and somatic cells. PLoS One. 2012;7:e37414. doi: 10.1371/journal.pone.0037414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.An JY, et al. UBR2 mediates transcriptional silencing during spermatogenesis via histone ubiquitination. Proc Natl Acad Sci U S A. 2010;107:1912–1917. doi: 10.1073/pnas.0910267107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu RG, et al. The N-end rule pathway as a nitric oxide sensor controlling the levels of multiple regulators. Nature. 2005;437:981–986. doi: 10.1038/nature04027. [DOI] [PubMed] [Google Scholar]
- 17.Hu RG, et al. The N-end rule pathway is a sensor of heme. Proc Natl Acad Sci U S A. 2008;105:76–81. doi: 10.1073/pnas.0710568105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee MJ, et al. Characterization of arginylation branch of N-end rule pathway in G-protein-mediated proliferation and signaling of cardiomyocytes. J Biol Chem. 2012;287:24043–24052. doi: 10.1074/jbc.M112.364117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee MJ, et al. RGS4 and RGS5 are in vivo substrates of the N-end rule pathway. Proc Natl Acad Sci U S A. 2005;102:15030–15035. doi: 10.1073/pnas.0507533102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Solomon V, et al. Rates of ubiquitin conjugation increase when muscles atrophy, largely through activation of the N-end rule pathway. Proc Natl Acad Sci U S A. 1998;95:12602–12607. doi: 10.1073/pnas.95.21.12602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Solomon V, et al. The N-end rule pathway catalyzes a major fraction of the protein degradation in skeletal muscle. J Biol Chem. 1998;273:25216–25222. doi: 10.1074/jbc.273.39.25216. [DOI] [PubMed] [Google Scholar]
- 22.Lecker SH, et al. Ubiquitin conjugation by the N-end rule pathway and mRNAs for its components increase in muscles of diabetic rats. J Clin Invest. 1999;104:1411–1420. doi: 10.1172/JCI7300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang G, et al. Signaling mechanism of tumor cell-induced up-regulation of E3 ubiquitin ligase UBR2. FASEB J. 2013;27:2893–2901. doi: 10.1096/fj.12-222711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.An JY, et al. Impaired neurogenesis and cardiovascular development in mice lacking the E3 ubiquitin ligases UBR1 and UBR2 of the N-end rule pathway. Proc Natl Acad Sci U S A. 2006;103:6212–6217. doi: 10.1073/pnas.0601700103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brower CS, et al. Neurodegeneration-associated protein fragments as short-lived substrates of the N-end rule pathway. Mol Cell. 2013;50:161–171. doi: 10.1016/j.molcel.2013.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park SE, et al. Control of mammalian G protein signaling by N-terminal acetylation and the N-end rule pathway. Science. 2015;347:1249–1252. doi: 10.1126/science.aaa3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cha-Molstad H, et al. Amino-terminal arginylation targets endoplasmic reticulum chaperone BiP for autophagy through p62 binding. Nat Cell Biol. 2015;17:917–929. doi: 10.1038/ncb3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Varshavsky A. The N-end rule pathway and regulation by proteolysis. Protein Sci. 2011;20:1298–1345. doi: 10.1002/pro.666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Varshavsky A. The ubiquitin system, an immense realm. Annu Rev Biochem. 2012;81:167–176. doi: 10.1146/annurev-biochem-051910-094049. [DOI] [PubMed] [Google Scholar]
- 30.Jiang Y, et al. Characterization of mammalian N-degrons and development of heterovalent inhibitors of the N-end rule pathway. Chem Sci. 2013;4:3339–3346. [Google Scholar]
- 31.Jiang Y, et al. A neurostimulant para-chloroamphetamine inhibits the arginylation branch of the N-end rule pathway. Sci Rep. 2014;4:6344. doi: 10.1038/srep06344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Varshavsky A. The N-end rule: functions, mysteries, uses. Proc Natl Acad Sci U S A. 1996;93:12142–12149. doi: 10.1073/pnas.93.22.12142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Prakash S, et al. An unstructured initiation site is required for efficient proteasome-mediated degradation. Nat Struct Mol Biol. 2004;11:830–837. doi: 10.1038/nsmb814. [DOI] [PubMed] [Google Scholar]
- 34.Varshavsky A. Naming a targeting signal. Cell. 1991;64:13–15. doi: 10.1016/0092-8674(91)90202-a. [DOI] [PubMed] [Google Scholar]
- 35.Hwang CS, et al. N-terminal acetylation of cellular proteins creates specific degradation signals. Science. 2010;327:973–977. doi: 10.1126/science.1183147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shemorry A, et al. Control of protein quality and stoichiometries by N-terminal acetylation and the N-end rule pathway. Mol Cell. 2013;50:540–551. doi: 10.1016/j.molcel.2013.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Crawford ED, Wells JA. Caspase substrates and cellular remodeling. Annu Rev Biochem. 2011;80:1055–1087. doi: 10.1146/annurev-biochem-061809-121639. [DOI] [PubMed] [Google Scholar]
- 38.Piatkov KI, et al. Calpain-generated natural protein fragments as short-lived substrates of the N-end rule pathway. Proc Natl Acad Sci U S A. 2014;111:E817–826. doi: 10.1073/pnas.1401639111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rao H, et al. Degradation of a cohesin subunit by the N-end rule pathway is essential for chromosome stability. Nature. 2001;410:955–959. doi: 10.1038/35073627. [DOI] [PubMed] [Google Scholar]
- 40.Arnesen T, et al. Proteomics analyses reveal the evolutionary conservation and divergence of N-terminal acetyltransferases from yeast and humans. Proc Natl Acad Sci U S A. 2009;106:8157–8162. doi: 10.1073/pnas.0901931106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kwon YT, et al. The mouse and human genes encoding the recognition component of the N-end rule pathway. Proc Natl Acad Sci U S A. 1998;95:7898–7903. doi: 10.1073/pnas.95.14.7898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kwon YT, et al. Construction and analysis of mouse strains lacking the ubiquitin ligase UBR1 (E3alpha) of the N-end rule pathway. Mol Cell Biol. 2001;21:8007–8021. doi: 10.1128/MCB.21.23.8007-8021.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kwon YT, et al. Female lethality and apoptosis of spermatocytes in mice lacking the UBR2 ubiquitin ligase of the N-end rule pathway. Mol Cell Biol. 2003;23:8255–8271. doi: 10.1128/MCB.23.22.8255-8271.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tasaki T, et al. Biochemical and genetic studies of UBR3, a ubiquitin ligase with a function in olfactory and other sensory systems. J Biol Chem. 2007;282:18510–18520. doi: 10.1074/jbc.M701894200. [DOI] [PubMed] [Google Scholar]
- 45.Tasaki T, Kwon YT. The mammalian N-end rule pathway: new insights into its components and physiological roles. Trends Biochem Sci. 2007;32:520–528. doi: 10.1016/j.tibs.2007.08.010. [DOI] [PubMed] [Google Scholar]
- 46.Choi WS, et al. Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases. Nat Struct Mol Biol. 2010;17:1175–1181. doi: 10.1038/nsmb.1907. [DOI] [PubMed] [Google Scholar]
- 47.Matta-Camacho E, et al. Structural basis of substrate recognition and specificity in the N-end rule pathway. Nat Struct Mol Biol. 2010;17:1182–1187. doi: 10.1038/nsmb.1894. [DOI] [PubMed] [Google Scholar]
- 48.Erbse A, et al. ClpS is an essential component of the N-end rule pathway in Escherichia coli. Nature. 2006;439:753–756. doi: 10.1038/nature04412. [DOI] [PubMed] [Google Scholar]
- 49.Roman-Hernandez G, et al. Molecular basis of substrate selection by the N-end rule adaptor protein ClpS. Proc Natl Acad Sci U S A. 2009;106:8888–8893. doi: 10.1073/pnas.0903614106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xie Y, Varshavsky A. The E2-E3 interaction in the N-end rule pathway: the RING-H2 finger of E3 is required for the synthesis of multiubiquitin chain. Embo J. 1999;18:6832–6844. doi: 10.1093/emboj/18.23.6832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee PC, et al. Alternative ubiquitin activation/conjugation cascades interact with N-end rule ubiquitin ligases to control degradation of RGS proteins. Mol Cell. 2011;43:392–405. doi: 10.1016/j.molcel.2011.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tasaki T, et al. A family of mammalian E3 ubiquitin ligases that contain the UBR box motif and recognize N-degrons. Mol Cell Biol. 2005;25:7120–7136. doi: 10.1128/MCB.25.16.7120-7136.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schnupf P, et al. Listeriolysin O secreted by Listeria monocytogenes into the host cell cytosol is degraded by the N-end rule pathway. Infect Immun. 2007;75:5135–5147. doi: 10.1128/IAI.00164-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Piatkov KI, et al. The N-end rule pathway counteracts cell death by destroying proapoptotic protein fragments. Proc Natl Acad Sci U S A. 2012;109:E1839–1847. doi: 10.1073/pnas.1207786109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yamano K, Youle RJ. PINK1 is degraded through the N-end rule pathway. Autophagy. 2013;9:1758–1769. doi: 10.4161/auto.24633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zenker M, et al. Deficiency of UBR1, a ubiquitin ligase of the N-end rule pathway, causes pancreatic dysfunction, malformations and mental retardation (Johanson-Blizzard syndrome) Nat Genet. 2005;37:1345–1350. doi: 10.1038/ng1681. [DOI] [PubMed] [Google Scholar]
- 57.Kwon YT, et al. An essential role of N-terminal arginylation in cardiovascular development. Science. 2002;297:96–99. doi: 10.1126/science.1069531. [DOI] [PubMed] [Google Scholar]
- 58.Kwon YT, et al. Bivalent inhibitor of the N-end rule pathway. J Biol Chem. 1999;274:18135–18139. doi: 10.1074/jbc.274.25.18135. [DOI] [PubMed] [Google Scholar]
- 59.De Vries L, et al. The regulator of G protein signaling family. Annu Rev Pharmacol Toxicol. 2000;40:235–271. doi: 10.1146/annurev.pharmtox.40.1.235. [DOI] [PubMed] [Google Scholar]
- 60.Tirziu D, et al. Myocardial hypertrophy in the absence of external stimuli is induced by angiogenesis in mice. J Clin Invest. 2007;117:3188–3197. doi: 10.1172/JCI32024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang J, et al. Genetic variations of regulator of G-protein signaling 2 in hypertensive patients and in the general population. J Hypertens. 2005;23:1497–1505. doi: 10.1097/01.hjh.0000174606.41651.ae. [DOI] [PubMed] [Google Scholar]
- 62.Uttara B, et al. Oxidative stress and neurodegenerative diseases: a review of upstream and downstream antioxidant therapeutic options. Curr Neuropharmacol. 2009;7:65–74. doi: 10.2174/157015909787602823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stadtman ER. Protein oxidation and aging. Free Radic Res. 2006;40:1250–1258. doi: 10.1080/10715760600918142. [DOI] [PubMed] [Google Scholar]
- 64.Ahmed EK, et al. Protein oxidative modifications and replicative senescence of WI-38 human embryonic fibroblasts. Ann N Y Acad Sci. 2007;1119:88–96. doi: 10.1196/annals.1404.020. [DOI] [PubMed] [Google Scholar]
- 65.David DC, et al. Proteasomal degradation of tau protein. J Neurochem. 2002;83:176–185. doi: 10.1046/j.1471-4159.2002.01137.x. [DOI] [PubMed] [Google Scholar]
- 66.Petrucelli L, et al. CHIP and Hsp70 regulate tau ubiquitination, degradation and aggregation. Hum Mol Genet. 2004;13:703–714. doi: 10.1093/hmg/ddh083. [DOI] [PubMed] [Google Scholar]
- 67.Shimura H, et al. CHIP-Hsc70 complex ubiquitinates phosphorylated tau and enhances cell survival. J Biol Chem. 2004;279:4869–4876. doi: 10.1074/jbc.M305838200. [DOI] [PubMed] [Google Scholar]
- 68.Shin Y, et al. The co-chaperone carboxyl terminus of Hsp70-interacting protein (CHIP) mediates alpha-synuclein degradation decisions between proteasomal and lysosomal pathways. J Biol Chem. 2005;280:23727–23734. doi: 10.1074/jbc.M503326200. [DOI] [PubMed] [Google Scholar]
- 69.Liani E, et al. Ubiquitylation of synphilin-1 and alpha-synuclein by SIAH and its presence in cellular inclusions and Lewy bodies imply a role in Parkinson's disease. Proc Natl Acad Sci U S A. 2004;101:5500–5505. doi: 10.1073/pnas.0401081101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lagier-Tourenne C, et al. TDP-43 and FUS/TLS: emerging roles in RNA processing and neurodegeneration. Hum Mol Genet. 2010;19:R46–64. doi: 10.1093/hmg/ddq137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lee EB, et al. Gains or losses: molecular mechanisms of TDP43-mediated neurodegeneration. Nat Rev Neurosci. 2012;13:38–50. doi: 10.1038/nrn3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Igaz LM, et al. Expression of TDP-43 C-terminal Fragments in Vitro Recapitulates Pathological Features of TDP-43 Proteinopathies. J Biol Chem. 2009;284:8516–8524. doi: 10.1074/jbc.M809462200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pesiridis GS, et al. A “two-hit” hypothesis for inclusion formation by carboxyl-terminal fragments of TDP-43 protein linked to RNA depletion and impaired microtubule-dependent transport. J Biol Chem. 2011;286:18845–18855. doi: 10.1074/jbc.M111.231118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ding WX, et al. Linking of autophagy to ubiquitin-proteasome system is important for the regulation of endoplasmic reticulum stress and cell viability. Am J Pathol. 2007;171:513–524. doi: 10.2353/ajpath.2007.070188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhu K, et al. Proteasome inhibitors activate autophagy as a cytoprotective response in human prostate cancer cells. Oncogene. 2010;29:451–462. doi: 10.1038/onc.2009.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Korolchuk VI, et al. Autophagy inhibition compromises degradation of ubiquitin-proteasome pathway substrates. Mol Cell. 2009;33:517–527. doi: 10.1016/j.molcel.2009.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tasaki T, et al. UBR box N-recognin-4 (UBR4), an N-recognin of the N-end rule pathway, and its role in yolk sac vascular development and autophagy. Proc Natl Acad Sci U S A. 2013;110:3800–3805. doi: 10.1073/pnas.1217358110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Prasad R, et al. A nucleus-based quality control mechanism for cytosolic proteins. Mol Biol Cell. 2010;21:2117–2127. doi: 10.1091/mbc.E10-02-0111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Heck JW, et al. Cytoplasmic protein quality control degradation mediated by parallel actions of the E3 ubiquitin ligases Ubr1 and San1. Proc Natl Acad Sci U S A. 2010;107:1106–1111. doi: 10.1073/pnas.0910591107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hershko A, et al. The protein substrate binding site of the ubiquitin-protein ligase system. J Biol Chem. 1986;261:11992–11999. [PubMed] [Google Scholar]
- 81.Reiss Y, et al. Specificity of binding of NH2-terminal residue of proteins to ubiquitin-protein ligase. Use of amino acid derivatives to characterize specific binding sites. J Biol Chem. 1988;263:2693–2698. [PubMed] [Google Scholar]
- 82.Bartel B, et al. The recognition component of the N-end rule pathway. Embo J. 1990;9:3179–3189. doi: 10.1002/j.1460-2075.1990.tb07516.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hershko A, et al. Role of the alpha-amino group of protein in ubiquitin-mediated protein breakdown. Proc Natl Acad Sci U S A. 1984;81:7021–7025. doi: 10.1073/pnas.81.22.7021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Baker RT, Varshavsky A. Inhibition of the N-end rule pathway in living cells. Proc Natl Acad Sci U S A. 1991;88:1090–1094. doi: 10.1073/pnas.88.4.1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sriram S, et al. Development and characterization of monomeric N-end rule inhibitors through in vitro model substrates. J Med Chem. 2013;56:2540–2546. doi: 10.1021/jm400046q. [DOI] [PubMed] [Google Scholar]
- 86.Gonda DK, et al. Universality and structure of the N-end rule. J Biol Chem. 1989;264:16700–16712. [PubMed] [Google Scholar]
- 87.Silk DB, et al. Protein digestion and amino acid and peptide absorption. Proc Nutr Soc. 1985;44:63–72. doi: 10.1079/pns19850011. [DOI] [PubMed] [Google Scholar]
- 88.Newey H, Smyth DH. Intracellular hydrolysis of dipeptides during intestinal absorption. J Physiol. 1960;152:367–380. doi: 10.1113/jphysiol.1960.sp006493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Craft IL, et al. Absorption and malabsorption of glycine and glycine peptides in man. Gut. 1968;9:425–437. doi: 10.1136/gut.9.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Alagramam K, et al. A recognition component of the ubiquitin system is required for peptide transport in Saccharomyces cerevisiae. Mol Microbiol. 1995;15:225–234. doi: 10.1111/j.1365-2958.1995.tb02237.x. [DOI] [PubMed] [Google Scholar]
- 91.Byrd C, et al. The N-end rule pathway controls the import of peptides through degradation of a transcriptional repressor. EMBO J. 1998;17:269–277. doi: 10.1093/emboj/17.1.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Turner GC, et al. Peptides accelerate their uptake by activating a ubiquitin-dependent proteolytic pathway. Nature. 2000;405:579–583. doi: 10.1038/35014629. [DOI] [PubMed] [Google Scholar]
- 93.Xia Z, et al. Amino acids induce peptide uptake via accelerated degradation of CUP9, the transcriptional repressor of the PTR2 peptide transporter. J Biol Chem. 2008;283:28958–28968. doi: 10.1074/jbc.M803980200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Nillegoda NB, et al. Ubr1 and Ubr2 function in a quality control pathway for degradation of unfolded cytosolic proteins. Mol Biol Cell. 2010;21:2102–2116. doi: 10.1091/mbc.E10-02-0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Du F, et al. Pairs of dipeptides synergistically activate the binding of substrate by ubiquitin ligase through dissociation of its autoinhibitory domain. Proc Natl Acad Sci U S A. 2002;99:14110–14115. doi: 10.1073/pnas.172527399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hondermarck H, et al. Dipeptide inhibitors of ubiquitin-mediated protein turnover prevent growth factor-induced neurite outgrowth in rat pheochromocytoma PC12 cells. Biochem Biophys Res Commun. 1992;189:280–288. doi: 10.1016/0006-291x(92)91555-5. [DOI] [PubMed] [Google Scholar]
- 97.Huskens J. Multivalent interactions at interfaces. Curr Opin Chem Biol. 2006;10:537–543. doi: 10.1016/j.cbpa.2006.09.007. [DOI] [PubMed] [Google Scholar]
- 98.Sabatini DM, et al. RAFT1: a mammalian protein that binds to FKBP12 in a rapamycin-dependent fashion and is homologous to yeast TORs. Cell. 1994;78:35–43. doi: 10.1016/0092-8674(94)90570-3. [DOI] [PubMed] [Google Scholar]
- 99.Banaszynski LA, et al. Characterization of the FKBP.rapamycin.FRB ternary complex. J Am Chem Soc. 2005;127:4715–4721. doi: 10.1021/ja043277y. [DOI] [PubMed] [Google Scholar]
- 100.Lee MJ, et al. Synthetic heterovalent inhibitors targeting recognition E3 components of the N-end rule pathway. Proc Natl Acad Sci U S A. 2008;105:100–105. doi: 10.1073/pnas.0708465105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lassen JB. The effect of p-chloroamphetamine on motility in rats after inhibition of monoamine synthesis, storage, uptake and receptor interaction. Psychopharmacologia. 1974;34:243–254. doi: 10.1007/BF00421965. [DOI] [PubMed] [Google Scholar]
- 102.Harvey JA, et al. P-Chloramphetamine: Selective neurotoxic action in brain. Science. 1975;187:841–843. doi: 10.1126/science.47181. [DOI] [PubMed] [Google Scholar]
- 103.Erdely HA, et al. Regional expression of RGS4 mRNA in human brain. Eur J Neurosci. 2004;19:3125–3128. doi: 10.1111/j.0953-816X.2004.03364.x. [DOI] [PubMed] [Google Scholar]
- 104.Sjogren B, Neubig RR. Thinking outside of the “RGS box”: new approaches to therapeutic targeting of regulators of G protein signaling. Mol Pharmacol. 2010;78:550–557. doi: 10.1124/mol.110.065219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Xie Y, et al. Breast cancer migration and invasion depend on proteasome degradation of regulator of G-protein signaling 4. Cancer Res. 2009;69:5743–5751. doi: 10.1158/0008-5472.CAN-08-3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Besche HC, et al. Isolation of mammalian 26S proteasomes and p97/VCP complexes using the ubiquitin-like domain from HHR23B reveals novel proteasome-associated proteins. Biochemistry. 2009;48:2538–2549. doi: 10.1021/bi802198q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Battaglia G, De Souza EB. Pharmacologic profile of amphetamine derivatives at various brain recognition sites: selective effects on serotonergic systems. NIDA Res Monogr. 1989;94:240–258. [PubMed] [Google Scholar]
- 108.Brower CS, Varshavsky A. Ablation of arginylation in the mouse N-end rule pathway: loss of fat, higher metabolic rate, damaged spermatogenesis, and neurological perturbations. PLoS One. 2009;4:e7757. doi: 10.1371/journal.pone.0007757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Saha S, et al. Small molecule inhibitors of arginyltransferase regulate arginylation-dependent protein degradation, cell motility, and angiogenesis. Biochem Pharmacol. 2012;83:866–873. doi: 10.1016/j.bcp.2012.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Yin SQ, et al. The development of MetAP-2 inhibitors in cancer treatment. Curr Med Chem. 2012;19:1021–1035. doi: 10.2174/092986712799320709. [DOI] [PubMed] [Google Scholar]
- 111.Ingber D, et al. Synthetic analogues of fumagillin that inhibit angiogenesis and suppress tumour growth. Nature. 1990;348:555–557. doi: 10.1038/348555a0. [DOI] [PubMed] [Google Scholar]
- 112.Kisselev AF, et al. Importance of the different proteolytic sites of the proteasome and the efficacy of inhibitors varies with the protein substrate. J Biol Chem. 2006;281:8582–8590. doi: 10.1074/jbc.M509043200. [DOI] [PubMed] [Google Scholar]
- 113.Broekman F, et al. Tyrosine kinase inhibitors: Multi-targeted or single-targeted? World J Clin Oncol. 2011;2:80–93. doi: 10.5306/wjco.v2.i2.80. [DOI] [PMC free article] [PubMed] [Google Scholar]