Abstract
Background
Division of double-membraned plastids (primary plastids) is performed by constriction of a ring-like division complex consisting of multiple plastid division proteins. Consistent with the endosymbiotic origin of primary plastids, some of the plastid division proteins are descended from cyanobacterial cell division machinery, and the others are of host origin. In several algal lineages, complex plastids, the “secondary plastids”, have been acquired by the endosymbiotic uptake of primary plastid-bearing algae, and are surrounded by three or four membranes. Although homologous genes for primary plastid division proteins have been found in genome sequences of secondary plastid-bearing organisms, little is known about the function of these proteins or the mechanism of secondary plastid division.
Results
To gain insight into the mechanism of secondary plastid division, we characterized two plastid division proteins, FtsZD-1 and FtsZD-2, in chlorarachniophyte algae. FtsZ homologs were encoded by the nuclear genomes and carried an N-terminal plastid targeting signal. Immunoelectron microscopy revealed that both FtsZD-1 and FtsZD-2 formed a ring-like structure at the midpoint of bilobate plastids with a projecting pyrenoid in Bigelowiella natans. The ring was always associated with a shallow plate-like invagination of the two innermost plastid membranes. Furthermore, gene expression analysis confirmed that transcripts of ftsZD genes were periodically increased soon after cell division during the B. natans cell cycle, which is not consistent with the timing of plastid division.
Conclusions
Our findings suggest that chlorarachniophyte FtsZD proteins are involved in partial constriction of the inner pair of plastid membranes, but not in the whole process of plastid division. It is uncertain how the outer pair of plastid membranes is constricted, and as-yet-unknown mechanism is required for the secondary plastid division in chlorarachniophytes.
Electronic supplementary material
The online version of this article (doi:10.1186/s12870-015-0662-7) contains supplementary material, which is available to authorized users.
Keywords: Chlorarachniophytes, Endosymbiosis, FtsZ, Nucleomorph, Plastid division
Background
Plastids are endosymbiotically derived organelles, and their evolutionary histories are complicated due to multiple endosymbiotic events. Plants and three algal groups (green algae, red algae, and glaucophytes) acquired plastids through a single primary endosymbiosis between a heterotrophic protist and a photosynthetic cyanobacterium, and we refer to the plastids originating from this process as primary plastids [1, 2]. In contrast, many other algae (e.g., dinoflagellates, heterokonts, haptophytes, cryptophytes, euglenophytes, and chlorarachniophytes) and apicomplexan parasites possess complex plastids. These are called secondary plastids, which evolved through endosymbiotic uptakes of red and green algae by distinct protists [3, 4]. Multiple secondary endosymbioses led to the phylogenetic diversity of the current plastid-bearing organisms [5]. In terms of structures, primary and secondary plastids are distinct in the number of envelope membranes. Primary plastids are surrounded by two membranes, whereas secondary plastids have one or two additional membranes; euglenophyte and dinoflagellate plastids are bounded by three membranes, and the others have four membranes [6]. In four membrane-bound secondary plastids, the two innermost membranes are regarded as descendants of the primary plastid membranes of the engulfed alga, and the two outermost membranes are suggested to be the former phagosomal membrane of the host and the relict plasma membrane of the endosymbiont, respectively [6, 7]. The outermost membrane is connected to the endoplasmic reticulum (ER) in some secondary plastids of red algal origin, and the recent ER-enclosure model has proposed that the outer pair of membranes are derived from the host ER [8]. These additional membranes should be a barrier for transport of nucleus-encoded plastid proteins as well as for metabolite exchange. Thus, secondary plastid-bearing organisms had to evolve transport mechanisms for breaking through the membranes [9, 10]. Furthermore, in terms of plastid division, new mechanisms would be necessary for constriction of the additional membranes in secondary plastids.
Primary plastid division is performed by the simultaneous constriction of the two envelope membranes at the division site, which is mediated by a ring-like division complex encompassing both the inside and the outside of the two membranes [11, 12]. Consistent with the origin of primary plastids, protein components of the division complex on the stromal side are typically descended from cyanobacterial cell division proteins, and several host-derived proteins comprise the cytoplasmic portion [13]. In plants and algae, prior to the onset of plastid division, the tubulin-like GTPase FtsZ assembles into a ring-like structure on the stromal surface of the inner plastid membrane at the division site [14–18]. In the model plant Arabidopsis thaliana, the FtsZ ring (Z ring) is tethered to the inner plastid membrane via an interaction with the integral membrane protein ARC6 [19], and the positioning of the Z ring is regulated by several stromal proteins, MinD [20], MinE [21], ARC3 [22], and MCD1 [23]. Subsequently, the stromal plastid division proteins directly or indirectly recruit cytoplasmic components of the division complex, including the outer membrane proteins PDV1 and PDV2 [24], and the dynamin-related GTPase DRP5B (also called ARC5) that assembles into a ring-like structure on the cytoplasmic portion [25]. Two self-assembling GTPases, FtsZ and DRP5B, are thought to generate contractile force for membrane constriction [26, 27]. In earlier electron microscopic studies, electron-dense ring structures, called the plastid-dividing (PD) rings, were observed at the plastid division site on both the stromal and the cytoplasmic surface of the plastid membranes in plants and algae [28]. A recent study showed that the outer PD ring of the primitive red alga Cyanidioschyzon merolae is composed of polyglucan filaments [29], and filament sliding mediated by DRP5B (CmDnm2) may generate a contractile force for membrane constriction [30]. The principal feature of the plant-like plastid division mechanism seems to be widely conserved in primary plastid-bearing algae that have homologs of plastid division proteins [31]. However, some plastid division proteins are depleted in certain algal lineages [13, 32]. For example, the glaucophyte Cyanophora paradoxa genome does not encode any known plastid division proteins of host origin, and constriction of the outer membrane appears to rely on the peptidoglycan hydrolyzing activity with DipM, as in cyanobacterial cell division [33]. The genome of the red alga C. merolae encodes only two plant-like plastid division proteins, FtsZ and DRP5B, and it remains unknown how positioning of the plastid division complex at the division site is regulated.
Several homologous genes of primary plastid division proteins have also been identified in whole genomes of secondary plastid-bearing organisms [13]. As shown in Table 1 (updated from [13, 18]), homologous genes for FtsZ, MinD, MinE, and/or DRP5B, were found in the heterokonts Thalassiosira pseudonana, Phaeodactylum tricornutum, and Aureococcus anophagefferens, the haptophyte Emiliania huxleyi, and the cryptophyte Guillardia theta based on BLAST surveys. These homologous genes are assumed to be derived from the engulfed red algae via secondary endosymbioses. Plastid division proteins are mostly encoded by the nuclear genomes as the result of endosymbiotic gene transfer, whereas the MinD and MinE of G. theta and the MinD of E. huxleyi are encoded by the plastid genomes [34, 35], and G. theta ftsZ gene is found in the nucleomorph genome that is a relict nuclear genome of the red algal endosymbiont [36]. Localization studies on secondary plastid division proteins are limited to a few organisms. Green fluorescent protein (GFP)-tagged experiments confirmed that FtsZ proteins of the diatom Chaetoceros negogracile carried an N-terminal plastid targeting signal [37], and immunofluorescence localization revealed that the T. pseudonana DRP5B formed a ring-like structure at the putative plastid division site [11]. Thus, constriction of the inner pair of secondary plastid membranes appears to be achieved by a portion of the primary plastid division complex that is derived from the engulfed endosymbiont. Although it is uncertain how the outer pair of secondary plastid membranes are divided, a study of apicomplexan parasites has shed light on this problem. Apicomplexans have non-photosynthetic secondary plastids (called apicoplasts) that are surrounded by four membranes. van Dooren et al. (2009) reported that a dynamin-related protein of Toxoplasma gondii (TgDrpA) localized to punctate regions on the apicoplast surface and it was essential for apicoplast division [38]. The drpA is an apicomplexan-specific gene that is phylogenetically distinct from the drp5B genes of primary plastid-bearing organisms. Interestingly, apicomplexans lack any homologous genes of primary plastid division proteins including FtsZ and DRP5B, suggesting that apicomplexans have lost the primary plastid division machinery and developed a new mechanism mediated by DrpA [38]. To date, few proteins have been characterized as a portion of the mechanism of secondary plastid division, furthermore, the whole picture of secondary plastid division is still unrevealed.
Table 1.
Distribution of homologous genes for plastid division proteins
| organisms | FtsZ | ARC6 | MinC | MinD | MinE | DRP5B/ARC5 (DrpA)a | PDV, ARC3, MCD1 | PDR1 |
|---|---|---|---|---|---|---|---|---|
| Synechocystis PCC6803 | BAA17496 | BAA10060 | BAA10664 | BAA10662 | BAA10661 | N/A | N/A | N/A |
| Cyanophora paradoxa | BAD99307 | Contig37232 | N/A | BAM09147 | BAM09148 | N/A | N/A | N/A |
| Cyanidioschyzon merolae | BAC87807, | N/A | N/A | N/A | N/A | BAC55068 | N/A | CMR358C |
| BAC87808 | ||||||||
| Chlamydomonas reinhardtii | XP_001702420, | XP_001690917 | XP_001692722 | XP_001697031 | XP_001697195 | XP_001702662 | N/A | N/A |
| XP_001700508 | ||||||||
| Arabidopsis thaliana | At2g36250, | At5g42480 | N/A | At5g24020 | At1g69390 | At3g19720 | At5g53280 (PDV) | N/A |
| At3g52750, | At2g16070 (PDV) | |||||||
| At5g55280 | At1g75010 (ARC3) | |||||||
| At1g20830 (MCD1) | ||||||||
| Thalassiosira pseudonana | XP_002296337, | N/A | N/A | XP_002291334 | N/A | XP_002290986 | N/A | N/A |
| XP_002294718, | ||||||||
| XP_002293505 | ||||||||
| Phaeodactylum tricornutum | XP_002182929, | N/A | N/A | N/A | N/A | XP_002181459 | N/A | N/A |
| XP_002182240, | ||||||||
| XP_002185124 | ||||||||
| Aureococcus anophagefferens | XP_009032511, | N/A | N/A | N/A | N/A | XP_009032466 | N/A | N/A |
| XP_009038601 | ||||||||
| Guillardia theta | XP_001713182Nm | N/A | N/A | NP_050687P | NP_050686P | N/A | N/A | N/A |
| Emiliania huxleyi | EOD19496, | N/A | N/A | YP_277381P | EOD04187 | EOD14983, | N/A | N/A |
| EOD06732 | EOD23222 | |||||||
| Toxoplasma gondii | N/A | N/A | N/A | N/A | N/A | (FJ264918)a | N/A | N/A |
| Bigelowiella natnas | JGI39262, | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
| JGI92991 |
The presence/absence of genes are represented by accession numbers of the NCBI GenBank, JGI, or the Cyanophora paradoxa Genome Project. N/A represents ‘not applicable’. The data is updated from [13, 18]
Nmnucleomorph-genome encoded
Pplastid-genome encoded
aapicomplexan-specific dynamin-related protein
To gain insight into secondary plastid division mechanisms, we investigated homologs of primary plastid division proteins in chlorarachniophyte algae that obtained secondary plastids by the uptake of a green alga. Chlorarachniophyte plastids are surrounded by four membranes. Each plastid possesses a relict nucleus (called nucleomorph) of the endosymbiont in the periplastidal compartment between the inner and the outer pair of membranes, and the existence of the nucleomorph would make plastid division more complicated [39]. Genome sequencing has been completed in the chlorarachniophyte Bigelowiella natans, including the nuclear genome [40], nucleomorph [41], and plastid [42]. A previous proteomic study identified two nucleus-encoded FtsZ homologs in isolated B. natans plastids [43]. In this study, we report detailed subcellular localization and periodic gene expression pattern for two FtsZ proteins, named FtsZD-1 and FtsZD-2, in the chlorarachniophyte B. natans. Our findings suggest that the FtsZD proteins are involved in partial constriction of the two innermost plastid membranes, but not in the whole process of plastid division. It is uncertain how the outer pair of plastid membranes is constricted, and as-yet-unknown components are required for secondary plastid division in chlorarachniophytes.
Results
Sequences of plastid division protein FtsZD in chlorarachniophytes
To identify homologous genes of primary plastid division proteins in chlorarachniophytes, we surveyed B. natans genomes by reciprocal BLAST with corresponding known sequences of Arabidopsis thaliana, Chlamydomonas reinhardtii, and Cyanidioschyzon merolae [12, 13]. Two homologous genes for FtsZ proteins were found in the B. natans nuclear genome as reported previously [43], but the others for ARC6, MinC, MinD, MinE, DRP5B, PDV, ARC3, MCD1, and PDR1 were not detected (Table 1). Two types of ftsZ genes, named ftsZD-1 and ftsZD-2, were also identified in RNAseq transcriptome data from four chlorarachniophytes, Amorphochlora amoebiformis, Chlorarachnion reptans, Gymnochlora sp. (CCMP2014), and Lotharella globosa. All of these FtsZD proteins were composed of an N-terminal leader, a highly conserved FtsZ domain, and a variable C-terminal region (Fig. 1a). The N-terminal leaders were predicted to be a typical plastid targeting signal consisting of an ER targeting signal peptide and a transit peptide-like sequence (Fig. 1a) by SignalP 3.0 [44] and ChloroP 1.1 [45] programs. To confirm the plastid targeting ability of the N-terminal leaders, we performed GFP-based experiments using the transient transformation system of A. amoebiformis. GFP fusion proteins with an N-terminal leader of A. amoebiformis FtsZD proteins (AaFtsZD1-N90 + GFP and AaFtsZD2-N92 + GFP) were imported into the plastid stroma (Fig. 1b, c). The C-terminal regions of FtsZD proteins were divergent in sequence and length, whereas their C-terminal ends were mostly conserved among FtsZD-1 and FtsZD-2 proteins; the C-terminal conserved motif of FtsZD-1 is ‘T-L-R/G-G-K-A-K-R-x-A-G-x-x-L-R-R/K-A/V-L-G’ and that of FtsZD-2 is ‘G-L-G-G-W-W-Y-W-W-R-N’ (Fig. 1a).
Fig. 1.
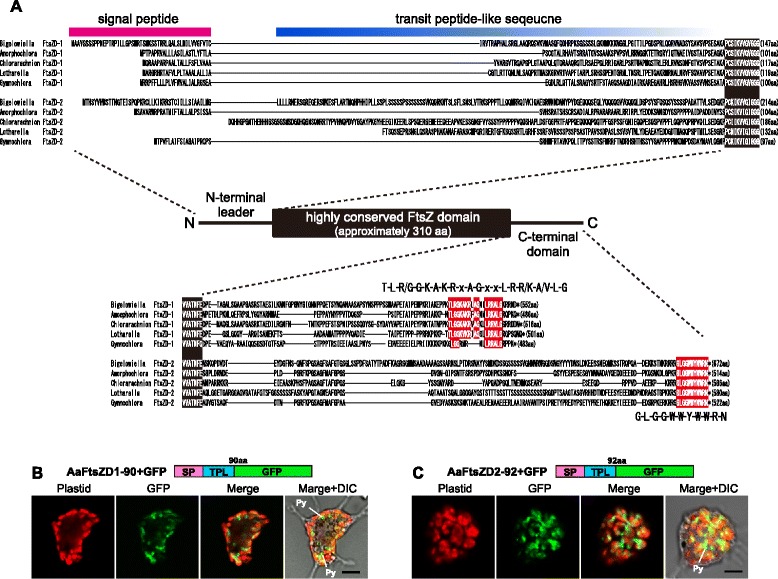
Alignment of N- and C-terminal sequences of chlorarachniophyte FtsZD proteins. a The N-terminal leaders are predicted to consist of a signal peptide and a transit peptide-like sequence. Identical amino acids within the highly conserved FtsZ domains are boxed in black, and the C-terminal conserved motifs are highlighted with red boxes. The FtsZD-2 sequences of Chlorarachnion and Lotharella lack their N-terminal ends. (b, c) Confocal images of Amorphochlora amoebiformis cells expressing GFP fused with the N-terminal leader of AaFtsZD-1 and AaFtsZD-2. The images show red chlorophyll autofluorescence of plastids and green fluorescence of GFP. The pyrenoid (Py) is a part of plastid stroma that has no chlorophyll autofluorescence. The scale bars represent 5 μm
Two previous phylogenetic studies reported that chlorarachniophyte ftsZ genes appear to be derived from a red algal lineage despite the green algal origin of chlorarachniophyte plastids [43, 46]. To examine the evolutionary process of the two types of ftsZ genes in chlorarachniophytes, we reconstructed a phylogenetic tree using prokaryotic and eukaryotic FtsZ sequences with eight chlorarachniophyte FtsZ sequences. The tree indicated the monophyly of chlorarachniophyte ftsZ genes, which were closely related to a sequence of the haptophyte Emiliania huxleyi bearing a red algal secondary plastid, and the ftsZD-1 and ftsZD-2 genes are robustly separated into two clades (Fig. 2). This result strongly suggested that the duplication of ftsZD-1 and ftsZD-2 occurred before the divergence of chlorarachniophyte species, whereas the red origin of chlorarachniophyte ftsZD genes was not well supported by our analyses.
Fig. 2.
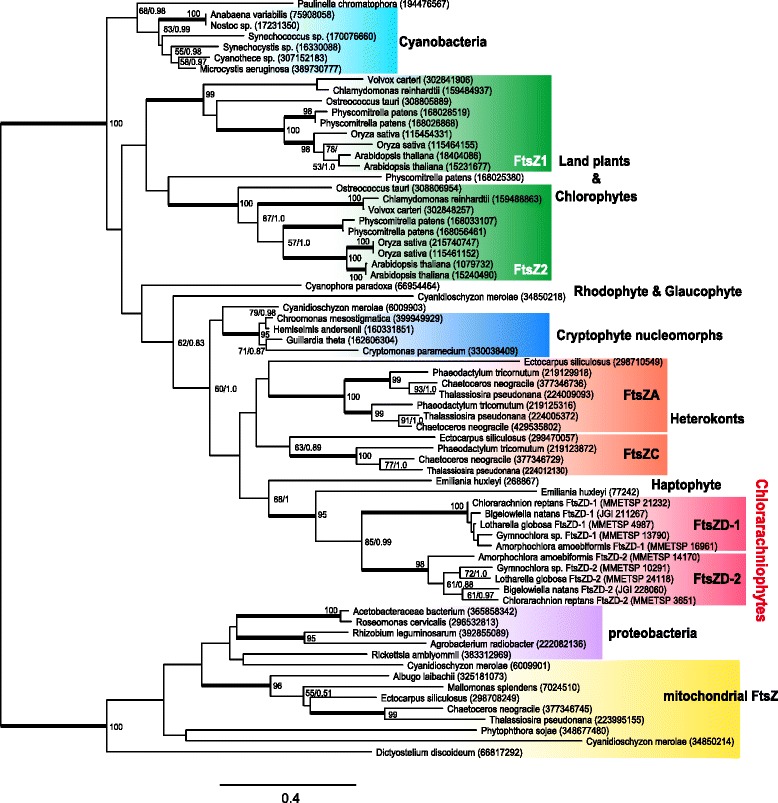
Unrooted phylogenetic tree representing the diversity of eukaryotic and prokaryotic FtsZ. The topology correspond to the best-scoring ML tree as obtained with RAxML. The values at nodes indicate the ML bootstrap supports (BP) and Bayesian posterior probabilities (PP) when they are higher than 50 % and 0.8, respectively. Bold lines correspond to ≥95 % BP and 1.0 PP. GenBank/JGI/MMETSP accession numbers are shown on the right side of species names. The scale bar represents the estimated number of amino acid substitutions per site
Subcellular localization of chlorarachniophyte FtsZD proteins
To determine the detailed subcellular localization of chlorarachniophyte FtsZD proteins, we first examined the full-length of AaFtsZD-1 (486 amino acids) and AaFtsZD-2 (514 amino acids) fused with GFP. Unfortunately their localization was ambiguous and we observed no obvious accumulation of GFP at the plastid division site, probably due to an artifact of high expression of the GFP tagged proteins. Therefore, we generated polyclonal antibodies against B. natans FtsZD proteins (anti-BnFtsZD-1 and anti-BnFtsZD-2) to perform immunolocalization experiments. B. natans typically has a single plastid in the tiny cell, which has an advantage in ultrastructural studies by electron microscopy. The B. natans plastids typically possess a projecting pyrenoid with a plate-like invagination of the two innermost membranes, and a nucleomorph is located near the pyrenoid base [47]. The specificity of two antibodies was examined by immunoblot analyses against the whole proteins extracted from B. natans cells; the detected bands were consistent with the predicted size of mature proteins of BnFtsZD-1 (44 kDa) and BnFtsZD-2 (50 kDa) (Fig. 3a, b). We first performed immunofluorescence microscopy. Fluorescein isothiocyanate (FITC) signals were observed at the midpoint of the bilobate plastids in both of the anti-BnFtsZD-1 and anti-BnFtsZD-2 (Fig. 3c, d), and cytoplasmic signals would be a nonspecific background, because such signals were also observed in negative control cells treated with pre-immune serums (Fig. 3c, d). Furthermore, we carried out immunoelectron microscopy using these two antibodies. In both cases, conjugated gold particles were observed near the tip of the shallow invagination of the two innermost plastid membranes within projecting pyrenoids (Fig. 4a, e, f), on opposite sides of the pyrenoid on horizontal sections (Fig. 4b, g), on the innermost membrane in the central narrow part of the bilobate plastids near the nucleomorph (Fig. 4c, h), and in a vertical line on the bottom of the bilobate plastids (Fig. 4d, i). Immunogold signals for both BnFtsZD proteins were almost always detected at the plastid midpoints in observed cells. Taken together, these findings indicated that BnFtsZD-1 and BnFtsZD-2 assembled into a gourd-shaped ring structure on the stromal side of the innermost membrane at the midpoint of bilobate plastids, and the ring always associates with a shallow plate-like invagination of the inner pair of plastid membranes (Fig. 4j). In our electron microscopic observation, electron-dense PD-rings were not detected at the putative plastid division sites where immunogold particles were located.
Fig. 3.
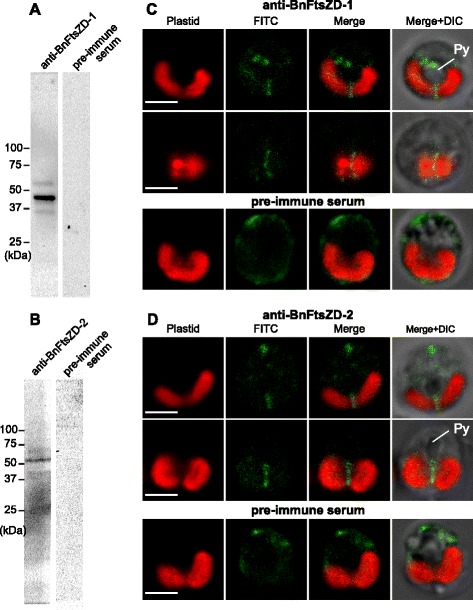
Immunoblot analyses and immunofluorescence microscopy of BnFtsZD-1 and BnFtsZD-2 proteins. Immunoblot analyses of anti-BnFtsZD-1 (a) and anti-BnFtsZD-2 (b) antibodies against the whole proteins extracted from B. natans cells. (c, d) Confocal images of immunofluorescence labeling of BnFtsZD-1 and BnFtsZD-2 with FITC (green) in B. natans cells. Pre-immune serums were used as negative controls. The chlorophyll autofluorescence of plastids is shown by red. Py, pyrenoid. The scale bars represent 2 μm
Fig. 4.
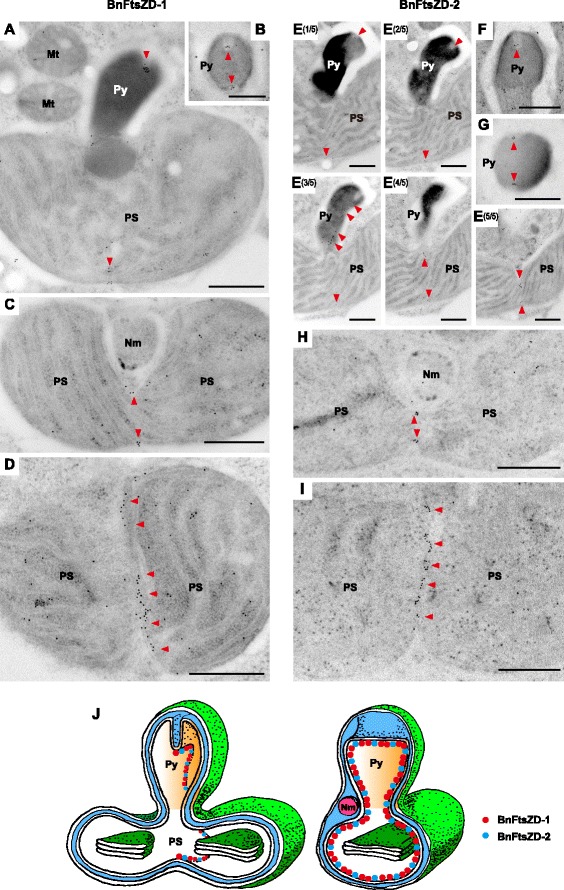
Immunoelectron micrographs of B. natans plastids showing localization of BnFtsZD proteins. Images present immunogold localization of BnFtsZD-1 (a to d) and BnFtsZD-2 (e to i). Conjugated gold particles (10 nm) are highlighted with red arrowheads. (a) Longitudinal section of plastid stroma (PS) with a projecting pyrenoid (Py). (b, g) Transverse sections of pyrenoids. (c, h) Mid-sections of plastid stroma with a nucleomorph (Nm). (d, i) Transverse sections at the bottom of plastid stroma. (e) Serial longitudinal sections of a plastid. (f) Longitudinal section of a pyrenoid. The scale bars represent 1 μm. (j) Schematic illustration of BnFtsZD localization in the plastid; red and blue circles correspond to BnFtsZD-1 and BnFtsZD-2, respectively
Transcription pattern of ftsZD genes during the cell cycle
In unicellular algae with a single plastid, plastid division is generally regulated by the host cell cycle, and transcription of nucleus-encoded plastid division proteins is generally restricted to the S, G2, or M phase, when plastids divide prior to cytokinesis. Transcripts of plastidal ftsZ genes are known to accumulate during the S phase of the red alga C. merolae [48] and a couple of green algae [18], and during the S/G2 phase of the secondary plastid-bearing diatom Seminavis robusta [49]. To clarify the relationship between the timing of ftsZ gene expression and host cell division in chlorarachniophytes, we examined the transcription levels of BnftsZD genes during the cell cycle using synchronized B. natans culture. Cell division was synchronized by pretreatment of continuous light deprivation for 36 h followed by a 12:12 h light:dark cycle; cell division occurred during the second dark phase (Fig. 5a). Total RNA was extracted at 4 h intervals during the second light and dark, and third light phase, and we calculated mRNA transcription levels of BnftsZD-1 and BnftsZD-2 using real-time quantitative PCR (RT-qPCR). Transcription levels of both genes increased soon after cell division in the late dark phase (Fig. 5b, c). The transcription level of BnftsZD-1 reached a peak at the end of the dark phase corresponding to the host M/G1 phase, whereas the peak of BnftsZD-2 was 4 h before that of BnftsZD-1. Both BnFtsZ genes appeared to be regulated by the host cell cycle. Unlike other unicellular algae, however, the transcription pattern of chlorarachniophyte ftsZD genes is not consistent with the timing of plastid division that occurs before the cytokinesis. When the mRNA transcription ratio between BnftsZD-1 and BnftsZD-2 was simply calculated by Ct values of RT-qPCR at their peaks (24 h and 20 h for BnftsZD-1 and 2, respectively), the maximum transcription level of BnftsZD-1 was 2.3-fold higher than that of BnftsZD-2. This suggests that two BnFtsZD proteins might exist in the plastids in different molecular amounts.
Fig. 5.
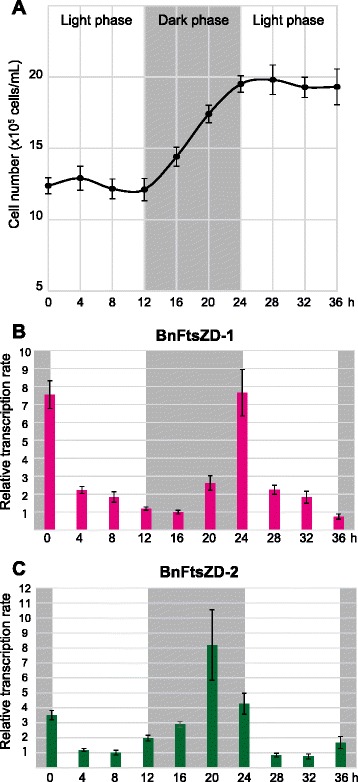
Transcription of BnftsZD genes during the cell cycle. a Cell densities (cells mL−1) in the synchronized B. natans culture determined by cell counting; error bars represent standard deviations calculated based on six independent measurements. (b, c) Relative transcription levels of BnftsZD-1 and BnftsZD-2 genes detected using real-time quantitative PCR for a total of 13 time points (4 h intervals for 36 h). Each transcription level was normalized against a transcription level of the reference gene 18S rRNA. Error bars represent standard deviations of triplicate experiments
Discussion
Prospective function of FtsZD proteins in chlorarachniophytes
One previous ultrastructural study of the chlorarachniophyte B. natans has shown that divisions of the plastid and the nucleomorph take place in an early stage of cell division, whereas partial bisection of the pyrenoid with an invagination of the two innermost membranes appears to start very soon after cell division [47]. Therefore, a shallow membrane invagination is almost always observed within the pyrenoids by electron microscopy. This infers that the partial membrane constriction would be achieved by a mechanism that is independent of plastid division. Our immunolocalization results indicate that the two FtsZD proteins of B. natans accumulate to form a gourd-shaped ring structure associated with the membrane invagination of projecting pyrenoids (Fig. 5j). This suggests that the FtsZD proteins are involved in the partial constriction of the inner pair of plastid membranes, which is supported by the finding that the expression patterns of BnftsZD genes are consistent with the timing of formation of the membrane invaginations. Interestingly, the membrane constriction somehow arrests on the way, and the FtsZD proteins are kept in a ring-like structure along the membrane invagination. However, it remains unknown how the membrane constriction is controlled and when the plastid stroma is completely divided.
The projecting pryrenoids of chlorarachniophyte plastids have a remarkable morphological diversity in membrane invaginations among genera, which is utilized for their taxonomy. The pyrenoids of Bigelowiella and Norrisiella have a shallow invagination of the two innermost plastid membrane (the depth is less than half the pyrenoid’s height) [50], whereas Lotharella, Amorphochlora, and Chlorarachnion pyrenoids possess a deep invagination that reaches the plastid stroma [51]. It is expected that variable degrees of Z ring constriction might cause the morphological diversity of chlorarachniophyte plastids. Unlike other chlorarachniophytes, Gymnochlora have many single-membraned tubular invaginations in the pyrenoids [52], implying that this genus might exhibit lineage-specific behavior of FtsZD localization.
Functional divergence of two FtsZ families
Although cyanobacteria possess a single gene for FtsZ protein, multiple types of plastidal FtsZ proteins have been reported in plants and algae; Viridiplantae (land plants and green algae) have two phylogenetically distinct types of FtsZ families named FtsZ1 and FtsZ2, red algae have FtsZA and FtsZB, and secondary plastid-bearing heterokonts possess FtsZA and FtsZC [53]. Two distinct plastidal FtsZ families emerged by multiple duplication events of an ftsZ gene originally derived from a cyanobacterium in each common ancestor of Viridiplantae, red algae, and heterokonts [53]. Green lineage FtsZ2 and red lineage FtsZA as well as cyanobacterial FtsZ contain a highly conserved sequence “D/E-I/V-P-x-F/Y-L” in their C-termini, the “C-terminal core domain”, whereas other FtsZ1, FtsZB, and FtsZC proteins lack this domain [53, 54]. This suggests that one of two FtsZ families have lost the C-terminal core domain after the duplication events. In the model plant A. thaliana, the C-terminal core domain of FtsZ2 mediates a specific interaction with the transmembrane protein ARC6 localized at the plastid division site to tether the Z ring to the inner plastid membrane [19]. We revealed that chlorarachniophytes also have two distinct FtsZ families, FtsZD-1 and FtsZD-2, that emerged by gene duplication in a common ancestor of chlorarachniophytes (Fig. 2). Both families carry chlorarachniophyte-specific C-terminal conserved motifs, T-L-R/G-G-K-A-K-R-x-A-G-x-x-L-R-R/K-A/V-L-G within FtsZD-1 and G-L-G-G-W-W-Y-W-W-R-N within FtsZD-2, which are different from the C-terminal core domain of FtsZ2 and FtsZA. This suggests that chlorarachniophyte FtsZD-1 and FtsZD-2 independently acquired the unique C-terminal motifs after gene duplication. Because of the absence of homologous gene for ARC6 in chlorarachniophyte genomes, the C-terminal conserved motifs are not involved in the ARC6-medicated membrane tethering, and the functional importance of the C-terminal conserved motifs remains uncertain.
Genetic complementation experiments demonstrated that Arabidopsis FtsZ1 and FtsZ2 were functionally non-redundant in plastid division [55]. Their functional divergence is partly explained by the facts that only FtsZ2 has a specific interaction with the transmembrane protein ARC6 [19] and turnover of the FtsZ1 filaments is faster than that of FtsZ2, suggesting that FtsZ1 facilitates Z ring remodeling [56]. Furthermore, it has been reported that Arabidopsis FtsZ1 and FtsZ2 maintain a constant 1:2 ratio in whole mature leaves [57], and that they are able to form heteropolymers at variable ratios in vitro [58]. In this study, we found that the two types of B. natans ftsZ genes, BnftsZD-1 and BnftsZD-2, showed slightly different transcription patterns in the cell cycle, and the maximum transcription level of BnftsZD-1 was predicted to be approximately 2-fold higher than that of BnftsZD-2. These findings lead us to infer that the two FtsZ families of chlorarachniophytes also exhibit functional divergence similar to Arabidopsis FtsZ families. The earlier transcription of BnftsZD-2 than BnftsZD-1 raise the possibility that BnFtsZD-2 plays a role in an early stage of Z ring assembly and positioning.
Evolution of the plastid division machinery in secondary plastids
In land plants, the Z ring assembly is restricted to the plastid midpoint by several division-site positioning factors, ARC3, MCD1, MinD, and MinE [54]. Homologous genes for cyanobacteria-derived Min proteins have also been found in a glaucophyte and green algae, as well as secondary plastid-bearing algae (Table 1). Therefore, a portion of the Z ring positioning mechanism appears to be widely conserved in both primary and secondary plastids. However, the chlorarachniophyte B. natans completely lacks all genes for the plant-like Z ring positioning factors, similar to the red alga C. melorae. Despite the absence of canonical positioning factors, FtsZ proteins are able to assemble at the plastid midpoint in both algae, suggesting that they have independently evolved a currently unknown mechanism for Z ring positioning.
The division complex of primary plastids generally consists of two different GTPase proteins, FtsZ and DRP5B, localizing to ring-like structures on the stromal side of the inner membrane and the cytoplasm side of the outer membrane, respectively. Homologous genes for FtsZ and DRP5B have been found in some secondary plastid-bearing algae, heterokonts and a haptophyte, and these proteins are assumed to function in the constriction of the inner pair of four plastid membranes [11]. However, no genes for DRP5B have been identified in genomes of B. natans and the cryptophyte Guillardia theta [32]. Previous studies have reported that drp5B (arc5) mutants of A. thaliana and the moss Physcomitrella patens exhibit defects in complete plastid constriction, resulting in enlarged, dumbbell-shaped plastids [25, 59, 60]. However, each cell of the drp5B mutants contains a small number of plastids, indicating that the plastids can still divide into daughter plastids without DRP5B. Therefore, it is possible that the two innermost membranes of secondary plastids are constricted by an FtsZ-dependent but DRP5B-independent mechanism in chlorarachniophytes and cryptophytes, like the plant drp5B mutants. Furthermore, the absence of DRP5B might be related to the partiality of membrane constriction in chlorarachniophyte plastids.
In some secondary plastids, constriction of the outer pair of plastid membranes takes place behind that of the inner pair [61]. Although it is not known how the outer pair of membranes divide, the constriction is performed by a mechanism that is distinct from the mechanism for the inner pair. In an apicomplexan parasite, the apicomplexan-specific dynamin-related protein DrpA presumably localizing on the surface of four membrane-bound apicoplasts is essential for apicoplast division [38]. It may be possible to infer that a lineage-specific dynamin-like protein is involved in constriction of the outer pair of plastid membranes in other secondary plastid-bearing organisms. The B. natans genome is found to encode two highly divergent dynamin-like proteins [GenBank ID: 113201831 and 113201829], and future studies on these proteins will provide further insights into our findings.
Conclusions
We characterized two plastid division proteins, named FtsZD-1 and FtsZD-2, in chlorarachniophyte algae possessing secondary plastids. These two distinct FtsZ families emerged by gene duplication in a common ancestor of chlorarachniophytes. Both FtsZD proteins assemble to a ring-like structure at the midpoint of bilobate plastids with a projecting pyrenoid. The ring was associated with a shallow plate-like invagination of the inner pair of plastid membranes. Transcription level of B. natans ftsZD genes was periodically increased soon after the cell division, which is consistent with the timing of the formation of the membrane invagination. Our findings suggest that chlorarachniophyte FtsZD proteins are involved in partial constriction of the inner pair of plastid membranes, but not in the whole process of plastid division. At present, it remains uncertain how the outer pair of plastid membranes is constricted in secondary plastid-bearing algae. We need further investigations to determine the whole system of secondary plastid division.
Methods
Chlorarachniophyte cultures
B. natans (CCMP2755) and A. amoebiformis (CCMP2058) cells were grown at 22 °C under white illumination (80–100 μmol photons m−2 s−1) on a 12:12 h light:dark cycle in ESM medium. Synchronization of B. natans cell division was achieved as previously described [62].
Homology searches of plastid division proteins
Genes for plastid division proteins (FtsZ, ARC6, MinC, MinD, MinE, DRP5B, PDV, ARC3, MCD1, and PDR1) were identified using whole genome data of land plants and several algae in previous studies [13, 18]. To update the information, homologous genes for plastid division proteins were searched in the whole genomes of secondary plastid-bearing algae (Thalassiosira pseudonana, Phaeodactylum tricornutum, Aureococcus anophagefferens, Emiliania huxleyi, Guillardia theta, Toxoplasma gondii, and Bigelowiella natans), based on reciprocal BLASTp with e-value cutoff of 1e−10 using corresponding known sequences of Arabidopsis thaliana, Chlamydomonas reinhardtii, and Cyanidioschyzon merolae. The genome sequences were obtained from the GenBank and the DOE Joint Genome Institute (JGI) site.
Phylogenetic analyses
Chlorarachniophyte ftsZ genes were identified from the genome sequence of B. natans [40] and Illumina-based transcriptomes of four chlorarachniophytes, A. amoebiformis, C. reptans, Gymnochlora sp., and L. globosa, using tBLASTn homology search with corresponding sequences of A. thaliana, C. reinhardtii, and C. merolae. The transcriptome analyses were performed by the Marine Microbial Eukaryote Transcriptome Sequencing Project [63], and the sequence data are available from the iMicrobe website (http://data.imicrobe.us/project/view/104) under sample IDs: MMETSP 0041, 0042, 0109, and 0110. Phylogenetic trees were constructed with 60 sequences of prokaryotic and eukaryotic FtsZ proteins retrieved from GenBank and 10 chlorarachniophyte sequences. The sequences automatically aligned by the L-INS-i method of the MAFFT package [64], and poorly aligned positions were manually inspected with MEGA6 [65]. Phylogenetic analyses were performed using Maximum-likelihood (ML) and Bayesian methods with the final dataset including 315 unambiguously aligned amino acid positions from 70 taxa. ML analyses were carried out using RAxML v8.1.15 [66] with the LG + GAMMA model selected as the best-fit model by Aminosan [67]. The best-scoring ML tree was determined in multiple searches using 10 distinct randomized maximum-parsimony trees, and statistical support was evaluated by 1,000 bootstrap replicates. Bayesian analyses were performed using MrBayes v3.1.2 under the LG + GAMMA model [68]. Metropolis-coupled Markov Chain Monte Carlo was run from a random starting tree for 1,000,000 generations, sampling every 1,000 cycles. Three heated and one cold chains were run simultaneously and the initial 380,000 cycles were discarded as burn-in. Node posterior probabilities were calculated from the remaining 620 trees.
Localization analyses of GFP fusion proteins
Total RNA was extracted from A. amoebiformis cells using TRIzol reagent (Invitrogen), and cDNA was synthesized using SuperScript II Reverse Transcriptase (Invitrogen) with an oligo (dT) primer. To construct plasmids encoding GFP fusion proteins, cDNA fragments encoding the N-terminal leaders of AaFtsZD-1 (90 amino acids) and AaFtsZD-2 (92 amino acids) were amplified by PCR with specific primer sets (see Additional file 1), and each fragment was inserted between HindIII and NcoI sites of pLaRGfp + mc vector [69] that carried the promoter of a rubisco small subunit gene. To analyze the subcellular localization of GFP fusion proteins, A. amoebiformis cells were transformed with each plasmid using a Biolistic PDS-1000/He particle delivery system (Bio-Rad) as described previously [70]. Twenty-four to 48 h after transformation, the transient transformed cells expressing reporter genes were observed under an inverted Zeiss LSM 510 laser scanning microscope (Carl Zeiss) with the single-track mode.
Preparation of antibodies
Polyclonal antibodies against B. natans FtsZD proteins were raised in rabbits using the respective recombinant proteins corresponding to their mature proteins of 210 amino acids (from 256 to 444 residue of BnFtsZD-1 or from 326 to 514 residue of BnFtsZD-2) by BioGate Co., Ltd (Gifu, Japan). Amplified cDNA fragments encoding these BnFtsZD mature proteins were cloned into pET28 expression vectors, and 6xHis fusion polypeptides were expressed in Rosetta 2 (DE3) Escherichia coli cells (Novagen), according to the manufacture’s instruction. The recombinant proteins were purified with His GraviTrap columns (GE Healthcare) under a denaturing condition with 8 M urea, followed by an overnight dialysis against phosphate-buffered saline (PBS). Polyclonal rabbit immunoglobulin G (IgG) antibodies were purified using HiTrap Protein A HP columns (GE Healthcare). The specificity of antibodies was tested by immunoblot analyses with the total proteins extracted from B. natans cells. The proteins were electrophoresed on a 12 % SDS polyacrylamide gel, and blotted to a PVDF membrane using a Trans-Blot Turbo Transfer System (Bio-Rad). Immunoblotting was performed by an iBind Western system (Life Technologies) with each of anti-BnFtsZD antibodies/pre-immune serums diluted 1:500, followed by horseradish peroxidase (HRP)-conjugated secondary antibodies at a dilution of 1:10,000. The signals were detected with ECL Prime Western Blotting Detection Reagent (GE Healthcare) and a ChemiDoc MP System (Bio-Rad).
Immunofluorescence and Immunoelectron microscopy
For immunofluorescence microscopy, B. natans cells were fixed for 1 h at room temperature in 4 % paraformaldehyde/0.25 M sucrose in PHEM buffer (60 mM PIPES, 25 mM HEPES, 10 mM EGTA, 2 mM MgCl2; pH 7.4). Fixed cells were transferred onto poly-L-lysine coated coverslips, and rinsed with PBS. Permeabilization of cell membranes was performed with 5 % tween 20 in PBS for 10 min; the detergent Triton X-100 was not useful because the chlorophyll autofluorescence was removed even in a low concentration of 0.01 %. After blocking with 3 % skim milk/1 % normal goat serum in PBS for 30 min, the cells were treated for 2 h at room temperature with respective anti-BnFtsZD antibodies/pre-immune serums diluted to 1:50 in PBS. The cells were washed by PBS, and then the primary antibodies were labeled for 1 h with a FITC-conjugated anti-rabbit IgG antibody (Sigma: F9887) diluted to 1:100 in PBS. The cells were mounted using a SlowFade Diamond antifade, and observed under an inverted Zeiss LSM 510 laser scanning microscope (Carl Zeiss).
For immunoelectron microscopy, specimens were prepared using a modified protocol of cryofixation and freeze substitution described previously [71]. Cryofixed B. natans cells were incubated in cooled 100 % ethanol (−80 °C) for 2 days, and finally embedded in LR White resin. Ultrathin sections of polymerized blocks were created by a Leica ultramicrotome EM UC6, and collected onto Formvar-coated copper grids. Immunogold labeling was performed with respective anti-BnFtsZD antibodies at a dilution of 1:10 in PBS, and a gold-conjugated anti-rabbit IgG secondary antibody (Sigma: G7402) diluted to 1:20. The ultrathin sections were stained with uranyl acetate for 10 min, and observed under a Hitachi H7650 transmission electron microscope at 80 kV.
Real-time quantitative PCR
B. natans cells were harvested from 100 mL synchronized cultures at 4 h intervals for 36 h, and the cell number was monitored with a Fuchs-Rosenthal hemocytometer. Total RNA was extracted from 13 time points using TRIzol reagent (Invitrogen), and cDNA was synthesized using a ReverTra Ace qPCR RT Kit (Toyobo) according to the manufacture’s protocols. Primers for quantification of BnftsZD-1 and BnftsZD-2 were designed using the Primer3Plus online software (primer sequences were listed in Additional file 1), and a previously reported primer set of 18S rRNA [62] was used for normalization. RT-qPCR was performed using a Thermal Cycler Dice Real Time System II (Takara) under the following conditions: 1 μL of cDNA, 0.4 μM of each primer, 12.5 μL of SYBR Premix Ex Taq II (Takara), and DNase/RNase-free water up to 25 μL. The cycling conditions comprised 3 min of denaturation at 95 °C, followed by 40 cycles of 10 s at 95 °C and 30 s at 60 °C, and a melting curve program for detection of nonspecific products. Relative changes in gene expression were calculated by the ΔΔCT method [72] in different time points.
Availability of supporting data
Nucleotide sequences used in this study can be obtained from GenBank (National Center for Biotechnology Information), the JGI genome portal site of Bigelowiella natans (http://genome.jgi.doe.gov/Bigna1/Bigna1.home.html), and the Cyanophora paradoxa Genome Project site (http://cyanophora.rutgers.edu/cyanophora/home.php), and the iMicrobe website (http://data.imicrobe.us/project/view/104).
Acknowledgements
We thank Mr. Takashi Shiratori (University of Tsukuba) for helping in the phylogenetic analyses, and Prof. Tetsuo Hashimoto (University of Tsukuba) for granting access to the ChemiDoc MP System. This work was supported by the Japan Society for the Promotion of Science (JSPS) KAKENHI Grant numbers: 23117004 and 15K18582.
Abbreviations
- ER
endoplasmic reticulum
- GFP
green fluorescent protein
- FITC
fluorescein isothiocyanate
- RT-qPCR
real-time quantitative PCR
- IgG
immunoglobulin G
Additional file
Primer sequences for plasmid construction (Table S1) and primer sequences for RT-qPCR experiments (Table S2). (PDF 120 kb)
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
YH designed this study, performed all experiments and analyses, and wrote the manuscript. KI conceived of the study, and helped to draft and revise the manuscript. Both authors read and approved the final manuscript.
Contributor Information
Yoshihisa Hirakawa, Email: hirakawa.yoshi.fp@u.tsukuba.ac.jp.
Ken-ichiro Ishida, Email: ken@biol.tsukuba.ac.jp.
References
- 1.Rodríguez-Ezpeleta N, Brinkmann H, Burey SC, Roure B, Burger G, Löffelhardt W, et al. Monophyly of primary photosynthetic eukaryotes: green plants, red algae, and glaucophytes. Curr Biol. 2005;15:1325–30. [DOI] [PubMed]
- 2.Price DC, Chan CX, Yoon HS, Yang EC, Qiu H, Weber APM, et al. Cyanophora paradoxa genome elucidates origin of photosynthesis in algae and plants. Science. 2012;335:843–7. [DOI] [PubMed]
- 3.Gould SB, Waller RF, McFadden GI. Plastid evolution. Annu Rev Plant Biol. 2008;59:491–517. doi: 10.1146/annurev.arplant.59.032607.092915. [DOI] [PubMed] [Google Scholar]
- 4.Keeling PJ. The endosymbiotic origin, diversification and fate of plastids. Philos Trans R Soc Lond B Biol Sci. 2010;365:729–748. doi: 10.1098/rstb.2009.0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burki F. The eukaryotic tree of life from a global phylogenomic perspective. Cold Spring Harb Perspect Biol. 2014;6:a016147. doi: 10.1101/cshperspect.a016147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cavalier-Smith T. Membrane heredity and early chloroplast evolution. Trends Plant Sci. 2000;5:174–182. doi: 10.1016/S1360-1385(00)01598-3. [DOI] [PubMed] [Google Scholar]
- 7.McFadden GI. Plastids and protein targeting. J Eukaryot Microbiol. 1999;46:339–346. doi: 10.1111/j.1550-7408.1999.tb04613.x. [DOI] [PubMed] [Google Scholar]
- 8.Gould SB, Maier UG, Martin WF. Protein import and the origin of red complex plastids. Curr Biol. 2015;25:R515–R521. doi: 10.1016/j.cub.2015.04.033. [DOI] [PubMed] [Google Scholar]
- 9.Bolte K, Bullmann L, Hempel F, Bozarth A, Zauner S, Maier UG. Protein targeting into secondary plastids. J Eukaryot Microbiol. 2009;56:9–15. doi: 10.1111/j.1550-7408.2008.00370.x. [DOI] [PubMed] [Google Scholar]
- 10.Sheiner L, Striepen B. Protein sorting in complex plastids. Biochim Biophys Acta. 1833;2013:352–359. doi: 10.1016/j.bbamcr.2012.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miyagishima S. Mechanism of plastid division: from a bacterium to an organelle. Plant Physiol. 2011;155:1533–1544. doi: 10.1104/pp.110.170688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Osteryoung KW, Pyke KA. Division and dynamic morphology of plastids. Annu Rev Plant Biol. 2014;65:443–472. doi: 10.1146/annurev-arplant-050213-035748. [DOI] [PubMed] [Google Scholar]
- 13.Miyagishima S, Kabeya Y. Chloroplast division: squeezing the photosynthetic captive. Curr Opin Microbiol. 2010;13:738–746. doi: 10.1016/j.mib.2010.10.004. [DOI] [PubMed] [Google Scholar]
- 14.Mori T, Kuroiwa H, Takahara M, Miyagishima S, Kuroiwa T. Visualization of an FtsZ ring in chloroplasts of Lilium longiflorum leaves. Plant Cell Physiol. 2001;42:555–9. [DOI] [PubMed]
- 15.Vitha S, McAndrew RS, Osteryoung KW. FtsZ ring formation at the chloroplast division site in plants. J Cell Biol. 2001;153:111–120. doi: 10.1083/jcb.153.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miyagishima S, Takahara M, Mori T, Kuroiwa H, Higashiyama T, Kuroiwa T. Plastid division is driven by a complex mechanism that involves differential transition of the bacterial and eukaryotic division rings. Plant Cell. 2001;13:2257–2268. doi: 10.1105/tpc.13.10.2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sato M, Nishikawa T, Kajitani H, Kawano S. Conserved relationship between FtsZ and peptidoglycan in the cyanelles of Cyanophora paradoxa similar to that in bacterial cell division. Planta. 2007;227:177–187. doi: 10.1007/s00425-007-0605-0. [DOI] [PubMed] [Google Scholar]
- 18.Miyagishima S, Suzuki K, Okazaki K, Kabeya Y. Expression of the nucleus-encoded chloroplast division genes and proteins regulated by the algal cell cycle. Mol Biol Evol. 2012;29:2957–2970. doi: 10.1093/molbev/mss102. [DOI] [PubMed] [Google Scholar]
- 19.Maple J, Aldridge C, Møller SG. Plastid division is mediated by combinatorial assembly of plastid division proteins. Plant J. 2005;43:811–823. doi: 10.1111/j.1365-313X.2005.02493.x. [DOI] [PubMed] [Google Scholar]
- 20.Colletti KS, Tattersall EA, Pyke KA, Froelich JE, Stokes KD, Osteryoung KW. A homologue of the bacterial cell division site-determining factor MinD mediates placement of the chloroplast division apparatus. Curr Biol. 2000;10:507–516. doi: 10.1016/S0960-9822(00)00466-8. [DOI] [PubMed] [Google Scholar]
- 21.Itoh R, Fujiwara M, Nagata N, Yoshida S. A chloroplast protein homologous to the eubacterial topological specificity factor minE plays a role in chloroplast division. Plant Physiol. 2001;127:1644–1655. doi: 10.1104/pp.010386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang M, Schmitz AJ, Kadirjan-Kalbach DK, Terbush AD, Osteryoung KW. Chloroplast division protein ARC3 regulates chloroplast FtsZ-ring assembly and positioning in arabidopsis through interaction with FtsZ2. Plant Cell. 2013;25:1787–1802. doi: 10.1105/tpc.113.111047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakanishi H, Suzuki K, Kabeya Y, Miyagishima S. Plant-specific protein MCD1 determines the site of chloroplast division in concert with bacteria-derived MinD. Curr Biol. 2009;19:151–156. doi: 10.1016/j.cub.2008.12.018. [DOI] [PubMed] [Google Scholar]
- 24.Miyagishima S, Froehlich JE, Osteryoung KW. PDV1 and PDV2 mediate recruitment of the dynamin-related protein ARC5 to the plastid division site. Plant Cell. 2006;18:2517–2530. doi: 10.1105/tpc.106.045484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gao H, Kadirjan-Kalbach D, Froehlich JE, Osteryoung KW. ARC5, a cytosolic dynamin-like protein from plants, is part of the chloroplast division machinery. Proc Natl Acad Sci U S A. 2003;100:4328–4333. doi: 10.1073/pnas.0530206100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sweitzer SM, Hinshaw JE. Dynamin undergoes a GTP-dependent conformational change causing vesiculation. Cell. 1998;93:1021–1029. doi: 10.1016/S0092-8674(00)81207-6. [DOI] [PubMed] [Google Scholar]
- 27.Osawa M, Anderson DE, Erickson HP. Reconstitution of contractile FtsZ rings in liposomes. Science. 2008;320:792–794. doi: 10.1126/science.1154520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuroiwa T, Kuroiwa H, Sakai A, Takahashi H, Toda K, Itoh R. The division apparatus of plastids and mitochondria. Int Rev Cytol. 1998;181:1–41. doi: 10.1016/S0074-7696(08)60415-5. [DOI] [PubMed] [Google Scholar]
- 29.Yoshida Y, Kuroiwa H, Misumi O, Yoshida M, Ohnuma M, Fujiwara T, et al. Chloroplasts divide by contraction of a bundle of nanofilaments consisting of polyglucan. Science. 2010;329:949–53. [DOI] [PubMed]
- 30.Yoshida Y, Kuroiwa H, Misumi O, Nishida K, Yagisawa F, Fujiwara T, et al. Isolated chloroplast division machinery can actively constrict after stretching. Science. 2006;313:1435–8. [DOI] [PubMed]
- 31.Miyagishima S. Origin and evolution of the chloroplast division machinery. J Plant Res. 2005;118:295–306. doi: 10.1007/s10265-005-0226-2. [DOI] [PubMed] [Google Scholar]
- 32.Miyagishima S, Nakamura M, Uzuka A, Era A. FtsZ-less prokaryotic cell division as well as FtsZ- and dynamin-less chloroplast and non-photosynthetic plastid division. Front Plant Sci. 2014;5:459. doi: 10.3389/fpls.2014.00459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miyagishima S, Kabeya Y, Sugita C, Sugita M, Fujiwara T. DipM is required for peptidoglycan hydrolysis during chloroplast division. BMC Plant Biol. 2014;14:57. doi: 10.1186/1471-2229-14-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Douglas SE, Penny SL. The plastid genome of the cryptophyte alga, Guillardia theta: complete sequence and conserved synteny groups confirm its common ancestry with red algae. J Mol Evol. 1999;48:236–244. doi: 10.1007/PL00006462. [DOI] [PubMed] [Google Scholar]
- 35.Sánchez Puerta MV, Bachvaroff TR, Delwiche CF. The complete plastid genome sequence of the haptophyte Emiliania huxleyi: A comparison to other plastid genomes. DNA Res. 2005;12:151–156. doi: 10.1093/dnares/12.2.151. [DOI] [PubMed] [Google Scholar]
- 36.Fraunholz MJ, Moerschel E, Maier UG. The chloroplast division protein FtsZ is encoded by a nucleomorph gene in cryptomonads. Mol Gen Genet. 1998;260:207–211. doi: 10.1007/s004380050887. [DOI] [PubMed] [Google Scholar]
- 37.Nishikawa T, Moriyama Y, Sato M, Sano T, Hasezawa S, Ota S, et al. Isolation of mitochondrial and plastid ftsZ genes and analysis of the organelle targeting sequence in the diatom Chaetoceros neogracile (Diatoms, Bacillariophyceae). Phycol Res. 2012;60:123–36.
- 38.van Dooren GG, Reiff SB, Tomova C, Meissner M, Humbel BM, Striepen B. A novel dynamin-related protein has been recruited for apicoplast fission in Toxoplasma gondii. Curr Biol. 2009;19:267–276. doi: 10.1016/j.cub.2008.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hirakawa Y. Complex plastids of chlorarachniophyte algae. Perspect Phycol. 2014;1:87–92. doi: 10.1127/pip/2014/0014. [DOI] [Google Scholar]
- 40.Curtis BA, Tanifuji G, Burki F, Gruber A, Irimia M, Maruyama S, et al. Algal genomes reveal evolutionary mosaicism and the fate of nucleomorphs. Nature. 2012;492:59–65. [DOI] [PubMed]
- 41.Gilson PR, Su V, Slamovits CH, Reith ME, Keeling PJ, McFadden GI. Complete nucleotide sequence of the chlorarachniophyte nucleomorph: nature’s smallest nucleus. Proc Natl Acad Sci U S A. 2006;103:9566–9571. doi: 10.1073/pnas.0600707103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rogers MB, Gilson PR, Su V, McFadden GI, Keeling PJ. The complete chloroplast genome of the chlorarachniophyte Bigelowiella natans: evidence for independent origins of chlorarachniophyte and euglenid secondary endosymbionts. Mol Biol Evol. 2007;24:54–62. doi: 10.1093/molbev/msl129. [DOI] [PubMed] [Google Scholar]
- 43.Hopkins JF, Spencer DF, Laboissiere S, Neilson JA, Eveleigh RJ, Durnford DG, et al. Proteomics reveals plastid- and periplastid-targeted proteins in the chlorarachniophyte alga Bigelowiella natans. Genome Biol Evol. 2012;4:1391–406. [DOI] [PMC free article] [PubMed]
- 44.Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 45.Emanuelsson O, Nielsen H, von Heijne G. ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 1999;8:978–984. doi: 10.1110/ps.8.5.978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang Y, Matsuzaki M, Takahashi F, Qu L, Nozaki H. Phylogenomic analysis of “red” genes from two divergent species of the “green” secondary phototrophs, the chlorarachniophytes, suggests multiple horizontal gene transfers from the red lineage before the divergence of extant chlorarachniophytes. PLoS One. 2014;9:e101158. doi: 10.1371/journal.pone.0101158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moestrup Ø, Sengco M. Ultrastructural studies on Bigelowiella natans, gen. et sp. nov., a chlorarachniophyte flagellate. J Phycol. 2001;37:624–646. doi: 10.1046/j.1529-8817.2001.037004624.x. [DOI] [Google Scholar]
- 48.Fujiwara T, Misumi O, Tashiro K, Yoshida Y, Nishida K, Yagisawa F, et al. Periodic gene expression patterns during the highly synchronized cell nucleus and organelle division cycles in the unicellular red alga Cyanidioschyzon merolae. DNA Res. 2009;16:59–72. [DOI] [PMC free article] [PubMed]
- 49.Gillard J, Devos V, Huysman MJ, De Veylder L, D’Hondt S, Martens C, et al. Physiological and transcriptomic evidence for a close coupling between chloroplast ontogeny and cell cycle progression in the pennate diatom Seminavis robusta. Plant Physiol. 2008;148:1394–411. [DOI] [PMC free article] [PubMed]
- 50.Ota S, Ueda K, Ishida K. Norrisiella sphaerica gen. et sp. nov., a new coccoid chlorarachniophyte from Baja California, Mexico. J Plant Res. 2007;120:661–670. doi: 10.1007/s10265-007-0115-y. [DOI] [PubMed] [Google Scholar]
- 51.Ishida K, Green B, Cavalier-Smith T. Diversification of a chimaeric algal group, the chlorarachniophytes: phylogeny of nuclear and nucleomorph small-subunit rRNA genes. Mol Biol Evol. 1999;16:321–331. doi: 10.1093/oxfordjournals.molbev.a026113. [DOI] [Google Scholar]
- 52.Ishida K, Nakayama T, Hara Y. Taxonomic studies on the Chlorarachniophyta. II. Generic delimitation of the chlorarachniophytes and description of Gymnochlora stellata gen. et sp. nov. and Lotharella gen. nov. Phycol Res. 1996;44:37–45. doi: 10.1111/j.1440-1835.1996.tb00036.x. [DOI] [Google Scholar]
- 53.Miyagishima S, Nozaki H, Nishida K, Nishida K, Matsuzaki M, Kuroiwa T. Two types of FtsZ proteins in mitochondria and red-lineage chloroplasts: the duplication of FtsZ is implicated in endosymbiosis. J Mol Evol. 2004;58:291–303. doi: 10.1007/s00239-003-2551-1. [DOI] [PubMed] [Google Scholar]
- 54.TerBush AD, Yoshida Y, Osteryoung KW. FtsZ in chloroplast division: structure, function and evolution. Curr Opin Cell Biol. 2013;25:461–470. doi: 10.1016/j.ceb.2013.04.006. [DOI] [PubMed] [Google Scholar]
- 55.Schmitz AJ, Glynn JM, Olson BJ, Stokes KD, Osteryoung KW. Arabidopsis FtsZ2-1 and FtsZ2-2 are functionally redundant, but FtsZ-based plastid division is not essential for chloroplast partitioning or plant growth and development. Mol Plant. 2009;2:1211–1222. doi: 10.1093/mp/ssp077. [DOI] [PubMed] [Google Scholar]
- 56.TerBush AD, Osteryoung KW. Distinct functions of chloroplast FtsZ1 and FtsZ2 in Z-ring structure and remodeling. J Cell Biol. 2012;199:623–637. doi: 10.1083/jcb.201205114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McAndrew RS, Olson BJ, Kadirjan-Kalbach DK, Chi-Ham CL, Vitha S, Froehlich JE, et al. In vivo quantitative relationship between plastid division proteins FtsZ1 and FtsZ2 and identification of ARC6 and ARC3 in a native FtsZ complex. Biochem J. 2008;412:367–78. [DOI] [PubMed]
- 58.Olson BJ, Wang Q, Osteryoung KW. GTP-dependent heteropolymer formation and bundling of chloroplast FtsZ1 and FtsZ2. J Biol Chem. 2010;285:20634–20643. doi: 10.1074/jbc.M110.122614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Robertson EJ, Rutherford SM, Leech RM. Characterization of chloroplast division using the Arabidopsis mutant arc5. Plant Physiol. 1996;112:149–159. doi: 10.1104/pp.112.1.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sakaguchi E, Takechi K, Sato H, Yamada T, Takio S, Takano H. Three dynamin-related protein 5B genes are related to plastid division in Physcomitrella patens. Plant Sci. 2011;180:789–795. doi: 10.1016/j.plantsci.2011.02.003. [DOI] [PubMed] [Google Scholar]
- 61.Hashimoto H. The ultrastructural features and division of secondary plastids. J Plant Res. 2005;118:163–172. doi: 10.1007/s10265-005-0214-6. [DOI] [PubMed] [Google Scholar]
- 62.Hirakawa Y, Burki F, Keeling PJ. Nucleus- and nucleomorph-targeted histone proteins in a chlorarachniophyte alga. Mol Microbiol. 2011;80:1439–1449. doi: 10.1111/j.1365-2958.2011.07643.x. [DOI] [PubMed] [Google Scholar]
- 63.Keeling PJ, Burki F, Wilcox HM, Allam B, Allen EE, Amaral Zettler LA, et al. The marine microbial eukaryote transcriptome sequencing project (MMETSP): illuminating the functional diversity of eukaryotic life in the oceans through transcriptome sequencing. PLoS Biol. 2014;12:e1001889. [DOI] [PMC free article] [PubMed]
- 64.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stamatakis A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tanabe AS. Kakusan4 and Aminosan: two programs for comparing nonpartitioned, proportional and separate models for combined molecular phylogenetic analyses of multilocus sequence data. Mol Ecol Resour. 2011;11:914–921. doi: 10.1111/j.1755-0998.2011.03021.x. [DOI] [PubMed] [Google Scholar]
- 68.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 69.Hirakawa Y, Nagamune K, Ishida K. Protein targeting into secondary plastids of chlorarachniophytes. Proc Natl Acad Sci U S A. 2009;106:12820–12825. doi: 10.1073/pnas.0902578106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hirakawa Y, Kofuji R, Ishida K. Transient transformation of a chlorarachniophyte alga, Lotharella amoebiformis (chlorarachniophyceae), with uidA and egfp reporter genes. J Phycol. 2008;44:814–820. doi: 10.1111/j.1529-8817.2008.00513.x. [DOI] [PubMed] [Google Scholar]
- 71.Nagasato C, Motomura T. Ultrastructural study on mitosis and cytokinesis in Scytosiphon lomentaria zygotes (Scytosiphonales, Phaeophyceae) by freeze-substitution. Protoplasma. 2002;219:140–149. doi: 10.1007/s007090200015. [DOI] [PubMed] [Google Scholar]
- 72.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]


