Abstract
Breast cancer is a global health issue, and as the tumor burden increases, we need to come up with newer, better technologies which are convenient, cheap, rapid, sensitive with a high specificity. Technological advancements in the field of cancer biomarker has led to the development of techniques such as mass spectrometric analysis and microarray analysis in which genes, proteins and hundreds and thousands of metabolites can be identified with the emergence of genomics, proteomics and metabolomics. This research is focused on finding biomarkers for diagnosis, prognosis, staging, treatment response and targets for chemotherapy, generating a panel of markers which provide better clinical information compared to a single marker in the panel. This review briefly summarizes application of genomics and proteomics followed by key concepts and applications of metabolomics in breast cancer, with the conclusion that an integration of the three “OMIC” technologies may hold the key to future biomarker discovery.
Sources of Data Study Selection:
The information for this review was collected by searching the Google Scholar and PubMed database for English articles published in the period from 2002 to 2015. The search terms included “biomarkers in breast cancer” along with the following search terms: “genomics”, “proteomics”, “metabolomics”, “breast cancer”, “mass spectrometry”, “molecular markers” and “cancer biomarker”. We have endeavored to quote only the primary sources. Titles and abstracts of retrieved studies were assessed first followed by selection and retrieval of selected full text articles.
KEY WORDS: Biomarker, Breast cancer, Genomics, Proteomics, Metabolomics
Abbreviations: ASCO – American Society of Clinical Oncology, ER – estrogen receptor, PgR – progesterone receptor, HER2 – human epidermal growth factor receptor 2, CA – cancer antigen, TAILORx – Trial Assigning Individualized Options for Treatment (Rx), MINDACT – Microarray InNode Negative Disease may Avoid ChemoTherapy using Mammaprint–, FDA – Federal Drug Authority, GC-TOF-MS – time of flight, LC/ESI-MS – electrospray ionization, PCho – phosphocholine, GPCho – glycerophosphocholine, Cdx-2 – caudal type homebox 2, KiSS1 – Kisspeptin 1, KAL1 – Kallman syndrome 1 sequence, NIST – National Institute of Standards and Technology, HMDB – Human metabolome database
INTRODUCTION
Breast cancer (BC) is a major health issue in women Worldwide as well as in Pakistan. However marked geographic variation has been noted in the incidence, natural course of the disease as well as survival statistics. This is reflected in the comparison between age standardized rates (ASR) and survival rates (SR) of North American women with that of Pakistani women i.e. ASR of 99.4 per 100,000 and SR of 80%1 for the former with ASR of 69.1 per 100,0002 and SR of less than 40% for the latter.1 The gravity of the situation further increases when it is noted that BC in Pakistan affects younger women with an advanced stage at the time of presentation.3
This puts an enormous burden on the resources of a poor country like Pakistan. Mammography for screening, histopathology and blood tests for diagnosis, prognosis and treatment are considered gold standards for breast cancer.4 According to 2007 recommendations of ASCO for tumor markers ER, PgR and HER2 expression in primary invasive breast cancer should be evaluated for diagnosis or recurrence especially as a guide for therapy, while increasing levels of CA 27.29 or CA 15-3 may indicate treatment failure.5 This however cannot be applied to all breast tumors leaving a wide gap in our understanding of this heterogeneous tumor. Hence development of new methods for exploring the molecular pathogenesis of this disease becomes imperative. Detection of malignancy by a sample blood test for identification of tumor markers has been explored thoroughly in medical field. These biomarkers are released by the tumor itself or by other tissues as a reaction to the tumor or inflammation occurring in response to tumor. An ideal tumor marker is easily measured, reliable, and cheap, with a high sensitivity and specificity. It should help not only in screening early cancer but also recurrence, vary with different stages of disease and has prognostic and predictive value.6
This review briefly covers the concepts of genomics and proteomics, followed by an in-depth analysis of the evolving field of metabolomics for biomarker discovery in breast cancer.
1. “Omics” in Breast Cancer
In the quest for identification of a suitable biomarker for breast cancer, novel and high-yield technologies like genomics, proteomics, and metabolomics have received a lot of attention in recent times, prompting the researchers to name this as the era of “Breast cancer – OMICS”.7 Studies have shown that malignant transformation of normal breast tissue and evolution of metastatic clone involves altered gene expression (altered transcription) or altered protein expression (altered translation).8 This has led to the development of promising technologies of genomics, proteomics (respectively) and metabolomics.
Gene expression profiles of certain predictive and prognostic markers of breast cancer have been developed and are available commercially such as molecular technologies for improvement in breast cancer diagnosis and treatment with the development of MammaPrint (Agendia, Amsterdam, The Netherlands) (a 70 gene microarray study for prediction of breast cancer relapse)9 and OncotypeDx (Genome Health, Redwood city, CA, USA) (a 21 gene expression profile by RT-PCR).10 These have been approved by FDA and are commercially available since 2007 and 2004 respectively. Their value in assessing individualized options for treatment in selection of an effective and appropriate chemotherapeutic agent for breast cancer patients is being explored in ongoing clinical trials: TAILORx11 and MINDACT,12 but their routine clinical use is not yet recommended.13
Recently researchers are interested in finding out whether other ‘omic’ technologies can also add to the information provided by genomics.14 Elevated levels of protein in biological fluid in cancer can be due to atypical secretion, shedding of membrane-associated proteins, change in cancer cells polarity, increased expression of proteases15 and single nucleotide polymorphism (SNP) of signal peptide,16 etc. HER2 is a cancer biomarker, and classic example of a membrane bound tyrosine kinase, that is shed into fluids. HER2 protein has an extracellular domain (ECD), a transmembrane domain and a cytoplasmic domain. The ECD of HER2 is cleaved by a protease from the receptor protein and can be detected as a biomarker in serum. Over expression of HER2 seen in some cases of breast cancer is an indicator of poor prognosis for these patients.5 Since 2000, HER2 test has been approved by FDA and is used in the management and follow-up of patients with metastatic breast cancer.
mRNA transcript does not reflect the function of proteins, hence different proteomic strategies in biomarker discovery have emerged as proteins in complex mixtures require systemic characterization by mass spectrometry (MS). Limitation of MS for proteomic approaches include improper sample collection and storage, inability to identify established serological biomarkers, bias in identification of high-abundance molecules within the serum, conflict in reporting of ms peaks reported by different research laboratories17,18 and possible artifacts in bioinformatics.19 Hence serum proteomic analysis and profiling is not currently recommended for clinical use by experts.13
Enzymes are proteins, and there should be good quantitative relationship between mRNA concentration and enzyme function. But on the contrary, metabolites which are downstream are better indicators of enzyme activity,20 and are more sensitive monitors of a change in biological system, represented by the genome (‘genomics’), transcriptome (‘transcriptomics’), proteome (‘proteomics’) and metabolome (‘metabolomics’). At the end of the spectrum, metabolome represents the phenotypic changes and even slight alterations in metabolites can be detected.21 Hence cancer researchers have renewed interest in the field of metabolomics for the discovery of specific biomarkers for use as diagnostic or prognostic markers.21
2. Metabolomics
Warburg effect (put forward by Otto Warburg in 1924)is characterized by an increase in glucose uptake by cancer cells converting it into lactate by glycolysis in spite of normal oxygen supply, hence also called ‘aerobic glycolysis’ or ‘aerobic fermentation’.22 Cancer cell is also shown to have an altered protein metabolism and altered lipid metabolism.23 Hence cancer is regarded as a disease with gene mutation resulting in changes in gene expression to produce a metabolic phenotype with altered glycolytic, amino acid, nucleotide and glycerophospholipid / lipid metabolism. This results in a cancer cell phenotype resulting in cancer cell growth, differentiation and survival.
Metabolomics technology involves identification of hundreds to thousands of metabolites and exploring multiple cellular pathways at the same time. Presence of small molecules in the body fluids or tissues contribute to the construction of a unique ‘fingerprint’, distinguishing between disease and health implying that metabolomics can distinguish between cancer and normal tissues. So metabolomics is emerging as a promising new ‘omics’ field7 a high throughput technology increasingly being used for breast cancer research especially for screening, diagnosis, cancer typing, staging and therapeutic intervention.21,24 Advantages of metabolomics include being cost-effective, high throughput, and being automated with sample analysis taking 10 to 30 minutes per sample approximately.23
Two terms are frequently used in metabolomics; ‘metabolic profiling’ and ‘metabolic fingerprinting’. Metabolic profiling refers to a measure of total number of individual metabolites in a sample while metabolic fingerprinting means measuring a group or class of metabolites or quantification of a limited number of metabolites to differentiate between different samples.25 In spite of recent advances in the field of metabolomics its application has been limited by technical problems.
2.1 Metabolomic Approaches
Most popular methods for metabolomics include mass spectrometry (MS) and nuclear magnetic resonance (NMR) spectroscopy,26 which are complementary to each other. Mass spectrometry can be coupled with gas chromatography (GC-MS) (identification of approximately 1000 metabolites), with liquid chromatography (LC-MS) (identification of hundreds of metabolites) or capillary electrophoresis (CE-MS). NMR MS identifies a variable number of metabolites depending on the nature of samples i.e. 20 – 40 metabolites in tissue samples and 100 – 200 in urine samples.20,25 GC-MS and LC-MS are commonly used techniques for cancer samples.21
Benefits of NMR include high reproducibility, ability to quantify metabolites in complex mixtures and metabolite detection in vivo, as well as in biological fluids and tissues without any prior preparation of the sample. Its main disadvantage is its low sensitivity.27 An improvement of NMR spectroscopic procedure is a technique called high resolution magic angle spinning (HR-MAS) NMR spectroscopy, which involves spinning of a biopsy sample at an angle to the magnetic field, to improve the spectrum resolution.28 Its advantage include simultaneous in situ assessment of both aqueous and lipid soluble metabolites.23 High resolution NMR (HR-NMR) and HR-MAS MRS can be used on biofluids and tissues as they do not cause destruction of samples making it possible to carry out parallel analysis.29
Benefits of GC-MS include high sensitivity, quantification of metabolites and ability to identify more compounds in comparison to other MS techniques. Its main limitation is complex and lengthy steps involved in sample preparation and interpretation of its spectra. Benefits of LC-MS is its use for non-volatile compounds, quantification of wide range a of metabolites and its complementary nature to GC-MS.21
CE-MS separates and identifies polar or ionic compounds in complex mixtures, has high resolution with no complex and laborious sample handling as for GC-MS, and has low sensitivity but high variability than that of LC-MS or GC-MS.21
2.2 Diagnosis, Prognosis & Treatment of Breast Cancer
Heterogeneity of breast cancer (BC) ranges from its morphology, to prognosis, to metastatic potential and to treatment response. Studies carried out for understanding breast cancer pathogenesis are aimed at identification of biomarkers and new targets for effective cancer chemotherapy.30 A GC-TOF MS based metabolomics study in breast cancer detected 368 metabolites that differentiated between cancer and normal tissues, a property that can be utilized for screening of breast cancer. The ratio of cytidine-5-monophosphate / pentadecanoic acid was the most specific and sensitive discriminator.31 In a research carried out to observe changes in lipid metabolism, an important feature of cancer, increased levels of sn-glycerol-3 phosphate was detected by GC-MS, and increased levels of phospholipids by LC-MS in breast cancer tissue.32
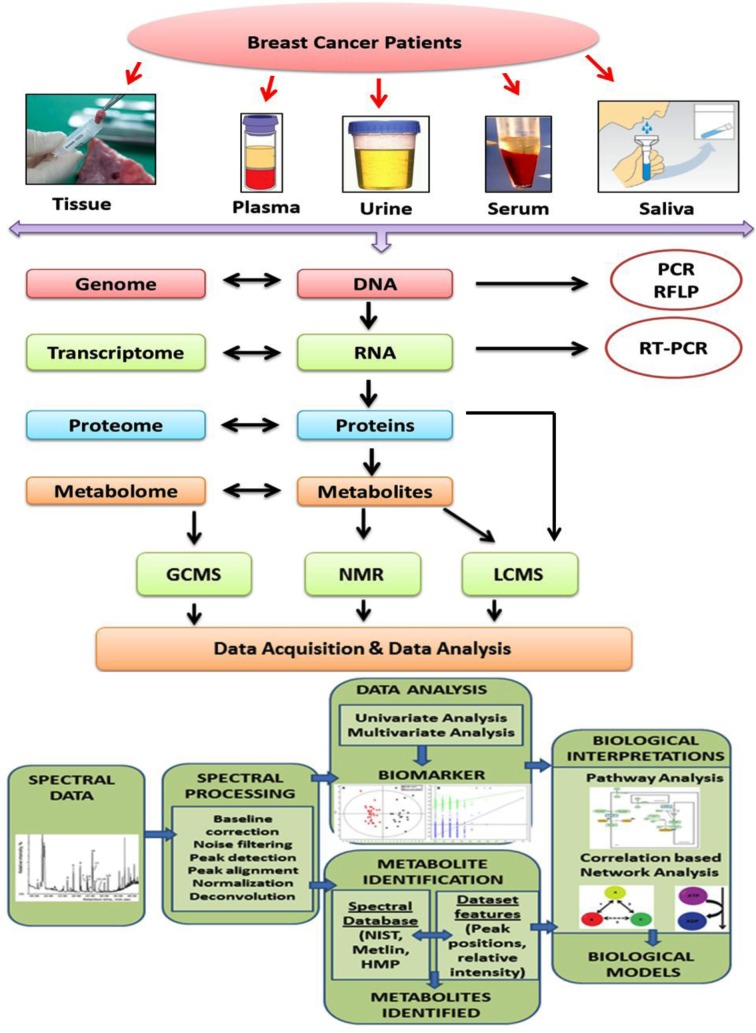
Fig.1.
Schematic representation of “-Omic” platforms in breast cancer biomarker discovery.
Breast cancer prognosis can be predicted by analyzing the metabolic profile and comparing with survival rates. Giske degård et al.33 analyzed breast cancer tissue by HR-MAS MRS and found high levels of glycine and lactate in a subgroup of ER positive breast cancer patients with lower survival rates and hence poor prognosis. The other subgroup of ER positive patients had better prognosis while these metabolic changes (elevated glycine and lactate) were not seen in ER negative patients. In another study it was reported that ER positive Luminal type A breast cancer has a variable response to hormone therapy, dividing this group into responders and non-responders. Metabolomic analysis of luminal type A identified three groups with the help of ‘a’ and ‘b’ glucose amino acids, myoinositol and lipid residues. Gene analysis of one of the group of luminal A subtype showed its relationship to cell cycle and DNA repair and most probably represented the non-responders to hormone therapy.34
Increased levels of total choline (t-Cho) containing compounds have been detected in breast cancer cells forming a basis for many metabolomics analysis carried out in these patients.35-37 For example, in a study carried out by Mimmi et al.(2011),36 breast tissue biopsies from normal subjects, patients with fibrocystic disease, benignlesions and breast cancer were analyzed by LC / ESI-MS for the presence of Cho, PCho and GPCho. Results showed raised levels of choline and its phosphorylated metabolites in subjects with benign and malignant tumors only. Metastasis in breast cancer has also been detected with the help of a distinct metabolic profile in the serum or urine of these patients.35,38 Similarly serum metabolic profile by NMR showed that metastatic breast cancer can be differentiated from early stage breast cancer with a prediction accuracy of 72%.35 Jobard et al.39 carried out serum metabolomics by HNMR to differentiate between localized early disease (EBC) and metastatic breast cancer (MBC) with respect to diagnosis, prognosis and management of these patients. He constructed a model with 9 differentiating metabolites which included histidine, acetoacetate, pyruvate, glycerol, glycoprotein (N-acetyl), glutamate, mannose and phenylalanine.
Asiago et al.38 studied recurrence of breast cancer by a combination of analytical (NMR, GC- MS) and multivariate statistical methods to identify 11 metabolites which can predict recurrence with a sensitivity of 86% and specificity of 84%. By combining NMR and LC- MS, four metabolites namely threonine, glutamine, isoleucine and linolenic acid were found to be predictors of response to neoadjuvant chemotherapy. Breast cancer patients were placed into three groups, i.e. with no response, partial response, or complete response. Altered metabolic profiles were seen for these four amino acid metabolites which distinguished between the different groups.40
Pharmacometabolomics, a promising and novel field can predict response to chemotherapeutic agents. In patients with metastatic breast cancer treated with paclitaxel and lapatinib, serum metabolic profile was analyzed before and during chemotherapy, and was found to have a positive correlation with patient survival and time to progression in HER2 positive patients. Metabolomics also helped in selecting the subset of HER2 positive breast cancer patients with metastatic disease who were responsive to this combination therapy.41
“Omics” in Pakistan: Present and Future
Different types and sites of BRCA 1 and 2 gene mutations, known risk factors for breast cancer, have been studied extensively in Pakistani women with BC (both sporadic as well as familial cases).42-44 Breast cancer research in recent times is focusing more on gene polymorphisms other than BRCA 1 and 2 as a possible explanation for racial differences in incidence, clinical presentations and prognosis of breast cancer. Prevalence of TP53 mutations in BRCA1 & 2 negative young Pakistani BC patients (≤ 30 years) was assessed, uncovering novel mutations which can account for a subset of cancers occurring in the younger age group.45 An association between vitamin D receptor Cdx-2 gene polymorphism and risk of breast cancer in premenopausal women revealed an increased risk of BC in young women with GG genotype,46 while another study reported a reduced expression of metastasis suppression genes (KiSS1 and KAL1) in Pakistani BC patients.47 Absence of FANCM c. 5101c >: T mutation in triple negative and BRCA 1 & 2 negative patients48 and insignificant role of RAD51C, a gene responsible for DNA repair and stability of genome, in BRCA 1 & 2 negative BC patients49 have also been reported.
Literature search regarding proteomics research in Pakistan yielded a single article reporting a distinct proteomic profile distinguishing between breast cancer, benign breast lesions and healthy controls, serving as diagnostic biomarkers. Sera of the three groups were analyzed by one-dimensional SDS polyacrylamide gel electrophoresis (PAGE) and protein identified through LC/MS/MS.50 Similarly in spite of global advances in field of biomarker discovery for BC by metabolomics, to the best of our knowledge no work has been carried out in Pakistan. Currently we are working on BC metabolomics biomarker project, which is in final stage of completion, in collaboration with H.E.J. research institute of chemistry, Dr. Panjwani Center for Molecular Medicine and Drug Research (PCMD), University of Karachi.
CONCLUSION
High-throughput “omic” technologies, especially metabolomics is a promising evolving field for advancing our knowledge and understanding of breast cancer pathogenesis, identification of diagnostic biomarkers, tumor typing and staging, and response to therapy. Extensive and widespread studies employing a large sample size are required for proper validation of these different biomarkers. Clinical application of “omics” approach can further be improved by integration of genomics, proteomics and metabolomics, thus exploring new frontiers in biomarker discovery for breast cancer.
Author’s Contribution
NIH: Conceived, designed and edited the manuscript.
QJ: Critically reviewed the content of the manuscript.
REFERENCES
- 1.Coleman MP, Quaresma F, Berrino J, Lutz JM, De Angelis R, Capocaccia R, et al. Cancer survival in five continents: a worldwide population based study (CONCORD) Lancet Oncol. 2008;9(8):730–756. doi: 10.1016/S1470-2045(08)70179-7. doi: 10.1016/S1470-2045(08)70179-7. [DOI] [PubMed] [Google Scholar]
- 2.Bhurgri Y, Bhurgri A, Nishter S, Ahmed A, Usman A, Pervez S, et al. Pakistan - country profile of cancer and cancer control 1995-2004. J Pak Med Assoc. 2006;56(3):124–130. [PubMed] [Google Scholar]
- 3.Khokher S, Mahmood S, Khan SA. Response to neoadjuvant chemotherapy in patients withadvanced breast cancer: A local hospital experience. Asian Pac J Cancer Prev. 2010;11(2):303–308. [PubMed] [Google Scholar]
- 4.Shi L. Racial differences in breast cancer patterns. Science. 2010;329(5987):32. doi: 10.1126/science.329.5987.32-a. doi: 10.1126/science.329.5987.32-a. [DOI] [PubMed] [Google Scholar]
- 5.Harris L, Fritsche H, Mennel R, Norton L, Ravdin P, Taube S, et al. American Society of Clinical Oncology 2007 update of recommendations for the use of tumor marker in breast cancer. JCO. 2007;24(33):5287–5312. doi: 10.1200/JCO.2007.14.2364. [DOI] [PubMed] [Google Scholar]
- 6.Malati Y. Tumor markers: An overview. Indian J Clin Biochem. 2007;22(2):17–31. doi: 10.1007/BF02913308. doi: 10.1007/BF02913308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang AH, Sun H, Qui S, Wang XJ. Metabolomics in noninvasive breast cancer. Clinica Chimica Acta. 2013;424:3–7. doi: 10.1016/j.cca.2013.05.003. doi: 10.1016/j.cca.2013.05.003. [DOI] [PubMed] [Google Scholar]
- 8.Nuyten DS, van de Vijver MJ. Using microarray analysis as a prognostic and predictive tool in oncology: Focus on breast cancer and normal tissue toxicity. Semin Radiat Oncol. 2008;18(2):105–114. doi: 10.1016/j.semradonc.2007.10.007. doi: 10.1016/j.semradonc.2007.10.007. [DOI] [PubMed] [Google Scholar]
- 9.van’t Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 10.Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351:2817–2826. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 11.Coates AS, Colleoni M, Goldhrisch A. Is adjuvant chemotherapy useful for women with luminal A breast cancer? JCO. 2012;30(12):1260–1263. doi: 10.1200/JCO.2011.37.7879. doi: 10.1200/JCO.2011.37.7879. [DOI] [PubMed] [Google Scholar]
- 12.Cardoso F, Van’t Veer L, Rutgers E, Loi S, Mook S, Piccart-Gebhart MJ. Clinical application of the 70 gene profile: The MINDACT trial. JCO. 2008;26(5):729–735. doi: 10.1200/JCO.2007.14.3222. doi: 10.1200/JCO.2007.14.3222. [DOI] [PubMed] [Google Scholar]
- 13.Sturgeon CM, Hoffman BR, Chan DW, Ch’ng SL, Hammond E, Hayes DF, et al. National Academy of Clinical Biochemistry Laboratory Medicine Practice Guidelines for use of tumor markers in clinical practice: quality requirements. Clin Chem. 2008;54:1–27. doi: 10.1373/clinchem.2007.094144. doi: 10.1373/clinchem.2007.094144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Howell A. Can metabolomics in addition to genomics add to prognostic and predictive information in breast cancer? BMC Medicine. 2010;8:73. doi: 10.1186/1741-7015-8-73. doi: 10.1186/1741-7015-8-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kulasingam V, Diamandis EP. Strategies for discovering novel cancer biomarkers through utilization of emerging technologies. Nat Clin Pract Oncol. 2008;5:588–599. doi: 10.1038/ncponc1187. doi: 10.1038/ncponc1187. [DOI] [PubMed] [Google Scholar]
- 16.Jarjanazi H, Savas S, Pabalan N, Dennis JW, Ozcelik H. Biological implications of SNPs in signal peptide domains of human proteins. Proteins. 2008;70(2):394–403. doi: 10.1002/prot.21548. [DOI] [PubMed] [Google Scholar]
- 17.Diamandis EP. Mass spectrometry as a diagnostic and a cancer biomarker discovery tool. Molecular & Cellular Proteomics. 2004;3:367–378. doi: 10.1074/mcp.R400007-MCP200. doi: 10.1074/mcp.R400007-MCP200. [DOI] [PubMed] [Google Scholar]
- 18.Karsan A, Eigl BJ, Flibotte S, Gelmon K, Switzer P, Hassell P, et al. Analytical and preanalytical biases in serum proteomic pattern analysis for breast cancer diagnosis. Clin Chem. 2005;51(8):1525–1528. doi: 10.1373/clinchem.2005.050708. doi: 10.1373/clinchem.2005.050708. [DOI] [PubMed] [Google Scholar]
- 19.Baggerly KA, Morris JS, Edmonson SR, Coombes ER. Signal in noise: evaluating reported reproducibility of serum proteomic tests for ovarian cancer. J Natl Cancer Inst. 2005;97(4):307–309. doi: 10.1093/jnci/dji008. doi: 10.1093/jnci/dji008. [DOI] [PubMed] [Google Scholar]
- 20.Spartlin JL, Serkova NJ, Eckhardt SG. Clinical applications of metabolomics in oncology: a review. Clin Cancer Res. 2009;15(2):431–440. doi: 10.1158/1078-0432.CCR-08-1059. doi: 10.1158/1078-0432.CCR-08-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Armitage EG, andBarbas C. Metabolomics in cancer biomarker discovery: Current trends and future perspectives. J Pharmaceutical Biomedical Analysis. 2014;87:1–11. doi: 10.1016/j.jpba.2013.08.041. doi: 10.1016/j.jpba.2013.08.041. [DOI] [PubMed] [Google Scholar]
- 22.Stricker TP, Kumar V. Growth promoting metabolic alterations: The Warburg Effect. In: Kumar V, Abbas AK, Aster JC, editors. In Neoplasia (Chapter 7). Robbins and Cotran pathologic basis of disease. (Ninth edition) Philadelphia, PA: Elsevier Saunders; 2015. pp. 265–340. ISBN: 978-1-4557-2613-4. [Google Scholar]
- 23.Griffin JL, Shockcor JP. Metabolic profiles of cancer cells. Nat Rev Cancer. 2004;4:551–556. doi: 10.1038/nrc1390. doi: 10.1038/nrc1390. [DOI] [PubMed] [Google Scholar]
- 24.Denkert C, Bucher E, Hilvo M, Salek R, Orešič M, Griffin J, et al. Metabolomics of human breast cancer: new approaches for tumor typing and biomarker discovery. Genome Med. 2012;4(4):37. doi: 10.1186/gm336. doi: 10.1186/gm336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dettmer K, Aronov PA, Hammock BD. Mass spectrometry-based metabolomics. Mass Spectrom Rev. 2007;26(1):51–78. doi: 10.1002/mas.20108. doi: 10.1002/mas.20108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arakaki AK, Skolnick J, McDonald JF. Marker metabolites can be therapeutic targets as well. Nature. 2008;456(7221):443. doi: 10.1038/456443c. doi: 10.1038/456443c. [DOI] [PubMed] [Google Scholar]
- 27.Van QN, Veenstra TD. How close is the bench to the bedside? Metabolic profiling in cancer research. Genome Med. 2009;1:5. doi: 10.1186/gm5. doi: 10.1186/gm5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sitter B, Bathen TF, Singstad TE, Fjosne HE, Lundgren S, Helgunset J, et al. Quantification of metabolites in breast cancer patients with different clinical prognosis using HR MAS MR spectroscopy. NMR Biomed. 2010;23(4):424–431. doi: 10.1002/nbm.1478. doi: 10.1002/nbm.1478. [DOI] [PubMed] [Google Scholar]
- 29.Choi JS, Baek H-M, Kim S, Kim MJ, Youk JH, Moon HJ, et al. HR-MAS MR Spectroscopy of Breast Cancer Tissue Obtained with Core Needle Biopsy: Correlation with Prognostic Factors. PLoS ONE. 2012;7(12):e51712. doi: 10.1371/journal.pone.0051712. doi: 10.1371/journal.pone.0051712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang A, Sun H, Wu X, Wang X. Urine metabolomics. Clin Chim Acta. 2012;414:65–69. doi: 10.1016/j.cca.2012.08.016. doi: 10.1016/j.cca.2012.08.016. [DOI] [PubMed] [Google Scholar]
- 31.Budczies J, Denkert C, Müller BM, Brockmöller SF, Klauschen F, Györffy B, et al. Remodeling of central metabolismin invasive breast cancer compared to normal breast tissue —a GC–TOFMS based metabolomics study. BMC Genomics. 2012;13:334. doi: 10.1186/1471-2164-13-334. doi: 10.1186/1471-2164-13-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brockmöller SF, Bucher E, Müller BM, Budczies J, Hilvo M, Griffin JL, et al. Integration of metabolomics and expression of glycerol-3-phosphate acyltransferase (GPAM) in breast cancer—link to patient survival, hormone receptor status, and metabolic profiling. J Proteome Res. 2012;11(2):850–860. doi: 10.1021/pr200685r. doi: 10.1021/pr200685r. [DOI] [PubMed] [Google Scholar]
- 33.Giskeødegård GF, Lundgren S, Sitter B, Fjøsne HE, Postma G, Buydens LM, et al. Lactate and glycine-potential MR biomarkers of prognosis in estrogen receptor-positive breast cancers. NMR Biomed. 2012;25(11):1271–1279. doi: 10.1002/nbm.2798. doi: 10.1002/nbm.2798. [DOI] [PubMed] [Google Scholar]
- 34.Borgan E, Sitter B, Lingjaerde OL, Johnsen H, Lundgren S, Bathen TF, et al. Merging transcriptomics and metabolomics - advances in breast cancer profiling. BMC Cancer. 2010;10:628. doi: 10.1186/1471-2407-10-628. doi: 10.1186/1471-2407-10-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oakman C, Tenori L, Claudino WM, Cappadona S, Nepi S, Battaglia A, et al. Identification of a serum-detectable metabolomic fingerprint potentially correlated with the presence of micrometastatic disease in early breast cancer patients at varying risks of disease relapse by traditional prognostic methods. Ann Oncol. 2011b;22(6):1295–1301. doi: 10.1093/annonc/mdq606. doi: 10.1093/annonc/mdq606. [DOI] [PubMed] [Google Scholar]
- 36.Mimmi MC, Picotti P, Corazza A, Betto E, Pucillo CE, Cesaratto L, et al. High-performance metabolic marker assessment in breast cancer tissue by mass spectrometry. Clin Chem Lab Med. 2011;49(2):317–324. doi: 10.1515/CCLM.2011.060. doi: 10.1515/CCLM.2011.060. [DOI] [PubMed] [Google Scholar]
- 37.Mimmi MC, Finato N, Pizzolato G, Beltrami CA, Fogolari F, Corazza A, et al. Absolute quantification of choline-related biomarkers in breast cancer biopsies by liquid chromatography electrospray ionization mass spectrometry. Anal Cell Pathol (Amst) 2013;36(3-4):71–83. doi: 10.3233/ACP-130082. doi: 10.3233/ACP-130082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Asiago VM, Alvarado LZ, Shanaiah N, Gowda GA, Owusu-Sarfo K, Ballas RA, et al. Early detection of recurrent breast cancer using metabolite profiling. Cancer Res. 2010;70(21):8309–8318. doi: 10.1158/0008-5472.CAN-10-1319. doi: 10.1158/0008-5472.CAN-10-1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jobard E, Pontoizeau C, Blaise BJ, Bachelot T, Elena-Herrmann B, Trédan O. A serum nuclear magnetic resonance-based metabolomic signature of advanced metastatic human breast cancer. Cancer Lett. 2014;343(1):33–41. doi: 10.1016/j.canlet.2013.09.011. doi: 10.1016/j.canlet.2013.09.011. [DOI] [PubMed] [Google Scholar]
- 40.Wei S, Liu L, Zhang J, Bowers J, Gowda GAN, Seeger H, et al. Metabolomics approach for predicting response to neoadjuvant chemotherapy for breast cancer. Mol Oncol. 2013;7(3):297–307. doi: 10.1016/j.molonc.2012.10.003. doi: 10.1016/j.molonc.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tenori L, Oakman C, Claudino WM, Bernini P, Cappadona S, Nepi S, et al. Exploration of serum metabolomic profiles and outcomes in women with metastatic breast cancer: A pilot study. Mol Oncol. 2012;6:437–444. doi: 10.1016/j.molonc.2012.05.003. doi: 10.1016/j.molonc.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rebbeck TR, Mitra N, Wan F, Sinilnikova OM, Healey S, McGuffog L, et al. Association of type and location of BRCA 1 and BRCA 2 mutations with risk of breast and ovarian cancer. JAMA. 2015;313(13):1347–1361. doi: 10.1001/jama.2014.5985. doi: 10.1001/jama.2014.5985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rashid MU, Muhammad N, Iqbal K, Yusuf HA, Hamann U. BRCA 1 gene testing in a Pakistani breast-ovarian cancer family with multiple consanguineous marriages. Clin Genet. 2015;88(2):198–199. doi: 10.1111/cge.12533. doi: 10.1111/cge.12533. [DOI] [PubMed] [Google Scholar]
- 44.Spurdle AB, Couch FJ, Parsons MT, McGuffog L, Barrowdale D, Bolla MK, et al. Refined histopathological predictors for BRCA1 and BRCA2 mutation status: a large scale analysis of breast cancer characteristics from the BCAC, CIMBA, and ENIGMA consortia. Breast Cancer Res. 2014;16(6):3419. doi: 10.1186/s13058-014-0474-y. doi: 10.1186/s13058-014-0474-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rashid MU, Gull S, Asghar K, Muhammad N, Amin A, Hamann U. Prevalence of TP53 germ line mutations in young Pakistani breast cancer patients. Fam Cancer. 2012;11(2):307–311. doi: 10.1007/s10689-012-9509-7. doi: 10.1007/s10689-012-9509-7. [DOI] [PubMed] [Google Scholar]
- 46.Iqbal MU, Khan TA, Maqbool SA. Vitamin D receptor cdx-2 polymorphism and premenopausal breast cancer risk in southern Pakistani patients. PLoS One. 2015;10(3):e0122657. doi: 10.1371/journal.pone.0122657. doi: 10.1371/journal.pone.0122657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mooez S, Malik FA, Kayani MA, Rashid R, Zahid A, Khan A. Expressional alterations and transcript isoforms of metastasis suppressor genes (KAL1 and KiSS1) in breast cancer patients. Asian Pac J. Can Prev. 2011;12(10):2785–2791. [PubMed] [Google Scholar]
- 48.Rashid MU, Muhammad N, Khan FA, Hamann U. Absence of the FANCM c.5101C >: T mutation in BRCA1 /2-negative triple-negative breast cancer patients from Pakistan. Breast Cancer Res Treat. 2015;152(1):229–230. doi: 10.1007/s10549-015-3457-5. doi: 10.1007/s10549-015-3457-5. [DOI] [PubMed] [Google Scholar]
- 49.Rashid MU, Muhammad N, Faisal S, Amin A, Hamann U. Deleterious RAD51C germline mutations rarely predispose to breast and ovarian cancer in Pakistan. Breast Cancer Res Treat. 2014;145(3):775–784. doi: 10.1007/s10549-014-2972-0. doi: 10.1007/s10549-014-2972-0. [DOI] [PubMed] [Google Scholar]
- 50.Nasim FU, Ejaz S, Ashraf M, Asif AR, Ollerich M, Ahmed G, et al. Potential biomarkers in the sera of breast cancer patients from Bahawalpur, Pakistan. Biomark Cancer. 2012;10(4):19–34. doi: 10.4137/BIC.S10502. doi: 10.4137/BIC.S10502. [DOI] [PMC free article] [PubMed] [Google Scholar]