Abstract
Background and Purpose
Diffusion tensor imaging tractography reconstruction of white matter pathways can help guide brain tumor resection. However, DTI tracts are complex mathematical objects and the validity of tractography-derived information in clinical settings has yet to be fully established. To address this issue, we initiated the DTI Challenge, an international working group of clinicians and scientists whose goal was to provide standardized evaluation of tractography methods for neurosurgery. The purpose of this empirical study was to evaluate different tractography techniques in the first DTI Challenge workshop.
Methods
Eight international teams from leading institutions reconstructed the pyramidal tract in four neurosurgical cases presenting with a glioma near the motor cortex. Tractography methods included deterministic, probabilistic, filtered, and global approaches. Standardized evaluation of the tracts consisted in the qualitative review of the pyramidal pathways by a panel of neurosurgeons and DTI experts and the quantitative evaluation of the degree of agreement among methods.
Results
The evaluation of tractography reconstructions showed a great inter-algorithm variability. Although most methods found projections of the pyramidal tract from the medial portion of the motor strip, only a few algorithms could trace the lateral projections from the hand, face, and tongue area. In addition, the structure of disagreement among methods was similar across hemispheres despite the anatomical distortions caused by pathological tissues.
Conclusions
The DTI Challenge provides a benchmark for the standardized evaluation of tractography methods on neurosurgical data. This study suggests that there are still limitations to the clinical use of tractography for neurosurgical decision-making.
Keywords: Diffusion MRI, Diffusion Tensor Imaging, Tractography, Brain Gliomas, Neurosurgery
Introduction
Diffusion MRI provides the first non-invasive window on the architecture of the brain white matter in vivo.1 The development of Diffusion Tensor Imaging (DTI) has opened up the possibility for investigating structural and anatomical features of white matter in human subjects non-invasively.2,3 From DTI data, tractography techniques have been proposed for extracting the trajectories of specific white matter pathways.4 In neurosurgical settings, DTI tractography reconstructions of peritumoral white matter anatomy have the potential for providing clinically relevant information during pre-operative planning and intra-operative mapping of brain tumor resection.5-6 In particular, the knowledge of the spatial relationship of a tumor with white matter tracts involved in motor, visual, or language function is essential to the neurosurgeon for preventing any post-operative disorder.7 Pre-operative tractography reconstructions can be generated interactively,8 and can be integrated into a neuronavigation workstation to help guide brain tumor surgery.9,10 However, tractography remains a challenging technology based on complex data acquisition and geometrical models that rely on many assumptions.11 In neurosurgical settings, tractography errors can give clinicians incorrect information on the location of critical structures, and thus present the risk of post-operative deficits for the patient.12
To overcome these limitations, the medical image computing community has developed a multitude of innovative tractography techniques.4 However, neurosurgeons face the challenge of selecting the appropriate tractography method in the absence of ground truth. The first attempt at comparing tractography algorithms on neurosurgical data was the 2010 IEEE visualization contest on multimodal visualization for neurosurgical planning.13 While this visualization contest provided relevant information on the performances of tractography algorithms on patient data, the tractography reconstructions were presented in different software environments and with different anatomical orientation making the comparative evaluation challenging.
The novelty of our work is a standardized evaluation of tractography algorithms on a common set of neurosurgical cases. The reconstructions were performed through the DTI Challenge, a publicly announced image processing contest that we organized at the 14th International conference on Medical Image Computing and Computer Assisted Intervention (MICCAI) in Toronto, Canada. This paper presents the methodology and results of the first DTI Challenge workshop that is, to the best of our knowledge, the first attempt at comparing neurosurgical data processed with different tractography algorithms in a standardized and interactive way.
Materials and methods
Anatomical objective
The pyramidal tract (PT) was chosen as targeted white matter structure due to its well-defined anatomical origin and termination, and its critical contribution to motor function. In addition, the crossing of the PT with the superior longitudinal fasciculus and corpus callosum was a representative of fiber-crossing regions which are often challenging for tractography techniques.
MRI data acquisition
Informed consent was obtained from all participants and the study was approved by Partners’ Institutional Review Board. MRI data were acquired on four patients presenting with a glioma near the motor cortex area. The cases included a recurrent/residual anaplastic oligoastrocytoma WHO grade III (patient 1), an anaplastic oligoastrocytoma WHO grade III (patient 2), an anaplastic oligodendroglioma WHO grade III (patient 3), and a glioblastoma WHO grade IV (patient 4). MR images were acquired on a GE Signa Excite 3T scanner (GE Medical Systems, Milwaukee, WI, USA) using an 8-channel head coil. Diffusion weighted images were acquired using a single-shot EPI sequence with 30 gradient-encoding directions. In total, 31 images were acquired, including one baseline image with no diffusion sensitization. This sequence represented the diffusion imaging data used in our neurosurgery clinic at the time of the study. The acquisition parameters were: b=1,000 s/mm2, TR=14000 ms, TE=30 ms, FOV=25.6 cm. Whole brain coverage was obtained by collecting 52 slices with 1.0 mm pixel size and 2.6 mm slice thickness. Anatomical imaging included a T1-weighted SPGR axial scan and a T2-weighted scan.
MRI data pre-processing
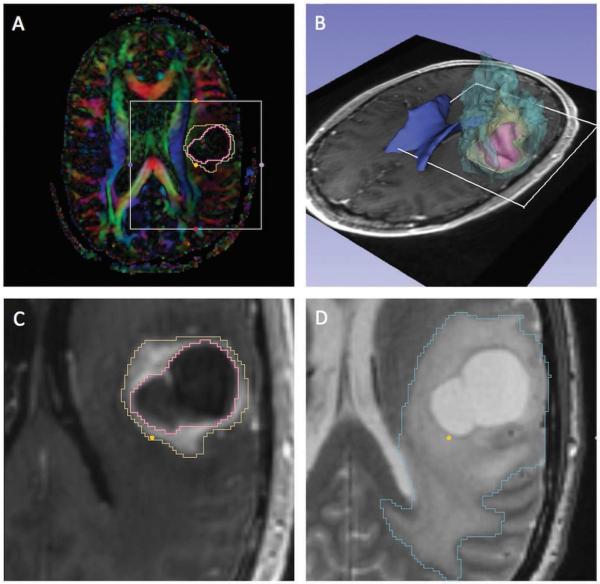
The pre-processing steps included image registration, segmentation, and tensor calculation. All steps were performed using the 3D Slicer open-source software.14 No distortion correction was performed on the DWI scans to provide original clinical datasets to the tractography teams. A diffusion tensor was estimated from the DWI volumes using a weighted least squares estimation algorithm. The anatomical scans were co-registered to the diffusion scan using rigid and non-rigid registration.15 For anatomical reference purpose, the tumor and edema were manually segmented from the registered anatomical images with input from a trained neuroradiologist. In patient 3, the outline of the surgical cavity resulting from previous resection was included in the segmentation. Surface models were generated from the segmented images using the Marching Cubes algorithm. Figure 1 shows the segmentation and 3D models of anatomical structures in patient 1.
Figure 1. Patient 1, pre-operative dataset.
The figure shows the pre-operative imaging data of patient 1 (anaplastic oligoastrocytoma WHO grade III). (A) Diffusion-Encoded Color map with superimposed segmentation of the tumor (yellow) and necrotic center (pink); (B) 3D view of surface models of the tumor (yellow), necrosis (pink), peri-tumoral edema (light blue), and lateral ventricles (dark blue) overlaid on an axial T1-weighted image; (C) T1-weighted image with segmented tumor (yellow) and necrosis (pink); (D) T2-weighted image with segmented peritumoral edema (light blue). The views presented in (C) and (D) correspond to the region of interest (white square) defined in (A) and (B).
Tractography reconstruction
The tractography reconstruction was performed through the DTI Challenge, an image processing challenge which consisted of an internet-based component and a physical meeting at the MICCAI conference.16 The teams reconstructed the pyramidal tract from the DWI datasets acquired on patient 1 and patient 2 in a 3-month period prior to the workshop. The DWI datasets of patient 3 and patient 4 were processed during a 5-hour on-site challenge to simulate clinical constraints in which post-processing time is limited. The tractography results consisted of the trajectories of the tracts represented as streamlines and the envelopes of the voxelized tracts represented as binary labelmaps.
Table 1 provides a short summary of the eight methods used by the teams.16 Each method is characterized by a diffusion model, a fiber tracking algorithm and a set of anatomical regions-of-interest (ROIs). The diffusion model describes the probability density function of the displacement of water molecules in brain tissues. The fiber tracking algorithm reconstructs the trajectory of the molecules based on the directional information given by the diffusion model and the anatomical ROIs. Fiber tracking methods can be divided into four categories: deterministic, probabilistic, filtered and global. Deterministic algorithms reconstruct the trajectory of a fascicle by following the principal direction of diffusion at each voxel.17 Probabilistic algorithms compute the distribution of fiber pathways emanating from a given seed point and assign a confidence level to a specific trajectory.18 Filtered tractography algorithms perform simultaneous multi-tensor estimation and fiber tractography within a Kalman filtering framework.19 Finally, energy-based global tractography methods reconstruct all the fibers simultaneously by optimizing a cost function that best fits the measured data.20
Table. 1.
Tractography methods
| Team ID | Diffusion Model | Fiber Tracking Algorithm | Regions-of-interest |
|---|---|---|---|
| Team1 | Single tensor | Deterministic | Precentral gyrus, postcentral gyrus, cerebral peduncles |
| Team 2 | Multi-compartment Diffusion Direction Imaging | Deterministic | Mesencephalon, internal capsule |
| Team 3 | Single tensor | Probabilistic | Somatosensory cortex, pons, temporal lobes |
| Team 4 | Mixture of two Watson directional functions | Filtered | Cerebral peduncles, pons, medulla |
| Team 5 | Two-tensor | Filtered | Cortex, brainstem |
| Team 6 | Regularized q-ball | Global | Pons, internal capsule, corpus callosum, pyramidal tract |
| Team 7 | Q-ball | Global | Cerebral peduncles, cingulum, pyramidal tract |
| Team 8 | Single tensor | Global | Precentral gyrus, cerebral peduncles |
The table lists the diffusion model, fiber tracking algorithm and regions-of-interest used by the teams.
Post-processing
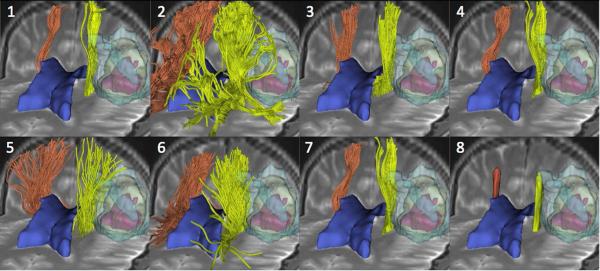
The tractography results submitted by the teams consisted of the trajectories of the tracts represented as streamlines, and the envelopes of the voxelized tracts represented as binary labelmaps. In order to enable a standardized review of the results, we loaded the tractography results in 3D Slicer and we created a series of 3D views of the tracts overlaid on the registered anatomical images along with 3D models of the tumor, edema, and ventricles (Figure 2). The tracts submitted by the different teams were given the same orientation and color for consistency, with tracts on the tumor side displayed in yellow and tracts on the contralateral side in orange. To avoid any observation bias, the names of the algorithms were blinded to the reviewers, and the teams were identified by a number ranging from 1 to 8.
Figure 2. Patient 1, tractography results.
The mosaic image shows 3D anterior views of the pyramidal tract reconstructed by the eight tractography teams on the patient 1 dataset. Each view presents the tracts (yellow: tumor side; orange: contralateral side) overlaid on an axial and a coronal T2-weighted image, along with 3D surface models of the tumor (light yellow), necrosis (pink), edema (light blue) and lateral ventricles (dark blue). The teams are identified by a number, from 1 to 8, in the top left corner of each view.
Qualitative evaluation
The evaluation of the tractography results was based on the qualitative review of the 3D views of the tracts by a panel of five judges. The panel was composed of two neurosurgeons experienced in the clinical use of DTI data and three DTI experts. The tractography results were evaluated based the presence of false-positive and false-negative tracts and anatomical accuracy of the reconstructed bundles. An important aspect of the DTI Challenge was the possibility for the jury to visualize the tractography results interactively. To that end, we provided the judges with a copy of the 3D Slicer software and an archive file that included, for each patient dataset, the eight tractography reconstructions of the pyramidal tract submitted by the teams along with the DTI volumes, anatomical scans, and 3D models of the anatomy. The judges could load and interactively rotate and zoom the data within the 3D Slicer software, so as to define the views that maximized their visual assessment of the tractography results.
Quantitative evaluation
The quantitative comparison of the different tractography reconstructions was performed on the basis of the estimation of the distance between tracts and percentage of affected regions visited by the tracts.
Distance between tracts
To evaluate the distances between two given fiber bundles, fx and fy generated by team x and y, respectively, we used the Average Mean Distance (AMD) and the Hausdorff distance, two metrics that have been proposed for the purpose of comparing tractography results.21 The AMD is defined as the average of closest distance between fiber fx and fiber fy; the Hausdorff distance is defined as the maximum of closest distance between fiber fx and fiber fy. We computed the AMD and Hausdorff distance for all pairs of fiber bundles reconstructed by the different teams on the tumor side and contralateral side, in all four patient datasets. We used box plots to display the summary statistics and distribution of distances.
Volumetric fraction of affected regions visited by tracts
To assess the capabilities of the tractography algorithms within and in the vicinity of the gliomas, we computed the volumetric fraction of tumor (ft) and volumetric fraction of edema (fe) visited by the tracts. In patient 3, we also calculated the volumetric fraction of surgical cavity (fc) visited by the tracts. For a given fiber bundle fx reconstructed by team x, fxt and fxe were defined as the volume of the intersection of the envelope of fx with the segmented tumor and edema region, respectively. In patient 3, fxc was defined as the volume of the intersection of the envelope of fx with the segmented cavity.
Results
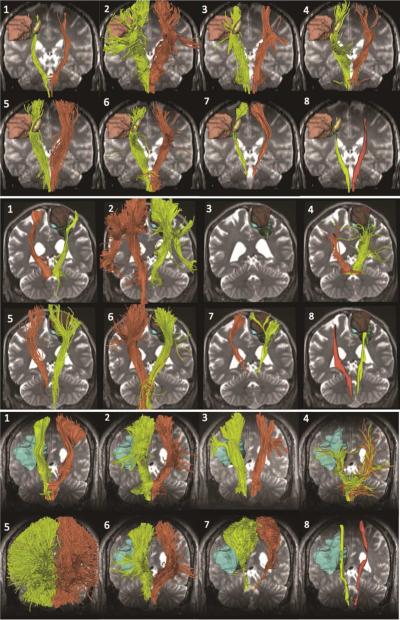
All tractography teams processed two neurosurgical cases prior to the workshop and seven teams processed two additional cases during the 5-hour on-site portion of the challenge. One team (team 3) failed to reconstruct the tracts on-site due to a hardware issue, and submitted their results for patient 4 after the challenge. The tractography results were visualized within 3D Slicer, which allowed for the comparison of tractography reconstructions in a non-ambiguous manner. Figure 2 shows the results obtained for patient 1. Figure 3 presents the analogous results for patients 2, 3, and 4.
Figure 3. Patient 2, 3 and 4: tractography results.
The figure shows the eight tractography reconstructions of the pyramidal tract for patient 2 (top), patient 3 (center), and patient 4 (bottom). The views include the following models: patient 2: tumor (green, light brown, yellow); patient 3: tumor (green), edema (blue) and contrast-enhanced surgical cavity (dark brown); patient 4: tumor (green) and edema (light blue).
Visual comparison of the tractography results showed large inter-algorithm variability in both tumor and unaffected hemisphere. Overall, the tractography results correctly identified the projection of the pyramidal tract from the foot motor area on both sides, but failed to include the fibers arising from the hand and face areas. In addition, most of the algorithms did not reconstruct any tract in edematous regions, which could represent false-negative tracts. Further problems included false-positive identification of frontal and parieto-occipital aspects of the corona radiata and reconstruction of tracts in the surgical cavity (Figure 4). The grades assigned by the judges ranged from A (excellent) to D (poor), and were averaged on the clinical criteria used for the review. The average grade for all clinical cases was B. We divided the teams into two groups: group 1 (teams 1, 3, 5, 6, and 7) achieved an overall score of B or higher; group 2 (teams 2, 4, and 8) achieved an overall score of B- or lower (Table 2). Overall, ratings of the tractography results showed a difference in performance between tractography algorithms that belong to the same fiber tracking family: for example, team 1 and team 2 (deterministic) belong to group 1 and group 2, respectively. Only the methods used by team 6 and team 7 (global fiber tracking) were both in the same group (group 1).
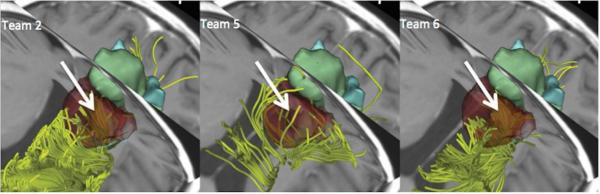
Figure 4. False-positive tractography reconstruction in a surgical cavity.
The figure shows a 3D superior view of the reconstruction of the pyramidal tract by team 2 (left), team 5 (center) and team 6 (right) in patient 3. The tracts (yellow) are presented overlaid on an axial and a coronal T1-weighted image, along with 3D surface models of the tumor (green) and edema (blue). The arrow points to the false-positive tractography reconstructions in the surgical cavity (dark brown).
Table 2.
Summary of qualitative review of the tractography results
| Team Id | Fiber Tracking Algorithm | Patient 1 | Patient 2 | Patient 3 | Patient 4 |
|---|---|---|---|---|---|
| Team1 | Deterministic | B+ | B+ | B− | B |
| Team 2 | Deterministic | B− | C+ | C+ | B− |
| Team 3 | Probabilistic | B− | B+ | N/A | N/A |
| Team 4 | Filtered | B− | B− | C | C+ |
| Team 5 | Filtered | A− | B+ | B− | B+ |
| Team 6 | Global | B+ | B+ | B | B |
| Team 7 | Global | B | B+ | B− | C+ |
| Team 8 | Global | C | C | D | D |
The table presents the average review score of the teams for each patient dataset (A: Excellent, B: Good, C: Fair, D: Poor), and the fiber tracking algorithm.
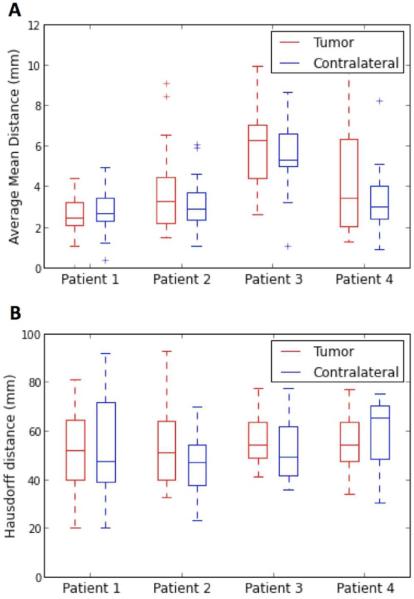
Figure 5 presents the graphs of the AMD and Hausdorff distance. The overlap of the boxes representing the tumor and contralateral side indicate that there is no significant difference across hemispheres, for both distances. The median AMD varied across patients between 2.4 mm and 6.3 mm on the tumor side, and between 2.7 mm and 5.3 mm on the contralateral side. In patients 2, 3, and 4, both the interquartile range and the distance between the two whiskers are larger in the tumor side than in the contralateral side, indicating a larger variability in AMD values on the pathological side. The median Hausdorff distance was relatively large, and varied across patients between 50.8 mm and 54.2 mm in the tumor side, and between 46.9 mm and 65.1 mm in the contralateral side. The interquartile range and distance between whiskers are comparable in the tumor and contralateral side, indicating that the presence of tumor or edema had little influence on the Hausdorff distance among the tracts reconstructed by the different methods.
Figure 5. Distances between tractography reconstructions.
The box plots represent summary statistics for the average mean distance and Hausdorff distance; each pair of box-and-whisker plots describes the distances between the eight tractography reconstructions of the pyramidal tract on the tumor and contralateral side, for a given patient dataset. The graphs show the median, interquartile range, minimum and maximum values of the distances. The shorter the distances, the closer the tracts reconstructed by the different teams should be. The overlap of the boxes representing the tumor and contralateral side indicates that there is no significant difference across hemispheres for both distances.
Table 3 presents the volumetric fraction of affected regions visited by the tracts. In patient 1 (anaplastic oligoastrocytoma grade III), all the tractography teams were in agreement with the absence of tracts in the tumor, which correlates with the cystic aspect of the lesion depicted on the T1-weighted image (Figure 1). In patient 2 (anaplastic oligoastrocytoma grade III) and patient 4 (glioblastoma grade IV), both the average and standard deviation of the fraction of tracts in the tumor regions were small (patient 2: 1.5±1.3%; patient 4: 0.5±0.9%), which is consistent with the type and grade of the tumor in the two cases. In patient 3 (anaplastic oligodendroglioma grade III), the volumetric fraction of the tumor visited by the tracts was higher, as expected from the infiltrative nature of the tumor and its location. While the tractography reconstructions are consistent with the type and location of the tumor, we observed a larger variability among results in the edematous regions. The mean and standard deviation of the volumetric fraction of edema visited by the tracts, which correspond to the percentage of edematous region that has tracts in it, was 5.3±5.3%, 5.6±6.8% and 7.1±7.9% in patients 1, 3, and 4, respectively. Finally, teams 2, 5, and 6 found false-positive tracts in the 13.3 cm3 fluid-filled cavity in patient 3.
Table 3.
Volumetric fraction of affected regions visited by tracts
| Patient | Patient 1 | Patient 2 | Patient 3 | Patient 4 | ||||
|---|---|---|---|---|---|---|---|---|
| f (%) | ft | fe | ft | ft | fe | fc | ft | fe |
| Team 1 | 0 | 2.9 | 1 | 22.8 | 17.3 | 0 | 0.02 | 12.7 |
| Team 2 | 0 | 17 | 4 | 12.3 | 1.5 | 21.1 | 0 | 5.4 |
| Team 3 | 0 | 3.5 | 2.2 | n/a | n/a | n/a | 0 | 0.01 |
| Team 4 | 0 | 1.9 | 0.9 | 1.8 | 0.1 | 0 | 0.2 | 0.8 |
| Team 5 | 0.06 | 2.7 | 1.4 | 3.1 | 4 | 8.2 | 2.8 | 20.4 |
| Team 6 | 0 | 8.4 | 2.4 | 18.3 | 3.1 | 23.0 | 0.9 | 15.4 |
| Team 7 | 0 | 5.6 | 0.01 | 10.5 | 13.4 | 0 | 0.2 | 2.6 |
| Team 8 | 0 | 0.5 | 0 | 0.02 | 0 | 0 | 0 | 0 |
| Mean | 0.007 | 5.3 | 1.5 | 9.8 | 5.6 | 7.5 | 0.5 | 7.1 |
| Max | 0.06 | 17 | 4 | 22.8 | 17.3 | 23.0 | 2.8 | 20.4 |
| Std.Dev | 0.02 | 5.3 | 1.3 | 8.7 | 6.8 | 10.3 | 0.9 | 7.9 |
The table shows the volumetric fraction of tumor (ft), edema (fe), and cavity (fc) regions visited by the tracts. Mean, maximum, and standard deviation are presented for each patient. Results show that the volumetric fractions are consistent with the type of tumor.
Discussion
DTI tractography is a research tool that holds promise for guiding to the location of critical white matter pathways during surgical planning of brain tumor resection. The technology can play a key role in the definition of resection boundaries by providing visual information on the presence of fibers near or within a lesion. However, DTI tractography suffers from a lack of standardization and the validity of tractography findings for neurosurgical decision-making needs to be fully established. Our study was designed to address this need using a novel approach based on the standardized comparison of tractography methods on patient data. Eight international teams reconstructed the pyramidal tract from a set of four neurosurgical datasets that we made available to the clinical and scientific community. A panel of experts reviewed the tractography reconstructions and the teams and jury members discussed the results at the first MICCAI DTI tractography challenge workshop. We reported the qualitative evaluation of tractography reconstructions by experts and the quantitative assessment of the agreement between methods.
The main findings of our study are: (1) that a large variability exists in the pyramidal tract reconstructed by different tractography algorithms in the clinical datasets provided; (2) most algorithms find projections of the pyramidal tract from the medial portion of the motor strip only, and few algorithms are able to trace the lateral projections from the hand, face and tongue area; and, (3) that the presence of tumoral tissues or edema has little influence on the pattern of disagreement among methods.
The tractography methods of this study illustrate the diversity of strategies for tracing white matter pathways proposed by leading academic teams. These methods include deterministic (teams 1 and 2), probabilistic (team 3), filtered (teams 4 and 5) and global (teams 6, 7, and 8) approaches. Because of that diversity, we chose not to provide seeding ROIs as, in general, such regions are a specific feature of a tractography tool and cannot be generalized to all approaches. The eight different methods present complementary strengths. For example, the multi-compartment model of team 2 and the global tractography approach of team 6 lead to the detection of some of the tracts emanating from the hand, face and tongue area in all neurosurgical cases. The deterministic streamline, filtered multi-tensor and energy-based global tractography approaches proposed by teams 1, 4, 7, and 8, respectively performed well by not producing false-positive tracts in the surgical cavity in patient 3. Finally, the boxplot representation of AMD and Hausdorff distance demonstrated that the structure of disagreement among methods was similar across the hemispheres despite the presence of alterations of neural architectures created by tumoral cells or edema.
This study has several limitations. First, the metrics employed to evaluate the variability of the results did not always capture the specificity of tractography data. We used voxelized tracts to calculate the volumetric fraction of affected regions visited by the fibers. There are several drawbacks in converting streamlines into a voxel grid, such as the influence of partial volume effects. Second, we did not take into account the observer variability. Overall, all judges agreed that most of algorithms failed to reconstruct the lateral projections of the pyramidal tract, and that tracts in the surgical cavity were clinically impossible. However, there were some discrepancies in the judging that did not appear in the overall grade as we averaged the ratings by all judges. This preliminary study was a first attempt to provide a standardized evaluation of tractography results by a team of experts. We plan to refine our review criteria and qualitative evaluation process by designing a survey questionnaire that will capture, in finer detail, the review by experts. In addition, we will include a test-retest study of the review of the pyramidal tracts by each judge, to evaluate intra-rater consistency. Finally, there was the absence of ground truth to which the tractography results could be compared for validation. While this issue affects the whole field of diffusion MR imaging, in general, our approach was to use the neuroanatomical knowledge of practicing neurosurgeons and DTI experts as a model of ground truth. We plan to add functional MRI to our study data to provide complementary information on the location of critical white matter pathways.
Conclusion
This report presents the results of an empirical research study on the evaluation of DTI tractography algorithms on a common set of clinical data from neurosurgical patients in the absence of ground truth. Our approach is based on the hypothesis that since all the different tractography methods aim at reconstructing the same anatomy, they should lead to the same results. Therefore, we focused our effort on collecting tractography data on neurosurgical cases and measuring a set of quantitative metrics and clinical criteria to evaluate them. Our study demonstrates considerable variability among different approaches despite the choice of a commonly known structure with a clear anatomical definition and an image processing challenge bringing together the most advanced algorithms. The difficulties in fully reconstructing the pyramidal pathway indicate that the potential to reconstruct less well-defined tracts could be even more challenging. Tracts associated with other critical functions such as the arcuate fasciculus and the optic radiation present additional difficulties due to the individual variability in the location of the language circuit as well as in the extent of the Meyer's loop.
It was not our goal to find out which method is the best, but to quantify the variability among tractography methods in the context of neurosurgical planning. This study is intended to serve as a first step towards the standardized evaluation of DTI tractography algorithms and, as a community-building effort, to address the highly multidisciplinary aspect of validation of DTI findings for clinical use. By bringing together a group of leading research teams in diffusion MRI who accepted run their tractography algorithms on a common set of clinical data, we were able to provide neurosurgeons an overview of the tractography results produced by advanced methods available in the field. By providing a review of the results by experts and a quantitative comparison of different methods, we provided algorithm developers with clues on the advantages and weaknesses of their individual approach.
This collaborative effort is intended to facilitate the exchange of knowledge and expertise among the scientists developing tractography algorithms tools and the neurosurgeons using tractography applications in the clinic. DTI tractography is a clinical research tool, and neurosurgeons and neuroradiologists should take into account the uncertainty associated with tractography reconstructions as different tractography algorithms give different results that can actually affect clinical outcomes. Future work includes the definition of ground truth model and a set of recommendations for the use of DTI tractography for neurosurgical decision-making.
Acknowledgments
This work was partially funded by the National Alliance for Medical Image Computing (NIH Grant U54EB005149), the Neuroimage Analysis Center (NIH Grant P41RR013218), and the National Center for Image-Guided Therapy (NIH Grant NCIGT P41EB015898). The authors would like to thank Nina Geller for providing editorial assistance with the manuscript.
Footnotes
Disclosure
All authors declare no financial conflicts-of-interest.
References
- 1.Le Bihan D, Breton E, Lallemand D, Grenier P, Cabanis E, Laval-Jeantet M. MR imaging of intravoxel incoherent motions: application to diffusion and perfusion in neurologic disorders. Radiology. 1986;161(2):401–7. doi: 10.1148/radiology.161.2.3763909. [DOI] [PubMed] [Google Scholar]
- 2.Basser PJ, Mattiello J, LeBihan D. MR diffusion tensor spectroscopy and imaging. Biophys J. 1994;66(1):259–67. doi: 10.1016/S0006-3495(94)80775-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pierpaoli C, Jezzard P, Basser PJ, Barnett A, Di Chiro G. Diffusion tensor MR imaging of the human brain. Radiology. 1996;201(3):637–48. doi: 10.1148/radiology.201.3.8939209. [DOI] [PubMed] [Google Scholar]
- 4.Behrens TEJ, Sotiropoulos SN, Jbabdi S. MR diffusion tractography. In: Johansen-Berg H, Berens TEJ, editors. Diffusion MRI: From quantitative measurements to in vivo neuroanatomy. Academic Press; Waltham, Ma: 2013. pp. 429–51. [Google Scholar]
- 5.Stieltjes B, Kaufmann WE, van Zijl PC, et al. Diffusion tensor imaging and axonal tracking in the human brainstem. Neuroimage. 2001;14(3):723–35. doi: 10.1006/nimg.2001.0861. [DOI] [PubMed] [Google Scholar]
- 6.Potgieser ARE, Wagemakers M, van Hulzen ALJ, de Jong BM, Hoving EW, Groen RJM. The role of diffusion tensor imaging in brain tumor surgery: a review of the literature. Clin Neurol Neurosurg. 2014;124:51–8. doi: 10.1016/j.clineuro.2014.06.009. [DOI] [PubMed] [Google Scholar]
- 7.McGirt MJ, Mukherjee DM, Chaichana KL, Than KD, Weingart JD, et al. Association of surgically acquired motor and language deficits on overall survival after resection of glioblastoma multiforme. Neurosurgery. 2009;65:463–70. doi: 10.1227/01.NEU.0000349763.42238.E9. [DOI] [PubMed] [Google Scholar]
- 8.Golby AJ, Kindlmann G, Norton I, Yarmarkovich A, Pieper S, Kikinis R. Interactive diffusion tensor tractography visualization for neurosurgical planning. Neurosurgery. 2011;68(2):496–505. doi: 10.1227/NEU.0b013e3182061ebb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coenen VA, Krings T, Mayfrank L, et al. Three-dimensional visualization of the pyramidal tract in a neuronavigation system during brain tumor surgery: first experiences and technical note. Neurosurgery. 2001;49(1):86–92. doi: 10.1097/00006123-200107000-00013. [DOI] [PubMed] [Google Scholar]
- 10.Nimsky C, Ganslandt O, Fahlbusch R. Implementation of fiber tract navigation. Neurosurgery. 2006;58:292–303. doi: 10.1227/01.NEU.0000204726.00088.6D. [DOI] [PubMed] [Google Scholar]
- 11.Jones DK, Knösche TR, Turner R. White matter integrity, fiber count, and other fallacies: the do's and don'ts of diffusion MRI. Neuroimage. 2013;73:239–54. doi: 10.1016/j.neuroimage.2012.06.081. [DOI] [PubMed] [Google Scholar]
- 12.Duffau H. The dangers of magnetic resonance imaging diffusion tensor tractography in brain surgery. World Neurosurg. 2014;81(1):56–8. doi: 10.1016/j.wneu.2013.01.116. [DOI] [PubMed] [Google Scholar]
- 13.Diepenbrock S, Prassni JS, Lindemann F, Bothe HW, Ropinski T. 2010 IEEE visualization contest winner: interactive planning for brain tumor resections. IEEE Comput Graph Appl. 2011;31(5):6–13. doi: 10.1109/mcg.2011.70. [DOI] [PubMed] [Google Scholar]
- 14.Pieper S, Halle M, Kikinis R. 3D Slicer.. Presented at the 2nd IEEE International Symposium on Biomedical Imaging: From Nano to Macro; Arlington, VA, USA.. April 15-18, 2004. [Google Scholar]
- 15.Magnotta VA, Harris G, Andreasen NC, O'Leary DS, Yuh WTC, Heckel D. Structural MR image processing using the brains2 toolbox. Comput Med Imaging Graph. 2002;26(4):251–264. doi: 10.1016/s0895-6111(02)00011-3. [DOI] [PubMed] [Google Scholar]
- 16.Proceedings of the first MICCAI DTI Challenge on Tractography for Neurosurgical Planning. Available at: http://www.namic.org/Wiki/images/3/35/DTIChallenge2011_Proceedings.pdf (accessed 23 June 2015)
- 17.Basser PJ, Pajevic S, Pierpaoli C, Duda J, Aldroubi A. In vivo fiber tractography using DT-MRI data. Magn Reson Med. 2000;44:625–32. doi: 10.1002/1522-2594(200010)44:4<625::aid-mrm17>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 18.Behrens TEJ, Johansen-Berg H, Woolrich MW, et al. Non-invasive mapping of connections between human thalamus and cortex using diffusion imaging. Nat Neurosci. 2003;6(7):750–7. doi: 10.1038/nn1075. [DOI] [PubMed] [Google Scholar]
- 19.Kalman RE. A new approach to linear filtering and prediction problems. Transactions of the ASME. Journal of Basic Engineering. 1960;82(D):35–45. [Google Scholar]
- 20.Mangin JF, Fillard P, Cointepas Y, Le Bihan D, Frouin V, Poupon C. Toward global tractography. Neuroimage. 2013;80:290–296. doi: 10.1016/j.neuroimage.2013.04.009. [DOI] [PubMed] [Google Scholar]
- 21.Goodlett B, Fletcher T, Gilmore J, Gerig G. Group Analysis of DTI fiber tract statistics with application to neurodevelopment. Neuroimage. 2009;45(1 Suppl):S133–S142. doi: 10.1016/j.neuroimage.2008.10.060. [DOI] [PMC free article] [PubMed] [Google Scholar]