Abstract
CRISPR/Cas systems act to protect the cell from invading nucleic acids in many bacteria and archaea. The bacterial immune protein Cas9 is a component of one of these CRISPR/Cas systems and has recently been adapted as a tool for genome editing. Cas9 is easily targeted to bind and cleave a DNA sequence via a complimentary RNA; this straightforward programmability has gained Cas9 rapid acceptance in the field of genetic engineering. While this technology has developed quickly, a number of challenges regarding Cas9 specificity, efficiency, fusion protein function, and spatiotemporal control within the cell remain. In this work, we develop a platform for constructing novel proteins to address these open questions. We demonstrate methods to either screen or select active Cas9 mutants and use the screening technique to isolate functional Cas9 variants with a heterologous PDZ domain inserted directly into the protein. As a proof of concept, these methods lay the groundwork for the future construction of diverse Cas9 proteins. Straightforward and accessible techniques for genetic editing are helping to elucidate biology in new and exciting ways; a platform to engineer new functionalities into Cas9 will help forge the next generation of genome modifying tools.
Keywords: CRISPR, Cas9, protein engineering, nuclease, genome editing, gene activation/repression, PDZ, synthetic biology
Acronyms: SpCas9, IPTG, sgRNA, RNA, DNA, ssDNA, crRNA, tracrRNA, PAM, PI, BH, EM, aTC, dCas9, FACS, GFP, RFP, LB, NHEJ, HR, WT Cas9
Introduction
The manipulation of gene sequence and expression is fundamental to unraveling the complexity of biological systems. However, our inability to make such manipulations across different organisms and cell types has limited the power of recombinant DNA technology to a handful of model systems. Consequently, numerous strategies for genome engineering – the ability to programmably disrupt or replace genomic loci - have emerged in recent years, yet there remains no universal solution to the problem (Carroll, 2014).
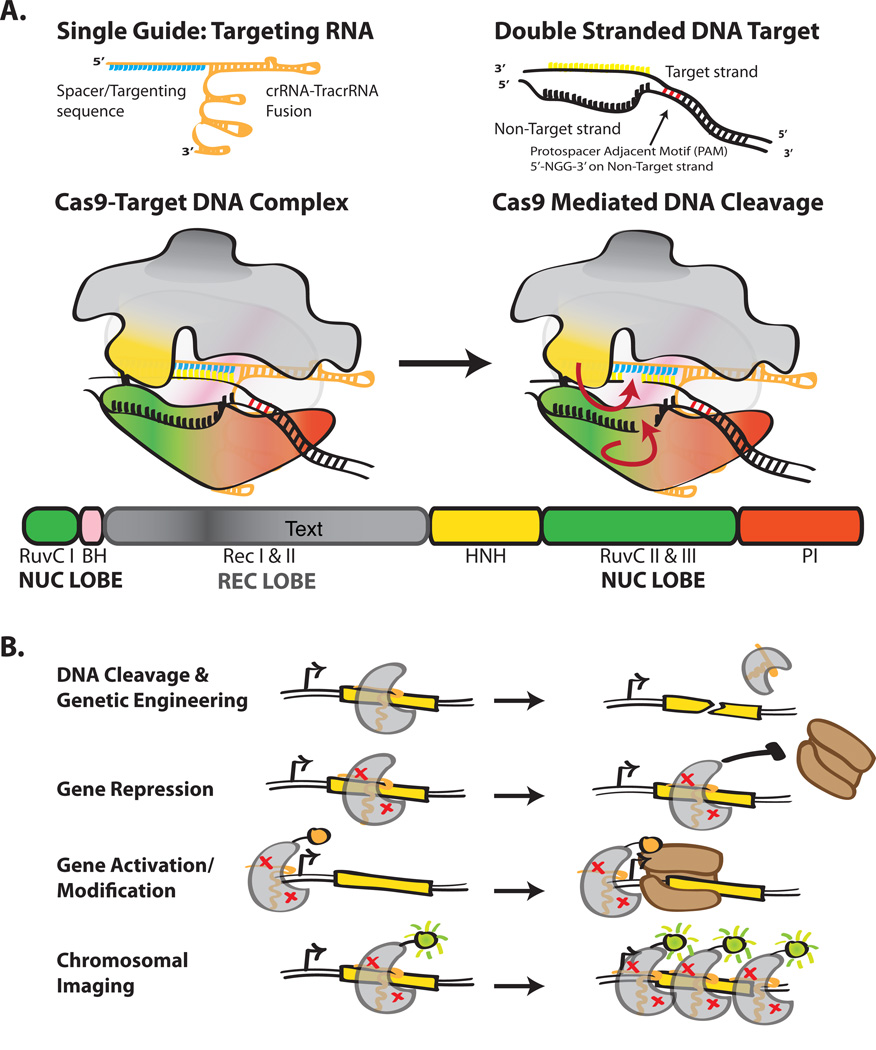
Investigation of bacterial adaptive immunity, known as the ‘clustered regularly interspaced short palindromic repeats’ (CRISPR) system, led to the discovery of the RNA-guided DNA nuclease Cas9, which has proven particularly potent for genome engineering (Barrangou et al., 2007; Deltcheva et al., 2011; Jinek et al., 2012; Wiedenheft, Sternberg, & Doudna, 2012). In its biological context, Cas9 is part of a Type II CRISPR interference system that functions to degrade pathogenic phage or plasmid DNA (Fig. 1A). Targeting of Cas9 is enabled by host-encoded CRISPR-RNAs (crRNAs), which recognize, through RNA:DNA hybridization, 20 bp of complementary target DNA sequence (referred to as a protospacer). The Cas9 protein itself also plays a role in target recognition by binding a short DNA sequence adjacent and opposite the protospacer, called the protospacer adjacent motif (PAM). Although there is significant variation in PAM specificity among Cas9 orthologs the commonly employed Cas9 from Streptococcus pyogenes (SpCas9) recognizes the PAM sequence 5'-NGG-3'. PAM binding is thought to prime Cas9 for target recognition by the crRNA sequence (Sternberg, Redding, Jinek, Greene, & Doudna, 2014). Upon target recognition, two nuclease domains, termed the RuvC and HNH domains because of their sequence similarity to other endonucleases, engage and cleave the separated strands of DNA between 3 and 4 bp upstream of the PAM site (Jinek et al., 2012). A second trans-activating RNA (tracrRNA), with partial complementarity to crRNA, is also required for crRNA maturation and activity. Doudna and colleagues have shown that the crRNA and tracrRNA can be fused together with a tetraloop insertion to form a single guide RNA (sgRNA or ‘guide’) (Jinek et al., 2012). Expression of Cas9 and this sgRNA is both necessary and sufficient for targeting DNA. Therefore, the rapid success of Cas9-based engineering has been driven by programmability – Cas9 can be targeted to any DNA locus by simply changing the sgRNA sequence.
Figure 1.
Holo Cas9 structure and its potential uses. (A) Single guide RNA, target dsDNA and Cas9 modeled. Domains of Cas9 are colored accordingly, RuvC-green, BH-pink, RecI-gray, RecII- dark grey, HNH-yellow, PI-red. (B) Common uses of Cas9 as a tool. Red x's in Cas9 represent a nuclease dead variant.
Intense interest in Cas9-based genomic engineering has already led to a number of directed alterations to change or improve Cas9 functionality (Fig. 1B). Based on sequence conservation of the RuvC and HNH nuclease domains, a number of point mutants were constructed to transform the normal endonuclease activity into either a nickase (for genome editing) or a catalytically dead mutant (dCas9) that can function as a transcription inhibitor (CRISPRi) (Cong et al., 2013; Jinek et al., 2012; Qi et al., 2013). As PAM recognition is critical to functionality but encoded by the protein, a number of efforts have identified Cas9 orthologs with minimal PAM requirements for use in conjunction with, or in place of, SpCas9 (Esvelt et al., 2013). Finally, a number of N- and C-terminal fusions to Cas9 have been used to recruit alternative factors to specific DNA loci, including RNA polymerase subunits to activate transcription and additional nuclease domains for improving the on-target specificity of genome editing (Bikard et al., 2013; Guilinger, Thompson, & Liu, 2014; Tsai et al., 2014). These advances show that, from an engineering perspective, Cas9 can be thought of as a unifying factor able to recruit any protein, RNA, and DNA together in the cell (Mali, Esvelt, & Church, 2013b). However, even with these recent improvements, there are a number of additional desirable that could be engineered into Cas9.
Enabling complex functions requires more elaborate protein engineering efforts than previously attempted. Fortunately, high-resolution structures of apo and holo Cas9 were recently solved and inform this process greatly (Jinek et al., 2014; Nishimasu et al., 2014). Thus, we begin with a discussion of structure, which frames the protein engineering effort. This is followed by a discussion of potentially feasible and advantageous manipulations of Cas9. Ultimately, the isolation of an enhanced Cas9 with new function will require assaying the activity of large libraries (> 106) of variants. Fortuitously, the basic function of Cas9, disrupting gene sequence or gene expression, facilitates the construction of a genetic screen and means that directed evolution methods can be employed in the same conditions functionality is ultimately desired. To this end, we present a detailed set of methods, employing either screening or selection, for the directed evolution of novel Cas9 proteins.
1. 1 The structure of Cas9
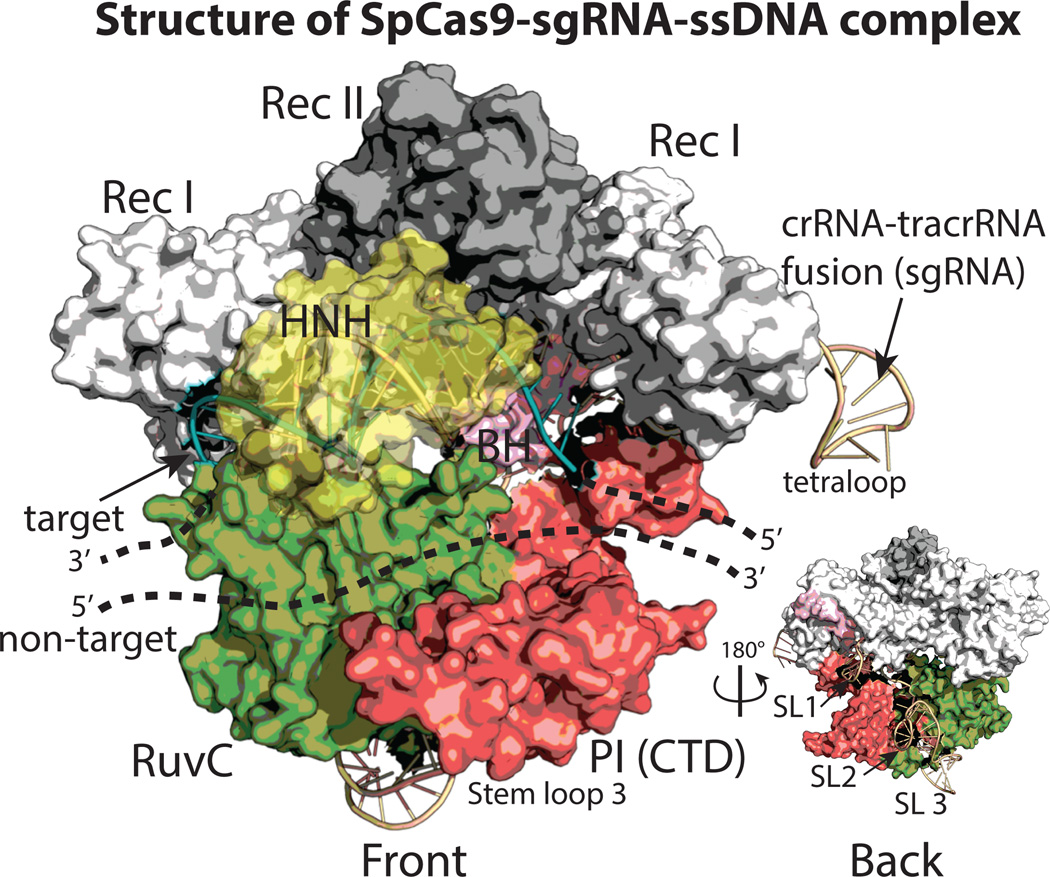
Recently published high-resolution structures of Cas9 serve as a useful starting point to inform the protein engineering process. Doudna and colleagues reported the high-resolution structure of apoSpCas9 using x-ray crystallography and a holo complex using electron microscopy (EM) (Jinek et al., 2014). Concurrently, Zhang and colleagues solved the x-ray structure of SpCas9 bound to sgRNA and single-stranded target DNA (ssDNA) (Nishimasu et al., 2014). Here, we summarize these structures in the context of engineering novel Cas9 variants.
Cas9 possesses a hand-shaped structure of size 100 Å × 100 Å × 50 Å and is composed of two major lobes, an N-terminal recognition (REC) lobe and a C-terminal lobe (NUC) possessing two endonuclease domains (Fig. 2). The REC lobe is composed of three segments: a small portion of the RuvC domain, an arginine-rich bridge helix (BH) which links the REC to the NUC lobe and an α-helical recognition segment with two subdomains (RecI and II). The NUC lobe possesses three domains: the RuvC endonuclease, the HNH endonuclease, and a PAM-interacting (PI) C-terminal domain. The sgRNA-DNA complex lies at the interface of the two lobes, with the BH and RecI domains making many of the primary interactions. It should be noted that the sgRNA-target DNA heteroduplex generally makes non-specific, sequence independent interactions with the protein, while the sgRNA repeat:anti-repeat region makes sequence-dependent interactions; this is consistent with the precise sgRNA recognition yet broad target flexibility of Cas9. Interestingly, although the sgRNA has extensive structural features – there are three stem loops in the tracrRNA sequence – stem loops 2 and 3 exit out the ‘back’ of the structure and make few sequence-specific contacts with Cas9 (inset Fig. 2).
Figure 2.
The structure of Cas9 in complex with sgRNA and a single-stranded target DNA (ssDNA) adapted from Nishimasu et al. 2014 (PDB code 4OO8). The structure is shown in a surface representation and colored as in Fig. 1. Note, the inset is rotated 180° and displays the location of the sgRNA stem loops (SLs).
The structures inform a number of rational protein engineering design strategies. Many publications have employed N and C-terminal fusions to Cas9 as a means of recruiting specific factors to a genomic locus, such as RNA polymerase for transcriptional activation (Bikard et al., 2013; Mali, Aach, et al., 2013a). The structures suggest the protein N-terminus is adjacent to the 3’ end of the target DNA exiting Cas9 while the C-terminus is adjacent to the 5’-end of the same DNA. The close proximity of DNA to protein termini explains probably explains why many simple fusions have been successful. Intriguingly, the N and C termini are in the same lobe and roughly 50 Å apart, suggesting it might be possible to make circularly permuted forms of Cas9 for more elaborate protein engineering uses. Moreover, the bi-lobed nature of the apo complex indicates it could be possible to construct split variants of Cas9 that are only active under conditions when both halves are recruited together. Such variants would be a useful tool in the construction of two-hybrid systems and for the engineering of allosterically controlled Cas9 derivatives, such as with optogenetic domains.
The sequence conservation of Cas9 orthologs mapped onto the structure also suggests domains that may prove malleable for engineering. Phylogenetic variation among Cas9 orthologs is significantly higher in the RecII domain of the REC lobe and the PI domain in the NUC lobe (Chylinski, Makarova, Charpentier, & Koonin, 2014; Nishimasu et al., 2014). The RecII domain makes very few contacts with the sgRNA or target DNA in the holo complex structure and Nishimasu et al. demonstrated that the domain (Δ175–307) can be completely eliminated yet still retain roughly 50% of editing activity. Conversely, two deletions (Δ97–150 and Δ312–409) in the highly conserved RecI domain were catalytically dead in the same assay. This is intriguing because Cas9 is very large; all known Cas9 proteins range from 984–1629 amino acids (Chylinski, Le Rhun, & Charpentier, 2013). Thus, a current goal in the community is to find or engineer smaller, but equally effective, Cas9 proteins and this may be possible through multiple domain deletions. Sequence diversity between PI domains also suggests a means to alter PAM specificity. Cross-linking experiments and structural evidence conclude that the C-terminal PI domain interacts directly with the PAM (Jinek et al., 2014). However, this specificity may in fact be modular. Domain swapping of the SpCas9 PI domain for that of the orthologous sequence from S. thermophilus imbues a preference for the S. thermophilus PAM sequence (TGGCG) (Nishimasu et al., 2014). Expanded domain swapping experiments, coupled with directed evolution, could therefore provide a means for creating orthogonal Cas9 variants from a single, well-characterized SpCas9 scaffold.
1.2 Current uses
Cas9 has rapidly established itself as a promising genome engineering technology in widely used model organisms (Friedland et al., 2013; Gratz et al., 2013; Guilinger et al., 2014; Hou et al., 2013; Hsu et al., 2013; Hwang et al., 2013; Nishimasu et al., 2014; Niu et al., 2014; Shan et al., 2013; Tsai et al., 2014; Wang et al., 2013). In these systems, Cas9 has been used to create both small genomic insertions and deletions (indels) via non-homologus end joining (NHEJ) and to facilitate larger sequence manipulation with homologous recombination (HR). Cas9 also allows for multiplexed genome engineering, and has been used to create large knock-out libraries in human cells, a feat both surprising in its simplicity and impressive in its efficacy (Shalem et al., 2014; Zhou et al., 2014). Decoupling the DNA binding activity of Cas9 proteins from cleavage activity has lead to a broader set of uses such as repression and activation of transcription (Gilbert et al., 2013). Finally, recent evidence suggests that Cas9 may even be able to manipulate RNA (Sampson, Saroj, Llewellyn, Tzeng, & Weiss, 2013). Although still nascent, the simple programmability and effectiveness of Cas9-based technology promises to democratize access to genome manipulation.
1.3 Initial engineering questions
As previously mentioned, there are a number of clear initial questions pertaining to Cas9 that are addressable using existing protein engineering tools. Namely, we believe that designing novel Cas9s with domain insertions and deletions will lead to the creation of a new family of synthetic orthologs whose outputs are manifold. For example, domain insertions could act to recruit a additional protein partners with desired activity onto Cas9-associated nucleic acids; domain deletions will reduce Cas9's size and increase its versatility.
Alternatively, improving N or C-terminal fusions with engineered linkers or creating Cas9s with new N and C-termini altogether, may greatly increase the efficacy of fusions. For example, to address issues of Cas9 targeting specificity dCas9 has been fused to FokI, an obligate dimeric sequence-independent nuclease (Guilinger et al., 2014; Tsai et al., 2014). This system requires the mutual on-target activity of two different FokI-dCas9 fusions to adjacent sites, a combined 40 bp of targeting, to catalyze a DNA cleavage event. Unfortunately, these FokI-dCas9 fusions are substantially less active than either WT Cas9 or the dual nickase strategy at inducing indels, rendering them a less attractive tool. Nevertheless, it is known that FokI protein fusions to other DNA binding domains can achieve cleavage efficiencies similar to that of WT Cas9 (Hwang et al., 2013; Mali, Yang, et al., 2013c). Therefore, lower activity of the current FokI-dCas9 is likely due to imperfect positioning of the FokI nuclease domain and further engineering the dCas9-FokI interface should yield an increase in activity.
Lastly, split proteins are known to function as switches or response elements in many different systems (Olson & Tabor, 2012). Splitting Cas9, as mentioned above, would be a simple method for engineering allosteric control and open the door to a number of uses including optogenetics, small-molecule dependence, or linking function to a cellular signal, such as a phosphorylation-dependent signal transduction cascade. Nevertheless, all of the previous engineered scenarios require that Cas9 is active despite the modifications introduced. Therefore, it is imperative that any engineering attempt start with an assay allowing for the separation of active mutants from the inactive.
Methods
To advance some of the aforementioned goals related to Cas9 protein engineering, we have developed a suite of protocols allowing for the rapid isolation of functional Cas9 proteins. These techniques rely on the ability to screen or select active Cas9 from a large pool of variants. As such, the methods may be applied equally to either rational or library-based approaches for engineering Cas9.
2.1 A note on applications
Although we present here a general method for directed evolution of Cas9, it is impossible to cover the myriad of nuances inherent to different applications. Instead, as a proof of concept, we demonstrate that Cas9 can be manipulated by domain insertion, a common event found in eukaryotic proteomes (Lander et al., 2001). Specifically, we created libraries of Cas9 in which the α-syntrophin PDZ domain was randomly inserted throughout the SpCas9 gene once per variant using in vitro transposition-based methods (Edwards, Busse, Allemann, & Jones, 2008). PDZ domains are small (~ 100 amino acids) proteins with adjacent N- and C-termini that mediate protein-protein interactions by specifically binding the C-terminal peptide of a cognate partner in the cell (Nourry, Grant, & Borg, 2003). These domains have been used extensively in synthetic biology applications as a tool for scaffolding proteins (Dueber, Mirsky, & Lim, 2007; Dueber, Yeh, Chak, & Lim, 2003). In the context of Cas9, PDZ insertion could recruit additional factors to a DNA-bound Cas9 in the cell, such as florescent tags, chromatin remodeling machinery and nuclease domains.
2.2 Electrocompetent E. coli preparation for library construction
Construction of libraries containing 106–109 diverse members is a basic step for engineering new functions into Cas9. A simple method for the creation of electrocompetent cells that consistently yields E. coli with transformation efficiencies of ≥108 per µg of plasmid is described. If necessary, cells can be pre-transformed with additional screening/selection plasmids, as required, to maximize library transformation efficiency.
Begin with the desired strain grown as single colonies on appropriate plates. For example, we use plasmid 44251: pgRNA-bacteria (addgene) with the RFP guide from Qi et al. (2013) transformed and grown on carbenicillin plates (100µg/mL) overnight at 37°C to single colonies.
Pick a single colony and inoculate into 5mL SOC (BD Difco 244310: Super Optimal Broth + 20 mL 20 % glucose per L) plus carbenicillin. Grow overnight at 37°C.
Inoculate 1 L of SOC + carbenicillin with the 5 mL overnight culture. Grow at 37°C for 2–4 hours to an OD600 between 0.55 and 0.65.
Rapidly cool the culture in an ice bath by swirling. Keep cells at during all subsequent steps.
Centrifuge cells at 4000g for 10 min. Wash the cell culture with 500 mL ice-cold water by gentle resuspension. Centrifuge at 4000g for 10 min. Repeat wash step twice.
Wash cells with 500mL of ice cold 10% glycerol. Repeat twice.
Perform a final spin and discard the supernatant. Resuspend the pellet in 2.75 mL of 10% glycerol and aliquot into cold microcentrifuge tubes, 80 µL each. Flash freeze.
Transforming cells: Thaw on ice, add library plasmid to 75 µL cells and vortex. Electroporate at 1800 V, 200ohms, 25uF in 0.1cm cuvettes, resuspending in warm SOC immediately afterward. Recover for one hour at 37°C, before adding antibiotics, as required.
2.3 Discovery of functional, engineered, variants of Cas9 proteins
After generating any library of Cas9 variants, it is necessary to have an platform which can separate active variants from the library with minimal effort. Two assays to probe this functionality are the coupling of dCas9 activity to either RFP expression or media-dependent cell growth. When devising these systems, it is important to keep the First Law of Directed Evolution (Schmidt-Dannert & Arnold, 1999) in mind: ‘you get what you screen for.’
2.4 Screening for Cas9
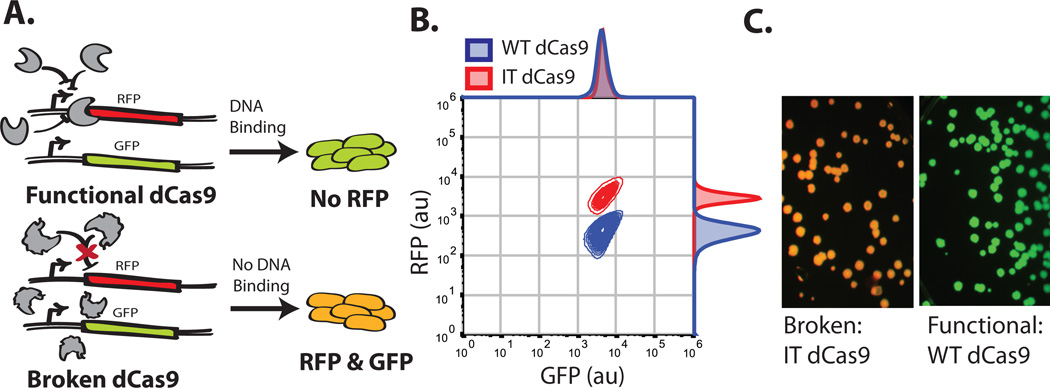
The catalytically dead version of Cas9, has a functional output that can be tied directly to transcription in E.coli; namely, it can repress transcription of a desired gene (Qi et al., 2013). Qi et al. previously demonstrated that dCas9 with a guide sequence of 5'-AACTTTCAGTTTAGCGGTCT-3' can target and repress a genome-encoded RFP while avoiding repression of a genome-encoded upstream GFP (Qi et al., 2013). In a screening context, this provides a simple output for assaying dCas9 functionality (i.e. RFP knock-down) while correcting for extrinsic noise in the population by monitoring GFP (Elowitz, Levine, Siggia, & Swain, 2002). The basic method of screening is schematized in Figure 3A. Briefly, cells containing functional dCas9s will repress RFP and express GFP while those with non-functional dCas9s will express both fluorescent proteins. This signal is easily distinguished using flow cytometry and florescence imaging (Fig.3B and 3C).
Figure 3.
Screen for functional Cas9s. (A) Schematic representation of the screen. (B) Flow cytometry data of the functional positive (WT dCas9) control in blue and negative 'Inactive Truncation' Cas9 (IT dCas9) control in Red, IT dCas9 contains only the C-terminal 250 amino acids. Both controls contain the sgRNA plasmid targeting RFP for repression. Samples were grown overnight in rich induction media. (C) Colony fluorescence of the functional (WT dCas9) and 'Broken' negative (IT dCas9) controls.
2.5 Selecting Cas9
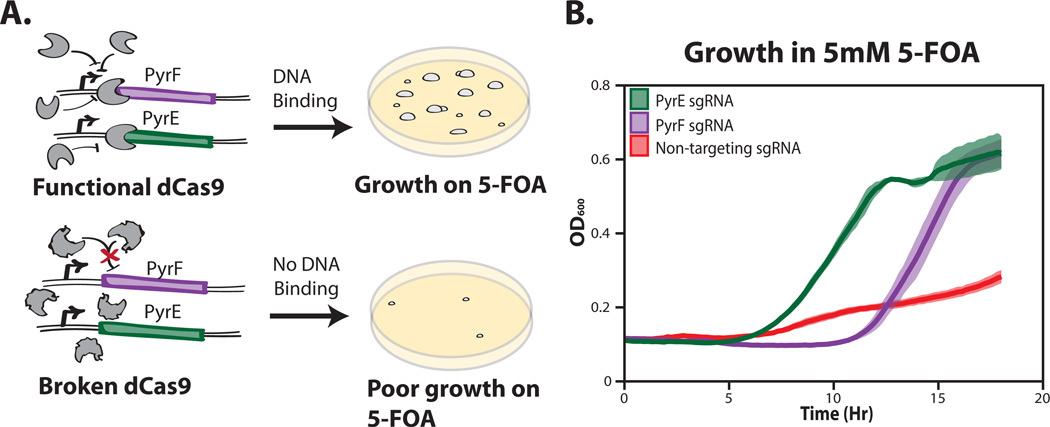
To complement the screening method and for use with larger libraries of Cas9 mutants, we have also developed a technique to select functional dCas9s using cellular growth. We fashioned a derivative of the classic yeast counter-selection method, which takes advantage of the toxicity of 5-fluoroorotic acid (5-FOA) in cells with the URA3 gene (Fig. 4A)(Boeke, Trueheart, Natsoulis, & Fink, 1987). In yeast, URA3 encodes orotidine-5’phosphate decarboxylase, which catalyzes 5-FOA into a highly toxic compound (Boeke, LaCroute, & Fink, 1984). The E.coli homolog of URA3, PyrF, is thought to act in a similar manner, and PyrF and the upstream gene PyrE are known to function as a selectable marker in other gram-negative bacteria (Galvao & de Lorenzo, 2005; Yano, Sanders, Catalano, & Daldal, 2005). Nevertheless, it was unclear whether dCas9 based repression would mimic the effects of these full gene knockouts in an E. coli system. To this end, we tested whether repression of either of PyrF and PyrE by dCas9 was sufficient to rescue a slow growth phenotype on 5-FOA. After creating a number of different sgRNA's to the start of PyrE and PyrF, we determined that the guides 5'-accttcttgatgatgggcac-3' for PyrF and 5'-taagcgcaaattcaataaac-3' for PyrE rescued growth in 5mM of 5-FOA (Fig. 4B).
Figure 4.
Functional Cas9 selection overview. (A) Schematic representation of the selection system. (B) Growth rate of a functional dCas9 + sgRNAs repressing the PyrF gene (purple), PyrE gene (green), and a no guide sequence control (red). Samples were grown in rich induction media +5mM 5-Fluoroorotic Acid. All measurements represent the average (line) and standard deviation (shading) of three biological replicates.
Ultimately, it is important to decide which approach, screening or selection, will be used to enrich for functional engineered Cas9 mutants. A primary determining factor is the theoretical library size. Screening systems can effectively cover libraries of sizes up to ~106, which is roughly the amount of E. coli that can conveniently be sorted 10× by flow cytometer in an hour. On the other hand, selection systems, which rely on repression/activation of a toxic/essential gene for growth, can screen libraries of random protein variants of up to ~109 in size (Persikov, Rowland, Oakes, Singh, & Noyes, 2014).
2.6 Screening for functional Cas9 variants
We have found that Fluorescence-activated cell sorting (FACS) is a convenient method for isolating functional Cas9 variants. This approach is somewhat more flexible than a selection – a gating strategy in FACS is easily manipulated while the growth constraints of a selection are not – yet still provides reasonable throughput. As a proof of concept, we demonstrate the FACS-based screening of functional dCas9 variants possessing the insertion of an α-syntrophin PDZ domain.
Obtain a dCas9 library containing ≤ 106 variants on an expression plasmid of choice. Here we used a tetracycline inducible expression plasmid, plasmid 44249: pdCas9-bacteria from addgene (Qi et al., 2013) to create a library which has a SNTA1 PDZ domain inserted across the whole dCas9 protein (Dueber et al., 2003). Based on the possible insertion sites and linkers, the size of this library is roughly equal to 106.
Transform electrocompetent E. coli expressing GFP and RFP with 1 µg of the library plasmid and 1 µg/µL of a sgRNA to RNA, if necessary. Here, the E. coli strain and guide RNA plasmid come from Qi et al. 2013 (plasmid is 44251: pgRNA-bacteria; addgene).
To ensure adequate coverage of the library the transformation efficiency should be at least 5–10× greater than the theoretical library size. To determine this plate 5 µl aliquots of serially-diluted transformants, and grow to colonies (overnight, 37°C) on double selection-media (chloramphenicol (50 µg/mL) to maintain the engineered dCas9 plasmids and carbenicillin (100 µg/mL) for maintenance of the guide RNA plasmid). Store the remaining transformed cells at 4 °C overnight.
Determine the volume of the transformation mixture needed to cover the theoretical library size 5–10× and inoculate into 5 mL of rich induction media: SOC, chloramphenicol, carbenicillin and 2 µM anhydrotetracycline (aTC). Concurrently, inoculate tubes of rich induction media with controls WT dCas9 and IT dCas9 with the RFP sgRNA. Grow at 37°C; we have found shaking ≥ 250 rpm is helpful for maximum RFP and GFP fluorescence.
After 8–12 hours of growth, centrifuge 500 µL of each sample, wash 2× with 1 mL 1× PBS and re-suspend 1:20 in 1× PBS for flow cytometry.
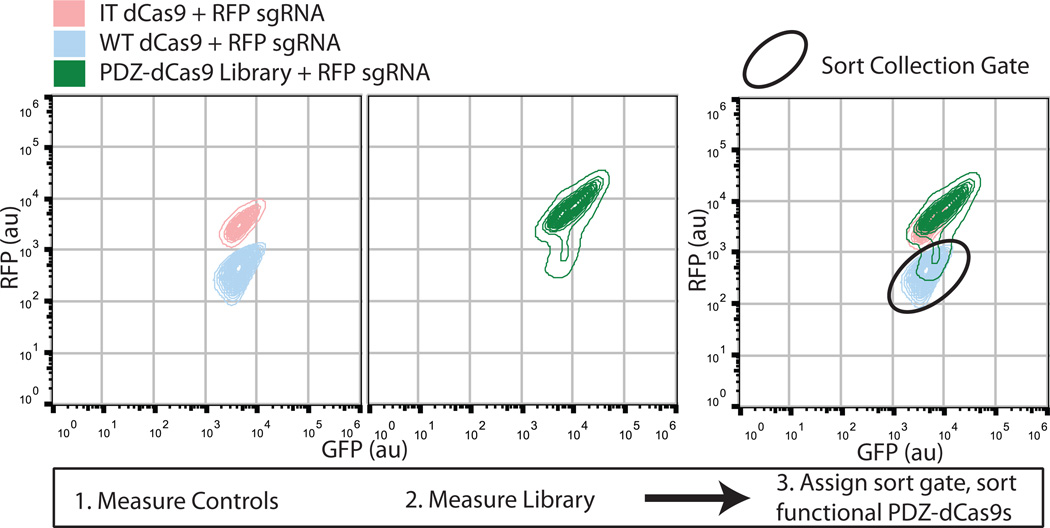
Run the controls on a FACS instrument to establish correct positive and negative gating (Fig 5.).
Screen the library dCas9's using FACS and collect the cells falling into the previously determined positive gate in rich media without antibiotic (Fig. 5). Screen at least 10× the library size to, as cellular viability post-FACS is often substantially less than 100%.
Recover sorted cells for 2 hours at 37 °C.
Depending on the library enrichment after a single round of sorting, repeating steps 4–8 may be necessary to further enrich for functional, engineered dCas9 clones.
Figure 5.
Cell sorting data from the GFP-RFP screen. The first panel depicts the RFP vs GFP measurements of WT dCas9 (blue) and IT dCas9 (pink) as run on a Sony SH800 cell sorter. The separation of these two controls into distinct populations is readily apparent. The second panel portrays the spread of the PDZ-Cas9 intercalation library (green). The last panel shows the overlay of all three FACS plots. It is evident that there are populations of functional and non-functional proteins within the single PDZ-dCas9 library and the third panel also provides a demonstration of a gate for isolation of functional PDZ- dCas9 intercalations.
2.7 Determining screening enrichment of PDZ-dCas9 domain insertions
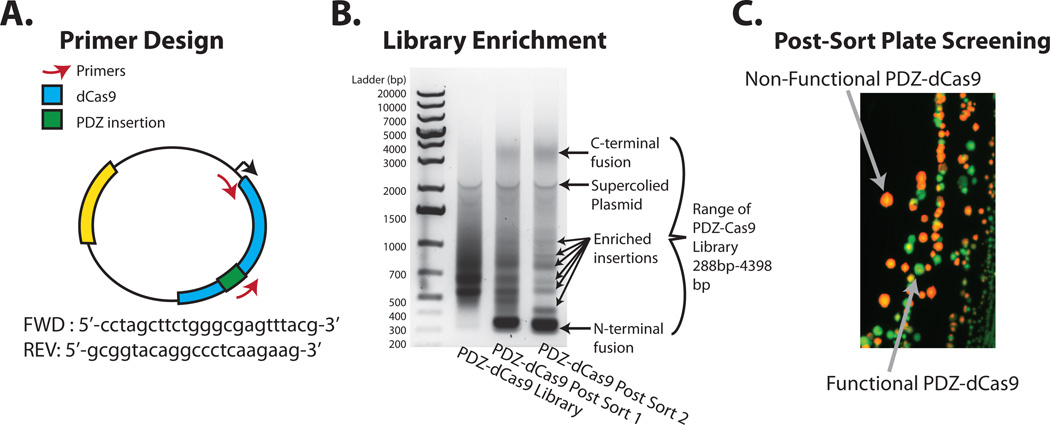
A successful round of screening with the PDZ-dCas9 library should enrich for functional PDZ-dCas9 insertion mutants (‘intercalations’). A straightforward method to check for enrichment is to PCR with a primer specific to the inserted domain and one external to the engineered Cas9 (Fig 6A). An amplified smear indicates a relatively ‘naïve’ library while specific bands indicate enriched library members. The following is a representative protocol for checking screening success.
Plate approximately 1000–10,000 of the sorted and recovered cells on rich induction plates with antibiotics and inducer. Add the remaining cells in liquid media with appropriate antibiotics.
Grow the induction plate(s) overnight at 37°C and allow 12 hrs at room temperature for RFP to fully mature. As described in section 2.8 below, this plate will be used to pick colonies with functionally intercalated PDZ-dCas9s.
Grow the liquid culture at 37°C overnight and prepare a glycerol stock of the sorted cells for future use by mixing 800 µL of culture with 400 µL of 50% glycerol in lysogeny broth (LB).
Centrifuge the remaining liquid culture and miniprep to recover plasmid DNA (Qiagen).
Perform a PCR on the original and the screened libraries with the primers described above (Fig. 6A). The screened library PCR should show enrichment bands (Fig. 6B). If bands are not evident, the library may require further rounds of screening (section 2.6, step 4). Alternatively, deep sequencing may be used to rigorously characterize the library.
Figure 6.
Checking success of a screen and picking final clones. (A) An overview of primer design for the PDZ-dCas9 library. (B) Gel electrophoresis of PCRs run on the original PDZ-dCas9 library and the first and second round of screening. The banding patterns that appear after the first and second sorts are indicative of library enrichment, representing the insertion sites of a PDZ domain. It is also evident that the N and C termini fusions to PDZ are also enriched. Since these fusions are expected to be functional this serves as an internal control. (C) Fluorescent image of the on-plate 'finishing' screen. Colonies that express only GFP are expected to have a functional PDZ-dCas9.
2.8 Identifying and testing PDZ-Cas9 clones from a screened library
Next, it is next necessary to isolate functional dCas9 clones with intercalated PDZ domains. This is done via a final plate-based screen. Once identified and isolated, it is then possible to collect, test, and verify unique PDZ-dCas9 clones in a secondary, discrete, screening method, such as repression of an alternative gene.
Set up a 96-well plate of 50 µL PCR reactions using the primers from section 2.7, step 5. In parallel, fill a 96-well plate with 100 µL of rich media per well.
From the induction plates grown in section 2.7, step 2, pick colonies expressing only GFP (Fig. 6C), as assayed via fluorescence imaging (Bio-Rad Chemidoc MP). Spot each colony into a well of the PCR plate by pipetting up and down 5× and then resuspend the colony in the corresponding well of the media plate.
Run the aforementioned PCR and sequence the amplicons. Store the resuspended colony plate at 4 °C.
Align sequences to the original plasmid map using appropriate software. Ensure variants are in-frame and determine unique clones.
Take 50 µL of the corresponding unique clones from the colony resuspension plate and grow overnight in 5 mL of rich media with antibiotics. Miniprep DNA to obtain a mix of both the engineered PDZ-dCas9 plasmid and the RFP guide plasmid for each isolate.
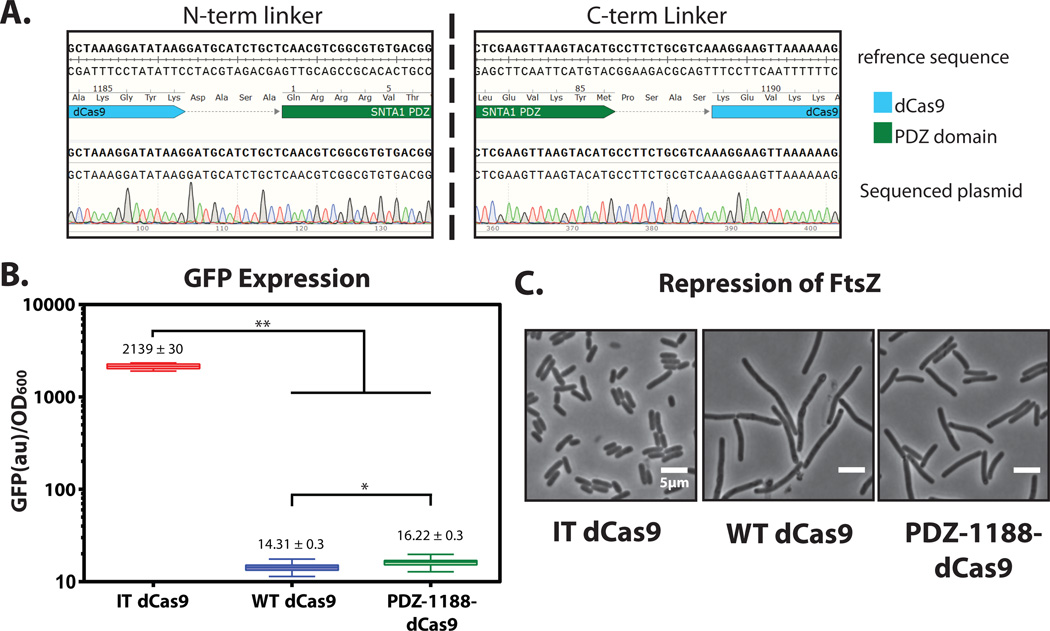
Using a primer upstream of the dCas9 insertion site, sequence the plasmid to determine the insertion site and linkers. (Fig 7. A)
Digest 5 µg of plasmid mixture with BsaI to remove the guide plasmid and clean up DNA (Qiagen) to remove restriction enzyme. (Digestion rxn occurs as follows 1. 37°C–60 min, 2. 50°C–60 min, 3. 80°C–10 min)
Transform the digested plasmid mixture with 200ng of new guide plasmids to examine the functionally of the intercalated PDZ-dCas9 on other genes and endogenous loci. Specifically, we transformed one of our PDZ-dCas9 intercalations with guides for GFP and FtsZ, an essential cell division protein (sequences 5'-atctaattcaacaagaatt-3', 5'-tcggcgtcggcggcggcgg-3' respectively).
Culture the bacteria with the PDZ-dCas9 intercalation isolates and the new guides. Grow the original dCas9 controls with these new guides, induce with 2µM aTC and measure the phenotypes accordingly.
Validate that the qualitative and quantitative phenotypes are within range of the WT Cas9 (Fig. 7 B–C).
Figure 7.
Validating functionality of engineered dCas9. (A) Sequence validation of the site 1188 PDZ intercalation. Sequence alignment via SnapGene. (B) Quantitative repression of GFP by the PDZ-1188 -dCas9 intercalation clone. Bulk florescence measurements of GFP expression levels over five hrs. Double asterisks represents a p value of < 0.0001 in a 1 way ANOVA. Single asterisk represents p values of < 0.0001 in an unpaired Student’s T-test. (C) Qualitative repression of ftsZ gene by PDZ-dCas9 and controls, scale bar is 5 µm.
3.1 Expanding Horizons
While the democratization of simple and multiplex genome engineering is central to the story of Cas9, the question of reliable specificity is paramount to its future use. Ideally, Cas9 would target and cleave one site in a complex genome yet leave other, similar, sites un-marred. Scarring the genome obscures the genotype-phenotype relationship, limiting basic science utility, and Cas9 cannot translate into the therapeutic arena if it is known to induce spurious mutations. Thus, how and when PAM and guide interactions integrate to provide specificity and activate Cas9-mediated cleavage is essential. Studies have shown that while the spCas9 PAM 5'-NGG-3' requirement is strict, only poorly tolerating one other target site (5'-NAG-3'), sgRNA:target-DNA hybridization may accept a number of mismatches, especially towards the 5' end of the sgRNA (Hsu et al., 2013; Mali et al., 2013a; Pattanayak et al., 2013). Accordingly, detrimental off-target binding and cleavage activity when using Cas9 is a pressing issue.
A number of reports have addressed this concern. Truncating the guide sequence appears to lessen the accepted number of mismatches in a given guide (Fu, Sander, Reyon, Cascio, & Joung, 2014). Alternatively, Cas9 nickases, which cleave only one strand, can be multiplexed to require mutual on-target activity of two Cas9's in order for editing to occur (Mali et al., 2013a; Ran et al., 2013). Finally, it has been shown that lowering the expression of Cas9 can lessen off-target effects (Hsu et al., 2013). Nevertheless, rigorous engineering may lead to superior solutions.
While the systems for isolating active Cas9 variants presented here are not designed to directly address specificity concerns, we envisage that with small changes our selection and screening platforms could separate mutant Cas9s with less specificity from those with more. For example, it should be possible to introduce high affinity off-target binding-sites in front of the fluorescent reporter not actively targeted, such that any binding to these mock sites would act as an internal counterscreen. We are motivated by the likelihood that, in the future, screens and selections of this vein may be used to engineer synthetic Cas9 proteins with can tolerate few, if any mismatches, in the guide and/or PAM sequence.
Conclusion
Cas9 has fundamentally altered the genome engineering landscape due to its simple programmability and overall effectiveness. Here, for the first time, we have delineated protein engineering-based methods for the directed evolution of Cas9 proteins with novel functions. We believe that such techniques will be critical for answering unresolved biochemical questions of protein structure and function. Moreover, directed evolution of Cas9 will allow for more refined improvement of this singular protein and the construction of next-generation tools for both interrogating the genome and biomedical therapies.
References
- Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science (New York, N.Y.) 2007;315(5819):1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- Bikard D, Jiang W, Samai P, Hochschild A, Zhang F, Marraffini LA. Programmable repression and activation of bacterial gene expression using an engineered CRISPR-Cas system. Nucleic Acids Research. 2013;41(15):7429–7437. doi: 10.1093/nar/gkt520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke JD, LaCroute F, Fink GR. A positive selection for mutants lacking orotidine-5'-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Molecular and General Genetics MGG. 1984;197(2):345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- Boeke JD, Trueheart J, Natsoulis G, Fink GR. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods in Enzymology. 1987;154:164–175. doi: 10.1016/0076-6879(87)54076-9. [DOI] [PubMed] [Google Scholar]
- Carroll D. Genome Engineering with Targetable Nucleases. Annual Review of Biochemistry. 2014;83(1):409–439. doi: 10.1146/annurev-biochem-060713-035418. [DOI] [PubMed] [Google Scholar]
- Chylinski K, Makarova KS, Charpentier E, Koonin EV. Classification and evolution of type II CRISPR-Cas systems. Nucleic Acids Research. 2014 doi: 10.1093/nar/gku241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, et al. Multiplex genome engineering using CRISPR/Cas systems. Science (New York, N.Y.) 2013;339(6121):819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, et al. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471(7340):602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dueber JE, Mirsky EA, Lim WA. Engineering synthetic signaling proteins with ultrasensitive input/output control. Nature Biotechnology. 2007;25(6):660–662. doi: 10.1038/nbt1308. [DOI] [PubMed] [Google Scholar]
- Dueber JE, Yeh BJ, Chak K, Lim WA. Reprogramming control of an allosteric signaling switch through modular recombination. Science (New York, N.Y.) 2003;301(5641):1904–1908. doi: 10.1126/science.1085945. [DOI] [PubMed] [Google Scholar]
- Edwards WR, Busse K, Allemann RK, Jones DD. Linking the functions of unrelated proteins using a novel directed evolution domain insertion method. Nucleic Acids Research. 2008;36(13):e78–e78. doi: 10.1093/nar/gkn363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elowitz MB, Levine AJ, Siggia ED, Swain PS. Stochastic gene expression in a single cell. Science (New York, N.Y.) 2002;297(5584):1183–1186. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- Esvelt KM, Mali P, Braff JL, Moosburner M, Yaung SJ, Church GM. Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nature Methods. 2013;10(11):1116–1121. doi: 10.1038/nmeth.2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedland AE, Tzur YB, Esvelt KM, Colaiácovo MP, Church GM, Calarco JA. Heritable genome editing in C. elegans via a CRISPR-Cas9 system. Nature Methods. 2013;10(8):741–743. doi: 10.1038/nmeth.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvao TC, de Lorenzo V. Adaptation of the Yeast URA3 Selection System to Gram-Negative Bacteria and Generation of a betCDE Pseudomonas putida Strain. Applied and Environmental Microbiology. 2005;71(2):883–892. doi: 10.1128/AEM.71.2.883-892.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154(2):442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratz SJ, Cummings AM, Nguyen JN, Hamm DC, Donohue LK, Harrison MM, et al. Genome Engineering of Drosophila with the CRISPR RNA-Guided Cas9 Nuclease. Genetics. 2013;194(4):1029–1035. doi: 10.1534/genetics.113.152710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilinger JP, Thompson DB, Liu DR. Fusion of catalytically inactive Cas9 to FokI nuclease improves the specificity of genome modification. Nature Biotechnology. 2014;32(6):577–582. doi: 10.1038/nbt.2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Z, Zhang Y, Propson NE, Howden SE, Chu L-F, Sontheimer EJ, Thomson JA. Efficient genome engineering in human pluripotent stem cells using Cas9 from Neisseria meningitidis. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(39):15644–15649. doi: 10.1073/pnas.1313587110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nature Biotechnology. 2013;31(9):827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, Sander JD, et al. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nature Biotechnology. 2013;31(3):227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science (New York, N.Y.) 2012;337(6096):816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Jiang F, Taylor DW, Sternberg SH, Kaya E, Ma E, et al. Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science (New York, N.Y.) 2014;343(6176):1247997. doi: 10.1126/science.1247997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Mali P, Aach J, Stranges PB, Esvelt KM, Moosburner M, Kosuri S, et al. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nature Biotechnology. 2013a;31(9):833–838. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Esvelt KM, Church GM. Cas9 as a versatile tool for engineering biology. Nature Methods. 2013b;10(10):957–963. doi: 10.1038/nmeth.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, et al. RNA-guided human genome engineering via Cas9. Science (New York, N.Y.) 2013c;339(6121):823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimasu H, Ran FA, Hsu PD, Konermann S, Shehata SI, Dohmae N, et al. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell. 2014;156(5):935–949. doi: 10.1016/j.cell.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu Y, Bin Shen, Cui Y, Chen Y, Wang J, Wang L, et al. Generation of Gene-Modified Cynomolgus Monkey via Cas9/RNA-Mediated Gene Targeting in One-Cell Embryos. Cell. 2014;156(4):836–843. doi: 10.1016/j.cell.2014.01.027. [DOI] [PubMed] [Google Scholar]
- Nourry C, Grant SGN, Borg J-P. PDZ domain proteins: plug and play! Science's STKE : Signal Transduction Knowledge Environment. 2003;2003(179):RE7. doi: 10.1126/stke.2003.179.re7. [DOI] [PubMed] [Google Scholar]
- Olson EJ, Tabor JJ. Post-translational tools expand the scope of synthetic biology. Current Opinion in Chemical Biology. 2012:1–7. doi: 10.1016/j.cbpa.2012.06.003. [DOI] [PubMed] [Google Scholar]
- Persikov AV, Rowland EF, Oakes BL, Singh M, Noyes MB. Deep sequencing of large library selections allows computational discovery of diverse sets of zinc fingers that bind common targets. Nucleic Acids Research. 2014;42(3):1497–1508. doi: 10.1093/nar/gkt1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152(5):1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson TR, Saroj SD, Llewellyn AC, Tzeng Y-L, Weiss DS. A CRISPR/Cas system mediates bacterial innate immune evasion and virulence. Nature. 2013;497(7448):254–257. doi: 10.1038/nature12048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt-Dannert C, Arnold FH. Directed evolution of industrial enzymes. Trends in Biotechnology. 1999;17(4):135–136. doi: 10.1016/s0167-7799(98)01283-9. [DOI] [PubMed] [Google Scholar]
- Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelsen TS, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science (New York, N.Y.) 2014;343(6166):84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan Q, Wang Y, Li J, Zhang Y, Chen K, Liang Z, et al. Multiplex and homologous recombination-mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nature Biotechnology. 2013;31(8):688–691. doi: 10.1038/nbt.2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg SH, Redding S, Jinek M, Greene EC, Doudna JA. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature. 2014;507(7490):62–67. doi: 10.1038/nature13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai SQ, Wyvekens N, Khayter C, Foden JA, Thapar V, Reyon D, et al. Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nature Biotechnology. 2014;32(6):569–576. doi: 10.1038/nbt.2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R. One-Step Generation of Mice Carrying Mutations in Multiple Genes by CRISPR/Cas-Mediated Genome Engineering. Cell. 2013;153(4):910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedenheft B, Sternberg SH, Doudna JA. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012;482(7385):331–338. doi: 10.1038/nature10886. [DOI] [PubMed] [Google Scholar]
- Yano T, Sanders C, Catalano J, Daldal F. sacB-5-Fluoroorotic Acid-pyrE-Based Bidirectional Selection for Integration of Unmarked Alleles into the Chromosome of Rhodobacter capsulatus. Applied and Environmental Microbiology. 2005;71(6):3014–3024. doi: 10.1128/AEM.71.6.3014-3024.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Zhu S, Cai C, Yuan P, Li C, Huang Y, Wei W. High-throughput screening of a CRISPR/Cas9 library for functional genomics in human cells. Nature. 2014;509(7501):487–491. doi: 10.1038/nature13166. [DOI] [PubMed] [Google Scholar]