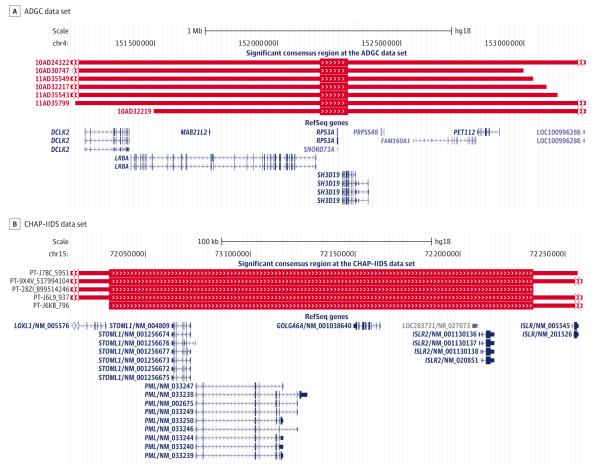
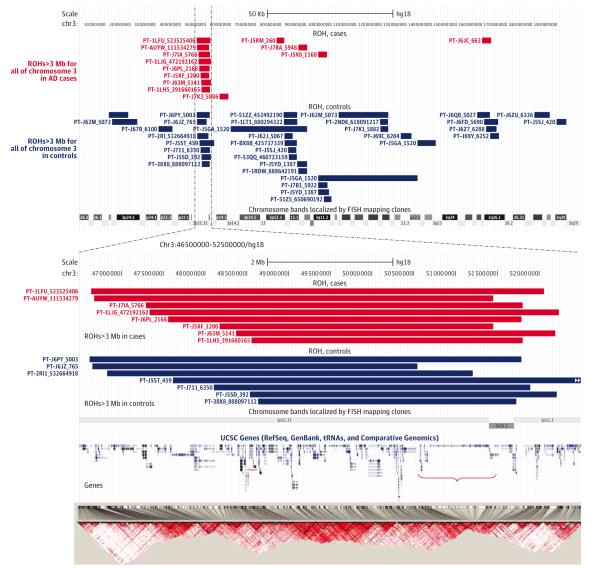
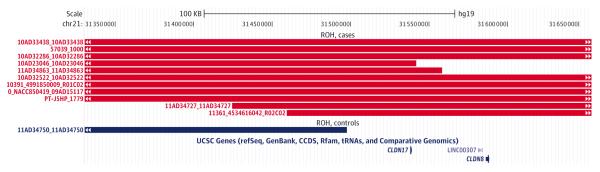
Mahdi Ghani
Mahdi Ghani, MD, PhD
1Tanz Centre for Research in Neurodegenerative Diseases, University of Toronto, Toronto, Ontario, Canada (Ghani, Sato, St. George-Hyslop, Rogaeva); Taub Institute for Research on Alzheimer’s Disease and the Aging Brain, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Cheng, Manly, Lee, Mayeux); Gertrude H. Sergievsky Center, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Vardarajan, Lee, Mayeux); Department of Neurology, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Manly, Mayeux); Department of Medicine (Biomedical Genetics), Boston University, Boston, Massachusetts (Jun, Logue, Baldwin, Farrer); Department of Biostatistics, Boston University, Boston, Massachusetts (Jun, Lunetta, Farrer); Department of Ophthalmology, Boston University, Boston, Massachusetts (Jun, Farrer); The John P. Hussman Institute for Human Genomics, University of Miami, Miami, Florida (Naj, Rajbhandary, Pericak-Vance); Department of Pathology and Laboratory Medicine, University of Pennsylvania Perelman School of Medicine, Philadelphia (Wang, Valladares, Lin, Cantwell, Schellenberg); Department of Medicine, University of Washington, Seattle (Larson, Crane); Group Health Research Institute, Group Health, Seattle, Washington (Larson); Department of Neuroscience, Mayo Clinic, Jacksonville, Florida (Graff-Radford, Ertekin-Taner); Department of Neurology, Mayo Clinic, Jacksonville, Florida (Graff-Radford, Ertekin-Taner); Rush Institute for Healthy Aging, Department of Internal Medicine, Rush University Medical Center, Chicago, Illinois (Evans); Program in Translational Neuropsychiatric Genomics, Department of Neurology, Brigham and Women’s Hospital, Boston, Massachusetts (De Jager); Harvard Medical School, Boston, Massachusetts (De Jager, Raj, Green); Program in Medical and Population Genetics, The Broad Institute, Cambridge, Massachusetts (De Jager); Department of Psychiatry, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Genetics and Genomics Sciences, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Neuroscience, Mount Sinai School of Medicine, New York, New York (Buxbaum); Friedman Brain Institute, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Medical and Molecular Genetics, Indiana University, Indianapolis (Murrell); Division of Genetics, Department of Medicine, Brigham and Women’s Hospital, Boston, Massachusetts (Green); Partners Center for Personalized Genetic Medicine, Brigham and Women’s Hospital, Boston, Massachusetts (Green); Department of Neurological Sciences, Rush University Medical Center, Chicago, Illinois (Barnes, Bennett); Department of Behavioral Sciences, Rush University Medical Center, Chicago, Illinois (Barnes, Foroud); Department of Epidemiology, Johns Hopkins University School of Public Health, Baltimore, Maryland (Fallin); School of Public Health, University of Alabama at Birmingham (Go); SABA University School of Medicine, SABA, Dutch Caribbean (Griffith); Division of Geriatrics, Howard University Hospital, Washington, DC (Obisesan); Department of Human Genetics, University of Pittsburgh, Pittsburgh, Pennsylvania (Kamboh); Alzheimer’s Disease Research Center, University of Pittsburgh, Pittsburgh, Pennsylvania (Kamboh, Lopez, Mayeux); Rush Alzheimer’s Disease Center, Rush University Medical Center, Chicago, Illinois (Bennett); Indiana University Center for Aging Research, Indianapolis (Hendrie); Department of Psychiatry, Indiana University School of Medicine, Indianapolis (Hendrie, Hall); Regenstrief Institute Inc, Indianapolis, Indiana (Hendrie); Hope Center Program on Protein Aggregation and Neurodegeneration, Department of Psychiatry, Washington University School of Medicine, St Louis, Missouri (Goate); Department of Biology, North Carolina A & T University, Greensboro (Byrd); National Alzheimer’s Coordinating Center, Department of Epidemiology, University of Washington, Seattle (Kukull); Vanderbilt Center for Human Genetics Research, Department of Molecular Physiology and Biophysics, Vanderbilt University, Nashville, Tennessee (Haines); Department of Neurology, Boston University, Boston, Massachusetts (Farrer); Department of Epidemiology, Boston University, Boston, Massachusetts (Farrer); Department of Epidemiology, College of Physicians and Surgeons, Columbia University, New York, New York (Lee); Department of Psychiatry, College of Physicians and Surgeons, Columbia University, New York, New York (Mayeux).
1,
Christiane Reitz
Christiane Reitz, MD, PhD
1Tanz Centre for Research in Neurodegenerative Diseases, University of Toronto, Toronto, Ontario, Canada (Ghani, Sato, St. George-Hyslop, Rogaeva); Taub Institute for Research on Alzheimer’s Disease and the Aging Brain, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Cheng, Manly, Lee, Mayeux); Gertrude H. Sergievsky Center, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Vardarajan, Lee, Mayeux); Department of Neurology, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Manly, Mayeux); Department of Medicine (Biomedical Genetics), Boston University, Boston, Massachusetts (Jun, Logue, Baldwin, Farrer); Department of Biostatistics, Boston University, Boston, Massachusetts (Jun, Lunetta, Farrer); Department of Ophthalmology, Boston University, Boston, Massachusetts (Jun, Farrer); The John P. Hussman Institute for Human Genomics, University of Miami, Miami, Florida (Naj, Rajbhandary, Pericak-Vance); Department of Pathology and Laboratory Medicine, University of Pennsylvania Perelman School of Medicine, Philadelphia (Wang, Valladares, Lin, Cantwell, Schellenberg); Department of Medicine, University of Washington, Seattle (Larson, Crane); Group Health Research Institute, Group Health, Seattle, Washington (Larson); Department of Neuroscience, Mayo Clinic, Jacksonville, Florida (Graff-Radford, Ertekin-Taner); Department of Neurology, Mayo Clinic, Jacksonville, Florida (Graff-Radford, Ertekin-Taner); Rush Institute for Healthy Aging, Department of Internal Medicine, Rush University Medical Center, Chicago, Illinois (Evans); Program in Translational Neuropsychiatric Genomics, Department of Neurology, Brigham and Women’s Hospital, Boston, Massachusetts (De Jager); Harvard Medical School, Boston, Massachusetts (De Jager, Raj, Green); Program in Medical and Population Genetics, The Broad Institute, Cambridge, Massachusetts (De Jager); Department of Psychiatry, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Genetics and Genomics Sciences, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Neuroscience, Mount Sinai School of Medicine, New York, New York (Buxbaum); Friedman Brain Institute, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Medical and Molecular Genetics, Indiana University, Indianapolis (Murrell); Division of Genetics, Department of Medicine, Brigham and Women’s Hospital, Boston, Massachusetts (Green); Partners Center for Personalized Genetic Medicine, Brigham and Women’s Hospital, Boston, Massachusetts (Green); Department of Neurological Sciences, Rush University Medical Center, Chicago, Illinois (Barnes, Bennett); Department of Behavioral Sciences, Rush University Medical Center, Chicago, Illinois (Barnes, Foroud); Department of Epidemiology, Johns Hopkins University School of Public Health, Baltimore, Maryland (Fallin); School of Public Health, University of Alabama at Birmingham (Go); SABA University School of Medicine, SABA, Dutch Caribbean (Griffith); Division of Geriatrics, Howard University Hospital, Washington, DC (Obisesan); Department of Human Genetics, University of Pittsburgh, Pittsburgh, Pennsylvania (Kamboh); Alzheimer’s Disease Research Center, University of Pittsburgh, Pittsburgh, Pennsylvania (Kamboh, Lopez, Mayeux); Rush Alzheimer’s Disease Center, Rush University Medical Center, Chicago, Illinois (Bennett); Indiana University Center for Aging Research, Indianapolis (Hendrie); Department of Psychiatry, Indiana University School of Medicine, Indianapolis (Hendrie, Hall); Regenstrief Institute Inc, Indianapolis, Indiana (Hendrie); Hope Center Program on Protein Aggregation and Neurodegeneration, Department of Psychiatry, Washington University School of Medicine, St Louis, Missouri (Goate); Department of Biology, North Carolina A & T University, Greensboro (Byrd); National Alzheimer’s Coordinating Center, Department of Epidemiology, University of Washington, Seattle (Kukull); Vanderbilt Center for Human Genetics Research, Department of Molecular Physiology and Biophysics, Vanderbilt University, Nashville, Tennessee (Haines); Department of Neurology, Boston University, Boston, Massachusetts (Farrer); Department of Epidemiology, Boston University, Boston, Massachusetts (Farrer); Department of Epidemiology, College of Physicians and Surgeons, Columbia University, New York, New York (Lee); Department of Psychiatry, College of Physicians and Surgeons, Columbia University, New York, New York (Mayeux).
1,
Rong Cheng
Rong Cheng, PhD
1Tanz Centre for Research in Neurodegenerative Diseases, University of Toronto, Toronto, Ontario, Canada (Ghani, Sato, St. George-Hyslop, Rogaeva); Taub Institute for Research on Alzheimer’s Disease and the Aging Brain, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Cheng, Manly, Lee, Mayeux); Gertrude H. Sergievsky Center, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Vardarajan, Lee, Mayeux); Department of Neurology, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Manly, Mayeux); Department of Medicine (Biomedical Genetics), Boston University, Boston, Massachusetts (Jun, Logue, Baldwin, Farrer); Department of Biostatistics, Boston University, Boston, Massachusetts (Jun, Lunetta, Farrer); Department of Ophthalmology, Boston University, Boston, Massachusetts (Jun, Farrer); The John P. Hussman Institute for Human Genomics, University of Miami, Miami, Florida (Naj, Rajbhandary, Pericak-Vance); Department of Pathology and Laboratory Medicine, University of Pennsylvania Perelman School of Medicine, Philadelphia (Wang, Valladares, Lin, Cantwell, Schellenberg); Department of Medicine, University of Washington, Seattle (Larson, Crane); Group Health Research Institute, Group Health, Seattle, Washington (Larson); Department of Neuroscience, Mayo Clinic, Jacksonville, Florida (Graff-Radford, Ertekin-Taner); Department of Neurology, Mayo Clinic, Jacksonville, Florida (Graff-Radford, Ertekin-Taner); Rush Institute for Healthy Aging, Department of Internal Medicine, Rush University Medical Center, Chicago, Illinois (Evans); Program in Translational Neuropsychiatric Genomics, Department of Neurology, Brigham and Women’s Hospital, Boston, Massachusetts (De Jager); Harvard Medical School, Boston, Massachusetts (De Jager, Raj, Green); Program in Medical and Population Genetics, The Broad Institute, Cambridge, Massachusetts (De Jager); Department of Psychiatry, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Genetics and Genomics Sciences, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Neuroscience, Mount Sinai School of Medicine, New York, New York (Buxbaum); Friedman Brain Institute, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Medical and Molecular Genetics, Indiana University, Indianapolis (Murrell); Division of Genetics, Department of Medicine, Brigham and Women’s Hospital, Boston, Massachusetts (Green); Partners Center for Personalized Genetic Medicine, Brigham and Women’s Hospital, Boston, Massachusetts (Green); Department of Neurological Sciences, Rush University Medical Center, Chicago, Illinois (Barnes, Bennett); Department of Behavioral Sciences, Rush University Medical Center, Chicago, Illinois (Barnes, Foroud); Department of Epidemiology, Johns Hopkins University School of Public Health, Baltimore, Maryland (Fallin); School of Public Health, University of Alabama at Birmingham (Go); SABA University School of Medicine, SABA, Dutch Caribbean (Griffith); Division of Geriatrics, Howard University Hospital, Washington, DC (Obisesan); Department of Human Genetics, University of Pittsburgh, Pittsburgh, Pennsylvania (Kamboh); Alzheimer’s Disease Research Center, University of Pittsburgh, Pittsburgh, Pennsylvania (Kamboh, Lopez, Mayeux); Rush Alzheimer’s Disease Center, Rush University Medical Center, Chicago, Illinois (Bennett); Indiana University Center for Aging Research, Indianapolis (Hendrie); Department of Psychiatry, Indiana University School of Medicine, Indianapolis (Hendrie, Hall); Regenstrief Institute Inc, Indianapolis, Indiana (Hendrie); Hope Center Program on Protein Aggregation and Neurodegeneration, Department of Psychiatry, Washington University School of Medicine, St Louis, Missouri (Goate); Department of Biology, North Carolina A & T University, Greensboro (Byrd); National Alzheimer’s Coordinating Center, Department of Epidemiology, University of Washington, Seattle (Kukull); Vanderbilt Center for Human Genetics Research, Department of Molecular Physiology and Biophysics, Vanderbilt University, Nashville, Tennessee (Haines); Department of Neurology, Boston University, Boston, Massachusetts (Farrer); Department of Epidemiology, Boston University, Boston, Massachusetts (Farrer); Department of Epidemiology, College of Physicians and Surgeons, Columbia University, New York, New York (Lee); Department of Psychiatry, College of Physicians and Surgeons, Columbia University, New York, New York (Mayeux).
1,
Badri Narayan Vardarajan
Badri Narayan Vardarajan, PhD
1Tanz Centre for Research in Neurodegenerative Diseases, University of Toronto, Toronto, Ontario, Canada (Ghani, Sato, St. George-Hyslop, Rogaeva); Taub Institute for Research on Alzheimer’s Disease and the Aging Brain, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Cheng, Manly, Lee, Mayeux); Gertrude H. Sergievsky Center, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Vardarajan, Lee, Mayeux); Department of Neurology, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Manly, Mayeux); Department of Medicine (Biomedical Genetics), Boston University, Boston, Massachusetts (Jun, Logue, Baldwin, Farrer); Department of Biostatistics, Boston University, Boston, Massachusetts (Jun, Lunetta, Farrer); Department of Ophthalmology, Boston University, Boston, Massachusetts (Jun, Farrer); The John P. Hussman Institute for Human Genomics, University of Miami, Miami, Florida (Naj, Rajbhandary, Pericak-Vance); Department of Pathology and Laboratory Medicine, University of Pennsylvania Perelman School of Medicine, Philadelphia (Wang, Valladares, Lin, Cantwell, Schellenberg); Department of Medicine, University of Washington, Seattle (Larson, Crane); Group Health Research Institute, Group Health, Seattle, Washington (Larson); Department of Neuroscience, Mayo Clinic, Jacksonville, Florida (Graff-Radford, Ertekin-Taner); Department of Neurology, Mayo Clinic, Jacksonville, Florida (Graff-Radford, Ertekin-Taner); Rush Institute for Healthy Aging, Department of Internal Medicine, Rush University Medical Center, Chicago, Illinois (Evans); Program in Translational Neuropsychiatric Genomics, Department of Neurology, Brigham and Women’s Hospital, Boston, Massachusetts (De Jager); Harvard Medical School, Boston, Massachusetts (De Jager, Raj, Green); Program in Medical and Population Genetics, The Broad Institute, Cambridge, Massachusetts (De Jager); Department of Psychiatry, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Genetics and Genomics Sciences, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Neuroscience, Mount Sinai School of Medicine, New York, New York (Buxbaum); Friedman Brain Institute, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Medical and Molecular Genetics, Indiana University, Indianapolis (Murrell); Division of Genetics, Department of Medicine, Brigham and Women’s Hospital, Boston, Massachusetts (Green); Partners Center for Personalized Genetic Medicine, Brigham and Women’s Hospital, Boston, Massachusetts (Green); Department of Neurological Sciences, Rush University Medical Center, Chicago, Illinois (Barnes, Bennett); Department of Behavioral Sciences, Rush University Medical Center, Chicago, Illinois (Barnes, Foroud); Department of Epidemiology, Johns Hopkins University School of Public Health, Baltimore, Maryland (Fallin); School of Public Health, University of Alabama at Birmingham (Go); SABA University School of Medicine, SABA, Dutch Caribbean (Griffith); Division of Geriatrics, Howard University Hospital, Washington, DC (Obisesan); Department of Human Genetics, University of Pittsburgh, Pittsburgh, Pennsylvania (Kamboh); Alzheimer’s Disease Research Center, University of Pittsburgh, Pittsburgh, Pennsylvania (Kamboh, Lopez, Mayeux); Rush Alzheimer’s Disease Center, Rush University Medical Center, Chicago, Illinois (Bennett); Indiana University Center for Aging Research, Indianapolis (Hendrie); Department of Psychiatry, Indiana University School of Medicine, Indianapolis (Hendrie, Hall); Regenstrief Institute Inc, Indianapolis, Indiana (Hendrie); Hope Center Program on Protein Aggregation and Neurodegeneration, Department of Psychiatry, Washington University School of Medicine, St Louis, Missouri (Goate); Department of Biology, North Carolina A & T University, Greensboro (Byrd); National Alzheimer’s Coordinating Center, Department of Epidemiology, University of Washington, Seattle (Kukull); Vanderbilt Center for Human Genetics Research, Department of Molecular Physiology and Biophysics, Vanderbilt University, Nashville, Tennessee (Haines); Department of Neurology, Boston University, Boston, Massachusetts (Farrer); Department of Epidemiology, Boston University, Boston, Massachusetts (Farrer); Department of Epidemiology, College of Physicians and Surgeons, Columbia University, New York, New York (Lee); Department of Psychiatry, College of Physicians and Surgeons, Columbia University, New York, New York (Mayeux).
1,
Gyungah Jun
Gyungah Jun, PhD
1Tanz Centre for Research in Neurodegenerative Diseases, University of Toronto, Toronto, Ontario, Canada (Ghani, Sato, St. George-Hyslop, Rogaeva); Taub Institute for Research on Alzheimer’s Disease and the Aging Brain, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Cheng, Manly, Lee, Mayeux); Gertrude H. Sergievsky Center, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Vardarajan, Lee, Mayeux); Department of Neurology, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Manly, Mayeux); Department of Medicine (Biomedical Genetics), Boston University, Boston, Massachusetts (Jun, Logue, Baldwin, Farrer); Department of Biostatistics, Boston University, Boston, Massachusetts (Jun, Lunetta, Farrer); Department of Ophthalmology, Boston University, Boston, Massachusetts (Jun, Farrer); The John P. Hussman Institute for Human Genomics, University of Miami, Miami, Florida (Naj, Rajbhandary, Pericak-Vance); Department of Pathology and Laboratory Medicine, University of Pennsylvania Perelman School of Medicine, Philadelphia (Wang, Valladares, Lin, Cantwell, Schellenberg); Department of Medicine, University of Washington, Seattle (Larson, Crane); Group Health Research Institute, Group Health, Seattle, Washington (Larson); Department of Neuroscience, Mayo Clinic, Jacksonville, Florida (Graff-Radford, Ertekin-Taner); Department of Neurology, Mayo Clinic, Jacksonville, Florida (Graff-Radford, Ertekin-Taner); Rush Institute for Healthy Aging, Department of Internal Medicine, Rush University Medical Center, Chicago, Illinois (Evans); Program in Translational Neuropsychiatric Genomics, Department of Neurology, Brigham and Women’s Hospital, Boston, Massachusetts (De Jager); Harvard Medical School, Boston, Massachusetts (De Jager, Raj, Green); Program in Medical and Population Genetics, The Broad Institute, Cambridge, Massachusetts (De Jager); Department of Psychiatry, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Genetics and Genomics Sciences, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Neuroscience, Mount Sinai School of Medicine, New York, New York (Buxbaum); Friedman Brain Institute, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Medical and Molecular Genetics, Indiana University, Indianapolis (Murrell); Division of Genetics, Department of Medicine, Brigham and Women’s Hospital, Boston, Massachusetts (Green); Partners Center for Personalized Genetic Medicine, Brigham and Women’s Hospital, Boston, Massachusetts (Green); Department of Neurological Sciences, Rush University Medical Center, Chicago, Illinois (Barnes, Bennett); Department of Behavioral Sciences, Rush University Medical Center, Chicago, Illinois (Barnes, Foroud); Department of Epidemiology, Johns Hopkins University School of Public Health, Baltimore, Maryland (Fallin); School of Public Health, University of Alabama at Birmingham (Go); SABA University School of Medicine, SABA, Dutch Caribbean (Griffith); Division of Geriatrics, Howard University Hospital, Washington, DC (Obisesan); Department of Human Genetics, University of Pittsburgh, Pittsburgh, Pennsylvania (Kamboh); Alzheimer’s Disease Research Center, University of Pittsburgh, Pittsburgh, Pennsylvania (Kamboh, Lopez, Mayeux); Rush Alzheimer’s Disease Center, Rush University Medical Center, Chicago, Illinois (Bennett); Indiana University Center for Aging Research, Indianapolis (Hendrie); Department of Psychiatry, Indiana University School of Medicine, Indianapolis (Hendrie, Hall); Regenstrief Institute Inc, Indianapolis, Indiana (Hendrie); Hope Center Program on Protein Aggregation and Neurodegeneration, Department of Psychiatry, Washington University School of Medicine, St Louis, Missouri (Goate); Department of Biology, North Carolina A & T University, Greensboro (Byrd); National Alzheimer’s Coordinating Center, Department of Epidemiology, University of Washington, Seattle (Kukull); Vanderbilt Center for Human Genetics Research, Department of Molecular Physiology and Biophysics, Vanderbilt University, Nashville, Tennessee (Haines); Department of Neurology, Boston University, Boston, Massachusetts (Farrer); Department of Epidemiology, Boston University, Boston, Massachusetts (Farrer); Department of Epidemiology, College of Physicians and Surgeons, Columbia University, New York, New York (Lee); Department of Psychiatry, College of Physicians and Surgeons, Columbia University, New York, New York (Mayeux).
1,
Christine Sato
Christine Sato, MSc
1Tanz Centre for Research in Neurodegenerative Diseases, University of Toronto, Toronto, Ontario, Canada (Ghani, Sato, St. George-Hyslop, Rogaeva); Taub Institute for Research on Alzheimer’s Disease and the Aging Brain, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Cheng, Manly, Lee, Mayeux); Gertrude H. Sergievsky Center, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Vardarajan, Lee, Mayeux); Department of Neurology, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Manly, Mayeux); Department of Medicine (Biomedical Genetics), Boston University, Boston, Massachusetts (Jun, Logue, Baldwin, Farrer); Department of Biostatistics, Boston University, Boston, Massachusetts (Jun, Lunetta, Farrer); Department of Ophthalmology, Boston University, Boston, Massachusetts (Jun, Farrer); The John P. Hussman Institute for Human Genomics, University of Miami, Miami, Florida (Naj, Rajbhandary, Pericak-Vance); Department of Pathology and Laboratory Medicine, University of Pennsylvania Perelman School of Medicine, Philadelphia (Wang, Valladares, Lin, Cantwell, Schellenberg); Department of Medicine, University of Washington, Seattle (Larson, Crane); Group Health Research Institute, Group Health, Seattle, Washington (Larson); Department of Neuroscience, Mayo Clinic, Jacksonville, Florida (Graff-Radford, Ertekin-Taner); Department of Neurology, Mayo Clinic, Jacksonville, Florida (Graff-Radford, Ertekin-Taner); Rush Institute for Healthy Aging, Department of Internal Medicine, Rush University Medical Center, Chicago, Illinois (Evans); Program in Translational Neuropsychiatric Genomics, Department of Neurology, Brigham and Women’s Hospital, Boston, Massachusetts (De Jager); Harvard Medical School, Boston, Massachusetts (De Jager, Raj, Green); Program in Medical and Population Genetics, The Broad Institute, Cambridge, Massachusetts (De Jager); Department of Psychiatry, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Genetics and Genomics Sciences, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Neuroscience, Mount Sinai School of Medicine, New York, New York (Buxbaum); Friedman Brain Institute, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Medical and Molecular Genetics, Indiana University, Indianapolis (Murrell); Division of Genetics, Department of Medicine, Brigham and Women’s Hospital, Boston, Massachusetts (Green); Partners Center for Personalized Genetic Medicine, Brigham and Women’s Hospital, Boston, Massachusetts (Green); Department of Neurological Sciences, Rush University Medical Center, Chicago, Illinois (Barnes, Bennett); Department of Behavioral Sciences, Rush University Medical Center, Chicago, Illinois (Barnes, Foroud); Department of Epidemiology, Johns Hopkins University School of Public Health, Baltimore, Maryland (Fallin); School of Public Health, University of Alabama at Birmingham (Go); SABA University School of Medicine, SABA, Dutch Caribbean (Griffith); Division of Geriatrics, Howard University Hospital, Washington, DC (Obisesan); Department of Human Genetics, University of Pittsburgh, Pittsburgh, Pennsylvania (Kamboh); Alzheimer’s Disease Research Center, University of Pittsburgh, Pittsburgh, Pennsylvania (Kamboh, Lopez, Mayeux); Rush Alzheimer’s Disease Center, Rush University Medical Center, Chicago, Illinois (Bennett); Indiana University Center for Aging Research, Indianapolis (Hendrie); Department of Psychiatry, Indiana University School of Medicine, Indianapolis (Hendrie, Hall); Regenstrief Institute Inc, Indianapolis, Indiana (Hendrie); Hope Center Program on Protein Aggregation and Neurodegeneration, Department of Psychiatry, Washington University School of Medicine, St Louis, Missouri (Goate); Department of Biology, North Carolina A & T University, Greensboro (Byrd); National Alzheimer’s Coordinating Center, Department of Epidemiology, University of Washington, Seattle (Kukull); Vanderbilt Center for Human Genetics Research, Department of Molecular Physiology and Biophysics, Vanderbilt University, Nashville, Tennessee (Haines); Department of Neurology, Boston University, Boston, Massachusetts (Farrer); Department of Epidemiology, Boston University, Boston, Massachusetts (Farrer); Department of Epidemiology, College of Physicians and Surgeons, Columbia University, New York, New York (Lee); Department of Psychiatry, College of Physicians and Surgeons, Columbia University, New York, New York (Mayeux).
1,
Adam Naj
Adam Naj, PhD
1Tanz Centre for Research in Neurodegenerative Diseases, University of Toronto, Toronto, Ontario, Canada (Ghani, Sato, St. George-Hyslop, Rogaeva); Taub Institute for Research on Alzheimer’s Disease and the Aging Brain, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Cheng, Manly, Lee, Mayeux); Gertrude H. Sergievsky Center, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Vardarajan, Lee, Mayeux); Department of Neurology, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Manly, Mayeux); Department of Medicine (Biomedical Genetics), Boston University, Boston, Massachusetts (Jun, Logue, Baldwin, Farrer); Department of Biostatistics, Boston University, Boston, Massachusetts (Jun, Lunetta, Farrer); Department of Ophthalmology, Boston University, Boston, Massachusetts (Jun, Farrer); The John P. Hussman Institute for Human Genomics, University of Miami, Miami, Florida (Naj, Rajbhandary, Pericak-Vance); Department of Pathology and Laboratory Medicine, University of Pennsylvania Perelman School of Medicine, Philadelphia (Wang, Valladares, Lin, Cantwell, Schellenberg); Department of Medicine, University of Washington, Seattle (Larson, Crane); Group Health Research Institute, Group Health, Seattle, Washington (Larson); Department of Neuroscience, Mayo Clinic, Jacksonville, Florida (Graff-Radford, Ertekin-Taner); Department of Neurology, Mayo Clinic, Jacksonville, Florida (Graff-Radford, Ertekin-Taner); Rush Institute for Healthy Aging, Department of Internal Medicine, Rush University Medical Center, Chicago, Illinois (Evans); Program in Translational Neuropsychiatric Genomics, Department of Neurology, Brigham and Women’s Hospital, Boston, Massachusetts (De Jager); Harvard Medical School, Boston, Massachusetts (De Jager, Raj, Green); Program in Medical and Population Genetics, The Broad Institute, Cambridge, Massachusetts (De Jager); Department of Psychiatry, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Genetics and Genomics Sciences, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Neuroscience, Mount Sinai School of Medicine, New York, New York (Buxbaum); Friedman Brain Institute, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Medical and Molecular Genetics, Indiana University, Indianapolis (Murrell); Division of Genetics, Department of Medicine, Brigham and Women’s Hospital, Boston, Massachusetts (Green); Partners Center for Personalized Genetic Medicine, Brigham and Women’s Hospital, Boston, Massachusetts (Green); Department of Neurological Sciences, Rush University Medical Center, Chicago, Illinois (Barnes, Bennett); Department of Behavioral Sciences, Rush University Medical Center, Chicago, Illinois (Barnes, Foroud); Department of Epidemiology, Johns Hopkins University School of Public Health, Baltimore, Maryland (Fallin); School of Public Health, University of Alabama at Birmingham (Go); SABA University School of Medicine, SABA, Dutch Caribbean (Griffith); Division of Geriatrics, Howard University Hospital, Washington, DC (Obisesan); Department of Human Genetics, University of Pittsburgh, Pittsburgh, Pennsylvania (Kamboh); Alzheimer’s Disease Research Center, University of Pittsburgh, Pittsburgh, Pennsylvania (Kamboh, Lopez, Mayeux); Rush Alzheimer’s Disease Center, Rush University Medical Center, Chicago, Illinois (Bennett); Indiana University Center for Aging Research, Indianapolis (Hendrie); Department of Psychiatry, Indiana University School of Medicine, Indianapolis (Hendrie, Hall); Regenstrief Institute Inc, Indianapolis, Indiana (Hendrie); Hope Center Program on Protein Aggregation and Neurodegeneration, Department of Psychiatry, Washington University School of Medicine, St Louis, Missouri (Goate); Department of Biology, North Carolina A & T University, Greensboro (Byrd); National Alzheimer’s Coordinating Center, Department of Epidemiology, University of Washington, Seattle (Kukull); Vanderbilt Center for Human Genetics Research, Department of Molecular Physiology and Biophysics, Vanderbilt University, Nashville, Tennessee (Haines); Department of Neurology, Boston University, Boston, Massachusetts (Farrer); Department of Epidemiology, Boston University, Boston, Massachusetts (Farrer); Department of Epidemiology, College of Physicians and Surgeons, Columbia University, New York, New York (Lee); Department of Psychiatry, College of Physicians and Surgeons, Columbia University, New York, New York (Mayeux).
1,
Ruchita Rajbhandary
Ruchita Rajbhandary, MPH
1Tanz Centre for Research in Neurodegenerative Diseases, University of Toronto, Toronto, Ontario, Canada (Ghani, Sato, St. George-Hyslop, Rogaeva); Taub Institute for Research on Alzheimer’s Disease and the Aging Brain, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Cheng, Manly, Lee, Mayeux); Gertrude H. Sergievsky Center, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Vardarajan, Lee, Mayeux); Department of Neurology, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Manly, Mayeux); Department of Medicine (Biomedical Genetics), Boston University, Boston, Massachusetts (Jun, Logue, Baldwin, Farrer); Department of Biostatistics, Boston University, Boston, Massachusetts (Jun, Lunetta, Farrer); Department of Ophthalmology, Boston University, Boston, Massachusetts (Jun, Farrer); The John P. Hussman Institute for Human Genomics, University of Miami, Miami, Florida (Naj, Rajbhandary, Pericak-Vance); Department of Pathology and Laboratory Medicine, University of Pennsylvania Perelman School of Medicine, Philadelphia (Wang, Valladares, Lin, Cantwell, Schellenberg); Department of Medicine, University of Washington, Seattle (Larson, Crane); Group Health Research Institute, Group Health, Seattle, Washington (Larson); Department of Neuroscience, Mayo Clinic, Jacksonville, Florida (Graff-Radford, Ertekin-Taner); Department of Neurology, Mayo Clinic, Jacksonville, Florida (Graff-Radford, Ertekin-Taner); Rush Institute for Healthy Aging, Department of Internal Medicine, Rush University Medical Center, Chicago, Illinois (Evans); Program in Translational Neuropsychiatric Genomics, Department of Neurology, Brigham and Women’s Hospital, Boston, Massachusetts (De Jager); Harvard Medical School, Boston, Massachusetts (De Jager, Raj, Green); Program in Medical and Population Genetics, The Broad Institute, Cambridge, Massachusetts (De Jager); Department of Psychiatry, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Genetics and Genomics Sciences, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Neuroscience, Mount Sinai School of Medicine, New York, New York (Buxbaum); Friedman Brain Institute, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Medical and Molecular Genetics, Indiana University, Indianapolis (Murrell); Division of Genetics, Department of Medicine, Brigham and Women’s Hospital, Boston, Massachusetts (Green); Partners Center for Personalized Genetic Medicine, Brigham and Women’s Hospital, Boston, Massachusetts (Green); Department of Neurological Sciences, Rush University Medical Center, Chicago, Illinois (Barnes, Bennett); Department of Behavioral Sciences, Rush University Medical Center, Chicago, Illinois (Barnes, Foroud); Department of Epidemiology, Johns Hopkins University School of Public Health, Baltimore, Maryland (Fallin); School of Public Health, University of Alabama at Birmingham (Go); SABA University School of Medicine, SABA, Dutch Caribbean (Griffith); Division of Geriatrics, Howard University Hospital, Washington, DC (Obisesan); Department of Human Genetics, University of Pittsburgh, Pittsburgh, Pennsylvania (Kamboh); Alzheimer’s Disease Research Center, University of Pittsburgh, Pittsburgh, Pennsylvania (Kamboh, Lopez, Mayeux); Rush Alzheimer’s Disease Center, Rush University Medical Center, Chicago, Illinois (Bennett); Indiana University Center for Aging Research, Indianapolis (Hendrie); Department of Psychiatry, Indiana University School of Medicine, Indianapolis (Hendrie, Hall); Regenstrief Institute Inc, Indianapolis, Indiana (Hendrie); Hope Center Program on Protein Aggregation and Neurodegeneration, Department of Psychiatry, Washington University School of Medicine, St Louis, Missouri (Goate); Department of Biology, North Carolina A & T University, Greensboro (Byrd); National Alzheimer’s Coordinating Center, Department of Epidemiology, University of Washington, Seattle (Kukull); Vanderbilt Center for Human Genetics Research, Department of Molecular Physiology and Biophysics, Vanderbilt University, Nashville, Tennessee (Haines); Department of Neurology, Boston University, Boston, Massachusetts (Farrer); Department of Epidemiology, Boston University, Boston, Massachusetts (Farrer); Department of Epidemiology, College of Physicians and Surgeons, Columbia University, New York, New York (Lee); Department of Psychiatry, College of Physicians and Surgeons, Columbia University, New York, New York (Mayeux).
1,
Li-San Wang
Li-San Wang, PhD
1Tanz Centre for Research in Neurodegenerative Diseases, University of Toronto, Toronto, Ontario, Canada (Ghani, Sato, St. George-Hyslop, Rogaeva); Taub Institute for Research on Alzheimer’s Disease and the Aging Brain, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Cheng, Manly, Lee, Mayeux); Gertrude H. Sergievsky Center, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Vardarajan, Lee, Mayeux); Department of Neurology, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Manly, Mayeux); Department of Medicine (Biomedical Genetics), Boston University, Boston, Massachusetts (Jun, Logue, Baldwin, Farrer); Department of Biostatistics, Boston University, Boston, Massachusetts (Jun, Lunetta, Farrer); Department of Ophthalmology, Boston University, Boston, Massachusetts (Jun, Farrer); The John P. Hussman Institute for Human Genomics, University of Miami, Miami, Florida (Naj, Rajbhandary, Pericak-Vance); Department of Pathology and Laboratory Medicine, University of Pennsylvania Perelman School of Medicine, Philadelphia (Wang, Valladares, Lin, Cantwell, Schellenberg); Department of Medicine, University of Washington, Seattle (Larson, Crane); Group Health Research Institute, Group Health, Seattle, Washington (Larson); Department of Neuroscience, Mayo Clinic, Jacksonville, Florida (Graff-Radford, Ertekin-Taner); Department of Neurology, Mayo Clinic, Jacksonville, Florida (Graff-Radford, Ertekin-Taner); Rush Institute for Healthy Aging, Department of Internal Medicine, Rush University Medical Center, Chicago, Illinois (Evans); Program in Translational Neuropsychiatric Genomics, Department of Neurology, Brigham and Women’s Hospital, Boston, Massachusetts (De Jager); Harvard Medical School, Boston, Massachusetts (De Jager, Raj, Green); Program in Medical and Population Genetics, The Broad Institute, Cambridge, Massachusetts (De Jager); Department of Psychiatry, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Genetics and Genomics Sciences, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Neuroscience, Mount Sinai School of Medicine, New York, New York (Buxbaum); Friedman Brain Institute, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Medical and Molecular Genetics, Indiana University, Indianapolis (Murrell); Division of Genetics, Department of Medicine, Brigham and Women’s Hospital, Boston, Massachusetts (Green); Partners Center for Personalized Genetic Medicine, Brigham and Women’s Hospital, Boston, Massachusetts (Green); Department of Neurological Sciences, Rush University Medical Center, Chicago, Illinois (Barnes, Bennett); Department of Behavioral Sciences, Rush University Medical Center, Chicago, Illinois (Barnes, Foroud); Department of Epidemiology, Johns Hopkins University School of Public Health, Baltimore, Maryland (Fallin); School of Public Health, University of Alabama at Birmingham (Go); SABA University School of Medicine, SABA, Dutch Caribbean (Griffith); Division of Geriatrics, Howard University Hospital, Washington, DC (Obisesan); Department of Human Genetics, University of Pittsburgh, Pittsburgh, Pennsylvania (Kamboh); Alzheimer’s Disease Research Center, University of Pittsburgh, Pittsburgh, Pennsylvania (Kamboh, Lopez, Mayeux); Rush Alzheimer’s Disease Center, Rush University Medical Center, Chicago, Illinois (Bennett); Indiana University Center for Aging Research, Indianapolis (Hendrie); Department of Psychiatry, Indiana University School of Medicine, Indianapolis (Hendrie, Hall); Regenstrief Institute Inc, Indianapolis, Indiana (Hendrie); Hope Center Program on Protein Aggregation and Neurodegeneration, Department of Psychiatry, Washington University School of Medicine, St Louis, Missouri (Goate); Department of Biology, North Carolina A & T University, Greensboro (Byrd); National Alzheimer’s Coordinating Center, Department of Epidemiology, University of Washington, Seattle (Kukull); Vanderbilt Center for Human Genetics Research, Department of Molecular Physiology and Biophysics, Vanderbilt University, Nashville, Tennessee (Haines); Department of Neurology, Boston University, Boston, Massachusetts (Farrer); Department of Epidemiology, Boston University, Boston, Massachusetts (Farrer); Department of Epidemiology, College of Physicians and Surgeons, Columbia University, New York, New York (Lee); Department of Psychiatry, College of Physicians and Surgeons, Columbia University, New York, New York (Mayeux).
1,
Otto Valladares
Otto Valladares, MS
1Tanz Centre for Research in Neurodegenerative Diseases, University of Toronto, Toronto, Ontario, Canada (Ghani, Sato, St. George-Hyslop, Rogaeva); Taub Institute for Research on Alzheimer’s Disease and the Aging Brain, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Cheng, Manly, Lee, Mayeux); Gertrude H. Sergievsky Center, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Vardarajan, Lee, Mayeux); Department of Neurology, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Manly, Mayeux); Department of Medicine (Biomedical Genetics), Boston University, Boston, Massachusetts (Jun, Logue, Baldwin, Farrer); Department of Biostatistics, Boston University, Boston, Massachusetts (Jun, Lunetta, Farrer); Department of Ophthalmology, Boston University, Boston, Massachusetts (Jun, Farrer); The John P. Hussman Institute for Human Genomics, University of Miami, Miami, Florida (Naj, Rajbhandary, Pericak-Vance); Department of Pathology and Laboratory Medicine, University of Pennsylvania Perelman School of Medicine, Philadelphia (Wang, Valladares, Lin, Cantwell, Schellenberg); Department of Medicine, University of Washington, Seattle (Larson, Crane); Group Health Research Institute, Group Health, Seattle, Washington (Larson); Department of Neuroscience, Mayo Clinic, Jacksonville, Florida (Graff-Radford, Ertekin-Taner); Department of Neurology, Mayo Clinic, Jacksonville, Florida (Graff-Radford, Ertekin-Taner); Rush Institute for Healthy Aging, Department of Internal Medicine, Rush University Medical Center, Chicago, Illinois (Evans); Program in Translational Neuropsychiatric Genomics, Department of Neurology, Brigham and Women’s Hospital, Boston, Massachusetts (De Jager); Harvard Medical School, Boston, Massachusetts (De Jager, Raj, Green); Program in Medical and Population Genetics, The Broad Institute, Cambridge, Massachusetts (De Jager); Department of Psychiatry, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Genetics and Genomics Sciences, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Neuroscience, Mount Sinai School of Medicine, New York, New York (Buxbaum); Friedman Brain Institute, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Medical and Molecular Genetics, Indiana University, Indianapolis (Murrell); Division of Genetics, Department of Medicine, Brigham and Women’s Hospital, Boston, Massachusetts (Green); Partners Center for Personalized Genetic Medicine, Brigham and Women’s Hospital, Boston, Massachusetts (Green); Department of Neurological Sciences, Rush University Medical Center, Chicago, Illinois (Barnes, Bennett); Department of Behavioral Sciences, Rush University Medical Center, Chicago, Illinois (Barnes, Foroud); Department of Epidemiology, Johns Hopkins University School of Public Health, Baltimore, Maryland (Fallin); School of Public Health, University of Alabama at Birmingham (Go); SABA University School of Medicine, SABA, Dutch Caribbean (Griffith); Division of Geriatrics, Howard University Hospital, Washington, DC (Obisesan); Department of Human Genetics, University of Pittsburgh, Pittsburgh, Pennsylvania (Kamboh); Alzheimer’s Disease Research Center, University of Pittsburgh, Pittsburgh, Pennsylvania (Kamboh, Lopez, Mayeux); Rush Alzheimer’s Disease Center, Rush University Medical Center, Chicago, Illinois (Bennett); Indiana University Center for Aging Research, Indianapolis (Hendrie); Department of Psychiatry, Indiana University School of Medicine, Indianapolis (Hendrie, Hall); Regenstrief Institute Inc, Indianapolis, Indiana (Hendrie); Hope Center Program on Protein Aggregation and Neurodegeneration, Department of Psychiatry, Washington University School of Medicine, St Louis, Missouri (Goate); Department of Biology, North Carolina A & T University, Greensboro (Byrd); National Alzheimer’s Coordinating Center, Department of Epidemiology, University of Washington, Seattle (Kukull); Vanderbilt Center for Human Genetics Research, Department of Molecular Physiology and Biophysics, Vanderbilt University, Nashville, Tennessee (Haines); Department of Neurology, Boston University, Boston, Massachusetts (Farrer); Department of Epidemiology, Boston University, Boston, Massachusetts (Farrer); Department of Epidemiology, College of Physicians and Surgeons, Columbia University, New York, New York (Lee); Department of Psychiatry, College of Physicians and Surgeons, Columbia University, New York, New York (Mayeux).
1,
Chiao-Feng Lin
Chiao-Feng Lin, PhD
1Tanz Centre for Research in Neurodegenerative Diseases, University of Toronto, Toronto, Ontario, Canada (Ghani, Sato, St. George-Hyslop, Rogaeva); Taub Institute for Research on Alzheimer’s Disease and the Aging Brain, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Cheng, Manly, Lee, Mayeux); Gertrude H. Sergievsky Center, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Vardarajan, Lee, Mayeux); Department of Neurology, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Manly, Mayeux); Department of Medicine (Biomedical Genetics), Boston University, Boston, Massachusetts (Jun, Logue, Baldwin, Farrer); Department of Biostatistics, Boston University, Boston, Massachusetts (Jun, Lunetta, Farrer); Department of Ophthalmology, Boston University, Boston, Massachusetts (Jun, Farrer); The John P. Hussman Institute for Human Genomics, University of Miami, Miami, Florida (Naj, Rajbhandary, Pericak-Vance); Department of Pathology and Laboratory Medicine, University of Pennsylvania Perelman School of Medicine, Philadelphia (Wang, Valladares, Lin, Cantwell, Schellenberg); Department of Medicine, University of Washington, Seattle (Larson, Crane); Group Health Research Institute, Group Health, Seattle, Washington (Larson); Department of Neuroscience, Mayo Clinic, Jacksonville, Florida (Graff-Radford, Ertekin-Taner); Department of Neurology, Mayo Clinic, Jacksonville, Florida (Graff-Radford, Ertekin-Taner); Rush Institute for Healthy Aging, Department of Internal Medicine, Rush University Medical Center, Chicago, Illinois (Evans); Program in Translational Neuropsychiatric Genomics, Department of Neurology, Brigham and Women’s Hospital, Boston, Massachusetts (De Jager); Harvard Medical School, Boston, Massachusetts (De Jager, Raj, Green); Program in Medical and Population Genetics, The Broad Institute, Cambridge, Massachusetts (De Jager); Department of Psychiatry, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Genetics and Genomics Sciences, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Neuroscience, Mount Sinai School of Medicine, New York, New York (Buxbaum); Friedman Brain Institute, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Medical and Molecular Genetics, Indiana University, Indianapolis (Murrell); Division of Genetics, Department of Medicine, Brigham and Women’s Hospital, Boston, Massachusetts (Green); Partners Center for Personalized Genetic Medicine, Brigham and Women’s Hospital, Boston, Massachusetts (Green); Department of Neurological Sciences, Rush University Medical Center, Chicago, Illinois (Barnes, Bennett); Department of Behavioral Sciences, Rush University Medical Center, Chicago, Illinois (Barnes, Foroud); Department of Epidemiology, Johns Hopkins University School of Public Health, Baltimore, Maryland (Fallin); School of Public Health, University of Alabama at Birmingham (Go); SABA University School of Medicine, SABA, Dutch Caribbean (Griffith); Division of Geriatrics, Howard University Hospital, Washington, DC (Obisesan); Department of Human Genetics, University of Pittsburgh, Pittsburgh, Pennsylvania (Kamboh); Alzheimer’s Disease Research Center, University of Pittsburgh, Pittsburgh, Pennsylvania (Kamboh, Lopez, Mayeux); Rush Alzheimer’s Disease Center, Rush University Medical Center, Chicago, Illinois (Bennett); Indiana University Center for Aging Research, Indianapolis (Hendrie); Department of Psychiatry, Indiana University School of Medicine, Indianapolis (Hendrie, Hall); Regenstrief Institute Inc, Indianapolis, Indiana (Hendrie); Hope Center Program on Protein Aggregation and Neurodegeneration, Department of Psychiatry, Washington University School of Medicine, St Louis, Missouri (Goate); Department of Biology, North Carolina A & T University, Greensboro (Byrd); National Alzheimer’s Coordinating Center, Department of Epidemiology, University of Washington, Seattle (Kukull); Vanderbilt Center for Human Genetics Research, Department of Molecular Physiology and Biophysics, Vanderbilt University, Nashville, Tennessee (Haines); Department of Neurology, Boston University, Boston, Massachusetts (Farrer); Department of Epidemiology, Boston University, Boston, Massachusetts (Farrer); Department of Epidemiology, College of Physicians and Surgeons, Columbia University, New York, New York (Lee); Department of Psychiatry, College of Physicians and Surgeons, Columbia University, New York, New York (Mayeux).
1,
Eric B Larson
Eric B Larson, MD, MPH
1Tanz Centre for Research in Neurodegenerative Diseases, University of Toronto, Toronto, Ontario, Canada (Ghani, Sato, St. George-Hyslop, Rogaeva); Taub Institute for Research on Alzheimer’s Disease and the Aging Brain, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Cheng, Manly, Lee, Mayeux); Gertrude H. Sergievsky Center, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Vardarajan, Lee, Mayeux); Department of Neurology, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Manly, Mayeux); Department of Medicine (Biomedical Genetics), Boston University, Boston, Massachusetts (Jun, Logue, Baldwin, Farrer); Department of Biostatistics, Boston University, Boston, Massachusetts (Jun, Lunetta, Farrer); Department of Ophthalmology, Boston University, Boston, Massachusetts (Jun, Farrer); The John P. Hussman Institute for Human Genomics, University of Miami, Miami, Florida (Naj, Rajbhandary, Pericak-Vance); Department of Pathology and Laboratory Medicine, University of Pennsylvania Perelman School of Medicine, Philadelphia (Wang, Valladares, Lin, Cantwell, Schellenberg); Department of Medicine, University of Washington, Seattle (Larson, Crane); Group Health Research Institute, Group Health, Seattle, Washington (Larson); Department of Neuroscience, Mayo Clinic, Jacksonville, Florida (Graff-Radford, Ertekin-Taner); Department of Neurology, Mayo Clinic, Jacksonville, Florida (Graff-Radford, Ertekin-Taner); Rush Institute for Healthy Aging, Department of Internal Medicine, Rush University Medical Center, Chicago, Illinois (Evans); Program in Translational Neuropsychiatric Genomics, Department of Neurology, Brigham and Women’s Hospital, Boston, Massachusetts (De Jager); Harvard Medical School, Boston, Massachusetts (De Jager, Raj, Green); Program in Medical and Population Genetics, The Broad Institute, Cambridge, Massachusetts (De Jager); Department of Psychiatry, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Genetics and Genomics Sciences, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Neuroscience, Mount Sinai School of Medicine, New York, New York (Buxbaum); Friedman Brain Institute, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Medical and Molecular Genetics, Indiana University, Indianapolis (Murrell); Division of Genetics, Department of Medicine, Brigham and Women’s Hospital, Boston, Massachusetts (Green); Partners Center for Personalized Genetic Medicine, Brigham and Women’s Hospital, Boston, Massachusetts (Green); Department of Neurological Sciences, Rush University Medical Center, Chicago, Illinois (Barnes, Bennett); Department of Behavioral Sciences, Rush University Medical Center, Chicago, Illinois (Barnes, Foroud); Department of Epidemiology, Johns Hopkins University School of Public Health, Baltimore, Maryland (Fallin); School of Public Health, University of Alabama at Birmingham (Go); SABA University School of Medicine, SABA, Dutch Caribbean (Griffith); Division of Geriatrics, Howard University Hospital, Washington, DC (Obisesan); Department of Human Genetics, University of Pittsburgh, Pittsburgh, Pennsylvania (Kamboh); Alzheimer’s Disease Research Center, University of Pittsburgh, Pittsburgh, Pennsylvania (Kamboh, Lopez, Mayeux); Rush Alzheimer’s Disease Center, Rush University Medical Center, Chicago, Illinois (Bennett); Indiana University Center for Aging Research, Indianapolis (Hendrie); Department of Psychiatry, Indiana University School of Medicine, Indianapolis (Hendrie, Hall); Regenstrief Institute Inc, Indianapolis, Indiana (Hendrie); Hope Center Program on Protein Aggregation and Neurodegeneration, Department of Psychiatry, Washington University School of Medicine, St Louis, Missouri (Goate); Department of Biology, North Carolina A & T University, Greensboro (Byrd); National Alzheimer’s Coordinating Center, Department of Epidemiology, University of Washington, Seattle (Kukull); Vanderbilt Center for Human Genetics Research, Department of Molecular Physiology and Biophysics, Vanderbilt University, Nashville, Tennessee (Haines); Department of Neurology, Boston University, Boston, Massachusetts (Farrer); Department of Epidemiology, Boston University, Boston, Massachusetts (Farrer); Department of Epidemiology, College of Physicians and Surgeons, Columbia University, New York, New York (Lee); Department of Psychiatry, College of Physicians and Surgeons, Columbia University, New York, New York (Mayeux).
1,
Neill R Graff-Radford
Neill R Graff-Radford, MD
1Tanz Centre for Research in Neurodegenerative Diseases, University of Toronto, Toronto, Ontario, Canada (Ghani, Sato, St. George-Hyslop, Rogaeva); Taub Institute for Research on Alzheimer’s Disease and the Aging Brain, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Cheng, Manly, Lee, Mayeux); Gertrude H. Sergievsky Center, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Vardarajan, Lee, Mayeux); Department of Neurology, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Manly, Mayeux); Department of Medicine (Biomedical Genetics), Boston University, Boston, Massachusetts (Jun, Logue, Baldwin, Farrer); Department of Biostatistics, Boston University, Boston, Massachusetts (Jun, Lunetta, Farrer); Department of Ophthalmology, Boston University, Boston, Massachusetts (Jun, Farrer); The John P. Hussman Institute for Human Genomics, University of Miami, Miami, Florida (Naj, Rajbhandary, Pericak-Vance); Department of Pathology and Laboratory Medicine, University of Pennsylvania Perelman School of Medicine, Philadelphia (Wang, Valladares, Lin, Cantwell, Schellenberg); Department of Medicine, University of Washington, Seattle (Larson, Crane); Group Health Research Institute, Group Health, Seattle, Washington (Larson); Department of Neuroscience, Mayo Clinic, Jacksonville, Florida (Graff-Radford, Ertekin-Taner); Department of Neurology, Mayo Clinic, Jacksonville, Florida (Graff-Radford, Ertekin-Taner); Rush Institute for Healthy Aging, Department of Internal Medicine, Rush University Medical Center, Chicago, Illinois (Evans); Program in Translational Neuropsychiatric Genomics, Department of Neurology, Brigham and Women’s Hospital, Boston, Massachusetts (De Jager); Harvard Medical School, Boston, Massachusetts (De Jager, Raj, Green); Program in Medical and Population Genetics, The Broad Institute, Cambridge, Massachusetts (De Jager); Department of Psychiatry, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Genetics and Genomics Sciences, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Neuroscience, Mount Sinai School of Medicine, New York, New York (Buxbaum); Friedman Brain Institute, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Medical and Molecular Genetics, Indiana University, Indianapolis (Murrell); Division of Genetics, Department of Medicine, Brigham and Women’s Hospital, Boston, Massachusetts (Green); Partners Center for Personalized Genetic Medicine, Brigham and Women’s Hospital, Boston, Massachusetts (Green); Department of Neurological Sciences, Rush University Medical Center, Chicago, Illinois (Barnes, Bennett); Department of Behavioral Sciences, Rush University Medical Center, Chicago, Illinois (Barnes, Foroud); Department of Epidemiology, Johns Hopkins University School of Public Health, Baltimore, Maryland (Fallin); School of Public Health, University of Alabama at Birmingham (Go); SABA University School of Medicine, SABA, Dutch Caribbean (Griffith); Division of Geriatrics, Howard University Hospital, Washington, DC (Obisesan); Department of Human Genetics, University of Pittsburgh, Pittsburgh, Pennsylvania (Kamboh); Alzheimer’s Disease Research Center, University of Pittsburgh, Pittsburgh, Pennsylvania (Kamboh, Lopez, Mayeux); Rush Alzheimer’s Disease Center, Rush University Medical Center, Chicago, Illinois (Bennett); Indiana University Center for Aging Research, Indianapolis (Hendrie); Department of Psychiatry, Indiana University School of Medicine, Indianapolis (Hendrie, Hall); Regenstrief Institute Inc, Indianapolis, Indiana (Hendrie); Hope Center Program on Protein Aggregation and Neurodegeneration, Department of Psychiatry, Washington University School of Medicine, St Louis, Missouri (Goate); Department of Biology, North Carolina A & T University, Greensboro (Byrd); National Alzheimer’s Coordinating Center, Department of Epidemiology, University of Washington, Seattle (Kukull); Vanderbilt Center for Human Genetics Research, Department of Molecular Physiology and Biophysics, Vanderbilt University, Nashville, Tennessee (Haines); Department of Neurology, Boston University, Boston, Massachusetts (Farrer); Department of Epidemiology, Boston University, Boston, Massachusetts (Farrer); Department of Epidemiology, College of Physicians and Surgeons, Columbia University, New York, New York (Lee); Department of Psychiatry, College of Physicians and Surgeons, Columbia University, New York, New York (Mayeux).
1,
Denis Evans
Denis Evans, MD
1Tanz Centre for Research in Neurodegenerative Diseases, University of Toronto, Toronto, Ontario, Canada (Ghani, Sato, St. George-Hyslop, Rogaeva); Taub Institute for Research on Alzheimer’s Disease and the Aging Brain, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Cheng, Manly, Lee, Mayeux); Gertrude H. Sergievsky Center, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Vardarajan, Lee, Mayeux); Department of Neurology, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Manly, Mayeux); Department of Medicine (Biomedical Genetics), Boston University, Boston, Massachusetts (Jun, Logue, Baldwin, Farrer); Department of Biostatistics, Boston University, Boston, Massachusetts (Jun, Lunetta, Farrer); Department of Ophthalmology, Boston University, Boston, Massachusetts (Jun, Farrer); The John P. Hussman Institute for Human Genomics, University of Miami, Miami, Florida (Naj, Rajbhandary, Pericak-Vance); Department of Pathology and Laboratory Medicine, University of Pennsylvania Perelman School of Medicine, Philadelphia (Wang, Valladares, Lin, Cantwell, Schellenberg); Department of Medicine, University of Washington, Seattle (Larson, Crane); Group Health Research Institute, Group Health, Seattle, Washington (Larson); Department of Neuroscience, Mayo Clinic, Jacksonville, Florida (Graff-Radford, Ertekin-Taner); Department of Neurology, Mayo Clinic, Jacksonville, Florida (Graff-Radford, Ertekin-Taner); Rush Institute for Healthy Aging, Department of Internal Medicine, Rush University Medical Center, Chicago, Illinois (Evans); Program in Translational Neuropsychiatric Genomics, Department of Neurology, Brigham and Women’s Hospital, Boston, Massachusetts (De Jager); Harvard Medical School, Boston, Massachusetts (De Jager, Raj, Green); Program in Medical and Population Genetics, The Broad Institute, Cambridge, Massachusetts (De Jager); Department of Psychiatry, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Genetics and Genomics Sciences, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Neuroscience, Mount Sinai School of Medicine, New York, New York (Buxbaum); Friedman Brain Institute, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Medical and Molecular Genetics, Indiana University, Indianapolis (Murrell); Division of Genetics, Department of Medicine, Brigham and Women’s Hospital, Boston, Massachusetts (Green); Partners Center for Personalized Genetic Medicine, Brigham and Women’s Hospital, Boston, Massachusetts (Green); Department of Neurological Sciences, Rush University Medical Center, Chicago, Illinois (Barnes, Bennett); Department of Behavioral Sciences, Rush University Medical Center, Chicago, Illinois (Barnes, Foroud); Department of Epidemiology, Johns Hopkins University School of Public Health, Baltimore, Maryland (Fallin); School of Public Health, University of Alabama at Birmingham (Go); SABA University School of Medicine, SABA, Dutch Caribbean (Griffith); Division of Geriatrics, Howard University Hospital, Washington, DC (Obisesan); Department of Human Genetics, University of Pittsburgh, Pittsburgh, Pennsylvania (Kamboh); Alzheimer’s Disease Research Center, University of Pittsburgh, Pittsburgh, Pennsylvania (Kamboh, Lopez, Mayeux); Rush Alzheimer’s Disease Center, Rush University Medical Center, Chicago, Illinois (Bennett); Indiana University Center for Aging Research, Indianapolis (Hendrie); Department of Psychiatry, Indiana University School of Medicine, Indianapolis (Hendrie, Hall); Regenstrief Institute Inc, Indianapolis, Indiana (Hendrie); Hope Center Program on Protein Aggregation and Neurodegeneration, Department of Psychiatry, Washington University School of Medicine, St Louis, Missouri (Goate); Department of Biology, North Carolina A & T University, Greensboro (Byrd); National Alzheimer’s Coordinating Center, Department of Epidemiology, University of Washington, Seattle (Kukull); Vanderbilt Center for Human Genetics Research, Department of Molecular Physiology and Biophysics, Vanderbilt University, Nashville, Tennessee (Haines); Department of Neurology, Boston University, Boston, Massachusetts (Farrer); Department of Epidemiology, Boston University, Boston, Massachusetts (Farrer); Department of Epidemiology, College of Physicians and Surgeons, Columbia University, New York, New York (Lee); Department of Psychiatry, College of Physicians and Surgeons, Columbia University, New York, New York (Mayeux).
1,
Philip L De Jager
Philip L De Jager, MD, PhD
1Tanz Centre for Research in Neurodegenerative Diseases, University of Toronto, Toronto, Ontario, Canada (Ghani, Sato, St. George-Hyslop, Rogaeva); Taub Institute for Research on Alzheimer’s Disease and the Aging Brain, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Cheng, Manly, Lee, Mayeux); Gertrude H. Sergievsky Center, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Vardarajan, Lee, Mayeux); Department of Neurology, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Manly, Mayeux); Department of Medicine (Biomedical Genetics), Boston University, Boston, Massachusetts (Jun, Logue, Baldwin, Farrer); Department of Biostatistics, Boston University, Boston, Massachusetts (Jun, Lunetta, Farrer); Department of Ophthalmology, Boston University, Boston, Massachusetts (Jun, Farrer); The John P. Hussman Institute for Human Genomics, University of Miami, Miami, Florida (Naj, Rajbhandary, Pericak-Vance); Department of Pathology and Laboratory Medicine, University of Pennsylvania Perelman School of Medicine, Philadelphia (Wang, Valladares, Lin, Cantwell, Schellenberg); Department of Medicine, University of Washington, Seattle (Larson, Crane); Group Health Research Institute, Group Health, Seattle, Washington (Larson); Department of Neuroscience, Mayo Clinic, Jacksonville, Florida (Graff-Radford, Ertekin-Taner); Department of Neurology, Mayo Clinic, Jacksonville, Florida (Graff-Radford, Ertekin-Taner); Rush Institute for Healthy Aging, Department of Internal Medicine, Rush University Medical Center, Chicago, Illinois (Evans); Program in Translational Neuropsychiatric Genomics, Department of Neurology, Brigham and Women’s Hospital, Boston, Massachusetts (De Jager); Harvard Medical School, Boston, Massachusetts (De Jager, Raj, Green); Program in Medical and Population Genetics, The Broad Institute, Cambridge, Massachusetts (De Jager); Department of Psychiatry, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Genetics and Genomics Sciences, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Neuroscience, Mount Sinai School of Medicine, New York, New York (Buxbaum); Friedman Brain Institute, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Medical and Molecular Genetics, Indiana University, Indianapolis (Murrell); Division of Genetics, Department of Medicine, Brigham and Women’s Hospital, Boston, Massachusetts (Green); Partners Center for Personalized Genetic Medicine, Brigham and Women’s Hospital, Boston, Massachusetts (Green); Department of Neurological Sciences, Rush University Medical Center, Chicago, Illinois (Barnes, Bennett); Department of Behavioral Sciences, Rush University Medical Center, Chicago, Illinois (Barnes, Foroud); Department of Epidemiology, Johns Hopkins University School of Public Health, Baltimore, Maryland (Fallin); School of Public Health, University of Alabama at Birmingham (Go); SABA University School of Medicine, SABA, Dutch Caribbean (Griffith); Division of Geriatrics, Howard University Hospital, Washington, DC (Obisesan); Department of Human Genetics, University of Pittsburgh, Pittsburgh, Pennsylvania (Kamboh); Alzheimer’s Disease Research Center, University of Pittsburgh, Pittsburgh, Pennsylvania (Kamboh, Lopez, Mayeux); Rush Alzheimer’s Disease Center, Rush University Medical Center, Chicago, Illinois (Bennett); Indiana University Center for Aging Research, Indianapolis (Hendrie); Department of Psychiatry, Indiana University School of Medicine, Indianapolis (Hendrie, Hall); Regenstrief Institute Inc, Indianapolis, Indiana (Hendrie); Hope Center Program on Protein Aggregation and Neurodegeneration, Department of Psychiatry, Washington University School of Medicine, St Louis, Missouri (Goate); Department of Biology, North Carolina A & T University, Greensboro (Byrd); National Alzheimer’s Coordinating Center, Department of Epidemiology, University of Washington, Seattle (Kukull); Vanderbilt Center for Human Genetics Research, Department of Molecular Physiology and Biophysics, Vanderbilt University, Nashville, Tennessee (Haines); Department of Neurology, Boston University, Boston, Massachusetts (Farrer); Department of Epidemiology, Boston University, Boston, Massachusetts (Farrer); Department of Epidemiology, College of Physicians and Surgeons, Columbia University, New York, New York (Lee); Department of Psychiatry, College of Physicians and Surgeons, Columbia University, New York, New York (Mayeux).
1,
Paul K Crane
Paul K Crane, MD, MPH
1Tanz Centre for Research in Neurodegenerative Diseases, University of Toronto, Toronto, Ontario, Canada (Ghani, Sato, St. George-Hyslop, Rogaeva); Taub Institute for Research on Alzheimer’s Disease and the Aging Brain, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Cheng, Manly, Lee, Mayeux); Gertrude H. Sergievsky Center, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Vardarajan, Lee, Mayeux); Department of Neurology, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Manly, Mayeux); Department of Medicine (Biomedical Genetics), Boston University, Boston, Massachusetts (Jun, Logue, Baldwin, Farrer); Department of Biostatistics, Boston University, Boston, Massachusetts (Jun, Lunetta, Farrer); Department of Ophthalmology, Boston University, Boston, Massachusetts (Jun, Farrer); The John P. Hussman Institute for Human Genomics, University of Miami, Miami, Florida (Naj, Rajbhandary, Pericak-Vance); Department of Pathology and Laboratory Medicine, University of Pennsylvania Perelman School of Medicine, Philadelphia (Wang, Valladares, Lin, Cantwell, Schellenberg); Department of Medicine, University of Washington, Seattle (Larson, Crane); Group Health Research Institute, Group Health, Seattle, Washington (Larson); Department of Neuroscience, Mayo Clinic, Jacksonville, Florida (Graff-Radford, Ertekin-Taner); Department of Neurology, Mayo Clinic, Jacksonville, Florida (Graff-Radford, Ertekin-Taner); Rush Institute for Healthy Aging, Department of Internal Medicine, Rush University Medical Center, Chicago, Illinois (Evans); Program in Translational Neuropsychiatric Genomics, Department of Neurology, Brigham and Women’s Hospital, Boston, Massachusetts (De Jager); Harvard Medical School, Boston, Massachusetts (De Jager, Raj, Green); Program in Medical and Population Genetics, The Broad Institute, Cambridge, Massachusetts (De Jager); Department of Psychiatry, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Genetics and Genomics Sciences, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Neuroscience, Mount Sinai School of Medicine, New York, New York (Buxbaum); Friedman Brain Institute, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Medical and Molecular Genetics, Indiana University, Indianapolis (Murrell); Division of Genetics, Department of Medicine, Brigham and Women’s Hospital, Boston, Massachusetts (Green); Partners Center for Personalized Genetic Medicine, Brigham and Women’s Hospital, Boston, Massachusetts (Green); Department of Neurological Sciences, Rush University Medical Center, Chicago, Illinois (Barnes, Bennett); Department of Behavioral Sciences, Rush University Medical Center, Chicago, Illinois (Barnes, Foroud); Department of Epidemiology, Johns Hopkins University School of Public Health, Baltimore, Maryland (Fallin); School of Public Health, University of Alabama at Birmingham (Go); SABA University School of Medicine, SABA, Dutch Caribbean (Griffith); Division of Geriatrics, Howard University Hospital, Washington, DC (Obisesan); Department of Human Genetics, University of Pittsburgh, Pittsburgh, Pennsylvania (Kamboh); Alzheimer’s Disease Research Center, University of Pittsburgh, Pittsburgh, Pennsylvania (Kamboh, Lopez, Mayeux); Rush Alzheimer’s Disease Center, Rush University Medical Center, Chicago, Illinois (Bennett); Indiana University Center for Aging Research, Indianapolis (Hendrie); Department of Psychiatry, Indiana University School of Medicine, Indianapolis (Hendrie, Hall); Regenstrief Institute Inc, Indianapolis, Indiana (Hendrie); Hope Center Program on Protein Aggregation and Neurodegeneration, Department of Psychiatry, Washington University School of Medicine, St Louis, Missouri (Goate); Department of Biology, North Carolina A & T University, Greensboro (Byrd); National Alzheimer’s Coordinating Center, Department of Epidemiology, University of Washington, Seattle (Kukull); Vanderbilt Center for Human Genetics Research, Department of Molecular Physiology and Biophysics, Vanderbilt University, Nashville, Tennessee (Haines); Department of Neurology, Boston University, Boston, Massachusetts (Farrer); Department of Epidemiology, Boston University, Boston, Massachusetts (Farrer); Department of Epidemiology, College of Physicians and Surgeons, Columbia University, New York, New York (Lee); Department of Psychiatry, College of Physicians and Surgeons, Columbia University, New York, New York (Mayeux).
1,
Joseph D Buxbaum
Joseph D Buxbaum, PhD
1Tanz Centre for Research in Neurodegenerative Diseases, University of Toronto, Toronto, Ontario, Canada (Ghani, Sato, St. George-Hyslop, Rogaeva); Taub Institute for Research on Alzheimer’s Disease and the Aging Brain, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Cheng, Manly, Lee, Mayeux); Gertrude H. Sergievsky Center, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Vardarajan, Lee, Mayeux); Department of Neurology, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Manly, Mayeux); Department of Medicine (Biomedical Genetics), Boston University, Boston, Massachusetts (Jun, Logue, Baldwin, Farrer); Department of Biostatistics, Boston University, Boston, Massachusetts (Jun, Lunetta, Farrer); Department of Ophthalmology, Boston University, Boston, Massachusetts (Jun, Farrer); The John P. Hussman Institute for Human Genomics, University of Miami, Miami, Florida (Naj, Rajbhandary, Pericak-Vance); Department of Pathology and Laboratory Medicine, University of Pennsylvania Perelman School of Medicine, Philadelphia (Wang, Valladares, Lin, Cantwell, Schellenberg); Department of Medicine, University of Washington, Seattle (Larson, Crane); Group Health Research Institute, Group Health, Seattle, Washington (Larson); Department of Neuroscience, Mayo Clinic, Jacksonville, Florida (Graff-Radford, Ertekin-Taner); Department of Neurology, Mayo Clinic, Jacksonville, Florida (Graff-Radford, Ertekin-Taner); Rush Institute for Healthy Aging, Department of Internal Medicine, Rush University Medical Center, Chicago, Illinois (Evans); Program in Translational Neuropsychiatric Genomics, Department of Neurology, Brigham and Women’s Hospital, Boston, Massachusetts (De Jager); Harvard Medical School, Boston, Massachusetts (De Jager, Raj, Green); Program in Medical and Population Genetics, The Broad Institute, Cambridge, Massachusetts (De Jager); Department of Psychiatry, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Genetics and Genomics Sciences, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Neuroscience, Mount Sinai School of Medicine, New York, New York (Buxbaum); Friedman Brain Institute, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Medical and Molecular Genetics, Indiana University, Indianapolis (Murrell); Division of Genetics, Department of Medicine, Brigham and Women’s Hospital, Boston, Massachusetts (Green); Partners Center for Personalized Genetic Medicine, Brigham and Women’s Hospital, Boston, Massachusetts (Green); Department of Neurological Sciences, Rush University Medical Center, Chicago, Illinois (Barnes, Bennett); Department of Behavioral Sciences, Rush University Medical Center, Chicago, Illinois (Barnes, Foroud); Department of Epidemiology, Johns Hopkins University School of Public Health, Baltimore, Maryland (Fallin); School of Public Health, University of Alabama at Birmingham (Go); SABA University School of Medicine, SABA, Dutch Caribbean (Griffith); Division of Geriatrics, Howard University Hospital, Washington, DC (Obisesan); Department of Human Genetics, University of Pittsburgh, Pittsburgh, Pennsylvania (Kamboh); Alzheimer’s Disease Research Center, University of Pittsburgh, Pittsburgh, Pennsylvania (Kamboh, Lopez, Mayeux); Rush Alzheimer’s Disease Center, Rush University Medical Center, Chicago, Illinois (Bennett); Indiana University Center for Aging Research, Indianapolis (Hendrie); Department of Psychiatry, Indiana University School of Medicine, Indianapolis (Hendrie, Hall); Regenstrief Institute Inc, Indianapolis, Indiana (Hendrie); Hope Center Program on Protein Aggregation and Neurodegeneration, Department of Psychiatry, Washington University School of Medicine, St Louis, Missouri (Goate); Department of Biology, North Carolina A & T University, Greensboro (Byrd); National Alzheimer’s Coordinating Center, Department of Epidemiology, University of Washington, Seattle (Kukull); Vanderbilt Center for Human Genetics Research, Department of Molecular Physiology and Biophysics, Vanderbilt University, Nashville, Tennessee (Haines); Department of Neurology, Boston University, Boston, Massachusetts (Farrer); Department of Epidemiology, Boston University, Boston, Massachusetts (Farrer); Department of Epidemiology, College of Physicians and Surgeons, Columbia University, New York, New York (Lee); Department of Psychiatry, College of Physicians and Surgeons, Columbia University, New York, New York (Mayeux).
1,
Jill R Murrell
Jill R Murrell, PhD
1Tanz Centre for Research in Neurodegenerative Diseases, University of Toronto, Toronto, Ontario, Canada (Ghani, Sato, St. George-Hyslop, Rogaeva); Taub Institute for Research on Alzheimer’s Disease and the Aging Brain, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Cheng, Manly, Lee, Mayeux); Gertrude H. Sergievsky Center, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Vardarajan, Lee, Mayeux); Department of Neurology, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Manly, Mayeux); Department of Medicine (Biomedical Genetics), Boston University, Boston, Massachusetts (Jun, Logue, Baldwin, Farrer); Department of Biostatistics, Boston University, Boston, Massachusetts (Jun, Lunetta, Farrer); Department of Ophthalmology, Boston University, Boston, Massachusetts (Jun, Farrer); The John P. Hussman Institute for Human Genomics, University of Miami, Miami, Florida (Naj, Rajbhandary, Pericak-Vance); Department of Pathology and Laboratory Medicine, University of Pennsylvania Perelman School of Medicine, Philadelphia (Wang, Valladares, Lin, Cantwell, Schellenberg); Department of Medicine, University of Washington, Seattle (Larson, Crane); Group Health Research Institute, Group Health, Seattle, Washington (Larson); Department of Neuroscience, Mayo Clinic, Jacksonville, Florida (Graff-Radford, Ertekin-Taner); Department of Neurology, Mayo Clinic, Jacksonville, Florida (Graff-Radford, Ertekin-Taner); Rush Institute for Healthy Aging, Department of Internal Medicine, Rush University Medical Center, Chicago, Illinois (Evans); Program in Translational Neuropsychiatric Genomics, Department of Neurology, Brigham and Women’s Hospital, Boston, Massachusetts (De Jager); Harvard Medical School, Boston, Massachusetts (De Jager, Raj, Green); Program in Medical and Population Genetics, The Broad Institute, Cambridge, Massachusetts (De Jager); Department of Psychiatry, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Genetics and Genomics Sciences, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Neuroscience, Mount Sinai School of Medicine, New York, New York (Buxbaum); Friedman Brain Institute, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Medical and Molecular Genetics, Indiana University, Indianapolis (Murrell); Division of Genetics, Department of Medicine, Brigham and Women’s Hospital, Boston, Massachusetts (Green); Partners Center for Personalized Genetic Medicine, Brigham and Women’s Hospital, Boston, Massachusetts (Green); Department of Neurological Sciences, Rush University Medical Center, Chicago, Illinois (Barnes, Bennett); Department of Behavioral Sciences, Rush University Medical Center, Chicago, Illinois (Barnes, Foroud); Department of Epidemiology, Johns Hopkins University School of Public Health, Baltimore, Maryland (Fallin); School of Public Health, University of Alabama at Birmingham (Go); SABA University School of Medicine, SABA, Dutch Caribbean (Griffith); Division of Geriatrics, Howard University Hospital, Washington, DC (Obisesan); Department of Human Genetics, University of Pittsburgh, Pittsburgh, Pennsylvania (Kamboh); Alzheimer’s Disease Research Center, University of Pittsburgh, Pittsburgh, Pennsylvania (Kamboh, Lopez, Mayeux); Rush Alzheimer’s Disease Center, Rush University Medical Center, Chicago, Illinois (Bennett); Indiana University Center for Aging Research, Indianapolis (Hendrie); Department of Psychiatry, Indiana University School of Medicine, Indianapolis (Hendrie, Hall); Regenstrief Institute Inc, Indianapolis, Indiana (Hendrie); Hope Center Program on Protein Aggregation and Neurodegeneration, Department of Psychiatry, Washington University School of Medicine, St Louis, Missouri (Goate); Department of Biology, North Carolina A & T University, Greensboro (Byrd); National Alzheimer’s Coordinating Center, Department of Epidemiology, University of Washington, Seattle (Kukull); Vanderbilt Center for Human Genetics Research, Department of Molecular Physiology and Biophysics, Vanderbilt University, Nashville, Tennessee (Haines); Department of Neurology, Boston University, Boston, Massachusetts (Farrer); Department of Epidemiology, Boston University, Boston, Massachusetts (Farrer); Department of Epidemiology, College of Physicians and Surgeons, Columbia University, New York, New York (Lee); Department of Psychiatry, College of Physicians and Surgeons, Columbia University, New York, New York (Mayeux).
1,
Towfique Raj
Towfique Raj, PhD
1Tanz Centre for Research in Neurodegenerative Diseases, University of Toronto, Toronto, Ontario, Canada (Ghani, Sato, St. George-Hyslop, Rogaeva); Taub Institute for Research on Alzheimer’s Disease and the Aging Brain, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Cheng, Manly, Lee, Mayeux); Gertrude H. Sergievsky Center, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Vardarajan, Lee, Mayeux); Department of Neurology, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Manly, Mayeux); Department of Medicine (Biomedical Genetics), Boston University, Boston, Massachusetts (Jun, Logue, Baldwin, Farrer); Department of Biostatistics, Boston University, Boston, Massachusetts (Jun, Lunetta, Farrer); Department of Ophthalmology, Boston University, Boston, Massachusetts (Jun, Farrer); The John P. Hussman Institute for Human Genomics, University of Miami, Miami, Florida (Naj, Rajbhandary, Pericak-Vance); Department of Pathology and Laboratory Medicine, University of Pennsylvania Perelman School of Medicine, Philadelphia (Wang, Valladares, Lin, Cantwell, Schellenberg); Department of Medicine, University of Washington, Seattle (Larson, Crane); Group Health Research Institute, Group Health, Seattle, Washington (Larson); Department of Neuroscience, Mayo Clinic, Jacksonville, Florida (Graff-Radford, Ertekin-Taner); Department of Neurology, Mayo Clinic, Jacksonville, Florida (Graff-Radford, Ertekin-Taner); Rush Institute for Healthy Aging, Department of Internal Medicine, Rush University Medical Center, Chicago, Illinois (Evans); Program in Translational Neuropsychiatric Genomics, Department of Neurology, Brigham and Women’s Hospital, Boston, Massachusetts (De Jager); Harvard Medical School, Boston, Massachusetts (De Jager, Raj, Green); Program in Medical and Population Genetics, The Broad Institute, Cambridge, Massachusetts (De Jager); Department of Psychiatry, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Genetics and Genomics Sciences, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Neuroscience, Mount Sinai School of Medicine, New York, New York (Buxbaum); Friedman Brain Institute, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Medical and Molecular Genetics, Indiana University, Indianapolis (Murrell); Division of Genetics, Department of Medicine, Brigham and Women’s Hospital, Boston, Massachusetts (Green); Partners Center for Personalized Genetic Medicine, Brigham and Women’s Hospital, Boston, Massachusetts (Green); Department of Neurological Sciences, Rush University Medical Center, Chicago, Illinois (Barnes, Bennett); Department of Behavioral Sciences, Rush University Medical Center, Chicago, Illinois (Barnes, Foroud); Department of Epidemiology, Johns Hopkins University School of Public Health, Baltimore, Maryland (Fallin); School of Public Health, University of Alabama at Birmingham (Go); SABA University School of Medicine, SABA, Dutch Caribbean (Griffith); Division of Geriatrics, Howard University Hospital, Washington, DC (Obisesan); Department of Human Genetics, University of Pittsburgh, Pittsburgh, Pennsylvania (Kamboh); Alzheimer’s Disease Research Center, University of Pittsburgh, Pittsburgh, Pennsylvania (Kamboh, Lopez, Mayeux); Rush Alzheimer’s Disease Center, Rush University Medical Center, Chicago, Illinois (Bennett); Indiana University Center for Aging Research, Indianapolis (Hendrie); Department of Psychiatry, Indiana University School of Medicine, Indianapolis (Hendrie, Hall); Regenstrief Institute Inc, Indianapolis, Indiana (Hendrie); Hope Center Program on Protein Aggregation and Neurodegeneration, Department of Psychiatry, Washington University School of Medicine, St Louis, Missouri (Goate); Department of Biology, North Carolina A & T University, Greensboro (Byrd); National Alzheimer’s Coordinating Center, Department of Epidemiology, University of Washington, Seattle (Kukull); Vanderbilt Center for Human Genetics Research, Department of Molecular Physiology and Biophysics, Vanderbilt University, Nashville, Tennessee (Haines); Department of Neurology, Boston University, Boston, Massachusetts (Farrer); Department of Epidemiology, Boston University, Boston, Massachusetts (Farrer); Department of Epidemiology, College of Physicians and Surgeons, Columbia University, New York, New York (Lee); Department of Psychiatry, College of Physicians and Surgeons, Columbia University, New York, New York (Mayeux).
1,
Nilufer Ertekin-Taner
Nilufer Ertekin-Taner, MD, PhD
1Tanz Centre for Research in Neurodegenerative Diseases, University of Toronto, Toronto, Ontario, Canada (Ghani, Sato, St. George-Hyslop, Rogaeva); Taub Institute for Research on Alzheimer’s Disease and the Aging Brain, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Cheng, Manly, Lee, Mayeux); Gertrude H. Sergievsky Center, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Vardarajan, Lee, Mayeux); Department of Neurology, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Manly, Mayeux); Department of Medicine (Biomedical Genetics), Boston University, Boston, Massachusetts (Jun, Logue, Baldwin, Farrer); Department of Biostatistics, Boston University, Boston, Massachusetts (Jun, Lunetta, Farrer); Department of Ophthalmology, Boston University, Boston, Massachusetts (Jun, Farrer); The John P. Hussman Institute for Human Genomics, University of Miami, Miami, Florida (Naj, Rajbhandary, Pericak-Vance); Department of Pathology and Laboratory Medicine, University of Pennsylvania Perelman School of Medicine, Philadelphia (Wang, Valladares, Lin, Cantwell, Schellenberg); Department of Medicine, University of Washington, Seattle (Larson, Crane); Group Health Research Institute, Group Health, Seattle, Washington (Larson); Department of Neuroscience, Mayo Clinic, Jacksonville, Florida (Graff-Radford, Ertekin-Taner); Department of Neurology, Mayo Clinic, Jacksonville, Florida (Graff-Radford, Ertekin-Taner); Rush Institute for Healthy Aging, Department of Internal Medicine, Rush University Medical Center, Chicago, Illinois (Evans); Program in Translational Neuropsychiatric Genomics, Department of Neurology, Brigham and Women’s Hospital, Boston, Massachusetts (De Jager); Harvard Medical School, Boston, Massachusetts (De Jager, Raj, Green); Program in Medical and Population Genetics, The Broad Institute, Cambridge, Massachusetts (De Jager); Department of Psychiatry, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Genetics and Genomics Sciences, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Neuroscience, Mount Sinai School of Medicine, New York, New York (Buxbaum); Friedman Brain Institute, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Medical and Molecular Genetics, Indiana University, Indianapolis (Murrell); Division of Genetics, Department of Medicine, Brigham and Women’s Hospital, Boston, Massachusetts (Green); Partners Center for Personalized Genetic Medicine, Brigham and Women’s Hospital, Boston, Massachusetts (Green); Department of Neurological Sciences, Rush University Medical Center, Chicago, Illinois (Barnes, Bennett); Department of Behavioral Sciences, Rush University Medical Center, Chicago, Illinois (Barnes, Foroud); Department of Epidemiology, Johns Hopkins University School of Public Health, Baltimore, Maryland (Fallin); School of Public Health, University of Alabama at Birmingham (Go); SABA University School of Medicine, SABA, Dutch Caribbean (Griffith); Division of Geriatrics, Howard University Hospital, Washington, DC (Obisesan); Department of Human Genetics, University of Pittsburgh, Pittsburgh, Pennsylvania (Kamboh); Alzheimer’s Disease Research Center, University of Pittsburgh, Pittsburgh, Pennsylvania (Kamboh, Lopez, Mayeux); Rush Alzheimer’s Disease Center, Rush University Medical Center, Chicago, Illinois (Bennett); Indiana University Center for Aging Research, Indianapolis (Hendrie); Department of Psychiatry, Indiana University School of Medicine, Indianapolis (Hendrie, Hall); Regenstrief Institute Inc, Indianapolis, Indiana (Hendrie); Hope Center Program on Protein Aggregation and Neurodegeneration, Department of Psychiatry, Washington University School of Medicine, St Louis, Missouri (Goate); Department of Biology, North Carolina A & T University, Greensboro (Byrd); National Alzheimer’s Coordinating Center, Department of Epidemiology, University of Washington, Seattle (Kukull); Vanderbilt Center for Human Genetics Research, Department of Molecular Physiology and Biophysics, Vanderbilt University, Nashville, Tennessee (Haines); Department of Neurology, Boston University, Boston, Massachusetts (Farrer); Department of Epidemiology, Boston University, Boston, Massachusetts (Farrer); Department of Epidemiology, College of Physicians and Surgeons, Columbia University, New York, New York (Lee); Department of Psychiatry, College of Physicians and Surgeons, Columbia University, New York, New York (Mayeux).
1,
Mark Logue
Mark Logue, PhD
1Tanz Centre for Research in Neurodegenerative Diseases, University of Toronto, Toronto, Ontario, Canada (Ghani, Sato, St. George-Hyslop, Rogaeva); Taub Institute for Research on Alzheimer’s Disease and the Aging Brain, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Cheng, Manly, Lee, Mayeux); Gertrude H. Sergievsky Center, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Vardarajan, Lee, Mayeux); Department of Neurology, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Manly, Mayeux); Department of Medicine (Biomedical Genetics), Boston University, Boston, Massachusetts (Jun, Logue, Baldwin, Farrer); Department of Biostatistics, Boston University, Boston, Massachusetts (Jun, Lunetta, Farrer); Department of Ophthalmology, Boston University, Boston, Massachusetts (Jun, Farrer); The John P. Hussman Institute for Human Genomics, University of Miami, Miami, Florida (Naj, Rajbhandary, Pericak-Vance); Department of Pathology and Laboratory Medicine, University of Pennsylvania Perelman School of Medicine, Philadelphia (Wang, Valladares, Lin, Cantwell, Schellenberg); Department of Medicine, University of Washington, Seattle (Larson, Crane); Group Health Research Institute, Group Health, Seattle, Washington (Larson); Department of Neuroscience, Mayo Clinic, Jacksonville, Florida (Graff-Radford, Ertekin-Taner); Department of Neurology, Mayo Clinic, Jacksonville, Florida (Graff-Radford, Ertekin-Taner); Rush Institute for Healthy Aging, Department of Internal Medicine, Rush University Medical Center, Chicago, Illinois (Evans); Program in Translational Neuropsychiatric Genomics, Department of Neurology, Brigham and Women’s Hospital, Boston, Massachusetts (De Jager); Harvard Medical School, Boston, Massachusetts (De Jager, Raj, Green); Program in Medical and Population Genetics, The Broad Institute, Cambridge, Massachusetts (De Jager); Department of Psychiatry, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Genetics and Genomics Sciences, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Neuroscience, Mount Sinai School of Medicine, New York, New York (Buxbaum); Friedman Brain Institute, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Medical and Molecular Genetics, Indiana University, Indianapolis (Murrell); Division of Genetics, Department of Medicine, Brigham and Women’s Hospital, Boston, Massachusetts (Green); Partners Center for Personalized Genetic Medicine, Brigham and Women’s Hospital, Boston, Massachusetts (Green); Department of Neurological Sciences, Rush University Medical Center, Chicago, Illinois (Barnes, Bennett); Department of Behavioral Sciences, Rush University Medical Center, Chicago, Illinois (Barnes, Foroud); Department of Epidemiology, Johns Hopkins University School of Public Health, Baltimore, Maryland (Fallin); School of Public Health, University of Alabama at Birmingham (Go); SABA University School of Medicine, SABA, Dutch Caribbean (Griffith); Division of Geriatrics, Howard University Hospital, Washington, DC (Obisesan); Department of Human Genetics, University of Pittsburgh, Pittsburgh, Pennsylvania (Kamboh); Alzheimer’s Disease Research Center, University of Pittsburgh, Pittsburgh, Pennsylvania (Kamboh, Lopez, Mayeux); Rush Alzheimer’s Disease Center, Rush University Medical Center, Chicago, Illinois (Bennett); Indiana University Center for Aging Research, Indianapolis (Hendrie); Department of Psychiatry, Indiana University School of Medicine, Indianapolis (Hendrie, Hall); Regenstrief Institute Inc, Indianapolis, Indiana (Hendrie); Hope Center Program on Protein Aggregation and Neurodegeneration, Department of Psychiatry, Washington University School of Medicine, St Louis, Missouri (Goate); Department of Biology, North Carolina A & T University, Greensboro (Byrd); National Alzheimer’s Coordinating Center, Department of Epidemiology, University of Washington, Seattle (Kukull); Vanderbilt Center for Human Genetics Research, Department of Molecular Physiology and Biophysics, Vanderbilt University, Nashville, Tennessee (Haines); Department of Neurology, Boston University, Boston, Massachusetts (Farrer); Department of Epidemiology, Boston University, Boston, Massachusetts (Farrer); Department of Epidemiology, College of Physicians and Surgeons, Columbia University, New York, New York (Lee); Department of Psychiatry, College of Physicians and Surgeons, Columbia University, New York, New York (Mayeux).
1,
Clinton T Baldwin
Clinton T Baldwin, PhD
1Tanz Centre for Research in Neurodegenerative Diseases, University of Toronto, Toronto, Ontario, Canada (Ghani, Sato, St. George-Hyslop, Rogaeva); Taub Institute for Research on Alzheimer’s Disease and the Aging Brain, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Cheng, Manly, Lee, Mayeux); Gertrude H. Sergievsky Center, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Vardarajan, Lee, Mayeux); Department of Neurology, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Manly, Mayeux); Department of Medicine (Biomedical Genetics), Boston University, Boston, Massachusetts (Jun, Logue, Baldwin, Farrer); Department of Biostatistics, Boston University, Boston, Massachusetts (Jun, Lunetta, Farrer); Department of Ophthalmology, Boston University, Boston, Massachusetts (Jun, Farrer); The John P. Hussman Institute for Human Genomics, University of Miami, Miami, Florida (Naj, Rajbhandary, Pericak-Vance); Department of Pathology and Laboratory Medicine, University of Pennsylvania Perelman School of Medicine, Philadelphia (Wang, Valladares, Lin, Cantwell, Schellenberg); Department of Medicine, University of Washington, Seattle (Larson, Crane); Group Health Research Institute, Group Health, Seattle, Washington (Larson); Department of Neuroscience, Mayo Clinic, Jacksonville, Florida (Graff-Radford, Ertekin-Taner); Department of Neurology, Mayo Clinic, Jacksonville, Florida (Graff-Radford, Ertekin-Taner); Rush Institute for Healthy Aging, Department of Internal Medicine, Rush University Medical Center, Chicago, Illinois (Evans); Program in Translational Neuropsychiatric Genomics, Department of Neurology, Brigham and Women’s Hospital, Boston, Massachusetts (De Jager); Harvard Medical School, Boston, Massachusetts (De Jager, Raj, Green); Program in Medical and Population Genetics, The Broad Institute, Cambridge, Massachusetts (De Jager); Department of Psychiatry, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Genetics and Genomics Sciences, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Neuroscience, Mount Sinai School of Medicine, New York, New York (Buxbaum); Friedman Brain Institute, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Medical and Molecular Genetics, Indiana University, Indianapolis (Murrell); Division of Genetics, Department of Medicine, Brigham and Women’s Hospital, Boston, Massachusetts (Green); Partners Center for Personalized Genetic Medicine, Brigham and Women’s Hospital, Boston, Massachusetts (Green); Department of Neurological Sciences, Rush University Medical Center, Chicago, Illinois (Barnes, Bennett); Department of Behavioral Sciences, Rush University Medical Center, Chicago, Illinois (Barnes, Foroud); Department of Epidemiology, Johns Hopkins University School of Public Health, Baltimore, Maryland (Fallin); School of Public Health, University of Alabama at Birmingham (Go); SABA University School of Medicine, SABA, Dutch Caribbean (Griffith); Division of Geriatrics, Howard University Hospital, Washington, DC (Obisesan); Department of Human Genetics, University of Pittsburgh, Pittsburgh, Pennsylvania (Kamboh); Alzheimer’s Disease Research Center, University of Pittsburgh, Pittsburgh, Pennsylvania (Kamboh, Lopez, Mayeux); Rush Alzheimer’s Disease Center, Rush University Medical Center, Chicago, Illinois (Bennett); Indiana University Center for Aging Research, Indianapolis (Hendrie); Department of Psychiatry, Indiana University School of Medicine, Indianapolis (Hendrie, Hall); Regenstrief Institute Inc, Indianapolis, Indiana (Hendrie); Hope Center Program on Protein Aggregation and Neurodegeneration, Department of Psychiatry, Washington University School of Medicine, St Louis, Missouri (Goate); Department of Biology, North Carolina A & T University, Greensboro (Byrd); National Alzheimer’s Coordinating Center, Department of Epidemiology, University of Washington, Seattle (Kukull); Vanderbilt Center for Human Genetics Research, Department of Molecular Physiology and Biophysics, Vanderbilt University, Nashville, Tennessee (Haines); Department of Neurology, Boston University, Boston, Massachusetts (Farrer); Department of Epidemiology, Boston University, Boston, Massachusetts (Farrer); Department of Epidemiology, College of Physicians and Surgeons, Columbia University, New York, New York (Lee); Department of Psychiatry, College of Physicians and Surgeons, Columbia University, New York, New York (Mayeux).
1,
Robert C Green
Robert C Green, MD, MPH
1Tanz Centre for Research in Neurodegenerative Diseases, University of Toronto, Toronto, Ontario, Canada (Ghani, Sato, St. George-Hyslop, Rogaeva); Taub Institute for Research on Alzheimer’s Disease and the Aging Brain, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Cheng, Manly, Lee, Mayeux); Gertrude H. Sergievsky Center, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Vardarajan, Lee, Mayeux); Department of Neurology, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Manly, Mayeux); Department of Medicine (Biomedical Genetics), Boston University, Boston, Massachusetts (Jun, Logue, Baldwin, Farrer); Department of Biostatistics, Boston University, Boston, Massachusetts (Jun, Lunetta, Farrer); Department of Ophthalmology, Boston University, Boston, Massachusetts (Jun, Farrer); The John P. Hussman Institute for Human Genomics, University of Miami, Miami, Florida (Naj, Rajbhandary, Pericak-Vance); Department of Pathology and Laboratory Medicine, University of Pennsylvania Perelman School of Medicine, Philadelphia (Wang, Valladares, Lin, Cantwell, Schellenberg); Department of Medicine, University of Washington, Seattle (Larson, Crane); Group Health Research Institute, Group Health, Seattle, Washington (Larson); Department of Neuroscience, Mayo Clinic, Jacksonville, Florida (Graff-Radford, Ertekin-Taner); Department of Neurology, Mayo Clinic, Jacksonville, Florida (Graff-Radford, Ertekin-Taner); Rush Institute for Healthy Aging, Department of Internal Medicine, Rush University Medical Center, Chicago, Illinois (Evans); Program in Translational Neuropsychiatric Genomics, Department of Neurology, Brigham and Women’s Hospital, Boston, Massachusetts (De Jager); Harvard Medical School, Boston, Massachusetts (De Jager, Raj, Green); Program in Medical and Population Genetics, The Broad Institute, Cambridge, Massachusetts (De Jager); Department of Psychiatry, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Genetics and Genomics Sciences, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Neuroscience, Mount Sinai School of Medicine, New York, New York (Buxbaum); Friedman Brain Institute, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Medical and Molecular Genetics, Indiana University, Indianapolis (Murrell); Division of Genetics, Department of Medicine, Brigham and Women’s Hospital, Boston, Massachusetts (Green); Partners Center for Personalized Genetic Medicine, Brigham and Women’s Hospital, Boston, Massachusetts (Green); Department of Neurological Sciences, Rush University Medical Center, Chicago, Illinois (Barnes, Bennett); Department of Behavioral Sciences, Rush University Medical Center, Chicago, Illinois (Barnes, Foroud); Department of Epidemiology, Johns Hopkins University School of Public Health, Baltimore, Maryland (Fallin); School of Public Health, University of Alabama at Birmingham (Go); SABA University School of Medicine, SABA, Dutch Caribbean (Griffith); Division of Geriatrics, Howard University Hospital, Washington, DC (Obisesan); Department of Human Genetics, University of Pittsburgh, Pittsburgh, Pennsylvania (Kamboh); Alzheimer’s Disease Research Center, University of Pittsburgh, Pittsburgh, Pennsylvania (Kamboh, Lopez, Mayeux); Rush Alzheimer’s Disease Center, Rush University Medical Center, Chicago, Illinois (Bennett); Indiana University Center for Aging Research, Indianapolis (Hendrie); Department of Psychiatry, Indiana University School of Medicine, Indianapolis (Hendrie, Hall); Regenstrief Institute Inc, Indianapolis, Indiana (Hendrie); Hope Center Program on Protein Aggregation and Neurodegeneration, Department of Psychiatry, Washington University School of Medicine, St Louis, Missouri (Goate); Department of Biology, North Carolina A & T University, Greensboro (Byrd); National Alzheimer’s Coordinating Center, Department of Epidemiology, University of Washington, Seattle (Kukull); Vanderbilt Center for Human Genetics Research, Department of Molecular Physiology and Biophysics, Vanderbilt University, Nashville, Tennessee (Haines); Department of Neurology, Boston University, Boston, Massachusetts (Farrer); Department of Epidemiology, Boston University, Boston, Massachusetts (Farrer); Department of Epidemiology, College of Physicians and Surgeons, Columbia University, New York, New York (Lee); Department of Psychiatry, College of Physicians and Surgeons, Columbia University, New York, New York (Mayeux).
1,
Lisa L Barnes
Lisa L Barnes, PhD
1Tanz Centre for Research in Neurodegenerative Diseases, University of Toronto, Toronto, Ontario, Canada (Ghani, Sato, St. George-Hyslop, Rogaeva); Taub Institute for Research on Alzheimer’s Disease and the Aging Brain, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Cheng, Manly, Lee, Mayeux); Gertrude H. Sergievsky Center, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Vardarajan, Lee, Mayeux); Department of Neurology, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Manly, Mayeux); Department of Medicine (Biomedical Genetics), Boston University, Boston, Massachusetts (Jun, Logue, Baldwin, Farrer); Department of Biostatistics, Boston University, Boston, Massachusetts (Jun, Lunetta, Farrer); Department of Ophthalmology, Boston University, Boston, Massachusetts (Jun, Farrer); The John P. Hussman Institute for Human Genomics, University of Miami, Miami, Florida (Naj, Rajbhandary, Pericak-Vance); Department of Pathology and Laboratory Medicine, University of Pennsylvania Perelman School of Medicine, Philadelphia (Wang, Valladares, Lin, Cantwell, Schellenberg); Department of Medicine, University of Washington, Seattle (Larson, Crane); Group Health Research Institute, Group Health, Seattle, Washington (Larson); Department of Neuroscience, Mayo Clinic, Jacksonville, Florida (Graff-Radford, Ertekin-Taner); Department of Neurology, Mayo Clinic, Jacksonville, Florida (Graff-Radford, Ertekin-Taner); Rush Institute for Healthy Aging, Department of Internal Medicine, Rush University Medical Center, Chicago, Illinois (Evans); Program in Translational Neuropsychiatric Genomics, Department of Neurology, Brigham and Women’s Hospital, Boston, Massachusetts (De Jager); Harvard Medical School, Boston, Massachusetts (De Jager, Raj, Green); Program in Medical and Population Genetics, The Broad Institute, Cambridge, Massachusetts (De Jager); Department of Psychiatry, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Genetics and Genomics Sciences, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Neuroscience, Mount Sinai School of Medicine, New York, New York (Buxbaum); Friedman Brain Institute, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Medical and Molecular Genetics, Indiana University, Indianapolis (Murrell); Division of Genetics, Department of Medicine, Brigham and Women’s Hospital, Boston, Massachusetts (Green); Partners Center for Personalized Genetic Medicine, Brigham and Women’s Hospital, Boston, Massachusetts (Green); Department of Neurological Sciences, Rush University Medical Center, Chicago, Illinois (Barnes, Bennett); Department of Behavioral Sciences, Rush University Medical Center, Chicago, Illinois (Barnes, Foroud); Department of Epidemiology, Johns Hopkins University School of Public Health, Baltimore, Maryland (Fallin); School of Public Health, University of Alabama at Birmingham (Go); SABA University School of Medicine, SABA, Dutch Caribbean (Griffith); Division of Geriatrics, Howard University Hospital, Washington, DC (Obisesan); Department of Human Genetics, University of Pittsburgh, Pittsburgh, Pennsylvania (Kamboh); Alzheimer’s Disease Research Center, University of Pittsburgh, Pittsburgh, Pennsylvania (Kamboh, Lopez, Mayeux); Rush Alzheimer’s Disease Center, Rush University Medical Center, Chicago, Illinois (Bennett); Indiana University Center for Aging Research, Indianapolis (Hendrie); Department of Psychiatry, Indiana University School of Medicine, Indianapolis (Hendrie, Hall); Regenstrief Institute Inc, Indianapolis, Indiana (Hendrie); Hope Center Program on Protein Aggregation and Neurodegeneration, Department of Psychiatry, Washington University School of Medicine, St Louis, Missouri (Goate); Department of Biology, North Carolina A & T University, Greensboro (Byrd); National Alzheimer’s Coordinating Center, Department of Epidemiology, University of Washington, Seattle (Kukull); Vanderbilt Center for Human Genetics Research, Department of Molecular Physiology and Biophysics, Vanderbilt University, Nashville, Tennessee (Haines); Department of Neurology, Boston University, Boston, Massachusetts (Farrer); Department of Epidemiology, Boston University, Boston, Massachusetts (Farrer); Department of Epidemiology, College of Physicians and Surgeons, Columbia University, New York, New York (Lee); Department of Psychiatry, College of Physicians and Surgeons, Columbia University, New York, New York (Mayeux).
1,
Laura B Cantwell
Laura B Cantwell, MPH
1Tanz Centre for Research in Neurodegenerative Diseases, University of Toronto, Toronto, Ontario, Canada (Ghani, Sato, St. George-Hyslop, Rogaeva); Taub Institute for Research on Alzheimer’s Disease and the Aging Brain, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Cheng, Manly, Lee, Mayeux); Gertrude H. Sergievsky Center, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Vardarajan, Lee, Mayeux); Department of Neurology, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Manly, Mayeux); Department of Medicine (Biomedical Genetics), Boston University, Boston, Massachusetts (Jun, Logue, Baldwin, Farrer); Department of Biostatistics, Boston University, Boston, Massachusetts (Jun, Lunetta, Farrer); Department of Ophthalmology, Boston University, Boston, Massachusetts (Jun, Farrer); The John P. Hussman Institute for Human Genomics, University of Miami, Miami, Florida (Naj, Rajbhandary, Pericak-Vance); Department of Pathology and Laboratory Medicine, University of Pennsylvania Perelman School of Medicine, Philadelphia (Wang, Valladares, Lin, Cantwell, Schellenberg); Department of Medicine, University of Washington, Seattle (Larson, Crane); Group Health Research Institute, Group Health, Seattle, Washington (Larson); Department of Neuroscience, Mayo Clinic, Jacksonville, Florida (Graff-Radford, Ertekin-Taner); Department of Neurology, Mayo Clinic, Jacksonville, Florida (Graff-Radford, Ertekin-Taner); Rush Institute for Healthy Aging, Department of Internal Medicine, Rush University Medical Center, Chicago, Illinois (Evans); Program in Translational Neuropsychiatric Genomics, Department of Neurology, Brigham and Women’s Hospital, Boston, Massachusetts (De Jager); Harvard Medical School, Boston, Massachusetts (De Jager, Raj, Green); Program in Medical and Population Genetics, The Broad Institute, Cambridge, Massachusetts (De Jager); Department of Psychiatry, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Genetics and Genomics Sciences, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Neuroscience, Mount Sinai School of Medicine, New York, New York (Buxbaum); Friedman Brain Institute, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Medical and Molecular Genetics, Indiana University, Indianapolis (Murrell); Division of Genetics, Department of Medicine, Brigham and Women’s Hospital, Boston, Massachusetts (Green); Partners Center for Personalized Genetic Medicine, Brigham and Women’s Hospital, Boston, Massachusetts (Green); Department of Neurological Sciences, Rush University Medical Center, Chicago, Illinois (Barnes, Bennett); Department of Behavioral Sciences, Rush University Medical Center, Chicago, Illinois (Barnes, Foroud); Department of Epidemiology, Johns Hopkins University School of Public Health, Baltimore, Maryland (Fallin); School of Public Health, University of Alabama at Birmingham (Go); SABA University School of Medicine, SABA, Dutch Caribbean (Griffith); Division of Geriatrics, Howard University Hospital, Washington, DC (Obisesan); Department of Human Genetics, University of Pittsburgh, Pittsburgh, Pennsylvania (Kamboh); Alzheimer’s Disease Research Center, University of Pittsburgh, Pittsburgh, Pennsylvania (Kamboh, Lopez, Mayeux); Rush Alzheimer’s Disease Center, Rush University Medical Center, Chicago, Illinois (Bennett); Indiana University Center for Aging Research, Indianapolis (Hendrie); Department of Psychiatry, Indiana University School of Medicine, Indianapolis (Hendrie, Hall); Regenstrief Institute Inc, Indianapolis, Indiana (Hendrie); Hope Center Program on Protein Aggregation and Neurodegeneration, Department of Psychiatry, Washington University School of Medicine, St Louis, Missouri (Goate); Department of Biology, North Carolina A & T University, Greensboro (Byrd); National Alzheimer’s Coordinating Center, Department of Epidemiology, University of Washington, Seattle (Kukull); Vanderbilt Center for Human Genetics Research, Department of Molecular Physiology and Biophysics, Vanderbilt University, Nashville, Tennessee (Haines); Department of Neurology, Boston University, Boston, Massachusetts (Farrer); Department of Epidemiology, Boston University, Boston, Massachusetts (Farrer); Department of Epidemiology, College of Physicians and Surgeons, Columbia University, New York, New York (Lee); Department of Psychiatry, College of Physicians and Surgeons, Columbia University, New York, New York (Mayeux).
1,
M Daniele Fallin
M Daniele Fallin, PhD
1Tanz Centre for Research in Neurodegenerative Diseases, University of Toronto, Toronto, Ontario, Canada (Ghani, Sato, St. George-Hyslop, Rogaeva); Taub Institute for Research on Alzheimer’s Disease and the Aging Brain, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Cheng, Manly, Lee, Mayeux); Gertrude H. Sergievsky Center, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Vardarajan, Lee, Mayeux); Department of Neurology, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Manly, Mayeux); Department of Medicine (Biomedical Genetics), Boston University, Boston, Massachusetts (Jun, Logue, Baldwin, Farrer); Department of Biostatistics, Boston University, Boston, Massachusetts (Jun, Lunetta, Farrer); Department of Ophthalmology, Boston University, Boston, Massachusetts (Jun, Farrer); The John P. Hussman Institute for Human Genomics, University of Miami, Miami, Florida (Naj, Rajbhandary, Pericak-Vance); Department of Pathology and Laboratory Medicine, University of Pennsylvania Perelman School of Medicine, Philadelphia (Wang, Valladares, Lin, Cantwell, Schellenberg); Department of Medicine, University of Washington, Seattle (Larson, Crane); Group Health Research Institute, Group Health, Seattle, Washington (Larson); Department of Neuroscience, Mayo Clinic, Jacksonville, Florida (Graff-Radford, Ertekin-Taner); Department of Neurology, Mayo Clinic, Jacksonville, Florida (Graff-Radford, Ertekin-Taner); Rush Institute for Healthy Aging, Department of Internal Medicine, Rush University Medical Center, Chicago, Illinois (Evans); Program in Translational Neuropsychiatric Genomics, Department of Neurology, Brigham and Women’s Hospital, Boston, Massachusetts (De Jager); Harvard Medical School, Boston, Massachusetts (De Jager, Raj, Green); Program in Medical and Population Genetics, The Broad Institute, Cambridge, Massachusetts (De Jager); Department of Psychiatry, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Genetics and Genomics Sciences, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Neuroscience, Mount Sinai School of Medicine, New York, New York (Buxbaum); Friedman Brain Institute, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Medical and Molecular Genetics, Indiana University, Indianapolis (Murrell); Division of Genetics, Department of Medicine, Brigham and Women’s Hospital, Boston, Massachusetts (Green); Partners Center for Personalized Genetic Medicine, Brigham and Women’s Hospital, Boston, Massachusetts (Green); Department of Neurological Sciences, Rush University Medical Center, Chicago, Illinois (Barnes, Bennett); Department of Behavioral Sciences, Rush University Medical Center, Chicago, Illinois (Barnes, Foroud); Department of Epidemiology, Johns Hopkins University School of Public Health, Baltimore, Maryland (Fallin); School of Public Health, University of Alabama at Birmingham (Go); SABA University School of Medicine, SABA, Dutch Caribbean (Griffith); Division of Geriatrics, Howard University Hospital, Washington, DC (Obisesan); Department of Human Genetics, University of Pittsburgh, Pittsburgh, Pennsylvania (Kamboh); Alzheimer’s Disease Research Center, University of Pittsburgh, Pittsburgh, Pennsylvania (Kamboh, Lopez, Mayeux); Rush Alzheimer’s Disease Center, Rush University Medical Center, Chicago, Illinois (Bennett); Indiana University Center for Aging Research, Indianapolis (Hendrie); Department of Psychiatry, Indiana University School of Medicine, Indianapolis (Hendrie, Hall); Regenstrief Institute Inc, Indianapolis, Indiana (Hendrie); Hope Center Program on Protein Aggregation and Neurodegeneration, Department of Psychiatry, Washington University School of Medicine, St Louis, Missouri (Goate); Department of Biology, North Carolina A & T University, Greensboro (Byrd); National Alzheimer’s Coordinating Center, Department of Epidemiology, University of Washington, Seattle (Kukull); Vanderbilt Center for Human Genetics Research, Department of Molecular Physiology and Biophysics, Vanderbilt University, Nashville, Tennessee (Haines); Department of Neurology, Boston University, Boston, Massachusetts (Farrer); Department of Epidemiology, Boston University, Boston, Massachusetts (Farrer); Department of Epidemiology, College of Physicians and Surgeons, Columbia University, New York, New York (Lee); Department of Psychiatry, College of Physicians and Surgeons, Columbia University, New York, New York (Mayeux).
1,
Rodney C P Go
Rodney C P Go, PhD
1Tanz Centre for Research in Neurodegenerative Diseases, University of Toronto, Toronto, Ontario, Canada (Ghani, Sato, St. George-Hyslop, Rogaeva); Taub Institute for Research on Alzheimer’s Disease and the Aging Brain, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Cheng, Manly, Lee, Mayeux); Gertrude H. Sergievsky Center, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Vardarajan, Lee, Mayeux); Department of Neurology, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Manly, Mayeux); Department of Medicine (Biomedical Genetics), Boston University, Boston, Massachusetts (Jun, Logue, Baldwin, Farrer); Department of Biostatistics, Boston University, Boston, Massachusetts (Jun, Lunetta, Farrer); Department of Ophthalmology, Boston University, Boston, Massachusetts (Jun, Farrer); The John P. Hussman Institute for Human Genomics, University of Miami, Miami, Florida (Naj, Rajbhandary, Pericak-Vance); Department of Pathology and Laboratory Medicine, University of Pennsylvania Perelman School of Medicine, Philadelphia (Wang, Valladares, Lin, Cantwell, Schellenberg); Department of Medicine, University of Washington, Seattle (Larson, Crane); Group Health Research Institute, Group Health, Seattle, Washington (Larson); Department of Neuroscience, Mayo Clinic, Jacksonville, Florida (Graff-Radford, Ertekin-Taner); Department of Neurology, Mayo Clinic, Jacksonville, Florida (Graff-Radford, Ertekin-Taner); Rush Institute for Healthy Aging, Department of Internal Medicine, Rush University Medical Center, Chicago, Illinois (Evans); Program in Translational Neuropsychiatric Genomics, Department of Neurology, Brigham and Women’s Hospital, Boston, Massachusetts (De Jager); Harvard Medical School, Boston, Massachusetts (De Jager, Raj, Green); Program in Medical and Population Genetics, The Broad Institute, Cambridge, Massachusetts (De Jager); Department of Psychiatry, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Genetics and Genomics Sciences, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Neuroscience, Mount Sinai School of Medicine, New York, New York (Buxbaum); Friedman Brain Institute, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Medical and Molecular Genetics, Indiana University, Indianapolis (Murrell); Division of Genetics, Department of Medicine, Brigham and Women’s Hospital, Boston, Massachusetts (Green); Partners Center for Personalized Genetic Medicine, Brigham and Women’s Hospital, Boston, Massachusetts (Green); Department of Neurological Sciences, Rush University Medical Center, Chicago, Illinois (Barnes, Bennett); Department of Behavioral Sciences, Rush University Medical Center, Chicago, Illinois (Barnes, Foroud); Department of Epidemiology, Johns Hopkins University School of Public Health, Baltimore, Maryland (Fallin); School of Public Health, University of Alabama at Birmingham (Go); SABA University School of Medicine, SABA, Dutch Caribbean (Griffith); Division of Geriatrics, Howard University Hospital, Washington, DC (Obisesan); Department of Human Genetics, University of Pittsburgh, Pittsburgh, Pennsylvania (Kamboh); Alzheimer’s Disease Research Center, University of Pittsburgh, Pittsburgh, Pennsylvania (Kamboh, Lopez, Mayeux); Rush Alzheimer’s Disease Center, Rush University Medical Center, Chicago, Illinois (Bennett); Indiana University Center for Aging Research, Indianapolis (Hendrie); Department of Psychiatry, Indiana University School of Medicine, Indianapolis (Hendrie, Hall); Regenstrief Institute Inc, Indianapolis, Indiana (Hendrie); Hope Center Program on Protein Aggregation and Neurodegeneration, Department of Psychiatry, Washington University School of Medicine, St Louis, Missouri (Goate); Department of Biology, North Carolina A & T University, Greensboro (Byrd); National Alzheimer’s Coordinating Center, Department of Epidemiology, University of Washington, Seattle (Kukull); Vanderbilt Center for Human Genetics Research, Department of Molecular Physiology and Biophysics, Vanderbilt University, Nashville, Tennessee (Haines); Department of Neurology, Boston University, Boston, Massachusetts (Farrer); Department of Epidemiology, Boston University, Boston, Massachusetts (Farrer); Department of Epidemiology, College of Physicians and Surgeons, Columbia University, New York, New York (Lee); Department of Psychiatry, College of Physicians and Surgeons, Columbia University, New York, New York (Mayeux).
1,
Patrick A Griffith
Patrick A Griffith, MD
1Tanz Centre for Research in Neurodegenerative Diseases, University of Toronto, Toronto, Ontario, Canada (Ghani, Sato, St. George-Hyslop, Rogaeva); Taub Institute for Research on Alzheimer’s Disease and the Aging Brain, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Cheng, Manly, Lee, Mayeux); Gertrude H. Sergievsky Center, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Vardarajan, Lee, Mayeux); Department of Neurology, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Manly, Mayeux); Department of Medicine (Biomedical Genetics), Boston University, Boston, Massachusetts (Jun, Logue, Baldwin, Farrer); Department of Biostatistics, Boston University, Boston, Massachusetts (Jun, Lunetta, Farrer); Department of Ophthalmology, Boston University, Boston, Massachusetts (Jun, Farrer); The John P. Hussman Institute for Human Genomics, University of Miami, Miami, Florida (Naj, Rajbhandary, Pericak-Vance); Department of Pathology and Laboratory Medicine, University of Pennsylvania Perelman School of Medicine, Philadelphia (Wang, Valladares, Lin, Cantwell, Schellenberg); Department of Medicine, University of Washington, Seattle (Larson, Crane); Group Health Research Institute, Group Health, Seattle, Washington (Larson); Department of Neuroscience, Mayo Clinic, Jacksonville, Florida (Graff-Radford, Ertekin-Taner); Department of Neurology, Mayo Clinic, Jacksonville, Florida (Graff-Radford, Ertekin-Taner); Rush Institute for Healthy Aging, Department of Internal Medicine, Rush University Medical Center, Chicago, Illinois (Evans); Program in Translational Neuropsychiatric Genomics, Department of Neurology, Brigham and Women’s Hospital, Boston, Massachusetts (De Jager); Harvard Medical School, Boston, Massachusetts (De Jager, Raj, Green); Program in Medical and Population Genetics, The Broad Institute, Cambridge, Massachusetts (De Jager); Department of Psychiatry, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Genetics and Genomics Sciences, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Neuroscience, Mount Sinai School of Medicine, New York, New York (Buxbaum); Friedman Brain Institute, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Medical and Molecular Genetics, Indiana University, Indianapolis (Murrell); Division of Genetics, Department of Medicine, Brigham and Women’s Hospital, Boston, Massachusetts (Green); Partners Center for Personalized Genetic Medicine, Brigham and Women’s Hospital, Boston, Massachusetts (Green); Department of Neurological Sciences, Rush University Medical Center, Chicago, Illinois (Barnes, Bennett); Department of Behavioral Sciences, Rush University Medical Center, Chicago, Illinois (Barnes, Foroud); Department of Epidemiology, Johns Hopkins University School of Public Health, Baltimore, Maryland (Fallin); School of Public Health, University of Alabama at Birmingham (Go); SABA University School of Medicine, SABA, Dutch Caribbean (Griffith); Division of Geriatrics, Howard University Hospital, Washington, DC (Obisesan); Department of Human Genetics, University of Pittsburgh, Pittsburgh, Pennsylvania (Kamboh); Alzheimer’s Disease Research Center, University of Pittsburgh, Pittsburgh, Pennsylvania (Kamboh, Lopez, Mayeux); Rush Alzheimer’s Disease Center, Rush University Medical Center, Chicago, Illinois (Bennett); Indiana University Center for Aging Research, Indianapolis (Hendrie); Department of Psychiatry, Indiana University School of Medicine, Indianapolis (Hendrie, Hall); Regenstrief Institute Inc, Indianapolis, Indiana (Hendrie); Hope Center Program on Protein Aggregation and Neurodegeneration, Department of Psychiatry, Washington University School of Medicine, St Louis, Missouri (Goate); Department of Biology, North Carolina A & T University, Greensboro (Byrd); National Alzheimer’s Coordinating Center, Department of Epidemiology, University of Washington, Seattle (Kukull); Vanderbilt Center for Human Genetics Research, Department of Molecular Physiology and Biophysics, Vanderbilt University, Nashville, Tennessee (Haines); Department of Neurology, Boston University, Boston, Massachusetts (Farrer); Department of Epidemiology, Boston University, Boston, Massachusetts (Farrer); Department of Epidemiology, College of Physicians and Surgeons, Columbia University, New York, New York (Lee); Department of Psychiatry, College of Physicians and Surgeons, Columbia University, New York, New York (Mayeux).
1,
Thomas O Obisesan
Thomas O Obisesan, MD
1Tanz Centre for Research in Neurodegenerative Diseases, University of Toronto, Toronto, Ontario, Canada (Ghani, Sato, St. George-Hyslop, Rogaeva); Taub Institute for Research on Alzheimer’s Disease and the Aging Brain, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Cheng, Manly, Lee, Mayeux); Gertrude H. Sergievsky Center, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Vardarajan, Lee, Mayeux); Department of Neurology, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Manly, Mayeux); Department of Medicine (Biomedical Genetics), Boston University, Boston, Massachusetts (Jun, Logue, Baldwin, Farrer); Department of Biostatistics, Boston University, Boston, Massachusetts (Jun, Lunetta, Farrer); Department of Ophthalmology, Boston University, Boston, Massachusetts (Jun, Farrer); The John P. Hussman Institute for Human Genomics, University of Miami, Miami, Florida (Naj, Rajbhandary, Pericak-Vance); Department of Pathology and Laboratory Medicine, University of Pennsylvania Perelman School of Medicine, Philadelphia (Wang, Valladares, Lin, Cantwell, Schellenberg); Department of Medicine, University of Washington, Seattle (Larson, Crane); Group Health Research Institute, Group Health, Seattle, Washington (Larson); Department of Neuroscience, Mayo Clinic, Jacksonville, Florida (Graff-Radford, Ertekin-Taner); Department of Neurology, Mayo Clinic, Jacksonville, Florida (Graff-Radford, Ertekin-Taner); Rush Institute for Healthy Aging, Department of Internal Medicine, Rush University Medical Center, Chicago, Illinois (Evans); Program in Translational Neuropsychiatric Genomics, Department of Neurology, Brigham and Women’s Hospital, Boston, Massachusetts (De Jager); Harvard Medical School, Boston, Massachusetts (De Jager, Raj, Green); Program in Medical and Population Genetics, The Broad Institute, Cambridge, Massachusetts (De Jager); Department of Psychiatry, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Genetics and Genomics Sciences, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Neuroscience, Mount Sinai School of Medicine, New York, New York (Buxbaum); Friedman Brain Institute, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Medical and Molecular Genetics, Indiana University, Indianapolis (Murrell); Division of Genetics, Department of Medicine, Brigham and Women’s Hospital, Boston, Massachusetts (Green); Partners Center for Personalized Genetic Medicine, Brigham and Women’s Hospital, Boston, Massachusetts (Green); Department of Neurological Sciences, Rush University Medical Center, Chicago, Illinois (Barnes, Bennett); Department of Behavioral Sciences, Rush University Medical Center, Chicago, Illinois (Barnes, Foroud); Department of Epidemiology, Johns Hopkins University School of Public Health, Baltimore, Maryland (Fallin); School of Public Health, University of Alabama at Birmingham (Go); SABA University School of Medicine, SABA, Dutch Caribbean (Griffith); Division of Geriatrics, Howard University Hospital, Washington, DC (Obisesan); Department of Human Genetics, University of Pittsburgh, Pittsburgh, Pennsylvania (Kamboh); Alzheimer’s Disease Research Center, University of Pittsburgh, Pittsburgh, Pennsylvania (Kamboh, Lopez, Mayeux); Rush Alzheimer’s Disease Center, Rush University Medical Center, Chicago, Illinois (Bennett); Indiana University Center for Aging Research, Indianapolis (Hendrie); Department of Psychiatry, Indiana University School of Medicine, Indianapolis (Hendrie, Hall); Regenstrief Institute Inc, Indianapolis, Indiana (Hendrie); Hope Center Program on Protein Aggregation and Neurodegeneration, Department of Psychiatry, Washington University School of Medicine, St Louis, Missouri (Goate); Department of Biology, North Carolina A & T University, Greensboro (Byrd); National Alzheimer’s Coordinating Center, Department of Epidemiology, University of Washington, Seattle (Kukull); Vanderbilt Center for Human Genetics Research, Department of Molecular Physiology and Biophysics, Vanderbilt University, Nashville, Tennessee (Haines); Department of Neurology, Boston University, Boston, Massachusetts (Farrer); Department of Epidemiology, Boston University, Boston, Massachusetts (Farrer); Department of Epidemiology, College of Physicians and Surgeons, Columbia University, New York, New York (Lee); Department of Psychiatry, College of Physicians and Surgeons, Columbia University, New York, New York (Mayeux).
1,
Jennifer J Manly
Jennifer J Manly, PhD
1Tanz Centre for Research in Neurodegenerative Diseases, University of Toronto, Toronto, Ontario, Canada (Ghani, Sato, St. George-Hyslop, Rogaeva); Taub Institute for Research on Alzheimer’s Disease and the Aging Brain, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Cheng, Manly, Lee, Mayeux); Gertrude H. Sergievsky Center, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Vardarajan, Lee, Mayeux); Department of Neurology, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Manly, Mayeux); Department of Medicine (Biomedical Genetics), Boston University, Boston, Massachusetts (Jun, Logue, Baldwin, Farrer); Department of Biostatistics, Boston University, Boston, Massachusetts (Jun, Lunetta, Farrer); Department of Ophthalmology, Boston University, Boston, Massachusetts (Jun, Farrer); The John P. Hussman Institute for Human Genomics, University of Miami, Miami, Florida (Naj, Rajbhandary, Pericak-Vance); Department of Pathology and Laboratory Medicine, University of Pennsylvania Perelman School of Medicine, Philadelphia (Wang, Valladares, Lin, Cantwell, Schellenberg); Department of Medicine, University of Washington, Seattle (Larson, Crane); Group Health Research Institute, Group Health, Seattle, Washington (Larson); Department of Neuroscience, Mayo Clinic, Jacksonville, Florida (Graff-Radford, Ertekin-Taner); Department of Neurology, Mayo Clinic, Jacksonville, Florida (Graff-Radford, Ertekin-Taner); Rush Institute for Healthy Aging, Department of Internal Medicine, Rush University Medical Center, Chicago, Illinois (Evans); Program in Translational Neuropsychiatric Genomics, Department of Neurology, Brigham and Women’s Hospital, Boston, Massachusetts (De Jager); Harvard Medical School, Boston, Massachusetts (De Jager, Raj, Green); Program in Medical and Population Genetics, The Broad Institute, Cambridge, Massachusetts (De Jager); Department of Psychiatry, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Genetics and Genomics Sciences, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Neuroscience, Mount Sinai School of Medicine, New York, New York (Buxbaum); Friedman Brain Institute, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Medical and Molecular Genetics, Indiana University, Indianapolis (Murrell); Division of Genetics, Department of Medicine, Brigham and Women’s Hospital, Boston, Massachusetts (Green); Partners Center for Personalized Genetic Medicine, Brigham and Women’s Hospital, Boston, Massachusetts (Green); Department of Neurological Sciences, Rush University Medical Center, Chicago, Illinois (Barnes, Bennett); Department of Behavioral Sciences, Rush University Medical Center, Chicago, Illinois (Barnes, Foroud); Department of Epidemiology, Johns Hopkins University School of Public Health, Baltimore, Maryland (Fallin); School of Public Health, University of Alabama at Birmingham (Go); SABA University School of Medicine, SABA, Dutch Caribbean (Griffith); Division of Geriatrics, Howard University Hospital, Washington, DC (Obisesan); Department of Human Genetics, University of Pittsburgh, Pittsburgh, Pennsylvania (Kamboh); Alzheimer’s Disease Research Center, University of Pittsburgh, Pittsburgh, Pennsylvania (Kamboh, Lopez, Mayeux); Rush Alzheimer’s Disease Center, Rush University Medical Center, Chicago, Illinois (Bennett); Indiana University Center for Aging Research, Indianapolis (Hendrie); Department of Psychiatry, Indiana University School of Medicine, Indianapolis (Hendrie, Hall); Regenstrief Institute Inc, Indianapolis, Indiana (Hendrie); Hope Center Program on Protein Aggregation and Neurodegeneration, Department of Psychiatry, Washington University School of Medicine, St Louis, Missouri (Goate); Department of Biology, North Carolina A & T University, Greensboro (Byrd); National Alzheimer’s Coordinating Center, Department of Epidemiology, University of Washington, Seattle (Kukull); Vanderbilt Center for Human Genetics Research, Department of Molecular Physiology and Biophysics, Vanderbilt University, Nashville, Tennessee (Haines); Department of Neurology, Boston University, Boston, Massachusetts (Farrer); Department of Epidemiology, Boston University, Boston, Massachusetts (Farrer); Department of Epidemiology, College of Physicians and Surgeons, Columbia University, New York, New York (Lee); Department of Psychiatry, College of Physicians and Surgeons, Columbia University, New York, New York (Mayeux).
1,
Kathryn L Lunetta
Kathryn L Lunetta, PhD
1Tanz Centre for Research in Neurodegenerative Diseases, University of Toronto, Toronto, Ontario, Canada (Ghani, Sato, St. George-Hyslop, Rogaeva); Taub Institute for Research on Alzheimer’s Disease and the Aging Brain, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Cheng, Manly, Lee, Mayeux); Gertrude H. Sergievsky Center, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Vardarajan, Lee, Mayeux); Department of Neurology, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Manly, Mayeux); Department of Medicine (Biomedical Genetics), Boston University, Boston, Massachusetts (Jun, Logue, Baldwin, Farrer); Department of Biostatistics, Boston University, Boston, Massachusetts (Jun, Lunetta, Farrer); Department of Ophthalmology, Boston University, Boston, Massachusetts (Jun, Farrer); The John P. Hussman Institute for Human Genomics, University of Miami, Miami, Florida (Naj, Rajbhandary, Pericak-Vance); Department of Pathology and Laboratory Medicine, University of Pennsylvania Perelman School of Medicine, Philadelphia (Wang, Valladares, Lin, Cantwell, Schellenberg); Department of Medicine, University of Washington, Seattle (Larson, Crane); Group Health Research Institute, Group Health, Seattle, Washington (Larson); Department of Neuroscience, Mayo Clinic, Jacksonville, Florida (Graff-Radford, Ertekin-Taner); Department of Neurology, Mayo Clinic, Jacksonville, Florida (Graff-Radford, Ertekin-Taner); Rush Institute for Healthy Aging, Department of Internal Medicine, Rush University Medical Center, Chicago, Illinois (Evans); Program in Translational Neuropsychiatric Genomics, Department of Neurology, Brigham and Women’s Hospital, Boston, Massachusetts (De Jager); Harvard Medical School, Boston, Massachusetts (De Jager, Raj, Green); Program in Medical and Population Genetics, The Broad Institute, Cambridge, Massachusetts (De Jager); Department of Psychiatry, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Genetics and Genomics Sciences, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Neuroscience, Mount Sinai School of Medicine, New York, New York (Buxbaum); Friedman Brain Institute, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Medical and Molecular Genetics, Indiana University, Indianapolis (Murrell); Division of Genetics, Department of Medicine, Brigham and Women’s Hospital, Boston, Massachusetts (Green); Partners Center for Personalized Genetic Medicine, Brigham and Women’s Hospital, Boston, Massachusetts (Green); Department of Neurological Sciences, Rush University Medical Center, Chicago, Illinois (Barnes, Bennett); Department of Behavioral Sciences, Rush University Medical Center, Chicago, Illinois (Barnes, Foroud); Department of Epidemiology, Johns Hopkins University School of Public Health, Baltimore, Maryland (Fallin); School of Public Health, University of Alabama at Birmingham (Go); SABA University School of Medicine, SABA, Dutch Caribbean (Griffith); Division of Geriatrics, Howard University Hospital, Washington, DC (Obisesan); Department of Human Genetics, University of Pittsburgh, Pittsburgh, Pennsylvania (Kamboh); Alzheimer’s Disease Research Center, University of Pittsburgh, Pittsburgh, Pennsylvania (Kamboh, Lopez, Mayeux); Rush Alzheimer’s Disease Center, Rush University Medical Center, Chicago, Illinois (Bennett); Indiana University Center for Aging Research, Indianapolis (Hendrie); Department of Psychiatry, Indiana University School of Medicine, Indianapolis (Hendrie, Hall); Regenstrief Institute Inc, Indianapolis, Indiana (Hendrie); Hope Center Program on Protein Aggregation and Neurodegeneration, Department of Psychiatry, Washington University School of Medicine, St Louis, Missouri (Goate); Department of Biology, North Carolina A & T University, Greensboro (Byrd); National Alzheimer’s Coordinating Center, Department of Epidemiology, University of Washington, Seattle (Kukull); Vanderbilt Center for Human Genetics Research, Department of Molecular Physiology and Biophysics, Vanderbilt University, Nashville, Tennessee (Haines); Department of Neurology, Boston University, Boston, Massachusetts (Farrer); Department of Epidemiology, Boston University, Boston, Massachusetts (Farrer); Department of Epidemiology, College of Physicians and Surgeons, Columbia University, New York, New York (Lee); Department of Psychiatry, College of Physicians and Surgeons, Columbia University, New York, New York (Mayeux).
1,
M Ilyas Kamboh
M Ilyas Kamboh, PhD
1Tanz Centre for Research in Neurodegenerative Diseases, University of Toronto, Toronto, Ontario, Canada (Ghani, Sato, St. George-Hyslop, Rogaeva); Taub Institute for Research on Alzheimer’s Disease and the Aging Brain, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Cheng, Manly, Lee, Mayeux); Gertrude H. Sergievsky Center, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Vardarajan, Lee, Mayeux); Department of Neurology, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Manly, Mayeux); Department of Medicine (Biomedical Genetics), Boston University, Boston, Massachusetts (Jun, Logue, Baldwin, Farrer); Department of Biostatistics, Boston University, Boston, Massachusetts (Jun, Lunetta, Farrer); Department of Ophthalmology, Boston University, Boston, Massachusetts (Jun, Farrer); The John P. Hussman Institute for Human Genomics, University of Miami, Miami, Florida (Naj, Rajbhandary, Pericak-Vance); Department of Pathology and Laboratory Medicine, University of Pennsylvania Perelman School of Medicine, Philadelphia (Wang, Valladares, Lin, Cantwell, Schellenberg); Department of Medicine, University of Washington, Seattle (Larson, Crane); Group Health Research Institute, Group Health, Seattle, Washington (Larson); Department of Neuroscience, Mayo Clinic, Jacksonville, Florida (Graff-Radford, Ertekin-Taner); Department of Neurology, Mayo Clinic, Jacksonville, Florida (Graff-Radford, Ertekin-Taner); Rush Institute for Healthy Aging, Department of Internal Medicine, Rush University Medical Center, Chicago, Illinois (Evans); Program in Translational Neuropsychiatric Genomics, Department of Neurology, Brigham and Women’s Hospital, Boston, Massachusetts (De Jager); Harvard Medical School, Boston, Massachusetts (De Jager, Raj, Green); Program in Medical and Population Genetics, The Broad Institute, Cambridge, Massachusetts (De Jager); Department of Psychiatry, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Genetics and Genomics Sciences, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Neuroscience, Mount Sinai School of Medicine, New York, New York (Buxbaum); Friedman Brain Institute, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Medical and Molecular Genetics, Indiana University, Indianapolis (Murrell); Division of Genetics, Department of Medicine, Brigham and Women’s Hospital, Boston, Massachusetts (Green); Partners Center for Personalized Genetic Medicine, Brigham and Women’s Hospital, Boston, Massachusetts (Green); Department of Neurological Sciences, Rush University Medical Center, Chicago, Illinois (Barnes, Bennett); Department of Behavioral Sciences, Rush University Medical Center, Chicago, Illinois (Barnes, Foroud); Department of Epidemiology, Johns Hopkins University School of Public Health, Baltimore, Maryland (Fallin); School of Public Health, University of Alabama at Birmingham (Go); SABA University School of Medicine, SABA, Dutch Caribbean (Griffith); Division of Geriatrics, Howard University Hospital, Washington, DC (Obisesan); Department of Human Genetics, University of Pittsburgh, Pittsburgh, Pennsylvania (Kamboh); Alzheimer’s Disease Research Center, University of Pittsburgh, Pittsburgh, Pennsylvania (Kamboh, Lopez, Mayeux); Rush Alzheimer’s Disease Center, Rush University Medical Center, Chicago, Illinois (Bennett); Indiana University Center for Aging Research, Indianapolis (Hendrie); Department of Psychiatry, Indiana University School of Medicine, Indianapolis (Hendrie, Hall); Regenstrief Institute Inc, Indianapolis, Indiana (Hendrie); Hope Center Program on Protein Aggregation and Neurodegeneration, Department of Psychiatry, Washington University School of Medicine, St Louis, Missouri (Goate); Department of Biology, North Carolina A & T University, Greensboro (Byrd); National Alzheimer’s Coordinating Center, Department of Epidemiology, University of Washington, Seattle (Kukull); Vanderbilt Center for Human Genetics Research, Department of Molecular Physiology and Biophysics, Vanderbilt University, Nashville, Tennessee (Haines); Department of Neurology, Boston University, Boston, Massachusetts (Farrer); Department of Epidemiology, Boston University, Boston, Massachusetts (Farrer); Department of Epidemiology, College of Physicians and Surgeons, Columbia University, New York, New York (Lee); Department of Psychiatry, College of Physicians and Surgeons, Columbia University, New York, New York (Mayeux).
1,
Oscar L Lopez
Oscar L Lopez, MD
1Tanz Centre for Research in Neurodegenerative Diseases, University of Toronto, Toronto, Ontario, Canada (Ghani, Sato, St. George-Hyslop, Rogaeva); Taub Institute for Research on Alzheimer’s Disease and the Aging Brain, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Cheng, Manly, Lee, Mayeux); Gertrude H. Sergievsky Center, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Vardarajan, Lee, Mayeux); Department of Neurology, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Manly, Mayeux); Department of Medicine (Biomedical Genetics), Boston University, Boston, Massachusetts (Jun, Logue, Baldwin, Farrer); Department of Biostatistics, Boston University, Boston, Massachusetts (Jun, Lunetta, Farrer); Department of Ophthalmology, Boston University, Boston, Massachusetts (Jun, Farrer); The John P. Hussman Institute for Human Genomics, University of Miami, Miami, Florida (Naj, Rajbhandary, Pericak-Vance); Department of Pathology and Laboratory Medicine, University of Pennsylvania Perelman School of Medicine, Philadelphia (Wang, Valladares, Lin, Cantwell, Schellenberg); Department of Medicine, University of Washington, Seattle (Larson, Crane); Group Health Research Institute, Group Health, Seattle, Washington (Larson); Department of Neuroscience, Mayo Clinic, Jacksonville, Florida (Graff-Radford, Ertekin-Taner); Department of Neurology, Mayo Clinic, Jacksonville, Florida (Graff-Radford, Ertekin-Taner); Rush Institute for Healthy Aging, Department of Internal Medicine, Rush University Medical Center, Chicago, Illinois (Evans); Program in Translational Neuropsychiatric Genomics, Department of Neurology, Brigham and Women’s Hospital, Boston, Massachusetts (De Jager); Harvard Medical School, Boston, Massachusetts (De Jager, Raj, Green); Program in Medical and Population Genetics, The Broad Institute, Cambridge, Massachusetts (De Jager); Department of Psychiatry, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Genetics and Genomics Sciences, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Neuroscience, Mount Sinai School of Medicine, New York, New York (Buxbaum); Friedman Brain Institute, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Medical and Molecular Genetics, Indiana University, Indianapolis (Murrell); Division of Genetics, Department of Medicine, Brigham and Women’s Hospital, Boston, Massachusetts (Green); Partners Center for Personalized Genetic Medicine, Brigham and Women’s Hospital, Boston, Massachusetts (Green); Department of Neurological Sciences, Rush University Medical Center, Chicago, Illinois (Barnes, Bennett); Department of Behavioral Sciences, Rush University Medical Center, Chicago, Illinois (Barnes, Foroud); Department of Epidemiology, Johns Hopkins University School of Public Health, Baltimore, Maryland (Fallin); School of Public Health, University of Alabama at Birmingham (Go); SABA University School of Medicine, SABA, Dutch Caribbean (Griffith); Division of Geriatrics, Howard University Hospital, Washington, DC (Obisesan); Department of Human Genetics, University of Pittsburgh, Pittsburgh, Pennsylvania (Kamboh); Alzheimer’s Disease Research Center, University of Pittsburgh, Pittsburgh, Pennsylvania (Kamboh, Lopez, Mayeux); Rush Alzheimer’s Disease Center, Rush University Medical Center, Chicago, Illinois (Bennett); Indiana University Center for Aging Research, Indianapolis (Hendrie); Department of Psychiatry, Indiana University School of Medicine, Indianapolis (Hendrie, Hall); Regenstrief Institute Inc, Indianapolis, Indiana (Hendrie); Hope Center Program on Protein Aggregation and Neurodegeneration, Department of Psychiatry, Washington University School of Medicine, St Louis, Missouri (Goate); Department of Biology, North Carolina A & T University, Greensboro (Byrd); National Alzheimer’s Coordinating Center, Department of Epidemiology, University of Washington, Seattle (Kukull); Vanderbilt Center for Human Genetics Research, Department of Molecular Physiology and Biophysics, Vanderbilt University, Nashville, Tennessee (Haines); Department of Neurology, Boston University, Boston, Massachusetts (Farrer); Department of Epidemiology, Boston University, Boston, Massachusetts (Farrer); Department of Epidemiology, College of Physicians and Surgeons, Columbia University, New York, New York (Lee); Department of Psychiatry, College of Physicians and Surgeons, Columbia University, New York, New York (Mayeux).
1,
David A Bennett
David A Bennett, MD
1Tanz Centre for Research in Neurodegenerative Diseases, University of Toronto, Toronto, Ontario, Canada (Ghani, Sato, St. George-Hyslop, Rogaeva); Taub Institute for Research on Alzheimer’s Disease and the Aging Brain, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Cheng, Manly, Lee, Mayeux); Gertrude H. Sergievsky Center, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Vardarajan, Lee, Mayeux); Department of Neurology, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Manly, Mayeux); Department of Medicine (Biomedical Genetics), Boston University, Boston, Massachusetts (Jun, Logue, Baldwin, Farrer); Department of Biostatistics, Boston University, Boston, Massachusetts (Jun, Lunetta, Farrer); Department of Ophthalmology, Boston University, Boston, Massachusetts (Jun, Farrer); The John P. Hussman Institute for Human Genomics, University of Miami, Miami, Florida (Naj, Rajbhandary, Pericak-Vance); Department of Pathology and Laboratory Medicine, University of Pennsylvania Perelman School of Medicine, Philadelphia (Wang, Valladares, Lin, Cantwell, Schellenberg); Department of Medicine, University of Washington, Seattle (Larson, Crane); Group Health Research Institute, Group Health, Seattle, Washington (Larson); Department of Neuroscience, Mayo Clinic, Jacksonville, Florida (Graff-Radford, Ertekin-Taner); Department of Neurology, Mayo Clinic, Jacksonville, Florida (Graff-Radford, Ertekin-Taner); Rush Institute for Healthy Aging, Department of Internal Medicine, Rush University Medical Center, Chicago, Illinois (Evans); Program in Translational Neuropsychiatric Genomics, Department of Neurology, Brigham and Women’s Hospital, Boston, Massachusetts (De Jager); Harvard Medical School, Boston, Massachusetts (De Jager, Raj, Green); Program in Medical and Population Genetics, The Broad Institute, Cambridge, Massachusetts (De Jager); Department of Psychiatry, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Genetics and Genomics Sciences, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Neuroscience, Mount Sinai School of Medicine, New York, New York (Buxbaum); Friedman Brain Institute, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Medical and Molecular Genetics, Indiana University, Indianapolis (Murrell); Division of Genetics, Department of Medicine, Brigham and Women’s Hospital, Boston, Massachusetts (Green); Partners Center for Personalized Genetic Medicine, Brigham and Women’s Hospital, Boston, Massachusetts (Green); Department of Neurological Sciences, Rush University Medical Center, Chicago, Illinois (Barnes, Bennett); Department of Behavioral Sciences, Rush University Medical Center, Chicago, Illinois (Barnes, Foroud); Department of Epidemiology, Johns Hopkins University School of Public Health, Baltimore, Maryland (Fallin); School of Public Health, University of Alabama at Birmingham (Go); SABA University School of Medicine, SABA, Dutch Caribbean (Griffith); Division of Geriatrics, Howard University Hospital, Washington, DC (Obisesan); Department of Human Genetics, University of Pittsburgh, Pittsburgh, Pennsylvania (Kamboh); Alzheimer’s Disease Research Center, University of Pittsburgh, Pittsburgh, Pennsylvania (Kamboh, Lopez, Mayeux); Rush Alzheimer’s Disease Center, Rush University Medical Center, Chicago, Illinois (Bennett); Indiana University Center for Aging Research, Indianapolis (Hendrie); Department of Psychiatry, Indiana University School of Medicine, Indianapolis (Hendrie, Hall); Regenstrief Institute Inc, Indianapolis, Indiana (Hendrie); Hope Center Program on Protein Aggregation and Neurodegeneration, Department of Psychiatry, Washington University School of Medicine, St Louis, Missouri (Goate); Department of Biology, North Carolina A & T University, Greensboro (Byrd); National Alzheimer’s Coordinating Center, Department of Epidemiology, University of Washington, Seattle (Kukull); Vanderbilt Center for Human Genetics Research, Department of Molecular Physiology and Biophysics, Vanderbilt University, Nashville, Tennessee (Haines); Department of Neurology, Boston University, Boston, Massachusetts (Farrer); Department of Epidemiology, Boston University, Boston, Massachusetts (Farrer); Department of Epidemiology, College of Physicians and Surgeons, Columbia University, New York, New York (Lee); Department of Psychiatry, College of Physicians and Surgeons, Columbia University, New York, New York (Mayeux).
1,
Hugh Hendrie
Hugh Hendrie, MB, ChB, DSc
1Tanz Centre for Research in Neurodegenerative Diseases, University of Toronto, Toronto, Ontario, Canada (Ghani, Sato, St. George-Hyslop, Rogaeva); Taub Institute for Research on Alzheimer’s Disease and the Aging Brain, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Cheng, Manly, Lee, Mayeux); Gertrude H. Sergievsky Center, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Vardarajan, Lee, Mayeux); Department of Neurology, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Manly, Mayeux); Department of Medicine (Biomedical Genetics), Boston University, Boston, Massachusetts (Jun, Logue, Baldwin, Farrer); Department of Biostatistics, Boston University, Boston, Massachusetts (Jun, Lunetta, Farrer); Department of Ophthalmology, Boston University, Boston, Massachusetts (Jun, Farrer); The John P. Hussman Institute for Human Genomics, University of Miami, Miami, Florida (Naj, Rajbhandary, Pericak-Vance); Department of Pathology and Laboratory Medicine, University of Pennsylvania Perelman School of Medicine, Philadelphia (Wang, Valladares, Lin, Cantwell, Schellenberg); Department of Medicine, University of Washington, Seattle (Larson, Crane); Group Health Research Institute, Group Health, Seattle, Washington (Larson); Department of Neuroscience, Mayo Clinic, Jacksonville, Florida (Graff-Radford, Ertekin-Taner); Department of Neurology, Mayo Clinic, Jacksonville, Florida (Graff-Radford, Ertekin-Taner); Rush Institute for Healthy Aging, Department of Internal Medicine, Rush University Medical Center, Chicago, Illinois (Evans); Program in Translational Neuropsychiatric Genomics, Department of Neurology, Brigham and Women’s Hospital, Boston, Massachusetts (De Jager); Harvard Medical School, Boston, Massachusetts (De Jager, Raj, Green); Program in Medical and Population Genetics, The Broad Institute, Cambridge, Massachusetts (De Jager); Department of Psychiatry, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Genetics and Genomics Sciences, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Neuroscience, Mount Sinai School of Medicine, New York, New York (Buxbaum); Friedman Brain Institute, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Medical and Molecular Genetics, Indiana University, Indianapolis (Murrell); Division of Genetics, Department of Medicine, Brigham and Women’s Hospital, Boston, Massachusetts (Green); Partners Center for Personalized Genetic Medicine, Brigham and Women’s Hospital, Boston, Massachusetts (Green); Department of Neurological Sciences, Rush University Medical Center, Chicago, Illinois (Barnes, Bennett); Department of Behavioral Sciences, Rush University Medical Center, Chicago, Illinois (Barnes, Foroud); Department of Epidemiology, Johns Hopkins University School of Public Health, Baltimore, Maryland (Fallin); School of Public Health, University of Alabama at Birmingham (Go); SABA University School of Medicine, SABA, Dutch Caribbean (Griffith); Division of Geriatrics, Howard University Hospital, Washington, DC (Obisesan); Department of Human Genetics, University of Pittsburgh, Pittsburgh, Pennsylvania (Kamboh); Alzheimer’s Disease Research Center, University of Pittsburgh, Pittsburgh, Pennsylvania (Kamboh, Lopez, Mayeux); Rush Alzheimer’s Disease Center, Rush University Medical Center, Chicago, Illinois (Bennett); Indiana University Center for Aging Research, Indianapolis (Hendrie); Department of Psychiatry, Indiana University School of Medicine, Indianapolis (Hendrie, Hall); Regenstrief Institute Inc, Indianapolis, Indiana (Hendrie); Hope Center Program on Protein Aggregation and Neurodegeneration, Department of Psychiatry, Washington University School of Medicine, St Louis, Missouri (Goate); Department of Biology, North Carolina A & T University, Greensboro (Byrd); National Alzheimer’s Coordinating Center, Department of Epidemiology, University of Washington, Seattle (Kukull); Vanderbilt Center for Human Genetics Research, Department of Molecular Physiology and Biophysics, Vanderbilt University, Nashville, Tennessee (Haines); Department of Neurology, Boston University, Boston, Massachusetts (Farrer); Department of Epidemiology, Boston University, Boston, Massachusetts (Farrer); Department of Epidemiology, College of Physicians and Surgeons, Columbia University, New York, New York (Lee); Department of Psychiatry, College of Physicians and Surgeons, Columbia University, New York, New York (Mayeux).
1,
Kathleen S Hall
Kathleen S Hall, PhD
1Tanz Centre for Research in Neurodegenerative Diseases, University of Toronto, Toronto, Ontario, Canada (Ghani, Sato, St. George-Hyslop, Rogaeva); Taub Institute for Research on Alzheimer’s Disease and the Aging Brain, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Cheng, Manly, Lee, Mayeux); Gertrude H. Sergievsky Center, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Vardarajan, Lee, Mayeux); Department of Neurology, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Manly, Mayeux); Department of Medicine (Biomedical Genetics), Boston University, Boston, Massachusetts (Jun, Logue, Baldwin, Farrer); Department of Biostatistics, Boston University, Boston, Massachusetts (Jun, Lunetta, Farrer); Department of Ophthalmology, Boston University, Boston, Massachusetts (Jun, Farrer); The John P. Hussman Institute for Human Genomics, University of Miami, Miami, Florida (Naj, Rajbhandary, Pericak-Vance); Department of Pathology and Laboratory Medicine, University of Pennsylvania Perelman School of Medicine, Philadelphia (Wang, Valladares, Lin, Cantwell, Schellenberg); Department of Medicine, University of Washington, Seattle (Larson, Crane); Group Health Research Institute, Group Health, Seattle, Washington (Larson); Department of Neuroscience, Mayo Clinic, Jacksonville, Florida (Graff-Radford, Ertekin-Taner); Department of Neurology, Mayo Clinic, Jacksonville, Florida (Graff-Radford, Ertekin-Taner); Rush Institute for Healthy Aging, Department of Internal Medicine, Rush University Medical Center, Chicago, Illinois (Evans); Program in Translational Neuropsychiatric Genomics, Department of Neurology, Brigham and Women’s Hospital, Boston, Massachusetts (De Jager); Harvard Medical School, Boston, Massachusetts (De Jager, Raj, Green); Program in Medical and Population Genetics, The Broad Institute, Cambridge, Massachusetts (De Jager); Department of Psychiatry, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Genetics and Genomics Sciences, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Neuroscience, Mount Sinai School of Medicine, New York, New York (Buxbaum); Friedman Brain Institute, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Medical and Molecular Genetics, Indiana University, Indianapolis (Murrell); Division of Genetics, Department of Medicine, Brigham and Women’s Hospital, Boston, Massachusetts (Green); Partners Center for Personalized Genetic Medicine, Brigham and Women’s Hospital, Boston, Massachusetts (Green); Department of Neurological Sciences, Rush University Medical Center, Chicago, Illinois (Barnes, Bennett); Department of Behavioral Sciences, Rush University Medical Center, Chicago, Illinois (Barnes, Foroud); Department of Epidemiology, Johns Hopkins University School of Public Health, Baltimore, Maryland (Fallin); School of Public Health, University of Alabama at Birmingham (Go); SABA University School of Medicine, SABA, Dutch Caribbean (Griffith); Division of Geriatrics, Howard University Hospital, Washington, DC (Obisesan); Department of Human Genetics, University of Pittsburgh, Pittsburgh, Pennsylvania (Kamboh); Alzheimer’s Disease Research Center, University of Pittsburgh, Pittsburgh, Pennsylvania (Kamboh, Lopez, Mayeux); Rush Alzheimer’s Disease Center, Rush University Medical Center, Chicago, Illinois (Bennett); Indiana University Center for Aging Research, Indianapolis (Hendrie); Department of Psychiatry, Indiana University School of Medicine, Indianapolis (Hendrie, Hall); Regenstrief Institute Inc, Indianapolis, Indiana (Hendrie); Hope Center Program on Protein Aggregation and Neurodegeneration, Department of Psychiatry, Washington University School of Medicine, St Louis, Missouri (Goate); Department of Biology, North Carolina A & T University, Greensboro (Byrd); National Alzheimer’s Coordinating Center, Department of Epidemiology, University of Washington, Seattle (Kukull); Vanderbilt Center for Human Genetics Research, Department of Molecular Physiology and Biophysics, Vanderbilt University, Nashville, Tennessee (Haines); Department of Neurology, Boston University, Boston, Massachusetts (Farrer); Department of Epidemiology, Boston University, Boston, Massachusetts (Farrer); Department of Epidemiology, College of Physicians and Surgeons, Columbia University, New York, New York (Lee); Department of Psychiatry, College of Physicians and Surgeons, Columbia University, New York, New York (Mayeux).
1,
Alison M Goate
Alison M Goate, PhD
1Tanz Centre for Research in Neurodegenerative Diseases, University of Toronto, Toronto, Ontario, Canada (Ghani, Sato, St. George-Hyslop, Rogaeva); Taub Institute for Research on Alzheimer’s Disease and the Aging Brain, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Cheng, Manly, Lee, Mayeux); Gertrude H. Sergievsky Center, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Vardarajan, Lee, Mayeux); Department of Neurology, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Manly, Mayeux); Department of Medicine (Biomedical Genetics), Boston University, Boston, Massachusetts (Jun, Logue, Baldwin, Farrer); Department of Biostatistics, Boston University, Boston, Massachusetts (Jun, Lunetta, Farrer); Department of Ophthalmology, Boston University, Boston, Massachusetts (Jun, Farrer); The John P. Hussman Institute for Human Genomics, University of Miami, Miami, Florida (Naj, Rajbhandary, Pericak-Vance); Department of Pathology and Laboratory Medicine, University of Pennsylvania Perelman School of Medicine, Philadelphia (Wang, Valladares, Lin, Cantwell, Schellenberg); Department of Medicine, University of Washington, Seattle (Larson, Crane); Group Health Research Institute, Group Health, Seattle, Washington (Larson); Department of Neuroscience, Mayo Clinic, Jacksonville, Florida (Graff-Radford, Ertekin-Taner); Department of Neurology, Mayo Clinic, Jacksonville, Florida (Graff-Radford, Ertekin-Taner); Rush Institute for Healthy Aging, Department of Internal Medicine, Rush University Medical Center, Chicago, Illinois (Evans); Program in Translational Neuropsychiatric Genomics, Department of Neurology, Brigham and Women’s Hospital, Boston, Massachusetts (De Jager); Harvard Medical School, Boston, Massachusetts (De Jager, Raj, Green); Program in Medical and Population Genetics, The Broad Institute, Cambridge, Massachusetts (De Jager); Department of Psychiatry, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Genetics and Genomics Sciences, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Neuroscience, Mount Sinai School of Medicine, New York, New York (Buxbaum); Friedman Brain Institute, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Medical and Molecular Genetics, Indiana University, Indianapolis (Murrell); Division of Genetics, Department of Medicine, Brigham and Women’s Hospital, Boston, Massachusetts (Green); Partners Center for Personalized Genetic Medicine, Brigham and Women’s Hospital, Boston, Massachusetts (Green); Department of Neurological Sciences, Rush University Medical Center, Chicago, Illinois (Barnes, Bennett); Department of Behavioral Sciences, Rush University Medical Center, Chicago, Illinois (Barnes, Foroud); Department of Epidemiology, Johns Hopkins University School of Public Health, Baltimore, Maryland (Fallin); School of Public Health, University of Alabama at Birmingham (Go); SABA University School of Medicine, SABA, Dutch Caribbean (Griffith); Division of Geriatrics, Howard University Hospital, Washington, DC (Obisesan); Department of Human Genetics, University of Pittsburgh, Pittsburgh, Pennsylvania (Kamboh); Alzheimer’s Disease Research Center, University of Pittsburgh, Pittsburgh, Pennsylvania (Kamboh, Lopez, Mayeux); Rush Alzheimer’s Disease Center, Rush University Medical Center, Chicago, Illinois (Bennett); Indiana University Center for Aging Research, Indianapolis (Hendrie); Department of Psychiatry, Indiana University School of Medicine, Indianapolis (Hendrie, Hall); Regenstrief Institute Inc, Indianapolis, Indiana (Hendrie); Hope Center Program on Protein Aggregation and Neurodegeneration, Department of Psychiatry, Washington University School of Medicine, St Louis, Missouri (Goate); Department of Biology, North Carolina A & T University, Greensboro (Byrd); National Alzheimer’s Coordinating Center, Department of Epidemiology, University of Washington, Seattle (Kukull); Vanderbilt Center for Human Genetics Research, Department of Molecular Physiology and Biophysics, Vanderbilt University, Nashville, Tennessee (Haines); Department of Neurology, Boston University, Boston, Massachusetts (Farrer); Department of Epidemiology, Boston University, Boston, Massachusetts (Farrer); Department of Epidemiology, College of Physicians and Surgeons, Columbia University, New York, New York (Lee); Department of Psychiatry, College of Physicians and Surgeons, Columbia University, New York, New York (Mayeux).
1,
Goldie S Byrd
Goldie S Byrd, PhD
1Tanz Centre for Research in Neurodegenerative Diseases, University of Toronto, Toronto, Ontario, Canada (Ghani, Sato, St. George-Hyslop, Rogaeva); Taub Institute for Research on Alzheimer’s Disease and the Aging Brain, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Cheng, Manly, Lee, Mayeux); Gertrude H. Sergievsky Center, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Vardarajan, Lee, Mayeux); Department of Neurology, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Manly, Mayeux); Department of Medicine (Biomedical Genetics), Boston University, Boston, Massachusetts (Jun, Logue, Baldwin, Farrer); Department of Biostatistics, Boston University, Boston, Massachusetts (Jun, Lunetta, Farrer); Department of Ophthalmology, Boston University, Boston, Massachusetts (Jun, Farrer); The John P. Hussman Institute for Human Genomics, University of Miami, Miami, Florida (Naj, Rajbhandary, Pericak-Vance); Department of Pathology and Laboratory Medicine, University of Pennsylvania Perelman School of Medicine, Philadelphia (Wang, Valladares, Lin, Cantwell, Schellenberg); Department of Medicine, University of Washington, Seattle (Larson, Crane); Group Health Research Institute, Group Health, Seattle, Washington (Larson); Department of Neuroscience, Mayo Clinic, Jacksonville, Florida (Graff-Radford, Ertekin-Taner); Department of Neurology, Mayo Clinic, Jacksonville, Florida (Graff-Radford, Ertekin-Taner); Rush Institute for Healthy Aging, Department of Internal Medicine, Rush University Medical Center, Chicago, Illinois (Evans); Program in Translational Neuropsychiatric Genomics, Department of Neurology, Brigham and Women’s Hospital, Boston, Massachusetts (De Jager); Harvard Medical School, Boston, Massachusetts (De Jager, Raj, Green); Program in Medical and Population Genetics, The Broad Institute, Cambridge, Massachusetts (De Jager); Department of Psychiatry, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Genetics and Genomics Sciences, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Neuroscience, Mount Sinai School of Medicine, New York, New York (Buxbaum); Friedman Brain Institute, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Medical and Molecular Genetics, Indiana University, Indianapolis (Murrell); Division of Genetics, Department of Medicine, Brigham and Women’s Hospital, Boston, Massachusetts (Green); Partners Center for Personalized Genetic Medicine, Brigham and Women’s Hospital, Boston, Massachusetts (Green); Department of Neurological Sciences, Rush University Medical Center, Chicago, Illinois (Barnes, Bennett); Department of Behavioral Sciences, Rush University Medical Center, Chicago, Illinois (Barnes, Foroud); Department of Epidemiology, Johns Hopkins University School of Public Health, Baltimore, Maryland (Fallin); School of Public Health, University of Alabama at Birmingham (Go); SABA University School of Medicine, SABA, Dutch Caribbean (Griffith); Division of Geriatrics, Howard University Hospital, Washington, DC (Obisesan); Department of Human Genetics, University of Pittsburgh, Pittsburgh, Pennsylvania (Kamboh); Alzheimer’s Disease Research Center, University of Pittsburgh, Pittsburgh, Pennsylvania (Kamboh, Lopez, Mayeux); Rush Alzheimer’s Disease Center, Rush University Medical Center, Chicago, Illinois (Bennett); Indiana University Center for Aging Research, Indianapolis (Hendrie); Department of Psychiatry, Indiana University School of Medicine, Indianapolis (Hendrie, Hall); Regenstrief Institute Inc, Indianapolis, Indiana (Hendrie); Hope Center Program on Protein Aggregation and Neurodegeneration, Department of Psychiatry, Washington University School of Medicine, St Louis, Missouri (Goate); Department of Biology, North Carolina A & T University, Greensboro (Byrd); National Alzheimer’s Coordinating Center, Department of Epidemiology, University of Washington, Seattle (Kukull); Vanderbilt Center for Human Genetics Research, Department of Molecular Physiology and Biophysics, Vanderbilt University, Nashville, Tennessee (Haines); Department of Neurology, Boston University, Boston, Massachusetts (Farrer); Department of Epidemiology, Boston University, Boston, Massachusetts (Farrer); Department of Epidemiology, College of Physicians and Surgeons, Columbia University, New York, New York (Lee); Department of Psychiatry, College of Physicians and Surgeons, Columbia University, New York, New York (Mayeux).
1,
Walter A Kukull
Walter A Kukull, PhD
1Tanz Centre for Research in Neurodegenerative Diseases, University of Toronto, Toronto, Ontario, Canada (Ghani, Sato, St. George-Hyslop, Rogaeva); Taub Institute for Research on Alzheimer’s Disease and the Aging Brain, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Cheng, Manly, Lee, Mayeux); Gertrude H. Sergievsky Center, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Vardarajan, Lee, Mayeux); Department of Neurology, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Manly, Mayeux); Department of Medicine (Biomedical Genetics), Boston University, Boston, Massachusetts (Jun, Logue, Baldwin, Farrer); Department of Biostatistics, Boston University, Boston, Massachusetts (Jun, Lunetta, Farrer); Department of Ophthalmology, Boston University, Boston, Massachusetts (Jun, Farrer); The John P. Hussman Institute for Human Genomics, University of Miami, Miami, Florida (Naj, Rajbhandary, Pericak-Vance); Department of Pathology and Laboratory Medicine, University of Pennsylvania Perelman School of Medicine, Philadelphia (Wang, Valladares, Lin, Cantwell, Schellenberg); Department of Medicine, University of Washington, Seattle (Larson, Crane); Group Health Research Institute, Group Health, Seattle, Washington (Larson); Department of Neuroscience, Mayo Clinic, Jacksonville, Florida (Graff-Radford, Ertekin-Taner); Department of Neurology, Mayo Clinic, Jacksonville, Florida (Graff-Radford, Ertekin-Taner); Rush Institute for Healthy Aging, Department of Internal Medicine, Rush University Medical Center, Chicago, Illinois (Evans); Program in Translational Neuropsychiatric Genomics, Department of Neurology, Brigham and Women’s Hospital, Boston, Massachusetts (De Jager); Harvard Medical School, Boston, Massachusetts (De Jager, Raj, Green); Program in Medical and Population Genetics, The Broad Institute, Cambridge, Massachusetts (De Jager); Department of Psychiatry, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Genetics and Genomics Sciences, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Neuroscience, Mount Sinai School of Medicine, New York, New York (Buxbaum); Friedman Brain Institute, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Medical and Molecular Genetics, Indiana University, Indianapolis (Murrell); Division of Genetics, Department of Medicine, Brigham and Women’s Hospital, Boston, Massachusetts (Green); Partners Center for Personalized Genetic Medicine, Brigham and Women’s Hospital, Boston, Massachusetts (Green); Department of Neurological Sciences, Rush University Medical Center, Chicago, Illinois (Barnes, Bennett); Department of Behavioral Sciences, Rush University Medical Center, Chicago, Illinois (Barnes, Foroud); Department of Epidemiology, Johns Hopkins University School of Public Health, Baltimore, Maryland (Fallin); School of Public Health, University of Alabama at Birmingham (Go); SABA University School of Medicine, SABA, Dutch Caribbean (Griffith); Division of Geriatrics, Howard University Hospital, Washington, DC (Obisesan); Department of Human Genetics, University of Pittsburgh, Pittsburgh, Pennsylvania (Kamboh); Alzheimer’s Disease Research Center, University of Pittsburgh, Pittsburgh, Pennsylvania (Kamboh, Lopez, Mayeux); Rush Alzheimer’s Disease Center, Rush University Medical Center, Chicago, Illinois (Bennett); Indiana University Center for Aging Research, Indianapolis (Hendrie); Department of Psychiatry, Indiana University School of Medicine, Indianapolis (Hendrie, Hall); Regenstrief Institute Inc, Indianapolis, Indiana (Hendrie); Hope Center Program on Protein Aggregation and Neurodegeneration, Department of Psychiatry, Washington University School of Medicine, St Louis, Missouri (Goate); Department of Biology, North Carolina A & T University, Greensboro (Byrd); National Alzheimer’s Coordinating Center, Department of Epidemiology, University of Washington, Seattle (Kukull); Vanderbilt Center for Human Genetics Research, Department of Molecular Physiology and Biophysics, Vanderbilt University, Nashville, Tennessee (Haines); Department of Neurology, Boston University, Boston, Massachusetts (Farrer); Department of Epidemiology, Boston University, Boston, Massachusetts (Farrer); Department of Epidemiology, College of Physicians and Surgeons, Columbia University, New York, New York (Lee); Department of Psychiatry, College of Physicians and Surgeons, Columbia University, New York, New York (Mayeux).
1,
Tatiana M Foroud
Tatiana M Foroud, PhD
1Tanz Centre for Research in Neurodegenerative Diseases, University of Toronto, Toronto, Ontario, Canada (Ghani, Sato, St. George-Hyslop, Rogaeva); Taub Institute for Research on Alzheimer’s Disease and the Aging Brain, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Cheng, Manly, Lee, Mayeux); Gertrude H. Sergievsky Center, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Vardarajan, Lee, Mayeux); Department of Neurology, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Manly, Mayeux); Department of Medicine (Biomedical Genetics), Boston University, Boston, Massachusetts (Jun, Logue, Baldwin, Farrer); Department of Biostatistics, Boston University, Boston, Massachusetts (Jun, Lunetta, Farrer); Department of Ophthalmology, Boston University, Boston, Massachusetts (Jun, Farrer); The John P. Hussman Institute for Human Genomics, University of Miami, Miami, Florida (Naj, Rajbhandary, Pericak-Vance); Department of Pathology and Laboratory Medicine, University of Pennsylvania Perelman School of Medicine, Philadelphia (Wang, Valladares, Lin, Cantwell, Schellenberg); Department of Medicine, University of Washington, Seattle (Larson, Crane); Group Health Research Institute, Group Health, Seattle, Washington (Larson); Department of Neuroscience, Mayo Clinic, Jacksonville, Florida (Graff-Radford, Ertekin-Taner); Department of Neurology, Mayo Clinic, Jacksonville, Florida (Graff-Radford, Ertekin-Taner); Rush Institute for Healthy Aging, Department of Internal Medicine, Rush University Medical Center, Chicago, Illinois (Evans); Program in Translational Neuropsychiatric Genomics, Department of Neurology, Brigham and Women’s Hospital, Boston, Massachusetts (De Jager); Harvard Medical School, Boston, Massachusetts (De Jager, Raj, Green); Program in Medical and Population Genetics, The Broad Institute, Cambridge, Massachusetts (De Jager); Department of Psychiatry, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Genetics and Genomics Sciences, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Neuroscience, Mount Sinai School of Medicine, New York, New York (Buxbaum); Friedman Brain Institute, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Medical and Molecular Genetics, Indiana University, Indianapolis (Murrell); Division of Genetics, Department of Medicine, Brigham and Women’s Hospital, Boston, Massachusetts (Green); Partners Center for Personalized Genetic Medicine, Brigham and Women’s Hospital, Boston, Massachusetts (Green); Department of Neurological Sciences, Rush University Medical Center, Chicago, Illinois (Barnes, Bennett); Department of Behavioral Sciences, Rush University Medical Center, Chicago, Illinois (Barnes, Foroud); Department of Epidemiology, Johns Hopkins University School of Public Health, Baltimore, Maryland (Fallin); School of Public Health, University of Alabama at Birmingham (Go); SABA University School of Medicine, SABA, Dutch Caribbean (Griffith); Division of Geriatrics, Howard University Hospital, Washington, DC (Obisesan); Department of Human Genetics, University of Pittsburgh, Pittsburgh, Pennsylvania (Kamboh); Alzheimer’s Disease Research Center, University of Pittsburgh, Pittsburgh, Pennsylvania (Kamboh, Lopez, Mayeux); Rush Alzheimer’s Disease Center, Rush University Medical Center, Chicago, Illinois (Bennett); Indiana University Center for Aging Research, Indianapolis (Hendrie); Department of Psychiatry, Indiana University School of Medicine, Indianapolis (Hendrie, Hall); Regenstrief Institute Inc, Indianapolis, Indiana (Hendrie); Hope Center Program on Protein Aggregation and Neurodegeneration, Department of Psychiatry, Washington University School of Medicine, St Louis, Missouri (Goate); Department of Biology, North Carolina A & T University, Greensboro (Byrd); National Alzheimer’s Coordinating Center, Department of Epidemiology, University of Washington, Seattle (Kukull); Vanderbilt Center for Human Genetics Research, Department of Molecular Physiology and Biophysics, Vanderbilt University, Nashville, Tennessee (Haines); Department of Neurology, Boston University, Boston, Massachusetts (Farrer); Department of Epidemiology, Boston University, Boston, Massachusetts (Farrer); Department of Epidemiology, College of Physicians and Surgeons, Columbia University, New York, New York (Lee); Department of Psychiatry, College of Physicians and Surgeons, Columbia University, New York, New York (Mayeux).
1,
Jonathan L Haines
Jonathan L Haines, PhD
1Tanz Centre for Research in Neurodegenerative Diseases, University of Toronto, Toronto, Ontario, Canada (Ghani, Sato, St. George-Hyslop, Rogaeva); Taub Institute for Research on Alzheimer’s Disease and the Aging Brain, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Cheng, Manly, Lee, Mayeux); Gertrude H. Sergievsky Center, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Vardarajan, Lee, Mayeux); Department of Neurology, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Manly, Mayeux); Department of Medicine (Biomedical Genetics), Boston University, Boston, Massachusetts (Jun, Logue, Baldwin, Farrer); Department of Biostatistics, Boston University, Boston, Massachusetts (Jun, Lunetta, Farrer); Department of Ophthalmology, Boston University, Boston, Massachusetts (Jun, Farrer); The John P. Hussman Institute for Human Genomics, University of Miami, Miami, Florida (Naj, Rajbhandary, Pericak-Vance); Department of Pathology and Laboratory Medicine, University of Pennsylvania Perelman School of Medicine, Philadelphia (Wang, Valladares, Lin, Cantwell, Schellenberg); Department of Medicine, University of Washington, Seattle (Larson, Crane); Group Health Research Institute, Group Health, Seattle, Washington (Larson); Department of Neuroscience, Mayo Clinic, Jacksonville, Florida (Graff-Radford, Ertekin-Taner); Department of Neurology, Mayo Clinic, Jacksonville, Florida (Graff-Radford, Ertekin-Taner); Rush Institute for Healthy Aging, Department of Internal Medicine, Rush University Medical Center, Chicago, Illinois (Evans); Program in Translational Neuropsychiatric Genomics, Department of Neurology, Brigham and Women’s Hospital, Boston, Massachusetts (De Jager); Harvard Medical School, Boston, Massachusetts (De Jager, Raj, Green); Program in Medical and Population Genetics, The Broad Institute, Cambridge, Massachusetts (De Jager); Department of Psychiatry, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Genetics and Genomics Sciences, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Neuroscience, Mount Sinai School of Medicine, New York, New York (Buxbaum); Friedman Brain Institute, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Medical and Molecular Genetics, Indiana University, Indianapolis (Murrell); Division of Genetics, Department of Medicine, Brigham and Women’s Hospital, Boston, Massachusetts (Green); Partners Center for Personalized Genetic Medicine, Brigham and Women’s Hospital, Boston, Massachusetts (Green); Department of Neurological Sciences, Rush University Medical Center, Chicago, Illinois (Barnes, Bennett); Department of Behavioral Sciences, Rush University Medical Center, Chicago, Illinois (Barnes, Foroud); Department of Epidemiology, Johns Hopkins University School of Public Health, Baltimore, Maryland (Fallin); School of Public Health, University of Alabama at Birmingham (Go); SABA University School of Medicine, SABA, Dutch Caribbean (Griffith); Division of Geriatrics, Howard University Hospital, Washington, DC (Obisesan); Department of Human Genetics, University of Pittsburgh, Pittsburgh, Pennsylvania (Kamboh); Alzheimer’s Disease Research Center, University of Pittsburgh, Pittsburgh, Pennsylvania (Kamboh, Lopez, Mayeux); Rush Alzheimer’s Disease Center, Rush University Medical Center, Chicago, Illinois (Bennett); Indiana University Center for Aging Research, Indianapolis (Hendrie); Department of Psychiatry, Indiana University School of Medicine, Indianapolis (Hendrie, Hall); Regenstrief Institute Inc, Indianapolis, Indiana (Hendrie); Hope Center Program on Protein Aggregation and Neurodegeneration, Department of Psychiatry, Washington University School of Medicine, St Louis, Missouri (Goate); Department of Biology, North Carolina A & T University, Greensboro (Byrd); National Alzheimer’s Coordinating Center, Department of Epidemiology, University of Washington, Seattle (Kukull); Vanderbilt Center for Human Genetics Research, Department of Molecular Physiology and Biophysics, Vanderbilt University, Nashville, Tennessee (Haines); Department of Neurology, Boston University, Boston, Massachusetts (Farrer); Department of Epidemiology, Boston University, Boston, Massachusetts (Farrer); Department of Epidemiology, College of Physicians and Surgeons, Columbia University, New York, New York (Lee); Department of Psychiatry, College of Physicians and Surgeons, Columbia University, New York, New York (Mayeux).
1,
Lindsay A Farrer
Lindsay A Farrer, PhD
1Tanz Centre for Research in Neurodegenerative Diseases, University of Toronto, Toronto, Ontario, Canada (Ghani, Sato, St. George-Hyslop, Rogaeva); Taub Institute for Research on Alzheimer’s Disease and the Aging Brain, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Cheng, Manly, Lee, Mayeux); Gertrude H. Sergievsky Center, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Vardarajan, Lee, Mayeux); Department of Neurology, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Manly, Mayeux); Department of Medicine (Biomedical Genetics), Boston University, Boston, Massachusetts (Jun, Logue, Baldwin, Farrer); Department of Biostatistics, Boston University, Boston, Massachusetts (Jun, Lunetta, Farrer); Department of Ophthalmology, Boston University, Boston, Massachusetts (Jun, Farrer); The John P. Hussman Institute for Human Genomics, University of Miami, Miami, Florida (Naj, Rajbhandary, Pericak-Vance); Department of Pathology and Laboratory Medicine, University of Pennsylvania Perelman School of Medicine, Philadelphia (Wang, Valladares, Lin, Cantwell, Schellenberg); Department of Medicine, University of Washington, Seattle (Larson, Crane); Group Health Research Institute, Group Health, Seattle, Washington (Larson); Department of Neuroscience, Mayo Clinic, Jacksonville, Florida (Graff-Radford, Ertekin-Taner); Department of Neurology, Mayo Clinic, Jacksonville, Florida (Graff-Radford, Ertekin-Taner); Rush Institute for Healthy Aging, Department of Internal Medicine, Rush University Medical Center, Chicago, Illinois (Evans); Program in Translational Neuropsychiatric Genomics, Department of Neurology, Brigham and Women’s Hospital, Boston, Massachusetts (De Jager); Harvard Medical School, Boston, Massachusetts (De Jager, Raj, Green); Program in Medical and Population Genetics, The Broad Institute, Cambridge, Massachusetts (De Jager); Department of Psychiatry, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Genetics and Genomics Sciences, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Neuroscience, Mount Sinai School of Medicine, New York, New York (Buxbaum); Friedman Brain Institute, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Medical and Molecular Genetics, Indiana University, Indianapolis (Murrell); Division of Genetics, Department of Medicine, Brigham and Women’s Hospital, Boston, Massachusetts (Green); Partners Center for Personalized Genetic Medicine, Brigham and Women’s Hospital, Boston, Massachusetts (Green); Department of Neurological Sciences, Rush University Medical Center, Chicago, Illinois (Barnes, Bennett); Department of Behavioral Sciences, Rush University Medical Center, Chicago, Illinois (Barnes, Foroud); Department of Epidemiology, Johns Hopkins University School of Public Health, Baltimore, Maryland (Fallin); School of Public Health, University of Alabama at Birmingham (Go); SABA University School of Medicine, SABA, Dutch Caribbean (Griffith); Division of Geriatrics, Howard University Hospital, Washington, DC (Obisesan); Department of Human Genetics, University of Pittsburgh, Pittsburgh, Pennsylvania (Kamboh); Alzheimer’s Disease Research Center, University of Pittsburgh, Pittsburgh, Pennsylvania (Kamboh, Lopez, Mayeux); Rush Alzheimer’s Disease Center, Rush University Medical Center, Chicago, Illinois (Bennett); Indiana University Center for Aging Research, Indianapolis (Hendrie); Department of Psychiatry, Indiana University School of Medicine, Indianapolis (Hendrie, Hall); Regenstrief Institute Inc, Indianapolis, Indiana (Hendrie); Hope Center Program on Protein Aggregation and Neurodegeneration, Department of Psychiatry, Washington University School of Medicine, St Louis, Missouri (Goate); Department of Biology, North Carolina A & T University, Greensboro (Byrd); National Alzheimer’s Coordinating Center, Department of Epidemiology, University of Washington, Seattle (Kukull); Vanderbilt Center for Human Genetics Research, Department of Molecular Physiology and Biophysics, Vanderbilt University, Nashville, Tennessee (Haines); Department of Neurology, Boston University, Boston, Massachusetts (Farrer); Department of Epidemiology, Boston University, Boston, Massachusetts (Farrer); Department of Epidemiology, College of Physicians and Surgeons, Columbia University, New York, New York (Lee); Department of Psychiatry, College of Physicians and Surgeons, Columbia University, New York, New York (Mayeux).
1,
Margaret A Pericak-Vance
Margaret A Pericak-Vance, PhD
1Tanz Centre for Research in Neurodegenerative Diseases, University of Toronto, Toronto, Ontario, Canada (Ghani, Sato, St. George-Hyslop, Rogaeva); Taub Institute for Research on Alzheimer’s Disease and the Aging Brain, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Cheng, Manly, Lee, Mayeux); Gertrude H. Sergievsky Center, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Vardarajan, Lee, Mayeux); Department of Neurology, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Manly, Mayeux); Department of Medicine (Biomedical Genetics), Boston University, Boston, Massachusetts (Jun, Logue, Baldwin, Farrer); Department of Biostatistics, Boston University, Boston, Massachusetts (Jun, Lunetta, Farrer); Department of Ophthalmology, Boston University, Boston, Massachusetts (Jun, Farrer); The John P. Hussman Institute for Human Genomics, University of Miami, Miami, Florida (Naj, Rajbhandary, Pericak-Vance); Department of Pathology and Laboratory Medicine, University of Pennsylvania Perelman School of Medicine, Philadelphia (Wang, Valladares, Lin, Cantwell, Schellenberg); Department of Medicine, University of Washington, Seattle (Larson, Crane); Group Health Research Institute, Group Health, Seattle, Washington (Larson); Department of Neuroscience, Mayo Clinic, Jacksonville, Florida (Graff-Radford, Ertekin-Taner); Department of Neurology, Mayo Clinic, Jacksonville, Florida (Graff-Radford, Ertekin-Taner); Rush Institute for Healthy Aging, Department of Internal Medicine, Rush University Medical Center, Chicago, Illinois (Evans); Program in Translational Neuropsychiatric Genomics, Department of Neurology, Brigham and Women’s Hospital, Boston, Massachusetts (De Jager); Harvard Medical School, Boston, Massachusetts (De Jager, Raj, Green); Program in Medical and Population Genetics, The Broad Institute, Cambridge, Massachusetts (De Jager); Department of Psychiatry, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Genetics and Genomics Sciences, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Neuroscience, Mount Sinai School of Medicine, New York, New York (Buxbaum); Friedman Brain Institute, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Medical and Molecular Genetics, Indiana University, Indianapolis (Murrell); Division of Genetics, Department of Medicine, Brigham and Women’s Hospital, Boston, Massachusetts (Green); Partners Center for Personalized Genetic Medicine, Brigham and Women’s Hospital, Boston, Massachusetts (Green); Department of Neurological Sciences, Rush University Medical Center, Chicago, Illinois (Barnes, Bennett); Department of Behavioral Sciences, Rush University Medical Center, Chicago, Illinois (Barnes, Foroud); Department of Epidemiology, Johns Hopkins University School of Public Health, Baltimore, Maryland (Fallin); School of Public Health, University of Alabama at Birmingham (Go); SABA University School of Medicine, SABA, Dutch Caribbean (Griffith); Division of Geriatrics, Howard University Hospital, Washington, DC (Obisesan); Department of Human Genetics, University of Pittsburgh, Pittsburgh, Pennsylvania (Kamboh); Alzheimer’s Disease Research Center, University of Pittsburgh, Pittsburgh, Pennsylvania (Kamboh, Lopez, Mayeux); Rush Alzheimer’s Disease Center, Rush University Medical Center, Chicago, Illinois (Bennett); Indiana University Center for Aging Research, Indianapolis (Hendrie); Department of Psychiatry, Indiana University School of Medicine, Indianapolis (Hendrie, Hall); Regenstrief Institute Inc, Indianapolis, Indiana (Hendrie); Hope Center Program on Protein Aggregation and Neurodegeneration, Department of Psychiatry, Washington University School of Medicine, St Louis, Missouri (Goate); Department of Biology, North Carolina A & T University, Greensboro (Byrd); National Alzheimer’s Coordinating Center, Department of Epidemiology, University of Washington, Seattle (Kukull); Vanderbilt Center for Human Genetics Research, Department of Molecular Physiology and Biophysics, Vanderbilt University, Nashville, Tennessee (Haines); Department of Neurology, Boston University, Boston, Massachusetts (Farrer); Department of Epidemiology, Boston University, Boston, Massachusetts (Farrer); Department of Epidemiology, College of Physicians and Surgeons, Columbia University, New York, New York (Lee); Department of Psychiatry, College of Physicians and Surgeons, Columbia University, New York, New York (Mayeux).
1,
Joseph H Lee
Joseph H Lee, PhD
1Tanz Centre for Research in Neurodegenerative Diseases, University of Toronto, Toronto, Ontario, Canada (Ghani, Sato, St. George-Hyslop, Rogaeva); Taub Institute for Research on Alzheimer’s Disease and the Aging Brain, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Cheng, Manly, Lee, Mayeux); Gertrude H. Sergievsky Center, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Vardarajan, Lee, Mayeux); Department of Neurology, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Manly, Mayeux); Department of Medicine (Biomedical Genetics), Boston University, Boston, Massachusetts (Jun, Logue, Baldwin, Farrer); Department of Biostatistics, Boston University, Boston, Massachusetts (Jun, Lunetta, Farrer); Department of Ophthalmology, Boston University, Boston, Massachusetts (Jun, Farrer); The John P. Hussman Institute for Human Genomics, University of Miami, Miami, Florida (Naj, Rajbhandary, Pericak-Vance); Department of Pathology and Laboratory Medicine, University of Pennsylvania Perelman School of Medicine, Philadelphia (Wang, Valladares, Lin, Cantwell, Schellenberg); Department of Medicine, University of Washington, Seattle (Larson, Crane); Group Health Research Institute, Group Health, Seattle, Washington (Larson); Department of Neuroscience, Mayo Clinic, Jacksonville, Florida (Graff-Radford, Ertekin-Taner); Department of Neurology, Mayo Clinic, Jacksonville, Florida (Graff-Radford, Ertekin-Taner); Rush Institute for Healthy Aging, Department of Internal Medicine, Rush University Medical Center, Chicago, Illinois (Evans); Program in Translational Neuropsychiatric Genomics, Department of Neurology, Brigham and Women’s Hospital, Boston, Massachusetts (De Jager); Harvard Medical School, Boston, Massachusetts (De Jager, Raj, Green); Program in Medical and Population Genetics, The Broad Institute, Cambridge, Massachusetts (De Jager); Department of Psychiatry, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Genetics and Genomics Sciences, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Neuroscience, Mount Sinai School of Medicine, New York, New York (Buxbaum); Friedman Brain Institute, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Medical and Molecular Genetics, Indiana University, Indianapolis (Murrell); Division of Genetics, Department of Medicine, Brigham and Women’s Hospital, Boston, Massachusetts (Green); Partners Center for Personalized Genetic Medicine, Brigham and Women’s Hospital, Boston, Massachusetts (Green); Department of Neurological Sciences, Rush University Medical Center, Chicago, Illinois (Barnes, Bennett); Department of Behavioral Sciences, Rush University Medical Center, Chicago, Illinois (Barnes, Foroud); Department of Epidemiology, Johns Hopkins University School of Public Health, Baltimore, Maryland (Fallin); School of Public Health, University of Alabama at Birmingham (Go); SABA University School of Medicine, SABA, Dutch Caribbean (Griffith); Division of Geriatrics, Howard University Hospital, Washington, DC (Obisesan); Department of Human Genetics, University of Pittsburgh, Pittsburgh, Pennsylvania (Kamboh); Alzheimer’s Disease Research Center, University of Pittsburgh, Pittsburgh, Pennsylvania (Kamboh, Lopez, Mayeux); Rush Alzheimer’s Disease Center, Rush University Medical Center, Chicago, Illinois (Bennett); Indiana University Center for Aging Research, Indianapolis (Hendrie); Department of Psychiatry, Indiana University School of Medicine, Indianapolis (Hendrie, Hall); Regenstrief Institute Inc, Indianapolis, Indiana (Hendrie); Hope Center Program on Protein Aggregation and Neurodegeneration, Department of Psychiatry, Washington University School of Medicine, St Louis, Missouri (Goate); Department of Biology, North Carolina A & T University, Greensboro (Byrd); National Alzheimer’s Coordinating Center, Department of Epidemiology, University of Washington, Seattle (Kukull); Vanderbilt Center for Human Genetics Research, Department of Molecular Physiology and Biophysics, Vanderbilt University, Nashville, Tennessee (Haines); Department of Neurology, Boston University, Boston, Massachusetts (Farrer); Department of Epidemiology, Boston University, Boston, Massachusetts (Farrer); Department of Epidemiology, College of Physicians and Surgeons, Columbia University, New York, New York (Lee); Department of Psychiatry, College of Physicians and Surgeons, Columbia University, New York, New York (Mayeux).
1,
Gerard D Schellenberg
Gerard D Schellenberg, PhD
1Tanz Centre for Research in Neurodegenerative Diseases, University of Toronto, Toronto, Ontario, Canada (Ghani, Sato, St. George-Hyslop, Rogaeva); Taub Institute for Research on Alzheimer’s Disease and the Aging Brain, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Cheng, Manly, Lee, Mayeux); Gertrude H. Sergievsky Center, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Vardarajan, Lee, Mayeux); Department of Neurology, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Manly, Mayeux); Department of Medicine (Biomedical Genetics), Boston University, Boston, Massachusetts (Jun, Logue, Baldwin, Farrer); Department of Biostatistics, Boston University, Boston, Massachusetts (Jun, Lunetta, Farrer); Department of Ophthalmology, Boston University, Boston, Massachusetts (Jun, Farrer); The John P. Hussman Institute for Human Genomics, University of Miami, Miami, Florida (Naj, Rajbhandary, Pericak-Vance); Department of Pathology and Laboratory Medicine, University of Pennsylvania Perelman School of Medicine, Philadelphia (Wang, Valladares, Lin, Cantwell, Schellenberg); Department of Medicine, University of Washington, Seattle (Larson, Crane); Group Health Research Institute, Group Health, Seattle, Washington (Larson); Department of Neuroscience, Mayo Clinic, Jacksonville, Florida (Graff-Radford, Ertekin-Taner); Department of Neurology, Mayo Clinic, Jacksonville, Florida (Graff-Radford, Ertekin-Taner); Rush Institute for Healthy Aging, Department of Internal Medicine, Rush University Medical Center, Chicago, Illinois (Evans); Program in Translational Neuropsychiatric Genomics, Department of Neurology, Brigham and Women’s Hospital, Boston, Massachusetts (De Jager); Harvard Medical School, Boston, Massachusetts (De Jager, Raj, Green); Program in Medical and Population Genetics, The Broad Institute, Cambridge, Massachusetts (De Jager); Department of Psychiatry, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Genetics and Genomics Sciences, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Neuroscience, Mount Sinai School of Medicine, New York, New York (Buxbaum); Friedman Brain Institute, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Medical and Molecular Genetics, Indiana University, Indianapolis (Murrell); Division of Genetics, Department of Medicine, Brigham and Women’s Hospital, Boston, Massachusetts (Green); Partners Center for Personalized Genetic Medicine, Brigham and Women’s Hospital, Boston, Massachusetts (Green); Department of Neurological Sciences, Rush University Medical Center, Chicago, Illinois (Barnes, Bennett); Department of Behavioral Sciences, Rush University Medical Center, Chicago, Illinois (Barnes, Foroud); Department of Epidemiology, Johns Hopkins University School of Public Health, Baltimore, Maryland (Fallin); School of Public Health, University of Alabama at Birmingham (Go); SABA University School of Medicine, SABA, Dutch Caribbean (Griffith); Division of Geriatrics, Howard University Hospital, Washington, DC (Obisesan); Department of Human Genetics, University of Pittsburgh, Pittsburgh, Pennsylvania (Kamboh); Alzheimer’s Disease Research Center, University of Pittsburgh, Pittsburgh, Pennsylvania (Kamboh, Lopez, Mayeux); Rush Alzheimer’s Disease Center, Rush University Medical Center, Chicago, Illinois (Bennett); Indiana University Center for Aging Research, Indianapolis (Hendrie); Department of Psychiatry, Indiana University School of Medicine, Indianapolis (Hendrie, Hall); Regenstrief Institute Inc, Indianapolis, Indiana (Hendrie); Hope Center Program on Protein Aggregation and Neurodegeneration, Department of Psychiatry, Washington University School of Medicine, St Louis, Missouri (Goate); Department of Biology, North Carolina A & T University, Greensboro (Byrd); National Alzheimer’s Coordinating Center, Department of Epidemiology, University of Washington, Seattle (Kukull); Vanderbilt Center for Human Genetics Research, Department of Molecular Physiology and Biophysics, Vanderbilt University, Nashville, Tennessee (Haines); Department of Neurology, Boston University, Boston, Massachusetts (Farrer); Department of Epidemiology, Boston University, Boston, Massachusetts (Farrer); Department of Epidemiology, College of Physicians and Surgeons, Columbia University, New York, New York (Lee); Department of Psychiatry, College of Physicians and Surgeons, Columbia University, New York, New York (Mayeux).
1,
Peter St George-Hyslop
Peter St George-Hyslop, MD
1Tanz Centre for Research in Neurodegenerative Diseases, University of Toronto, Toronto, Ontario, Canada (Ghani, Sato, St. George-Hyslop, Rogaeva); Taub Institute for Research on Alzheimer’s Disease and the Aging Brain, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Cheng, Manly, Lee, Mayeux); Gertrude H. Sergievsky Center, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Vardarajan, Lee, Mayeux); Department of Neurology, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Manly, Mayeux); Department of Medicine (Biomedical Genetics), Boston University, Boston, Massachusetts (Jun, Logue, Baldwin, Farrer); Department of Biostatistics, Boston University, Boston, Massachusetts (Jun, Lunetta, Farrer); Department of Ophthalmology, Boston University, Boston, Massachusetts (Jun, Farrer); The John P. Hussman Institute for Human Genomics, University of Miami, Miami, Florida (Naj, Rajbhandary, Pericak-Vance); Department of Pathology and Laboratory Medicine, University of Pennsylvania Perelman School of Medicine, Philadelphia (Wang, Valladares, Lin, Cantwell, Schellenberg); Department of Medicine, University of Washington, Seattle (Larson, Crane); Group Health Research Institute, Group Health, Seattle, Washington (Larson); Department of Neuroscience, Mayo Clinic, Jacksonville, Florida (Graff-Radford, Ertekin-Taner); Department of Neurology, Mayo Clinic, Jacksonville, Florida (Graff-Radford, Ertekin-Taner); Rush Institute for Healthy Aging, Department of Internal Medicine, Rush University Medical Center, Chicago, Illinois (Evans); Program in Translational Neuropsychiatric Genomics, Department of Neurology, Brigham and Women’s Hospital, Boston, Massachusetts (De Jager); Harvard Medical School, Boston, Massachusetts (De Jager, Raj, Green); Program in Medical and Population Genetics, The Broad Institute, Cambridge, Massachusetts (De Jager); Department of Psychiatry, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Genetics and Genomics Sciences, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Neuroscience, Mount Sinai School of Medicine, New York, New York (Buxbaum); Friedman Brain Institute, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Medical and Molecular Genetics, Indiana University, Indianapolis (Murrell); Division of Genetics, Department of Medicine, Brigham and Women’s Hospital, Boston, Massachusetts (Green); Partners Center for Personalized Genetic Medicine, Brigham and Women’s Hospital, Boston, Massachusetts (Green); Department of Neurological Sciences, Rush University Medical Center, Chicago, Illinois (Barnes, Bennett); Department of Behavioral Sciences, Rush University Medical Center, Chicago, Illinois (Barnes, Foroud); Department of Epidemiology, Johns Hopkins University School of Public Health, Baltimore, Maryland (Fallin); School of Public Health, University of Alabama at Birmingham (Go); SABA University School of Medicine, SABA, Dutch Caribbean (Griffith); Division of Geriatrics, Howard University Hospital, Washington, DC (Obisesan); Department of Human Genetics, University of Pittsburgh, Pittsburgh, Pennsylvania (Kamboh); Alzheimer’s Disease Research Center, University of Pittsburgh, Pittsburgh, Pennsylvania (Kamboh, Lopez, Mayeux); Rush Alzheimer’s Disease Center, Rush University Medical Center, Chicago, Illinois (Bennett); Indiana University Center for Aging Research, Indianapolis (Hendrie); Department of Psychiatry, Indiana University School of Medicine, Indianapolis (Hendrie, Hall); Regenstrief Institute Inc, Indianapolis, Indiana (Hendrie); Hope Center Program on Protein Aggregation and Neurodegeneration, Department of Psychiatry, Washington University School of Medicine, St Louis, Missouri (Goate); Department of Biology, North Carolina A & T University, Greensboro (Byrd); National Alzheimer’s Coordinating Center, Department of Epidemiology, University of Washington, Seattle (Kukull); Vanderbilt Center for Human Genetics Research, Department of Molecular Physiology and Biophysics, Vanderbilt University, Nashville, Tennessee (Haines); Department of Neurology, Boston University, Boston, Massachusetts (Farrer); Department of Epidemiology, Boston University, Boston, Massachusetts (Farrer); Department of Epidemiology, College of Physicians and Surgeons, Columbia University, New York, New York (Lee); Department of Psychiatry, College of Physicians and Surgeons, Columbia University, New York, New York (Mayeux).
1,
Richard Mayeux
Richard Mayeux, MD
1Tanz Centre for Research in Neurodegenerative Diseases, University of Toronto, Toronto, Ontario, Canada (Ghani, Sato, St. George-Hyslop, Rogaeva); Taub Institute for Research on Alzheimer’s Disease and the Aging Brain, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Cheng, Manly, Lee, Mayeux); Gertrude H. Sergievsky Center, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Vardarajan, Lee, Mayeux); Department of Neurology, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Manly, Mayeux); Department of Medicine (Biomedical Genetics), Boston University, Boston, Massachusetts (Jun, Logue, Baldwin, Farrer); Department of Biostatistics, Boston University, Boston, Massachusetts (Jun, Lunetta, Farrer); Department of Ophthalmology, Boston University, Boston, Massachusetts (Jun, Farrer); The John P. Hussman Institute for Human Genomics, University of Miami, Miami, Florida (Naj, Rajbhandary, Pericak-Vance); Department of Pathology and Laboratory Medicine, University of Pennsylvania Perelman School of Medicine, Philadelphia (Wang, Valladares, Lin, Cantwell, Schellenberg); Department of Medicine, University of Washington, Seattle (Larson, Crane); Group Health Research Institute, Group Health, Seattle, Washington (Larson); Department of Neuroscience, Mayo Clinic, Jacksonville, Florida (Graff-Radford, Ertekin-Taner); Department of Neurology, Mayo Clinic, Jacksonville, Florida (Graff-Radford, Ertekin-Taner); Rush Institute for Healthy Aging, Department of Internal Medicine, Rush University Medical Center, Chicago, Illinois (Evans); Program in Translational Neuropsychiatric Genomics, Department of Neurology, Brigham and Women’s Hospital, Boston, Massachusetts (De Jager); Harvard Medical School, Boston, Massachusetts (De Jager, Raj, Green); Program in Medical and Population Genetics, The Broad Institute, Cambridge, Massachusetts (De Jager); Department of Psychiatry, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Genetics and Genomics Sciences, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Neuroscience, Mount Sinai School of Medicine, New York, New York (Buxbaum); Friedman Brain Institute, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Medical and Molecular Genetics, Indiana University, Indianapolis (Murrell); Division of Genetics, Department of Medicine, Brigham and Women’s Hospital, Boston, Massachusetts (Green); Partners Center for Personalized Genetic Medicine, Brigham and Women’s Hospital, Boston, Massachusetts (Green); Department of Neurological Sciences, Rush University Medical Center, Chicago, Illinois (Barnes, Bennett); Department of Behavioral Sciences, Rush University Medical Center, Chicago, Illinois (Barnes, Foroud); Department of Epidemiology, Johns Hopkins University School of Public Health, Baltimore, Maryland (Fallin); School of Public Health, University of Alabama at Birmingham (Go); SABA University School of Medicine, SABA, Dutch Caribbean (Griffith); Division of Geriatrics, Howard University Hospital, Washington, DC (Obisesan); Department of Human Genetics, University of Pittsburgh, Pittsburgh, Pennsylvania (Kamboh); Alzheimer’s Disease Research Center, University of Pittsburgh, Pittsburgh, Pennsylvania (Kamboh, Lopez, Mayeux); Rush Alzheimer’s Disease Center, Rush University Medical Center, Chicago, Illinois (Bennett); Indiana University Center for Aging Research, Indianapolis (Hendrie); Department of Psychiatry, Indiana University School of Medicine, Indianapolis (Hendrie, Hall); Regenstrief Institute Inc, Indianapolis, Indiana (Hendrie); Hope Center Program on Protein Aggregation and Neurodegeneration, Department of Psychiatry, Washington University School of Medicine, St Louis, Missouri (Goate); Department of Biology, North Carolina A & T University, Greensboro (Byrd); National Alzheimer’s Coordinating Center, Department of Epidemiology, University of Washington, Seattle (Kukull); Vanderbilt Center for Human Genetics Research, Department of Molecular Physiology and Biophysics, Vanderbilt University, Nashville, Tennessee (Haines); Department of Neurology, Boston University, Boston, Massachusetts (Farrer); Department of Epidemiology, Boston University, Boston, Massachusetts (Farrer); Department of Epidemiology, College of Physicians and Surgeons, Columbia University, New York, New York (Lee); Department of Psychiatry, College of Physicians and Surgeons, Columbia University, New York, New York (Mayeux).
1,
Ekaterina Rogaeva
Ekaterina Rogaeva, PhD
1Tanz Centre for Research in Neurodegenerative Diseases, University of Toronto, Toronto, Ontario, Canada (Ghani, Sato, St. George-Hyslop, Rogaeva); Taub Institute for Research on Alzheimer’s Disease and the Aging Brain, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Cheng, Manly, Lee, Mayeux); Gertrude H. Sergievsky Center, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Vardarajan, Lee, Mayeux); Department of Neurology, College of Physicians and Surgeons, Columbia University, New York, New York (Reitz, Manly, Mayeux); Department of Medicine (Biomedical Genetics), Boston University, Boston, Massachusetts (Jun, Logue, Baldwin, Farrer); Department of Biostatistics, Boston University, Boston, Massachusetts (Jun, Lunetta, Farrer); Department of Ophthalmology, Boston University, Boston, Massachusetts (Jun, Farrer); The John P. Hussman Institute for Human Genomics, University of Miami, Miami, Florida (Naj, Rajbhandary, Pericak-Vance); Department of Pathology and Laboratory Medicine, University of Pennsylvania Perelman School of Medicine, Philadelphia (Wang, Valladares, Lin, Cantwell, Schellenberg); Department of Medicine, University of Washington, Seattle (Larson, Crane); Group Health Research Institute, Group Health, Seattle, Washington (Larson); Department of Neuroscience, Mayo Clinic, Jacksonville, Florida (Graff-Radford, Ertekin-Taner); Department of Neurology, Mayo Clinic, Jacksonville, Florida (Graff-Radford, Ertekin-Taner); Rush Institute for Healthy Aging, Department of Internal Medicine, Rush University Medical Center, Chicago, Illinois (Evans); Program in Translational Neuropsychiatric Genomics, Department of Neurology, Brigham and Women’s Hospital, Boston, Massachusetts (De Jager); Harvard Medical School, Boston, Massachusetts (De Jager, Raj, Green); Program in Medical and Population Genetics, The Broad Institute, Cambridge, Massachusetts (De Jager); Department of Psychiatry, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Genetics and Genomics Sciences, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Neuroscience, Mount Sinai School of Medicine, New York, New York (Buxbaum); Friedman Brain Institute, Mount Sinai School of Medicine, New York, New York (Buxbaum); Department of Medical and Molecular Genetics, Indiana University, Indianapolis (Murrell); Division of Genetics, Department of Medicine, Brigham and Women’s Hospital, Boston, Massachusetts (Green); Partners Center for Personalized Genetic Medicine, Brigham and Women’s Hospital, Boston, Massachusetts (Green); Department of Neurological Sciences, Rush University Medical Center, Chicago, Illinois (Barnes, Bennett); Department of Behavioral Sciences, Rush University Medical Center, Chicago, Illinois (Barnes, Foroud); Department of Epidemiology, Johns Hopkins University School of Public Health, Baltimore, Maryland (Fallin); School of Public Health, University of Alabama at Birmingham (Go); SABA University School of Medicine, SABA, Dutch Caribbean (Griffith); Division of Geriatrics, Howard University Hospital, Washington, DC (Obisesan); Department of Human Genetics, University of Pittsburgh, Pittsburgh, Pennsylvania (Kamboh); Alzheimer’s Disease Research Center, University of Pittsburgh, Pittsburgh, Pennsylvania (Kamboh, Lopez, Mayeux); Rush Alzheimer’s Disease Center, Rush University Medical Center, Chicago, Illinois (Bennett); Indiana University Center for Aging Research, Indianapolis (Hendrie); Department of Psychiatry, Indiana University School of Medicine, Indianapolis (Hendrie, Hall); Regenstrief Institute Inc, Indianapolis, Indiana (Hendrie); Hope Center Program on Protein Aggregation and Neurodegeneration, Department of Psychiatry, Washington University School of Medicine, St Louis, Missouri (Goate); Department of Biology, North Carolina A & T University, Greensboro (Byrd); National Alzheimer’s Coordinating Center, Department of Epidemiology, University of Washington, Seattle (Kukull); Vanderbilt Center for Human Genetics Research, Department of Molecular Physiology and Biophysics, Vanderbilt University, Nashville, Tennessee (Haines); Department of Neurology, Boston University, Boston, Massachusetts (Farrer); Department of Epidemiology, Boston University, Boston, Massachusetts (Farrer); Department of Epidemiology, College of Physicians and Surgeons, Columbia University, New York, New York (Lee); Department of Psychiatry, College of Physicians and Surgeons, Columbia University, New York, New York (Mayeux).
1,
for the Alzheimer’s Disease Genetics Consortium