Abstract
Despite availability of a universal vaccine, hepatitis B virus (HBV) infection has a huge impact on public health worldwide. Accurate and timely diagnosis of HBV infection is needed. Rapid developments have been made in the diagnostic and monitoring methods for HBV infection, including serological and molecular assays. In clinical practice, qualitative hepatitis B surface antigen (HBsAg) testing has long served as a diagnostic marker for individuals infected with HBV. More recently, HBsAg level has been used to predict treatment outcome when determined early during treatment or at baseline. However, identification of HBV DNA positive cases that do not have detectable HBsAg has encouraged the application of molecular tests. Hence, combination of quantitative detection of HBV DNA and HBsAg can be used to discriminate patients during the course of HBV infection and to monitor therapy. This article reviews the most commonly used quantitative methods for HBsAg and HBV DNA.
Keywords: Hepatitis B virus, Biosensor, Polymerase chain reaction, Isothermal amplification methods, Quantitative assay
Core tip: The combination of quantitative detection of hepatitis B surface antigen (HBsAg) and hepatitis B virus (HBV) DNA can be used to classify individuals during the course of HBV infection and to monitor therapy. The most popular platforms for HBsAg detection are based on chemiluminescent microparticle immunoassay, while polymerase chain reaction based methods are widely used for HBV DNA assay. Recently, isothermal amplification and biosensors offered a lower cost and more rapid alternative for HBV quantification. This article reviews the most commonly used quantitative methods for HBV.
INTRODUCTION
Hepatitis B virus (HBV) is an enveloped virus with a small (3.2 kb) partially double-stranded DNA genome that causes acute and chronic infections[1]. The impact of HBV infection on public health is enormous, with an estimated prevalence of 2 billion infected and 360 million chronically infected[2]. The diagnosis of HBV infection relies heavily on serological and molecular tests. Serological tests use serum-based blood tests that can be analyzed by enzyme immunoassays either qualitatively or quantitatively[3]. The tests identify virus-encoded antigens and their corresponding antibodies: hepatitis B surface antigen (HBsAg), anti-HBs, hepatitis B e antigen (HBeAg), anti-HBe, and antibodies to hepatitis core antigen (anti-HBc). In contrast, the molecular tests focus on quantitative viral load, genotyping, drug resistance mutations, and core promotion/pre-core mutation assays.
In clinical practice, qualitative assays for HBsAg have served as diagnostic markers for patients with HBV infection. More recently, quantitative methods for HBsAg have been used to predict treatment outcome when determined early during treatment or at baseline[4]. However, identification of HBV-DNA-positive cases, which do not have detectable HBsAg, is greatly encouraging for the application of molecular testing. This article reviews the most commonly used quantitative detection methods for HBsAg and HBV DNA (Figure 1).
Figure 1.

Quantitative methods for hepatitis B virus detection. 1These methods are qualitative or semi-quantitative for HBsAg. HBsAg: Hepatitis B surface antigen.
QUANTITATIVE METHODS FOR HBsAg
Since the discovery of HBsAg in 1965[5], it has served as a biomarker for the diagnosis of HBV infection. Methods for HBsAg detection were first described in the 1970s using electron microscopy, radioimmunoassay, and enzyme immunoassays[6-8], which were cumbersome, labor intensive, and restricted to a research setting. Since then, various diagnostic methods have been developed for quantitative HBV detection.
In the early 1990s, HBsAg quantification was considered to be a simple, promising, and inexpensive method to monitor viral replication in chronic hepatitis B (CHB) patients who were receiving interferon (IFN) therapy[9]. HBsAg quantification is associated with the concentration of covalently closed circular DNA (cccDNA), the persistent intrahepatic form of HBV DNA[10,11]. The amount of circulating HBsAg is hypothesized to be predictive for response to antiviral therapy. Currently, standard quantitative HBsAg assays have been developed, which are fully automated and have high throughput. Two commercially available assays are briefly introduced here.
The Architect HBsAg QT (Abbott Laboratories, Abbott Park, IL, United States) is an automated chemiluminescent microparticle immunoassay method, which is the most widely used assay in clinical practice[12]. The Architect HBsAg QT assay is a two-step immunoassay with flexible assay protocols, referred to as Chemiflex, for quantitatively measuring HBsAg concentrations in serum and plasma[13]. The Architect HBsAg system can detect as low as 0.2 ng/mL HBsAg with a dynamic range of 0.05-250.0 IU/mL (1 IU/mL is equivalent to 1-10 ng/mL HBsAg)[14]. The assay is capable of processing up to 800 tests per hour.
Elecsys HBsAg II (Roche Diagnostics, Indianapolis, IN, United States) is another popular assay for HBsAg quantification[15]. Elecsys HBsAg II is a “sandwich” assay with a total testing time of 18 min. The results are reported as a cutoff index (signal sample/cutoff). The sample is considered nonreactive when the index is < 0.9, while samples with index value > 1.0 are interpreted as reactive[15].
These two methods can be confidently used for HBsAg quantification for the most prevalent HBV genotypes. These tests are easy to use, inexpensive, and have a rapid turnaround time; the analytical performance of the assays is generally satisfactory.
QUANTITATIVE METHODS FOR HBV DNA
With the increased prevalence of serologically negative HBV infections (HBeAg negative CHB and occult HBV infection) and the rapid advent of diagnostic escape mutants, the detection of HBV DNA has gained more attention in clinical medicine[16]. The detection of HBV DNA in peripheral blood is a reliable marker of HBV activity, while high levels of HBV DNA are associated with a higher incidence of hepatocellular carcinoma (HCC) and more rapid progression to cirrhosis[17]. In addition, HBV DNA detection is beneficial in routine clinical practice to identify individuals who need anti-viral treatment and provide them the most suitable therapy[18]. Nowadays, a variety of molecular technologies have been used in HBV DNA quantification, such as ultraviolet (UV) spectrophotometry, real-time polymerase chain reaction (PCR), digital PCR, isothermal amplification methods, and biosensors[19-22]. Here, we review the rapid and quantitative methods that have been used for HBV DNA detection.
UV spectrophotometry
The aromatic rings of the bases absorb UV light with a maximum peak at 260 nm. When a beam of UV light shines through a sample containing DNA or RNA, the amount of UV absorption by the sample depends on the DNA or RNA concentration. The amount of UV light absorbed by a series of standard DNA amounts is measured to calibrate the technique, and the ratio of UV light absorbed by the unknown sample is measured and plotted on the calibration curve to deduce the concentration of DNA or RNA.
NanoDrop instruments (Thermo Scientific, Wilmington, DE, United States) are based on UV spectrophotometry and utilize a patented sample-retention system that allows the quantification of DNA or RNA from 1-2 μL samples[23]. The usable concentration ranges are 0.4-15000 ng/μL. No specific sample preparations are needed for NanoDrop instruments[23].
Real-time PCR
Since developed in 1983 by Kary Mullis[24], PCR has become an essential tool for molecular biologists, and its application in nucleic acids detection system has revolutionized the quantitative analysis of DNA and RNA. PCR techniques have rapidly evolved over the last few years, and their quantitative applications have favored the development of real-time PCR. Real-time PCR follows the general principle of conventional PCR, and its key feature is that the amplified DNA is detected as the reaction progresses in real time. This new approach has a broader dynamic range compared to conventional PCR. The most commonly used reagents for real-time PCR are TaqMan probes[25]. TaqMan probes are hydrolysis probes that were designed in 1991 to increase the specificity of quantitative PCR[26]. Figure 2 outlines the reaction mechanism of real-time PCR based on TaqMan probes.
Figure 2.

Reaction mechanism of real-time polymerase chain reaction based on TaqMan probe technology. TaqMan probe is an oligo-nucleotide probe that has a fluorescent reporter at the 5’ end and a quencher attached to the 3’ end. Once hybridized to the target sequence during annealing, TaqMan probe is cleaved by DNA polymerase, which separates the fluorescent reporter from the quencher. Once they are separated, the signal is emitted and detected in the real-time machine. The intensity of fluorescence is proportional to the amount of PCR product produced. FRET: Fluorescent resonance energy transfer.
In the study by Abe et al[27], a sensitive, accurate, and reproducible assay for HBV DNA quantification based on real-time PCR using TaqMan probes was reported. Their results demonstrated that the limit of detection was as few as 10 copies/reaction, with a linear standard curve between 10 and 108 DNA copies/reaction. The coefficient of variation for both inter- and intra-experimental variability indicated remarkable reproducibility.
Sitnik et al[28] developed a real-time PCR assay for HBV DNA quantification with TaqMan and minor groove binder (MGB) probes. In this assay, primers and probes were designed using an alignment of sequences from all HBV genotypes in order to amplify all the genotypes equally. The assay had a dynamic range from 50 to 108 IU/mL.
Although many laboratories have developed in-house real-time PCR-based assays, all of them had a lack of quality control and standardization[3], which restricted their application in clinical diagnosis. The major diagnostics companies also provide commercial quantitative assays. These assays vary in their detection limit and dynamic range: the first version suffered from a narrow dynamic range from 102 to 105 copies/mL, while the detection limit of the newer assay is lower than 50 IU/mL with a dynamic range of approximately 108 IU/mL[29-32]. Recently, the World Health Organization has established a universal standard for HBV DNA quantification measured in IU/mL, with the purpose of correlating different results using a single reference unit[33]. However, significant variability in quantification among different assays can occur randomly in spite of the standardization of reporting units and the generally good correlation between different assays[34,35]. Hence, patients are suggested to be monitored by a single assay[1].
Digital PCR
Digital PCR, developed by Vogelstein et al[36], is a refinement of conventional PCR methods that can be used to quantify and clonally amplify nucleic acids directly, including DNA, cDNA, and RNA. The key difference between digital and conventional PCR lies in the measuring method; digital PCR is a more precise method than conventional PCR[37]. A sample is partitioned in digital PCR so that individual nucleic acid molecules within the sample are localized and concentrated within many separate regions. The partitioning of the sample allows for estimation of the number of molecules, by assuming that the molecular population follows a Poisson distribution. Hindson et al[38] developed a fundamentally distinct method of partitioning, droplet digital PCR, which partitions a sample into 20000 droplets and provides digital counting of nucleic acids.
High-sensitivity techniques distinguish differences in the number of HBV copies among samples, especially in cases of low copy number (e.g., nucleic acids extracted from tissues). Huang et al[39] applied droplet digital PCR to measure the number of HBV copies in formalin-fixed paraffin-embedded (FFPE) HCC tissue. A total of 131 HCC FFPE samples with different tumor stages and clinical features were classified by their serological tests. The number of HBV copies were successfully determined by droplet digital PCR for all FFPE tissues, with copy numbers ranging from 1.1 to 175.5 copies/μL. These results showed that droplet digital PCR improved the analytical sensitivity and specificity of nucleic acids measurement to a single-molecule level and was suitable for HBV DNA quantification.
Isothermal amplification methods
PCR-based assays are the most widely used methods for HBV DNA quantification; however, they need a thermo-cycling machine to separate DNA strands and amplify the fragments[21]. Isothermal amplification methods are carried out at a constant temperature, and do not require a thermal cycler. The isothermal amplification methods have been developed according to new findings in the molecular biology of DNA/RNA synthesis and in vitro nucleic acid amplification function of some accessory proteins. Here, we describe several isothermal amplification methods that have been used to quantify HBV DNA.
Loop-mediated isothermal amplification
Loop-mediated isothermal amplification (LAMP) is a single-tube technique for DNA amplification[40]. LAMP may be combined with a reverse transcription step to allow the detection of RNA[41]. In LAMP, the target sequence is amplified at a constant temperature of 60 °C-65 °C using either two or three sets of primers and a polymerase with high activity of strand displacement and replication activity. An additional pair of loop primers can accelerate the amplification[42]. Due to the specific nature of the primers, the amount of DNA products in LAMP is considerably higher than in PCR-based amplification[42]. Compared with PCR, LAMP was less sensitive to inhibitors in complex samples, such as blood, due to the use of a different DNA polymerase. However, complex primer design has been considered as a weakness of LAMP, and it may limit its application in some aspects of molecular biology[21,41].
The original article on LAMP, which was published in 2000 by Notomi et al[40], showed that 600 and 6000 copies of HBV DNA were detected at 13 and 11 min, respectively, which indicated a high specificity and efficiency for HBV detection. Cai et al[43] developed an accurate and rapid real-time fluorogenic LAMP protocol to quantify HBV. Their study demonstrated a dynamic range of eight orders of magnitude, a lower detection limit of 210 copies/mL, low inter-assay and intra-assay variability (4.24%-12.11%), and excellent correlation with real-time PCR (R2 = 0.96). Similar LAMP assays have been reported by others, indicating that LAMP may be useful in the future as a low-cost alternative for HBV DNA quantification[44,45].
Transcription-mediated amplification
Transcription-mediated amplification (TMA) is an isothermal amplification technique used in molecular biology research and in clinical laboratories for the rapid diagnosis of infections. In contrast to PCR, this method involves RNA transcription (via RNA polymerase) and DNA synthesis (via reverse transcriptase). There are several more differences between TMA and PCR: (1) TMA is isothermal; (2) TMA produces RNA rather than DNA amplicons. Since RNA is more labile in a laboratory environment, this reduces the possibility of carry-over contamination; and (3) TMA produces 100-1000 copies per cycle (PCR only produces two copies per cycle), which results in a 10 billion-fold increase in nucleic acid products within 15-30 min[46].
Kamisango et al[47] developed a sensitive and quantitative assay using TMA and hybridization protection assay for the detection of HBV DNA in serum. The assay achieved a detection range of 5 × 103 to 5 × 108 genome equivalents/mL. It takes about 5 h to complete a moderately sized manual assay. Ide et al[48] noted that TMA was more useful in understanding the changes of HBV DNA level than branched DNA signal amplification assay in lamivudine-treated CHB patients. Kubo et al[49] examined the usefulness of TMA for evaluation of the active degree of hepatitis and estimation of recurrence after resection of HBV-related HCC.
Nucleic acid sequence-based amplification
Nucleic acid sequence-based amplification (NASBA) is similar to TMA, which was developed by Compton[50] in 1991 to amplify RNA sequences. This technique can also be used for amplification of DNA with modifications in the basic method, such as primer design, sample extraction, and template denaturation[16]. Compared with PCR, major advantages of NASBA are: (1) it works under isothermal conditions - usually at a constant temperature of 41 °C; and (2) it is more rapid and sensitive than PCR in medical diagnostics[51].
Yates et al[52] developed an HBV DNA quantification system based on amplification with NASBA and real-time detection with molecular beacon technology. The detection range of the assay is 103-109 copies/mL in plasma or serum, with good reproducibility and precision. Deiman et al[53] used NASBA, including a restriction enzyme digestion for HBV DNA amplification, and found that the sensitivity of normal NASBA was improved 100-1000 times when restriction enzyme digestion was performed prior to amplification. The limit of detection was 10 IU/mL with a dynamic detection range of 102-109 IU/mL.
Rolling circle amplification
Rolling circle amplification (RCA) is a simple, reliable, and isothermal amplification method, which is driven by DNA polymerase to generate a long tandem repeat product based on a circular DNA template[54]. This technique does not require advanced laboratory equipment or experimental expertise. Compared with PCR, the main advantages of RCA include: (1) it is resistant to inhibitors present in clinical samples and requires almost no assay optimization[16]; and (2) it can amplify targets on solid support or in solution, offering opportunity for microarray and biosensor application[55] (Figure 3).
Figure 3.
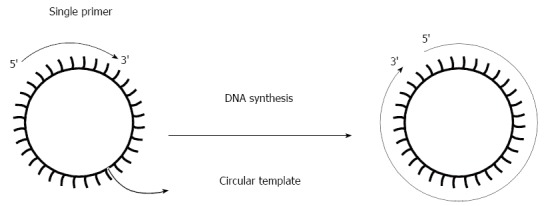
Scheme of the rolling circle amplification reaction. RCA rapidly synthesizes multiple copies of a single circular template with use of a single primer. RCA: Rolling circle amplification.
The property of a circular template for RCA makes it ideal for detection of HBV DNA, especially cccDNA. Margeridon et al[56] reported that cccDNA from liver biopsies could be amplified from as few as 13 copies using RCA. Martel et al[57] developed a method using in vitro completion/ligation of plus-strand HBV relax circular DNA and amplification using RCA. The method can amplify complete HBV genomes from serum with viral loads ranging from 103 to 108 IU/mL.
The isothermal amplification methods mentioned above offer several advantages over PCR, most importantly, they do not require an expensive and cost-intensive thermal cycler. Comparisons between isothermal amplification methods and conventional PCR are shown in Table 1.
Table 1.
Characteristics of isothermal amplification methods and polymerase chain reaction reviewed in this article
| Method | Temperature requirement (°C) | No. of enzymes | Primer design | Product detection method | Rapid detection possibility | Tolerance to biological components |
| PCR | 55-95 | 1 | Simple | GE, ELISA, Real-time | Yes | No |
| IAM | ||||||
| LAMP | 60-65 | 1 | Complex | GE, turbidity, Real-time | Yes | Yes |
| TMA | 50-60 | 2 | Simple | GE, ELISA, Real-time, ECL | Yes | No |
| NASBA | 37-41 | 2 or 3 | Simple | GE, ELISA, Real-time, ECL | Yes | No |
| RCA | 37 | 1 | Simple | GE, Real-time | Yes | No |
ECL: Electrochemiluminescence; GE: Gel electrophoresis; PCR: Polymerase chain reaction; IAM: Isothermal amplification methods; LAMP: Loop-mediated isothermal amplification; TMA: Transcription-mediated amplification; NASBA: Nucleic acid sequence-based amplification; RCA: Rolling circle amplification.
Biosensors
Biosensors are analytical devices used for detection, which combine a biological component with a physicochemical detector[22]. Recently, an increasing number of biosensors have been used in clinical research; a common example is the blood glucose biosensor, which uses glucose oxidase to break blood glucose down[58,59]. Most of the clinical research on biosensors was based on immunological reactions or DNA hybridization, and the biosensors always yielded rapid results with high sensitivity[60-62].
Electrochemical biosensors: Electrochemical biosensors work by detecting current or potential changes caused by binding reactions that occur on or near the electrode surface[63]. Recently, electrochemical biosensors have offered sensitivity, selectivity, and low-cost detection for DNA sequences and have attracted considerable attention. Ding et al[64] described a label-free electrochemical biosensor for the detection of oligonucleotides related to HBV sequences via the interactions of DNA with the redox-active complex 2,9-dimethyl-1,10-phenantroline cobalt[Co(dmp)(H2O)(NO3)2]. The experiment was performed by hybridizing 21-mer DNA probes modified on glassy carbon electrode with target DNA and cobalt[Co(dmp)(H2O)(NO3)2], whose sizes were comparable to the small groove of native double-helix DNA that was used as an electrochemical indicator. Under optimal conditions, the electrical signal had a linear relationship, with the concentration of target DNA ranging from 3.96 × 10-7 to 1.32 × 10-6 mol/L, and the detection limit was 1.94 × 10-8 mol/L.
Zhang et al[65] developed an electrochemical biosensor using diaquabis[N-(2-pyridinylmethyl)benzamide-kappa N-2,O]-cadmium(II) dinitrate as a new electroactive indicator for the detection of human HBV DNA. The hybridization between the probe and its complementary single-stranded DNA was determined by differential pulse voltammetry. Experiments with non-complementary oligonucleotides were carried out to assess the selectivity of the developed electrochemical DNA biosensor. HBV DNA could be quantified in a range from 1.01 × 10-8 to 1.62 × 10-6 mol/L with good linearity (γ = 0.9962). The detection limit was 7.19 × 10-9 mol/L.
Quartz crystal microbalance biosensors: Piezoelectric materials, typically crystals, generate an electrical potential in response to a mechanical force[19]. Piezoelectric biosensors are mass-sensitive, and the additional mass to the sensor causes a detectable change in the resonance frequency of the crystal. The most common type of piezoelectric biosensor is quartz crystal microbalance (QCM). QCM biosensors have gained increasing attention in recent years because of their high sensitivity, good specificity, low-cost, label-free detection, and rapid response[66].
Zhou et al[67] developed a highly sensitive piezoelectric HBV DNA biosensor based on the sensitive mass-transducing function of the QCM and the specificity of nucleic acid hybridization reaction. HBV nucleic acid probes were immobilized onto the gold electrodes of a 9 MHz AT-cut piezoelectric quartz crystal via the polyethyleneimine adhesion, glutaraldehyde cross-linking method or the physical adsorption method. The frequency shifts of hybridization have a good linear relationship with the amount of HBV DNA, when the amount was 0.02-0.14 mg/mL.
Peptide nucleic acid (PNA)-based piezoelectric biosensor for real time monitoring of hybridization of HBV genomic DNA was constructed by Yao et al[68]. PNA probes can combine target sequences more effectively and specifically than DNA probes. The PNA probes were designed and immobilized on the surface of the biosensor to be a substitute for the conventional DNA probes for direct detection of HBV genomic DNA without previous amplification by PCR. The hybridization assay was completed in 50 min. The detection limit was 8.6 pg/L, and the clinical specificity was 94.44% in comparison with real-time PCR.
Microcantilever biosensors: In the last two decades, microcantilevers have emerged as a sensitive tool for the detection of chemicals and bio-organisms. Because of their light weight, small size, and high surface-to-volume ratio, microcantilever sensors improve the detection and identification of biological agents by several orders of magnitude[69-71]. An HBV DNA detection method using a silica-nanoparticle-enhanced dynamic microcantilever biosensor was developed by Cha et al[72], with a 243-mer nucleotide of HBV DNA pre-core/core region used as the target DNA. In this study, the capture probe immobilized on the microcantilever surface and the detection probe conjugated with silica nanoparticles were designed specifically for the target DNA. HBV DNA was detected using a silica-nanoparticle-enhanced microcantilever biosensor with a concentration of 23.1 fmol/L to 2.3 nmol/L, which was obtained from the PCR procedure. The HBV target DNA of 243-mer was detected up to the picomolar level without nanoparticle enhancement and up to the femtomolar level using a nanoparticle-based signal amplification process.
Surface plasmon resonance biosensors: Surface plasmon resonance (SPR) biosensors are optical sensors that exploit special electromagnetic waves, surface plasmon polaritons, to probe interactions between analytes and biomolecular recognition elements immobilized on the SPR sensor surface[73] (Figure 4). SPR biosensors are a label-free, real-time analytical technology for the detection of biological analytes and the analysis of biomolecular interactions.
Figure 4.
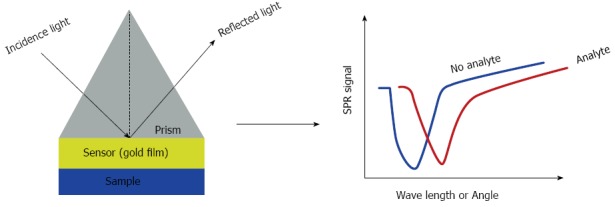
Scheme of the surface plasmon resonance biosensor. The incidence light causes the changes in wavelength or angle. Changes were detected in real time monitor. SPR: Surface plasmon resonance.
Chuang et al[45] constructed a simple, low-cost, SPR-sensing cartridge based on the LAMP method for the on-site detection of HBV. The HBV template mixed in 10 μL LAMP solution could be detected by the new system in 17 min, even at the detection-limited concentration of 2 fg/mL. They also analyzed the correlation coefficients between the initial concentrations of HBV DNA templates and the system response at varying amplification times to establish an optimum amplification time endpoint of 25 min (R2 = 0.98).
All rapid and quantitative methods for HBV detection mentioned above are summarized in Table 2.
Table 2.
Quantitative detection methods for hepatitis B virus discussed in this review
| Method | Target | Detection range | Ref. |
| Architect | HBsAg | 0.05-250.0 IU/mL | [12] |
| Elecsys | HBsAg | NA | [15] |
| UV spectrophotometry | HBV DNA | 0.4-15000 ng/µL | [23] |
| In-house assays based on real-time PCR | HBV DNA | 101-108 copies/reaction | [27] |
| HBV DNA | 50-108 IU/mL | [19] | |
| Digital PCR | HBV DNA | Single copy | [39] |
| IAM | |||
| LAMP | HBV DNA | 48-108 IU/mL | [43] |
| TMA | HBV DNA | 5 × 103-5 × 108 GE/mL | [47] |
| NASBA | HBV DNA | 103-109 copies/mL | [52] |
| HBV DNA | 102-109 IU/mL | [53] | |
| RCA | HBV DNA | 103-108 IU/mL | [57] |
| Biosensors | |||
| Electrochemical biosensors | HBV DNA | 3.96 × 10-7-1.32 × 10-6 mol/L | [65] |
| HBV DNA | 1.01 × 10-8-1.62 × 10-6 mol/L | [66] | |
| QCM biosensors | HBV DNA | 0.02-0.14 mg/mL | [67] |
| HBV DNA | 8.6 pg/L1 | [68] | |
| Microcantilever biosensors | HBV DNA | 23.1 fmol/L-2.31 nmol/L | [69] |
| SPR biosensors | HBV DNA | 2 fg/mL1 | [45] |
Only the limits of detection were given in these two papers. HBsAg: Hepatitis B surface antigen; HBV: Hepatitis B virus; NA: Not available; LAMP: Loop-mediated isothermal amplification; TMA: Transcription-mediated amplification; NASBA: Nucleic acid sequence-based amplification; RCA: Rolling circle amplification; QCM: Quartz crystal microbalance; SPR: Surface plasmon resonance; IAM: Isothermal amplification methods.
FUTURE PERSPECTIVES
The level of HBsAg has been suggested as a marker for the amount of cccDNA or infected liver mass. Although commercial assays are now available for HBsAg quantification, their dynamic range is narrow, and manual dilution is required if HBsAg concentration exceeds the dynamic range. More sensitive assays with broader detection ranges are needed for rapid and quantitative detection of HBV in clinical practice.
Our review describes the most assays that have been used for quantification of HBV DNA, along with their merits and limitations. PCR is the most widely used method in HBV DNA quantification, but it is also known to have erroneous results caused by its hybridization mechanism and false-negative results because of low HBV copy numbers. Additionally, PCR requires efficient control of thermal cycles and thus is instrument intensive. The development of isothermal amplification methods makes the amplification of HBV DNA without a thermal cycler possible, which also offers an opportunity for biosensor miniaturization. In spite of isothermal amplification and high sensitivity, isothermal amplification techniques have remained less utilized for developing commercial detection assays for HBV quantification. Biosensors have major advantages, such as speed, sensitivity, and low cost. Detection and quantification of viral components and viruses using biosensors is a new concept and is still in its infancy[63]. Tests developed previously are now being integrated into other novel platforms, such as the combination of LAMP and biosensors[45].
Development of rapid quantification detection for HBV is progressing towards procedural simplicity, tolerance to crude samples, high sensitivity, specific amplification, and robust reliable performance. With the development of technologies, quantification of HBV may be moved away from centralized laboratory facilities to other locations, such as emergency rooms, physician’s office, and even the family. Point-of-care and point-of-patient testing may be the mainstream in the future[19,20].
Footnotes
Supported by National Natural Science Foundation of China, No. 81371885.
Conflict-of-interest statement: The authors declare that there is no conflict of interest in this study.
Open-Access: This article is an open-access article which was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/
Peer-review started: April 24, 2015
First decision: July 13, 2015
Article in press: September 14, 2015
P- Reviewer: Kim K, Slomiany BL S- Editor: Ma YJ L- Editor: Filipodia E- Editor: Liu XM
References
- 1.Valsamakis A. Molecular testing in the diagnosis and management of chronic hepatitis B. Clin Microbiol Rev. 2007;20:426–439, table of contents. doi: 10.1128/CMR.00009-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arzumanyan A, Reis HM, Feitelson MA. Pathogenic mechanisms in HBV- and HCV-associated hepatocellular carcinoma. Nat Rev Cancer. 2013;13:123–135. doi: 10.1038/nrc3449. [DOI] [PubMed] [Google Scholar]
- 3.Gish RG, Locarnini SA. Chronic hepatitis B: current testing strategies. Clin Gastroenterol Hepatol. 2006;4:666–676. doi: 10.1016/j.cgh.2006.03.017. [DOI] [PubMed] [Google Scholar]
- 4.Fung J, Lai CL, Yuen MF. Hepatitis B virus DNA and hepatitis B surface antigen levels in chronic hepatitis B. Expert Rev Anti Infect Ther. 2010;8:717–726. doi: 10.1586/eri.10.45. [DOI] [PubMed] [Google Scholar]
- 5.Blumberg BS, Sutnick AI, London WT, Millman I. Australia antigen and hepatitis. N Engl J Med. 1970;283:349–354. doi: 10.1056/NEJM197008132830707. [DOI] [PubMed] [Google Scholar]
- 6.Engvall E, Perlmann P. Enzyme-linked immunosorbent assay (ELISA). Quantitative assay of immunoglobulin G. Immunochemistry. 1971;8:871–874. doi: 10.1016/0019-2791(71)90454-x. [DOI] [PubMed] [Google Scholar]
- 7.Wolters G, Kuijpers L, Kacaki J, Schuurs A. Solid-phase enzyme-immunoassay for detection of hepatitis B surface antigen. J Clin Pathol. 1976;29:873–879. doi: 10.1136/jcp.29.10.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dane DS, Cameron CH, Briggs M. Virus-like particles in serum of patients with Australia-antigen-associated hepatitis. Lancet. 1970;1:695–698. doi: 10.1016/s0140-6736(70)90926-8. [DOI] [PubMed] [Google Scholar]
- 9.Martinot-Peignoux M, Lapalus M, Asselah T, Marcellin P. HBsAg quantification: useful for monitoring natural history and treatment outcome. Liver Int. 2014;34 Suppl 1:97–107. doi: 10.1111/liv.12403. [DOI] [PubMed] [Google Scholar]
- 10.Thompson AJ, Nguyen T, Iser D, Ayres A, Jackson K, Littlejohn M, Slavin J, Bowden S, Gane EJ, Abbott W, et al. Serum hepatitis B surface antigen and hepatitis B e antigen titers: disease phase influences correlation with viral load and intrahepatic hepatitis B virus markers. Hepatology. 2010;51:1933–1944. doi: 10.1002/hep.23571. [DOI] [PubMed] [Google Scholar]
- 11.Chan HL, Wong VW, Tse AM, Tse CH, Chim AM, Chan HY, Wong GL, Sung JJ. Serum hepatitis B surface antigen quantitation can reflect hepatitis B virus in the liver and predict treatment response. Clin Gastroenterol Hepatol. 2007;5:1462–1468. doi: 10.1016/j.cgh.2007.09.005. [DOI] [PubMed] [Google Scholar]
- 12.Deguchi M, Yamashita N, Kagita M, Asari S, Iwatani Y, Tsuchida T, Iinuma K, Mushahwar IK. Quantitation of hepatitis B surface antigen by an automated chemiluminescent microparticle immunoassay. J Virol Methods. 2004;115:217–222. doi: 10.1016/j.jviromet.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 13.Lee JM, Ahn SH. Quantification of HBsAg: basic virology for clinical practice. World J Gastroenterol. 2011;17:283–289. doi: 10.3748/wjg.v17.i3.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chudy M, Scheiblauer H, Hanschmann KM, Kress J, Nick S, Wend U, Schüttler C, Nübling CM, Gerlich WH. Performance of hepatitis B surface antigen tests with the first WHO international hepatitis B virus genotype reference panel. J Clin Virol. 2013;58:47–53. doi: 10.1016/j.jcv.2013.06.011. [DOI] [PubMed] [Google Scholar]
- 15.Mühlbacher A, Weber B, Bürgisser P, Eiras A, Cabrera J, Louisirirotchanakul S, Tiller FW, Kim HS, v Helden J, Bossi V, et al. Multicenter study of a new fully automated HBsAg screening assay with enhanced sensitivity for the detection of HBV mutants. Med Microbiol Immunol. 2008;197:55–64. doi: 10.1007/s00430-007-0059-9. [DOI] [PubMed] [Google Scholar]
- 16.Datta S, Chatterjee S, Veer V. Recent advances in molecular diagnostics of hepatitis B virus. World J Gastroenterol. 2014;20:14615–14625. doi: 10.3748/wjg.v20.i40.14615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen CJ, Yang HI, Su J, Jen CL, You SL, Lu SN, Huang GT, Iloeje UH. Risk of hepatocellular carcinoma across a biological gradient of serum hepatitis B virus DNA level. JAMA. 2006;295:65–73. doi: 10.1001/jama.295.1.65. [DOI] [PubMed] [Google Scholar]
- 18.Chevaliez S, Pawlotsky JM. Diagnosis and management of chronic viral hepatitis: antigens, antibodies and viral genomes. Best Pract Res Clin Gastroenterol. 2008;22:1031–1048. doi: 10.1016/j.bpg.2008.11.004. [DOI] [PubMed] [Google Scholar]
- 19.O’Connor L, Glynn B. Recent advances in the development of nucleic acid diagnostics. Expert Rev Med Devices. 2010;7:529–539. doi: 10.1586/erd.10.22. [DOI] [PubMed] [Google Scholar]
- 20.Craw P, Balachandran W. Isothermal nucleic acid amplification technologies for point-of-care diagnostics: a critical review. Lab Chip. 2012;12:2469–2486. doi: 10.1039/c2lc40100b. [DOI] [PubMed] [Google Scholar]
- 21.Gill P, Ghaemi A. Nucleic acid isothermal amplification technologies: a review. Nucleosides Nucleotides Nucleic Acids. 2008;27:224–243. doi: 10.1080/15257770701845204. [DOI] [PubMed] [Google Scholar]
- 22.Ziegler C, Göpel W. Biosensor development. Curr Opin Chem Biol. 1998;2:585–591. doi: 10.1016/s1367-5931(98)80087-2. [DOI] [PubMed] [Google Scholar]
- 23.Desjardins P, Conklin D. NanoDrop microvolume quantitation of nucleic acids. J Vis Exp. 2010;(45):pii 2565. doi: 10.3791/2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mullis KB, Faloona FA. Specific synthesis of DNA in vitro via a polymerase-catalyzed chain reaction. Methods Enzymol. 1987;155:335–350. doi: 10.1016/0076-6879(87)55023-6. [DOI] [PubMed] [Google Scholar]
- 25.Fitzmaurice J, Glennon M, Duffy G, Sheridan JJ, Carroll C, Maher M. Application of real-time PCR and RT-PCR assays for the detection and quantitation of VT 1 and VT 2 toxin genes in E. coli O157: H7. Mol Cell Probes. 2004;18:123–132. doi: 10.1016/j.mcp.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 26.Holland PM, Abramson RD, Watson R, Gelfand DH. Detection of specific polymerase chain reaction product by utilizing the 5’----3’ exonuclease activity of Thermus aquaticus DNA polymerase. Proc Natl Acad Sci USA. 1991;88:7276–7280. doi: 10.1073/pnas.88.16.7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abe A, Inoue K, Tanaka T, Kato J, Kajiyama N, Kawaguchi R, Tanaka S, Yoshiba M, Kohara M. Quantitation of hepatitis B virus genomic DNA by real-time detection PCR. J Clin Microbiol. 1999;37:2899–2903. doi: 10.1128/jcm.37.9.2899-2903.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sitnik R, Paes A, Mangueira CP, Pinho JR. A real-time quantitative assay for hepatitis B DNA virus (HBV) developed to detect all HBV genotypes. Rev Inst Med Trop Sao Paulo. 2010;52:119–124. doi: 10.1590/s0036-46652010000300001. [DOI] [PubMed] [Google Scholar]
- 29.Cai SH, Lv FF, Zhang YH, Jiang YG, Peng J. Dynamic comparison between Daan real-time PCR and Cobas TaqMan for quantification of HBV DNA levels in patients with CHB. BMC Infect Dis. 2014;14:85. doi: 10.1186/1471-2334-14-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qiu N, Li R, Yu JG, Yang W, Zhang W, An Y, Li T, Liu XE, Zhuang H. Comparison of Abbott and Da-an real-time PCR for quantitating serum HBV DNA. World J Gastroenterol. 2014;20:11762–11769. doi: 10.3748/wjg.v20.i33.11762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yeh ML, Huang CF, Hsieh MY, Huang JF, Dai CY, Yu ML, Chuang WL. Comparison of the Abbott RealTime HBV assay with the Roche Cobas AmpliPrep/Cobas TaqMan HBV assay for HBV DNA detection and quantification. J Clin Virol. 2014;60:206–214. doi: 10.1016/j.jcv.2014.04.008. [DOI] [PubMed] [Google Scholar]
- 32.Krajden M, Minor J, Cork L, Comanor L. Multi-measurement method comparison of three commercial hepatitis B virus DNA quantification assays. J Viral Hepat. 1998;5:415–422. doi: 10.1046/j.1365-2893.1998.00129.x. [DOI] [PubMed] [Google Scholar]
- 33.Baylis SA, Heath AB, Chudy M, Pisani G, Klotz A, Kerby S, Gerlich W. An international collaborative study to establish the 2nd World Health Organization International Standard for hepatitis B virus DNA nucleic acid amplification technology-based assays. Vox Sang. 2008;94:358–362. doi: 10.1111/j.1423-0410.2008.01023.x. [DOI] [PubMed] [Google Scholar]
- 34.Germer JJ, Qutub MO, Mandrekar JN, Mitchell PS, Yao JD. Quantification of hepatitis B virus (HBV) DNA with a TaqMan HBV analyte-specific reagent following sample processing with the MagNA pure LC instrument. J Clin Microbiol. 2006;44:1490–1494. doi: 10.1128/JCM.44.4.1490-1494.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Konnick EQ, Erali M, Ashwood ER, Hillyard DR. Evaluation of the COBAS amplicor HBV monitor assay and comparison with the ultrasensitive HBV hybrid capture 2 assay for quantification of hepatitis B virus DNA. J Clin Microbiol. 2005;43:596–603. doi: 10.1128/JCM.43.2.596-603.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vogelstein B, Kinzler KW. Digital PCR. Proc Natl Acad Sci USA. 1999;96:9236–9241. doi: 10.1073/pnas.96.16.9236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sanders R, Huggett JF, Bushell CA, Cowen S, Scott DJ, Foy CA. Evaluation of digital PCR for absolute DNA quantification. Anal Chem. 2011;83:6474–6484. doi: 10.1021/ac103230c. [DOI] [PubMed] [Google Scholar]
- 38.Hindson BJ, Ness KD, Masquelier DA, Belgrader P, Heredia NJ, Makarewicz AJ, Bright IJ, Lucero MY, Hiddessen AL, Legler TC, et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal Chem. 2011;83:8604–8610. doi: 10.1021/ac202028g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang JT, Liu YJ, Wang J, Xu ZG, Yang Y, Shen F, Liu XH, Zhou X, Liu SM. Next generation digital PCR measurement of hepatitis B virus copy number in formalin-fixed paraffin-embedded hepatocellular carcinoma tissue. Clin Chem. 2015;61:290–296. doi: 10.1373/clinchem.2014.230227. [DOI] [PubMed] [Google Scholar]
- 40.Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, Hase T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28:E63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parida M, Sannarangaiah S, Dash PK, Rao PV, Morita K. Loop mediated isothermal amplification (LAMP): a new generation of innovative gene amplification technique; perspectives in clinical diagnosis of infectious diseases. Rev Med Virol. 2008;18:407–421. doi: 10.1002/rmv.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nagamine K, Hase T, Notomi T. Accelerated reaction by loop-mediated isothermal amplification using loop primers. Mol Cell Probes. 2002;16:223–229. doi: 10.1006/mcpr.2002.0415. [DOI] [PubMed] [Google Scholar]
- 43.Cai T, Lou G, Yang J, Xu D, Meng Z. Development and evaluation of real-time loop-mediated isothermal amplification for hepatitis B virus DNA quantification: a new tool for HBV management. J Clin Virol. 2008;41:270–276. doi: 10.1016/j.jcv.2007.11.025. [DOI] [PubMed] [Google Scholar]
- 44.Nagamine K, Watanabe K, Ohtsuka K, Hase T, Notomi T. Loop-mediated isothermal amplification reaction using a nondenatured template. Clin Chem. 2001;47:1742–1743. [PubMed] [Google Scholar]
- 45.Chuang TL, Wei SC, Lee SY, Lin CW. A polycarbonate based surface plasmon resonance sensing cartridge for high sensitivity HBV loop-mediated isothermal amplification. Biosens Bioelectron. 2012;32:89–95. doi: 10.1016/j.bios.2011.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chevaliez S, Rodriguez C, Pawlotsky JM. New virologic tools for management of chronic hepatitis B and C. Gastroenterology. 2012;142:1303–1313.e1. doi: 10.1053/j.gastro.2012.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kamisango K, Kamogawa C, Sumi M, Goto S, Hirao A, Gonzales F, Yasuda K, Iino S. Quantitative detection of hepatitis B virus by transcription-mediated amplification and hybridization protection assay. J Clin Microbiol. 1999;37:310–314. doi: 10.1128/jcm.37.2.310-314.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ide T, Kumashiro R, Hino T, Murashima S, Ogata K, Koga Y, Sata M. Transcription-mediated amplification is more useful in the follow-up of patients with chronic hepatitis B treated with lamivudine. Hepatol Res. 2001;21:76–84. doi: 10.1016/s1386-6346(01)00081-x. [DOI] [PubMed] [Google Scholar]
- 49.Kubo S, Hirohashi K, Tanaka H, Shuto T, Takemura S, Yamamoto T, Uenishi T, Kinoshita H, Nishiguchi S. Usefulness of viral concentration measurement by transcription-mediated amplification and hybridization protection as a prognostic factor for recurrence after resection of hepatitis B virus-related hepatocellular carcinoma. Hepatol Res. 2003;25:71–77. doi: 10.1016/s1386-6346(02)00167-5. [DOI] [PubMed] [Google Scholar]
- 50.Compton J. Nucleic acid sequence-based amplification. Nature. 1991;350:91–92. doi: 10.1038/350091a0. [DOI] [PubMed] [Google Scholar]
- 51.Schneider P, Wolters L, Schoone G, Schallig H, Sillekens P, Hermsen R, Sauerwein R. Real-time nucleic acid sequence-based amplification is more convenient than real-time PCR for quantification of Plasmodium falciparum. J Clin Microbiol. 2005;43:402–405. doi: 10.1128/JCM.43.1.402-405.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yates S, Penning M, Goudsmit J, Frantzen I, van de Weijer B, van Strijp D, van Gemen B. Quantitative detection of hepatitis B virus DNA by real-time nucleic acid sequence-based amplification with molecular beacon detection. J Clin Microbiol. 2001;39:3656–3665. doi: 10.1128/JCM.39.10.3656-3665.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Deiman B, Jay C, Zintilini C, Vermeer S, van Strijp D, Venema F, van de Wiel P. Efficient amplification with NASBA of hepatitis B virus, herpes simplex virus and methicillin resistant Staphylococcus aureus DNA. J Virol Methods. 2008;151:283–293. doi: 10.1016/j.jviromet.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 54.Fire A, Xu SQ. Rolling replication of short DNA circles. Proc Natl Acad Sci USA. 1995;92:4641–4645. doi: 10.1073/pnas.92.10.4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fakruddin M, Mannan KS, Chowdhury A, Mazumdar RM, Hossain MN, Islam S, Chowdhury MA. Nucleic acid amplification: Alternative methods of polymerase chain reaction. J Pharm Bioallied Sci. 2013;5:245–252. doi: 10.4103/0975-7406.120066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Margeridon S, Carrouée-Durantel S, Chemin I, Barraud L, Zoulim F, Trépo C, Kay A. Rolling circle amplification, a powerful tool for genetic and functional studies of complete hepatitis B virus genomes from low-level infections and for directly probing covalently closed circular DNA. Antimicrob Agents Chemother. 2008;52:3068–3073. doi: 10.1128/AAC.01318-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Martel N, Gomes SA, Chemin I, Trépo C, Kay A. Improved rolling circle amplification (RCA) of hepatitis B virus (HBV) relaxed-circular serum DNA (RC-DNA) J Virol Methods. 2013;193:653–659. doi: 10.1016/j.jviromet.2013.07.045. [DOI] [PubMed] [Google Scholar]
- 58.Quershi A, Gurbuz Y, Kang WP, Davidson JL. A novel interdigitated capacitor based biosensor for detection of cardiovascular risk marker. Biosens Bioelectron. 2009;25:877–882. doi: 10.1016/j.bios.2009.08.043. [DOI] [PubMed] [Google Scholar]
- 59.Qureshi A, Niazi JH, Kallempudi S, Gurbuz Y. Label-free capacitive biosensor for sensitive detection of multiple biomarkers using gold interdigitated capacitor arrays. Biosens Bioelectron. 2010;25:2318–2323. doi: 10.1016/j.bios.2010.03.018. [DOI] [PubMed] [Google Scholar]
- 60.Haasnoot W, Gerçek H, Cazemier G, Nielen MW. Biosensor immunoassay for flumequine in broiler serum and muscle. Anal Chim Acta. 2007;586:312–318. doi: 10.1016/j.aca.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 61.Kalogianni DP, Koraki T, Christopoulos TK, Ioannou PC. Nanoparticle-based DNA biosensor for visual detection of genetically modified organisms. Biosens Bioelectron. 2006;21:1069–1076. doi: 10.1016/j.bios.2005.04.016. [DOI] [PubMed] [Google Scholar]
- 62.Yao CY, Fu WL. Biosensors for hepatitis B virus detection. World J Gastroenterol. 2014;20:12485–12492. doi: 10.3748/wjg.v20.i35.12485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Drummond TG, Hill MG, Barton JK. Electrochemical DNA sensors. Nat Biotechnol. 2003;21:1192–1199. doi: 10.1038/nbt873. [DOI] [PubMed] [Google Scholar]
- 64.Ding C, Zhao F, Zhang M, Zhang S. Hybridization biosensor using 2,9-dimethyl-1,10-phenantroline cobalt as electrochemical indicator for detection of hepatitis B virus DNA. Bioelectrochemistry. 2008;72:28–33. doi: 10.1016/j.bioelechem.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 65.Zhang SS, Tan QQ, Li F, Zhang XR. Hybridization biosensor using diaquabis[N-(2-pyridinylmethyl)benzamide-kappa N-2,O]-cadmium(II) dinitrate as a new electroactive indicator for detection of human hepatitis B virus DNA. Sensor Actuat B-Chem. 2007;124:290–296. [Google Scholar]
- 66.March C, Manclús JJ, Jiménez Y, Arnau A, Montoya A. A piezoelectric immunosensor for the determination of pesticide residues and metabolites in fruit juices. Talanta. 2009;78:827–833. doi: 10.1016/j.talanta.2008.12.058. [DOI] [PubMed] [Google Scholar]
- 67.Zhou X, Liu L, Hu M, Wang L, Hu J. Detection of hepatitis B virus by piezoelectric biosensor. J Pharm Biomed Anal. 2002;27:341–345. doi: 10.1016/s0731-7085(01)00538-6. [DOI] [PubMed] [Google Scholar]
- 68.Yao C, Zhu T, Tang J, Wu R, Chen Q, Chen M, Zhang B, Huang J, Fu W. Hybridization assay of hepatitis B virus by QCM peptide nucleic acid biosensor. Biosens Bioelectron. 2008;23:879–885. doi: 10.1016/j.bios.2007.09.003. [DOI] [PubMed] [Google Scholar]
- 69.Hansen KM, Thundat T. Microcantilever biosensors. Methods. 2005;37:57–64. doi: 10.1016/j.ymeth.2005.05.011. [DOI] [PubMed] [Google Scholar]
- 70.Buchapudi KR, Huang X, Yang X, Ji HF, Thundat T. Microcantilever biosensors for chemicals and bioorganisms. Analyst. 2011;136:1539–1556. doi: 10.1039/c0an01007c. [DOI] [PubMed] [Google Scholar]
- 71.Ji HF, Gao H, Buchapudi KR, Yang X, Xu X, Schulte MK. Microcantilever biosensors based on conformational change of proteins. Analyst. 2008;133:434–443. doi: 10.1039/b713330h. [DOI] [PubMed] [Google Scholar]
- 72.Cha BH, Lee SM, Park JC, Hwang KS, Kim SK, Lee YS, Ju BK, Kim TS. Detection of Hepatitis B Virus (HBV) DNA at femtomolar concentrations using a silica nanoparticle-enhanced microcantilever sensor. Biosens Bioelectron. 2009;25:130–135. doi: 10.1016/j.bios.2009.06.015. [DOI] [PubMed] [Google Scholar]
- 73.Homola J. Present and future of surface plasmon resonance biosensors. Anal Bioanal Chem. 2003;377:528–539. doi: 10.1007/s00216-003-2101-0. [DOI] [PubMed] [Google Scholar]


